Abstract
This study contains synthesis, antimicrobial activity, density functional modelling and molecular docking studies of benzoxazole derivative: 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole. The synthetic procedure of investigated compound is given in detail. The newly synthesized benzoxazole compound and standard drugs were evaluated for their antimicrobial activity against some Gram-positive, Gram-negative bacteria and fungus C. albicans and their drug-resistant isolates. The benzoxazole compound has been characterized by using 1H-NMR, IR and MASS spectrometry and elemental analysis techniques. The molecular structure of the compound in the ground state has been modelling using density functional theory (DFT) with B3LYP/6-311++g(d,p) level. The molecular docking of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with COVID-19 main protease has been also performed by using optimized geometry and the experimentally determined dimensional structure of the main protease (M-pro) of COVID-19.
Keywords: Benzoxazoles, Density functional theory, Dpectroscopy, Molecular docking, Covid-19
Graphical abstract
1. Introduction
During the last decades, a better comprehension of viral replication and disease states caused by viral infections have led to the development of newer antiviral agents with enhanced activity and better tolerability. However, because of limited efficacy of treatment and treatment-emergent antiviral resistance in infections there are needed urgently new compounds. Especially nowadays a great number of deaths and various public health problems are occurring throughout the globe because of COVID-19 which has been designated as SARS-CoV-2 around the globe [1,2]. The scientists around the world have been struggling to understand SARS-CoV-2 and investigate the pathophysiology of this disease to find out potential treatment and discover effective therapeutic drug candidate. Benzoxazole derivatives have acquired a lot of importance in the past few years because of their use in intermediates for new biological materials. Many known drugs are available having benzoxazole as core active moiety like, nonsteroidal anti-inflammatory drug; flunoxaprofen, benoxaprofen, muscle relaxant-chloroxazone and an antibiotic; calcimycin. Benzoxazoles are important materials in medicinal chemistry due to their wide spectrum of different biological activities such as antimicrobial [3], [4], [5], anticancer [6], anti-inflammatory [7], antimicrobacterial [8], antihistaminic [9], antiparkinson [10], antiviral [11] and Rho-kinase inhibition [12]. Our research group has been intensively studying the synthesis of benzoxazoles and their molecular properties because of the importance of the benzoxazole derivatives [[3], [4], [5], 8, 13,14]. Prompted by the above findings in the present study, we hereby report the synthesis, spectroscopic, antimicrobial activity, Density Functional Theory (DFT) calculations studies of the investigated new benzoxazole derivative. The molecular docking studies have become efficient tool for drug discovery and development. While traditional methods of drug discovery can take many years with high costs, the possible medications are investigated by using molecular docking studiesin a short time and with very low costs. The experimentally determined dimensional structure of the main protease (M-pro) of COVID-19 which plays a pivotal role in mediating viral replication and transcription are available in the Protein Data Bank (PDB) [15]. This protease represents a potential target for the inhibition of COVID-19 replication [15]. Therefore, we have also performed the molecular docking of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with COVID-19 main protease.
2. Materials and methods
2.1. Reagents and techniques
The analytical grade chemicals procured from Sigma-Aldrich Co. (Taufkirchen, Munich Germany) and Fisher Scientific (Pittsburgh, PA, USA) were used as such without further purification. Thin-layer chromatography on 0.3 mm silica gel (Merck) plates was performed for monitoring the progress of reaction, using chloroform and methanol as mobile phase in ratio of 3:1. Melting point was recorded on a Stuart Scientific SMP 1 (Bibby Scientific Limited, Stone, Staffordshire, UK) and are uncorrected. Schimadzu spectrometer was used for recording infrared spectrum. Varian Mercury 400 MHz NMR spectrometer was used for recording 1H spectra in appropriate deuterated solvent and are expressed in parts per million (ppm) downfield from tetramethylsilane (internal standard). NMR data are given as multiplicity (s, singlet; d, doublet; t, triplet; m, multiplet) and number of protons. LECO 932 CHNS (St. Joseph, MI, USA) instrument C, H and N analyzer was utilized for the elemental analysis of the new synthesized compound. Mass spectra were obtained on Waters ZQ Micromass LC-MS spectrometer (Milford, MA, USA) using the ESI(+) method. Infrared absorption spectra of the investigated compound were obtained from Shimadzu IR Affinity-1 model FT-IR spectrometer and were reported in cm−1 units. The FT-IR spectrum of the investigated compound (3) are given in Fig. S1 (Supplementary material).
2.2. Procedure for synthesis of benzoxazole derivatives
The method to synthesize the designed benzoxazole derivative is given in Fig. 1. Initially, 5-amino-2-(p-chlorobenzyl)-benzoxazole (1) was synthesized by the reaction of ortho aminophenol, p-chlorophenylacetic acid in polyphosphoric acid. 5-[3-chloropropanamido]-2-(p-chlorobenzyl)-benzoxazole (2) was synthesized by the reaction of 3-chloropropionylchloride with 5-amino-2-(p-chlorobenzyl)-benzoxazole in diethyl eter-water-sodium bicarbonate solution in ice-cooled condition. Finally, reaction of (2) with N-ethyl-piperazine gave the title compound with yield of %61 (3) (Fig. 1 ). Its melting point was 110-112°C. The molecular structures of the synthesized compound (3) were determined by IR (cm−1), 1H/13C-NMR (DMSO-d6, 400 MHz, ppm), mass spectra and elemental analysis studies. The synthetic procedure of investigated compound (3) is given following step by step:
| (1) |
Fig. 1.
The synthesize diagrams.
5-Amino-2-(p-chlorobenzyl)-benzoxazole was synthesized by heating (0.02 mol) 2,4-diaminophenol-hydrochloride with (0.02 mol) p-chlorophenyl acetic acid in 25 g polyphosphoric acid (PPA) and stirring for about 1 h at 160 °C. Then the residue was poured over ice, and the solution was neutralized with 10% NaOH. The resulting precipitate was filtered and was crystallized in ethanol [2].
| (2) |
Chloropropionyl chloride (0.02 mol) was added over a period of 1 h to a stirred, ice-cooled mixture of 5-amino-2-(p-chloro-benzyl)-benzoxazole (0.02 mol) (1), sodium bicarbonate (0.02 mol), diethyl ether (40 ml), and water (20 ml). The mixture was stirred overnight. The precipitate formed was filtered off, washed with water, and dissolved in ethanol. Elemental analyses result was ± 0.04% of theoretical values for compounds (1), (2) and given as Supplementary material (Table S1).
| (3) |
Then, (0.002 mol) 5-(3-chloropropionamido)-2-(p-chloro-benzyl)-benzoxazole (2) was added to (0.002 mol) N-ethyl-piperazine 2 ml of triethylamine solution in 3 ml of N,N-dimethylformamide (DMF) and 2 ml of ethanol. The mixture was stirred at room temperature for 24 h. At the end of the reaction time, the mixture was poured over ice, an equal volume of 5% (w/v) of aqueous NaOH solution was added, and the mixture extracted with chloroform. The solvent was evaporated under reduced pressure, and the resulting crude product was purified by column chromatography using chloroform as mobile phase. Finally, the chloroform fractions were collected, the solvent evaporated, and crystallization was achieved by dissolving the residue in chloroform and adding petroleum ether. The crystalline material was dried in vacuo. The investigated compound 3 was prepared as original product. The structure of its was confirmed by spectral data and the analysis results agree with its of the proposed structure. Yield: %61, Melting Point:110-112°C, 1H-NMR (DMSO-d6 δ ppm): 0.954-0.990 (3H, t, CH 3CH2-), 2.256-2.516 (12H, m, piperazine -CH2, CH3 CH 2 -, CH 2-CO), 2.608-2.643 (2H, t, CH 2-CH2CO), 4.334 (2H, s, benzyl CH2), 7.415-7.441 (5H, m, phenyl-H, benzoxazole H-6), 7.571-7.594 (1H, d, Jo=9.2, benzoxazole H-7), 8.018-8.022 (1H, d, Jm=1.6 benzoxazole H-4), 10.217 (1H, s). MS (m/z): 427(100, M+H) 429(33). C23H27ClN4O2 Calculated C: 64.70 H: 6.37 N: 13.12, Observed C: 64.80 H: 6.55 N: 12.95
2.3. Screening for antimicrobial activities
Standard powders that were obtained Sigma, were used as standard antimicrobial agents. Isolates were; P. aeruginosa isolate [resistant to gentamycine], E. coli isolate [has Extended Spectrum Beta Lactamase (ESBL) enzyme], E. faecalis isolate [resistant to vancomycin (VRE)], and S. aureus isolate [resistant to methicilline (MRSA)]. Standard strains were; Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212 and Candida albicans ATCC 10231 standard strains and clinical isolates provided from Gazi University Hospital Microbiology Laboratory (Ankara, Turkey) were used in this research.
2.4. Calculation details
2.4.1. DFT calculations
The molecular geometry of the compound is modelled by the Gauss-view visualization program [16] and the geometry optimization is performed by the Gaussian 09 software package program [17] for the DFT calculations with Becke's three parameters hybrid exchange–correlation functional (B3LYP) at 6–311++G(d,p) basis set [18]. MEP surface was evaluated by using B3LYP/6-311++G(d,p) method to investigate the reactive sites and to identify sites of intra- and intermolecular interactions of the compound. The total energy, frontier molecular orbitals energies, band gaps and chemical parameters such as total molecular dipole moments (μ), the absolute electro negativity (χ), the absolute hardness (η) and softness (σ) are calculated by the B3LYP/6–311++G(d,p).
2.4.2. Molecular docking
We have performed molecular docking calculations to examine the state of three dimensional structure of the main protease (M-pro) of COVID-19 settlement of investigated 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole (3) compound. Thus to implement the molecular docking study, 3D molecular structure of the main protease (M-pro) in complex with an inhibitor N3 of COVID-19 virus which PDB ID number is 6LU7 was taken from the protein data bank [15]. The molecular geometry of the (3) was directly taken from the optimization results from output of Gaussian 09 software package program. The molecular docking simulations between inhibitor (3) and the SARS-CoV-2 main protease were performed using the AutoDock-Vina code, running 3-ways multithreading, Lamarkian Genetic Algorithm and the feasible region center_x = -26.283, center_y = 12.599, center_z = 58.965, size_x = 50, size_y = 60, size_z = 60, spacing = 1 and exhaustiveness = 8 [19]. All residues were removed and polar hydrogens were added, producing favorable protonation states for molecular docking by using Discover Studio Visualizer 4.0 software [20]. The active site of the SARS-CoV-2 main protease was defined to include residues of active site within the grid size of 50 × 60 × 60 Å for 6LU7. Between molecular docking executions were performed, the most favorable ones being represented by the lowest free-bond energy (ΔG) [21]. The other criteria is the Root Mean Square Deviation (RMSD) values which is lower than 2 Å [22]. The interactions figures were drafted using Discover Studio Visualizer 4.0 software and PyMol [20, 23].
3. Results and discussions
3.1. Spectroscopic studies
The presence of IR absorption respectively band at 1682 cm−1 and 1618 cm−1 in the spectral data of synthesized derivative (I) corresponds to the group NH-C=O (amide I band and amide II band). Oxazole ring vibration of benzoxazole compounds shows band around 1476 cm−1. In case of halogen group Ar–Cl vibration appears at 760–799 cm−1(Fig. S1, Supplementary material). DMSO-d6 was used for recording the 1H-NMR spectra of benzoxazole derivative. In case of 1H-NMR spectra the presence of multiplet signals between 7.42 and 7.44 ppm reflected the presence of phenyl and 6th position of benzoxazole ring protons in synthesized derivative. Also 7th position of benzoxazole ring proton and 4th position of benzoxazole ring proton appeared respectively at 7.571-7.594 ppm (1H, d, Jo=9.2Hz) and, 8.018-8.022 (1H, d, Jm=1.6 Hz). The compound showed singlet at 10.22 ppm because of the presence of NH of NH-C=O. The appearance of singlet at 4.33 ppm, is due to the existence of benzylic CH2 group. Compound 3 showed triplet around 2.61-2.64 ppm and 0.95-0.99 ppm respectively due to existence of CH 2-CH2CO group at CH2 protons and CH3 protons of ethyl group. Also compound (3) showed that multiplet at range of 2.26-2.52 ppm as 12 H due to presence of piperazine CH2, CH2 protons of ethyl group and CH2 protons of CH 2-CO group. According to Mass spectral data, its M+H peak was observed that the spectral signals and proposed molecular structure of the prepared compound showed good agreement. Finally, elemental analyses result was ± 0.04% of theoretical values.
3.2. Optimized molecular structure
The molecular structure of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole (3) compound has been modelled by the Gauss-view visualization program [16] and optimized by the Gaussian 09 software package program [17]. The bond lengths, bond angles, and dihedral angles of the title compound were calculated by using density functional theory (DFT) with the functional B3LYP using the 6-311++G(d,p) basis set. The selected bond distances, bond angles and torsion angles are given in the Table 1 . The optimized molecular geometry with atom label of the investigated compound is given in Fig. 2 .
Table 1.
Optimized structural parameters using DFT/B3LYP with 6-311++G(d,p) basis set of the title compound in the ground state. Bond distances (Å) and angles (o).
| Bond distances (Å) | B3LYP6-311++G(d,p) | Bond angles (0) | B3LYP6-311++G(d,p) | Selected torsion angles (o) | B3LYP6-311++G(d,p) |
|---|---|---|---|---|---|
| C1-C2 | 1.5276 | C1-C2-N1 | 113.799 | C1-C2-N1-C3 | -164.005 |
| C2-N1 | 1.4633 | C2-N1-C3 | 111.392 | C1-C2-N1-C6 | 70.446 |
| N1-C3 | 1.4881 | N1-C3-C4 | 111.305 | N1-C6-C5-N2 | -28.290 |
| N1-C6 | 1.4582 | C3-C4-N2 | 110.204 | N1-C3-C4-N2 | -30.675 |
| C3-C4 | 1.5416 | C4-N2-C5 | 110.685 | N2-C7-C8-C9 | -64.293 |
| C4-N2 | 1.4621 | N2-C5-C6 | 111.058 | C3-C4-N2-C7 | -165.825 |
| N2-C5 | 1.4729 | C5-C6-N1 | 109.830 | C6-C5-N2-C7 | -162.835 |
| C5-C6 | 1.5406 | C6-N1-C2 | 114.558 | C4-N2-C7-C8 | 163.516 |
| N2-C7 | 1.4661 | C6-N1-C3 | 109.924 | C6-N2-C7-C8 | -69.044 |
| C7-C8 | 1.5375 | C4-N2-C7 | 113.525 | C8-C9-N3-C10 | 177.749 |
| C8-C9 | 1.5333 | C5-N2-C7 | 113.382 | C7-C8-C9-O1 | -143.052 |
| C9-O1 | 1.2218 | N2-C7-C8 | 113.916 | C9-N3-C10-C15 | 4.790 |
| C9-N3 | 1.3695 | C7-C8-C9 | 117.346 | C9-N3-C10-C11 | -175.371 |
| N3-C10 | 1.4102 | C8-C9-N3 | 114.315 | N3-C10-C15-C14 | 179.627 |
| C10-C11 | 1.4157 | C9-N3-C10 | 128.882 | N3-C10-C11-C12 | 179.449 |
| C11-C12 | 1.3909 | N3-C10-C11 | 115.888 | C15-C14-N4-C16 | 179.749 |
| C12-C13 | 1.3838 | N3-C10-C15 | 123.265 | C12-C13-O2-C16 | 179.984 |
| C13-C14 | 1.3953 | C10-C11-C12 | 122.100 | N4-C14-C13-C12 | 179.960 |
| C14-C15 | 1.3945 | C11-C12-C13 | 115.960 | O2-C13-C14-C15 | 179.702 |
| C15-C10 | 1.3997 | C12-C13-C14 | 123.073 | C13-O2-C16-C17 | -178.664 |
| C13-O2 | 1.3753 | C13-C14-C15 | 121.182 | C14-N4-C16-C17 | 178.528 |
| O2-C16 | 1.3756 | C14-C15-C10 | 116.837 | N4-C16-C17-C18 | -104.230 |
| C16-N4 | 1.2910 | C15-C10-C11 | 120.847 | O2-C16-C17-C18 | 74.397 |
| N4-C14 | 1.3995 | C12-C13-O2 | 129.219 | C16-C-17-C18-C23 | 66.554 |
| C16-C17 | 1.4929 | C13-O2-C16 | 103.978 | C16-C-17-C18-C19 | -113.590 |
| C17-C18 | 1.5216 | O2-C16-N4 | 115.272 | C19-C20-C21-Cl1 | 179.936 |
| C18-C19 | 1.3955 | C16-N4-C14 | 104.755 | C23-C22-C21-Cl1 | 179.920 |
| C19-C20 | 1.3942 | N4-C14-C13 | 108.287 | ||
| C20-C21 | 1.3894 | O2-C16-C17 | 116.838 | ||
| C21-Cl1 | 1.7602 | N4-C16-C17 | 127.878 | ||
| C21-C22 | 1.3921 | C16-C17-C18 | 113.060 | ||
| C22-C23 | 1.3915 | C17-C18-C19 | 121.054 | ||
| C23-C18 | 1.3988 | C18-C19-C20 | 121.125 | ||
| C19-C20-C21 | 119.041 | ||||
| C20-C21-Cl1 | 119.491 | ||||
| Cl1-C21-C22 | 119.451 | ||||
| C20-C21-C22 | 121.058 | ||||
| C21-C22-C23 | 119.138 | ||||
| C22-C23-C18 | 121.014 | ||||
| C23-C18-C19 | 118.624 |
Fig. 2.
Optimized geometry of investigated compound.
As an optimized geometry, whole of moieties is not nearly planar. The torsion angles for 2-(p-chloro-benzyl) ring Ph1 with the benzoxazole Ring3 are C19-C18-C17-C16= -113.59, C18-C17-C16-O2=74.40 and C18-C17-C16-N4=-104.23o. The propionamido group is titled from the phenyl ring Ph2 which is evident from the torsion angles C11-C10-N3-C9= -175.37 and C8-C9-N3-C10=177.75o. The torsion angle N2-C7-C8-C9 value between with piperazynl ring and propionamido group is -64.29o. The angles between 2-(p-chloro-benzyl) ring Ph1, benzoxazole Ring3 and the phenyl ring Ph2 are 76.75 and 76.67o respectively.
The bond distances, bond angles and torsion angles which are theoretically determinedby DFT with the functional B3LYP using the 6-311++G(d,p) basis set in the investigated compound are good consistent with experimentally (X-ray structure analysis) corresponding values of similar benzoxazoles derivatives in the literature [13,14, 24].
3.6. Molecular electrostatic potential surface
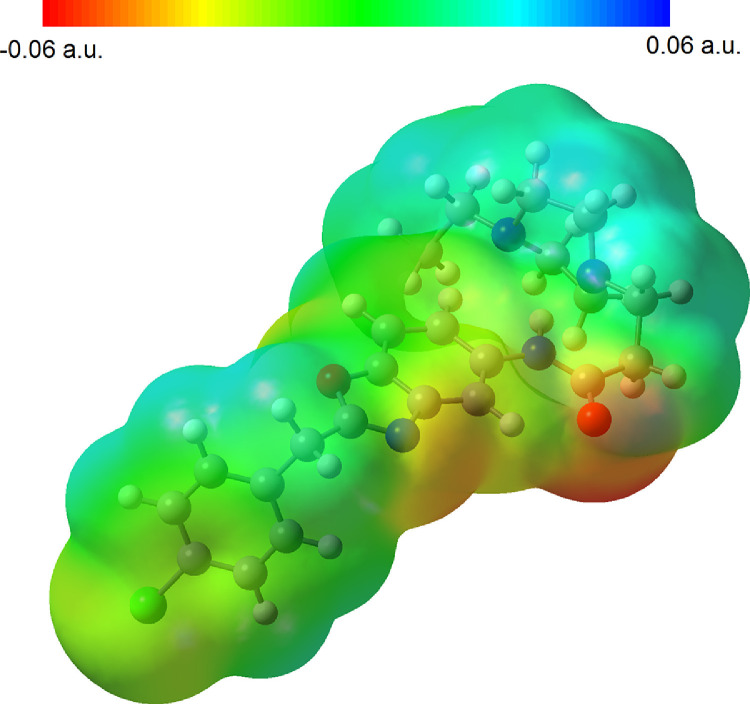
The electrostatic potential is well suited for analyzing processes based on the ‘‘recognition’’ of one molecule by another, as in drug-receptor, and enzyme–substrate interactions, because it is through their potentials that the two species first ‘‘see’’ each other [25, 26]. To investigate the regions of the MEP for the title compound was composed by DFT calculation using the optimized geometry at the B3LYP/6-311++G(d,p). The MEP surface, electrophilic reactivity (negative regions: red and yellow colours) and nucleophilic reactivity (positive regions: blue colours) shown in Fig. 3 . As can be seen from Fig. 3, the compound has three possible sites for electrophilic attack. The O1, N1 and Cl1 atoms have negative region. The negative molecular electrostatic potential value for is the O1, N1 and Cl1 atoms 0.060, 0.041 and 0.016 a.u, respectively. The main regions for nucleophilic reactivity in the whole molecule is 2-(p-chloro-benzyl) ring Ph1, the propionamido group and the piperazynl ring. These groups have positive region between 0.013-0.024 a.u.
Fig. 3.
Molecular electrostatic potential surface of the investigated compound.
3.7. HOMO-LUMO band gap and chemical parameters
The determining highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) are key to calculating chemical parameters and very important for chemical stability because of these orbitals play an important role in the electric properties and determine the way the molecule interacts with other species [27, 28]. In the other words, HOMO and LUMO are the main orbitals for the chemical reactions.
The HOMO represents the ability to donate an electron, whereas LUMO represents the ability to obtain an electron. Thus, the HOMO is directly related to the ionization potential (IP), while LUMO is directly related to the electron affinity (EA). According to the Koopman's theorem [29] for closed-shell molecules, IP and EA can be expressed as follows in terms of HOMO and LUMO energies:
| (5) |
| (6) |
If the values of IP and EA are predicted, HOMO-LUMO band gap and the global chemical reactivity descriptors of molecules such as chemical hardness (η), electronegativity (χ), softness (σ), chemical potential (µp), and electrophilicity index (ω) as well as local reactivity can determine according to the Koopman's theorem.
The total molecular energies, HOMO and LUMO energies were predicted by the B3LYP/6-311++G(d,p) level for investigating. The values of total energy, HOMO-LUMO band gap and chemical parameters of the investigated compound were listed in Table 2 . The frontier molecular orbital distributions and energy levels of the HOMO and LUMO which is mostly the π-antibonding type molecular orbitals of the title compound are shown in Fig. 4 . Except for the propionamido group and the piperazynl ring in the structure, the LUMO are mainly localized on the whole structure, whereas the HOMO is localized only on the 2-(p-chloro-benzyl) ring Ph1 with the benzoxazole Ring3 and phenyl ring Ph2, as seen from Fig. 4. The HOMO-LUMO gap was calculated to be 4.8795 eV. The large HOMO-LUMO band gap means a hard molecule whereas small band gap means a soft molecule. They are very important relation between HOMO-LUMO band gap and light-emitting properties of the benzoxazole derivatives [13].
Table 2.
Calculated chemical parameters of the title compound at B3LYP/6-311++G(d,p) level.
| Basis Set | B3LYP/6-311G++(d,p) |
|---|---|
| ETOTAL(Hartree) | -1722.7301 |
| EHOMO(eV) | -5.9492 |
| ELUMO (eV) | -1.0697 |
| aΔEGap (eV) | 4.8795 |
| I (eV) | 5.9492 |
| A (eV) | 1.0697 |
| μ(Debye) | 7.4052 |
| η(eV) | 2.4398 |
| χ(eV) | 3.5095 |
| σ(eV) | 0.4099 |
| μp(eV) | -3.5095 |
| ω | 15.0245 |
ETOTAL: Total energy
EHOMO and ELUMO: Energy values of HOMO and LUMO
µ: Total molecular dipole moments,
I: Ionization potential,
A: Electron affinity,
η: Absolute hardness, (I-A)/2,
χ: Electronegativity, (I+A)/2,
σ: Softness, 1/η,
μp:Chemical potential, -(I+A)/2
ω: Electrophilicity, μp2/2η.
ΔΕGap= (ELUMO – EHOMO): gap of energy,
Fig. 4.
Molecular orbital surfaces and energy levels given in parentheses for the HOMO and LUMO orbitals of the title compound computed at 6-311++G(d,p) level.
3.8. Antimicrobial activity
Antimicrobial susceptibility testing was performed through CLSI M100-S18 and CLSI M27-A3 directions [12, 13]. Mueller Hinton Agar (MHA) (Merck), Mueller Hinton Broth (MHB) (Merck), Sabouraud Dextrose Agar (SDA) (Merck), Sabouraud Liquid Medium (SLM) (Merck) and RPMI-1640 medium (Sigma) with L-glutamine buffered pH 7 with 3-[N-morpholino]-propansulfonic acid (MOPS) (Sigma) were used for microbial cultures. Stock solutions of the test compounds were prepared in DMSO (Merck). Ampicillin was prepared in phosphate buffer solution and other antibiotic solutions were prepared in sterile distilled water according to the guideline of CLSI) M100-S25. Bacterial isolates were subcultured in Mueller Hinton Agar (MHA) plates and incubated over night at 37 °C and C. albicans was subcultured in Sabouraud Dextrose Agar (SDA) plates at 35 °C for 24-48 h. Pure colonies were transferred to MHB and SLM for bacteria and fungi, respectively. They were incubated in the appropriate conditions overnight. After incubation, the bacterial suspensions used for inoculation were prepared at 105 cfu/ml by diluting fresh cultures at MacFarland 0.5 density (108 cfu/ml). Yeast suspensions were also prepared according to McFarland 0.5 density and a working suspension was made by a 1:100 dilution followed by a 1:20 dilution of the stock suspension (2.5 × 103 CFU/ml). Susceptibility testing was performed with MHB for bacteria and RPMI-1640 medium with L-glutamine buffered pH 7 with 3-[N-morpholino]-propansulfonic acid (MOPS) for fungi. The solution of the newly synthesized compounds and standard drugs were prepared at 512, 256, 128, 64, 32, 16, 8, 4 µg/mL and 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.06, 0.03, 0.015, 0.0078 µg/mL concentrations, respectively by diluting the stock concentrations in a microdilution tray with a multichannel pipette. After dilution, a 10 µl bacterial or fungal inoculum was added to each well of the microdilution trays. The trays were incubated at 37 °C for bacteria and 35 °C for fungi, in a humid chamber and MIC endpoints were read after 24 h of incubation. The lowest concentration of the compound that completely inhibits macroscopic growth was determined and minimum inhibitory concentrations (MICs) were reported.
All organisms were tested in triplicate in each run of the experiments. Solvents, pure microorganisms and pure media were used as control wells. The data on the antimicrobial activity of the compound and the control drugs as MIC values (µg/mL) are given in Table 3 . Based on these data, the antimicrobial effect of this benzoxazole compound against various microorganisms has been detected in a broad spectrum.
Table 3.
In vitro antibacterial and antifungal MIC values (μg/mL) of the new compound (3) and reference antimicrobial drugs.
| Gram-negative bacteria |
Gram-positive bacteria |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Comp. No | E.c. | E.c.* | P.a. | P.a* | S.a. | S.a.* | E.f. | E.f* | C.a. |
| 3 | 64 | 64 | 64 | 64 | 128 | 256 | 64 | 32 | 128 |
| Vancomycin | n.d | n.d | n.d | n.d | 1 | 1 | 1 | 32 | n.d |
| Ampicillin | 2 | 128 | n.d | n.d | 2 | 64 | 2 | 2 | n.d |
| Ofloxacin | ˂0,0625 | 64 | 8 | 64 | 0,25 | 0,25 | 1 | 4 | n.d |
| Gentamycin | 0.5 | ˃ 512 | 0.5 | ˃512 | 0.125 | 32 | 4 | 32 | n.d |
| Amphotericin B | n.d | n.d | n.d | n.d | n.d | n.d | n.d | n.d | 0,25 |
| Fluconazole | n.d | n.d | n.d | n.d | n.d | n.d | n.d | n.d | 1 |
nd: not determined
When the benzoxazole ring system's chemical structure is investigated, it is thought that the nucleic acids are analog to the adenine and guanine bases in their structure and can show their antimicrobial effects by inhibiting nucleic acid synthesis [30,31]. So that, studies on the benzoxazole derivatives have been increased in recent years [3, [32], [33], [34]].Antimicrobial activities of some benzoxazole derivatives obtained were observed equal or more effective than reference drugs. In previous studies, some derivatives containing p-(substituted)phenyl/benzyl) at position 2 and 6-membered rings attached to the amide side chain at position 5 were synthesized, and promising results were obtained by examining their antimicrobial effects [35], [36], [37], [38].
3.9. Molecular docking studies of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with COVID-19 main protease
The study of molecules employing molecular docking has become increasingly relevant to predict bond modes to understanding of receptor-binder interactions. Benzoxazoles are important materials in medicinal chemistry due to especially their antimicrobial and antiviral inhibition [[3], [4], [5], 11]. A new coronavirus which is named COVID-19 has spread worldwide and the World Health Organization (WHO) is declared a pandemic [1,2].
With the onset of the COVID-19 epidemic, studies have started on interactions of some Antiviral molecules with CoV-2 main protease with molecular docking simulations. Molecular modeling studies of this type are available on some quinoline and indole compounds with a long history as antiviral agents [39, 40]. Benzoxazoles, benzimidazoles and benzothiazoles are isosteres of indoles that indicate potent antiviral activity.A23187 is also known as Calcimycin that is a benzoxazole compound has potent anti-influenza activity [41, 42]. Based on this information, this study paved the way for further experimental evaluation of the benzoxazole molecule, which is thought to have potential antiviral effects against SARS-CoV-2.
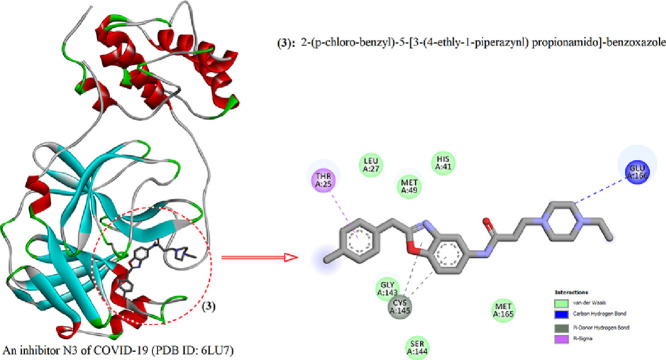
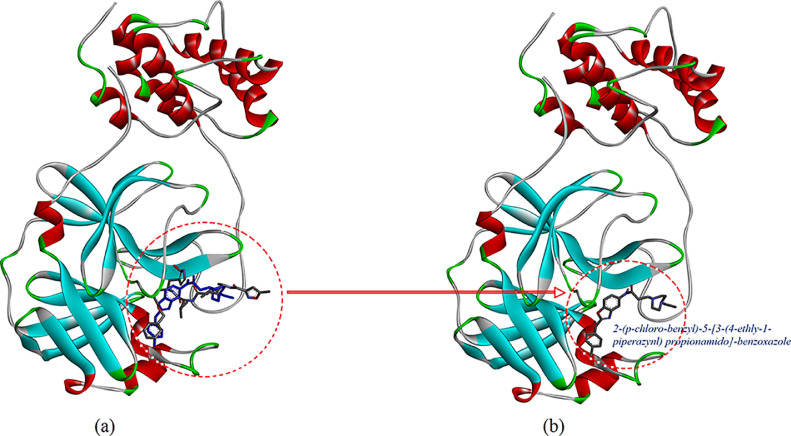
To implement the molecular docking study, 3D molecular structure of the main protease (M-pro) in complex of COVID-19 virus (PDB ID: 6LU7). The generated bonding energy and RMSD values as a result of molecular docking are given in Table 4 . The docking protocol was tested by removing co-crystallized inhibitor from the main protease (M-pro) and then docking it at the same site as seen in Fig. 5 a. Analysis of the molecular docking simulations showed that investigated new benzoxazole molecule are linked with relative binding energy value is -6.1 kcal/mol in site of the main protease (M-pro) in complex of COVID-19 virus. Energetically most favorable docked structures obtained from the rigid molecular docking of the compound 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with 6LU7 are shown in Fig. 5b. The interaction of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with the protease showed a high affinity interaction in the main protease (M-pro) as the ligand fits inside the core pocket region of the protease (Fig. 5b). Analysis of interactions with 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole showed that the molecular fitting simulation of the inhibitor (Fig. 6 ) resulted in the formation of four interactions type with the enzyme, one of the carbon hydrogen bonding between the C3 atom of the piperazynl ring of investigated benzoxazole ligand with GLU166 (3.57 Å), a Pi-sigma bonding between the ph1 group of the ligand with THR25 (3.74 Å), two Pi-donor hydrogen bonding between the Rng3 and Ph2 groups of the benzoxazole ligand with CYS145 (3.73 and 4.01 Å). Some of the van der Waals interaction of the investigated new benzoxazole ligand with LEU27, MET49, HIS41, GLY143 and MET165 has been observed (Fig. 6). These results draw us to the conclusion that the investigated benzoxazole ligand might exhibit an inhibitor activity. But biological tests need to be done to validate the computational predictions. Docking calculations look for a suitable in site of the main protease (M-pro) in complex of COVID-19 virus and perform the interaction energy calculations using the topology of the site chosen beforehand.
Table 4.
Binding affinity of different poses of the investigated ligand (3) as predicted Autodock Vina.
| Compound-inhibitor | Mode Affinity Distance from best mode(kcal/mol)RMSD l.b.RMSD u.b. | |||
|---|---|---|---|---|
| (3)- (6LU7) | ||||
| 1 | -6.1 | 0.000 | 0.000 | |
| 2 | -6.0 | 22.045 | 25.624 | |
| 3 | -6.0 | 27.555 | 31.601 | |
| 4 | -6.0 | 26.621 | 30.749 | |
| 5 | -5.9 | 27.359 | 31.497 | |
| 6 | -5.9 | 29.657 | 33.283 | |
| 7 | -5.9 | 22.296 | 26.690 | |
| 8 | -5.8 | 22.722 | 24.648 | |
| 9 | -5.8 | 28.833 | 33.034 | |
(3): 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole
Fig. 5.
(a) Co-crystallized molecule shown in grey color and the docked conformation of L (blue) as predicted by the Autodock Vina show very low RMSD value. (b) Representation of docking results of investigated a new benzoxazole compound embedded into the main protease (M-pro) in complex of COVID-19 virus (PDB ID: 6LU7). (For interpretation of the references to color in this figure legend, the reader is referred to web version of this article).
Fig. 6.
(a) The ligand binds at the active site of the main protease (M-pro) in complex of COVID-19 virus (PDB ID: 6LU7). (b) 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole and 6LU7 interaction (2D).
4. Conclusion
In this study, initially synthesis, spectroscopic (NMR, FTIR) and elemental analysis of a new benzoxazole compound titled 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole (compound 3) have been presented. Then, it was further evaluated for antibacterial, antifungal activity and it was observed that thecompound 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole (compound 3) displayed the moderate activity against various microbial species in comparison to reference drugs. Also, The molecular structure of the compound 3 in the ground state has been molecular modelling using density functional theory (DFT) with B3LYP/6-311++g(d,p) level. The theoretical 3D-geometric parameters values of the new benzoxazol were quite agreement with the experimental values of the similar compound in literature. MEP surface was evaluated by using B3LYP/6-311++G(d,p) method to predicted the reactive sites and to identify sites of intra- and intermolecular interactions of the compound. The O1, N1 and Cl1 atoms have negative region. The total energy, frontier molecular orbitals energies, band gaps and chemical parameters such as total molecular dipole moments (μ), the absolute electro negativity (χ), the absolute hardness (η) and softness (σ) are calculated by the B3LYP/6–311++G(d,p). The HOMO-LUMO gap was calculated to be 4.8795 eV. All organisms were tested in triplicate in each run of the experiments. Solvents, pure microorganisms and pure media were used as control wells. The data on the antimicrobial activity of the compound and the control drugs as MIC values (µg/mL) are given in this study. The molecular docking of 2-(p-chloro-benzyl)-5-[3-(4-ethly-1-piperazynl) propionamido]-benzoxazole with COVID-19 main protease has been also performed by using optimized geometry and the experimentally determined dimensional structure of the main protease (M-pro) of COVID-19. The molecular fitting simulation of the inhibitor resulted in the formation of four interactions type with the enzyme;the carbon hydrogen bonding, Pi-sigma bonding, Pi-donor hydrogen bonding and van der Waals interaction. A virtual screening based on molecular docking emerges as an important tool for obtaining new antiviral molecules, where researchers can use this tool as a complementary approach so that the synthesis of new compounds or the repositioning of drugs can be assigned.
Author statement
Celal Tuğrul Zeyrek: Conceptualization, Methodology, Software, Molecular modelling, Visualization, Validation, Writing - original draft
Özlem Temiz Arpacı: Investigation, Resources, Writing- Original draft preparation, Synthesis and structure elucidation
Mustafa Arısoy: Investigation, Resources, Writing- Original draft preparation, Synthesis and structure elucidation
Fatma Kaynak Onurdağ: Investigation, Resources, Writing- Original draft preparation, Antimicrobial activity
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.130413.
Appendix. Supplementary materials
References
- 1.Zhou P., Yang X-L., X-G Wang, Hu B., Zhang L., Zhang W., Si H-R., Zhu Y., Li B., Huang C.L., Chen H-D., Chen J., Luo Y., Guo H., Jiang R-D., Liu M.-Q., Chen Y., Shen X-R., Wang X., Zheng X-S., Zhao K., Chen Q-J., Deng F., Liu L-L, Yan B., Zhan F-X, Wang Y-Y., Xiao G-F., Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby PW., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395:470–4733. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arisoy M., Temız-Arpaci O., Yıldız I., Kaynak-Onurdag F., Akı E., Yalcın I., Abbasoglu U. Synthesis, antimicrobial activity and QSAR studies of 2,5-disubstituted benzoxazoles. SAR and QSAR Environ. Res.19. 2008;5–6:589–612. doi: 10.1080/10629360802348738. [DOI] [PubMed] [Google Scholar]
- 4.Temiz-Arpacı O., Cıfcıoglu-Goztepe B.E., Kaynak-Onurdag F., Ozgen S., Senol F.S., Erdogan-Orhan I. Synthesis and Different Biological Activities of Novel Benzoxazoles. Acta Biologica Hungarica. 2013;64(2):249–261. doi: 10.1556/ABiol.64.2013.2.10. [DOI] [PubMed] [Google Scholar]
- 5.Tasci M., Temız-Arpaci O., Kaynak-Onurdag F., Okten S. Syntesis and antimicrobial evaluation of 2-(p-tert-butylphenyl)benzoxazoles. Ind. J. Chem. 2018;57B:385–389. [Google Scholar]
- 6.Aiello S., Wells G., Stone E.L., Kadri H., Bazzi R., Bell D.R., Stevens M.F.G., Matthews C.S.T., Bradshaw D., Westwell A.D. Synthesis and biological properties of benzothiazole, benzoxazole, and chromen-4-one analogues of the potent antitumor Agent 2-(3,4-dimethoxyphenyl)-5-fluorobenzothiazol. J. Med. Chem. 2008;51(16):5135–5139. doi: 10.1021/jm800418z. [DOI] [PubMed] [Google Scholar]
- 7.Sondhi S.M., Singh N., Kumar A., Lozach O., Meijer L. Synthesis, anti-inflammatory, analgesic and kinase (CDK-1, CDK-5 and GSK-3) inhibition activity evaluation of benzimidazole/benzoxazole derivatives and some Schiff's bases. Bioorg. Med. Chem. 2006;14(11):3758–3765. doi: 10.1016/j.bmc.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 8.Arisoy M., Temız-Arpaci O., Kaynak-Onurdag F., Ozgen S. Synthesis and antimicrobial activity of novel benzoxazoles. Z. Naturforsch. 2012;67C:466–472. doi: 10.1515/znc-2012-9-1004. [DOI] [PubMed] [Google Scholar]
- 9.Katsura Y., Inoue Y., Nishino S., Tomoi M., Itoh H., Takasugi H. Studies on antiulcer drugs. III. Synthesis and antiulcer activities of imidazo[1,2-a]pyridinylethylbenzoxazoles and related compounds. A novel class of histamine H2-receptor antagonists. Chem. Pharm. Bull. 1992;40(6):1424–1438. doi: 10.1248/cpb.40.1424. [DOI] [PubMed] [Google Scholar]
- 10.Benazzouz A., Boraud T., Dubedat P., Boireau A., Stutzmann J.M., Gross C. Riluzole prevents MPTP-induced parkinsonism in the rhesus monkey: a pilot study. Eur. J. Pharmacol. 1995;284(3):299–307. doi: 10.1016/0014-2999(95)00362-o. [DOI] [PubMed] [Google Scholar]
- 11.Smith P., Ward D.N. Heterocyclic benzoxazole compositions as inhibitors of hepatitis c virüs, Publication Number WO/2011/047390 Publication Date 21.04.2011 International Application No. PCT/US2010/053085 International Filing Date 18. 2010;10 [Google Scholar]
- 12.Sessions E.H., Yin Y., Bannister T.D., Weiser A., Griffin E., Pocas J., Cameron M.D., Ruiz C., Lin L., Schürer S.C., Schröter T., LoGrasso P., Feng Y. Benzimidazole- and benzoxazole-based inhibitors of Rho kinase. Bioorg. Med. Chem. Lett. 2008;18(24):6390–6393. doi: 10.1016/j.bmcl.2008.10.095. [DOI] [PubMed] [Google Scholar]
- 13.Zeyrek C.T., Unver H., Temiz- Arpaci O., Polat K., Iskeleli N.O., Yildiz M. Experimental and theoretical characterization of the 2-(4-bromobenzyl)-5-ethylsulphonyl-1,3- benzoxazole. J. Mol. Struct. 2015;1081:22–37. [Google Scholar]
- 14.Zeyrek C.T., Boyacioglu B., Temiz-Arpaci O., Unver H., Elmali A. Spectroscopic, quantum mechanical and molecular docking studies of a new benzoxazole compound with an oxidoreductase enzyme and DNA. J. Mol. Struct. 2017;1136:112–126. [Google Scholar]
- 15.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 16.Dennington R., Keith T., Millam J. Semichem, Inc., Shawnee Mission; KS: 2009. GaussView, Version 5. [Google Scholar]
- 17.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., P. J.E., Jr, Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas €O., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision D.01. [Google Scholar]
- 18.Petersson G.A., Bennett A., Tensfeldt T.G., Allaham M.A., Shirley W.A., Mantzaris J. A complete basis set model chemistry .1. The total energies of closed-shell atoms and hydrides of the 1st-row elements. J. Chem. Phys. 1988;89(4):2193–2218. [Google Scholar]
- 19.Trott O., Olson A.J. Software news and update autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Accelrys Software Inc; SanDiego: 2013. Accelrys Software Inc Discovery Studio Modelling Environment. Relaser 4.0. [Google Scholar]
- 21.Gurung A.B., Ali M.A., Bhattacharjee A., Abul Farah M., Al-Hemaid F., Abou-Tarboush F.M., Al-Anazi K.M., Al-Anazi F.S.M., Lee J. Molecular docking of the anticancer bioactive compound proceraside with macromolecules involved in the cell cycle and DNA replication. Genet. Mol. Res. 2016 doi: 10.4238/gmr.15027829. [DOI] [PubMed] [Google Scholar]
- 22.Kramer B., Rarey M., Lengauer T. Evaluation of the FLEXX Incremental Construction Algorithm for Protein–Ligand Docking. Proteins. 1999;37(2):228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.DeLano W.L. San Carlos; CA, USA: 2016. The PyMOL Molecular Graphics System. [Google Scholar]
- 24.Sivakumar M., Rajavelu K.Saravanan K., Rajakumar P., Aravindhan S. Chemical Data Collections Synthesis, structural exploration and Hirshfeld surface analysis of a novel bioactive heterocycle: (4-(6-Fluorobenzo[d]isoxazol-3-yl) piperidin-1-yl)(morpholino)methanone. Chemical Data Collections. 2018;15–16:161–169. [Google Scholar]
- 25.Scrocco E., Tomasi J. Vol. 7. Springer Berlin; 1973. (Topics in Current Chem). [Google Scholar]
- 26.Fleming I. John Wiley &Sons; New York: 1976. Frontier Orbitals and Organic Chemical Reactions; p. 182. [Google Scholar]
- 27.Govindarajan M., Periandy S., Carthigayen K. Spectrochim. Acta A, FT-IR and FT-Raman spectra, thermo dynamical behavior, HOMO and LUMO, UV, NLO properties, computed frequency estimation analysis and electronic structure calculations on α-bromotoluene. Spectrochim. Acta Part A Mol. Biomol. Spectros. 2012;97:411–422. doi: 10.1016/j.saa.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Ravikumar C., Joe I.H., Jayakumar V.S. Charge transfer interactions and nonlinear optical properties of push–pull chromophore benzaldehyde phenylhydrazone: A vibrational approach. Chem. Phys. Lett. 2008;460:552–558. [Google Scholar]
- 29.Koopmans T. Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den Einzelnen Elektronen Eines Atoms. Physica. 1933;1:104–113. [Google Scholar]
- 30.Rida S.M., Ashour F.A., El-Hawash S.A., El Semary M.M., Badr M.H., Shalaby M.A. Synthesis of some novel benzoxazole derivatives as anticancer, anti-HIV-1 and antimicrobial agents. Eur. J. Med. Chem. 2005;40:949–959. doi: 10.1016/j.ejmech.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 31.Oehlers L., Mazzitelli C.L., Brodbelt J.S., Rodriguez M., Kerwin S. Evaluation of complexes of DNA duplexes and novel benzoxazoles or benzimidazoles by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1593–1603. doi: 10.1016/j.jasms.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Ertan T., Yildiz I., Tekiner-Gulbas B., Bolelli K., Temiz-Arpaci O., Ozkan S., Kaynak F., Yalcin I., Aki E. Synthesis, biological evaluation and 2D-QSAR analysis of benzoxazoles as antimicrobial agents. Eur. J. Med. Chem. 2009;44:501–510. doi: 10.1016/j.ejmech.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Tekiner-Gulbas B., Temiz-Arpaci O., Yildiz I., Altanlar N. Synthesis and in vitro antimicrobial activity of new 2-[p-substituted-benzyl]-5-[substituted-carbonylamino]benzoxazoles. Eur. J. Med. Chem. 2007;42:1293–1299. doi: 10.1016/j.ejmech.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Kuroyanagi J.-i., Kanai K., Horiuchi T., Takeshita H., Kobayashi S., Achiwa I., Yoshida K., Nakamura K., Kawakami K. Structure-activity relationships of 1,3-benzoxazole-4-carbonitriles as novel antifungal agents with potent in vivo efficacy. Chem. Pharm. Bull. 2011;59:341. doi: 10.1248/cpb.59.341. [DOI] [PubMed] [Google Scholar]
- 35.Temiz-Arpacı Ö., Ozdemir A., Yalçın İ., Yıldız İ., Akı-Şener E., Altanlar N. Synthesis and Antimicrobial Activity of Some 5-[2-(Morpholin-4-yl)acetamido] and/or 5-[2-(4-Substituted piperazin-1-yl)acetamido]-2-(p-substituted phenyl)benzoxazoles. Arch. Pharm. Chem. Life Sci. 2005;338:105–111. doi: 10.1002/ardp.200400923. [DOI] [PubMed] [Google Scholar]
- 36.Temiz-Arpacı Ö., Yıldız İ., Özkan S., Kaynak F., Akı-Şener E., Yalçın İ. Synthesis and biological activity of some new benzoxazoles. Eur. J. Med. Chem. 2008;43:1423–1431. doi: 10.1016/j.ejmech.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 37.Arisoy M., Temiz-Arpaci O., Kaynak-Onurdag F., Ozgen S. Synthesis and antimicrobial evaluation of 2-(p-substituted phenyl)-5-[(4-substituted piperazin-l-yl)acetamido]-benzoxazoles. Z. Naturforsch C J. Biosci. 2014;69:368–374. doi: 10.5560/znc.2014-0024. [DOI] [PubMed] [Google Scholar]
- 38.Arisoy M., Temiz-Arpaci O., Kaynak-Onurdag F., Ozgen S. Synthesis of some piperazinobenzoxazole derivatives and their antimicrobial properties. Indian J. Chem. B. 2016;55:240–247. [Google Scholar]
- 39.I.Celı̇k, A.Onay-Besı̇kcı̇, G.Ayhan-Kilcigı̇l,cApproach to the mechanism of action of hydroxychloroquine on SARS-CoV-2: a molecular docking study, (2020) 1-13.
- 40.Llanes A., Cruz H., Nguyen V.D., Larionov O.V., Fernández P.L. A computational approach to explore the interaction of semisynthetic nitrogenous heterocyclic compounds with the SARS-CoV-2 main protease. Biomolecules. 2021;11:18–31. doi: 10.3390/biom11010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francesconi V., Cichero E., Schenone S., Naesens L., Tonelli M. Synthesis and biological evaluation of novel (thio)semicarbazone-based benzimidazoles as antiviral agents against human respiratory viruses. Molecules. 2020;25(7):1487–1508. doi: 10.3390/molecules25071487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jantaratrirat S., Boonarkart C., Ruangrung K., Suptawiwat O., Auewarakul P. Microparticle release from cell lines and its anti-influenza activity. Viral Immunol. 2018;31(6):447–456. doi: 10.1089/vim.2017.0201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.