Abstract
Background & Aims
Defining the genetic heterogeneity of intrahepatic biliary epithelial cells (BECs) is challenging, and tools for identifying BEC subpopulations are limited. Here, we characterize the expression of a Sox9EGFP transgene in the liver and demonstrate that green fluorescent protein (GFP) expression levels are associated with distinct cell types.
Methods
Sox9EGFP BAC transgenic mice were assayed by immunofluorescence, flow cytometry, and gene expression profiling to characterize in vivo characteristics of GFP populations. Single BECs from distinct GFP populations were isolated by fluorescence-activated cell sorting, and functional analysis was conducted in organoid forming assays. Intrahepatic ductal epithelium was grown as organoids and treated with a Yes-associated protein (Yap) inhibitor or bile acids to determine upstream regulation of Sox9 in BECs. Sox9EGFP mice were subjected to bile duct ligation, and GFP expression was assessed by immunofluorescence.
Results
BECs express low or high levels of GFP, whereas periportal hepatocytes express sublow GFP. Sox9EGFP+ BECs are differentially distributed by duct size and demonstrate distinct gene expression signatures, with enrichment of Cyr61 and Hes1 in GFPhigh BECs. Single Sox9EGFP+ cells form organoids that exhibit heterogeneous survival, growth, and HNF4A activation dependent on culture conditions, suggesting that exogenous signaling impacts BEC heterogeneity. Yap is required to maintain Sox9 expression in biliary organoids, but bile acids are insufficient to induce BEC Yap activity or Sox9 in vivo and in vitro. Sox9EGFP remains restricted to BECs and periportal hepatocytes after bile duct ligation.
Conclusions
Our data demonstrate that Sox9EGFP levels provide readout of Yap activity and delineate BEC heterogeneity, providing a tool for assaying subpopulation-specific cellular function in the liver.
Keywords: Biliary Epithelium, Cholangiocytes, Sox9, Yap
Abbreviations used in this paper: BDL, bile duct ligation; BEC, biliary epithelial cell; DCA, deoxycholic acid; DMEM, Dulbecco modified Eagle medium; EGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; K19, cytokeratin 19; LSEC, liver sinusoidal endothelial cell; PBS, phosphate-buffered saline; RT-qPCR, reverse transcriptase quantitative polymerase chain reaction; TNF, tumor necrosis factor; WGA, wheat germ agglutinin; Yap, Yes-associated protein
Graphical abstract
Summary.
We characterize expression of a Sox9EGFP transgene in the mouse liver. Differential expression of EGFP facilitates identification and isolation of peribiliary hybrid hepatocytes and two biliary epithelial populations with distinct gene expression signatures.
Biliary epithelial cells (BECs) line intrahepatic bile ducts and are responsible for modifying and transporting bile during homeostasis. After acute or chronic liver injury, BECs undergo a proliferative response, termed ductular reaction, that is associated with liver repair and modeled in rodents through chemical injury or surgical ligation of the common bile duct. BECs are also impacted by cholangiopathies, which can result in liver failure and have few therapeutic interventions.1 Evidence suggests a significant degree of functional heterogeneity among BECs with relevance to physiology and liver disease. For example, BEC secretory function is modulated by hormones, peptides, and neurotransmitters, many of which act on a subpopulation of ductal epithelium.1 BECs also demonstrate differential proliferation after ductal injury, and some BECs are capable of transdifferentiating into hepatocytes after severe or chronic liver injury.2, 3, 4 However, assigning potentially heterogeneous responses to specific cell “types” is complicated by the fact that BEC subpopulations lack the clear molecular and genetic definitions that have facilitated a deeper understanding of cell biology in other epithelial tissues. New and accessible tools for interrogating BEC heterogeneity are needed to define and dissect context-dependent roles of BEC subpopulations in liver physiology and disease.
BEC heterogeneity has been defined relative to BEC size, with “small” cholangiocytes residing in proximal ductules near the canals of Hering and “large” cholangiocytes in large ducts.5 Although isolated bile ducts and immortalized small and large cholangiocyte cell lines have provided significant insight into biliary physiology, size-based definitions complicate isolation of BECs from genetically or pharmacologically challenged livers for direct, in vivo insight. Recent single cell transcriptomic studies have shown that BECs demonstrate variable levels of Yes-associated protein (Yap) activity.6 Yap activity is increased in bile ducts relative to hepatocytes, and transgenic activation of Yap in hepatocytes drives transdifferentiation to a ductal phenotype.7 The Yap pathway and its downstream effectors may present an opportunity for studying BEC heterogeneity from a genetic perspective.
Studies from our lab and others have used a Sox9EGFP BAC transgene to isolate stem, progenitor, and differentiated epithelial cells from mouse intestine and colon.8, 9, 10 Because Sox9EGFP is expressed broadly and at distinct levels in these tissues, isolation of cells based on green fluorescent protein (GFP) expression provides a single transgene approach to studying cellular heterogeneity. In the liver, Sox9 is a BEC biomarker that is activated during hepatoblast specification into BEC precursors, where it is required for proper timing of biliary differentiation during development.11 We hypothesized that Sox9EGFP could facilitate isolation of BEC subpopulations, similar to previous work in the luminal gastrointestinal tract. Here, we examine Sox9EGFP transgene expression in intrahepatic bile ducts and exploit differential GFP expression levels to isolate distinct cellular subpopulations. Our results demonstrate that Sox9EGFP expression levels facilitate dissection of BEC heterogeneity.
Results
Sox9EGFP Is Expressed in Intrahepatic BECs and Periportal Hepatocytes
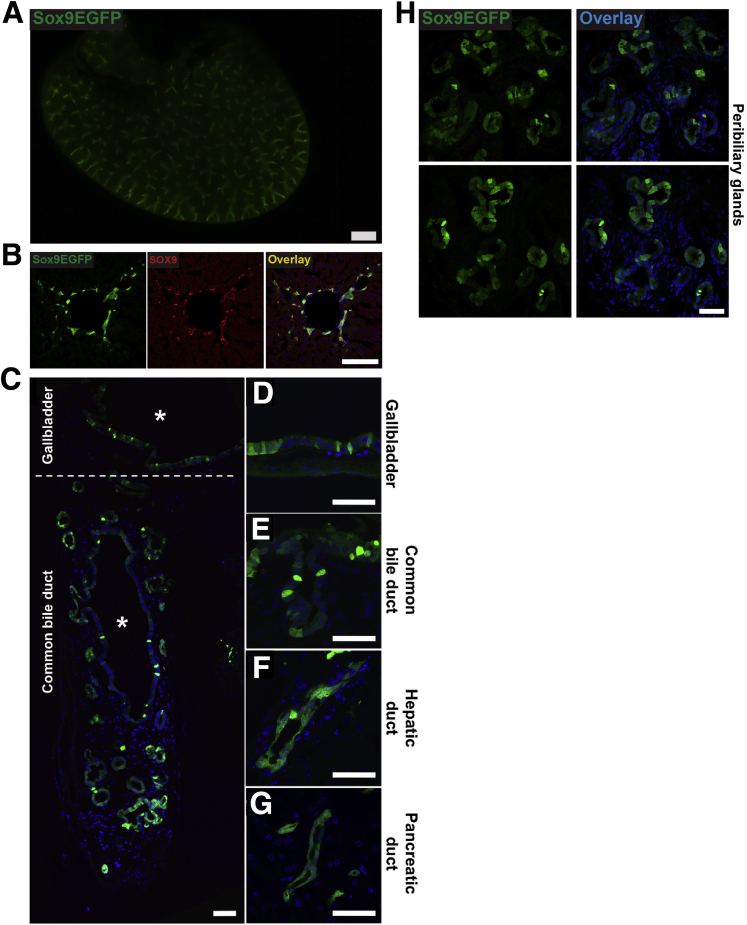
We sought to determine whether the Sox9EGFP BAC transgene, previously established as a stem/progenitor cell marker in intestinal and colonic epithelium, accurately labels known Sox9+ populations in the liver. Low magnification, epifluorescent imaging of whole liver lobes demonstrated robust GFP signal in branching patterns consistent with intrahepatic bile ducts (Figure 1A). Examination of histologic sections revealed GFP+ cells with typical ductal morphology confined to the portal area and co-localized with endogenous SOX9 (Figure 1B). We also observed Sox9EGFP expression throughout the biliary tree, including the gallbladder, extrahepatic bile duct, and pancreatic duct, consistent with known expression patterns of Sox9 in these tissues (Figure 1C–G). In addition, we noted expression of GFP throughout the epithelium of peribiliary glands in the extrahepatic bile duct, which have been previously reported to express Sox9 and are implicated in regeneration after injury (Figure 1H).12 Because of independent developmental origins and significant differences in gene expression of biliary epithelium in extrahepatic tissue, we focused the present study on intrahepatic ducts.13,14
Figure 1.
A Sox9EGFPtransgene is expressed at variable levels throughout the biliary tree. (A) Low magnification imaging of Sox9EGFP expression throughout the intrahepatic biliary tree (11-week-old female mouse, right lobe; scale bar = 1 mm). (B) Sox9EGFP is expressed in intrahepatic bile ducts and co-localizes with SOX9 (scale bar = 100 μm). (C) Epithelial cells of the (D) gallbladder, (E) common bile duct, (F) hepatic ducts, and (G) pancreatic ducts express Sox9EGFP at variable levels (∗ indicate lumens; scale bars = 50 μm). (H) Peribiliary glands of the extrahepatic duct also express Sox9EGFP (scale bar = 50 μm).
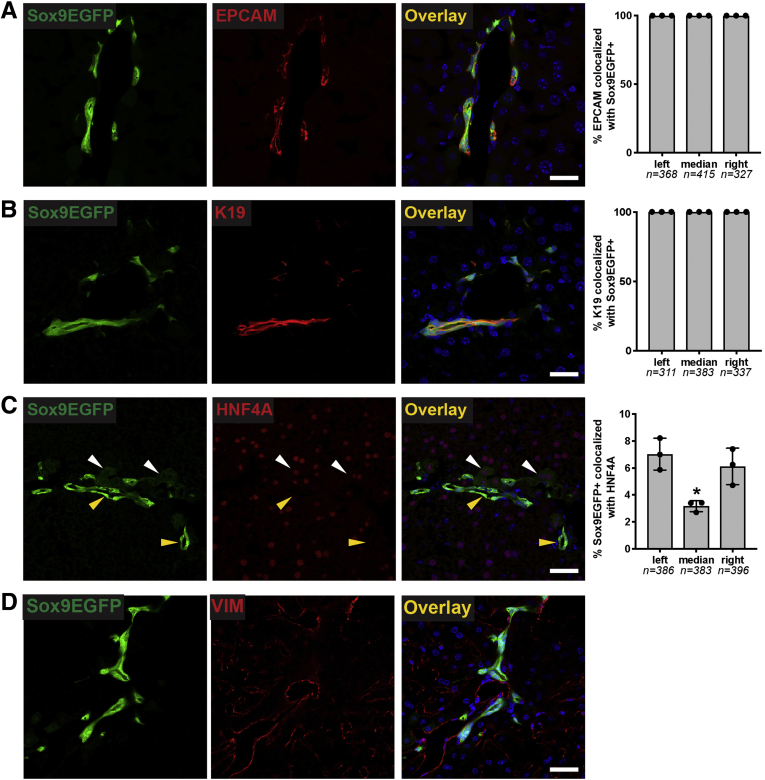
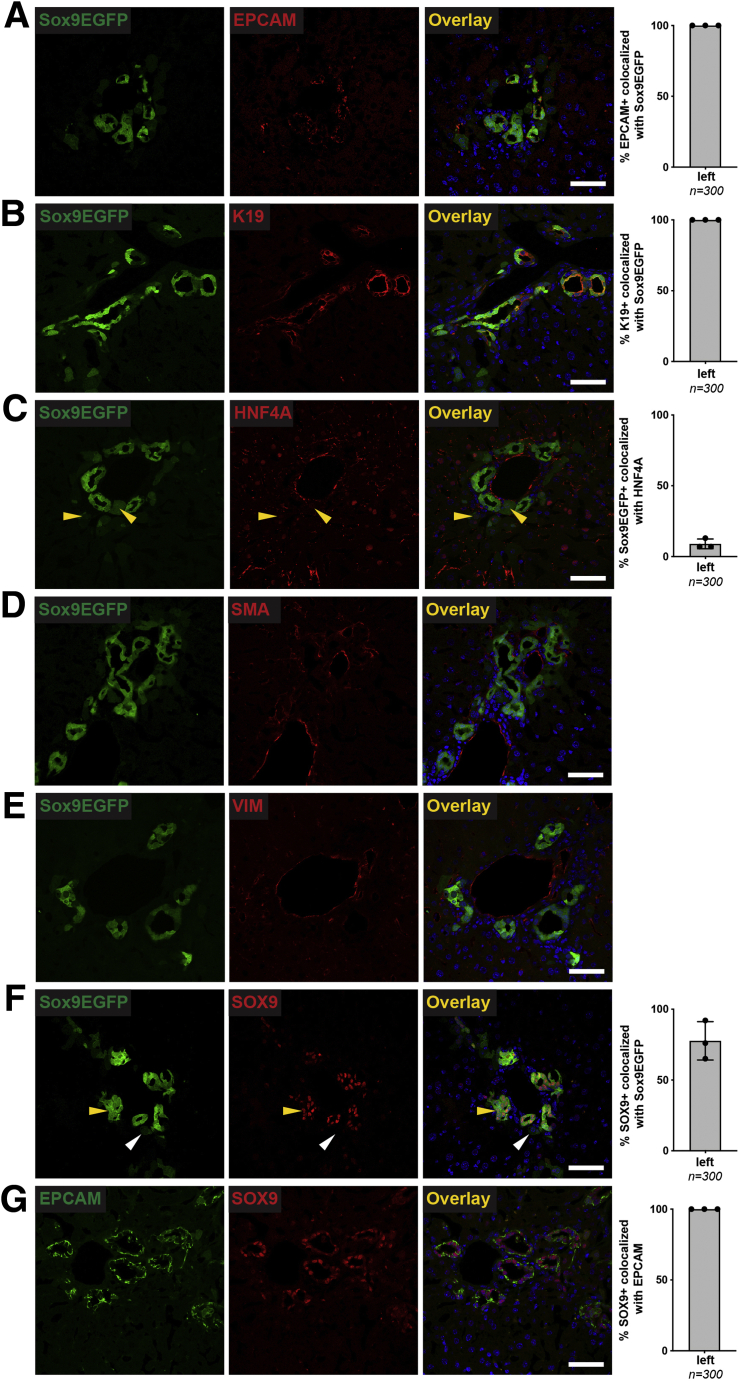
To determine whether Sox9EGFP expression accurately labels all BECs and whether Sox9EGFP is restricted to BECs, we next examined enhanced GFP (EGFP) expression relative to the independent BEC markers EPCAM and cytokeratin 19 (K19) by immunofluorescence. In both cases, we found that 100% of BECs identified by EPCAM or K19 were also positive for GFP in left, median, and right lobes (Figure 2A and B). Previous reports have shown that a subpopulation of periportal hepatocytes co-express BEC markers, including Sox9.15 To test whether Sox9EGFP is also expressed in hybrid hepatocytes, we co-localized GFP+ cells with hepatocyte transcription factor HNF4A. We found that a small percentage of Sox9EGFP cells are HNF4A+ (Figure 2C). These cells were observed to have typical hepatocyte morphology and express very low levels of the GFP transgene (Figure 2C, white arrowheads), in contrast to GFP+ cells with BEC morphology, which were appreciably brighter (Figure 2C, yellow arrowheads). We did not observe any co-localization between Sox9EGFP and vimentin, which is expressed on mesenchymal, endothelial, and stellate cells (Figure 2D).16
Figure 2.
Sox9EGFPis expressed in intrahepatic bile ducts and peribiliary hepatocytes. Immunofluorescence demonstrates that EGFP is co-expressed in (A) 100% of EPCAM+ and (B) K19+ positive BECs across left, median, and right lobes. (C) Rare peribiliary cells expressing very low levels of EGFP co-localize with HNF4A and are morphologically consistent with hepatocytes (white arrows indicate EGFP+/HNF4A+; yellow arrows indicate EGFP+/HNF4A–). (D) Sox9EGFP does not co-localize with mesenchymal marker, vimentin (VIM) (scale bar = 50 μm).
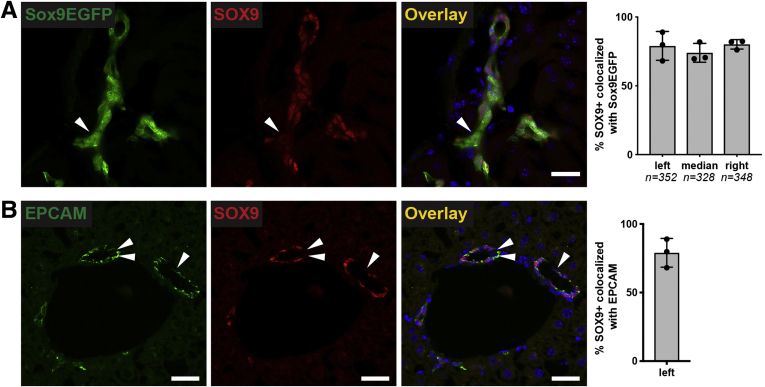
Finally, we co-localized Sox9EGFP with endogenous SOX9. Surprisingly, we found that some cells expressing the GFP transgene did not co-express endogenous SOX9 protein (Figure 3A, arrowheads). The number of GFP+/SOX9– cells was not significantly variable between biological replicates. Because SOX9 is considered a pan-biliary marker, we independently confirmed the presence of SOX9– BECs by co-localizing SOX9 with EPCAM by immunofluorescence. We found that SOX9 co-localized with EPCAM+ BECs (79.0% ± 10.5%) at approximately the same rate as EGFP+ BECs (77.7% ± 7.1%) (Figure 3B). Collectively, our immunofluorescence analyses confirm that Sox9EGFP is expressed ubiquitously in BECs, including BECs that lack endogenous SOX9 protein. In addition, very low levels of GFP are expressed in a subpopulation of periportal hepatocytes, consistent with known expression of endogenous Sox9 in hybrid hepatocytes.
Figure 3.
Rare Sox9EGFP-positive BECs do not express SOX9 protein. (A) SOX9 is expressed in most, but not all, Sox9EGFP+ cells (arrowheads indicate EGFP+/SOX9–) (scale bar = 50 μm; ∗ indicates P < .05, one-way analysis of variance and Tukey test). (B) Co-localization of SOX9 with EPCAM independently confirms the presence and occurrence of SOX9- BECs (left lobe, arrowheads indicate EPCAM+/SOX9– cells, scale bars = 50 μm).
GFPhigh Cells Are More Plentiful in Smaller Ducts
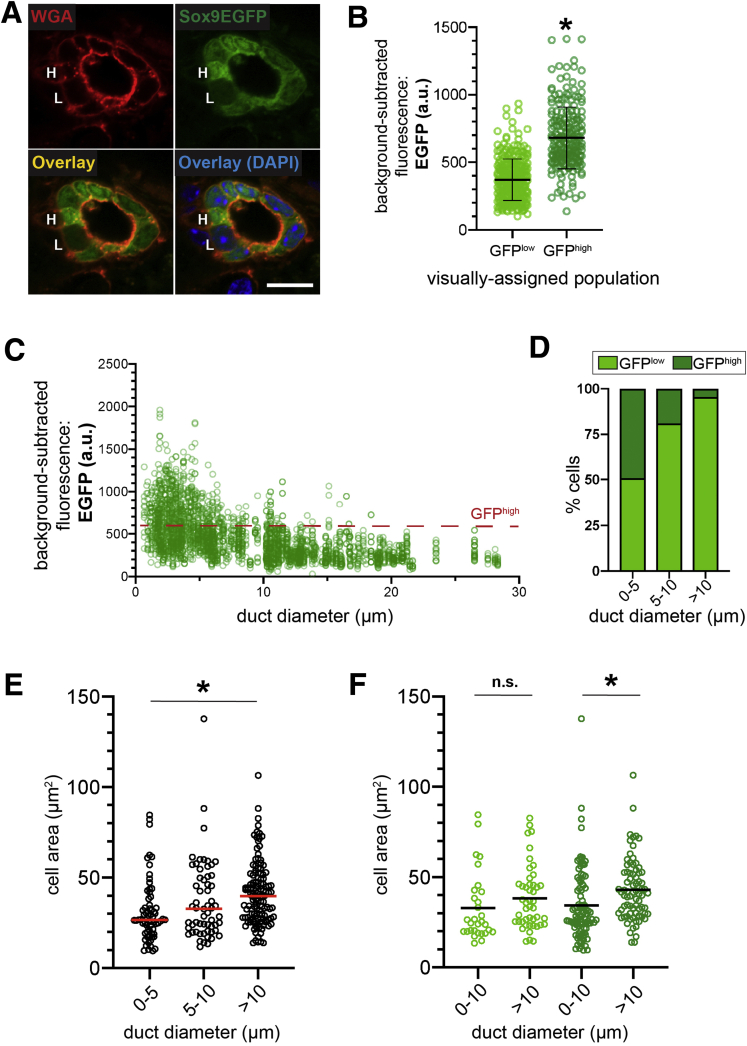
Although qualitative observation demonstrated variable Sox9EGFP expression in intrahepatic bile ducts, we sought to quantify ductal GFP at the single cell level and determine whether different levels of expression correlate with anatomic localization. We used semiquantitative confocal microscopy and measured GFP in individual cells by using wheat germ agglutinin (WGA) to delineate cell membranes. To avoid artifacts associated with antibody detection, all experiments measured endogenous GFP. First, we visually categorized BECs as GFPlow or GFPhigh and asked whether qualitatively identified Sox9EGFP populations demonstrated quantitatively discernible differences in GFP intensity (Figure 2A). We found that BECs identified as GFPhigh had significantly higher fluorescence intensity relative to those identified as GFPlow, validating our ability to resolve Sox9EGFP populations by histology (Figure 4B).
Figure 4.
GFPhighBECs are enriched in small intrahepatic ductules. (A) WGA labels cell membranes and facilitates quantification of Sox9EGFP cells qualitatively identified as high (H) and low (L) (scale bar = 10 μm). (B) Qualitatively identified GFPhigh cells are significantly brighter than GFPlow cells by quantification of confocal images (n = 307 GFPlow, 215 GFPhigh; ∗ indicates P < .001, unpaired t test; a.u. = arbitrary units). (C) Quantification of EGFP intensity relative to duct diameter demonstrates distribution of Sox9EGFP populations across the intrahepatic biliary tree (n = 2589 cells). (D) GFPhigh BECs are most abundant in small ductules and rare with increasing duct size (n = 954 cells 0–5 μm, 523 cells 5–10 μm, 1112 cells >10 μm). (E) BEC size increases with increasing duct diameter, and BECs located in ducts with luminal diameter ≥10 μm are significantly larger than BECs located in the smallest ductules (n = 954 cells 0–5 μm, 523 cells 5–10 μm, 1112 cells >10 μm; ∗ indicates P < .001, one-way analysis of variance and Tukey test). (F) GFPlow BECs located in ductules and ducts do not differ significantly in size, whereas GFPhigh BECs located in ducts are significantly larger than GFPhigh BECs located in ductules (∗ indicates P = .01, one-way analysis of variance and Tukey test).
Because BEC heterogeneity has been classically described relative to cell size and location in small or large ducts, we next quantified GFP expression in individual BECs relative to duct diameter.5 GFP fluorescence and duct diameter of the resident bile duct were measured for 2589 BECs, and we noted a clear inverse relationship between the number of GFPhigh cells and duct diameter (Figure 4C). To quantify the percentage of GFPhigh BECs in different-sized ducts, we arbitrarily defined GFPhigh BECs as having mean fluorescence intensity ≥600, which is based on the upper limit of fluorescence in a majority of GFPlow cells (Figure 4B). Next, we adopted duct size definitions used in recent functional studies of murine intrahepatic biliary epithelium; ducts are defined by a luminal diameter ≥10 μm, and ductules are defined by a luminal diameter of <10 μm.2 We determined that the smallest ductules (0–5 μm luminal diameter) are composed of 49.16% GFPhigh and 50.84% GFPlow cells, larger ductules (5–10 μm luminal diameter) are 18.93% GFPhigh and 81.07% GFPlow cells, and ducts (>10 μm luminal diameter) are composed of 4.50% GFPhigh and 95.50% GFPlow cells (Figure 4D). To ask whether cells in the same Sox9EGFP population varied by size relative to their resident duct, we first confirmed that BECs in ducts are larger than BECs in ductules, consistent with previous observations (Figure 4E).17 Interestingly, cell area was not significantly different between smaller (0–5 μm) and larger (5–10 μm) ductules (Figure 4E). Grouping cells by both GFP level and resident duct type revealed that GFPhigh BECs in ducts are significantly larger than GFPhigh BECs in ductules, whereas the size of GFPlow BECs in ducts and ductules was not significantly different (Figure 4F).
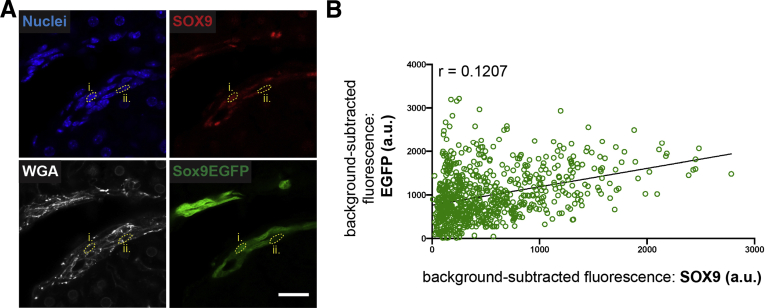
Sox9EGFP is driven by upstream transcriptional regulators of Sox9, and GFP levels accurately predict endogenous SOX9 protein levels in GFP+ cells of the small intestine and colon.8,10 We quantified 747 BECs by using WGA and bisbenzimide as membrane and nuclear markers for GFP and SOX9 immunofluorescence, respectively (Figure 5A). GFP level was a poor predictor of endogenous SOX9 protein level, and we did not observe specific association of SOX9– cells with low or high GFP expression (Figure 5B). Interestingly, some BECs expressing the highest observed level of GFP expressed little or no appreciable SOX9. Together, these data demonstrate that Sox9EGFP BECs are compartmentalized by duct size, with GFPhigh cells present in increased numbers in ductules. Furthermore, GFPhigh BECs located in ducts are larger than GFPhigh BECs in ductules. We also find that Sox9EGFP level is not predictive of SOX9 protein expression, in contrast with observations in the small intestine and colon.
Figure 5.
EGFP levels are not predictive of endogenous SOX9 levels. (A) Bisbenzimide and WGA label nuclei and cell membranes, respectively, for quantification of SOX9 and EGFP in single cells (roman numerals denote cells across multiple channels; scale bar = 25 μm). (B) SOX9 correlates poorly with EGFP levels (n = 747 cells). a.u. = arbitrary units.
Sox9EGFP Expression Levels Facilitate Isolation of Cellular Subpopulations
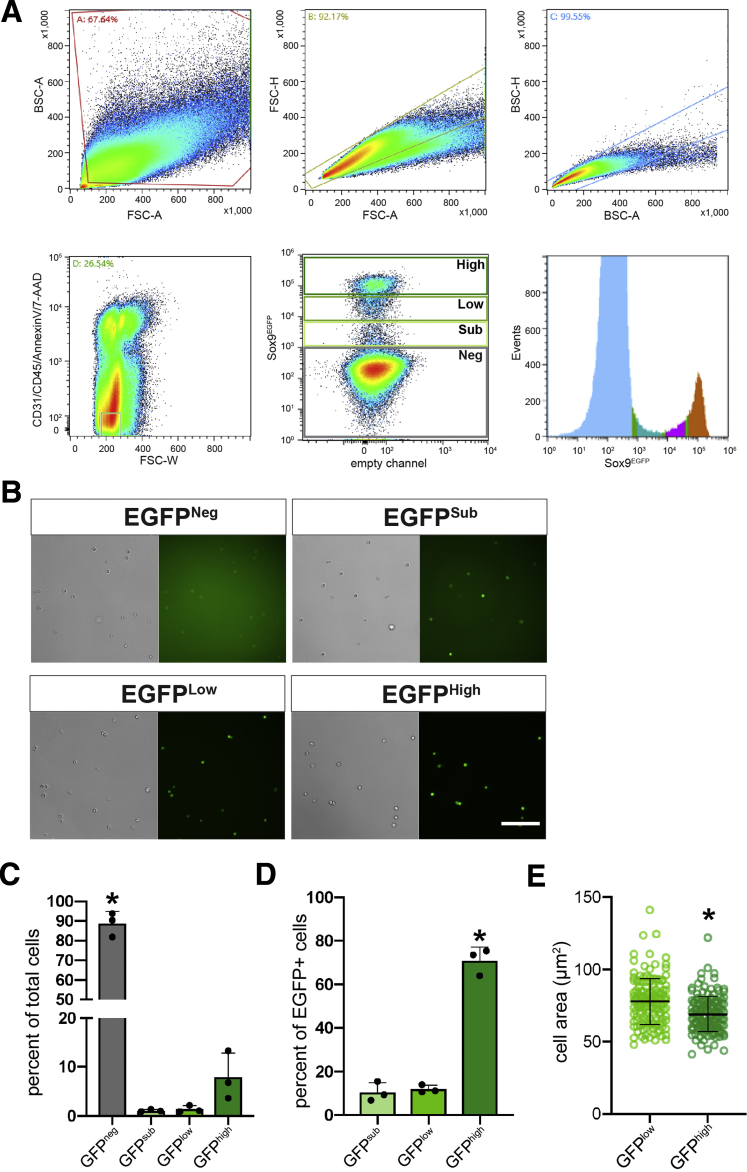
To further quantify relative levels of Sox9EGFP expression, we analyzed intrahepatic GFP by flow cytometry. Livers were dissociated by using a protocol optimized to obtain single BECs at the expense of hepatocyte viability.18 Flow cytometry revealed that a majority of cells in our single cell isolations were negative for the GFP transgene (88.7% ± 6.1%) (Figure 6A and C). Within the GFP+ fraction, we defined 3 distinct subpopulations: GFPsublow (GFPsub), GFPlow, and GFPhigh (Figure 6A and B). Of GFP+ populations, GFPhigh cells (71.0% ± 6.1%) were significantly more abundant than GFPlow (11.9% ± 1.8%) and GFPsub (10.6% ± 4.4%) cells (Figure 6D). These data demonstrate that intrahepatic Sox9EGFP is expressed at distinct levels that can be resolved by flow cytometry.
Figure 6.
FACS isolation of Sox9EGFPpopulations. (A) FACS isolation strategy for Sox9EGFP populations demonstrating that Sox9EGFP is divided into 4 populations by flow analysis. (B) FACS-isolated cells from Sox9EGFP populations exhibit increasing EGFP intensity (scale bar = 100 μm). (C) GFPneg cells are the most abundant population present in single cell preps after exclusion of CD31/CD45/Annexin V/7-AAD+ cells, and (D) GFPhigh cells are the most abundant of GFP+ populations (∗ indicates significance, P < .05, one-way analysis of variance and Tukey test). (E) Cell area measurements demonstrate that isolated GFPhigh cells are significantly smaller than GFPlow cells (n = 150 GFPlow, 150 GFPhigh; ∗ indicates P < .001, unpaired t test).
On the basis of our histologic assays, we reasoned that GFPlow and GFPhigh populations were most likely to represent cells of the intrahepatic bile ducts, whereas GFPsub were likely to represent periportal hepatocytes. To determine whether the size of sorted GFPlow and GFPhigh BECs was consistent with what we observed in vivo, we measured the area of fluorescence-activated cell sorting (FACS) isolated Sox9EGFP cells from the corresponding gates. We found that, on average, isolated GFPhigh cells were significantly smaller than GFPlow cells (Figure 6E). This could be explained by a greater representation of small, ductule-derived BECs in our liver prep or by changes to cell area after dissociation of epithelial tissues.
Sox9EGFP Populations Exhibit Differential Gene Expression Patterns
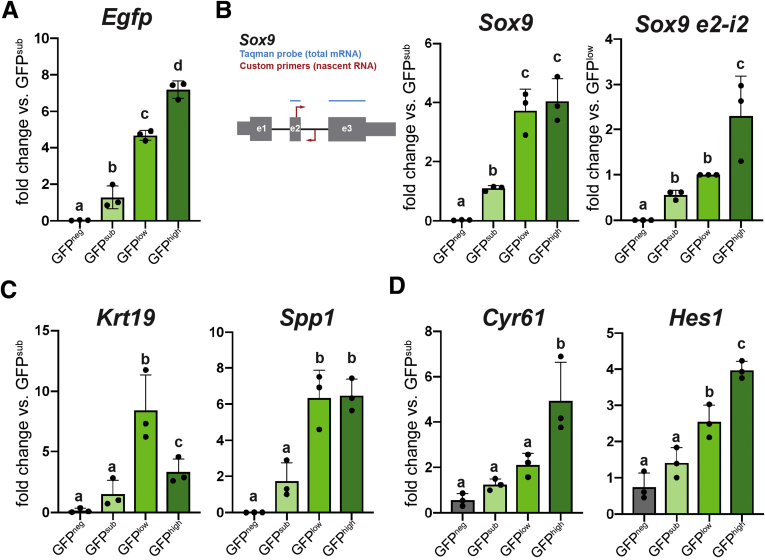
We next analyzed gene expression in FACS-isolated Sox9EGFP BEC populations. As expected, Egfp was differentially expressed across GFPneg, GFPsub, GFPlow, and GFPhigh populations (Figure 7A). Sox9 was enriched as expected between (1) GFPneg and GFPsub and (2) GFPsub and GFPlow/high (Figure 7B). However, we observed no difference in Sox9 expression between GFPlow and GFPhigh populations. We reasoned that differences in post-transcriptional regulation could lead to differential Egfp expression without differential Sox9 expression. To test this hypothesis, we designed reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) primers spanning the second exon and intron of Sox9 to detect nascent Sox9 RNA and complement our Taqman assay, which spanned exons 2 and 3 of Sox9 and is specific to mRNA (Figure 7B). We found that GFPhigh cells express significantly higher levels of nascent Sox9 RNA relative to GFPlow and GFPsub populations (Figure 7B). These data demonstrate that Sox9EGFP levels provide readout of Sox9 transcription, which is highest in GFPhigh BECs despite equivalent amounts of Sox9 mRNA between GFPlow and GFPhigh populations. This also suggests that Sox9 undergoes post-transcriptional regulation in BEC subpopulations.
Figure 7.
BEC genes demonstrate differential expression across Sox9EGFPpopulations. (A) Egfp expression is significantly different between GFPneg, sub, low, and high populations. (B) RT-qPCR probes detecting Sox9 mRNA show enrichment between GFPneg and sub populations but not GFPlow and high. Primers against nascent Sox9 RNA demonstrate up-regulation in GFPhigh. (C) Canonical BEC biomarkers Krt19 and Spp1 are differentially expressed in Sox9EGFP populations. (D) Cyr61 and Hes1 are most significantly enriched in GFPhigh cells, suggesting increased Yap activity (letters indicate grouping by significance, P < .05, one-way analysis of variance and Tukey test).
Next, we examined the expression of canonical BEC genes in Sox9EGFP populations. Spp1, which encodes pan-biliary marker osteopontin, was significantly enriched in both GFPlow and GFPhigh populations (Figure 7C). Interestingly, although both GFPlow and GFPhigh cells expressed significantly higher levels of Krt19 relative to GFPneg and GFPsub populations, we observed enrichment of Krt19 in GFPlow relative to GFPhigh (Figure 7C). To test whether Sox9EGFP populations capture recently reported Yap-associated heterogeneity, we assayed expression of Cyr61 and Hes1, which have been previously correlated with increased Yap activity.6,7 Both genes were significantly up-regulated in GFPhigh cells relative to GFPlow cells, and Hes1 was also enriched in GFPlow relative to GFPsub (Figure 7D). These data support histologic evidence that GFPlow and GFPhigh cells represent BEC populations, imply distinct transcriptional identities for GFPlow and GFPhigh BECs, and suggest that Sox9EGFP expression levels capture previously described heterogeneity relative to ductal YAP activity.
Unique Transcriptomic Signatures Define Intrahepatic Sox9EGFP Populations
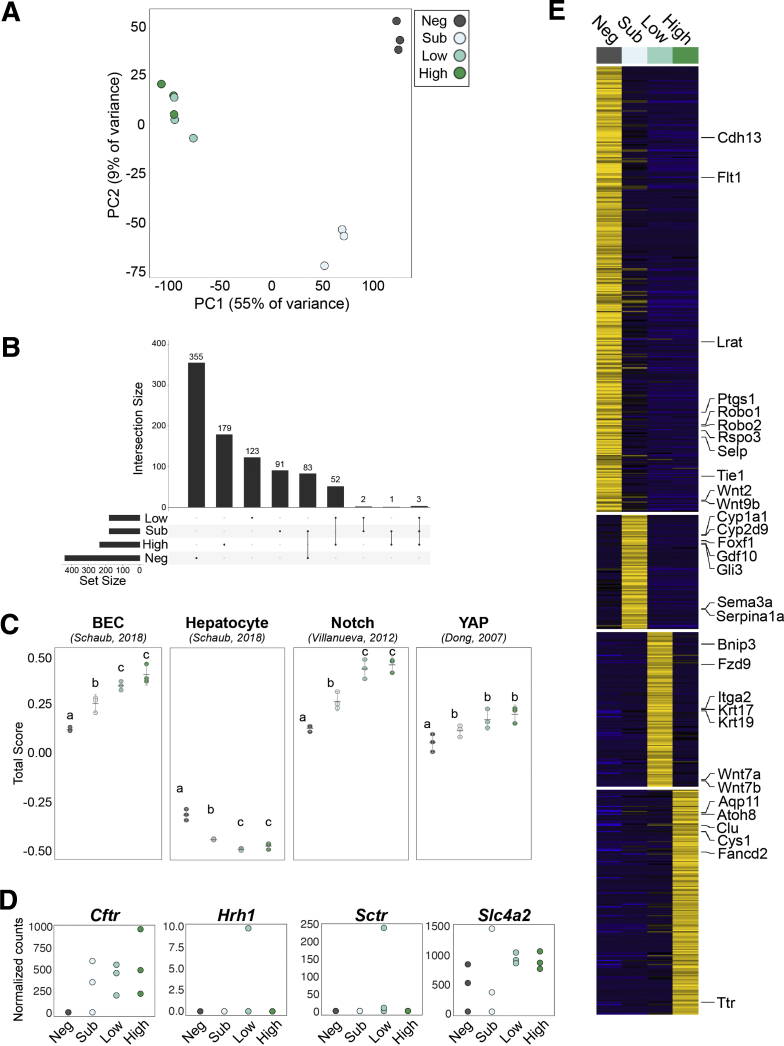
To determine gene expression signatures of Sox9EGFP subpopulations, we conducted RNA-seq on FACS-isolated GFPneg, GFPsub, GFPlow, and GFPhigh populations. Clustering by principal components analysis reinforced expected relationships predicted by histology and RT-qPCR. GFPlow and GFPhigh populations, which are both consistent with BEC identity, clustered together, whereas GFPneg and GFPsub samples demonstrated more significant differences (Figure 8A). Differential gene expression analysis identified genes unique to and shared between Sox9EGFP populations, with the largest shared gene sets consisting of genes shared between GFPneg and GFPsub populations, followed by genes shared between GFPlow and GFPhigh populations (Figure 8B). By comparison, very few genes were shared between GFPlow/high and GFPsub, and no genes were shared between GFPlow/high and GFPneg.
Figure 8.
Sox9EGFPpopulations have distinct transcriptomes. (A) GFPneg and sub populations form distinct clusters by principal components analysis, whereas GFPlow and high cluster similarly. (B) Genes shared between multiple populations reinforce similarities between GFPlow and high. (C) Gene signature analysis demonstrates significant enrichment of BEC, Notch, and Yap gene sets in GFPlow and high populations relative to GFPneg and sub, whereas all populations are depleted for hepatocyte genes (error bars represent 95% confidence interval; letters indicate grouping by significance, P < .05, one-way analysis of variance and Tukey test). (D) Expression of small and large cholangiocyte markers in Sox9EGFP populations by RNA-seq. (E) Heatmap represents genes uniquely up-regulated in a single Sox9EGFP population.
We next analyzed gene expression signatures of Sox9EGFP populations against published transcriptomic datasets using rank-based gene set scoring.19 Gene signature analysis demonstrated that GFPlow and GFPhigh were more enriched for intrahepatic BEC-associated genes than GFPneg and GFPsub (Figure 8C).20 All 4 populations were depleted for genes associated with hepatocytes, affirming that our single cell isolation and collection protocol selects against GFPneg hepatocytes. To examine enrichment of known biliary regulatory pathways, we assayed Notch and Yap target genes and again observed significant enrichment in GFPlow and GFPhigh populations (Figure 4C).21,22 Although our qPCR data demonstrated differential expression of specific YAP target genes, transcriptome-scale analyses did not reveal a significant difference between GFPlow and GFPhigh populations for either Yap or Notch gene signature scores (Figure 8C). We also examined expression of genes previously associated with functional heterogeneity in small (Hrh1) and large (Cftr, Sctr, Slc4a2) cholangiocytes. Surprisingly, both Hrh1 and Sctr were not highly expressed in any Sox9EGFP population from most biological replicates examined (Figure 8D). While Cftr and Slc4a2 were expressed, we did not detect significantly different levels of expression between GFPlow and GFPhigh BECs, consistent with the localization of both populations in small ductules as well as larger ducts (Figure 8D).
To more stringently assay transcriptomic differences between Sox9EGFP populations, we identified genes that were significantly up-regulated in a single population (Figure 8E, Supplementary Tables 1 and 2). Many of the genes uniquely enriched in the GFPneg population were consistent with pericentral liver sinusoidal endothelial cells (LSECs), including Rspo3, Cdh13, Flt1, Ptgs1, Selp, Wnt2, and Wnt9b (Figure 8E).23 Although pericentral LSECs express CD31, it remains restricted to the cytoplasm, explaining why this population would persist in our isolation strategy despite negative selection against CD31 by FACS24 (Figure 6A). The hepatic stellate cell-specific gene, Lrat, was also up-regulated in GFPneg cells.25 Although a majority of genes specific to individual GFP+ populations have no established function in the liver, we observed gene expression patterns consistent with hybrid hepatocyte and BEC identities. Hepatocyte-associated metabolic enzymes Cyp1a1 and Serpina1a were both up-regulated in GFPsub cells, along with the Hedgehog signaling target Gli3. Consistent with our qPCR data, Krt19 was significantly enriched in GFPlow BECs, along with Krt17 and WNT pathway genes Fzd9, Wnt7a, and Wnt7b. GFPhigh BECs expressed Aqp11, previously shown to be enriched in developing bile ducts, and Clu, which was recently identified as a marker and functional mediator of facultative stem cells in the small intestine (Figure 8E).26,27 Together, our transcriptomic data reinforce the broad cellular identities of Sox9EGFP populations defined by histology and RT-qPCR and identify unique gene expression signatures that differentiate GFPsub hybrid hepatocytes and GFPlow and GFPhigh BECs.
Sox9EGFP Populations Exhibit Distinct Phenotypic Responses to Single Cell Organoid Culture
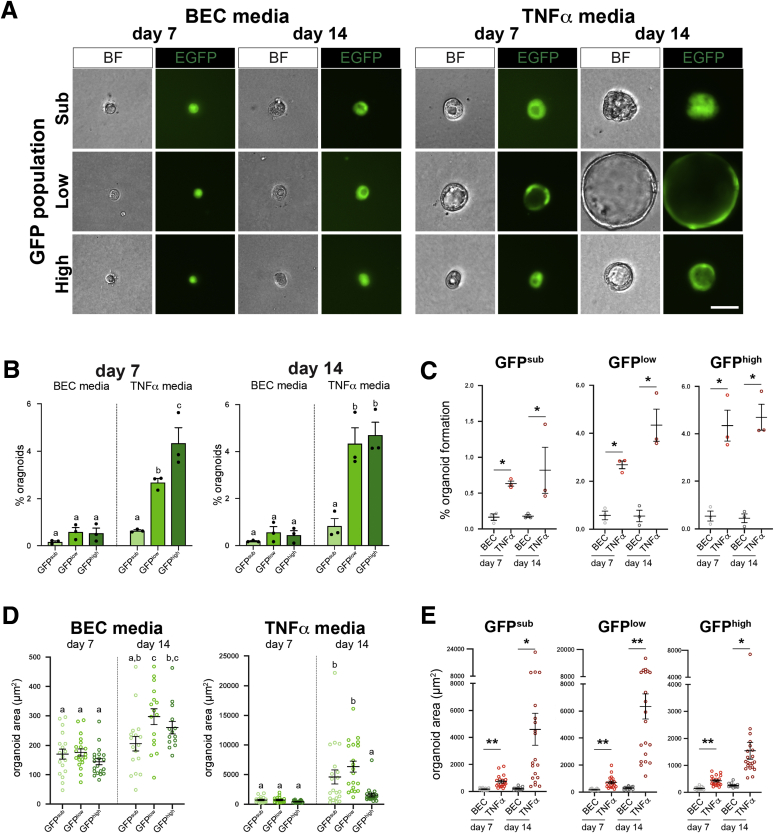
To assay functional properties of proliferation and cell fate in Sox9EGFP populations, we turned to organoid assays. Liver organoids were initially reported to be formed by BECs, with subsequent studies describing media conditions supportive of hepatocyte organoids.28,29 Because the Sox9EGFP transgene is expressed in both a subpopulation of hepatocytes (GFPsub) as well as BECs (GFPlow and GFPhigh), we used conditions developed for BECs (BEC media) as well as conditions developed for hepatocyte organoid culture (tumor necrosis factor [TNF]α media). All GFP+ populations formed organoids in both medium conditions, whereas GFPneg cells never formed organoids (Figure 9A). Consistent with previous reports, organoids with clear lumens or spherical morphology took approximately 5–6 days to form, and we quantified organoid formation rates at 7 and 14 days of culture.30 Whereas all GFP+ populations formed organoids at approximately the same rate in BEC media, single GFPlow and GFPhigh cells grown in TNFα media formed significantly more organoids than GFPsub cells at both 7 and 14 days of culture (Figure 9B). We observed an increase in GFPlow-derived organoids between days 7 and 14, suggesting an extended delay in the ability of some GFPlow BECs to form morphologically appreciable organoids in TNFα media (Figure 9B). Furthermore, TNFα media increased organoid formation relative to BEC media in organoids grown from the same GFP+ population (Figure 9C).
Figure 9.
Functional differences in organoids derived from single GFP+cells are dependent on medium conditions. (A) Single cells from Sox9EGFP populations form organoids in medium conditions developed for BECs (BEC media) and hepatocytes (TNFα media) (scale bar = 25 μm). (B and C) TNFα media significantly increase organoid formation relative to BEC media in all Sox9EGFP populations and are required for significantly different organoid formation rates between GFPsub, GFPlow, and GFPhigh (B): letters indicate grouping by significance, P < .05, one-way analysis of variance and Tukey test; (C) ∗ indicates significance, P < .01, unpaired t test. (D and E) TNFα media also drive more significant increases in organoid size between 7 and 14 days of culture and result in significantly larger GFPsub- and GFPlow-derived organoids at day 14 relative to GFPhigh-derived organoids at the same time point (D): letters indicate grouping by significance, P < .05, one-way analysis of variance and Tukey test; (E): ∗ indicates significance, P < .002; ∗∗ indicates significance, P ≤ .0001, unpaired t test.
We quantified organoid area and found that relative to organoids grown in BEC media, organoids grown in TNFα media demonstrated a greater increase in average area between days 7 and 14 in culture, suggesting more robust proliferation induced by TNFα media (Figure 9D). Interestingly, GFPsub- and GFPlow-derived organoids were significantly larger than GFPhigh-derived organoids at day 14 in TNFα media (Figure 9D). Finally, organoids grown in TNFα media were significantly larger than organoids grown in BEC media at both time points and across Sox9EGFP populations (Figure 9E).
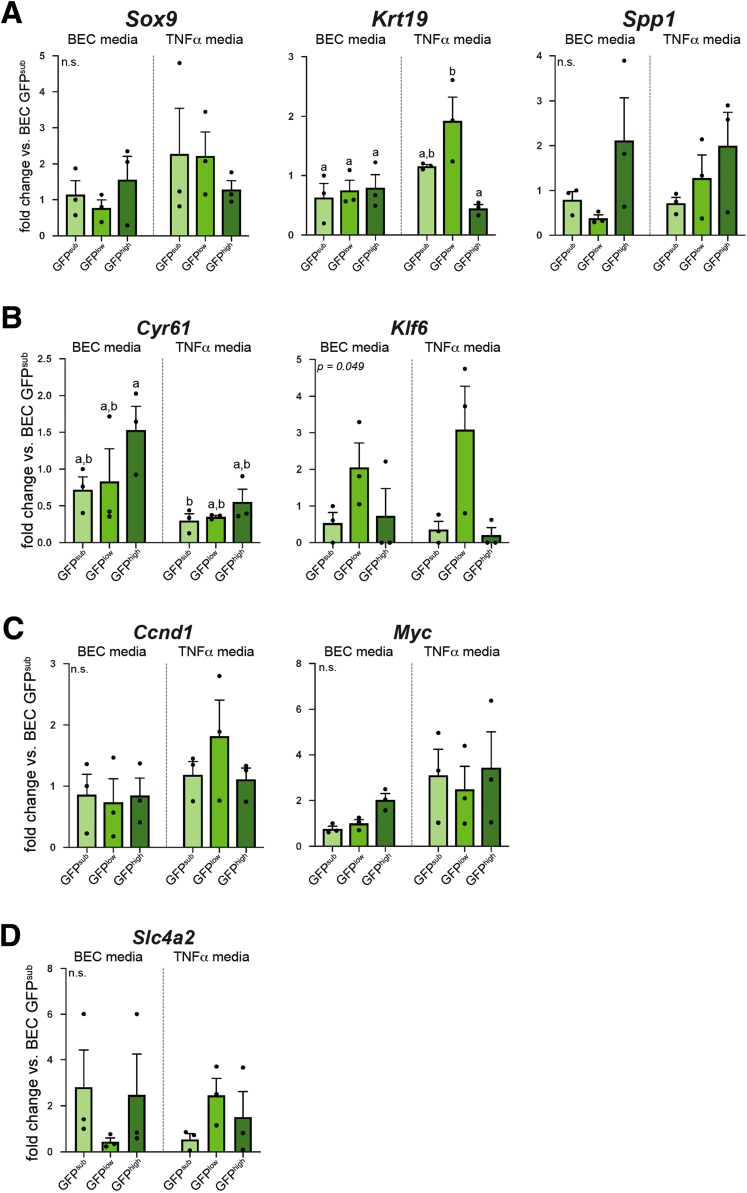
To determine whether organoids derived from different Sox9EGFP populations exhibited distinct gene expression profiles, we conducted qPCR on organoids grown in BEC and TNFα media for 7 days. Although we observed some trends in expression for most genes examined, variability between biological replicates resulted in few significant differences between Sox9EGFP populations or medium conditions. Whereas Sox9 and Spp1 were not differentially expressed, Krt19 remained up-regulated in GFPlow-derived organoids grown in TNFα media relative to all organoids grown in BEC media and GFPhigh-derived organoids grown in TNFα media (Figure 10A). Cyr61 was enriched in GFPhigh-derived BEC media organoids relative to GFPsub-derived TNFα media but was not significantly different between other populations and conditions (Figure 10B). In addition, Klf6 trended toward enrichment in GFPlow-derived organoids grown in both medium conditions (Figure 10B). Despite increased organoid size and the use of WNT agonist CHIR99021 in TNFα media, Wnt targets Ccnd1 and Myc were not significantly up-regulated in organoids grown in TNFα media, consistent with recent reports of WNT-independence in biliary organoids (Figure 10C).31 To determine whether BECs isolated from distinct Sox9EGFP populations produced organoids enriched for small or large cholangiocyte-associated genes, we examined expression of Cftr, Hrh1, Sctr, and Slc4a2. Of these, only Slc4a2 was detected by RT-qPCR. Slc4a2 demonstrated trends in expression between different Sox9EGFP populations and medium conditions, but no significant differences were observed, likely because of variability between biological replicates (Figure 10D). Notably, recent studies of biliary organoid development from single cells also demonstrated repression of some biliary markers at early time points, suggesting that culture conditions may impact expression of some functional cholangiocyte genes.30
Figure 10.
Biliary genes exhibit similar expression across organoids derived from different Sox9EGFPsubpopulations. (A) Sox9 and Spp1 are not differentially expressed in BEC or TNFα media, but Krt19 is enriched in GFPlow-derived organoids relative to GFPhigh-derived organoids in TNFα media only, exhibiting an expression pattern similar to in vivo results. (B) Cyr61 is significantly up-regulated in GFPhigh-derived BEC media organoids relative to GFPsub-derived TNFα media. Although Klf6 is not significantly regulated between populations and conditions, it trends toward increased expression in GFPlow-derived organoids in both BEC and TNFα media. (C) WNT target genes Myc and Ccnd1 are not differentially expressed between populations and conditions (letters indicate grouping by significance, P < .05, one-way analysis of variance and Tukey test; n.s. = not significant).
Our results demonstrate that organoid-forming capacity is restricted to Sox9EGFP populations but suggest that GFPsub, GFPlow, and GFPhigh cells perform similarly in organoid assays in terms of survival and gene expression. Our data also demonstrate that the most pronounced differences in organoid survival and size were found between medium conditions rather than Sox9EGFP populations. Furthermore, significant interpopulation differences between organoid survival, size, and Krt19 expression were found exclusively in TNFα media, suggesting that functional heterogeneity of BECs may be driven by exogenous conditions.
Sox9EGFP Populations Demonstrate Different Rates of Transdifferentiation
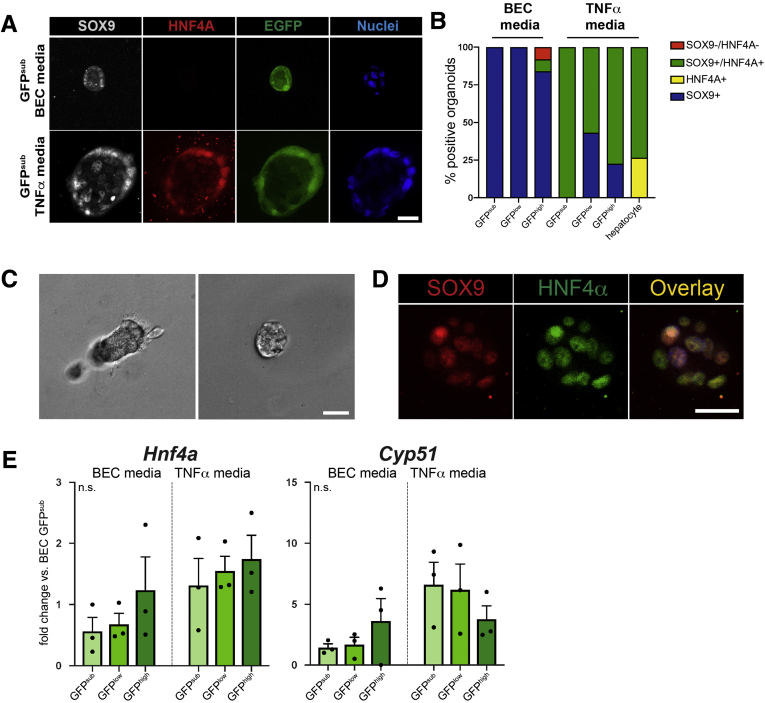
Previous reports have indicated that BECs are capable of transdifferentiating into hepatocytes, and vice versa, in defined organoid culture systems.28,32 Because TNFα media conditions were developed for long-term hepatocyte organoid culture, we reasoned that BEC vs hepatocyte fate decisions might differ between BEC media and TNFα media. To assay cellular identity, we examined protein expression of SOX9 and hepatocyte-specific transcription factor HNF4A by whole-mount immunofluorescence in single cell-derived Sox9EGFP organoids after 7 days of culture in BEC or TNFα media (Figure 11A). Hepatocytes were isolated by collagenase perfusion and grown in TNFα media as positive controls for hepatocyte identity and HNF4A expression (Figure 11C and D). Organoids produced by single GFPsub and GFPlow cells expressed SOX9 exclusively in BEC media, whereas a small number of GFPhigh-derived organoids (8.0%) co-expressed SOX9 and HNF4A (Figure 11B, Table 1). Interestingly, rare GFPhigh-derived organoids were also observed to be negative for both SOX9 and HNF4A, consistent with SOX9–/GFP+ BECs observed in vivo.
Figure 11.
HNF4A expression in single cell derived organoids is dependent on culture conditions and Sox9EGFPpopulation. (A) Representative images of SOX9 and HNF4A immunofluorescence in organoids derived from single Sox9EGFP populations (scale bar = 25 μm). (B) SOX9 and HNF4A expression is dependent on both medium conditions and Sox9EGFP population, with TNFα media driving increased expression of HNF4A in GFPsub, low, and high-derived organoids, but at distinct rates (n per group in Table 1). (C) Representative images of hepatocyte organoids after 7 days in culture (scale bar = 25 μm). (D) A majority of hepatocyte organoids express both SOX9 and HNF4A in vitro (scale bar = 25 μm). (E) Hnf4a and Cyp51 gene expression is not significantly different in organoids derived from different Sox9EGFP populations or between medium conditions (P > .05, one-way analysis of variance and Tukey test; n.s. = not significant).
Table 1.
Organoid Quantification for SOX9/HNF4A Co-localization (Related to Figure 11B)
| Media condition | |||||||
|---|---|---|---|---|---|---|---|
| BEC |
TNF |
||||||
| Sox9EGFP population |
Hepatocytes | ||||||
| GFPsub | GFPlow | GFPhigh | GFPsub | GFPlow | GFPhigh | ||
| SOX9–/HNF4A– | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| SOX9+/HNF4A+ | 0 | 0 | 4 | 27 | 25 | 41 | 27 |
| SOX9+/HNF4A– | 5 | 23 | 42 | 0 | 19 | 12 | 0 |
| SOX9–/HNF4A+ | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| Total organoids | 5 | 23 | 50 | 27 | 44 | 53 | 30 |
Single cells grown in TNFα media produced organoids that were more likely to express HNF4A relative to single cells grown in BEC media, regardless of which Sox9EGFP population they were derived from (Figure 5E). Strikingly, although GFPsub-derived organoids did not express HNF4A in BEC media, 100% co-expressed SOX9 and HNF4A in TNFα media. SOX9 and HNF4A were also co-expressed in a larger proportion of GFPhigh-derived organoids (77.4%) in TNFα media versus GFPlow (56.8%) (Figure 11B). Only collagenase-isolated hepatocytes produced organoids expressing HNF4A in the absence of SOX9 (Figure 11B). Next, we examined gene expression of Hnf4a and Cyp51, which were previously reported to be up-regulated in Lgr5EGFP+ liver organoids in hepatocyte media conditions.28 In contrast to HNF4A protein expression, neither gene was differentially expressed across Sox9EGFP populations or medium conditions (Figure 11E). These data demonstrate that organoids derived from Sox9EGFP populations exhibit varying potential to express HNF4A and that HNF4A expression is enhanced in all Sox9EGFP populations by TNFα media conditions. Our gene expression data suggest that HNF4A activation in TNFα media is regulated at the post-transcriptional level.
Sox9 Expression Is Maintained by Yap but Is not Modulated by Bile Acids
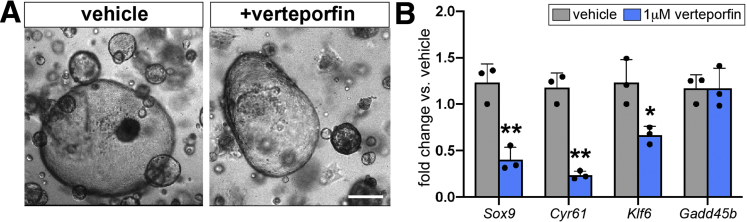
Sox9 is known to be activated downstream of Yap activity in diverse cellular contexts, including developing and regenerating hepatocytes and esophageal cancer cells.33,34 Because we observed up-regulation of Cyr61 and Hes1 in GFPhigh cells, we hypothesized that Sox9EGFP may serve as readout for Yap in BECs. To test whether biliary Sox9 expression is Yap-dependent, we treated organoids isolated from whole bile ducts with verteporfin, a small molecule inhibitor of Yap.35 Verteporfin treatment did not result in appreciable changes to organoid morphology (Figure 12A). As expected, we observed a significant decrease in the expression of Yap target genes Cyr61 and Klf6, as well as Sox9 (Figure 12B). Gadd45b, another known target of Yap signaling, remained unchanged, suggesting differential regulation of canonical Yap targets in biliary epithelium (Figure 12B).
Figure 12.
Sox9 expression in BECs is maintained by Yap. (A) Verteporfin treatment does not impact organoid morphology (scale bar = 50 μm) but (B) results in significant down-regulation of Sox9 and established BEC Yap targets, Cyr61 and Klf6, whereas Gadd45b is unaffected (∗ indicates significance, P < .05; ∗∗ indicates significance, P ≤ .01, unpaired t test).
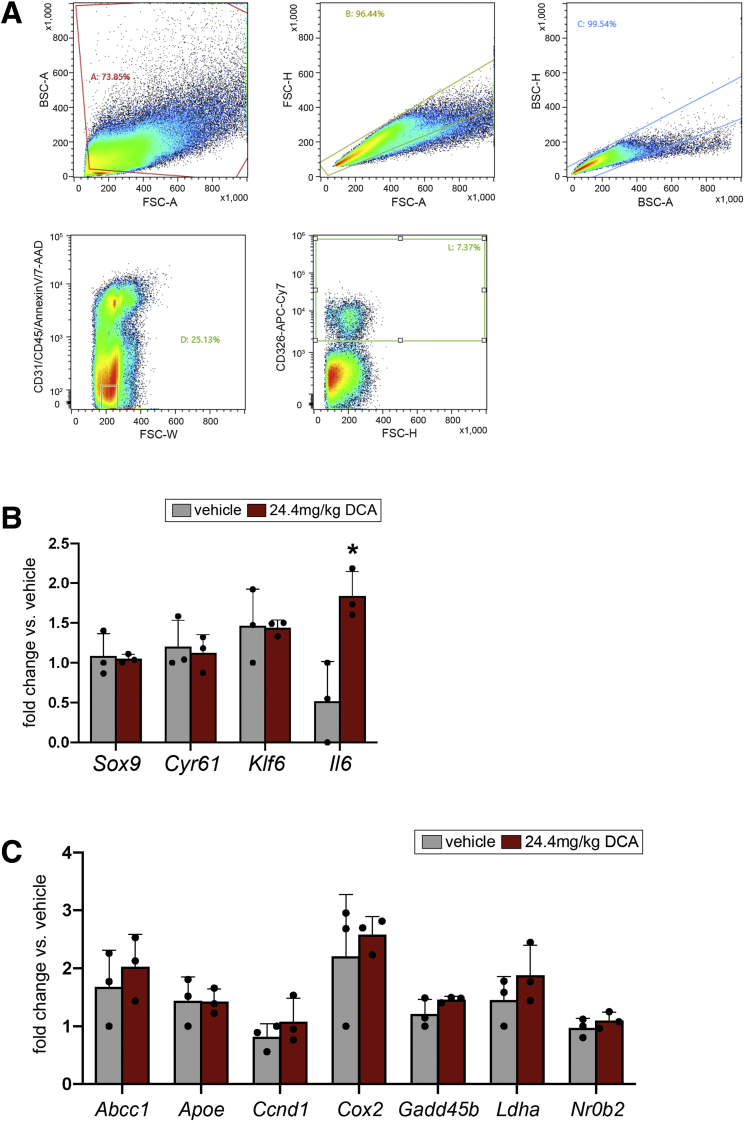
Recent studies have shown that bile acids induce Yap in both hepatocytes and BECs.6,36 To test whether biliary Sox9 is also induced by bile acids upstream of Yap, we administered deoxycholic acid (DCA) to wild-type mice via a single intraperitoneal injection and analyzed gene expression in FACS-isolated EPCAM+ BECs 24 hours later (Figure 13A).37 We found that neither Sox9 nor Yap targets Cyr61 or Klf6 were responsive to DCA in vivo (Figure 13B). To identify a BEC-specific positive control gene and validate DCA treatment, we assayed a panel of putative targets of farnesoid X receptor that were identified by published farnesoid X receptor ChIP-seq and expressed in GFPlow or GFPhigh BECs in our RNA-seq.38 Although most candidate target genes were unresponsive to DCA, we observed significant up-regulation of Il6 in DCA-treated mice relative to vehicle controls, consistent with previous reports of nuclear factor kappa B induction of Il6 in BECs after bile acid treatment (Figure 13B and C).39
Figure 13.
Sox9 expression is not up-regulated by bile acids in vivo. (A) FACS isolation strategy for EPCAM+ BECs from vehicle and DCA-treated mice. (B) Sox9 and Yap target genes are not differentially expressed in EPCAM+ cells from mice treated with DCA for 24 hours, but Il6 is significantly up-regulated. (C) Candidate DCA targets that were not differentially expressed in isolated BECs after DCA administration in vivo (∗ indicates significance, P < .05, unpaired t test).
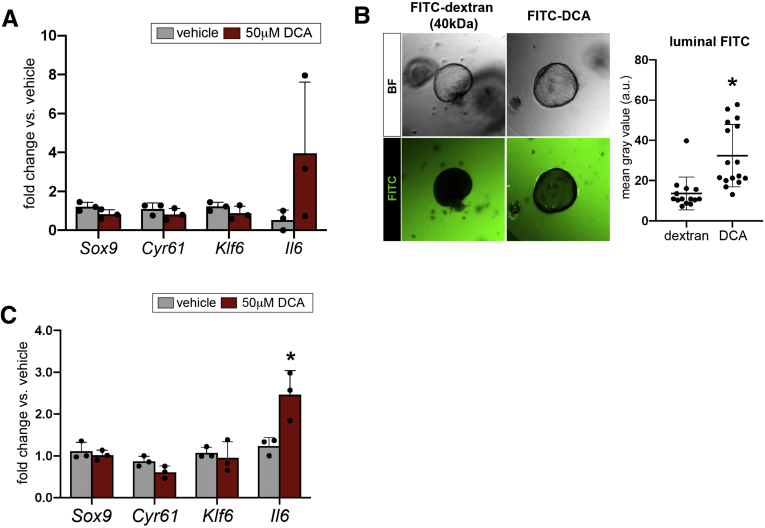
We reasoned that transcriptional responses to bile acid fluctuations in vivo might be subtle and next assayed BEC-autonomous responses to DCA by treating organoids in vitro. We did not observe a change in expression of Sox9 or Yap target genes at 24 hours of treatment, despite induction of Il6 in 2 of 3 replicates (Figure 14A). Because DCA-induced Yap was previously shown to be dependent on apical sodium-dependent bile acid transporter (ASBT), which is found on the apical membrane of BECs, we confirmed that DCA was interacting with apical cell surfaces via 2 independent experiments.6 First, we treated biliary organoids with fluorescein isithiocyanate (FITC)-conjugated DCA and measured luminal fluorescence 9 hours after treatment. Organoids treated with FITC-DCA exhibited significantly increased luminal fluorescence relative to controls treated with FITC-dextran, indicating that DCA is capable of crossing the basolateral membrane and entering organoid lumens (Figure 14B). Next, we passaged biliary organoids into monolayers, so apical membranes interact directly with overlaid media. As observed in our in vivo and organoid experiments, DCA failed to induce Cyr61, Klf6, or Sox9 in biliary monolayers, but Il6 was significantly up-regulated (Figure 14C). These data demonstrate that biliary Sox9 expression is maintained by Yap, but that both Yap activity and Sox9 are unaffected by DCA.
Figure 14.
Sox9 expression is not up-regulated by bile acids in vitro. (A) Biliary organoids treated with DCA for 24 hours reproduce in vivo results, failing to up-regulate Sox9, Cyr61, and Klf6 and demonstrating heterogeneous response in terms of Il6 up-regulation. (B) FITC-conjugated DCA was found in organoid lumens after 9 hours of treatment, whereas FITC-dextran was excluded (n = 16 FITC-dextran treated organoids, 14 FITC-DCA treated organoids; ∗ indicates significance, P = .004, unpaired t test). (C) Primary biliary monolayers treated with DCA in vitro also up-regulated Il6 but failed to demonstrate significantly different expression of Sox9, Cyr61, or Klf6 (∗ indicates significance, P < .05; ∗∗ indicates significance, P ≤ .01, unpaired t test).
Sox9EGFP Is Expressed in BECs and Peribiliary Hepatocytes After Cholestatic Injury
As proof-of-concept for the utility of the Sox9EGFP transgene in studying BECs after liver injury, we examined EGFP expression by histology 7 days after BDL. BDL is a well-established model of cholestatic injury that is associated with ductular reaction and activation of BEC phenotype in some peribiliary hepatocytes.40,41 We observed distinct levels of EGFP expression, along with expansion of EGFP+ BECs in portal fields and increase in GFPsub hepatocytes, consistent with ductular reaction to cholestasis (Figure 15). As in uninjured livers, all EPCAM+ and K19+ BECs co-expressed the EGFP transgene (Figure 15A and B), whereas a portion of EGFP+ cells with hepatocyte morphology and relatively low levels of EGFP co-expressed HNF4A (Figure 15C). Because Sox9 is known to be expressed in activated stellate cells and is associated with development of fibrosis after injury, we examined expression of alpha-smooth muscle actin and vimentin.42,43 We did not observe any co-localization between either marker and the Sox9EGFP transgene (Figure 15D and E, ≥10 portal fields from each of 3 mice 7 days after BDL). Finally, we assayed co-localization between EGFP and endogenous SOX9 protein. Although the overall number of SOX9-expressing EGFP+ cells was similar to uninjured livers (77.6% ± 13.6% after BDL vs 77.7% ± 7.1% uninjured), we noticed that SOX9–/EGFP+ cells appeared to exclusively demonstrate hepatocyte morphology (Figure 15F, yellow vs white arrowheads). We confirmed that all EPCAM+ BECs co-express SOX9 protein, which suggests that the only EGFP+ cells that do not express SOX9 after BDL are hepatocytes (Figure 15G). Together, these data demonstrate that Sox9EGFP expression levels are preserved after cholestatic liver injury induced by BDL, and that the EGFP transgene is expressed in similar cell populations in homeostasis and in the setting of injury/regeneration.
Figure 15.
Sox9EGFPexpression labels BECs and peribiliary hepatocytes in cholestatic injury after BDL. Immunofluorescence 7 days after BDL demonstrates that all (A) EPCAM+ and (B) K19+ BECs associated with typical ductular reactions co-express EGFP. (C) Consistent with results in uninjured livers, HNF4A is found in a subpopulation of Sox9EGFP+ peribiliary hepatocytes after BDL (yellow arrowheads indicate EGFP+/HNF4A+). BDL does not result in Sox9EGFP activation in (D) SMA+ or (E) VIM+ cells. (F) Although some Sox9EGFP+ cells do not express SOX9 after BDL (white arrowhead), those with ductal morphology appear SOX9+ (yellow arrowhead). (G) Independent co-localization of SOX9+ with EPCAM indicates that all BECs co-express SOX9 7 days after BDL (n = 100 cells from left lobes of each of 3 biological replicates per co-localization study; scale bars = 50 μm).
Discussion
Dissecting cellular heterogeneity is important for understanding how subpopulations of cells contribute to tissue homeostasis and disease. Although advances in single cell genomics provide tools for rapid and unbiased cataloguing of cell types, models that facilitate identification and manipulation of cell populations in intact tissues are still critical for understanding cellular function. Here, we characterize Sox9EGFP expression in the liver and establish a tool for understanding heterogeneity in BECs. Sox9EGFP is expressed at variable levels that are associated with phenotypically distinct cell populations in the small intestine, colon, and pancreatic duct.8,10,44 Our data demonstrate variability in Sox9EGFP expression levels throughout the intrahepatic bile ducts and periportal hepatocytes and confirm unique anatomic distribution, gene expression, and functional properties characteristic of each Sox9EGFP population. In the present study, we show that EGFP is not expressed in non-epithelial cells during homeostasis or at 7 days after BDL. However, the known activation of Sox9 in hepatic stellate cells in the setting of chronic fibrosis warrants confirmation of EGFP expression in other injury models.42,43 We also observe distinct Sox9EGFP expression levels in the extrahepatic bile ducts and gallbladder, suggesting that the transgene may be a useful biomarker for understanding cellular heterogeneity in these tissues as well. Notably, Sox9EGFP is expressed at variable levels in peribiliary glands, which have been proposed to house BECs with stem cell-like properties.12 Although the ubiquitous expression of Sox9EGFP and near-ubiquitous expression of endogenous SOX9 in EPCAM+ and K19+ ductal epithelium suggest that Sox9 itself is unlikely to be a specific biomarker of any one BEC subpopulation, distinct EGFP expression levels may be indicative of cellular heterogeneity throughout the biliary tree.
Unlike previous reports in the intestine, we find that Sox9EGFP does not accurately report endogenous SOX9 protein or mRNA levels. Instead, the highest levels of Egfp expression are associated with increased nascent Sox9 RNA. In addition, some GFP+ BECs do not express SOX9 protein during homeostasis. Because SOX9 is considered a ubiquitous marker of BECs and is required for BEC specification during development, this observation was unexpected but confirmed by co-localization of EPCAM and SOX9, demonstrating a similar number of SOX9- BECs.11 Interestingly, all BECs express SOX9 after BDL, suggesting that pan-biliary translation of Sox9 mRNA may play an important role in response to injury. The mechanistic significance of the regulation of Sox9 mRNA and protein expression, as well as its impact on adult BEC identity, function, and regeneration, warrants further investigation.
In contrast to other epithelial tissues, the biliary epithelium lacks clearly defined cell populations with compartmentalized functional properties (eg, stem, enteroendocrine, and absorptive cells in intestinal epithelium; type I and type II pneumocytes in alveolar epithelium). This lack of defined populations and general ambiguity of distinctions between cell types in the biliary epithelium complicate defining cell types captured within each Sox9EGFP population, as well as intrapopulation heterogeneity. The clearest distinction found in the present study is between GFPsub hepatocytes and GFPlow/high BECs. Sox9 expressing hybrid hepatocytes have been previously defined by their peribiliary anatomic location and co-expression of hepatocyte and BEC markers.15 GFPsub cells are localized to the peribiliary niche, co-express HNF4A in vivo, and demonstrate enrichment of BEC genes by transcriptomic signature analysis, consistent with hybrid hepatocytes. Interestingly, GFPsub-derived organoids exclusively expressed SOX9 in BEC media and always co-expressed SOX9 and HNF4A in TNFα media. Although the low rate of organoid formation precluded functional assays in the present study, these data suggest bipotency of GFPsub cells and their progeny. We also observed an increase in GFPsub hepatocytes after BDL, consistent with previous reports of Sox9 activation in peribiliary hepatocytes after injury to the bile ducts.41 Accordingly, the Sox9EGFP transgene may be a useful tool for differentially assessing BEC and hybrid hepatocyte phenotypes in models of liver injury and regeneration.
Transcriptomic differences are present between GFPlow and GFPhigh BEC populations but are more subtle relative to those observed between GFPsub and GFPlow/high populations. Interestingly, we find that GFPlow BECs express significantly higher levels of Krt19, a pan-biliary marker that has been associated with BEC maturation in induced pluripotent stem cell models.45 However, it is unclear whether differential Krt19 expression in vivo is indicative of different stages of hierarchical cell fate or simply heterogeneity across distinct, mature cell types. Our RNA-seq analysis also uncovered Wnt7a and Wnt7b, which were recently described as specific to a subpopulation of BECs in a DDC damage model, as uniquely enriched in GFPlow cells.6 Although we did not detect a significant difference in a previously published Yap gene signature between GFPlow and GFPhigh populations, we observed increased expression of Cyr61 and Hes1 in GFPhigh vs GFPlow BECs by RT-qPCR.21 Cyr61 and Hes1 have previously been identified as Yap-regulated markers of BEC heterogeneity.6 Taken in context of our verteporfin experiments demonstrating dependence of Sox9 expression on Yap signaling in organoids, these results suggest that high levels of Sox9EGFP expression in BECs are associated with increased Yap activity.
In organoid-forming assays using medium conditions developed for biliary organoids, both GFPlow and GFPhigh BECs behaved similarly in terms of organoid size and survival.28 It is known that despite heterogeneous Yap activity in vivo, BEC media conditions drive broad activation of Yap in vitro, which could lead to homogenization of organoid phenotypes.6 Compellingly, we find that TNFα media conditions result in organoid size differences between GFPlow- and GFPhigh-derived organoids, as well as increased initial organoid formation from GFPhigh cells. This may indicate that exogenous factors drive or repress functional heterogeneity in BECs. Like GFPsub cells, GFPlow and GFPhigh cells also demonstrated differential activation of HNF4A in TNFα media. Interestingly, GFPhigh-derived organoids were more likely to express HNF4A, tempting speculation that different subpopulations of BECs may be more likely to undergo transdifferentiation to hepatocytes. Because organoid culture conditions impact multiple signaling pathways and fail to perfectly recapitulate the in vivo environment, further work will be required to expand insight into how in vitro conditions affect BEC subpopulations.
BEC heterogeneity has been classically defined relative to cell size, with small cholangiocytes residing in small ducts and large cholangiocytes residing in large ducts.5 In this study, we classified ductules as biliary structures with an inner diameter <10 μm and ducts as ≥10 μm, as defined in recent three-dimensional studies of ductal morphology.2 Although we demonstrate that GFPhigh BEC numbers are greater in small ductules relative to larger ducts, our data also show that GFPhigh BECs in ducts are significantly larger than those in ductules. Together, this suggests that the GFPhigh population consists of BECs that could be classified as small or large cholangiocytes by classical definitions. Although we do not observe size differences in GFPlow BECs located in ductules vs ducts, it is possible that such heterogeneity exists between these smaller structures and rare large ducts leading to the extrahepatic biliary tree, which were not specifically examined in the current study because of their relative scarcity in histologic sections. Future studies may benefit from small cytoplasmic RNA-seq to further tease apart heterogeneity within Sox9EGFP populations. However, the lack of distinct clustering in BEC small cytoplasmic RNA-seq by t-Distributed Stochastic Neighbor Embedding suggests that deep sequencing would be required to detect differences between subpopulations driven by low-expressed genes.6,31 Our organoid studies, which demonstrate increased HNF4A protein expression despite no change in Hnf4a gene expression, suggest that post-transcriptional regulation may significantly impact biliary identity. Along with our observation that some BECs are SOX9–, this result points to the potential of proteomic characterization to further resolve BEC heterogeneity. We also show that interpopulation differences in organoid size and survival are found in TNFα media conditions but not in BEC media. This suggests that BEC functional heterogeneity, which may be associated with dynamic transcriptomic and proteomic heterogeneity, could be context-dependent and driven by changes to the biliary “niche”. The Sox9EGFP model presented here will provide a platform for further exploration of such heterogeneity in biliary homeostasis, injury, and regeneration.
Methods
Mice
Sox9EGFP mice were previously developed and characterized.8,46 All Sox9EGFP mice were heterozygous for the EGFP transgene and maintained on the C57Bl/6 background. All experiments were carried out on mice between 8 and 24 weeks of age. Male mice were used for gene expression studies, and male and female mice were used for immunofluorescence and organoid experiments. Mice were fed Tekland Global Soy Protein-Free Extruded Rodent Diet (Envigo, Indianapolis, IN; 2020SX) and received water ad libitum. For gene expression studies, mice were fasted overnight (12–14 hours) before tissue harvest. For in vivo DCA administration, male C57Bl/6 mice were given 24.4 mg/kg DCA in phosphate-buffered saline (PBS) (pH 7.4) or PBS only via a single intraperitoneal injection, as previously described.37 DCA- and vehicle-treated mice were fasted 16 hours and euthanized 24 hours after treatment. Sox9EGFP mice were phenotyped by observing EGFP expression in tail clippings by epifluorescent microscopy.8 The Institutional Animal Care and Use Committees of the University of North Carolina at Chapel Hill and Emory University reviewed and approved all animal use protocols.
Bile Duct Ligation
BDLs were carried out by the UNC Animal Surgery Core and the Emory University Pediatric Animal Physiology Core in accordance with Institutional Animal Care and Use Committee approvals, as previously described.47 Briefly, 8- to 24-week-old Sox9EGFP mice were anesthetized with 1%–3% isoflurane in 100% oxygen and subjected to ∼2-cm midline laparotomy using sterile surgical scissors. The common bile duct was exposed by gentle lifting of liver and caudal movement of the gut by using a sterile swab moistened with sterile 0.9% sodium chloride. The common bile duct was dissected from the portal vein using microserrated forceps and ligated with nonabsorbable 5-0 polyester suture, with a second cranial ligation placed in the same manner to thoroughly occlude the duct. The peritoneal cavity was rinsed with sterile 0.9% sodium before closing the peritoneum and skin with separate running sutures (6-0 monofilament, nonabsorbable). Sham-operated mice underwent the same procedure without ligation of the common bile duct. BDL and sham mice were allowed to recover on heating pads and monitored twice daily for 2 days after surgery and then once daily for the remainder of the study. No adverse effects or unexpected mortality were observed in either group. Analgesia consisted of 0.1 mg/kg buprenorphine given once perioperatively and twice daily postoperatively for 2 days after surgery. Mice were euthanized 7 days after BDL, and livers were processed for histology by intracardiac perfusion of 4% paraformaldehyde (PFA) in PBS, as described below.
Immunofluorescence
Livers were fixed by intracardiac perfusion of 4% PFA in PBS, followed by incubation overnight in 4% PFA in PBS at 4°C. Tissue was transferred to 30% sucrose and incubated overnight at 4°C. Livers were dissected into individual lobes, embedded in Tissue-Tek Optimal Cutting Temperature media (Sakura Finetek USA, Torrance, CA), frozen, and cut into 10-μm sections. Sections were stored at –80°C until use.
Immunofluorescence was carried out as previously described, and all steps were carried out at room temperature unless otherwise noted.9 Sections were permeabilized with 0.3% Triton X-100 in PBS for 20 minutes and then blocked in 1X Animal-Free Blocking Solution (Cell Signaling Technology, Danvers, MA; 15019) for 30 minutes. Primary antibodies were applied in PBS for 2 hours at room temperature or overnight at 4°C. Slides were rinsed in PBS 3 times for 5 minutes. Secondary antibodies were applied in PBS for 45 minutes, and slides were rinsed in PBS 3 times for 5 minutes. Nuclei were stained with bisbenzimide (Sigma-Aldrich; B2883), diluted 1:1000 in PBS for 10 minutes, before a final wash in PBS 3 times for 5 minutes. Slides were mounted to coverslips using Hydromount Mounting Media (Electron Microscopy Sciences, Hatfield, PA; 17966). For semiquantitative confocal analysis of EGFP, slides were mounted using ProLong Gold Mounting Media (Thermo Fisher, Waltham, MA; P36934). For mouse-on-mouse detection, tissue sections were incubated in 0.13 mg/mL goat anti-mouse Fab fragments for 1 hour at room temperature or overnight at 4°C immediately before primary antibody incubation. Antibodies used in this article are listed in Supplementary Table 4. For co-labeling with WGA, tissue was incubated for 10 minutes in 1 mg/mL WGA-AlexaFluor647 (Thermo Fisher; W32466) in Hanks’ balanced salt solution before permeabilization. After WGA incubation, tissue was washed in Hanks’ balanced salt solution twice for 5 minutes.
Images were acquired by using an Olympus IX-81 (Tokyo, Japan) widefield epifluorescent microscope or Zeiss LSM700 (Carl Zeiss, Oberkochen, Germany) and Leica SP8 (Wetzlar, Germany) confocal microscopes. Confocal images of tissue were acquired as 1 μmol/L optical sections; confocal images of organoids were acquired as 5 μmol/L optical sections. Low magnification image of Sox9EGFP right lobe (Figure 1A) was acquired by tile scan imaging of fresh tissue immediately after dissection by using Olympus IX-81 widefield epifluorescent microscope fitted with 4× objective lens. Tile scanned images were merged by image stitching in Metamorph (Molecular Devices, San Jose, CA). Confocal imaging was carried out in the Microscopy Services Laboratory at UNC Chapel Hill and the Integrated Cellular Imaging Core at Emory University. Quantification of confocal images was carried out in ImageJ (National Institutes of Health, Bethesda, MA) with subtraction of background fluorescence. Statistical analyses were carried out in Prism 8.4.2 (GraphPad Software, San Diego, CA).
Intrahepatic Bile Duct Isolation
To isolate single BECs from intrahepatic bile ducts, we dissociated liver tissue as previously described with minor modifications.18 Briefly, liver lobes were dissected without removing extrahepatic duct and gallbladder to avoid contamination of intrahepatic biliary preps with extrahepatic biliary tissue. The tissue was rinsed in sterile Dulbecco PBS (Gibco, Gaithersburg, MD), minced (<0.5 cm2 pieces) with a razor blade, and transferred to 50-mL conical tube. Liver tissue pieces were rinsed twice with Liver Wash Buffer (DMEM-H [Gibco], 1% fetal bovine serum, 1% Glutamax [Gibco], 1% penicillin/streptomycin [Gibco]) by resuspending and allowing tissue to sediment before removing supernatant. After the second rinse, tissue was centrifuged at 600g at 4°C for 5 minutes to remove residual Liver Wash Buffer. The supernatant was discarded, and liver tissue pieces were resuspended in 10 mL of Dissociation buffer (Liver Wash Buffer with 0.15 mg/mL collagenase type XI [Sigma-Aldrich, St Louis, MO; C9407], 0.3 U/mL Dispase [Corning, Corning, NY; 354235], 200 μg/mL DNase [Sigma-Aldrich; DN25]), pre-warmed to 37°C. The tube was transferred to a rocking platform (60 rpm) and incubated at 37°C for 90 minutes with continuous shaking. The tube was retrieved every 30 minutes and vigorously pipetted 50 times using a P1000 micropipette set to 1 mL to aid in dissociation of bile ducts. After 90 minutes of digest, dissociated tissue was rinsed twice with 10 mL Liver Wash Buffer, followed by centrifugation at 200g at 4°C for 5 minutes. Supernatant was discarded after each wash by careful decanting so as not to disturb the cell pellet. The pellet was resuspended in 5 mL Red Blood Cell Lysis Buffer (BioLegend, San Jose, CA; 420301) diluted in sterile Molecular Grade Water (Corning), as per manufacturer protocol, and incubated on ice for 5 minutes with repeated agitation. Ten milliliters Dulbecco PBS (Gibco) was added to quench the lysis buffer, and the tube was centrifuged at 600g at 4°C for 5 minutes. Supernatant was discarded, and ductal fragments were resuspended in 500 μL Advanced Dulbecco modified Eagle medium (DMEM) (Gibco) for whole duct organoid culture or used for single cell dissociation.
Single BEC Dissociation and Flow Cytometry/FACS
Ductal fragments were resuspended in 1 mL TrypLE (Gibco) with 100 μg/mL Y-27632 (Selleck Chemicals, Houston, TX; S1049) for single cell dissociation. The tube was incubated at 37°C for 12 minutes with vigorous pipetting every 2 minutes to aid in dissociation of isolated ductal fragments. Ten milliliters Advanced DMEM (Gibco) was added to quench the TrypLE and stop the dissociation. Dissociated cells were passed through a 40-μm cell strainer into a new 50-mL conical tube and centrifuged at 600g at 4°C for 5 minutes. The supernatant was discarded, and cells were resuspended in 500 μL Sort Media (Advanced DMEM with 500 μL B27 supplement without vitamin A [Thermo Fisher], 250 μL N2 supplement [Thermo Fisher], 250 μL Glutamax [Gibco], 250 μL Hepes Buffer Solution [Gibco], 250 μL penicillin/streptomycin [Gibco], 0.1% Y-27632 100 μL/mL [Selleck Chemicals; S1049], 0.1% DNase [Sigma-Aldrich; DN25).
Single BECs were stained with anti-CD31-APC (1:100) (BioLegend) and anti-CD45-APC (1:100) (BioLegend) and incubated, covered, on ice for 45 minutes. Cells were rinsed with 3 mL Advanced DMEM (Gibco) and centrifuged at 600g at 4°C for 5 minutes. After decanting the supernatant, the cells were resuspended in 1.5 mL Sort Media. Five microliters of 7AAD (BioLegend; 420404) and 5 μL AnnexinV-APC (BioLegend; 640941) were added to resuspended cells to distinguish dead and dying cells, respectively. Cells were analyzed and collected by using a Sony SH800 fluorescence-activated cell sorter. Gating schemes are shown in Figures 6A and 13A. For gene expression experiments, cells were collected directly into 500 μL RNA Lysis Buffer (Ambion RNAqueous Micro Kit, Austin, TX; AM1931); for organoid culture experiments, cells were collected directly into Sort Media.
RNA Isolation and RT-qPCR
FACS-isolated cells and organoids for gene expression studies were lysed in RNA Lysis Buffer, and RNA was isolated using the RNAqueous Micro Kit (Ambion) following manufacturer instructions. cDNA was generated with the iScript cDNA Synthesis Kit (BioRad, Hercules, CA; 1708891) and diluted 1:10 in molecular grade H2O (Corning) for RT-qPCR. RT-qPCR was carried out using Taqman probes and SsoAdvanced Universal Probes Supermix (BioRad; 1725280). To detect nascent Sox9 RNA, primers were designed spanning the junction of Sox9 exon 2-intron 2 using Primer-BLAST.48 Primers were validated to have efficiency between 90% and 110% and produced a single PCR product that was validated by Sanger sequencing (Eurofins Genomics, Luxembourg). RT-qPCR for nascent Sox9 was carried out using SsoAdvanced Universal SYBR Green Supermix (BioRad; 1725270). Relative fold change of gene expression was calculated using the delta-delta CT method, with 18S as the internal reference gene.49 Statistical analyses were carried out in Prism 8.4.2 (GraphPad). Taqman assay IDs and Sox9 primer sequences used in this article are listed in Supplementary Table 5.
RNA-seq and Analysis
Libraries for RNA-seq were prepared as previously described.9 Twelve thousand cells per Sox9EGFP population were collected directly into 500 μL RNA Lysis Buffer (Ambion RNAqueous Micro Kit), and RNA was isolated as described above, with an adjustment to the protocol so that final volume of eluate is equivalent to 15 mL. cDNA was prepared from 5 mL RNA, which was used to validate FACS isolation by RT-qPCR for Egfp (Life Technologies, Carlsbad, CA) using 18S as the internal reference gene. One microliter of RNA from each sample was used to assess RNA integrity number by Bioanalyzer (Agilent, Santa Clara, CA) using the RNA Pico assay. RIN values were ≥7.0 for all RNA-seq samples. Libraries were prepared from 8 mL RNA using the SMARTer Stranded Total RNA-seq Pico kit v2 (Clontech, Mountain View, CA) per manufacturer instructions. Libraries were pooled in equimolar ratios using concentrations determined by Qubit 3.0 High Sensitivity DNA assay (Life Technologies) and Bioanalyzer (Agilent) using the High Sensitivity DNA Analysis kit. Libraries were sequenced on a NextSeq500 (Illumina, San Diego, CA) with 75 base pair single-end reads, v2 chemistry.
Transcript abundances were estimated using Salmon (1.1.0) indexed to Gencode mouse annotations v24 and collapsed to gene counts using tximport (1.14.2).50,51 Differential expression analysis was performed using DESeq2 (1.26.0).52 Heatmaps were generated using ComplexHeatmap (2.2.0) and the Z-scored normalized count values. Signature scores were derived using the singscore R package (1.6.0)19; data.20, 21, 22 P-values for gene set signature scores are presented in Supplementary Table 3. All code used in these analyses available at https://doi.org/10.5281/zenodo.3858321. RNA-seq data are deposited in GEO under series number: GSE151387.
Primary Hepatocyte Isolation
Hepatocyte isolation was adapted from a previously published protocol.53 Briefly, mice weighing 20–35 g were anesthetized with Nembutal (40–60 mg/kg, intraperitoneal). Abdominal cavity was opened, and portal vein was cannulated (24-gauge × ¾-inch Terumo Surflo ETFE IV catheter; Fisher Scientific, Hampton, NH). The catheter was connected to a perfusion system for cell isolation from mouse organs (Cat #73-3659; Harvard Apparatus, Holliston, MA). Liver was perfused with 50 mL of 37°C sterile buffer I solution (50 mmol/L EGTA, 1 mol/L glucose, 1% penicillin/streptomycin in calcium- and magnesium-free Hank’s balanced salt solution) at a rate of 7 mL/min. At this time, inferior vena cava was cut. Subsequently, liver was digested with 40 mL of 37°C sterile buffer II solution (1 mol/L CaCl2, 1 mol/L glucose, 1% penicillin/streptomycin, and 3600 U Collagenase IV in calcium- and magnesium-free Hank’s balanced salt solution) at a rate of 5 mL/min. Livers were surgically removed, and hepatocytes were released in cold isolation medium (1X DMEM, 1% penicillin/streptomycin) by removing the Glisson’s capsule using sterile tweezers. Hepatocytes were isolated by size exclusion using 100-mm and 70-mm filters, respectively. Cells were washed at 120g for 5 minutes at 4°C. Live/dead cell exclusion was performed by Percoll gradient (1:1 1X Percoll/isolation medium) and centrifugation at 120g for 5 minutes at 4°C. Hepatocytes were resuspended in isolation medium.
Biliary and Hepatocyte Organoid Culture
Whole duct culture and bile acid treatment
Ten microliter aliquots from ductal preps were examined by light microscopy to qualitatively determine cellular density. Five to ten microliters of ductal prep was diluted in Advanced DMEM/F12 (Gibco), and Cultrex Type II Growth Factor Reduced extracellular matrix (R&D Biosystems, Minneapolis, MN; 3533-010-02) was added to a final concentration of 66% Cultrex. Cultrex-duct suspensions were plated as 40-μL droplets per well in pre-warmed 48-well plates and allowed to polymerize at 37°C for 20 minutes. After polymerization, droplets were overlaid with 200 μL Biliary Expansion Media (50% Advanced DMEM/F12 [Gibco], 40% WNT3A-conditioned media, 10% RSPO1-conditioned media, B27 supplement without vitamin A [Thermo Fisher], N2 supplement [Thermo Fisher], Glutamax [Gibco], 10 mmol/L HEPES [Gibco], penicillin/streptomycin [Gibco], 50 ng/mL recombinant murine EGF [Gibco; PMG8043], 100 ng/mL recombinant human Noggin [Peprotech, Rocky Hill, NJ; 120-10C], 100 ng/mL recombinant human FGF10 [Peprotech; 100-26], 10 μmol/L recombinant human gastrin [Sigma-Aldrich; G9145], 50 ng/mL recombinant human HGF [Peprotech 100-39H], 10 mmol/L nicotinamide, and 10 μmol/L Y-27632 [Selleck Chemicals]). Media were replaced every other day. Starting on day 4, WNT3A-conditioned medium was removed and replaced with Advanced DMEM/F12, and Noggin and Y-27632 were withdrawn from culture.
For bile acid and verteporfin organoid experiments, ductal organoids were passaged twice before treatment. Organoids were grown for 7 days before each passage and passaged by removing medium and adding 250 μL TrypLE (Gibco). Cultrex droplets were mechanically dissociated in TrypLE by pipetting and then incubated at 37°C for 3 minutes. TrypLE was quenched by adding 250 μL Advanced DMEM/F12, and organoid fragments were pelleted at 6000g for 5 minutes at room temperature. Organoid fragments were re-plated at 1:2 ratio as 40-μL droplets (66% Cultrex: 34% Advanced DMEM/F12) in pre-warmed 48-well plates, as described above. For monolayer experiments, organoid fragments were passaged onto 48-well plates coated with 10% Cultrex at passage 2. After each passage, organoids and monolayers were initially grown in Biliary Expansion Media with WNT3A, Noggin, and Y27632 for 4 days. WNT3A, Noggin, and Y27632 were removed from culture for 48 hours before treatment. Samples were treated with 50 μmol/L deoxycholic acid (Sigma-Aldrich), 1 μmol/L verteporfin (Tocris Bioscience, Bristol, UK; 5305), or an equivalent volume of dimethyl sulfoxide for 24 hours before lysis in 500 μL of RNA Lysis Buffer (Ambion RNAqueous Micro Kit). To determine bile acid presence in organoid lumens, organoids were treated with 50 μmol/L FITC-conjugated DCA (E. Mash, University of Arizona) or 40 kDa FITC-conjugated dextran (Sigma-Aldrich; FD40S).54 Organoid images were acquired as 1 μmol/L optical sections on a Zeiss LSM700 confocal microscope 9 hours after treatment with DCA or dextran.
Single cell culture
FACS-isolated BECs were resuspended in Cultrex Type II Growth Factor Reduced extracellular matrix (R&D Biosystems). For organoid survival and whole-mount immunofluorescence experiments, Cultrex cell suspensions were plated as 40-μL droplets per well in pre-warmed 48-well plates. For gene expression experiments, Cultrex cell suspensions were plated as 10-μL droplets per well to pre-warmed 96-well plates. All Cultrex droplets were allowed to polymerize at 37°C for 20 minutes. After polymerization, BEC or TNFα medium was overlaid: 200 μL per well for 48-well plates or 100 μL per well for 96-well plates. BECs were plated at density of 5000 cells per 40-μL Cultrex droplet per well (48-well plate) or 1200 cells per 10-μL Cultrex droplet per well (96-well plate). Primary hepatocytes were plated at a density of 10,000 cells per 40-μL Cultrex droplet per well (48-well plate).
Organoids grown in BEC media were initially overlaid with Biliary Expansion Media. Medium was replaced every other day. Starting on day 4, WNT3A-conditioned medium was removed and replaced with Advanced DMEM/F12, and Noggin and Y-27632 were withdrawn from culture. Organoids grown in TNFα conditions were overlaid with TNFα media (Advanced DMEM/F12 [Gibco], B27 supplement without vitamin A [Thermo Fisher], N2 supplement [Thermo Fisher], Glutamax [Gibco], 10 mmol/L HEPES [Gibco], penicillin/streptomycin [Gibco], 25 ng/mL recombinant murine EGF [Gibco], 50 ng/mL recombinant human HGF [Peprotech], 10 μmol/L Y-27632 [Selleck Chemicals], 1 μmol/L A8301 [Tocris; 2939], 3 μmol/L CHIR99021 [Selleck Chemicals; S1263], and 100 ng/mL recombinant murine TNFα [Peprotech; 315-01A]. Medium was replaced every other day.
Whole Mount Immunofluorescence
Organoids grown from single cells were fixed 7 days after plating as follows. All steps were carried out at room temperature unless noted. Overlay medium was removed, and 200 μL of 4% paraformaldehyde (VWR 41678) in Dulbecco PBS (pre-warmed at 37°C) was added to each well for 15 minutes to fix the organoids. Four percent paraformaldehyde was removed, and the organoids were rinsed with 200 μL PBS (Gibco) twice for 5 minutes. The organoids were permeabilized with 200 μL 0.5% Triton (Sigma-Aldrich; T8787) in PBS for 20 minutes and then rinsed with 200 μL 100 mmol/L glycine in PBS twice for 15 minutes. Organoids were incubated with 250 μL of Blocking Buffer (10% Normal Donkey Serum [Jackson Immunoresearch Laboratories, West Grove, PA; 017-000-121] in Immunofluorescence Buffer [Dulbecco PBS- 0.1% bovine serum albumin, 0.2% Triton, 0.05% Tween-20]) for 90 minutes. Primary antibodies were applied in 250 μL of Blocking Buffer and incubated overnight at 4°C.
Primary antibodies were removed, and organoids were washed with 250 μL Immunofluorescence Buffer 3 times for 20 minutes. Secondary antibodies were applied in 250 μL of Blocking Buffer and incubated for 2 hours. Secondary antibodies were removed, and organoids were washed with 250 μL Immunofluorescence Buffer 3 times for 20 minutes. Nuclei were stained with bisbenzimide (Sigma-Aldrich) diluted 1:1000 in Immunofluorescence Buffer for 30 minutes. Organoids were washed with Dulbecco PBS for 5 minutes 3 times and immediately quantified. SOX9 and HNF4A expression was observed by using an Olympus IX-81 inverted epifluorescent microscope.
Acknowledgments
The authors thank Drs Bailey Zwarcyz, Scott Magness, Susan Henning, Paul Dawson, and Terry Magnuson and members of the Magnuson Lab (UNC) for helpful discussion and feedback. FITC-DCA was a gift of Dr Eugene A. Mash (University of Arizona). Dr Joshua Maxwell and Ming Shen provided assistance with bile duct ligation surgery via the Children’s Healthcare of Atlanta and Emory University’s Pediatric Animal Physiology Core.
CRediT Authorship Contributions
Deepthi Y. Tulasi (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Visualization: Lead; Writing – review & editing: Supporting)
Diego Martinez Castaneda (Formal analysis: Supporting; Investigation: Supporting Visualization: Supporting; Writing – review & editing: Supporting)
Kortney Wager (Formal analysis: Supporting; Investigation: Supporting Methodology: Supporting; Visualization: Supporting)
Connor B. Hogan (Formal analysis: Supporting; Investigation: Supporting)
Karel P. Alcedo (Investigation: Supporting; Methodology: Supporting Writing – review & editing: Supporting)
Jesse R. Raab (Formal analysis: Supporting; Investigation: Supporting; Visualization: Supporting; Writing – review & editing: Supporting)
Adam David Gracz, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Supervision: Lead; Validation: Equal; Visualization: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the American Gastroenterological Association (Research Scholar Award to A.D.G.), the National Institutes of Diabetes and Digestive and Kidney Diseases (P30 DK34987 to R. Sandler, pilot award to A.D.G.), and the Emory University Department of Medicine (start-up funds to A.D.G.). K.P.A was supported by the National Cancer Institute (T32 CA071341). The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Supplementary Material
References
- 1.Maroni L., Haibo B., Ray D., Zhou T., Wan Y., Meng F., Marzioni M., Alpini G. Functional and structural features of cholangiocytes in health and disease. Cell Mol Gastroenterol Hepatol. 2015;1:368–380. doi: 10.1016/j.jcmgh.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamimoto K., Kaneko K., Kok C.Y., Okada H., Miyajima A., Itoh T. Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. Elife. 2016;5 doi: 10.7554/eLife.15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manco R., Clerbaux L.A., Verhulst S., Bou Nader M., Sempoux C., Ambroise J., Bearzatto B., Gala J.L., Horsmans Y., van Grunsven L., Desdouets C., Leclercq I. Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J Hepatol. 2019;70:1180–1191. doi: 10.1016/j.jhep.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O'Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., Kotelevtsev Y., Ffrench-Constant C., Boulter L., Forbes S.J. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpini G., Roberts S., Kuntz S.M., Ueno Y., Gubba S., Podila P.V., LeSage G., LaRusso N.F. Morphological, molecular, and functional heterogeneity of cholangiocytes from normal rat liver. Gastroenterology. 1996;110:1636–1643. doi: 10.1053/gast.1996.v110.pm8613073. [DOI] [PubMed] [Google Scholar]
- 6.Pepe-Mooney B.J., Dill M.T., Alemany A., Ordovas-Montanes J., Matsushita Y., Rao A., Sen A., Miyazaki M., Anakk S., Dawson P.A., Ono N., Shalek A.K., van Oudenaarden A., Camargo F.D. Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and gegeneration. Cell Stem Cell. 2019 doi: 10.1016/j.stem.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raab J.R., Tulasi D.Y., Wager K.E., Morowitz J.M., Magness S.T., Gracz A.D. Quantitative classification of chromatin dynamics reveals regulators of intestinal stem cell differentiation. Development. 2020;147(1) doi: 10.1242/dev.181966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramalingam S., Daughtridge G.W., Johnston M.J., Gracz A.D., Magness S.T. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol. 2012;302:G10–G20. doi: 10.1152/ajpgi.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B.Z., Jacquemin P., Pierreux C.E., Clotman F., Lemaigre F.P. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardinale V., Wang Y., Carpino G., Cui C.B., Gatto M., Rossi M., Berloco P.B., Cantafora A., Wauthier E., Furth M.E., Inverardi L., Dominguez-Bendala J., Ricordi C., Gerber D., Gaudio E., Alvaro D., Reid L. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology. 2011;54:2159–2172. doi: 10.1002/hep.24590. [DOI] [PubMed] [Google Scholar]
- 13.Rimland C.A., Tilson S.G., Morell C.M., Tomaz R.A., Lu W.Y., Adams S.E., Georgakopoulos N., Otaizo-Carrasquero F., Myers T.G., Ferdinand J.R., Gieseck R.L., 3rd, Sampaziotis F., Tysoe O.C., Wesley B., Muraro D., Oniscu G.C., Hannan N.R., Forbes S.J., Saeb-Parsy K., Wynn T.A., Vallier L. Regional differences in human biliary tissues and corresponding in vitro derived organoids. Hepatology. 2020 doi: 10.1002/hep.31252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong Y., Stanger B.Z. Molecular mechanisms of liver and bile duct development. Wiley Interdiscip Rev Dev Biol. 2012;1:643–655. doi: 10.1002/wdev.47. [DOI] [PubMed] [Google Scholar]
- 15.Font-Burgada J., Shalapour S., Ramaswamy S., Hsueh B., Rossell D., Umemura A., Taniguchi K., Nakagawa H., Valasek M.A., Ye L., Kopp J.L., Sander M., Carter H., Deisseroth K., Verma I.M., Karin M. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troeger J.S., Mederacke I., Gwak G.Y., Dapito D.H., Mu X., Hsu C.C., Pradere J.P., Friedman R.A., Schwabe R.F. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology. 2012;143:1073–1083 e22. doi: 10.1053/j.gastro.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser S.S., Gaudio E., Rao A., Pierce L.M., Onori P., Franchitto A., Francis H.L., Dostal D.E., Venter J.K., DeMorrow S., Mancinelli R., Carpino G., Alvaro D., Kopriva S.E., Savage J.M., Alpini G.D. Morphological and functional heterogeneity of the mouse intrahepatic biliary epithelium. Lab Invest. 2009;89:456–469. doi: 10.1038/labinvest.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.K., Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- 19.Foroutan M., Bhuva D.D., Lyu R., Horan K., Cursons J., Davis M.J. Single sample scoring of molecular phenotypes. BMC Bioinformatics. 2018;19:404. doi: 10.1186/s12859-018-2435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaub J.R., Huppert K.A., Kurial S.N.T., Hsu B.Y., Cast A.E., Donnelly B., Karns R.A., Chen F., Rezvani M., Luu H.Y., Mattis A.N., Rougemont A.L., Rosenthal P., Huppert S.S., Willenbring H. De novo formation of the biliary system by TGFbeta-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villanueva A., Alsinet C., Yanger K., Hoshida Y., Zong Y., Toffanin S., Rodriguez-Carunchio L., Sole M., Thung S., Stanger B.Z., Llovet J.M. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660–1669 e7. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halpern K.B., Shenhav R., Massalha H., Toth B., Egozi A., Massasa E.E., Medgalia C., David E., Giladi A., Moor A.E., Porat Z., Amit I., Itzkovitz S. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36:962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLeve L.D., Wang X., Hu L., McCuskey M.K., McCuskey R.S. Rat liver sinusoidal endothelial cell phenotype is maintained by paracrine and autocrine regulation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G757–G763. doi: 10.1152/ajpgi.00017.2004. [DOI] [PubMed] [Google Scholar]
- 25.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.P., Schwabe R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayyaz A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J., Chan K., Wrana J.L., Gregorieff A. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 2019;569:121–125. doi: 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- 27.Poling H.M., Mohanty S.K., Tiao G.M., Huppert S.S. A comprehensive analysis of aquaporin and secretory related gene expression in neonate and adult cholangiocytes. Gene Expr Patterns. 2014;15:96–103. doi: 10.1016/j.gep.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., Haft A., Vries R.G., Grompe M., Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng W.C., Logan C.Y., Fish M., Anbarchian T., Aguisanda F., Alvarez-Varela A., Wu P., Jin Y., Zhu J., Li B., Grompe M., Wang B., Nusse R. Inflammatory cytokine TNFalpha promotes the long-term expansion of primary hepatocytes in 3D culture. Cell. 2018;175:1607–1619 e15. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aloia L., McKie M.A., Vernaz G., Cordero-Espinoza L., Aleksieva N., van den Ameele J., Antonica F., Font-Cunill B., Raven A., Aiese Cigliano R., Belenguer G., Mort R.L., Brand A.H., Zernicka-Goetz M., Forbes S.J., Miska E.A., Huch M. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nature Cell Biology. 2019;21:1321–1333. doi: 10.1038/s41556-019-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planas-Paz L., Sun T., Pikiolek M., Cochran N.R., Bergling S., Orsini V., Yang Z., Sigoillot F., Jetzer J., Syed M., Neri M., Schuierer S., Morelli L., Hoppe P.S., Schwarzer W., Cobos C.M., Alford J.L., Zhang L., Cuttat R., Waldt A., Carballido-Perrig N., Nigsch F., Kinzel B., Nicholson T.B., Yang Y., Mao X., Terracciano L.M., Russ C., Reece-Hoyes J.S., Gubser Keller C., Sailer A.W., Bouwmeester T., Greenbaum L.E., Lugus J.J., Cong F., McAllister G., Hoffman G.R., Roma G., Tchorz J.S. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell. 2019;25:39–53 e10. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Hu H., Gehart H., Artegiani B., C LO-I, Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J., van den Born M., Zou C., Quirk C., Chiriboga L., Rice C.M., Ma S., Rios A., Peters P.J., de Jong Y.P., Clevers H. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606 e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Song S., Ajani J.A., Honjo S., Maru D.M., Chen Q., Scott A.W., Heallen T.R., Xiao L., Hofstetter W.L., Weston B., Lee J.H., Wadhwa R., Sudo K., Stroehlein J.R., Martin J.F., Hung M.C., Johnson R.L. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014;74:4170–4182. doi: 10.1158/0008-5472.CAN-13-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi J., Lu L., Yanger K., Wang W., Sohn B.H., Stanger B.Z., Zhang M., Martin J.F., Ajani J.A., Chen J., Lee J.S., Song S., Johnson R.L. Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology. 2016;64:1757–1772. doi: 10.1002/hep.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu-Chittenden Y., Huang B., Shim J.S., Chen Q., Lee S.J., Anders R.A., Liu J.O., Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anakk S., Bhosale M., Schmidt V.A., Johnson R.L., Finegold M.J., Moore D.D. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 2013;5:1060–1069. doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paolini M., Pozzetti L., Montagnani M., Potenza G., Sabatini L., Antelli A., Cantelli-Forti G., Roda A. Ursodeoxycholic acid (UDCA) prevents DCA effects on male mouse liver via up-regulation of CYP [correction of CXP] and preservation of BSEP activities. Hepatology. 2002;36:305–314. doi: 10.1053/jhep.2002.34939. [DOI] [PubMed] [Google Scholar]
- 38.Thomas A.M., Hart S.N., Kong B., Fang J., Zhong X.B., Guo G.L. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai J., Wang H., Dong Y., Zhang Y., Wang J. Bile acids affect the growth of human cholangiocarcinoma via NF-kB pathway. Cancer Invest. 2013;31:111–120. doi: 10.3109/07357907.2012.762781. [DOI] [PubMed] [Google Scholar]
- 40.Gadd V.L., Aleksieva N., Forbes S.J. Epithelial plasticity during liver injury and regeneration. Cell Stem Cell. 2020;27:557–573. doi: 10.1016/j.stem.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athwal V.S., Pritchett J., Llewellyn J., Martin K., Camacho E., Raza S.M., Phythian-Adams A., Birchall L.J., Mullan A.F., Su K., Pearmain L., Dolman G., Zaitoun A.M., Friedman S.L., MacDonald A., Irving W.L., Guha I.N., Hanley N.A., Piper Hanley K. SOX9 predicts progression toward cirrhosis in patients while its loss protects against liver fibrosis. EMBO Mol Med. 2017;9:1696–1710. doi: 10.15252/emmm.201707860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanley K.P., Oakley F., Sugden S., Wilson D.I., Mann D.A., Hanley N.A. Ectopic SOX9 mediates extracellular matrix deposition characteristic of organ fibrosis. J Biol Chem. 2008;283:14063–14071. doi: 10.1074/jbc.M707390200. [DOI] [PubMed] [Google Scholar]
- 44.Rezanejad H., Ouziel-Yahalom L., Keyzer C.A., Sullivan B.A., Hollister-Lock J., Li W.C., Guo L., Deng S., Lei J., Markmann J., Bonner-Weir S. Heterogeneity of SOX9 and HNF1beta in pancreatic ducts is dynamic. Stem Cell Reports. 2018;10:725–738. doi: 10.1016/j.stemcr.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sampaziotis F., de Brito M.C., Madrigal P., Bertero A., Saeb-Parsy K., Soares F.A.C., Schrumpf E., Melum E., Karlsen T.H., Bradley J.A., Gelson W.T., Davies S., Baker A., Kaser A., Alexander G.J., Hannan N.R.F., Vallier L. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 47.Tag C.G., Sauer-Lehnen S., Weiskirchen S., Borkham-Kamphorst E., Tolba R.H., Tacke F., Weiskirchen R. Bile duct ligation in mice: induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J Vis Exp. 2015;96:52438. doi: 10.3791/52438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim N.D., Moon J.O., Slitt A.L., Copple B.L. Early growth response factor-1 is critical for cholestatic liver injury. Toxicol Sci. 2006;90:586–595. doi: 10.1093/toxsci/kfj111. [DOI] [PubMed] [Google Scholar]
- 54.Jean-Louis S., Akare S., Ali M.A., Mash E.A., Jr., Meuillet E., Martinez J.D. Deoxycholic acid induces intracellular signaling through membrane perturbations. J Biol Chem. 2006;281:14948–14960. doi: 10.1074/jbc.M506710200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.