Graphical abstract
Keywords: Data management, Phenotype, Phenomics, Controlled terminologies and vocabularies
Abbreviations: COVID-19, Coronavirus disease 2019; EHR, Electronic health record; RD, Research Derivative; OMOP, Observational Medical Outcomes Partnership; ICD, International Classification of Disease; CPT, Current Procedural Terminology
Abstract
From the start of the coronavirus disease 2019 (COVID-19) pandemic, researchers have looked to electronic health record (EHR) data as a way to study possible risk factors and outcomes. To ensure the validity and accuracy of research using these data, investigators need to be confident that the phenotypes they construct are reliable and accurate, reflecting the healthcare settings from which they are ascertained. We developed a COVID-19 registry at a single academic medical center and used data from March 1 to June 5, 2020 to assess differences in population-level characteristics in pandemic and non-pandemic years respectively. Median EHR length, previously shown to impact phenotype performance in type 2 diabetes, was significantly shorter in the SARS-CoV-2 positive group relative to a 2019 influenza tested group (median 3.1 years vs 8.7; Wilcoxon rank sum P = 1.3e-52). Using three phenotyping methods of increasing complexity (billing codes alone and domain-specific algorithms provided by an EHR vendor and clinical experts), common medical comorbidities were abstracted from COVID-19 EHRs, defined by the presence of a positive laboratory test (positive predictive value 100%, recall 93%). After combining performance data across phenotyping methods, we observed significantly lower false negative rates for those records billed for a comprehensive care visit (p = 4e-11) and those with complete demographics data recorded (p = 7e-5). In an early COVID-19 cohort, we found that phenotyping performance of nine common comorbidities was influenced by median EHR length, consistent with previous studies, as well as by data density, which can be measured using portable metrics including CPT codes. Here we present those challenges and potential solutions to creating deeply phenotyped, acute COVID-19 cohorts.
1. Introduction
The emergence of coronavirus disease 2019 (COVID-19) has raised urgent questions about the susceptibility of vulnerable populations, effectiveness of public health interventions, and efficacy of new prophylaxis and treatment strategies. The rapid emergence of this disease has mobilized a large number of researchers to study the impact of the pandemic through observational cohort studies and randomized clinical trials.[1], [2], [3] Many are leveraging electronic health records (EHRs) to rapidly identify patients diagnosed with COVID-19 and to track the clinical course and outcomes with routinely collected data.[4], [5]
Conducting EHR-based research requires clear definitions of cohorts and the ability to accurately and reliably identify relevant comorbidities. Both laboratory results and billing codes can be used to identify patients affected by COVID-19, but the relative performance of these variables has not been studied. For identifying comorbidities and outcomes, researchers may use existing or previously published algorithms developed to extract research-grade phenotypes from the EHR.[6], [7] However, it is unknown how these approaches perform in the context of a rapidly shifting pandemic, during which patterns of patient engagement with the healthcare system as well as healthcare delivery itself may be seriously altered.[8], [9] Furthermore, researchers need frameworks that guide them in assessing performance in both static and rapidly evolving situations.
In this analysis, we explore the challenges of phenotyping during the COVID-19 pandemic. First, we examine different approaches to identifying populations diagnosed with COVID-19. Next, we examine the characteristics of a SARS-CoV-2 tested cohort and a non-pandemic cohort of patients tested for influenza in 2019, with a focus on differences in available data prior to testing. Finally, we measure the performance of established phenotyping methods to find comorbidities present prior to SARS-CoV-2 testing in order to study the interaction between data availability and algorithm performance. Through this work, we have identified key variables that may help researchers better characterize cohorts in a rapidly emerging and shifting situation.
2. Background and significance
The first case of the novel coronavirus that causes COVID-19 was reported in the United States on January 20, 2020.[10] Six weeks later, on March 5th, the first reported case in the state of Tennessee was diagnosed through polymerase chain reaction (PCR) testing completed by Vanderbilt University Medical Center (VUMC). VUMC continues to test a large number of patients and thus has accumulated a significant amount of data on both testing and clinical processes in COVID-19. As part of an institutional effort to make COVID-19 related data broadly available to researchers, a de-identified registry of patients tested for SARS-CoV-2 was created and populated with structured data from our linked institutional EHR, Epic Systems. Phenotyping efforts were initiated to identify pre-existing co-morbidities and exposures, characterize disease progression and severity of COVID-19, and monitor long-term outcomes.
Creation of a COVID-19 registry was intended to accelerate COVID-19 informatics research by utilizing robust systems, such as the Research Derivative (RD) in place at VUMC to analyze near real-time EHR data. The RD is a database of clinical information curated from the EHR and made available for research.[11] Output may include structured data points, such as billing codes and encounter dates, semi-structured data such as laboratory tests and results, or unstructured data such as physician progress reports. Researchers have the option of coupling EHR data with DNA biorepositories such as BioVU, an opt-out biobank that currently has close to 250,000 DNA samples.[12]. These tools have been successful in producing replicable identification of genetic variants that modulate risk for human disease.[13], [14]
2.1. Known performance and risks of ePhenotyping
Phenotyping has used successfully to identify genetic variants of significance and to provide targeted clinical decision support.[15], [16] Billing codes are often used to identify patient cohorts, but risk losing the clinical processes or sets of contextual events from which they were ascertained. Adding classes of data (e.g., medications, labs) to billing codes can improve phenotyping performance but may overfit for local, institution-specific algorithms.[17] Algorithm development in a single EHR system also relies on data cleaning for incomplete or highly complex data. As others have acknowledged, local phenotyping execution necessitates anticipating data quality issues,[18] defined value sets,[19] and an explicit study of bias.[20]
3. Materials and methods
3.1. COVID-19 data sources - healthcare systems processes
Beginning in March, EHR data for patients tested for COVID-19 were collected in a data repository updated daily using the Observational Medical Outcomes Partnership (OMOP) structure. The OMOP database included many of the data points contained in the operational EHR, but some COVID-19 specific data elements were missing. Paper intake forms were created for clinicians to collect COVID-19 related symptoms, duration of illness and patient-reported vaccination history among other data (Appendix Fig. A1) to assist in the triage of growing numbers of patients needing SARS-CoV-2 testing at designated “COVID-19” outpatient clinics. These forms were later scanned into the medical record, with structured data abstracted using optical character recognition and manual processes for use in future EHR-based research. Because of a shifting knowledge-base, dynamic changes to clinical decision support alerts, patient flags, and order sets occurred as the pandemic evolved and clinicians expressed needs for changing tools and support.
3.2. Creation of a COVID-19 data registry
All patients with PCR testing for SARS-CoV-2 after March 1 were included in the COVID-19 registry. Following a positive test, patient labs, comorbidities, and disease progression were integrated using standardized International Classification of Disease, Tenth revision, Clinical Modification codes (ICD-10), Current Procedural Terminology (CPT) codes, and phecodes, groups of ICD-9 and ICD-10 codes developed for the purpose of phenome-wide association studies.[21] Members of the data team monitored and normalized incoming labs (e.g., duplicate lab names, conflicting units), and developed a chronology of incoming raw data streams with respect to the SARS-CoV-2 PCR test, or “time zero,” in the RD (Fig. 1 ). Data cleaning was recorded in a data dictionary.
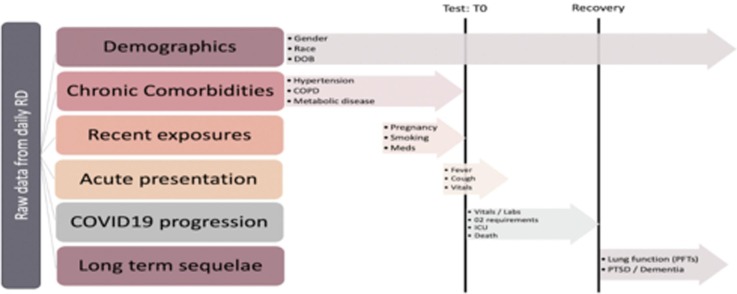
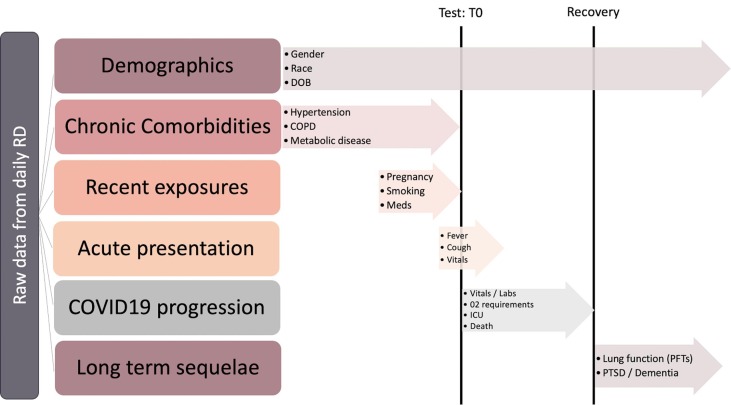
Fig. 1.
Chronology of coronavirus disease 2019 (COVID-19) registry data types. “Test: T0” indicates the timestamp of a positive PCR test and defines the acute phase of disease in our registry. As depicted, T0 is critical for distinguishing between risk factors (e.g., history of DVT/PE in the pre-infection phase prior to T0) and sequelae of disease (e.g., acute DVT/PE in the acute or recovery phase). RD: Research Derivative, a database of clinical information curated from the EHR at Vanderbilt University Medical Center and restructured for research; T0: chronology of incoming raw data streams ordered with respect to a SARS-CoV-2 PCR test; DOB: Date of birth; COPD: Chronic obstructive pulmonary disease; Meds: Medications; O2: Oxygen; ICU: Intensive Care Unit; PFTs: Pulmonary function tests; PTSD: Post traumatic stress disorder.
3.3. COVID-19 case definition
Fig. 2 depicts the data workflow following registry creation. In late February, the Centers for Disease Control released official coding guidelines for patients testing positive for COVID-19, effective April 1.[22] Previously, coders were prompted to use a set of nonspecific billing codes (e.g., J22, “Unspecified acute lower respiratory infection”) to bill for COVID-19 cases. In our registry, only 10% of SARS-CoV-2 PCR positive patients before the month of April had billing codes according to these recommendations, likely due to uncertainty around classifying “possible,” “suspected,” or “probable” cases. Consequently, we chose to interrogate the relevance of three COVID-19 case definitions at our institution: presence of one or more U07.1 ICD-10 billing codes, laboratory testing, or both. COVID-19 case definition algorithms queried from OMOP on May 15, 2020 were validated against a manual chart review of a portion of these cases (n = 140) performed by a clinician reviewer blinded to the algorithm’s billing or lab status. Differences in billing practices at inpatient versus outpatient centers and for those tests performed at outside facilities were not accounted for on this initial review. To assess timeliness of billing code availability in the RD, a second unblinded review recorded time between test date and billing date in our system. In the case of multiple COVID-19 PCR tests with the same result, SARS-CoV-2 test date was recorded as the date of the first positive or last negative test.
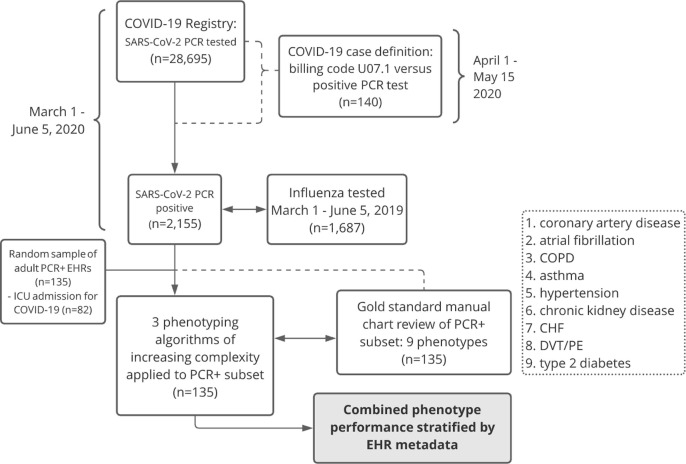
Fig. 2.
Data workflow. Registry created from individuals with at least one SARS-CoV-2 positive PCR test at any of our 18 care sites across the Mid-South between March 1 and June 5, 2020. COVID-19 case definition validated on a subset of EHRs dated after billing guidelines issued April 1. Random sampling of adult inpatient and outpatient EHRs selected for phenotyping. Double arrows indicate comparison between cohorts. Dotted lines indicate processes that contributed to decision making in the methods workflow (solid lines). COVID-19: coronavirus disease 2019; PCR: polymerase chain reaction; ICD-10: International classification of diseases, Tenth revision, Clinical modification; EHR: electronic health record; ICU: intensive care unit; COPD: chronic obstructive pulmonary disease; CHF: congestive heart failure; DVT/PE: deep venous thrombosis/pulmonary embolism.
3.4. Defining phenotyping metadata
Pandemics cause individuals to seek medical care, many not previously known to the healthcare system, and require changes to healthcare systems processes to accommodate this influx, thus impacting research conducted with the data. Although less granular than the data they represent, metadata attempt to stratify for possible influences on data fitness, grouping records by data available to all EHRs subject to similar healthcare processes. Because validated phenotypes are developed on longitudinal cohorts, selecting for median EHR length, we hypothesized that new patients with incomplete or absent retrospective records could increase false negative rates among algorithms as demonstrated by Wei et al. in a longitudinal Mayo clinic cohort.[23], [24]. Adjusting for metadata, studying its effect on phenotyping performance specifically, is a local effort to acknowledge those pandemic-associated changes in how data elements are recorded,[25] including scanned paper COVID-19 patient intake forms unavailable for use in phenotyping algorithms at the time of study. To ascertain the extent of the quantity and quality of data available for individual patients, we studied four categories of metadata (Table 1 ) available in most EHR systems (e.g., CPT codes), excepting data density categorized by center-specific visit identifiers.
Table 1.
Metadata studied within a COVID-19 cohort.
| Metadata (Data type) | Description | Data Reference | |
|---|---|---|---|
| Median EHR length (Years) | Difference in years between the first recorded test date (either influenza or SARS-CoV-2) and first recorded visit, any type. | Data quantity | |
| Missingness (Count) | “Unknown” demographic(s) (i.e., any incomplete age, self-reported race, gender) data element in the RD. | Data quantity | |
| Data density (Categorical, institution-specific) | No Visits | Individuals with no visit(s) billed prior to the week before the first test date. | Data quality |
| No Primary Care Visit | Non-primary care visit(s) billed before the first test date. | ||
| Medical Home | At least one primary care visit, identified by local site IDs, billed before the first test date. | ||
| Data density (Binary, not institution-specific) | Presence of a CPT code that indicates a ‘Comprehensive history’ was taken prior to or on the day of the first SARS-CoV-2 test (Appendix Table A4) | Data quality | |
COVID-19: Coronavirus disease 2019; EHR: Electronic health record; RD: RD: Research Derivative, a database of clinical information curated from the EHR at Vanderbilt University Medical Center and restructured for research; CPT: Current procedural terminology.
3.5. Evaluation of an early COVID-19 registry
We selected four cohorts of adult patients (age greater than or equal to 18 years) to describe features of our COVID-19 cohort for the study period of March 1 to June 5, 2020. (1) SARS-CoV-2 tested: Adults tested for SARS-CoV-2 within the 2020 study period; (2) Influenza tested 2019: Adults tested for influenza in the equivalent 2019 study period; (3) SARS-CoV-2 positive: Adults testing positive for SARS-CoV-2 at least once in the 2020 study period; (4) SARS-CoV-2 negative: Adults testing negative at least once for SARS-CoV-2 with no positive testing in the 2020 study period. A portion of SARS-CoV-2 positive individuals also had negative SARS-CoV-2 tests but were not included in the SARS-CoV-2 negative cohort by definition. The test date was defined as either the first positive test for SARS-CoV-2 positive patients, or the last test for patients who only tested negative. Testing site(s) was not limited to our institution and included those individuals with COVID-19 transferred to our facility for higher level of care. We calculated descriptive statistics for all four cohorts, including summary statistics for demographics and metadata. Wilcoxon rank sum test was used to describe differences in median EHR length, otherwise Fisher’s exact test was used to study statistical difference among metadata categories. Age was calculated at the start of the study period.
3.6. Defining standard phenotypes in the COVID-19 population
We created a gold standard phenotype set by manual chart review of a random sample of inpatient and outpatient charts of patients who tested positive for COVID-19, stratified by admission to the intensive care unit (n = 82, 61%). Practicing physicians reviewed the charts of 135 SARS-CoV-2 PCR-tested individuals to identify commonly encountered comorbidities in inpatient and outpatient populations including: presence of Type 2 diabetes mellitus (chart diagnosis), chronic kidney disease (chart diagnosis and manual review of estimated glomerular filtration rate <60 mL/min for >3 months where available), essential hypertension (chart diagnosis, including evidence of at least one blood pressure lowering medication in history where available and excluding diagnosis of elevated blood pressure only without evidence of outpatient medication use), congestive heart failure (chart diagnosis with or without echocardiogram evidence of diastolic or systolic dysfunction), history of atrial fibrillation (chart diagnosis), coronary artery disease (chart diagnosis). Pulmonary-specific comorbidities included chronic obstructive pulmonary disease (chart diagnosis, emphysema or chronic bronchitis unspecified), asthma (requiring at least one asthma medication on historical medication list), history of pulmonary embolism and/or deep venous thrombosis (chart diagnosis). Although additional phenotypes were identified through manual chart review, we selected comorbid phenotypes in the COVID-19 population based, in part, on existing and emerging phenotyping algorithms developed using the common data model.
3.7. Selection of phenotyping algorithms
Validated phenotyping algorithms were applied to the same 135 SARS-CoV-2 tested records, including ICD-based phecodes and more complex algorithms shared by the EHR vendor Epic-systems. Data elements used in phenotype definitions are shown in Appendix Table A1. We used standard phenotyping metrics, including sensitivity, specificity and F-score to evaluate the performance of selected standard phenotypes in our local COVID-19 population.
3.8. Validation of standard phenotypes in the COVID-19 population
Comorbidities identified by each high-throughput phenotyping method were corroborated with manual chart review to formulate a gold standard set. In cases where manual chart review results disagreed with results of selected phenotyping algorithms, the same subject that previously performed chart review returned to the EHR to provide final input on the presence or absence of comorbidities and identify sources of false positive or false negative results. A major source of false negatives was hypothesized to be comorbid phenotypes identified by scanned paper COVID-19 patient intake forms, used in the outpatient setting only. Specific notation was used for phenotypes recorded from scanned paper intake forms on manual review.
4. Results
4.1. COVID-19 case definition
We found significant variation in positive predictive value (PPV) and recall of case definitions applied between April 1 and May 15, 2020 (Table 2 ). Although the highest recall among algorithms was achieved for those records with both an ICD-10 code and laboratory test, we desired maximum return of true positive cases. Thus, laboratory testing only (PPV 100%) was chosen for identifying COVID-19 cases for chart review. Sufficient recall (93%) was achieved through positive laboratory testing only. The average days between the earliest confirmed lab result and ICD-10 diagnosis in our system was 7 days. Differences in inpatient versus outpatient billing practices may have contributed to delays between laboratory and billing results.
Table 2.
Comparison of COVID-19 case definitions between April 1 and May 15, 2020.
| Phenotype Definition | PPV | Recall |
|---|---|---|
| ICD-10 Only | 90.6% | 46.4% |
| Laboratory testing Only | 100% | 93.0% |
| ICD-10 or Laboratory testing | 95.4% | 100% |
| ||
4.2. Characterization of COVID-19 cohort
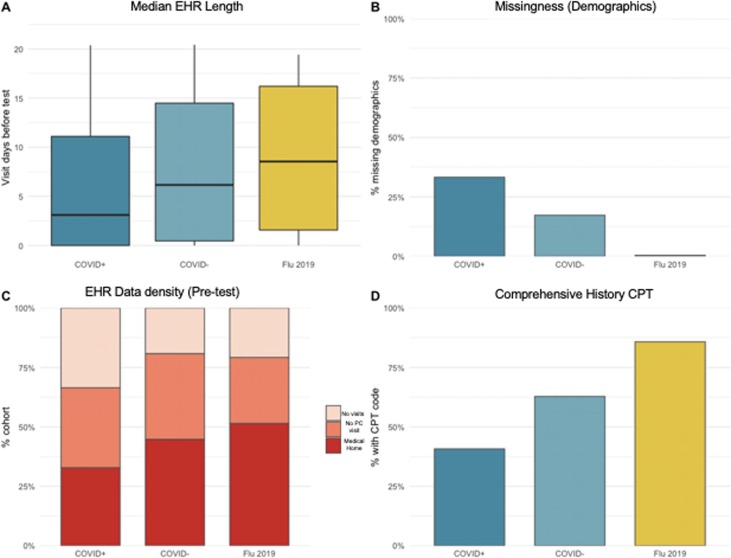
Of 28,695 patients with SARS-CoV-2 test results in the VUMC COVID-19 registry from March 1 through June 5, 2020, 8% (n = 2,155) were SARS-CoV-2 PCR positive. Breakdown of age, gender, and self-reported race is provided in Appendix Table A2. High numbers of missing testing demographics in the SARS-CoV-2 tested group resulted in a limited distribution of self-reported race. Reasons for incomplete demographics include the way data was recorded at designated “COVID-19” testing sites and absence of these check boxes on paper COVID-19 patient intake forms. Average age was lower in our early SARS-CoV-2 tested population than an influenza tested cohort tested in 2019 (40 years versus 52 years in COVID-19 positive and influenza tested respectively; standard deviation[SD] 16 and 19 years). The proportion of individuals who identify as female was consistent for both tests 2019–2020.
4.3. Comparison of influenza tested and SARS-CoV-2 tested cohorts
More than half of those with SARS-CoV-2 testing results in our EHR had no visits at any of our 18 care sites across the Mid-South prior to testing (Table 3 ). Median EHR length was significantly shorter in the SARS-CoV-2 positive group than in the influenza tested group from the same time period in the year prior (3.1 years(interquartile range[IQR] 0–11.1) vs. 8.7 years (IQR 1.6–16.2)). One-third (n = 717) of the COVID-19 individuals missing demographic information (age, race, and/or gender). In the 2019 influenza tested group, seven individuals (0.4%) lacked a complete demographic profile. These influenza-tested individuals were more likely to be “Medical Home” patients, having at least one prior primary care visit recorded by center specific visit identifiers, or have a comprehensive history recorded in their EHR, as suggested by the presence of the corresponding CPT code.
Table 3.
Metadata results for SARS-CoV-2 tested cohort between March 1 through June 5, 2020 and influenza tested March 1 through June 5, 2019.
| SARS-CoV-2 Positive | SARS-CoV-2 Negative | Influenza tested 2019 | ||
|---|---|---|---|---|
| (n = 2155) | (n = 26,540) | (n = 1687) | ||
| Missing demographic data elements (any age, race, gender reported “Unknown”) | 717 (33%) | 4585 (17%) | 7 (0.4%) | |
| Median EHR length (median years + IQR) | 3.10 [0.0–11.1] | 6.16 [0.5–14.5] | 8.65 [1.6–16.2] | |
| Data density (institution-specific) | No Visits | 723 (34%) | 5108 (19%) | 351 (21%) |
| No PC Visit | 726 (34%) | 9559 (36%) | 469 (28%) | |
| Medical Home | 706 (33%) | 11,873 (45%) | 867 (51%) | |
| Data density (comprehensive history CPT code) | 878 (41%) | 16,665 (63%) | 1,447 (86%) | |
EHR: Electronic health record; Medical Home: At least one primary care visit before the first test date as defined using clinic location identifiers; No PC visit: Only non-primary care visit(s) before the first test date; No visits: No billing dates prior to the week before the first test date; IQR: Interquartile range; CPT: Common procedural terminology.
Phenotyping data quantity was lowest in all four metadata categories for SARS-CoV-2 positive individuals (Fig. 2). This group had fewest available records for phenotyping prior to testing. Individuals in the COVID-19 cohort were also less likely to have a comprehensive CPT code billed prior to testing relative to non-pandemic, influenza-tested EHRs (OR 0.1, 95% Confidence Interval[CI] 0.09–0.13). Odds of having complete demographic variables (age, race, gender) were also lowest in this group (OR 0.008, 95% CI 0.02–0.003).
4.4. Comparison of different phenotyping choices
Comorbidities extracted from 135 COVID-19 patient charts by three phenotyping methods (i.e., ICD-based, EHR Vendor and expert algorithm) that differed from manual chart review resulted in 129 comorbid phenotypes for additional review, many comorbidities occurring in the same EHR. Phenotyping identified 67 false observations (e.g., high blood pressure, pulmonary hypertension) and 14 true comorbidities not observed on chart review. There was no statistical difference among false negative or false positive rates between methods. Scanned forms, both transfer records from outside hospitals on inpatient admission and the aforementioned COVID-19 intake forms used in the outpatient setting, accounted for 11 false negative comorbidities. Results of phenotyping from nine commonly encountered comorbidities in the adult population from 135 records are described in Appendix Table A3.
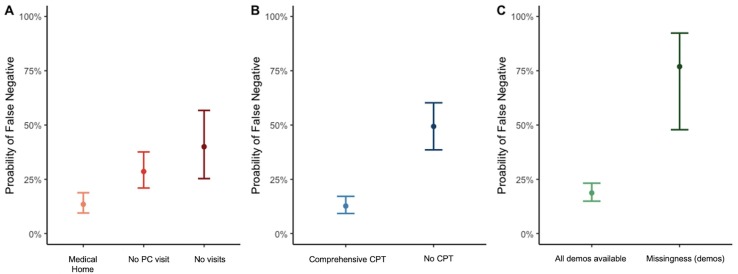
Combining phenotyping methods, significantly lower false negative rates were observed for those records with phenotyping data available from a comprehensive history, identified by the presence of a CPT code, and complete demographics reported in the EHR (Fig. 3 ). SARS-CoV-2 positive individuals with 1 of 9 comorbid phenotypes were 6 times more likely to be missed by phenotyping algorithms if a comprehensive care visit CPT (Appendix Table A4) was never recorded in the EHR (p = 4e-11, 95% CI 3.8–11.9). Increasing data density using institution-specific clinic site identifiers (e.g., “Medical Home” records with history of visit(s) at a primary care clinic site) resulted in the lowest probability of a false negative phenotype (Fig. 3A). Highest rates of false negative phenotypes were seen in those records with any missing demographic(s) (Fig. 3C).
Fig. 3.
EHR: Electronic health record; COVID+: Patients testing positive for SARS-CoV-2 during the study period; COVID-: Patients testing negative for SARS-CoV-2 during the study period; Flu 2019: Patients testing positive for influenza during the study period equivalent dates March 1 through June 5, 2019. CPT: Common procedural terminology.
Accounting for data density, either by history of a visit to a local clinic site or presence of a comprehensive CPT code, and missingness resulted in higher F-scores when phenotyping methods were combined. Increasing data density resulted in a statistically significant decrease in false negatives for the phecodes method only (No history of a primary care visit p = .001; No visits p = 3e-4). A stepwise increase was observed in false negative rates of patient comorbidities returned from the EHR from the phecodes methods along data density categories (14% for EHRs in the “Medical Home” group, having at least one primary care visit; 29% for those with some records but no primary care visits; 60% false negatives for those with any prior records at our institution). We hypothesize that this pattern was not observed for more complex algorithms due to access to additional data types including natural language processing or medications data, even for EHRs with few encounters. It is also possible that billing practices at acute care testing encounters did not preference chronic comorbid phenotypes (e.g., hypertension), thus having the largest impact on the phecodes algorithm (i.e., ICD-10 codes only).
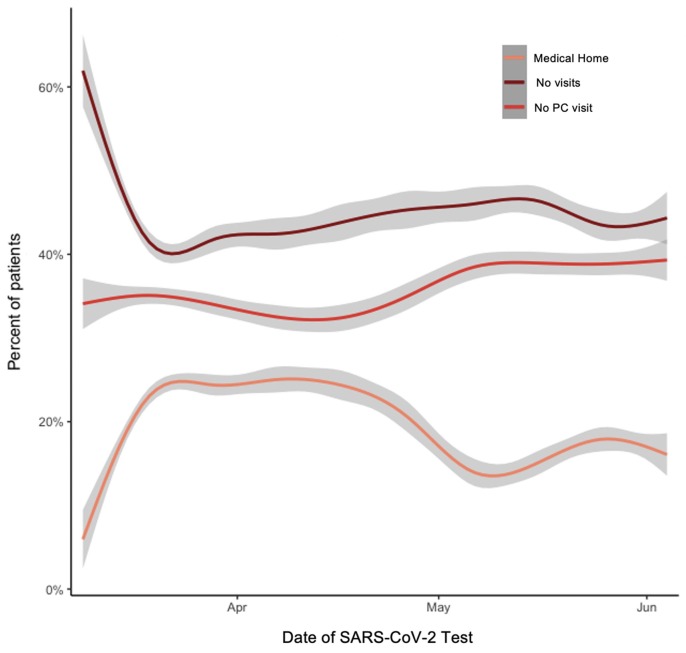
Since March, we have seen decreasing numbers of individuals with any primary care visits in our medical system (Fig. 4 ) suggesting that individuals who typically receive their medical care elsewhere are interacting with VUMC for testing for COVID-19 and treatment specifically. Early in the pandemic, testing was reserved for symptomatic individuals in our system. In May, however, screening was started at select inpatient sites (e.g., oncology clinics) as testing became more available. We hypothesize that the spike in screening tests and corresponding decrease in “Medical Home” population may be in part due to the reintroduction of elective surgeries in our system, during which time VUMC instituted testing before undergoing elective surgery (see Fig. 5 ).
Fig. 4.
Probability of false negative results for 9 comorbid phenotypes across 3 phenotyping algorithms (ICD-10 based and ICD-10 plus domain-specific algorithms provided by an EHR vendor and clinical experts) among an early COVID-19 population, March 1 through June 5, 2020. Medical Home: At least one primary care visit before the first test date as defined using clinic location identifiers; No PC visit: Only non-primary care visit(s) before the first test date; No visits: No billing dates prior to the week before the first test date; Comprehensive CPT: patients with comprehensive history CPT code; No CPT: patients without comprehensive history CPT code; All demos available: EHRs with complete age, self-reported race, and gender data elements; Missingness (demos): at least one missing demographic variable in the EHR.
Fig. 5.
Percent of total individuals tested between March 1 and June 5, 2020 grouped by data density categorized by center-specific visit identifiers. Since March, we have seen fewer individuals with any primary care visits in our medical system. Medical Home: At least one primary care visit before the first test date as defined using clinic location identifiers; No PC visit: Only non-primary care visit(s) before the first test date; No visits: No billing dates prior to the week before the first test date.
5. Discussion
The emergence of COVID-19 in the U.S. represented a public health crisis that required an urgent scientific response to understand the risks and outcomes of afflicted patients. While EHRs represent a broadly available source of observational data, the ability to extract reproducible and meaningful scientific data from them depends on an understanding of the flow of information from which the data was recorded. Methods exist for extracting research grade phenotypes from EHR data, but these approaches have been developed in relatively static circumstances. The ability of validated phenotypes to perform in the context of a rapidly changing pandemic was unknown. Lessons learned from phenotyping COVID-19 (Table 4 ) will have implications not only for the current pandemic, but for any circumstance with such rapidly shifting contexts.
Table 4.
Lessons learned1 from early phenotyping efforts during the coronavirus disease 2019 (COVID-19) pandemic.
| Domain | Challenges to Phenotyping acute patient cohorts | Description | Potential Solutions |
|---|---|---|---|
| Data | Data Availability (Completeness) | Longitudinal records may not be available for all patients. | Anticipate data quality issues with available data types including electronic health record (EHR) metadata. Consider sources of metadata indirectly related to EHR data types (e.g., geospatial and Census Block data or other “community vital signs”)[26]) to interpolate various systems processes not captured in the clinical record. |
| Data Management (Timeliness) | Discordant temporality of data streams (e.g., from operational to structured data). | Evaluate time from event to data pull; create automated systems to accommodate differences. Exploit missingness to interpolate time between variables.[27] |
|
| Data Validation (Correctness) | Patient histories may rely on data from limited visit(s) and visit types. | Evaluate ways data is gathered and recorded in your healthcare system.[25] Identify essential population and database characteristics,[28] including the degree to which a given variable tends to over or underestimate a feature or change over time. Target novel data sources and note types (e.g., clinical communications) to validate narrative or structured elements.[9], [29] |
|
| Authoring | Defined Cohorts | No reliable billing code available to identify cohorts. | Validate local testing practices (i.e., presence of laboratory testing). Assign probability of known disease;[30] evaluate data driven selection of cases or controls such as a maximum likelihood approach.[31] |
| Defined Logic | Data use requires knowledge of data cleaning processes. | Build a data dictionary documenting representation of data elements (e.g., Boolean, temporal) as well as cleaning methods. |
elements of the table were adapted with permission from Rasmussen, et al. 2019.[19]
In an early COVID-19 dataset, we found that phenotyping performance of nine common comorbidities was subject to data quantity and quality, measured using portable metrics such as missingness and CPT codes. It is important to consider that circumstances that create an increase in patient encounters, such as for SARS-CoV-2 testing, may inflate the number of EHRs available for study, but without a corresponding contribution of comprehensive data. To the degree that these patients do not reflect the existing patient population, using EHR data in observational research without accounting for data density risks mischaracterizing the population being studied.
5.1. Impact of ‘data fitness’ on phenotyping accuracy
The primary goal of this paper was to highlight healthcare experiences at our institution during the COVID-19 pandemic that have challenged assumptions present in previously validated methods of characterizing patient cohorts. As minimizing false positive rates and timeliness were important early in data collection, the average delay between the earliest confirmed lab date and ICD-10 date in our system being 7 days, defining COVID-19 cohorts was best addressed in our system by laboratory results only; however, these methods may differ among other systems with unique healthcare processes in place. In contrast to the multi-site efforts undertaken by The National COVID Cohort Collaborative to identify those tested for SARS-CoV-2 (both positive and negative) via laboratory codes (or LOINC), we sought to validate COVID-19 case definitions specific to our institution’s concurrent healthcare practices.[32], [33] Based on our experience, we suggest evaluations of and updates to case definition algorithms occur as often as healthcare processes change at your site.
We have identified sources of information bias, specifically missingness and data density, within our own COVID-19 cohort that significantly impacted phenotype performance. New data types (e.g., Respiratory Therapy notes, ventilator flow sheets) present new challenges but also new opportunities to increase certainty among reported medical histories in the EHR. It is likely that similar biases exist within COVID-19 registries at other academic and tertiary care sites. We hope that our attempts to address these findings might be instructive in developing approaches and frameworks for performing research in new healthcare environments. An unexpected finding was that those metadata identified as having a varying impact on phenotyping performance have changed since data collection.
Researchers interested in using EHR data to study COVID-19 should evaluate algorithms for identifying patient cohorts at their institution(s) and the impact of metadata, including consideration of how these characteristics change over time. Maintained logic and defined cohort selection are the means by which thoughtful data management is bridged to implementation. An important consideration in our COVID-19 registry was that not all EHR visit types provide equal amounts of data for phenotyping. Other institutions might replicate data density categories (i.e., No visits to “Medical Home”) defined using internal clinic location identifiers using natural language processing or additional CPT code types.
5.2. Best practices for phenotyping
Phenotyping an emerging infectious disease requires early characterization of risks and outcomes without knowledge of a static exposure. We continue to partner with our colleagues involved in observational cohort studies and randomized clinical trials to guide phenotyping targets. For example, work on COVID-19 cardiovascular outcomes, including the incidence of deep venous thromboses, is well underway. As before the pandemic, we validate recorded patient information (e.g., diagnoses, past medical history) using additional data sources (e.g., laboratory results, medication data), especially for those with limited or patient reported histories in our system. Assigning probabilities of certainty to phenotypes and data types within COVID-19 registries is a potential solution to minimize bias and may be aided by metadata.[28] Registry maintenance using data dictionaries tracking data cleaning methods is key to preserving defined logic over time.
5.3. Method considerations for phenotyping for acute events
Although we used chart review as a gold standard validation of our efforts, other methods may attempt data reuse to account for missingness. We envision increasing features of metadata that can be used to further stratify data, recent exposures including Census Block data and socioeconomic variables. OMOP queries have provided new data elements such as ventilator flow sheets, previously not available to researchers in the RD. Perl scripts, in development, will anticipate outcomes of interest in the COVID-19 population such as changes to lung function and post-traumatic stress disorder.
6. Limitations
The key limitation of our study is that it reflects the experience of one medical center, which, as an academic medical center, may not reflect the SARS-CoV-2 tested patient population at other sites. The fact that our clinical center draws complex patients in our region may have unduly over-populated our patient population with those high complexity patients in whom we saw the highest rate of false positive phenotypes (e.g., sickle cell disease, polypharmacy). Furthermore, our study at present fails to consider the predictive value of data types (e.g., ICD-10) with respect to desired phenotypes. For example, it is possible that more sensitive ICD-10 codes (e.g., congestive heart failure) returned fewer false positives than those less specific (e.g., hypertension) and we did not study this difference. It is important to note that due to limited sample size of phenotypes derived from manual review, a comparison between phenotypes derived from increasingly complex algorithms is underpowered and that the records used to create a more complex phenotyping algorithm from billing codes (EHR vendor) do not work off of the common data model. Thus, perceived difference among algorithm results in part due to underlying differences in methodology may be misleading. As for all EHR-based research, accuracy of phenotypes is limited to accuracy of data collection. Among metadata, our local assessment of data density (i.e., “Medical Home” per history of a primary care note) assumes that primary care notes record complete and accurate medical histories, and this may not be the case. For those records with only a COVID-19 related visit type and no previous medical history, phenotyping research must rely on self-reported data, which may be incomplete or inaccurate.
Future systems might consider ways to exploit missingness to interpolate time between variables.[27] Major sources of false negative results included data from scanned COVID-19 intake forms, and limited retrospective histories especially for episodic events (e.g., history of DVT). Future work will distinguish acute and ‘history of’ events computationally. These systems should consider methods of converting manual to automated steps, training successful queries in techniques such as active learning. Our assessment of this early registry is a limited window into a dynamic process. It is likely that COVID-19 cohorts will change over time as the virus reaches new populations, perhaps necessitating new approaches to combat bias and ascertain missingness. More sensitive ascertainment of these cohorts might be identified by a maximum likelihood approach (e.g., temperature and positive laboratory data combinations). Furthermore, targeting records with history of any outpatient note in our EHR system may yield higher reported sensitivities for local algorithms.
7. Conclusion
COVID-19 represents a phenotyping challenge that exposes many of the known difficulties of EHR phenotype development. At our institution, testing for SARS-CoV-2 infection has generated an influx of new patient encounters, EHRs rich prospectively but incomplete or absent retrospectively. Using a COVID-19 cohort validated at our institution, we found that phenotyping performance of common comorbidities was significantly impacted by data density, that we describe using institution-specific identifiers and CPT codes. We pose challenges and potential solutions to ascertaining accurate, high throughput COVID-19 phenotypes in an evolving clinical landscape. Those in EHR-based data science have enormous opportunity and responsibility to make contributions in COVID-19 research via new or existing data sources.
8. Financial Disclosure
No financial disclosure necessary.
9. Funding sources
This work was supported by Beyond PheWAS: Recognition of Phenotype Patterns for Discovery and Translation 2 R01 LM010685-09 and U01 HG011166-01S1 from NIH/NIGMS. This sponsor had no role in the processes of study design; collection, analysis and interpretation of the data; writing of the report; or the decision to submit for publication.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
AUTHOR’S ROLES:
1) Research project: A. Conception, B. Organization, C. Execution; 2) Statistical analysis: A. Design, B. Execution, C. Review and critique; 3) Manuscript: A. writing of the first draft, B. Review and critique
Lisa Bastarache: 1A, 1B, 1C, 2A, 2B, 2C, 3B
Josh Peterson: 1A, 1B, 1C, 2A, 2C, 3A, 3B
Melissa McPheeters: 1A, 1B, 2C, 3B
Quinn Wells: 1A, 1B, 1C, 3B
Eric Farber-Eger: 1B, 1C
Cosmin A. Bejan: 1A, 1B, 2C, 3B
S. Trent Rosenbloom: 1A, 1B, 2C, 3B
Kevin Johnson: 1A, 1B, 3B
Dan Roden: 1A, 1B, 2C
Daniel Fabbri: 1A
Sarah Bland: 1A, 1B, 1C, 3B
Janey Wang: 1B, 2C
Sarah DeLozier: 1A, 1B, 1C, 2A, 2C, 3A, 3B
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103777.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Reference
- 1.Guan W., Ni Z., Hu Y., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao B., Wang Y., Wen D., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J.J. Reeves, H.M. Hollandsworth, F.J. Torriani, et al., Rapid response to COVID-19: health informatics support for outbreak management in an academic health system, J. Am. Med. Inform. Assoc. Published online April 27, 2020. doi:10.1093/jamia/ocaa037. [DOI] [PMC free article] [PubMed]
- 5.Grange E.S., Neil E.J., Stoffel M., et al. Responding to COVID-19: the UW medicine information technology services experience. Appl. Clin. Inform. 2020;11(02):265–275. doi: 10.1055/s-0040-1709715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman O., Tromp G., Faucett W.A., et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet. Med. 2013;15(10):761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirby J.C., Speltz P., Rasmussen L.V., et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J. Am. Med. Inform. Assoc. 2016;23(6):1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathak J., Kho A.N., Denny J.C. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J. Am. Med. Inform. Assoc. 2013;20(e2):e206–e211. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng C., Shah N.H., Hripcsak G. Deep phenotyping: Embracing complexity and temporality—Towards scalability, portability, and interoperability. J. Biomed. Inform. 2020;105 doi: 10.1016/j.jbi.2020.103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holshue M.L., DeBolt C., Lindquist S., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danciu I., Cowan J.D., Basford M., et al. Secondary use of clinical data: the Vanderbilt approach. J. Biomed. Inform. 2014;52:28–35. doi: 10.1016/j.jbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roden D., Pulley J., Basford M., et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny J.C., Crawford D.C., Ritchie M.D., et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011;89(4):529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny J.C., Bastarache L., Ritchie M.D., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31(12):1102–1111. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho A.N., Pacheco J.A., Peissig P.L., et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci. Transl. Med. 2011;3(79) doi: 10.1126/scitranslmed.3001807. 79re1-79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deisseroth C.A., Birgmeier J., et al. ClinPhen extracts and prioritizes patient phenotypes directly from medical records to expedite genetic disease diagnosis. Genet. Med. 2019;21(7):1585–1593. doi: 10.1038/s41436-018-0381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W.-Q., Teixeira P.L., Mo H., Cronin R.M., Warner J.L., Denny J.C. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. 2016;23(e1):e20–e27. doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huckvale K., Venkatesh S., Christensen H. Toward clinical digital phenotyping: a timely opportunity to consider purpose, quality, and safety. npj Digit Med. 2019;2:88. doi: 10.1038/s41746-019-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen L., Brandt P., Jiang G., et al. Considerations for improving the portability of electronic health record-based phenotype algorithms. AMIA Annu Symp Proc. Published online. 2020:755–764. [PMC free article] [PubMed] [Google Scholar]
- 20.Hripcsak G., Albers D.J. High-fidelity phenotyping: richness and freedom from bias. J. Am. Med. Inform. Assoc. 2018;25(3):289–294. doi: 10.1093/jamia/ocx110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W.-Q. Wei, L.A. Bastarache, R.J. Carroll, et al., Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. Rzhetsky A, ed., PLOS ONE. 12(7) (2017) e0175508. doi:10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed]
- 22.New ICD-10-CM code for the 2019 Novel Coronavirus (COVID-19), April 1, 2020. Published online February 20, 2020. https://www.cdc.gov/nchs/data/icd/Announcement-New-ICD-code-for-coronavirus-3-18-2020.pdf.
- 23.Wei W.-Q., Leibson C.L., Ransom J.E., et al. The absence of longitudinal data limits accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects. Int. J. Med. Inform. 2013;82(4):239–247. doi: 10.1016/j.ijmedinf.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei W.-Q., Leibson C.L., Ransom J.E., et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J. Am. Med. Inform. Assoc. 2012;19:219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hripcsak G., Albers D.J. Next-generation phenotyping of electronic health records. J. Am. Med. Inform. Assoc. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazemore A.W., Cottrell E.K., Gold R., et al. “Community vital signs”: incorporating geocoded social determinants into electronic records to promote patient and population health. J. Am. Med. Inform. Assoc. 2016;23(2):407–412. doi: 10.1093/jamia/ocv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J.-H., Haug P.J. Exploiting missing clinical data in Bayesian network modeling for predicting medical problems. J. Biomed. Inform. 2008;41(1):1–14. doi: 10.1016/j.jbi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Sagreiya H., Altman R.B. The utility of general purpose versus specialty clinical databases for research: warfarin dose estimation from extracted clinical variables. J. Biomed. Inform. 2010;43(5):747–751. doi: 10.1016/j.jbi.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tracy J.M., Özkanca Y., Atkins D.C., Hosseini Ghomi R. Investigating voice as a biomarker: Deep phenotyping methods for early detection of Parkinson’s disease. J. Biomed. Inform. 2020;104 doi: 10.1016/j.jbi.2019.103362. [DOI] [PubMed] [Google Scholar]
- 30.N.S. Zheng, Q. Feng, V.E. Kerchberger, et al., PheMap: a multi-resource knowledge base for high-throughput phenotyping within electronic health records, J. Am. Med. Inform. Assoc. Published online September 24, 2020:ocaa104. doi:10.1093/jamia/ocaa104. [DOI] [PMC free article] [PubMed]
- 31.Zhang L., Ding X., Ma Y., et al. A maximum likelihood approach to electronic health record phenotyping using positive and unlabeled patients. J. Am. Med. Inform. Assoc. 2020;27(1):119–126. doi: 10.1093/jamia/ocz170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.H. Melissa, C. Christopher, G. Kenneth, The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J. Am. Med. Inform. Assoc. 2020 Aug 17:ocaa196. doi: 10.1093/jamia/ocaa196. Epub ahead of print. PMID: 32805036; PMCID: PMC7454687. [DOI] [PMC free article] [PubMed]
- 33.Brat G.A., Weber G.M., Gehlenborg N., et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. npj Digit Med. 2020;3:109. doi: 10.1038/s41746-020-00308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.