Abstract
Transcription is an elaborate process that is required to establish and maintain the identity of the more than two hundred cell types of a metazoan organism. Strict regulation of gene expression is therefore vital for tissue formation and homeostasis. An accumulating body of work found that ubiquitylation of histones, transcription factors, or RNA polymerase II is crucial for ensuring that transcription occurs at the right time and place during development. Here, we will review principles of ubiquitin‐dependent control of gene expression and discuss how breakdown of these regulatory circuits leads to a wide array of human diseases.
Keywords: histone modification, RNA polymerase II, transcription, ubiquitin
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Post-translational Modifications, Proteolysis & Proteomics
This review discusses the principles of ubiquitin‐dependent control of gene expression and how the dysfunction of these regulatory circuits results in diverse disease conditions.
Glossary
- βTrCP
beta‐transducin repeat‐containing protein
- APC/C
anaphase‐promoting complex/cyclosome
- ASCL
achaete‐scute homolog
- AXR
auxin‐resistance protein
- BAP
BRCA1‐associated protein
- BCL
B‐cell lymphoma
- BCOR
BCL6 co‐repressor
- BRD
bromodomain‐containing protein
- BRE
brefeldin A sensitivity
- BTB
Broad‐Complex, Tramtrack and Bric a brac
- CBP
CREB‐binding protein
- CDK
cyclin‐dependent kinase
- CK
casein kinase
- CRBN
protein cereblon
- CRY
cryptochrome
- CUL
cullin
- DDB
DNA damage binding
- DCAF
DDB1 and CUL4 associated factor
- DDI
DNA damage inducible homolog
- DEF
RNA polymerase II degradation factor
- DOT
disrupter of telomere silencing protein
- dTAG
degradation tag
- DUB
deubiquitylase
- ER
endoplasmic reticulum
- FACT
facilitates chromatin transcription
- FBXL
F‐box and leucine‐rich repeat protein
- FBXW
F‐box and WD‐40 domain‐containing protein
- GATA
GATA‐binding factor
- GID
gibberellin‐insensitive dwarf protein
- Gli
glioma‐associated oncogene
- GSK
glycogen‐synthase kinase
- HIF
hypoxia‐inducible factor
- HUWE
HECT, UBA and WWE domain‐containing protein
- IAA
indoleacetic acid‐induced protein
- IκBα
inhibitor of nuclear factor kappa B, alpha
- KANSL
KAT8 regulatory NSL complex
- KEAP
kelch‐like ECH‐associated protein
- LGE
Large cells
- MDM
mouse double minute homolog
- MGA
multicopy suppressor of Gam1
- MLL
mixed lineage leukemia
- MSL
male‐specific lethal
- MYC
myelocytomatosis
- NMYC
v‐myc avian myelocytomatosis viral oncogene neuroblastoma‐derived homolog
- MYSM
Myb‐like, SWIRM, and MPN domain‐containing protein
- NCoR
nuclear receptor co‐repressor
- NEDD
neural precursor cell expressed developmentally downregulated protein
- NF‐κB
nuclear factor of kappa light polypeptide gene enhancer in B‐cells
- NRF
nuclear respiratory factor
- PAF
RNA polymerase II‐associated factor
- PRC
polycomb repressive complex
- PROTAC
proteolysis‐targeting chimera
- RPB1
RNA polymerase II subunit B1
- RBX
RING‐box protein
- RING
really interesting new gene
- RNA Pol II
RNA polymerase II
- RNF
RING finger protein
- RPN
regulatory particle non‐ATPase
- RSF
remodeling and spacing factor
- RSP
reverses SPT‐phenotype protein
- SALL
Sal‐like protein
- SCF
Skp, Cullin, F‐box containing
- SETDB
SET domain bifurcated
- SIAH
seven in absentia homolog
- SMAD
Mothers against decapentaplegic homolog
- SMRT
silencing mediator for retinoid or thyroid‐hormone receptors
- SMURF
SMAD ubiquitination regulatory factor
- SOX
SRY (Sex determining region Y)‐box
- SPT
suppressor of Ty
- STAT
signal transducer and activator of transcription
- TBP
TATA‐box binding protein
- TC‐NER
transcription‐coupled nucleotide excision repair
- TCF/LEF
T‐cell factor/lymphoid enhancer‐binding factor
- TGF‐β
transforming growth factor‐β
- TIR
transport inhibitor response
- TLE
transducin‐like enhancer
- TP
tumor protein
- UBR
ubiquitin protein ligase E3 component n‐recognin
- USP
ubiquitin specific peptidase
- VCP
valosin‐containing protein
- VHL
von Hippel Lindau disease tumor suppressor
- WDR
WD repeat‐containing protein
- WNT
wingless‐related MMTV integration site 1
- WWP
WW domain‐containing protein
Introduction
Human bodies contain more than two hundred cell types that perform specialized functions and allow us to react to developmental or environmental challenges. Cell identity is established by the expression of tissue‐specific subsets of genes, which requires that differentiating cells guide RNA polymerase II (RNA Pol II) to select genomic loci (Djebali et al, 2012). While loss of transcription disrupts tissue formation and is incompatible with life, the erroneous execution of gene expression programs can lead to a plethora of developmental abnormalities and is often found at the heart of tumorigenesis (Lee & Young, 2013).
Accurate gene expression relies on transcription factors that bind to specific sequences in gene promoters or enhancers (Zaret & Carroll, 2011). These proteins can collaborate with co‐repressors and limit transcription by maintaining chromatin in a compact state (Venkatesh & Workman, 2015). Conversely, when gene expression needs to be stimulated, transcription factors attract co‐activators or chromatin remodelers that increase the accessibility of RNA Pol II for the respective genomic loci (Bulynko & O'Malley, 2011). After initiation of RNA synthesis, a distinct set of elongation factors helps RNA Pol II traverse histones within the coding region to allow production of a full‐length transcript (Chen et al, 2018a). Proteins with roles in transcription termination dissociate RNA Pol II from chromatin, which sets the complex free for another round of gene expression at a distinct locus (Proudfoot, 2016).
Ensuring that this sequence of events occurs at the right time and place, organisms tightly control chromatin architecture and fine‐tune the availability and function of transcription factors or RNA Pol II. Central for the underlying regulatory circuits is ubiquitylation, a posttranslational modification that was first documented for a histone with roles in transcriptional control (Goldknopf et al, 1975). Several aspects of its biochemistry and biology allow ubiquitylation to exert precise control over gene expression networks: cells can assemble several ubiquitin conjugates of different topologies that elicit distinct responses and hence provide an opportunity to employ similar chemistry for multiple transcriptional outputs (Fig 1A; Yau & Rape, 2016; Haakonsen & Rape, 2019). While, for example, attachment of a single ubiquitin typically changes protein interactions, polymers linked through K11 or K48 of one ubiquitin and the carboxy‐terminus of another trigger protein degradation (Chau et al, 1989; Jin et al, 2008). Cells can further adjust ubiquitin signaling to their needs by synthesizing more complex conjugates, such as branched ubiquitin chains used to dismantle stable protein complexes (Meyer & Rape, 2014; Yau et al, 2017; Oh et al, 2020). Ubiquitin conjugates can also be edited to modulate their output (Newton et al, 2008; Ohtake et al, 2018), or disassembled (Komander, 2009; Sahtoe & Sixma, 2015), which creates a cellular language for the dynamic control of gene expression in changing environments.
Figure 1. The ubiquitin modification system.
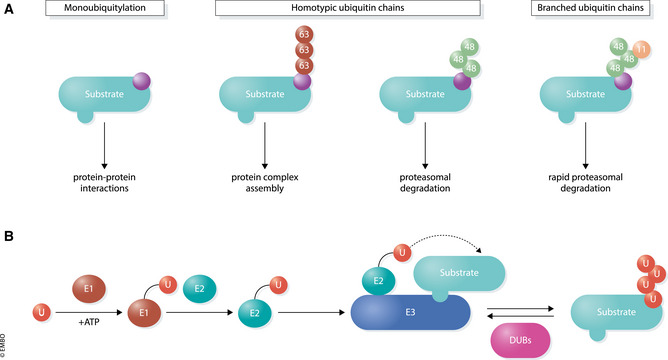
(A) Various structures and functions of different types of ubiquitin conjugates. Monoubiquitylation (left) involves transfer of a single ubiquitin to a substrate. E3 ligases can also connect several ubiquitin molecules together using the C‐terminus of one subunit and one of seven lysine (K) residues (K6, K11, K27, K29, K33, K48, K63) or the N‐terminal methionine (M1) on the other. Different ubiquitin topologies adopt distinct structural conformations which direct a wide array of substrate outcomes. (B) Ubiquitylation occurs through an enzymatic cascade. E1 enzymes use ATP to form a high‐energy thioester bond to ubiquitin through an active site cysteine residue. Charged E1s transfer ubiquitin to one of ~ 40 E2 ubiquitin‐conjugating enzymes, which then bind to one of ~ 600 E3 ubiquitin ligases that facilitate the transfer of ubiquitin onto a specific substrate. Approximately 100 DUBs remove ubiquitin from substrates to reverse the ubiquitylation process.
In humans, the writers of this ubiquitin code are ~ 600 E3 ligases that are counteracted by ~ 100 deubiquitylases (DUBs) (Fig 1B). Multiple effectors with ubiquitin‐binding domains decipher the type of ubiquitylation and couple the modification to its intended biological outcome. Many E3 ligases, DUBs, and effectors control gene expression, and their mutation is often deleterious for organismal development. Conversely, hijacking the ubiquitylation machinery with small molecules is beginning to be used to alter gene expression for therapeutic benefit. Here, we will review principles of ubiquitin‐dependent control of transcription and discuss how our increasing understanding of these processes is laying the foundation for therapeutic approaches to modulate gene expression in diseases of aberrant cell specification.
Ubiquitin‐dependent control of chromatin architecture
Transcription occurs within the context of chromatin whose basic unit is the nucleosome: an octamer of the core histones H2A, H2B, H3, and H4, wrapped around 147 base pairs of DNA. As nucleosomes can restrict the accessibility of the transcription machinery to promoter or enhancer sequences, their position on chromatin needs to be tightly regulated. While transcription factors can destabilize the interaction of genomic DNA with nucleosomes (Michael et al, 2020), they often recruit protein complexes with histone‐modifying or chromatin‐remodeling activities. As an essential histone modification, ubiquitylation plays a crucial role in the regulatory processes that shape chromatin architecture and, thereby, gene expression.
Histone H2A monoubiquitylation maintains transcriptional silencing
It is befitting to begin a review about ubiquitin‐dependent control of gene expression by discussing the modification of histone H2A, the first protein known to be ubiquitylated (Goldknopf et al, 1975). Monoubiquitylation of H2A at K119 is catalyzed by polycomb repressive complex 1 (PRC1) (Blackledge et al, 2014; Aranda et al, 2015) and plays a critical role in restricting gene expression at tissue‐specific loci, at the inactive female X chromosome, or at sites of DNA damage (Uckelmann & Sixma, 2017; Brockdorff, 2017). Underscoring the importance of this modification, loss of PRC1 is incompatible with life (Voncken et al, 2003), and mutations that impede its activity cause microcephaly or learning disabilities (Pierce et al, 2018). Aberrant regulation of PRC1 also results in tumors of the brain, liver, colon, breast, lung, prostate or lymphatic system (Grossniklaus & Paro, 2014; Wang et al, 2015).
PRC1 installs H2A monoubiquitylation at the proper sites by detecting CpG‐rich promoter sequences or chromatin‐associated noncoding RNAs (Brockdorff, 2017), which sets in motion a series of events leading to chromatin compaction (Bantignies et al, 2011; Denholtz et al, 2013; Isono et al, 2013) (Fig 2A): The monoubiquitylated H2A is recognized by PRC2 complexes, which methylate K27 on histone H3 (Kalb et al, 2014). This recruits SETDB1 to trimethylate K9 in histone H3 (Zhao et al, 2016), which serves as a landing platform for chromatin compactors, like HP1 (Nakagawa et al, 2008; Machida et al, 2018). Trimethylated H3K27 also attracts more PRC1 complexes to seed further H2A modification for stable long‐range signaling (Blackledge et al, 2014; Tamburri et al, 2020; Blackledge et al, 2020). Moreover, monoubiquitylated H2A can engage the linker histone H1 to establish repressive higher‐order chromatin folding (Jason et al, 2005; Fyodorov et al, 2018), a reaction that may involve the chromatin remodeler RSF1 (Zhang et al, 2017).
Figure 2. Transcriptional effects of histone ubiquitylation.
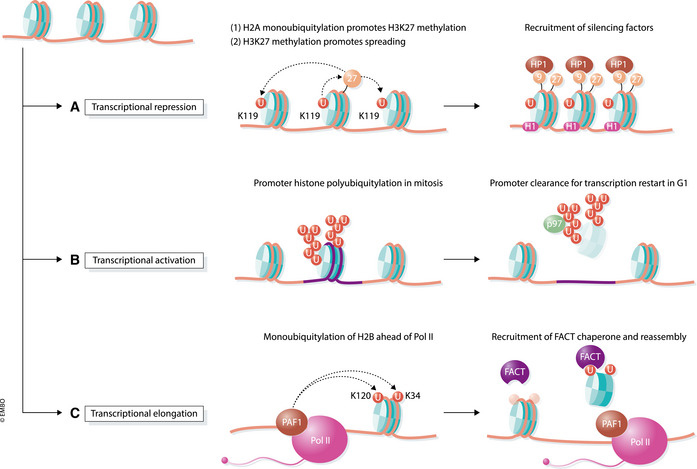
(A) Monoubiquitylation of histone H2A at K119 by PRC1 promotes transcriptional silencing. This modification recruits PRC2 to trimethylate histone H3 at K27, a mark which in turn recruits PRC1 to ubiquitylate additional H2A histones, resulting in spreading. H3K27me3 also leads to H3K9 trimethylation, which together with ubiquitylated H2A, recruit factors that compact chromatin and silence transcription. (B) Histone polyubiquitylation promotes transcriptional reactivation following cell division. During mitosis, the APC/C E3 ligase is recruited to specific promoters to polyubiquitylate histones, leading to their extraction by p97/VCP and proteasomal degradation. This action licenses these promoters for rapid transcriptional reactivation when cells enter G1. (C) Monoubiquitylation of histone H2B at K120 and K34 mediates transcriptional elongation. As part of the PAF1 elongation complex, the E3 ligases RNF20/RNF40 and MSL1/MSL2 travel with RNA Pol II and ubiquitylate H2B on nucleosomes obstructing RNA Pol II’s path. H2B ubiquitylation promotes nucleosome remodeling, through the FACT histone chaperone, to allow polymerase passage. Ubiquitin is then removed by H2B DUBs to reestablish chromatin behind RNA Pol II.
To allow for precise gene activation during development, the ubiquitylation of H2A needs to be reversible and multiple deubiquitylases target H2A to counteract PRC1‐mediated gene silencing. Most H2A‐directed deubiquitylases are subunits of co‐activators (Belle & Nijnik, 2014; Chittock et al, 2017), such as MYSM1 that associates with the histone acetyltransferase p300/CBP and helps release histone H1 from chromatin (Zhu et al, 2007). The H2A deubiquitylases USP16, BAP1 and MYSM1 derepress different sets of lineage‐specific genes in stem cells (Scheuermann et al, 2010; Yang et al, 2014; Gu et al, 2016; Campagne et al, 2019), while USP21 has a role in liver regeneration (Nakagawa et al, 2008). In addition to activating tissue‐specific genes, removal of ubiquitin from H2A increases the expression of cell cycle inhibitors to limit stem cell proliferation (Gatzka et al, 2015; Wilms et al, 2018). Aberrant H2A deubiquitylation can thus deplete stem cell populations, as observed in Down’s syndrome and its triplication of USP16 (Adorno et al, 2013). By contrast, inactivation of H2A deubiquitylases can set in place a differentiation block associated with more aggressive cancers and worsened patient outcomes (Carbone et al, 2013).
H2B monoubiquitylation facilitates transcription elongation
Contrary to the effects of H2A modification, monoubiquitylation of the neighboring nucleosome subunit, histone H2B, promotes gene expression (Fig 2C; Weake & Workman, 2008; Minsky et al, 2008). H2B monoubiquitylation at K120 is carried out by RNF20/RNF40, which is recruited to RNA Pol II as part of the polymerase‐associated factor 1 (PAF1) complex. The yeast homolog of RNF20/RNF40, BRE1, uses its partner LGE1 and liquid‐liquid phase separation to increase the local concentration of H2B for efficient modification (Gallego et al, 2020). H2B can also be monoubiquitylated at K34 by MSL1/MSL2 (Wu et al, 2011; Wu et al, 2014), which is similarly recruited to RNA Pol II by the PAF1 complex.
H2B monoubiquitylation stimulates transcription by multiple means. It disrupts higher‐order nucleosome assembly by forming a wedge between histone surfaces to prevent their tight packing (Fierz et al, 2011; Debelouchina et al, 2017). The modification of H2B may also destabilize the nucleosome itself (Krajewski et al, 2018), a function that is in part mediated by the FACT histone chaperone that transiently displaces H2A/H2B dimers (Pavri et al, 2006; Wang et al, 2018; Chen et al, 2018b). In addition, monoubiquitylated H2B acts as a platform to attract over ninety effectors, many of which are histone modifiers and chromatin remodelers (Shema‐Yaacoby et al, 2013). One intriguing effector is the histone H3 methyltransferase, DOT1L, which forms part of a histone crosstalk cascade (Worden et al, 2019; Worden et al, 2020). Monoubiquitylation of H2B stimulates DOT1L methylation of K79 of histone H3, which in turn promotes H3K4 methylation by MLL to recruit transcriptional activators (Wu et al, 2011; Fuchs & Oren, 2014; Levendosky et al, 2016; Valencia‐Sanchez et al, 2019). In this arrangement, H2B ubiquitylation nucleates downstream events that make chromatin more permissive for transcription elongation.
As observed with H2A, the modification of H2B is reversible, and several deubiquitylases remove ubiquitin from H2B to stabilize chromatin in its original state (Zhang et al, 2008; Zhao et al, 2008; Atanassov et al, 2016; Nune et al, 2019). For example, the deubiquitylase USP44 inhibits transcript elongation within the NCoR co‐repressor (Lan et al, 2016), and downregulation of USP44 accounts for an increase in H2B ubiquitylation at lineage‐specific genes during stem cell differentiation (Fuchs et al, 2012; Karpiuk et al, 2012; Chen et al, 2012). Conversely, USP22 opposes RNF20/RNF40 during differentiation of regulatory T cells to limit the immune system’s anti‐tumor activity (Cortez et al, 2020). While defects in H2B monoubiquitylation block cell fate specification, altered levels of ubiquitylated H2B are observed in many cancers (Wang et al, 2013; Fuchs & Oren, 2014; Dickson et al, 2016; Tarcic et al, 2016). Together, these observations underscore that the site of ubiquitylation can be critical for its effects on transcription: Although H2A and H2B are in close proximity within the nucleosome, their modification with ubiquitin has opposite consequences, i.e., inhibition or activation of gene expression.
Histone polyubiquitylation restarts gene expression after mitosis
While monoubiquitylation of histones has long been recognized to control chromatin architecture, histone polyubiquitylation has recently emerged as a means of transcriptional regulation during the cell cycle. As gene expression is downregulated during mitosis (Palozola et al, 2017), cells must restart their transcriptional programs every time they enter a new division cycle (Hsiung & Blobel, 2016). This process is facilitated by factors referred to as “mitotic bookmarks” that are deposited at active genes during the G2 phase and persist through the rigors of mitosis into the next G1 (Palozola et al, 2019). Despite chromatin condensation during mitosis, bookmarked loci remain accessible to the transcriptional machinery (Martinez‐Balbas et al, 1995; Michelotti et al, 1997; Teves et al, 2018).
Early proteins proposed to serve as mitotic bookmarks were transcription factors such as BRD4 (Dey et al, 2009), GATA1 (Kadauke et al, 2012), or SOX2 (Teves et al, 2016; Deluz et al, 2016). These included the TATA‐box binding protein TBP (Teves et al, 2018), which during interphase teams up with WDR5 to recruit RNA Pol II to actively transcribed genes (Tjian, 1996; Wysocka et al, 2005; Thomas et al, 2015; Guarnaccia & Tansey, 2018). As cells enter mitosis, WDR5 and TBP replace RNA Pol II with the E3 ligase anaphase‐promoting complex (APC/C) (Oh et al, 2020; Fig 2B). The TBP‐ and WDR5‐bound APC/C modifies histones at transcription start sites with K11/K48‐branched ubiquitin chains (Oh et al, 2020), a conjugate that recruits the p97/VCP segregase and proteasomes to degrade proteins found in stable complexes (Meyer & Rape, 2014; Yau et al, 2017). Accordingly, histone polyubiquitylation clears nucleosomes off promoters for rapid reactivation of transcription in G1. Because the APC/C also triggers mitotic exit (Glotzer et al, 1991; Peters et al, 1996), this circuit coordinates gene expression with cell division, a role that is particularly important in stem cells (Pilaz et al, 2016; Oh et al, 2020) (Box 1).
Box 1. In need of answers.
How specific are ubiquitylation enzymes and effectors that control gene expression? Many E3 ligases and DUBs control transcription, yet the whole spectrum of their substrates is typically unknown. As E3 ligases often target multiple proteins, ubiquitylating more than one transcription factor might provide novel ways of coordinating gene expression programs.
How do E3 ligases collaborate to control gene expression? Multiple E3 ligases can target the same transcription factor in response to distinct inputs or to modify it with complex ubiquitin conjugates containing multiple linkages. How such coordination between ubiquitylation enzymes is established and regulated needs to be addressed.
How does ubiquitin‐dependent control of gene expression intersect with other roles of the ubiquitylation machinery? Addressing this question is of particular importance in understanding the relationship between transcription regulation and ubiquitin‐dependent quality control, both of which preserve cell identity and are affected in aging or neurodegenerative disease.
How does ubiquitin‐dependent control of protein complex composition affect transcriptional outputs? As most transcription factors act within dynamic multisubunit complexes, the ability of the ubiquitin system to detect and eliminate aberrant complex subunits is likely crucial for ensuring proper gene expression. Determining transcription factor complex composition on a global scale and discovering new examples of quality control of transcription factor complex composition will be critical.
How broadly can small molecules be used to re‐wire gene expression in development and disease? Can molecular glues or PROTACs be used to alter cell fate decisions during development? How important is the proper timing, duration, and efficiency of such therapeutic approaches?
As seen for monoubiquitylation of H2A or H2B, histone polyubiquitylation is likely part of extensive crosstalk between posttranslational marks. In addition to the APC/C, WDR5 binds the histone methyltransferase MLL, which is also required for transcriptional reactivation upon mitotic exit (Blobel et al, 2009) and may increase binding of WDR5 to promoters (Wysocka et al, 2005). Mitotic WDR5 also interacts with histone acetyl transferases that target residues in histone H3 and H4 (Oh et al, 2020). Histone acetyl transferases likely act upstream of MLL, as acetylation of H4K16 is required for H3K4 methylation at a subset of human genes (Katoh et al, 2011; Zhou et al, 2014), as well as for maintaining a pluripotent state (Spedale et al, 2012; Sheikh et al, 2019). Revealing another principle of ubiquitin‐dependent control of transcription, different types of ubiquitin conjugates collaborate with histone methylation or acetylation to fine‐tune gene expression to the needs of cell division and differentiation.
Ubiquitin‐dependent control of transcription factor stability and function
Key to regulating access of RNA Pol II to its target sequences are the ~ 1,600 transcription factors that are encoded in the human genome (Juven‐Gershon & Kadonaga, 2010; Magnani et al, 2011; Lambert et al, 2018). A single transcription factor can regulate multiple genes and act in concert with other transcription factors, thereby controlling the expression of specific gene sets during differentiation. Both abundance and activity of transcription factors therefore have to be tightly controlled during development, a task that has often been delegated to ubiquitylation.
Rapid response times through constitutive transcription factor degradation
In its simplest iteration, ubiquitin‐dependent degradation eliminates transcription factors that have fulfilled their functions. However, many transcription factors are intrinsically unstable proteins that are constantly synthesized, yet immediately degraded, if they are not needed. While seemingly wasteful, this setup allows cells to stall degradation as a means to translate a developmental signal into rapid transcription factor accumulation. Radial glial cells of the hippocampus, for example, synthesize the transcription factor ASCL1, yet use constitutive ubiquitin‐dependent degradation to keep its concentration low. This prevents quiescent precursor cells from premature cell cycle entry, which would deplete the stem cell pool (Blomfield et al, 2019). When these stem cells have to undergo self‐renewal, however, they simply inhibit ASCL1 degradation, without having to set in motion the machinery for ASCL1 transcription and translation. Rapid response times are also important for stress responses that detect the many adverse conditions faced by developing organisms. Accordingly, transcription factors responding to hypoxia (HIF1α), oxidative stress (NRF2), or DNA damage (TP53), are constantly turned over under homeostatic conditions, yet instantly stabilized upon stress (Salceda & Caro, 1997; Giaccia & Kastan, 1998; Ohh et al, 2000; Ma, 2013).
As ubiquitylation can be quantitatively tuned, inhibition of constitutive degradation is used to translate a certain amount of a developmental signal into a distinct transcription factor level and gene expression program. This is illustrated by β‐catenin, a WNT‐responsive transcription factor that determines pluripotency and tissue specification (Fig 3A). In the absence of WNT, β‐catenin is phosphorylated by CK1 and GSK3β kinases, which are part of a destruction complex that includes the scaffold proteins Axin and adenomatous polyposis coli. Its phosphorylation marks β‐catenin for ubiquitylation by the E3 ligase complex SCFβTrCP and proteasomal degradation. Binding of WNT to receptors inactivates the destruction complex, which disrupts β‐catenin ubiquitylation and dependent on WNT levels, initiates the synthesis of proteins that govern specific cell fate decisions (Nguyen et al, 2009; Vermeulen et al, 2010; Yu et al, 2012; Nusse & Clevers, 2017; Tammela et al, 2017). Cells monitor differences in β‐catenin abundance before and after signaling, a mechanism of fold‐change detection that helps establish robust gene expression responses despite stochastic variation in cellular protein expression (Goentoro & Kirschner, 2009; Goentoro et al, 2009).
Figure 3. Ubiquitin‐dependent regulation of transcriptional machinery.
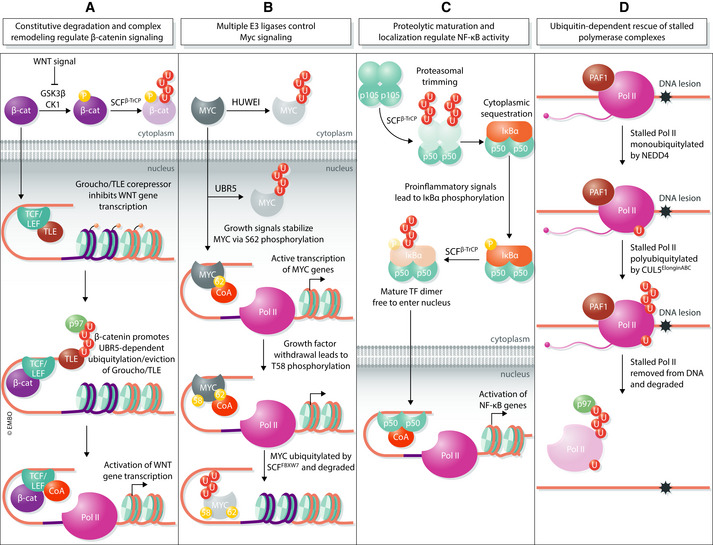
(A) In the absence of WNT signal, the β‐catenin transcription factor (β‐cat) is constitutively degraded by a destruction complex consisting of kinases GSK3β and CK1 as well as the ubiquitin ligase SCFβTrCP. The presence of WNT ligand stabilizes β‐catenin by inhibiting its phosphorylation, thus allowing it to translocate into the nucleus. At TCF/LEF transcriptional complexes, β‐catenin facilitates the exchange of co‐repressors for co‐activators (CoA) through UBR5‐mediated ubiquitylation and p97/VCP‐dependent extraction of the repressive Groucho/TLE subunit. (B) MYC levels are kept low in the cytoplasm and nucleus by the E3 ligases HUWE1 and UBR5, respectively. Upstream growth signals stabilize and activate MYC through phosphorylation of the S62 residue. This primes MYC for subsequent phosphorylation at nearby T58, which occurs upon growth factor withdrawal. T58‐phosphorylated MYC is recognized by the SCFFBXW7 E3 ligase and degraded to prevent subsequent re‐initiation of transcription. (C) The proinflammatory transcription factor NF‐κB is synthesized as a 105‐kDa precursor (p105) that is ubiquitylated by the SCFβTrCP E3 ligase and partially processed by the proteasome to yield a mature 50‐kDa form (p50). Mature NF‐κB dimers are sequestered in the cytoplasm by an inhibitor, IκBα, until cellular signals lead to degradation of IκBα and liberation of mature NF‐κB. Only then can mature NF‐κB enter the nucleus and initiate transcription of target genes. (D) RNA Pol II complexes that have stalled, such as at a DNA lesion, are first monoubiquitylated by the E3 ligase NEDD4. A second E3, CUL5ElonginABC, adds K48‐linked polyubiquitin chains to monoubiquitylated RNA Pol II leading to p97/VCP‐dependent removal of the stalled complex from DNA.
While rapid and quantitative signaling is highly advantageous for cells that need to adapt to changing environments, the circuits described above come at considerable risk. Mutations that prevent transcription factor degradation uncouple developmental inputs from gene expression outputs, with devastating consequences for the organism. Indeed, mutations that prevent E3 ligases from recognizing transcription factors or aberrant activity of E3 ligases often drive tumorigenesis. Inactivation of the destruction complex or the β‐catenin phosphodegron is associated with most cases of familial and sporadic colon cancers (Shang et al, 2017; Gao et al, 2018), whereas mutation or aberrant expression of VHL, KEAP1, or MDM2, the E3 ligases responsible for HIF1α, NRF2, and TP53 degradation, are among the most frequent events across all cancers (Lawrence et al, 2014; Gossage et al, 2015; Karni‐Schmidt et al, 2016).
Negative feedback regulation of transcription factor activity
In addition to regulating the speed or amplitude by which developmental signals are transmitted, transcription factor turnover can set the duration of a gene expression response. This function is often built around negative feedback loops, in which a transcription factor induces the expression of its own E3 ligase. In an example of particular importance for disease, TP53 drives the synthesis of the E3 MDM2, which in turn ubiquitylates TP53 to induce its proteasomal degradation (Karni‐Schmidt et al, 2016). The resulting negative feedback plays a role in establishing the pulsatile nature of DNA repair signaling, when cells take the persistence of DNA damage as basis for their decision to exit the cell cycle (Purvis & Lahav, 2013). The balance between TP53 and MDM2 is skewed in cancer, as MDM2 is amplified in a large number of hematological and solid tumors (Dembla et al, 2018). Elevated levels of MDM2 suppress wild‐type TP53 and hamper a cell’s response to genotoxic damage, which increases mutagenesis rates to drive tumor evolution (Manfredi, 2010).
While negative feedback regulation is a common principle of ubiquitin‐dependent control of gene expression, it comes in several twists. In addition to producing its own E3 ligase, SCFβTrCP, β‐catenin also drives the synthesis of Axin, the rate‐limiting component of the destruction complex that marks β‐catenin for ubiquitylation and proteasomal degradation (Jho et al, 2002; Lee et al, 2003). Thus, β‐catenin accelerates maturation of its degron to increase rates of ubiquitylation. Conversely, yeast RPN4 promotes expression of proteasomal subunits, in part to provide efficient responses to unfolded proteins accumulating in the cytoplasm (Schmidt et al, 2019). Once RPN4 has accumulated and proteasomes have been assembled, the proteolytic machinery degrades the transcription factor (Xie & Varshavsky, 2001).
Signal integration through multiple E3 ligases
Differentiating cells not only need to respond quickly to signals received from their environment, they also have to integrate multiple inputs at a time to make the appropriate fate choice. Ubiquitylation provides a unique opportunity to combine developmental signals into a coherent response, as a single transcription factor can be targeted by several E3 ligases that react to distinct inputs. A striking example is provided by MYC and MYCN, two transcription factors that play crucial roles in relaying anabolic signals to the cell cycle machinery (Fig 3B).
MYC and MYCN release paused RNA Pol II to increase expression of many genes at a time (Baluapuri et al, 2020). Ensuring that MYC is activated to the right extent during cell division, its levels are kept in check by phosphorylation of two residues, T58 and S62, and subsequent ubiquitylation by SCFFBXW7 (Welcker et al, 2004; Welcker & Clurman, 2008). Phosphorylated T58 also mediates binding of MYC by a distinct E3 ligase composed of SCFFBXL3 and its co‐adaptor CRY2 (Huber et al, 2016). Rather than connecting MYC activity to the cell cycle, this regulation establishes proper transcription factor activity in the context of the circadian clock. Mutation of T58 of MYC, which is observed in a high percentage of Burkitt’s lymphoma patients (Bahram et al, 2000; Gregory & Hann, 2000), thus disrupts control by both the cell cycle and circadian clock. In a similar manner, mutation of the analogous residue in MYCN can lead to neuroblastoma, medulloblastoma, or glioblastoma (Rickman et al, 2018).
As the most frequently amplified human oncogene, the levels of MYC must not exceed a threshold that would trigger cell death (McMahon, 2014). MYC and MYCN levels are additionally restricted by the E3 ligase UBR5, which triggers degradation independently of T58 or SCFFBXW7 (Qiao et al, 2020; Schukur et al, 2020). UBR5 thus sets a cap for MYC activity, which is exploited by cancer cells that overexpress UBR5 together with stabilized MYC. As MYC controls the transcription of many ribosomal genes (van Riggelen et al, 2010), downregulation of MYC by UBR5 decreases protein synthesis by limiting ribosomal biogenesis. Interestingly, UBR5 also eliminates aberrant translation products through quality control of nascent peptides (Yau et al, 2017). Another E3 ligase, HUWE1, controls both MYCN levels (Adhikary et al, 2005) and excess ribosomal proteins (Sung et al, 2016), underscoring the importance of coordinating MYC activity with protein synthesis in dividing cells. Cells therefore implement at least four E3 ligases to establish the appropriate levels of MYC or MYCN under a wide range of conditions.
Transcription factor regulation by multiple E3 ligases is a common theme in development, as also illustrated by members of the SMAD family that control transforming growth factor‐β (TGF‐β) signaling (Budi et al, 2017). Binding of TGF‐β growth factors to plasma membrane receptors induces phosphorylation and nuclear translocation of receptor‐regulated SMADs, allowing them to promote transcription of genes involved in cell proliferation, migration, and apoptosis (David & Massague, 2018). While nuclear SMADs are ubiquitylated by the E3 NEDD4‐L to prevent aberrant re‐initiation of transcription (Gao et al, 2009), constitutive modification of cytoplasmic SMADs by the E3s SMURF1 and SMURF2 sharpens a cell’s sensitivity toward changing TGF‐β levels and helps establish rapid response times (Zhu et al, 1999; Lin et al, 2000; Zhang et al, 2001). This integrated control of SMADs in multiple compartments is likely pivotal for setting the dynamics of TGFβ‐signaling that drive tissue patterning (Sorre et al, 2014).
Ubiquitin‐dependent processing of transcription factors
While degradation is an established consequence of ubiquitylation, this modification can also be used to rapidly activate transcription factors. This surprising role of ubiquitylation has emerged for inactive precursors of transcription factors, which are cleaved, rather than degraded, by the proteasome (Fig 3C). Providing an early example, NF‐κB transcription factors are synthesized as cytosolic p100 or p105 precursors that are converted into active p50 or p52 subunits through ubiquitin‐ and proteasome‐dependent cleavage (Palombella et al, 1994; Skaug et al, 2009; Yilmaz et al, 2014). Proteasomal processing occurs in multiple organisms and distinct contexts: proteasomal cleavage of the yeast transcription factors SPT23 and MGA2 modulates lipid composition at the endoplasmic reticulum (ER) membrane (Hoppe et al, 2000), yet processing of Drosophila Cubitus interruptus controls Hedgehog‐dependent transcription during development (Jiang, 2006). Proteasomal processing also regulates gene expression through targets other than transcription factors. In response to transcriptional stress, trimming of the DEF1 protein generates the substrate receptor subunit of an E3 ligase that, in turn, downregulate levels of RNA Pol II (Wilson et al, 2013b).
To prevent complete degradation, precursors of proteasomal processing are often tethered to another protein or even an organelle (Hoppe et al, 2001; Piwko & Jentsch, 2006; Yilmaz et al, 2014). This provides a physical impediment to the proteasome, which in turn can initiate, but not complete, degradation to yield a processed target. Following proteasome action, processed transcription factors need to be extracted from their protective partners, which led to the discovery that the AAA‐ATPase p97/VCP is a ubiquitin‐selective segregase (Rape et al, 2001). Although p97/VCP mostly collaborates with proteasomes, it also allows for the ubiquitin‐dependent, but proteasome‐independent, activation of the transcription factor NRF1 (Radhakrishnan et al, 2014). NRF1 is synthesized as a precursor that is embedded into the ER membrane yet constitutively degraded. In cells experiencing proteasome inhibition, NRF1 evades degradation and is instead cleaved by the ubiquitin‐selective rhomboid protease DDI2. P97/VCP then liberates cleaved NRF1 from the ER membrane and allows it to enter the nucleus for renewed synthesis of proteasomal subunits (Radhakrishnan et al, 2014; Koizumi et al, 2016; Sha & Goldberg, 2016; Vangala et al, 2016; Lehrbach & Ruvkun, 2016; Dirac‐Svejstrup et al, 2020).
Ubiquitin‐dependent remodeling of transcription factor complexes
Most transcription factors operate within multisubunit assemblies that define their specificity for particular loci or set the extent of transcriptional activation, and the formation of these complexes needs to be tightly monitored for development. The majority of the ~ 120 transcription factors with BTB domains, for example, form homodimers to engage their proper effector proteins impacting transcription. The transcription repressor BCL6, which is mutated in B‐cell lymphomas (Yang & Green, 2019), binds to a SMRT, BCOR, and NCoR co‐repressor through each of its BTB subunits (Ahmad et al, 2003; Ghetu et al, 2008). BCL6, however, also forms aberrant heterodimers that are unable to engage co‐repressors and impede gene regulation (Mena et al, 2018). A protective pathway referred to as dimerization quality control selectively detects and eliminates the inactive heterodimers to ensure proper BCL6 complex formation (Mena et al, 2018; Mena et al, 2020).
While dimerization quality control constitutively regulates transcription factor interactions, development or environmental inputs often elicit signal‐dependent changes in binding partners to alter gene expression (Mottis et al, 2013). This is illustrated by WNT‐mediated cell specification, which relies on remodeling of a complex around TCF/LEF family transcription factors (Fig 3A). In the absence of WNT, TCF/LEF is kept in check by the Groucho/TLE co‐repressor, but when WNT has been bound by cell membrane receptors, the stabilized β‐catenin directs the E3 ligase UBR5 to polyubiquitylate Groucho/TLE (Flack et al, 2017). Groucho/TLE ubiquitylation signals p97/VCP‐dependent extraction from the TCF/LEF complex to relieve chromatin compaction and initiate efficient transcription.
A frequent consequence of ubiquitin‐dependent changes in complex composition is the re‐localization of transcription factors in or out of the nucleus. Processed NF‐κB, for example, is held back in the cytosol by its inhibitor IκBα, until proinflammatory signals elicit IκBα degradation and nuclear entry of NF‐κB (Fig 3C; Winston et al, 1999). The NF‐κB‐dependent resynthesis of IκBα re‐exports NF‐κB out of the nucleus, thereby establishing pulsatile gene expression (Ashall et al, 2009). Such regulation might also occur in the absence of degradation: it has been proposed that monoubiquitylation of TP53 masks its recognition by importin‐α3 and thereby prevents it from entering the nucleus (Lohrum et al, 2001; Marchenko et al, 2010). Conversely, monoubiquitylation may block recognition of export sequences within transcription factors to promote nuclear accumulation (van der Horst et al, 2006; Trotman et al, 2007).
Together, these examples illustrate the diverse nature of ubiquitin‐dependent control of transcription factors. It ranges from fine‐tuning protein levels to ensuring that stimulus‐induced changes in their activity or location are set in place for the necessary duration. Ubiquitin‐dependent control of transcription factor activity and abundance can be established rapidly and with high precision, making it a powerful tool for regulatory circuits that integrate the many inputs received by gene expression networks during development.
Ubiquitin‐dependent regulation of transcription elongation
While initiation is often the rate‐limiting step in transcription, the entire message must be produced to generate a stable transcript that can be translated into a functional protein. Premature abortion of transcription produces mRNA species that are turned over by nucleases, in part because they would encode truncated and likely misfolded proteins. Despite the dangerous consequences of inefficient transcription, elongation of the message is not a smooth process and RNA Pol II frequently stalls due to topological constraints of chromatin or damaged templates (Selth et al, 2010; Wilson et al, 2013a). While transcription‐coupled nucleotide excision repair (TC‐NER) can remove DNA lesions in the path of RNA Pol II (Gaillard & Aguilera, 2013), the polymerase itself is evicted from DNA if repair fails (Woudstra et al, 2002; Anindya et al, 2007).
Recent studies showed that the largest subunit of RNA Pol II, RPB1, is a central target for ubiquitin‐dependent control of transcription elongation. While UV‐induced DNA damage broadly downregulates transcription, it is followed by recovery of gene expression once damage has been cleared. During this process, RPB1 is ubiquitylated on a specific Lys residue, K1268, which leads to its rapid proteasomal degradation (Wilson et al, 2013a; Tufegdzic Vidakovic et al, 2020) (Fig 3D). Proteolytic clearance of stalled RNA Pol II prevents inappropriate re‐initiation of transcription for short genes when DNA damage persists. A failure to ubiquitylate RNA Pol II, and its resulting aberrant transcription, yields a Cockayne syndrome‐like phenotype characterized by premature aging and neurodegeneration (Anindya et al, 2010; Nakazawa et al, 2020).
Several E3 ligases act on RNA Pol II to decorate it with distinct ubiquitin conjugates. RPB1 is monoubiquitylated by the E3 ligase NEDD4 (Anindya et al, 2007; Harreman et al, 2009), while CUL5ElonginABC attaches K48‐linked chains for p97/VCP‐dependent extraction and proteasomal degradation (Yasukawa et al, 2008; Harreman et al, 2009). NEDD4 and CUL5ElonginABC likely act on different Lys residues in RPB1 (Somesh et al, 2007; Tufegdzic Vidakovic et al, 2020). Elongin ABC functions as a transcription elongation factor independent of CUL5 and only recruits the catalytic CUL5‐RBX1 module upon stalling (Yasukawa et al, 2012; Weems et al, 2015). Finally, the E3 ligase WWP2 can ubiquitylate RNA Pol II for extraction at double‐strand breaks (Caron et al, 2019), which is co‐opted by MYCN to alleviate transcriptional stalling in neuroblastoma (Herold et al, 2019). Akin to transcription factor regulation by multiple E3 ligases, these findings suggest that RNA Pol II ubiquitylation plays a similar role in coordinating the cellular response to multiple sources of transcriptional stalling.
Regulation of transcription by small molecules targeting ubiquitylation
As highlighted in this review, ubiquitylation plays a pivotal role in controlling gene expression in development and disease. This is dramatically underscored by mutations in the degron motifs of transcription factors, such as TP53, NRF2, β‐catenin, or MYC, that drive many cancers. Given the prevalence of aberrant gene expression in disease, it has been a goal of many drug discovery programs to alter the stability of transcription factors for therapeutic benefit. As most E3 ligases and transcription factors lack obvious pockets for small molecules (Gu et al, 2018), it only recently became apparent how protein stability can be altered to modulate gene expression. Complementing the physiological regulation of transcription by ubiquitin, our final paragraphs will therefore discuss how the ubiquitylation machinery can be hijacked by small molecules to modulate the transcriptional networks that control development and prevent disease.
E3 ligase inhibition to target unstable transcription factors
A straightforward way to affect gene expression by small molecules are inhibitors of E3 ligases that target transcription factors (Huang & Dixit, 2016), as spearheaded for the tumor suppressor TP53 (Sarek & Ojala, 2007) (Fig 4A). While TP53 is mutated in ~ 50% of cancers, many tumors downregulate TP53 by overexpression of its E3 MDM2. In these cases, inhibition of MDM2 could reactivate TP53‐dependent gene expression and help eliminate malignant cells. The first class of MDM2 inhibitors were “Nutlins”, which prevent MDM2 from interacting with TP53 (Vassilev et al, 2004). Other compounds directly inhibit the E3 ligase activity of MDM2 or destabilize MDM2 by shutting off its protective deubiquitylase USP7 (Turnbull et al, 2017; Kategaya et al, 2017; Sanz et al, 2019) (Fig 4A).
Figure 4. Regulation of transcription by small molecules targeting ubiquitin components.
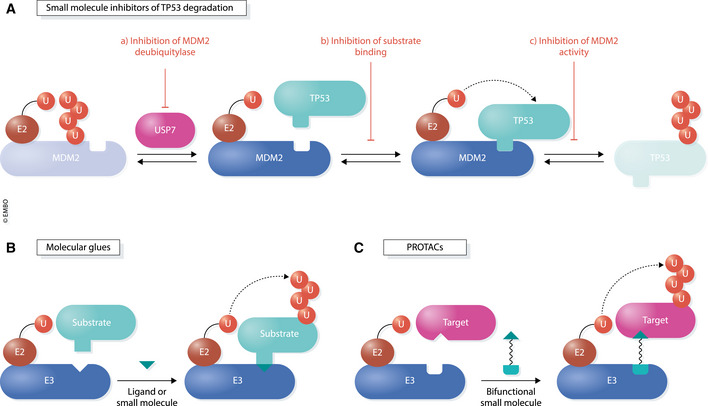
(A) Mechanisms of small molecules that prevent degradation of the transcription factor and tumor suppressor, TP53, by its E3 ubiquitin ligase, MDM2. (a) Inhibitors of the USP7 deubiquitylase lead to increased autoubiquitylation and destabilization of MDM2. (b) Small molecules, such as the Nutlins, block TP53 degradation by disrupting the substrate‐ligase interaction surface. (c) Compounds that block the E3 ligase activity of MDM2, but not substrate binding, also result in accumulation of TP53. (B) Molecular glues are small molecules that alter the surface of proteins, namely an E3 ligase and substrate, to promote their association. These compounds often display low binding affinities for each interactor but simultaneously bind both components to enhance their interaction. (C) PROTACs (proteolysis‐targeting chimeras) are heterobifunctional molecules that consist of two small molecules tethered by a linker. One of the small molecules binds a target protein while the other binds an E3 ligase. In this manner, the cell’s endogenous ubiquitin–proteasome system can be used to rapidly and selectively eliminate any given protein target‐of‐interest.
Small molecule inhibitors of E3 ligases are most effective against transcription factors that are constitutively turned over. In addition to TP53, this is illustrated by the antioxidant transcription factor NRF2 (Hayes & Dinkova‐Kostova, 2014), whose transient stabilization is of interest for the treatment of neurodegeneration or diabetes (Schmoll et al, 2017). NRF2 can be regulated by targeting reactive cysteine residues in its E3, CUL3KEAP1 (Zhang et al, 2004), using compounds, such as dimethyl fumarate, which has been approved for the treatment of multiple sclerosis (Dodson et al, 2019). However, inhibition of NRF2 has to be carefully monitored, as persistent stabilization of this transcription factor induces a gene expression program that triggers reductive stress and prevents muscle stem cell differentiation (Manford et al, 2020).
Induced protein degradation to control gene expression
Providing an alternative approach, small molecules can alter gene expression by eliciting the turnover of transcription factors. Such induced protein degradation was first naturally observed in plants, where hormones of the auxin family bridge an E3 ligase, SCFTIR1, with the transcriptional repressors AXR2/IAA7 and AXR3/IAA17 (Sakamoto et al, 2001; Dharmasiri et al, 2005; Kepinski & Leyser, 2005). In a similar manner, the hormone gibberellin triggers complex formation between the E3 SCFGID2 and the transcriptional repressor DELLA (Hauvermale et al, 2012). As molecular glues between an E3 ligase and a protein, these hormones induce the ubiquitylation of transcription regulators and control gene expressions programs important for plant development (Fig 4B).
Synthetic molecular glues are now used to modulate transcription factor stability in human cells, as illustrated by the immunomodulatory drug thalidomide that was once prescribed as sleep medication and now is prescribed to combat hematological malignancies (Lasagna, 1960; Vargesson, 2009). Thalidomide and its derivatives direct the E3 CUL4CRBN to degrade zinc finger proteins, many of which are transcription factors (Lu et al, 2014; Sievers et al, 2018). One transcription factor, SALL4, is essential for limb development, and its induced degradation during early pregnancy likely accounts for the drastic thalidomide‐induced birth defects (Donovan et al, 2018; Matyskiela et al, 2020). While thalidomide was introduced into the clinic without knowledge of its mechanism, the first prospective molecular glue to connect an E3 ligase to a pathological protein was recently reported (Simonetta et al, 2019). This compound tethered unphosphorylated β‐catenin to the E3 SCFβTrCP to induce degradation of this transcription factor in colon cancer cells carrying mutations in the destruction complex. Molecular glues can also elicit transcription factor degradation by means other than directly attracting an E3 ligase: Compounds that bind BCL6, for example, trigger formation of BCL6 fibers that recruit the E3 ligase SIAH1 and counteract BCL6‐dependent gene repression in lymphoma cells (Kerres et al, 2017; Slabicki et al, 2020b). Moreover, molecular glues can affect the activity of RNA Pol II itself, by controlling the stability of a cyclin‐dependent kinase that regulates transcript elongation (Mayor‐Ruiz et al, 2020; Lv et al, 2020; Slabicki et al, 2020a).
Understanding thalidomide’s mechanism of action led to the development of bifunctional molecules composed of two warheads, one that binds an E3 ligase and the other that recognizes the target (Winter et al, 2015). Having been proposed to elicit degradation almost two decades earlier (Sakamoto et al, 2001; Sakamoto et al, 2003), such proteolysis‐targeting chimeras (PROTACs) induce the recruitment of pathological proteins to E3 ligases to trigger proteasomal degradation of the disease‐causing agent. PROTACs have been widely applied to alter gene expression, as they can eliminate transcription factors, such as STAT3 (Bai et al, 2019), histone modifiers, such as the PRC2 complex (Potjewyd et al, 2020), or readers of histone modifications, such as BRD4 (Winter et al, 2015) (Fig 4C). In 2019, the first PROTAC entered clinical trials for the treatment of castration‐resistant prostate cancer, targeting the androgen receptor and its transcriptional networks (Wang et al, 2020). Degradation tags (dTAGs) (Nabet et al, 2018) are PROTACs that target the FKBP12F36V domain which, when fused to a transcriptional regulator, can be a powerful way to acutely change gene expression in the laboratory (Erb et al, 2017). Hijacking E3 ligases thus offers multiple opportunities to alter gene expression networks that have been notoriously difficult to target by traditional means.
To conclude, accurate transcription is critical for guiding tissue formation and homeostasis. The failure to initiate gene expression at the right time and place or problems in resolving transcriptional stress can lead to cell death and disease. It is clear that cells use ubiquitylation in proteolytic and nonproteolytic ways to control nearly every step of transcription—from modulating chromatin architecture over limiting transcription factor abundance to ensuring transcript elongation. The past years have witnessed groundbreaking improvements in our understanding of ubiquitin‐dependent control of gene expression, which led to novel therapeutic strategies that may help patients with cancer and other ailments. It is now possible to mobilize the ubiquitylation machinery to selectively degrade transcription factors that were once deemed “undruggable”. As clinical trials are underway, whether these compounds live up to their promise will soon become apparent. If they do, ubiquitin‐dependent control of gene expression will be a textbook example of how fundamental research in understanding principles of signal transduction can have a profound impact on human health.
Conflict of interest
M.R. is a co‐founder and consultant to Nurix and scientific advisory board member to Monte Rosa.
Acknowledgements
We apologize to our colleagues whose work we were unable to include due to space constraints. We thank D. Haakonsen and E. Oh for critical reading of this manuscript, and we are grateful to all members of our laboratory for their ideas and discussions regarding the role of ubiquitin in transcription. K.G.M. is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award F32GM120956. M.R. is an Investigator of the Howard Hughes Medical Institute.
EMBO Reports (2021) 22: e51078.
See the Glossary for abbreviations used in this article.
Contributor Information
Kevin G Mark, Email: markk@berkeley.edu.
Michael Rape, Email: mrape@berkeley.edu.
References
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S et al (2005) The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 [DOI] [PubMed] [Google Scholar]
- Adorno M, Sikandar S, Mitra SS, Kuo A, Nicolis Di Robilant B, Haro‐Acosta V, Ouadah Y, Quarta M, Rodriguez J, Qian D et al (2013) Usp16 contributes to somatic stem‐cell defects in Down's syndrome. Nature 501: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG (2003) Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell 12: 1551–1564 [DOI] [PubMed] [Google Scholar]
- Anindya R, Aygun O, Svejstrup JQ (2007) Damage‐induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 28: 386–397 [DOI] [PubMed] [Google Scholar]
- Anindya R, Mari PO, Kristensen U, Kool H, Giglia‐Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ (2010) A ubiquitin‐binding domain in Cockayne syndrome B required for transcription‐coupled nucleotide excision repair. Mol Cell 38: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Mas G, di Croce L (2015) Regulation of gene transcription by Polycomb proteins. Sci Adv 1: e1500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS et al (2009) Pulsatile stimulation determines timing and specificity of NF‐kappaB‐dependent transcription. Science 324: 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, McIvor E, Li W, Zhang Y, Florens L et al (2016) ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell 62: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahram F, von der Lehr N, Cetinkaya C, Larsson LG (2000) c‐Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome‐mediated turnover. Blood 95: 2104–2110 [PubMed] [Google Scholar]
- Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, Chen J, Yang CY, Liu Z, Wang M et al (2019) A potent and selective small‐molecule degrader of STAT3 achieves complete tumor regression in vivo . Cancer Cell 36: 498–511.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluapuri A, Wolf E, Eilers M (2020) Target gene‐independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol 21: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G (2011) Polycomb‐dependent regulatory contacts between distant Hox loci in Drosophila . Cell 144: 214–226 [DOI] [PubMed] [Google Scholar]
- Belle JI, Nijnik A (2014) H2A‐DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. Int J Biochem Cell Biol 50: 161–174 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y et al (2014) Variant PRC1 complex‐dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157: 1445–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ (2020) PRC1 catalytic activity is central to polycomb system function. Mol Cell 77: 857–874.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR (2009) A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell 36: 970–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield IM, Rocamonde B, Masdeu MDM, Mulugeta E, Vaga S, van den Berg DL, Huillard E, Guillemot F, Urban N (2019) Id4 promotes the elimination of the pro‐activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. Elife 8: e48561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N (2017) Polycomb complexes in X chromosome inactivation. Philos Trans R Soc Lond B Biol Sci 372: 20170021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi EH, Duan D, Derynck R (2017) Transforming growth factor‐beta receptors and smads: regulatory complexity and functional versatility. Trends Cell Biol 27: 658–672 [DOI] [PubMed] [Google Scholar]
- Bulynko YA, O'Malley BW (2011) Nuclear receptor coactivators: structural and functional biochemistry. Biochemistry 50: 313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne A, Lee MK, Zielinski D, Michaud A, le Corre S, Dingli F, Chen H, Shahidian LZ, Vassilev I, Servant N et al (2019) BAP1 complex promotes transcription by opposing PRC1‐mediated H2A ubiquitylation. Nat Commun 10: 348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G (2013) BAP1 and cancer. Nat Rev Cancer 13: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P, Pankotai T, Wiegant WW, Tollenaere MAX, Furst A, Bonhomme C, Helfricht A, de Groot A, Pastink A, Vertegaal A et al (2019) WWP2 ubiquitylates RNA polymerase II for DNA‐PK‐dependent transcription arrest and repair at DNA breaks. Genes Dev 33: 684–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short‐lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Chen S, Li J, Wang DL, Sun FL (2012) Histone H2B lysine 120 monoubiquitination is required for embryonic stem cell differentiation. Cell Res 22: 1402–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, Shilatifard A (2018a) Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19: 464–478 [DOI] [PubMed] [Google Scholar]
- Chen P, Dong L, Hu M, Wang YZ, Xiao X, Zhao Z, Yan J, Wang PY, Reinberg D, Li M et al (2018b) Functions of FACT in breaking the nucleosome and maintaining its integrity at the single‐nucleosome level. Mol Cell 71: 284–293.e4 [DOI] [PubMed] [Google Scholar]
- Chittock EC, Latwiel S, Miller TC, Muller CW (2017) Molecular architecture of polycomb repressive complexes. Biochem Soc Trans 45: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez JT, Montauti E, Shifrut E, Gatchalian J, Zhang Y, Shaked O, Xu Y, Roth TL, Simeonov DR, Zhang Y et al (2020) CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582: 416–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Massague J (2018) Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol 19: 419–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelouchina GT, Gerecht K, Muir TW (2017) Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat Chem Biol 13: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluz C, Friman ET, Strebinger D, Benke A, Raccaud M, Callegari A, Leleu M, Manley S, Suter DM (2016) A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev 30: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembla V, Somaiah N, Barata P, Hess K, Fu S, Janku F, Karp DD, Naing A, Piha‐Paul SA, Subbiah V et al (2018) Prevalence of MDM2 amplification and coalterations in 523 advanced cancer patients in the MD Anderson phase 1 clinic. Oncotarget 9: 33232–33243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K (2013) Long‐range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13: 602–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K (2009) Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20: 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F‐box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dickson KA, Cole AJ, Gill AJ, Clarkson A, Gard GB, Chou A, Kennedy CJ, Henderson BR, Australian Ovarian Cancer Study , Fereday S et al (2016) The RING finger domain E3 ubiquitin ligases BRCA1 and the RNF20/RNF40 complex in global loss of the chromatin mark histone H2B monoubiquitination (H2Bub1) in cell line models and primary high‐grade serous ovarian cancer. Hum Mol Genet 25: 5460–5471 [DOI] [PubMed] [Google Scholar]
- Dirac‐Svejstrup AB, Walker J, Faull P, Encheva V, Akimov V, Puglia M, Perkins D, Kumper S, Hunjan SS, Blagoev B et al (2020) DDI2 Is a ubiquitin‐directed endoprotease responsible for cleavage of transcription factor NRF1. Mol Cell 79: 332–341.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F et al (2012) Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M, de la Vega MR, Cholanians AB, Schmidlin CJ, Chapman E, Zhang DD (2019) Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol 59: 555–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KA, An J, Nowak RP, Yuan JC, Fink EC, Berry BC, Ebert BL, Fischer ES (2018) Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife 7: e38430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, Souza A, Roberts JM, Dastjerdi S, Buckley DL et al (2017) Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543: 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar‐Dagan M, Raleigh DP, Muir TW (2011) Histone H2B ubiquitylation disrupts local and higher‐order chromatin compaction. Nat Chem Biol 7: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flack JE, Mieszczanek J, Novcic N, Bienz M (2017) Wnt‐Dependent Inactivation of the Groucho/TLE Co‐repressor by the HECT E3 Ubiquitin Ligase Hyd/UBR5. Mol Cell 67: 181–193.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj‐Yahya M et al (2012) RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 46: 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Oren M (2014) Writing and reading H2B monoubiquitylation. Biochim Biophys Acta 1839: 694–701 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y (2018) Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 19: 192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Aguilera A (2013) Transcription coupled repair at the interface between transcription elongation and mRNP biogenesis. Biochim Biophys Acta 1829: 141–150 [DOI] [PubMed] [Google Scholar]
- Gallego LD, Schneider M, Mittal C, Romanauska A, Gudino Carrillo RM, Schubert T, Pugh BF, Kohler A (2020) Phase separation directs ubiquitination of gene‐body nucleosomes. Nature 579: 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument‐Bromage H, Tempst P, Massague J (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF‐beta signaling. Mol Cell 36: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wang Y, Broaddus R, Sun L, Xue F, Zhang W (2018) Exon 3 mutations of CTNNB1 drive tumorigenesis: a review. Oncotarget 9: 5492–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzka M, Tasdogan A, Hainzl A, Allies G, Maity P, Wilms C, Wlaschek M, Scharffetter‐Kochanek K (2015) Interplay of H2A deubiquitinase 2A‐DUB/Mysm1 and the p19(ARF)/p53 axis in hematopoiesis, early T‐cell development and tissue differentiation. Cell Death Differ 22: 1451–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG (2008) Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell 29: 384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12: 2973–2983 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Goentoro L, Kirschner MW (2009) Evidence that fold‐change, and not absolute level, of beta‐catenin dictates Wnt signaling. Mol Cell 36: 872–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentoro L, Shoval O, Kirschner MW, Alon U (2009) The incoherent feedforward loop can provide fold‐change detection in gene regulation. Mol Cell 36: 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H (1975) Isolation and characterization of protein A24, a "histone‐like" non‐histone chromosomal protein. J Biol Chem 250: 7182–7187 [PubMed] [Google Scholar]
- Gossage L, Eisen T, Maher ER (2015) VHL, the story of a tumour suppressor gene. Nat Rev Cancer 15: 55–64 [DOI] [PubMed] [Google Scholar]
- Gregory MA, Hann SR (2000) c‐Myc proteolysis by the ubiquitin‐proteasome pathway: stabilization of c‐Myc in Burkitt's lymphoma cells. Mol Cell Biol 20: 2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Paro R (2014) Transcriptional silencing by polycomb‐group proteins. Cold Spring Harb Perspect Biol 6: a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Jones AE, Yang W, Liu S, Dai Q, Liu Y, Swindle CS, Zhou D, Zhang Z, Ryan TM et al (2016) The histone H2A deubiquitinase Usp16 regulates hematopoiesis and hematopoietic stem cell function. Proc Natl Acad Sci USA 113: E51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Cui D, Chen X, Xiong X, Zhao Y (2018) PROTACs: an emerging targeting technique for protein degradation in drug discovery. BioEssays 40: e1700247 [DOI] [PubMed] [Google Scholar]
- Guarnaccia AD, Tansey WP (2018) Moonlighting with WDR5: a cellular multitasker. J Clin Med 7: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haakonsen DL, Rape M (2019) Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol 29: 704–716 [DOI] [PubMed] [Google Scholar]
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW et al (2009) Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci USA 106: 20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova‐Kostova AT (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39: 199–218 [DOI] [PubMed] [Google Scholar]
- Herold S, Kalb J, Buchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C et al (2019) Recruitment of BRCA1 limits MYCN‐driven accumulation of stalled RNA polymerase. Nature 567: 545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S (2000) Activation of a membrane‐bound transcription factor by regulated ubiquitin/proteasome‐dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S (2001) Membrane‐bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol 13: 344–348 [DOI] [PubMed] [Google Scholar]
- van der Horst A, De Vries‐Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073 [DOI] [PubMed] [Google Scholar]
- Hsiung CC, Blobel GA (2016) A new bookmark of the mitotic genome in embryonic stem cells. Nat Cell Biol 18: 1124–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Dixit VM (2016) Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res 26: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AL, Papp SJ, Chan AB, Henriksson E, Jordan SD, Kriebs A, Nguyen M, Wallace M, Li Z, Metallo CM et al (2016) CRY2 and FBXL3 cooperatively degrade c‐MYC. Mol Cell 64: 774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, Koseki H (2013) SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 26: 565–577 [DOI] [PubMed] [Google Scholar]
- Jason LJ, Finn RM, Lindsey G, Ausio J (2005) Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J Biol Chem 280: 4975–4982 [DOI] [PubMed] [Google Scholar]
- Jho E, Zhang T, Domon C, Joo CK, Freund JN, Constantini F (2002) Wnt/beta‐catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J (2006) Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 5: 2457–2463 [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin‐chain formation by the human anaphase‐promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven‐Gershon T, Kadonaga JT (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA (2012) Tissue‐specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J (2014) Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571 [DOI] [PubMed] [Google Scholar]
- Karni‐Schmidt O, Lokshin M, Prives C (2016) The roles of MDM2 and MDMX in cancer. Annu Rev Pathol 11: 617–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, Snaidero N, Vogel T, Shchebet A, Begus‐Nahrmann Y et al (2012) The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell 46: 705–713 [DOI] [PubMed] [Google Scholar]
- Kategaya L, di Lello P, Rouge L, Pastor R, Clark KR, Drummond J, Kleinheinz T, Lin E, Upton JP, Prakash S et al (2017) USP7 small‐molecule inhibitors interfere with ubiquitin binding. Nature 550: 534–538 [DOI] [PubMed] [Google Scholar]
- Katoh H, Qin ZS, Liu R, Wang L, Li W, Li X, Wu L, Du Z, Lyons R, Liu CG et al (2011) FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU‐1. Mol Cell 44: 770–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F‐box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kerres N, Steurer S, Schlager S, Bader G, Berger H, Caligiuri M, Dank C, Engen JR, Ettmayer P, Fischerauer B et al (2017) Chemically induced degradation of the oncogenic transcription factor BCL6. Cell Rep 20: 2860–2875 [DOI] [PubMed] [Google Scholar]
- Koizumi S, Irie T, Hirayama S, Sakurai Y, Yashiroda H, Naguro I, Ichijo H, Hamazaki J, Murata S (2016) The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. Elife 5: e18357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Krajewski WA, Li J, Dou Y (2018) Effects of histone H2B ubiquitylation on the nucleosome structure and dynamics. Nucleic Acids Res 46: 7631–7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT (2018) The human transcription factors. Cell 175: 598–599 [DOI] [PubMed] [Google Scholar]
- Lan X, Atanassov BS, Li W, Zhang Y, Florens L, Mohan RD, Galardy PJ, Washburn MP, Workman JL, Dent SYR (2016) USP44 Is an integral component of N‐CoR that contributes to gene repression by deubiquitinating histone H2B. Cell Rep 17: 2382–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagna L (1960) Thalidomide–a new nonbarbiturate sleep–inducing drug. J Chronic Dis 11: 627–631 [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW (2003) The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 1: E10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA (2013) Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Ruvkun G (2016) Proteasome dysfunction triggers activation of SKN‐1A/Nrf1 by the aspartic protease DDI‐1. Elife 5: e17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levendosky RF, Sabantsev A, Deindl S, Bowman GD (2016) The Chd1 chromatin remodeler shifts hexasomes unidirectionally. Elife 5: e21356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome‐dependent degradation of Smad2 in transforming growth factor‐beta signaling. J Biol Chem 275: 36818–36822 [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH (2001) C‐terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol 21: 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG Jr (2014) The myeloma drug lenalidomide promotes the cereblon‐dependent destruction of Ikaros proteins. Science 343: 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Chen P, Cao L, Li Y, Zeng Z, Cui Y, Wu Q, Li J, Wang JH, Dong MQ et al (2020) Discovery of a molecular glue promoting CDK12‐DDB1 interaction to trigger cyclin K degradation. Elife 9: e59994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53: 401–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Takizawa Y, Ishimaru M, Sugita Y, Sekine S, Nakayama JI, Wolf M, Kurumizaka H (2018) Structural basis of heterochromatin formation by human HP1. Mol Cell 69: 385–397.e8 [DOI] [PubMed] [Google Scholar]
- Magnani L, Eeckhoute J, Lupien M (2011) Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet 27: 465–474 [DOI] [PubMed] [Google Scholar]
- Manford AG, Rodriguez‐Perez F, Shih KY, Shi Z, Berdan CA, Choe M, Titov DV, Nomura DK, Rape M (2020) A cellular mechanism to detect and alleviate reductive stress. Cell 183: 46–61.e21 [DOI] [PubMed] [Google Scholar]
- Manfredi JJ (2010) The Mdm2‐p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24: 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM (2010) Stress‐mediated nuclear stabilization of p53 is regulated by ubiquitination and importin‐alpha3 binding. Cell Death Differ 17: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C (1995) Displacement of sequence‐specific transcription factors from mitotic chromatin. Cell 83: 29–38 [DOI] [PubMed] [Google Scholar]
- Matyskiela ME, Clayton T, Zheng X, Mayne C, Tran E, Carpenter A, Pagarigan B, McDonald J, Rolfe M, Hamann LG et al (2020) Crystal structure of the SALL4‐pomalidomide‐cereblon‐DDB1 complex. Nat Struct Mol Biol 27: 319–322 [DOI] [PubMed] [Google Scholar]
- Mayor‐Ruiz C, Bauer S, Brand M, Kozicka Z, Siklos M, Imrichova H, Kaltheuner IH, Hahn E, Seiler K, Koren A et al (2020) Rational discovery of molecular glue degraders via scalable chemical profiling. Nat Chem Biol 16: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB (2014) MYC and the control of apoptosis. Cold Spring Harb Perspect Med 4: a014407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena EL, Kjolby RAS, Saxton RA, Werner A, Lew BG, Boyle JM, Harland R, Rape M (2018) Dimerization quality control ensures neuronal development and survival. Science 586: 452–456 [DOI] [PubMed] [Google Scholar]
- Mena EL, Jevtic P, Greber BJ, Gee CL, Lew BG, Akopian D, Nogales E, Kuriyan J, Rape M (2020) Structural basis for dimerization quality control. Nature 586: 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, Bunker RD, Schenk AD, Graff‐Meyer A, Pathare GR et al (2020) Mechanisms of OCT4‐SOX2 motif readout on nucleosomes. Science 368: 1460–1465 [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D (1997) Marking of active genes on mitotic chromosomes. Nature 388: 895–899 [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Mottis A, Mouchiroud L, Auwerx J (2013) Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev 27: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A et al (2018) The dTAG system for immediate and target‐specific protein degradation. Nat Chem Biol 14: 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M et al (2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans‐histone cross‐talk with H3K4 di‐ and trimethylation. Genes Dev 22: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hara Y, Oka Y, Komine O, van den Heuvel D, Guo C, Daigaku Y, Isono M, He Y, Shimada M et al (2020) Ubiquitination of DNA damage‐stalled RNAPII promotes transcription‐coupled repair. Cell 180: 1228–1244.e24 [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA et al (2008) Ubiquitin chain editing revealed by polyubiquitin linkage‐specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J (2009) WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nune M, Morgan MT, Connell Z, McCullough L, Jbara M, Sun H, Brik A, Formosa T, Wolberger C (2019) FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics. Elife 8: e40988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Clevers H (2017) Wnt/beta‐catenin signaling, disease, and emerging therapeutic modalities. Cell 169: 985–999 [DOI] [PubMed] [Google Scholar]
- Oh E, Mark KG, Mocciaro A, Watson ER, Prabu JR, Cha DD, Kampmann M, Gamarra N, Zhou CY, Rape M (2020) Gene expression and cell identity controlled by anaphase‐promoting complex. Nature 579: 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG (2000) Ubiquitination of hypoxia‐inducible factor requires direct binding to the beta‐domain of the von Hippel‐Lindau protein. Nat Cell Biol 2: 423–427 [DOI] [PubMed] [Google Scholar]
- Ohtake F, Tsuchiya H, Saeki Y, Tanaka K (2018) K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci USA 115: E1401–E1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T (1994) The ubiquitin‐proteasome pathway is required for processing the NF‐kappa B1 precursor protein and the activation of NF‐kappa B. Cell 78: 773–785 [DOI] [PubMed] [Google Scholar]
- Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, Zaret KS (2017) Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358: 119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palozola KC, Lerner J, Zaret KS (2019) A changing paradigm of transcriptional memory propagation through mitosis. Nat Rev Mol Cell Biol 20: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Peters JM, King RW, Hoog C, Kirschner MW (1996) Identification of BIME as a subunit of the anaphase‐promoting complex. Science 274: 1199–1201 [DOI] [PubMed] [Google Scholar]
- Pierce SB, Stewart MD, Gulsuner S, Walsh T, Dhall A, McClellan JM, Klevit RE, King MC (2018) De novo mutation in RING1 with epigenetic effects on neurodevelopment. Proc Natl Acad Sci USA 115: 1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, Silver DL (2016) Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron 89: 83–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W, Jentsch S (2006) Proteasome‐mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol 13: 691–697 [DOI] [PubMed] [Google Scholar]
- Potjewyd F, Turner AW, Beri J, Rectenwald JM, Norris‐Drouin JL, Cholensky SH, Margolis DM, Pearce KH, Herring LE, James LI (2020) Degradation of polycomb repressive complex 2 with an EED‐targeted bivalent chemical degrader. Cell Chem Biol 27: 47–56.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2016) Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G (2013) Encoding and decoding cellular information through signaling dynamics. Cell 152: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Liu Y, Prada ML, Mohan AK, Gupta A, Jaiswal A, Sharma M, Merisaari J, Haikala HM, Talvinen K et al (2020) UBR5 Is coamplified with MYC in Breast tumors and encodes an ubiquitin ligase that limits MYC‐dependent apoptosis. Cancer Res 80: 1414–1427 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, den Besten W, Deshaies RJ (2014) p97‐dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife 3: e01856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane‐tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin‐selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Rickman DS, Schulte JH, Eilers M (2018) The expanding world of N‐MYC‐driven tumors. Cancer Discov 8: 150–163 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Sahtoe DD, Sixma TK (2015) Layers of DUB regulation. Trends Biochem Sci 40: 456–467 [DOI] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ (2001) Protacs: chimeric molecules that target proteins to the Skp1‐Cullin‐F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA 98: 8554–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Verma R, Ransick A, Stein B, Crews CM, Deshaies RJ (2003) Development of Protacs to target cancer‐promoting proteins for ubiquitination and degradation. Mol Cell Proteomics 2: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Salceda S, Caro J (1997) Hypoxia‐inducible factor 1alpha (HIF‐1alpha) protein is rapidly degraded by the ubiquitin‐proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox‐induced changes. J Biol Chem 272: 22642–22647 [DOI] [PubMed] [Google Scholar]
- Sanz G, Singh M, Peuget S, Selivanova G (2019) Inhibition of p53 inhibitors: progress, challenges and perspectives. J Mol Cell Biol 11: 586–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Ojala PM (2007) p53 reactivation kills KSHV lymphomas efficiently in vitro and in vivo: new hope for treating aggressive viral lymphomas. Cell Cycle 6: 2205–2209 [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, De Ayala Alonso AG, Oktaba K, Ly‐Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J (2010) Histone H2A deubiquitinase activity of the Polycomb repressive complex PR‐DUB. Nature 465: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RM, Schessner JP, Borner GH, Schuck S (2019) The proteasome biogenesis regulator Rpn4 cooperates with the unfolded protein response to promote ER stress resistance. Elife 8: e43244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll D, Engel CK, Glombik H (2017) The Keap1‐Nrf2 protein‐protein interaction: a suitable target for small molecules. Drug Discov Today Technol 24: 11–17 [DOI] [PubMed] [Google Scholar]
- Schukur L, Zimmermann T, Niewoehner O, Kerr G, Gleim S, Bauer‐Probst B, Knapp B, Galli GG, Liang X, Mendiola A et al (2020) Identification of the HECT E3 ligase UBR5 as a regulator of MYC degradation using a CRISPR/Cas9 screen. Sci Rep 10: 20044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ (2010) Transcript elongation by RNA polymerase II. Annu Rev Biochem 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Sha Z, Goldberg AL (2016) Reply to Vangala et al: complete inhibition of the proteasome reduces new proteasome production by causing Nrf1 aggregation. Curr Biol 26: R836–R837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S, Hua F, Hu ZW (2017) The regulation of beta‐catenin activity and function in cancer: therapeutic opportunities. Oncotarget 8: 33972–33989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh BN, Guhathakurta S, Akhtar A (2019) The non‐specific lethal (NSL) complex at the crossroads of transcriptional control and cellular homeostasis. EMBO Rep 20: e47630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema‐Yaacoby E, Nikolov M, Haj‐Yahya M, Siman P, Allemand E, Yamaguchi Y, Muchardt C, Urlaub H, Brik A, Oren M et al (2013) Systematic identification of proteins binding to chromatin‐embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep 4: 601–608 [DOI] [PubMed] [Google Scholar]
- Sievers QL, Petzold G, Bunker RD, Renneville A, Slabicki M, Liddicoat BJ, Abdulrahman W, Mikkelsen T, Ebert BL, Thoma NH (2018) Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362: eaat0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Taygerly J, Boyle K, Basham SE, Padovani C, Lou Y, Cummins TJ, Yung SL, von Soly SK, Kayser F et al (2019) Prospective discovery of small molecule enhancers of an E3 ligase‐substrate interaction. Nat Commun 10: 1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ (2009) The role of ubiquitin in NF‐kappaB regulatory pathways. Annu Rev Biochem 78: 769–796 [DOI] [PubMed] [Google Scholar]
- Slabicki M, Kozicka Z, Petzold G, Li YD, Manojkumar M, Bunker RD, Donovan KA, Sievers QL, Koeppel J, Suchyta D et al (2020a) The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585: 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabicki M, Yoon H, Koeppel J, Nitsch L, Roy Burman SS, Di Genua C, Donovan KA, Sperling AS, Hunkeler M, Tsai JM et al (2020b). Small‐molecule‐induced polymerization triggers degradation of BCL6. Nature 588: 164–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh BP, Sigurdsson S, Saeki H, Erdjument‐Bromage H, Tempst P, Svejstrup JQ (2007) Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell 129: 57–68 [DOI] [PubMed] [Google Scholar]
- Sorre B, Warmflash A, Brivanlou AH, Siggia ED (2014) Encoding of temporal signals by the TGF‐beta pathway and implications for embryonic patterning. Dev Cell 30: 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Timmers HT, Pijnappel WW (2012) ATAC‐king the complexity of SAGA during evolution. Genes Dev 26: 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ (2016) Ribosomal proteins produced in excess are degraded by the ubiquitin‐proteasome system. Mol Biol Cell 27: 2642–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburri S, Lavarone E, Fernandez‐Perez D, Conway E, Zanotti M, Manganaro D, Pasini D (2020) Histone H2AK119 mono‐ubiquitination is essential for polycomb‐mediated transcriptional repression. Mol Cell 77: 840–856.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T, Sanchez‐Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft RA et al (2017) A Wnt‐producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545: 355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H, Boocholez H, Hubert A, Rotkopf R, Baniyash M et al (2016) RNF20 links histone H2B ubiquitylation with inflammation and inflammation‐associated cancer. Cell Rep 14: 1462–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R (2016) A dynamic mode of mitotic bookmarking by transcription factors. Elife 5: e22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Bhargava‐Shah A, Xie L, Darzacq X, Tjian R (2018) A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife 7: e35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LR, Wang Q, Grieb BC, Phan J, Foshage AM, Sun Q, Olejniczak ET, Clark T, Dey S, Lorey S et al (2015) Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol Cell 58: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R (1996) The biochemistry of transcription in eukaryotes: a paradigm for multisubunit regulatory complexes. Philos Trans R Soc Lond B Biol Sci 351: 491–499 [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya‐Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon‐Cardo C, Erdjument‐Bromage H et al (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128: 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufegdzic Vidakovic A, Mitter R, Kelly GP, Neumann M, Harreman M, Rodriguez‐Martinez M, Herlihy A, Weems JC, Boeing S, Encheva V et al (2020) Regulation of the RNAPII pool is integral to the DNA damage response. Cell 180: 1245–1261.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull AP, Ioannidis S, Krajewski WW, Pinto‐Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR et al (2017) Molecular basis of USP7 inhibition by selective small‐molecule inhibitors. Nature 550: 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann M, Sixma TK (2017) Histone ubiquitination in the DNA damage response. DNA Repair (Amst) 56: 92–101 [DOI] [PubMed] [Google Scholar]
- Valencia‐Sanchez MI, de Ioannes P, Wang M, Vasilyev N, Chen R, Nudler E, Armache JP, Armache KJ (2019) Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. Mol Cell 74: 1010–1019.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangala JR, Sotzny F, Kruger E, Deshaies RJ, Radhakrishnan SK (2016) Nrf1 can be processed and activated in a proteasome‐independent manner. Curr Biol 26: R834–R835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargesson N (2009) Thalidomide‐induced limb defects: resolving a 50‐year‐old puzzle. BioEssays 31: 1327–1336 [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C et al (2004) In vivo activation of the p53 pathway by small‐molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL (2015) Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16: 178–189 [DOI] [PubMed] [Google Scholar]
- Vermeulen L, de Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H et al (2010) Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 12: 468–476 [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M (2003) Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA 100: 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Kawaoka S, Yu M, Shi J, Ni T, Yang W, Zhu J, Roeder RG, Vakoc CR (2013) Histone H2B ubiquitin ligase RNF20 is required for MLL‐rearranged leukemia. Proc Natl Acad Sci USA 110: 3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Li CL, Cui J, Jiao M, Wu T, Jing LI, Nan KJ (2015) BMI‐1, a promising therapeutic target for human cancer. Oncol Lett 10: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Liu Y, Edwards G, Krzizike D, Scherman H, Luger K (2018) The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci Alliance 1: e201800107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang X, Feng F, Liu W, Sun H (2020) Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B 10: 207–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Weems JC, Slaughter BD, Unruh JR, Hall SM, McLaird MB, Gilmore JM, Washburn MP, Florens L, Yasukawa T, Aso T et al (2015) Assembly of the elongin A ubiquitin ligase is regulated by genotoxic and other stresses. J Biol Chem 290: 15030–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation‐dependent c‐Myc protein degradation. Proc Natl Acad Sci USA 101: 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8: 83–93 [DOI] [PubMed] [Google Scholar]
- Wilms C, Krikki I, Hainzl A, Kilo S, Alupei M, Makrantonaki E, Wagner M, Kroeger CM, Brinker TJ, Gatzka M (2018) 2A‐DUB/Mysm1 regulates epidermal development in part by suppressing p53‐mediated programs. Int J Mol Sci 19: 687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Harreman M, Svejstrup JQ (2013a) Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta 1829: 151–157 [DOI] [PubMed] [Google Scholar]
- Wilson MD, Harreman M, Taschner M, Reid J, Walker J, Erdjument‐Bromage H, Tempst P, Svejstrup JQ (2013b) Proteasome‐mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell 154: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer‐Romero P, Chu CY, Elledge SJ, Harper JW (1999) The SCFbeta‐TRCP‐ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta‐catenin and stimulates IkappaBalpha ubiquitination in vitro . Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe‐Paganon S, Bradner JE (2015) DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348: 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden EJ, Hoffmann NA, Hicks CW, Wolberger C (2019) Mechanism of Cross‐talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell 176: 1490–1501.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden EJ, Zhang X, Wolberger C (2020) Structural basis for COMPASS recognition of an H2B‐ubiquitinated nucleosome. Elife 9: e53199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument‐Bromage H, Tempst P, Svejstrup JQ (2002) A Rad26‐Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415: 929–933 [DOI] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y (2011) The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell 43: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Li L, Zhou B, Qin Z, Dou Y (2014) H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol Cell 54: 920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci USA 98: 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lee YH, Jones AE, Woolnough JL, Zhou D, Dai Q, Wu Q, Giles KE, Townes TM, Wang H (2014) The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat Commun 5: 3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Green MR (2019) Epigenetic programing of B‐cell lymphoma by BCL6 and its genetic deregulation. Front Cell Dev Biol 7: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T (2008) Mammalian Elongin A complex mediates DNA‐damage‐induced ubiquitylation and degradation of Rpb1. EMBO J 27: 3256–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Bhatt S, Takeuchi T, Kawauchi J, Takahashi H, Tsutsui A, Muraoka T, Inoue M, Tsuda M, Kitajima S et al (2012) Transcriptional elongation factor elongin A regulates retinoic acid‐induced gene expression during neuronal differentiation. Cell Rep 2: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Yau R, Rape M (2016) The increasing complexity of the ubiquitin code. Nat Cell Biol 18: 579–586 [DOI] [PubMed] [Google Scholar]
- Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez‐Martin N, Matsumoto ML, Dixit VM et al (2017) Assembly and function of heterotypic ubiquitin chains in cell‐cycle and protein quality control. Cell 171: 918–933.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz ZB, Kofahl B, Beaudette P, Baum K, Ipenberg I, Weih F, Wolf J, Dittmar G, Scheidereit C (2014) Quantitative dissection and modeling of the NF‐kappaB p100–p105 module reveals interdependent precursor proteolysis. Cell Rep 9: 1756–1769 [DOI] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ et al (2012) RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487: 510–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M (2004) Keap1 is a redox‐regulated substrate adaptor protein for a Cul3‐dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB (2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell‐cycle progression. Mol Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati‐Brivanlou A, Derynck R (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA 98: 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones AE, Wu W, Kim J, Kang Y, Bi X, Gu Y, Popov IK, Renfrow MB, Vassylyeva MN et al (2017) Role of remodeling and spacing factor 1 in histone H2A ubiquitination‐mediated gene silencing. Proc Natl Acad Sci USA 114: E7949–E7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, le Guezennec X, Stunnenberg HG, Krasnov A et al (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
- Zhao D, Zhang X, Guan H, Xiong X, Shi X, Deng H, Li H (2016) The BAH domain of BAHD1 is a histone H3K27me3 reader. Protein Cell 7: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu J, Li B, Liu D, Yu J, Yan X, Zheng S, Wang J, Zhang L, Zhang L et al (2014) PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia 28: 1436–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400: 687–693 [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument‐Bromage H, Tempst P, Glass CK, Rosenfeld MG (2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 27: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]


