Abstract
A comprehensive understanding of the changes in gene expression in cell types involved in idiopathic pulmonary fibrosis (IPF) will shed light on the mechanisms underlying the loss of alveolar epithelial cells, and development of honeycomb cysts and fibroblastic foci. We sought to understand changes in IPF lung cell transcriptomes and gain insight into innate immune aspects of pathogenesis. We investigated IPF pathogenesis using single cell RNA-sequencing of fresh lung explants, comparing human IPF fibrotic lower lobes reflecting late disease, upper lobes reflecting early disease and normal lungs. IPF lower lobes showed increased fibroblasts, and basal, ciliated, goblet and club cells, but decreased alveolar epithelial cells, and marked alterations in inflammatory cells. We found three discrete macrophage subpopulations in normal and fibrotic lungs, one expressing monocyte markers, one highly expressing FABP4 and INHBA (FABP4hi), and one highly expressing SPP1 and MERTK (SPP1hi). SPP1hi macrophages in fibrotic lower lobes showed highly upregulated SPP1 and MERTK expression. Low-level local proliferation of SPP1hi macrophages in normal lungs was strikingly increased in IPF lungs. Co-localization and causal modeling supported the role for these highly proliferative SPP1hi macrophages in activation of IPF myofibroblasts in lung fibrosis. These data suggest SPP1hi macrophages contribute importantly to lung fibrosis in IPF, and that therapeutic strategies targeting MERTK and macrophage proliferation may show promise for treatment of this disease.
Introduction
The complex pathogenesis of idiopathic pulmonary fibrosis (IPF) involves multiple cell types [1]: injury and loss of alveolar epithelial cells [2, 3]; formation of honeycomb cysts lined by several epithelial cell lineages: p63+, goblet and ciliated cells [4]; expansion of reparative basal, stem cells [5–7]; and development of fibroblastic foci composed of myofibroblasts acting as fibrotic effector cells [8].
In murine bleomycin-induced pulmonary fibrosis, several studies suggest that inflammation and monocyte-derived macrophages drive fibrosis, through overactive reparative responses to alveolar cell injury. CCR2 deletion depletes bleomycin-induced pulmonary macrophages and fibrosis [9]. Similarly, deletion of CD11b-expressing monocyte-derived macrophages ameliorates bleomycin-induced lung fibrosis [10]. A recent study has indicated that monocyte-derived alveolar macrophages drive bleomycin induced lung fibrosis [11].
Recent studies show that in most tissues resident macrophages are derived from embryonic progenitors. Alveolar macrophages develop from embryonic progenitors originating in the yolk sac and fetal liver [12, 13]. Interstitial macrophages also derive from yolk sac and fetal liver macrophages, although a second group of interstitial monocyte/macrophages appears to derive from circulating monocytes [13, 14]. Thus, in normal adult mouse lungs most resident macrophages are derived from embryonic progenitors, but a subset of interstitial cells derives from monocytes. Multiple studies have shown that murine macrophages are of capable self-renewal. Tissue resident macrophages have a long life span and proliferate under the influence of M-CSF and GM-CSF [15, 16], and IL-4 stimulates resident macrophage proliferation during nematode infections [17]. M-CSF is required for proliferation of both inflammatory bone marrow derived macrophages and resident macrophage populations in zymosan-induced peritonitis [18]. Whether proliferation of resident macrophages contributes to human IPF is less studied.
We used single cell RNA-sequencing (scRNA-seq) to provide a comprehensive view of altered cell numbers and transcriptomes associated with IPF. As a criterion for IPF disease progression we examined changes in upper (early disease) and lower (late fibrotic disease) lobes compared to normal lungs. We identify subpopulations of macrophages in normal lungs that in IPF show increased proliferation and highly upregulated expression of SPP1, suggesting these cells play an important role in IPF pathogenesis.
Methods
Healthy control lungs were obtained under a protocol approved by the University of Pittsburgh, Committee for Oversight of Research and Clinical Training Involving Decedents, following rejection as candidate donors for transplant. IPF lung tissue was obtained under a protocol approved by the University of Pittsburgh, Institutional Review Board, during transplantation surgery. Detailed methods are included in the Online Supplement. ScRNA-seq data can be accessed at the Gene Expression Omnibus: GSE128033.
Results
Macrophage subpopulations in normal lungs.
We analyzed single cell transcriptomes of 24,220 cells from seven normal lung samples by scRNA-seq, including one technical replicate together by t-Distributed Stochastic Neighbor Embedding (t-SNE) and colored by groups of cells (Table S1). This analysis showed 16 different clusters of cells (Figure 1A), using previously described markers, including inflammatory, epithelial, vascular and mesenchymal cell types, as indicated (Figures S1, S2, Table S2 and Supplemental Results). Cells identified by the subject of origin ensured that clusters included cells from each sample (Figure 1B).
Figure 1.
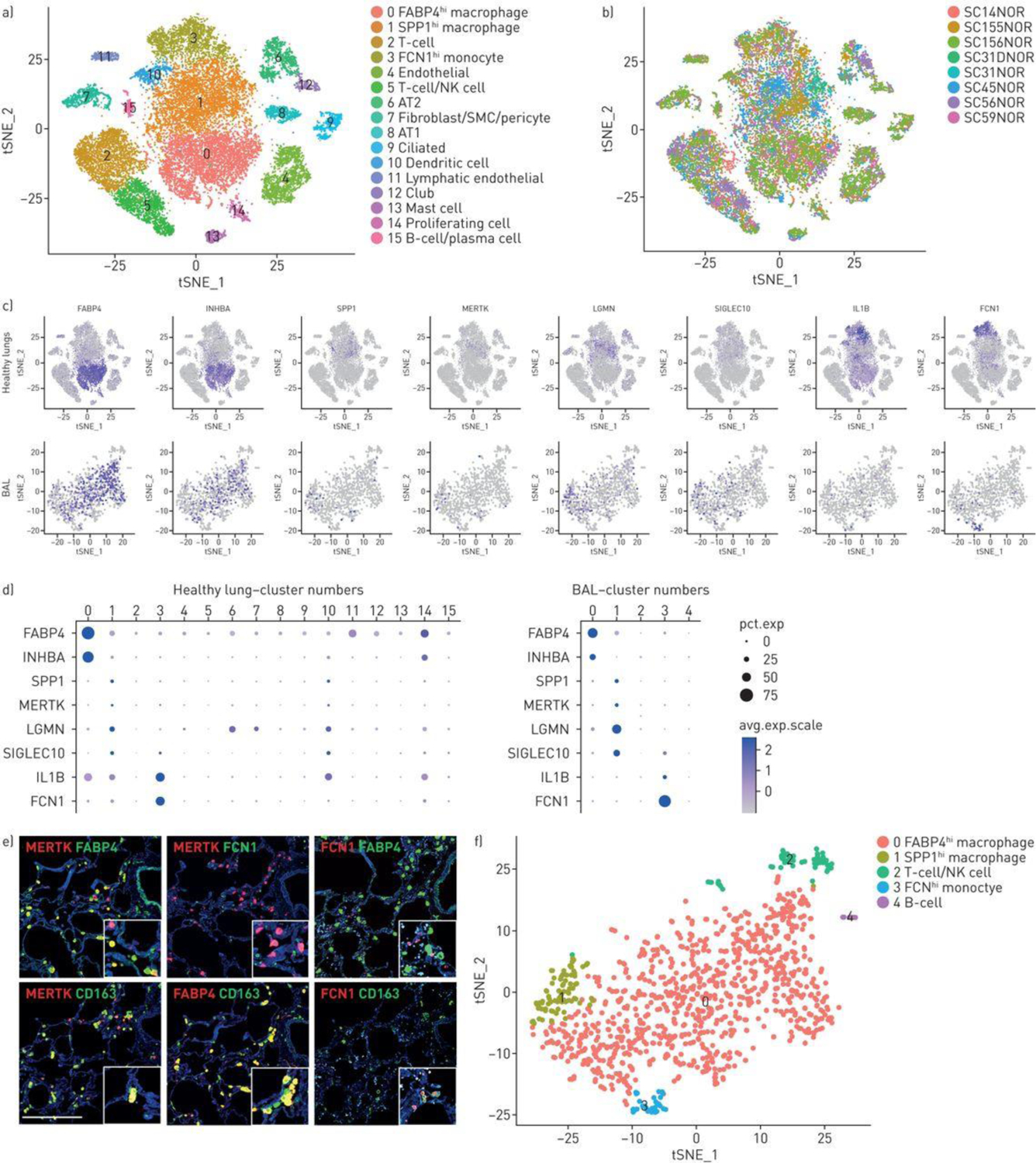
Characterisation of normal lung macrophages. T-distributed stochastic neighbour embedding (T-SNE) of normal lung cells indicates 10 cell types including a large central grouping of macrophage/monocytes/dendritic cells composed of clusters 0, 1, 3 and 10 (panel a) All four of these clusters express AIF1 and all but the dendritic cell cluster (cluster 10) express CD163 (supplementary figure S1). The subject of origin for each of the samples in indicated by a different colour in panel b. The three different macrophage/monocyte clusters (clusters 0, 1 and 3) can be distinguished in feature plots (panel c, purple intensity indicates level of gene expression) and dot plots (panel d, dot size indicates percent of cells expressing and the intensity of purple the level of expression) by expression of FABP4 and INHBA (FABP4hi macrophages, cluster 0); SPP1, MERTK, LGMN and SIGLEC10 (SPP1hi macrophages, cluster 1); or IL1B and FCN1 (FCN1hi monocyte/macrophages, cluster 3). Dendritic cells, expressing CD1C, are found in cluster 10 (see also supplementary figure S1). Macrophage monocyte cell types in bronchoalveolar lavage (BAL) clusters can be distinguished by gene expression (panels c and d), showing FABP4 and INHBA (FABP4hi macrophages, cluster 0); SPP1, MERTK, LGMN and SIGLEC10 (SPP1hi macrophages, cluster 1); or IL1B and FCN1 (FCN1hi monocyte/macrophages, cluster 3). Double immunoflorescent staining for MERTK, FABP4 and FCN1 encoded proteins in serial sections show mostly discrete staining for these three macrophage populations, all three co-stain for CD163. Scale bar=500 μm (panel e). BAL cells from a healthy subject clustered by t-SNE show discrete clusters of macrophages (panel f).
Normal control lungs showed three discrete populations of monocyte/macrophages, all of which expressed high levels of CD163 and AIF1 (Figures S1 and 1A, clusters 0, 1 and 3). The first group of macrophages (FABP4hi macrophages, Figures 1A–D, cluster 0) expressed high levels of FABP4 and INHBA, and relatively low levels of SPP1, MERTK, LGMN and SIGLEC10. A second group of macrophages (SPP1hi, Figure 1A–D, cluster 1) expressed relatively high levels of SPP1, as well as MERTK, LGMN and SIGLEC10 and relatively low levels of FABP4 and INHBA. A third macrophage population (FCN1hi monocyte/macrophages) expressed relatively high levels of FCN1, as well as several marker genes associated with monocytes: CD14, IL1B, INSIG1, OSM, IL1R2 and THBS1 [14], and low to no expression of FABP4, INHBA, MERTK and SPP1 (Figure 1A–D, cluster 3, and data not shown). Previously defined flow cytometry markers did not distinguish these subsets well (Figure S3). A fourth population of cells adjacent to the macrophage populations discretely expressed CD1C and therefore most likely represented dendritic cells (Figure 1A, S1 and S2; cluster 10). Re-clustering only the myeloid cells showed the same cell subsets (Figures S4 and S5).
Immunofluorescent (IF) staining of normal lungs showed that discrete cells co-stained for CD163, and either FABP4 or SPP1/osteopontin (not shown). Although SPP1-expressing cells were detected, staining of SPP1/osteopontin, a matricellular protein, also stained extracellular matrix. IF staining of MERTK better identified the SPP1hi subset of macrophages that was generally discrete from FABP4 staining of FABP4hi macrophages, although some overlap in protein expression of each of these markers was seen as was also seen a the level of MERTK and FABP4 gene expression (Figure 1E and S6). The staining for these markers was more discrete in IPF tissues, as described below. FCN1 staining showed smaller cells, representing FCN1hi macrophages. All three macrophages populations could be found in the interstitial space in normal lungs samples.
Normal alveolar macrophages are composed primarily of FABP4hi macrophages.
Since morphologic evaluation of IF staining of macrophages in normal lung tissues did not provide a clear localization of macrophage subsets between alveolar and interstitial spaces, we also analyzed cells from normal lung bronchoalveolar lavage (BAL, Figure 1F). The cell types identified by marker genes were all inflammatory cells, mostly macrophages, expressing CD163 and AIF1, but also discrete clusters that included both T cells (expressing CD3D) and NK cells (expressing KLRF1, GNLY, and NKG7, cluster 2), and B cells (expressing MS4A1/CD20, cluster 4; Figures S7 and S8). As seen in normal lung tissue three macrophage populations could be distinguished. Most of the macrophages showed gene expression features of FABP4hi macrophages, expressing relatively high levels of FABP4 and INHBA, and lower levels of SPP1, MERTK, SIGLEC10, LGMN, IL1B and FCN1 (Figure 1C and 1D, cluster 0). However, minor populations of SPP1hi and FCN1hi macrophages were also seen in BAL (Figure 1C D and F-, clusters 1 and 3). Clustering of the macrophages from this healthy BAL with macrophages from a second normal BAL with brushing showed essentially the same results with a predominance of FABP4hi macrophages (Figure S9 and S10).
Consistent with these observations, when the BAL cells were analyzed with the normal lungs cells by t-SNE, most of the cells clustered with the FABP4hi macrophages (Figure S11). However, on this combined analysis, consistent with t-SNE clustering of the BAL cells alone, some of the BAL macrophages clustered with the SPP1hi and others with the FCN1hi macrophages (Figure S10 and S11). Thus, alveolar macrophages were composed primarily of FABP4hi macrophages, but all three macrophage populations, as well as lymphocyte subsets, were found in BAL.
Lung samples from patients with IPF show alterations in cellular composition compared to healthy controls.
In order to better understand the changes in populations and gene expression of cell types associated with the development of IPF, we analyzed freshly digested lung explants from three patients with IPF, and three healthy controls showing the least changes on pathology and analyzed using the same chemistry (V2 10X Genomics) by scRNA-seq (Table 1 and Table S1). Since fibrotic changes in IPF lungs typically are first evident at the lung bases and the disease then progresses apically, we analyzed samples from both the right upper and lower lobes to capture disease at different points in evolution. Lower lobe pathology showed classical changes associated with IPF, including honeycomb cysts and fibroblastic foci (Figure 2A), as well as smooth muscle actin staining myofibroblasts and collagen deposition (Figures S12A and S12B). Two of the three IPF upper lobes showed markedly less fibrosis, and similar or more inflammation.
Table 1.
Lung sample analysed by single-cell RNA sequencing, demographics and associated review of pathology
| Sample ID | Single Cell ID | Tissue Type | Sex | Age Years | Number of Cells Analyzed | Pathologic Assessment of Adjacent Lung Tissue |
|---|---|---|---|---|---|---|
| 2017-025-NOR | SC56 | Control Lung | FEMALE | 57 | 4334 | Good normal. No evidence of abnormal tissue |
| 2017-029-NOR | SC59 | Control Lung | MALE | 18 | 3155 | Healthy tissue a bit of bronchitis likely due to vent. Some of the small airways are prominent and more usclular than expecated which may indicate this patients had allergies or some asthma. |
| 2017-105-NOR | SC155 | Control Lung | FEMALE | 23 | 4122 | Normal healthy lung |
| SC156 | 5620 | |||||
| 2017-064-IPF | SC87 | IPF Lung Lower Lobe | FEMALE | 70 | 10479 | UIP: acute exacerbation of UIP, diffuse alveolar damage and inflammation |
| SC88 | IPF Lung Upper Lobe | 4717 | Mostly UIP; more diffuse than lower lobe. Some mixed fibrotic and cellular NSIP (acute). Some honeycomb cysts. | |||
| 2017-067-IPF | SC93 | IPF Lung Lower Lobe | MALE | 69 | 5920 | Classic end stage lung. Honeycomb cysts, bronchial metaplastic cells, some smooth muscle metaplasia. Pathologists may call this “honeycomb lung.” |
| SC94 | IPF Lung Upper Lobe | 2146 | Scarring around the airways, smoker, much better than the lower lobe. Some of the scarring may be from previous acute exacerbations. No UIP or fibrosis, maybe some COPD. | |||
| 2017-100-IPF | SC153 | IPF Lung Lower Lobe | FEMALE | 69 | 3259 | complete replacement of the lung architecture by scar tissue with honeycomb change with end stage lung disease. |
| SC154 | IPF Lung Upper Lobe | 4019 | complete replacement of the lung architecture by scar tissue with honeycomb change with end stage lung disease. |
IPF: idiopathic pulmonary fibrosis; UIP: usual interstitial pneumonia; NSIP: nonspecific interstitial pneumonia; COPD: chronic obstructive pulmonary disease.
Figure 2.
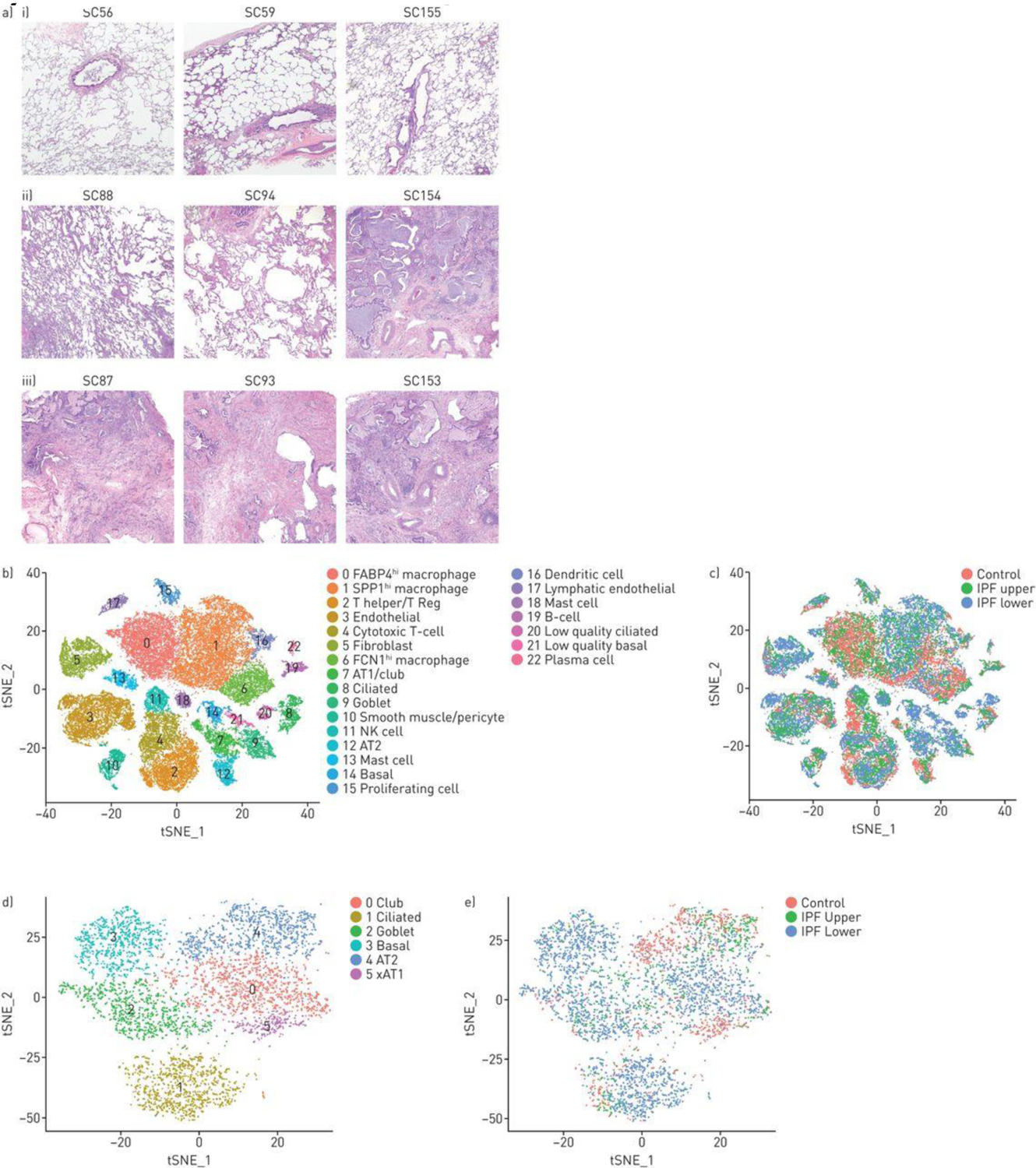
Combined t-distributed stochastic neighbour embedding (t-SNE) analysis of single-cell transcriptomes from three normal, three idiopathic pulmonary fibrosis (IPF) upper and three IPF lower lobes. a) Tissue sections of i) three control lung samples show preserved lung architecture with no significant pleural, subpleural or interstitial fibrosis and no increase in inflammatory cells. Tissue sections from patients with IPF showed iii) usual interstitial pneumonia lower lobes with a predominant cicatricial process and complete replacement of the lung architecture by scar tissue with honeycomb change, increased inflammatory cells and fibroblastic foci. ii) In comparison, the upper lobes showed relatively uninvolved lung tissue with preserved architecture and only mild interstitial organisation and mildly increased interstitial mononuclear cells suggestive of acute/subacute inflammation in samples SC88 and SC94 with more extensive fibrosis in SC154. Scale bar=1 mm. b) The t-SNE plot shows 20 clusters, with cell types identified by marker genes (supplementary figures S13 and S14). c,e) The tissue origin of the cells stratified by the type of tissue (control, IPF upper or IPF lower lobes) is indicated by different colouring of the cells. d) Reclustering of epithelial cells, showing the discrete clusters of club, ciliated (cluster 1) goblet and AT1 and AT2 cells, as identified by marker genes (supplementary figure S15).
Different cell types in normal and IPF lungs revealed by scRNA-seq.
Single cell transcriptomes of 47,771 cells, representing 17,231 cells from healthy lungs and 30,540 cells from IPF lungs from the different lung samples were analyzed together by t-SNE and colored by groups of cells (Figure 2B), showing 23 different cell clusters. Cells were also identified by the subject of origin to ensure that clusters represented cell types found in all samples (Figure S13). Cells were also identified by the disease status (normal, upper lobes and lower lobes) to show the change in patterns of cell types with disease (Figure 2C). Using previously described markers most clusters were identified as discrete cell types, including inflammatory, epithelial, vascular and mesenchymal cell types (see Supplemental Results and Figures S14–S16; Table S3). Epithelial cell types (cluster 7, 8, 9, 12, 14, 20, 21) were reanalyzed and low quality cells filtered, allowing for discrimination of each epithelial cell type (Figures 2D, 2E, S17).
Macrophages and dendritic cells identified by AIF1 and CD163 expression were found in five different clusters (Figures 2B, S15 and S16; clusters 0, 1, 6, 15 and 16). Cells in cluster 0 expressed FABP4 most highly and represented the same FABP4hi macrophage population described above in normal lungs. Cells in cluster 1 expressed SPP1 most highly and SPP1hi healthy control macrophages fell into this cluster, which also expressed the highest levels of MERTK, LGMN and SIGLEC10. Cells in cluster 6 expressed FCN1 most highly as well as CD14, IL1B, INSIG1, OSM, IL1R2 and THBS1 the other markers shown above to be associated with the FCN1hi monocyte/macrophage subset in normal lungs. Cluster 15 represented proliferating macrophages analyzed further below. Cluster 16 represented CD1C-expressing, dendritic cells, again as also seen in normal lungs. Thus, this combined analysis recapitulated the macrophage/monocyte/dendritic cell subsets seen in healthy lungs.
Increased and decreased proportions of cell types changed in a graded fashion between normal, upper lobe IPF and lower lobe IPF.
The proportion of different cell populations changed in IPF compared to normal lungs. In almost all cases changes in lower lobes were more dramatic than changes in upper lobes, compared to normal lungs (Figure 3A and 3B). Several inflammatory cell populations decreased in this graded fashion, including FABP4hi and FCN1hi macrophages, and NK cells. In contrast, SPP1hi macrophages trended toward modestly increased proportion in IPF lower lobes. The proportion of T cells, B cells and plasma cells showed little difference between normal, IPF upper and IPF lower lobes.
Figure 3.
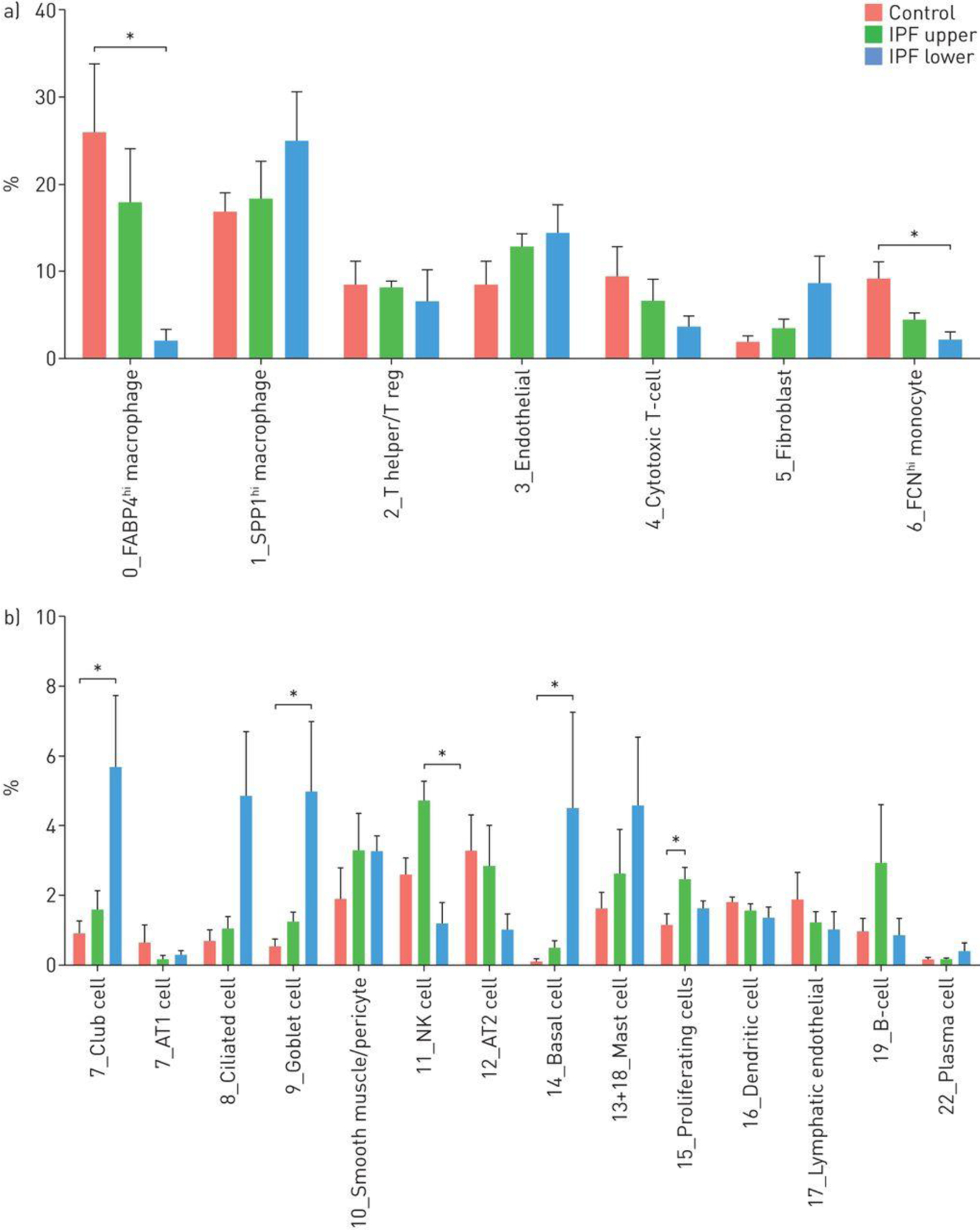
Percentages of cell types captured in control, upper idiopathic pulmonary fibrosis (IPF) and lower IPF lung samples. The average proportion of cell subgroups as a percentage of total cells analysed, in control, upper IPF lobes and lower IPF lobes are shown for each cluster as shown in figure 2a and subclusters as shown in figure 2d. *: p<0.05.
MUC5+(MUC5AC and MUC5B), FoxJ1+(FOXJ1) and p63+(TP63) expressing cells, which make up honeycomb cysts [19] associated with, respectively, goblet cells (cluster 9), ciliated cell (cluster 8) and basal cells (cluster 14), as well as club cells were all highly increased in IPF lungs (Figure 3B). Consistent with their marked expansion, we detected increased numbers of cells with club and basal cell markers that co-expressed markers of cell proliferation (Table S4). AGER-expressing AT1 and SFTPC-expressing AT2 cells declined in fibrotic lower lobes though not reaching statistical significance (Figure 3B), and we did not detect proliferating AT2 cells, though this might represent the relative paucity of AT2 cells surviving digestion [20]. We did see rare bipolar cells described to have makers of both AT1 and AT2 cells in normal or IPF lungs (Figure S18), however there were no distinctive markers for these cells and thus we are uncertain whether these represent cellular transitions or doublet cells.
The proportion of alveolar and bronchiolar epithelial cells in normal lungs appeared lower than anticipated. Overall gene expression between bulk tissue RNA-seq, RNA-seq of digested cells and combined scRNA-seq gene expression correlated well between these three groups (data not shown). However, expression of selected marker genes of cells after digestion indicated a selective loss of mesenchymal and alveolar epithelial cells compared to inflammatory cells (Table S5). This skewing of cell types associated with cell survival during digestion was consistent between two control and two IPF samples analyzed in this manner, indicating that although the absolute proportions of cells represented reflect the loss, the relative changes between samples are accurate.
Our results are generally consistent with a previous report (Figure S19) in which EPCAM purified cells in normal lungs were mainly AT2 cells [21]. Ciliated, goblet, club and basal cells showed large increases in proportion of total cells in IPF. Fibroblasts, representing only 2.0% of cell in normal lungs increased to 9.0% of cells in fibrotic lower lobe. Pericytes, endothelial cells and lymphatic endothelial cells showed little difference in cells numbers comparing IPF to control samples (Figure 3A and 3B).
Macrophage subpopulations in IPF lungs.
IPF upper lobes showed relatively few SPP1hi, macrophages, and many FABP4hi macrophages, similar to control lungs (Figures 4A and 4B). Macrophages expressing high levels of SPP1 and FABP4 were in large part mutually exclusive (Figure 4A, 4C). IF staining showed the same pattern of expression with mainly FABP4-staining cells in the upper lobes, low level SPP1-staining and few MERTK-staining cells (Figure 4D and S21). IPF lower lobes showed higher numbers of SPP1-expressing cells compared to control or upper lobe IPF (Figures 4A, 4B and S21). IF staining of IPF lower lobes showed much brighter diffuse staining for SPP1 as well as cells embedded in the SPP1 matrix, many MERTK-staining cells, and relatively few FABP4-staining cells (Figure 4C, S21). Staining of serial sections of lower lobes confirmed that the three macrophage populations were distinct and co-stained with CD163. (Figure S22).
Figure 4.
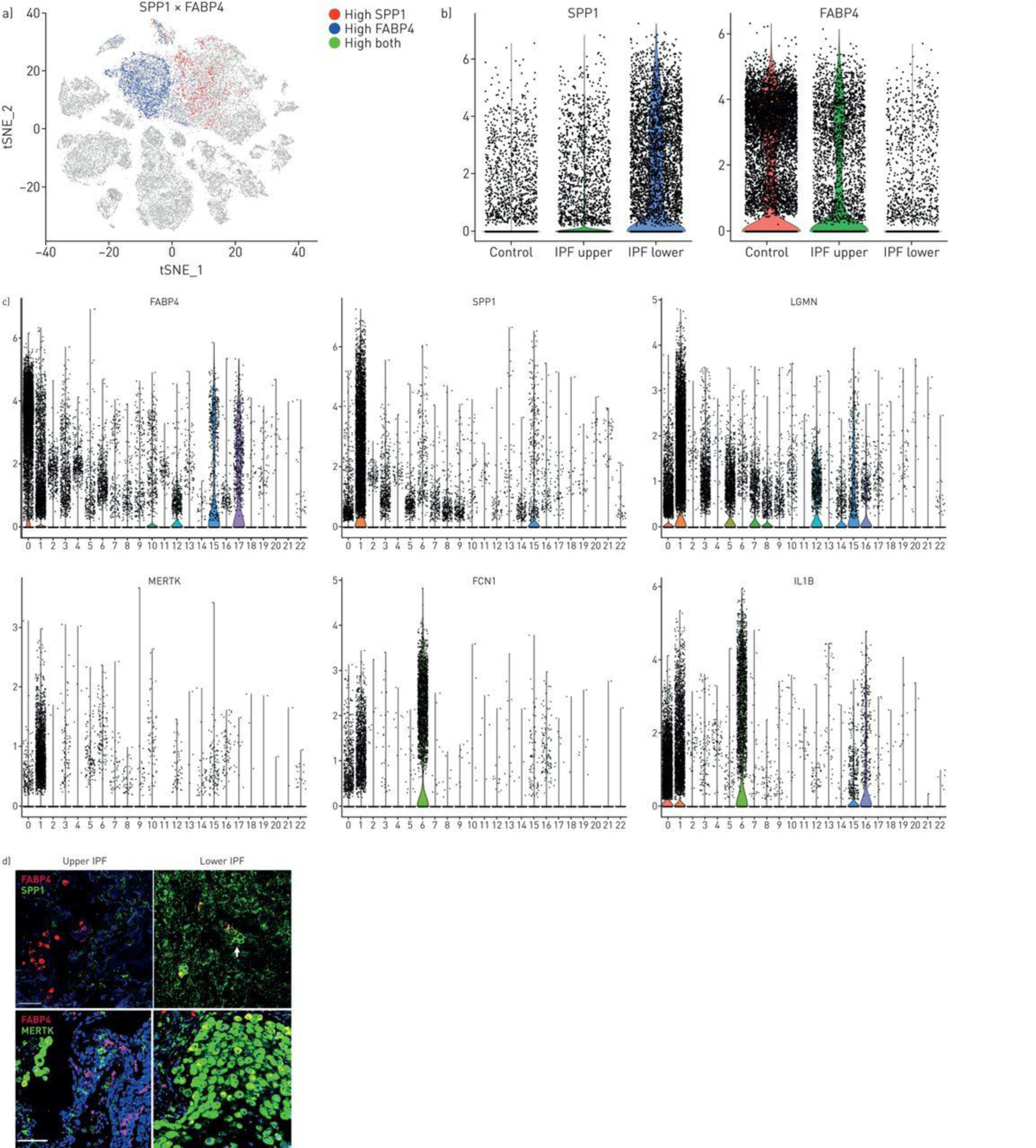
Macrophage populations in idiopathic pulmonary fibrosis (IPF) lungs. a) SPP1 and FABP4 expression define two discrete populations in IPF and normal lungs. b) (and supplementary figure S18) SPP1hi macrophages express more SPP1 and make up a higher percentage of cells in lower lobes, while FABP4hi macrophages are a higher percentage of the cells in healthy and IPF upper lobes. c) Violin plots of combined IPF/control lung data show expression of FABP4, SPP1, LGMN, MERTK, FCN1 and IL1B limited mainly to macrophage populations (clusters 0, FABP4hi macrophages; cluster 1, SPP1hi macrophages and cluster 6, FCN1hi monocyte/macrophages; figure 2). d) (and supplementary figure S19) FABP4 expression is increased in FABP4hi macrophages, although it is also expressed at lower levels in SPP1hi macrophages; SPP1, MERTK and LGMN show highly increased expression in SPP1hi macrophages; FCN1 and IL1B are most highly expressed in FCN1hi monocyte/macrophages. Immunofluorescent staining for SPP1 shows macrophages embedded in the SPP1/osteopontin matrix; while staining for MERTK and FABP4 reveals these largely two discrete macrophage populations with increased FABP4-staining macrophages in upper lobes and increased MERTK macrophages in lower lobes. Scale bar=100 μm.
SPP1 stained much more highly in IPF lower lobe than upper lobe, staining most intensely surrounding and within fibroblastic foci (Figure 5A, S23). MERTK discretely stained macrophages surrounding and within fibroblastic foci.
Figure 5.
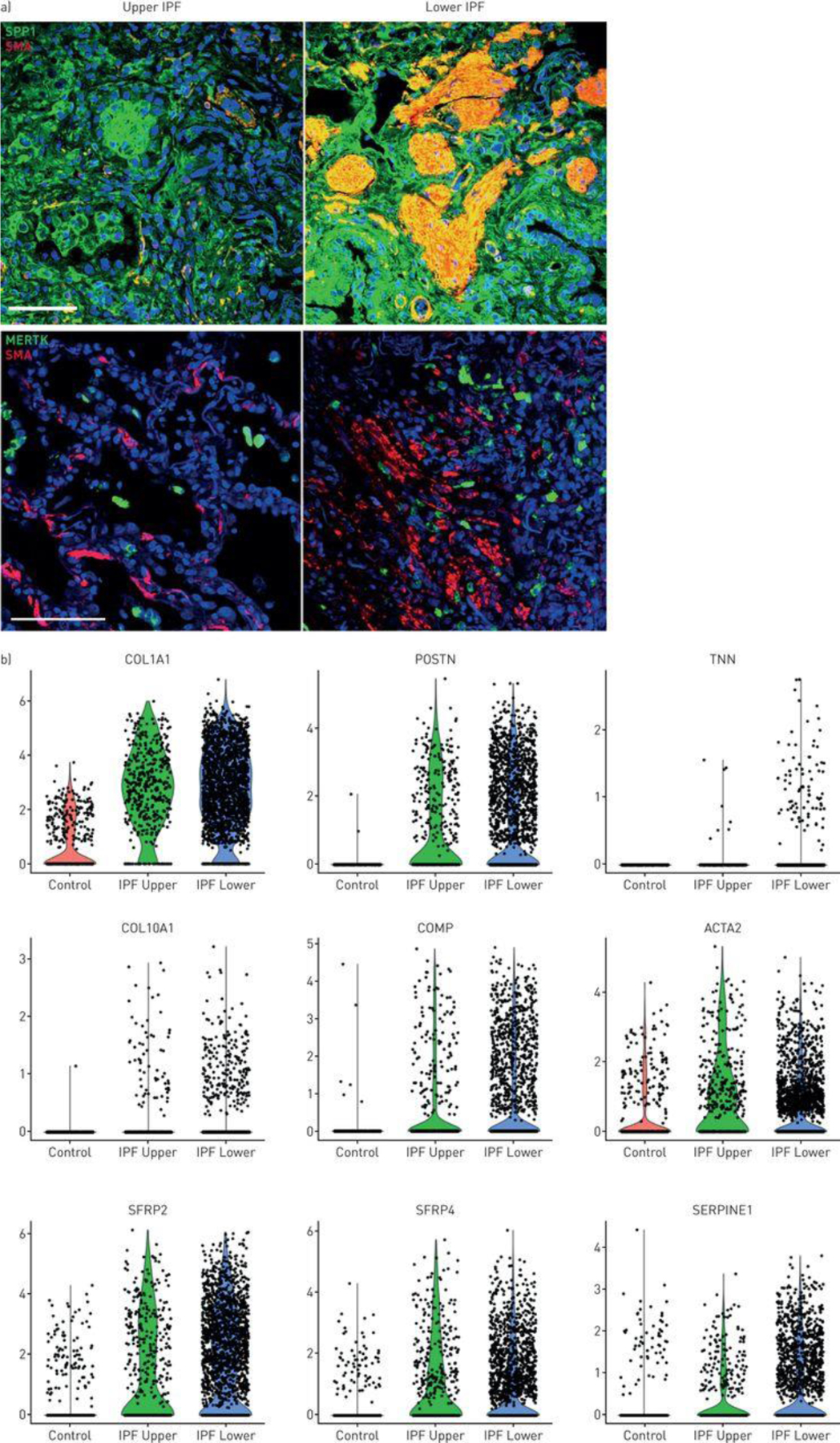
SPP1 macrophages and myofibroblasts in idiopathic pulmonary fibrosis (IPF) lungs. a) Fibroblastic foci in IPF lungs were stained with α-smooth muscle actin (SMA; red). Macrophages were stained with SPP1 or MERTK as indicated (green). b) Violin plots, indicating expression of several genes associated with fibrosis in control, IPF upper and IPF lower lungs.
Gene regulation in IPF fibroblasts.
Altered average gene expression was associated with each cluster in IPF upper and lower lobes and healthy lungs (Tables S6A and S6B; see: ERJ website or https://dom.pitt.edu/rheum/centers-institutes/scleroderma/systemicsclerosiscenter/database/). IPF fibroblasts showed many highly upregulated matrix genes, including TNN (no denominator, nd), COL10A1 (612-fold), COMP (17-fold), POSTN (470-fold), COL1A1 (170-fold). Wnt-related genes that are expressed by dermal fibroblast subtypes [22] were also upregulated, SFRP4 (5.1-fold) and SFRP2 (8.8-fold) (Figure 5B).
Genes upregulated in SPP1 macrophages in IPF lungs.
To highlight genes regulated by SPP1 macrophages in IPF patients, we ranked genes by fold-change between IPF and normal SPP1hi macrophages. To limit detection of ambient RNA, we selected genes that were more highly expressed by lower lobe IPF SPP1hi macrophages than any other cell type, and low level expressed genes (expressed at an average level of 0.01 or less by SPP1hi lower lobe macrophages) were excluded (Table S7). The most highly upregulated genes in lower lobe IPF compared to control lungs included LEP (nd), KCNJ5 (15.76-fold), HS3ST2 (15.58-fold), SPP1 (7.56-fold), SIGLEC15 (7.43-fold), ATP6V0D2 (7.12-fold), LGMN (6.07-fold), MERTK (4.96-fold) and MMP9 (3.81-fold). LEP, HS3ST2, SPP1, SIGLEC15, LGMN, MERTK, LGMN and MMP9 were most specifically upregulated in IPF SPP1hi and proliferating macrophages (Figure S24). Many other genes that were highly upregulated in SPP1hi IPF macrophages showed a graded increase in gene expression comparing control macrophages to upper lobe IPF macrophages to lower lobe IPF macrophages, suggesting a continuum in differentiation of these cells throughout disease progression (Table S8)
We examined IL4, IL13, MRC1 and TMG2 expression, the latter two genes markers of human M2 macrophages [23, 24]. Although expressed at very low levels, IL4 was expressed and upregulated mainly T cells in IPF compared to controls, whereas IL13 expression was down regulated in mast cells (Figure S25). TGM2 was modestly upregulated in all IPF macrophage subsets, though much more highly expressed in non-macrophage cell types. MRC1 was modestly down-regulated in all macrophage cell types.
Proliferating cells in IPF lungs.
We found in cluster 15 from Control/IPF lung clustering (Figure 2B) cells expressing highest levels of G2/M markers and specific markers of cell proliferation, including MKI67 (i.e. Ki67), KIAA0101/PCLAF (PCNA-associated factor), BIRC5 (Survivin) and UBE2C/UBCH10 Ubiquitin-conjugating enzyme E2 C (Figure 6B, 6D). Both FAPB4hi and SPP1hi macrophages were proliferating. More FAPB4hi macrophages were proliferating in the IPF upper lobe, but when adjusted for the paucity of these cells in lower lobes, the percent of proliferating cells was similar (10.59% compared to 16.95%, Table S4). SPP1hi macrophages were rarely proliferating in healthy lungs (0.072%) but showed dramatically increased proliferation in the IPF upper (2.01% of SPP1hi macrophages) and lower lobes (3.50% of SPP1hi macrophages; Figure 6C, 6E and Table S4), representing the most common proliferating macrophage in IPF lower lobes (2.8% compared to 1.7% and 0.3% proliferating FABP4 and FCN1, respectively, of total macrophages, Table S4). Epithelial cells were also proliferating. Low numbers of proliferating AT1, AT2, club and ciliated cells did not clearly trend toward altered rates of proliferation in IPF (Table S4, Figures S26 and S27). However KRT5+ basal cells showed a clear trend toward increased proliferation with control, IPF upper and IPF lower lungs showing, respectively, 0 (0%), 1 (1.49%) and 33 (4.64%) of basal cells proliferating. These cells also showed highly upregulated expression of TP63 (p63, Figure S28).
Figure 6.
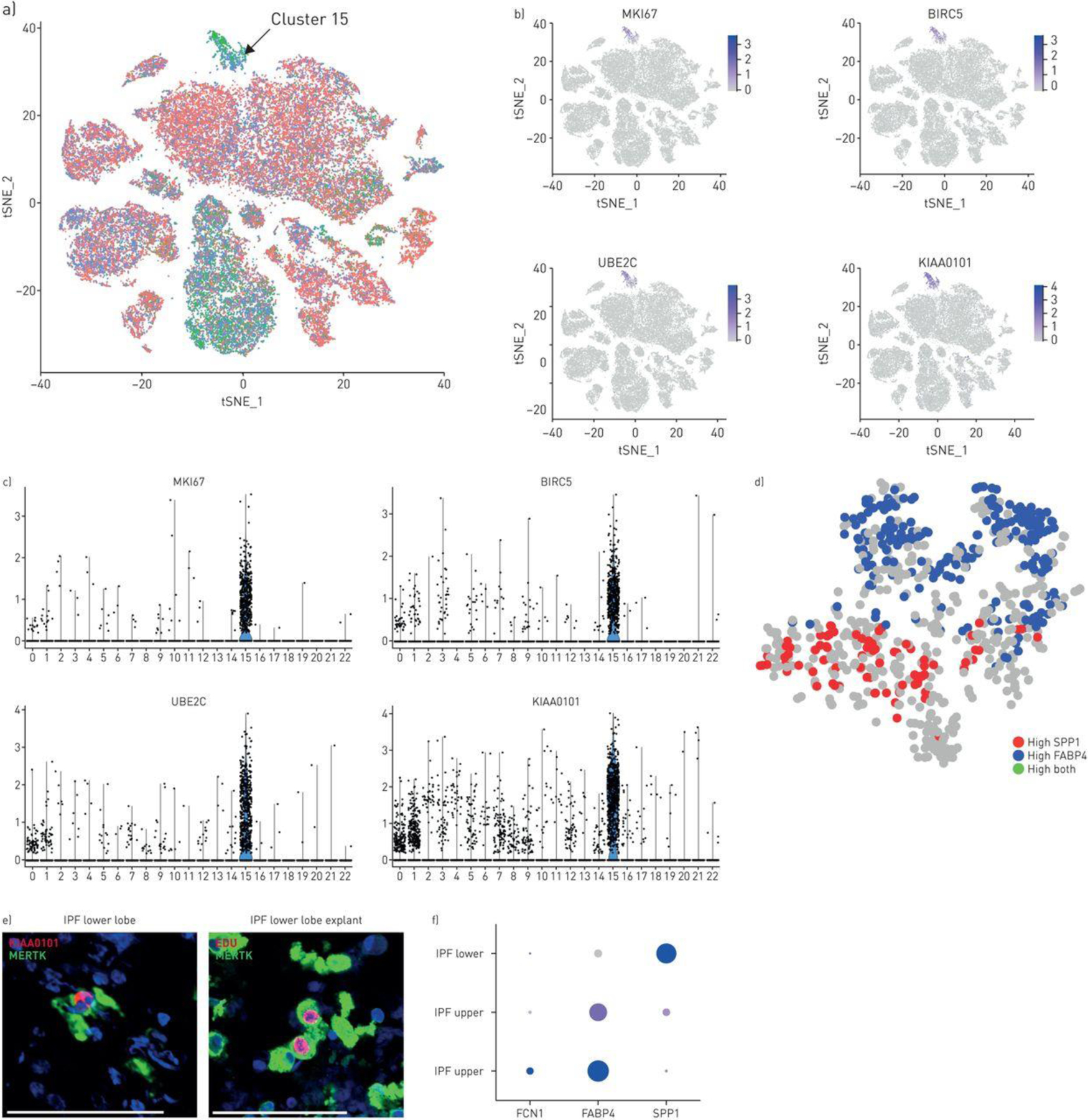
Proliferating cells in idiopathic pulmonary fibrosis (IPF) and normal lungs. a) Proliferating cells from control and IPF lungs form a distinct cluster (cluster 15, green G2/M phase cells marked by arrow; see also figure 2b, cluster 15). b, c) Cluster 15 cells highly express MKI67 (Ki-67), BIRC5, UBE2C and KIAA0101. d) Reclustering of cluster 15 cells shows macrophages expressing either FABP4 or SPP1. Using KIAA as a marker for proliferating cells, both FABP4 and SPP1hi cells are proliferating in this cluster. e) MERTK-expressing macrophages from IPF lower lobe lung explant co-stained with MERTK and proliferation marker KIAA0101. Macrophages labelled in vitro by incubation with EdU (red), co-stained by immunofluorescent with MERTK (green). f) Examining gene expression by only the proliferating macrophage subset shows low level proliferation of FCN1/(IL1B-expressing) macrophages in control lungs; primarily proliferation of FCN1/(IL1B-expressing) macrophages and FABP4hi macrophages in IPF upper lobes and primarily proliferation of SPP1hi macrophages in IPF lower lobes (size of dot indicates the proportion of cells and intensity of purple the relative level of gene expression). Scale bar=100 μm.
Proliferating cells were identified by KIAA0101/PCLAF staining by IF. KIAA0101/PCLAF cells co-expressed SPP1 in lower lobes and FABP4 in the upper lobes (Figure 6F, S14B). Proliferating IPF cells in explant culture, incorporating EdU (5-ethynyl-2’-deoxyuridine), co-stained with CD163, confirming proliferating macrophages (Figure 6F). Expression of CSF1, whose gene product CSF-1 is known to stimulate monocyte/macrophage proliferation, was strongly upregulated in IPF mast cells (Figure S25).
Connectivity maps of cell subtypes.
We built a graphical model to observe the direct relationships between differentially expressed genes for SPP1/MERTK macrophages, fibroblasts, and various epithelial cell types. The resulting network showed the most densely connected gene groups were between SPP1 macrophages and fibroblasts, suggesting a causal relationship between these two cell types (Figure S29, Table S9 and see Supplemental results).
We also examined gene expression associated with each of the macrophage populations. Gene Ontology (GO) analysis. FABP4hi macrophages expressed genes involved in lipid metabolism more highly, SPP1hi macrophages express genes involved in the stress response more highly; and FCN1hi macrophages express genes associated with the immune response more highly (Table S10). Additionally we compared GO terms activated in IPF compared to control IPF macrophages, showing upregulated expression of genes associated with extracellular matrix organization
Discussion
SPP1/osteopontin is a consistently observed marker of IPF [25]. Our results show that SPP1 is a highly selective marker for an expanded subpopulation of macrophages found in human IPF. SPP1 is selectively expressed by a subgroup of macrophages found in normal healthy lungs but seen rarely in BAL, indicating that they are part of the interstitial macrophage compartment. In contrast to another recent description of scRNA-seq data in IPF [26], our data indicate that SPP1hi macrophages do not arise as a new macrophage population in IPF, but are present in normal lungs as well. In our reclustering of the data from this previous study [26], we found similar populations of SPP1hi, FABP4hi and FCN1hi macrophages in normal lungs, indicating that SPP1hi cells in IPF likely arise from a macrophage population already present in normal lungs. However, SPP1 expression increased dramatically in this macrophage subset in IPF, particularly in the fibrotic IPF lower lobe. The SPP1 gene product, osteopontin, was strikingly deposited in fibrotic IPF lower lobes, where it was associated with fibroblastic foci. Osteopontin also supports monocyte/macrophage proliferation [27], suggesting that macrophage secretion of this matrix protein might support SPP1 macrophage proliferation in IPF. Deletion of SPP1 in bleomycin-induced lung fibrosis reduces upregulated expression of collagen type 1 and MMP2 [28, 29]. SPP1 deletion also ameliorates bleomycin-induced dermal fibrosis and carbon nanotube-induced lung fibrosis [30, 31]. Osteopontin is also increased in the serum of patients with systemic sclerosis [30], and has been implicated in renal cardiac and bone marrow fibrosis, suggesting that SPP1 macrophages may have a more general role in promoting fibrosis [32–35]. We propose that SPP1hi macrophages represent a profibrotic macrophage population in IPF lungs.
Highly increased co-expression of SPP1 and MERTK by SPP1hi macrophages may be key to a role for these cells in IPF tissue repair and fibrosis. MERTK is a receptor for complexes of Gas6 or Protein S bound to phosphatidylserine, exposed on apoptotic cells [36, 37], and the main apoptotic cell receptor on macrophages [38, 39]. Alveolar cell apoptosis is a feature of IPF [40], and blocking Gas6 inhibits markers of fibroblast activation [41]. Macrophage efferocytosis suppresses inflammation, in part through upregulated TGFβ, pGE2 and PAF [42]. In addition, MERTK engagement by apoptotic cells stimulates macrophage production of pro-resolving lipid mediators [43]. Supporting the notion that SPP1hi macrophages are reparative macrophages, MERTK macrophages aid tissue repair after cardiac or liver injury [44, 45]. As inhibiting MERTK leads to apoptosis of myeloid cells [46], MERTK inhibitors under clinical development [46–48] might deplete profibrotic macrophage and fibroblast activation in IPF lungs. In the central nervous system microglial cells not only clear apoptotic cells, but also target live, damaged cells [49], referred to as phagoptosis [50, 51]. Thus, macrophages expressing increased MERTK might phagocytose live, damaged or senescent alveolar epithelial cells.
FABP4hi macrophages represented a second subset of lung macrophages, making up most of the macrophages in alveoli showed dramatically decreased numbers in IPF in lower lobes. FABP4 expression by macrophages induces inflammation associated with obesity and atherosclerosis [52, 53]. Deletion of macrophage FABP4 is associated with increased intracellular fatty acid and increased unfolded protein response [54]. In other studies FABP4 expression in macrophages is associated with pro-inflammatory macrophages and IL1β secretion [55].
FCN1hi macrophages represented a third population of macrophages with marker closely related to monocytes. Although these could include true intravascular monocytes, they were not consistently seen in blood vessels in IHC; the normal healthy lungs were always on ex vivo lung perfusion prior to harvest, likely washing out most or all blood associated monocytes; and a small fraction were captured in BAL. Thus, they likely represent a third macrophage subpopulation found primarily in the interstitial compartment. Complementary expression of several marker genes suggests that SPP1hi, FABP4hi and FCN1hi macrophages represent discrete macrophage subsets.
Both FAPB4hi and SPP1hi macrophage subsets increased their rate of proliferation in IPF lungs, SPP1hi macrophages particularly in fibrotic lower lobes. FCN1hi monocyte/macrophages showed lower rates of proliferation that were relatively constant between healthy and IPF lungs (~2%). Observed basal proliferation of both FCN1hi and FABP4hi macrophages supports some degree of self-renewal of these populations in healthy lungs.
Tissue resident macrophages self-renew under the influence of M-CSF and/or GM-CSF [15, 16, 18, 56]. We found increased expression of CSF1 in lung mast cells, suggesting this might contribute to increased macrophage proliferation in IPF. IL-4 also stimulates resident macrophage proliferation, suggesting proliferation might be a feature of M2 macrophages [17]. IL4/IL13 have been implicated in murine lung fibrosis [57, 58]. We saw increased IL4 mRNA expression by IPF T cells, though low-level expression of IL4 and IL13 make these results uncertain. In IPF lungs, SPP1hi macrophages upregulated DC-SIGN (CD209), LGMN and CHI3L1, all regulated by IL-4 and/or IL-13 [59–62]. Together these results are consistent with possible CSF-1, IL-4 and/or IL-13 induced local macrophage proliferation.
Despite increased proliferation of FABP4hi macrophages, the total percentage of these cells decreased particularly in IPF lower lobes. Thus, proliferating FAPB4hi macrophages appear to be either dying or changing phenotype. We speculate that FAPB4hi macrophages, which are the primary cell type in alveoli, might die in fibrotic IPF lower lobes. Alternatively these cells might transition into SPP1hi macrophages in IPF lungs. In summary, our data show striking changes in cell populations in IPF that include marked increases in local proliferation of two populations of macrophages, one of which is highly associated with IPF diseased lungs.
Supplementary Material
Take Home Message.
By single cell RNA-sequencing we identify three discrete pulmonary macrophage subsets, including one expressing highly upregulated SPP1 and proliferating in fibrotic IPF lower lobes, accompanied by marked deposition of osteopontin in the matrix.
Acknowledgements.
The authors acknowledge the assistance of William Horne for helpful discussions and technical support for RNA-sequencing.
Support:
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases under award number 2P50 AR060780; National Institute for Heart, Lung and Blood INstitue under award number R01 HL123766 and U01 HL137159; and National Librarbay of Medicine under award R01 LM012087 of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Blackwell TS, Tager AM, Borok Z, Moore BB, Schwartz DA, Anstrom KJ, Bar-Joseph Z, Bitterman P, Blackburn MR, Bradford W, Brown KK, Chapman HA, Collard HR, Cosgrove GP, Deterding R, Doyle R, Flaherty KR, Garcia CK, Hagood JS, Henke CA, Herzog E, Hogaboam CM, Horowitz JC, King TE Jr., Loyd JE, Lawson WE, Marsh CB, Noble PW, Noth I, Sheppard D, Olsson J, Ortiz LA, O’Riordan TG, Oury TD, Raghu G, Roman J, Sime PJ, Sisson TH, Tschumperlin D, Violette SM, Weaver TE, Wells RG, White ES, Kaminski N, Martinez FJ, Wynn TA, Thannickal VJ, Eu JP. Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. American journal of respiratory and critical care medicine 2014: 189(2): 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sisson TH, Mendez M, Choi K, Subbotina N, Courey A, Cunningham A, Dave A, Engelhardt JF, Liu X, White ES, Thannickal VJ, Moore BB, Christensen PJ, Simon RH. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. American journal of respiratory and critical care medicine 2010: 181(3): 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoz DF, Lawson WE, Blackwell TS. Idiopathic pulmonary fibrosis: a disorder of epithelial cell dysfunction. The American journal of the medical sciences 2011: 341(6): 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans CM, Fingerlin TE, Schwarz MI, Lynch D, Kurche J, Warg L, Yang IV, Schwartz DA. Idiopathic Pulmonary Fibrosis: A Genetic Disease That Involves Mucociliary Dysfunction of the Peripheral Airways. Physiological reviews 2016: 96(4): 1567–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell stem cell 2011: 8(6): 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America 2009: 106(31): 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 2015: 517(7536): 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni T, O’Reilly P, Antony VB, Gaggar A, Thannickal VJ. Matrix Remodeling in Pulmonary Fibrosis and Emphysema. American journal of respiratory cell and molecular biology 2016: 54(6): 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuma T, Terasaki Y, Kaikita K, Kobayashi H, Kuziel WA, Kawasuji M, Takeya M. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. The Journal of pathology 2004: 204(5): 594–604. [DOI] [PubMed] [Google Scholar]
- 10.McCubbrey AL, Barthel L, Mohning MP, Redente EF, Mould KJ, Thomas SM, Leach SM, Danhorn T, Gibbings SL, Jakubzick CV, Henson PM, Janssen WJ. Deletion of c-FLIP from CD11b(hi) Macrophages Prevents Development of Bleomycin-induced Lung Fibrosis. American journal of respiratory cell and molecular biology 2017. [DOI] [PMC free article] [PubMed]
- 11.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen CI, Anekalla KR, Joshi N, Williams KJN, Abdala-Valencia H, Yacoub TJ, Chi M, Chiu S, Gonzalez-Gonzalez FJ, Gates K, Lam AP, Nicholson TT, Homan PJ, Soberanes S, Dominguez S, Morgan VK, Saber R, Shaffer A, Hinchcliff M, Marshall SA, Bharat A, Berdnikovs S, Bhorade SM, Bartom ET, Morimoto RI, Balch WE, Sznajder JI, Chandel NS, Mutlu GM, Jain M, Gottardi CJ, Singer BD, Ridge KM, Bagheri N, Shilatifard A, Budinger GRS, Perlman H. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. The Journal of experimental medicine 2017: 214(8): 2387–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. The Journal of experimental medicine 2013: 210(10): 1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012: 336(6077): 86–90. [DOI] [PubMed] [Google Scholar]
- 14.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, van Rooijen N, Grainger JR, Belkaid Y, Ma’ayan A, Riches DW, Yokoyama WM, Ginhoux F, Henson PM, Randolph GJ. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 2013: 39(3): 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, Garcia-Sastre A, Stanley ER, Ginhoux F, Frenette PS, Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013: 38(4): 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies LC, Rosas M, Smith PJ, Fraser DJ, Jones SA, Taylor PR. A quantifiable proliferative burst of tissue macrophages restores homeostatic macrophage populations after acute inflammation. European journal of immunology 2011: 41(8): 2155–2164. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 2011: 332(6035): 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nature immunology 2013: 14(10): 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, Schwarz MI, Schwartz DA, Reynolds SD. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PloS one 2013: 8(3): e58658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014: 509(7500): 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl AT, Funari VA, Gokey JJ, Stripp BR, Whitsett JA. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI insight 2016: 1(20): e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabib T, Morse C, Wang T, Chen W, Lafyatis R. SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. The Journal of investigative dermatology 2017. [DOI] [PMC free article] [PubMed]
- 23.Christmann RB, Hayes E, Pendergrass S, Padilla C, Farina G, Affandi AJ, Whitfield ML, Farber HW, Lafyatis R. Interferon and alternative activation of monocyte/macrophages in systemic sclerosis-associated pulmonary arterial hypertension. Arthritis and rheumatism 2011: 63(6): 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, Thomas B, Fabbri M, Crawshaw A, Ho LP, Ten Hacken NH, Cobos Jimenez V, Kootstra NA, Hamann J, Greaves DR, Locati M, Mantovani A, Gordon S. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood 2013: 121(9): e57–69. [DOI] [PubMed] [Google Scholar]
- 25.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, Kaminski N. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS medicine 2005: 2(9): e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, Verma R, Abdala-Valencia H, Nam K, Chi M, Han S, Gonzalez-Gonzalez FJ, Soberanes S, Watanabe S, Williams KJN, Flozak AS, Nicholson TT, Morgan VK, Winter DR, Hinchcliff M, Hrusch CL, Guzy RD, Bonham CA, Sperling AI, Bag R, Hamanaka RB, Mutlu GM, Yeldandi AV, Marshall SA, Shilatifard A, Amaral LAN, Perlman H, Sznajder JI, Argento AC, Gillespie CT, Dematte J, Jain M, Singer BD, Ridge KM, Lam AP, Bharat A, Bhorade SM, Gottardi CJ, Budinger GRS, Misharin AV. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. American journal of respiratory and critical care medicine 2018. [DOI] [PMC free article] [PubMed]
- 27.Tardelli M, Zeyda K, Moreno-Viedma V, Wanko B, Grun NG, Staffler G, Zeyda M, Stulnig TM. Osteopontin is a key player for local adipose tissue macrophage proliferation in obesity. Molecular metabolism 2016: 5(11): 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman JS, Serlin D, Li X, Whitley G, Hayes J, Rishikof DC, Ricupero DA, Liaw L, Goetschkes M, O’Regan AW. Altered bleomycin-induced lung fibrosis in osteopontin-deficient mice. American journal of physiology Lung cellular and molecular physiology 2004: 286(6): L1311–1318. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi F, Takahashi K, Okazaki T, Maeda K, Ienaga H, Maeda M, Kon S, Uede T, Fukuchi Y. Role of osteopontin in the pathogenesis of bleomycin-induced pulmonary fibrosis. American journal of respiratory cell and molecular biology 2001: 24(3): 264–271. [DOI] [PubMed] [Google Scholar]
- 30.Wu M, Schneider DJ, Mayes MD, Assassi S, Arnett FC, Tan FK, Blackburn MR, Agarwal SK. Osteopontin in systemic sclerosis and its role in dermal fibrosis. The Journal of investigative dermatology 2012: 132(6): 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong J, Ma Q. Osteopontin enhances multi-walled carbon nanotube-triggered lung fibrosis by promoting TGF-beta1 activation and myofibroblast differentiation. Particle and fibre toxicology 2017: 14(1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins AR, Schnee J, Wang W, Kim S, Fishbein MC, Bruemmer D, Law RE, Nicholas S, Ross RS, Hsueh WA. Osteopontin modulates angiotensin II-induced fibrosis in the intact murine heart. Journal of the American College of Cardiology 2004: 43(9): 1698–1705. [DOI] [PubMed] [Google Scholar]
- 33.Merszei J, Wu J, Torres L, Hicks JM, Bartkowiak T, Tan F, Lou YH. Osteopontin overproduction is associated with progression of glomerular fibrosis in a rat model of anti-glomerular basement membrane glomerulonephritis. American journal of nephrology 2010: 32(3): 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruberti S, Bianchi E, Guglielmelli P, Rontauroli S, Barbieri G, Tavernari L, Fanelli T, Norfo R, Pennucci V, Fattori GC, Mannarelli C, Bartalucci N, Mora B, Elli L, Avanzini MA, Rossi C, Salmoiraghi S, Zini R, Salati S, Prudente Z, Rosti V, Passamonti F, Rambaldi A, Ferrari S, Tagliafico E, Vannucchi AM, Manfredini R. Involvement of MAF/SPP1 axis in the development of bone marrow fibrosis in PMF patients. Leukemia 2017. [DOI] [PMC free article] [PubMed]
- 35.Zahradka P Novel role for osteopontin in cardiac fibrosis. Circulation research 2008: 102(3): 270–272. [DOI] [PubMed] [Google Scholar]
- 36.Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL, Earp HS, Matsushima GK. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 2001: 411(6834): 207–211. [DOI] [PubMed] [Google Scholar]
- 37.van der Meer JH, van der Poll T, van ‘t Veer C. TAM receptors, Gas6, and protein S: roles in inflammation and hemostasis. Blood 2014: 123(16): 2460–2469. [DOI] [PubMed] [Google Scholar]
- 38.Sather S, Kenyon KD, Lefkowitz JB, Liang X, Varnum BC, Henson PM, Graham DK. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 2007: 109(3): 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. Journal of immunology 2007: 178(9): 5635–5642. [DOI] [PubMed] [Google Scholar]
- 40.Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proceedings of the American Thoracic Society 2006: 3(4): 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Espindola MS, Habiel DM, Narayanan R, Jones I, Coelho AL, Murray LA, Jiang D, Noble PW, Hogaboam CM. Targeting of TAM Receptors Ameliorates Fibrotic Mechanisms in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine 2018: 197(11): 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. The Journal of clinical investigation 1998: 101(4): 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai B, Thorp EB, Doran AC, Subramanian M, Sansbury BE, Lin CS, Spite M, Fredman G, Tabas I. MerTK cleavage limits proresolving mediator biosynthesis and exacerbates tissue inflammation. Proceedings of the National Academy of Sciences of the United States of America 2016: 113(23): 6526–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeBerge M, Yeap XY, Dehn S, Zhang S, Grigoryeva L, Misener S, Procissi D, Zhou X, Lee DC, Muller WA, Luo X, Rothlin C, Tabas I, Thorp EB. MerTK Cleavage on Resident Cardiac Macrophages Compromises Repair After Myocardial Ischemia Reperfusion Injury. Circulation research 2017: 121(8): 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triantafyllou E, Pop OT, Possamai LA, Wilhelm A, Liaskou E, Singanayagam A, Bernsmeier C, Khamri W, Petts G, Dargue R, Davies SP, Tickle J, Yuksel M, Patel VC, Abeles RD, Stamataki Z, Curbishley SM, Ma Y, Wilson ID, Coen M, Woollard KJ, Quaglia A, Wendon J, Thursz MR, Adams DH, Weston CJ, Antoniades CG. MerTK expressing hepatic macrophages promote the resolution of inflammation in acute liver failure. Gut 2017. [DOI] [PMC free article] [PubMed]
- 46.Koda Y, Itoh M, Tohda S. Effects of MERTK Inhibitors UNC569 and UNC1062 on the Growth of Acute Myeloid Leukaemia Cells. Anticancer research 2018: 38(1): 199–204. [DOI] [PubMed] [Google Scholar]
- 47.Wu J, Frady LN, Bash RE, Cohen SM, Schorzman AN, Su YT, Irvin DM, Zamboni WC, Wang X, Frye SV, Ewend MG, Sulman EP, Gilbert MR, Earp HS, Miller CR. MerTK as a therapeutic target in glioblastoma. Neuro-oncology 2017. [DOI] [PMC free article] [PubMed]
- 48.Yi JH, Jang J, Cho J, Do IG, Hong M, Kim ST, Kim KM, Lee S, Park SH, Park JO, Park YS, Kang WK, Lim HY, Lee J. MerTK is a novel therapeutic target in gastric cancer. Oncotarget 2017: 8(57): 96656–96667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fourgeaud L, Traves PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagorska A, Rothlin CV, Nimmerjahn A, Lemke G. TAM receptors regulate multiple features of microglial physiology. Nature 2016: 532(7598): 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown GC, Neher JJ. Eaten alive! Cell death by primary phagocytosis: ‘phagoptosis’. Trends in biochemical sciences 2012: 37(8): 325–332. [DOI] [PubMed] [Google Scholar]
- 51.Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nature reviews Neuroscience 2014: 15(4): 209–216. [DOI] [PubMed] [Google Scholar]
- 52.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nature medicine 2001: 7(6): 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hotamisligil GS, Bernlohr DA. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nature reviews Endocrinology 2015: 11(10): 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu H, Hertzel AV, Steen KA, Wang Q, Suttles J, Bernlohr DA. Uncoupling lipid metabolism from inflammation through fatty acid binding protein-dependent expression of UCP2. Molecular and cellular biology 2015: 35(6): 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steen KA, Xu H, Bernlohr DA. FABP4/aP2 Regulates Macrophage Redox Signaling and Inflammasome Activation via Control of UCP2. Molecular and cellular biology 2017: 37(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonov AS, Munn DH, Kolodgie FD, Virmani R, Gerrity RG. Aortic endothelial cells regulate proliferation of human monocytes in vitro via a mechanism synergistic with macrophage colony-stimulating factor. Convergence at the cyclin E/p27(Kip1) regulatory checkpoint. The Journal of clinical investigation 1997: 99(12): 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huaux F, Liu T, McGarry B, Ullenbruch M, Phan SH. Dual roles of IL-4 in lung injury and fibrosis. Journal of immunology 2003: 170(4): 2083–2092. [DOI] [PubMed] [Google Scholar]
- 58.Lee CG, Homer RJ, Zhu Z, Lanone S, Wang X, Koteliansky V, Shipley JM, Gotwals P, Noble P, Chen Q, Senior RM, Elias JA. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1). The Journal of experimental medicine 2001: 194(6): 809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Relloso M, Puig-Kroger A, Pello OM, Rodriguez-Fernandez JL, de la Rosa G, Longo N, Navarro J, Munoz-Fernandez MA, Sanchez-Mateos P, Corbi AL. DC-SIGN (CD209) expression is IL-4 dependent and is negatively regulated by IFN, TGF-beta, and anti-inflammatory agents. Journal of immunology 2002: 168(6): 2634–2643. [DOI] [PubMed] [Google Scholar]
- 60.Soilleux EJ, Morris LS, Leslie G, Chehimi J, Luo Q, Levroney E, Trowsdale J, Montaner LJ, Doms RW, Weissman D, Coleman N, Lee B. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. Journal of leukocyte biology 2002: 71(3): 445–457. [PubMed] [Google Scholar]
- 61.Lee CG, Hartl D, Lee GR, Koller B, Matsuura H, Da Silva CA, Sohn MH, Cohn L, Homer RJ, Kozhich AA, Humbles A, Kearley J, Coyle A, Chupp G, Reed J, Flavell RA, Elias JA. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. The Journal of experimental medicine 2009: 206(5): 1149–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakazaki Y, Hoshino T, Takei S, Sawada M, Oda H, Takenaka S, Imaoka H, Matsunaga K, Ota T, Abe Y, Miki I, Fujimoto K, Kawayama T, Kato S, Aizawa H. Overexpression of chitinase 3-like 1/YKL-40 in lung-specific IL-18-transgenic mice, smokers and COPD. PloS one 2011: 6(9): e24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


