Abstract
The coronavirus (COVID-19) is currently the most common contagious disease which is prevalent all over the world. The main challenge of this disease is the primary diagnosis to prevent secondary infections and its spread from one person to another. Therefore, it is essential to use an automatic diagnosis system along with clinical procedures for the rapid diagnosis of COVID-19 to prevent its spread. Artificial intelligence techniques using computed tomography (CT) images of the lungs and chest radiography have the potential to obtain high diagnostic performance for Covid-19 diagnosis. In this study, a fusion of convolutional neural network (CNN), support vector machine (SVM), and Sobel filter is proposed to detect COVID-19 using X-ray images. A new X-ray image dataset was collected and subjected to high pass filter using a Sobel filter to obtain the edges of the images. Then these images are fed to CNN deep learning model followed by SVM classifier with ten-fold cross validation strategy. This method is designed so that it can learn with not many data. Our results show that the proposed CNN-SVM with Sobel filter (CNN-SVM + Sobel) achieved the highest classification accuracy, sensitivity and specificity of 99.02%, 100% and 95.23%, respectively in automated detection of COVID-19. It showed that using Sobel filter can improve the performance of CNN. Unlike most of the other researches, this method does not use a pre-trained network. We have also validated our developed model using six public databases and obtained the highest performance. Hence, our developed model is ready for clinical application.
Keywords: Image processing, Data mining, Machine learning, Deep learning, Feature extraction, Covid-19, Sobel operator, SVM, CNN
1. Introduction
Coronavirus disease 2019 (COVID-19) has been spreading unprecedentedly across the globe from the beginning of 2020. The clinical characteristics of COVID-19 include respiratory symptoms, fever, cough, dyspnea, pneumonia, and fatigue during early stages [[1], [2], [3]]. The COVID-19 also affects the cardiovascular and respiratory systems and may lead to multiple organ failure or acute respiratory distress in critical cases and is highly contagious [[3], [4], [5], [6], [7]]. Therefore, COVID-19 infections are a crucial healthcare challenge around the world and has become a global threat [8].
The World Health Organization (WHO) declared the outbreak a “public health emergency of international concern” on 30th January 2020. Reverse-transcription polymerase chain reaction (RT-PCR) is generally used to confirm the incidence of COVID-19 [9]. Contrary to RT-PCR, chest X-ray is found to be low cost, fast, and widely available for the early diagnosis and screening of COVID-19 [10]. It is economical and can be made available in most of the clinical settings, even in third world countries. This non-invasive imaging modality can help to detect specific characteristic manifestations in the lung related to the COVID-19 [11]. Indeed, it is especially useful (and viable) for symptomatic patients. One of the bottlenecks of the system is that expert radiologists are required to interpret the radiography images. As such, computer-aided diagnostic systems (CAD) can help the radiologists to detect COVID-19 cases accurately and rapidly. There are few deep learning (DL)-based techniques proposed for such automated detection using X-ray radiographs [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]]. These techniques have exhibited great success by extracting relevant hidden signatures (features) from the medical images using deep learning [22].
The main contributions of this work are listed as follows:
-
•
New private database collected by the authors is used.
-
•
Proposed model is tested using six public databases and the results are found to be better than most of the existing state of the art methods.
-
•
Sobel filter is found to improve the performance of CNN.
-
•
Obtained highest classification performance for all databases.
Nowadays, machine learning (ML) methods are widely used for Covid-19. These methods can improve the diagnosis accuracy of clinicians. However, there are few limitations in these methods. For example, feature extraction is a challenging step in almost all ML methods. So, automatic feature extraction is a great improvement in this field. Among the different ML methods, deep learning (DL) can solve this challenge. It can do feature extraction automatically. In addition, when there are large amount of data, its performance is better than other ML methods. Consequently, nowadays DL is used to diagnose different diseases [[23], [24], [25], [26], [27], [28], [29]] such as COVID-19 [21,30]. An overview of the works done on automated detection of COVID-19 using DL is presented in Table 1 . In this table, the recently published DL works on COVID-19 detection using X-ray and CT scan images are listed. However, almost all of them used pre-trained networks using public databases.
Table 1.
Summary of works done on automated detection of COVID-19 using DL techniques with X-ray and CT images.
| Study | Modality | Number of Cases (or Images) | Network |
|---|---|---|---|
| Wang et al. [31] | X-ray | 13,975 images | Deep CNN |
| Hall et al. [11] | X-ray | 455 images | VGG-16 and ResNet-50 |
| Farooq et al. [12] | X-ray | 5941 images | ResNet-50 |
| Hemdan et al. [14] | X-ray | 50 images | DesnseNet, VGG16, MobileNet v2.0 etc. |
| Abbas et al. [15] | X-ray | 196 images | CNN with transfer learning |
| Minaee et al. [16] | X-ray | 5000 images | DenseNet-121, SqueezeNet, ResNet50, ResNet18 |
| Zhang et al. [17] | X-ray | 213 images | ResNet, EfficientNet |
| Apostolopoulos et al. [19] | X-ray | 3905 images | MobileNet v2.0 |
| Narin et al. [20] | X-ray | 100 images | InceptionResNetV2, InceptionV3, ResNet50 |
| Luz et al. [32] | X-ray | 13, 800 images | EfficientNet |
| Brunese et al. [33] | X-ray | 6523 images | VGG-16 and transfer learning |
| Ozturk et al. [34] | X-ray | Two publically available databases were used where images were updated regularly. | Darknet-19 |
| Khan et al. [35] | X-ray | 1251 images | CNN |
| Silva et al. [36] | CT scans | 2482 images | A slice voting-based approach extending the Efficient Net Family of deep artificial neural networks |
| Luz et al. [32] | X-ray | 13, 800 images | Efficient Net |
| Ozturk et al. [34] | X-ray | Two publically available databases were used where images were updated regularly. | Darknet-19 |
| Khan et al. [35] | X-ray | 1251 images | CNN |
| Haghanifar et al. [37] | X-ray | 7700 images | DenseNet-121 |
| U-Net | |||
| Oh et al. [38] | X-ray | 502 images | DenseNet |
| U-Net | |||
| Tartaglione et al. [39] | X-ray | 5 different databases | ResNet |
| Rahimzadeh et al. [40] | X-ray | 11,302 images | Xception and ResNet50V2 |
| Jamil et al. [41] | X-ray | 14,150 images | Deep CNN |
| Horry et al. [42] | X-ray | 60,798 images | VGG, Inception, Xception, and Resnet |
| Elasnaoui et al. [43] | X-ray | 6087 images | inception_Resnet_V2 and Densnet201 |
| And CT | |||
| Ardakani et al. [44] | CT | 1020 | ResNet-101, ResNet-50, ResNet-18, GoogleNet, SqueezeNet, VGG-19, AlexNet |
This paper is organized as follows. The computer aided diagnosis (CAD) based on the proposed deep learning to detect COVID-19 is described in Section 2. The results obtained is presented and discussed in Section 3. Finally, the paper concludes with brief summary in Section 4.
2. CADS based COVID-19 diagnosis using 2D-CNN
Nowadays, many CAD systems have been developed using deep learning techniques to detect various diseases, including COVID-19, have attracted the attention of many researchers. The CAD based deep learning methods require huge database to yield highest performance.
2.1. X-ray database
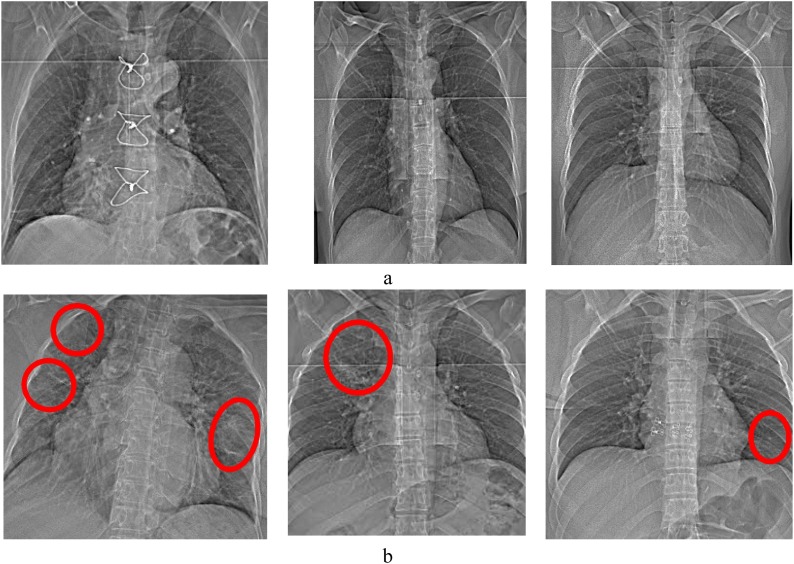
In this study, 333 chest X-ray images comprising of 77 images of COVID-19 patients and 256 images of normal subjects were recorded at Omid Hospital in Tehran. They are collected from February 2020 to April 2020. The mean and standard deviation of their age are 49.5 ± 18.5 years old. 55% of cases are female. Three radiologists checked each image and determined whether a case has Covid-19 or not. Ethical approval of these data was also obtained. Some examples of these data can be seen in Fig. 1 . They show the typical X-ray images of normal and COVID-19 patients.
Fig. 1.
Sample X-ray images: a) healthy subjects and b) COVID-19 patients. The marked region indicates the infected parts.
2.2. Proposed method
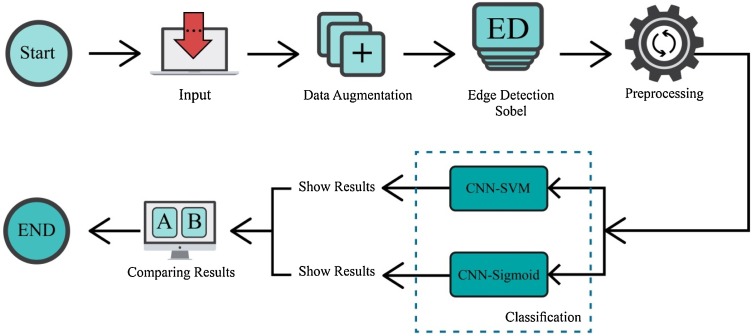
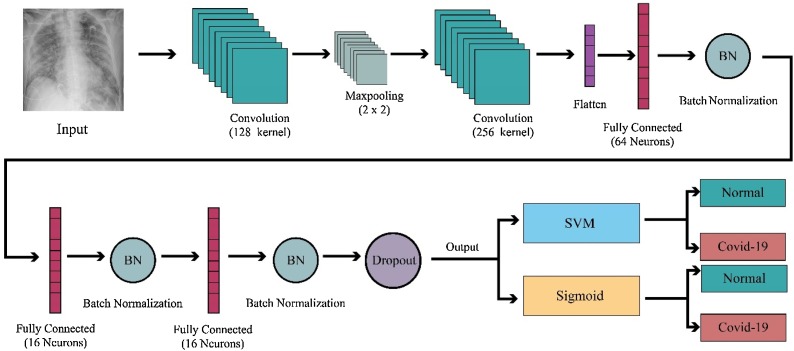
This paper proposes a novel 2D-CNN architecture to detect COVID-19 using X-ray images. The 2D-CNN with a number of convolutional layers, max-pooling, and fully connected (FC) layers are used. In our methodology, support vector machine (SVM) is used instead of the sigmoid activation function in fully connected layers to obtain highest classification performance. The proposed CAD system is shown in Fig. 2 .
Fig. 2.
Proposed methodology used for the automated detection of COVID-19 patients using X-ray images.
As shown in Fig. 2, X-ray images are fed to our CAD system. The data augmentation technique is adopted to prevent the overfitting and increase the input data size. Each of the existing images in the dataset undergoes augmentation to obtain multiple new ones. The number of generated images differs depending on the class of the original image. Width and height shift, changing brightness and rotation were used with parameter {0.10, 0.15}. These eight transformation methods were used to generate eight new examples for each image in positive class. Two or three of the eight possible transformations are used randomly to generate new examples for each image in negative class. As the number of positive and negative examples was not the same, we applied data augmentation more on positive cases to reduce the bias in the performance. At the end of this step, about 45% of cases were positive and about 55% were negative.
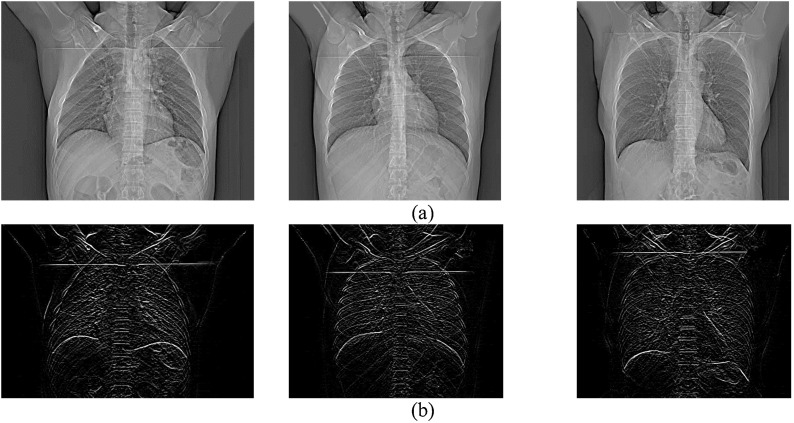
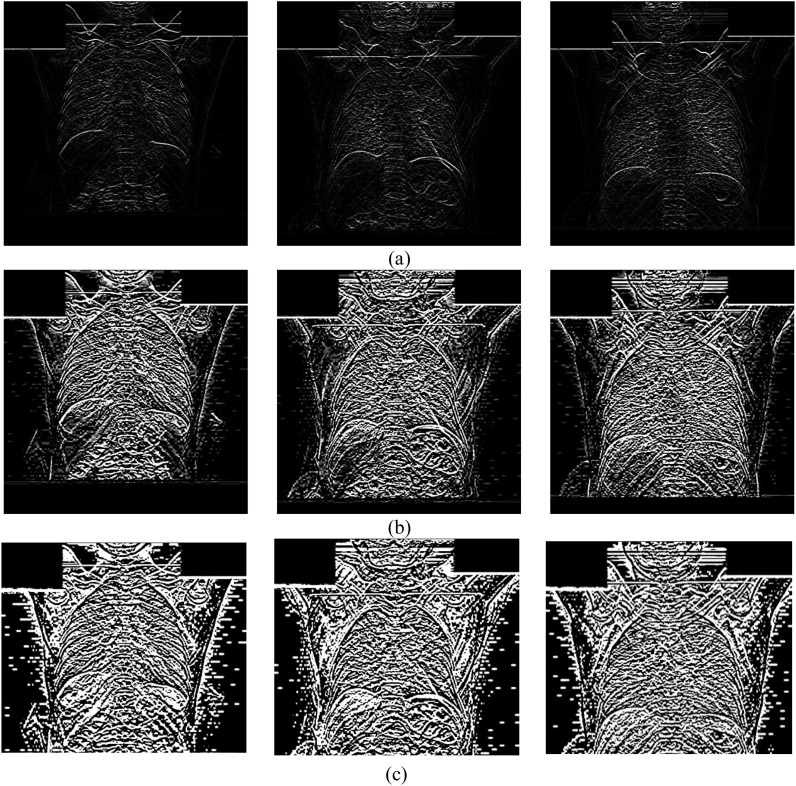
In the next step, we performed edge detection on the images to improve the deep networks classification performance. Sobel and Canny are among popular edge detection methods [45]. The computational complexity of Canny is higher compared to Sobel [46]. In our experiments, Sobel was able to provide us with acceptable results. Therefore, we chose Sobel over Canny to reduce computational complexity without sacrificing performance. Sobel employs two separate filters to extract vertical and horizontal edges. Using just one of the vertical or horizontal filters leads to much noise on the results. Therefore, we employed both of them simultaneously to avoid such an issue. An example of applying Sobel filter on the images is shown in Fig. 3 . The noise enhancement of Sobel is controlled by setting its filter size appropriately. The effect of different filter sizes is demonstrated in Fig. 4 . It is evident that the appropriate size is 3 (Fig. 4.a) which has reasonable noise level and extracts useful edge information. The filter sizes of 5 (Fig. 4.b) and 7 (Fig. 4.c) have flooded the image with noisy pixels which impair CNN training.
Fig. 3.
Sample images: (a) original (b) after applying Sobel filter.
Fig. 4.
Results of applying various filter sizes of: (a) 3, (b) 5 and (c) 7.
Using images with original dimensions (512 512) increases the training complexity of the 2D-CNN network without noticeable performance gain. Moreover, data normalization almost always leads to better training results. Therefore, as a pre-processing step, the images are resized to 100 100 and normalized to the interval [0,1]. The preprocessed images are fed to the convolutional layers of the 2D-CNN network to extract the features. Then, the classification operation is accomplished by FC layers using two methods: (i) sigmoid and (ii) SVM separately. The generated results are compared to select the best performing method. The motivation of employing SVM in the last layer of the 2D-CNN is improvement of classification performance. SVM is a robust binary classifier which draws a decision boundary between samples of the two classes such that the gap between them is maximized.
2.3. CNN architecture
Nowadays, 2D-CNN networks are employed in many medical applications, including the diagnosis of COVID-19 using X-ray images [47]. These deep learning networks consists of three main layers, convolutional layers, pooling, and fully connected (FC) layers [25]. The convolutional layers are responsible for extracting features from images. Max-pooling layers are often applied to reduce the features in CNN architectures. The last part of 2D-CNN is FC, and in the previous layer, there is an activation function that is responsible for classification. Usually, the Softmax function is employed. The Sigmoid activation function has been proved to perform efficiently in binary classification problems in this deep learning architecture. In 2D-CNNs, replacing Sigmoid with support vector machine (SVM) may lead to better results. Such achievement using SVM has been reported in some of the prior works such as [48].
In this work, at first, the number of data is increased using data augmentation algorithm. Data augmentation is done by using width shift range, height shift range, and rotation techniques. Using this method, the data is increased from 333 to 1332 images. Then, a 2D-CNN with sigmoid activation function is used to classify X-ray images. In addition, binary SVM is also used in the 2D-CNN network for classification. The hinge error function is used to obtain best results when using SVM in 2D-CNN. More details about the proposed 2D-CNN architecture is shown in Table 2 and Fig. 5 .
Table 2.
Details of parameters used in the proposed CNN architecture.
| Number of Kernels related to first and second connection | Size of the convolution kernels | Size of the max pooling kernels | Number of neurons in the Fully Connected layer | Number of neurons in the output layer | Size of the Dropout layer | Number of batch size | Number of epochs | Value of validation data | Optimizer function | Activator function | Loss function for CNN + Sigmoid | Loss function for CNN + SVM | SVM function kernel | Output layer classifiers |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 128 and 256 | 3*3 | 2*2 | 64, 32 and 16 | 2(health and sick) | 0.2 | 32 | 100 | 0.3 and 0.2 | Adam | ReLU | binary cross entropy | Hinge | Linear | Sigmoid and SVM |
Fig. 5.
Proposed CNN architecture for the automated detection of COVID-19 patients using X-ray images.
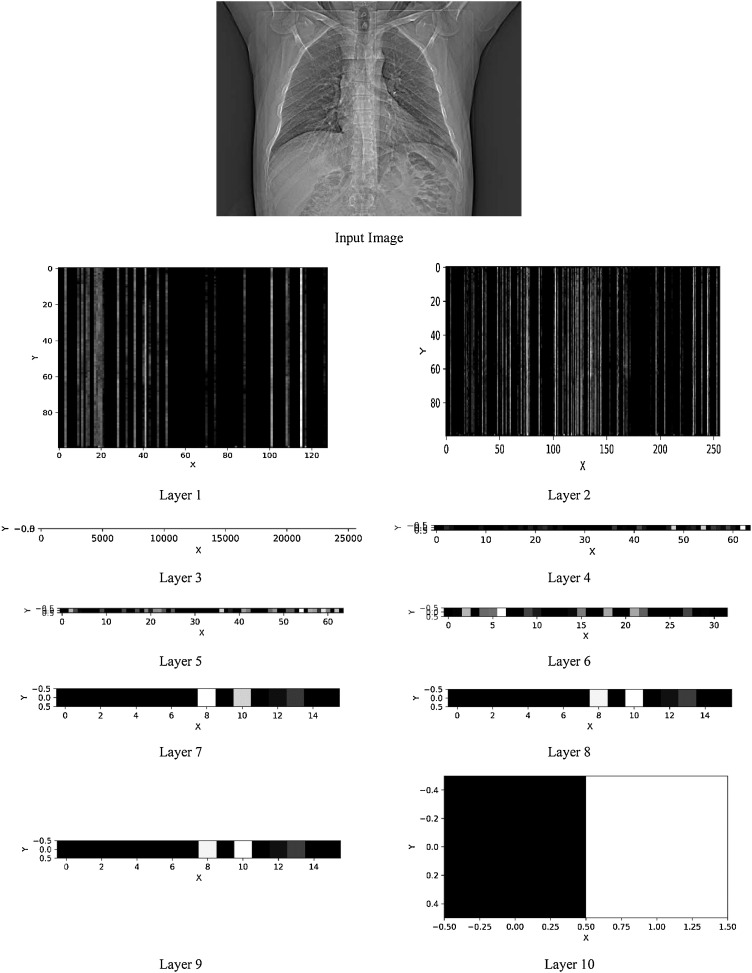
As CNN operates as a black box, its training steps are not clear. So, when using this algorithm, one of the important challenges is checking the correct training process. To verify that the CNN training is sound, the outputs of its layers have been inspected. In each generated image, the x axis shows the input data and y axis shows the output of a layer. The output obtained at the different layers of CNN are shown in Fig. 6 . The size of input data in first and second layer (convolutional layers) is equal to the number of filters in that layer. The size of input to the third layer onwards (fully-connected layers) is equal to the number of neurons in the layer. In these images, the brighter regions indicate more weight and more features can be extracted from them.
Fig. 6.
Output obtained at different layers of CNN.
2.4. Performance measures
In this study, to evaluate the performance of proposed methods, various evaluation metrics have been used and they are given below:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
In these equations, true positive (TP) is the correct classification of positive class. False-negative (FN) is the incorrect prediction of the positive case. True negative (TN) is the correct classification of the samples in the negative class. False-positive (FP) is the incorrect prediction of the negative case. In this work, positive class is symptom of COVID-19 and normal class is negative class.
3. Results and discussion
In this section, the results of our proposed CNN-SVM and CNN-Sigmoid methods and its combination with Sobel filter are provided. All simulations are done using Keras library have been conducted with back-end TensorFlow. The COVID-19 X-ray images database is obtained from Omid Hospital, Tehran, Iran. In this work, total number of 1332 (total images number is 333, which is increased to 1332 after the data augmentation operation) images are used. The results are obtained in two modes: (i) CNN network with sigmoid output layer and (ii) CNN network with SVM output layer with 10-fold cross-validation strategy. In order to validate the proposed method, we have tested with another public database named as augmented COVID-19 X-ray images database [49]. The experiment results are presented in Figs. 7 to 14.
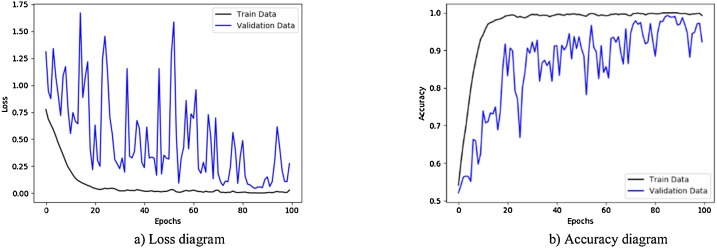
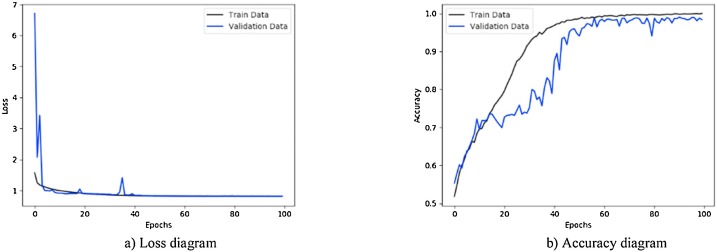
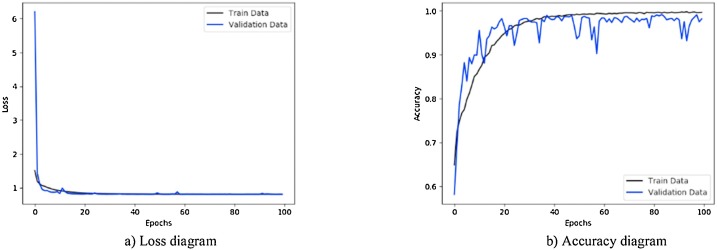
Fig. 7.
Performance metrics of CNN-sigmoid method using private database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
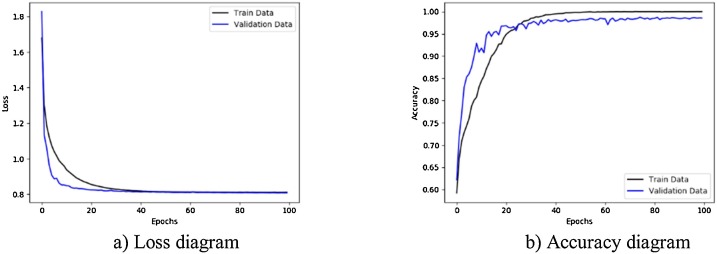
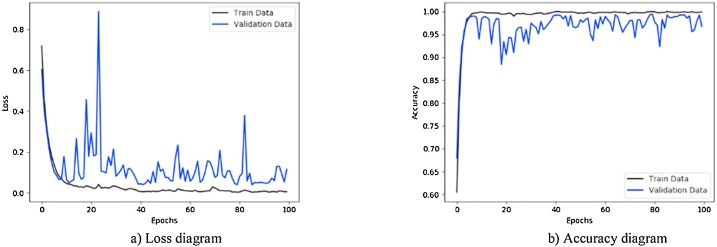
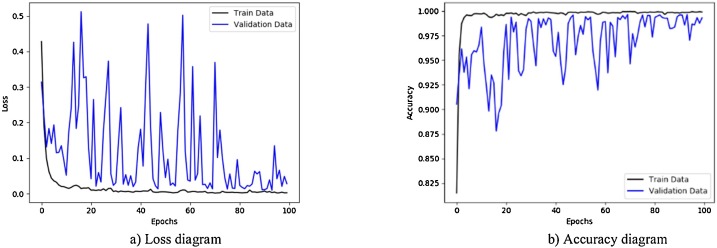
Fig. 14.
Performance metrics of CNN-SVM method with Sobel operator using augmented COVID-19 X-ray images database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
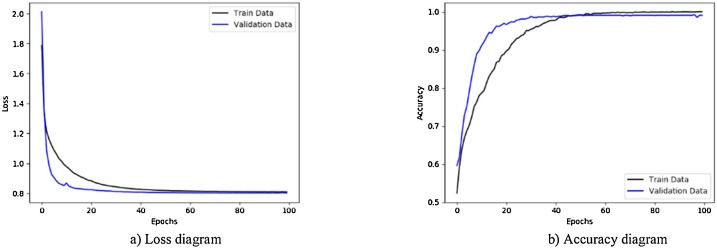
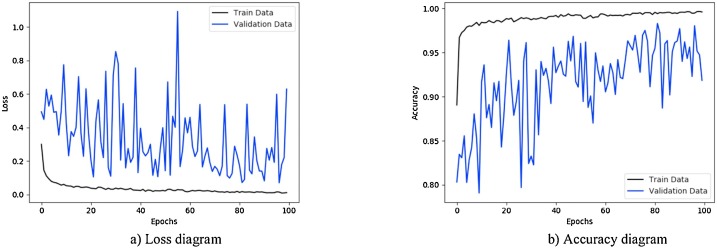
Fig. 7 illustrates the results obtained using private database with CNN-sigmoid method with 10-fold cross-validation. Figs. 8 to 10, shows the private database results obtained by applying CNN-SVM, CNN-sigmoid with Sobel operator, and CNN-SVM with Sobel operator, respectively with 10-fold cross-validation.
Fig. 8.
Performance metrics of CNN-SVM method using private database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
Fig. 10.
Performance metrics of CNN-SVM with Sobel operator method using private database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
Figs. 11 to 14, show the results obtained by applying CNN-Sigmoid, CNN-SVM, CNN-sigmoid with Sobel, and CNN-SVM with Sobel operator respectively with 10-fold cross-validation strategy using augmented COVID-19 X-ray images database.
Fig. 11.
Performance metrics of CNN-sigmoid method using augmented COVID-19 X-ray images database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
As can be seen in Figs. 7 to 14, during the learning, the loss of the algorithms is decreased and their accuracy is increased. These figures make it possible to observe the effect of using Sobel and data augmentation with CNN-Sigmoid and CNN-SVM algorithms. Fig. 7, Fig. 8 belong to CNN-Sigmoid and CNN-SVM methods, respectively. The performance of these methods accompanied with Sobel operator is illustrated in Fig. 9, Fig. 10. Careful comparison of Figs. 7 with 9 and 8 with 10 reveals that Sobel has accelerated the convergence of both methods.
Fig. 9.
Performance metrics of CNN-sigmoid with Sobel operator method using private database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
As a further enhancement, the data augmentation was added to the four experiments mentioned above. Fig. 11, Fig. 12, Fig. 13, Fig. 14 are related to scenarios CNN-Sigmoid + data-augmentation, CNN-SVM + data-augmentation, CNN-Sigmoid + Sobel + data-augmentation, and CCN-SVM + Sobel + data-augmentation. Comparing Figs. 7 with 11, 8 with 12, 9 with 13 and 10 with 14 shows that data augmentation leads to performance improvement just like Sobel.
Fig. 12.
Performance metrics of CNN-SVM method using augmented COVID-19 X-ray images database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
Fig. 13.
Performance metrics of CNN-sigmoid method with Sobel operator using augmented COVID-19 X-ray images database: (a) loss function curve, and b) accuracy curve with 10-fold cross-validation strategy.
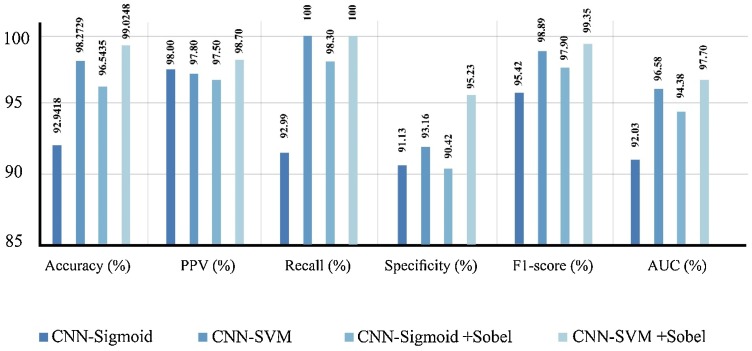
Table 3, Table 4 depict the results obtained using various combination of networks with private database and augmented COVID-19 X-ray images database, respectively. Table 3 clearly shows the effect of using sigmoid or SVM classifiers and Sobel Filter when we used in our proposed method with our database.
Table 3.
Various performance measures obtained using different combination of methods.
| Methods | Accuracy (%) |
PPV (%) | Recall (%) | Specificity (%) | F1-score (%) | Loss | AUC | ||
|---|---|---|---|---|---|---|---|---|---|
| AVG | Min | Max | |||||||
| CNN-Sigmoid | 92.9418 | 89.3256 | 95.1267 | 98.00 | 92.99 | 91.13 | 95.42 | 0.2327 | 0.9203 |
| CNN-SVM | 98.2729 | 96.2564 | 99.0224 | 97.80 | 100 | 93.16 | 98.89 | 0.8088 | 0.9658 |
| CNN-Sigmoid + Sobel | 96.5435 | 93.1657 | 98.9652 | 97.50 | 98.30 | 90.42 | 97.90 | 0.1368 | 0.9438 |
| CNN-SVM + Sobel | 99.0248 | 97.9521 | 100 | 98.70 | 100 | 95.23 | 99.35 | 0.8031 | 0.9770 |
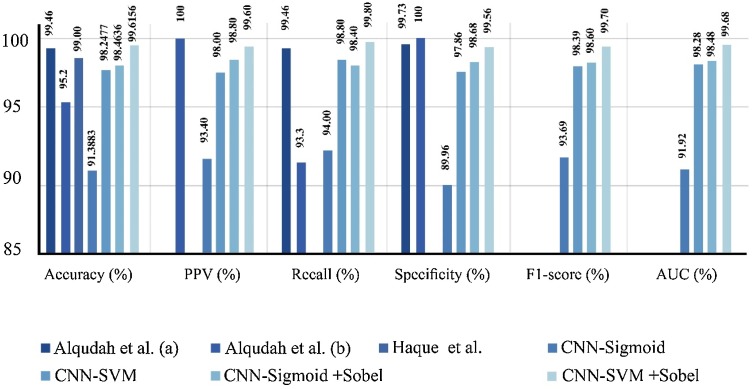
Table 4.
Evaluation performance measures obtained by applying different algorithms and combination of our methods using augmented COVID-19 X-ray images database.
| Methods | Accuracy (%) | PPV (%) | Recall (%) | Specificity (%) | F1-score (%) | Loss | AUC |
|---|---|---|---|---|---|---|---|
| Alqudah et al. (a) [50] | 99.46 | NA | 99.46 | 99.73 | NA | NA | NA |
| Alqudah et al. (b) [51] | 95.2 | 100 | 93.3 | 100 | NA | NA | NA |
| Haque et al. [52] | 99.00 | NA | NA | NA | NA | NA | NA |
| CNN-Sigmoid | 91.3883 | 93.40 | 94.00 | 89.96 | 93.69 | 0.6894 | 0.9192 |
| CNN-SVM | 98.2477 | 98.00 | 98.80 | 97.86 | 98.39 | 0.8044 | 0.9828 |
| CNN-Sigmoid + Sobel | 98.4636 | 98.80 | 98.40 | 98.68 | 98.60 | 0.0100 | 0.9848 |
| CNN-SVM + Sobel | 99.6156 | 99.60 | 99.80 | 99.56 | 99.70 | 0.8047 | 0.9968 |
It can be noted from Table 3 that, SVM classifier experiences loss values higher than Sigmoid classifier. At first glance this observation might suggest that Sigmoid classifier is superior to SVM. However, SVM has clearly outperformed Sigmoid classifier. This counterintuitive scenario is due to the difference between loss function of SVM and Sigmoid classifier. Comparing the first with third row and second with forth row show that Sobel filter can be used to improve the performance of classification.
Table 4 shows the evaluation performance measures obtained by applying different algorithms and combination of our methods using augmented COVID-19 X-ray images database.
It can be noted from Table 4 that Sobel operator improved the performance of CNN-Sigmoid and CNN-SVM approaches in detecting COVID-19 significantly.
For better comparison between the achieved results in Table 3, Table 4, the results are also illustrated in Fig. 15, Fig. 16 . They show the impact of using Sobel filtering in our algorithms. As SVM is a more robust classifier, when it is used in our algorithms, the performance has improved.
Fig. 15.
Performance obtained using different methods with our private database for COVID-19 diagnosis.
Fig. 16.
Performance obtained using different methods with augmented COVID-19 X-ray images database for COVID-19 diagnosis.
Our proposed method is also tested with six public databases to evaluate the performance of our developed model. The public database can be accessed from this link: https://www.kaggle.com [[53], [54], [55], [56], [57], [58]]. The details of the database and results obtained using our database are provided in Table 5 . It can be noted from this table that using the Sobel filter can improve the performance of our algorithm. In all tests, using Sobel filter has a positive impact on the results. Also, CNN-SVM + Sobel performed better than others combinations. For all databases, CNN-Sigmoid + Sobel performed better than the rest of the combinations.
Table 5.
Evaluation metrics obtained for our proposed method using different public databases.
| Accuracy (%) |
Other Performance Measurement Factors |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Database | Collected from | Number of cases | Method | AVG | Min | Max | PPV (%) | Recall (%) | Specificity (%) | F1-score (%) | Loss | AUC |
| [53] | Bangladesh | 1820 | CNN-Sigmoid | 91.39 | 90.02 | 92.56 | 93.40 | 94.00 | 89.93 | 93.70 | 0.69 | 0.92 |
| CNN-SVM | 98.25 | 96.35 | 99.06 | 98.00 | 98.80 | 97.87 | 98.40 | 0.80 | 0.98 | |||
| CNN-Sigmoid + Sobel | 98.46 | 96.25 | 99.63 | 98.80 | 98.40 | 98.68 | 98.60 | 0.01 | 0.98 | |||
| CNN-SVM + Sobel | 99.61 | 97.98 | 100 | 99.60 | 99.80 | 99.57 | 99.70 | 0.80 | 0.99 | |||
| [54] | India | 1160 | CNN-Sigmoid | 96.47 | 93.56 | 97.98 | 96.00 | 100 | 92.86 | 97.96 | 0.20 | 0.96 |
| CNN-SVM | 97.82 | 94.89 | 99.28 | 97.10 | 100 | 95.46 | 98.53 | 0.80 | 0.98 | |||
| CNN-Sigmoid + Sobel | 99.56 | 98.67 | 100 | 99.30 | 100 | 99.26 | 99.65 | 0.01 | 0.99 | |||
| CNN-SVM + Sobel | 99.98 | 97.59 | 100 | 99.95 | 100 | 99.97 | 99.97 | 0.79 | 0.99 | |||
| [55] | Italy | 1550 | CNN-Sigmoid | 85.92 | 83.62 | 89.65 | 87.30 | 86.20 | 83.75 | 86.75 | 1.44 | 0.85 |
| CNN-SVM | 86.60 | 82.78 | 89.63 | 97.30 | 87.30 | 70.99 | 92.03 | 0.84 | 0.84 | |||
| CNN-Sigmoid + Sobel | 94.97 | 93.04 | 97.05 | 97.80 | 96.50 | 85.07 | 97.15 | 0.15 | 0.91 | |||
| CNN-SVM + Sobel | 96.86 | 94.67 | 98.14 | 96.80 | 99.70 | 78.56 | 98.23 | 0.82 | 0.89 | |||
| [56] | India | 1120 | CNN-Sigmoid | 97.54 | 95.63 | 99.04 | 96.60 | 99.40 | 95.81 | 97.98 | 0.07 | 0.97 |
| CNN-SVM | 99.10 | 97.46 | 100 | 99.50 | 98.80 | 99.37 | 99.15 | 0.80 | 0.99 | |||
| CNN-Sigmoid + Sobel | 99.46 | 98.73 | 99.98 | 98.90 | 100 | 99.05 | 99.45 | 0.01 | 0.99 | |||
| CNN-SVM + Sobel | 99.92 | 98.91 | 100 | 99.80 | 100 | 99.84 | 99.90 | 0.80 | 0.99 | |||
| [57] | Singapore | 460 | CNN-Sigmoid | 89.67 | 86.07 | 92.56 | 92.90 | 92.70 | 83.42 | 92.80 | 0.33 | 0.88 |
| CNN-SVM | 97.61 | 94.92 | 99.46 | 99.70 | 96.50 | 99.33 | 98.07 | 0.80 | 0.98 | |||
| CNN-Sigmoid + Sobel | 98.04 | 96.35 | 99.79 | 99.10 | 98.70 | 98 | 98.90 | 0.05 | 0.98 | |||
| CNN-SVM + Sobel | 99.35 | 97.43 | 100 | 99.10 | 100 | 98 | 99.55 | 0.79 | 0.99 | |||
| [58] | Unknown | 1930 | CNN-Sigmoid | 97.50 | 95.68 | 99.63 | 98.10 | 99.00 | 73.32 | 98.55 | 0.10 | 0.86 |
| CNN-SVM | 97.30 | 94.57 | 98.79 | 97.90 | 99.30 | 66.69 | 98.60 | 0.82 | 0.83 | |||
| CNN-Sigmoid + Sobel | 98.18 | 95.76 | 98.67 | 97.90 | 100 | 71.64 | 98.94 | 0.15 | 0.86 | |||
| CNN-SVM + Sobel | 98.07 | 96.38 | 99.46 | 97.90 | 99.90 | 71.64 | 98.89 | 0.81 | 0.86 | |||
Meanwhile, in Table 6 , the results of the proposed method applied on our database are compared with other researches who used different databases. Accordingly, the performance of our proposed method is better than other researches.
Table 6.
Comparison of proposed CNN-SVM + Sobel method using private database with other methods in detecting COVID-19 using X-ray images from different private databases.
| Study | Number of Cases | Network | Train-Test | Evaluation Metrics |
|---|---|---|---|---|
| Hall et al. [11] | 455 images | VGG-16 and ResNet-50 | 10-fold | AUC: 0.997 |
| Hemdan et al. [14] | 50 images | DesnseNet, VGG16, MobileNet v2.0 etc. | 80–20% | F1 score: 91% |
| Abbas et al. [15] | 196 images | CNN with transfer learning | 70–30% | Accuracy: 95.12% |
| Sensitivity: 97.91% | ||||
| Specificity: 91.87% | ||||
| PPV: 93.36% | ||||
| Zhang et al. [17] | 213 images | ResNet, EfficientNet | 5-fold | Sensitivity: 71.70% |
| AUC: 0.8361 | ||||
| Narin et al. [20] | 100 images | ResNet50 | 10-fold | Accuracy: 98% |
| Ozturk et al. [34] | 625 images | Darknet-19 | 5-fold | Accuracy: 98.08% |
| Khan et al. [35] | 1251 images | CNN | 4-fold | Accuracy: 89.6% |
| Sensitivity: 98.2% | ||||
| PPV: 93% | ||||
| Iwendi et al. [59] | NA | Random Forest algorithm | NA | Accuracy: 94% |
| boosted by the AdaBoost algorithm | F1-score: 86% | |||
| Haghanifar et al. [37] | 780 images | DenseNet-121 | 75–25% | Accuracy: 87.21% |
| U-Net | ||||
| Oh et al. [38] | 502 images | DenseNet | 80–20% | Accuracy: 91.9% |
| U-Net | ||||
| Tartaglione et al. [39] | 137 images | ResNet | 70–30% | Accuracy: 85% |
| Proposed Method | 1332 images | CNN-SVM + Sobel | 10-fold | Accuracy: 99.02% |
| Sensitivity: 100% | ||||
| Specificity: 95.23% | ||||
| AUC: 0.9770 |
Fig. 15 shows the performance obtained using different proposed methods with our private database for automated detection of COVID-19 patients using X-ray images. Fig. 16 shows the performance obtained using various proposed methods with augmented COVID-19 X-ray images database for COVID-19 diagnosis. Fig. 15, Fig. 16 clearly show that our proposed CNN-SVM + Sobel model has performed better than rest of the methods on our database and augmented COVID-19 X-ray images database respectively. Our proposed method has performed better even using six public databases.
Advantages of our proposed method are as follows:
-
1.
We collected a new database to validate our developed model.
-
2.
Our proposed method is also tested on six public databases and showed excellent performance.
-
3.
Data augmentation is used to enable it works with small databases.
-
4.
Sobel filter is used to improve the performance of our method
Limitations of our proposed method are as follows:
-
1.
Computational cost of different deep learning algorithm is high.
-
2.
Limitation of input data is another weakness of our algorithm.
4. Conclusion
COVID-19 is currently one of the most life-threatening diseases endangering the health of many people globally. One of the main features of this disease is its rapid prevalence among people in the community. In this work, we have developed a novel COVID-19 detection system using X-ray images. In this work, we have used 333 X-ray images (77 COVID-19 + 256 normal) from Omid Hospital, Tehran to develop the model. First the images are subjected to Sobel filter to obtain the contours of the images and then fed to CNN model followed by SVM classifier. Our method is able to detect the COVID-19 cases correctly with an accuracy of 99.02%. The developed model has also yielded highest detection accuracy using six public databases. Hence, this justifies that our developed model is robust and accurate. For better evaluation of our proposed method, the final model (the best model created during 10-fold validation) was tested on some new data which their labels were determined by two experts. It could classify these new data precisely. In future, we intend to use this model to detect other chest related diseases like cancer, pneumonia, cystic fibrosis, infection, and chronic obstructive pulmonary disease (COPD).
CRediT authorship contribution statement
Danial Sharifrazi: Conceptualization, Methodology, Software, Validation, Investigation, Data curation, Visualization. Roohallah Alizadehsani: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing, Visualization. Mohamad Roshanzamir: Conceptualization, Methodology, Writing - review & editing. Javad Hassannataj Joloudari: Conceptualization, Methodology, Writing - review & editing. Afshin Shoeibi: Conceptualization, Methodology, Investigation, Resources, Writing - review & editing. Mahboobeh Jafari: Conceptualization, Methodology, Writing - review & editing. Sadiq Hussain: Conceptualization, Methodology, Writing - review & editing. Zahra Alizadeh Sani: Writing - original draft, Writing - review & editing, Supervision. Fereshteh Hasanzadeh: Investigation, Data curation, Writing - original draft. Fahime Khozeimeh: Writing - original draft, Writing - review & editing, Investigation. Abbas Khosravi: Writing - original draft, Writing - review & editing, Supervision. Saeid Nahavandi: Writing - original draft, Writing - review & editing, Project administration, Funding acquisition. Maryam Panahiazar: Writing - original draft, Writing - review & editing. Assef Zare: Writing - original draft, Writing - review & editing. Sheikh Mohammed Shariful Islam: Writing - original draft, Writing - review & editing, Project administration. U. Rajendra Acharya: Writing - original draft, Writing - review & editing, Project administration.
Declaration of Competing Interest
The authors have no competing interests to declare.
References
- 1.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alizadehsani R., et al. Risk factors prediction, clinical outcomes, and mortality of COVID-19 patients. medRxiv. 2020 doi: 10.1002/jmv.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. 2020/02/15/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holshue M.L., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S., et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) MedRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes J.C., et al. IKONOS: an intelligent tool to support diagnosis of COVID-19 by texture analysis of X-ray images. Res. Biomed. Eng. 2020 doi: 10.1007/s42600-020-00091-7. 2020/09/03. [DOI] [Google Scholar]
- 11.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv. 2020;2004:02060. [Google Scholar]
- 12.Farooq M., Hafeez A. Covid-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv. 2020;2003:14395. [Google Scholar]
- 13.Chowdhury M.E., et al. Can AI help in screening viral and COVID-19 pneumonia? arXiv preprint arXiv. 2020;2003:13145. [Google Scholar]
- 14.Hemdan E.E.-D., Shouman M.A., Karar M.E. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv. 2020;2003:11055. [Google Scholar]
- 15.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint arXiv. 2020;2003:13815. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. arXiv preprint arXiv. 2020;2004:09363. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., Xie Y., Li Y., Shen C., Xia Y. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv. 2020;2003:12338. [Google Scholar]
- 18.Wang L., Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv preprint arXiv. 2020;2003:09871. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv. 2020;2003:10849. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoeibi A., et al. Automated detection and forecasting of COVID-19 using deep learning techniques: a review. arXiv preprint arXiv. 2020;2007:10785. [Google Scholar]
- 22.Ye H., et al. Precise diagnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network. Eur. Radiol. 2019;29(11):6191–6201. doi: 10.1007/s00330-019-06163-2. 2019/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gozes O., et al. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv preprint arXiv. 2020;2003:05037. [Google Scholar]
- 24.Alizadehsani R., et al. Handling of uncertainty in medical data using machine learning and probability theory techniques: A review of 30 years (1991-2020) arXiv preprint arXiv. 2020;2008:10114. doi: 10.1007/s10479-021-04006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharifrazi D., et al. 2020. CNN-KCL: Automatic Myocarditis Diagnosis Using Convolutional Neural Network Combined With K-means Clustering. [DOI] [PubMed] [Google Scholar]
- 26.Shoeibi A., et al. A comprehensive comparison of handcrafted features and convolutional autoencoders for epileptic seizures detection in EEG signals. Expert Syst. Appl. 2021;163:113788. doi: 10.1016/j.eswa.2020.113788. [DOI] [Google Scholar]
- 27.Khodatars M., et al. Deep learning for neuroimaging-based diagnosis and rehabilitation of autism Spectrum disorder: a review. arXiv preprint arXiv. 2020;2007:01285. doi: 10.1016/j.compbiomed.2021.104949. [DOI] [PubMed] [Google Scholar]
- 28.Shoeibi A., et al. Epileptic seizure detection using deep learning techniques: a Review. arXiv preprint arXiv. 2020;2007:01276. [Google Scholar]
- 29.Alizadehsani R., et al. A database for using machine learning and data mining techniques for coronary artery disease diagnosis. Sci. Data. 2019;6(1):227. doi: 10.1038/s41597-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asgharnezhad H., et al. Objective evaluation of deep uncertainty predictions for COVID-19 detection. arXiv preprint arXiv. 2020;2012:11840. doi: 10.1038/s41598-022-05052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthimopoulos M., Christodoulidis S., Ebner L., Christe A., Mougiakakou S. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans. Med. Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 32.Luz E., Silva P.L., Silva R., Moreira G. Towards an efficient deep learning model for covid-19 patterns detection in x-ray images. arXiv preprint arXiv. 2020;2004:05717. [Google Scholar]
- 33.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva P., et al. 2020. Efficient Deep Learning Model for COVID-19 Detection in Large CT Images Datasets: a Cross-dataset Analysis. [Google Scholar]
- 37.Haghanifar A., Majdabadi M.M., Ko S. COVID-CXNet: detecting COVID-19 in frontal chest X-ray images using deep learning. arXiv preprint arXiv. 2020;2006:13807. doi: 10.1007/s11042-022-12156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 39.Tartaglione E., Barbano C.A., Berzovini C., Calandri M., Grangetto M. Unveiling COVID-19 from Chest X-ray with deep learning: a hurdles race with small data. arXiv preprint arXiv. 2020;2004:05405. doi: 10.3390/ijerph17186933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahimzadeh M., Attar A. A new modified deep convolutional neural network for detecting COVID-19 from X-ray images. arXiv preprint arXiv. 2020;2004:08052. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamil M., Hussain I. Automatic detection of COVID-19 infection from chest X-ray using deep learning. medRxiv. 2020 [Google Scholar]
- 42.Horry M.J., Paul M., Ulhaq A., Pradhan B., Saha M., Shukla N. 2020. X-Ray Image Based COVID-19 Detection Using Pre-trained Deep Learning Models. [Google Scholar]
- 43.El Asnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. 2020/06/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas R., Sil J. An improved canny edge detection algorithm based on type-2 fuzzy sets. Procedia Technol. 2012;4:820–824. [Google Scholar]
- 46.Katiyar S.K., Arun P. Comparative analysis of common edge detection techniques in context of object extraction. arXiv preprint arXiv. 2014;1405:6132. [Google Scholar]
- 47.Alizadehsani R., et al. Uncertainty-aware semi-supervised method using large unlabelled and limited labeled COVID-19 data. arXiv preprint arXiv. 2021;2102:06388. [Google Scholar]
- 48.Sun X., Liu L., Li C., Yin J., Zhao J., Si W. Classification for remote sensing data with improved CNN-SVM method. IEEE Access. 2019;7:164507–164516. doi: 10.1109/ACCESS.2019.2952946. [DOI] [Google Scholar]
- 49.Alqudah A.M., Qazan S. Augmented COVID-19 X-ray images dataset. Mendeley Data. 2020;4 doi: 10.17632/2fxz4px6d8.4. doi: 10.17632/2fxz4px6d8. [DOI] [Google Scholar]
- 50.Alqudah A.M., Qazan S., Alqudah A. 2020. Automated Systems for Detection of COVID-19 Using Chest X-ray Images and Lightweight Convolutional Neural Networks. [Google Scholar]
- 51.Alqudah A.M., Qazan S., Alquran H., Qasmieh I.A., Alqudah A. COVID-19 detection from X-ray images using different artificial intelligence hybrid models. Jordan J. Electr. Eng. 2020;6(6):168. [Google Scholar]
- 52.Haque A.B., Rahman M. 2021. Augmented COVID-19 X-ray Images Dataset (Mendely) Analysis Using Convolutional Neural Network and Transfer Learning. [Google Scholar]
- 53."https://www.kaggle.com/amanullahasraf/covid19-pneumonia-normal-chest-xray-pa-dataset.".
- 54."https://www.kaggle.com/fusicfenta/chest-xray-for-covid19-detection.".
- 55."https://www.kaggle.com/tawsifurrahman/covid19-radiography-database.".
- 56."https://www.kaggle.com/tarandeep97/covid19-normal-posteroanteriorpa-xrays.".
- 57."https://www.kaggle.com/khoongweihao/covid19-xray-dataset-train-test-sets.".
- 58."https://www.kaggle.com/darshan1504/covid19-detection-xray-dataset.".
- 59.Iwendi C., et al. COVID-19 patient health prediction using boosted random forest algorithm. Front. Public Health. 2020;8:357. doi: 10.3389/fpubh.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]