Soft extracellular matrix protects breast cancer cells from apoptosis and reduces the efficacy of chemo- and radiation therapies through enhanced NF-κB signaling and diminished proapoptotic JNK activity. Therapeutic targeting of the NF-κB–JNK axis may thwart treatment resistance in breast cancer.
Abstract
Triple-negative breast cancers (TNBCs) are associated with poor survival mediated by treatment resistance. TNBCs are fibrotic, yet little is known regarding how the extracellular matrix (ECM) evolves following therapy and whether it impacts treatment response. Analysis revealed that while primary untreated TNBCs are surrounded by a rigid stromal microenvironment, chemotherapy-resistant residual tumors inhabit a softer niche. TNBC organoid cultures and xenograft studies showed that organoids interacting with soft ECM exhibit striking resistance to chemotherapy, ionizing radiation, and death receptor ligand TRAIL. A stiff ECM enhanced proapoptotic JNK activity to sensitize cells to treatment, whereas a soft ECM promoted treatment resistance by elevating NF-κB activity and compromising JNK activity. Treatment-resistant residual TNBCs residing within soft stroma had elevated activated NF-κB levels, and disengaging NF-κB activity sensitized tumors in a soft matrix to therapy. Thus, the biophysical properties of the ECM modify treatment response, and agents that modulate stiffness-dependent NF-κB or JNK activity could enhance therapeutic efficacy in patients with TNBC.
Introduction
Neoadjuvant chemotherapy (NAC) is increasingly used in the treatment of invasive breast cancer, providing the opportunity to understand the relative sensitivity or resistance of tumors while still in situ (Abt et al., 2014). Many studies have demonstrated the more aggressive nature of hormone receptor–negative, HER2-negative breast cancer (triple-negative breast cancer [TNBC]) compared with other phenotypes, and TNBCs have been repeatedly correlated with increased local recurrence, distant recurrence, and worse overall survival (von Minckwitz et al., 2012). Despite significant advances in targeted therapies for hormone receptor–positive and HER2-positive disease, cytotoxic chemotherapy remains the standard of care for most TNBC cases, followed by radiation in selected cases.
TNBC tumors that are completely eradicated in the breast with NAC, known as a pathological complete response (pCR), are associated with an excellent patient prognosis, significantly better than for patients whose tumors lack such a response (von Minckwitz et al., 2012; Symmans et al., 2017; Cortazar et al., 2014). However, this response is heterogeneous, and TNBC patients continue to have worse overall outcomes than patients with breast tumors of other subtypes. Currently, there are no reliable methods to identify patients whose disease is likely to respond well and those whose disease is likely to be chemotherapy resistant. The ability to improve predicted response would facilitate personalization of treatment and patient outcomes. There is also an urgent need for a deeper understanding of the biological basis for chemotherapy response in TNBC that will inform treatment decisions and the development of effective strategies to overcome therapy resistance.
In addition to their highly aggressive phenotype, TNBCs have a characteristically high degree of extracellular matrix (ECM) remodeling and stiffening (Acerbi et al., 2015; Maller et al., 2020). ECM stiffness is well known to heavily influence cell behavior and has been shown to extrinsically modulate drug responses via blood vessel occlusion that limits transport. ECM stiffness can also regulate cell-intrinsic behaviors that affect therapy response, including proliferation, survival, and growth factor signaling (Pickup et al., 2014; Piersma et al., 2020). Despite the important role the ECM plays in regulating cell behavior, it is unclear whether the stiffer ECM measured in TNBC contributes to the underlying mechanisms whereby the evolving ECM may impact the development of therapy resistance.
NF-κB signaling is critically important to ECM-mediated cell survival. The apoptosis resistance phenotype of three-dimensional (3D) mammary epithelial tissue–like structures in a reconstituted basement membrane (rBM) depended upon the activation of NF-κB, but NF-κB activity per se failed to protect mammary epithelial cells (MECs) from apoptosis when cells were plated as 2D monolayers (Weaver et al., 2002; Zahir et al., 2003). Importantly, NF-κB activity can drive resistance to chemotherapy and radiation therapy in many tumor types by modifying the expression and stability of apoptosis regulators, including the stress-activated protein kinase JNK (Baldwin, 2001; Baeuerle and Baltimore, 1996; Tergaonkar et al., 2002; Javelaud and Besançon, 2001; Tang et al., 2001). JNK permits apoptosis induction in a variety of cell types in response to assorted cell stress stimuli, including paclitaxel, TNF-related apoptosis-inducing ligand (TRAIL), and UV and ionizing radiation (IR) exposure (Sunters et al., 2006; Hu et al., 1999; Chen et al., 1996). Recently, an in vivo screen also implicated JNK as a key mediator of chemotherapeutic efficacy (Ashenden et al., 2017). Importantly, JNK signaling is responsive to mechanical stimuli such as substrate strain and mediates mechanical stretch–induced apoptosis of renal epithelial cells (Katsumi et al., 2005; Nguyen et al., 2006). Moreover, Rho-GTPases that are elevated in MECs cultured on stiff matrices also activate JNK (Marinissen et al., 2004; Paszek et al., 2005). Thus, substrate stiffness could enhance JNK activation to sensitize MECs to antitumor treatment.
Here we show that residual disease from NAC-treated tumors localizes within a soft, remodeled ECM niche. We then demonstrate that MECs and tumor cells assembled into organoid-like structures in soft ECMs can efficiently engage NF-κB signaling to resist apoptosis induction. Conversely, cells interacting with a stiff substrate show reduced NF-κB signaling and increased JNK activity that sensitize them to several apoptosis-inducing agents. Consistent with this mechanism, we show human breast tumors that failed to achieve pCR in response to NAC were enriched for an NF-κB gene expression signature, and NF-κB levels were highly elevated in residual disease of NAC-treated TNBC tumors. This study implicates ECM stiffness as an important mediator of response to chemotherapy and radiation in TNBC. It has important clinical implications for understanding the heterogeneous response to therapy in TNBC and for making rational, informed decisions in selecting effective therapies.
Results
Residual triple-negative breast tumors are surrounded by a soft interstitial stroma
TNBCs are highly fibrotic and evolve within a stiffened, cross-linked, collagen-rich ECM (Acerbi et al., 2015; Egeblad et al., 2010; Maller et al., 2020). A stiff ECM promotes cell growth, survival, and migration and can foster resistance to receptor tyrosine kinase inhibitors (Levental et al., 2009; Pickup et al., 2014; Hanker et al., 2017).
Here we explored the role of ECM stiffness in the chemotherapy responsiveness of human TNBC. To begin, we assessed the organization and biophysical properties of the ECM associated with primary human TNBCs and compared these stromal features with the ECM of residual tumor tissue of NAC-treated tumors (Table S1). Histological analysis of a cohort of primary untreated TNBC biopsies demonstrated the presence of an interstitial stroma composed of abundant ECM proteins that was infiltrated by numerous tumor epithelial cells (Fig. 1 A). Picrosirius red (PS) staining and polarized light imaging revealed that the ECM in the primary, stage-matched, human TNBCs (n = 9) contained abundant quantities of fibrillar type collagens (Fig. 1, B and C), which atomic force microscopy (AFM) indentation showed was significantly stiffer than that measured previously in the normal human breast (Fig. 1, F and G; Acerbi et al., 2015). By contrast, hematoxylin and eosin staining indicated that the residual tumor epithelium in the NAC-treated TNBCs (n = 7) was dramatically reorganized into epithelial aggregates, many of which were reminiscent of tubular carcinoma (compare Fig. 1 A). Polarized light imaging of PS-stained tissue further revealed that the residual TNBC tumors were surrounded by large quantities of a remodeled, collagen-rich stroma (Fig. 1, B and C). However, second harmonic generation (SHG) microscopy showed that the collagen matrix adopted strikingly different architectures between primary and residual TNBC tumors. Collagen in primary TNBC predominantly was organized into long, linearized fibers, whereas the collagen matrix in residual tumors was significantly less aligned and was often organized isotropically in pericellular regions and within the regions surrounding tumor cell clusters (Fig. 1, D and E). AFM analysis revealed that the collagen-rich stroma associated with residual tumor was significantly softer than that measured in the primary TNBCs (Fig. 1, F and G; and Fig. S1). The findings indicate that chemotherapy treatment not only significantly reduces the density of the tumor epithelium but also induces significant remodeling and reorganization of the ECM stroma that substantially decreases its stiffness. The data also raise the intriguing possibility that the softened, remodeled ECM might be causally linked to the pathogenesis of the treatment-resistant residual tumor tissue.
Figure 1.
Treatment-resistant residual disease localizes within a soft ECM niche. (A) Representative hematoxylin and eosin–stained tissue sections of untreated human primary TNBC or residual disease from NAC-treated TNBC. (B) Representative images of brightfield and polarized light PS-stained primary tumor or residual disease tissue sections. (C) Scatterplot showing mean ± SEM of polarized light PS signal for primary tumor (n = 9) and residual disease (n = 7) from three to five fields of view per sample. (D) Representative images of SHG microscopy of tissue sections from human primary and chemotherapy-treated residual TNBC. (E) Scatterplot showing mean ± SEM of collagen coherence calculated from SHG microscopy images of primary tumor (n = 5) and residual TNBC (n = 4). (F) Scatterplot showing mean and SD of AFM elasticity measurements of tumor cell–adjacent ECM from primary tumor (n = 9) and residual disease (n = 7). Values reflect measurements from three to five elasticity maps per sample. (G) Histogram showing distribution of the top 10% of AFM elasticity measurements in each group. All statistical analyses were performed using two-tailed Mann-Whitney U test (*, P < 0.05; ***, P < 0.001). All scale bars are 200 µm.
Figure S1.
Treatment-resistant residual disease localizes within a soft ECM niche. Scatterplot showing mean ± SEM with individual values of the elastic modulus of primary human TNBC (n = 7) and chemotherapy-treated residual disease (n = 7) samples measured by AFM.
ECM stiffness regulates treatment responsiveness of human mammary tumors
To determine if a remodeled, compliant ECM could compromise the chemotherapy responsiveness of mammary tumors and whether any changes in response vary with tumor stage, we tested the impact of ECM stiffness on the apoptosis response of premalignant and malignant mammary epithelial organoids to a standard chemotherapy agent. We generated proliferating organoid structures of estrogen receptor (ER)–negative and HER2-negative, premalignant human MEC MCF10A, and malignant HMT-3522 T4-2 cells (T4-2) and TNBC-derived HCC70 cells to model early- and late-stage TNBCs. Mammary cells were plated on rBM-conjugated polyacrylamide (PA) substrates calibrated to recapitulate the low and high ends of the elasticity (E) we measured in the normal (E = 150 Pa) and tumor stroma (E = 5,000 Pa; Paszek et al., 2005; Johnson et al., 2007). The mammary tumor cells were overlaid with growth media containing 2% rBM to create a 3D ECM microenvironment. The tumor cells were then grown for 5 d until they formed small, uniformly proliferating, tissue-like colonies (Paszek et al., 2005; Johnson et al., 2007). Notably, the colonies grown on soft and stiff substrates had a similar percentage of cells in direct contact with the basal substrate, with those on stiff substrate containing a slightly higher percentage (Fig. S2, A and B). The organoids were then treated with an increasing dose of the chemotherapeutic drug paclitaxel. Intriguingly, the treated premalignant colonies cultured on the stiff PA substrate displayed a significant, dose-dependent loss of metabolic activity, as measured by 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay, and a concomitant increase in apoptosis, as measured by positive staining for cleaved caspase 3 (Fig. 2, A and B). By contrast, the premalignant structures cultured on the soft substrate showed only a modest reduction in metabolic activity, and apoptosis levels were significantly lower than those quantified in the same proliferating structures plated on a stiff ECM substrate, despite exposure to the same dosages of paclitaxel (Fig. 2, A and B; and Fig. S2 C). To investigate whether ECM stiffness influences tumor treatment efficacy by modifying drug uptake or tumor cell accessibility, we next examined the impact of ECM stiffness on IR responsiveness, a standard therapeutic modality used to treat TNBCs that is not influenced by multidrug resistance pumps or drug transport limitations. Interestingly, once again, experiments showed that premalignant mammary organoids cultured on a stiff substrate exhibited a significant reduction in metabolic activity and displayed greater numbers of cleaved caspase 3–positive cells in response to increasing doses of IR (Fig. 2 C and Fig. S2 D). Moreover, a clonogenic survival assay revealed that a significantly higher proportion of the 3D premalignant mammary cell colonies irradiated on a soft substrate not only survived better but also were able to regrow into viable colonies when plated on a soft ECM, as compared with parallel cultures of premalignant cell colonies irradiated and replated on stiff substrates (Fig. 2 D). These data suggest that the observed apoptosis resistance by cells cultured on soft substrates is not attributable to any potential differences in paclitaxel diffusion or accessibility but is rather a result of differences in apoptotic signaling induced by substrate stiffness.
Figure S2.
Stiff ECM enhances apoptosis in response to treatment. (A) Representative maximum-intensity projections and basal slice images of HCC70 cell cultured on soft and stiff substrates treated with DMSO vehicle stained with DAPI (gray). (B) Bar graph showing mean ± SEM of the percentage of HCC70 cells in contact with the basal substrate when cultured on soft (n = 4) or stiff (n = 4) substrates. (C) Representative images of MCF10A cells cultured on soft and stiff substrates and treated with 10 nM paclitaxel or DMSO vehicle stained for cleaved caspase 3 (green) and with DAPI (gray). (D) Bar graph showing mean ± SEM of loss of metabolic activity by MTT assay after IR treatment in MCF10A cells cultured on soft or stiff substrates (n = 2). (E) Representative images of HCC70 cells cultured on soft and stiff substrates and treated with 100 nM paclitaxel or DMSO vehicle stained for cleaved caspase 3 (green) and with DAPI (gray). All scale bars are 100 μm.
Figure 2.
Stiff ECM enhances apoptosis in response to treatment. (A) Bar graph showing mean ± SEM with individual values of loss of metabolic activity by MTT assay after paclitaxel treatment in MCF10A cells cultured on soft or stiff substrates. (B) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cultures on soft and stiff substrates with increasing doses of paclitaxel. At least 300 cells on soft and 900 cells on stiff substrates were counted in each experiment. (C) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cultures on soft and stiff substrates with increasing doses of IR. At least 300 cells on soft and 500 cells on stiff substrates were counted in each experiment (n = 4 or 5). (D) Bar graph showing mean ± SEM with individual values of the percentage of MCF10A cells cultured on a soft or stiff substrate that are capable of reforming colonies after IR. (E) Representative maximum-intensity projection of T4-2 cells cultured on soft and stiff substrates and treated with 10 nM paclitaxel or DMSO vehicle stained for cleaved caspase 3 (green) and with DAPI (gray). (F) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from T4-2 cultures on soft and stiff substrates with increasing doses of paclitaxel. At least 900 cells on soft and stiff substrates were counted in each experiment. (G) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from T4-2 cultures on soft and stiff substrates with increasing doses of IR. At least 800 T4-2 cells on soft substrate and 1,000 T4-2 cells on stiff substrate were counted in each experiment (n = 4). (H) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from HCC70 cultures on soft and stiff substrates with increasing doses of paclitaxel. At least 750 cells on soft and stiff substrates were counted in each experiment (n = 3 or 4). (I) Bar graph showing mean ± SEM of the change in tumor volume after 14 d of paclitaxel treatment of animals with orthotopically injected T4-2 cells in soft (n = 7) or stiff (n = 8) collagen gels. (J) Representative images of FFPE tumor tissue sections stained for cleaved caspase 3 from orthotopically injected T4-2 tumors in soft or stiff collagen after 14 d of paclitaxel treatment. (K) Bar graph showing mean ± SEM with individual values of the change in tumor volume after 21 d of paclitaxel treatment of animals with orthotopically injected HCC70 cells expressing empty vector control (control; n = 6) or V737N β1 integrin mutant (n = 6). Statistical analyses were performed using two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (L) Bar graph showing mean ± SEM with individual values of the time to paclitaxel resistance of animals with orthotopically injected HCC70 cells expressing empty vector control (control; n = 9) or V737N β1 integrin mutant (n = 7). (M) Bar graph showing mean ± SEM with individual values of the time to paclitaxel resistance of animals with orthotopically implanted BCM2665 triple-negative PDX pieces in soft (n = 7) or stiff (n = 7) collagen gels. Statistical analyses of time to resistance were performed using two-tailed Mann-Whitney test (*, P < 0.05). Unless otherwise noted, n = 3 for all experiments. All scale bars are 100 µm.
We repeated these experiments in two triple-negative mammary tumor cell lines, the HMT-3522–derived ER-, progesterone receptor–, and HER2-negative T4-2 and the TNBC-derived HCC70, to determine whether stiff ECM also sensitizes malignant cells to apoptosis-inducing therapies. Colonies from both malignant cell lines plated on the compliant ECM substrates confirmed a striking apoptosis resistance in response to both paclitaxel chemotherapy and IR, as indicated by significantly lower levels of cleaved caspase 3 following paclitaxel treatment than in those malignant colonies grown on the stiffer ECM substrate (Fig. 2, E–H; and Fig. S2 E). Importantly, malignant tumor colonies of T4-2 embedded within stiff ribose cross-linked collagen gels and injected into the fat pads of immunocompromised mice were also significantly more sensitive to chemotherapy treatment. Indeed, these xenografted tumor colonies shrank significantly more (Fig. 2 I) and displayed higher levels of apoptosis in response to paclitaxel treatment than the orthotopically implanted tumors injected within the soft collagen gels (Fig. 2 J). Cell ligation to stiffened ECM increases focal adhesion–mediated signaling, which is a major mechanism by which ECM stiffness regulates cell behavior (Paszek et al., 2005; Levental et al., 2009; Leight et al., 2017). To test whether increased integrin signaling could sensitize tumor cells to chemotherapy, we orthotopically injected TNBC-derived HCC70 cells into the fat pads of immunocompromised mice engineered to express an activating V737N β1 integrin mutant or an empty vector control (Paszek et al., 2005; Levental et al., 2009). Paclitaxel-treated mice bearing V737N β1 mutant tumors showed increased paclitaxel treatment sensitivity compared with control tumors, as indicated initially by a greater reduction in tumor volume and chronically by a significantly delayed onset of chemoresistance (Fig. 2, K and L). These data indicate that increased integrin signaling likely contributes to stiffness-mediated sensitivity to chemotherapy. To further explore these findings, we orthotopically implanted the human TNBC patient-derived xenograft (PDX) BCM2665, which recapitulates the phenotype and cellular heterogeneity of a human TNBC, into the fat pads of immunocompromised mice within either a soft or stiffened ribose–cross-linked collagen and assayed for chemotherapy responsiveness. Consistently, those PDXs implanted in the stiff collagen were more acutely sensitive to paclitaxel treatment and had significantly delayed development of paclitaxel resistance compared with the same TNBCs within the soft ECMs (Fig. 2 M). These findings argue that a stiff ECM sensitizes breast tumors to treatment both in culture and in vivo.
ECM stiffness does not sensitize organoids to treatment by enhancing cell proliferation
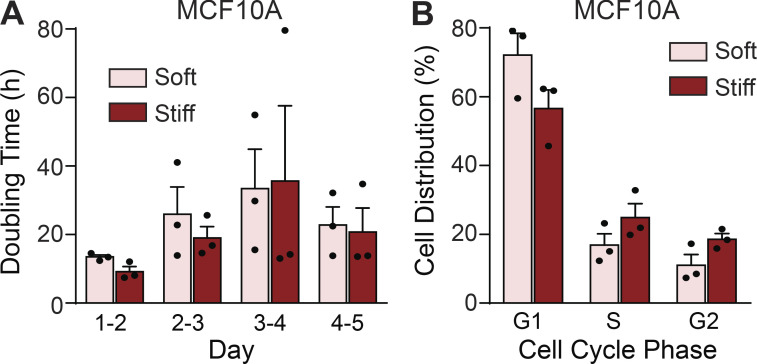
Chemotherapy and radiation efficacy are due in part to exploitation of the rapid division of cancer cells to induce apoptosis. A stiff ECM can enhance growth factor–dependent signaling and elevate cell growth (Paszek et al., 2005; Levental et al., 2009; Pickup et al., 2014; Northey et al., 2017). This enhanced growth phenotype raises the possibility that the differential tumor sensitivity incurred by interaction with a stiff ECM substrate to both the chemotherapy and the radiation treatment reflected an increased rate of cell proliferation. However, all culture and in vivo experiments were initiated when the tumor colonies were actively proliferating. Thus, all treatments were started at days 4 and 5 of culturing, when the premalignant and malignant organoids were proliferating at similar levels on both the soft and stiff ECM substrates. Indeed, quantification of phosphorylated histone H3 (Ser10) staining confirmed that the level of proliferation was equally high in both the premalignant and malignant tumor 3D colonies plated on the soft and stiff substrates between days 4 and 5, when the antitumor treatments were initiated (Fig. 3, A and B). FACS analysis of propidium iodide (PI)–labeled cells further revealed no change in the doubling time and only a modest, nonsignificant increase in the proportion of cells in S and G2 phases of the cell cycle in cells cultured on stiff ECM substrates at the time of treatment (Fig. S3, A and B). Accordingly, the findings rule out the possibility that significant differences in cell proliferation or cell cycle status explain the differential apoptosis sensitivity exhibited by the premalignant and malignant tumor colonies on the soft versus stiff ECM substrates.
Figure 3.
ECM stiffness does not sensitize organoids to treatment by enhancing cell proliferation. (A) Representative images of MCF10A, T4-2, and HCC70 cells at day 5 of culture on soft or stiff substrates stained for phosphorylated histone H3 at Ser10 (red) and DAPI (blue). (B) Bar graph showing mean ± SEM with individual values of the percentage of phosphorylated histone H3 (Ser10)–positive MCF10A, T4-2, and HCC70 cells on day 5 of culture on soft or stiff substrates. (C–E) Bar graphs showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from (C) MCF10A (n = 3 or 4), (D) T4-2, and (E) HCC70 (n = 3 or 4) cultures on soft and stiff substrates with increasing doses of TRAIL. At least 300 cells on soft substrate and 700 cells on stiff substrate were counted in each experiment (n = 3 or 4). All statistical analyses were performed using two-tailed t test (*, P < 0.05; **, P < 0.01). Unless otherwise noted, n = 3 for all experiments. All scale bars are 100 µm.
Figure S3.
ECM stiffness does not sensitize organoids to treatment by enhancing cell proliferation. (A) Bar graph showing mean ± SEM with individual values of the doubling time of MCF10A cells cultured on soft or stiff substrates each day over 5 d of culture (n = 3). (B) Bar graph showing mean ± SEM with individual values of the percentage of MCF10A cells in each cell cycle phase at day 5 of culture on soft or stiff substrates (n = 3).
Nonetheless, to provide further evidence to rule out the possibility that the stiffness-mediated sensitization of premalignant and malignant colonies to apoptosis-inducing agents could simply be explained by differences in cell proliferation, we treated the two types of tumor colonies with TRAIL, a cytokine that binds to the death receptors DR4 and DR5. Importantly, TRAIL is induced in tumors in response to chemotherapy and radiation treatment, and the cytokine induces tumor cell apoptosis through a pathway that is independent of cell growth (Srivastava, 2001; Nagane et al., 2001). Consistent with our conclusion that the differential sensitivity of mammary tumors to antitumor treatment is independent of cell growth, we noted that the proliferating premalignant MCF10A and the malignant T4-2 and HCC70 colonies interacting with soft ECM substrates were all protected from TRAIL-induced apoptosis. Thus, we observed a dose-dependent increase in apoptosis, as indicated by cleaved caspase 3, in the TRAIL-treated tumor colonies cultured on stiff as compared with soft ECM substrates (Fig. 3, C–E). Accordingly, these data demonstrate that the sensitivity of the tumors to these anticancer treatments is likely not simply due to differences in cell proliferation. Instead, the findings suggest that differences in tumor treatment sensitivity likely reflect a cell-intrinsic phenotype conferred on the tumors by the compliance of the ECM substrate.
A stiff ECM sensitizes tumor colonies to antitumor treatment by enhancing JNK activation
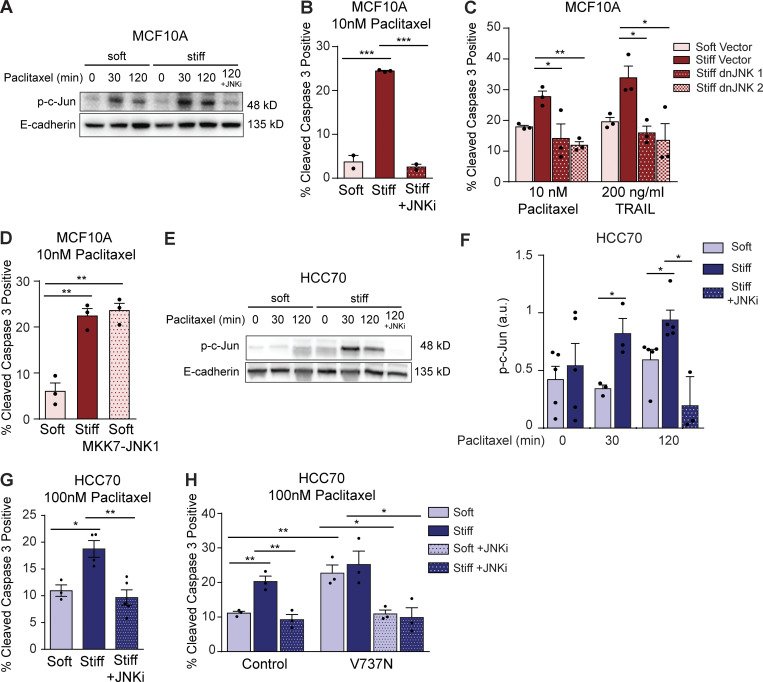
Cell survival in response to an antitumor treatment is regulated by stress response pathways whose activity is controlled by a comparatively small number of key molecular regulators or signaling nodes (Fulda and Debatin, 2006; Wong, 2011). JNKs compose one major signaling node that can dictate whether a cell survives or undergoes apoptosis in response to an exogenous stress (Dhanasekaran and Reddy, 2008; Sui et al., 2014). JNKs phosphorylate c-Jun, which is a component of the Fos/Jun AP-1 transcriptional complex that regulates the expression of key cell growth and survival genes (Dhanasekaran and Reddy, 2008). AP-1 levels and transcriptional activity can be induced by a stiff ECM, possibly because a stiff ECM increases RhoA–Rho-associated protein kinase signaling (Marinissen et al., 2004). Elevated JNK activity has also been implicated in both the intrinsic apoptosis pathway triggered in response to stress-inducing stimuli such as UV radiation or chemotherapeutic agents, as well as via the extrinsic apoptosis pathway stimulated in response to death receptor signaling (Dhanasekaran and Reddy, 2008). Accordingly, we reasoned that a stiff ECM substrate could sensitize mammary tumors to antitumor treatment by enhancing JNK activity. Consistently, we observed a significant and sustained increase in JNK activity following paclitaxel treatment in both the premalignant and malignant tumor colonies when they were cultured on the stiff as compared with the soft ECM substrate (Fig. 4, A and E; and Fig. S4, C and D). Furthermore, and importantly, treating tumor cells with a pharmacological inhibitor of JNK activity, SP600125, significantly protected the premalignant tumor colonies plated on the stiff ECM substrate from apoptosis induction. Indeed, inhibiting JNK reduced apoptosis, as indicated by the percentage of activated caspase 3–positive cells induced in the tumor colonies in response to paclitaxel treatment when they were cultured on the stiff ECM substrates as compared with those cultured on a soft ECM substrate (Fig. 4 B). Furthermore, to more directly implicate JNK in the stiffness-mediated sensitization of tumor tissue to anticancer treatment, we engineered the MCF10A MECs to express either a dominant-negative JNK1 or JNK2 construct that was designed to prevent kinase activation (Wojtaszek et al., 1998). Consistent with our JNK hyperactivation hypothesis, both dominant-negative JNK constructs rendered the MCF10A MECs interacting with the stiff ECM substrates resistant to apoptosis induction following treatment with either paclitaxel or TRAIL (Fig. 4 C). Moreover, to further implicate stiffness-mediated modulation of JNK activity in tumor treatment responsiveness, we next tested whether a gain of JNK activity could sensitize the tumor tissue interacting with a soft ECM substrate to antitumor treatment. The MCF10A MECs were engineered to constitutively activate JNK by expressing a MKK7-JNK1 fusion protein that renders the JNK1 protein constitutively active (Takada et al., 2005; Han et al., 2002). Importantly, increasing JNK activity sensitized the tumor tissue to paclitaxel treatment and enhanced the level of apoptosis induced, as indicated by greater numbers of activated caspase 3–positive cells, even when the cells were cultured on a soft ECM substrate (Fig. 4 D). Notably, the level of apoptosis induced in the MKK7-JNK1–expressing premalignant tumor cells cultured on the soft ECM substrate was virtually identical to that observed in the vector-expressing tumor tissue cultured on the stiff ECM substrate. The data functionally implicate JNK activity as one key mechanism whereby a stiff ECM substrate can sensitize mammary tumors to anticancer apoptosis-inducing agents and suggest that overcoming JNK suppression can abrogate the treatment resistance conferred by a soft ECM.
Figure 4.
Enhanced JNK activity in cells on stiff ECM sensitizes cells to chemotherapy. (A) Representative Western blot showing bands for phosphorylated c-Jun (Ser63) and E-cadherin in MCF10A cells cultured on soft or stiff substrates at indicated time points after 10 nM paclitaxel treatment or paclitaxel treatment with 5 µM JNK inhibitor SP600125. (B) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cultured on soft and stiff substrates treated with 10 nM paclitaxel or 10 nM paclitaxel plus 5 µM of JNK inhibitor SP600125 (n = 2 or 3). (C) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cells cultured on soft and stiff substrates and stably transduced with dominant-negative JNK1 or JNK2 constructs or the empty vector control and treated with 10 nM paclitaxel. (D) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cells cultured on soft and stiff substrates and stably transduced with an inducible, constitutively active MKK7–JNK1 fusion protein and treated with 10 nM paclitaxel. (E) Representative Western blot showing bands for phosphorylated c-Jun (Ser63) and E-cadherin in HCC70 cells cultured on soft or stiff substrates at indicated time points after 100 nM paclitaxel treatment or paclitaxel treatment with 5 µM JNK inhibitor SP600125. (F) Bar graph showing mean ± SEM with individual values of p-c-Jun (Ser63) levels in HCC70 cultured on soft or stiff substrates at indicated time points after 100 nM paclitaxel treatment or paclitaxel treatment with 5 µM JNK inhibitor SP600125 quantified from Western blots by pixel density and normalized to E-cadherin (n = 3–5). (G) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from HCC70 cultured on soft and stiff substrates treated with 100 nM paclitaxel or 100 nM paclitaxel plus 5 µM JNK inhibitor SP600125 (n = 3 or 4). (H) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from HCC70 transduced with empty vector (control) or V737N β1 integrin mutant cultured on soft and stiff substrates treated with 100 nM paclitaxel or 100 nM paclitaxel plus 5 µM JNK inhibitor SP600125. Unless otherwise noted, n = 3 for all experiments. All statistical analyses were performed using two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Figure S4.
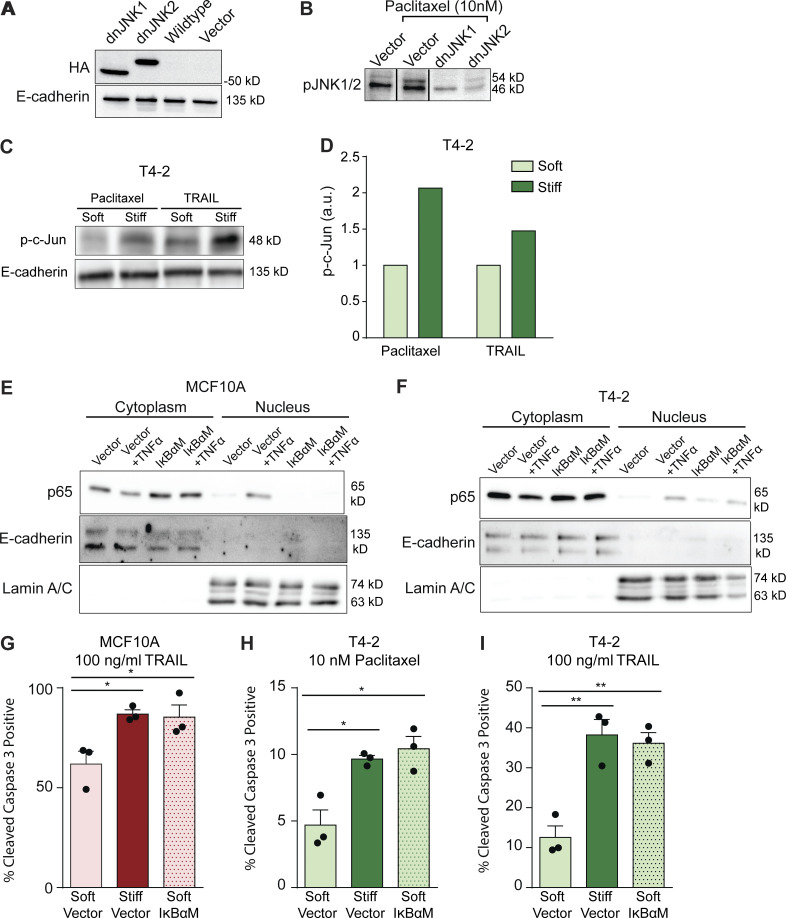
A soft ECM increases NF-κB signaling to regulate JNK activity and apoptosis. (A) Western blot validating dominant-negative (dn)JNK construct expression in MCF10A cells. (B) Western blot of phosphorylated JNK1/2 in vector and dnJNK-transduced MCF10A cells treated with 10 nM paclitaxel. (C) Western blot of p-c-Jun (Ser63) and E-cadherin of T4-2 cells cultured on soft and stiff gels after 60-min incubations with 10 nM paclitaxel or 200 ng/ml TRAIL. (D) Bar graph quantifying p-c-Jun (Ser63) relative to E-cadherin from Western blot in C using pixel density (n = 1). (E) Western blot for p65 in cytoplasmic and nuclear protein extracts from MCF10A containing the IκBαM construct and treated with recombinant human TNFα. (F) Western blot for p65 in cytoplasmic and nuclear protein extracts from MCF10A containing the IκBαM construct and treated with recombinant human TNFα. (G) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cells expressing a vector control or dominant-negative IκBαM mutant cultured on soft or stiff substrates and treated with 100 ng/ml TRAIL (n = 3). (H) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from T4-2 cells expressing a vector control or dominant-negative IκBαM mutant cultured on soft or stiff substrates and treated with 10 nM paclitaxel (n = 3). (I) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from T4-2 cells expressing a vector control or dominant-negative IκBαM mutant cultured on soft or stiff substrates and treated with 100 ng/ml TRAIL (n = 3). All statistical analyses were performed using two-tailed t test (*, P < 0.05; **, P < 0.01).
The results were confirmed using the malignant TNBC cell line HCC70. Paclitaxel treatment induced significantly greater levels and prolonged JNK activity in HCC70 cells cultured on stiff substrates compared with those cultured on soft substrates, and this effect was entirely nullified upon treatment with the JNK inhibitor SP600125 (Fig. 4, E and F). JNK inhibition also rendered HCC70 cells cultured on stiff substrates resistant to paclitaxel-induced apoptosis, as shown by the reduction in the percentage of cleaved caspase 3–positive cells to levels similar to those in cells cultured on soft substrates (Fig. 4 G). To determine whether stiffness-induced apoptosis sensitivity was regulated by increased JNK activity due to enhanced integrin signaling, HCC70 expressing the V737N β1 integrin mutant or vector control were seeded on soft and stiff substrates and treated with paclitaxel or paclitaxel plus JNK inhibitor. V737N-expressing cells cultured on soft substrates had significantly higher levels of cleaved caspase 3 staining than control cells, while the percentage of cleaved caspase 3–positive cells between V737N mutant cells cultured on soft and stiff substrates remained unchanged (Fig. 4 H). These data implicate focal adhesion signaling in stiffness-mediated sensitivity to apoptosis. Importantly, when cells expressing the V737N mutant were treated with paclitaxel plus JNK inhibitor, both cells cultured on soft and stiff substrates showed a significant reduction in cleaved caspase 3–positive cells (Fig. 4 H). These data suggest that substrate stiffness regulates apoptosis sensitivity by increasing focal adhesion signaling, which in turn increases proapoptotic JNK activity in response to chemotherapy.
NF-κB represses JNK activity to promote tumor treatment resistance
We next assessed whether ECM stiffness could modulate the levels and/or activity of JNK regulators previously implicated in tumor treatment resistance. Elevated tumor levels of nuclear NF-κB have been implicated in cancer aggression and poor patient outcome (Yamamoto and Gaynor, 2001; Zeligs et al., 2016). Experimental models have also shown that NF-κB activity enhances tumor cell survival and resistance to antitumor treatment both in culture and in vivo (Weaver et al., 2002; Zahir et al., 2003; Friedland et al., 2007; Ahmed et al., 2013; Murray et al., 2012). We therefore tested whether ablating NF-κB activity could sensitize mammary tumor organoids to antitumor treatment and if this was linked to enhanced JNK activation. We engineered premalignant MCF10A mammary tumor cells with a dominant-negative inhibitor of NF-κB using an IκBα mutant (IκBαM) that sequesters NF-κB in the cytosol. Consistent with prior studies, we noted that ablating NF-κB activity significantly increased JNK activity, even in tumor organoids cultured on the soft ECM substrate (Fig. 5, A and B). Furthermore, and importantly, inhibiting NF-κB significantly sensitized MCF10A, T4-2, and HCC70 colonies to paclitaxel and TRAIL treatment, even when they were grown on a soft ECM substrate (Fig. 5, C and D; and Fig. S4, G–I). These findings not only implicate NF-κB activity as a key regulator of JNK-dependent apoptosis signaling but also reveal an unappreciated potential link between ECM stiffness and NF-κB and JNK activity in the pathogenesis of treatment-resistant breast tumors.
Figure 5.
A soft ECM increases NF-κB signaling to regulate JNK activity and apoptosis. (A) Western blot showing bands for phosphorylated c-Jun (Ser63) in MCF10A cells stably transduced with a dominant-negative IκBαM mutant or the vector backbone and cultured on soft or stiff substrates at indicated time points after 10 nM paclitaxel treatment. (B) Bar graph showing mean ± SEM with individual values of Western blot quantifications of phosphorylated c-Jun (Ser63) by pixel density normalized to E-cadherin loading control in MCF10A cells stably transduced with a dominant-negative IκBαM mutant or the vector backbone and cultured on soft or stiff substrates at indicated time points after 10 nM paclitaxel treatment (n = 4). (C) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from MCF10A cells expressing a dominant-negative IκBαM mutant cultured on soft or stiff substrates and treated with 10 nM paclitaxel or vehicle. (D) Bar graph showing mean ± SEM with individual values of the percentage of cleaved caspase 3–positive cells from HCC70 cells expressing a dominant-negative IκBαM mutant cultured on soft or stiff substrates and treated with 100 nM paclitaxel, 100 nM paclitaxel with 5 µM SP600125, or vehicle. Unless otherwise noted, n = 3 for all experiments. All statistical analyses were performed using two-tailed t test (*, P < 0.05; **, P < 0.01).
NF-κB activity predicts pathological response to NAC, modulates antitumor treatment response, and is elevated in residual TNBCs
We next explored the relevance of NF-κB activity in the breast cancer patient response to therapy. Analysis of Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis (I-SPY) trial patient datasets from NAC-treated patients showed that patients who failed to achieve a pCR had primary tumors that were significantly enriched for an NF-κB–dependent gene signature with a normalized enrichment score of 1.799 (Fig. 6 A; Table S2; Magbanua et al., 2015). The findings are consistent with prior studies suggesting that NF-κB target genes modulate tumor response to chemotherapy.
Figure 6.
NF-κB activity antagonizes tumor therapy. (A) Gene set enrichment analysis for NF-κB target genes in human breast cancer samples before treatment that failed to achieve a pCR (n = 138 total patients). Normalized enrichment score (NES) is 1.799 (P = 0.0034). (B) Bar graph showing mean ± SEM of tumor volume of s.c. injected T4-2 tumors treated with IR or radiation in combination with NF-κB inhibitor JSH-23 (n = 7 or 8 animals per group). Cartoon shows experimental design. (C) Representative images of p65 IHC in untreated primary human triple-negative breast tumors or in residual disease of chemotherapy-treated human triple-negative breast tumors (n = 6 primary, n = 8 residual disease). (D) Bar graph showing mean ± SEM of the percentage of p65-positive cells in primary human triple-negative breast tumors or in residual disease of chemotherapy-treated human triple-negative breast tumors (n = 4 primary, n = 7 residual disease). Statistical analyses were performed using two-tailed t test (B) or two-tailed Mann-Whitney U test (D; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Scale bars are 100 µm.
Nonetheless, to directly test whether NF-κB activity could antagonize antitumor treatment responsiveness, we injected malignant T4-2 cells s.c. into the rear flanks of immunocompromised nude mice. After 2 wk, the mice were injected either with vehicle or the NF-κB inhibitor JSH-23, and thereafter the tumors were subjected to IR (4 Gy) treatment. Tumor response rates were assessed by monitoring changes in tumor growth. We observed that NF-κB inhibition potentiated therapy response, and those tumors whose NF-κB activity had been inhibited with JSH-23 grew significantly more slowly following radiation treatment than the vehicle-treated tumors did (Fig. 6 B). These findings further implicate elevated NF-κB activity in the treatment resistance phenotype of mammary tumors.
To explore the clinical relevance of links between ECM substrate stiffness, elevated NF-κB activity, and treatment resistance, we next examined NF-κB levels in the neoadjuvant treated residual TNBCs surrounded by the highly compliant, remodeled ECM stroma (Table S3). Consistent with a compliant ECM enhancing NF-κB activity to potentially impair the response of TNBCs to anticancer treatment, we quantified a significant increase in NF-κB (p65) in the residual epithelium in the chemotherapy-treated TNBC patients that were surrounded by the remodeled soft stroma. In contrast, the level of NF-κB was markedly lower in the tumor epithelium in a cohort of untreated TNBC patients in whom the associated and surrounding ECM was significantly stiffer (Fig. 1, F and G; and Fig. 6, C and D). These studies imply that a remodeled, compliant ECM could contribute to the treatment resistance of TNBCs to preclude a pCR. The data also implicate antiapoptotic NF-κB activity in the pathogenesis of this residual tumor phenotype.
Discussion
In this study, we identified ECM stiffness as a critical modulator of antitumor treatment response. Using cultured 3D organoids of premalignant and malignant tumors, xenografted malignant tumors, and a PDX model, we determined that a stiff ECM substrate broadly sensitizes tumor cells to the chemotherapeutic agent paclitaxel and to IR and that it did so regardless of the cell cycle status or level of cell proliferation. Surprisingly, we found that the tumor tissue interacting with a soft ECM was able to efficiently activate NF-κB to resist even high doses of chemotherapy, IR, and death receptor signaling. By contrast, we observed that a stiff ECM substrate sensitized tumor organoids to antitumor therapies by promoting therapy-induced stress signaling through JNK to abrogate prosurvival NF-κB signaling. Critically, we determined that breast cancer patients whose primary tumors had high NF-κB activity also had a higher rate of treatment resistance and after treatment were enriched for NF-κB activity. Consistently, residual chemotherapy-resistant TNBC tissue that was surrounded by a highly remodeled soft ECM also showed high levels of nuclear NF-κB. These findings are consistent with prior studies implicating NF-κB activity in the treatment resistance of tumors and highlight the importance of tissue context in regulating antitumor treatment response. These data also suggest that therapies designed to inhibit NF-κB could be used to improve the treatment response of TNBCs to enhance the long-term survival of these patients.
Consistent with an association between a soft ECM and treatment resistance, we observed a profoundly remodeled ECM in post-treatment human TNBCs and showed that this modified ECM was softer than that measured in primary TNBCs. Manipulations using 3D organoid models in which the substrate biomechanical properties could be precisely manipulated provided direct evidence to show that a soft ECM protects, whereas a stiff ECM sensitizes, tumor cells to antitumor treatment. The findings are consistent with prior studies that showed that 3D mammary organoids grown within soft rBM gels exhibit improved survival following chemotherapy and radiation therapy by activating NF-κB (Weaver et al., 2002; Zahir et al., 2003; Friedland et al., 2007; Ahmed et al., 2013). Critically, these and other reports established a link between the 3D culture format and multicellularity in generating the treatment resistance phenotype of tumors (Sutherland, 1988; Durand and Sutherland, 1972; Weaver et al., 2002; Zahir et al., 2003). Our experiments, which rigorously controlled for confounding variables, including hypoxia, proliferation, and drug accessibility, not only confirmed those results but also extended those earlier observations to unequivocally demonstrate a role for ECM stiffness and multicellular architecture in modulating the treatment response of tumors (Schrader et al., 2011). Our studies also imply that the organoid models used to study treatment resistance in culture and to screen for novel antitumor agents need to consider the biophysical properties of the matrix when interpreting data and designing assays.
A stiff ECM promotes epidermal growth factor receptor signaling to increase the activity of phosphoinositide 3-kinase (PI3K) and ERK that in turn enhance tumor cell growth and survival (Levental et al., 2009; Pickup et al., 2014). Not surprisingly, a stiff tissue microenvironment was shown to promote the resistance of melanomas to targeted kinase inhibitors both in culture and in vivo, presumably by potentiating receptor tyrosine kinase activity to increase net prosurvival signaling (Hirata et al., 2015). Those prior studies addressed the impact of therapies that target a specific defined receptor tyrosine kinase signaling pathway whose activity is significantly potentiated by ECM stiffness. However, our studies addressed the impact of ECM stiffness on agents and treatments that trigger apoptosis by inducing a general cellular stress response. To this end, we found that a stiff ECM permits an unencumbered chronic elevation in JNK activity in response to an exogenous stress such as chemotherapy, and we provided evidence that this effect compromises tumor cell survival. Consistent with a reduced stress response in cells interacting with a soft ECM, we showed that premalignant MECs treated with 6–12 Gy of IR not only failed to die but also showed a significantly enhanced clonogenic survival only when replated on a soft ECM, an observation also made with chemotherapy-treated hepatic cancer cells (Schrader et al., 2011). Intriguingly, a set of studies conducted in monolayer 2D cultures implicated higher stress signaling through JNK as a protective mechanism whereby cultured breast and hepatic cancer cells plated on a stiffer ECM substrate acquired greater resistance to the multikinase inhibitor sorafenib (Nguyen et al., 2014). In marked contrast to these monolayer culture studies, our work was conducted in cancer organoids in which we recapitulated the 3D tissue-like structures of in vivo tumor tissue. Moreover, unlike many 2D studies, our culture studies were faithfully reproduced in mouse models and patient samples (Lee et al., 2019; Riedl et al., 2017). Indeed, while cells grown as 2D monolayers show an absolute requirement for PI3K for survival and for ERK activation to progress through G1, cells grown as 3D organoids depend less upon PI3K and ERK and instead require Rac-GTPase for both their growth and survival (Zahir et al., 2003; Fournier et al., 2008).
We and others have identified NF-κB as a key regulator of treatment resistance (Weaver et al., 2002; Zahir et al., 2003; Ahmed et al., 2013; Xia et al., 2014). NF-κB directly mediates the transcription of several antiapoptotic genes and alters angiogenesis, metabolism, immune cell interactions, and metastasis (Xia et al., 2014; Lin et al., 2010). We noted an increase in NF-κB in the residual tumor of chemotherapy-treated patients, and, mechanistically, we determined that the failure of tumor organoids in a stiff 3D ECM to robustly activate NF-κB in response to therapy permitted a high degree of JNK activation, which induced apoptosis in these cells. These findings are consistent with work by others showing that JNK stress signaling reduces tumor viability and compromises survival in response to therapy (Chen et al., 1996). Conversely, JNK activation in some contexts, such as cells plated on a stiff 2D substrate, prevents apoptosis to promote drug resistance, consistent with its pleiotropic role in apoptosis and stress response (Nguyen et al., 2014; Dhanasekaran and Reddy, 2008). Nevertheless, compelling in vivo evidence identified JNK as a key mediator of chemotherapy sensitivity, and, furthermore, human TNBC datasets linked a high JNK activity gene expression signature with improved chemotherapy response, implicating JNK in chemosensitivity (Ashenden et al., 2017). Our findings further support the physiological relevance of JNK in chemotherapy response and demonstrate that the soft remodeled ECM niche contributes to the ability of residual tumor cells to resist apoptosis via NF-κB–JNK signaling. These data suggest that the targeting of additional pathways, such as the NF-κB–JNK axis, may sensitize or potentially resensitize chemotherapy-resistant tumor cells to enhance treatment efficacy and eradicate residual tumor cells.
Resistance to chemotherapy and radiation continue to be significant barriers to overcome, particularly in TNBC. Many TNBC patients respond only partially to NAC, with as much as 60–70% of these cancers showing some level of residual disease (von Minckwitz et al., 2012). Importantly, an incomplete pathological response to therapy is associated with worse overall patient survival (von Minckwitz et al., 2012). Indeed, primary TNBCs that fail to respond well to chemotherapy frequently recur with metastatic disease that is refractory to nearly all therapies, with less than 30% of patients surviving beyond 5 yr. Targeting the prosurvival molecular effectors activated in cells within a soft ECM niche will very likely be a more tractable and efficacious strategy to improve chemotherapy response than modulating ECM mechanics directly. Unfortunately, despite promise in many tumor types, systemic NF-κB inhibitors have proved immensely toxic due to lack of tumor cell selectivity and immune suppression (Baud and Karin, 2009). Targeting NF-κB–regulated antiapoptotic proteins has shown more promise. A recent clinical trial using LCL161, an antagonist of inhibitor of apoptosis proteins, in combination with paclitaxel found that the rate of pCR more than doubled in this group when patients had a high TNFα gene expression signature (Bardia et al., 2018). However, this combination treatment was also associated with a higher incidence of serious adverse events. Recently, a small molecule selectively targeting NF-κB–JNK signaling by disruption of an inactivating interaction between NF-κB–regulated GADD45β and MKK7 in multiple myeloma enabled MKK7 to phosphorylate JNK and sensitized these cells to apoptosis (Tornatore et al., 2014). While this mechanism has not been demonstrated in breast cancer, it serves as a proof of principle that selective targeting of the NF-κB–JNK pathway may be of considerable benefit to patients. Other approaches to indirectly modulate NF-κB activity may also mitigate therapy resistance. For example, chemotherapy administered at the maximum tolerated dose can activate NF-κB in stromal fibroblasts, triggering the secretion of potent cytokines that induce the formation of highly drug-resistant tumor-initiating cells. Low-dose metronomic chemotherapy similarly abrogated stromal NF-κB activation, thus potentiating tumor response to systemic chemotherapy (Chan et al., 2016). Such ongoing efforts are needed to clarify the nature of the mechanisms that mediate treatment resistance and to develop effective, selective, and safe therapeutics.
Materials and methods
Human breast tissue acquisition and processing
Human breast tumor specimens were either embedded in an optimum cutting temperature (OCT) aqueous embedding compound (Tissue-Plus, catalog no. 4583; Scigen) within a disposable plastic base mold (catalog no. 22-363-554; Fisher Scientific) and snap frozen by direct immersion into liquid nitrogen and kept at −80°C in a freezer until cryosectioning for analysis, or were fixed in formalin and embedded in paraffin (FFPE). All human breast specimens were collected from prospective patients undergoing surgical resection at the University of California, San Francisco (UCSF), or Duke University Medical Center between 2010 and 2014. The selected specimens were de-identified, stored, and analyzed according to the procedures described in institutional review board (IRB) protocols 10-03832 and 10-05046, approved by the UCSF Committee of Human Resources and the Duke IRB (Pro00034242; Acerbi et al., 2015).
Cell culture
Human MEC lines MCF10A and HMT-3522 T4-2 were grown and maintained as previously described (Paszek et al., 2005). HCC70 was purchased from the American Type Culture Collection (ATCC CRL-2315) and grown according to American Type Culture Collection guidelines. Growth medium was supplemented to a final concentration of 160 µg/ml rBM 24 h after plating. Growth medium was changed 1 d after plating and every 2 d thereafter. All assays were performed between days 4 and 5 after seeding.
Mouse experiments
5-wk-old nude mice were orthotopically injected with 1 million T4-2 cells in soft or stiff ribose–cross-linked collagen gels. Collagen gels were prepared as previously described (Levental et al., 2009). Briefly, collagen was incubated with 500 mM L-ribose for at least 4 wk at 4°C before neutralization and gelation. Paclitaxel was dissolved in a 1:1 ratio of ethanol and Kolliphor EL, diluted in PBS, and injected i.p. (10 mg/kg) every 2 d starting 7 wk after tumor cell injection, coinciding with the emergence of measurable tumors. Mice were sacrificed and their tissue was harvested 14 d following the start of paclitaxel injections.
5-wk-old nude mice were injected with 5 million T4-2 cells s.c. in collagen gels. Mice were given three i.p. injections with NF-κB inhibitor JSH-23 (4.8 mg/kg) or DMSO vehicle, with the final injection coinciding with 4 Gy of IR to the tumor. Mice were sacrificed and their tissue was harvested 2 wk after irradiation.
6-wk-old nonobese diabetic/severe combined immunodeficiency mice were administered orthotopic injections with 3 million HCC70 cells transduced with mutant V737N β1 integrin or an empty vector control. Mice were fed doxycycline-supplemented chow (200 mg/kg) starting 8 d after tumor cell injection. Paclitaxel (10 mg/kg) was administered i.p. twice weekly after tumors reached an average of 7 mm in size. Mice were sacrificed at 10 d and 38 d following the start of treatment.
Equal sized pieces of human triple-negative PDX BCM2665 (Zhang et al., 2013) were inserted into the mammary glands of 8-wk-old nonobese diabetic/severe combined immunodeficiency mice in soft or ribose-stiffened collagen gels (Levental et al., 2009). Paclitaxel injections (10 mg/kg) began twice weekly 3.5 wk after PDX implantation following the emergence of tumors with an average size of 9 mm. Mice were sacrificed 21 d following the start of treatment.
Tumor resistance was calculated as the day at which tumor size increased greater than or equal to a 1.4-fold change followed by no reduction in size at the next measurement and no substantial size reductions for the remainder of chemotherapy treatment. In the case of HCC70 tumors, tumor resistance was calculated as this criterion or a 10% size increase for greater than or equal to three consecutive measurements, whichever came first. This alternative methodology was used for HCC70 because these tumors grew very slowly relative to the others and thus did not always achieve a 1.4-fold change despite not responding to paclitaxel.
All mouse studies were maintained under specific pathogen–free conditions and performed in accordance with the guidelines of the institutional animal care and use committee and the Laboratory Animal Research Center at UCSF.
Lentiviral infection and vectors
MCF10A cells were transduced with the dominant-negative forms of JNK (HA-JNK1-APF and HA-JNK2-APF) and used as previously described (L. Heasley, University of Colorado, Denver, CO; Wojtaszek et al., 1998). MCF10A cells were transduced with the constitutively active JNK1, the MKK7-JNK1α fusion protein, as previously described (Takada et al., 2005; Han et al., 2002).
MCF10A, T4-2, and HCC70 cells were electroporated to introduce a transposon (PiggyBac) expressing a doxycycline-inducible IκBαM and PiggyBac transposase (Van Antwerp et al., 1996). Cells were selected in G418 (500 µg/ml) for purity.
HCC70 cells were electroporated to introduce a transposon (PiggyBac) expressing mEmerald and a doxycycline-inducible V737N β1 integrin mutant or an empty vector control. Cells were selected in G418 (500 µg/ml) for purity.
PA gel preparation
Basement membrane–conjugated PA gels were prepared as described previously (Przybyla et al., 2016). Soft gels with measured elastic moduli of 140 Pa were prepared with 3% (wt/vol) acrylamide and 0.04% (wt/vol) bis-acrylamide, and stiff gels with elastic moduli of 5,000 Pa were prepared with 5.5% (wt/vol) acrylamide and 0.15% (wt/vol) bis-acrylamide. Gels were conjugated with 10 µg/ml rBM and washed overnight with basal medium plus antibiotic.
Apoptosis assays
MECs grown on soft or stiff PA gels for 4–5 d were treated with either paclitaxel (5, 10, 50, or 100 nM), TRAIL (100 or 200 ng/ml), or IR (4–12 Gy). Apoptosis was directly assayed at 24 h after treatment via immunofluorescence (IF) staining of cleaved caspase 3. The percentage of apoptosis was calculated as the number of cells positive for activated caspase 3 expressed as a percentage of the total number of cells using 300–1,600 cells per condition in each experiment counted from several fields of view. Maximum-intensity projections of z-stacks were used for analysis of cleaved caspase 3–positive cells.
IF and immunohistochemistry (IHC)
Cells were briefly rinsed in PBS and directly fixed using 2–4% paraformaldehyde for 20 min at room temperature. Cells were washed with IF buffer (32.5 mM NaCl, 3.3 mM Na2HPO4, 76 µM KH2PO4, 1.92 mM NaN3, 0.1% [wt/vol] BSA, 0.2% [vol/vol] Triton X-100, 0.05% [vol/vol] Tween 20) and then incubated with the blocking solution (3% BSA, 5% normal goat serum in IF buffer; 1 h; room temperature). Cells were incubated with primary antibodies in a humidified chamber overnight at 4°C. Samples were washed with IF buffer (three times for 10 min while rocking) and incubated with Alexa Fluor 488– or Alexa Fluor 555–conjugated secondary antibodies (1 h, room temperature). Samples were washed again with IF buffer (three times for 15 min while rocking), and nuclei were counterstained with DAPI (1 µg/ml in PBS, 5 min, room temperature). Cells on gels were inverted onto a number 1 coverglass immediately before imaging.
Fluorescence images were acquired on a spinning disk confocal Nikon Eclipse Ti microscope at 20× magnification with a 0.75 NA objective. Cells on PA gels were inverted directly onto a number 1 coverslip and imaged immediately at room temperature. The images were acquired using an Andor Zyla scientific complementary metal–oxide–semiconductor camera and MetaMorph acquisition software. Images were processed in ImageJ.
FFPE slides were deparaffinized in xylene and rehydrated in graded ethanol solutions. Heat-mediated antigen retrieval was performed in 10 mM citrate buffer, pH 6.0, and endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide. Tissues were incubated with primary antibody overnight at 4°C and with biotin-conjugated secondary antibody for 1 h at room temperature. The Vectastain Elite ABC Kit (Vector Laboratories; PK-6100) and a 3,3′-diaminobenzidine peroxidase substrate kit (Vector Laboratories; SK-4100) were used for signal detection according to the manufacturer’s instructions.
IHC images were acquired on a Nikon Eclipse TE2000-U inverted microscope at 20× magnification with a 0.75 NA objective. Images were acquired with a Hamamatsu ORCA-Flash4.0 LT camera using NIS-Elements software.
Protein lysates
Protein lysates were prepared using radioimmunoprecipitation assay (RIPA; 50 mM Tris-HCl, 150 mM NaCl, 0.25% Na-deoxycholate, 0.1% SDS, and 1.0% IGEPAL CA-630 [NP-40]; pH 7.4 at room temperature) or Laemmli lysis buffer (50 mM Tris-HCl, 2% SDS, and 5 mM EDTA; pH 7.4 at room temperature). Immediately before cell lysis, a cocktail of protease inhibitors was added to the buffer (1.2 µg leupeptin, 1.2 µg pepstatin, 2.4 µg aprotinin, 12 µg E-64, 0.5 mM benzamidine, 50 mM NaF, and 1.2 µg Pefabloc) and the phosphatase inhibitor Na-orthovanadate (1 mM activated with 1.5% H2O2). RIPA lysates were carried out on ice, while Laemmli lysates were performed at room temperature due to SDS precipitation at cold temperatures. After rinsing the dishes with PBS, lysis buffer was added and cells were scraped off the dish with a cell scraper. RIPA lysates were sonicated three times, each for 10 s, while Laemmli lysates were passed through a fine pipette tip (p200) several times. Both were centrifuged for 10 min at 20,817 relative centrifugal force to pellet any internal organelles and cellular debris. Supernatant was collected and fast frozen on dry ice. Protein concentration was determined using a bicinchoninic acid assay kit (Promega) following the manufacturer’s protocol.
Cytoplasmic and nuclear protein extracts were obtained with a kit using the manufacturer’s instructions (78833; Thermo Fisher Scientific).
Western blotting
Cells were lysed as described above, and equal amounts of protein were separated on reducing SDS- PA gels, immunoblotted, and detected with the ECL Plus System. Samples were boiled for 5 min (95°C) and loaded onto the SDS-PA gel, and protein was separated at 120 constant volts.
The protein was transferred onto a prewet polyvinylidene difluoride membrane (100% methanol, 1 min), either at 300 mA for 2 h or at 25 constant volts overnight (4°C). The polyvinylidene difluoride membrane was rinsed with Tris-buffered saline with Tween 20 (TBST), and nonspecific binding was blocked with 5% nonfat dry milk dissolved in TBST. The membrane was then incubated with the primary antibody overnight at 4°C, washed with TBST, incubated with HRP-conjugated secondary antibody (1 h; room temperature; dilution, 1:5,000), washed with TBST, and detected with the chemiluminescence system Quantum HRP substrate (K-12042; Advansta). Quantification was performed using ImageJ.
Metabolic activity assay
MECs were washed with 1× PBS and incubated with MTT working solution (5 mg/ml [wt/vol] MTT stock solution in PBS, diluted 10-fold in DMEM/F12 for a final MTT concentration of 500 µg/ml) for 2 h at 37°C. MTT was solubilized with DMSO for 10 min at 37°C. The plate was tapped vigorously to extract all of the MTT from the cells, and, once the solution was an even purple color, a quantity of 100 µl was transferred to a 96-well plate. The absorbance was measured directly using a plate reader set at 550 nm and background subtraction set at 630 nm. Absorbance values were normalized to the untreated controls to determine relative metabolic activity.
Irradiation
All cell culture and mouse experiments used a JL Shepherd Mark I model 20 137Cs source irradiator.
Proliferation assay
To measure the proliferation rate of MECs in log phase of growth on soft (E = 140 Pa) and stiff (E = 5,000 Pa) PA gels, cells were trypsinized and counted at 24-h intervals. The growth rate constant, k, was derived using the equation N/No = ekt, where No is the initial absorbance or cell number and N is the absorbance or cell number at time t. The doubling time, td, was calculated using the derived k and setting N/No = 2 in the following equation: td = (ln 2)/k.
To assess the proliferation rate of MECs in 3D rBM, cultures was fixed and stained at different time points with anti-phosphorylated histone H3. Proliferating cells or colonies (positive phH3 staining) were scored using a fluorescence microscope. A minimum of 250 cells per condition were scored from several fields of view.
Colony size was determined by measuring the diameter of MEC colonies after 10–14 d in 3D rBM or 3D collagen/rBM. The colony diameters were measured using a precalibrated eyepiece on a light microscope. At least 50 colonies per sample were scored from different fields of view in the center of each gel.
Cell cycle analysis
The percentage of cells in each stage of the cell cycle was determined by PI staining and FACS analysis for DNA content (FACSCalibur; BD Biosciences). MECs were grown on soft and stiff PA gels and processed on days 4 and 5 in order to determine cell cycle status. Briefly, MECs were trypsinized, counted, and washed with ice-cold 1% BSA/Dulbecco’s PBS. MECs were fixed with a 1:4 dilution of 1% BSA/Dulbecco’s PBS in 96% ethanol. Fixed MECs were centrifuged at 259 relative centrifugal force, washed with ice-cold PBS, and incubated in PI staining solution (50 µg/ml PI in PBS supplemented with 3.8 mM sodium citrate and 500 µg/ml RNase A; 500 µl per 2 × 106 cells; 3 h on ice). DNA content was determined using the FACSCalibur set to low flow, and data were acquired for 20,000 cells within the gated region of cells in G0/G1, S, and G2/M phases of the cell cycle.
Clonogenic survival assay
MECs were grown for 4 d on soft and stiff PA gels and treated with apoptosis inducers on day 4 as described. Cells were trypsinized and replated on soft or stiff PA gels at low density on day 4 (500 cells in a 60-mm gel; triplicates), grown for 7–10 d, and assayed for colony formation. Briefly, cells were rinsed with 1× PBS, fixed (ice-cold 100% methanol, 10 min, −20°C), rinsed with 1× PBS, stained with crystal violet (0.5% [wt/vol] solution, 10 min, room temperature), rinsed with 1× PBS, rinsed with double-distilled H2O, and air dried by inverting onto a paper towel (overnight).
Plates were scanned with a conventional flatbed scanner (grayscale, 150 dots per inch, TIF format). Colonies were counted using a MATLAB script. First, a representative positive colony was chosen for calibration, where a positive colony contained 50–100 cells (∼10 square pixels). The program output was the number of colonies greater than or equal to the size indicated for a positive colony. Any fused colonies in the program output were accounted for by adding 1 to the final colony number for each fusion.
PS staining and quantification
FFPE tissue sections were stained using 0.1% PS (Direct Red 80, catalog no. 365548, and picric acid solution, catalog no. P6744; Sigma-Aldrich) and counterstained with Weigert’s hematoxylin (catalog no. CM3951; Cancer Diagnostics). Brightfield and polarized light images were acquired using an Olympus IX81 microscope fitted with an analyzer (U-ANT) and a polarizer (U-POT, Olympus) oriented parallel or orthogonal to each other, respectively. The percentage of PS signal per field of view was quantified in polarized light images using ImageJ.
SHG microscopy
20-µm-thick OCT-embedded tissue sections were thawed in room temperature PBS and fixed in 4% paraformaldehyde. Nuclei were stained with PI. SHG was performed using a custom-built two-photon laser scanning microscope using both 18-W Coherent Chameleon Vision II and 8-W Mai Tai Ti:Sapphire lasers and a Prairie Technology Ultima System with an Olympus BX-51 fixed-stage microscope equipped with a 25× (NA 1.05) water immersion objective. Images were acquired using µMagellan software.
SHG images of collagen fibers were analyzed using the ImageJ plugin OrientationJ (Fonck et al., 2009; Clemons et al., 2018). Fiber coherency was measured in the dominant fiber direction as determined by OrientationJ for each image.
AFM data acquisition
AFM measurements were performed as previously described (Acerbi et al., 2015). Briefly, 20-µm-thick OCT-embedded frozen human breast tumor tissue sections were thawed by immersion in room temperature PBS. Thawed sections were immersed in PBS containing phosphatase inhibitor (genDEPOT Xpert; P3200-001), protease inhibitor cocktail (genDEPOT Xpert; P3100-001), and PI (catalog no. 440300250; ACROS Organics) and placed on the stage for AFM measurements. AFM indentations were performed using an MFP3D-BIO inverted optical AFM (Asylum Research) mounted on a Nikon TE2000-U inverted fluorescence microscope. Silicon nitride cantilevers were used with an approximate spring constant of 0.06 N m−1 and a borosilicate glass spherical tip with 5-µm diameter (Novascan Technologies). The cantilever was calibrated using the thermal oscillation method before each experiment. Specimens were indented at a 1 μm s−1 loading rate. Force maps were obtained in tumor-adjacent ECM as a raster series of indentations using the FMAP function of the IGOR PRO build supplied by Asylum Research. Elastic properties of ECM were determined by fitting data with the Hertz model. A Poisson’s ratio of 0.5 was used in the calculation of Young’s elastic modulus.
Antibodies and reagents
We used commercial ECM (Matrigel; Collaborative Research) for the rBM assays. Primary antibodies used were as follows: cleaved caspase 3 (1:400, 9661; Cell Signaling Technology), phospho-c-Jun (serine 63; 1:1,000, 3270; Cell Signaling Technology), JNK1/JNK2 (1:1,000, 554285; BD PharMingen), E-cadherin (1:1,000, 3195; Cell Signaling Technology), phospho-JNK (G-7) mAb (1:100, sc-6254; Santa Cruz Biotechnology), HA.11 clone 16B12 (1:2,000; BabCo), and p65 (1:1,000 Western blot, 1:800 IHC, 8242; Cell Signaling Technology). The secondary antibodies used were as follows: HRP-conjugated anti-rabbit and anti-mouse (Amersham Biosciences) and Alexa Fluor 488– and Alexa Fluor 555–conjugated anti-mouse and anti-rabbit IgGs (Molecular Probes). The following reagents were used in these studies: PI, DAPI, MTT, and Weigert’s iron hematoxylin (all from Sigma-Aldrich); N-tetramethyl ethylenediamine, 30% acrylamide/bis solution (37.5:1; 2.6% C), 40% acrylamide solution, and 2% bis solution (Bio-Rad Laboratories); TRAIL (100–200 ng/ml), paclitaxel (5–100 nM), and the JNK inhibitor SP600125 (5 µM, A4604; APExBIO); recombinant human TNFα (20 ng/ml, 68-8786-63; Invitrogen); and NF-κB inhibitor JSH-23 (481408; MilliporeSigma).
Gene set enrichment analysis
Gene set enrichment analysis was performed to assess differential expression of NF-κB target genes in patients who achieved or failed to achieve a pCR to NAC, using publicly available software maintained by the Broad Institute (Subramanian et al., 2005). Publicly available gene expression and treatment response data from the I-SPY-1 trial were used (Gene Expression Omnibus accession no. GSE32603; Magbanua et al., 2015). The NF-κB gene signature gene set was taken from publicly available chromatin immunoprecipitation sequencing data from the Cistrome data browser. The gene set consisted of the top 100 targets from a p65 (RELA) chromatin immunoprecipitation sequencing experiment in the MCF-7 MEC line treated with TNFα compared with vehicle-treated cells (Stender et al., 2017).
Statistics
Statistical tests are noted in each figure legend. All tests used a significance level of 0.05. Data were analyzed using GraphPad Prism software All data reported herein displays one star denoting a P value ≤ 0.05, two stars denoting P ≤ 0.01, and three stars denoting P ≤ 0.001. For all data presented, the number of biological replicate experiments conducted is denoted as n.
Study approval
All human breast specimens were collected from prospective patients undergoing surgical resection at UCSF or Duke University Medical Center between 2010 and 2014. Informed consent was received from patients before tissue collection. The selected specimens were de-identified, stored, and analyzed according to the procedures described in IRB protocols 10-03832 and 10-05046, approved by the UCSF Committee of Human Resources and the Duke IRB (Pro00034242; Acerbi et al., 2015). All mouse studies were maintained under specific pathogen–free conditions and performed in accordance with the guidelines of the institutional animal care and use committee and the Laboratory Animal Research Center at UCSF.
Online supplemental material
Fig. S1 shows AFM analysis for each primary tumor and residual disease sample. Fig. S2 shows additional data supporting stiffness-induced apoptosis sensitivity and representative images of MCF10A and HCC70 treated with paclitaxel. Fig. S3 shows MCF10A cell cycle data on soft and stiff substrates. Fig. S4 shows dnJNK and IκBαM construct validation and additional apoptosis sensitivity data in MCF10A and T4-2 cells treated with paclitaxel and TRAIL with substrate stiffness and with loss of NF-κB activity. Table S1 shows clinical data for patient samples used in AFM, SHG, and PS analysis. Table S2 shows clinical data for patient samples used in the gene set enrichment analysis. Table S3 shows clinical data for patient samples used in p65 IHC analysis.
Supplementary Material
shows human triple-negative breast tumor patient data for AFM, SHG, and PS analysis.
shows human breast tumor patient data for gene set enrichment analysis.
shows human triple-negative breast tumor patient data for p65 IHC.
Acknowledgments
We thank Josette Northcott for writing the picrosirius quantification macro and Connor Stashko for assistance in gene expression data set analysis. We also thank Lynn Heasley for generously providing dnJNK1/2 and MKK7-JNK1 constructs. We thank Kelvin Tsai for writing the colony quantification script. We are grateful to the UCSF Biological Imaging Development Center for maintenance and assistance with the two-photon microscope.
This work was supported by investigator grants through the US Department of Defense Breast Cancer Research Program (BC122990 to V.M. Weaver), the National Cancer Institute (R01 CA174929 to C.C. Park and V.M. Weaver; R01 CA192914, R01 CA222508, and R35 CA242447 to V.M. Weaver), and the Breast Cancer Research Foundation (A132292 to V.M. Weaver).
Author contributions: V.M. Weaver, C.C. Park, A.P. Drain, and N. Zahir conceived the project, and V.M. Weaver, C.C. Park, A.P. Drain, and N. Zahir prepared figures and wrote the manuscript. A.P. Drain performed AFM in human tissues, and O. Maller performed and quantified PS staining. A.P. Drain, H. Zhang, N. Zahir, and J.L. Leight performed 3D cell culture experiments and cleaved caspase 3 staining, imaging, and quantification. A.P. Drain designed and A.P. Drain, J.J. Northey, H. Zhang, and X. Yu conducted in vivo experiments with paclitaxel and radiation treatments. N. Zahir performed proliferation and clonogenic survival assays. J.N. Lakins, N. Zahir, and A.P. Drain generated dominant-negative JNK, MKK7-JNK1, IκBαM, and β1 integrin transduced cell lines. N. Zahir, B.P. Alston-Mills, and A.P. Drain performed apoptosis assays in gain/loss of JNK function experiments. A.P. Drain and H. Zhang performed p-c-Jun Western blot experiments. E.S. Hwang and Y.-Y. Chen provided primary treatment-naive and chemotherapy-treated human breast tumor samples. A.P. Drain conducted gene set enrichment analysis on human patient gene expression from the I-SPY-1 trial. P.-J. Huang and J.J. Northey performed p65 staining and quantification on primary human tumor and residual disease tissues.
References
- Abt, N.B., Flores J.M., Baltodano P.A., Sarhane K.A., Abreu F.M., Cooney C.M., Manahan M.A., Stearns V., Makary M.A., and Rosson G.D.. 2014. Neoadjuvant chemotherapy and short-term morbidity in patients undergoing mastectomy with and without breast reconstruction. JAMA Surg. 149:1068–1076. 10.1001/jamasurg.2014.1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acerbi, I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., and Weaver V.M.. 2015. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 7:1120–1134. 10.1039/c5ib00040h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, K.M., Zhang H., and Park C.C.. 2013. NF-κB regulates radioresistance mediated by β1-integrin in three-dimensional culture of breast cancer cells. Cancer Res. 73:3737–3748. 10.1158/0008-5472.CAN-12-3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenden, M., van Weverwijk A., Murugaesu N., Fearns A., Campbell J., Gao Q., Iravani M., and Isacke C.M.. 2017. An in vivo functional screen identifies JNK signaling as a modulator of chemotherapeutic response in breast cancer. Mol. Cancer Ther. 16:1967–1978. 10.1158/1535-7163.MCT-16-0731 [DOI] [PubMed] [Google Scholar]
- Baeuerle, P.A., and Baltimore D.. 1996. NF-κ B: ten years after. Cell. 87:13–20. 10.1016/S0092-8674(00)81318-5 [DOI] [PubMed] [Google Scholar]
- Baldwin, A.S. Jr. 2001. Series introduction: the transcription factor NF-kappaB and human disease. J. Clin. Invest. 107:3–6. 10.1172/JCI11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardia, A., Parton M., Kümmel S., Estévez L.G., Huang C.S., Cortés J., Ruiz-Borrego M., Telli M.L., Martin-Martorell P., López R., et al. 2018. Paclitaxel with inhibitor of apoptosis antagonist, LCL161, for localized triple-negative breast cancer, prospectively stratified by gene signature in a biomarker-driven neoadjuvant trial. J. Clin. Oncol. 36:JCO2017748392. 10.1200/JCO.2017.74.8392 [DOI] [PubMed] [Google Scholar]
- Baud, V., and Karin M.. 2009. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 8:33–40. 10.1038/nrd2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, T.-S., Hsu C.-C., Pai V.C., Liao W.-Y., Huang S.-S., Tan K.-T., Yen C.-J., Hsu S.-C., Chen W.-Y., Shan Y.-S., et al. 2016. Metronomic chemotherapy prevents therapy-induced stromal activation and induction of tumor-initiating cells. J. Exp. Med. 213:2967–2988. 10.1084/jem.20151665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-R., Wang X., Templeton D., Davis R.J., and Tan T.-H.. 1996. The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and γ radiation. Duration of JNK activation may determine cell death and proliferation. J. Biol. Chem. 271:31929–31936. 10.1074/jbc.271.50.31929 [DOI] [PubMed] [Google Scholar]
- Clemons, T.D., Bradshaw M., Toshniwal P., Chaudhari N., Stevenson A.W., Lynch J., Fear M.W., Wood F.M., and Iyer K.S.. 2018. Coherency image analysis to quantify collagen architecture: implications in scar assessment. RSC Advances. 8:9661–9669. 10.1039/C7RA12693J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar, P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. 2014. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 384:164–172. 10.1016/S0140-6736(13)62422-8 [DOI] [PubMed] [Google Scholar]
- Dhanasekaran, D.N., and Reddy E.P.. 2008. JNK signaling in apoptosis. Oncogene. 27:6245–6251. 10.1038/onc.2008.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, R.E., and Sutherland R.M.. 1972. Effects of intercellular contact on repair of radiation damage. Exp. Cell Res. 71:75–80. 10.1016/0014-4827(72)90265-0 [DOI] [PubMed] [Google Scholar]
- Egeblad, M., Rasch M.G., and Weaver V.M.. 2010. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 22:697–706. 10.1016/j.ceb.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonck, E., Feigl G.G., Fasel J., Sage D., Unser M., Rüfenacht D.A., and Stergiopulos N.. 2009. Effect of aging on elastin functionality in human cerebral arteries. Stroke. 40:2552–2556. 10.1161/STROKEAHA.108.528091 [DOI] [PubMed] [Google Scholar]
- Fournier, A.K., Campbell L.E., Castagnino P., Liu W.F., Chung B.M., Weaver V.M., Chen C.S., and Assoian R.K.. 2008. Rac-dependent cyclin D1 gene expression regulated by cadherin- and integrin-mediated adhesion. J. Cell Sci. 121:226–233. 10.1242/jcs.017012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland, J.C., Lakins J.N., Kazanietz M.G., Chernoff J., Boettiger D., and Weaver V.M.. 2007. α6β4 integrin activates Rac-dependent p21-activated kinase 1 to drive NF-kappaB-dependent resistance to apoptosis in 3D mammary acini. J. Cell Sci. 120:3700–3712. 10.1242/jcs.03484 [DOI] [PubMed] [Google Scholar]
- Fulda, S., and Debatin K.M.. 2006. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 25:4798–4811. 10.1038/sj.onc.1209608 [DOI] [PubMed] [Google Scholar]
- Han, S.Y., Kim S.H., and Heasley L.E.. 2002. Differential gene regulation by specific gain-of-function JNK1 proteins expressed in Swiss 3T3 fibroblasts. J. Biol. Chem. 277:47167–47174. 10.1074/jbc.M204270200 [DOI] [PubMed] [Google Scholar]
- Hanker, A.B., Estrada M.V., Bianchini G., Moore P.D., Zhao J., Cheng F., Koch J.P., Gianni L., Tyson D.R., Sánchez V., et al. 2017. Extracellular matrix/integrin signaling promotes resistance to combined inhibition of HER2 and PI3K in HER2+ breast cancer. Cancer Res. 77:3280–3292. 10.1158/0008-5472.CAN-16-2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, E., Girotti M.R., Viros A., Hooper S., Spencer-Dene B., Matsuda M., Larkin J., Marais R., and Sahai E.. 2015. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell. 27:574–588. 10.1016/j.ccell.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y.-L., Li S., Shyy J.Y.-J., and Chien S.. 1999. Sustained JNK activation induces endothelial apoptosis: studies with colchicine and shear stress. Am. J. Physiol. 277:H1593–H1599. 10.1152/ajpheart.1999.277.4.H1593 [DOI] [PubMed] [Google Scholar]
- Javelaud, D., and Besançon F.. 2001. NF-κ B activation results in rapid inactivation of JNK in TNF α-treated Ewing sarcoma cells: a mechanism for the anti-apoptotic effect of NF-κ B. Oncogene. 20:4365–4372. 10.1038/sj.onc.1204570 [DOI] [PubMed] [Google Scholar]
- Johnson, K.R., Leight J.L., and Weaver V.M.. 2007. Demystifying the Effects of a Three-Dimensional Microenvironment in Tissue Morphogenesis. Methods Cell Biol. 83:547–583. 10.1016/S0091-679X(07)83023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumi, A., Naoe T., Matsushita T., Kaibuchi K., and Schwartz M.A.. 2005. Integrin activation and matrix binding mediate cellular responses to mechanical stretch. J. Biol. Chem. 280:16546–16549. 10.1074/jbc.C400455200 [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Chang J.K., Dominguez A.A., Lee H.P., Nam S., Chang J., Varma S., Qi L.S., West R.B., and Chaudhuri O.. 2019. YAP-independent mechanotransduction drives breast cancer progression. Nat. Commun. 10:1848. 10.1038/s41467-019-09755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leight, J.L., Drain A.P., and Weaver V.M.. 2017. Extracellular matrix remodeling and stiffening modulate tumor phenotype and treatment response. Annu. Rev. Cancer Biol. 1:313–334. 10.1146/annurev-cancerbio-050216-034431 [DOI] [Google Scholar]
- Levental, K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F.T., Csiszar K., Giaccia A., Weninger W., et al. 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139:891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., Bai L., Chen W., and Xu S.. 2010. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin. Ther. Targets. 14:45–55. 10.1517/14728220903431069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua, M.J.M., Wolf D.M., Yau C., Davis S.E., Crothers J., Au A., Haqq C.M., Livasy C., Rugo H.S., Esserman L., et al. I-SPY 1 TRIAL Investigators . 2015. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res. 17:73. 10.1186/s13058-015-0582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller, O., Drain A.P., Barrett A.S., Borgquist S., Ruffell B., Zakharevich I., Pham T.T., Gruosso T., Kuasne H., Lakins J.N., et al. 2020. Tumour-associated macrophages drive stromal cell-dependent collagen crosslinking and stiffening to promote breast cancer aggression. Nat. Mater. 10.1038/s41563-020-00849-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen, M.J., Chiariello M., Tanos T., Bernard O., Narumiya S., and Gutkind J.S.. 2004. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell. 14:29–41. 10.1016/S1097-2765(04)00153-4 [DOI] [PubMed] [Google Scholar]
- Murray, S., Briasoulis E., Linardou H., Bafaloukos D., and Papadimitriou C.. 2012. Taxane resistance in breast cancer: mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 38:890–903. 10.1016/j.ctrv.2012.02.011 [DOI] [PubMed] [Google Scholar]
- Nagane, M., Huang H.J.S., and Cavenee W.K.. 2001. The potential of TRAIL for cancer chemotherapy. Apoptosis. 6:191–197. 10.1023/A:1011336726649 [DOI] [PubMed] [Google Scholar]
- Nguyen, H.T., Hsieh M.H., Gaborro A., Tinloy B., Phillips C., and Adam R.M.. 2006. JNK/SAPK and p38 SAPK-2 mediate mechanical stretch-induced apoptosis via caspase-3 and -9 in NRK-52E renal epithelial cells. Nephron, Exp. Nephrol. 102:e49–e61. 10.1159/000088401 [DOI] [PubMed] [Google Scholar]
- Nguyen, T.V., Sleiman M., Moriarty T., Herrick W.G., and Peyton S.R.. 2014. Sorafenib resistance and JNK signaling in carcinoma during extracellular matrix stiffening. Biomaterials. 35:5749–5759. 10.1016/j.biomaterials.2014.03.058 [DOI] [PubMed] [Google Scholar]
- Northey, J.J., Przybyla L., and Weaver V.M.. 2017. Tissue force programs cell fate and tumor aggression. Cancer Discov. 7:1224–1237. 10.1158/2159-8290.CD-16-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek, M.J., Zahir N., Johnson K.R., Lakins J.N., Rozenberg G.I., Gefen A., Reinhart-King C.A., Margulies S.S., Dembo M., Boettiger D., et al. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pickup, M.W., Mouw J.K., and Weaver V.M.. 2014. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15:1243–1253. 10.15252/embr.201439246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma, B., Hayward M.K., and Weaver V.M.. 2020. Fibrosis and cancer: A strained relationship. Biochim. Biophys. Acta Rev. Cancer. 1873:188356. 10.1016/j.bbcan.2020.188356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla, L., Lakins J.N., Sunyer R., Trepat X., and Weaver V.M.. 2016. Monitoring developmental force distributions in reconstituted embryonic epithelia. Methods. 94:101–113. 10.1016/j.ymeth.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl, A., Schlederer M., Pudelko K., Stadler M., Walter S., Unterleuthner D., Unger C., Kramer N., Hengstschläger M., Kenner L., et al. 2017. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 130:203–218. 10.1242/jcs.188102 [DOI] [PubMed] [Google Scholar]
- Schrader, J., Gordon-Walker T.T., Aucott R.L., van Deemter M., Quaas A., Walsh S., Benten D., Forbes S.J., Wells R.G., and Iredale J.P.. 2011. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 53:1192–1205. 10.1002/hep.24108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, R.K. 2001. TRAIL/Apo-2L: mechanisms and clinical applications in cancer. Neoplasia. 3:535–546. 10.1038/sj.neo.7900203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender, J.D., Nwachukwu J.C., Kastrati I., Kim Y., Strid T., Yakir M., Srinivasan S., Nowak J., Izard T., Rangarajan E.S., et al. 2017. Structural and molecular mechanisms of cytokine-mediated endocrine resistance in human breast cancer cells. Mol. Cell. 65:1122–1135.e5. 10.1016/j.molcel.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, X., Kong N., Ye L., Han W., Zhou J., Zhang Q., He C., and Pan H.. 2014. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 344:174–179. 10.1016/j.canlet.2013.11.019 [DOI] [PubMed] [Google Scholar]
- Sunters, A., Madureira P.A., Pomeranz K.M., Aubert M., Brosens J.J., Cook S.J., Burgering B.M.T., Coombes R.C., and Lam E.W.F.. 2006. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 66:212–220. 10.1158/0008-5472.CAN-05-1997 [DOI] [PubMed] [Google Scholar]
- Sutherland, R.M. 1988. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 240:177–184. 10.1126/science.2451290 [DOI] [PubMed] [Google Scholar]
- Symmans, W.F., Wei C., Gould R., Yu X., Zhang Y., Liu M., Walls A., Bousamra A., Ramineni M., Sinn B., et al. 2017. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J. Clin. Oncol. 35:1049–1060. 10.1200/JCO.2015.63.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, E., Shimo K., Hata K., Abiake M., Mukai Y., Moriyama M., Heasley L., and Mizuguchi J.. 2005. Interferon-β-induced activation of c-Jun NH2-terminal kinase mediates apoptosis through up-regulation of CD95 in CH31 B lymphoma cells. Exp. Cell Res. 304:518–530. 10.1016/j.yexcr.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Tang, G., Minemoto Y., Dibling B., Purcell N.H., Li Z., Karin M., and Lin A.. 2001. Inhibition of JNK activation through NF-kappaB target genes. Nature. 414:313–317. 10.1038/35104568 [DOI] [PubMed] [Google Scholar]
- Tergaonkar, V., Pando M., Vafa O., Wahl G., and Verma I.. 2002. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 1:493–503. 10.1016/S1535-6108(02)00068-5 [DOI] [PubMed] [Google Scholar]
- Tornatore, L., Sandomenico A., Raimondo D., Low C., Rocci A., Tralau-Stewart C., Capece D., D’Andrea D., Bua M., Boyle E., et al. 2014. Cancer-selective targeting of the NF-κB survival pathway with GADD45β/MKK7 inhibitors. Cancer Cell. 26:495–508. 10.1016/j.ccr.2014.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerp, D.J., Martin S.J., Kafri T., Green D.R., and Verma I.M.. 1996. Suppression of TNF-α-induced apoptosis by NF-kappaB. Science. 274:787–789. 10.1126/science.274.5288.787 [DOI] [PubMed] [Google Scholar]
- von Minckwitz, G., Untch M., Blohmer J.U., Costa S.D., Eidtmann H., Fasching P.A., Gerber B., Eiermann W., Hilfrich J., Huober J., et al. 2012. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 30:1796–1804. 10.1200/JCO.2011.38.8595 [DOI] [PubMed] [Google Scholar]
- Weaver, V.M., Lelièvre S., Lakins J.N., Chrenek M.A., Jones J.C.R., Giancotti F., Werb Z., and Bissell M.J.. 2002. β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2:205–216. 10.1016/S1535-6108(02)00125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek, P.A., Heasley L.E., Siriwardana G., and Berl T.. 1998. Dominant-negative c-Jun NH2-terminal kinase 2 sensitizes renal inner medullary collecting duct cells to hypertonicity-induced lethality independent of organic osmolyte transport. J. Biol. Chem. 273:800–804. 10.1074/jbc.273.2.800 [DOI] [PubMed] [Google Scholar]
- Wong, R.S.Y. 2011. Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30:87. 10.1186/1756-9966-30-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Shen S., and Verma I.M.. 2014. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2:823–830. 10.1158/2326-6066.CIR-14-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y., and Gaynor R.B.. 2001. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J. Clin. Invest. 107:135–142. 10.1172/JCI11914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir, N., Lakins J.N., Russell A., Ming W., Chatterjee C., Rozenberg G.I., Marinkovich M.P., and Weaver V.M.. 2003. Autocrine laminin-5 ligates α6β4 integrin and activates RAC and NFkappaB to mediate anchorage-independent survival of mammary tumors. J. Cell Biol. 163:1397–1407. 10.1083/jcb.200302023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeligs, K.P., Neuman M.K., and Annunziata C.M.. 2016. Molecular pathways: the balance between cancer and the immune system challenges the therapeutic specificity of targeting nuclear factor-κB signaling for cancer treatment. Clin. Cancer Res. 22:4302–4308. 10.1158/1078-0432.CCR-15-1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Claerhout S., Prat A., Dobrolecki L.E., Petrovic I., Lai Q., Landis M.D., Wiechmann L., Schiff R., Giuliano M., et al. 2013. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 73:4885–4897. 10.1158/0008-5472.CAN-12-4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows human triple-negative breast tumor patient data for AFM, SHG, and PS analysis.
shows human breast tumor patient data for gene set enrichment analysis.
shows human triple-negative breast tumor patient data for p65 IHC.