Abstract
Pathogenic Escherichia coli is an important cause of diarrhea, edema disease, and septicemia in swine. In Japan, the volume of antimicrobial drugs used for animals is highest in swine, but information about the prevalence of antimicrobial-resistant bacteria is confined to apparently healthy animals. In the present study, we determined the O serogroups, virulence factors, and antimicrobial resistance of 360 E. coli isolates from swine that died of disease in Kagoshima Prefecture, Japan, between 1999 and 2017. The isolates of the predominant serogroups O139, OSB9, O149, O8, and O116 possessed virulence factor genes typically found in diarrheagenic E. coli. We further found five strains of third-generation cephalosporin-resistant E. coli that each produced an extended-spectrum β-lactamase encoded by blaCTX-M-14, blaCTX-M-15, blaCTX-M-24, blaCTX-M-61, or blaSHV-12. In 218 swine with a clear history of antimicrobial drug use, we further analyzed associations between the use of antimicrobials for the treatment of diseased swine and the isolation of resistant E. coli. We found significant associations between antimicrobial use and selection of resistance to the same class of antimicrobials, such as the use of ceftiofur and resistance to cefotaxime, cefazolin, or ampicillin, the use of aminoglycosides and resistance to streptomycin, and the use of phenicols and resistance to chloramphenicol. A significant association between antimicrobial use and the resistance of E. coli isolates to structurally unrelated antimicrobials, such as the use of ceftiofur and resistance to chloramphenicol, was also observed.
Keywords: antimicrobial resistance, pathogenic Escherichia coli, swine
Pathogenic Escherichia coli is an important cause of several diseases in swine worldwide, including neonatal diarrhea, postweaning diarrhea, edema disease, and septicemia. In particular, diarrhea and edema disease result in significant economic losses due to high morbidity and mortality, decreased weight gain, and the high cost of treatment [1, 10]. In swine, enterotoxigenic E. coli (ETEC) causes diarrhea by producing one or more enterotoxins, including heat-labile enterotoxin (LT), heat-stable enterotoxins (STa and STb), and enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1), as virulence factors (VFs) [11]. Shiga toxin (Stx)-producing E. coli (STEC) causes edema disease by producing a subtype of Stx (Stx2e) as the main VF, and some STEC strains produce enterotoxins in addition to Stx2e [2, 26]. Swine ETEC and STEC typically have specific types of fimbriae, including F4 (K88), F5 (K99), F6 (987P), F18, and F41, that mediate bacterial colonization of the epithelial surface of the swine intestine [2, 11]. Serogroups O8, O138, O139, O141, O147, O149, and O157 are most frequently reported in swine pathogenic E. coli worldwide [11, 13]. In Japan, we previously reported that recent isolates of swine ETEC and STEC contained a greater diversity of O serogroups than isolates from the 1990s (O139 and O149 were nearly the sole serogroups), and O116 and OSB9 have emerged in the last 10 to 15 years as the third and fourth major O serogroups, respectively [26]. Although the four major serogroups, O139, O149, O116, and OSB9, represented the majority of the strains in most prefectures investigated, the proportions of each O serogroup varied remarkably among prefectures [26].
Antimicrobial resistance of pathogenic E. coli also impacts the swine industry because of limited treatment options and growing public health concerns due to the potential transfer of antimicrobial resistance genes into the food chain [10]. In Japan, the use of antimicrobial drugs in farm animals, which has been calculated from the sales volume of veterinary medical products, is greater than that of antimicrobial feed additives [17], and antimicrobial drug use is highest in swine, followed by poultry and cattle [22]. According to the national surveillance of antimicrobials and resistant bacteria isolated from apparently healthy farm animals in Japan, the prevalence of resistant E. coli was closely associated with the use of antimicrobials through not only direct selection of the related resistance but also indirect selection via cross-resistance and coresistance [17, 19, 28]. Therefore, swine are considered important farm animals involved in the emergence and dissemination of antimicrobial-resistant bacteria, but there is little information about associations between antimicrobial use and resistance in pathogenic bacteria isolated from diseased swine.
In the present study, we investigated the O serogroups, VF gene profiles, and antimicrobial resistance of E. coli strains isolated from swine that died of disease in Kagoshima Prefecture, Japan, between April 1999 and March 2017. Kagoshima is known as the prefecture with the greatest numbers of both swine and swine farms in Japan; in 2018, there were 1.3 million swine in this prefecture, accounting for 14% of the total swine in the country (47 prefectures), as reported by the Ministry of Agriculture, Forestry and Fisheries [27]. We also investigated the history of antimicrobial drug use on farms and analyzed the associations between the antimicrobials used for the treatment of diseased swine and the resistance of E. coli isolated from the same swine.
MATERIALS AND METHODS
Bacterial strains and culture conditions
We investigated 360 E. coli strains isolated from swine that were aged from one day to two years old and died of diarrhea, edema disease, or septicemia on 137 farms in Kagoshima Prefecture, Japan, between April 1999 and March 2017 (detailed information on serogroup, year, anonymized farm ID, disease, and antimicrobial use for all the isolates is provided in Supplementary Table 1 in the supplemental material). All of these strains were not included in our previous study [26]. The bacterial strains were isolated from the organs (e.g., the heart, lung, liver, kidney, spleen, mesenteric lymph nodes, spinal cord, or brain) or intestinal contents of the diseased swine using 5% sheep blood agar and DHL agar (Becton, Dickinson and Co., Sparks, MD, USA) and were identified as E. coli through biochemical tests using the API20E system (Sysmex-bioMerieux, Tokyo, Japan). The O serogroups of the strains were determined through agglutination tests using antisera obtained from the Statens Serum Institute (Copenhagen, Denmark) according to the manufacturer’s instructions. All strains were grown in Mueller-Hinton broth (Becton, Dickinson and Co.) for 18 hr at 37°C.
PCR-based VF gene profiling of E. coli strains
The presence of genes encoding various VFs found in diarrheagenic E. coli (DEC), namely, LT, STa, STb, EAST1, Stx1, Stx2, Stx2e, F4, F5, F6, F18, F41, and intimin, was examined through PCR using the primers described by Vu-Khac et al. [38]. Template DNA for PCR was prepared using a previously described alkaline-boiling method [35]. PCR was performed in a 50-µl reaction mixture containing template DNA, 0.2 µM each primer, 0.2 mM deoxynucleoside triphosphates (dNTPs), PCR buffer, and 1.25 U of Ex Taq DNA polymerase (TaKaRa Bio Inc., Kusatsu, Japan) using 30 amplification cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min.
Antimicrobial susceptibility and extended-spectrum β-lactamase (ESBL) production tests
The Kirby-Bauer disk diffusion test was performed using Sensi-Disc Susceptibility Disks (Becton, Dickinson and Co.) according to the recommendation of the Clinical and Laboratory Standards Institute (CLSI) [6]. First, 0.5 McFarland suspensions of the test strains were prepared using the Prompt System (Becton, Dickenson and Co.). The following antimicrobials were tested: ampicillin (AMP), piperacillin (PIP), cefazolin (CFZ), cefuroxime (CXM), cefotaxime (CTX), cefepime (FEP), cefoxitin (FOX), moxalactam (MOX), aztreonam (ATM), imipenem (IPM), meropenem (MEM), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), tetracycline (TET), chloramphenicol (CHL), nalidixic acid (NAL), ciprofloxacin (CIP), levofloxacin (LVX), gatifloxacin (GAT) and trimethoprim-sulfamethoxazole (SXT). The E. coli strains that showed resistance to third- and/or fourth-generation cephalosporins (i.e., CTX and FEP in this study) were further examined for ESBL production using the double-disk synergy test (DDST) described by Jarlier et al. [23].
Analysis of β-lactamase genes
DDST-positive E. coli strains were investigated for the presence of the blaTEM, blaSHV, and blaCTX-M ESBL genes by PCR, as described by Kojima et al. [25] and Mena et al. [30]. Amplicons were sequenced for subtyping the ESBL genes using a BigDye Terminator Cycle Sequencing Kit and a 3130xl sequencer (Applied Biosystems, Inc., Foster City, CA, USA) according to the manufacturer’s instructions. For the ESBL-negative CTX-resistant strains, the presence of several β-lactamase genes, including blaTEM, blaSHV, blaCMY-1, blaCMY-2, and blaFOX, was investigated by PCR as described by Kojima et al. [25].
Collection of general information about swine
We collected the following information about swine that died due to pathogenic E. coli infection: age, sex, and breed of the swine; histories of feeding, vaccination, and antimicrobial drug use; course of the symptoms and the treatment record; autopsy findings; and the rearing scale, facilities, and hygienic conditions of the farm. Although the collected information included a mixture of the history of antimicrobial drug use for each swine and for each herd of swine containing a diseased individual, the cases showing the use of antimicrobials at least for diseased individuals were used for the analysis of associations between the use of antimicrobials and resistance of E. coli isolates.
Analysis of the associations between the use of antimicrobials in swine and the isolation of resistant E. coli from swine
We classified the antimicrobials used for swine on farms in Kagoshima Prefecture into seven groups, i.e., penicillins, ceftiofur, aminoglycosides, tetracyclines, phenicols, fluoroquinolones, and sulfamethoxazole-trimethoprim, because many different antimicrobials are used on farms, even antimicrobials in the same drug class. Ceftiofur (a third-generation cephalosporin) was the only cephalosporin antimicrobial used in this study. For the 218 strains, which were isolated from 218 swine with a clear history of antimicrobial drug use, we calculated the statistical significance of the associations between the antimicrobial use of the seven groups and the resistance of E. coli isolates to 16 antimicrobials belonging to the drug classes approved for the treatment of animals in Japan, i.e., penicillins (AMP and PIP), cephalosporins (CFZ, CXM, CTX, and FEP), aminoglycosides (GEN, KAN, and STR), tetracyclines (TET), phenicols (CHL), quinolones (NAL), fluoroquinolones (CIP, LVX, and GAT), and sulfamethoxazole-trimethoprim (SXT). We organized the antimicrobial resistance patterns of the 218 isolates according to the antimicrobials used. The association between antimicrobial use and resistance was tested by univariate analysis using the odds ratio with a 95% confidence interval, which was calculated by R software. For the calculation of P-values, Fisher’s exact test was performed when at least one of the four cells had an expected frequency of less than five in the χ2 test.
RESULTS
O serogroups and VF gene profiles of E. coli isolates
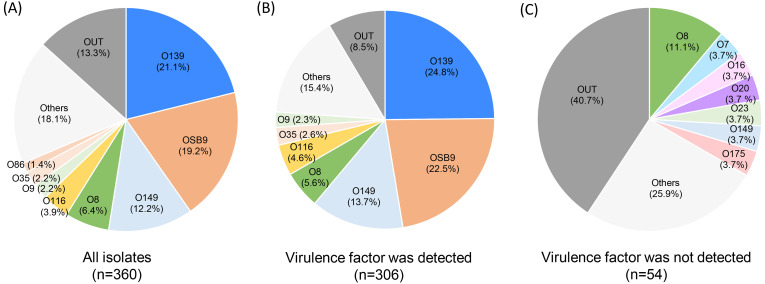
Among the 360 E. coli strains analyzed in the present study, 312 (86.7%) were classified into 45 O serogroups, and 48 (13.3%) were untypable (OUT). O139 (21.1%), OSB9 (19.2%), O149 (12.2%), O8 (6.4%), and O116 (3.9%) were predominant among the 45 serogroups, and these five serogroups represented 62.8% of the 360 strains examined (Fig. 1A). Almost all strains of the five major O serogroups were isolated from swine with diarrhea or edema disease and possessed the VF genes typically found in DEC (Fig. 1B and Table 1). In contrast, 54 strains possessing no VF gene were isolated mainly from swine with septicemia and consisted mainly of minor O serogroups (Fig. 1C).
Fig. 1.
Prevalence of O serogroups in 360 strains of all Escherichia coli isolates examined (A) and in 306 or 54 strains in which typical virulence factor genes of diarrheagenic E. coli were detected (B) or not detected (C), respectively. The proportions of each O serogroup are indicated in different colors.
Table 1. Results of O-serogrouping and virulence factor-profiling.
| O serogroup |
Virulence factor profile |
||||
|---|---|---|---|---|---|
| O serogroup | No. of isolates (%) | Toxin | No. of isolates (%) | Fimbriae | No. of isolates (%) |
| O139 | 76 (21.1) | Stx2e | 76 (100) | F18 | 74 (97.4) |
| STa | 6 (7.9) | ||||
| STb | 8 (10.5) | ||||
| OSB9 | 69 (19.2) | Stx2e | 69 (100) | F18 | 69 (100) |
| LT | 69 (100) | ||||
| STa | 69 (100) | ||||
| STb | 58 (84.1) | ||||
| EAST1 | 33 (47.8) | ||||
| O149 | 44 (12.2) | LT | 41 (93.2) | F4 | 39 (88.6) |
| STa | 33 (75.0) | F18 | 1 (2.3) | ||
| STb | 40 (90.9) | ||||
| EAST1 | 41 (93.2) | ||||
| Stx2e | 1 (2.3) | ||||
| O8 | 23 (6.4) | LT | 8 (34.8) | F4 | 2 (8.7) |
| STa | 6 (26.1) | F5 | 1 (4.4) | ||
| STb | 13 (56.5) | F18 | 8 (34.8) | ||
| EAST1 | 14 (60.9) | ||||
| Stx2e | 5 (21.7) | ||||
| O116 | 14 (3.9) | LT | 13 (92.9) | F18 | 14 (100) |
| STa | 14 (100) | ||||
| STb | 14 (100) | ||||
| EAST1 | 2 (14.3) | ||||
| Stx2e | 14 (85.7) | ||||
Stx2e, Shiga toxin 2e; STa, heat-stable enterotoxin a; STb, heat-stable enterotoxin b; LT, heat-labile enterotoxin; EAST1, enteroaggregative Escherichia coli heat-stable enterotoxin 1.
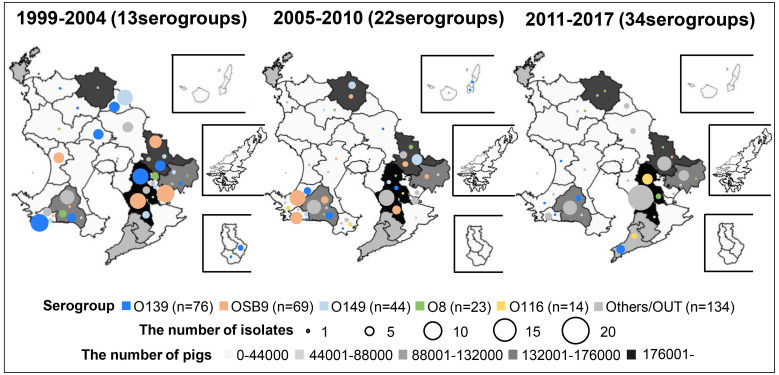
Figure 2 shows the changes in the temporal and geographical distributions of O serogroups of swine-pathogenic E. coli isolates between 1999 and 2017. As the numbers of healthy swine on farms in each area are also shown on the map of Kagoshima Prefecture, it can be seen that many swine farms are concentrated in the southeastern area. Overall, the diversity of isolated serogroups has been increasing recently: 13, 22, and 34 serogroups were isolated in 1999−2004, 2005−2010, and 2011−2017, respectively. Strains of the predominant serogroups O139, OSB9, O149, O8, and O116 first appeared in 1999, 2000, 2000, 1999, and 2008, respectively. The proportions of the five major O serogroups have decreased over time, i.e., the proportions of these serogroups isolated in 1999−2004, 2005−2010, and 2011−2016 were 80.3%, 67.0%, and 31.6%, respectively. Instead, strains of minor O serogroups and OUT strains have been isolated increasingly frequently in the last decade, especially in the southeastern area of the prefecture (Fig. 2). Notably, in the same area, isolation of O116 strains has also increased in recent years, although their prevalence is still low (4.3% and 9.3% in 2005−2010 and 2011−2016, respectively).
Fig. 2.
Temporal changes in O serogroups in Escherichia coli isolated from diseased swine in each area of Kagoshima Prefecture, Japan. The prevalence of O serogroups between 1999 and 2004, 2005 and 2010, and 2011 and 2017 was compared, and the number of O serogroups detected in each period is shown in parentheses. The size of the circle in each area represents the number of isolates. The O serogroups O139, OSB9, O149, O8, and O116 are indicated in blue, pink, light blue, green, and yellow, respectively, and the other O serogroups (other than O139, OSB9, O149, O8, and O116) and OUT isolates are indicated in gray. The number of healthy swine in each area is represented on the map with color gradation from light gray to black.
Antimicrobial susceptibility
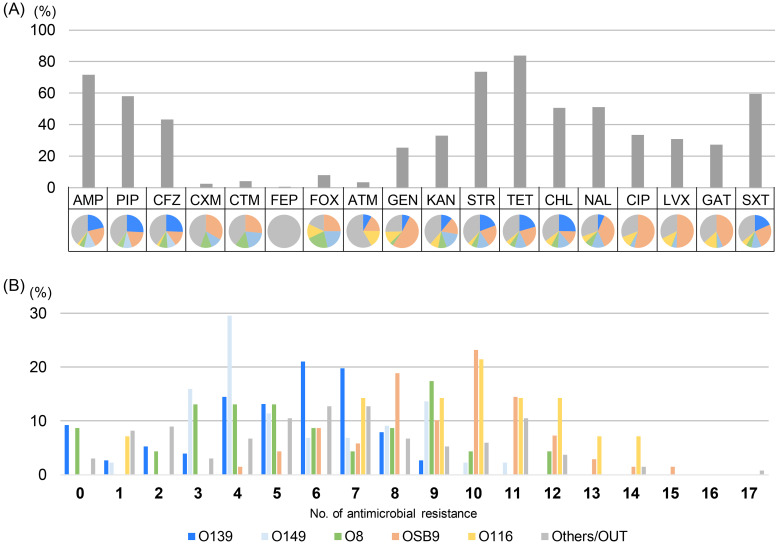
All the strains were susceptible to MOX, IPM, and MEM, and 354 of the 360 strains (98.3%) were resistant to one or more of the other 18 antimicrobials (Fig. 3). Many of the strains were resistant to AMP, STR, and TET (the proportions of resistant strains were 71.7%, 73.6%, and 83.9%, respectively). There were similar proportions of the five major O serogroups among the strains that were resistant to AMP, PIP, CFZ, KAN, STR, TET, CHL, and SXT (the proportions of resistant strains were 60.9%, 60.3%, 59.6%, 62.2%, 62.6%, 64.6%, 65.4%, and 64.5%, respectively). In contrast, OSB9 accounted for half of the strains that were resistant to GEN and fluoroquinolones (the proportions of strains resistant to CIP, LVX, and GAT were 53.3%, 50.5%, and 43.9%, respectively) (Fig. 3A). As shown in Fig. 3B, the OSB9 and O116 strains were resistant to a wider range of antimicrobials (with medians of 10 in both serogroups) than O139, O149, and O8 (with medians of 6, 5, and 5, respectively). Notably, 15 of the 360 strains (4.2%) showed resistance to a third-generation cephalosporin (CTX) and two of these 15 strains were also resistant to a fourth-generation cephalosporin (FEP).
Fig. 3.
(A) Comparison of the prevalence of O serogroups and the rate of resistance to each antimicrobial. O serogroups are indicated by the following colors: blue, O139; pink, OSB9; light blue, O149; green, O8; yellow, O116; and gray, other O serogroups and OUT isolates. AMP, ampicillin; PIP, piperacillin; CFZ, cefazolin; CXM, cefuroxime; CTX, cefotaxime; FEP, cefepime; FOX, cefoxitin; ATM, aztreonam; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; TET, tetracycline; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; GAT, gatifloxacin; SXT, trimethoprim-sulfamethoxazole. (B) Comparisons of multidrug resistance levels among O serogroups. The horizontal axis shows the number of antimicrobials to which strains in the O serogroup are resistant.
As a result of DDSTs and subsequent ESBL genotyping, we found that five of the 15 CTX and/or FEP-resistant strains produced various types of ESBLs encoded by blaCTX-M-14, blaCTX-M-15, blaCTX-M-24, blaCTX-M-61, or blaSHV-12 (Table 2). The two FEP-resistant strains were both OUT strains isolated from swine with diarrhea and possessed blaCTX-M-14 or blaCTX-M-15 in addition to blaTEM-1. Among the 10 strains of ESBL-negative and CTX-resistant E. coli, six strains possessed an AmpC β-lactamase gene, blaCMY-2. The other four strains were E. coli OSB9, but the CTX resistance gene remained unclear.
Table 2. Detail of double-disk synergy test-positive isolates.
| Strain name | Year isolated | O serogroup | Virulence factors | β-lactamase | Resistance | Use of antimicrobial |
|---|---|---|---|---|---|---|
| E2499 | 2005 | UT | STa, STb, F4 | TEM-1, CTX-M-15 |
AMP, PIP, CFZ, CXM, CTX, FEP, ATM, GEN, KAN, STR, TET, CHL, NAL, CIP, LVX, GAT, SXT | Unknown |
| E2525 | 2006 | UT | EAST1 | TEM-1, CTX-M-14 |
AMP, PIP, CFZ, CXM, CTX, FEP, GEN, STR, TET, NAL, CIP, LVX, GAT, SXT | Ampicillin, Lincomycin |
| E2619 | 2010 | 34 | - | CTX-M-24 | AMP, PIP, CFZ, CXM, CTX, ATM, STR, TET, CHL, NAL, CIP, LVX, GAT, SXT | Not used |
| E2642 | 2013 | 23 | - | TEM-1, SHV-12 |
AMP, PIP, CFZ, CTX, ATM, STR, TET | Unknown |
| E2646 | 2013 | 8 | EAST1 | CTX-M-61 | AMP, PIP, CFZ, CXM, CTX, CHL, SXT | Ceftiofur, Fluoroquinolone |
AMP, ampicillin; PIP, piperacillin; CFZ, cefazolin; CXM, cefuroxime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; TET, tetracycline; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; GAT, gatifloxacin; SXT, trimethoprim-sulfamethoxazole.
Associations between the use of antimicrobials and isolation of resistant E. coli
Of the 360 E. coli strains analyzed in the present study, 218 strains were isolated from 218 diseased swine for which the history of antimicrobial use was available: 146 were treated with one or more antimicrobials, while 72 died without antimicrobial treatment. For the remaining 142 swine, there was a lack of definite data on the individual treatment. Table 3 shows the history of antimicrobial use for the swine and the resistance patterns of the 218 strains. Multidrug-resistant E. coli strains were highly prevalent, even with use of individual or no antimicrobial drugs. Among the 72 strains isolated from swine without antimicrobial treatment, 70 strains (97.2%) showed resistance to at least one antimicrobial, and the majority of the strains were resistant to AMP, PIP, STR, TET, CHL, NAL, and SXT (the resistance rates were 65.3%, 56.9%, 63.9%, 87.5%, 58.3%, 61.1%, and 65.3%, respectively).
Table 3. Association between the use of antimicrobials and the resistance pattern of the isolates.
| Antimicrobials used | Antimicrobial resistance pattern of the isolatesa) | No. of isolates |
|---|---|---|
| Penicillins | AMP-PIP-CFZ-FOX-GEN-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-CFZ-CXM-CTX-FEP-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-CFZ-FOX-GEN-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-CXM-CTX-FOX-GEN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-GEN-KAN-STR-TET-CHL-NAL-CIP-LVX-SXT | 1 | |
| AMP-CFZ-FOX-KAN-STR-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 4 | |
| AMP-PIP-CFZ-FOX-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-GEN-TET-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-PIP-GEN-KAN-STR-TET-CHL-NAL-CIP-SXT | 1 | |
| AMP-PIP-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-SXT | 3 | |
| GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-GEN-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-GEN-KAN-STR-TET-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL | 1 | |
| AMP-PIP-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-KAN-STR-TET-CHL-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET-SXT | 1 | |
| AMP-PIP-GEN-STR-TET-NAL | 1 | |
| AMP-PIP-CFZ-STR-TET | 1 | |
| AMP-PIP-CFZ-TET-CHL | 1 | |
| AMP-PIP-KAN-STR-TET | 1 | |
| KAN-TET-CHL-NAL-SXT | 1 | |
| CFZ-FOX-CHL-NAL | 2 | |
| KAN-TET-CHL-SXT | 1 | |
| TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-TET | 1 | |
| KAN-TET-SXT | 2 | |
| STR-TET | 2 | |
| -b) | 2 | |
| Ceftiofur | AMP-PIP-CFZ-CTX-FOX-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-CFZ-CTX-FOX-GEN-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-CTX-FOX-STR-TET-CHL | 1 | |
| AMP-PIP-CFZ-FOX-STR-TET-CHL | 3 | |
| Aminoglycoside | AMP-PIP-KAN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-KAN-STR-TET-CHL-SXT | 1 | |
| AMP-PIP-GEN-KAN-STR-TET-NAL | 1 | |
| AMP-PIP-KAN-STR-TET-CHL | 1 | |
| AMP-PIP-STR-TET-SXT | 1 | |
| -b) | 1 | |
| Tetracyclines | AMP-PIP-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-CFZ-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| GEN-STR-TET-NAL-CIP-LVX-GAT | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL-SXT | 3 | |
| AMP-PIP-CFZ-STR-TET-CHL | 1 | |
| AMP-PIP-STR-TET-CHL-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET | 1 | |
| TET | 1 | |
| -b) | 1 | |
| Phenicols | AMP-PIP-CFZ-GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-CFZ-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL-SXT | 3 | |
| KAN-STR-TET-CHL-NAL-SXT | 1 | |
| TET-CHL-NAL | 1 | |
| STR-TET | 1 | |
| Fluoroquinolones | AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-GEN-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-PIP-CFZ-GEN-STR-TET-NAL-CIP-LVX-SXT | 1 | |
| AMP-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| GEN-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-KAN-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| CFZ-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-FOX-STR-TET-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL | 2 | |
| AMP-CFZ-FOX-GEN-STR-TET-NAL | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL | 2 | |
| AMP-PIP-STR-NAL-SXT | 1 | |
| AMP-PIP-STR-TET-NAL | 1 | |
| AMP-PIP-CFZ-NAL | 1 | |
| STR-TET | 2 | |
| CHL | 1 | |
| TET | 1 | |
| Sulfamethoxazole-trimethoprim | AMP-CXM-GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-GEN-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| Penicillins + Aminoglycoside | AMP-PIP-CFZ-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 2 |
| AMP-PIP-GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-GEN-KAN-STR-TET-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-GEN-STR-TET-CHL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-FOX-KAN-STR-TET-CHL-NAL | 2 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-SXT | 1 | |
| AMP-PIP-KAN-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-GEN-KAN-STR-TET-CIP-LVX-GAT | 1 | |
| AMP-PIP-CFZ-STR-TET | 1 | |
| CFZ-KAN-STR-TET | 1 | |
| KAN-TET-CHL-SXT | 1 | |
| STR-TET | 1 | |
| Penicillins + Tetracyclines | AMP-PIP-CFZ-STR-TET-CHL-SXT | 1 |
| Penicillins + Phenicols | AMP-PIP-CFZ-GEN-KAN-TET-CHL-NAL | 1 |
| AMP-PIP-CFZ-STR-TET-CHL-SXT | 3 | |
| AMP-KAN-STR-CHL-SXT | 1 | |
| Penicillins + Fluoroquinolones | AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| Penicillins + Sulfamethoxazole-trimethoprim | NAL-SXT | 1 |
| Ceftiofur + Fluoroquinolones | AMP-CFZ-FOX-ATM-KAN-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-CFZ-FOX-KAN-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-CXM-CTX-CHL-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-NAL | 1 | |
| Fluoroquinolones + Sulfamethoxazole-trimethoprim | AMP-PIP-CFZ-GEN-KAN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-GEN-STR-TET-NAL-CIP-SXT | 2 | |
| AMP-PIP-CFZ-GEN-KAN-STR-TET | 1 | |
| AMP-PIP-CFZ-GEN-KAN-STR | 1 | |
| AMP-PIP-STR-TET-NAL | 1 | |
| Penicillins + Ceftiofur + Aminoglycoside | AMP-PIP-KAN-STR-TET-CHL-SXT | 1 |
| Penicillins + Aminoglycoside + Tetracyclines | AMP-PIP-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-CFZ-STR-TET-NAL-CIP-LVX-GAT | 2 | |
| TET | 1 | |
| Penicillins + Aminoglycoside + Fluoroquinolones | AMP-CFZ-FOX-GEN-STR-TET-NAL-CIP | 1 |
| Penicillins + Aminoglycoside + Sulfamethoxazole-trimethoprim | AMP-GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| GEN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| CFZ-GEN-STR-TET-NAL-CIP | 1 | |
| Penicillins + Tetracyclines + Phenicols + Fluoroquinolones | AMP-PIP-CFZ-STR-TET-NAL | 1 |
| Unused | AMP-PIP-CFZ-CTX-FOX-ATM-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 |
| AMP-PIP-CFZ-CXM-CTX-ATM-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-CFZ-CXM-CTX-FOX-ATM-GEN-KAN-STR-TET-NAL-CIP-LVX | 1 | |
| AMP-CFZ-CXM-CTX-FOX-GEN-STR-TET-NAL-CIP-LVX-GAT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-CXM-CTX-FOX-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-KAN-STR-TET-CHL-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-CFZ-FOX-KAN-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-GEN-STR-TET-NAL-CIP-LVX-GAT-SXT | 2 | |
| AMP-CFZ-CTX-FOX-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-ATM-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-CFZ-GEN-KAN-STR-TET-CHL-SXT | 1 | |
| AMP-PIP-CFZ-KAN-STR-TET-CHL-NAL-SXT | 2 | |
| AMP-PIP-GEN-STR-TET-NAL-CIP-LVX-SXT | 1 | |
| AMP-GEN-STR-TET-NAL-CIP-LVX-SXT | 1 | |
| AMP-KAN-TET-CHL-NAL-CIP-LVX-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL-NAL-SXT | 1 | |
| AMP-PIP-GEN-KAN-STR-TET-CHL-SXT | 2 | |
| AMP-PIP-TET-NAL-CIP-LVX-GAT-SXT | 3 | |
| AMP-PIP-CFZ-STR-TET-CHL-SXT | 2 | |
| AMP-PIP-GEN-STR-TET-NAL-CIP | 1 | |
| AMP-PIP-STR-TET-CHL-NAL-SXT | 4 | |
| GEN-STR-TET-NAL-CIP-LVX-GAT | 1 | |
| STR-CHL-NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CFZ-STR-TET-CHL | 1 | |
| AMP-PIP-CFZ-TET-CHL-SXT | 1 | |
| AMP-PIP-KAN-STR-TET-CHL | 1 | |
| AMP-PIP-STR-TET-CHL-SXT | 3 | |
| GEN-STR-TET-NAL-CIP-LVX | 4 | |
| AMP-PIP-CFZ-TET-CHL | 3 | |
| AMP-PIP-CFZ-TET-SXT | 1 | |
| AMP-PIP-KAN-STR-TET | 1 | |
| AMP-PIP-STR-TET-SXT | 1 | |
| GEN-STR-TET-NAL-CIP | 1 | |
| NAL-CIP-LVX-GAT-SXT | 1 | |
| AMP-PIP-CHL-SXT | 1 | |
| AMP-PIP-TET-CHL | 1 | |
| GEN-STR-TET-NAL | 1 | |
| KAN-STR-CHL-SXT | 1 | |
| KAN-STR-TET-SXT | 1 | |
| KAN-TET-CHL-SXT | 1 | |
| TET-CHL-NAL-SXT | 5 | |
| STR-TET | 1 | |
| NAL | 2 | |
| TET | 3 | |
| -b) | 2 | |
a) AMP, ampicillin; PIP, piperacillin; CFZ, cefazolin; CXM, cefuroxime; CTX, cefotaxime; FEP, cefepime; FOX, cefoxitin; ATM, aztreonam; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; TET, tetracycline; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; GAT, gatifloxacin; SXT, trimethoprim-sulfamethoxazole. b) Susceptible to all antimicrobials tested.
Table 4 shows the results of univariate analysis, in which the P-values were less than 0.2, for associations between the seven groups of antimicrobials used for swine and the resistance of E. coli isolates to 16 antimicrobials belonging to the drug classes approved for the treatment of animals in Japan (see Supplementary Table 2 in the supplemental material for the full list of results). Significant associations between antimicrobial use and resistance to the same class of antimicrobials were observed: the use of ceftiofur and resistance to CTX (P<0.01), CFZ (P<0.01), or AMP (P<0.05); the use of aminoglycosides and resistance to STR (P<0.05); and the use of phenicols and resistance to CHL (P<0.05). Associations between antimicrobial use and structurally unrelated antimicrobial resistance were also observed: the use of penicillins and resistance to GEN (P<0.05); the use of ceftiofur and resistance to CHL (P<0.05); and the use of sulfamethoxazole-trimethoprim and resistance to GEN (P<0.001) or NAL (P<0.05). Moreover, a negative association between the use of tetracyclines and resistance to KAN (odds ratio <0.23, P<0.05) was observed.
Table 4. Association between the use of antimicrobials and antimicrobial resistance (P-values <0.2).
| Antimicrobials used | Resistance outcomea) | Exposed | Not exposed | Odds ratio | P-value | ||
|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | Resistant | Susceptible | ||||
| Penicillins | CFZ | 40 | 35 | 59 | 84 | 1.63 (0.93–2.86) | 0.0890 |
| GEN* | 31 | 44 | 37 | 106 | 2.02 (1.12–3.65) | 0.0193 | |
| GAT | 27 | 48 | 39 | 104 | 1.50 (0.82–2.73) | 0.1828 | |
| Ceftiofur | AMP* | 11 | 0 | 149 | 58 | 9.0 (0.52–155.21) | 0.0394b) |
| CFZ** | 10 | 1 | 89 | 118 | 13.26 (1.67–105.49) | 0.0030b) | |
| CTX** | 4 | 7 | 8 | 199 | 14.21 (3.45–58.65) | 0.0014b) | |
| GEN | 1 | 10 | 67 | 140 | 0.21 (0.03–1.67) | 0.1791b) | |
| KAN | 6 | 5 | 69 | 138 | 2.40 (0.71–8.14) | 0.1936b) | |
| TET | 8 | 3 | 185 | 22 | 0.32 (0.08–1.28) | 0.1183b) | |
| CHL* | 10 | 1 | 113 | 94 | 8.32 (1.05–66.17) | 0.0253b) | |
| Aminoglycosides | GEN | 14 | 17 | 54 | 133 | 2.03 (0.93–4.40) | 0.0699 |
| KAN | 14 | 17 | 61 | 126 | 1.70 (0.79–3.68) | 0.1734 | |
| STR* | 28 | 3 | 135 | 52 | 3.60 (1.05–12.34) | 0.0314 | |
| CHL | 13 | 18 | 110 | 77 | 0.51 (0.23–1.09) | 0.0790 | |
| CIP | 16 | 15 | 67 | 120 | 1.91 (0.89–4.11) | 0.0937 | |
| LVX | 14 | 17 | 62 | 125 | 1.66 (0.77–3.59) | 0.1939 | |
| GAT | 14 | 17 | 52 | 135 | 2.14 (0.98–4.65) | 0.0514 | |
| Tetracyclines | GEN | 2 | 15 | 66 | 135 | 0.27 (0.06–1.23) | 0.0718 |
| KAN* | 2 | 15 | 73 | 128 | 0.23 (0.05–1.05) | 0.0407 | |
| NAL | 7 | 10 | 123 | 78 | 0.44 (0.16–1.21) | 0.1063 | |
| SXT | 8 | 9 | 131 | 70 | 0.47 (0.18–1.29) | 0.1357 | |
| Phenicols | CFZ* | 10 | 4 | 89 | 115 | 3.23 (0.98–10.64) | 0.0433 |
| CHL* | 12 | 2 | 111 | 93 | 5.03 (1.10–23.03) | 0.0223 | |
| NAL | 6 | 8 | 124 | 80 | 0.48 (0.16–1.45) | 0.1860 | |
| CIP | 2 | 12 | 81 | 123 | 0.25 (0.06–1.16) | 0.0581 | |
| LVX | 2 | 12 | 74 | 130 | 0.29 (0.06–1.34) | 0.1457b) | |
| Fluoroquinolones | AMP | 31 | 6 | 129 | 52 | 2.08 (0.82–5.29) | 0.1166 |
| KAN | 17 | 20 | 58 | 123 | 1.8 (0.88–3.70) | 0.1048 | |
| TET | 30 | 7 | 163 | 18 | 0.47 (0.18–1.23) | 0.1528b) | |
| CHL | 16 | 21 | 107 | 74 | 0.53 (0.26–1.08) | 0.0760 | |
| CIP | 18 | 19 | 65 | 116 | 1.69 (0.83–3.45) | 0.1460 | |
| SXT | 20 | 17 | 119 | 62 | 0.61 (0.30–1.25) | 0.1777 | |
| Sulfamethoxazole-trimethoprim | GEN*** | 12 | 3 | 56 | 147 | 10.5 (2.86–38.61) | 0.0001b) |
| STR | 14 | 1 | 149 | 54 | 5.07 (0.65–39.51) | 0.1229b) | |
| NAL* | 13 | 2 | 117 | 86 | 4.78 (1.05–21.73) | 0.0270 | |
| CIP | 9 | 6 | 74 | 129 | 2.61 (0.90–7.64) | 0.0699 | |
a) *P<0.05, **P<0.01, ***P<0.001. b) The P-value is based on Fisher’s exact test. AMP, ampicillin; CFZ, cefazolin; CTX, cefotaxime; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; TET, tetracycline; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; GAT, gatifloxacin; SXT, trimethoprim-sulfamethoxazole.
DISCUSSION
Through an analysis of swine-pathogenic E. coli strains isolated over the past 18 years in Kagoshima Prefecture, Japan, we show that O139, OSB9, O149, O8, and O116 are the predominant serogroups in this prefecture (Fig. 1), and the recently isolated strains contain a greater diversity of O serogroups than previously isolated strains (Fig. 2). Most of the STEC O139 or ETEC O149 strains were isolated from swine with edema disease or diarrhea, respectively. These strains had already been disseminated throughout the prefecture before 1999 and produced diseases in swine over a long period. Almost all strains of OSB9 and O116 possessed multiple VF genes for both ETEC and STEC (Table 1) and showed resistance to many antimicrobials, including GEN and fluoroquinolones (Fig. 3). These results for DEC are consistent with our previous study on ETEC and STEC strains isolated from diseased swine in 23 Japanese prefectures other than Kagoshima [26]. In the present study, we isolated E. coli from not only swine with edema disease or diarrhea but also swine with other diseases (mainly septicemia) and found O8 in both DEC and non-DEC strains (Fig. 1). E. coli O8 has been isolated from both diseased swine and healthy swine [11, 13, 15, 26] and from cattle, poultry, and humans with extraintestinal infections [7, 29] worldwide.
The swine-pathogenic E. coli OSB9 and O116 serogroups are considered to belong to the same or closely related lineages: these strains were classified into a single sequence type (ST88) and a single fingerprinting cluster (Cluster III) by multilocus sequence typing and pulsed-field gel electrophoresis, respectively [26]. In several prefectures in Japan, O116 has been the most predominant serogroup of swine-pathogenic E. coli isolates in the last decade [14, 26]. In Kagoshima Prefecture, OSB9 strains first appeared in 2000 and were isolated until 2013, but they have not been isolated from diseased swine since then. On the other hand, O116 strains first appeared in 2008 and have been continuously isolated from swine with diarrhea until now, specifically in the southeast area of the prefecture in which many swine farms are concentrated (Fig. 2). Therefore, careful monitoring and prevention of the dissemination of swine-pathogenic E. coli O116 on farms is required, although the prevalence in the prefecture is relatively low at this time (9.3% in 2011−2016).
Importantly, approximately half of the cephalosporin-resistant strains were dominated by minor O serogroups (other than O139, OSB9, O149, O8, and O116) or OUT strains, of which some strains showed high multidrug resistance (Fig. 3). As shown in Table 2, most of the ESBL-producing strains found in the present study belonged to the minor O serogroups or OUT, and the two OUT strains were resistant to a fourth-generation cephalosporin (FEP) in addition to a third-generation cephalosporin (CTX). The emergence of ESBL-producing bacteria has become a serious concern because ESBLs confer resistance to clinically important third- and fourth-generation cephalosporins to humans. In recent years, ESBL-producing E. coli has increasingly been isolated from humans, animals, food, and the environment worldwide [3, 4, 8]. The prevalence of ESBL-producing E. coli in farm animals has subsequently been investigated thoroughly, and this prevalence in broilers and retail chicken meat has significantly increased in Japan [20, 21, 24]. Therefore, careful attention should be paid even to minor O serogroups and OUT strains for the control of antimicrobial-resistant E. coli on farms.
The two FEP-resistant OUT strains possessed blaCTX-M-14 or blaCTX-M-15 (Table 2), and both ESBL genes have been found in E. coli isolated from humans and animals worldwide [4, 8, 9, 25]. Although the prevalence of ESBL genes originating from swine and cattle is reported much less frequently than that from poultry in Japan, Norizuki et al. isolated 22 strains of ESBL-producing E. coli isolated from swine and showed that two, two, 12, and six of these strains possessed blaCTX-M-3, blaCTX-M-14, blaCTX-M-15, and blaCTX-M-55, respectively [32]. Moreover, Ohnishi et al. isolated three and seven strains of CTX-resistant E. coli possessing blaCTX-M-14 and blaCTX-M-15 from cattle, respectively, and reported that each of these groups included some FEP-resistant strains [34]. Therefore, certain strains of E. coli possessing blaCTX-M-14 or blaCTX-M-15 are resistant to fourth-generation cephalosporins. In the present study, the FEP-resistant strains possessed blaTEM-1 in addition to blaCTX-M-14 or blaCTX-M-15 (Table 2). In a study of ESBL-producing E. coli isolated from animals in Switzerland, Geser et al. found several isolates possessing the combination blaCTX-M-14/blaTEM-1 or blaCTX-M-15/blaTEM-1 but reported that all of these isolates were susceptible to FEP [16]. Further studies are required to clarify the mechanism responsible for the resistance to fourth-generation cephalosporins in E. coli possessing blaCTX-M-14 or blaCTX-M-15.
In the present study, the associations between the use of antimicrobial drugs and antimicrobial resistance in E. coli isolated from diseased swine are demonstrated, but there are some limitations: a univariate analysis was performed, multiple causes of cross-resistance and coresistance were not considered, and data on the period between antimicrobial drug use and the isolation of E. coli were not fully collected. Improvements in the data collection and analysis system for the investigation of pathogenic bacteria isolated from dead animals are required to clarify these issues.
Significant associations between antimicrobial use and selection of resistance to the same class of antimicrobials were observed between the use of ceftiofur and CTX, CFZ, or AMP resistance, the use of aminoglycosides and STR resistance, and the use of phenicols and CHL resistance (Table 4). Associations between the use of β-lactams, aminoglycosides, or phenicols and the resistance of E. coli isolates to structurally related antimicrobials were also observed in a study on healthy swine [28]. However, in this study, there were few significant associations that suggested direct selection or cross-selection in most other combinations of antimicrobial use and resistance of E. coli isolates. In the case of swine colibacillosis, pathogenic E. coli was already dominant in the intestine when the symptoms appeared and was selected by the antimicrobial drugs used in therapy. As shown in Fig. 3 and Table 3, most of the pathogenic E. coli strains showed high multidrug resistance even though they were isolated from swine without antimicrobial treatment. In terms of the time of treatment and sampling, Cavaco et al. reported that selection of CTX-resistant E. coli occurred from one day to 22 days after treatment with ceftiofur [5]. Although full data on the period between the use of antimicrobials and the isolation of E. coli were not collected in this study, there could be slight selection pressure imposed by antimicrobials in bacterial samples that were collected before treatment or shortly after treatment. For reasons related to multidrug resistance and time of sampling, the resistance of E. coli isolates may be considered to have little association with direct selection or cross-selection by most of the antimicrobial drugs used for swine with symptoms.
We also observed statistically significant associations between antimicrobial use and resistance of pathogenic E. coli isolates to structurally unrelated antimicrobials, such as the use of ceftiofur and CHL resistance, the use of penicillins and GEN resistance, and the use of sulfamethoxazole-trimethoprim and GEN or NAL resistance (Table 4). According to Harada et al. [18], the use of antimicrobials other than CHL contributed considerably to the coselection of CHL-resistant E. coli isolated from diseased cattle and swine. Indeed, recent reports have demonstrated that CHL resistance genes (floR, cmlA, and cat) were found on some plasmids encoding an ESBL or the AmpC β-lactamase in E. coli isolated from humans and animals [12, 31, 33, 37]. In this study, there was no swine treated with both ceftiofur and phenicols. Therefore, the association of ceftiofur usage with the isolation of CHL-resistant E. coli may be a result of coselection. However, it is unlikely that the associations of penicillins and sulfamethoxazole-trimethoprim usage with the isolation of GEN- and NAL-resistant E. coli have been a result of coselection, because associations of these antimicrobial uses and the same classes of resistance cannot be found (Table 4). The detected associations of penicillins and sulfamethoxazole-trimethoprim usage should be due to the multidrug resistance of pathogenic E. coli.
In conclusion, we identified and characterized the five major serogroups of swine-pathogenic E. coli isolated from Kagoshima Prefecture and analyzed associations between the use of antimicrobials for the treatment of diseased swine and the isolation of resistant E. coli. Although there were few significant associations for most combinations of antimicrobial use and resistance of E. coli isolates because of high multidrug resistance, we observed a clear association (P<0.01) between the use of ceftiofur and resistance to a third-generation cephalosporin (CTX) and other β-lactams. In this prefecture, the prevalence of CTX-resistant isolates is low (5.5%) at present. However, heavy use of antimicrobials is known to be an important risk factor [36]. Therefore, careful monitoring of the use of third-generation cephalosporins and the prevalence of resistant E. coli are required.
POTENTIAL CONFLICTS OF INTEREST
The authors have nothing to disclose.
Supplementary
Acknowledgments
This study was conducted under the research project on “Regulatory Research Projects for Food Safety, Animal Health and Plant Protection (JPJ008617.17935699)” funded by the Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.Abubakar R. H., Madoroba E., Adenubi O., Morar-Leather D., Fasina F. O.2017. Bacterial pathogens of pigs with particular reference to Escherichia coli: a systematic review and meta-analysis. J. Vet. Med. Anim. Health 9: 159–185. [Google Scholar]
- 2.Blanco M., Blanco J. E., Gonzalez E. A., Mora A., Jansen W., Gomes T. A., Zerbini L. F., Yano T., de Castro A. F., Blanco J.1997. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli strains belonging to different O:K:H serotypes: relationship with toxic phenotypes. J. Clin. Microbiol. 35: 2958–2963. doi: 10.1128/JCM.35.11.2958-2963.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K., Bradford P. A.2020. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 33: e00047–e19. doi: 10.1128/CMR.00047-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantón R., Novais A., Valverde A., Machado E., Peixe L., Baquero F., Coque T. M.2008. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14 Suppl 1: 144–153. doi: 10.1111/j.1469-0691.2007.01850.x [DOI] [PubMed] [Google Scholar]
- 5.Cavaco L. M., Abatih E., Aarestrup F. M., Guardabassi L.2008. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrob. Agents Chemother. 52: 3612–3616. doi: 10.1128/AAC.00354-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement, Document M100-S2, Clinical and Laboratory Standards Institute, Wayne.
- 7.Contrepois M., Bertin Y., Girardeau J. P., Picard B., Goullet P.1993. Clonal relationships among bovine pathogenic Escherichia coli producing surface antigen CS31A. FEMS Microbiol. Lett. 106: 217–222. doi: 10.1111/j.1574-6968.1993.tb05962.x [DOI] [PubMed] [Google Scholar]
- 8.Dorado-García A., Smid J. H., van Pelt W., Bonten M. J. M., Fluit A. C., van den Bunt G., Wagenaar J. A., Hordijk J., Dierikx C. M., Veldman K. T., de Koeijer A., Dohmen W., Schmitt H., Liakopoulos A., Pacholewicz E., Lam T. J. G. M., Velthuis A. G., Heuvelink A., Gonggrijp M. A., van Duijkeren E., van Hoek A. H. A. M., de Roda Husman A. M., Blaak H., Havelaar A. H., Mevius D. J., Heederik D. J. J.2018. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother. 73: 339–347. doi: 10.1093/jac/dkx397 [DOI] [PubMed] [Google Scholar]
- 9.Ewers C., Bethe A., Semmler T., Guenther S., Wieler L. H.2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18: 646–655. doi: 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- 10.Fairbrother J. M., Gyles C. L.2012. Colibacillosis, pp. 723–749. In: Diseases of Swine, 10th ed. (Zimmerman, J. J., Karriker, L. A., Ramirez, A., Schwartz, K. J. and Stevenson, G. W. eds.), Wiley-Blackwell, Hoboken. [Google Scholar]
- 11.Fairbrother J. M., Nadeau E., Gyles C. L.2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6: 17–39. doi: 10.1079/AHR2005105 [DOI] [PubMed] [Google Scholar]
- 12.Freitag C., Michael G. B., Kadlec K., Hassel M., Schwarz S.2017. Detection of plasmid-borne extended-spectrum β-lactamase (ESBL) genes in Escherichia coli isolates from bovine mastitis. Vet. Microbiol. 200: 151–156. doi: 10.1016/j.vetmic.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 13.Frydendahl K.2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85: 169–182. doi: 10.1016/S0378-1135(01)00504-1 [DOI] [PubMed] [Google Scholar]
- 14.Fujii Y., Tanabe H., Nishino H., Otani Y., Tsuzuku S., Oouchi Y., Akiba M., Kusumoto M.2017. Comparative analysis of pathogenic Escherichia coli isolated from diseased swine in Ibaraki prefecture, Japan: Analysis of the most prevalent serogroup O116. Nippon Juishikai Zasshi 70: 643–649. [Google Scholar]
- 15.Garabal J. I., González E. A., Vázquez F., Blanco J., Blanco M., Blanco J. E.1996. Serogroups of Escherichia coli isolated from piglets in Spain. Vet. Microbiol. 48: 113–123. doi: 10.1016/0378-1135(95)00150-6 [DOI] [PubMed] [Google Scholar]
- 16.Geser N., Stephan R., Hächler H.2012. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8: 21. doi: 10.1186/1746-6148-8-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada K., Asai T.2010. Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J. Biomed. Biotechnol. 2010: 180682. doi: 10.1155/2010/180682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada K., Asai T., Kojima A., Ishihara K., Takahashi T.2006. Role of coresistance in the development of resistance to chloramphenicol in Escherichia coli isolated from sick cattle and pigs. Am. J. Vet. Res. 67: 230–235. doi: 10.2460/ajvr.67.2.230 [DOI] [PubMed] [Google Scholar]
- 19.Harada K., Asai T., Ozawa M., Kojima A., Takahashi T.2008. Farm-level impact of therapeutic antimicrobial use on antimicrobial-resistant populations of Escherichia coli isolates from pigs. Microb. Drug Resist. 14: 239–244. doi: 10.1089/mdr.2008.0836 [DOI] [PubMed] [Google Scholar]
- 20.Hiroi M., Harada T., Kawamori F., Takahashi N., Kanda T., Sugiyama K., Masuda T., Yoshikawa Y., Ohashi N.2011. A survey of β-lactamase-producing Escherichia coli in farm animals and raw retail meat in Shizuoka Prefecture, Japan. Jpn. J. Infect. Dis. 64: 153–155. [PubMed] [Google Scholar]
- 21.Hiroi M., Yamazaki F., Harada T., Takahashi N., Iida N., Noda Y., Yagi M., Nishio T., Kanda T., Kawamori F., Sugiyama K., Masuda T., Hara-Kudo Y., Ohashi N.2012. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 74: 189–195. doi: 10.1292/jvms.11-0372 [DOI] [PubMed] [Google Scholar]
- 22.Hosoi Y., Asai T., Koike R., Tsuyuki M., Sugiura K.2013. Use of veterinary antimicrobial agents from 2005 to 2010 in Japan. Int. J. Antimicrob. Agents 41: 489–490. doi: 10.1016/j.ijantimicag.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 23.Jarlier V., Nicolas M. H., Fournier G., Philippon A.1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10: 867–878. doi: 10.1093/clinids/10.4.867 [DOI] [PubMed] [Google Scholar]
- 24.Kawamura K., Goto K., Nakane K., Arakawa Y.2014. Molecular epidemiology of extended-spectrum β-lactamases and Escherichia coli isolated from retail foods including chicken meat in Japan. Foodborne Pathog. Dis. 11: 104–110. doi: 10.1089/fpd.2013.1608 [DOI] [PubMed] [Google Scholar]
- 25.Kojima A., Ishii Y., Ishihara K., Esaki H., Asai T., Oda C., Tamura Y., Takahashi T., Yamaguchi K.2005. Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49: 3533–3537. doi: 10.1128/AAC.49.8.3533-3537.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusumoto M., Hikoda Y., Fujii Y., Murata M., Miyoshi H., Ogura Y., Gotoh Y., Iwata T., Hayashi T., Akiba M.2016. Emergence of a multidrug-resistant Shiga toxin-producing enterotoxigenic Escherichia coli lineage in diseased swine in Japan. J. Clin. Microbiol. 54: 1074–1081. doi: 10.1128/JCM.03141-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MAFF. 2018. The 93rd statistical yearbook of ministry of agriculture, forestry and fisheries, https://www.maff.go.jp/e/data/stat/93th/index.html [accessed on March 6, 2020].
- 28.Makita K., Goto M., Ozawa M., Kawanishi M., Koike R., Asai T., Tamura Y.2016. Multivariable analysis of the association between antimicrobial use and antimicrobial resistance in Escherichia coli isolated from apparently healthy pigs in Japan. Microb. Drug Resist. 22: 28–39. doi: 10.1089/mdr.2014.0311 [DOI] [PubMed] [Google Scholar]
- 29.Maluta R. P., Logue C. M., Casas M. R., Meng T., Guastalli E. A., Rojas T. C., Montelli A. C., Sadatsune T., de Carvalho Ramos M., Nolan L. K., da Silveira W. D.2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9: e105016. doi: 10.1371/journal.pone.0105016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mena A., Plasencia V., García L., Hidalgo O., Ayestarán J. I., Alberti S., Borrell N., Pérez J. L., Oliver A.2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44: 2831–2837. doi: 10.1128/JCM.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meunier D., Jouy E., Lazizzera C., Doublet B., Kobisch M., Cloeckaert A., Madec J. Y.2010. Plasmid-borne florfenicol and ceftiofur resistance encoded by the floR and blaCMY-2 genes in Escherichia coli isolates from diseased cattle in France. J. Med. Microbiol. 59: 467–471. doi: 10.1099/jmm.0.016162-0 [DOI] [PubMed] [Google Scholar]
- 32.Norizuki C., Kawamura K., Wachino J. I., Suzuki M., Nagano N., Kondo T., Arakawa Y.2018. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum beta-lactamases from pigs in Aichi prefecture, Japan, between 2015 and 2016. Jpn. J. Infect. Dis. 71: 33–38. doi: 10.7883/yoken.JJID.2017.206 [DOI] [PubMed] [Google Scholar]
- 33.Novais A., Baquero F., Machado E., Cantón R., Peixe L., Coque T. M.2010. International spread and persistence of TEM-24 is caused by the confluence of highly penetrating enterobacteriaceae clones and an IncA/C2 plasmid containing Tn1696:Tn1 and IS5075-Tn21. Antimicrob. Agents Chemother. 54: 825–834. doi: 10.1128/AAC.00959-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnishi M., Okatani A. T., Esaki H., Harada K., Sawada T., Murakami M., Marumo K., Kato Y., Sato R., Shimura K., Hatanaka N., Takahashi T.2013. Herd prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2 β-lactamases among Japanese dairy farms. J. Appl. Microbiol. 115: 282–289. doi: 10.1111/jam.12211 [DOI] [PubMed] [Google Scholar]
- 35.Ojima T., Hirano K., Honda K., Kusumoto M.2016. Development of a multiplex PCR assay for rapid virulence factor profiling of extraintestinal pathogenic Escherichia coli isolated from cattle. J. Microbiol. Methods 128: 31–33. doi: 10.1016/j.mimet.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 36.Paterson D. L., Bonomo R. A.2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18: 657–686. doi: 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehman M. A., Yin X., Lepp D., Laing C., Ziebell K., Talbot G., Topp E., Diarra M. S.2017. Genomic analysis of third generation cephalosporin resistant Escherichia coli from dairy cow manure. Vet. Sci. 4: 57. doi: 10.3390/vetsci4040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu-Khac H., Holoda E., Pilipcinec E., Blanco M., Blanco J. E., Dahbi G., Mora A., López C., González E. A., Blanco J.2007. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhoea in Slovakia. Vet. J. 174: 176–187. doi: 10.1016/j.tvjl.2006.05.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.