Abstract
Metastasis is the main cause of cancer-related death and the major challenge in cancer treatment. Cancer cells in circulation are termed circulating tumor cells (CTCs). Primary tumor metastasis is likely due to CTCs released into the bloodstream. These CTCs extravasate and form fatal metastases in different organs. Analyses of CTCs are clarifying the biological understanding of metastatic cancers. These data are also helpful to monitor disease progression and to inform the development of personalized cancer treatment-based liquid biopsy. However, CTCs are a rare cell population with 1–10 CTCs per ml and are difficult to isolate from blood. Numerous approaches to detect CTCs have been developed based on the physical and biological properties of the cells. The present review summarizes the progress made in detecting CTCs.
Keywords: circulating tumor cells, separation technologies, label-free methods, label-based methods
1. Introduction
Globally in 2018, there were an estimated 18.1 million new cancer cases and 9.6 million cancer-related deaths. Globally, ~20% of people aged ≤75 years are at risk of developing cancer and the mortality for these people is 10% (1,2). Cancer morbidity and mortality are rapidly increasing worldwide (3). It is estimated that by 2035, the annual number of new cancer cases and cancer-related deaths globally will be 24 million and 14.5 million, respectively (3). Since patients with cancer can display atypical symptoms at an early clinical stage for example, early gastric cancer is asymptomatic or only manifests as nausea, vomiting and other symptoms, and it is difficult to distinguish early gastric cancer from benign dyspepsia (4). If the patient doesn't pay enough attention the disease gradually deteriorates (5). The disease develops to the late stage showing obvious abdominal pain, anorexia, dyspepsia, weight loss, gastrointestinal bleeding and other symptoms before diagnosis (6). Metastasis is the major cause of cancer-related mortality, accounting for ~90% of treatment failure and cancer-related deaths (7). The development of reliable technologies to detect tumorigenesis and tumor metastasis at an earlier stage could enable the timely excision of tumors, which would undoubtedly reduce cancer-related mortality (5).
Metastasis is a multi-stage process, and cancer cells are initially released into the bloodstream from a primary tumor (8). Cancer cells in circulation are termed circulating tumor cells (CTCs) (9). Some CTCs from the primary tumor or metastatic cells survive in circulation and extravasate into the surrounding tissues and grow at the new site as single or cluster cells (10). The immune system and tumor microenvironment are important in tumor progression (11). Thousands of tumor cells are released into the blood from the primary tumor each day (11). However, on average, only a very small number of CTCs can be detected (12). Most tumor cells die during transmission to the blood (12). Biological and physical factors, such as shear stress and immune surveillance cause death of CTCs (12). Only a minor subset of CTCs (0.01%) survive (12), and their survival time is short, with a half-life from 1.0–2.4 h (13). It is currently estimated that there are typically 0–100 individual CTCs and ~0-5 CTCs clusters in 10 ml of blood sampled from the peripheral circulation of a patient with metastatic cancer, while red blood cells (RBCs), white blood cells (WBCs) and platelets in these blood samples are ~50×109, 80×109, and 3×109, respectively (14). These estimates can vary greatly according to cancer type, blood collection site and treatment stage (14).
Tumors have been traditionally diagnosed primarily based on tumor tissue biopsy (15). However, this means of diagnosis is limited by some inherent factors, such as the patient being unsuitable for surgery, inconvenient location of the tumor, clinical risks involved in tissue biopsy, and tumor heterogeneity (15). Compared with traditional tissue biopsy, liquid biopsy has notable advantages (15). Liquid biopsy was originally used in the analysis of CTCs and then extended to the analysis of cell-free DNA, cell-free RNA, micro RNA, extracellular vesicles, and tumor-derived metabolites found in blood, urine, cerebrospinal fluid, and other fluids, such as plueral fluid and ascites in patients with cancer (16). Liquid biopsy is non-invasive, hence it can be used to indicate the real-time status of disease, including information on how the tumor evolves and tumor heterogeneity (17). CTCs are integral cells that break away from the tumor and undergo metastasis (10). In contrast to other liquid biopsies, CTCs can be used to analyze the levels of DNA, RNA and protein (15,18). The functional cellular characteristics of CTCs can also be analyzed, which can provide information on cancer biology (19). The existence of CTCs has been confirmed in many cancer types, including lung (20), prostate (21), liver (22), breast (23), pancreas (24), colon (25), ovarian (26) cancers and so on. Its development includes early cancer diagnosis, prognosis monitoring, treatment efficacy, drug resistance analysis, and therapeutic targeting of CTCs to prevent metastasis, hence enabling personalized therapy (27,28). The potential value of harnessing CTCs as a non-invasive means of understanding the biology of tumor cells has been recognized by the global oncology research community (29).
CTCs can be distinguished from whole blood based on physical properties, such as size, deformability, density, adhesion, and dielectric properties (8,15). However, because of the low numbers, CTCs are very difficult to detect and capture at a satisfactorily high efficiency and purity (15). From a technical point of view, the heterogeneity of tumor cells has become another challenge in technology (8). Different tumor types can display histologic differences, which can manifest as the expression of different proteins on the cell surface, resulting in tumor cell heterogeneity (15). In addition, the same type of tumor occurring in different patients may also display different patterns and levels of protein expression on the surface of the cancer cell (30). Furthermore, cancer cells must be released from the primary tumor to disseminate. This goal necessitates the transition of epithelial tumor cells in the form of epithelial-to-mesenchymal transition (EMT) (31). During EMT, surface markers of epithelial cells, such as epithelial cell adhesion molecule (EpCAM) are downregulated, while markers of mesothelial cells are upregulated, such as vimentin (30). These alterations allow tumor cells to sever their intercellular adhesions, hence achieving their variability and invasiveness. The tumor cells invade the bloodstream and become CTCs (30). After extravasation, CTCs can invade a new organ and undergo mesenchymal-to-epithelial transition (MET) (31). MET enables CTCs that have undergone EMT to reverse and restore the epithelial phenotype, regain the ability to adhesion and metastasize (31). As tumor cells are heterogeneous, CTCs can be detected and captured depending on the targeting molecule, although this is also technically challenging (10).
There is an urgency to effectively and reliably detect and isolate CTCs (32). During recent decades, various methods have been developed to solve these technological problems (32). The aim of the present review is to summarize the latest advances in the methodology for improved enrichment and detection of CTCs.
2. Novel materials for enrichment and detection of CTCs
A number of novel materials and devices have been recently developed to enrich and detect CTCs (33,34). These include the most widely used nanomaterials and microfluidic devices (33,34).
Application of nanomaterials for isolation and detection of CTCs
Development of nanotechnology has included the adaptation of numerous state-of-the-art nanoscale materials, such as metals, metal oxides, semiconductors, liposomes, graphene and graphene oxide, into nanoparticles, nanowires, nanofibers, nanopillars, and nanotubes (18,35–40). These nanomaterials have been successfully incorporated into platforms capable of detecting CTCs. The numerous advantages of nanoparticles include their small size, ease of modification with different ligands and high surface-to-volume ratios (41). The size, shape and surface characteristics of nanoparticles enable their use in detecting biomarkers that are present in low levels and indirectly amplify the signal to enhance the detection rate, even in the early stage of cancer (42). Nanomaterials can be combined with a number of reactive functional groups to further interact with antibodies, polypeptides, aptamers, and other molecules to produce multifunctional hybrid nanomaterials for biological targeted cancer therapies, multiple detectors, biosensors, and other applications (33,41).
A recent emerging novel engineering strategy is the fabrication of biomimetic nanoparticles that combine synthetic nanomaterials with natural biomaterials, such as the membranes of leukocytes (43), RBCs (44) and platelets (45). A fluidic and multivalent engineered nanointerface decorating a microfluidic chip with aptamer-functionalized leukocyte membrane nanovesicles has been described (46). Platelet-leukocyte hybrid membrane-coated magnetic nanoparticles were designed with cell membranes modified with anti-EpCAM for the high-performance isolation of CTCs (45). The combination of nanoparticles and cell membranes significantly decreases the nonspecific adsorption and scavenging probability of nanoparticles (45). In addition, placing a layer of soft yet flexible nanovesicles between the cell and capture substrate, which serves as a cushion, can minimize cell damage resulting from interfacial collision (46).
Application of microfluidics technology for isolation and detection of CTCs
Conventional chemical and biological approaches can be integrated onto a microfluidic chip to achieve a number of basic operations, such as reaction, separation, detection, cell culture, and others (47). This variety of applications has been recognized by the description of the microfluidic chip as a lab-on-a-chip (47,48). The chip can consist of different kinds of materials, including glass, silicon, polymethylmethacrylate and polydimethylsiloxane (49–52). By manipulating the fluids at the microscale, microfluidics can be used to minimize sample consumption, enhance sensitivity and purity, and allow high-speed and high-throughput analysis (47). In addition, microfluidic platforms can be integrated into other technologies, such as nanotechnology, to enhance the efficacy of the assay in detecting CTCs (53). CTCs detection can be automated to a greater degree using microfluidic technology, as can cell culture and molecular analysis of CTCs (54). Thus, the microfluidics approach provides versatile platforms for liquid biopsy (47). For these reasons, this technology has become important in enriching and detecting CTCs (47). Numerous microfluidic devices have been developed for the separation and analysis of CTCs, such as ApoStream™ (ApoCell), system CellSearch® system (Veridex; Menarini Silicon Biosystems) and CellCollector® (Gilupi).
3. Novel technologies for isolation and detection of CTCs
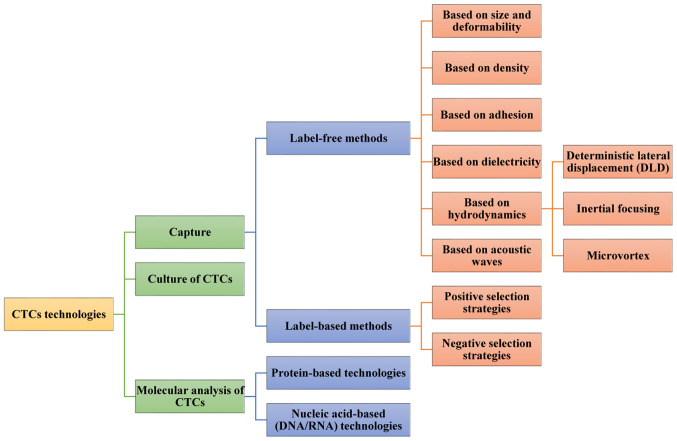
CTCs technologies mainly consist of two steps (15,55). They can be used to identify and enumerate these rare cells (8). They can also be used to better understand their molecular features (56). The first stage is detection or capture. In this stage, some methods is used to increase the concentration of CTCs and/or deplete surrounding normal blood cells (15,55). In the second stage, the retrieved CTCs are characterized by various molecular techniques (15,55). In this new age of cancer treatment, the ex vivo culture of captured CTCs also provides exceptional reagents to study cancer metastasis, as well as perform individualized preclinical testing for drug susceptibility (57) (Fig. 1).
Figure 1.
Methods for CTCs analysis. CTCs analysis technologies mainly include CTCs capture, culture and molecular analysis. CTCs are mainly detected and separated using label-free and label-based methods. Label-free methods mainly depend on physical properties of cells, which include size, deformability, density, adhesion, and dielectric properties. Label-based methods are used for targeting specific markers for selective CTCs enrichment and leukocyte depletion and divided into two sub-categories according to the target cells: Negative selection and positive selection. Two main approaches have been adopted according to current technologies for molecular characterization of CTCs: Protein-based technologies and nucleic acid-based (DNA/RNA) technologies. CTCs, circulating tumor cells.
Isolation technologies for CTCs
CTCs are mainly detected and separated using label-free and label-based methods (34). Label-free methods mainly depend on physical properties of cells, which include size, deformability, density, adhesion, and dielectric properties (15,34). Label-based methods are mainly based on the binding affinity between unique surface proteins expressed on CTCs, such as EpCAM and synthetic molecular probes (15,34). Aptamers are synthetic oligonucleotide ligands with high affinity and specificity for targets compared to an antibody/antigen interaction (18). They can be selected using systematic evolution of ligands by exponential enrichment technology. Aptamers can be used specifically to recognize numerous kinds of targets, including small metal ions, proteins, organic molecules, and whole cells (58). A number of aptamers have been developed over the past decades to target cancer cell biomarkers, such as prostate-specific membrane antigen (59), mucin 1 (60), cell surface vimentin (61), and EpCAM (41). Compared to antibodies, aptamers can easily be synthesized in large quantities and modified with different chemical groups (62). If captured, cells can be released gently by using nucleases or a complementary strand of aptamers (62). Peptides can serve as surrogates for antibodies because short peptides at the contacting interfaces participate in the molecular recognition between the antigens and the antibodies (63). Since the peptides are small and stable, they can be synthesized easily (64). They are promising probes for biological detection (63). Numerous peptides with high affinity have been designed for the detection of CTCs, and high capture efficiency has been achieved compared to antibodies (63). For instance, a microfluidic chip was developed to lithographically pattern silicon nanowires functionalized with the specific CKAAKN peptide to capture CTCs in patients with pancreatic cancer. The recovery rate exceeded 95.6%, and after enzymatic release, the purity and cell viability of the obtained CTCs was 28.5 and 93.5%, respectively (63). Capture of CTCs may be impeded because the tumor cells are heterogeneous (65). To solve this problem, two or more probes could be made in response to cancer cells for wide use in cancer diagnosis (65).
Indicators for CTCs isolation technologies
A total of six technical indicators are typically used to compare the performance of isolation technologies: i) Recovery rate; ii) purity; iii) throughput; iv) sensitivity; v) specificity; and vi) biocompatibility (32). Recovery rate, also known as capture efficiency or capture rate, refers to the proportion of captured CTCs from the total number of CTCs in the blood sample, which is used to indicate the assay efficiency in isolating CTCs. However, the recovery rate is almost always measured by recovering the cells from the estimated cells when the total number of cancer cells is already known. It is imperative to translate the aforementioned measurements into clinical samples. Purity is the percentage of isolated CTCs in the total isolated cells. The purity may be heavily biased because of different experimental designs or because of clinical samples at different disease stages. Throughput is used to demonstrate how much blood is required for CTCs analysis. While 5–10 ml volume of blood was once required for detection of CTCs, only 1–2 ml blood is now required owing to the development in CTCs isolation technology. Clinical sensitivity and specificity are used to indicate whether the technology can be used to correctly identify patients with cancer and avoid false positives. Biocompatibility refers to cells that maintain their integrity even after sequential processing (34,66). Since the actual number of CTCs in the clinical samples from patients is unknown, blood samples from healthy donors spiked with known numbers of tumor cells from cancer cell lines are used for evaluating a system's performance for these parameters (67). An optimal technology should have high recovery and purity of CTCs, high-throughput for sample processing, and the ability to retain heterogeneous CTCs populations for further downstream analysis to aid in providing options for clinical management.
Label-free methods
Methods based on size and deformability of CTCs
Size and deformability of cells are the foundation of the physical separation approaches for CTCs enrichment in peripheral blood (8). In some studies, CTCs have been described as being larger and stiffer compared with hematopoietic cells (68). Numerous technologies have been developed based on these physical differences to increase recovery rates in CTCs enrichment (34). Size-based filtration is the most common enrichment strategy for CTCs separation (69). The commercially available ISET system was one of the earliest technologies (70). In this system, blood is diluted and then filtered with a 8-µm pore size filter (70). CTCs are retained by the filter as they are larger in size, while RBCs and leukocytes can pass through the randomly distributed pores as their sizes are smaller than the pore diameter (70). The variety of CTCs separation methods depends solely on the size difference (69). Since the sizes of CTCs and leukocytes overlap, the efficiency of the size-dependent filtration system to capture target cells is limited (68). Therefore, a strategy to increase cell size by utilizing modified microbeads that specifically bind to CTCs before cell filtration was proposed to improve capture efficiency (71). Up to 91% of target cells were isolated from whole blood samples using this microfluidic capture system at a flow rate of 1 ml/min (71).
Another possible solution to this limitation is to harness the features of CTCs (72). Cells that are more deformable or smaller can more easily pass through the channel, while higher critical pressure is required to help rigid and larger cells pass through the channel; the channel will be blocked if the pressure provided is insufficient (72). A proposed mechanical low-pass filtering technique based on microfluidic constriction has a microfluidic device with consecutive constriction channels of different sizes and an electrical current-sensing detector for detection of novel CTCs in whole blood without any specific antigen-antibody interaction or biochemical modification of the cell surface (73). The ionic current is reduced when cells pass through the constriction channel. The intensity of the ionic current at the wide constriction reflects the volume of the migrating cell, while the residence time of the migrating cell at the narrow constriction depends on the deformability of the cell (73). This consecutive constriction device has been applied to distinguish CTCs from blood cells with an accuracy rate of almost 95% (73).
Cell clogging reduces the efficiency of filtration (66). To resolve this problem, an electromagnetic vibration-based filtration (eVBF) device was developed (74). The eVBF device can be used to generate an electromagnetic force with a periodic vibration that prevents cells from clogging (74). High-throughput (1 ml/min) is achieved without the need for antibodies or chemical reagents in the continuous isolation of CTCs or pretreatment processes (74). In addition, the eVBF device has been optimized with a high-amplitude vibration, and its recovery rate has increased to 80–90% for whole blood sampled with 100 or 1,000 gastric cancer cells/ml (74).
Caution and precision are required in the determination of pore size and in measurement based on cell deformability (75). In clinical cases, CTCs from patients can be small and similar in size to leukocytes (69). CTCs that have undergone EMT and stem cell-like CTCs are capable of considerable deformation and can be missed with deformability-based separation technologies (69). Therefore, physical isolation of CTCs using rational design and performance analyses based on cell size and deformability may be unreliable (75).
Methods based on density of CTCs
Density gradient centrifugation can be used to separate CTCs from other blood cells based on the different densities of different cell types (19,68). As erythrocytes and granulocytes have a higher density compared with the separation medium, they precipitate at the bottom of the pipe when centrifugation is completed (19,68). Monocytes and lymphocytes have lower densities compared with the separation medium (19,68). Hence, they remain at the surface of the medium or suspended in the medium. CTCs are mainly deposited in the monocyte enrichment layer, allowing for their convenient collection and analysis (19,68). Ficoll-Hypaque® (Cytiva) and OncoQuick® (Greiner Bio-One) density gradient centrifugation technologies are the most popular in preclinical and clinical research (8). Ficoll-Hypaque® (Cytiva) is based on a co-polymer of sucrose and epichlorohydrin (8). It is mainly used to recover mononuclear cells from peripheral blood (8). Ficoll-Hypaque® (Cytiva) has been used in laboratories for a long time (67). Nonetheless, the use of Ficoll-Hypaque® (Cytiva) remains hampered by limitations (67). For instance, the tumor cells may migrate to the plasma fraction or to the bottom of the gradient owing to the formation of aggregates (8). This can result in the loss of CTCs (8). Unlike the simple Ficoll-Hypaque separation, OncoQuick® (Greiner Bio-One) incorporates a porous barrier during the separation process to prevent separated cells from mixing (76). This approach also has disadvantages. For example, large CTCs and clusters of CTCs fall to the bottom, making it impossible to eliminate leukocytes and difficult to obtain a pure preparation of CTCs (77). To overcome these limitations, it is necessary to combine centrifugation with another enrichment method (8). The RosetteSep™ CTCs Enrichment Cocktail (StemCell Technologies Inc.) in combination with Ficoll-Hypaque® (Cytiva) separation improves the enrichment and purity of CTCs (77). The system utilizes tetrameric antibody complexes that crosslink CD45-expressing leukocytes to RBCs (77). This artificially increases the density of the labeled leukocytes, which gather at the bottom of the density gradient (78). The aforementioned system has successfully detected CTCs in 77% of patients with prostate cancer and 90% of metastatic epithelial ovarian cancer (79).
The sedimentation rate of a particle in suspension is determined by the size of the particle and the difference in density between the particle and the surrounding solution (68). The sedimentation rate increases dramatically as the size and density of the particles increase (68). Selective density amplification of CTCs using antibody-coated microbeads is another strategy to separate CTCs based on density (34). The microbeads specifically bind to CTCs to maximize the difference in sedimentation rates between CTCs and normal blood cells, such as leukocytes and erythrocytes in blood samples (34). Huang et al (68) proposed a novel density gradient centrifugation method that used biodegradable gelatin nanoparticle-coated silicon microbeads (SiO2@Gel MBs) coated with anti-EpCAM and anti-CD146 antibodies to enhance the size and density of mesenchymal CTCs. The authors reported high rates of recovery (>80%) and purity (>85%). Degradation of the gelatin coating by matrix metalloproteinase-9 enzyme enabled high rates of CTCs release (94%) and viability (92.5%) (68). The authors also demonstrated the unparalleled robustness of this method in downstream CTCs analyses, such as the detection of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α mutations (68). The efficiency and versatility of the multifunctional density microbeads approach provides new opportunities for personalized cancer diagnostics and treatments.
Methods based on adhesion of CTCs
Compared to normal blood cells, a simple and effective strategy was reported by Chen et al (80). The authors used the preferential adhesion of cancer cells to surfaces that were rough on the nanoscale to capture CTCs regardless of their physical size and without using capture antibody. The reactive ion etching method was used to treat bare glass surfaces to produce different degrees of roughness at nanoscale resolution (80). On average, the recovery rates were 88.7±3.0% and 93.3±1.5% for MCF-7 cells mixed with peripheral blood mononuclear cells and spiked in lysed blood, respectively, while for MD-MB-231 cells, the recovery rates were 94.9±2.4% and 95.4±2.2% for the peripheral blood mononuclear cells and lysed blood samples, respectively (80). The aforementioned results confirmed the efficiency of nanorough substrates to capture cancer cells (80).
Methods based on dielectricity of CTCs
In addition to size, density, and deformability, the dielectricity of CTCs can also be harnessed to distinguish CTCs from other blood cells including RBCs, platelets and WBCs (81). Dielectrophoresis-based enrichment technologies rely on the different levels of polarization between cells generated using a non-uniform alternative electric field (82). The cells are also isolated when they move toward electric fields that have different intensities (82). The polarizability of a cell depends on its composition, morphology, and frequency of the applied electric field (82). Different cell populations can be isolated when they move in opposite directions if the electrical excitation frequency is set between the cross-over frequencies for different cells (81). Dielectrophoretic field-flow fractionation has been utilized in the commercial ApoStream™ (ApoCell) system that enables the label-free isolation of viable cancer cells (83). The system utilizes a 45–85 kHz AC signal that is in the range of the cross-over frequencies of cancer cells (~30-40 kHz) and peripheral blood cells (90–140 kHz) (84). Cancer cells move toward electrodes and force blood cells to the center of the channel (85,86). The cancer cells are enriched and collected at the product collection port, while blood cells exit from the chamber via the waste outlet (85,86). The system was used to obtain recovery rates of 75.4±3.1% and 71.2±1.6% for SKOV3 ovarian cancer cells and MDA-MB-231 breast cancer cells, respectively. In addition, the viability of these enriched cancer cells exceeded 97.1% (84). The continuous-flow design of the ApoStream™ allows the processing of a 7.5-ml sample within 1 h, which is faster compared with most dielectrophoresis-based CTCs enrichment technologies (84).
Methods based on hydrodynamics
A novel approach used to enrich CTCs relies on unique hydrodynamics and is used in microfluidic channels (87–89). The hydrodynamics method is primarily based on the different movements of cells with different sizes in response to the hydrodynamic force (90). There are three main types, which are separately detailed below: Deterministic lateral displacement (DLD), inertial focusing and micro-vortex (90).
Methods based on DLD
DLD is a technology in which the microcolumns in a chip are designed and controlled to have spacings smaller or larger compared with the critical particle diameter (69). This microcolumn array can influence the displacement of the suspended particles according to hydrodynamic forces created by the flow conditions (69). A parameter termed the critical particle diameter defined in these devices predicts the displacement of suspended particles as they pass through the array of microcolumns (69,88). If CTCs are larger compared with the critical diameter of the device, they are laterally displaced when they collide with the microcolumn and move to one side (following the bumped mode) (69,88). In contrast, blood cells that are smaller than the critical diameter maintain their original path without being laterally displaced after collision with the microcolumn (following the zigzag mode) (69,88). The array of microcolumns in DLD devices is arranged so that the trajectory of the particles is precisely controlled and particles larger or smaller compared with the critical diameter are more rapidly separated (69,88). A DLD array device was designed by Loutherback et al (91) to separate CTCs at a flow rate of 10 ml/min. The capture efficiency of this device exceeded 85% without affecting cell viability (91). The DLD device can become clogged with cells if the detected clinical samples are too sticky or contain large amounts of debris (91).
Methods based on inertial focusing
The inertial focusing method used in microfluidic devices exploits the advantage of the hydraulic phenomenon, in which particles/cells of different sizes occupy different equilibrium positions in the cross-section of the microfluidic channel because of the balance between the inertial lift force and the Dean drag force (87). As a result, when flowing in a spiral channel, larger cells move toward the inner wall, while smaller cells move toward the outer wall under the influences of these forces (69). CTCs separate from blood cells (87). The device can be operated at a wide range of flow rates from 5 µl/min-8 ml/min (87). Hydraulic-based separation has been further exploited by constructing a cascaded microfluidic device consisting of two spiral channels and one zigzag channel, to simultaneously isolate different types of CTCs from human blood (87). Recovery rates of 80.75% for A549 human lung cancer cells and 73.75% for MCF-7 human breast cancer cells have been reported (87).
Methods based on microvortex
Microvortex trapping is another hydrodynamics-based CTCs enrichment technology that enriches CTCs in the microvortex formed in a microfluidic device. The method is based on the high flow rate that produces microvortices when cells flow through regions of expanded and contracted volume. Smaller cells flow smoothly through the main channel, while larger cells swirl in the vortex and remain in the chamber. Finally, the flow rate is reduced to release the captured cells from the vortex (89,92,93). The microfluidic vortex chip was used by Rennier et al (94) to isolate CTCs from patients with advanced prostate cancer. CTCs were isolated with a purity of 1.74–37.59% (94). A disadvantage of the method is that some microfluidic vortex chips may produce bubbles inside the channel (94).
Methods based on acoustic waves
In 2014, Ding et al (95) proposed the use of acoustic waves to separate pathological cells from normal blood cells. Other studies have also addressed the use of acoustic waves to separate CTCs (96). Cells that are sustained by acoustic force with different magnitudes depending on the cell sizes, density, and deformation are translated by the acoustic force to the nodes with zero periodic pressure variations or maximum pressure to reach a balance point (95). An acoustic-based microfluidic device was developed by Wu et al (96) using tilted-angle surface acoustic waves to form multiple regions of slanted nodes or antinodes inside the microfluidic channel. Cells transiting these regions experience different levels of acoustic forces, which may change their positions inside the channel (90). A constructed high-throughput platform integrating acoustics and microfluidics has enabled the separation of CTCs from peripheral blood (96). The separated cells remain structurally, biologically, and functionally intact (96). The system can separate cancer cells from leukocytes at a throughput of up to 7.5 ml h−1 with an 86% recovery rate and no effect on cell proliferation (96). Using acoustic waves to separate cells is a relatively mild method that can maintain the original state, phenotype, and genotype of CTCs to the greatest extent (96). However, this method is greatly influenced by environmental factors (96).
Label-based methods
Specific cancer biomarkers are indispensable in most biological detection methods (47). Cancer biomarkers refer to the molecular changes that are measurable and occur between normal and cancerous tissues of patients (47). Specific molecular characteristics and pathological changes occur in each cancer during the transition from normal to cancerous cells (47). It is important to identify these biomarkers so that they can be further applied in capturing and isolating CTCs that are commonly described to express EpCAM and cytokeratins (CKs), and to be nucleated (they are identified by staining with a nuclear dye like 4′,6-diamidino-2-phenylindole) (67). These biomarkers are not used to express the leukocyte surface marker CD45 (67). Other biomarkers commonly used to specifically detect CTCs include human epidermal growth factor receptor 2, epidermal growth factor receptor, vimentin, and CD44 (47). In addition, ‘negative markers’ may be used to identify and eliminate questionable cells (47). These markers include the leukocyte markers CD45, CD66, and CD15, platelet marker CD61, and apoptosis marker M30 (97). Label-based methods can be further divided into two sub-categories according to the target cells: Negative selection and positive selection (97).
Positive selection strategies
CTCs are regarded as target cells in positive sorting methods (98). EpCAM is used as a specific biomarker in most of these methods (19). Positive selection to isolate CTCs is performed based on ligand-labeled beads, columns, or other devices (8). The CellSearch® system (Veridex; Menarini Silicon Biosystems) is a commonly accepted method that automatically detects EpCAM- and keratin-positive CTCs (32). This is followed by immunostaining of the captured cells and semi-automated fluorescence microscopy to evaluate the immunofluorescence (19). The aforementioned system has been approved by the United States Food and Drug Administration for analysis of blood samples from patients with metastatic breast, prostate, and colorectal cancer (99). The capture step is characterized by high sensitivity, but may lack specificity. In another innovative method marketed as MagSweeper by Illumina Inc., blood samples are diluted and prelabeled with EpCAM-coated magnetic particles (8). The cells are subsequently captured with a magnetic rod that is swept through the sample (56). The magnetic rod is then placed in a washing well, and the labeled cells are released by an external magnetic field in a buffer solution (56). The released cells remain viable and can be successfully transferred and cultured (56).
All the aforementioned methods have limitations because they rely on the enrichment of EpCAM-positive cells. Hence, they can only detect the EpCAM-positive subpopulation of CTCs. However, once CTCs undergo EMT, these EpCAM-based methods may be ineffective because EMT reduces EpCAM expression. CTCs with low or no EpCAM expression can remain undetected and their capture rate is greatly decreased (30).
Negative selection strategies
Leukocytes are regarded as target cells in negative sorting (34). Anti-CD45 is the biomarker used most often, reflecting its binding affinity with leukocytes (19). The EasySep® leukocyte depletion kit marketed by Stem Cells Technologies Inc. allows the enrichment of CTCs through leukocyte depletion using CD45-labeled magnetic beads (100). The CTCs remain viable for use in further experiments after leukocyte depletion (100). Leukocyte depletion methods reportedly reduce purity compared to preparations obtained by positive CTCs selection. Hence, negative selection strategies are often used in combination with other enrichment technologies (34).
Combination of label-free and label-based methods
Label-free isolation does not vary with the level of expression of the selected marker on the cell surface (90). Instead, the approach is used according to the physical differences between CTCs and blood cells (90). In addition, a label-free isolation method can be realized simply, rapidly, and at low cost compared with label-based isolation (90). However, label-free isolation has low specificity due to the overlap in size, density, rigidity, and other parameters between CTCs and leukocytes (68). Numerous researchers have sought to combine label-free and label-based methods to increase the efficiency of the isolation process, as both the label-free methods and label-based methods have limitations (101,102). The novel Herringbone (HB)-Chip is designed such that the microvortex increases the number of interactions between target CTCs and the antibody-coated chip surface (101). This system has been used to successfully isolate CTCs in patients with metastatic disease for different cancers, such as metastatic prostate and breast cancer (103), as well as to isolate CTCs clusters (104).
In vivo isolation of CTCs
Ex vivo CTCs isolation technologies have relatively low sensitivity as limited volume of blood is sampled from patients (47). A total of 1–10 ml blood is required for CTCs detection (90). However, the amount of CTCs in blood is very small with 1–10 CTCs/ml (90). The detection systems used also have limitations. CellCollector® marketed by Gilupi GmbH isolates CTCs based on EpCAM recognition. It is the first in vivo CTCs isolation product with CE approval (CE abbreviation of French phrase ‘Conformité Européene’ which literally means ‘European Conformity’) (67). Specifically, CellCollector® (Gilupi) is a medical wire coated with anti-EpCAM antibodies. The wire is directly placed in the bloodstream of the patient through a permanent catheter. The wire remains in the vein of the arm for 30 min, where it contacts a larger volume of blood and captures CTCs in vivo during this period (105,106). CTCs that have not been detected in healthy volunteers could be isolated in early-stage cancer patients not yet diagnosed with distant metastases (75). However, it is not simple to implement this technique in the clinic as manual selection is required to detect CTCs (67).
Culture of CTCs
Culture of viable CTCs is an attractive option for the study of captured CTCs (107). Once a CTCs line is established in cell culture, it can be used for direct testing of drug sensitivity or can be implanted into immunosuppressed mice to create xenograft models, permitting further drug testing and genetic profiling studies (57,107). Additionally, the metastatic mechanism can be further understood by using experimental platforms that mimic the tumor microenvironment in vitro (108). A three-dimensional porous poly(ε-caprolactone) scaffold-based method was established by De et al (108) to mimic more closely the native cellular in vivo environment and allow the deposition of extracellular matrix. Culture of CTCs from RBC-depleted nucleated cell pellets of patients with advanced breast cancer under hypoxic conditions without any prior enrichment allowed the detection of CK-positive and CD45-negative CTCs in 12 of 16 patient samples (108). However, as the captured CTCs were removed from their host environment and the immune system, they may be altered, precluding meaningful conclusions regarding disease evolution or treatment resistance (108). Hence, it is necessary to establish robust and reliable culture conditions for CTCs.
Molecular analysis of CTCs
It has been suggested in some studies that the enumeration of CTCs is not sufficient to guide therapeutic decisions (109). Rather, molecular analysis of CTCs should be performed to determine the design of the target therapy (110). Two main approaches have been adopted according to current technologies for molecular characterization of CTCs: Protein-based technologies and nucleic acid-based (DNA/RNA) technologies (10). These are reviewed in detail elsewhere (10,15,98,110) and not discussed in the present review.
4. Discussion
CTCs detection has numerous advantages compared to conventional invasive biopsy (41). For example, with approval of the patients, blood samples can be easily collected. CTCs harbor considerable information on tumorigenesis, tumor progression, metastasis, and drug resistance (41). This information increases the knowledge and treatment of tumor diseases (15). The new approach of employing CTCs has paved the way for next-generation liquid biopsy diagnostics, especially in tumors that are difficult to biopsy and in metastatic lesions (109).
There is great anticipation for label-free and label-based methods for CTCs isolation (90). Each method has value, and each aims to overcome the pitfalls involving sensitivity, specificity, throughput, and/or purity (90). Numerous reliable CTCs isolation platforms are available (90). However, the methodology that is superior and that would be the most suitable for the end user requirements remains unclear (82). These questions can be answered based on accrued biological knowledge (82).
The only two types of CTCs assays that are presently used are CellSearch® system (Veridex; Menarini Silicon Biosystems) and CellCollector® (Gilupi) (67). Both technologies have been limited in their clinical application, largely due to their low sensitivity (33). The technologies need to satisfy the following demands to meet the clinical requirements of CTCs detection (90). First, the assays must fully consider cell heterogeneity to distinguish different subpopulations of CTCs, and to achieve more efficient isolation of the target cells (111). This goal may require new technologies or a combination of several technologies (10). Second, high-throughput is conducive for the rapid recovery of a sufficient amount of cancer cells collected from a large volume. A high-throughput system that is more suited to clinical applications is required (34). Third, a shorter separation time would help preserve cell freshness, viability, and integrity and would maximize the potential to establish in vitro cultured CTCs lines (108). These advancements would make it possible to perform tasks such as investigation of CTCs phenotypic and biomarker heterogeneity or isolation of CTCs clusters that are not possible with slower processing methods due to the degradation of CTCs (108). Fourth, specific verification strategies to prove that the isolated targeted cells are real CTCs are important for clinical applications (8). Fifth, applying automated devices as much as possible is desirable to minimize intervention by human operators (98). Sixth, standardization is crucial to yield convincing and reproducible results and ensure the generalized use of the approach (42). Finally, clinical demonstration of the prowess of this technology in clinical trials is important (112). Most of the detection technologies have not yet been fully applied in clinical practice, and there is a demand for large-scale clinical validation (32). Moving the aforementioned techniques from the bench to the clinic is becoming a hot topic of research in the CTCs field.
It is hypothesized that there are two main directions for the development of CTCs in the future. One is the development of CTCs detection technology with higher sensitivity and specificity. The other is functional assessments of CTCs, especially the decoding of molecular pathological characteristics such as genes, proteins, and methylation expression profiles. Although great progress has been made, there is a long way to go before CTCs-based liquid biopsy is widely used as a routine test in clinical applications.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CTCs
circulating tumor cells
- RBCs
red blood cells
- WBCs
white blood cells
- EMT
epithelial-to-mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- MET
mesenchymal-to-epithelial transition
- eVBF
electromagnetic vibration-based filtration
- DLD
deterministic lateral displacement
Funding Statement
The present study was funded by the Maternal and Child Health Research Project of Jiangsu Province in 2019 (grant no. F201952) and Huai'an Health and Scientific Research Project in 2019 (grant no. 20).
Funding
The present study was funded by the Maternal and Child Health Research Project of Jiangsu Province in 2019 (grant no. F201952) and Huai'an Health and Scientific Research Project in 2019 (grant no. 20).
Availability of data and materials
Data sharing is not applicable to this article, as no datasets were generated or analyzed during the current study.
Authors' contributions
XH, XZ and YL all contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Cheng SJ, Hsieh KY, Chen SL, Chen CY, Huang CY, Tsou HI, Kumar PV, Hsieh JC, Chen GY. Microfluidics and nanomaterial-based technologies for circulating tumor cell isolation and detection. Sensors (Basel) 2020;20:1875. doi: 10.3390/s20071875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maconi G, Manes G, Porro GB. Role of symptoms in diagnosis and outcome of gastric cancer. World J Gastroenterol. 2008;14:1149–1155. doi: 10.3748/wjg.14.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–2044. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 7.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 8.Gabriel MT, Calleja LR, Chalopin A, Ory B, Heymann D. Circulating tumor cells: A review of non-EpCAM-based approaches for cell enrichment and isolation. Clin Chem. 2016;62:571–581. doi: 10.1373/clinchem.2015.249706. [DOI] [PubMed] [Google Scholar]
- 9.Prasanna BK, Balakrishnan A, Kumar P. Circulating tumor cell clusters and circulating tumor cell-derived explant models as a tool for treatment response. Biotechniques. 2020;69:362–363. doi: 10.2144/btn-2020-0029. [DOI] [PubMed] [Google Scholar]
- 10.Tellez-Gabriel M, Heymann MF, Heymann D. Circulating tumor cells as a tool for assessing tumor heterogeneity. Theranostics. 2019;9:4580–4594. doi: 10.7150/thno.34337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leone K, Poggiana C, Zamarchi R. The interplay between circulating tumor cells and the immune system: From immune escape to cancer immunotherapy. Diagnostics (Basel) 2018;8:59. doi: 10.3390/diagnostics8030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tayoun T, Faugeroux V, Oulhen M, Aberlenc A, Pawlikowska P, Farace F. CTC-derived models: A window into the seeding capacity of circulating tumor cells (CTCs) Cells. 2019;8:1145. doi: 10.3390/cells8101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 14.Au SH, Edd J, Haber DA, Maheswaran S, Stott SL, Toner M. Clusters of circulating tumor cells: A biophysical and technological perspective. Curr Opin Biomed Eng. 2017;3:13–19. doi: 10.1016/j.cobme.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiele JA, Bethel K, Králíčková M, Kuhn P. Circulating tumor cells: Fluid surrogates of solid tumors. Annu Rev Pathol. 2017;12:419–447. doi: 10.1146/annurev-pathol-052016-100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mocan T, Simão AL, Castro RE, Rodrigues CMP, Słomka A, Wang B, Strassburg C, Wöhler A, Willms AG, Kornek M. Liquid biopsies in hepatocellular carcinoma: Are we winning? J Clin Med. 2020;9:1541. doi: 10.3390/jcm9051541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Zhang J, Li H, Xu W, Zhang X. The potential of liquid biopsies in gastrointestinal cancer. Clin Biochem. 2020;84:1–12. doi: 10.1016/j.clinbiochem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Zhang P, Dou L, Wang Y, Sun K, Zhang X, Song G, Zhao C, Li K, Bai Y, et al. Detection of circulating tumor cells in breast cancer patients by nanopore sensing with aptamer-mediated amplification. ACS Sens. 2020;5:2359–2366. doi: 10.1021/acssensors.9b02537. [DOI] [PubMed] [Google Scholar]
- 19.Yap K, Cohen EN, Reuben JM, Khoury JD. Circulating tumor cells: State-of-the-art update on technologies and clinical applications. Curr Hematol Malig Rep. 2019;14:353–357. doi: 10.1007/s11899-019-00531-x. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, Yang X, Li Y, Zhao P, Fu R, Ren T, Hu P, Wu Y, Yang H, Guo N. Clinical significance of circulating tumor cells and metabolic signatures in lung cancer after surgical removal. J Transl Med. 2020;18:243. doi: 10.1186/s12967-020-02401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapatero A, Gómez-Caamaño A, Cabeza Rodriguez MÁ, Muinelo-Romay L, Martin de Vidales C, Abalo A, Calvo Crespo P, Leon Mateos L, Olivier C, Vega Piris LV. Detection and dynamics of circulating tumor cells in patients with high-risk prostate cancer treated with radiotherapy and hormones: A prospective phase II study. Radiat Oncol. 2020;15:137. doi: 10.1186/s13014-020-01577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun YF, Wang PX, Cheng JW, Gong ZJ, Huang A, Zhou KQ, Hu B, Gao PT, Cao Y, Qiu SJ, et al. Postoperative circulating tumor cells: An early predictor of extrahepatic metastases in patients with hepatocellular carcinoma undergoing curative surgical resection. Cancer Cytopathol. 2020;128:733–745. doi: 10.1002/cncy.22304. [DOI] [PubMed] [Google Scholar]
- 23.Deutsch TM, Stefanovic S, Feisst M, Fischer C, Riedel F, Fremd C, Domschke C, Pantel K, Hartkopf AD, Sutterlin M, et al. Cut-off analysis of CTC change under systemic therapy for defining early therapy response in metastatic breast cancer. Cancers (Basel) 2020;12:1055. doi: 10.3390/cancers12041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimitrov-Markov S, Perales-Patón J, Bockorny B, Dopazo A, Muñoz M, Baños N, Bonilla V, Menendez C, Duran Y, Huang L, et al. Discovery of new targets to control metastasis in pancreatic cancer by single-cell transcriptomics analysis of circulating tumor cells. Mol Cancer Ther. 2020;19:1751–1760. doi: 10.1158/1535-7163.MCT-19-1166. [DOI] [PubMed] [Google Scholar]
- 25.Pan X, Zhang X. Utility of circulating tumor cells and DNA in the management of advanced colorectal cancer. Future Oncol. 2020;16:1289–1299. doi: 10.2217/fon-2020-0073. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Lim M, Kim JY, Shin SJ, Cho YK, Cho CH. Circulating tumor cells enumerated by a centrifugal microfluidic device as a predictive marker for monitoring ovarian cancer treatment: A pilot study. Diagnostics (Basel) 2020;10:249. doi: 10.3390/diagnostics10040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: What we know and what we expect (Review) Int J Oncol. 2016;49:2206–2216. doi: 10.3892/ijo.2016.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabel L, Proudhon C, Gortais H, Loirat D, Coussy F, Pierga JY, Bidard FC. Circulating tumor cells: Clinical validity and utility. Int J Clin Oncol. 2017;22:421–430. doi: 10.1007/s10147-017-1105-2. [DOI] [PubMed] [Google Scholar]
- 29.Aoki M, Shoji H, Kashiro A, Takeuchi K, Shimizu Y, Honda K. Prospects for comprehensive analyses of circulating tumor cells in tumor biology. Cancers (Basel) 2020;12:1135. doi: 10.3390/cancers12051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown HK, Tellez-Gabriel M, Cartron PF, Vallette FM, Heymann MF, Heymann D. Characterization of circulating tumor cells as a reflection of the tumor heterogeneity: Myth or reality? Drug Discov Today. 2019;24:763–772. doi: 10.1016/j.drudis.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Lowes LE, Allan AL. Circulating tumor cells and implications of the epithelial-to-mesenchymal transition. Adv Clin Chem. 2018;83:121–181. doi: 10.1016/bs.acc.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Lin E, Cao T, Nagrath S, King MR. Circulating tumor cells: Diagnostic and therapeutic applications. Annu Rev Biomed Eng. 2018;20:329–352. doi: 10.1146/annurev-bioeng-062117-120947. [DOI] [PubMed] [Google Scholar]
- 33.Hong S, Wang AZ. Nanotechnology enabling the use of circulating tumor cells (CTCs) as reliable cancer biomarkers. Adv Drug Deliv Rev. 2018;125:1–2. doi: 10.1016/j.addr.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Zou D, Cui D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol Med. 2018;15:335–353. doi: 10.20892/j.issn.2095-3941.2018.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y, Peng Y, Tang S, Shen H, Sheng S, Wang Y, Wang T, Cai J, Xie G, Feng W. PdIrBP mesoporous nanospheres combined with superconductive carbon black for the electrochemical determination and collection of circulating tumor cells. Mikrochim Acta. 2020;187:216. doi: 10.1007/s00604-020-4213-z. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Peng J, Lai Y, Wu B, Sun L, Weng J. Ultrasensitive label-free detection of circulating tumor cells using conductivity matching of two-dimensional semiconductor with cancer cell. Biosens Bioelectron. 2019;142:111520. doi: 10.1016/j.bios.2019.111520. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Chen L, Du S, Wu J, Quan M, Yin H, Wu Y, Ye X, Liang X, Jiang H. High sensitive detection of circulating tumor cell by multimarker lipid magnetic nanoparticles and clinical verifications. J Nanobiotechnology. 2019;17:116. doi: 10.1186/s12951-019-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dou B, Xu L, Jiang B, Yuan R, Xiang Y. Aptamer-functionalized and gold nanoparticle array-decorated magnetic graphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumor cells in whole blood. Anal Chem. 2019;91:10792–10799. doi: 10.1021/acs.analchem.9b02403. [DOI] [PubMed] [Google Scholar]
- 39.Chen SL, Chen CY, Hsieh JC, Yu ZY, Cheng SJ, Hsieh KY, Yang JW, Kumar PV, Lin SF, Chen GY. Graphene oxide-based biosensors for liquid biopsies in cancer diagnosis. Nanomaterials (Basel) 2019;9:1725. doi: 10.3390/nano9121725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, Zou C, Zhang J, Jiang W, Guan F, Tang K, Li S, Li G, Wang J, Ke Z. dynamically monitoring the clonal evolution of lung cancer based on the molecular characterization of circulating tumor cells using aptamer cocktail-modified nanosubstrates. ACS Appl Mater Interfaces. 2020;12:5671–5679. doi: 10.1021/acsami.9b22234. [DOI] [PubMed] [Google Scholar]
- 41.Ding P, Wang Z, Wu Z, Zhou Y, Sun N, Pei R. Natural biointerface based on cancer cell membranes for specific capture and release of circulating tumor cells. ACS Appl Mater Interfaces. 2020;12:20263–20270. doi: 10.1021/acsami.0c03355. [DOI] [PubMed] [Google Scholar]
- 42.Li W, Wang H, Zhao Z, Gao H, Liu C, Zhu L, Wang C, Yang Y. Emerging nanotechnologies for liquid biopsy: The detection of circulating tumor cells and extracellular vesicles. Adv Mater. 2019;31:e1805344. doi: 10.1002/adma.201970318. [DOI] [PubMed] [Google Scholar]
- 43.Chen M, Liu A, Chen B, Zhu DM, Xie W, Deng FF, Ji LW, Chen LB, Huang HM, Fu YR, et al. Erythrocyte-derived vesicles for circulating tumor cell capture and specific tumor imaging. Nanoscale. 2019;11:12388–12396. doi: 10.1039/C9NR01805K. [DOI] [PubMed] [Google Scholar]
- 44.Xiong K, Wei W, Jin Y, Wang S, Zhao D, Wang S, Gao X, Qiao C, Yue H, Ma G, Xie HY. Biomimetic immuno-magnetosomes for high-performance enrichment of circulating tumor cells. Adv Mater. 2016;28:7929–7935. doi: 10.1002/adma.201601643. [DOI] [PubMed] [Google Scholar]
- 45.Rao L, Meng QF, Huang Q, Wang Z, Yu GT, Li A, Ma W, Zhang N, Guo SS, Zhao XZ, et al. Platelet-leukocyte hybrid membrane-coated immunomagnetic beads for highly efficient and highly specific isolation of circulating tumor cells. Adv Funct Mater. 2018;28:1803531. doi: 10.1002/adfm.201870241. [DOI] [Google Scholar]
- 46.Wu L, Ding H, Qu X, Shi X, Yang J, Huang M, Zhang J, Zhang H, Song J, Zhu L, et al. Fluidic multivalent membrane nanointerface enables synergetic enrichment of circulating tumor cells with high efficiency and viability. J Am Chem Soc. 2020;142:4800–4806. doi: 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
- 47.Gribko A, Künzel J, Wünsch D, Lu Q, Nagel SM, Knauer SK, Stauber RH, Ding GB. Is small smarter? Nanomaterial-based detection and elimination of circulating tumor cells: Current knowledge and perspectives. Int J Nanomedicine. 2019;14:4187–4209. doi: 10.2147/IJN.S198319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmanogli A, Gokcen D. Identification of circulating tumor cells using plasmonic resonance effect: Lab-on-a-chip analysis and modelling. J Nanosci Nanotechnol. 2020;20:1341–1350. doi: 10.1166/jnn.2020.17163. [DOI] [PubMed] [Google Scholar]
- 49.Al-Halhouli A, Al-Faqheri W, Alhamarneh B, Hecht L, Dietzel A. Spiral microchannels with trapezoidal cross section fabricated by femtosecond laser ablation in glass for the inertial separation of microparticles. Micromachines (Basel) 2018;9:171. doi: 10.3390/mi9040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao N, Nie Y, Tadimety A, Shen T, Zhang JXJ. Microfluidics-enabled rapid manufacturing of hierarchical silica-magnetic microflower toward enhanced circulating tumor cell screening. Biomater Sci. 2018;6:3121–3125. doi: 10.1039/C8BM00851E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson JM, Witek MA, Hupert ML, Brady C, Pullagurla S, Kamande J, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ, Soper SA. UV activation of polymeric high aspect ratio microstructures: Ramifications in antibody surface loading for circulating tumor cell selection. Lab Chip. 2014;14:106–117. doi: 10.1039/C3LC50618E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan X, Jia C, Yang J, Li G, Mao H, Jin Q, Zhao J. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens Bioelectron. 2015;71:380–386. doi: 10.1016/j.bios.2015.04.080. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Zhao D, Zhang Y, Huang L, Huang H, Guo Q, Zhang W, Hou C, Wang H, Zhang Q, et al. Facile synthesis of 3D hierarchical micro-/nanostructures in capillaries for efficient capture of circulating tumor cells. J Colloid Interface Sci. 2020;575:108–118. doi: 10.1016/j.jcis.2020.04.087. [DOI] [PubMed] [Google Scholar]
- 54.Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat Protoc. 2018;13:34–58. doi: 10.1038/nprot.2017.125. [DOI] [PubMed] [Google Scholar]
- 55.Harigopal M, Kowalski D, Vosoughi A. Enumeration and molecular characterization of circulating tumor cells as an innovative tool for companion diagnostics in breast cancer. Expert Rev Mol Diagn. 2020;20:815–828. doi: 10.1080/14737159.2020.1784009. [DOI] [PubMed] [Google Scholar]
- 56.Deng G, Krishnakumar S, Powell AA, Zhang H, Mindrinos MN, Telli ML, Davis RW, Jeffrey SS. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maheswaran S, Haber DA. Ex vivo culture of CTCs: An emerging resource to guide cancer therapy. Cancer Res. 2015;75:2411–2415. doi: 10.1158/0008-5472.CAN-15-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun D, Lu J, Zhang L, Chen Z. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Anal Chim Acta. 2019;1082:1–17. doi: 10.1016/j.aca.2019.07.054. [DOI] [PubMed] [Google Scholar]
- 59.Fan X, Guo Y, Wang L, Xiong X, Zhu L, Fang K. Diagnosis of prostate cancer using anti-PSMA aptamer A10-3.2-oriented lipid nanobubbles. Int J Nanomedicine. 2016;11:3939–3950. doi: 10.2147/IJN.S112951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin X, Chen B, He M, Hu B. A multifunctional platform for the capture, release, and enumeration of circulating tumor cells based on aptamer binding, nicking endonuclease-assisted amplification, and inductively coupled plasma mass spectrometry detection. Anal Chem. 2020;92:10308–10315. doi: 10.1021/acs.analchem.0c00276. [DOI] [PubMed] [Google Scholar]
- 61.Zheng Y, Zhang J, Huang M, Wang T, Qu X, Wu L, Song J, Wang W, Song Y, Yang C. Selection of aptamers against vimentin for isolation and release of circulating tumor cells undergoing epithelial mesenchymal transition. Anal Chem. 2020;92:5178–5184. doi: 10.1021/acs.analchem.9b05690. [DOI] [PubMed] [Google Scholar]
- 62.Ding P, Wang Z, Wu Z, Zhu W, Liu L, Sun N, Pei R. Aptamer-based nanostructured interfaces for the detection and release of circulating tumor cells. J Mater Chem B. 2020;8:3408–3422. doi: 10.1039/C9TB02457C. [DOI] [PubMed] [Google Scholar]
- 63.Shen Q, Yang H, Peng C, Zhu H, Mei J, Huang S, Chen B, Liu J, Wu W, Cao S. Capture and biological release of circulating tumor cells in pancreatic cancer based on peptide-functionalized silicon nanowire substrate. Int J Nanomedicine. 2018;14:205–214. doi: 10.2147/IJN.S187892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang SG, Zhang B, Li CG, Zhu JQ, Sun BS, Wang CL. Sorting and gene mutation verification of circulating tumor cells of lung cancer with epidermal growth factor receptor peptide lipid magnetic spheres. Thorac Cancer. 2020;11:2887–2895. doi: 10.1111/1759-7714.13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Y, Jiang L, Huang Y, Yang Y, He Y, Lu C, Yang H. DNA-mediated reversible capture and release of circulating tumor cells with a multivalent dual-specific aptamer coating network. Chem Commun (Camb) 2019;55:5387–5390. doi: 10.1039/C9CC02365H. [DOI] [PubMed] [Google Scholar]
- 66.Magbanua MJ, Das R, Polavarapu P, Park JW. Approaches to isolation and molecular characterization of disseminated tumor cells. Oncotarget. 2015;6:30715–30729. doi: 10.18632/oncotarget.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa C, Dávila-Ibáñez AB. Methodology for the isolation and analysis of CTCs. Adv Exp Med Biol. 2020;1220:45–59. doi: 10.1007/978-3-030-35805-1_4. [DOI] [PubMed] [Google Scholar]
- 68.Huang Q, Wang FB, Yuan CH, He Z, Rao L, Cai B, Chen B, Jiang S, Li Z, Chen J, et al. Gelatin nanoparticle-coated silicon beads for density-selective capture and release of heterogeneous circulating tumor cells with high purity. Theranostics. 2018;8:1624–1635. doi: 10.7150/thno.23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hao SJ, Wan Y, Xia YQ, Zou X, Zheng SY. Size-based separation methods of circulating tumor cells. Adv Drug Deliv Rev. 2018;125:3–20. doi: 10.1016/j.addr.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 70.Magbanua MJ, Park JW. Isolation of circulating tumor cells by immunomagnetic enrichment and fluorescence-activated cell sorting (IE/FACS) for molecular profiling. Methods. 2013;64:114–118. doi: 10.1016/j.ymeth.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 71.Sun N, Li X, Wang Z, Li Y, Pei R. High-purity capture of CTCs based on micro-beads enhanced isolation by size of epithelial tumor cells (ISET) method. Biosens Bioelectron. 2018;102:157–163. doi: 10.1016/j.bios.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 72.Aghaamoo M, Zhang Z, Chen X, Xu J. Deformability-based circulating tumor cell separation with conical-shaped microfilters: Concept, optimization, and design criteria. Biomicrofluidics. 2015;9:034106. doi: 10.1063/1.4922081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki T, Kaji N, Yasaki H, Yasui T, Baba Y. Mechanical low-pass filtering of cells for detection of circulating tumor cells in whole blood. Anal Chem. 2020;92:2483–2491. doi: 10.1021/acs.analchem.9b03939. [DOI] [PubMed] [Google Scholar]
- 74.Xiang A, Xue M, Ren F, Wang L, Ye Z, Li D, Ji Q, Ji G, Lu Z. High-throughput and continuous flow isolation of rare circulating tumor cells and clusters in gastric cancer from human whole blood samples using electromagnetic vibration-based filtration. Oncol Rep. 2020;43:1975–1985. doi: 10.3892/or.2020.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lei KF. A review on microdevices for isolating circulating tumor cells. Micromachines (Basel) 2020;11:531. doi: 10.3390/mi11050531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Z, Luo G, Du W, Kong T, Liu C, Liu Z. Recent advances in microfluidic platforms applied in cancer metastasis: Circulating tumor cells' (CTCs) isolation and tumor-on-a-chip. Small. 2020;16:e1903899. doi: 10.1002/smll.201903899. [DOI] [PubMed] [Google Scholar]
- 77.Drucker A, Teh EM, Kostyleva R, Rayson D, Douglas S, Pinto DM. Comparative performance of different methods for circulating tumor cell enrichment in metastatic breast cancer patients. PLoS One. 2020;15:e0237308. doi: 10.1371/journal.pone.0237308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soler A, Cayrefourcq L, Mazel M, Alix-Panabières C. EpCAM-independent enrichment and detection of viable circulating tumor cells using the EPISPOT assay. Methods Mol Biol. 2017;1634:263–276. doi: 10.1007/978-1-4939-7144-2_22. [DOI] [PubMed] [Google Scholar]
- 79.He W, Kularatne SA, Kalli KR, Prendergast FG, Amato RJ, Klee GG, Hartmann LC, Low PS. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. Int J Cancer. 2008;123:1968–1973. doi: 10.1002/ijc.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RH, Macoska JA, Merajver SD, Fu J. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano. 2013;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waheed W, Alazzam A, Mathew B, Christoforou N, Abu-Nada E. Lateral fluid flow fractionation using dielectrophoresis (LFFF-DEP) for size-independent, label-free isolation of circulating tumor cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1087-1088:133–137. doi: 10.1016/j.jchromb.2018.04.046. [DOI] [PubMed] [Google Scholar]
- 82.Murlidhar V, Rivera-Báez L, Nagrath S. Affinity versus label-free isolation of circulating tumor cells: Who wins? Small. 2016;12:4450–4463. doi: 10.1002/smll.201601394. [DOI] [PubMed] [Google Scholar]
- 83.Nasiri R, Shamloo A, Ahadian S, Amirifar L, Akbari J, Goudie MJ, Lee K, Ashammakhi N, Dokmeci MR, Di Carlo D, Khademhosseini A. Microfluidic-based approaches in targeted cell/particle separation based on physical properties: Fundamentals and applications. Small. 2020;16:e2000171. doi: 10.1002/smll.202000171. [DOI] [PubMed] [Google Scholar]
- 84.Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, Davis DW. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shim S, Gascoyne P, Noshari J, Hale KS. Dynamic physical properties of dissociated tumor cells revealed by dielectrophoretic field-flow fractionation. Integr Biol (Camb) 2011;3:850–862. doi: 10.1039/c1ib00032b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdulla A, Liu W, Gholamipour-Shirazi A, Sun J, Ding X. High-throughput isolation of circulating tumor cells using cascaded inertial focusing microfluidic channel. Anal Chem. 2018;90:4397–4405. doi: 10.1021/acs.analchem.7b04210. [DOI] [PubMed] [Google Scholar]
- 88.Aghilinejad A, Aghaamoo M, Chen X. On the transport of particles/cells in high-throughput deterministic lateral displacement devices: Implications for circulating tumor cell separation. Biomicrofluidics. 2019;13:034112. doi: 10.1063/1.5092718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin MX, Hyun KA, Moon HS, Sim TS, Lee JG, Park JC, Lee SS, Jung HI. Continuous labeling of circulating tumor cells with microbeads using a vortex micromixer for highly selective isolation. Biosens Bioelectron. 2013;40:63–67. doi: 10.1016/j.bios.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 90.Xu X, Jiang Z, Wang J, Ren Y, Wu A. Microfluidic applications on circulating tumor cell isolation and biomimicking of cancer metastasis. Electrophoresis. 2020;41:933–951. doi: 10.1002/elps.201900402. [DOI] [PubMed] [Google Scholar]
- 91.Loutherback K, D'Silva J, Liu L, Wu A, Austin RH, Sturm JC. Deterministic separation of cancer cells from blood at 10 ml/min. AIP Adv. 2012;2:42107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khojah R, Stoutamore R, Di Carlo D. Size-tunable microvortex capture of rare cells. Lab Chip. 2017;17:2542–2549. doi: 10.1039/C7LC00355B. [DOI] [PubMed] [Google Scholar]
- 93.Sollier E, Go DE, Che J, Gossett DR, O'Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N, et al. Size-selective collection of circulating tumor cells using vortex technology. Lab Chip. 2014;14:63–77. doi: 10.1039/C3LC50689D. [DOI] [PubMed] [Google Scholar]
- 94.Renier C, Pao E, Che J, Liu HE, Lemaire CA, Matsumoto M, Triboulet M, Srivinas S, Jeffrey SS, Rettig M, et al. Label-free isolation of prostate circulating tumor cells using vortex microfluidic technology. NPJ Precis Oncol. 2017;1:15. doi: 10.1038/s41698-017-0015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding X, Peng Z, Lin SC, Geri M, Li S, Li P, Chen Y, Dao M, Suresh S, Huang T. Cell separation using tilted-angle standing surface acoustic waves. Proc Natl Acad Sci U S A. 2014;111:12992–12997. doi: 10.1073/pnas.1413325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu M, Huang PH, Zhang R, Mao Z, Chen C, Kemeny G, Li P, Lee AV, Gyanchandani R, Armstrong AJ, et al. Circulating tumor cell phenotyping via high-throughput acoustic separation. Small. 2018;14:e1801131. doi: 10.1002/smll.201801131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castro-Giner F, Aceto N. Tracking cancer progression: From circulating tumor cells to metastasis. Genome Med. 2020;12:31. doi: 10.1186/s13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rawal S, Yang YP, Cote R, Agarwal A. Identification and quantitation of circulating tumor cells. Annu Rev Anal Chem (Palo Alto Calif) 2017;10:321–343. doi: 10.1146/annurev-anchem-061516-045405. [DOI] [PubMed] [Google Scholar]
- 99.Riethdorf S, O'Flaherty L, Hille C, Pantel K. Clinical applications of the CellSearch platform in cancer patients. Adv Drug Deliv Rev. 2018;125:102–121. doi: 10.1016/j.addr.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 100.Awasthi NP, Kumari S, Neyaz A, Gupta S, Agarwal A, Singhal A, Husain N. EpCAM-based flow cytometric detection of circulating tumor cells in gallbladder carcinoma cases. Asian Pac J Cancer Prev. 2017;18:3429–3437. doi: 10.22034/APJCP.2017.18.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen K, Dopico P, Varillas J, Zhang J, George TJ, Fan ZH. Integration of lateral filter arrays with immunoaffinity for circulating tumor cell isolation. Angew Chem Int Ed Engl. 2019;58:7606–7610. doi: 10.1002/anie.201901412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang S, Thomas A, Lee E, Yang S, Cheng X, Liu Y. Highly efficient and selective isolation of rare tumor cells using a microfluidic chip with wavy-herringbone micro-patterned surfaces. Analyst. 2016;141:2228–2237. doi: 10.1039/C6AN00236F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dizdar L, Fluegen G, van Dalum G, Honisch E, Neves RP, Niederacher D, Neubauer H, Fehm T, Rehders A, Krieg A, et al. Detection of circulating tumor cells in colorectal cancer patients using the GILUPI CellCollector: Results from a prospective, single-center study. Mol Oncol. 2019;13:1548–1558. doi: 10.1002/1878-0261.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He Y, Shi J, Shi G, Xu X, Liu Q, Liu C, Gao Z, Bai J, Shan B. Using the new CellCollector to capture circulating tumor cells from blood in different groups of pulmonary disease: A cohort study. Sci Rep. 2017;7:9542. doi: 10.1038/s41598-017-09284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Que Z, Luo B, Zhou Z, Dong C, Jiang Y, Wang L, Shi Q, Tian J. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019;19:21. doi: 10.1186/s12935-019-0735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De T, Goyal S, Balachander G, Chatterjee K, Kumar P, Babu KG, Rangarajan A. A novel ex vivo system using 3d polymer scaffold to culture circulating tumor cells from breast cancer patients exhibits dynamic E-M phenotypes. J Clin Med. 2019;8:1473. doi: 10.3390/jcm8091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hwang WL, Pleskow HM, Miyamoto DT. Molecular analysis of circulating tumors cells: Biomarkers beyond enumeration. Adv Drug Deliv Rev. 2018;125:122–131. doi: 10.1016/j.addr.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 110.Rossi E, Zamarchi R. Single-cell analysis of circulating tumor cells: How far have we come in the-omics era? Front Genet. 2019;10:958. doi: 10.3389/fgene.2019.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pramani KA, Jones S, Gao Y, Sweet C, Vangara A, Begum S, Ray PC. Multifunctional hybrid graphene oxide for circulating tumor cell isolation and analysis. Adv Drug Deliv Rev. 2018;125:21–35. doi: 10.1016/j.addr.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 112.Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, Telekes A. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:48. doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article, as no datasets were generated or analyzed during the current study.