Radial meiotic initiation in the fetal ovary is patterned by Tex14-dependent germ cell bridges that hinder the loss of Dppa3.
Abstract
Meiosis is critical to generating oocytes and ensuring female fertility; however, the mechanisms regulating the switch from mitotic primordial germ cells to meiotic germ cells are poorly understood. Here, we implicate intercellular bridges (ICBs) in this state transition. We used three-dimensional in toto imaging to map meiotic initiation in the mouse fetal ovary and revealed a radial geometry of this transition that precedes the established anterior-posterior wave. Our studies reveal that appropriate timing of meiotic entry across the ovary and coordination of mitotic-meiotic transition within a cyst depend on the ICB component Tex14, which we show is required for functional cytoplasmic sharing. We find that Tex14 mutants more rapidly attenuate the pluripotency transcript Dppa3 upon meiotic initiation, and Dppa3 mutants undergo premature meiosis similar to Tex14. Together, these results lead to a model that ICBs coordinate and buffer the transition from pluripotency to meiosis through dilution of regulatory factors.
INTRODUCTION
Meiosis is critical to generating haploid gametes for reproduction and shuffling genetic information to generate diversity. This unique cell division starts in utero in females, arrests, and resumes after months (for mice) or years (for humans). Although oocyte maturation and the resumption of meiosis in adult ovaries have been investigated in great detail, little is known about early events of meiosis in the fetal ovary. In mice, meiosis occurs asynchronously starting at embryonic day 13.5 (E13.5) in the anterior region of the ovary and disseminates to the posterior in a wave-like pattern in response to a morphogen, retinoic acid (RA), which is produced in the mesonephros and the ovary (1–6). Although the function of RA in meiosis has been demonstrated, the persistence of meiosis in mice deficient in RA receptors (RARs) suggests that additional mechanisms, such as other extrinsic inducers or germ cell–intrinsic factors, participate in this multifaceted developmental milestone (7–9).
To date, much of our knowledge of meiotic entry is based on RNA in situ hybridization or immunostaining in histological sections. While whole-mount RNA in situ hybridization allows three-dimensional (3D) analysis of intact samples, the transcript level and localization do not necessarily reflect protein expression. While histological section analysis of the ovary reveals protein localization, it is labor intensive (10) and could lead to misinterpretation of 3D structure or overcounting of objects because the result is based on sampling rather than evaluating the entire structure. Recent advances in confocal imaging and 3D analysis enabled quantitative analysis of ovarian follicles and their geography in the mouse ovary (11–13) but have yet to be applied toward spatiotemporal dynamics of early female meiosis.
Here, we develop an algorithm to analyze meiotic initiation in the intact mouse fetal ovary with confocal imaging and a novel 3D quantitative analysis. This reveals earlier meiotic onset and a new, radial, pattern of meiotic initiation that precedes the anterior-posterior (A-P) wave of meiosis in mouse fetal ovaries. We show that the onset of meiosis in the ovary propagates via intercellular bridges (ICBs) that transiently connect germ cells during fetal development. In Tex14 mutant (Tex14−/−) mice, which are deficient in ICBs (14), we demonstrate the absence of functional cytoplasmic sharing and find that meiosis initiates and advances prematurely along the AP axis but without a radial wave. Upon entering meiosis, Tex14−/− germ cells more rapidly extinguish pluripotency-associated transcripts including Dppa3, and Dppa3 mutant ovaries likewise show earlier meiotic onset. These studies suggest that cytoplasmic sharing via ICBs slows and coordinates the cell-state transition from pluripotency to meiosis, which, we suggest, occurs by a dilution effect of regulatory factor related to epigenetic reprogramming. Thus, we uncover a role for ICBs in female meiotic initiation by coordinating and buffering gene expression changes across sister cells in a germline cyst.
RESULTS
Generating a 3D map of meiosis in mouse fetal ovaries
To investigate the spatiotemporal dynamics of meiosis in fetal ovaries, we performed immunofluorescence in toto with markers that distinguish nonmeiotic and meiotic germ cells. Following confocal microscopy and 3D reconstruction, we determined the localization of labeled germ cells by creating spots, each demarcating an individual germ cell. To quantify the distribution of labeled germ cells within the fetal ovary, we used polar coordinates to divide the ovary into seven segments of equal thickness in the sagittal plane along the longitudinal [medial-lateral (M-L)] axis, as well as seven segments of equal thickness in the transverse plane along the A-P axis. Comparative analysis of the germ cell population in each segment generated a 3D map of cell distribution in fetal mouse ovaries (Fig. 1A, fig. S1A, and movie S1).
Fig. 1. 3D mapping of meiosis in mouse fetal ovaries.

(A) Schematic summarizing 3D analysis method for ovaries. Total germ cell population is illustrated in green, while meiotic germ cells are depicted in red and magenta. Following whole-mount immunofluorescence and 3D visualization of embryonic ovaries, individual germ cells were identified. The ovary was divided into seven segments based on distance and angle from origin, respectively. In M-L distribution graph, bins 1 and 7 are most lateral, and 3 to 5 are medial (toward the middle) regions of the ovary. In A-P distribution graph, bin 1 is anterior, and 7 is posterior. (B) Oct4-GFP revealed homogeneous distribution of germ cells at E14.5, whereas (C) OCT4 protein showed mild posterior skewing (P = 0.0274). STRA8-expressing (D) and SYCP3-expressing (F) cells were homogeneously distributed, indicating that most germ cells completed premeiotic DNA replication and started synaptonemal complex assembly. Dashed rectangle in (C) shows borders of zoom in (Bi). (Cii), (Diii) to (iv), and dashed yellow line defines borders of the OCT4-expressing cell cluster. (G) SYCP1 expression was detected in very few cells at E14.5. Dashed rectangle in (G) shows borders of zoom in (i) and (ii). (E) Quantification of OCT4+, STRA8+, SYCP3+, and SYCP1+ germ cells in whole-mount at E14.5. Adjacent to each immunostaining is the graph of M-L (top) and A-P (bottom) distribution with identical color scheme. Primary y axes (on the left) indicate germ cell numbers in each bin, dashed lines represent individual ovaries, and color-coded thick lines show mean value for each antibody. Secondary y axes (on the right and colored in gray) present the percentage of OCT4, STRA8, and SYCP1 cells (normalized to total germ cells) in each bin. At least n = 4 ovaries (from two litters and two embryos) were analyzed for each time point.
We applied this new method for 3D analysis to the dynamic transition between pluripotent primordial germ cells (PGCs) and meiotic prophase at E14.5. Previous studies showed that meiosis in mouse fetal ovaries initiates in a wave, as transcripts of synaptonemal complex protein 3 (Sycp3) appear in an A-P gradient, and concurrently, transcripts of the pluripotency marker Oct4 recede in a reciprocal pattern (2). While the distribution of green fluorescent protein-positive (GFP+) germ cells in Pou5f1-ΔPE-eGFP (Oct4-GFP) E14.5 ovaries was homogeneous regardless of meiotic status (Fig. 1B, Bi), we observed that cells expressing endogenous OCT4 were skewed toward the posterior (Fig. 1C, Cii), in agreement with previous studies (1–3). Almost all Oct4-GFP+ germ cells were STRA8+ (Stimulated by retinoic acid 8) and SYCP3+ at E14.5, suggesting that premeiotic DNA replication and recruitment of the lateral elements of the synaptonemal complex already occurred [Fig. 1, D (Diii) and F]. In addition, we found that very few germ cells in the E14.5 ovary stained positive for synaptonemal complex protein 1 (SYCP1), a protein normally localized to the synaptonemal complex at the late zygotene stage (Fig. 1G, i and ii). In addition to the spatial distribution of OCT4+, STRA8+, SYCP3+, and SYCP1+ germ cell subsets, we scored total numbers of each in E14.5 ovaries (Fig. 1E). These results validate our new method for interrogating a large number of cells, as well as rare populations such as SYCP1+ germ cells in E14.5, to generate a 3D map of meiosis.
3D analysis reveals earlier meiotic onset and radial meiotic wave in mice
To uncover the spatiotemporal dynamics of the mitotic-meiotic transition, we examined E12.5 and E13.5 ovaries, which predominantly consist of pluripotent PGCs (Fig. 2). The germ cell marker TRA98 confirmed that germ cells are located throughout the E12.5 ovary in an even distribution (Fig. 2A). While almost all germ cells expressed OCT4 at E12.5 (and that not all express TRA98; Fig. 2B), wholemount imaging revealed very small populations of STRA8+ and SYCP3+ cells (Fig. 2, C and D, and fig. S1, B and C) (~18 and 5%, respectively) (Fig. 2E), making this the earliest time point at which these meiotic proteins have been detected and rigorously quantified thus far. In contrast to the previously established AP wave of meiotic initiation, the distribution of the earliest STRA8+ germ cells did not show a strong AP bias as there is no statistically significant difference between the three anterior bins (1 to 3 on x axis of A-P distribution graph) and posterior (bins 5 to 7) in E12.5 ovaries (Fig. 2C). The earliest SYCP3+ germ cells were localized in clusters (fig. S1C) and biased in their radial distribution, with a preference for the anterior and medial [located in the middle or interior; 3 to 5 on x axis of M-L distribution graph] region of the ovary at E12.5 (Fig. 2D, i and ii). By E13.5, the clustering behavior of SYCP3+ cells became more prominent (Fig. 2I, i to iii, and fig. S2B, i to iv), and although still medial, SYCP3+ germ cell clusters were not confined to the anterior tip but arrayed along the entire length of the ovary with overall bias toward the anterior. To investigate whether this medial expression pattern differs by genetic background, we analyzed SYCP3 expression in embryonic ovaries from pure C57BL/6 mice. Distinct from mixed background mice in Fig. 2D, there was no SYCP3 expression in E12.5, as meiosis initiates about a day later (around E13.5) in C57BL/6 ovaries with the first SYCP3+ cells (~1%) similarly located in the middle of the ovary toward the anterior (fig. S2C). In mixed background, we compared the distribution of SYCP3+ cells on the dorsal-ventral axis at E13.5 and observed that the number of SYCP3+ cells in the dorsal region of the ovary was twice that of the ventral region (Fig. 2, J and K).
Fig. 2. Earlier meiotic initiation and a new radial meiotic wave in embryonic mouse ovaries.
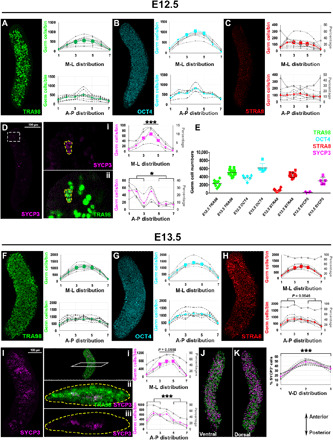
Detection of TRA98 (A), OCT4 (B), STRA8 (C), and SYCP3 (D) proteins in E12.5 ovaries by wholemount immunofluorescence. Sparse STRA8+ and SYCP3+ cells were identified as early as E12.5. White dashed rectangle in (D) indicates zoom at right. (Di) SYCP3+ meiotic germ cells at E12.5 are delineated by yellow dash with TRA98 stain in (Dii). Graphs show anterior skew of SYCP3+ (P = 0.0404) but not STRA8+ cells [(C); P = 0.8070] at E12.5. More SYCP3+ cells were detected in the medial part (segments 3 to 5) than lateral parts (1, 2, 6, and 7) (P = 0.0002) at E12.5. (E) Quantification of germ cells with each marker at E12.5 and E13.5. Wholemount immunolocalization of TRA98 (F), OCT4 (G), STRA8 (H), and SYCP3 (I) at E13.5. Uptick of STRA8+ and SYCP3+ germ cells was confirmed at E13.5, with STRA8+ cells enriched in the anterior (P = 0.0546). (J) SYCP3+ cells congregated in clusters in the anterior-medial part of the ovary [shown in transverse optical section view in (Ii) to (Iiii)] with significant A-P bias (P = 0.0001), although clusters were also noted near the posterior tip. (J) and (K) show SYCP3+ spots in the ventral and dorsal views of the E13.5 ovary, respectively, with graph showing the highest numbers in the middle segment and increased numbers in the dorsal compared to ventral region (P = 0.0003). Primary y axes (on the left of each graph) indicate germ cell numbers in each bin, dashed lines represent individual ovaries, and color-coded thick lines show mean value for each antibody. Secondary y axes (on the right and colored in gray) present percentage of STRA8 and SYCP3 cells (normalized to total germ cells) in each bin. At least n = 3 ovaries (from two litters and three to four embryos) were analyzed for each time point. ***P < 0.0001.
To confirm the radial wave of meiotic initiation observed with SYCP3, we used γH2AX, a marker of double-strand breaks (DSBs), which are initiated during the leptotene stage of meiotic prophase. Similar to SYCP3, γH2AX+ germ cells (validated by Oct4-GFP staining) mainly localized in the anterior-medial region of the ovary (fig. S2E). We colabeled with cleaved poly(adenosine 5′-diphosphate–ribose) polymerase (cPARP) to distinguish DSBs formed during homologous recombination from those induced by apoptotic DNA fragmentation. In contrast to the anterior-radial distribution of γH2AX+ cells, cPARP+ cells were localized in a spatially unbiased manner at E13.5, with only 5.8% of γH2AX+ GFP+ cells also staining positive for cPARP (fig. S2, F and J, top). To further validate spatial distribution of germ cell apoptosis, we analyzed expression of active caspase-3, which is another biochemical marker of apoptosis upstream of PARP cleavage [reviewed in (15)]. The distribution of active caspase-3+ germ cells was similar to cPARP+ (fig. S2, H and I), as ~75% of germ cells were double positive (cPARP+; active caspase-3+), while only ~25% of cells expressed cPARP without active caspase-3 immunoreaction (cPARP+; active caspase-3−) (fig. S2J, bottom). Meiotic but nonapoptotic γH2AX+ GFP+ and cPARP− germ cells, by contrast, were more likely to be located in the anterior core of the E13.5 ovary (fig. S2G). These experiments corroborate our observation with SYCP3 that a radial meiotic wave appears earlier than the AP meiotic wave in the mouse ovary. This pattern is similar to meiotic initiation in human ovaries, in which pluripotent, nonmeiotic germ cells are found in the peripheral region of the human ovary, while germ cells in meiosis or expressing late germ cell markers are located further toward the interior (16). As RA is known to diffuse in an anterior to posterior direction, and the newly demonstrated radial wave in mouse ovaries does not begin entirely at the anterior extreme of the ovary or the region adjacent to mesonephros, we reasoned that other regulatory mechanisms apart from RA signaling must be present during meiotic initiation.
ICBs regulate the radial meiotic wave in developing female germ cells
A potential mechanism underlying the first radial wave of meiosis is suggested by the clustered distribution of the earliest SYCP3+ cells. This clustering is reminiscent of germ cell cysts, which are formed through rounds of incomplete cytokinesis. Uniquely, in germ cells, the expression of TEX14 during mitosis renders the midbody into a stable ICB that connects the sister cells (fig. S3A) (14, 17). To test the role of ICBs in meiotic initiation, we turned to a mutant in Tex14 that has been shown to lack ICBs at birth (14, 18). The number and distribution of Oct4-GFP+ or TRA98 germ cells within the ovary were similar between Tex14 wild type (WT) and mutant (Tex14−/−) at E13.0 to E13.5, although it should be noted that they only mark most of the germ cells at this time point (Fig. 3, A to C, and fig. S3, C and D). However, the number and frequency of SYCP3+ cells were higher in Tex14−/− compared to WT ovaries; whereas the first SYCP3+ cells were concentrated at the core of the WT ovary at E13.0 (Fig. 3, A, D, and E, and fig. S3C) and remained absent near the surface at E13.5, age-matched mutant ovaries had more SYCP3+ cells that were distributed equally between the core and surface regions (Fig. 3, B, D, and E, and fig. S3D). SYCP3 expression in Tex14−/− advanced in an AP pattern, but the earlier radial meiotic wave was absent (Fig. 3B). This difference in the geography of meiotic entry did not result from differences in developmental timing in the mutant, as SYCP3 was absent at E12.5, and germ cells were evenly distributed throughout Tex14−/− ovaries (fig. S3B). By contrast, the progression of meiosis at E15.5 to 16.5 was comparable between WT and Tex14−/− as evaluated by 3D imaging and meiotic spreads (Fig. 3, F and G, and fig. S3E), suggesting that the defect is limited to earlier meiotic onset and the absence of a radial wave of meiotic initiation.
Fig. 3. ICBs orchestrate meiotic initiation and the radial meiotic wave in differentiating female germ cells.
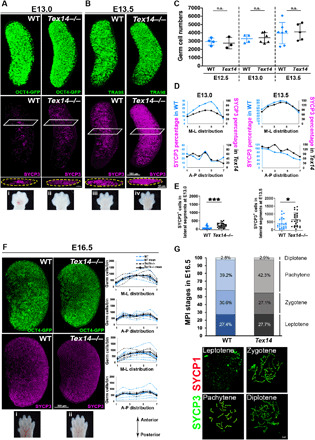
(A) Increased numbers and scattering of SYCP3+ cells were observed in Tex14 mutant (Tex14−/−) at E13.0 compared to SYCP3+ clusters in the core of WT ovaries. (B) At E13.5, SYCP3+ cells were increased in number in Tex14−/− ovaries and more advanced toward the posterior, compared to WT. (C) Similar germ cell numbers were observed in WT and Tex14 mutant in E12.5, E13.0, and E13.5 ovaries using TRA98 or Oct4-GFP. n.s., not significant. (D) Percentage of SYCP3+ cells in each ovary segment at E13.0 and E13.5. WT indicated in blue and Tex14−/− in black. (E) An increased number of SYCP3+ cells was detected in lateral segments (1 to 3 and 5 to 7) of Tex14−/− ovaries compared to WT at E13.0 (P = 0.0003) and E13.5 (P = 0.0335) supporting the absence of the radial wave in Tex14−/−. (F) Similar SYCP3 expression pattern was seen E16.5 WT and Tex14−/− ovaries, suggesting that meiosis progressed normally in later stages. Graphs corresponding to each immunostaining are adjacent, with WT indicated in blue and Tex14−/− in black. Dashed lines represent individual ovaries, and thick lines show the mean value for each genotype. (G) Analysis of different stages in MPI between genotypes showed that meiotic progression is comparable at E16.5. Insets (i) to (iv) in (A) and (i) and (ii) in (F) are forelimbs collected from WT and Tex14−/− mutants for accurate staging and age-matched comparison of embryos. WT includes genetically WT or heterozygous animals. MPI, Meiotic prophase I. At least n = 4 ovaries (from two litters and three embryos) were analyzed for each genotype and time point. *P < 0.05, ***P < 0.0001. ns, not significant.
Tex14 mutant ovaries lack cytoplasmic sharing
Although the absence of ICB structures in Tex14 mutants was observed by transmission electron microscopy at E13.5 (fig. S4A), it remained unclear whether functional cytoplasmic sharing between germ cells occurs in Tex14−/− ovaries. To test the capacity for sharing of transcripts and/or proteins between sister germ cells, we used an inducible multicolor reporter system. Rosa26-Rainbow;Tex14+/− females were crossed with Pou5f1-CreERT2;Tex14+/− males, and recombination in germ cells was induced coincident with colonization of the gonads at ~E11.5 by a single dose of tamoxifen at E10.5 (Fig. 4A). When cytoplasmic sharing occurs, separate recombination events within each nucleus of two-cell cysts could result in the stable production of two different fluorescent proteins (FPs) that transverse ICBs (Fig. 4B). These bicolored clones were detected at E13.5 in Rosa26-Rainbow; Pou5f1-CreERT2;Tex14+/+ ovaries. At a low frequency of labeling such that we could distinguish clones, we observed that approximately 2.3% of clones were bicolored among 4904 labeled cells analyzed (Fig. 4, C and E; fig. S4, B and D; and movie S2) on the basis of a mean clone size of 8 (Fig. 4D and fig. S4C). In Tex14 heterozygotes, a slightly lower frequency of bicolored clones was observed (fig. S4D). However, among the almost 7500 cells examined from intact E13.5 Rosa26-Rainbow; Pou5f1-CreERT2;Tex14−/− ovaries, no bicolored clones were found (P < 0.0001 by Fisher’s exact test; Fig. 4E and fig. S4D). Examination of Rosa26-Rainbow; Pou5f1-CreERT2;Tex14+/+ ovaries at E15.5 facilitated identification of bicolored cell groups; however, it was difficult to ascribe cyst membership to single labeled cells at this late time point because of increased cyst fragmentation (fig. S4E). Rainbow labeling further revealed a profound difference in the structure of clones between WT and Tex14 mutants; whereas germ cells of the same color “huddled” in close proximity in WT (Fig. 4C, fig. S4B, and movie S2), clones were far more dispersed and interdigitated in Tex14 mutant ovaries (Fig. 4C and movie S3). While bicolored clones do not measure the frequency of cysts with cytoplasmic exchange because they depend on stochastic and different recombination events, these data do provide functional evidence that cytoplasmic sharing of transcripts and/or proteins occurs within germ cell cysts of WT fetal oocytes. Most critically, the absence of bicolored clones in Tex14 mutants argues that such sharing requires TEX14-dependent structures, whereas the notable decrease in bicolored clones in Tex14 heterozygotes could suggest that functional connections within cysts are smaller.
Fig. 4. Cytoplasmic sharing through ICBs is absent in Tex14−/− fetal ovaries.
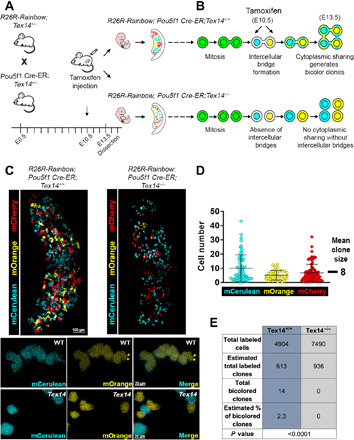
(A) Experimental approach to detect germ cell cytoplasmic sharing using recombination-induced multicolor clonal labeling. (B) Schematized formation of bicolored germ cell clones with R26R-Rainbow;Pou5f1Cre-ER in the presence and absence of ICBs through cytoplasmic sharing of FPs in WT and Tex14−/− genetic backgrounds, respectively. (C) Endogenous expression of FPs in E13.5 R26R-Rainbow;Pou5f1Cre-ER;Tex14+/+ and R26R-Rainbow;Pou5f1Cre-ER;Tex14−/− ovaries reveals divergent clone organization. Magnified image below shows a bicolor clone positive for mCerulean and mOrange in a R26R-Rainbow;Pou5f1Cre-ER;Tex14+/+ E13.5 ovary. Although germ cells expressing different FPs were closely juxtaposed in some cases, bicolored clones were not observed in E13.5 R26R-Rainbow;Pou5f1Cre-ER;Tex14−/− ovaries. (D) The mean clone size in 214 clones from E13.5 R26R-Rainbow;Pou5f1Cre-ER;Tex14+/+ ovaries was 8 (mCerulean, 64 clones; mOrange, 33 clones; mCherry, 177 clones). (E) Table of total labeled cells, estimated total labeled clones (based on mean clone size of 8), total bicolored clones, and estimated percentage of bicolored clones in R26R-Rainbow;Pou5f1Cre-ER;Tex14+/+ (n = 3) and R26R-Rainbow;Pou5f1Cre-ER;Tex14−/− (n = 4) ovaries at E13.5 (see fig. S4D for the breakdown of total number of cells per genotype).
Pluripotency transcripts are depleted in Tex14−/− meiotic germ cells
Although TEX14 is expressed exclusively in germ cells (14, 18, 19), given the role of extrinsic factors in meiotic initiation, we considered both somatic and germ cell defects in the Tex14−/− phenotype. To compare the transcriptional signature of germ cells during meiotic initiation and their neighboring somatic cells, we performed droplet-based single-cell RNA sequencing (scRNA-seq) of E13.5 Tex14+/− and Tex14−/− ovaries, using the reporter Pou5f1-ΔPE-eGFP to enrich germ cells (Fig. 5A). We profiled 14,715 (9368 germ cells and 5347 somatic cells) from mutant, with an average of 2675 genes per cell, and 12,260 cells (7590 germ cells and 4670 somatic cells) from Tex14 heterozygotes, with an average of 2824 genes per cell. By t-distributed stochastic neighborhood embedding (tSNE) from Tex14+/− and Tex14−/−, we identified 22 transcriptionally distinct clusters (10 germ cell and 12 somatic cell clusters), distinguishing germ cell clusters on the basis of expression of Dazl and six other germ cell markers (see Materials and Methods for the list of germ cell genes; Fig. 5, B and C). Considering the role of Dazl in licensing for meiosis (20) and its function in translational regulation of TEX14 in cultured mouse embryonic ovaries (21), we compared Dazl expression and confirmed that levels were comparable between Tex14+/− and Tex14−/− (fig. S5A). Premeiotic germ cells demarcated by Pou5f1high expression were transcriptionally distinct from germ cells in meiotic prophase that were Sycp3high (Fig. 5C). Computational integration of the two datasets using the scTransform algorithm (22) revealed that transcriptomes of Tex14+/− and Tex14−/− ovaries were highly similar (fig. S5B). One possible mechanism underlying premature meiosis could be the altered expression of RARs and/or retinoid X receptors (RXRs) in Tex14−/− germ cells. Increased levels of receptor transcripts could render Tex14−/− germ cells more sensitive to RA signaling and lead to earlier meiotic entry. However, analysis of RARs (Rara, Rarb, and Rarg) and RXRs (Rxra, Rxrb, and Rxrg) revealed comparable transcript levels between the two genotypes, suggesting that Tex14+/− and Tex14−/− germ cells respond to RA signaling similarly (fig. S6A). To examine subtle transcriptional differences between mutant and heterozygous cells, we applied an additional algorithmic approach; using the scAlign package (23), we trained a decoder network to project cells of the Tex14−/− dataset into the alignment space of the Tex14+/− dataset. This revealed distinct populations of the two samples that may have been merged together because of overfitting of the scTransform alignment paradigm. scAlign confirmed overlap between somatic cells of different genotypes but identified a small subpopulation of germ cells that diverged between Tex14+/− and Tex14−/− (fig. S5C, arrows). Upon examining these areas of divergence between genotypes, we identified three germ cell subclusters defined by expression of either Pou5f1 or Sycp3 (fig. S5D) and then determined genes that most define those subclusters (fig. S5, E to G, and table S1).
Fig. 5. Dissecting the transcriptional signature of Tex14+/− and Tex14−/− ovaries during the time of meiotic initiation.
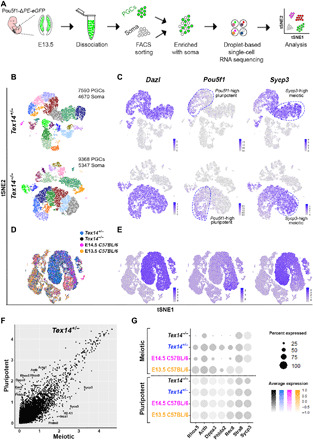
(A) Experimental design schematic. (B) Visualization of single-cell clustering by tSNE shows 10 germ cell and 12 somatic cell clusters in Tex14+/− and Tex14−/− ovaries represented by different colors. FACS, fluorescence-activated cell sorting. (C) Expression of germ cell marker Dazl identifies germ cell clusters. Pluripotent germ cells are defined by expression of Pou5f1 transcript; meiotic germ cells are defined by Sycp3 transcript. Dashed lines indicate cells with higher levels of Pou5f1 or Sycp3. (D) Integration of previously published E13.5 and E14.5 C57BL/6 datasets (24) with E13.5 Tex14+/− and Tex14−/− datasets. (E) Dazl identifies germ cell clusters, pluripotent germ cells are defined by expression of Pou5f1, and meiotic germ cells are defined by Sycp3 transcript. (F) Scatter plot of differentially expressed genes between pluripotent and meiotic clusters in Tex14+/−. (G) Dot plot graph of highly expressed genes (Rhox5, Actb, Dppa3, Phlda2, and Rec8) in the pluripotent clusters compared to meiotic clusters in E13.5 C57BL/6, E14.5 C57BL/6, E13.5 Tex14+/−, and Tex14−/− integrated datasets. Dot size indicates percentage of cells expressing the transcript, while colors denote expression level in cells. Expression of genes associated with the pluripotent cluster was the lowest in Tex14 mutant germ cells, whereas expression of meiotic markers Stra8 and Sycp3 was similar in E14.5 C57BL/6, E13.5 Tex14+/−, and Tex14−/−.
To corroborate our observations, we integrated our E13.5 Tex14+/− and Tex14−/− datasets with previously published scRNA-seq datasets of WT (C57BL/6) embryonic mouse ovaries (24). As meiosis in C57BL/6 mouse ovaries starts slightly later compared to a mixed genetic background, we integrated both E13.5 and E14.5 C57BL/6 datasets with our mixed background E13.5 data. Germ cells were identified on the basis of expression of Dazl (as well as six germ cell genes listed in Material and Methods) and totaled 782 and 2140 in the E13.5 and E14.5 C57/BL6 datasets, respectively (Fig. 5D). While we confirmed that Pou5f1high premeiotic germ cells were transcriptionally distinct from Sycp3high germ cells in meiotic prophase (Fig. 5E), the comparatively few Sycp3-expressing germ cells in the E13.5 C57BL/6 ovary indicate that the mixed-background Tex14+/− germ cells at E13.5 interpolate between the transcriptome of E13.5 and E14.5 C5BL/6 germ cells (fig. S6D). Next, in light of the divergent germ cell subclusters between Tex14+/− and Tex14−/− as well as the premature meiosis phenotype in Tex14−/−, we focused our transcriptional analysis on the transition between pluripotency and meiosis. In Seruat v3, we identified the most highly expressed genes (Rhox5, Actb, Dppa3, and others in table S2) that were more than 0.25-fold changed between the pluripotent (Pou5f1high) and the early meiotic (Sycp3high) clusters of Tex14+/− germ cells (Fig. 5F). Among the most differentially expressed genes between equivalently staged germ cell populations in mutant and heterozygote, those most abundant in the pluripotent (Pou5f1high) cluster (Fig. 5F) including Rhox5, Actb, Dppa3, and Phlda2 were down-regulated in Sycp3high meiotic germ cells in Tex14−/− compared to all controls (Tex14+/−, E13.5 and E14.5 C57BL/6 germ cells; Fig. 5G). This difference was specific to highly expressed transcripts in the pluripotent cluster, as meiotic genes such as Stra8 and Sycp3 were unchanged between Tex14+/−, Tex14−/−, and E14.5 C57BL/6 meiotic populations. A number of the same pluripotency cluster genes depleted in Tex14 mutant meiotic germ cells were also identified in divergent subclusters by scAlign (fig. S5E). Besides the most highly expressed genes in the Tex14+/− pluripotent cluster (Fig. 5E), we found that genes implicated in the pluripotency program such as Sox2 and Pou5f1 were also down-regulated in Sycp3high meiotic germ cells in Tex14−/− (fig. S6C). Such selective loss of a specific group of transcripts subset in Tex14−/−, which lacks cytoplasmic bridges, argues against the possibility that severed bridges during dissociation alter the transcriptome indiscriminately. Collectively, these observations indicate that the transcriptional signature of E13.5 Tex14+/− germ cells is similar to WT (and developmentally interpolates between E13.5 and E14.5 germ cells from a pure C57BL/6 background), and that the absence of Tex14 does not alter the transcriptional composition of ovarian somatic cells but specifically attenuates the level of pluripotency-associated transcripts as germ cells enter meiosis.
Dynamic expression and knockout implicate DPPA3 in meiotic onset in the ovary
Dppa3, also known as Stella or PGC7, is a developmentally regulated gene first expressed in PGCs at E7.5. DPPA3 expression decreases in fetal oocytes around E14.5 and rises again in postnatal oocytes (25). Functional studies have demonstrated a role for Dppa3 in early embryonic development as a maternal effect gene (26, 27), although, unexpectedly, no alterations in germ cell development have been identified in Dppa3 mutant mice (Dppa3−/−) (28). The emergence of Dppa3 as a transcript that is dynamically regulated between pluripotent and meiotic germ cells as well as down-regulated in Tex14 mutant meiotic germ cells inspired closer examination of the protein expression during this transition. Wholemount immunofluorescence revealed mutually exclusive protein expression of DPPA3 and SYCP3 in germ cells from WT (Fig. 6A) and Tex14−/− ovaries (Fig. 6B) at E13.5. To uncover the role of DPPA3 in the mitotic-meiotic transition, we analyzed the expression of SYCP3 in Dppa3−/− ovaries. We detected an elevated fraction of SYCP3+ germ cells and a more advanced meiotic transition in Dppa3−/− compared to WT ovaries (Fig. 6, C and D, and fig. S7, A and B). This result implicates DPPA3 as a regulator of the entry into meiosis in the fetal ovary and demonstrates that Dppa3−/− phenocopies earlier meiotic onset seen in Tex14−/− ovaries.
Fig. 6. Reciprocal expression of SYCP3 and DPPA3, precocious meiosis in Dppa3 mutants, and methylation changes in Tex14 mutants link ICBs, the epigenetic reprogramming during meiotic initiation.
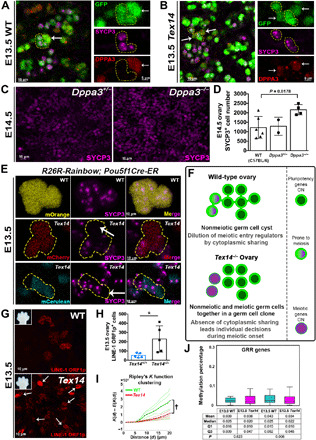
(A) WT germ cell cluster and (B) Tex14 mutant germ cell cluster (yellow outlines) expressing SYCP3 (magenta) at E13.5 lack DPPA3 (red), whereas SYCP3− cells express DPPA3 (arrows). (C) SYCP3 expression in Dppa3+/− and Dppa3−/− ovaries at E14.5, with quantification in (D) (n = 6 for WT C57BL/6, n = 2 for Dppa3+/−, and n = 4 for Dppa3−/− ovaries). (E) mOrange+ clone with uniform SYCP3 (magenta) in R26R-Rainbow;Pou5f1 Cre-ER;Tex14+/+ ovary. R26R-Rainbow;Pou5f1 Cre-ER;Tex14−/− exemplary mCherry and mCerulean clones (yellow outlines) do not homogeneously express SYCP3 (white arrows indicate SYCP3−cells). (F) Model for coordination of meiotic initiation through ICBs in E13.0 fetal germ cells depicts dilution of regulatory factors in mitotic-meiotic transition. Cytoplasmic sharing may synchronize the pluripotent state of germline cysts in WT by increasing cytoplasmic volume, as meiotic transcripts must be expressed by a quorum to initiate meiosis. Without ICBs, germ cells expressing meiotic transcripts enter meiosis individually, leading to earlier meiotic onset in Tex14−/−. (G) LINE-1 ORF1p expression, as a proxy of DNA demethylation, shows increased scattering (white arrows) and frequency (H) in Tex14−/− compared to Tex14+/+ ovaries at E13.5. (I) Ripley’s K function analysis shows the clustered distribution of LINE-1 ORF1p+ germ cells in WT compared to Tex14 mutant (†P = 0.002). Embryo forelimbs in (G) compare ages. n = 5 ovaries were analyzed in both cases. (J) Methylation of GRR genes is similar between Tex14−/− female germ cells and Tex14+/− or Tex14+/+ controls at E13.0 (n = 1 knockout, n = 2 control) but reduced at E13.5 (n = 2 knockout, n = 2 control) by WGBS. WT includes genetically WT and heterozygous animals in (J).
The timing of meiotic initiation depends on cytoplasmic continuity between germ cells
The precocious meiosis phenotypes in Dppa3 and Tex14 mutants together with the depletion of Dppa3 transcript in the latter raise the question of how ICBs affect the timing of meiotic initiation and mRNA levels of Dppa3. Corroborating a previous report using single-color clonal labeling (29), we observed similar SYCP3 levels between adjacent germ cells expressing the same Rainbow fluorophore, which confirms that cells in a cyst synchronously enter meiosis (Fig. 6E). However, the lack of cytoplasmic sharing that we have demonstrated between clonally related germ cells in Tex14 mutants provides the opportunity to ask whether the timing of meiotic entry is synchronous within clones because of direct cytoplasmic sharing or alternatively, due to similar cytoplasmically inherited determinants from a common precursor. We thus examined SYCP3-expression in Rosa26-Rainbow; Pou5f1-CreERT2;Tex14−/− ovaries at E13.5. Approximately 5.7% of all labeled Tex14 mutant clones contained both SYCP3− cells and adjacent sister cells (within 1 cell diameter) bearing the same FP that were SYCP3+. In comparison, the absolute coordination between clones in WT (from 4874/4874 labeled cells examined in Fig. 6E and fig. S7G) indicates that the transition to meiosis is synchronized between cells of a clone only when connected by ICBs, but not because of other cell-heritable entities. Together, these results suggest that direct cytoplasmic sharing within germ cell cysts via ICBs orchestrates the timing and geography of meiotic initiation in mouse ovaries. The underlying mechanism must involve the sharing of cytoplasmic factors that regulate meiosis.
A model that connects cytoplasmic sharing to meiotic initiation relies on the dilution of key regulatory factors to coordinate developmental transitions across the cyst until a threshold is reached (Fig. 6F). Asymmetries in gene expression that arise between cells within a germ cell cyst (whether stochastically or from differential stimulation by exogenous factors) would be diluted and equalized across ICBs. Such dilution accounts for the synchronization and the delay in meiotic initiation in WT compared to Tex14; the absence of dilution in mutants without ICBs would enable more discrete and rapid transitions, exemplified by the faster decline of transcripts that promote pluripotency such as Dppa3 and Pou5f1 in Tex14−/− germ cells that have initiated meiosis (Fig. 5G and fig. S6, B and C).
A candidate cell-intrinsic process that has been linked to meiotic initiation is DNA demethylation. Loss of the maintenance methyltransferase DNA methyltransferase 1 (Dnmt1) in germ cells leads to premature DNA demethylation and premature meiotic entry (30). By contrast, the absence of the enzyme Tet1 prevents active demethylation of promoters for a specific subset of germline reprogramming-responsive (GRR) genes involved in gamete formation and meiosis, which leads to a delay in meiotic initiation (31). The involvement of Dppa3 in DNA demethylation in the zygote (32) together with the similarity of Dppa3 and Tex14 phenotypes to the Dnmt1 germ cell knockout raises the possibility that DNA demethylation is delayed by cytoplasmic sharing. Whole-genome bisulfite sequencing (WGBS) on E13.0 and E13.5 germ cells from Tex14−/− ovaries and age-matched WT and heterozygous littermates revealed a significant decrease specifically in the methylation of GRR genes but not in CpG sites across the genome at E13.5 (Fig. 6J and fig. S7, E and H). Given that the expression of the LINE-1 (L1)–transposable element is silenced by methylation at promoters (33), we also examined methylation in repeat regions and observed a decrease in L1 MdA genes in mutants compared to controls (fig. S7F). Alleviating the dilution of epigenetic regulators between germ cell cysts in Tex14 mutants could lead to premature demethylation of L1 promoters and expression of L1 open reading frame 1 (ORF1p) that is both premature and desynchronized between cells within a clone. We observed that the onset of L1 ORF1p expression was clustered in WT at E13.5, whereas in Tex14 ovaries, L1 ORF1p expression was more abundant in germ cells and more dispersed according to Ripley’s K function (Fig. 6, G to I, and fig. S7, C and D). This result is consistent with a relationship between ICBs, epigenetic reprogramming, and the transition between pluripotent PGCs and meiosis.
DISCUSSION
Here, we describe a new mechanism by which ICBs coordinate a state transition during germ cell development in the fetal ovary by cytoplasmic sharing. In addition to the known gradient of meiotic initiation that sweeps from anterior to posterior, we identified an earlier onset of meiosis that occurs in interconnected cysts that begins in the medial region of the embryonic ovary and radiates outward. We found that cytoplasmic sharing through Tex14-dependent ICBs is crucial not only for the radial meiotic wave but also for appropriate timing of meiotic initiation. Upon initiation of meiosis, Tex14 mutant germ cells more abruptly deplete transcripts associated with pluripotency such as Dppa3. This finding, together with the lack of coordinated meiotic initiation between sister cells when ICBs are absent, suggests that the transition from pluripotency to meiosis involves the sharing of cell state regulatory factors across ICBs; sharing synchronizes differentiation across cysts but also slows it through dilution of transcripts that are unequally expressed across members of an interconnected cyst. We showed that meiotic entry correlates with the down-regulation of DPPA3 protein and that Dppa3 knockout causes premature meiosis in the fetal ovary. The resemblance of Dppa3 and Tex14 meiosis phenotypes to that of Dnmt1 germ cell knockout raises the possibility that key meiotic genes are prematurely demethylated in the absence of ICBs as in the absence of the DNA methyltransferase Dnmt1. We provide evidence that Tex14-deficient germ cells have reduced DNA methylation including LINE-1, which supports this model.
The involvement of ICBs in meiotic initiation reveals a new function of cytoplasmic sharing in regulating cell state transitions. As structures that provide cytoplasmic continuity between cells in a cyst, ICBs are deeply conserved in evolution and yet unique to germ cells (19). The connection of more than two cells through different mechanisms such as membrane fusion (e.g., skeletal muscle and placenta) (34) or multiple nuclear divisions without cytokinesis (e.g., hepatocytes) is an important feature that varies by cell type–specific function (35). In Drosophila ovaries, ICBs allow the transfer of oocyte-specific proteins, mRNAs, and organelles from supporting “nurse” cells to the dominant oocyte (19). Mammalian oocytes similarly develop in cyst structures, and the accumulation of organelles in later stages of embryonic germ cell development has been demonstrated (36). In mice, Tex14 was identified as an essential component of the ICB and fertility in males, as Tex14 knockouts lack physical connections between neonatal germ cells and succumb to apoptosis during meiotic prophase (18). By contrast, in female germ cells, ICBs exist more transiently during fetal development; Tex14 mice have fewer oocytes starting from postnatal development and bear fewer litters (14). A recent study of Tex14 mutants finds elevated apoptosis and decreased numbers of female germ cells starting at E12.5 (37); although we did not observe a parallel decrease in germ cell numbers, it is noteworthy that we maintain Tex14−/− mice on a mixed genetic background rather than pure C57BL/6. Membrane connections between germ cells in Tex14−/− ovaries were observed in the prior study by ultrastructure at E14.5 (37); however, our extensive analysis of Rainbow multicolored germ cells labeled from E10.5 to E13.5 did not reveal any clones bearing two fluorophores in Tex14 as occurs in WT (Fig. 4). A likely interpretation is that the concurrent or subsequent membrane fragmentation described in Tex14−/− germ cells by Nuzhat et al. (37) does not enable sharing of cytoplasmic contents. Our approach involved drug-induced recombination-based labeling of a subset of germ cells to distinguish individual clones in the ovary. The timing and dose of tamoxifen balanced clone identification with the relatively low probability of producing simultaneous but separate recombination events with different outcomes in two cell cysts. Thus, the 2.3% fraction of bicolored clones observed in WT does not represent the frequency of cytoplasmic sharing because not all germ cells were labeled in our system, and only three colors are possible. However, the presence of bicolored clones, which arises through the movement of fluorophore transcripts and/or proteins across ICBs in vivo, is an entirely novel and functional demonstration of cytoplasmic sharing, and it is significant that bicolored clones are absent in Tex14 mutants. The perdurance of pluripotency-associated transcripts in WT germ cells that have entered meiosis in scRNA-seq data, in contrast to Tex14 mutants, is a likely by-product of cytoplasmic sharing and signifies a new link between ICBs and cellular state transitions. Further investigation into the function of ICBs will be facilitated by generating mouse knockouts of additional components of the bridges (38) and performing functional tests of cytoplasmic sharing.
A novel insight from this study is the implication of a cellular structure in the regulation of meiosis. We used Tex14 mutants to show that ICBs prevent precocious meiosis in across the entire ovary (Fig. 3) and coordinate meiotic entry across cysts, as evidenced by the loss of synchrony between Tex14 clones identified by Rainbow labeling (Fig. 6). The identity of the extrinsic and cell-intrinsic drivers of meiotic initiation in mammals is an ongoing question. Analysis of the ovary in toto revealed an earlier cohort of meiotic cells in the medial region rather than the anterior tip of the ovary. Our robust 3D technique manifested natural variation in the number of germ cells between ovaries in the same developmental stage. Previous studies in mice show that meiosis initiates in the ovary anterior at E13.5 and sweeps toward the posterior in response to RA, which diffuses from mesonephros and stimulates the expression of meiotic genes (1, 2, 4–6). Challenging this model, genetic ablation of an RA synthesis enzyme (Aldh1a2−/−) (7) or all RAR isotypes (9) did not prevent the expression of Stra8 in addition to other meiotic markers in germ cells. Although SYCP3 onset has been detected before E13.5 in cultured ovaries (39), our rigorous demonstration of the first meiotic entrants and their 3D distribution at E12.5 in vivo revises our knowledge about the timing of mitotic to meiotic transition. Furthermore, our analysis revealed that the earliest STRA8+ germ cells are localized throughout the ovary at E12.5 without strong spatial distribution. Given prior evidence of AP meiotic initiation in response to external diffusible RA signaling, the unbiased localization of STRA8+ germ cells before meiotic onset raises the possibility that intrinsic factor(s) could be responsible for premeiotic DNA replication in germ cells at E12.5. Localization of the earliest SYCP3+ cells in the ovary core further supports the presence of other meiosis-inducing factors that precede RA function. A detailed description of this earliest wave of meiosis in RAR triple-knockout mice would provide further insight. An additional geographic feature of meiosis that emerged from our wholemount analysis was the relatively lower number of SYCP3+ cells in the ventral region of the ovary compared to middle and dorsal regions. By contrast, a prior study in histologic sections reports that STRA8+ cells skew toward the ventral region of the ovary at E13.5 (3). This discrepancy underscores the difference between STRA8 and SYCP3 and lends credence to the idea that premeiotic DNA replication (as read out by STRA8) may be regulated by a different mechanism, such as cell-intrinsic factors, as compared to the formation of the synaptonemal complex (as read out by SYCP3).
Recent work has established a link between meiosis and epigenetic reprogramming (30, 31, 40); this study reinforces and expands that relationship to include ICBs. Our model that ICBs coordinate the transition from pluripotency to meiosis through the cytoplasmic dilution of epigenetic regulators was informed by scRNA-seq. In stride with recent studies (24, 41), we showed that transcriptional progression accompanies the transition from mitosis to meiosis. An enrichment strategy allowed us to interrogate the largest number of E13.5 germ cells at single-cell level to date. Integration of our datasets with previously published datasets (24) confirmed that the transcriptomic signature of Tex14+/− germ cells is similar to WT. We used two different algorithms to interrogate gene expression differences associated with premature meiosis in Tex14−/− and validated scAlign (23) as a powerful tool for data integration and determination of subtle variations in different populations. A nuanced analysis of differentially expressed genes in pluripotent and advanced meiotic clusters revealed specific depletion of pluripotency genes in Tex14 meiotic cells compared to WT. On the basis of this observation, we hypothesize that germ cell cysts in WT retain pluripotency transcripts longer after initiating meiosis because they travel across ICBs. Reinforcing this hypothesis is the correlation between the decline of DPPA3 and appearance of SYCP3, which suggests a function of Dppa3 to preserve the pluripotent state before meiotic initiation; premature meiosis in Dppa3 mutants is the first phenotype described in developing germ cells. Dppa3 plays a role in ensuring appropriate methylation of oocytes (32) and preventing untimely demethylation in early embryos (27, 42). In the oocyte and preimplantation embryo, DPPA3 sequesters the E3 ubiquitin ligase UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1), preventing its nuclear localization and association with the DNA methyltransferase DNMT1 (32, 43). This mechanism does not extend to PGCs or gonocytes because UHRF1 is not expressed. Our finding that DNA methylation across the genome is not affected in Tex14 mutant germ cells, but that promoters of GRR genes and younger L1 genes are less methylated, poses new questions about precise Tex14- or ICB-dependent epigenetic mechanisms and the connection to Dppa3. Further studies of the specific function of Dppa3 in PGCs may elucidate a novel mechanism for this regulator of naïve pluripotency.
In conclusion, ICBs globally coordinate the earliest meiotic initiation across the embryonic ovary while also synchronizing cell-state transitions within germ cell cysts, potentially by diluting regulatory factors of epigenetic reprogramming. This raises questions about the impact of cytoplasmic sharing on oocyte competence and the potential for cellular connections in other cell lineages such as the placenta or cancer cells to regulate state transitions.
MATERIALS AND METHODS
Mice
CD1 female mice were mated with Oct4-ΔPE-eGFP (Oct4-GFP) male mice (MGI:3057158) to label germ cells with GFP in embryonic ovaries. Tex14+/− mice (MGI:3623684, provided by M. Matzuk, Baylor College of Medicine) were maintained on a mixed genetic background including C57Bl/6, CD1, and CBA and crossed with Oct4-GFP mice. C57BL/6 mice were purchased from the Jackson Laboratory and interbred. For clonal labeling experiments, mice from Tex14 colony were mated with R26R-Rainbow mice (MGI:5441200, a gift from I. Weissman, Stanford University) or Pou5f1 Cre-ER/+ (MGI:5049897) to obtain R26R-Rainbow;Tex14+/− females and Pou5f1Cre-ER/+;Tex14+/− males, which were further crossed to each other. Oct4-GFP, Tex14, R26R-Rainbow, and Pou5f1Cre-ER/+ genotypes were determined by polymerase chain reaction (PCR). Day of mating plug was identified as E0.5, and female mice were euthanized at different stages of pregnancy (E12.5, E13.0, E13.5, E14.5, E15.5, and E16.5); embryos were dissected and staged according to their morphological landmarks. All mouse work was performed under the University of California, San Francisco (UCSF), Institutional Animal Care and Use Committee guidelines in an approved facility of the Association for Assessment and Accreditation of Laboratory Animal Care International.
To induce clonal labeling in germ cells, R26R-Rainbow; Tex14+/− females and Pou5f1Cre-ER/+; Tex14+/− males were crossed, 0.8 mg of tamoxifen (Sigma-Aldrich, stock concentration of 5 mg/ml in sunflower seed oil) per 40 g of R26R-Rainbow; Tex14+/− mouse was intraperitoneally injected at E10.5. Embryonic ovaries were collected at E13.5 and processed further for wholemount immunofluorescence staining.
Genotyping
DNA was extracted from tail samples in alkaline lysis reagent comprising 25 mM NaOH and 0.2 mM EDTA, heated to 95°C for 45 min, and cooled to 4°C, and 40 mM tris-HCl was added in 1:1 ratio for neutralization. Primer sets used in the study were listed (table S3). Tex14 PCR was performed at 95°C for 2 min, followed by 35 cycles of 94°C for 20 s, 62.5°C for 20 s, and 72°C for 1 min. To determine eGFP (enhanced GFP) transgene, PCR was carried out as follows: 93°C for 1 min, followed by 30 cycles of 93°C for 20 s, 68°C for 3 min, and 72°C for 10 min. Pou5f1Cre-ER/+ reaction was performed at 95°C for 1 min, followed by 32 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 2 min. PCR products were electrophoretically separated in 2% agarose gel in tris/borate/EDTA buffer, and genotypes of adult mice and embryos were identified according to size of PCR products.
Whole-mount immunofluorescence
Mesonephros-attached mouse fetal ovaries for wholemount staining were dissected in 0.4% bovine serum albumin (BSA) in 1× phosphate-buffered saline (PBS) and transferred into 1.5-ml Eppendorf tubes, and subsequent steps were carried out while rocking. Fetal ovaries were fixed with 4% paraformaldehyde (PFA) in PBS at +4°C for 2 hours. After fixation, ovaries were washed three times with 0.2% BSA in PBS for 10 min each and blocked with 2% BSA and 0.1% Triton X-100 in PBS for 3 hours at room temperature. Primary antibodies (table S3) were diluted in 0.2% BSA and 0.1% Triton X-100 in PBS, and ovaries were incubated in primary antibodies at +4°C for 2 nights. Samples were washed four times with 0.1% Triton X-100 in PBS for 15 min each at room temperature and incubated with Alexa Fluor–conjugated secondary antibodies in 0.2% BSA and 0.1% Triton X-100 in PBS at +4°C overnight. Ovaries were washed three times with 0.2% BSA and 0.1% Triton X-100 in PBS for 30 min each, dehydrated with methanol:PBS series (25 to 50 to 75 to 100%) for 10 min each (only 100% twice) at room temperature and incubated with 3% H2O2 in methanol overnight at 4°C. The following day, ovaries were incubated in 100% methanol for 30 min twice, transferred to the 10-mm-long glass cylinders (ACE Glass 3865-10) mounted onto coverslips (Fisherfinest Premium Cover Glass 12–548-5P), and incubated in benzyl alcohol:benzyl benzoate (1:2) at +4°C overnight. Mouse ovaries were oriented as anatomically anterior part of ovary is at the top, and the posterior part of ovary is located at the bottom of field of view. Samples were imaged by using a white-light Leica TCS SP8 converted confocal microscope with a HC PL APO CS 10×/0.40 dry objective, 1.5× optic zoom, and 1024 × 1024 pixel resolution, and stacks were acquired at system-optimized z steps between optical sections (z step size, 1 μm).
For clonal labeling analysis, ovaries were cleared in Scale CUBIC Reagent 1 overnight at 4°C and imaged with by using a Fluotar VISIR 25×/0.95 water objective on a Leica TCS SP8 converted confocal microscope with 0.75× optic zoom, 1024 × 1024 pixel resolution and z stacks that were 2 μm apart. E13.5 Rainbow;Pou5f1Cre-ER/+; Tex14 ovaries were imaged using 3 × 1 tile scans, and tiles were merged using Leica software.
Image analysis for whole-mount stained ovaries
Image analysis was performed using Imaris v8.3.1 (Bitplane). After files were imported to Surpass mode, voxel size Z of the original images was reduced to increase the power of analysis. Anterior and posterior tip of the ovary was defined according to orientation of mesonephros, a surface was created manually on the ovary, mesonephros was removed by masking the channels corresponded to different antibody staining. Thus, the mesonephros was removed from the ovary to more accurately analyze the ovary. Fluorescently labeled germ cells were selected by using the Spot detection module. To label the total germ cell population, both TRA98 and OCT4-GFP were used on the basis of antibody compatibility and availability of the GFP transgene in embryos. The two markers showed an approximate 70% overlap in expression at E13.5. To count the most accurate number of germ cells, two different object sizes were applied for different antibody staining patterns (XY diameters of 5 and 2.5 μm for GFP; 4 and 3 μm for TRA98, OCT4, STRA8 and SYCP1, cPARP, active caspase-3, and γH2AX; and 4 and 2.6 μm for SYCP3).
For each marker, a new channel was created with a standard voxel value inside the surface by masking larger–object size spots from the original channel. Another spot population was created with smaller object size. To avoid overcounting, smaller–object size spots were filtered by a previously created larger–object size spot channel on the basis of intensity median value of the larger object channel. Threshold for filtering was applied by moving the left slider to the far left and right slider to just before the middle of the scale. After initial check that only smaller-sized spots were selected and bigger-sized spots were eliminated, this spot population was duplicated by clicking the duplicate selection to the new spots. Thus, spots were picked by using both (two different) object size and double-counted spots were eliminated by using filter function of the software. A new folder was created on Imaris to combine the germ cell counts, larger and filtered smaller–object size spots were merged under the new folder, and positive germ cell numbers for each marker were imported to Microsoft Office Excel 2007.
Clone size and bicolor clone analysis
Mean clone size at E13.5 was determined by analyzing Rainbow; Pou5f1Cre-ER/+; Tex14+/+ ovaries that have extremely low clone densities (an example presented in fig. S4C) to accurately identify clones as described previously (44). Surface module was used to detect individual clones, and clone size was identified by dividing the volume of surface (represents the volume of clone) by the volume of single germ cells in the ovary.
The Spot module was also used to analyze bicolored clones in Rainbow; Pou5f1Cre-ER/+; Tex14 E13.5 ovaries. Spots with XY diameters of 10 and 6 μm were created for each channel (mCerulean, mOrange, and mCherry), and a new channel was created with standard voxel value by masking 10-μm size spots from the original channel. To avoid overcounting, 6-μm spots were filtered by a previously created 10-μm spot channel on the basis of the intensity median value. Ten- and selected 6-μm spots were merged as described above. Double-positive germ cell spots for two different channels (mCerulean+; mOrange+, mCerulean+; mCherry+, and mOrange+; mCherry+) were determined by filtering feature of spots. Double-positive spots were also manually checked in original image whether all cells in the clone express two colors to be defined as a bicolored clone, and numbers were imported to Microsoft Office Excel 2007. To determine estimated clone size, total labeled cell number was divided by the mean clone size.
Transforming coordinate systems to quantify angular and radial distributions of cells within the mouse ovary
The 2D Cartesian coordinates of the objects created by the Imaris Spots module, which represent the centroid of cells, were recorded. In addition, in Cartesian coordinates, the geometric center and the end points (top and bottom) of the Imaris surface representing the ovarian outer surface were noted. Using the geometric center and two end points, the radius of curvature was calculated, and all spots object coordinates were offset so as to shift the circle described by the radius of curvature to the Cartesian origin. These coordinates were then transformed from Cartesian to polar coordinates by using a custom MATLAB script available at https://github.com/BIDCatUCSF/Angular-Radial-Position-Distribution.
If the mesonephros was on the left side of the ovary, then all polar coordinates were rotated by −180° so that the center of the ovary corresponded to an angle of 0°. To match the orientation of the ovary, such that dorsal is in the positive angular directions, the y-component of the Cartesian coordinates was multiplied by −1 before transforming in the polar coordinate system. If the mesonephros was on the right side of the ovary, then no rotations or inverses were necessary.
Clustering analysis
Spatial analysis of clustering was carried out on the basis of Ripley’s K function using the RipleyGUI (45) platform in MATLAB. Following the wholemount staining with OCT4-GFP and LINE-1 ORF1p antibodies, total germ cell population and LINE-1 ORF1p–positive cells were identified by the Spot module of Imaris software. Coordinates of objects (positive cells) for each marker were imported as text file, and analysis was performed as described previously (44).
Preparation of chromosomal spreads and immunofluorescent staining
Chromosomal spreads were prepared as previously described (46) with some modifications. E16.5 mouse ovaries were dissected in 0.4% BSA in PBS, and each pair of ovaries was digested in 0.025% trypsin, collagenase (2.5 mg/ml), and deoxyribonuclease I (DNase I)–containing dissociation buffer (0.1 mg/ml) at 37°C for 30 min (with pipette trituration every 10 min). To quench the reaction, an equal volume of fetal bovine serum (FBS) was added. After adding an equal volume of hypotonic buffer [30 mM tris (pH 8.2), 50 mM sucrose, 17 mM sodium citrate, 5 mM EDTA, 0.5 mM dithiothreitol (DTT), and 0.5 mM phenylmethanesulfonyl fluoride], samples were incubated for 30 min at room temperature and centrifuged for 10 min at 1000 rpm. Supernatant was removed from each tube; cells were resuspended in 100 mM sucrose. A rectangle-shaped barrier was drawn on precleaned (in 70% ethanol) positively charged glass microscope slides by a hydrophobic pen, a desired volume of fixative solution [1% PFA, 0.15% Triton X-100, and 3 mM DTT (pH 9.2)] was added in that bordered area. An equal volume of cell suspension was pipetted from a height. Slides were dried at room temperature and then submerged in 0.4% Kodak Photoflo in H2O for 2 min twice. Last, slides were air-dried and stored at −80°C until staining.
Samples were incubated in 0.1% Trition X-100 in PBS for 10 min at room temperature, washed three times with PBS for 5 min each, and blocked with 5% BSA in PBS for 1 hour at room temperature. After overnight incubation at 4°C with SYCP3 and SYCP1 primary antibodies (table S3) in 5% BSA in PBS, samples were washed three times with PBS for 5 min each and were incubated with secondary antibodies for 1 hour at room temperature. Samples were washed three times with PBS for 5 min each, and slides were mounted with VECTASHIELD and imaged on a Leica TCS SP5 confocal microscope by using an HCX PL APO CS 63×/1.4 oil objective.
Electron microscopy
E13.5 mouse ovaries were dissected in 0.4% BSA in PBS, fixed with 2.5% glutaraldehyde for 2 hours at +4°C, washed three times with 0.1 M Sorenson’s phosphate buffer at +4°C for 10 min each, and then postfixed with 1% OsO4 for 2 hours at +4°C. After postfixation, the washing step was repeated, and ovaries were dehydrated in 30, 50, and 70% ethanol series at +4°C. Samples were incubated in 1% uranyl acetate for 1 hour at +4°C, and dehydration was completed by incubating the samples in a gradient series from 80 to 100% ethanol at +4°C. After dehydration, samples were incubated two times with propylene oxide for 10 min each at +4°C, incubated in araldite/propylene oxide (1:1) solution for 8 hours at room temperature on a rotator, followed by pure araldite resin incubation overnight at room temperature on a rotator. Last, samples were transferred to embedding capsules containing araldite resin CY212 and polymerized at 60°C for 48 hours.
Sections (300 nm thick) were cut, stained with toluidine blue, and examined on a light microscope to determine the specific area for preparing ultrathin sections. Thin sections (70 nm) were cut on a Leica Ultracut UCT125 and contrasted with uranyl acetate and lead citrate. Sections were examined on a LEO 906E (Oberkochen, Germany) transmission electron microscope, and electron micrographs were captured for further analysis.
Dissociation and flow cytometry of embryonic ovaries and 10X library prep
E13.5 mouse fetuses were dissected in cold 0.4% BSA in PBS, and each pair of ovaries was digested in 0.25% trypsin-EDTA–containing 1.5-ml Eppendorf tubes at 37°C for 20 min; subsequently DNase I (1 mg/ml) was added, and the digest was incubated for 10 min with pipette trituration every 10 min. To quench the reaction, an equal volume of FBS was added; dissociated cells were passed through a 35-μm cell strainer and stained with Sytox Blue for live/dead cell discrimination. A BD FACSAria II was used to sort GFP+ germ and GFP− somatic cells into 1.5-ml Eppendorf tubes containing 0.1% BSA in PBS. We pooled three Tex14+/− and two Tex14−/− E13.5 ovaries for each group. A total of 43,310 Tex14+/− and 46,422 Tex14−/− germ cells were collected. Before loading cells onto a 10X Genomics platform, sorted GFP+ germ cell suspension was enriched with somatic cells; 42,000 and 38,000 somatic cells were loaded from Tex14+/− and Tex14−/−, respectively. Samples were processed for 10× sequencing by the UCSF Institute for Human Genetics. Briefly, the Chromium Single-Cell 3′ Reagent Version 2 Kit (10X Genomics) was used to capture individual cells producing gel bead-in-emulsions (GEMS). Reverse transcription of the GEMS allowed for barcoding of single cells and further complementary DNA amplification. Sample prep was followed according to 10X Protocol. Samples were sequenced on one lane of the HiSeq 4000 (Illumina) with paired-end sequencing parameters: Read1, 98 cycles; Index1, 14 cycles; Index2, 8 cycles; and Read2, 10 cycles.
scRNA-seq analysis: Seurat
Initial processing of the v2 10× libraries was done through CellRanger v2.1.0. Libraries for the Tex14+/−, and Tex14−/− samples were aligned to the mouse genome (10× Genomics prebuilt mm10 reference genome) with all cellranger default parameters for demultiplexing and aligning. Our libraries were sequenced at a depth of 23,567 read pairs per cell for the Tex14+/− sample and 19,735 read pairs per cell for the Tex14−/− sample. The resulting gene by cell matrices was analyzed with the R package Seurat v 3.0. We selected high-quality cells by filtering on the following metrics: genes expressed in at least three cells, fewer than 25,000 unique fragments (to exclude cell doublets), greater than 200 genes expressed (to exclude GEMS with no cells), and cells with 7.0% or less mitochondrial gene expression. We also excluded blood cells by filtering cells whose gene expression was 99.5% hemoglobin genes. The Tex14+/− and Tex14−/− libraries were log-normalized, and then variable genes were selected using the FindVariableFeatures Seurat function with the vst method. To compare the Tex14+/− and Tex14−/− libraries to one another, both libraries were integrated; the Seurat FindIntegrationAnchors and IntegrateData functions were run on both libraries using 1:30 dimensions. Last, the integrated libraries were scaled, and principle components were computed to determine sources of variability between the two datasets. Using the jackstraw statistical test, principle components 1 through 16 were determined to be statistically significant and were used for dimensional reduction analysis. The Seurat FindNeighbors and FindClusters functions [based on Louvain clustering; (47)] were used to identify cells most similar to one another and conduct dimensional reduction. Last, tSNE and UMAP (uniform manifold approximation and projection) were computed with a resolution of 1.0 and a perplexity of 30. We visualized the clustering using the RuntSNE and RunUMAP functions from Seurat. The differentially expressed genes of each cluster were determined by the FindAllMarkers function using a Wilcoxon rank sum test to determine genes expressed with at least a 0.5-fold difference (log scale) between the groups of cells; this test also calculates adjusted P values for multiple comparisons. We required genes to be expressed by a minimum fraction of 0.25 of cells of a cluster to be considered a marker. Last, to identify clusters that are made up of germ cells, we curated a list of the following germ cell markers: Dazl, Ddx4, Sycp1, Sycp3, Dppa3, Pou5f1, and Stra8. Using these seven markers, we were able to identify cluster numbers associated with these germ cell–expressing genes. To subset and create PGC-only datasets to further interrogate, we subset using the cluster numbers for cells highly expressing our seven germ cell markers. In this way, our PGC subset was derived without being influenced by developmental expression of one specific marker. We identified 7590 Tex14+/− and 9368 Tex14−/− germ cells. The integrated germ cells were subset and further analyzed as mentioned above. Jackstaw analysis determined the first 11 principle components to contain meaningful variability in the dataset. The first 11 principle components were used to FindNeighbors and FindClusters, with a resolution of 0.4.
To integrate our Tex14+/− and Tex14−/− libraries with previously published C57BL/6 E13.5 and E14.5 mouse ovaries (24), we normalized all four libraries and selected 2000 variable features. We used dimensions 1:30 for the Seurat functions FindIntegrationAnchors and IntegrateData. We then scaled the integrated Seurat objected and ran principal components analysis. We used principle components 1:15 to calculate TSNE and UMAP dimensionality reduction, as well as find cell neighbors. We then used the FindClusters command with a resolution of 1.0. Germ cells were subset as described above.
scRNA-seq analysis: scAlign
For 10X Single Cell 3′ v2 libraries, the recommend sequencing depth is 50,000 read pairs per cell. To achieve deeper sequencing on our samples, we resubmitted our same Tex14+/− and Tex14−/− libraries for sequencing; our more deeply sequenced libraries totaled 55,744 read pairs per cell for Tex14+/− library and 41,117 read pairs per cell for the Tex14−/− library. We confirmed that the Seurat analysis of the more deeply sequenced libraries mirrored the analysis from the shallowly sequenced libraries. However, we used these more deeply sequenced versions of our libraries for scAlign analysis, to ensure that we were able to detect subtle heterogonies, which may have been missed in the more shallowly sequenced iterations.
We selected high-quality cells by filtering on the following metrics: fewer than 30,000 unique fragments (to exclude cell doublets), greater than 1000 genes expressed (to exclude GEMS with no cells), and cells with 5.0% or less mitochondrial gene expression. We also excluded blood cells by filtering cells whose gene expression was 99.75% hemoglobin genes. After normalizing and scaling the data (using the Seurat v3.0 package), we used Seurat’s FindVariableFeatures function to select 3000 highly variable genes from Tex14+/− and Tex14−/− datasets. A total of 2347 genes present in both datasets were used as input to the encoder aligment algorithm. We used the log counts of both datasets input to the decoder, which projects cells from Tex14−/− genotypes into the alignment space of the Tex14+/− dataset. scAlign was then trained for 5000 steps using the architecture = “small” neural network option available in the scAlign options function. The resulting 2390 dimensions were input back into Seurat for further analysis as the Tex14−/− into Tex14+/− reduction; we used FindNeighbors, FindClusters, and RunTSNE on the 2210 dimensions of this reduction. Observing that the somatic populations did not differ in alignment space, we subset out germ cells only, on the basis of clusters marked with Dazl. We repeated this protocol on germ cells only, this time with 2334 highly variable genes. After converting the dataset back into a Seurat object and calculating FindNeighbors, FindClusters, and RunTSNE on the basis of the 2210 dimensions of the Tex14−/− into Tex14+/− germ cells–only reduction, we used the CellSelector Seurat function to manually gate and select cells that appear spatially distinct. We were then able to use the FindMarkers function to identify genes of interest in these selected populations.
WGBS by postbisulfite adapter tagging
E13.0 and E13.5 ovaries were dissected and digested, and Oct4-GFP+ female germ cells from single Tex14−/−, Tex14+/−, or Tex14+/+ embryos were purified by fluorescence-activated flow cytometry as described above. Sorted GFP+ germ cells (8611 cells from Tex14+/+, 9944 cells from Tex14+/−, 4371 cells from Tex14−/− at E13.0; 3224 and 8606 cells from Tex14+/− and 3710 and 10,371 cells from Tex14−/− at E13.5) were pelleted and lysed using the EZ DNA Methylation-DirectTM Kit (Zymo, D5020). To assess bisulfite conversion rate, 60 fg of unmethylated phage lamba (Promega, D1521) was spiked into each sample. We performed bisulfite conversion and library preparation using the Pico Methyl-Seq Library Prep Kit (Zymo, D5455). We implemented the purification steps as described in sections 3, 4, and 5 of the manufacturer’s protocols using a 5:1 ratio of binding buffer:sample. We amplified seven different libraries through nine PCR cycles (see section 4 of the manufacturer’s protocol). The concentration and purity of each library were assessed using a NanoDrop, and the quality of the libraries was evaluated using Agilent BioAnalyzer high-sensitivity DNA chips. The libraries were sequenced on NovaSeq 6000 S2 flow cells yielding to 50–base pair paired-end reads.
Bisulfite sequencing analysis
We performed the bisulfite sequencing analysis using the following pipeline. We trimmed 10 nt off the front of both mates of the raw sequences. We filtered the trimmed reads by removing reads with a mean Phred quality score of less than 20. Adapters were removed using cutadapt (v1.12). We used Bismark (v0.14.3) to align the filtered sequences to the mouse genome assembly GRC38/mm10 with the nondirectional parameter and used Bowtie (v1.2) within Bismark. We only kept one read pair when multiple reads pairs have both mates align to the same genomic location. CpG methylation was calculated at each CpG location in the genome by counting the number of reads showing methylation and the number showing unmethylation. We estimated the bisulfite conversion rate by calculating the percentage of unmethylated counts for CpGs on the phage lambda genome.
To assess CpG methylation at GRR promoters, we calculated the average weighted methylation score for each GRR promoter. The weighted methylation score takes into account variable coverage, and it is calculated by summing the methylated counts and the unmethylated counts of all CpGs in a region; the methylation percentage is then derived from these summed counts. This calculates the average methylation of multiple CpGs but adds more weight to the CpGs with more coverage. For GRR promoters, we used CpGs in the region from 1000 nt upstream of the TSS (transcription start site) to 500 nt downstream of the TSS, as defined by the GENCODE annotation (vM24). For L1 and IAP (Intracisternal A particle) elements, we used CpGs within the repeat regions, as defined by the repeat masker annotation. DNA methylation levels were visualized as box-and-whisker plots.
Statistical analysis
Built-in statistical function in RipleyGUI was used for clustering analysis. Methylation levels were compared by calculating P values with the two-sided paired Wilcoxon rank sum test. All other analyses were performed using Prism software. Fisher’s exact test was used for analyzing the bicolor clones. t test was used for analyzing germ cell numbers in anterior versus posterior part of ovaries and germ cell numbers between two genotypes.
Acknowledgments
We thank M. Matzuk for Tex14 mice, I. L. Weissman for Rainbow mice, B. Zhu and L. Yingfeng for Dppa3 ovaries, the Biological Imaging Development Center at UCSF for feedback on data analysis, and the Electron Microscopy Image Analysis Unit at Akdeniz University School of Medicine for electron microscopy analysis. Comments on the manuscript were provided by L. Afonso, S. A. Cincotta, and other members of the Laird laboratory. Funding: B.S. is supported by the International Research Fellowship Program 2214/A from Scientific and Technological Research Council of Turkey and in part by grant number GCRLE-1620 from the Global Consortium for Reproductive Longevity and Equality at the Buck Institute, made possible by the Bia-Echo Foundation. R.G.J. is supported by NIH F31HD096840. G.H. and S.M. are supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences Z01ES102745; R.A. is supported by a Basil O’Connor Starter Scholar Award #5-FY20-209 from the March of Dimes Foundation, 1 July 2020 to 30 June 2022, #5-FY20-209, “Non-steroidal anti-inflammatory drugs influence uterine 3D patterning and pregnancy outcomes”; and D.J.L. is supported by P30-ES030284, R01ES028212, and R01GM122902, the W.M. Keck Foundation, and the UCSF Program for Breakthrough Biomedical Research. Author contributions: B.S., R.A., and D.J.L. conceived the project. B.S. performed the experiments. B.S., R.A., and A.F. performed the image analysis. R.G.J. performed bioinformatics analysis. D.H.N. established the framework for clonal labeling experiments. S.M. and G.H. performed WGBS experiment and analysis. B.S. and D.J.L. analyzed the data and wrote the manuscript. R.A., N.D., and D.J.L. provided feedback. Competing interests: The authors declare that they have no competing interests. Data availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. WGBS data have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE165010. scRNA-seq data can be found under accession code GSE166121.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/15/eabc6747/DC1
REFERENCES AND NOTES
- 1.Menke D. B., Koubova J., Page D. C., Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol. 262, 303–312 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Bullejos M., Koopman P., Germ cells enter meiosis in a rostro-caudal wave during development of the mouse ovary. Mol. Reprod. Dev. 68, 422–428 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Cordeiro M. H., Kim S.-Y., Ebbert K., Duncan F. E., Ramalho-Santos J., Woodruff T. K., Geography of follicle formation in the embryonic mouse ovary impacts activation pattern during the first wave of folliculogenesis. Biol. Reprod. 93, 88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles J., Feng C.-W., Miles K., Ineson J., Spiller C., Koopman P., ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nat. Commun. 7, 10845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M. J., Rossant J., Hamada H., Koopman P., Retinoid signaling determines germ cell fate in mice. Science 312, 596–600 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Koubova J., Menke D. B., Zhou Q., Capel B., Griswold M. D., Page D. C., Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 2474–2479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S., Chatzi C., Brade T., Cunningham T. J., Zhao X., Duester G., Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat. Commun. 2, 151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora R., Abby E., Ross A. D. J., Cantu A. V., Kissner M. D., Castro V., Ho H.-Y. H., Livera G., Laird D. J., Meiotic onset is reliant on spatial distribution but independent of germ cell number in the mouse ovary. J. Cell Sci. 129, 2493–2499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernet N., Condrea D., Mayere C., Féret B., Klopfenstein M., Magnant W., Alunni V., Telentin M., Souali-Crespo S., Nef S., Mark M., Ghyselinck N. B., Meiosis occurs normally in the fetal ovary of mice lacking all retinoic acid receptors. Sci. Adv. 6, eaaz1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilly J. L., Ovarian follicle counts – not as simple as 1, 2, 3. Reprod. Biol. Endocrinol. 1, 11 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faire M., Skillern A., Arora R., Nguyen D. H., Wang J., Chamberlain C., German M. S., Fung J. C., Laird D. J., Follicle dynamics and global organization in the intact mouse ovary. Dev. Biol. 403, 69–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malki S., Tharp M. E., Bortvin A., A whole-mount approach for accurate quantitative and spatial assessment of fetal oocyte dynamics in mice. Biol. Reprod. 93, 113 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Feng Y., Cui P., Lu X., Hsueh B., Möller Billig F., Zarnescu Yanez L., Tomer R., Boerboom D., Carmeliet P., Deisseroth K., Hsueh A. J. W., CLARITY reveals dynamics of ovarian follicular architecture and vasculature in three-dimensions. Sci. Rep. 7, 44810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenbaum M. P., Iwamori N., Agno J. E., Matzuk M. M., Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol. Reprod. 80, 449–457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aitken R. J., Findlay J. K., Hutt K. J., Kerr J. B., Apoptosis in the germ line. Reproduction 141, 139–150 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Anderson R. A., Fulton N., Cowan G., Coutts S., Saunders P. T. K., Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev. Biol. 7, 136 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepling M. E., Spradling A. C., Female mouse germ cells form synchronously dividing cysts. Development 125, 3323–3328 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Greenbaum M. P., Yan W., Wu M.-H., Lin Y.-N., Agno J. E., Sharma M., Braun R. E., Rajkovic A., Matzuk M. M., TEX14 is essential for intercellular bridges and fertility in male mice. Proc. Natl. Acad. Sci. U.S.A. 103, 4982–4987 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenbaum M. P., Iwamori T., Buchold G. M., Matzuk M. M., Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 3, a005850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill M. E., Hu Y.-C., Lin Y., Page D. C., Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc. Natl. Acad. Sci. U.S.A. 108, 7443–7448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosario R., Crichton J. H., Stewart H. L., Childs A. J., Adams I. R., Anderson R. A., Dazl determines primordial follicle formation through the translational regulation of Tex14. FASEB J. 33, 14221–14233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen N., Quon G., scAlign: A tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 20, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge W., Wang J.-J., Zhang R.-Q., Tan S.-J., Zhang F.-L., Liu W.-X., Li L., Sun X.-F., Cheng S.-F., Dyce P. W., De Felici M., Shen W., Dissecting the initiation of female meiosis in the mouse at single-cell resolution. Cell. Mol. Life Sci. 78, 695–713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato M., Kimura T., Kurokawa K., Fujita Y., Abe K., Masuhara M., Yasunaga T., Ryo A., Yamamoto M., Nakano T., Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 113, 91–94 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Payer B., Saitou M., Barton S. C., Thresher R., Dixon J. P. C., Zahn D., Colledge W. H., Carlton M. B. L., Nakano T., Surani M. A., Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13, 2110–2117 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S., Shiota K., Nakano T., PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Bortvin A., Goodheart M., Liao M., Page D. C., Dppa3 / Pgc7 / stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev. Biol. 4, 2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei L., Spradling A. C., Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development 140, 2075–2081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargan-Calvopina J., Taylor S., Cook H., Hu Z., Lee S. A., Yen M.-R., Chiang Y.-S., Chen P.-Y., Clark A. T., Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev. Cell 39, 75–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill P. W. S., Leitch H. G., Requena C. E., Sun Z., Amouroux R., Roman-Trufero M., Borkowska M., Terragni J., Vaisvila R., Linnett S., Bagci H., Dharmalingham G., Haberle V., Lenhard B., Zheng Y., Pradhan S., Hajkova P., Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 555, 392–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Zhang Z., Chen J., Liu W., Lai W., Liu B., Li X., Liu L., Xu S., Dong Q., Wang M., Duan X., Tan J., Zheng Y., Zhang P., Fan G., Wong J., Xu G. L., Wang Z., Wang H., Gao S., Zhu B., Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564, 136–140 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M. A., Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Huppertz B., Tews D. S., Kaufmann P., Apoptosis and syncytial fusion in human placental trophoblast and skeletal muscle. Int. Rev. Cytol. 205, 215–253 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Margall-Ducos G., Celton-Morizur S., Couton D., Brégerie O., Desdouets C., Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J. Cell Sci. 120, 3633–3639 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Lei L., Spradling A. C., Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352, 95–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuzhat N., Ikami K., Abbott H., Tanner H., Spradling A. C., Lei L., Altered germline cyst and oocyte differentiation in Tex14 mutant mice reveal a new mechanism underlying female reproductive life-span. bioRxiv , 814384 (2019). [Google Scholar]
- 38.Iwamori T., Iwamori N., Matsumoto M., Imai H., Ono E., Novel localizations and interactions of intercellular bridge proteins revealed by proteomic profiling. Biol. Reprod. 102, 1134–1144 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Chuma S., Nakatsuji N., Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev. Biol. 229, 468–479 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Zamudio N., Barau J., Teissandier A., Walter M., Borsos M., Servant N., Bourc’his D., DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayère C., Neirijnck Y., Sararols P., Rands C. M., Stévant I., Kühne F., Chassot A.-A., Chaboissier M.-C., Dermitzakis E. T., Nef S., Single cell transcriptomics reveal temporal dynamics of critical regulators of germ cell fate during mouse sex determination. bioRxiv, 747279 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Nakamura T., Liu Y.-J., Nakashima H., Umehara H., Inoue K., Matoba S., Tachibana M., Ogura A., Shinkai Y., Nakano T., PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486, 415–419 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Du W., Dong Q., Zhang Z., Liu B., Zhou T., Xu R.-M., Wang H., Zhu B., Li Y., Stella protein facilitates DNA demethylation by disrupting the chromatin association of the RING finger-type E3 ubiquitin ligase UHRF1. J. Biol. Chem. 294, 8907–8917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen D. H., Soygur B., Peng S. P., Malki S., Hu G., Laird D. J., Apoptosis in the fetal testis eliminates developmentally defective germ cell clones. Nat. Cell Biol. 22, 1423–1435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansson K., Jafari-Mamaghani M., Krieger P., RipleyGUI: Software for analyzing spatial patterns in 3D cell distributions. Front. Neuroinform. 7, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malki S., van der Heijden G. W., O’Donnell K. A., Martin S. L., Bortvin A., A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell 29, 521–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E., Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/15/eabc6747/DC1


