SUMMARY
There is an increasing appreciation for the heterogeneity of myeloid lineages in the lung, but relatively little is known about populations specifically associated with the conducting airways. We use single-cell RNA sequencing, flow cytometry, and immunofluorescence to characterize myeloid cells of the mouse trachea during homeostasis and epithelial injury/repair. We identify submucosal macrophages, similar to lung interstitial macrophages, and intraepithelial macrophages. Following injury, there are early increases in neutrophils and submucosal macrophages, including M2-like macrophages. Intraepithelial macrophages are lost after injury and later restored by CCR2+ monocytes. We show that repair of the tracheal epithelium is impaired in Ccr2-deficient mice. Mast cells and group 2 innate lymphoid cells are sources of interleukin-13 (IL-13) that polarize macrophages and directly influence basal cell behaviors. Their proximity to the airway epithelium establishes these myeloid populations as potential therapeutic targets for airway disease.
Graphical Abstract

In Brief
Engler et al. identify molecularly unique populations of monocytes and macrophages associated with the tracheal epithelium. The composition of the tracheal myeloid compartment changes upon injury, and repair is delayed in the absence of Ccr2+ monocytes. Injury-associated myeloid cells and type 2 cytokines directly affect basal cell behaviors.
INTRODUCTION
The respiratory system can be divided into at least two functional zones: the conducting airways and the alveoli. Each contains distinct epithelial and mesenchymal lineages with unique functions (Rock and Hogan, 2011). Alveolar macrophages (AMs) are considered tissue-resident macrophages of the lung, and there is growing evidence for ontological, molecular, and functional heterogeneity in this population (Misharin et al., 2013; Tan and Krasnow, 2016). We have shown that CCR2+ monocytes and M2-like macrophages promote alveolar regeneration by type 2 epithelial cells in a mouse model of partial pneumonectomy (Lechner et al., 2017).
Interstitial macrophages (IMs), molecularly distinct from AMs, associate with the submucosa of the airways (Gibbings et al., 2017). Previous studies have shown that these myeloid cells increase in number after epithelial injury (Tadokoro et al., 2014), and Th2 cytokines influence the cellular composition of the airway epithelium (Danahay et al., 2015; Kuperman et al., 2002). Recent data implicate alternatively activated macrophages as critical components of the regenerative niche after injury of the airway epithelium (Dagher et al., 2020).
Here, we used flow cytometry and single-cell RNA sequencing (SCRNA-seq) to characterize the myeloid populations in the mouse trachea at steady state and during injury/repair of the epithelium. We provide evidence for airway-associated macrophages that are distinct from AMs and show a shift toward M2-like polarization during epithelial repair. Some of these M2-like macrophages originate from a Ccr2+ precursor, and regeneration of the tracheal epithelium is impaired in the absence of Ccr2 expression. Lastly, we identify a unique macrophage subset that is located in close proximity to airway basal cells within the tracheal epithelium. Our data suggest that these cells are lost along with the tracheal epithelium after chemically induced injury and are replaced by CCR2+ cells during epithelial repair.
RESULTS
Homeostatic Tracheal Macrophages
The mouse trachea provides a well-characterized model of the pseudostratified airway epithelium. We used flow cytometry to compare myeloid cells of dissociated tracheas to those of dissociated lung parenchyma. Approximately 38% of the CD45+ cells in the trachea were MerTK+. Most of these expressed CD11bhi and CD11c (Figure S1B), similar to IMs in the distal lung (Gibbings et al., 2017). CD45+;F4/80+ macrophages in the trachea did not express the AM marker SiglecF (Figure S1A). In contrast, the majority of CD45+;MerTK+ cells in the lung are Cd11bneg/lo;CD11chi AMs with a minor population of MerTK+;CD11bhi IMs (Figure S1B).
Tracheal Macrophage Dynamics during Injury and Repair of the Tracheal Epithelium
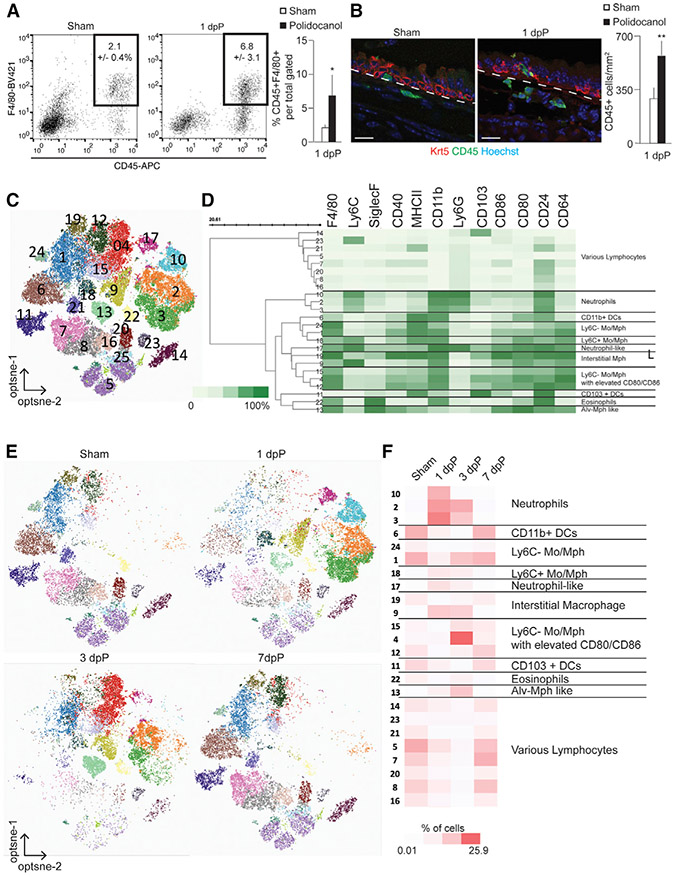
We injured the tracheal epithelium with polidocanol (Borthwick et al., 2001) to determine how the tracheal macrophage populations respond. Tracheas were analyzed 1, 3, and 7 days post-polidocanol (dpP) and compared to PBS-treated controls (Figure S1C). Polidocanol results in the death and sloughing of suprabasal cells of the pseudostratified epithelium, largely sparing basal cells that repair the epithelium within a week (Figure S1C). We observed increased numbers of CD45+ and CD45+F4/80+ cells in the mesenchyme beneath the injured epithelium 1 dpP (Figures 1A and 1B). Next, we performed flow cytometry with a 15-color myeloid immunophenotyping panel (Table S1). Opt-SNE dimensionality reduction and PhenoGraph unsupervised clustering of live, extravascular CD45+ cells from sham and 1, 3, and 7 dpP tracheas showed 24 distinct clusters (Figures 1C and S1D), of which 16 were categorized as myeloid populations (Figure 1D). The proportion of cells with a neutrophil cell surface phenotype increased 1 dpP compared to sham (Figures 1E and 1F, clusters 10, 2, and 3). We observed increased numbers of Ly6C+ IMs (Cluster 9) and Ly6C− monocytes and macrophages that showed elevated levels of CD80 and CD86 (cluster 4) at 3 dpP. Within 7 dpP, cellular composition had returned to baseline (Figures 1E and 1F).
Figure 1. Characterization of Tracheal Macrophages.
(A) FACS analysis of CD45+;F4/80+ myeloid cells after 1 dpP; (n = 4).
(B) Immunofluorescence and quantifications of CD45+ cells (green) near basal cells (KRT5, red) following polidocanol-induced injury. Scale bar, 25 μm.
(C) Unsupervised clustering of all cells combined from sham and from 1, 3, and 7 dpP based on 15-marker myeloid panel; (n = 1/time point, consistent of 2 animals).
(D) Heatmap of marker expression data, scaled from 0% to 100% of normalized, arcsin value of fluorescent intensity.
(E) Cells isolated on indicated days post-injury, visualized by cluster assignment.
(F) Relative abundance of cells from 24 distinct clusters across injury and repair, scaled 0.01% to 25.9% of total cells in analysis. See also Figure S1 and Table S1.
Transcriptional Changes during Injury and Repair
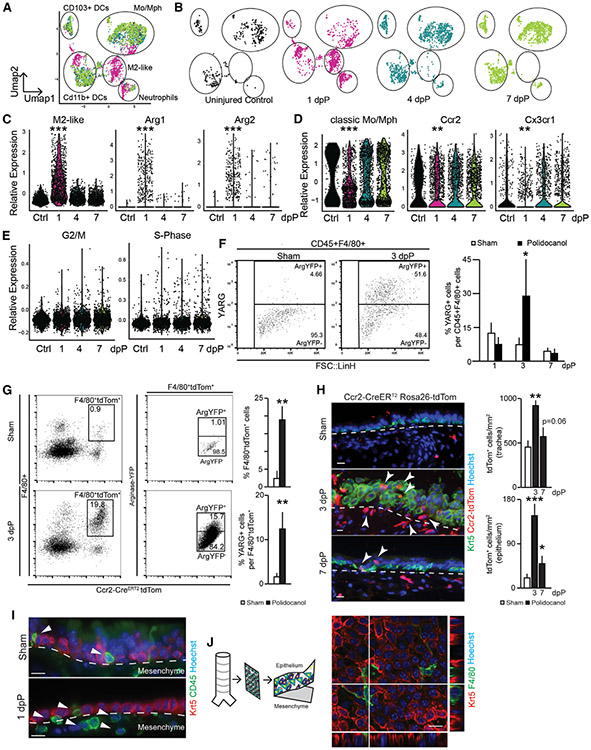
As an unbiased complimentary approach, we performed SCRNA-seq of CD45+ cells from dissociated tracheas 1, 4, and 7 dpP and uninjured controls. We performed post-processing filtering to focus our analyses on CD45+;F4/80+ myeloid cells (Figure S2A). Uniform manifold approximation and projection (UMAP) visualization (Figures 2A and 2B) and analysis of differential gene expression (Figure S2B; Table S2) showed unique features of myeloid populations at 1dpP compared to the largely overlapping 4 and 7 dpP tracheas that were similar to controls (Figures 2A and 2B). Upon injury, the number of cells expressing transcripts associated with alternatively activated M2-like macrophages, such as Arg1 and Arg2, were increased (Figure 2C; Figure S2B; Tables S2 and S4). Concomitantly, the expression of transcripts associated with classical monocytes and macrophages, including Ccr2+ and Cx3cr1+, was rapidly reduced, but returned to baseline 4 dpP (Figure 2D; Table S4). We did not observe changes in transcripts associated with cell cycle (Figure 2E; Table S4).
Figure 2. Dynamic Features of Tracheal Myeloid Populations after Injury.
(A) UMAP visualization of all SCRNA-seq data across airway injury and repair overlaid (3 animals pooled into 1 sample for each); captured cells: ctrl = 458, 1 dpP = 1,625, 4 dpP = 1,507, 7 dpP = 1,717.
(B) UMAP visualization of SCRNA-seq data on indicated days post-injury.
(C) Violin plots of M2-like macrophage gene signature, Arg1 and Arg2.
(D) Violin plots of classic monocyte/macrophage gene signature, Ccr2 and Cx3cr1.
(E) Violin plots of cell cycle genes associated with G2/M and S phase.
(F) Representative FACS plot and quantification of YARG expression gated on CD45+;F4/80+ (n = 3 mice per time point).
(G) FACS analysis and quantification of Ccr2-CreERT2;ROSA-tdTom+ cells from YARG animals after tamoxifen administration and sham or polidocanol treatment (n = 3, mice per time point).
(H) Immunofluorescence analysis of Ccr2-CreERT2;ROSA-tdTom animals; Krt5 (basal cells, green), Ccr2-CreERT2;ROSA-tdTom lineage trace (red), and Höechst nuclear stain (blue); scale bar, 15 μm.
(I) Histology of wild-type animals; Krt5 (red), CD45 (green), nuclear stain Höechst, scale bar, 15 μm.
(J) Confocal image of peeled epithelial sheet showing F4/80+ myeloid cells (green) and Krt5+ epithelial basal cells (red), Höechst nuclear stain. See also Figure S2 and Tables S2 and S4.
Increased M2-like Macrophages following Polidocanol-Induced Injury
We used an arginase-YFP reporter mouse (YARG; Reese et al., 2007) to quantify M2-like macrophages in the trachea by flow cytometry (Figure S2D). As described above (Figure 1), CD45+;F4/80+ myeloid cells increased at 1 and 3 dpP and returned to baseline by 7 dpP (Figure S2C). YARG+ cells were uniquely increased at 3 dpP compared to sham and other intervals post-injury (Figure 2F). This translated surrogate for arginase expression increased slightly later than Arg1 transcripts (Figure 2B).
Next, we crossed Ccr2-CreERT2 mice to ROSA-tdTom and YARG reporter mice to lineage trace CCR2+ monocytes (Figure S2D) and determine whether they gave rise to M2-like macrophages after injury. When we administered tamoxifen at the time of polidocanol (Figure S2D’), an average of 13.6% ± 3.6% of the YARG+ cells carried the CCR2 lineage trace 3 dpP (Figure 2G). In contrast, only 0.7% of cells were CCR2-lineage traced and YARG+ when polidocanol injury was performed 2 weeks after tamoxifen administration (Figure S2D”).
Next, we localized CCR2 lineage-traced cells on tissue sections 1, 3, and 7 dpP (Figure S2E). As expected, we observed Tom+ cells in the submucosa beneath the tracheal epithelium in sham and injured tracheas at all times examined. We also observed an increased number of Tom+ cells within the regenerating epithelial layer of the trachea at 3 and 7 dpP (Figure 2H). Few of these lineage-traced cells in sham Ccr2-CreERT2;ROSA-tdTom+ mice were found in the epithelial layer (Figures 2H and S2E”). This unexpected finding led us to confirm the existence of CD45+ leukocytes within the pseudostratified tracheal epithelium of sham and injured wild-type mice (Figure 2I).
A Population of Myeloid Cells Is Found within the Pseudostratified Tracheal Epithelium
We peeled sheets of epithelium away from the underlying mesenchyme following an enzymatic digestion (Rock et al., 2009). Analysis of isolated sheets revealed CD45+ (Figure S2F), F4/80+ (Figure 2J), CD64, and MerTK (Figures S2G and S2H) myeloid cells within the epithelial layer in close proximity to KRT5+ basal cells. We similarly analyzed the tracheas of transgenic mice that express GFP under the control of the CSF1R locus, a broad marker of myeloid lineages (cFMS::eGFP; Figure S2I) (Sasmono et al., 2003). Immunofluorescence showed that the intraepithelial CD45+ and F4/80+ cells express cFMS::eGFP+ (Figure S2J). We called these cells intraepithelial airway macrophages (IAMs).
Intraepithelial Myeloid Cells Are Molecularly Distinct from Other Pulmonary Myeloid Lineages
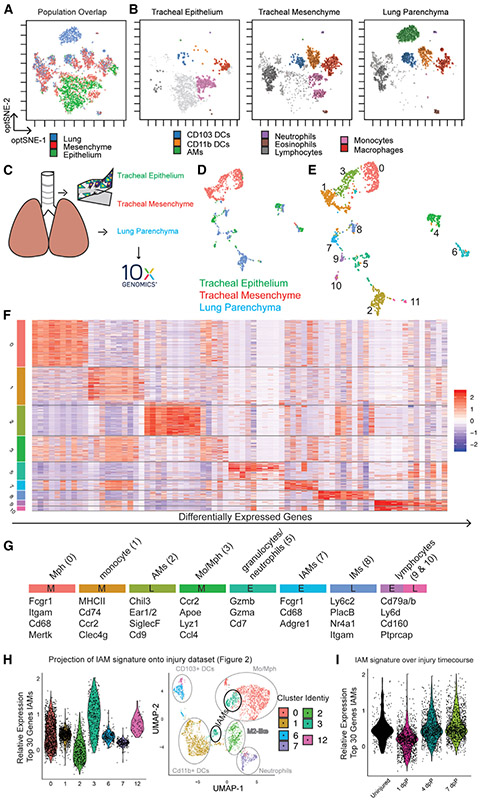
We used the 15-color myeloid fluorescence-activated cell sorting (FACS) panel (Table S1) to compare IAMs to cells from the tracheal mesenchyme and lung parenchyma. We performed opt-SNE dimensionality reduction on equal numbers of live, extravascular CD45+ cells from each of the 3 tissues (Figure 3A; Figures S3A and S3B). Cell identities were assigned (Figure 3B) based on marker expression (Table S1; Misharin et al., 2013). Because the gating strategy was developed for the lung parenchyma, the majority of cells from this tissue were contained within the conventional gates (Figure 3B). In contrast, the majority of myeloid cells from the tracheal epithelium fell outside these gates, suggesting novel myeloid populations or cell states associated with the trachea under homeostasis (Figures S3A and S3B; light gray cells in Figure 3B). The cells from the tracheal epithelium that fell within traditional gates were mostly macrophages and monocytes (Figure 3B) and a few were dendritic cells (DCs). The tracheal mesenchyme and lung parenchyma contained abundant CD103+ and CD11b+ DCs. It is interesting to note that some cell types, such as macrophages and DCs, were observed in all 3 tissues but exhibited slight differences in cell surface profiles represented as small shifts in opt-SNE space (Figure 3B).
Figure 3. Myeloid Cells Isolated from the Epithelium, Mesenchyme, and Lung Parenchyma Have Distinct Transcriptional and Translational Identities.
(A) Unsupervised clustering of 15-marker myeloid FACS panel, overlay (n = 5/tissue pooled).
(B) Visualization of cells isolated from epithelium (left), mesenchyme (middle), and lung (right) after pooled unsupervised clustering.
(C) Schematic representation of samples isolated for SCRNA-seq (n = 5/tissue animals pooled).
(D) UMAP colored for tissue of origin.
(E) Unsupervised Louvaine clustering at resolution 0.25 of SCRNA-seq data.
(F) Heatmap of differentially expressed genes in myeloid clusters.
(G) Cluster assignment based on selected transcripts and annotation of tissue of origin (E, epithelium; M, mesenchyme; L, lung).
(H) Projection of IAM gene signature (cluster 7; Figure 3F) onto single-cell injury dataset (Figures 2A-2E).
(I) Projection of IAM gene signature onto injury time points. See also Figure S3 and Tables S1, S3, and S4.
IAMs, IMs, and AMs Are Molecularly Distinct from Each Other
Because the conventional gating above pointed to unique myeloid populations in the tracheal epithelium and mesenchyme, we performed unbiased SCRNA-seq of CD45+ leukocytes isolated from the tracheal epithelium and CD45+;F4/80+ cells from the tracheal mesenchyme and lung parenchyma as described above (Figures 3C and S3C). Unsupervised clustering visualized in UMAP space showed that the majority of cells from each tissue formed distinct clusters (Figure 3D). Louvain clustering (resolution 0.25) of these data resulted in 12 populations (Figure 3E). Clusters 4, 6, and 11 were characterized as lymphocytes, fibroblasts, and lung epithelium, respectively, and excluded from further analysis (Figure S3D, crossed out; data included in Table S3). We characterized the remaining clusters by identifying the top 10 differentially expressed genes for each (Figures 3F and 3G; Figure S3E; Table S3).
Clusters 0, 1, and 3 were from the tracheal mesenchyme and could be identified as classic macrophages, monocytes, and cells with an intermediate signature, respectively (Figures 3E-3G; Table S3). These three populations share features with IMs from the lung (Gibbings et al., 2017; Ural et al., 2020), including the expression of Mertk, Fcgr1, and Cd68; variable expression of H2-ab1, H2-aa, and Retnla; and lack of expression of SiglecF and Marco. A similar signature was observed for cluster 8 that originated from the lung, presumably representing lung IMs.
Clusters 5, 7, and 9 were isolated from the tracheal epithelium. Of particular interest was cluster 7 that contained IAMs; macrophages enriched for the expression of Fcgr1, Cd68, and Adgre1 (Figure 2G); and undescribed markers such as Tgfβr1 and Scimp (cluster 7; Table S3). Importantly, the transcription profile of these cells was distinct from those reported for IMs (Gibbings et al., 2017). IAMs were also negative for Cd169, recently associated with a subpopulation of macrophages localized in close proximity to airway neurons (Ural et al., 2020). Cluster 5 expressed genes associated with neutrophils, and cluster 9 expressed markers of lymphocytes.
Consistent with the flow cytometry data presented above, the cells isolated from the lung parenchyma expressed genes associated with AMs (cluster 2), a smaller population of IMs (cluster 8), and a small population of lymphocytes (cluster 10) (Figure 3G; Table S3).
We were not able to peel the epithelium (and IAMs) away from the mesenchyme 1 dpP, precluding direct analysis of this population after injury. We therefore generated a transcriptional signature for the IAM cluster (cluster 7; Table S4) and back traced this signature into the SCRNA-seq injury dataset described in Figure 2. Cells enriched for the IAM signature fell into cluster 3 of the injury dataset. The IAM gene signature was lowest at 1 dpP, consistent with immunofluorescence data showing that most and IAMs are sloughed after injury. The IAM signature returned to baseline by 7 dpP (Figure 3I; Figure S3F), supporting a model in which CCR2+ monocytes replace IAMs after injury.
Type 2 Cytokines Modulate Basal Cell Behaviors In Vitro
To determine whether myeloid cells influence the behaviors of basal epithelia stem cells, we cocultured basal cells as tracheospheres (Rock et al., 2009) with CCR2+ monocytes or YARG+ cells isolated 3 dpP. Coculture with either population caused an increase in tracheosphere size compared to those cocultured with CCR2+ cells from control tracheas (Figure 4A; Figure S4A).
Figure 4. CCR2+ Myeloid Cells Promote Basal-Cell-Mediated Epithelial Repair.
(A) Cocultures (n = 4) of basal cells and CCR2+ cells or Arg1+ cells isolated from sham or polidocanol conditions.
(B) FACS analysis of IL-4Rα expression on basal cells (n = 3).
(C) Schematic representation of basal cell isolation, infection, and manipulation.
(D and E) Quantification (D) and bright field (E) of culture of IL-4Rα+/+ (n = 3) and IL-4Rα−/− (n = 3) basal cells in the presence of IL-4 or IL-13 and quantification of sphere size after 10 days of culture.
(F) Epithelial repair in CCR2-deficient mice and heterozygous controls, 3 dpP and 7 dpP. Heterozygote, sham: n = 2, injury: n = 3; homozygote sham: n = 4, injury: n = 3. Scale bars, 25 μm. Quantification of acetylated α-tubulin+ ciliated cells per mm2 as a metric of repair. See also Figure S4.
Type 2 cytokines (interleukin-4 [IL-4], IL-5, and IL-13) are required for the polarization of M2-like macrophages (Wynn et al., 2013). Our SCRNA-seq of CD45+ cells following tracheal injury identified mast cells and group 2 innate lymphoid cells as sources of Il13 after injury (data not shown). We sought to determine whether these cytokines might directly affect basal epithelial cells during repair in addition to their known role in the polarization of M2 macrophages. We cultured basal cells in a tracheosphere assay and supplemented the medium with cytokines associated with lung injury, including IL-1β, IL-2, IL-4, IL-5, and IL-13 (Lechner et al., 2017; Tadokoro et al., 2014; Figure S4B). As previously described (Danahay et al., 2015), the addition of either IL-4 or IL-13 resulted in an increase in average sphere size (Figure S4B). IL-4 and IL-13 signal by a common receptor subunit, IL-4Rα. Flow cytometry on isolated basal cells demonstrated that ~13% of basal cells express IL-4Rα (Figure 4B). We infected adherent cultures of IL-4Rαflox/flox basal cells with an adenovirus-expressing Cre-GFP fusion protein to delete the floxed Il4rα receptor subunit (Figure 4C). Ad5CMVCre-eGFP-infected cells, but not AdCMV-GFP controls, no longer expressed IL-4Rα 3 days later (Figure S4C). When grown as tracheospheres, the IL-4Rα-deficient cells did not respond to IL-4 or IL-13 like the control cells (Figures 4D and 4E).
Myeloid Cells Promote Repair of the Pseudostratified Epithelium
To assess the role of immune cells on tracheal repair in vivo, we injured CIAE-NOG animals that lack adaptive immunity and have dysfunctional innate immunity. Three days after polidocanol-induced injury, basal cells had not undergone extensive self-renewal and temporary stratification seen during normal repair (compare Figure S4D to Figure S1C). However, this defect is temporary, as the epithelium of immunodeficient mice had completely regenerated within 3 weeks (Figure S4F).
Since CCR2 lineage-traced cells accumulate within the regenerating tracheal epithelium (Figure 2H), we reasoned that monocytes and their progeny modulate epithelial repair. We bred mice homozygous for a Ccr2-RFP allele that lack Ccr2 expression and do not have functioning CCR2+ myeloid cells (Saederup et al., 2010; Figure S4G). Importantly, there were no developmental abnormalities of the tracheal epithelium of Ccr2-RFP homozygotes. Histology and immunofluorescence demonstrated incomplete epithelial repair in Ccr2-deficient mice compared to heterozygous littermate controls (Figure 4F), characterized by a lack of multiciliated cells 7 dpP.
DISCUSSION
The mouse trachea is a well characterized model to study airway biology that shares many morphological and histological features with small human airways (Rock et al., 2010). We used this model to study airway myeloid cells and their roles in homeostasis and injury/repair. Under steady-state conditions we identified macrophages in the tracheal submucosa that are similar to recently described IMs (Gibbings et al., 2017). We unexpectedly identified macrophages within the tracheal epithelium that are molecularly distinct from IMs and AMs. These IAMs are also distinct from DCs that have been described within the epithelium (Sertl et al., 1986).
Our flow cytometry and SCRNA-seq data of CD45+;F4/80+ cells revealed several dynamic properties of trachea-associated myeloid populations in response to epithelial injury. For example, at 1 and 3 dpP, we detected an increase in neutrophils, monocytes, and macrophages. At 1 dpP, we observed a shift toward alternatively activated M2-like macrophages and confirmed the accumulation of arginase-expressing M2-like macrophages after epithelial injury by using transgenic reporter mice.
Our lineage-tracing data show that some M2-like macrophages come from a Ccr2-expressing precursor. The accumulation of CCR2-derived cells within the epithelium concomitant with the re-emergence of the IAM transcriptional signature 4 and 7 dpP suggests that CCR2+ monocytes replenish IAMs lost with injury. Similar to previous reports, we observed Cx3cr1-expressing monocytes in homeostatic tracheas and in response to injury (Joshi et al., 2020; Ural et al., 2020). Whether or not these cells also give rise to M2-like macrophages and IAMs remains unknown.
Monocyte-derived, alternatively activated M2-like macrophages are crucial for development, regeneration, and remodeling in disease (Byrne et al., 2015; Lechner et al., 2017). Our previous work demonstrated the importance of M2-like macrophages for the compensatory growth of alveoli after pneumonectomy (Lechner et al., 2017). We used parallel strategies to determine whether leukocytes also promote the repair of the tracheal epithelium. First, we performed polidocanol injury of the airway epithelium in immunodeficient mice and demonstrated a delay in basal cell proliferation and differentiation 3 days post-injury. Second, we showed that tracheal repair is incomplete in CCR2-deficient mice, characterized by failure to generate a pseudostratified epithelium with multiciliated cells 3 and 7 dpP.
Our SCRNA-seq data revealed that IL-4 and IL-13 are expressed by group 2 innate lymphoid cells and mast cells in the trachea (data not shown). One function of these cytokines is the polarization of alternatively activated M2-like macrophages. Previous data suggest that IL-4 and IL-13 also directly affect epithelial cells, including basal cells. (Danahay et al., 2015; Kuperman et al., 2002). Indeed, treatment of wild-type basal cells with IL-4 or IL-13 caused an increased size of tracheospheres. Finally, coculture of basal cells with CCR2+ monocytes or alternatively activated M2-like YARG+ macrophages isolated from injured tracheas caused a similar increase in tracheosphere size. Together, these data demonstrate that inflammation is a key component of the epithelial repair process.
This work and recent work by others (Chakarov et al., 2019; Gibbings et al., 2017; Lechner et al., 2017; Misharin et al., 2013; Ural et al., 2020) illustrate the striking heterogeneity within the myeloid compartment of the respiratory system. With modern, single-cell technologies, we can slowly appreciate the high complexity of tissue -resident monocytes and macrophages that will be key for the development of future, targeted technologies and therapies (Klichinsky et al., 2020).
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by the Lead Contact, Jason R. Rock (rockjr@bu.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets generated during this study are available. FlowRepository: http://flowrepository.org/id/RvFrhTPb7LK2nAIF366KA2ta9q5awmTMo5zYwWCtyRqXotLW4DPmsd2coPsjMgaG; Single Cell Sequencing Data are deposited under GSE152501; Data related to Figure 3: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/macrophages/macrophages; Data related to Figure 2: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/19_09_18_alex_merge_sctransform/19_09_18_alex_merge_sctransform
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experiments were conducted as sex unbiased, with a minimum of three animals per experimental group. All mice were bred and maintained in a specific-pathogen-free barrier facility with free access to food and water. Both male and female mice were used between 8-12 weeks of age for all experiments. All studies were approved and performed according to the guidelines issued by Boston University’s (BU) Institutional Animal Care and Use Committees (IACUC) under license numbers PROTO201900002. Animals were commercially acquired via Jackson Laboratories. Tg (Csf1r-EGFP)1Hume/J (also referred to as Csf1r-GFP) mice (Sasmono et al., 2003), Ccr2tm2.1Ifc (also referred to as Ccr2RFP/+, [Saederup et al., 2010]) mice were used for macrophage and monocyte studies and Ccr2RFP/RFP (pure-bred C57BL/6 strain) mice were used to assess the contribution of Ccr2+ cells after Polidocanol mediated injury. Arg1tm1Lky/J ([Reese et al., 2007] also referred to as YARG) mice were used to assess M2-like polarization of macrophages and Il4ratm1Sz mice were used to isolate basal cells for culture assays.
METHOD DETAILS
Tamoxifen Administration
Tamoxifen (Sigma-Aldrich, catalog #T5648) was dissolved in corn oil to a final concentration of 20mg/mL and administered to mice via intraperitoneal injection at 0.25 mg of Tamoxifen per kg of mouse weight. A single full dose of TMX was given to each animal. Further analysis and additional experiments were performed earliest 4-days after last Tamoxifen dosage (early time-points were used for replenishment studies) and for progenitor analysis additional experiments were performed after full Tamoxifen washout after 14 days (YARG coexpression experiments.
Polidocanol Induced Injury
Polidocanol-induced injury was performed as previously described (Borthwick et al., 2001). Mice were anesthetized in an Isoflurane chamber and delivered one dose of 15 μL freshly prepared, 2% Polidocanol or PBS sham control by oropharyngeal aspiration to induce injury. Tracheas were harvested 1-, 3- or 7-days following injury for scRNA-Seq, FACS Analysis or immunohistochemistry.
Tissue Preparation
Mice were euthanized, chest cavity was opened and tracheas exposed. For scRNA-seq experiments the pulmonary vasculature was perfused via the right heart ventricle. Uninjured tracheas were peeled as previously described (Rock et al., 2009), in short, tracheas were dissected, cleaned, cut in smaller pieces and predigested in Dispase (15 U/mL, Sigma-Aldrich) for maximally 25 minutes and the epithelium was thereafter peeled from the mesenchyme. The epithelium was further digested in trypsin (0.25%) for 15 minutes.
Whole tracheas, leftover mesenchyme and lung were digested with 1.5 mg/mL Collagenase A (Roche), 0.4 mg/mL DNase I (Roche), and 2 U/mL Dispase (Sigma–Aldrich) in RPMI base medium at 37°C for 30-45 minutes. Reaction was stopped using FBS to a concentration of 10%.
For tissue collection for immunohistochemistry, whole throats were dissected and fixed for maximally 4 hours in 4% PFA. PFA was washed out of tissue using PBS and the trachea was dissected out of the throat. Tracheas were either embedded for Paraffin histology, by moving through ascending Ethanol (50%–100%), Xylene and Paraffin in a vacuum oven and sectioned at 5-7 μm on a microtome (Leica) or were prepared for frozen sectioning, by cryopreservation in 30% Sucrose for 48 hours and embedding in OCT medium and freezing on dry ice. Frozen organs were sectioned on Cryostat (Leica) at 10-12 μm thickness within one month and sectioned tissue was stored at −20°C.
Tracheosphere Culture
Basal Cells were cultured as previously described (Rock et al., 2009). In brief, basal cells were suspended in mouse tracheal epithelial cells (MTEC)/plus medium mixed at a 1:1 ratio with growth factor-reduced Matrigel (BD Biosciences), and seeded at 500 cells/90 μL droplet. Medium was changed every other day. For co-cultures media was adapted to contain 20% RPMI base media and 50ng/mL SCF, 10 ng/mL MCSF and 10 ng/mL IL3. Cells were seeded at 1:1 ratio (basal cell to macrophage) at 1000 cells/100 μL pure factor-reduced matrigel. For IL4Ra deletion, primary cells from IL4Raflox/flox animals were isolated and infected as pure, adherent basal cell cultures using MOI 3, Ad-CMV-GFP (Control) or Ad-CMV-Cre-GFP (Cre) virus (Viral Vectors were provided by the University of Iowa Viral Vector Core (http://www.medicine.uiowa.edu/vectorcore). 3-days after infection cells were dissociated from plastic and analyzed for deletion by FACS and plated as previously described for differentiation as tracheosphere.
Immunohistochemistry
Paraffin slides were rehydrated by consecutive, descending processing through Xylene and Ethanol (100%–50%). Paraffin slides underwent citrate-based retrieval (Vector Unmasking), for frozen sections only select panels were citrate retrieved.
Tissue sections were blocked and permeabilized for 30 – 60 minutes in 10% NDS, 4% BSA and 0.5% TritonX. Primary antibodies (Krt5 (rb) Covance (Prb-160P) 1:1000; F4/80 (rt) Biorad (MCA497GA) 1:250; RFP (gt) Abcam (ab25877) 1:500; acet.Tub (ms) Sigma (T7451) 1:2000; CD45 (gt) R&D (AF114) 1:250) were diluted in primary block solution (2.5% NDS, 1% BSA, 0.125% TritonX) and incubated overnight at 4°C in a humidified chamber. All fluorophore-conjugated secondary antibodies were diluted in secondary block solution (5% NDS, 2% BSA, 0.25% TX) at 1:500 containing Höechst nuclear stain dye. EdU staining was performed according to manufacturer recommendations (Invitrogen). Images of sections were captured on a Nikon Eclipse NiE or a Leica SP6.
Flow Cytometry
Whole tissue digests to obtain single cell suspension (as described in tissue preparation) were kept on ice and stained in flow cytometry/FACS buffer (1% BSA in PBS without Ca2+ or Mg2+). For live cell staining and sorting, a cell viability dye (DAPI or Calcein Blue) was added before sample acquisition. Flow cytometry analysis was performed on a Stratadigm (S1000EXI) or FACSAria II (BD Biosciences) and cell sorting was performed on Moflo Astrios (Beckman Coulter). Compensation was performed with single-stained UltraComp compensation beads (Thermo Fisher) or single-stained immune cells prepared from spleen or lung. To determine gating controls for reporter gene expression, mice lacking the reporter or protein were used. Initial data cleanup and expert-driven analysis were performed using FlowJo (BD Biosciences) and Cytobank cloud-based analysis software (Beckman Coulter). For the 15-color myeloid cell immunophenotyping, intravascular immune cells were labeled by tail vein injection (Anderson et al., 2014) of 2 μg of CD45-BUV737 (BD Biosciences) diluted in 100 μL of sterile saline and loaded into a 28.5 gauge insulin syringe. For tail vein injection animals were lightly anesthesized, injected and kept under controlled, anesthesized conditions, before right ventricle transcardial perfusion 3 minutes post injection.
Automated analysis of data generated from representative samples was performed in Omiq.ai cloud-based analysis platform. Representative files were deposited in FlowRepository (http://flowrepository.org/id/RvFrhTPb7LK2nAIF366KA2ta9q5awmTMo5zYwWCtyRqXotLW4DPmsd2coPsjMgaG) CD45+ live single cells phenotyped across 15 markers were clustered with Phenograph unsupervised clustering algorithm (Levine et al., 2015) and projected into opt-SNE space (Belkina et al., 2019). Median fluorescence intensities of each marker across Phenograph-found populations were plotted on a hierarchically cluster heatmap to allow expert annotation of specific populations.
Single Cell Sequencing
Tissue was prepared as described above for fluorescent activated cell sorting (FACS). For mesenchymal and lung samples viable CD45+;F4/80+ cells were sorted, for peeled tracheal epithelium and tracheas in Polidocanol injury experiments viable bulk CD45+ cells were sorted and brought to appropriate concentration, according to 10X recommendation for capture of up to 2000 cells/ lane. Single cells were captured for sequencing library preparation using a 10X Chromium (10X Genomics, Pleasanton, CA) instrument at the BUMC Single Cell Core. Single-cell RNA-seq libraries were prepared according to the Single Cell 3′ v2 Reagent Kits User Guide (10X Genomics). Sequence libraries were constructed using the Chromium Single-Cell 3′ Library Kit (10X Genomics). Sequencing libraries were loaded on a NextSeq500 (Illumina) to obtain a sequencing depth of~200K reads per cell. 329 cells from the tracheal epithelium, 721 cells from the tracheal mesenchyme and 460 cells from whole lung parenchyma were captured and sequenced. For the epithelium 143,824 mean reads per cell were obtained corresponding to 2133 UMI counts and 908 genes expressed per cell. For the mesenchyme 190,186 reads per cell were obtained corresponding to 5383 UMI counts and 1891 genes expressed per cell. For the lung parenchyma, 320,071 mean reads per cell were obtained corresponding to 3969 UMI counts per cell and 1636 genes expressed per cell. Data are deposited under GSE152501.
Single Cell Analysis
Single cell reads were mapped to the mouse genome reference (ENSEMBL, GRCm38) and pre-processed with Cell Ranger v.2.0.1 to obtain the matrix of UMI counts per gene per cell. We used Seurat v.3 to normalize, scale and regress out unwanted sources of variation (like cell degradation), and subsequently identified highly variable genes for linear dimensionality reduction with principal component analysis (PCA). The principal components were then used for clustering (using the Louvain algorithm). Further non-linear dimensionality reduction was done with Uniform Manifold Approximation and Projection (UMAP) for visualization purposes. Differential gene expression was tested using MAST (Finak et al., 2015). Clusters identified with the Louvain method were annotated based on their differentially expressed genes (DEGs).
QUANTIFICATION AND STATISTICAL ANALYSIS
Stained sections were analyzed with a Nikon Eclipse NiE or a Leica SP6. Images were converted with Fiji/ImageJ. For quantification of cells, at least 3 randomly selected, non-consecutive regions per trachea were imaged and counted per animal. Images were analyzed and quantified with ImageJ/FIJI (version 2.0.0, NIH). Statistical significance was determined by two-tailed Student’s t test on mean values, and percentages and proportions were arcsine square root transformed before statistical testing. Significance was determined as *p < 0.05, **p < 0.01, and ***p < 0.001; Deviance from the mean is displayed as standard deviation.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45, BUV395 (clone 30-F110) | BD Biosciences | Cat#564279; RRID:AB_2651134 |
| CD19, BUV737 (clone ID3) | BD Biosciences | Cat#612781; RRID:AB_2870110 |
| CD64, BV421 (clone X54-5/7.1) | Biolegend | Cat#139309; RRID:AB_2562694 |
| CD24, BV510 (clone M1/69) | Biolegend | Cat#101831; RRID:AB_2563894 |
| CD80, BV605 (clone 16-10a1) | Biolegend | Cat#104729; RRID:AB_11126141 |
| CD86, BV650 (clone 24F) | BD Biosciences | Cat#743214; RRID:AB_2741350 |
| MHC II (I-A/I-E), FITC (clone M5/114.15.2) | Biolegend | Cat#107606; RRID:AB_313321 |
| CD40, PE (clone 3/23) | BD Biosciences | Cat#561846; RRID:AB_10896482 |
| Siglec F, PE-CF594 (clone E50-2440) | BD Biosciences | Cat#562757; RRID:AB_2687994 |
| Ly6C, PerCp5.5 (clone HK1.4) | Biolegend | Cat#128012; RRID:AB_1659241 |
| F4/80, PE-Cy7 (clone BM8) | Biolegend | Cat#123114; RRID:AB_893478 |
| CD103, APC (clone 2E7) | Biolegend | Cat#121432; RRID:AB_2566552 |
| Ly6G, AF700 (clone 1A8) | Biolegend | Cat#127622; RRID:AB_10643269 |
| CD11b, APCFire (clone M1/70) | Biolegend | Cat#101262; RRID:AB_2572122 |
| CD45, APC (clone 30-F11) | Biolegend | Cat#103112; RRID:AB_312977 |
| CD45, FITC (clone 30-F11) | Biolegend | Cat#103108; RRID:AB_312973 |
| CD45, BV421 (clone 30-F11) | Biolegend | Cat#103134; RRID:AB_2562559 |
| CD45.2, BUV737 (clone 104) | BD Biosciences | Cat#612778; RRID:AB_2870107 |
| F4/80, APC (clone BM8) | Biolegend | Cat#123116; RRID:AB_893481 |
| F4/80, BV421 (clone BM8) | Biolegend | Cat#123132; RRID:AB_11203717 |
| CD11b, FITC (clone MI/70) | Biolegend | Cat#101206; RRID:AB_312789 |
| CD11c, PeCy7 (clone N418) | Biolegend | Cat#117317; RRID:AB_493569 |
| SiglecF, Alexa647 (clone E50-2440) | BD Biosciences | Cat#562680; RRID:AB_2687570 |
| CD64, PE (clone X54-5/7.1) | Biolegend | Cat#139303; RRID:AB_10613467 |
| Ly6G, PECy7 (clone 1A8) | Biolegend | Cat#127617; RRID:AB_1877262 |
| IL4Ra, APC (clone I015F8) | Biolegend | Cat#144807; RRID:AB_2750451 |
| Krt5 (rabbit) | Covance | Cat#Prb-160P; RRID:AB_10063444 |
| Acetylated tubulin (mouse) | Sigma | Cat#T7451; RRID:AB_609894 |
| F4/80 (rat) | Biorad | Cat#MCA497GA; RRID:AB_323806 |
| RFP (goat) | MyBioSource | Cat#MBS448122 |
| CD45 (goat) | R and D Systems | Cat#AF114; RRID:AB_442146 |
| Bacterial and Virus Strains | ||
| Ad-CMV-GFP | University of Iowa Viral Vector Core | Ad5CMVeGFP |
| Ad-CMV-Cre-GFP | University of Iowa Viral Vector Core | Ad5CMVCre-eGFP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma Aldrich | Cat#T5648 |
| Polidocanol | Sigma Aldrich | Cat#88315-100G |
| “Isothesia” Isoflurane | Henry Schein | Cat#029405 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat# 017-000-121 |
| Bovine Serum Albumin | Fisher Chemical | Cat#BP1600-100 |
| Triton-X | Fisher BioReagents | Cat#BP151-100 |
| EdU (5-ethynyl-20-deoxyuridine) | Invitrogen | Cat#C10340 |
| Collagenase A (1.5 mg/mL) | Roche | Cat#10103586001 |
| DNase I (0.4 mg/mL) | Roche | Cat#10104159001 |
| Dispase | Corning | Cat#354235 |
| RPMI 1640 Media | GIBCO | Cat#11875093 |
| Hoechst 33342p | Invitrogen | Cat#H1399 |
| Calcein Blue, AM | Invitrogen | Cat#C1429 |
| D-Sucrose | Fisher Bioreagents | Cat#BP220-1 |
| OCT Compound | Fisher Healthcare | Cat#4585 |
| Paraffin | Fisher Chemical | Cat#T565 |
| Xylenes | Fisher Chemical | Cat#X3S-4 |
| Paraformaldehyde | Fisher Chemical | Cat#O4042-500 |
| Antigen Unmasking Solution, Citric Acid Based | Vector Labs | Cat#H-3300 |
| DMEM/F-12 | GIBCO | Cat#11330-057 |
| HEPES | Fisher Bioreagents | Cat#BP299-1 |
| Sodium Bicarbonate (7.5%) | Sigma-Aldrich | Cat#S8761 |
| L-Glutamine | GIBCO | Cat#25030164 |
| Pen-Strep | GIBCO | Cat# 15140-122 |
| Amphotericin B | GIBCO | Cat#152-90-018 |
| Insulin-Transferrin-Selenium | GIBCO | Cat#41400045 |
| Cholera Toxin | Sigma-Aldrich | Cat#C8052 |
| Recombinant Mouse Epidermal Growth Factor | R&D Systems | Cat#2028-EG-200 |
| Bovine Pituitary Extract | GIBCO | Cat#13028-014 |
| Trypsin (0.25%) | GIBCO | Cat#25200114 |
| Retinoic Acid | Sigma-Aldrich | Cat#R2625 |
| GFR 3D Matrigel | Corning | Cat#356231 |
| Recombinant Mouse Il1b Protein | R&D Systems | Cat#401-ML-010 |
| Recombinant Mouse IL2 Protein | R&D Systems | Cat#402-ML-020 |
| Recombinant Mouse IL4 Protein | R&D Systems | Cat#404-ML-010 |
| Recombinant Mouse IL5 Protein | R&D Systems | Cat#405-ML-005 |
| Recombinant Mouse IL13 Protein | R&D Systems | Cat#413-ML-005 |
| Recombinant Mouse SCF Protein | R&D Systems | Cat#455-MC-050 |
| Recombinant Mouse MCSF Protein | R&D Systems | Cat#416-ML-010 |
| Recombinant Mouse IL3 Protein | R&D Systems | Cat#403-ML-010 |
| Deposited Data | ||
| Single Cell Sequencing Raw Data | NCBI GEO | GSE152501 |
| Flow Cytometry Representative Data | FlowRepository | FR-FCM-Z2MU |
| Experimental Models: Organisms/Strains | ||
| Tg(Csf1r-EGFP)1Hume/J mouse | The Jackson Laboratory, made by Sasmono et al., 2003 | Stock#018549 |
| Ccr2tm2.1Ifc/J mouse, heterozygous and homozygous | The Jackson Laboratory, made by Saederup et al., 2010 | Stock#017586 |
| B6.129S4-Arg1tm1Lky/J mouse | The Jackson Laboratory, made by Reese et al., 2007 | Stock#015857 |
| Il4ratm1Sz/J mouse | Received from NIH, made by (Noben-Trauth et al., 1997) | Stock#003514 |
| Software and Algorithms | ||
| Fiji/ImageJ | Fiji | https://fiji.sc/ |
| NIS-Elements | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Leica Application Suite X | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| FlowJo | BD Biosciences | https://www.fiowjo.com/ |
| Cytobank | Cytobank | https://www.cytobank.org/ |
| Cell Ranger | 10X Genomics | v.2.0.1 |
| Seurat | Satija Lab | v3 |
| OMIQ | OMIQ | https://www.omiq.ai |
| Other | ||
| Cryostat | Leica | CM1950 |
| Inverted fluorescent microscope | Nikon | Eclipse NiE |
| Confocal | Leica | SP6 |
| Flow Cytometer (Sorting) | Beckman Coulter | MoFlo Astrios |
| Flow Cytometer (Analysis) | BD Biosciences | Aria II |
| Stratedigm | Stratedigm | S1000EXI |
Highlights.
The myeloid compartment composition in the trachea changes upon injury
Monocytes and macrophages promote tracheal epithelial repair in vivo
Monocytes and macrophages support basal cell growth in vitro
Tracheal and lung macrophages are distinct at the transcript and protein level
ACKNOWLEDGMENTS
We thank Drs. Darrell Kotton, Brigid Hogan, and Joseph Mizgerd for helpful comments on the manuscript. We would like to thank the following people for excellent technical assistance: Brian Tilton (BUSM Flow Cytometry core), Dr. Yuriy Alekseyev, Ashley LeClerc (BUSM Sequencing core) and Dr. Michael T. Kirber (BUSM Cellular Imaging core). This research was funded by the National Institutes of Health grant numbers R01 HL127002, U01 HL148692, and U01 HL134766 (JRR); the Swiss National Science Foundation (172221); and by a sponsored research agreement from Janssen Pharmaceuticals (S.A.M.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108553.
DECLARATION OF INTERESTS
S.A.M. received sponsored research funding from Janssen Pharmaceuticals.
REFERENCES
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, and Masopust D (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Ciccolella CO, Anno R, Halpert R, Spidlen J, and Snyder-Cappione JE (2019). Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat. Commun 10, 5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, and Randell SH (2001). Evidence for stem-cell niches in the tracheal epithelium. Am. J. Respir. Cell Mol. Biol 24, 662–670. [DOI] [PubMed] [Google Scholar]
- Byrne AJ, Mathie SA, Gregory LG, and Lloyd CM (2015). Pulmonary macrophages: key players in the innate defence of the airways. Thorax 70, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang X-M, Foo S, Nakamizo S, Duan K, et al. (2019). Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, 80. [DOI] [PubMed] [Google Scholar]
- Dagher R, Copenhaver AM, Besnard V, Berlin A, Hamidi F, Maret M, Wang J, Qu X, Shrestha Y, Wu J, et al. (2020). IL-33-ST2 axis regulates myeloid cell differentiation and activation enabling effective club cell regeneration. Nat. Commun 11, 4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahay H, Pessotti AD, Coote J, Montgomery BE, Xia D, Wilson A, Yang H, Wang Z, Bevan L, Thomas C, et al. (2015). Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 10, 239–252. [DOI] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, Leach SM, Bratton DL, Henson PM, Janssen WJ, and Jakubzick CV (2017). Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol 57, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Watanabe S, Verma R, Jablonski RP, Chen CI, Cheresh P, Markov NS, Reyfman PA, McQuattie-Pimentel AC, Sichizya L, et al. (2020). A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur. Respir. J 55, 1900646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, Schmierer M, Gabrusiewicz K, Anderson NR, Petty NE, et al. (2020). Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol 38, 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, and Erle DJ (2002). Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med 8, 885–889. [DOI] [PubMed] [Google Scholar]
- Lechner AJ, Driver IH, Lee J, Conroy CM, Nagle A, Locksley RM, and Rock JR (2017). Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration following Pneumonectomy. Cell Stem Cell 21, 120–134.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GRS, and Perlman H (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol 49, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth N, Shultz LD, Brombacher F, Urban JF, Gu H, and Paul WE (1997). An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc. Natl. Acad. Sci. U. S. A 94, 10838–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, and Locksley RM (2007). Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, and Hogan BLM (2011). Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol 27, 493–512. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells asstem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Randell SH, and Hogan BLM (2010). Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech 3, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL, Ransohoff RM, and Charo IF (2010). Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One 5, e13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR, and Hume DA (2003). A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101, 1155–1163. [DOI] [PubMed] [Google Scholar]
- Sertl K, Takemura T, Tschachler E, Ferrans VJ, Kaliner MA, and Shevach EM (1986). Dendritic cells with antigen-presenting capability reside in airway epithelium, lung parenchyma, and visceral pleura. J. Exp. Med 163, 436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Wang Y, Barak LS, Bai Y, Randell SH, and Hogan BLM (2014). IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA 111, E3641–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SYS, and Krasnow MA (2016). Developmental origin of lung macrophage diversity. Development 143, 1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ural BB, Yeung ST, Damani-Yokota P, Devlin JC, de Vries M, Vera-Licona P, Samji T, Sawai CM, Jang G, Perez OA, et al. (2020). Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci. Immunol 5, eaax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, and Pollard JW (2013). Macrophage biology in development, homeostasis and disease. Nature 496, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available. FlowRepository: http://flowrepository.org/id/RvFrhTPb7LK2nAIF366KA2ta9q5awmTMo5zYwWCtyRqXotLW4DPmsd2coPsjMgaG; Single Cell Sequencing Data are deposited under GSE152501; Data related to Figure 3: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/macrophages/macrophages; Data related to Figure 2: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/19_09_18_alex_merge_sctransform/19_09_18_alex_merge_sctransform