Abstract
Background
South Africa was the African country with the most recorded cases of SARS-CoV-2 during 2020, experiencing 2 waves of infection. During the first wave, diagnostics were largely based on reverse transcription-linked PCR (RT-PCR). The Abbott PanBio antigen test was deployed during the 2nd wave which may have been driven by emergence of the B.1.351 variant. At the time of evaluation in mid-November 2020, B.1.351 was the dominant circulating virus in Nelson Mandela Bay, in the Eastern Cape Province.
Methods
Used PanBio antigen swabs (collected from patients with genetically characterised virus) were first validated as suitable for PCR. A prospective study was then undertaken to evaluate assay performance in the field. Testing was conducted at mobile community testing centres on 677 ambulant patients. Used swabs were kept and tested by RT-PCR.
Results
During initial validation, used swabs in proprietary lysis buffer were found to be suitable for PCR and secondly, the PB assay reliably detected patients infected with B.1.351. In the field study, of 146 RT-PCR positive individuals, 101 were RTD positive in the clinic. The RTD had a sensitivity of 69.2% (95%CI 61.4, 75.8) and specificity of 99.0% (95%CI 98.8, 99.3). Sensitivity was dependent on the amount of viral RNA in clinical samples, as reflected by the PCR cycle threshold (CT) value.
Conclusions
The assay reliably detected B.1.351 infections in ambulatory ill patients during initial validation and in field testing. In the field, assay sensitivity was >90% in patients with high viral loads who are expected to be most infectious. Negative and positive predictive values were also >90%.
Keywords: SARS-CoV-2, 501Y.v2, B.1.351, Rapid antigen test, Point of care, COVID-19
Background
During 2020 South Africa was the African country with the most recorded cases of SARS-CoV-2 with more than 1 380 000 laboratory confirmed cases and 83 918 excess deaths [1]. During this period, the country experienced 2 waves of infection [2]. Provision of an effective diagnostic service proved to be challenging. RT-PCR is the gold standard assay for SARS-CoV-2 diagnosis, however, in the context of high disease prevalence, laboratory systems may easily become overwhelmed. Rapid diagnostics such as antigen tests that can be performed at point of care provide a welcome solution. Their main drawback is lower sensitivity [3]. The WHO advises that assays that meet minimum performance requirements (>80% sensitivity, >97% specificity in the first 7 days of symptoms) can be used in contexts where nucleic acid-based testing is unavailable, or where turn-around times are prolonged [4]. The Abbott PanBio rapid SARS-CoV-2 antigen assay has fulfilled these criteria in evaluations in several studies [5,6]
This assay was deployed during the 2nd wave in South Africa which first became apparent in the Eastern Cape Province in October/November 2020. Increased disease activity was associated with emergence of a new variant, namely B1.351 [7]. This variant first detected in October 2020 rapidly became the predominant virus, across the country, potentially due to higher transmissibility [8]. At the time of evaluation in mid-November 2020, it was the dominant circulating virus, responsible for around 84% of infections in Nelson Mandela Bay, estimate based on genomes submitted to global initiative for sharing all influenza data (GISAID) over this time-period [9]. This prospective diagnostic evaluation study was designed to evaluate the field performance of the PanBio assay, but also provides evidence on its performance in individuals infected with B.1.351. Another novel aspect is that RT-PCR was performed on the same swab used for antigen testing, which obviated the need to collect further samples from patients and provided a more direct comparison with the antigen result.
Methods and results
Prospective diagnostic evaluation study in Nelson Mandela Bay municipality, Eastern Cape South Africa during a period of high disease prevalence, using nasopharyngeal swabs to determine the accuracy of Abbott PanBio COVID-19 antigen RTD.
Verification that used antigen swabs are suitable for PCR
46 paired swabs were collected from symptomatic patients, one nylon tip, standard issue swab for PCR and the flocked antigen swab from the PanBio test kit. SARSCoV-2 RT-PCR was done on used antigen and matched nylon swabs.
The antigen swab was prepared for PCR as follows: 1 mL saline was added to the swab container using a filter tip. The sample was vortexed and allowed to stand 2 min. The bottom cap was opened, and fluid bled into a sterile vial. The matching PCR swab was snipped into a vial containing 1.5 ml normal saline and vortexed.
Paired samples were extracted on the NucliSENS® easyMag® (bioMerieux, France) platform. RT-PCR was done using the Allplex™ 2019-nCoV (Seegene, South Korea) assay with amplification on CFX Real-Time PCR instrument (Bio-Rad, USA). For PCR positive swabs, mean CT values of the 3 assay targets were compared.
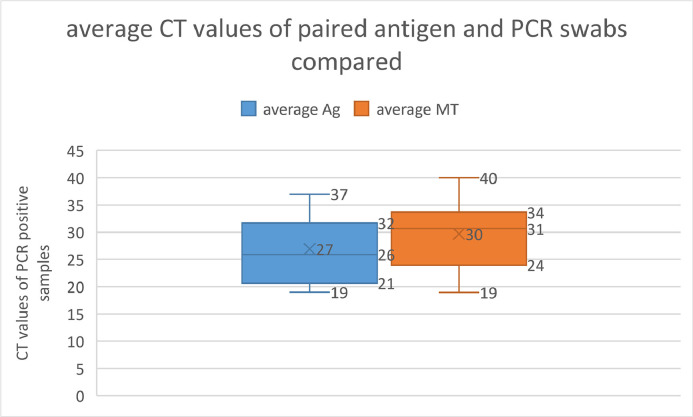
Of the 46 paired samples, 15 were antigen positive. 25 were concordantly PCR negative, 2 samples were PCR positive on the antigen swab, but negative on the regular swab and 19 samples were concordantly PCR positive. When comparing mean CT values of the paired swabs, the antigen swab had values 2 CTs lower than the swab collected for PCR. Wilcoxon signed-rank test p0.0073. (Fig. 1 )
Fig. 1.
Average CT values of PCR positive paired antigen vs regular swabs are compared: The range of CT values from PCR of the antigen swab were on average 2 CTs lower than for those from the standard swabs. Wilcoxon signed-rank test p0.0073.
Verification that PB assay detects 501Y.v2 infections
Whole genome sequencing was performed on 15 of the 19 PCR positive validation samples. All 15 were confirmed to be lineage B1.351. PB assay was positive in 13 of these 15 samples. Data on the 46 validation samples are given in Table S1.
Study protocol
Between 17 and 20 November 2020 mobile clinics ran community testing campaigns at 6 sites in Nelson Mandela Bay. Symptomatic patients were invited to undergo antigen testing. Nasopharyngeal (NP) swabs were tested using the PanBio SARS-CoV-2 RTD. Results were communicated to patients immediately. The used swabs were sent for RT-PCR which was performed as per the verification.
A total of 677 patients from 6 mobile clinics were tested by both antigen and PCR. Patients were ambulant and seeking COVID-19 testing. They ranged in age range from 3 to 85 years; 59% were female.
Of these, 101 (14.9%) were antigen positive in the clinic. With RT-PCR, 146 samples (21.4%) were reported as positive, 19 (2.8%) as inconclusive (single target positive, CT>38) and 509 (75.2%) were negative for both tests. Inconclusive samples were excluded from analysis as their significance was unresolved.
Antigen test performance
Using RT-PCR as the reference standard, the antigen test had an overall sensitivity (positive percent agreement) of 69.17% (95%CI 61.44, 75.80) and specificity (negative percent agreement) of 99.02% (95%CI 98.78, 99.26) in this clinical context.
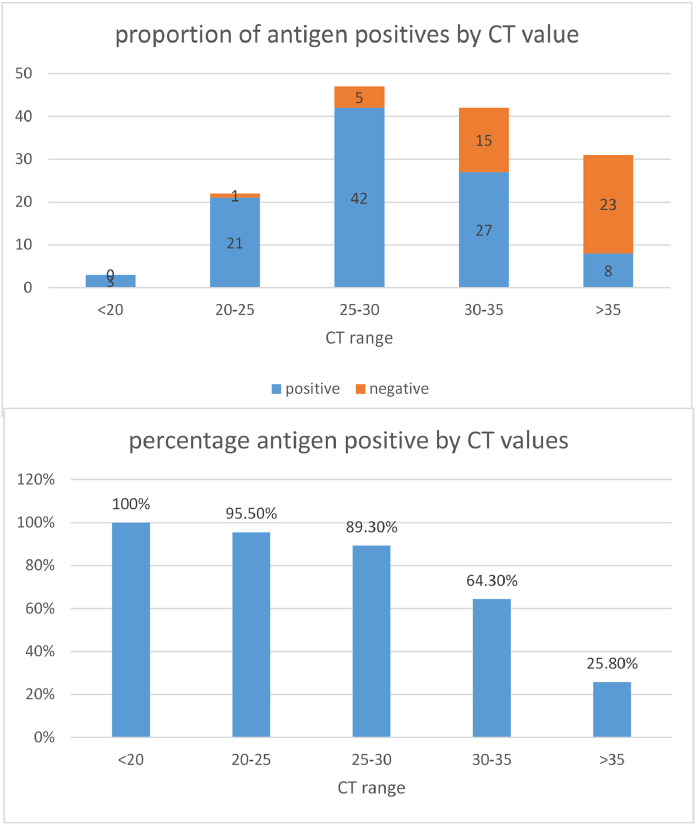
Sensitivity was dependent on the quantity of viral RNA in clinical samples, as reflected by the CT value, with 100% detection by the antigen test in samples where the CT was <20, 95.5% with CT between 20 and 25, 89.3% with CT between 26 and 30 and 64,3% when CT was 31–35. The CT values of antigen positive and negative samples are shown (Figs. 2 a, b)
Fig. 2.
(a) Compares the number of antigen positive and negative samples according to CT category values obtained in PCR, (b) Compares the percentage of antigen positive samples according to virus levels, as reflected by the mean CT value.
The antigen assay was positive in 3 RT-PCR negative patients. Given the prevalence of infection of 21% (as determined by RT-PCR) the predictive value of a negative test was 91.9% and that of a positive test was 97.12%.
Characteristics of PCR positive samples
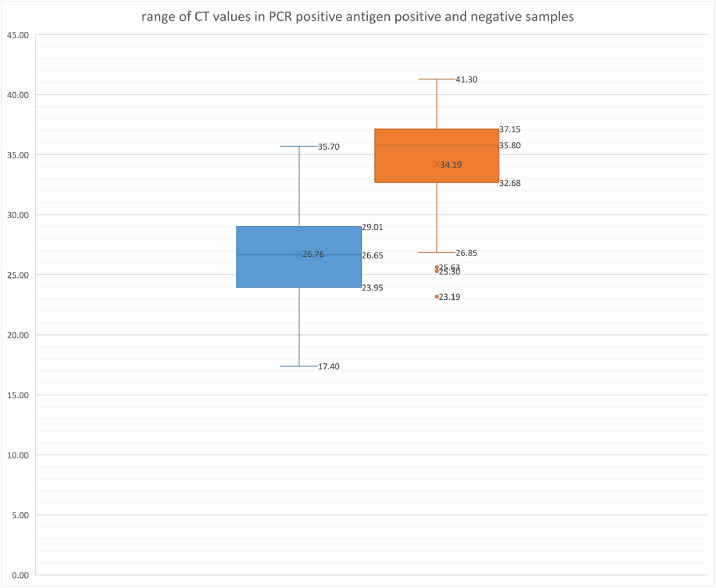
In the 146 PCR positive patients, CT values ranged from 17.4 to 41.3, median 30.1. As expected, the median CT and interquartile range (IQR) was lower in antigen positive samples at 28.7 (IQR 25.3–31.3). In comparison, the median and IQR of CT of antigen negatives was 35.8 (IQR 32.7–37.1) (Fig. 3 ).
Fig. 3.
Compares the range of CT values obtained on antigen positive and negative samples by reference PCR. Median and IQR values are given. CT values in antigen positive and negative samples were significantly different when compared with the Mann–Whitney rank sum test, p < 0.00001.
Discussion
This study took place during a period of high community transmission associated with emergence of the B.1.351 variant in the Eastern Cape. RT-PCR was performed directly on the antigen swab after testing allowing direct comparison of antigen reactivity and RT-PCR on the same sample. Initial verification confirmed that the RTD reliably detected B.1.351. In the field, the RTD had a sensitivity of 69.17% and specificity of 99.0%. The sensitivity is below the 80% WHO benchmark [4]. However, context is key. Testing was performed on unselected symptomatic individuals who requested testing, irrespective of symptom duration. This probably accounts for the fact that 50% of PCR positive patients had CT values > 30, indicating that on average sampling may have occurred later during infection than recommended for maximum performance. Nonetheless, performance exceeded benchmark in patients with CT<30. In this range, sensitivity was 91.3%. This concordance (using distinctly different technologies) suggests that B.1.351 was reliably detected by the RTD at clinically relevant RNA copy numbers [9,10].
Assay specificity was similarly good at 99% and the predictive value of a positive test was 95%. This fulfils the WHO benchmark specificity requirements for deployment of this assay [4].
Two factors could compromise detection of B.1.351; namely amino acid changes in the region of the nucleocapsid protein targeted by kit antibodies, or reduced virus shedding in respiratory tract samples. The potential higher infectivity of the variant makes the latter explanation unlikely and preliminary evidence does not support it. B.1.351 has a single amino acid change in the linker region of the nucleocapsid protein, namely N205I [11]. As this amino acid is located in an unstructured region [12], it should not affect antibody binding. This region is not targeted by capture antibodies in the RTD, according to the manufacturers.
The main limitation was that it was not feasible to confirm B.1.351 infection in positive cases. This was inferred from the fact that resurgence in this district was overwhelmingly due to B.1.351, based on contemporaneous genomes submitted to GISAID [11].
Conclusion
The assay reliably detected B.1.351 infection in ambulatory ill patients. Sensitivity was >90% in patients with high viral loads who are expected to be most infectious. To optimise the use of antigen RDTs in different and changing circumstances, clinical predictors and the epidemiological context should be considered when deciding on assay deployment.
Ethics
Ethical approval was obtained from the University of Cape Town Human Research Ethics Committee (UCT HREC 862.2020).
Informed consent
Patients were willing participants and gave verbal consent to diagnostic testing. Data was anonymised and delinked.
Author contributions
Study concept and design: DH, OL
Conduct of study, sample testing: OL, KS, GM
Manuscript write up: DH, OL, KS, GM
All authors approved the final manuscript
Funding
No funding was given for this study.
Declaration of Competing Interest
The authors report no competing interests.
Acknowledgments
We thank Deelan Doolabh, Arash Iranzadeh, Nei-Yuan Hsiao and Carolyn Williamson for providing access to whole genome sequence data on SARS-CoV-2 positive samples used in the assay validation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2021.100013.
Appendix. Supplementary materials
References
- 1.SAMRC, Report on weekly deaths in South Africa, South Africa Med. Res. Counc. 2021 (2020) 1. https://www.samrc.ac.za/reports/report-weekly-deaths-south-africa.
- 2.Waasila Jassat1 L.B. Cheryl Cohen2, Caroline Mudara1, The first and second wave of COVID-19 In three districts of South Africa, COVID-19 Spec. Public Heal. Surveill. Bull. COVID-19 Spec. PUBLIC Heal. Surveill. Bull. 2021;18:1–24. [Google Scholar]
- 3.Cunningham J., Beese S., Dretzke J., Im H., Mj P., Im H., Mj P., Hoo L., Mmg L., Spijker R. 2020. Diagnosis of SARS-CoV-2 Infection (Review)www.cochranelibrary.com V.D.B. A. [DOI] [Google Scholar]
- 4.WHO, Antigen-detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays Interim guidance, 11 September 2020, World Health Organisation (2020). https://apps.who.int/iris/handle/10665/334253.
- 5.Linares M., Pérez-Tanoira R., Carrero A., Romanyk J., Pérez-García F., Gómez-Herruz P., Arroyo T., Cuadros J. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J. Clin. Virol. 2020;133:3–6. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mühlemann B., Zuchowski M., Karen W., Lei J. Comparison of seven commercialSARS-CoV-2 rapid point of care antigen assays. MedRxiv. 2020 doi: 10.1101/2020.11.12.20230292. [DOI] [Google Scholar]
- 7.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Wibmer K., Sewell B.T., Lourenço J., Carlos L., Alcantara J., Kosakovsky S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., De Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Arghavan Alisoltani-Dehkordi. 2020;10 doi: 10.1101/2020.12.21.20248640. 2020.12.21.20248640. [DOI] [Google Scholar]
- 8.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M. BioRxiv Prepr; 2021. SARS-CoV-2 501Y . V2 Escapes Neutralization by South African COVID-19 Donor Plasma. doi: [DOI] [PubMed] [Google Scholar]
- 9.Manzulli V., Scioscia G., Giganti G., Capobianchi M.R., Lacedonia D., Pace L., Cipolletta D., Tondo P., De Nittis R., Rondinone V., Serrecchia L., Parisi A., Galante D., Lo Caputo S., Santantonio T.A., Moschetta D., Dattoli V., Fasanella A., Foschino Barbaro M.P. Real time PCR and culture-based virus isolation test in clinically recovered patients: is the subject still infectious for SARS-CoV2? J. Clin. Med. 2021;10:309. doi: 10.3390/jcm10020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:8–13. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.https://corona-maps.live/gisaid/ 2021
- 12.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M. Biochemica characterisation of SARS-VoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.