Abstract
The low affinity Fcγ receptor, FcγRIIA, harbors a common missense mutation, rs1801274 (G>A, Arg131His) that modifies binding affinity to human IgG2 and mouse IgG1 antibodies and is associated with increased risk of autoimmune disease. Despite the important role of the Arg131His variant, little is understood about heterozygous genotype effects on global gene expression and cytokine production during an FcγR-dependent response. To address this gap in knowledge, we treated human whole blood samples from 130 individuals with mouse IgG1 anti-CD3 and anti-CD28 antibodies, and characterized the genome-wide gene expression profiles and cytokine production among individuals stratified by rs1801274 genotype. Our analysis revealed that the levels of four cytokines (IFNγ, IL-12, IL-2, TNFα) and global gene expression patterns differed between all three genotype classes. Surprisingly, the heterozygotes showed suboptimal T cell activation compared to cells from individuals homozygous for the higher-affinity FcγRIIA allele (GG; Arg/Arg). The results of this study demonstrate IgG response varies among all rs1801274 genotype classes and results in profound differences in both cytokine responses and gene expression patterns in blood leukocytes. Because even heterozygotes showed dampened global responses, our data may provide insight into the heterogeneity of outcomes in cytokine release assays and immunotherapy efficacy.
Introduction
Fc receptors (FcRs) bind to the Fc region of immunoglobins (Ig) M, IgG, IgE, and IgA, which are secreted by B cells. FcRs play a critical role in activating neutrophils and NK cells to kill infected cells during the process of antibody-dependent cell-mediated cytotoxicity (ADCC), and in inducing basophil and mast cell degranulation1. The class of human FcRs that bind to IgG, FcγR, can be activating (FcγRI, FcγRIIA, FcγRIIC, FcγRIIIA) or inhibitory (FcγRIIB), and are further classified by their affinity to different classes of IgG antibodies. The low affinity receptor FcγRIIA is broadly expressed on monocytes, macrophages, dendritic cells, eosinophils, and neutrophils, and contains a missense mutation (G>A, rs1801274) in the fourth exon of the FCGR2A gene encoding FcγRIIA1. This polymorphism results in an arginine to histidine (Arg131His) amino acid substitution within the binding interface, resulting in increased affinity of the A (His) allele for the monomeric human IgG2 isotype2, 3 and an increase in phagocytosis of opsonized bacteria in granulocytes from individuals with AA (His/His) genotype4. The prevalence of this common mutation varies widely around the world (Figure S1), with the frequency of the A (His) allele ranging from 0.81 in Japanese to 0.39 in East Africans5. The prevalence and functional significance of rs1801274 is underscored by numerous genome-wide association studies (GWAS) reporting associations between the G allele and increased risk of autoimmune and inflammatory diseases, such as ulcerative colitis6, Kawasaki disease7, systemic lupus erythematosus8, and periodontitis9.
In addition to modifying affinity to human IgG2, this polymorphism changes the binding of human cells to mouse IgG1, an antibody frequently used in in vitro studies10. In contrast to binding of human IgG2, the G (Arg) allele has higher affinity to the mouse IgG1 antibody compared to the A (His) allele. The effects of this polymorphism on peripheral blood mononuclear cell (PBMC) responses to soluble mouse IgG1 anti-CD3 antibody was reported over 30 years ago when OKT3 (mouse anti-human CD3 IgG1 antibody) treatment failed to induce a mitogenic response in blood cells from five of eight members of a single family due to a defect in Fc receptor binding to OKT311, 12. Despite these observations, mouse IgG1 antibodies, and specifically anti-CD3 and/or anti-CD28 antibodies, continue to be used in studies of cytokine responses in stimulated blood cells13, 14. Most notably, these antibodies are commonly used as a positive control for many cytokine release assays, despite the fact that rs1801274 genotype will introduce substantial variation in cytokine release, depending on genetic composition of the study subjects15. According to a recent survey of cytokine release assay use and practices, some, but not all, laboratories take rs1801274 genotype into account, leading to wide variation in control levels of release15.
In spite of the experimental importance of the Arg131His genotype on human immune responses to different classes of mouse and human IgG, relatively little is known of the effect of this genotype on broad cytokine or gene expression responses. Notably, heterozygous (AG, His/Arg) individuals are typically grouped with the homozygote class not of interest to the study (e.g., see ref.16), assuming that only the homozygous genotype is functionally affected by this variant. As a result, the effect of heterozygosity at this variant site on response is largely unknown.
To address these questions, we conducted a study of cytokine and global gene expression responses of whole blood leukocytes to anti-CD3 and anti-CD28 mouse IgG1 antibodies (anti-CD3/CD28), in which we observed a dramatic impact of this polymorphism on a broad panel of cytokine responses as well as on global gene expression responses. Importantly, our studies demonstrate that individuals with the AG (His/Arg) heterozygous genotype respond differently to treatment with mouse anti-CD3/CD28 antibodies compared to either the AA (His/His) or GG (Arg/Arg) homozygotes at both the gene expression and cytokine levels. These observations yield insight into the gene regulation of anti-CD3/CD28 response, and challenge the assumption of equivalent responses in individuals with the AA (His/His) and AG (His/Arg) genotypes, as assumed in previous studies13, 16.
Results
rs1801274 genotype effect on cytokine responses in whole blood
We used mouse IgG1 anti-CD3 and IgG1 anti-CD28 antibodies, as prepared in the TruCulture tubes (Myriad RBM, Austin, Texas), to stimulate T cells for studies of cytokine and gene expression responses. The co-activation of the T cell receptor by these soluble antibodies is enhanced by FcγR binding, and therefore, in this study, T cell receptor activation provides a read-out of FcγR binding. First, we tested for association of rs1801274 with the levels of 21 cytokines detected in our sample. Overall, levels of 17 of the 21 cytokines were reduced in AA (His/His) homozygotes at a Bonferroni-corrected threshold of significance (P<2.4×10−3) (Table 1, Figure 1, S2A). Of these 17 cytokines, eight had highly significant (P<5×10−8) associations between rs1801274 genotype and cytokine level (Table 1; Figure 1). Among the remaining four cytokines (IL-15, IL-21, IL-23, and IL-25) no differences were observed by rs1801274 genotype (Table 1, Figure S2B). Three of these cytokines (IL-15, IL-23, IL-25) had a minimal response (<5 pg/ml) to anti-CD3/CD28 treatment across all rs1801274 genotypes, while IL-21 levels were more robust but not significantly different across all three genotype classes.
Table 1. Summary table of results of association studies of rs1801274 (ch1:161479745) at the FCGR2A locus and cytokines.
Cytokines with highly significant (P<5×10−8), or Bonferroni-corrected (P<2.4×10−3) associations are denoted with * or §, respectively. The beta values are shown for the minor allele (A, MAF=0.365).
| Cytokine | P-value | Beta | Standard error (Beta) |
|---|---|---|---|
| IL-2 | 7.35×10−19 * | −0.865 | 0.083 |
| IFNγ | 7.56×10−18 * | −0.873 | 0.087 |
| TNFα | 4.38×10−12 * | −0.723 | 0.095 |
| GM-CSF | 5.33×10−11 * | −0.619 | 0.086 |
| IL-1β | 3.02×10−10 * | −0.688 | 0.100 |
| IL-13 | 5.91×10−10 * | −0.578 | 0.086 |
| IL-10 | 3.06×10−9 * | −0.691 | 0.108 |
| IL-22 | 3.81×10−8 * | −0.289 | 0.049 |
| IL-5 | 5.25×10−8 § | −0.607 | 0.104 |
| IL-4 | 2.13×10−7 § | −0.559 | 0.102 |
| IL-17 | 2.19×10−7 § | −0.576 | 0.105 |
| MIP-3α | 2.95×10−7 § | −0.582 | 0.107 |
| IL-9 | 1.16×10−6 § | −0.529 | 0.103 |
| IL-6 | 3.02×10−6 § | −0.582 | 0.119 |
| IL-12p70 | 4.91×10−5 | −0.365 | 0.087 |
| TNFβ | 2.01×10−4 | −0.368 | 0.096 |
| IL-17F | 1.09×10−3 | −0.387 | 0.116 |
| IL-21 | 0.013 | −0.238 | 0.095 |
| IL-23 | 0.023 | −0.140 | 0.061 |
| IL-15 | 0.057 | −0.170 | 0.089 |
| IL-25 | 0.204 | −0.093 | 0.073 |
Figure 1. Cytokine levels following treatment with anti-CD3/CD28 antibodies in human whole blood by rs1801274 genotype in the FCGR2A gene.
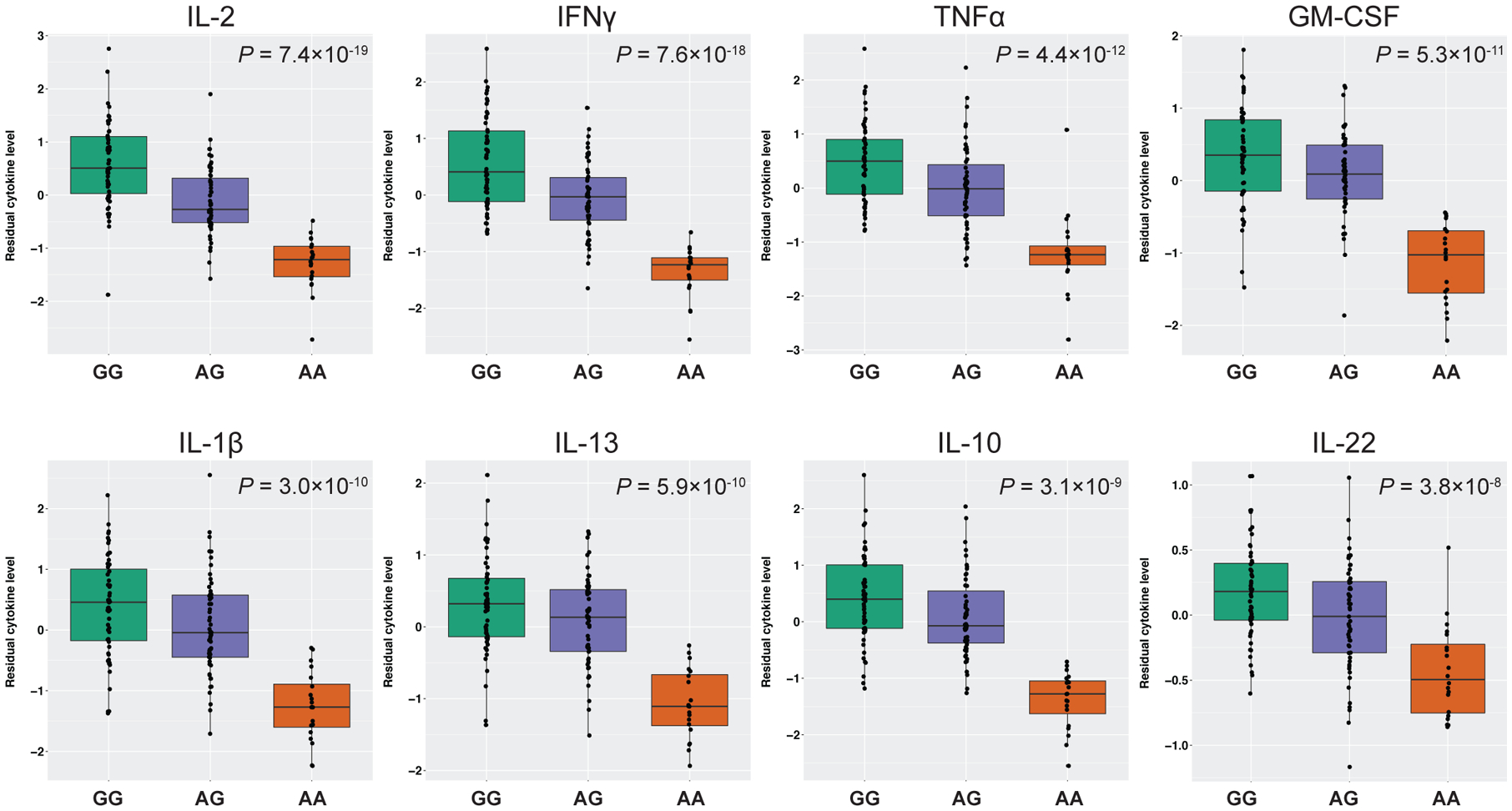
Boxes indicate the interquartile ranges, whiskers represent the 95% confidence intervals. The eight cytokines shown are associated with genotype at the FCGR2A locus at a P<5×10−8. For all other cytokines, see Supplementary Figure S2.
Four of the 17 cytokines were also significantly reduced in the AG (His/Arg) heterozygotes compared to the GG (Arg/Arg) homozygotes (IFNγ, IL-12, IL-2, and TNFα at P=6.3×10−5, 1.7×10−3, 6.7×10−7, and 2.3×10−3 respectively; Wilcoxon rank sum test), suggesting an additive effect of rs1801274 genotype. For the remaining 13 cytokines, the AG (His/Arg) heterozygotes had similar levels to the GG (Arg/Arg) homozygotes, consistent with a dominant/recessive effect.
Gene expression profiles of whole blood cells and rs1801274 genotype
The known effects of rs1801274 genotype on immunoglobulin binding and subsequent activation of T cells via crosslinking of their receptors2 are in line with the cytokine response patterns described above. However, the downstream effects of this variant on global gene expression have not previously been investigated. To directly assess this, we measured gene expression from the same anti-CD3/CD28 antibody-treated samples used in the cytokine studies along with the untreated samples from the same individuals. A principal components analysis (PCA) of genome-wide gene expression showed clear separation of treated and untreated samples along the first PC (PC1), which explained 63.2% of the variance (Figure 2A), although subsets of the treated samples—those with AA (His/His) genotype at rs1801274—clustered intermediate to or even within the untreated sample cluster.
Figure 2. Principal components plots of gene expression profiles in whole blood.

(A) PC plot of the PCs 1 and 2 (x- and y-axis, respectively) of untreated and anti-CD3/CD28 treated samples colored by rs1801274 genotype. Shape indicates untreated (open triangles) or anti-CD3/CD28-treated samples (filled circles). (B) Boxplot of PC1 by rs1801274 genotype in anti-CD3/CD28-treated samples only. (C) PC plot of PCs 1 and 2 (x- and y-axis, respectively) of anti-CD3/CD28 treated samples with AG or GG rs1801274 genotype. (D) Boxplot of PC1 by rs1801274 genotype.
In a PCA of anti-CD3/CD28 treated samples alone, the non-responding AA homozygotes cluster apart from samples with AG (His/Arg) and GG (His/His) genotypes along PC1, accounting for 41.6% of the variance in global gene expression (Figure 2B). The distributions of PC1 values by genotype additionally revealed a more subtle separation between samples with GG (Arg/Arg) and AG (His/Arg) genotype (Figure 2B). To better visualize gene expression differences between samples with the GG (Arg/Arg) and AG (His/Arg) genotypes, we removed individuals with the AA (His/His) genotype and repeated the PCA. In this analysis, genotype at rs1801274 was still significantly correlated with PC1, which now accounted for 17.2% of the variance in global gene expression response to anti-CD3/CD28 treatment (Figure 2C–D). Overall, these results reveal global alterations in gene expression response to anti-CD3/CD28 antibodies in whole blood cells from individuals carrying one or two copies of the A (His) allele at this SNP.
To investigate these apparent patterns of gene expression at a more granular level, we tested the interaction effect of rs1801274 genotype and treatment in the paired anti-CD3/CD28 treated samples and untreated controls using a linear model that included an interaction term. Of 10,328 genes tested, over 70% (7,359 genes) had a significant (false discovery rate [FDR] of 5%) rs1801274 genotype-by-treatment interaction effect (Figure S3). This gene set with significant interaction effects includes genes encoding cytokine receptors or cytokines that were measured in the supernatant, illustrating that differences in response exist at both the transcriptional and protein levels (Figure 3, S4, S5). The mean log fold-change in response to anti-CD3/CD28 treatment is smallest in individuals with genotype AA (His/His) in 8,139 genes, 79% (6,416) of which are also significant for a treatment by genotype interaction effect (Supplemental File 1). These results support the broad pattern of a lack of response in AA indivduals to anti-CD3/CD28 treatment also observed in the PCA results.
Figure 3. Gene expression of cytokine and cytokine receptor genes in untreated control and anti-CD3/CD28-treated samples, by rs1801274 genotype.
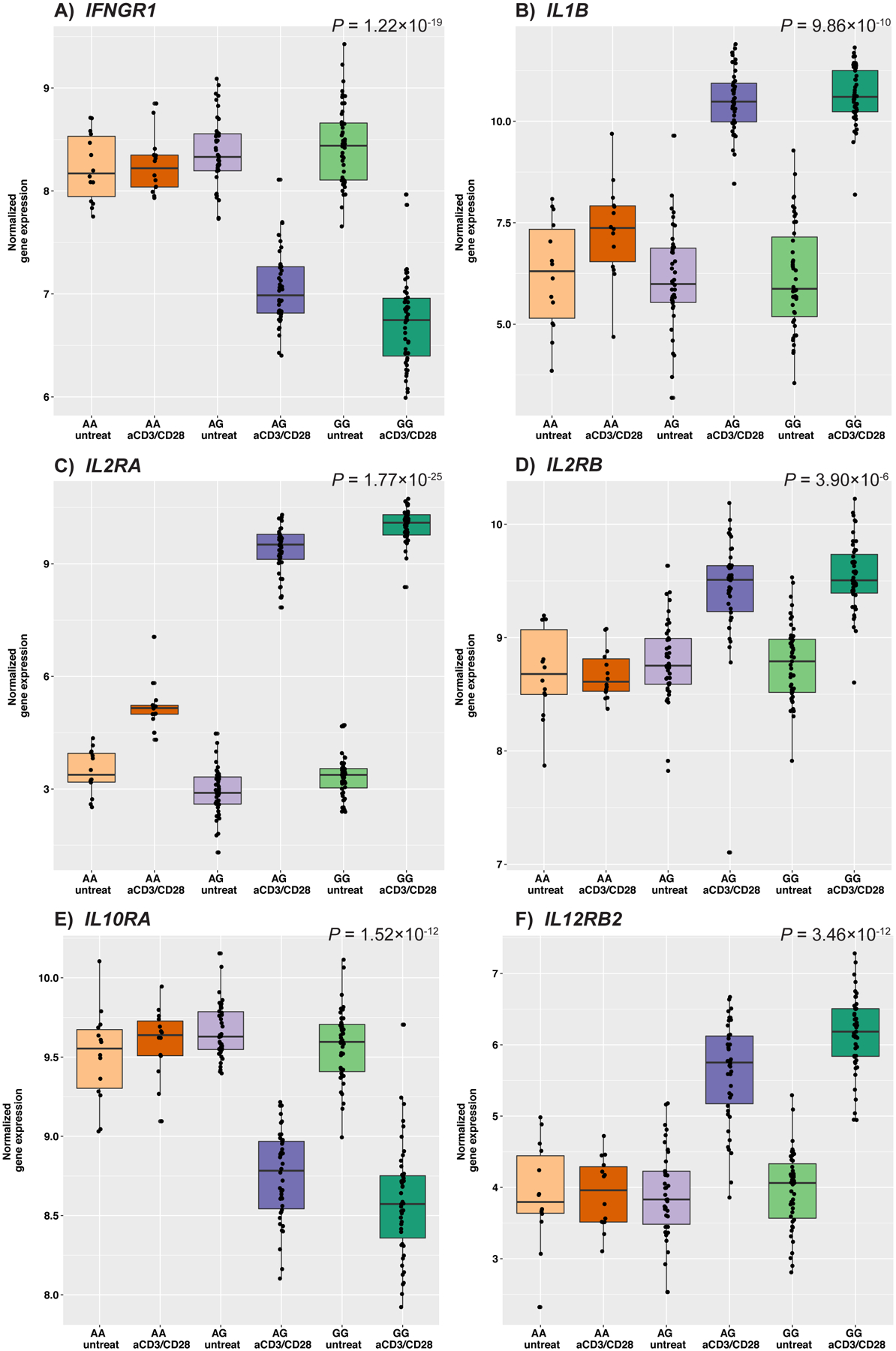
Boxes indicate the interquartile ranges, whiskers represent the 95% confidence intervals. P-values represent the genotype by treatment interaction effect, all genes shown are significant at FDR 5%. (A) IFNGR1, Interferon gamma receptor 1; (B) IL1B, Interleukin 1 beta; (C) IL2RA, Interleukin 2 receptor subunit alpha; (D) IL2RB, Interleukin 2 receptor subunit beta; (E) IL10RA, Interleukin 10 receptor subunit alpha; (F) IL12RB2, Interleukin 12 receptor beta 2 subunit.
To specifically identify the set of genes whose response to anti-CD3/CD28 treatment differed between individuals with AG (His/Arg) and GG (Arg/Arg) genotypes, we tested for differential expression in response to anti-CD3/CD28 treatment compared to untreated control cells in paired samples. At an FDR of 5%, 87% (9,288) and 85% (9,092) of genes were differentially expressed following anti-CD3/CD28 treatment compared to untreated controls in individuals with GG (Arg/Arg; n=47) and AG (His/Arg; n=42) genotype, respectively (Figure 4A–B). Of these genes, 8,789 were differentially expressed in both genotype classes. In addition, approximately 25% (2,197) of these genes also showed significant differences between AG (His/Arg) and GG (Arg/Arg) individuals in response (response in GG – response in AG ≠ 0) to anti-CD3/CD28 (Figure 4C). Nearly all (2,150/2,197) of these genes with significant differences in response have a greater magnitude of upregulation or downregulation in response to anti-CD3/CD28 treatment in GG (Arg/Arg) individuals compared to AG (His/Arg) or AA (His/His) individuals (Figure 4D). These differential response genes are enriched for transcription and RNA processing GO terms, suggesting that levels of transcriptional activation may differ by genotype class (Table S1). A further 91 genes had significant response to anti-CD3/CD28 treatment in GG (Arg/Arg) individuals, and no response in AG (His/Arg) individuals (Figure 4C), and are significantly enriched in immune processes and stimulated immune cells (Table S2). In contrast, the 41 genes with differential expression following treatment with anti-CD3/CD28 in AG (His/Arg) individuals only are enriched for gene sets associated with down regulation of unstimulated immune cells, highlighting that gene expression differences are driven by a less robust activation of T cell receptors in AG individuals compared to GG (Arg/Arg) homozygotes (Table S3).
Figure 4. Differences in gene expression response to anti-CD3/CD28-treated whole blood between individuals with the AG (Arg/His) and GG (His/His) genotype at rs1801274 in the FCGR2A gene.
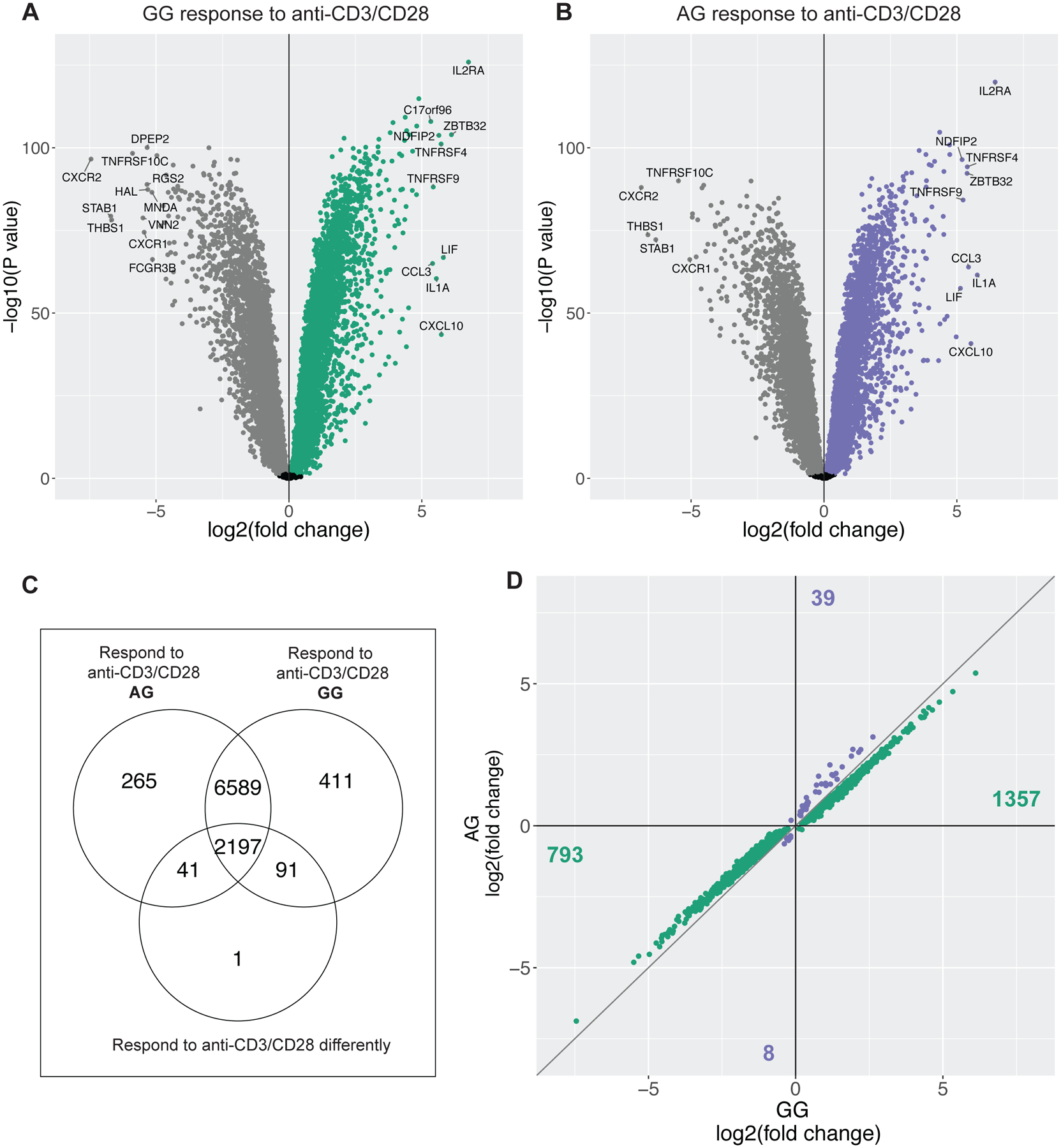
Volcano plot of gene expression response to anti-CD3/CD28 in individuals GG (A) and AG (B) at rs1801274. Each point represents a gene, and genes in gray have significantly decreased expression (FDR 5%) after anti-CD3/CD28 treatment, while genes in green or purple have significantly increased expression following treatment compared to the untreated control. Genes with a log2 fold change <5 or >−5 are labeled in each plot. (C) Venn diagram summarizing the number of genes differentially expressed following anti-CD3/CD28 treatment in GG homozygotes (top left circle), AG heterozygotes (top right circle), and genes that respond differently to treatment by genotype class in the bottom circle ([GG anti-CD3/CD28 - GG untreated] - [AG anti-CD3/CD28 – AG untreated] ≠ 0). (D) Genes that respond differently by genotype class plotted by fold change in GG homozygotes (x-axis) and AG heterozygotes (y-axis). Genes in green have a higher magnitude of response in GG individuals, genes colored purple have a higher magnitude of response in AG individuals. Number of genes listed in each quadrant.
Discussion
In this study of the effect of rs1801274 on whole blood leukocytes treated with mouse IgG1 anti-CD3 and anti-CD28 antibodies, we observed a striking lack of overall activation of immune response, as measured by cytokine and gene expression profiles, in individuals homozygous for the A (131His) allele. Though the absence of cytokine responses to anti-CD3/CD28 antibodies is in agreement with previous studies of rs180127413, 16, 17, differences in cytokine responses between AG heterozygotes (His/Arg) and GG homozygotes (Arg/Arg) have not been previously appreciated. In particular, our study identified IFNγ, IL-12, TNFα, and IL-2 cytokine responses to treatment were significantly different across all three genotype classes. Notably, these cytokines are associated with Th1 responses, and illustrate that efficacy of anti-CD3/CD28 treatment in whole blood affects Th1-associated cytokines differently across each rs1801274 genotype class. In contrast, Th2- and some Th17-associated cytokines, such as IL-13, IL-4, IL-5 (Th2), and IL-17, show decreased cytokine levels in AA (His/His) individuals, but little difference between AG (His/Arg) and GG (Arg/Arg) individuals. Still another facet of rs1801274 genotype effect on anti-CD3/CD28 treatment reveals that some cytokines associated with Th17 and follicular helper T (Tfh) cells (IL-23 and IL-21, respectively) show no differences by genotype. Taken together, these data suggest that the effect of rs1801274 on anti-CD3/CD28 treated whole blood cytokine response is greatest—and most distinctive between genotypes—in Th1-associated associated cytokine and consequent cell-mediated immune responses, and heterozygote (AG) effects are less pronounced among cytokines associated with other T cell subsets.
At the level of gene expression response to anti-CD3/CD28 treatment, genotype differences between AG (His/Arg) and GG (Arg/Arg) individuals had a broader impact. Over 20% of genes expressed had a different response to anti-CD3/CD28 treatment, with a decreased magnitude of response to treatment in AG heterozygotes compared to GG homozygotes. These results suggest a suboptimal activation of T cell receptors in AG (His/Arg) heterozygotes compared to GG (Arg/Arg) homozygotes. That these differences may be relevant beyond laboratory studies using these antibodies is evidenced by GWAS implicating genotype at rs1801274 in susceptibility to autoimmune and inflammatory diseases6–9, 18. In this context, the G allele is associated with risk for several autoimmune diseases because this it has a lower binding affinity to human IgG2. Thus, in vivo responses to stimuli may be skewed more or less toward immune activation and autoimmunity depending on genotype, and impact risk or protection from a variety of immune-mediated conditions.
Beyond the anti-CD3 and anti-CD28 IgG antibodies used in this study, this broad difference in gene expression by rs1801274 genotype suggest that measurement of gene expression before and during in vitro studies of therapeutic antibodies could be an important tool for identifying differential response to treatment by rs1801274 genotype, analogous to the more frequently employed cytokine release assays. Study of a more proximal phenotype (i.e., gene expression) may also help to dissect the inconsistent associations of the His131Arg polymorphism in the FCGR2A gene and tumor response and/or progression free survival reported in earlier studies16, 17, 19–26.
Generalizing the results of this study comes with several caveats. Most importantly, we studied the effect of rs1801274 in response to mouse IgG1 anti-CD3 and mouse IgG1 anti-CD28 antibodies, which bind to the His allele in the FcγRIIA receptors with lower affinity than to the Arg allele. In contrast, human IgG2 antibodies bind to Arg allele in the FcγRIIA receptors with lower affinity than to the His allele. While we can infer that treatment of whole blood leukocytes with human IgG2 anti-CD3/CD28 antibodies would reveal similar genotype differences in immune response, this has not been confirmed experimentally in our study or by others.
We also did not measure peripheral blood cell composition in the individuals studied. Inter-individual differences in cell proportion likely modulate response to anti-CD3/CD28 antibodies, which could introduce greater variability into the cytokine and gene expression studies. Despite this additional noise, we nevertheless observe striking differences in gene expression profile and cytokine levels by rs1801274 genotype. Finally, while there are other FcγR polymorphisms, such as rs396991 in FCGR3A, that were not examined in this study, the binding affinity to human IgG2 and IgG1 is much lower compared to either allele of rs1801274, and is expressed on a more narrow set of cell types (NK cells and monocytes)27.
Regardless, the results of this study provide clear evidence that individual responses to anti-CD3 and/or anti-CD28 antibodies differ by rs1801274 genotype. While differences between individuals with the low-affinity homozygous genotype class (AA [His/His] for mouse IgG1, or GG [Arg/Arg] for human IgG22) and the higher-affinity heterozygote and homozygous individuals have long been known, differences between the higher-affinity FcγRIIA homozygote and heterozygote have not previously been explored, but have potentially important implications for future studies of response to these stimuli, such as cytokine release assays. In vitro studies can side-step this issue with the use of antibody beads, which act as a steric hindrance to the FcγRIIA-Fc binding interface. Future studies should avoid study designs in which antibody beads cannot be used (e.g., Myriad RBM TruCulture tubes), unless genotype at rs1801274 is taken into account when interpreting results. Moving towards a more nuanced understanding of the spectrum of rs1801274 effects on gene expression and immune responses may contribute to a greater understanding of risk for immune-mediated conditions. Assessment of genotype effects for this polymorphism in more general clinical contexts may even broaden further the relevance of this functional variant to precision medicine.
Materials and Methods
Study population
This study was conducted in 153 Hutterites (7–76 years old) who are a subset of the >1,400 Hutterites who have participated in our population-based studies of complex phenotypes28–30. The Hutterites are of central European ancestry, and the participants in our studies live on communal farms in South Dakota and are related to each other through multiple lines of descent in a single 3,671-member pedigree with 64 founders. Written consent for these studies was obtained from the adult participants and parents of children under 18; written assent was obtained from all children. This study was approved by the University of Chicago Institutional Review Board.
Collection of Whole blood samples
One milliliter of whole blood was drawn into each of three TruCulture (Myriad RBM; Austin, Texas) tubes containing either proprietary TruCulture media alone or media + 0.4 μl/ml mouse IgG1 anti-CD3 (UCHT-1) + 0.33 μl/ml mouse IgG1 anti-CD28 (CD28.2) antibodies without prior knowledge of rs1801274 genotype. Samples were incubated upright in a dry heat block at 37C for 30 hours. Following incubation, supernatants were separated from the cell pellets, and then aliquoted and flash-frozen on dry ice. The remaining samples were washed twice with Buffer EL (Qiagen; Hilden, Germany) and the cell pellets were resuspended in 350 μl RLT Buffer (Qiagen) and frozen on dry ice. Frozen samples were shipped on dry ice overnight to Chicago and stored at −80°C. DNA and RNA were extracted from thawed cell pellets using AllPrep DNA/RNA Mini Kits (Qiagen).
Cytokine measurement and processing
Supernatants from 130 anti-CD3/CD28 treated samples were submitted to the Human Immunology Core at the University of Chicago for measurement of 25 cytokines using the Milliplex Th17 Magnetic Bead Panel (Millipore; Burlington, Massachusetts). Each sample was run in duplicate. Results from duplicate samples were averaged; in 10 cases one duplicate did not yield a detectable result and only a single measurement was kept. After removing cytokines with measurements outside the range of detection in >30% of individuals, 21 cytokines from anti-CD3/CD28-treated samples were retained for downstream analysis. Cytokine measurements were quantile normalized; and principal components analysis (PCA) identified bead panel “plate” as a confounding technical covariate, which was adjusted using ComBat31.
Genotyping and cytokine association with rs1801274
Genotype of rs1801274 was determined using a validated TaqMan assay (ThermoFisher; Waltham, Massachusetts). Associations between this variant and cytokine levels were calculated using GEMMA32, and included a kinship matrix based on identity by descent (IBD) to correct for the relatedness between Hutterites in the sample. In all analyses, age and sex were included as covariates; we used a Bonferroni-corrected P-value threshold (P=2.3×10−3) to correct for the 21 tests. A Wilcoxon rank-sum test was used to determine if cytokines differed between individuals AG or GG at rs1801274.
RNA sequencing
RNA samples from all TruCulture tubes were used to create RNA-seq libraries using the TruSeq Library kit (Illumina; San Diego, California); quality and concentration of libraries were assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies; Santa Clara, California) and quantitative PCR using the Kapa library quantification kit (Kapa Biosystems; Wilmington, Massachusetts). Samples were sequenced in pools of 16–18 samples across three flow cells of an Illumina HiSeq 2500; 119 samples with low read count were re-sequenced on two flow cells on the same machine. VerifyBamID33 was used to detect one sample swap, and these two samples were retained in downstream analyses. Read quality was assessed with multiQC34, and mapped to hg19 and genes were counted using STAR v2.5.235. Read counts for the same sample sequenced multiple times were summed together, and samples with >7 million uniquely mapped reads underwent Trimmed Means of M-values normalization (TMM) and a voom transformation to correct for differences in library sizes, resulting in log2 counts per million (CPM) values36. Confounding technical effects were assessed in the normalized expression data using principal components analysis (PCA), and sequencing pool was adjusted for plotting using the function RemoveBatchEffect from the R package limma37. PCA was also used to visualize differences in overall patterns of gene expression variation according to rs1801274 genotype in three analyses: (1) untreated vs. anti-CD3/CD28-treated samples; (2) within anti-CD3/CD28-treated samples; and (3) within anti-CD3/CD28-treated samples after removing individuals homozygous AA at rs1801274.
Analysis of Gene Expression
Differential gene expression in anti-CD3/CD28-treated and untreated control samples by rs1801274 genotypes was examined using limma37, with age, sex, and sequencing pool included as covariates. All gene expression analyses were conducted in paired samples; each individual (rs1801274 genotype: 14 AA, 42 AG, 47 GG) had RNAseq completed in both the untreated control and anti-CD3/CD28 treated whole blood samples. Two separate models were tested (shown below, T = treatment, SNP= rs1801274 genotype, TAG= treatment in individuals with genotype AG, TGG= treatment effects in individuals with genotype GG).
In the first, we identified genes with a significant genotype by treatment response interaction across all individuals, and in the second, we tested differences in response in individuals with genotype GG and AG only.
Differential expression was assessed using limma in order to test for treatment, genotype and genotype by treatment interaction effects while accounting for the paired nature of the data. As a consequence, relatedness between individuals was included in this model, potentially leading to some inflation of P-values. To to confirm that our conclusions regarding gene expression differences between rs1801274 genotype groups were not due to this omission, we used the modified linear model shown below to test for an rs1801274 genotype effect on gene expression differences between anti-CD3/CD28-treated and untreated control samples using GEMMA32, which allows inclusion of a kinship matrix to adjust for relatedness between individuals (SNP= rs1801274 genotype, model was tested across all three genotype classes). Gene expression profiles were corrected for sequencing pool using removeBatchEffect() function in limma.
In this approach, 91.7% of the genes with significant genotype by treatment interaction effects (FDR 5%) also had a significant genotype effect in the modified GEMMA model, and the P-values of all genes from the two models were significantly correlated (Spearman’s rho= 0.96, P=<2.2×10−16). Thus, overall our results are robust to effects of relatedness.
Significance was assessed using the methods of Benjamini and Hochberg38. Genes differentially expressed at a false discovery rate (FDR) of 5% were analyzed with Gene Set Enrichment Analysis (GSEA)39, 40. Overlaps were computed using Gene Ontology (GO), KEGG, and immunologic signatures gene sets.
Code availability
The analysis scripts for this project are available at https://github.com/mstein3/FcR_scripts.
Supplementary Material
Acknowledgments
The authors would like to thank Christine Billstrand and Raluca Nicolae for making RNAseq sample libraries and Catherine Igartua and Mark Abney for helpful comments and statistical advice. This work was supported by a grant from the National Institutes of Health (R01 HL085197).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcgamma receptors in dendritic cells and macrophages. Nat Rev Immunol 2014; 14(2): 94–108. [DOI] [PubMed] [Google Scholar]
- 2.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009; 113(16): 3716–25. [DOI] [PubMed] [Google Scholar]
- 3.Ivan E, Colovai AI. Human Fc receptors: critical targets in the treatment of autoimmune diseases and transplant rejections. Human immunology 2006; 67(7): 479–91. [DOI] [PubMed] [Google Scholar]
- 4.Bredius RG, Fijen CA, De Haas M, Kuijper EJ, Weening RS, Van de Winkel JG et al. Role of neutrophil Fc gamma RIIa (CD32) and Fc gamma RIIIb (CD16) polymorphic forms in phagocytosis of human IgG1- and IgG3-opsonized bacteria and erythrocytes. Immunology 1994; 83(4): 624–30. [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus JH, Novembre J. Visualizing the geography of genetic variants. Bioinformatics 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47(9): 979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet 2011; 43(12): 1241–6. [DOI] [PubMed] [Google Scholar]
- 8.Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015; 47(12): 1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song GG, Lee YH. Associations between FCGR2A rs1801274, FCGR3A rs396991, FCGR3B NA1/NA2 polymorphisms and periodontitis: a meta-analysis. Molecular biology reports 2013; 40(8): 4985–93. [DOI] [PubMed] [Google Scholar]
- 10.Stewart R, Hammond SA, Oberst M, Wilkinson RW. The role of Fc gamma receptors in the activity of immunomodulatory antibodies for cancer. Journal for ImmunoTherapy of Cancer 2014; 2(1): 29. [Google Scholar]
- 11.Ceuppens JL, Bloemmen FJ, Van Wauwe JP. T cell unresponsiveness to the mitogenic activity of OKT3 antibody results from a deficiency of monocyte Fc gamma receptors for murine IgG2a and inability to cross-link the T3-Ti complex. Journal of immunology 1985; 135(6): 3882–6. [PubMed] [Google Scholar]
- 12.Ceuppens JL, Meurs L, Van Wauwe JP. Failure of OKT3 monoclonal antibody to induce lymphocyte mitogenesis: a familial defect in monocyte helper function. Journal of immunology 1985; 134(3): 1498–502. [PubMed] [Google Scholar]
- 13.Duffy D, Rouilly V, Libri V, Hasan M, Beitz B, David M et al. Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity 2014; 40(3): 436–50. [DOI] [PubMed] [Google Scholar]
- 14.Mueller SC, Marz R, Schmolz M, Drewelow B. Intraindividual long term stability and response corridors of cytokines in healthy volunteers detected by a standardized whole-blood culture system for bed-side application. BMC medical research methodology 2012; 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finco D, Grimaldi C, Fort M, Walker M, Kiessling A, Wolf B et al. Cytokine release assays: current practices and future directions. Cytokine 2014; 66(2): 143–55. [DOI] [PubMed] [Google Scholar]
- 16.Davila-Fajardo CL, van der Straaten T, Baak-Pablo R, Medarde Caballero C, Cabeza Barrera J, Huizinga TW et al. FcGR genetic polymorphisms and the response to adalimumab in patients with rheumatoid arthritis. Pharmacogenomics 2015; 16(4): 373–81. [DOI] [PubMed] [Google Scholar]
- 17.Avila-Pedretti G, Tornero J, Fernandez-Nebro A, Blanco F, Gonzalez-Alvaro I, Canete JD et al. Variation at FCGR2A and functionally related genes is associated with the response to anti-TNF therapy in rheumatoid arthritis. PloS one 2015; 10(4): e0122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sole-Violan J, Garcia-Laorden MI, Marcos-Ramos JA, de Castro FR, Rajas O, Borderias L et al. The Fcgamma receptor IIA-H/H131 genotype is associated with bacteremia in pneumococcal community-acquired pneumonia. Critical care medicine 2011; 39(6): 1388–93. [DOI] [PubMed] [Google Scholar]
- 19.Wilson NS, Yang B, Yang A, Loeser S, Marsters S, Lawrence D et al. An Fcgamma receptor-dependent mechanism drives antibody-mediated target-receptor signaling in cancer cells. Cancer cell 2011; 19(1): 101–13. [DOI] [PubMed] [Google Scholar]
- 20.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2003; 21(21): 3940–7. [DOI] [PubMed] [Google Scholar]
- 21.Kjersem JB, Skovlund E, Ikdahl T, Guren T, Kersten C, Dalsgaard AM et al. FCGR2A and FCGR3A polymorphisms and clinical outcome in metastatic colorectal cancer patients treated with first-line 5-fluorouracil/folinic acid and oxaliplatin +/− cetuximab. BMC cancer 2014; 14: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2008; 26(11): 1789–96. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Shimizu C, Hojo T, Akashi-Tanaka S, Kinoshita T, Yonemori K et al. FcgammaR2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology 2011; 22(6): 1302–7. [DOI] [PubMed] [Google Scholar]
- 24.Ghesquieres H, Cartron G, Seymour JF, Delfau-Larue MH, Offner F, Soubeyran P et al. Clinical outcome of patients with follicular lymphoma receiving chemoimmunotherapy in the PRIMA study is not affected by FCGR3A and FCGR2A polymorphisms. Blood 2012; 120(13): 2650–7. [DOI] [PubMed] [Google Scholar]
- 25.Paez D, Pare L, Espinosa I, Salazar J, del Rio E, Barnadas A et al. Immunoglobulin G fragment C receptor polymorphisms and KRAS mutations: are they useful biomarkers of clinical outcome in advanced colorectal cancer treated with anti-EGFR-based therapy? Cancer science 2010; 101(9): 2048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurvitz SA, Betting DJ, Stern HM, Quinaux E, Stinson J, Seshagiri S et al. Analysis of Fcgamma receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2012; 18(12): 3478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 2012; 119(24): 5640–9. [DOI] [PubMed] [Google Scholar]
- 28.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 2008; 358(16): 1682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ober C, Nord AS, Thompson EE, Pan L, Tan Z, Cusanovich D et al. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. Journal of lipid research 2009; 50(5): 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao TC, Du G, Han L, Sun Y, Hu D, Yang JJ et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol 2014; 133(1): 248–55 e1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. biostatistics.oxfordjournals.org. [DOI] [PubMed]
- 32.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012; 44(7): 821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. American Journal of Human Genetics 2012; 91(5): 839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016; 32(19): 3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology 2014; 15(2): R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 2015; 43(7): e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg, Yosef. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society 1995; 57(1): 11. [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102(43): 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34(3): 267–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


