Abstract
Here we present and describe data on homozygous deletions (HD) of human CDKN2 A and neighboring regions on the p arm of Chromosome 9 from cancer genome sequences deposited on the online Catalogue of Somatic Mutations in Cancer (COSMIC) database. Although CDKN2 A HDs have been previously described in many cancers, this is a pan-cancer report of these aberrations with the aim to map the distribution of the breakpoints. We find that HDs of this locus have a median range of 1,255,650bps. When the deletion breakpoints were mapped on both the telomere and centromere proximal sides of CDKN2A, most of the telomere proximal breakpoints concentrate to a narrow region of the chromosome which includes the gene MTAP.. The centromere proximal breakpoints of the deletions are distributed over a wider chromosomal region. Furthermore, gene expression analysis shows that the deletions that include the CDKN2A region also include the MTAP region and this observation is tissue independent. We propose a model that may explain the origin of the telomere proximal CDKN2A breakpoints Finally, we find that HD distributions for at least three other loci, RB1, SMAD4 and PTEN are also not random.
Keywords: CDKN2A, Tumor suppressor, Homozygous deletion
1. Introduction
Cancers are in general characterized by the lifelong accumulation of thousands of mutations and other chromosomal aberrations resulting from inappropriately repaired DNA damage. These mutations do not accumulate linearly throughout the life of the individual. Rather, mutations of certain key genes serve as “primers” for rapid mutation accumulation [1]. This DNA damage may be caused by exogenous agents, but a vast majority occurs during DNA replication, accumulating particularly as a result of lagging strand synthesis [2,3].
In addition to simple base pair substitutions and small insertions and deletions (InDels) most cancer cells are also characterized by a high level of numerical instability (nCIN) that leads to aneuploidy [4] as well as structural chromosomal instability (sCIN) which represents large deletions, duplications and translocations [5]. These large chromosomal aberrations are the result of improperly rescued stalled or collapsed replication forks [6–8]. In at least one analysis of mice livers it was revealed that accumulation of chromosomal instability accelerates later in the life of the tissue [9]. We and others have previously shown that mutations in key genes that control the fidelity of replication and facilitate accurate DNA damage repair increase the numbers of sCINs [10–12].
It has been long appreciated that the most represented mutations in cancer cells are of genes that control the cell cycle and those involved in DNA damage repair. For example, p53 mutations are identified in over 50% of cancers [13]. The second most represented genetic aberration in cancers is in the INK4-ARF locus which is found on Ch9p21 [14–16]. This locus has two cyclin dependent kinase inhibitors: CDKN2 A which encodes p16 and the splicing variants p12 and p14 (ARF), and CDKN2B which encodes p15. p15 and p16 are inhibitors of Cyclin D/CDK4/6 while p14(ARF) promotes p53 stabilization. p12 is a splicing variant of p16 expressed only in pancreatic tissues. A non-coding RNA known as ANRIL required for regulation of transcription of the CDKN2 A/B loci is also encoded.
Mutations that inactivate CDKN2A/B locus have been studied for over 20 years [17,18] and from the beginning attempts have been made to catalogize the type of mutations observed in cancer cells [19–21]. The types of chromosomal aberrations observed at this locus in cancer cells fall into three general categories, simple mutations (amino acid substitution, insertion or deletion), promoter methylation, and homozygous deletions larger than 100 bp [22–24]. Remarkably, in some cancers, over half of the chromosomal aberrations that span the CDKN2A locus are large homozygous deletions (HDs) [25–28].
CDKN2 A HDs have been reported in several cancers (we only name a few) [29–33,66] including a recent pan-cancer distribution of of CDKN2A homozygous deletions. Here, we built on these previous studies and asked whether the breakpoints of these deletions distrubute equally on either side of the CDKN2A locus and attempted to identify a potential reason for the braks. We queried the COSMIC database which reports homozygous deletions in a variety of cancers using an Affymetrix SNP6.0 array [34–36]. The major finding of this study is that the CDKN2 A HDs telomere proximal breakpoints originate in a small chromosomal region (Chr9:20000000–22000000) where the gene MTAP is located. The centromere proximal breakpoints spread over a larger region. We propose a model which suggest that these homozygous deletions may require two events: loss of heterozygosity followed by loss of both copies (homozygous deletion).
2. Materials and methods
2.1. Homozygous deletion breakpoint data analysis
For this analysis, we relied on the data presented on Catalogue of Somatic Mutations in Cancer (COSMIC) website [34] using the copy number variation CONAN tool. The data deposited on COSMIC CONAN were derived from an Affymetrix SNP 6.0 Array and analyzed with PICNIC [37] and ASCAT [38]. Note that this database is a repository for many different studies. Therefore, it does not rely on data from only one study (please see results section). Deletion breakpoints were downloaded from the COSMIC CONAN database present on the same website (https://cancer.sanger.ac.uk/cosmic/conan/search). All aberration breakpoints for CDKN2A, RB1, SMAD4 and PTEN were downloaded as an Excel file. For the Chromosome 9 breakpoint distribution the search term was “9:1-40000000” (Genomic Region). For the genes breakpoint data the search terms were “CDKN2A, RB1, SMAD4, PTEN” (HGNC Gene Symbol). All data were exported as .csv format. Graphs of the breakpoints were analyzed in either SPSS or Excel.
2.2. Gene expression analysis
Two methods were used to extract the relevant gene expression data from the TCGA cancer samples analyzed for HDs. Note that only some of the TCGA samples also reported gene expression data. (1) A ‘PERL’ script was generated to load the raw Gene Expression Data as a .tsv file and the TCGA sample list. Then every sample of the Gene Expression Data was scanned and matched with TCGA sample list and gene name. All the matches were then written out to separate “per gene name” files. (2) The Linux ‘grep’ built in command was used to scan the Gene Expression Data.tsv file, extract all samples that match any of the TCGA patients and output results to file. Then the same ‘grep’ command was used to load this output file and match it with the gene name, then output matches to separate “per gene” files. The “per gene” name output files from both methods were compared using a PERL script and found to be identical.
2.3. Raw data availability
The data can be accessed on COSMIC CONAN using the prompts described in the above section. The files can be downloaded in .csv format. Each file contains 8 columns. The “# Sample” column lists the cancer subject identifier. The “Tissue” column lists the cancer tissues analyzed. The “Segment start” and “Segment end” columns list the chromosomal coordinates for the aberrations. These coordinates are at the resolution of the array. The “Total copy number” column lists the number of alleles identified. The “Minor allele” column represents the copies of the least frequent allele. The “Classification” column lists the type of aberration: HD = homozygous deletion, AMP = amplification, LOH = loss of heterozygosity. The gene expression data was downloaded as a .tsv file.
3. Results and discussion
3.1. A concentration of breakpoints in a narrow region on Chromosome 9
The COSMIC (Catalogue of Somatic Mutations in Cancer) database deposits cancer genome data from different sources, including the ICGC (International Cancer Genome Consortium), the cell line project (https://cancer.sanger.ac.uk/cell_lines/about), and TCGA (The Cancer Genome Atlas). To understand whether Chromosome 9 is more prone to breaking in certain regions, we first analyzed the distribution of the breakpoints in all cancer genomes reported on COSMIC. We used data deposited on the Copy Number Analysis (CONAN) database that catalogues only the copy number variations (CNVs) [39] acquired with an Affymetrix SNP6.0 array and reports CNV segment start and segment end for all breakpoints at probe resolution. The coordinates give the minimal region of the deletion. When we generated scatter plots of both the left and right breakpoints of the p arm of Chromosome 9, we found that many breakpoints concentrate in a narrow region between coordinates 2000000020,000,000 and 25,000,000 (Supplementary Fig. S1). Note that this pattern is unique to this region and is not found anywhere on the p arm of Chromosome 9.
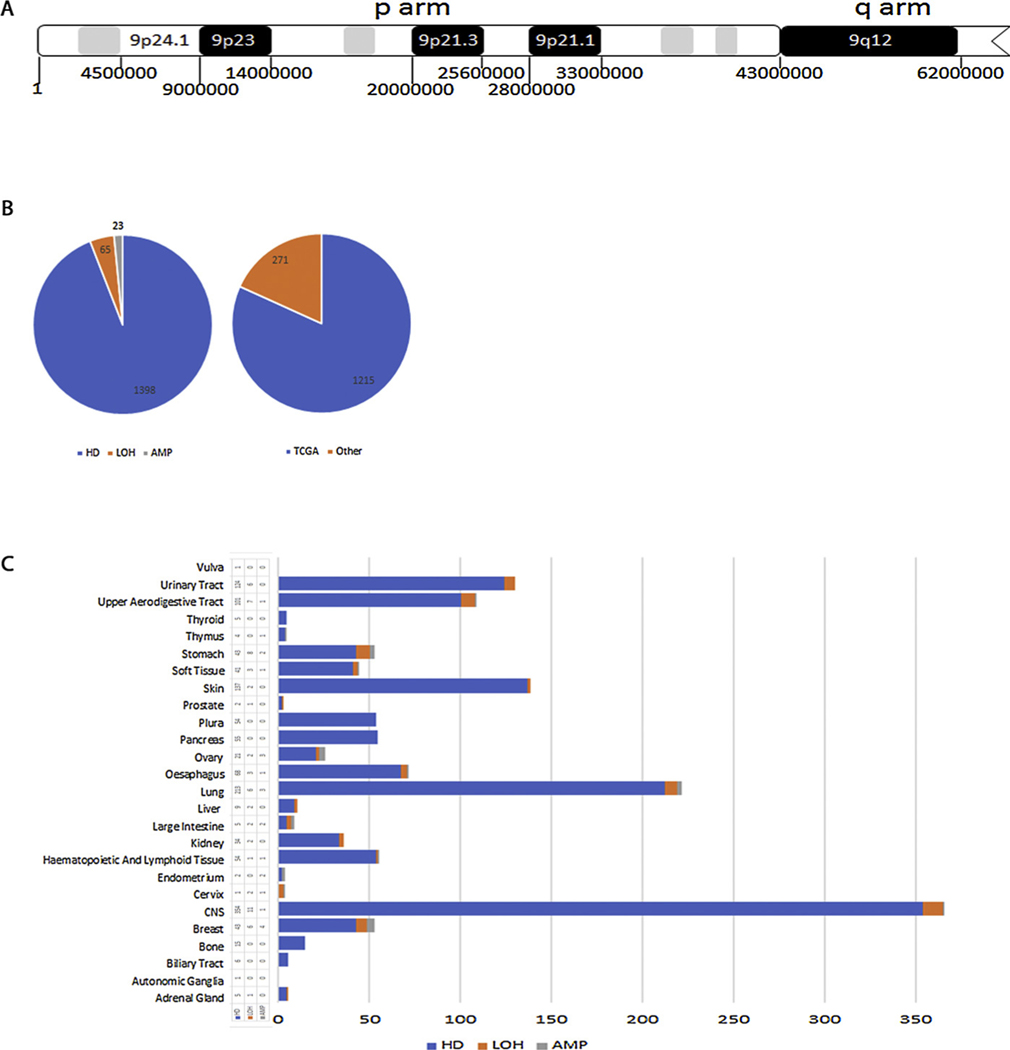
The concentration of breakpoints found in this narrow region of Chromosome 9 suggests that the chromosome is prone to breaking in this region which includes the CDKN2A locus but was not unexpected because the Chromosome 9 p21.1 and p21.2 (Fig. 1A) regions have been previously reported to be hotspots for large genomic aberrations [26,40,66]. We analyzed all the aberrations (1398) that include the CDKN2 A locus which fall into three categories: homozygous deletions (loss of both copies), loss of heterozygosity (loss of one copy) and amplification (gain of more than two copies) (Fig. 1B). 1215 of these samples are from TCGA while 271 are deposited from various other sources including the cell lines project and some have been described previously [66]. A significant number of these CDKN2A aberrations were reported in cancers of the central nervous system (CNS) but there was good representation of a variety of cancers, particularly lung (Fig. 1C). The high representation of aberrations in CNS is discussed later.
Fig. 1. Distribution of homozygous deletions that include the CDKN2A locus.
A. Map of the p-arm of chromosome 9 indicating some genomic regions and nucleotide coordinates. B. Pie charts showing the number of the three CDKN2 A aberrations (LOH, HD, AMP) analyzed in this study and their sources. C. Cancer tissue distribution of the three types of CDKN2 A aberrations.
3.2. Mapping breakpoints of CDKN2A aberrations
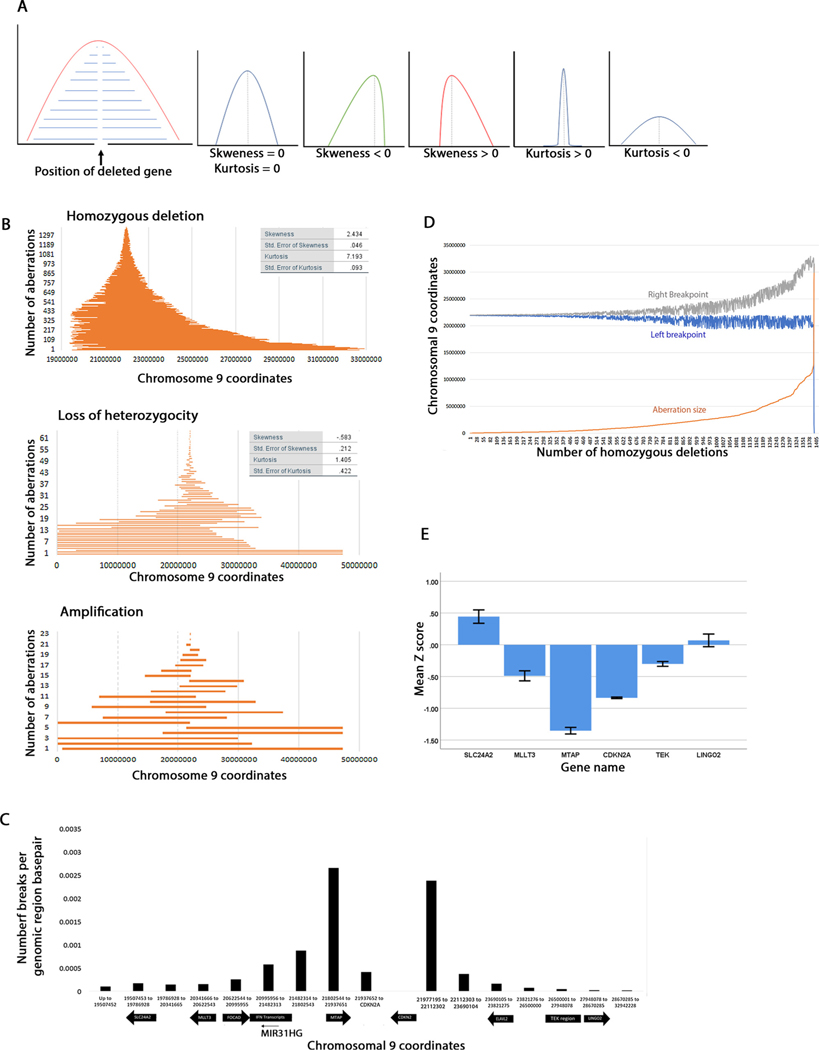
We hypothesized that if enough homozygous deletions are mapped, the breakpoints should have an equal distribution two the left and right of CDKN2A (Fig. 2A) and should form a perfect bell curve with zero skewness. However, if the breakpoints should concentrate in a narrower region on one side than on the other, the skewness would be negative (skewed to the left) or positive (skewed to the right).This would mean that breaks are more likely to occur on one side than the other. Kurtosis is a measure of how spread out the tails of a distribution are. Positive kurtosis distributions have small tails while negative kurtosis have long tails. This statistic will inform whether the breakpoints are really close to each other (short aberrations) or far away (long aberrations).
Fig. 2. Maps of chromosomal aberrations that include the CDKN2A locus.
A. Diagrams showing the interpretation of skewness and kurtosis statistics used in this study. We assume that for any deleted gene the breakpoints should map with equal frequency to the left and to the right. The skewness and kurtosis of such a curve is 0. A distribution skewed to the left will have a negative statistic while a distribution skewed to the right will have a positive statistic. Likewise, a sharp curve will have a positive kurtosis and a curve with wide tails will have a negative kurtosis. The kurtosis statistic will inform whether the breakpoints map close to the deleted gene (short deletions) or far (wide deletions). B. Graphs of homozygous deletions, loss of heterozygosity and amplification organized by size. Each line shows the span of an aberration. C. Density of CDKN2 A homozygous deletion breakpoint distribution to the left and right of CDKN2 A. Note that the graph is not to scale, and more loci are shown for the left breakpoint than for the right breakpoint. The coordinates were chosen arbitrarily between some loci. Chromosomal 9 coordinates are given for the indicated genes and the regions between the genes. D. Graph showing correlation between the length of homozygous deletion (orange), its left breakpoint (blue) and its right breakpoint (grey). Note that the left breakpoints are clustered, and the deletions size increase is dependent on the position of the right breakpoint. E. Gene expression levels of CDKN2 A and some loci flanking CDKN2 A. Shown are mean Z-scores and standard error for each gene.
When we mapped all of the reported HDs for the CDNK2A locus, the skewness was 19.712 and kurtosis was 736.15 (Supplementary Figure S2). However, a visual inspection showed that three homozygous deletions that extend over larger regions were likely to affect this statistic. When these homozygous deletions (coordinates 46587-31532212 CNS, 1627292-29951546 pancreas and 46587-141091394 kidney) were excluded from this graph, the skeweness was 2.434 and kurtosis 7.193 but both statistics were still positive (Fig. 2B). This indicates that the distribution is skewed to the right and most HDs (about 50%) are short. An analysis of CDKN2A deletions in cancer cell lines also found a shorter and a longer cluster [66]. A scatter graph also showed that most breakpoints concentrate just to the left and right of CDKN2A with no significant correlation between the position of the left and the right breakpoints (Pearson’s r= −.325) (Supplementary Figure S3A). Note that this is not a consequence of graphing method because in Fig. 2B each line shows position start and position end of the homozygous deletion. Clearly, the left breakpoints are more clustered that the right breakpoints (Supplementary Figure S3B,C). These data show that the breakpoints spread over a larger region on the centromere proximal side of CDKN2A. The deletions median range is 1,255,650bps. When we mapped the LOH aberrations we found that the breakpoints are moderately skewed to the left (skewness = −0.583, kurtosis = 1.405). These differences in skewness may have potentially explain how these how these homozygous deletions arise (discussed later). Remarkably the CDKN2A shows both small and large LOH events suggesting that it behaves like a fragile site as described in [67]. The amplifications map equally to the right and left of CDKN2A.
3.3. Analysis of the chromosomal region spanning the left and right breakpoints
We wanted to analyze in more detail the positions of the HD breakpoints in the regions flanking CDKN2 A. Fig. 2C shows the breakpoint density as number of breaks per genomic base pair with some of the genomic loci represented on the X-axis using information from the UCSC genome browser. Chromosomal coordinates were chosen to represent these loci or regions in between these loci. The left breakpoints clusters within 2.5Mbs (chromosomal coordinates 19400000-22000000) while the right breakpoints span over a region of about 10.96Mbs (Ch9: 21977195-32942228, approx. 10965033bp) which is about four times larger than the region spanning the left breakpoint.
The break interval where most left breakpoints occur includes the MTAP, the IFN genes, FOCAD, MLLT3 and SLC24 A2 genes, while the right breakpoints include ELAVL2, the TEK genes and LINGO2. Clearly most of the left breakpoints are concentrated in the MTAP region (Fig. 2C). We calculated the percentage of breakpoints in each region. Most left break points (1098/1398, 78.5%) occur approximately between coordinates 20995956-21937651, which is just under 1Mb. A smaller percentage of breakpoints (204/1398, 14.6%) occur in the region of the FOCAD, MLLT3, and SLC24A2 loci. Few breakpoints (96/1398, 6.9%) occur between CDKN2A and the end of MTAP. Within the right region we find that 70.7% of the breakpoints occur between CDKN2A and ELAVL2 (short deletions), 1.6% occur in the ELAVL2 region, 15.4% occur between ELAVL2 and the TEK region and 1.36% occur in the TEK region. The rest of the breakpoints are to the right of the TEK region. The deletion size increase is determined primarily by the position of the right breakpoint (Fig. 2D). Note that the left breakpoints remain concentrated in a small region while the right breakpoints move further and further to the right and this correlates with the increase in deletion size.
3.4. Analysis of gene expression in the region harboring homozygous deletions
We asked whether gene expression levels correlate with loss of CDKN2A as well as other loci flanking CDKN2A. However, only some of the HD samples acquired through the TCGA study also report gene expression and we checked gene expression for these ones. Gene expression should not be possible when both alleles are deleted. We note that gene expression levels for CDKN2A and MTAP regions where most of the breakpoints concentrate is low (Fig. 2E). This agrees with the data in Fig. 2B which shows that these HDs include both CDKN2A and MTAP. Moreover, loci farther to the left and right of the CDKN2A locus have increased levels of gene expression. The graph shows mean gene expression in all the samples reported to have CDKN2A HDs and is consistent with the spread of the breakpoints of these deletions (e.g. not all HDs include MLLT3 or the TEK region, so a percentage of the samples may show normal gene expression).
3.5. Tissue distribution of homozygous deletion breakpoints
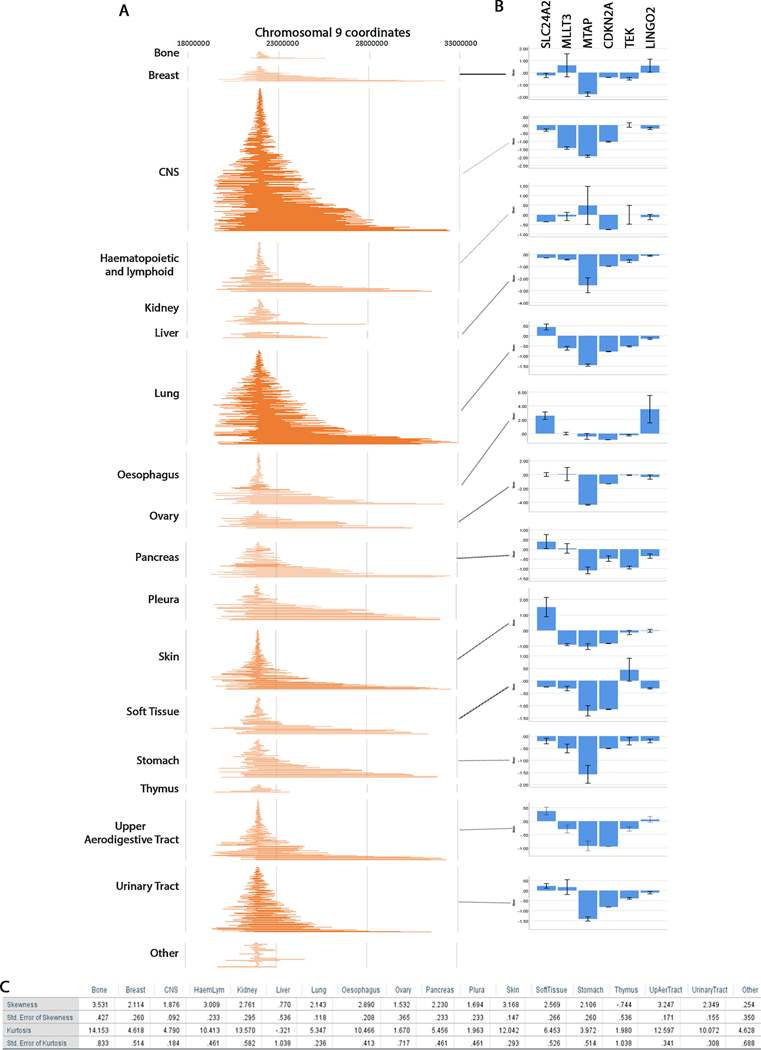
To understand whether the distribution of breakpoints is different in the various tissues analyzed, we graphed the breakpoints by tissue (Fig. 3A). We find that generally the distribution of breakpoints is similar for all tissues. We also plotted gene expression profiles for most tissues and compared with the breakpoint distribution (Fig. 3B). For some tissues gene expression was not available. The most important observation is that for most tissues MTAP is always co-deleted with CDKN2A. The exception was the haematopoietic and lymphoid tissues where some samples show normal expression of MTAP resulting in larger error. The skewness of this tissue is 3.009 with suggests that the breakpoints are highly skewed to the right (Fig. 3C). In this tissue, some of the left breakpoints may not incorporate MTAP.
Fig. 3. Tissue distribution of CDKN2A HD breakpoints and correlation with gene expression.
A. Maps of homozygous deletions by tissue similar to Fig. 2B. B. Gene expression per tissue similar to Fig. 2E. C. Skewness and kurtosis statistics for all maps in A.
We find that generally the skewness and kurtosis of the tissues correlated with gene expression; those with higher skewness have higher expression for the genes telomere proximal of CDKN2A (SLC24A2, MLLT3, MTAP) and lower gene expression of the genes centromere proximal to CDKN2A (TEK, LINGO2). For example, breast tissue has a high skewness but also high kurtosis. This suggests that some of the homozygous deletions extend far to the left of CDKN2A but most of them are short. Indeed, we see that MLLT3 and SLC24A2 are not always co-deleted with CDKN2A. Likewise, LINGO2 which is far to the right of CDKN2A is also not deleted in all samples because only one or two homozygous deletions extend to that region. This is also true for CNS and lung, the two tissues with the most homozygous deletions. Note that for CNS (skewness = 1.876, kurtosis = 4.790) most homozygous deletions spread as far as TEK and LINGO2 and expression of these genes is not as decreased as CDKN2A and MTAP while for lung (skeweness = 2.143, kurtosis = 5.347) TEK and LINGO2 show decrease expression because the high skewness and kurtosis indicates that most deletions are longer and spread farther to the right. Overall, these tissue independent analyses show that skewness and kurtosis correlate with levels of gene expression in the various tissues. COSMIC TCGA samples give Z-scores for gene expression in diploid tumor samples. Often genes that have high expression will show a more marked change (e.g. MTAP) However, CDKN2A/p16 expression in diploid tumor tissues is low (Z = −0.115 ± 0.127SEM for 20 diploid samples). A decrease from this value to a value of Z = −1.5is indicative of complete loss of gene expression.
3.6. Chromosome and replication origins analysis in CDKN2A neighboring regions
When we analyzed the nature of the repetitive elements in the region where aberrations breakpoints are found, we did see a high concentration of segmental duplications that localize within the IFNA transcripts region (Supplementary Figure S4C), where about 25% of the left breakpoints are found and within the TEK region where only 1.36% of right breakpoints are found (data not shown). Segmental duplications have been proposed to arise from repair of damaged DNA replication forks [8,41]. Although a correlation between the other approximately 74% of the breakpoints and segmental duplications cannot be made this still suggests that breaks may arise from rescued stalled or collapsed replication forks.
We next asked if there is anything unique about this region of the chromosome that make it prone to breakage. A snapshot of the human genome from UCSC genome browser [42] revealed that the telomere proximal region of the CDKN2 A locus is very gene rich but the centromere proximal region is not (data not shown). Histone H3K27 acetylation marks are associated with actively transcribed regions [43,44]. Indeed, the telomere proximal CDK2NA region is characterized by high levels of H3K27 acetylation indicating that these genes are highly transcribed (Supplementary Figure S4A). Further, the DNaseI sensitivity pattern also supports the observation that this region has relaxed chromatin characteristic of highly transcribed regions. The centromere proximal CDKN2A region has neither H3K27 acetylation marks nor is it sensitive to DNaseI typical of non-transcribed regions.
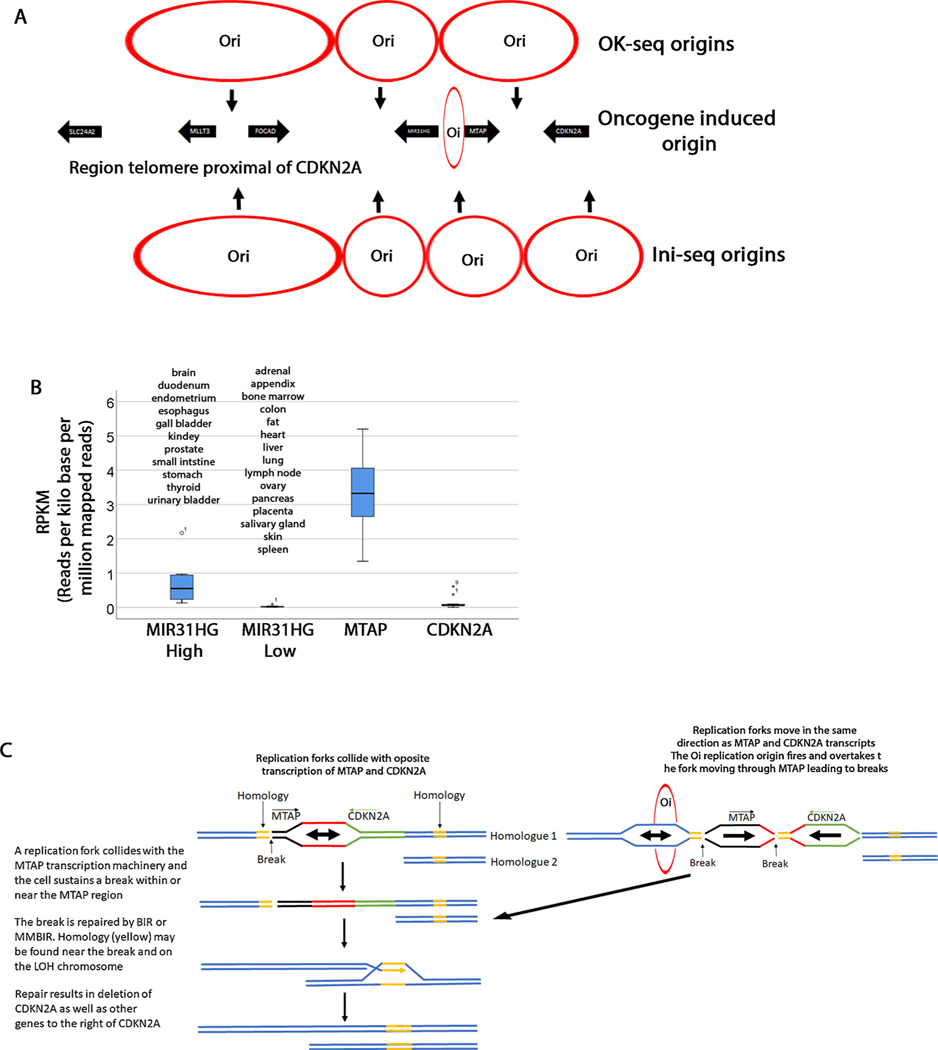
Transcription can cause chromosomal breaks when it interferes with DNA replication. Actively transcribed genes can act as replication fork barriers that can cause fork stalling and potential breaks [45]. To understand whether replication replication thorugh highly transcribed regions may be causing these breaks, we investigated the positions of origins of replications and fork directionality in this region. Petryk et al. have mapped all origins of replication and fork directionality by OK-seq [3]. We took a snapshot of the replication landscape in this region with the same chromosomal coordinates as the genome browser snapshot (Supplementary Figure S4). In the Petryk et al. analysis the presence of the Okazaki fragments was monitored (red and blue dots). A replication origin is determined by a shift between the blue dots and red dots with blue on the left and red on the right. A sudden shift represents an efficient origin. We identified 4 origins of replication within this region. Most genome wide replication forks have been shown to be oriented in the same direction as transcribed genes [3]. Here we found that at least in some cells one origin of replication may fire in between MTAP and CDKN2A while another one is right in the proximity of the MIR31HG gene (Fig. 4A). We hypothesized that the increase breakpoint frequency in this region may be due to collisions between replication forks and transcription machinery. However, a different study (Ini-seq origins) identified the same origins of replication in the FOCAD/MLLT3 and MIR31HG regions but places the other origins of replication on either side of MTAP and CDKN2A suggesting that that there is no collision [70]. Finally, one study showed that certain intragenic origins termed “oncogene induced origins (Oi)” are activated in cells with short G1 cycles. One such origin maps between MIR3HG and MTAP (Chr9: 21672000) [68]. Replication forks from these Oi origins may increase instability whether they are co-directional or head on with transcription [69]. Centromere proximal to CDKN2A we only mapped one other origin besides those shown in Fig. 4A between CDKN2A and ELAVL2 where most of the breakpoints are found using ini-seq data. This origin is found at position 22446404 next to DMRTA1 (data not shown). Forks from this origin advance in the same direction as DMRTA1 transcription. Other orirings were found further towards the centromere after coordinates 25350000.
Fig. 4. Speculative model of CDKN2a homozygous deletions.
A. Three origins of replication are shown (OK-seq) that fire at the position of the arrows. Data from Petryk et al. [3]. The replication forks converge where the circles touch. Four origins of replication (Ini-seq) [70] are shown on the bottom of the diagram. An oncogene induced origin is also shown. B. Expression level of MIR31HG, MTAP and CDKN2A from NCBI [46]. MIR31HG is differentially expressed in tissues and this graph groups all tissues with 0.135RPKM as high and those below 0.135RPKM as low. MTAP is ubiquitously expressed and the graph shows expression in all tissues. However, the pancreas has significantly lower expression levels (0.74RPKM) but it is represented in this graph. Although there is some differential expression in various tissues, when compared to MTAP, CDKN2A expression is overall lower (highest expression is 0.6RPKM in fat). C. Model explaining how CDKN2A deletions are generated. In this model we propose that the HDs arise in two steps. In the first step, one of the homologues (Homologue 2) may sustain a deletion (the whole arm or interstitial) leading to a LOH. A second break may occur due to interference between replication forks and MTAP or other genes (MIR31HG, CDKN2A). This break may be repaired by BIR or MMBIR from the LOH homologous chromosome. The outcome is that the CDKN2A and neighboring regions are deleted.
We next checked the level of transcription of these genes using data from NCBI (Fig. 4B) [46]. These data are reported as reads per kilo base per million mapped reads (RPKM). We also checked tissue distribution of these transcripts. Although, there is some differential expression of CDKN2A in various tissues, we find biquitous lower transcription when compared to MTAP which shows generally ubiqutous higher transcription. This observation was quite striking because the concentration of breakpoints in the MTAP region is not tissue specific suggesting that it this replication-transcription collision that may be responsible for the breaks. The MIR31HG expression is higher in brain, urinary and digestive systems and thyroid and lower in all other tissues. MIR41HG may contribute to the breaks in these tissues.
3.7. Speculative model for CDKN2A homozygous deletions
We propose a speculative model for the skewed distributions of these CDKN2 A HDs on the base of higher density of genes and origions on the telomere proximal side of CDKN2A (Fig. 4C). In this model a homozygous deletion may require two events. The first is an LOH event that leads to hemizygous deletion in one of the homologues. We note that the loss of heterozygosity distribution is skewed to the left (Fig. 2B) showing that segments of the left arm of chromosome 9 are preferentially lost in these samples. Note also that some of these LOH events spread to about coordinates 33,000,000. The second event involves a break within the MTAP or neighboring regions perhaps due to interference between replication and transcription. This break may then be repaired from the homologous chromosome by break induced replication (BIR). BIR has been shown to be the predominant mechanism for repair of DNA damage arising from stalled or collapsed replication forks [47]. Major repetitive sequences or passage through the centromere is not necessary because microhomology mediated BIR (MMBIR) has been identified which may require as few as 3–5 base pair homology and involves template switching [48]. This model proposes that this form of repair will result in homozygous deletion of CDKN2A and other genes between the break and wherever homology is found on the LOH chromosome. This also does not suggest that the window of homozygous deletion is identical for both chromosomes. In fact previous studies have shown that homozygous deletions may involve one longer deletion on one chromosome and a shorter one on the other [66]. Furthermore the CONAN coordinates predict the window of homozygous deletion with high confidence but does not exlude the posibility that the deletion on one of the chromosomes may be longer than the other; only that between the coordinates somes regions on both chromosomes are lost We realize that this is only a model and will require subsequent wet laboratory testing, but this is beyond the scope of this publication. This model also does not exclude the possibility of breaks occurring due to replication and transcription interference in the centromere proximal region of CDKN2A but this is less likely because both gene and origin density is lower. However, we did identify three Oi origins between CDKN2A and ELAVL2 that may collide with the oppositely transcribed CDKN2B (data not shown).
3.8. Functions of genes co-deleted with CDKN2A
We realize that large homozygous deletions should only be tolerated if the deleted genes are not essential. Therefore, there should be some form of selection for regions that do not include essential genes. The window of CDKN2A homozygous deletions lies between the ribosomal protein RPS6 (chr9:33025201-33039906) on the telomere side and and an enzyme essential for the trichloroacetic acid cycle (ACO1, chr9: 32384603-32450832) on the centromere side. These two genes could act as selectors for the HD window. The breakpoints of these deletions have been analyzed with ASCAT and PICNIC which should predict breakpoints with high accuracy at the resolution of the probe [37,38]. We investigated the functions and tissue-dependent expression of some of the co-deleted genes right and left of the CDKN2A (Table 1). Not unexpectedly none of these genes are essential and therefore dispensable for viability.
Table 1.
Function and tissue distribution of genes flanking the GDKN2A locus.
| Telomere proximal genes |
Centromere proximal genes |
||||
|---|---|---|---|---|---|
| Gene Name | Function | Tissue specificity | Gene Name | Function | Tissue specificity |
| SLC24A2 | Calcium/Cation Antiporter | Brain | ELAVL2 | Neural-specific RNA binding protein, metalloprotease stabilization | Brain |
| MLLT3 | Superelongation complex subunit, Mixed Lineage Leukemia family | Ubiquitous | TUSC 1 | Tumor supressor, possible role in lung tumors | N/A |
| FOCAD | Focal adehesion protein, tumor supressor function in gliomas | Ubiquitous | CAAP 1 | Apoptosis Inhibitor | Ubiquitous |
| MIR31HG | Small non-coding RNA Cellclar pluripotency and differentiation | Digestive system Urinary System Brain | TEK | Receptor Tyrosine Kinase | Ubiquitous |
| IFN | Proteins released by host cells in response to pathogens | Secreted | MOB3B | Mps 1 inter actor, mitotic checkpoint regulation | Ubiquitous Highest in lung |
| MTAP | Polyamine metabolism, adenine and methionine salvage | Ubiquitous | LING02 | Leucine rich repeat and Ig domain, associated with Parkinson’s disease | High in brain, endometrium, testis, thyroid |
The telomere proximal region includes several genes commonly altered in cancer cells. Inactivation of the MLL (Mixed Lineage Leukemia) and related genes have been identified in different forms of leukemias [49]. Many of these genes are inactivated by various translocations which produces chimeric mRNAs. The most famous is the Philadelphia chromosome translocation which produces a fusion between ABL1 and BCR1 [50]. Further analysis of translocation in leukemia cancers led to identification of a plethora of other genes which were named after the point of translocation. This includes MLLT3 (Multiple Lineage Leukemia Translocated to Ch. 3) [51]. MLLT3 characterized by tri-nucleotide repeats which may facilitate these form of translocations [52]. Several non-reciprocal translocations between MLLT3 and other chromosome loci have been identified [53]. The presence of an MLLT3 gene in the vicinity of CDKN2 A may explain why some of the breakpoints localize in this region but does not explain most of the breakpoints which occur in the MTAP region or between MTAP and MIR31HG. Remarkably, MTAP is the only transcript that collides with CDKN2 A which raises the possibility that collisions between transcriptional machineries may lead to a higher incidence of breaks in this region. However, in this analysis we do not have any data for this conclusion.
MIR31HG encodes a long-non-coding RNA with oncogenic properties that represses expression of p16 [54]. MIR31HG dysregulation has been identified in many cancers including pancreatic [54–56]. Thus, it appears that the homozygous deletion events seen here simultaneously inactivate both CDKN2A and its regulators. MTAP encodes the enzyme methylthioadenosine phosphorylase which is required early in the purine biosynthesis pathway [57]. Deletion of MTAP has been identified in many forms of cancers and is usually co-deleted with the CDKN2A locus [58].
The centromere proximal region includes three other cell cycle regulators (TUSC1, TEK, MOB3B) as well as an apoptosis regulator (CAAP1). TUSC1 is intriguing because it has been shown to have a possible role as a tumor suppressor in lung tumors [59] and some glioblastomas [60]. The TEK receptor tyrosine kinase functions in angiogenesis and it has been shown to be mutated particularly in lymphatic cancers [61]. The function of Mps1 in spindle checkpoint and its connection to cancer has also been actively studied [62,63]. Decrease in MOB3B transcriptional levels have also been associated with prostate cancers. CAAP1 has been shown to function in regulating apoptosis in response to double strand breaks arising from topoisomerase 2 errors [64,65]. This genomic analysis of the function of these genes shows the concentration of so many cell cycle regulators in this region allows for inactivation of several of them in one event.
Remarkably, we also found several brain specific genes (SLC24A2, ELAVL2, LINGO2). These observations suggest that in the CNS there may be selective pressure to also delete these genes and might explain why so many HDs appear in this tissue. We compared the tissue distributions of all genome-wide homozygous deletions with CDKN2a deletions and found a statistically significant increase in CDKN2A HDs in CNS (p < 0.0001, chi square test).
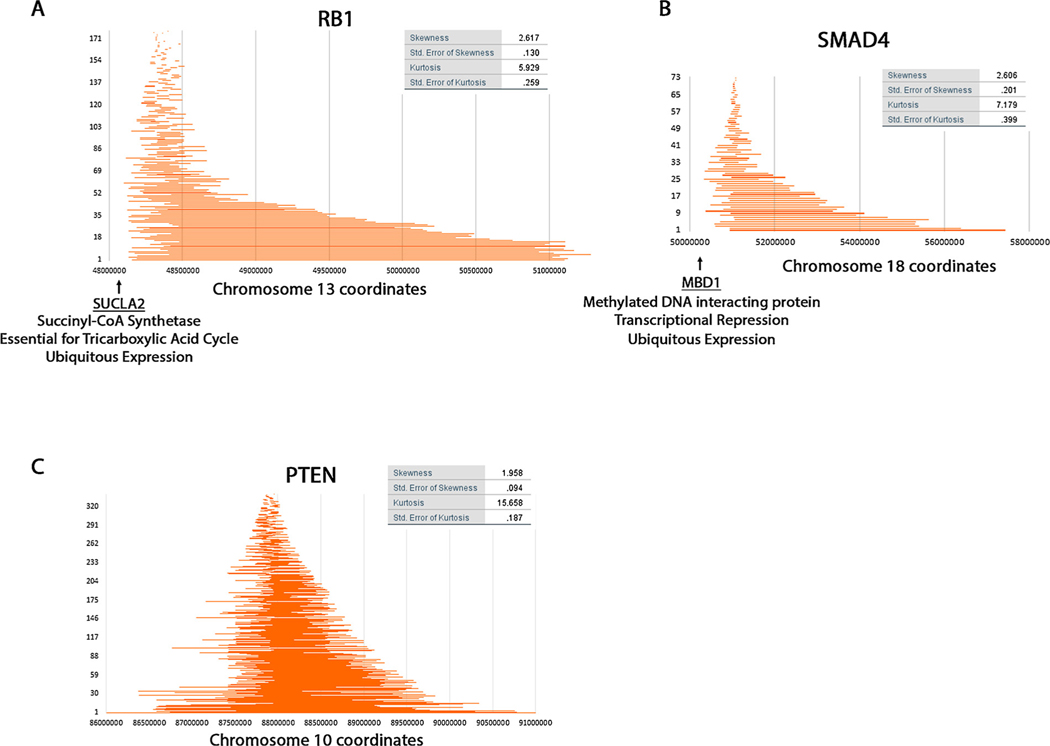
3.9. Distribution of homozygous deletions for other tumor supressor genes
We wanted to check whether other loci characterized by homozygous deletions have such skewed distributions. We investigated seven previously identified loci (Supplementary Figure S5) [66,67] and found that all these loci have a combination of both short and long deletions. Additionally, some of these loci appeared to show some skewness. In order analyze them similarly to CDKN2A we removed some of the longer deletions and identified 3 that show some pronounced skewness and kurtosis (RB1, SMAD4, PTEN (all deletions shown for PTEN)) (Fig. 5). We investigated the reason for this skewness in these genes but could not correlate it with transcription or replication as we did for CDKN2A. However, we identified SUCLA2 (Succinyl-CoA Synthetase) at position 48,000,000 immediately centromere proximal of RB1. Because this gene is essential for the tricarboxylic acid cycle it is possible that there is selection to retain the function of this gene though clearly some deletions include it (Supplementary Fig. S5).
Fig. 5. RB1, SMAD4, and PTEN breakpoint distribution of homozygous deletions.
Maps were generated for the three genes RB1 (A), SMAD4 (B) and PTEN (C) similarly to those shown in Fig. 2B.
We also found the gene MBD1 on one side of SMAD4. The MBD1 protein interacts with methylated DNA and is involved in transcriptional repression. Promoter methylation leading to changes in gene expression is a characteristic of cancer cells and is possible that this protein is required globally for this process. Therefore, there may be selection to preserve it.
We could not find a reason for the moderate skewness of PTEN. TGFBR2 also showed negative skewness (−2.309) when considering only the small deletions but we did not characterize it further because there were few data points and we are not confident in this statistic.
4. Conclusion
Previous analyses have shown that some cancers are characterized by CDKN2A deletions. Here we show that the telomere proximal breakpoints of these homozygous deletions are not random but concentrate in a region just left of CDKN2A. These aberrations are not cancer specific but are probably related to the structure or other molecular transactions in the chromosomal region in which they occur. We hypothesize that it is interference between replication and transcription that produces these concentrated breakpoints. Furthermore, this study agrees with previous studies showing that the CDKN2A homozygous deletions include the MTAP region [71]. Such skewed distributions of homozygous deletions were also found in other genes but the model is not immediately obvious.
Supplementary Material
Acknowledgements
We thank Dr. James and Ellen Bazzoli for their monetary sponsorship of our laboratory. We also thank Melissa Petreaca, Wayne Miles, and Rick Fishel for critical comments and scientific input.
Funding
This work was supported in part by the National Institutes of Health [grant number RO3 CA223545-01]. Other support from The Ohio State University startup funds.
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.mrfmmm.2019.04.002.
References
- [1].Kennedy SR, Loeb LA, Herr AJ, Somatic mutations in aging, cancer and neurodegeneration, Mech. Ageing Dev 133 (2012) 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Preston BD, Albertson TM, Herr AJ, DNA replication fidelity and cancer, Semin. Cancer Biol 20 (2010) 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, et al. , Replication landscape of the human genome, Nat. Commun 7 (2016) 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McClelland SE, Role of chromosomal instability in cancer progression, Endocr. Relat. Cancer 24 (2017) T23–T31. [DOI] [PubMed] [Google Scholar]
- [5].Venkatesan S, Natarajan AT, Hande MP, Chromosomal instability–mechanisms and consequences, Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793 (2015) 176–184. [DOI] [PubMed] [Google Scholar]
- [6].Li W, Vijg J, Measuring genome instability in aging - a mini-review, Gerontology 58 (2012) 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lambert S, Carr AM, Impediments to replication fork movement: stabilisation, reactivation and genome instability, Chromosoma 122 (2013) 33–45. [DOI] [PubMed] [Google Scholar]
- [8].Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, et al. , Break-induced replication repair of damaged forks induces genomic duplications in human cells, Science 343 (2014) 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J, Rapid accumulation of genome rearrangements in liver but not in brain of old mice, Nat. Genet 17 (1997) 431–434. [DOI] [PubMed] [Google Scholar]
- [10].Li PC, Petreaca RC, Jensen A, Yuan JP, Green MD, Forsburg SL, Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin, Cell Rep. 3 (2013) 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hromas R, Williamson E, Lee SH, Nickoloff J, Preventing the chromosomal translocations that cause Cancer, Trans. Am. Clin. Climatol. Assoc 127 (2016) 176–195. [PMC free article] [PubMed] [Google Scholar]
- [12].Bhattacharjee S, Nandi S, Choices have consequences: the nexus between DNA repair pathways and genomic instability in cancer, Clin. Transl. Med 5 (2016) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bieging KT, Mello SS, Attardi LD, Unravelling mechanisms of p53-mediated tumour suppression, Nat. Rev. Cancer 14 (2014) 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ortega S, Malumbres M, Barbacid M, Cyclin D-dependent kinases, INK4 inhibitors and cancer, Biochim. Biophys. Acta 1602 (2002) 73–87. [DOI] [PubMed] [Google Scholar]
- [15].Kim WY, Sharpless NE, The regulation of INK4/ARF in cancer and aging, Cell 127 (2006) 265–275. [DOI] [PubMed] [Google Scholar]
- [16].Poi MJ, Knobloch TJ, Sears MT, Warner BM, Uhrig LK, Weghorst CM, et al. , Alterations in RD(INK4/ARF) -mediated en bloc regulation of the INK4-ARF locus in human squamous cell carcinoma of the head and neck, Mol. Carcinog 54 (2015) 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Serrano M, The tumor suppressor protein p16INK4a, Exp. Cell Res 237 (1997) 7–13. [DOI] [PubMed] [Google Scholar]
- [18].LaPak KM, Burd CE, The molecular balancing act of p16(INK4a) in cancer and aging, Mol. Cancer Res 12 (2014) 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Smith-Sorensen B, Hovig E, CDKN2A (p16INK4A) somatic and germline mutations, Hum. Mutat 7 (1996) 294–303. [DOI] [PubMed] [Google Scholar]
- [20].Foulkes WD, Flanders TY, Pollock PM, Hayward NK, The CDKN2A (p16) gene and human cancer, Mol Med. 3 (1997) 5–20. [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao R, Choi BY, Lee MH, Bode AM, Dong Z, Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer, EBioMedicine. 8 (2016) 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. , The mutational landscape of head and neck squamous cell carcinoma, Science. 333 (2011) 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J, KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic Cancer, Cancers (Basel) 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mountzios G, Rampias T, Psyrri A, The mutational spectrum of squamous-cell carcinoma of the head and neck: targetable genetic events and clinical impact, Ann. Oncol 25 (2014) 1889–1900. [DOI] [PubMed] [Google Scholar]
- [25].Gast A, Scherer D, Chen B, Bloethner S, Melchert S, Sucker A, et al. , Somatic alterations in the melanoma genome: a high-resolution array-based comparative genomic hybridization study, Genes Chromosomes Cancer 49 (2010) 733–745. [DOI] [PubMed] [Google Scholar]
- [26].Lee B, Yoon K, Lee S, Kang JM, Kim J, Shim SH, et al. , Homozygous deletions at 3p22, 5p14, 6q15, and 9p21 result in aberrant expression of tumor suppressor genes in gastric cancer, Genes Chromosomes Cancer 54 (2015) 142–155. [DOI] [PubMed] [Google Scholar]
- [27].Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, et al. , Mutation spectrum revealed by breakpoint sequencing of human germline CNVs, Nat. Genet 42 (2010) 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Frazao L, do Carmo Martins M, Nunes VM, Pimentel J, Faria C, Miguens J, et al. , BRAF V600E mutation and 9p21: CDKN2A/B and MTAP co-deletions - Markers in the clinical stratification of pediatric gliomas, BMC Cancer 18 (2018) 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Adati N, Huang MC, Suzuki T, Suzuki H, Kojima T, High-resolution analysis of aberrant regions in autosomal chromosomes in human leukemia THP-1 cell line, BMC Res. Notes 2 (2009) 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Raschke S, Balz V, Efferth T, Schulz WA, Florl AR, Homozygous deletions of CDKN2A caused by alternative mechanisms in various human cancer cell lines, Genes Chromosomes Cancer 42 (2005) 58–67. [DOI] [PubMed] [Google Scholar]
- [31].Florl AR, Schulz WA, Peculiar structure and location of 9p21 homozygous deletion breakpoints in human cancer cells, Genes Chromosomes Cancer 37 (2003) 141–148. [DOI] [PubMed] [Google Scholar]
- [32].Xie H, Rachakonda PS, Heidenreich B, Nagore E, Sucker A, Hemminki K, et al. , Mapping of deletion breakpoints at the CDKN2A locus in melanoma: detection of MTAP-ANRIL fusion transcripts, Oncotarget. 7 (2016) 16490–16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Norris AL, Kamiyama H, Makohon-Moore A, Pallavajjala A, Morsberger LA, Lee K, et al. , Transflip mutations produce deletions in pancreatic cancer, Genes Chromosomes Cancer (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. http://cancer.sanger.ac.uk/cosmic.
- [35].Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. , COSMIC: somatic cancer genetics at high-resolution, Nucleic Acids Res. 45 (2017) D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Forbes SA, Beare D, Bindal N, Bamford S, Ward S, Cole CG, et al. , COSMIC: high-resolution Cancer genetics using the catalogue of somatic mutations in Cancer, Curr. Protoc. Hum. Genet 91 (2016) 10 1 1–1 37. [DOI] [PubMed] [Google Scholar]
- [37].Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, et al. , PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data, Biostatistics 11 (2010) 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. , Allele-specific copy number analysis of tumors, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Forer L, Schonherr S, Weissensteiner H, Haider F, Kluckner T, Gieger C, et al. , CONAN: copy number variation analysis software for genome-wide association studies, BMC Bioinformatics 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sasaki S, Kitagawa Y, Sekido Y, Minna JD, Kuwano H, Yokota J, et al. , Molecular processes of chromosome 9p21 deletions in human cancers, Oncogene 22 (2003) 3792–3798. [DOI] [PubMed] [Google Scholar]
- [41].Payen C, Koszul R, Dujon B, Fischer G, Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms, PLoS Genet. 4 (2008) e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. http://genome.ucsc.edu/.
- [43].Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kimura H, Histone modifications for human epigenome analysis, J. Hum. Genet 58 (2013) 439–445. [DOI] [PubMed] [Google Scholar]
- [45].Castan A, Hernandez P, Krimer DB, Schvartzman JB, The abundance of Fob1 modulates the efficiency of rRFBs to stall replication forks, Nucleic Acids Res. 45 (2017) 10089–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. , Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics, Mol. Cell Proteomics 13 (2014) 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anand RP, Lovett ST, Haber JE, Break-induced DNA replication, Cold Spring Harb. Perspect. Biol 5 (2013) a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hastings PJ, Ira G, Lupski JR, A microhomology-mediated break-induced replication model for the origin of human copy number variation, PLoS Genet. 5 (2009) e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Winters AC, Bernt KM, MLL-Rearranged Leukemias-An Update on Science and Clinical Approaches, Front. Pediatr 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, et al. , The Philadelphia chromosome in leukemogenesis, Chin. J. Cancer 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, et al. , Mllt3 gene on 9p22 involved in t(9–11) leukemia encodes a serine proline-rich protein homologous to Mllt1 on 19p13, Oncogene 8 (1993) 3085–3092. [PubMed] [Google Scholar]
- [52].Walker GJ, Walters MK, Palmer JM, Hayward NK, The Mllt3 gene maps between D9s156 and D9s171 and contains an unstable polymorphic trinucleotide repeat, Genomics 20 (1994) 490–491. [DOI] [PubMed] [Google Scholar]
- [53].Meyer C, Hofmann J, Burmeister T, Groger D, Park TS, Emerenciano M, et al. , The MLL recombinome of acute leukemias in 2013, Leukemia 27 (2013) 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, et al. , The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence, Nat. Commun 6 (2015). [DOI] [PubMed] [Google Scholar]
- [55].Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu S, et al. , Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b, Oncogene 35 (2016) 3647–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nie FQ, Ma SJ, Xie M, Liu YW, De W, Liu XH, Decreased long noncoding RNA MIR31HG is correlated with poor prognosis and contributes to cell proliferation in gastric cancer, J. Immunother. Emphasis Tumor Immunol 37 (2016) 7693–7701. [DOI] [PubMed] [Google Scholar]
- [57].Bertino JR, Waud WR, Parker WB, Lubin M, Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity Current strategies, Cancer Biol. Ther 11 (2011) 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen ZH, Zhang HY, Savarese TM, Gene deletion chemoselectivity: Codeletion of the genes for p16(INK4) methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy, Cancer Res. 56 (1996) 1083–1090. [PubMed] [Google Scholar]
- [59].Shan Z, Shakoori A, Bodaghi S, Goldsmith P, Jin J, Wiest JS, TUSC1, a putative tumor suppressor gene, reduces tumor cell growth in vitro and tumor growth in vivo, PLoS One 8 (2013) e66114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang R, Yu W, Liang G, Jia Z, Chen Z, Zhao L, et al. , Tumor suppressor candidate 1 suppresses cell growth and predicts better survival in glioblastoma, Cell. Mol. Neurobiol 37 (2017) 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Eklund L, Kangas J, Saharinen P, Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems, Clin Sci (Lond.) 131 (2017) 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW, Targeting mitosis in Cancer: emerging strategies, Mol. Cell 60 (2015) 524–536. [DOI] [PubMed] [Google Scholar]
- [63].Pachis ST, Kops G, Leader of the SAC: molecular mechanisms of Mps1/TTK regulation in mitosis, Open Biol. 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Aslam MA, Alemdehy MF, Pritchard CEJ, Song JY, Muhaimin FI, Wijdeven RH, et al. , Towards an understanding of C9orf82 protein/CAAP1 function, PLoS One 14 (2019) e0210526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wijdeven RH, Pang B, van der Zanden SY, Qiao X, Blomen V, Hoogstraat M, et al. , Genome-wide identification and characterization of novel factors conferring resistance to topoisomerase II poisons in Cancer, Cancer Res. 75 (2015) 4176–4187. [DOI] [PubMed] [Google Scholar]
- [66].Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, et al. , Signatures of mutation and selection in the cancer genome, Nature 463 (2010) 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Cheng, et al. , Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors, Nature Communications 8 (2017) 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Macheret Halzonetis, Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress, Nature 555 (7694) (2018) 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kotsantis, et al. , Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place, Cancer Discov. 8 (5) (2018) 537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Langley, et al. , Genome-wide identification and characterisation of human DNA replication origins by initiation site sequencing (ini-seq), Nucleic Acids Res. 44 (21) (2016) 10230–10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang, et al. , Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines, Cancer Genet Cytogenet 86 (1) (1996) 22–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data can be accessed on COSMIC CONAN using the prompts described in the above section. The files can be downloaded in .csv format. Each file contains 8 columns. The “# Sample” column lists the cancer subject identifier. The “Tissue” column lists the cancer tissues analyzed. The “Segment start” and “Segment end” columns list the chromosomal coordinates for the aberrations. These coordinates are at the resolution of the array. The “Total copy number” column lists the number of alleles identified. The “Minor allele” column represents the copies of the least frequent allele. The “Classification” column lists the type of aberration: HD = homozygous deletion, AMP = amplification, LOH = loss of heterozygosity. The gene expression data was downloaded as a .tsv file.