Abstract
Multiple primary lung cancer (MPLC) remains a tough challenge to diagnose and treat. Although neoadjuvant immunotherapy has shown promising results in early stage non-small cell lung cancer, whether such modality can benefit all primary lesions remains unclear. Herein, we performed integrated multiomics analysis in one patient with early stage MPLC with remarkable tumor shrinkage in a solid nodule and no response in two subsolid nodules after treatment with three cycles of neoadjuvant pembrolizumab. Genomic heterogeneity was observed among responding nodules with high levels of infiltrating CD8+ and CD68+ immune cells. Substantially downregulated human leukocyte antigen (HLA)-related genes and impaired T lymphocyte function were observed in non-responding nodules. A larger proportion of infiltrating tissue resident memory T cells (Trm) along with high T cell receptor repertoire clonality in responding nodules were validated as predictive and prognostic biomarkers in multiple cancer types using external public datasets. These results suggested that neoadjuvant programmed death 1 (PD-1)/programmed death ligand 1 inhibitors alone may not be an optimal therapeutic strategy for MPLC due to disparities in genomic alterations and immune microenvironment among different lesions. Additionally, we postulate that increased infiltration of Trm may be a unique marker of early immune responses to PD-1 blockade.
Keywords: lung neoplasms, immunotherapy, translational medical research, tumor biomarkers, tumor microenvironment
Background
Multiple primary lung cancer (MPLC) is a lung cancer subtype with a unique diagnosis and therapeutic strategy. Distinguishing a second primary lung cancer from intrapulmonary metastasis can be clinically challenging and thus, influences subsequent treatment.1 2 A previous study showed extremely low incidence of intrapulmonary metastasis among ground-glass or subsolid nodules.3 Although surgery is considered the optimal choice for MPLC treatment, the extent of resection and the potential of other local treatment approaches has not been fully determined.4–6 Immune checkpoint inhibitors blocking negative key regulator of T cells have shifted the management of multiple advanced cancers7 8; however, only a fraction of patients with advanced disease respond to immunotherapy aside from long-term benefits.9 10
Unlike advanced disease, early stage disease may harbor distinct tumor microenvironments. Immunoediting, a concept used to illustrate tumor immunogenicity over time,11 12 has demonstrated correlations between disease progression and metastasis.13 14 Initial disease, known as precancerous lesions, is accompanied by early immune sensing through the activation of resident immune cells and the activation of innate and adaptive immune responses.15 However, very low expression of programmed death 1/programmed death ligand 1 (PD-1/PD-L1) protein in either tumor cells or immune cells during precancerous and early stage disease can indicate the existence of other mechanisms of immune escape beyond PD-1/PD-L1 modulation.16 Conversely, relatively high PD-L1 expression has been observed in late-stage lung cancer.17 18 Since the development of neoadjuvant immunotherapy for early stage non-small cell lung cancer (NSCLC),19 multiple trials have been initiated to evaluate the efficacy of different checkpoint blockades and modalities and have shown encouraging preliminary results. However, no reported trials have assessed the feasibility of neoadjuvant immunotherapy in MPLC. Here, we report an integrated multiomics analysis in one patient with MPLC receiving three cycles of neoadjuvant pembrolizumab, and showing mixed response.
Case presentation
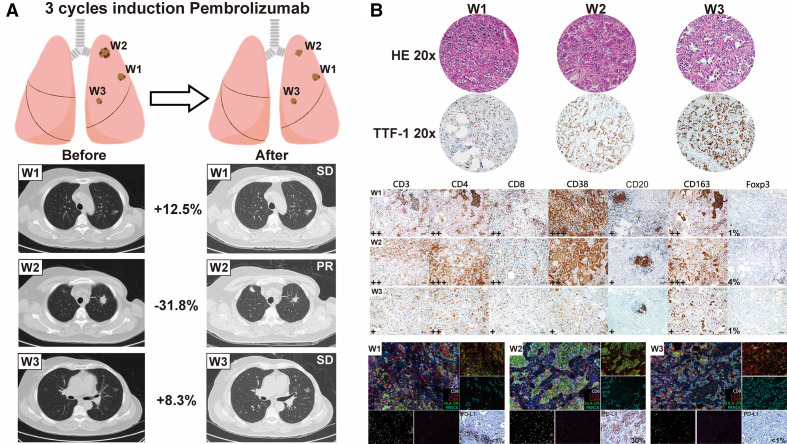
A woman aged 72 years admitted to outer hospital was accidentally found with multiple pulmonary nodules on a CT scan. One nodule in the left upper lobe was a solid lesion, whereas all others were mixed or pure ground-glass nodules. PET scan showed no distant metastasis or enlarged lymph nodes with increased uptake values. Percutaneous lung biopsy of the major solid nodule confirmed lung adenocarcinoma. Next-generation sequencing showed pan-negative results for all common driver mutations. Immunohistochemistry (IHC) using the Dako 22C3 assay revealed a PD-L1 expression level of 40%. However, due to the presence of bilateral pulmonary nodules, intrapulmonary metastasis and multiple primary lung cancer were indistinguishable. Given the negative results for driver mutations and the relatively high PD-L1 expression, the patient was prescribed the ‘first-line’ pembrolizumab, with informed consent. After three cycles (21 days/cycle) of pembrolizumab treatment, the patient was scheduled for radical treatment at our hospital. Chest CT showed significant shrinkage (31.8%) of the solid nodule; however, mixed and pure ground-glass nodules remained stable or slightly increased in size (figure 1A). Surgical resection of the major nodule and other nodules in the upper left lobe was considered due to the potential of early stage disease and radiological surveillance was recommended for mixed or pure ground-glass nodules in other lobes. IHC and multiplex immunohistochemistry (mIHC) revealed significantly high levels of infiltrating CD8+ lymphocytes and activated CD68+HLA-DR+ macrophages in responding nodules (W2). Notably, CD20 staining showed tertiary lymphoid structures (TLS) formation in all three nodules, which has been verified its potential of predicting immunotherapy efficacy.20 As expected, the extent of TLS observed in non-responding nodules (W1/W3) was smaller than that in W2 (figure 1B).
Figure 1.
Patient clinical and pathological evaluation following neoadjuvant immunotherapy. (A) Radiological evaluation of three resected nodules after three cycles of neoadjuvant immunotherapy showing notable tumor shrinkage of the solid nodule; however, the other nodules remained stable. (B) Comprehensive pathological evaluation integrated with mIHC of three surgically resected nodules. The extent of staining and the percentage of Foxp3-positive cells with nucleus staining are listed on the lower left corner of each slide. mIHC and PD-L1 images using the Dako 22C3 assay of nodules are obtained. mIHC panels are displayed in color on the lower right corner of the IHC slides: CD8 (white), CD56 (purple), CD68 (red), HLA-DR (green); PANCK (cyan). The expression level of PD-L1 is listed on the lower right corner of IHC slides. CNV, copy number variation; HLA, human leukocyte antigen; IC, immune cell: mIHC, multiplex immunohistochemistry; PD-L1, programmed death ligand 1; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors; SD, stable disease; TMB, tumor mutation burden; TTF-1, thyroid transcription factor 1.
Heterogeneous genomic features and T cell receptor repertoire profile among nodules
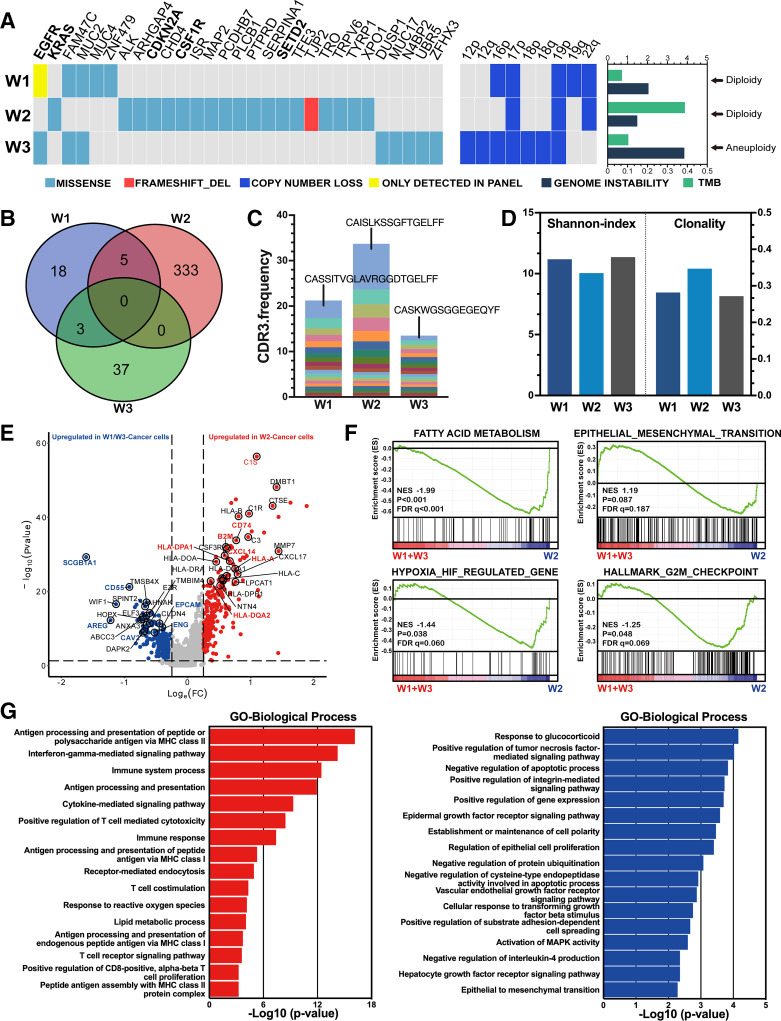
Multiomics sequencing was performed on three resected nodules as described in the online supplemental materials. Whole exome sequencing (WES) and T cell receptor repertoire (TCR) in resected nodules identified highly heterogenous genomic features among different nodules. Both W1 and W3, which were unresponsive to immunotherapy, harbored the EGFR exon 21 L858R mutation, whereas W2 harbored an oncogenic driver mutation in KRAS. Arm-level somatic copy number alterations indicated relatively fewer events in W1 and W2, likely leading to lower immune evasion.21 The tumor mutation burden of W2 was notably higher but showed the lowest genomic instability (figure 2A). Additionally, HLA phenotypes predicted nearly separate neoantigen distribution, indicating that the different nodules induced a heterogenous microenvironment (figure 2B). TCR-beta chain sequencing identified a markedly amplified complementarity-determining region 3 (CDR3) alignment (CAISLKSSGFTGELFE) in the W2 cell type, contributing to over 10% of the total repertoire. In contrast, the most predominant CDR3 clonotypes in W1 and W3 accounted for only 3.9% and 1.1%, respectively, suggesting a specific T cell proliferation in W2 (figure 2C). In line with the presence of a dominant clone, TCR clonality was relatively higher in W2; however, the W2 clonotype had lower TCR diversity compared with the other two nodules even under the same systemic immunity (figure 2D) (original data of mIHC, WES as well as HLA phenotype was summarized in online supplemental file 2).
Figure 2.
Heterogeneity of multiple primary lung cancer (MPLC) and single-cell RNA sequencing (scRNA-seq) analysis of differentially enriched pathways. (A) Genomic alteration of MPLC and somatic copy number variants. Bar chart on the right shows genomic instability, tumor mutation burden (TMB) and ploidy for each corresponding lesion. (B) A Venn diagram showing highly inconsistent human leukocyte antigen (HLA) types among nodules. (C) Stacked column chart of CDR3 frequency among nodules highlighting high frequency of CDR3 alignment in responding nodules. (D) Shannon index and CDR3 clonality of T cell receptor repertoire. (E) A volcano plot of differentially expressed genes between W2 and W1/W3 cancer cells (EPCAM+) with immune modulation (red) and tumor growth (blue) associated genes highlighted in corresponding color. (F) Gene set enrichment analysis (GSEA) between W2 and W1/W3 cancer cells among four cancer-related pathways, including fatty acid metabolism, epithelial mesenchymal transition, hypoxia-regulated genes and G2/M checkpoint. (G) Gene Ontology (GO) analysis of differentially enriched biological processes between W2 (red) and W1/W3 (blue) cancer cells.
jitc-2020-002312supp001.pdf (111KB, pdf)
jitc-2020-002312supp002.xlsx (1.2MB, xlsx)
Differentially enriched pathways and major histocompatibility complex expression modulation
We then analyzed gene expression in tumor cells selected using InferCNV from single-cell RNA sequencing (scRNA-seq) and determined differentially expressed genes in W2 and W1/W3 cancer cells. Genes correlated with HLA, complement activation and immune modulation were significantly upregulated in W2 cancer cells, whereas genes related to cell growth and adhesion were upregulated in W1/W3 cancer cells (figure 2E). We further examined several gene signatures that could influence the efficacy of antitumor treatment. The fatty acid metabolism gene signature was significantly enriched in W2 (p<0.001, false discovery rate (FDR) q<0.001). Both hypoxia-related (p=0.038, FDR q=0.060) and G2/M checkpoint signatures (p=0.048, FDR q=0.069) were markedly enriched in the W2. However, there was no significant enrichment in the epithelial mesenchymal transition signature (p=0.087, FDR q=0.187) among nodules (figure 2F). W2 cancer cells were significantly involved in antigen processing and presentation through major histocompatibility complex class II (p<0.001, FDR <0.001). Additionally, W2 cancer cells were involved in positive regulation of T cell-mediated cytotoxicity and other T cell activation processes. In contrast, W1/3 cancer cells were significantly involved in growth factor receptor signaling (p<0.001, FDR=0.058) and cell proliferation (p<0.001, FDR=0.060) (figure 2G), which could partially explain the limited response to immunotherapy.
Impaired lymphocyte immunity with fewer infiltrating tissue resident memory T cells
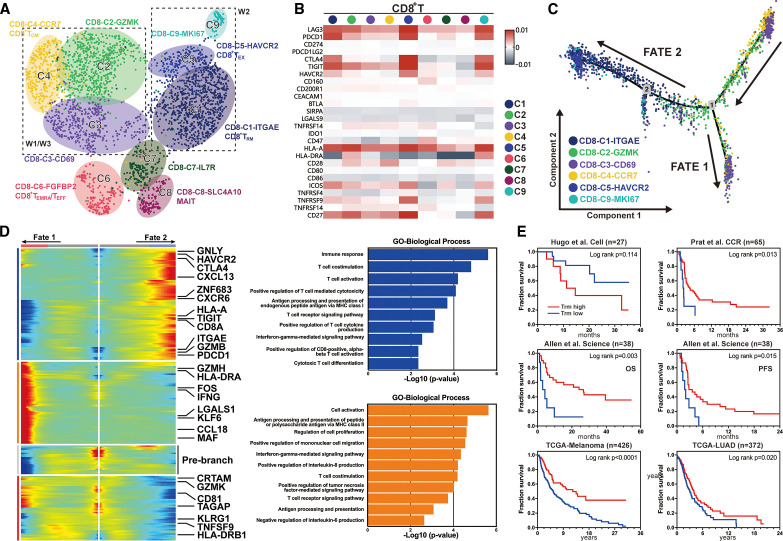
Given the major therapeutic mechanism of PD-1 checkpoint inhibitors on T lymphocytes, we evaluated differential T cell interactions between responding and non-responding nodules. T cell subtypes were distinguished by specific markers in CD4 (CD3+CD8+CD45+) and CD8 (CD3+CD8+CD45+) lymphocytes (figure 3A), respectively. To investigate how immunotherapy affected the expression of checkpoints and costimulator molecules in CD8 T lymphocytes, we examined the expression of 26 relevant genes in different cell types. CD8-C1-ITGAE, CD8-C5-HAVCR2 and CD8-C9-MKI67 showed relatively high expression of immune checkpoint proteins, such as LAG3, PDCD1, cytotoxic T-lymphocyte antigen 4 (CTLA-4), TIGIT and HLA-A, whereas other cell types had weak or no expression of these proteins (figure 3B). To map the evolutionary trajectory of CD8+ lymphocytes under immunotherapy, we performed pseudo-time analysis and identified two fates. W2 cells tended toward fate 2, characterized by high expression of HAVCR2, TIGIT, PDCD1, GNLY, HLA-A and GZMB, which correlated with T cell exhaustion and cytotoxic signals (figure 3C). W1/W3 cells tended toward fate 1, which was characterized by the high expression of GZMH, HLA-DRA and IFNG (figure 3C). These results were corroborated by Gene Ontology analysis, showing that fate 2 was enriched for the induction of antigen-specific cytotoxicity in CD8 lymphocytes. However, CD8 lymphocytes in fate 1 also exhibited activated T cell functions, such as T cell costimulation and TCR signaling (figure 3D). Trm, previously reported to reside in epithelia, connective tissues and other tissues,22 23 have been associated with superior long-term prognosis in various cancers.24 25 In this study, Trm subsets were prominently enriched in W2 cancer cells, accounting for nearly 50% of T lymphocytes. Therefore, we established a Trm signature by screening specific genes using the Wilcoxon rank-sum test, as described in the online supplemental materials. Using three studies26–28 and two datasets from The Cancer Genome Atlas (https://gdc.cancer.gov/) as external validation cohorts to assess the predictive immunotherapy and prognostic value of Trm, we found that cells with high Trm infiltration showed significantly prolonged overall survival in both lung adenocarcinoma and melanoma. Except in one study, patients with high Trm signature treated with either PD-1 or CTLA-4 inhibitors showed appreciable survival compared with patients with low Trm signature indicating that Trm was both a predictive and prognostic factor for immune infiltration in multiple cancer types (figure 3E).
Figure 3.
Diverse differentiation trajectories of CD8+ T lymphocytes and prognostic value of tissue resident memory T cells (Trm). (A) UMAP projection of 2755 CD8+ T lymphocytes from three nodules clustered into nine groups by feature markers. (B) Heatmap of average gene expression value of immune checkpoint and costimulation-related genes among different CD8 T clusters. (C) Pseudo-time analysis of six major CD8+ T lymphocyte clusters showing two differentiation trajectory fates. (D) Radiation plot showing significant differential expression for each fate. Highly expressed genes are colored in red and genes with low expression in blue. Bar charts on the right indicate highly involved processes in fate 1 (orange) and fate 2 (blue), respectively. (E) Confirmation of the predictive and prognostic value of the Trm signature using external validation cohorts including two TCGA datasets and three retrospective studies. Log-rank p value is presented for each independent survival analysis. TCGA, The Cancer Genome Atlas; UMAP, Uniform manifold approximation and projection.
Discussion
In this study, we showed that neoadjuvant immunotherapy unlikely enhances systemic antitumor immunity against all lesions in patients with early stage MPLC. A recent study revealed that 45% of advanced NSCLC had mixed progression after resistance to immunotherapy29 despite moderate tumor heterogeneity. Genomics analyses have shown that unlike metastatic lesions, multiple primary lesions are much more heterogeneous,30 and may explain the diverse HLA-phenotypes and TCR expansion observed in this study. Therefore, a single biopsy of one lesion may not be sufficient as a guide for subsequent treatment of the remaining lesions. Furthermore, multiple subsolid lesions revealed impaired antigen presentation and cytotoxicity induction by CD4 and CD8 lymphocytes, which might be a potential mechanism of insensitivity to immunotherapy, given their involvement in early phase antitumor immunity.31 Notably, impaired T lymphocyte immunity was not driven by the expression of PD-1/PD-L1 or other immune checkpoint proteins, as shown by our findings that non-responding nodules were negative for PD-L1 and exhibited low expression of immune checkpoints in lymphocytes. These results might be explained by differential distribution of Trm in responding and non-responding lesions of immune-modulated and immune-surveillant CD8 T cell types,32 indicating effective durable stimulation by tumor antigens. In contrast, CD8 T lymphocytes of non-responding nodules preferentially displayed naïve or early activated T cell genotypes, concordant with early stage disease. The TCR repertoire showed higher clonality in responding nodules with a specific amplified CDR3 alignment (CAISLKSSGFTGELFE). Furthermore, survival analysis demonstrated the prognostic value of Trm with or without immunotherapy in advanced-stage disease. We propose, therefore, that Trm might be a more sophisticated marker as an early response signal for immunotherapy than CD8 infiltrating T cells.
There are several caveats to our study. First, despite our multiomics approach, this is a case-based study; thus, the results should be carefully interpreted and a larger sample size study is warranted to establish clinical efficacy. Second, it should be noted that pseudo-progression might also explain the diverse responses in different lesions. However, two to four cycles of neoadjuvant treatment may not be sufficient to determine whether the diverse responses can be attributed to pseudo-progression. Besides, we have performed in-depth pathological evaluation including pathological regression showing relatively consistent results with radiological shrinkage (pathological regression −80% for W2 and −25%/−10% for W1/W3, respectively), which may in a way suggest unlikelihood of pseudo-progression. Third, due to the lack of pretreated specimens, changes in different infiltrating immune cells after immunotherapy could not be reliably determined. Moreover, the acquisition of all small nodules in MPLC, especially for subsolid lesions, is technically impracticable before surgery. Nevertheless, by comparing nodules with different responses to a specific treatment, it may be possible to analyze how immunotherapy influences the tumor immune microenvironment, leading to diverse clinical outcomes.
In summary, our results revealed that early stage MPLC harbored diverse genomic phenotypes and exhibited distinct neoantigen peptides and downregulated HLA genes, suggesting lower immunogenicity in subsolid nodules. Moreover, we showed that impaired T lymphocyte immunity could be attributed to an inferior response to immunotherapy within subsolid nodules, which accounted for the largest proportion of multiple pulmonary nodules. Several trials (NCT04047186, NCT04026841) evaluating the efficacy of neoadjuvant immunotherapy for MPLC are still ongoing. Recently, a retrospective study showed limited efficacy of PD-1/PD-L1 checkpoint blockade alone in multiple ground-glass nodules of advanced NSCLC.33 Altogether, these data suggested that neoadjuvant immunotherapy alone might not be an ideal option for patients with MPLC due to insufficient antitumor response for all subsolid lesions. However, given the genomic and immune microenvironment heterogeneity among MPLCs, neoadjuvant immunotherapy for MPLC with subsolid nodules should be deliberately considered for future clinical trials. In addition, Trm might be incorporated as an extensive immune biomarker in the design of future immunotherapy trials.
Acknowledgments
The authors would like to thank Geneseeq technology for providing high-throughput whole exome sequencing plus T cell receptor repertoire sequencing and NovelBio for providing scRNA-seq and bioinformatics analysis support. The authors would like to thank Editage (www.editage.cn) for English language editing. The authors would also like to thank the patient and her family for consenting to our publishing her clinical case.
Footnotes
CZ, KY and S-YL contributed equally.
Contributors: Conceptualization: CZ, W-ZZ, Y-LW, X-NY. Resources: CZ, W-ZZ and S-YL. Methodology: CZ, S-YL, JS, L-XY and X-CZ. Bioinformatic analysis: CZ, S-YL, KY. Writing—original draft: CZ and S-YL. Writing—review and editing: CZ, S-YL, KY, W-ZZ, X-NY and Y-LW. All authors approved the submission of the manuscript.
Funding: This study was supported by National Natural Science Foundation (grant 81872510 to W-ZZ) and High-level Hospital Construction Project (grant DFJH201801 to W-ZZ).
Competing interests: W-ZZ has received honoraria from AstraZeneca and Roche outside the submitted work; Y-LW has received honoraria from AstraZeneca, Eli Lilly, Roche, Pierre Fabre, Pfizer and Sanofi; consulting or advisory tole with AstraZeneca, Roche, Merck and Boehringer Ingelheim and Roche outside the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Single-cell RNA sequencing datasets generated in this study are available on the GEO database under the accession number GSE146100. Detailed mIHC data, WES MAF files, and HLA-phenotype data are summarized in supplemental data. External datasets used in this study were accessed using the accession numbers: GEO, GSE93157 (Prat et al.); GEO, GSE78220 (Hugo et al.). All other relevant data are available from the corresponding author of this study (Xue-Ning Yang, yangxuening@gdph.org.cn) upon reasonable request.
Ethics statements
Patient consent for publication
Obtained.
Ethics approval
Written informed consent of tumor acquisition for research has been obtained before surgery and approved by internal review board from Guangdong Lung Cancer Institute and Guangdong Provincial People’s Hospital (Guangzhou, China, IRB approved protocol number GDREC2016175H).
References
- 1. Girard N, Deshpande C, Lau C, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. Am J Surg Pathol 2009;33:1752–64. 10.1097/PAS.0b013e3181b8cf03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Detterbeck FC, Franklin WA, Nicholson AG, et al. The IASLC lung cancer staging project: background data and proposed criteria to distinguish separate primary lung cancers from metastatic foci in patients with two lung tumors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol 2016;11:651–65. 10.1016/j.jtho.2016.01.025 [DOI] [PubMed] [Google Scholar]
- 3. Suh YJ, Lee H-J, Sung P, et al. A novel algorithm to differentiate between multiple primary lung cancers and intrapulmonary metastasis in multiple lung cancers with multiple pulmonary sites of involvement. J Thorac Oncol 2020;15:203-215. 10.1016/j.jtho.2019.09.221 [DOI] [PubMed] [Google Scholar]
- 4. Lee BE, Port JL, Stiles BM, et al. Tnm stage is the most important determinant of survival in metachronous lung cancer. Ann Thorac Surg 2009;88:1100–5. 10.1016/j.athoracsur.2009.06.098 [DOI] [PubMed] [Google Scholar]
- 5. Ishikawa Y, Nakayama H, Ito H, et al. Surgical treatment for synchronous primary lung adenocarcinomas. Ann Thorac Surg 2014;98:1983–8. 10.1016/j.athoracsur.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 6. De Leyn P, Moons J, Vansteenkiste J, et al. Survival after resection of synchronous bilateral lung cancer. Eur J Cardiothorac Surg 2008;34:1215–22. 10.1016/j.ejcts.2008.07.069 [DOI] [PubMed] [Google Scholar]
- 7. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hargadon KM, Johnson CE, Williams CJ. Immune checkpoint blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors. Int Immunopharmacol 2018;62:29–39. 10.1016/j.intimp.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 9. Schadendorf D, Hodi FS, Robert C, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol 2015;33:1889–94. 10.1200/JCO.2014.56.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garon EB, Hellmann MD, Rizvi NA, et al. Five-year overall survival for patients with advanced Non‒Small-Cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol 2019;37:2518–27. 10.1200/JCO.19.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn GP, Bruce AT, Ikeda H, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3:991–8. 10.1038/ni1102-991 [DOI] [PubMed] [Google Scholar]
- 12. Mittal D, Gubin MM, Schreiber RD, et al. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol 2014;27:16–25. 10.1016/j.coi.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Angelova M, Mlecnik B, Vasaturo A, et al. Evolution of metastases in space and time under immune selection. Cell 2018;175:751–65. 10.1016/j.cell.2018.09.018 [DOI] [PubMed] [Google Scholar]
- 14. Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482:400–4. 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mascaux C, Angelova M, Vasaturo A, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature 2019;571:570–5. 10.1038/s41586-019-1330-0 [DOI] [PubMed] [Google Scholar]
- 16. Zhang C, Zhang J, Xu F-P, et al. Genomic landscape and immune microenvironment features of preinvasive and early invasive lung adenocarcinoma. J Thorac Oncol 2019;14:1912–23. 10.1016/j.jtho.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsunoda A, Morikawa K, Inoue T, et al. A prospective observational study to assess PD-L1 expression in small biopsy samples for non-small-cell lung cancer. BMC Cancer 2019;19:546. 10.1186/s12885-019-5773-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu H, Boyle TA, Zhou C, et al. Pd-L1 expression in lung cancer. J Thorac Oncol 2016;11:964–75. 10.1016/j.jtho.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med 2018;378:1976–86. 10.1056/NEJMoa1716078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Helmink BA, Reddy SM, Gao J, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355. 10.1126/science.aaf8399. [Epub ahead of print: 20 Jan 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar BV, Ma W, Miron M, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep 2017;20:2921–34. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 2016;16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- 24. Djenidi F, Adam J, Goubar A, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol 2015;194:3475–86. 10.4049/jimmunol.1402711 [DOI] [PubMed] [Google Scholar]
- 25. Savas P, Virassamy B, Ye C, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018;24:986–93. 10.1038/s41591-018-0078-7 [DOI] [PubMed] [Google Scholar]
- 26. Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prat A, Navarro A, Paré L, et al. Immune-related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res 2017;77:3540–50. 10.1158/0008-5472.CAN-16-3556 [DOI] [PubMed] [Google Scholar]
- 28. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osorio JC, Arbour KC, Le DT, et al. Lesion-level response dynamics to programmed cell death protein (PD-1) blockade. J Clin Oncol 2019;37:3546–55. 10.1200/JCO.19.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asmar R, Sonett JR, Singh G, et al. Use of oncogenic driver mutations in staging of multiple primary lung carcinomas: a single-center experience. J Thorac Oncol 2017;12:1524–35. 10.1016/j.jtho.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 31. Galon J, Bruni D, Immunology T. Tumor immunology and tumor evolution: intertwined histories. Immunity 2020;52:55–81. 10.1016/j.immuni.2019.12.018 [DOI] [PubMed] [Google Scholar]
- 32. van der Leun AM, Thommen DS, Schumacher TN. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer 2020;20:218–32. 10.1038/s41568-019-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu F, Li W, Zhao W. Synchronous ground-glass nodules showed limited response to anti-PD-1/PD-L1 therapy in patients with advanced lung adenocarcinoma. Ciln Transl Med 2020;10:e149. 10.1002/ctm2.149 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002312supp001.pdf (111KB, pdf)
jitc-2020-002312supp002.xlsx (1.2MB, xlsx)
Data Availability Statement
Single-cell RNA sequencing datasets generated in this study are available on the GEO database under the accession number GSE146100. Detailed mIHC data, WES MAF files, and HLA-phenotype data are summarized in supplemental data. External datasets used in this study were accessed using the accession numbers: GEO, GSE93157 (Prat et al.); GEO, GSE78220 (Hugo et al.). All other relevant data are available from the corresponding author of this study (Xue-Ning Yang, yangxuening@gdph.org.cn) upon reasonable request.