Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Main protease, Ebsulfur, Inhibitor
Abstract
The emerging COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has raised a global catastrophe. To date, there is no specific antiviral drug available to combat this virus, except the vaccine. In this study, the main protease (Mpro) required for SARS-CoV-2 viral replication was expressed and purified. Thirty-six compounds were tested as inhibitors of SARS-CoV-2 Mpro by fluorescence resonance energy transfer (FRET) technique. The half-maximal inhibitory concentration (IC50) values of Ebselen and Ebsulfur analogs were obtained to be in the range of 0.074–0.91 μM. Notably, the molecules containing furane substituent displayed higher inhibition against Mpro, followed by Ebselen 1i (IC50 = 0.074 μM) and Ebsulfur 2k (IC50 = 0.11 μM). The action mechanism of 1i and 2k were characterized by enzyme kinetics, pre-incubation and jump dilution assays, as well as fluorescent labeling experiments, which suggested that both compounds covalently and irreversibly bind to Mpro, while molecular docking suggested that 2k formed an S—S bond with the Cys145 at the enzymatic active site. This study provides two very potent scaffolds Ebsulfur and Ebselen for the development of covalent inhibitors of Mpro to combat COVID-19.
1. Introduction
A highly infectious viral disease (COVID-19) caused by SARS-CoV-2 became a global epidemic since its outbreak in December 2019 [1]. In February 2020, the World Health Organization (WHO) has elevated its risk of COVID-19 to ‘very high’ globally [2]. The basic reproduction number (R0) of this viral disease was estimated at the range of 2.24–3.18 [3], indicating that SARS-CoV-2 has high transmission efficiency. As of December 11, 2020, there are more than 1,500,000 deaths out of 68,000,000 confirmed cases in 220 countries (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). SARS-CoV-2, as the name indicates, is close to SARS-CoV and MERS-CoV, which belong to the beta lineage of the coronavirus [4]. The genome of SARS-CoV-2 contains the largest single-stranded, positive-sense RNA with approximately 30,000 nucleotides. The viral genome consists of 14 open reading frames, which mainly encode several non-structural, structural, and accessory proteins [5]. The low toxic compounds that target any of these viral proteins may be the potential antiviral drug candidates [6]. Based on the evidence from the clinical studies, the Remdesivir has been used to treat COVID-19, but the therapeutic effect remains very limited [7]. As a consequence, safe and effective therapy is urgently needed.
The main protease (Mpro), also called the chymotrypsin-like protease (3CLpro), is considered one of the most characteristic drug targets in coronavirus. It is composed of three domains that could form a functional dimer. Residues 8–101 (Domains I) and residues 102–184 (Domain II) are the catalytic domains. Residues 201–303 (Domain III) are responsible for the enzyme dimerization [8], [9]. The Mpro, a cysteine protease with an unconventional Cys catalytic residue, plays an essential role in coronavirus replication and transcription by digesting the polyproteins at more than 11 conserved sites. However, unlike other Cys (or Ser) hydrolases, it has a catalytic Cys-His (Cys145 and His41) dyad in the gap rather than a canonical Cys (Ser)-His-Asp (Glu) triad [10]. In addition, it is known to have recognition sequence of Leu-Gln (Ser, Ala, Gly) at the P1 site (↓ marks the cleavage site). Notably, the Gln residue is virtually always required in the P1 position [8], [11]. However, no homolog of Mpro exists in humans, therefore, Mpro is the ideal target for antiviral therapy [12].
To date, several approaches including high-throughput screening, virtual screening, drug design, and drug repurposing have been used to develop the potential inhibitors of SARS-CoV-2. Based on the fact that similar therapeutic regimens are commonly used for viral diseases associated with SARS-CoV-2, drug repurposing is now attracting attention with the advantages of lower cost and greater safety, especially medicines where pre-clinical safety studies have been conducted [13]. Several drugs, such as azithromycin [14], lopinavir/ritonavir [15], remdesivir [16], and favipiravir [17], have been currently suggested as antiviral drug candidates. However, due to the limited therapeutic efficacy of these drugs and the toxic side effects at high doses, only remdesivir has so far been approved by the FDA.
Given currently urgent need, the high-throughput and virtual screening may become a rapid and effective means for drug discovery. An in-silico attempt has been made to perform a large-scale virtual screening of enzyme inhibitors [18], these enzyme include Mpro [19], [20], [21], taste receptors (TR) [22], cyclin-dependent kinases (CDKs) [23], dual-specificity tyrosine-phosphorylation regulated kinase-1A (DYRK1A) [24], and checkpoint kinase 1 (CHK1) [25]. To develop anti-viral medicines with clinical potential, a large amount of Mpro inhibitors were reported, such as aldehyde peptidomimetics [26], α-ketoamide [27], benzotriazole [28], and decahydroisoquinoline [29], GC-376 analogs [30], aldehydes [31]. Recently, Brul et al reported that the Ebselen and its analogs to be potent covalent inhibitors of papain-like protease [32]. In this study, a fluorescence resonance energy transfer (FRET)-based enzymatic assay was employed to screen the inhibitors of Mpro, and the Ebselen and Ebsulfur derivatives were found to be very highly promising scaffolds for targeting the enzyme. This study provides additional insights to Ebselen and Ebsulfur derivatives. The action mechanism was also characterized by enzymatic kinetics and fluorescent labeling. The interaction of Mpro and the identified inhibitor was further investigated by molecular docking studies.
2. Results and discussion
2.1. FRET-based assay of Mpro
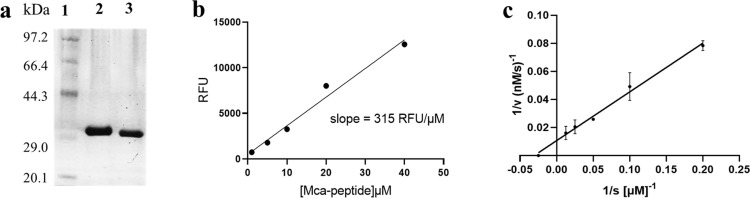
The Mpro gene was inserted into the PGEX-6P-1 vector and expressed in BL21 E. coli. The recombinant Mpro with native C and N termini was purified through Ni-NTA and HiTrap Q FF column (Fig. 1 a). The enzymatic activity was assayed by time-dependent kinetics using Mca–AVLQ/SGFRK(Dnp)K as fluorescent substrate [33]. A Mca-standard curve was produced to convert the relative fluorescence unit (RFU) to the amount of substrate depletion (Fig. 1b), and then the activity of Mpro was tested through measurement of K m and V max values. The Mpro (0.1 µM) was mixed with different concentrations of substrate (1–80 µM), the initial velocities were determined and plotted against substrate concentration. The data were fitted to the Line-weaver Burk equation with K m and V max of 39.1 ± 0.8 µM and 106.3 ± 6 nM/s, respectively (Fig. 1c). The catalytic efficiency was calculated to be 27,186 s−1 M−1, which was similar to the previously reported value (k cat/K m = 28,500 s−1 M−1) [34].
Fig. 1.
Expression and characterization of SARS-CoV-2 Mpro. (a) SDS–PAGE of Mpro. Lane 1, protein marker; lane 2, Mpro before cleavage with HRV 3C-protease; lane 3, Mpro with native C and N termini. (b) the standard curve of fluorescent substrate Mca–AVLQSGFRK(Dnp)K for converting RFU to the amount of the cleaved substrate. (c) Line-weaver Burk plot of 0.1 µM Mpro with various concentrations of substrate.
2.2. Screening of Mpro inhibitor
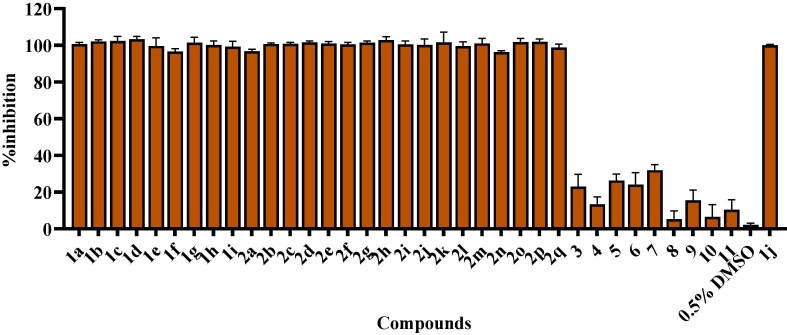
The fluorescence resonance energy transfer (FRET) experiments were employed to identify the potential inhibitors of Mpro [35]. Thirty-six compounds (Table 1 ) were screened, including Ebselen, Ebsulfur derivatives and compounds 3–11. These compounds were synthesized in our lab and characterized by NMR and mass spectrometry and evaluated to be the effective Metallo-β-lactamase inhibitors [36], [37], [38], [39], [40], [41], [42]. The compounds 3–11 have been identified to inhibit NDM-1 (cysteine protease). Mpro is also a cysteine protease, so we were interested in verifying whether these compounds have an inhibitory effect on Mpro. The percentage inhibition of Mpro with compounds is shown in Fig. 2 , which was calculated from enzyme activity without inhibitor (100%) minus residual activity with inhibitor. The Ebselen 1j has been reported to be the inhibitor of Mpro with an IC50 value of 0.67 μM [34]. In this work, we used 50 µM 1j as the positive control and 0.5% DMSO was used as the negative control in the absence of inhibitor. Encouragingly, it is clearly observed that both Ebselen and Ebsulfur exhibited more than 95% inhibition against Mpro, but compounds 3–11 had less than 40% inhibition. In this case, the following work would focus on the studies of Ebselen and Ebsulfur derivatives.
Table 1.
The structures of protease inhibitors tested against SARS-CoV-2 Mpro.
 |
Fig. 2.
Percent inhibition of Ebselen 1a-j, Ebsulfur 2a-q and compounds 3–11 (50 µM) against Mpro determined by FRET assays. Mca–AVLQSGFRK(Dnp)K (20 µM) was used as the fluorescent substrate, and the enzymatic concentration was 0.2 µM. Ebselen 1j was used as the positive control and 0.5% DMSO was used as the negative control.
2.3. Determination of IC50
Based on the encouraging results of the preliminary screening, the half-maximal inhibitory concentrations (IC50) of all Ebselen and Ebsulfur derivatives against Mpro were determined in the assay buffer using Mca–AVLQSGFRK(Dnp)K as substrate [43]. The concentrations of substrate and enzyme were 20 and 0.2 μM, respectively, and the concentration of inhibitor was varied in the range of 0 and 2 μM.
The collected IC50 data (Table 2 ) shown that all of these compounds had excellent inhibitory efficacy against Mpro, exhibiting IC50 values in the range of 0.074–0.91 μM. The inhibitors 1i and 2k were also found to be the most potent inhibitors with IC50 values of 0.074 and 0.11 μM, respectively. It should be noted that all these Ebselen and Ebsulfur derivatives tested had better inhibitory efficacy than the Ebselen molecule recently reported on Mpro (IC50 = 0.67 μM), except 2b-c and 2p, the molecule inhibited SARS-CoV-2 replication with an EC50 value of 4.67 μM [34]. Clearly, both Ebselen and Ebsulfur are highly promising scaffolds for the development of antiviral reagents.
Table 2.
The inhibitory activities (IC50, μM) of Ebselen and Ebsulfur against Mpro.
| Compd | IC50 | Compd | IC50 | Compd | IC50 |
|---|---|---|---|---|---|
| 1a | 0.65 | 2a | 0.66 | 2k | 0.11 |
| 1b | 0.41 | 2b | 0.78 | 2L | 0.41 |
| 1c | 0.35 | 2c | 0.73 | 2m | 0.49 |
| 1d | 0.13 | 2d | 0.41 | 2n | 0.36 |
| 1e | 0.15 | 2e | 0.48 | 2o | 0.23 |
| 1f | 0.26 | 2f | 0.63 | 2p | 0.91 |
| 1g | 0.24 | 2g | 0.13 | 2q | 0.38 |
| 1h | 0.33 | 2h | 0.17 | ||
|
1i 1j |
0.074 0.52 |
2i 2j |
0.33 0.39 |
2.4. Inhibition mode assays
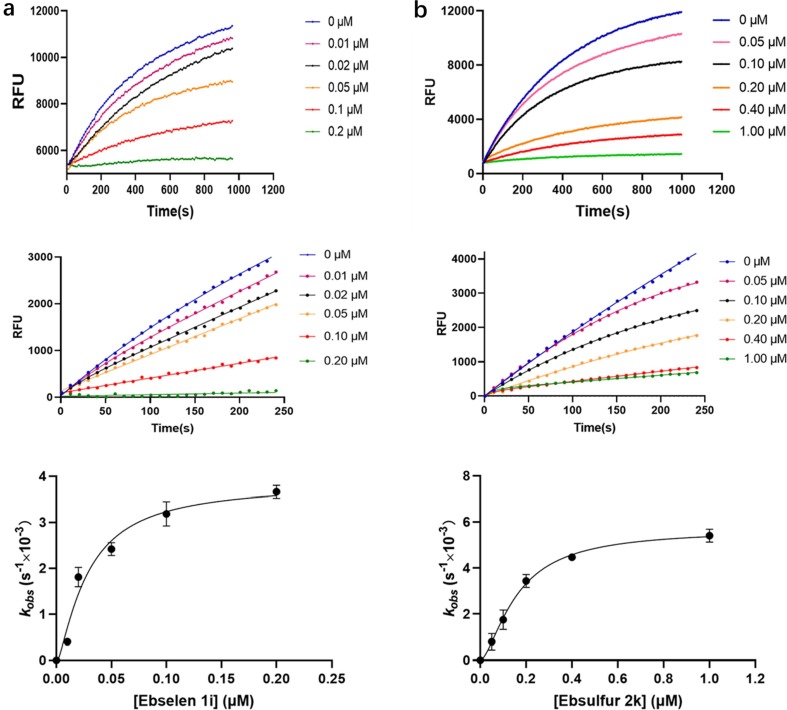
To further identify the inhibition mode of the Ebselen and Ebsulfur against Mpro, the enzyme kinetic parameters were determined. The concentrations of the inhibitors 1i and 2k were varied between 0 and 1 μM, and the FRET substrate concentration was 20 µM. After proteolysis proceeded for about 16 min, it can be observed that a large amount of substrate was consumed (Fig. 3 , upper column). As shown in Fig. 3 middle column, it is observed that the both inhibitors exhibited a concentration- and time-dependent inhibition pattern against Mpro, and the kinetic progression curves for the first 250 s showed a biphasic character, indicating the rate of inactivation follows pseudo-first-order rate kinetics, which implied that 1i and 2k covalently inhibit Mpro [34]. The observed first-order inhibition rate constant (Kobs) were fitted against inhibitor concentration by nonlinear regression to determine the inactivation kinetic parameters (Fig. 3, bottom column), to give a K i value of 31 ± 1.2 and 78 ± 3.5 nM, a k inact value of 0.0035 ± 0.0011 and 0.0058 ± 0.0002 s−1, and a k inact/K i value of 1.13x105 and 7.44x104 M−1s−1 for 1i and 2k, respectively.
Fig. 3.
Monitoring of fluorescent substrate Mca–AVLQSGFRK(Dnp)K hydrolysis catalyzed by Mpro in the presence of Ebselen 1i and Ebsulfur 2k. The concentrations of substrate and enzyme were 20 and 0.1 μM, respectively. The upper column shows the reaction progression up to 16 min; the middle column shows the progression curves for the first 250 s, which were used for curve fitting to generate the plot shown in the down column. (a), Ebselen 1i; (b), Ebsulfur 2k.
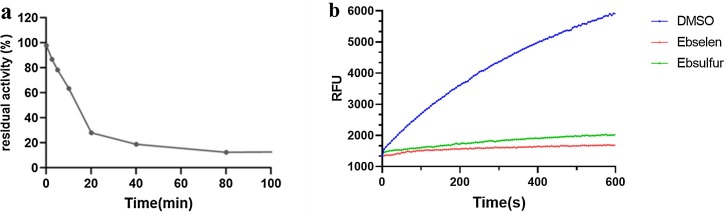
Given the excellent potency of Ebselen 1i and Ebsulfur 2k, the time-dependent inhibition of Mpro was assayed. The concentrations of enzyme and inhibitor were 0.2 and 1.0 μM, respectively. It could be observed (Fig. 4 a) that the residual activity of Mpro decreased progressively with increasing incubation time between the enzyme and inhibitor. After 60 min of incubation, the inhibitor 2k exhibited about 90% inhibition, suggesting that the Ebsulfur is time-dependent inhibitor.
Fig. 4.
The residual activity of Mpro (0.2 µM) incubation with 1 μM Ebsulfur 2k for different period (a), Jump dilution: Mpro was soaked with 1i and 2k (a concentration of 30 × IC50) for 2 h, and then 100-fold diluted with enzymatic substrate solution and monitored for 10 min (b). 0.5% DMSO was as the black control.
Furthermore, a jump dilution assay was performed to evaluate the reversibility of 1i and 2k binding to the Mpro [44]. The recombinant Mpro was incubated with a high concentration of inhibitors (equivalent to 30 × IC50) so that the inhibitor could fully occupy enzymatic active sites, and the resulting mixtures were diluted 100-fold with the fluorogenic substrate solution, it is clear to be observed that neither 1i nor 2k had significant activity recovery (Fig. 4b), implying that both compounds irreversibly inhibited the target enzyme.
2.5. Fluorescent labeling of Mpro
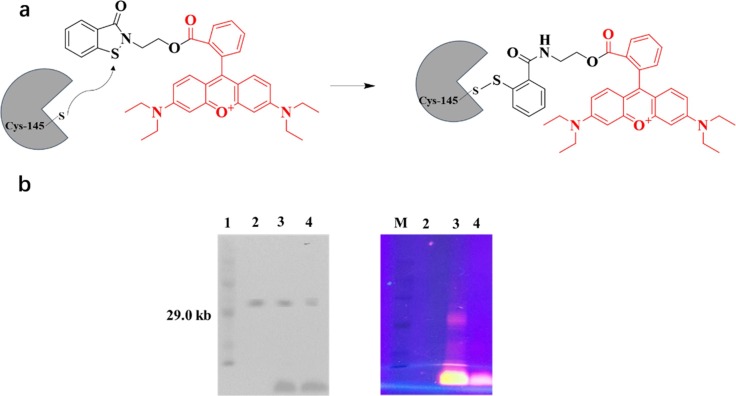
A fluorescent inhibitor Ebs-R was employed to further investigate the covalent binding of Ebsulfur to Mpro by SDS-PAGE [45]. The purified Mpro sample was incubated with Ebs-R in 10 mM Tris, pH 6.8, at 25 °C for 1 h, and then the gel was irradiated with UV light at 365 nm to visualize the labeled proteins. The scheme of Mpro labeling with Ebs-R is shown in Fig. 5 . A red protein band of approximately 33kDa was observed, which was confirmed by Coomassie Brilliant Blue (CBB) staining experiments to be consistent with Mpro (Fig. 5b, lane 3). In contrast, no red protein band was observed when Mpro was treated with rhodamine B (Fig. 5b, lane 4), indicating that rhodamine B could not label Mpro, suggesting that the Ebsulfur is capable of covalently binding to the active site of Mpro and thus exhibit potent inhibitory activity.
Fig. 5.
Structure and mechanism (a) and Gel electrophoresis analysis (b) of the fluorescent reagent Ebs-R labeling/binding Mpro. Lane 1: marker; lane 2: 10 μM purified Mpro; lane 3: 10 μM Mpro and 10 μM Ebs-R incubated; lane 4: 10 μM Mpro and 10 μM Rhodamine B incubated; Right column was observed with UV light irradiating the gel at 365 nm.
2.6. DTT-dependent assays
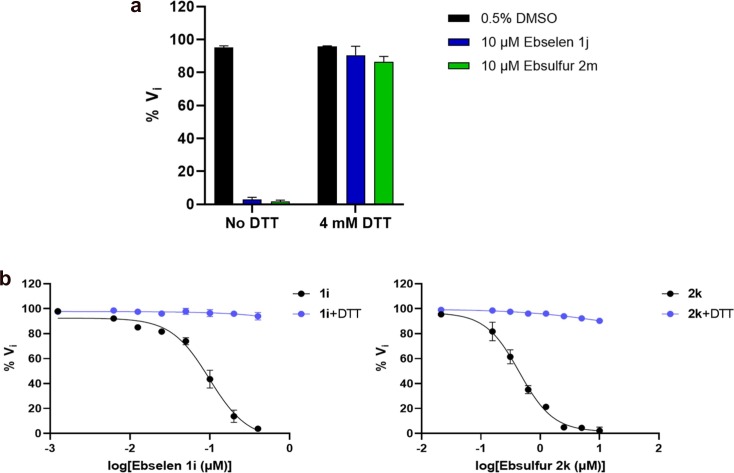
To investigate the effect of DTT on Mpro inhibition by Ebselen and Ebsulfur, the enzymatic activity assays with and without DTT (4 mM) were performed. The concentrations of Mpro and substrate were 0.2 and 20 μM, respectively. As shown in Fig. 6 a, it was found that compounds exhibited strong inhibition on Mpro in the absence of DTT, and the results are generally consistent with previous conclusion [46]. However, in the presence of DTT (4 mM), both compounds did not exhibit a potent inhibitory effect on Mpro.
Fig. 6.
Inhibition (a) and dose-dependent inhibition (b) of Ebselen and Ebsulfur on Mpro in the presence and absence of DTT. The Mpro (0.2 μM) was incubated with the compounds in the absence and presence of DTT (4 mM) buffer for 30 min. The enzymatic reaction was initiated by the addition of 20 μM substrate and the initial rate was determined. The reaction buffer without inhibitor (0.5% DMSO) was used as control.
Also, we performed dose-dependent inhibition of Mpro in the absence and presence of DTT. As shown in Fig. 6b, in the absence of DTT, the residual activity of Mpro decreased with the increase of inhibitor concentration from 0 to 10 μM. However, in the presence of DTT, 1i and 2k were unable to effectively inhibit the enzymatic activity, suggesting that inhibition of the Ebselen and Ebsulfur on Mpro was abolished probably due to that DTT disrupted the binding of inhibitors to enzyme, this case is similar to that previously reported [46], [47]. These assays indicate that both Ebselen and Ebsulfur are promiscuous cysteine protease inhibitors.
In addition, Ebselen has been reported to have potent antiviral activity against SARS-CoV-2 [34]. So, it is reasonable to believe that the Ebselen and Ebsulfur derivatives also have cellular antiviral activity, although the antiviral activity may not have a direct correlation with the in vitro enzymatic inhibition against Mpro in the absence of DTT.
2.7. Molecular modeling
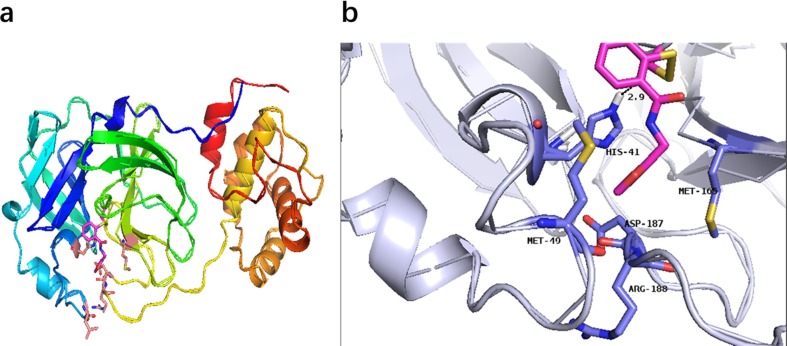
According to the reported crystal structure (PDB ID: 6LU7), the three-dimensional structure of Mpro was employed to simulate the binding mode of the most effective Ebsulfur 2k [34]. It has been reported that Cys145 is a key residue in the active site of Mpro, which makes the residue an attractive target for covalent bonding [48], [49]. The conformation shown in Fig. 7 is the lowest energy conformations with a binding energy of −5.34kcal mol−1. In this binding mode, in which the furan group of 2k forms hydrophobic interactions with Met165, Arg188, Asp187, and Met49, increasing the affinity of this substructure for the pocket. Besides, 2k also interacted with His41 residue through H‐bond, showing its effect on the catalytic coordination residues of Mpro. 2k also forms an S—S bond with Cys145 in the Mpro active site.
Fig. 7.
Lowest-energy conformations of 2k docked with Mpro (PDB code 6LU7). (a) Overview of the structure of 2k-bound Mpro. Protein is shown in cartoon representation and inhibitor is shown in the stick model. (b) Interactions formed between inhibitor and surrounding residues. The inhibitor and key residues are in the stick model and then colored by elements (N, blue; O, red; S, yellow). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Materials and methods
3.1. Materials
The HisTrapFF column and DEAE-Sepharose Fast Flow column were provided by Cytiva Technology Co (Shanghai, China). Fluorogenic substrate MCA-AVLQSGFR-Lys (Dnp)-Lys-NH2 (over 95% purity) was purchased from NJPeptide Ltd (Nanjing, China). PGEX-6P-1 vector and the SARS-CoV-2 Mpro gene were obtained from GENERAY (Shanghai, China). The other chemical reagents were purchased from Aladdin Chemical Co. (Shanghai, China) and Sigma–Aldrich (St. Louis, MO, USA).
3.2. Cloning, expression and purification of Mpro
The gene encoding the SARS-CoV-2 Mpro (NC_045512) with E. coli codon usage was optimized and synthesized by GENERAY. To obtain Mpro with authentic C and N terminals, the construct contains the cleavage site at the N-terminus (SAVLQ↓SGFR; ↓ indicates cleavage site), while eight amino acids (GPHHHHHH) was inserted into the C-terminus [33]. The amplified PCR product was cloned to the PGEX-6P-1 vector and then transformed into E. coli BL21-Gold (DE3) strain.
The signal clone was pre-cultured at 37 °C in 40 ml Luria Broth (LB) medium with ampicillin (100 μg/ml) overnight, and then incubated culture was inoculated into 4L LB medium with 100 μg/ml ampicillin. The cells are allowed to grow to an OD600 value in the range of 0.6–0.8, and protein expression was induced by adding 1.0 mM isopropyl-d-thiogalactoside (IPTG) at 18 °C. After 12 h, the cells were harvested, resuspended in buffer A (20 mM Tris, pH 7.8, 150 mM NaCl), and lysed by sonication on ice. Cell debris was removed by centrifugation at 3000g, 4 °C for 20 min. The supernatants were loaded onto a Ni-NTA affinity column and washed with buffer B (20 mM Tris, pH 7.8, 150 mM NaCl, 20 mM imidazole). The His-tagged Mpro was eluted with buffer C (20 mM Tris, pH 7.8, 150 mM NaCl, 300 mM imidazole), using a linear gradient of imidazole concentration from 0 to 100%. The eluted fraction was collected after dialysis and then human rhinovirus (HRV) 3C protease was added to cleave the C-terminal hexahistidine-tag at 4˚C overnight. The cleaved protein was loaded onto a His-Trap column to separate the His-tag and protein. The purity of the recombinant protein was confirmed by 15% SDS-PAGE and stored at −80 °C. The protein sequence for the native SARS-CoV-2 Mpro is SGFRKMAFPS GKVEGCMVQVTCGTTTLNGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHMELPTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQCSGVTFQ.
3.3. Enzymatic activity assay
The fluorescence resonance energy transfer (FRET) assay was employed to test the activity of Mpro as previously reported method [50], using Mca–AVLQSGFR-K(Dnp)K as substrate and monitoring at 405 nm (emission wavelength, the excitation wavelength was 320 nm). The assay mixture contains 0.1 µM Mpro and various concentrations of substrate (0.5–40 µM). The reaction progress of substrate hydrolysis was monitored on Hitachi F-4500 fluorescence spectrophotometer until the final signals reached a plateau, at which point the whole substrate was deemed to be digested by the enzyme. The end-point fluorescence signals were plotted against the substrate concentrations using a linear regression function to offer a Mca-standard curve.
For the measurement of Km/Vmax, Mpro was mixed with different concentrations of substrate in 200 μL of assay buffer (20 mM Tris, pH 6.5, 0.4 mM EDTA, 20% glycerol, 120 mM NaCl), in which the concentration of protein was 0.1 µM, and concentration of substrate was in the range of 1–80 μM. The reaction was monitored every 2 s for 16 min. The initial rate for the first 4 min was calculated by linear regression function in Graphpad Prism 5 and then plotted against the substrate concentration using the Michaelis-Menten equation.
3.4. Enzymatic inhibition assay
Enzymatic inhibition was assayed through the determination of percent inhibition and IC50 values of inhibitors on Mpro. The compounds were dissolved in a small volume of DMSO, diluted with assay buffer (see above), and then mixed with the enzyme samples. The final concentrations of DMSO in inhibition experiments were below 0.5%, and control experiments verified that 0.5% of DMSO had no inhibitory effect against Mpro.
For determination of the percent inhibition, the inhibitors dissolved in 200 μL of assay buffer were incubated with Mpro at 30 °C for 30 min, the substrate was then added and monitored every 2 s at 405 nm on a Hitachi F-4500 fluorescence spectrophotometer. The concentrations of protein, substrate and inhibitors were 0.2, 20 and 50 μM, respectively.
To determine the IC50, the inhibitors 1a-j and 2a-q were assayed at six different concentrations between 0 and 2 µM, and concentrations of enzyme and substrate were 0.2 and 20 µM, respectively. The hydrolysis of the substrate was monitored at 405 nm, and the rate of hydrolysis was determined in the linear range. All experiments were performed in triplicate. The IC50 values were calculated by plotting the average percentage inhibition against inhibitor concentration and fitting the data in Prism 5 [50].
3.5. Inhibition mode assays
Ebselen 1i and Ebsulfur 2k with effective inhibition were identified in the preliminary inhibitory experiments, and its kinetic parameters were determined as previously described [50]. Briefly, an enzyme sample was added into 200 μL of assay buffer with different concentrations of inhibitor, then the substrate was added and the reaction was monitored at 405 nm for about 16 min. The concentrations of enzyme and substrate were 20 nM and 20 µM, respectively. The continuously detected data were used for the enzyme kinetic analysis in GraphPad Prism 5. The time-dependent inhibition progress curves were fitted to a standard exponential (equation (1)) to give an observed first-order inhibition rate constant (kobs) [51], [52].
| (1) |
where [P] is the product fluorescence signal; D is the background signal at data collection start; t is time and V0 is the initial velocity. The best-fit kobs value was plotted against the inhibitor concentration to fit equation (2).
| (2) |
where [I] is inhibitor concentration; K i is the concentration of inactivator at the half-maximum inactivation rate constant, and k inact is the rate constant of inactivation.
3.6. SDS-PAGE analysis of labeled Mpro
Ebs-R and Mpro were firstly dissolved in buffer (10 mM Tris, pH 6.8, 0.5% DMSO) and incubated at room temperature for 2 h. Then an equal volume of the labeled protein was dissolved in 2 × SDS gel loading buffer (100 mM Tris, pH 6.8, 20% glycerol, 2.5% SDS) and analyzed by SDS-PAGE. The Gels were observed under 365 nm UV light and finally stained with Coomassie Brilliant Blue.
3.7. Docking study
To further investigate the binding interaction of the inhibitor 2k with Mpro, AutoDock 4.2 was used to predict their binding poses [53], [54]. The crystal structure of the Mpro complex with the inhibitor N3 (coded 6LU7) was obtained from the Protein Data Bank [34]. Before docking, AutoDockTools were applied to prepare protein by removing all water molecules and then adding hydrogen atoms and charges that modify the PDB files of ligand and receptor. The covalent bond between the enzyme and N3 was also removed. The center coordinates are set according to the center point of N3 (-10.36, 12.46, 68.7) and the grid box size was set to 35 × 35 × 35 grid points. The docking pose of the protein to the ligand was identified through comparison of the root mean square deviation (RMSD). All other parameters were kept at default values and no constraints were employed. Based on visual examination, the structure with low binding free energy was selected. The minimized energy of 2k obtained from the docking study was − 5.34kcal/mol.
4. Conclusions
The main protease (Mpro) that the SARS-CoV-2 viral replication employed was expressed and purified, and K m and Vmax were determined to be 39.1 ± 0.8 µM and 106.3 ± 6 nM/s, respectively. The inhibitory activity of Ebselen and Ebsulfur analogs against Mpro was identified using FRET assay with an IC50 value in the range of 0.074–0.91 µM. Several compounds displayed more potent enzymatic inhibition than the standard Ebselen, where Ebselen 1i and Ebsulfur 2k showed the highest inhibition on Mpro with an IC50 value of 0.074 and 0.11 µM, and a K i value of 0.031 and 0.078 µM, respectively. The pre-incubation and jump dilution assays and fluorescent labeling experiments showed that the compounds covalently and irreversibly bind to Mpro. DTT-dependent inhibition assays indicate that Ebselen and Ebsulfur are promiscuous cysteine protease inhibitors of Mpro. Docking studies further suggested that 2k formed an S—S bond with the Cys145 at the enzymatic active site. We believe that Ebselen and Ebsulfur derivatives still have clinical potential for the treatment of coronaviruses. This study found that both Ebsulfur and Ebselen derivatives are very potent scaffolds for the development of the covalent inhibitors of Mpro to combat COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the grants (22077100 and 2019KW-068) from the National Natural Science Foundation of China and Shanxi Province International Cooperation Project and the grant (201710010013) from the Science and Technology Program of Guangzhou, and the grant (17JS007) from the Shaanxi Education Commission.
References
- 1.Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., Zhang L., Fan G.H., Xu J.Y., Gu X.Y., Cheng Z.S., Yu T., Xia J.A., Wei Y., Wu W.J., Xie X.L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J.G., Wang G.F., Jiang R.M., Gao Z.C., Jin Q., Wang J.W., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2019;395(2020):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lei L., Huang X.M., Zhang S., Yang J.R., Yang L., Xu M. Comparison of prevalence and associated factors of anxiety and depression among people affected by versus people unaffected by quarantine during the COVID-9 epidemic in Southwestern China. Med. Sci. Monit. 2020;26:12. doi: 10.12659/msm.924609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Gayle A.A., Wilder Smith A., Rocklov J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020;27:1–4. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viswanathan T., Arya S., Chan S.H., Qi S., Dai N., Misra A., Park J.G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G.D., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2019;19(2020):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 7.Bhavana V., Thakor P., Singh S.B., Mehra N.K. COVID-19: Pathophysiology, treatment options, nanotechnology approaches, and research agenda to combating the SARS-CoV2 pandemic. Life Sci. 2020;261:118336. doi: 10.1016/j.lfs.2020.118336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grottesi A., Besker N., Emerson A., Manelfi C., Beccari A.R., Frigerio F., Lindahl E., Cerchia C., Talarico C. Computational Studies of SARS-CoV-2 3CLpro: Insights from MD Simulations. Int. J. Mol. Sci. 2020;21:5346. doi: 10.3390/ijms21155346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya A.E., Snijder E.J. Viral cysteine proteinases. Perspect. Drug Discovery Des. 1996;6(1):64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27(6):529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 12.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert H., Tanya H., Nigel W.H., Paul N.S., Georgios V.G., Michel D. Mouse model phenotypes provide information about human drug targets. Bioinformatics. 2013;5:719–725. doi: 10.1093/bioinformatics/btt613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippo A., Federica F., Alessia G., Pierluigi F., Anna G., Chiara P., Danilo D., Alessandra S., Marco M., Elena M., Giuseppe N. Impact of azithromycin and/or hydroxychloroquine on hospital mortality in COVID-19. J. Clin. Med. 2020;9:2800. doi: 10.3390/jcm9092800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D’Arminio Monforte A., Ismail S., Kato H., Lapadula G., L’Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe COVID-19. N Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou Y., Liu L., Yao H., Hu X., Su J., Xu K., Luo R., Yang X., He L., Lu X., Zhao Q., Liang T., Qiu Y. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: an exploratory randomized, controlled trial. Eur. J. Pharm. Sci. 2021;157:105631. doi: 10.1016/j.ejps.2020.105631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoichet B.K. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346:128933. doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj V., Singh R., Singh P., Purohit R., Kumar S. Elimination of bitter-off taste of stevioside through structure modification and computational interventions. J. Theor. Biol. 2020;486:110094. doi: 10.1016/j.jtbi.2019.110094. [DOI] [PubMed] [Google Scholar]
- 23.Singh R., Bhardwaj V., Das P., Purohit R. Natural analogues inhibiting selective cyclin-dependent kinase protein isoforms: a computational perspective. J. Biomol. Struct. Dyn. 2020;38(17):5126–5135. doi: 10.1080/07391102.2019.1696709. [DOI] [PubMed] [Google Scholar]
- 24.Bhardwaj V.K., Singh R., Sharma J., Das P., Purohit R. Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Comput Meth. Programs Biomed. 2020;194:105494. doi: 10.1016/j.cmpb.2020.105494. [DOI] [PubMed] [Google Scholar]
- 25.Singh Rahul, Bhardwaj Vijay Kumar, Sharma Jatin, Das Pralay, Purohit Rituraj. Discovery and in silico evaluation of aminoarylbenzosuberene molecules as novel checkpoint kinase 1 inhibitor determinants. Genomics. 2021;113(1):707–715. doi: 10.1016/j.ygeno.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011;92(2):204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sydnes M.O., Hayashi Y., Sharma V.K., Hamada T., Bacha U., Barrila J., Freire E., Kiso Y. Synthesis of glutamic acid and glutamine peptides possessing a trifluoromethyl ketone group as SARS-CoV 3CL protease inhibitors. Tetrahedron. 2006;62(36):8601–8609. doi: 10.1016/j.tet.2006.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K., Yakushiji F., Akaji K., Chen S.E., Naser Tavakolian A., Schon A., Freire E., Hayashi Y. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: Design, synthesis, biological evaluation, and docking studies. Eur. J. Med. Chem. 2013;68:372–384. doi: 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23(4):876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mproand cathepsin L. Sci. Adv. 2020;6:1–15. doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng Xi, Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weglarz-Tomczak E., Tomczak J.M., Talma M., Burda-Grabowska M., Giurg M., Brul S. Identification of ebselen and its analogues as potent covalent inhibitors of papain-like protease from SARS-CoV-2. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-83229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M-pro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L.L., Lin D.Z., Kusov Y., Nian Y., Ma Q.J., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 36.Zhai L., Zhang Y.-L., Kang J.S., Oelschlaeger P., Xiao L., Nie S.-S., Yang K.-W. Triazolylthioacetamide: A valid scaffold for the development of New Delhi Metallo-β-Lactmase-1 (NDM-1) Inhibitors. ACS Med. Chem. Lett. 2016;7(4):413–417. doi: 10.1021/acsmedchemlett.5b00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Y., Chen C., Wang W.-M., Xu L.-W., Yang K.-W., Oelschlaeger P., He Y. Rhodanine as a potent scaffold for the development of broad-spectrum Metallo-β-lactamase Inhibitors. ACS Med. Chem. Lett. 2018;9(4):359–364. doi: 10.1021/acsmedchemlett.7b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang Y.-N., Xiang Y., Zhang Y.-J., Wang W.-M., Chen C., Oelschlaeger P., Yang K.-W. Carbamylmethyl mercaptoacetate thioether: a novel scaffold for the development of L1 metallo-β-lactamase Inhibitors. ACS Med. Chem. Lett. 2017;8(5):527–532. doi: 10.1021/acsmedchemlett.7b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Chen C., Sun L.Y., Gao H., Zhen J.B., Yang K.-W. Meta-substituted benzenesulfonamide: A potent scaffold for the development of metallo-beta-lactamase ImiS inhibitors. RSC Med. Chem. 2020;11:259–267. doi: 10.1039/c9md00455f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y.L., Yang K.-W., Zhou Y.J., LaCuran A.E., Oelschlaeger P., Crowder M.W. Diaryl-substituted azolylthioacetamides: Inhibitor discovery of New Delhi Metallo-beta-Lactamase-1 (NDM-1) ChemMedChem. 2014;9:2445–2448. doi: 10.1002/cmdc.201402249. [DOI] [PubMed] [Google Scholar]
- 41.Chen C., Liu Y., Zhang Y.J., Ge Y., Lei J.E., Yang K.-W. The assemblage of covalent and metal binding dual functional scaffold for cross-class metallo-beta-lactamases inhibition. Future Med. Chem. 2019;11:2381–2394. doi: 10.4155/fmc-2019-0008. [DOI] [PubMed] [Google Scholar]
- 42.Su J.P., Liu J.Y., Chen C., Zhang Y.J., Yang K.W. Ebsulfur as a potent scaffold for inhibition and labelling of New Delhi metallo-beta-lactamase-1 in vitro and in vivo. Bioorg. Chem. 2019;84:192–201. doi: 10.1016/j.bioorg.2018.11.035. [DOI] [PubMed] [Google Scholar]
- 43.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z., Bjorkman P. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copeland R.A., Basavapathruni A., Moyer M., Scott M.P. Impact of enzyme concentration and residence time on apparent activity recovery in jump dilution analysis. Anal. Biochem. 2011;416(2):206–210. doi: 10.1016/j.ab.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Mizukami S., Watanabe S., Hori Y., Kikuchi K. Covalent protein labeling based on noncatalytic beta-Lactamase and a designed FRET substrate. J. Am. Chem. Soc. 2009;131:5016–5017. doi: 10.1021/ja8082285. [DOI] [PubMed] [Google Scholar]
- 46.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin are nonspecific promiscuous SARS-CoV-2 main protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020;3(6):1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moretto M.B., Thomazi A.P., Godinho G., Roessler T.M., Nogueira C.W., Souza D.O., Wofchuk S., Rocha J.B.T. Ebselen and diorganylchalcogenides decrease in vitro glutamate uptake by RAT brain slices: Prevention by DTT and GSH. Toxicol. In Vitro. 2007;21(4):639–645. doi: 10.1016/j.tiv.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Yoshino R., Yasuo N., Sekijima M. Identification of key interactions between SARS-CoV-2 main protease and inhibitor drug candidates. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-69337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shitrit A., Zaidman D., Kalid O., Bloch I., Doron D., Yarnizky T., Buch I., Segev I., Ben Zeev E., Segev E., Kobiler O. Conserved interactions required for inhibition of the main protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-77794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Yu, Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison J.F., Walsh C.T. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 52.Musharrafieh R., Ma C.L., Zhang J.T., Hu Y.M., Diesing J.M., Marty M.T., Wang J. Validating enterovirus D68–2A(pro) as an antiviral drug target and the discovery of telaprevir as a potent D68–2A(pro) Inhibitor. J. Virol. 2019;93:1–16. doi: 10.1128/jvi.02221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J., Macip G., Saldivar-Espinoza B., Cereto-Massagué A., Pujadas G., Garcia-Vallvé S. Garcia Vallve, Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int. J. Mol. Sci. 2020;21(11):3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bianco G., Forli S., Goodsell D.S., Olson A.J. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 2016;25:295–301. doi: 10.1002/pro.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]