Abstract
Breast cancer is the most common cancer in women and is one of the three most common malignancies worldwide. Serum microRNAs (miRNAs/miRs) are ideal biomarkers for tumor diagnosis and prognosis due to their specific biological characteristics. In several different types of cancer, miRNAs are associated with cell migration and invasion. In the present study, miR-25-3p expression levels were detected in tissue and serum samples derived from patients with breast cancer, and the diagnostic and prognostic value of miR-25-3p in breast cancer was evaluated. Cellular function assays were performed to evaluate the role of miR-25-3p in breast cancer. Moreover, dual-luciferase reporter assays and western blotting were performed to investigate the target of miR-25-3p. miR-25-3p expression was upregulated in breast cancer tissue and serum samples compared with normal breast tissue and serum samples. Patients with breast cancer with high serum miR-25-3p levels were more likely to have lymph node metastasis compared with those with low serum miR-25-3p levels. The area under the curve for miR-25-3p in the diagnosis of breast cancer was 0.748, with 57.1% sensitivity and 95.0% specificity. Moreover, the Kaplan-Meier survival curves demonstrated that patients with breast cancer with a low expression of serum miR-25-3p had a higher overall survival rate compared with patients with a high serum miR-25-3p expression. miR-25-3p knockdown suppressed breast cancer cell proliferation and invasion, and transducer of ERBB2, 1 (TOB1) was identified as a potential target gene regulated by miR-25-3p. Therefore, the present study suggested that miR-25-3p regulated cellular functions via TOB1 in breast cancer; therefore, miR-25-3p may serve as a breast cancer biomarker.
Keywords: microRNA, microRNA-25-3p, breast cancer, biomarker
Introduction
Breast cancer is the most common cancer in women and is one of the three most common malignancies worldwide (1). In 2018, an estimated 2,088,849 new cases of breast cancer were diagnosed worldwide, and it was predicted that 626,679 patients would die from the disease in the same year (2). The incidence of breast cancer has increased in the majority of countries over the past few decades (3). Although multidisciplinary synthetic compound therapies have used for the treatment of breast cancer, the morbidity and mortality rates remain high in developing countries and the prognosis of breast cancer remains poor, particularly in patients with triple-negative breast cancer (4,5). Early diagnosis and correct prognostic judgment can aid medical experts to implement individualized treatment strategies for patients, resulting in a significant improvement to their quality of life and disease prognosis (6).
MicroRNAs (miRNAs/miRs) are short non-coding RNAs, ~22-nucleotides in length, that form an indispensable and important aspect of gene regulation (7,8). Circulating miRNAs have received increasing attention in the field of tumor biomarkers over the past two decades (9). Serum miRNAs are ideal biomarkers for tumor diagnosis and prognosis due to their low cost, easy application, high stability and minimal invasiveness (8,10,11). Previous studies have reported the potential of serum miRNAs as biomarkers for early cancer diagnosis and prognosis, including breast cancer (12–14). Moreover, miRNAs are associated with cell migration and invasion in several different types of cancer, including breast cancer (15,16).
Transducer of ERBB2, 1 (TOB1) acts as a tumor suppressor in certain types of tumors. Previous findings have indicated that TOB1 is downregulated in breast, pancreatic, thyroid and stomach cancer (17). Moreover, it has been reported that TOB1 inhibits cancer cell proliferation, migration and invasion, and induces cancer cell apoptosis. In lung and gastric cancer cells, TOB1 knockdown promoted tumor cell migration and invasion (18,19), and in breast cancer cells, TOB1 overexpression induced cell apoptosis (20). miR-25-3p, which is one of the most extensively studied miRNAs, belongs to the miR-25 family and is a member of the 93 and 106b cluster (21). Increasing evidence has demonstrated that miR-25-3p can be used as a prognosis biomarker and that it can regulate cell functions in numerous types of cancer (22), including epithelial ovarian cancer (23), as well as hepatocellular and esophageal squamous cell carcinoma (24,25). One study analyzed tumor tissue miRNA expression and patient survival collecting data from The Cancer Genome Atlas, which used miRNA sequencing and overall survival datasets of 759 breast cancer patients to demonstrate that increased expression of miR-25 predicts improved breast cancer survival (26). A previous study demonstrated that miR-25-3p functions as an oncogene and promotes proliferation via BTG anti-proliferation factor 2 induction in breast cancer (27). Analysis of RNA in serum of patients with breast cancer also showed that miR-25-3p is highly overexpressed (28). However, the diagnostic and prognostic roles of miR-25-3p, as well as the mechanisms underlying miR-25-3p during breast cancer are not completely understood. Therefore, the present study aimed to investigate the diagnostic and prognostic role of miR-25-3p, as well as its molecular mechanism in breast cancer progression.
Materials and methods
Tissue specimens and serum samples
Tumor tissue samples and corresponding non-tumor breast tissue samples (>3 cm from tumor) were obtained from 25 female patients with breast cancer (median age, 51 years; age range, 37–68 years) in Tianjin Medical University Cancer Institute and Hospital from January 2017 to January 2018. Tissue specimens were verified and classified by an experienced pathologist with regard to their origin and type. Tissue specimens were immediately immersed in RNAlater reagent (Qiagen GmbH) for 30 min at room temperature and stored in liquid nitrogen until further analysis.
Blood samples were collected from 88 female patients with breast cancer prior to surgery, the aforementioned 25 female patients with breast cancer on day 1 and at 1 month-post surgery and 40 cancer-free blood donor female volunteers. All the blood samples were collected from Tianjin Medical University Cancer Institute and Hospital from January 2017 to January 2018. The blood samples were processed within 3 h of collection. To obtain serum samples, the blood samples were centrifuged at 1,200 × g for 10 min at 4°C. A second centrifugation was performed at 10,000 × g for 10 min at 4°C to remove residual cellular debris. Subsequently, serum samples were stored in liquid nitrogen until further analysis.
The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Written informed consent was obtained from each participant. The clinical characteristics of the patients with breast cancer are listed in Table I.
Table I.
Association between serum miR-25-3p expression and clinicopathological characteristics of breast cancer.
| Clinical characteristics | n | Serum miR-25-3p expression | P-value |
|---|---|---|---|
| Age (years) | 0.882 | ||
| ≤50 | 45 | 0.397±0.065 | |
| >50 | 43 | 0.401±0.062 | |
| Clinical stage | 0.112 | ||
| I, II | 60 | 0.391±0.066 | |
| III, IV | 28 | 0.417±0.053 | |
| Lymph node metastasis | 0.002 | ||
| Positive | 31 | 0.424±0.053 | |
| Negative | 57 | 0.385±0.064 | |
| ER | 0.563 | ||
| Positive | 36 | 0.404±0.059 | |
| Negative | 52 | 0.395±0.066 | |
| PR | 0.912 | ||
| Positive | 47 | 0.400±0.067 | |
| Negative | 41 | 0.398±0.059 | |
| HER2 | 0.523 | ||
| Positive | 51 | 0.394±0.060 | |
| Negative | 37 | 0.405±0.068 |
miR, microRNA; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
RNA extraction
Total RNA was extracted from tissue samples and cell lines (SUM-159PT and MCF-7 cells) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was extracted from serum samples (2.5 ml) using the Qiagen miRNeasy Mini Kit (Qiagen GmbH), according to the manufacturer's protocol. RNA quantity and purity were determined using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Inc.). RNA samples with an optical density ratio (260/280) of 1.8–2.0 were used for subsequent experiments. RNA samples were stored at −80°C until further analysis.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA (0.1 µg) was reverse transcribed into cDNA using the miScript Reverse Transcription kit II (Qiagen GmbH) with the following thermocycling conditions: 37°C for 60 min and 95°C for 5 min. Subsequently, qPCR was performed using the miScript SYBR® Green PCR kit (Qiagen GmbH) and the Roche Lightcycler 480 Real-Time PCR system (Roche Diagnostics). The following thermocycling conditions were used for qPCR: 95°C for 15 min; followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec. The following primer pairs were used for qPCR: miR-25-3p forward, 5′-CATTGCACTTGTCTCGGTCTGA-3′ and reverse, 5′-GCTGTCAACGATACGCTACGTAACG-3′. The miR-25-3p expression levels were quantified using the 2−ΔΔCq method (29) and normalized to the internal reference genes cel-miR-39 for blood samples (forward, 5′-UAAGGUGCAUCUAGUGCAGAUAG-3′ and reverse, 5′-AUCUGCACUAGAUGCACCUUAUU-3′) or U6 for tissue samples (forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-ACGCTTCACGAATTTGCGT-3′).
Cell culture and transfection
The normal breast cell line MCF-10A and the breast cancer cell lines SUM-159PT and MCF-7 used in the present study were purchased from American Type Culture Collection. Cells were cultured in complete medium consisting of DMEM (Gibco; Thermo Fisher Scientific), FBS (Invitrogen; Thermo Fisher Scientific), 100 µl/ml penicillin, 100 mg/ml streptomycin sulfates and glutamine. Cells were incubated at 37°C with 5% CO2.
SUM-159PT and MCF-7 cells (4×106) were transfected with miR-25-3p inhibitor (100 pmol; 5′-CUAUCAGACUAGAUCGACCUUA-3′; Shanghai GenePharma Co., Ltd.) or the negative control (NC; 5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma Co., Ltd.) using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific) according to the manufacturer's protocol. To confirm transfection efficiency, RT-qPCR was performed 24 h after transfection.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay (US Everbright Inc.) was performed to assess SUM-159PT and MCF-7 cell proliferation. Transfected cells were seeded (3×103 cells/well) into 96-well plates and cultured for 24, 48 or 72 h. Subsequently, 10 µl CCK-8 solution was added to each well and incubated for 1 h at 36.5°C. The optical density of each well at a wavelength of 450 nm was measured every 24 h for 72 h using an ELISA microplate reader (model 680; Bio Rad Laboratories, Inc.).
Cell invasion assay
The Transwell assay was conducted to assess breast cancer cell migration. Transwell membranes were precoated with Matrigel® at 36.5°C for 12 h. Transfected cells with serum-free DMEM (3×103) were seeded into the upper chambers of Transwell plates. DMEM (450 µl) supplemented with 50 µl FBS was plated into the lower chambers of Transwell plates to act as a chemoattractant. Following incubation for 36 h, cells on the upper surface of the Transwell membrane were washed with sterile water and removed. Invading cells were stained using 1% crystal violet at 36.5°C for 1 h. Stained cells were observed in three random fields of view using a light microscope (×200 magnification; Olympus Corporation).
Bioinformatic analysis
The potential target gene regulated by miR-25-3p was predicted using three miRNA-binding site prediction databases: TargetScan (www.targetscan.org/vert_72), miRWalk (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) and starBase (starbase.sysu.edu.cn). TOB1 expression levels were explored using the University of Alabama Cancer Database (UALCAN; ualcan.path.uab.edu). The correlation between TOB1 and miR-25-3p was explored using LinkedOmics (www.linkedomics.org).
Dual-luciferase reporter assay
The dual-luciferase reporter assay was performed to measure the translation coupling efficiency of recoding mechanisms and to evaluate gene expression regulation. The full-length wild-type (WT) TOB1−3′-untranslated region (UTR) containing the predicted binding site of miR-25-3p and its mutant (MT) fragment were cloned into the luciferase reporter vector (Syngene). Cells were co-transfected with WT or MT luciferase reporter vector and miR-25-3p mimic or NC using Lipofectamine® 3000. Luciferase activities were detected using the Dual-Luciferase Reporter assay system (Promega Corporation). Firefly luciferase activities were normalized to Renilla luciferase activities (30,31).
Western blotting
Transfected cells were washed using PBS. Subsequently, total protein was extracted from the cells using ice-cold RIPA buffer. Following homogenization, centrifugation (3,000 × g, 4°C, 60 min) was performed and the supernatant was collected. Protein determination was performed via BCA assay. Total protein (50 µg) was separated via 12% SDS-PAGE and transferred to PVDF membranes, which were blocked with 5% skimmed milk (room temperature, 60 min). Subsequently, the membranes were incubated at 4°C for 24 h with the following primary antibodies: Anti-TOB1 (1:1,000; ProteinTech Group, Inc.) and an anti-GAPDH (1:2,000; ProteinTech Group, Inc.). Following primary incubation, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:1,000; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. Protein bands were visualized using enhanced chemiluminescence substrates (EMD Millipore). Protein expression was quantified using Bio-Rad Gel Doc XR+ system (Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS (version 21.0; IBM Corp.) or GraphPad Prism (version 7; GraphPad Software, Inc.) software. Data are presented as the mean ± SD or SEM (n≥3). Differences among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. The correlation between miR-25-3p expression and clinicopathological variables was assessed using the χ2 test. Receiver-operating characteristics (ROC) and area under the curve (AUC) calculations were performed to assess the diagnostic value of serum miRNAs for the discrimination between normal control subjects and patients with breast cancer. The overall survival curve was drawn using the Kaplan-Meier method. Multivariate Cox regression analyses were performed to evaluate the potential prognostic factors for breast cancer. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-25-3p expression in tissue and serum samples
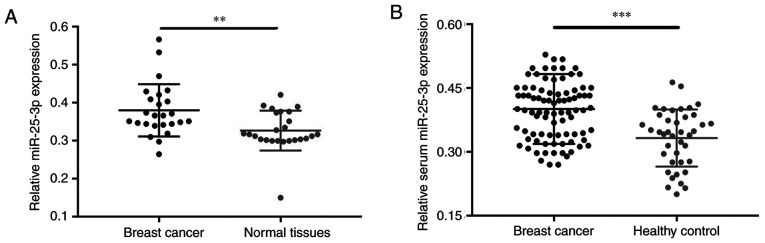
miR-25-3p expression levels were assessed in 50 tissue samples (25 breast cancer and 25 normal tissues) and 176 serum samples (86 samples obtained from patients with breast cancer, 25 paired samples obtained from patients with breast cancer 1 day and 1 month post-surgery, and 40 healthy control samples). The association between age and breast cancer was not significant (Table I). The RT-qPCR results demonstrated that miR-25-3p expression(± SD) was upregulated in breast cancer tissues compared with corresponding non-tumor tissues (Fig. 1A; P=0.005). Moreover, miR-25-3p expression levels were significantly increased in serum samples of patients with breast cancer compared with healthy control subjects (Fig. 1B; P<0.001).
Figure 1.
miR-25-3p expression in breast cancer and normal tissues. (A) Relative expression of miR-25-3p in breast cancer and normal tissues. (B) Relative expression of miR-25-3p in serum samples obtained from patients with breast cancer and healthy controls. **P<0.01 and ***P<0.001, as indicated. miR, microRNA.
Association between serum miR-25-3p expression and clinicopathological characteristics of patients with breast cancer
Subsequently, the association between the expression levels of serum miR-25-3p and the clinicopathological characteristics of patients with breast cancer was analyzed. Patients with breast cancer (n=88) were split into two groups and the median expression level of serum miR-25-3p (44 low serum miR-25-3p expression and 44 high serum miR-25-3p expression) was used as the cut-off point. Serum miR-25-3p expression levels were not significantly associated with age, clinical stage, estrogen receptor, progesterone receptor or human epidermal growth factor receptor 2 levels (Table I). By contrast, serum miR-25-3p expression levels were significantly associated with lymph node metastasis (Table I; P=0.002). The results suggested that patients with breast cancer with high serum miR-25-3p expression levels were more likely to display lymph node metastasis.
Serum miR-25-3p may serve as a promising diagnostic biomarker for breast cancer
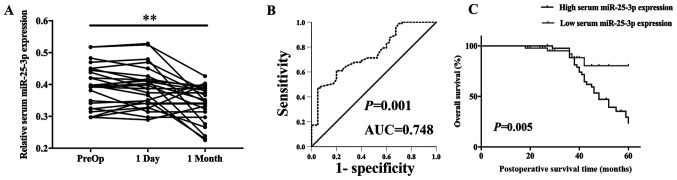
Serum miR-25-3p expression levels of 25 postoperative patients with breast cancer were detected. Serum miR-25-3p expression levels were not significantly different in samples obtained on day 1 post-surgery (P>0.05). However, miR-25-3p expression levels were significantly decreased in samples obtained at 1month post-surgery (P=0.002) compared with preoperative samples (Fig. 2A). Moreover, ROC curve analysis was performed to evaluate the diagnostic value of miR-25-3p in breast cancer (88 patients with breast cancer and 40 cancer-free blood donor volunteers). The AUC for miR-25-3p was 0.748, with 57.1% sensitivity and 95.0% specificity (Fig. 2B; P=0.001). The ROC curve corresponded to the diagnostic value of miR-25-3p for breast cancer.
Figure 2.
Serum miR-25-3p may serve as a promising diagnostic biomarker for breast cancer. (A) Serum miR-25-3p expression levels in preoperative, day 1 post-surgery and 1 month post-surgery samples. (B) ROC curve demonstrated that serum miR-25-3p expression levels successfully discriminated patients with breast cancer from healthy controls (AUC=0.748; 95% confidence interval, 0.660–836; P=0.001) with 57.1% sensitivity and 95.0% specificity. (C) The Kaplan-Meier survival curves demonstrated that patients with breast cancer with low expression of serum miR-25-3p displayed a higher overall survival rate. **P<0.01. miR, microRNA; ROC, receiver operating characteristic curve; AUC, area under curve; PreOp, preoperative.
Serum miR-25-3p may serve as a prognostic biomarker in breast cancer
The association between serum miR-25-3p expression levels and breast cancer prognosis was analyzed. The multivariate Cox proportional hazard regression analysis suggested that clinical stage [hazard ratio (HR)=2.235; 95% confidence interval (CI)=1.094–5.853; P=0.047], lymph node metastasis (HR=4.974; 95% CI=1.786–9.843; P=0.011) and serum miR-25-3p expression levels (HR=6.683; 95% CI=3.343–9.789; P=0.007) were potential factors associated with the overall survival of patients with breast cancer (Table II). The Kaplan-Meier survival curves indicated that patients with breast cancer with low expression of serum miR-25-3p displayed a higher overall survival rate compared with patients with breast cancer with high expression of serum miR-25-3p (P=0.005; Fig. 2C). The results demonstrated that serum miR-25-3p may serve as a promising prognostic biomarker for breast cancer.
Table II.
Univariate and multivariate survival analysis of 88 patients with breast cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (≤50 years vs. >50 years) | 0.844 | 0.417–1.709 | 0.638 | – | – | – |
| Clinical stage (I, II vs. III, IV) | 5.423 | 2.741–8.658 | 0.012 | 2.235 | 1.094–5.853 | 0.047 |
| Lymph node metastasis (positive vs. negative) | 10.356 | 4.404–24.353 | 0.001 | 4.974 | 1.786–9.843 | 0.011 |
| ER (positive vs. negative) | 1.172 | 0.578–2.378 | 0.660 | – | – | – |
| PR (positive vs. negative) | 0.828 | 0.409–1.678 | 0.601 | – | – | – |
| HER2 (positive vs. negative) | 0.799 | 0.393–1.622 | 0.534 | – | – | – |
| Serum miR-25-3p expression | 8.333 | 2.900–23.944 | 0.001 | 6.683 | 3.343–9.789 | 0.007 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; miR, microRNA; HR, hazard ratio; CI, confidence interval; -, not applicable.
miR-25-3p expression levels in breast cancer and normal cell lines
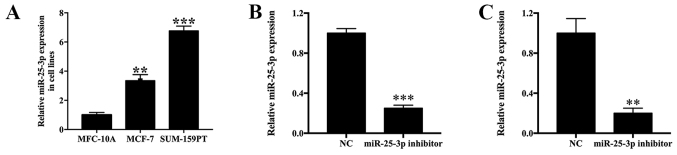
The expression levels of miR-25-3p in MCF-7 and SUM-159PT cells were measured by RT-qPCR. In addition, alterations to the expression levels of miR-25-3p following transfection with miR-25-3p inhibitor, miR-25-3p mimic or NC were also monitored. miR-25-3p expression levels were 3.45- and 6.67-fold higher in MCF-7 (P<0.01) and SUM-159PT (P<0.001) cells compared with MCF-10A cells (Fig. 3A). Moreover, miR-25-3p inhibitor-transfected SUM-159PT cells displayed a 0.249-fold decrease in miR-25-3p expression levels compared with the NC group (P<0.001; Fig. 3B). miR-25-3p inhibitor-transfected MCF-7 cells displayed a 0.201-fold decrease in miR-25-3p expression levels compared with the NC group (P<0.01; Fig. 3C). By contrast, miR-25-3p mimic-transfected SUM-159PT and MCF-7 cells displayed a 12.49- (Fig. S1A; P<0.001) and 13.21-fold (Fig. S1B; P<0.001) increase in miR-25-3p expression levels compared with the mimic NC group, respectively. The results indicated that the transfections were successful.
Figure 3.
miR-25-3p expression in breast cancer cell lines. (A) Relative expression of miR-25-3p in breast cancer cell lines. Transfection efficiency of miR-25-3p inhibitor in (B) SUM-159-PT and (C) MCF-7 cells. **P<0.01, ***P<0.001 vs. NC. miR, microRNA; NC, negative control.
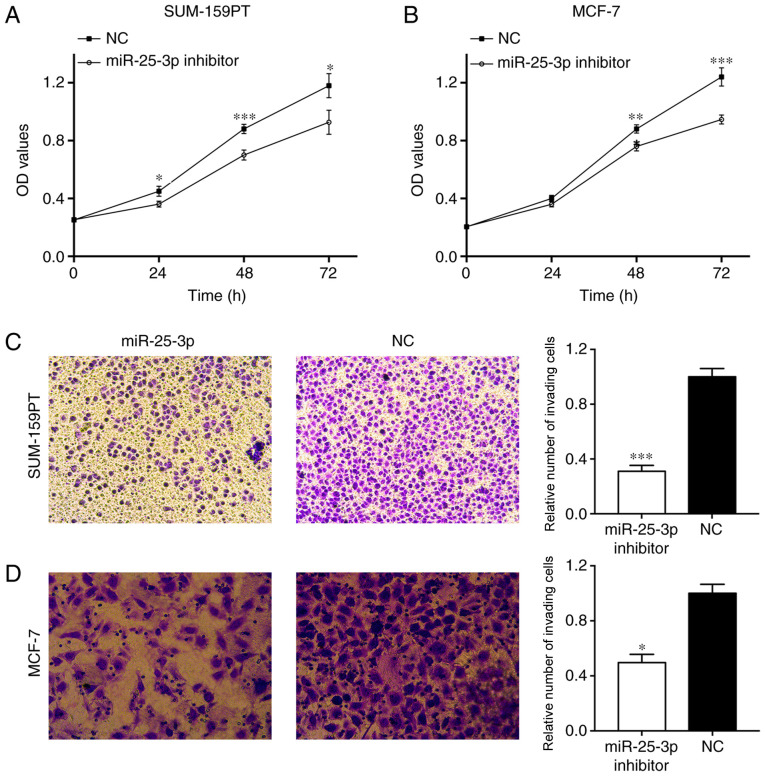
miR-25-3p knockdown suppresses cell proliferation and invasion
The CCK-8 assay indicated that SUM-159PT cell proliferation was significantly inhibited following miR-25-43p knockdown compared with the NC group (Fig. 4A). A similar result was observed in MCF-7 cells (Fig. 4B). In addition, the Transwell assay indicated that SUM-159PT cell invasion was significantly inhibited by miR-25-3p knockdown compared with the NC group (P<0.001; Fig. 4C). Similarly, MCF-7 cell invasion was also significantly decreased by miR-25-3p knockdown compared with the NC group (P<0.05; Fig. 4D). The results indicated that miR-25-3p knockdown suppressed cell proliferation and invasion.
Figure 4.
Effects of miR-25-3p on breast cancer cell function. (A) SUM-159-PT and (B) MCF-7 cell proliferation following miR-25-3p knockdown. (C) SUM-159-PT and (D) MCF-7 cell invasion following miR-25-3p knockdown (magnification, ×100). *P<0.05, **P<0.01 and ***P<0.001 vs. NC. miR, microRNA; NC, negative control; OD, optical density.
TOB1 is a target gene regulated by miR-25-3p
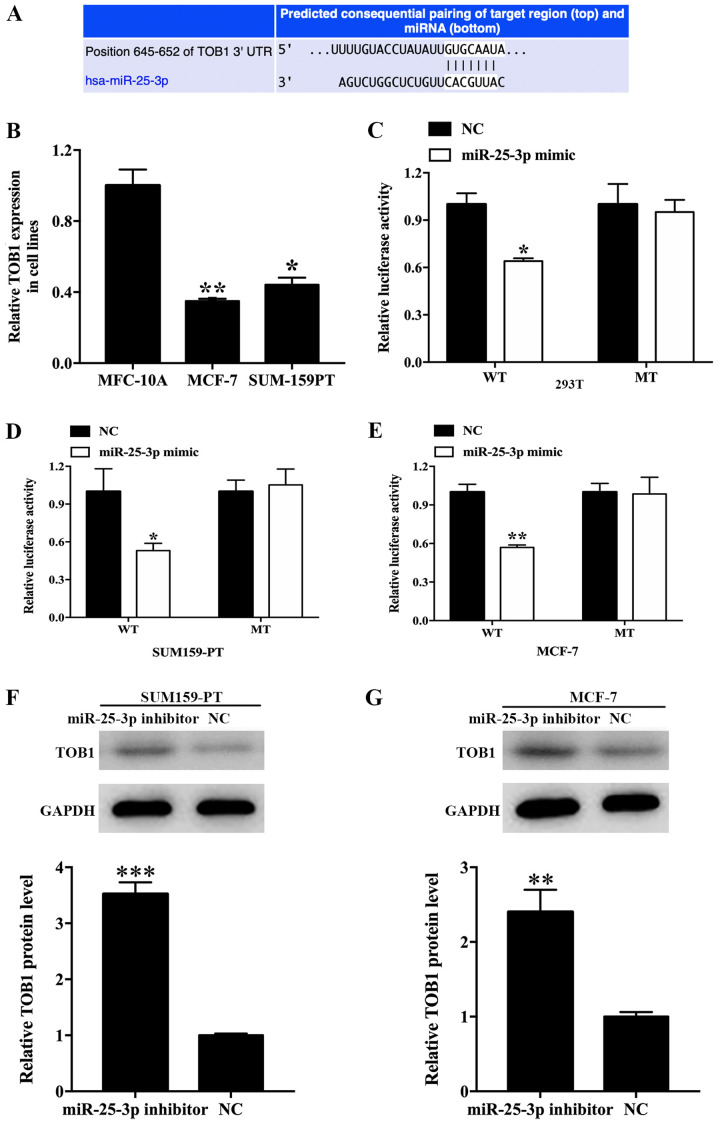
In the present study, three miRNA-binding site prediction databases (TargetScan, miRWalk and starBase) were utilized to predict the potential target gene regulated by miR-25-3p. TOB1 was identified as a target gene. The binding sequence between miR-25-3p and TOB1 is presented in Fig. 5A. TOB1 expression levels were 0.35- (P<0.01) and 0.44- (P<0.05) fold lower in MCF-7 and SUM-159PT cells compared with MCF-10A cells (Fig. 5B). In addition, the UALCAN analysis revealed that TOB1 expression levels were significantly decreased in breast cancer tissues compared with normal tissues (P=8.85×10−9; Fig. S2A). Furthermore, a negative association between TOB1 and miR-25-3p expression levels was identified (P=9.38×10−8; Fig. S2B). The results suggested that TOB1 is a potential target gene regulated by miR-25-3p. Subsequently, a dual-luciferase reporter assay was performed to verify the binding of miR-25-3p to TOB1. The luciferase activity of the TOB1-UTR-WT group in 293T cells was significantly inhibited by miR-25-3p overexpression compared with the NC group (P<0.05). By contrast, the luciferase activity of the TOB1-UTR-MT group was not altered by miR-25-3p expression (Fig. 5C). To confirm the specificity of TOB1 in breast cancer, a dual-luciferase reporter assay was also performed in breast cancer cell lines, and similar results were obtained in SUM159-PT (P<0.05; Fig. 5D) and MCF-7 cells (P<0.05; Fig. 5E). Western blotting was performed for target gene validation. In SUM-159PT cells, TOB1 protein expression levels in the miR-25-3p inhibitor group were significantly increased compared with the inhibitor NC group (P<0.001; Fig. 5F). Moreover, TOB1 protein expression levels in the miR-25-3p inhibitor group were significantly increased compared with the inhibitor NC group (P<0.01; Fig. 5G). Collectively, the results demonstrated that TOB1 was a potential target gene regulated by miR-25-3p in breast cancer.
Figure 5.
Validation of TOB1 as a direct target of miR-25-3p. (A) The binding sites between miR-25-3p and TOB1 3′-UTR. (B) Relative expression of TOB1 in cell lines. *P<0.05, **P<0.01 vs. MFC-10A. (C) Luciferase activities in the WT and MT groups treated with miR-25-3p mimic or NC in (C) 293T, (D) SUM159-PT and (E) MCF-7 cells. TOB1 protein expression levels following miR-25-3p knockdown in (D) SUM-159-PT and (E) MCF-7 cells. Relative TOB1 protein expression in (F) SUM159-PT and (G) MCF7 cells after treatment with miR-25-3p compared with NC group. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. TOB1, transducer of ERBB2, 1; miR, microRNA; 3′-UTR, 3′-untranslated region; WT, wild-type; MT, mutant; NC, negative control.
Discussion
miRNAs are a distinct class of small non-coding RNAs that display various effects in post-transcriptional gene silencing of target mRNAs (32). miRNAs are involved in numerous biological processes, including cell proliferation, invasion, migration, apoptosis, cell cycle control, differentiation, metabolism, immunity, neuronal patterning and stress responses (33–37). Previous findings have revealed that miRNAs exist in the 12 body fluids, which includes serum, urine, saliva, peritoneal fluid, pleural fluid, seminal fluid, tears, amniotic fluid, breast milk, bronchial lavage, cerebrospinal fluid and colostrum (38). Mitchell et al (39) initially described serum miRNAs and demonstrated that they remain in a stable form that is not degraded by endogenous RNase enzymes. It has also been suggested that serum miRNAs may serve as predictive prognostic biomarkers in various malignancies, including breast cancer (39–41).
miR-25-3p is a member of the of miR-25-93-106b cluster, which commonly act as oncogenes, is involved in carcinogenesis and is often upregulated in various malignancies (42). Wang et al (43) reported that miR-25 expression was significantly upregulated in hepatocellular carcinoma tissues, where it promoted cancer cell growth, migration and invasion via Rho GDP dissociation inhibitor-1 (43). miR-25 upregulation was also observed in esophageal squamous cell carcinoma cells and miR-25 knockdown significantly inhibited cell migration and invasion (44). Moreover, it has been reported that miR-25-3p may serve as a biomarker for numerous types of cancer, such as osteosarcoma and pancreatic cancer (45,46). Previous findings demonstrated that miR-25-3p expression was upregulated in breast cancer and that the abnormal expression levels promoted breast cancer cell proliferation (27). Therefore, the present study aimed to investigate the diagnostic and prognostic role of miR-25-3p in breast cancer.
In the present study, miR-25-3p expression levels were increased in breast cancer tissue and serum samples compared with normal breast tissue and serum samples. Moreover, serum miR-25-3p expression levels were not significantly different on day 1 post-surgery compared with presurgery; however, at 1 month post-surgery, miR-25-3p expression levels were significantly decreased compared with prior to surgery, which indicated that miR-25-3p downregulation was associated with the removal of the tumor and not due to surgical stress. Furthermore, the results indicated that serum miR-25-3p expression levels could successfully discriminate patients with breast cancer from healthy subjects. The AUC, sensitivity and specificity of serum miR-25-3p for the diagnosis of breast cancer were 0.748, 57.1 and 95.0%, respectively. The Kaplan-Meier survival curves demonstrated that patients with breast cancer with low serum miR-25-3p expression showed a higher overall survival rate compared with patients with breast cancer with high serum miR-25-3p expression. Therefore, the results demonstrated that serum miR-25-3p may serve as an alternative biomarker for the diagnosis and prognosis of breast cancer.
Previous studies revealed that serum miRNAs could be used as diagnostic biomarkers in breast cancer. For example, serum miR-103 expression levels were significantly increased in patients with breast cancer compared with healthy control subjects. Additionally, for the diagnosis of breast cancer, serum miR-103 displayed 84% sensitivity and 70% specificity, suggesting that serum miR-103 was a promising diagnostic marker for breast cancer detection (47). Serum miR-195 downregulation was observed in patients with breast cancer, and serum miR-195 displayed 69.0% sensitivity and 89.2% specificity for breast cancer detection. Furthermore, serum miR-195 was used to distinguish patients with breast cancer from healthy subjects, indicating that serum miR-195 was a potential tumor biomarker for breast cancer diagnosis (48). A meta-analysis was performed with 438 patients and 228 healthy subjects, and the results suggested that serum miR-21 displayed 0.79% sensitivity, 0.85% specificity and 0.89 AUC in the diagnosis of breast cancer; therefore, the study indicated that miR-21 was a novel biomarker for early detection of breast cancer (49).
Similarly, previous studies have indicated that serum miRNAs have the potential to be used as prognostic biomarkers in breast cancer. A meta-analysis of 1,629 cases demonstrated that patients with breast cancer with elevated miR-21 expression displayed a poor overall survival. miR-21 expression levels were significantly associated with lymph node metastasis, suggesting that circulating miR-21 could be used as a prognostic biomarker in patients with breast cancer (50). Hsieh et al (51) reported that serum miR-125a-5p expression was negatively associated with tumor grade, lymph-node status and tumor size. Moreover, low miR-125a-5p expression was an independent biomarker for the prediction of poor prognosis in patients with breast cancer. Another study indicated that serum miR-99a expression levels were significantly decreased in patients with breast cancer compared with healthy subjects. Furthermore, it was reported that patients with breast cancer with lower miR-99a expression levels displayed a poor overall survival, and that serum miR-99a expression is an independent risk factor for breast cancer, indicating that serum miR-99a was a tumor suppressor and a prognostic biomarker for breast cancer (52).
TOB1 is a transducer of ErbB-2 that is ubiquitously expressed in human adult tissues (53). The TOB1 gene is located on chromosome 17q21 and codes for a 45 kDa protein (54). Previous studies have revealed that TOB1 is associated with tumor cell proliferation and invasion (54,55). In the present study, TOB1 was downregulated in breast cancer cells compared with normal cells, and miR-25-3p knockdown suppressed breast cancer cell proliferation and invasion by regulating TOB1 expression.
The main limitation of the present study was that the sample size was small; therefore, a future study employing a larger sample size should be performed to verify the results of the present study. Moreover, the relevant signaling pathways and targets of miR-25-3p in breast cancer, as well as the function of TOB1 in breast cancer were not clarified in the present study.
In conclusion, the present study demonstrated that miR-25-3p, which regulated breast cancer cellular functions via TOB1, may serve as a breast cancer biomarker.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Natural Science Foundation of Tianjin (grant no. 18JCYBJC94100) and Key Task Project of Tianjin Health and Family Planning Commission (grant no. 2015KZ089).
Funding
The present study was supported by the Natural Science Foundation of Tianjin (grant no. 18JCYBJC94100) and Key Task Project of Tianjin Health and Family Planning Commission (grant no. 2015KZ089).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TZ, XP and LH designed the experiments and analyzed the data. TZ, WM, LG, YC and XY performed the experiments. SS recruited and examined participants. TZ and WM wrote the manuscript. All authors discussed the results. All authors confirm the authenticity of all the raw data and read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6:229–239. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Pogoda K, Niwińska A, Murawska M, Pieńkowski T. Analysis of pattern, time and risk factors influencing recurrence in triple-negative breast cancer patients. Med Oncol. 2013;30:388. doi: 10.1007/s12032-012-0388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang F, Liang H, Li Z, Huang P. JAK3 is a potential biomarker and associated with immune infiltration in kidney renal clear cell carcinoma. Int Immunopharmacol. 2020;86:106706. doi: 10.1016/j.intimp.2020.106706. [DOI] [PubMed] [Google Scholar]
- 7.Halvorsen AR, Helland Å, Gromov P, Wielenga VT, Talman MM, Brunner N, Sandhu V, Børresen-Dale AL, Gromova I, Haakensen VD. Profiling of microRNAs in tumor interstitial fluid of breast tumors-a novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol Oncol. 2017;11:220–234. doi: 10.1002/1878-0261.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madhavan D, Cuk K, Burwinkel B, Yang R. Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Genet. 2013;4:116. doi: 10.3389/fgene.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christou N, Meyer J, Popeskou S, David V, Toso C, Buchs N, Liot E, Robert J, Ris F, Mathonnet M. Circulating tumour cells, circulating tumour DNA and circulating tumour miRNA in blood assays in the different steps of colorectal cancer management, a review of the evidence in 2019. Biomed Res Int. 2019;2019:5953036. doi: 10.1155/2019/5953036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Wu X, Wang J, Lopez J, Zhou W, Yang L, Wang SE, Raz DJ, Kim JY. Circulating miRNA profile in esophageal adenocarcinoma. Am J Cancer Res. 2016;6:2713–2721. [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Zhang H, Qin S, Wang Q, Yang X, Wang K. Optical fiber amplifier for quantitative and sensitive point-of-care testing of myoglobin and miRNA-141. Biosens Bioelectron. 2019;129:87–92. doi: 10.1016/j.bios.2018.12.056. [DOI] [PubMed] [Google Scholar]
- 12.Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27:1879–1886. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 13.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, Dahiya R, Tanaka Y, Saini S. microRNA-1246 Is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018;78:1833–1844. doi: 10.1158/0008-5472.CAN-17-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Zhou L, Lin C, Pan X, Xie J, Zhao L, Quan J, Xu J, Guan X, Xu W, et al. miR-302b regulates cell functions and acts as a potential biomarker to predict recurrence in bladder cancer. Life Sci. 2018;209:15–23. doi: 10.1016/j.lfs.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C, Ling X, Li X, Hou X, Zhao D. MicroRNA-138-5p inhibits cell migration, invasion and EMT in breast cancer by directly targeting RHBDD1. Breast Cancer. 2019;26:817–825. doi: 10.1007/s12282-019-00989-w. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS, Kundu J, Kim RN, Shin YK. Transducer of ERBB2.1 (TOB1) as a tumor suppressor: A mechanistic perspective. Int J Mol Sci. 2015;16:29815–29828. doi: 10.3390/ijms161226203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu J, Wahab SM, Kundu JK, Choi YL, Erkin OC, Lee HS, Park SG, Shin YK. Tob1 induces apoptosis and inhibits proliferation, migration and invasion of gastric cancer cells by activating Smad4 and inhibiting β-catenin signaling. Int J Oncol. 2012;41:839–848. doi: 10.3892/ijo.2012.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao Y, Sun KK, Zhao L, Xu JY, Wang LL, Fan SJ. Suppression of human lung cancer cell proliferation and metastasis in vitro by the transducer of ErbB-2.1 (TOB1) Acta Pharmacol Sin. 2012;33:250–260. doi: 10.1038/aps.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Zhou W, Wang S, Zhou Z, Wang S, Chen L. Tob1 enhances radiosensitivity of breast cancer cells involving the JNK and p38 pathways. Cell Biol Int. 2015;39:1425–1430. doi: 10.1002/cbin.10545. [DOI] [PubMed] [Google Scholar]
- 21.Cioffi M, Trabulo SM, Vallespinos M, Raj D, Kheir TB, Lin ML, Begum J, Baker AM, Amgheib A, Saif J, et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609–21625. doi: 10.18632/oncotarget.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sárközy M, Kahán Z, Csont T. A myriad of roles of miR-25 in health and disease. Oncotarget. 2018;9:21580–21612. doi: 10.18632/oncotarget.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Meng X, Li H, Liu W, Shen S, Gao Z. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin Transl Oncol. 2014;16:954–958. doi: 10.1007/s12094-014-1178-6. [DOI] [PubMed] [Google Scholar]
- 24.Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137:1679–1690. doi: 10.1002/ijc.29544. [DOI] [PubMed] [Google Scholar]
- 25.Jia Y, Lu H, Wang C, Wang J, Zhang C, Wang F, Zhang C. miR-25 is upregulated before the occurrence of esophageal squamous cell carcinoma. Am J Transl Res. 2017;9:4458–4469. [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JT, Wang F, Chapin W, Huang RS. Identification of MicroRNAs as breast cancer prognosis markers through the cancer genome atlas. PLoS One. 2016;11:e0168284. doi: 10.1371/journal.pone.0168284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Pan H, Qian Y, Zhou W, Liu X. miR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17:4. doi: 10.1186/s12943-017-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou X, Xia T, Li M, Wang T, Liu P, Zhou X, Huang Z, Zhu W. MicroRNA profiling in serum: Potential signatures for breast cancer diagnosis. Cancer Biomark. 2021;30:41–53. doi: 10.3233/CBM-201547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 31.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 32.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mens MMJ, Ghanbari M. Cell cycle regulation of stem cells by MicroRNAs. Stem Cell Rev Rep. 2018;14:309–322. doi: 10.1007/s12015-018-9808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi Y, Wang X, Kong X, Zhai J, Fang Y, Guan X, Wang J. Expression signatures and roles of microRNAs in inflammatory breast cancer. Cancer Cell Int. 2019;19:23. doi: 10.1186/s12935-018-0709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Lin C, Zhao L, Zhou L, Pan X, Quan J, Peng X, Li W, Li H, Xu J, et al. Oncogene miR-187-5p is associated with cellular proliferation, migration, invasion, apoptosis and an increased risk of recurrence in bladder cancer. Biomed Pharmacother. 2018;105:461–469. doi: 10.1016/j.biopha.2018.05.122. [DOI] [PubMed] [Google Scholar]
- 36.Olejniczak M, Kotowska-Zimmer A, Krzyzosiak W. Stress-induced changes in miRNA biogenesis and functioning. Cell Mol Life Sci. 2018;75:177–191. doi: 10.1007/s00018-017-2591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh SK, Pal Bhadra M, Girschick HJ, Bhadra U. MicroRNAs-micro in size but macro in function. FEBS J. 2008;275:4929–4944. doi: 10.1111/j.1742-4658.2008.06624.x. [DOI] [PubMed] [Google Scholar]
- 38.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61–81. doi: 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;10:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Liu M, Ma F, Luo Y, Cai R, Wang L, Xu N, Xu B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS One. 2014;9:e104870. doi: 10.1371/journal.pone.0104870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen L, Wan Z, Ma Y, Wu L, Liu F, Zang H, Xin S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumour Biol. 2015;36:1993–2005. doi: 10.1007/s13277-014-2806-z. [DOI] [PubMed] [Google Scholar]
- 42.Gruszka R, Zakrzewska M. The oncogenic relevance of miR-17-92 cluster and its paralogous miR-106b-25 and miR-106a-363 clusters in brain tumors. Int J Mol Sci. 2018;19:879. doi: 10.3390/ijms19030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Wang X, Su Z, Fei H, Liu X, Pan Q. miR-25 promotes hepatocellular carcinoma cell growth, migration and invasion by inhibiting RhoGDI1. Oncotarget. 2015;6:36231–36244. doi: 10.18632/oncotarget.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua Y, Zhao K, Tao G, Dai C, Su Y. miR-25 promotes metastasis via targeting FBXW7 in esophageal squamous cell carcinoma. Oncol Rep. 2017;38:3030–3038. doi: 10.3892/or.2017.5995. [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8:33375–33392. doi: 10.18632/oncotarget.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng T, Yuan Y, Zhang C, Zhang C, Yao W, Wang C, Liu R, Ba Y. Identification of circulating miR-25 as a potential biomarker for pancreatic cancer diagnosis. Cell Physiol Biochem. 2016;39:1716–1722. doi: 10.1159/000447872. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Wu X, Yan L, Shao J. Serum miR-103 as a potential diagnostic biomarker for breast cancer. Nan Fang Yi Ke Da Xue Xue Bao. 2012;32:631–634. (In Chinese) [PubMed] [Google Scholar]
- 48.Zhao FL, Dou YC, Wang XF, Han DC, Lv ZG, Ge SL, Zhang YK. Serum microRNA-195 is down-regulated in breast cancer: A potential marker for the diagnosis of breast cancer. Mol Biol Rep. 2014;41:5913–5922. doi: 10.1007/s11033-014-3466-1. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Yang X, Yang J, Zhen J, Zhang D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: A systematic review and meta-analysis. Clin Exp Med. 2016;16:29–35. doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 50.Jinling W, Sijing S, Jie Z, Guinian W. Prognostic value of circulating microRNA-21 for breast cancer: A systematic review and meta-analysis. Artif Cells Nanomed Biotechnol. 2017;45:1–6. doi: 10.1080/21691401.2016.1216856. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai CY, Hou MF, Lee JN, Wu DC, Wang SC, Tsai EM. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2015;6:494–509. doi: 10.18632/oncotarget.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Song ZJ, Wang YY, Yin Y, Liu Y, Nan X. Low levels of serum miR-99a is a predictor of poor prognosis in breast cancer. Genet Mol Res. 2016:15. doi: 10.4238/gmr.15038338. doi: 10.4238/gmr.15038338. [DOI] [PubMed] [Google Scholar]
- 53.Lin S, Zhu Q, Xu Y, Liu H, Zhang J, Xu J, Wang H, Sang Q, Xing Q, Fan J. The role of the TOB1 gene in growth suppression of hepatocellular carcinoma. Oncol Lett. 2012;4:981–987. doi: 10.3892/ol.2012.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Malley S, Su H, Zhang T, Ng C, Ge H, Tang CK. TOB suppresses breast cancer tumorigenesis. Int J Cancer. 2009;125:1805–1813. doi: 10.1002/ijc.24490. [DOI] [PubMed] [Google Scholar]
- 55.Guo H, Ji F, Zhao X, Yang X, He J, Huang L, Zhang Y. MicroRNA-371a-3p promotes progression of gastric cancer by targeting TOB1. Cancer Lett. 2019;443:179–188. doi: 10.1016/j.canlet.2018.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.