Abstract
Hepatitis B virus (HBV) has infected one-third of world population and 240 million people are chronic carriers, to whom a curative therapy is still not available. Similar to other viruses, persistent HBV infection relies on the virus to exploit host cell functions to support its replication and efficiently evade host innate and adaptive antiviral immunity. Understanding HBV replication and concomitant host cell interactions is thus instrumental for development of therapeutics to disrupt the virus-host interactions critical for its persistence and cure chronic hepatitis B. Although the currently available cell culture systems of HBV infection are refractory to genome-wide high throughput screening of key host cellular factors essential for and/or regulating HBV replication, classic one-gene (or pathway)-at-a-time studies in last several decades have already revealed many aspects of HBV-host interactions. An overview of the landscape of HBV-hepatocyte interaction indicates that in addition to more tightly suppressing viral replication by directly targeting viral proteins, disruption of key viral-host cell interactions to eliminate or inactivate the covalently closed circular (ccc) DNA, the most stable HBV replication intermediate that exists as an episomal minichromosome in the nucleus of infected hepatocyte, is essential to achieve a functional cure of chronic hepatitis B. Moreover, therapeutic targeting of integrated HBV DNA and their transcripts may also be required to induce HBsAg seroclearance and prevent liver carcinogenesis.
As obligate intracellular parasites, successful colonization of cells by viruses depends on strategies evolved to exploit cellular functions to support their replication and capability to overcome cellular antiviral defenses. It is the dynamic virus-host interaction that determines the pathogenesis and outcomes of a viral infection. Therefore, identification of host cellular factors that support or restrict viral replication and elucidation of the underlying mechanisms are important to understand viral pathogenesis and develop antiviral therapeutics to treat viral diseases1. The advent of omics technologies in the last decades allows for investigation of virus-host interaction at the levels of genome, transcriptome, proteome and metabolome and identification of host cellular proviral and restriction factors through genome-wide genetic screenings2. These new technologies have profoundly shaped the landscape of virus-cell interaction research and revealed potential molecular targets for development of antiviral therapeutics3.
Hepatitis B virus (HBV) is a member of Hepadnaviridae family and contains a 3.2 kb, partially double-stranded, relaxed circular (rc) DNA genome4. Hepatocytes are the primary host cells of HBV. As depicted in Fig. 1, HBV infection begins by binding to its cellular receptor, sodium taurocholate cotransporting polypeptide (NTCP), on the surface of hepatocytes and delivering viral nucleocapsids into the cytoplasm via endocytosis. The viral rcDNA genome in nucleocapsid is then transported into the nucleus and converted into episomal covalently closed circular (ccc) DNA to serve as a template for transcription of viral RNA. Unlike other DNA viruses, HBV replicates its genomic DNA via viral DNA polymerase self-primed reverse transcription of a RNA pregenome in the cytoplasmic nucleocapsid. Briefly, binding of viral DNA polymerase to the stem-loop structure at the 5’ terminus of pregenomic (pg) RNA initiates their packaging by 120 copies of core protein dimers to form a nucleocapsid. Inside the nucleocapsid, viral DNA polymerase converts the pgRNA first to a single-stranded DNA and then to rcDNA. The rcDNA-containing mature nucleocapsids can either acquire an envelope and be secreted out of cells as infectious virions or deliver the rcDNA into the nucleus to amplify cccDNA pool, the sole transcription template to support HBV replication5.
Figure 1. Schematic representation of HBV replication cycle.
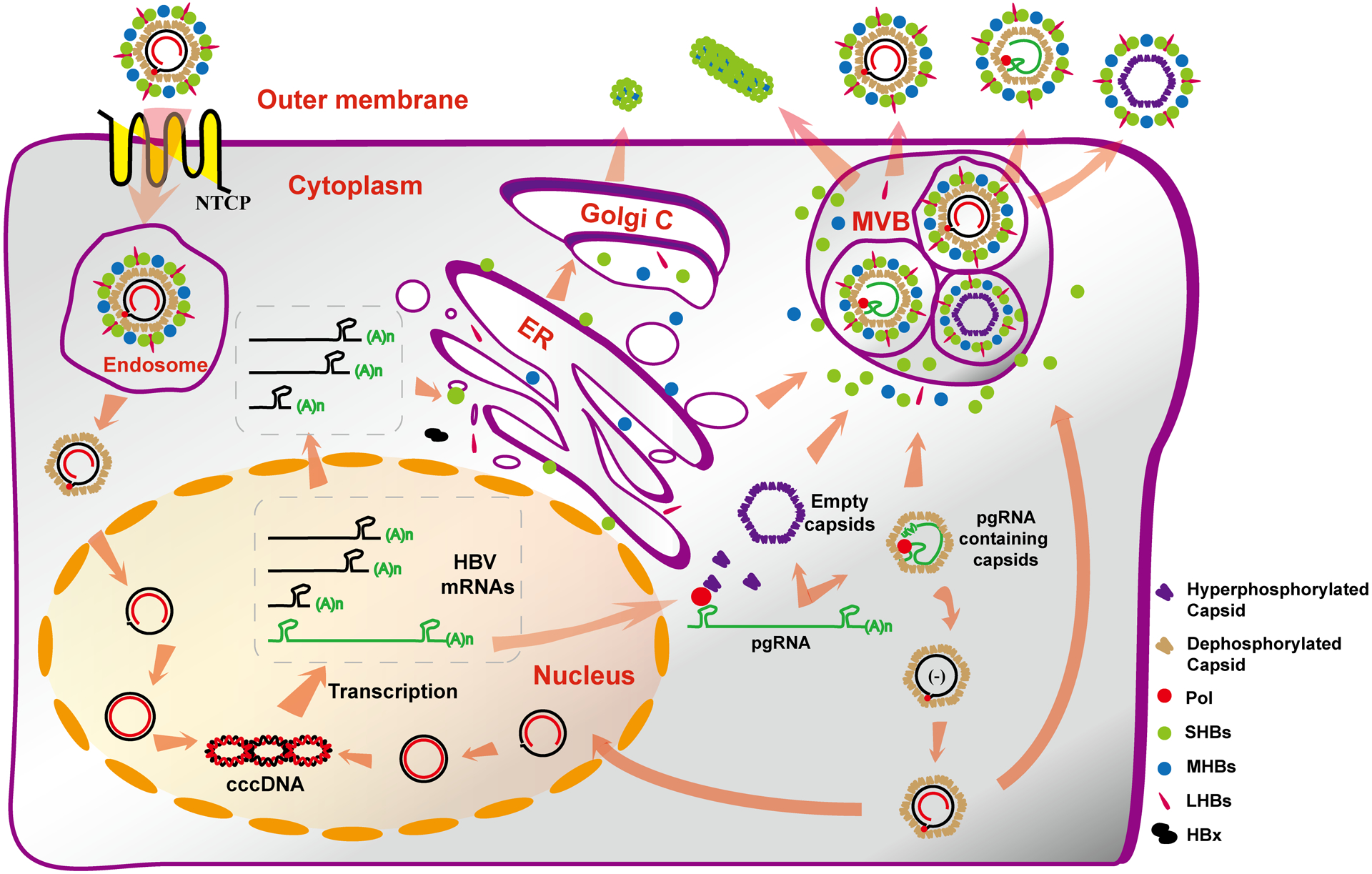
HBV binds to its cellular receptor NTCP to trigger endocytosis that internalizes virion into endosome where the nucleocapsid is released into the cytoplasm. The viral genomic DNA in the nucleocapsid is transported into the nucleus and converted into cccDNA, which serves as the template to transcribe viral RNAs. Viral DNA polymerase binds to pgRNA and is subsequently assembled by core proteins to form nucleocapsids where the pgRNA is reverse transcribed into DNA. Core protein also assembles into empty capsids. Both viral DNA- (to a lesser extent, RNA-) containing or empty capsids can be enveloped and secreted as virions or empty virions. Meanwhile, viral envelope proteins also assemble and secrete large amounts of small sphere and filament subviral particles. See text for more detailed explanation.
While vast majority of adulthood HBV infection is resolved within six months by host antiviral immune responses, particularly the vigorous polyclonal cytotoxic T lymphocyte response against multiple viral proteins, infection at early childhood usually fails to induce a sufficient immune response to clear the virus and results in a life-long persistent infection6. Chronic HBV infection is associated with high risk of severe liver diseases, including cirrhosis, hepatocellular carcinoma (HCC) and liver failure7. The currently available antiviral therapeutics, including alpha-interferon (IFN-α) that regulates host antiviral immune response and six nucleos(t)ide analogues (NUCs) that inhibit HBV DNA polymerase, can potently suppress viral replication and prevent the progression of liver diseases, but fail to cure the viral infection8. This is due primarily to their inability to eliminate cccDNA and restore the dysfunctional antiviral immune response against HBV5, 9. Investigation of HBV-host cell interaction will reveal cellular and viral functions that are essential for the stable maintenance and proper function of cccDNA to support the persistent infection and can thus be pharmacologically targeted for elimination or functional inactivation of cccDNA and ultimate cure of chronic hepatitis B. Although the currently available cell culture systems supporting HBV infection are not suitable for genome-wide high throughput screening of key host cellular factors essential for and/or regulating HBV replication, studies of HBV virology in the last several decades have already revealed many aspects of HBV-hepatocyte interaction. Integration of the existing knowledge should serve as a starting point toward construction of a comprehensive molecular network of HBV-hepatocyte interaction and establishment of a knowledge base for continuing search and validation of therapeutic targets for the cure of chronic HBV infection.
1. HBV replication depends on and is regulated by hundreds of host cellular proteins
Although numerous studies have shown that HBV does not induce an overt pattern recognition receptor-mediated innate immune response in infected hepatocytes10, classical virology studies have already revealed a very comprehensive molecular landscape of HBV-hepatocyte interaction and identified hundreds of host cellular genes that are required for, or regulate, HBV infection of hepatocytes at every step of the viral life cycle.
Entry: delivery of nucleocapsids into the cytoplasm
Three cellular proteins, heparan sulfate proteoglycan (HSPG), sodium taurocholate cotransporting polypeptide (NTCP, encoded by SLC10A1) and glypican 5, have been identified as attachment factor, receptor and co-receptor for the infectious entry of HBV into hepatocytes, respectively. The large envelope protein (LHBs) on the surface of HBV virions interacts with HSPG to attach virion particles on the surface of hepatocytes11. The myristoylated N-terminal preS1 domain of LHBs subsequently binds NTCP, bona fide receptor for HBV12. HBV entry can be efficiently blocked, in vitro and in vivo, by Myrcludex B, a synthetic N-acylated preS1 lipopeptide13. In spite of its requirement for HBV infection of HepG2-NTCP cells, the mechanism of glypican 5 in HBV entry remains elusive14. Moreover, whether HBV is internalized into hepatocytes via clathrin-mediated or caveolin-dependent endocytosis remains controversial15. Following endocytosis, HBV must travel through the complex network of the endocytic pathway and release nucleocapsids. While the dependence of HBV infection on Rab5 and Rab7 expression suggests that HBV transport from early to mature endosomes is required for infectious entry, where and how the viral envelope-host cellular membrane fusion and release of nucleocapsid into the cytoplasm take place remain to be determined16.
Nucleocapsid transport and viral genomic DNA nuclear import
Upon reaching the cytoplasm, nucleocapsids have to be transported through the viscous cytoplasm to nuclear pore complexes (NPCs). Nucleocapsids disassemble at the basket of NPCs and viral genomic DNA are released into the karyoplasm where it is converted into cccDNA by cellular DNA repair machinery17. As depicted in Fig. 1, in addition to the incoming virion DNA, cccDNA can also be synthesized from intracellular progeny nucleocapsid DNA18. However, although HBV cccDNA intracellular amplification pathway works efficiently in hepatoma cell lines supporting HBV replication19, it is less efficient in HBV-infected hepatoma cells or primary human hepatocytes17, 20. Mechanism of this discrepancy remains to be investigated. Regardless of their origins, the intra-cytoplasmic transportation and viral genomic DNA nuclear import of nucleocapsids ought to take place in a coordinated manner. First, a recent study showed that both double-stranded DNA-containing mature nucleocapsids and empty capsids, but not pgRNA-containing capsids, can be actively transported through the cytoplasm towards the nucleus21. It was further demonstrated that dynein light chain LL1 functions as an interaction partner linking capsids to the dynein motor complex and retrograde transport capsids along microtubules towards the NPCs21. Second, carboxyl terminal domain (CTD) of core protein contains two overlapping bipartite nuclear localization signals (NLS) and their exposure on the exterior of capsids is modulated by genomic DNA maturation or CTD phosphorylation. The exposed NLS interacts with nuclear import receptors and initiates nuclear transport of capsids22. Interestingly, while empty capsids interact with the nuclear import receptor importin-β directly, mature nucleocapsids need the adaptor molecule importin-α, most likely due to the differences in exposure of core protein CTD and/or its phosphorylation statu22a, 22b, 23. Third, premature or enhanced nucleocapsid uncoating (or disassembly) induced by core protein allosteric modulator (CpAM) treatment or core protein mutations results in reduced amounts of cytoplasmic mature HBV double-stranded DNA, but increased amounts of nuclear deproteinized (DP)-rcDNA and cccDNA19a, 24. A plausible explanation is that the premature/enhanced nucleocapsid uncoating increases core protein CTD exposure and the nucleocapsids are thus imported into nuclei via the more efficient importin-β pathway. This hypothesis is currently under investigation in our laboratory. Finally, NPCs are composed of ~30 different proteins collectively called nucleoporins (Nups)25. Interaction with the NLS on capsids changes the structure of import receptors, allowing interaction with Nup358 at the cytoplasmic face of NPC. The capsid is then translocated through the nuclear pore and disassociates from importins in the nuclear basket and directly binds to Nup153, followed by capsid disassembly and diffusion of rcDNA into the karyoplasm26.
Conversion of viral genomic DNA into cccDNA
It is generally acknowledged that the conversion of viral genomic DNA to cccDNA takes place in the nucleus and is catalyzed by host cellular DNA repair proteins. The strict requirement of Ku-80 and ligase IV strongly suggests that formation of cccDNA from double-stranded linear (dsl) DNA, a minor species of HBV genome derived from in situ priming of positive strand DNA, is through non-homologous end joining (NHEJ) DNA repair pathway27. However, only a few biochemical reactions essential for cccDNA synthesis from rcDNA precursor have been investigated. First, removal of viral DNA polymerase covalently linked to the 5’ terminus of rcDNA minus strand is an essential step to convert rcDNA into cccDNA. Tyrosol-DNA phosphodiesterase 2 (TDP2), an enzyme responsible for resolving topoisomerase II-DNA adduct28, has been demonstrated to cleave the tyrosyl-minus strand DNA linkage in vitro29. However, TDP2 gene knockout did not inhibit cccDNA formation during HBV infection of NTCP-expressing HepG2 cells30 and only delayed intracellular amplification of duck hepatitis virus (DHBV) cccDNA29, suggesting that additional cellular function(s) can process the 5’ terminus of rcDNA minus strand. Second, failure of viral DNA polymerase inhibitors to block cccDNA formation during de novo HBV infection of hepatocytes suggests that the completion of positive strand of rcDNA from viral particles is not catalyzed by viral DNA polymerase, but cellular DNA polymerase(s). A genetic screening in HBV infection of NTCP-expressing HepG2 cells identified cellular DNA polymerase ƙ, and to a lesser extent, DNA polymerase λ, are required for de novo cccDNA synthesis20. DNA polymerases ƙ and λ play a role in translesion DNA synthesis to bypass base damage in DNA and NHEJ DNA repair pathway, respectively. Exact biochemical reactions catalyzed by those cellular DNA polymerases, involving the elongation of positive strand DNA or filling the gaps in positive and negative strands after end processing, remain to be determined. It is also interesting to know how those error-prone DNA polymerases perfectly repair rcDNA to cccDNA. Third, it is obvious that ligation of both strands of viral DNA is essential for cccDNA synthesis. Recent identification of rcDNA species with covalently closed negative strand in hepatoma cells supporting HBV DNA replication suggests that this negative-strand closed rcDNA is possibly the intermediate of cccDNA formation from rcDNA precursor and argues that repairing of negative and positive strands of rcDNA sequentially takes place31. Moreover, a recent genetic study demonstrated that knocking out either DNA ligase I or ligase III significantly compromised HBV and DHBV cccDNA synthesis from de novo infection and intracellular amplification pathways27b, suggesting that both the ligases are required for cccDNA synthesis from rcDNA. Finally, it was reported recently that activation of retinoid X receptor alpha (RXRα) inhibited de novo HBV cccDNA synthesis in HBV infected human hepatocytes through inducing expression of the genes in arachidonic acid/eicosanoids biosynthesis pathways and production of arachidonic acid32. Obviously, further investigation is required to determine the biochemical reaction in cccDNA synthesis that is inhibited by this cellular metabolism pathway.
cccDNA minichromosome and transcription regulation
As depicted in Fig. 2, expression of the four open reading frames (ORFs) of HBV genome are regulated by four promoters (basal core, preS1, preS2/S, and X) and two enhancers (Enh I and Enh II) to transcribe four major species (3.5, 2.4, 2.1 and 0.7 kb) of mRNA. While 2.4 and 0.7 kb mRNA specify LHBs and HBx protein, respectively, 2.1 kb mRNA translate middle (MHBs) and small (SHBs) envelope proteins. The 3.5 kb mRNA contains pre-C mRNA and pgRNA. While the pre-C mRNA translates pre-core protein that is subsequently processed and secreted as e antigen (HBeAg), pgRNA translates core and polymerase proteins, in addition to serving as a template for reverse transcriptional DNA replication4. Similar to host cellular DNA, cccDNA in the nuclei of infected hepatocytes associate with nucleosomes to assemble into minichromosomes33. Transcription of cccDNA is regulated by ordered recruitment of specific chromatin modifiers and transcription factors as well as basal RNA polymerase II transcriptional complex.
Figure 2. HBV genome structure and transcription.
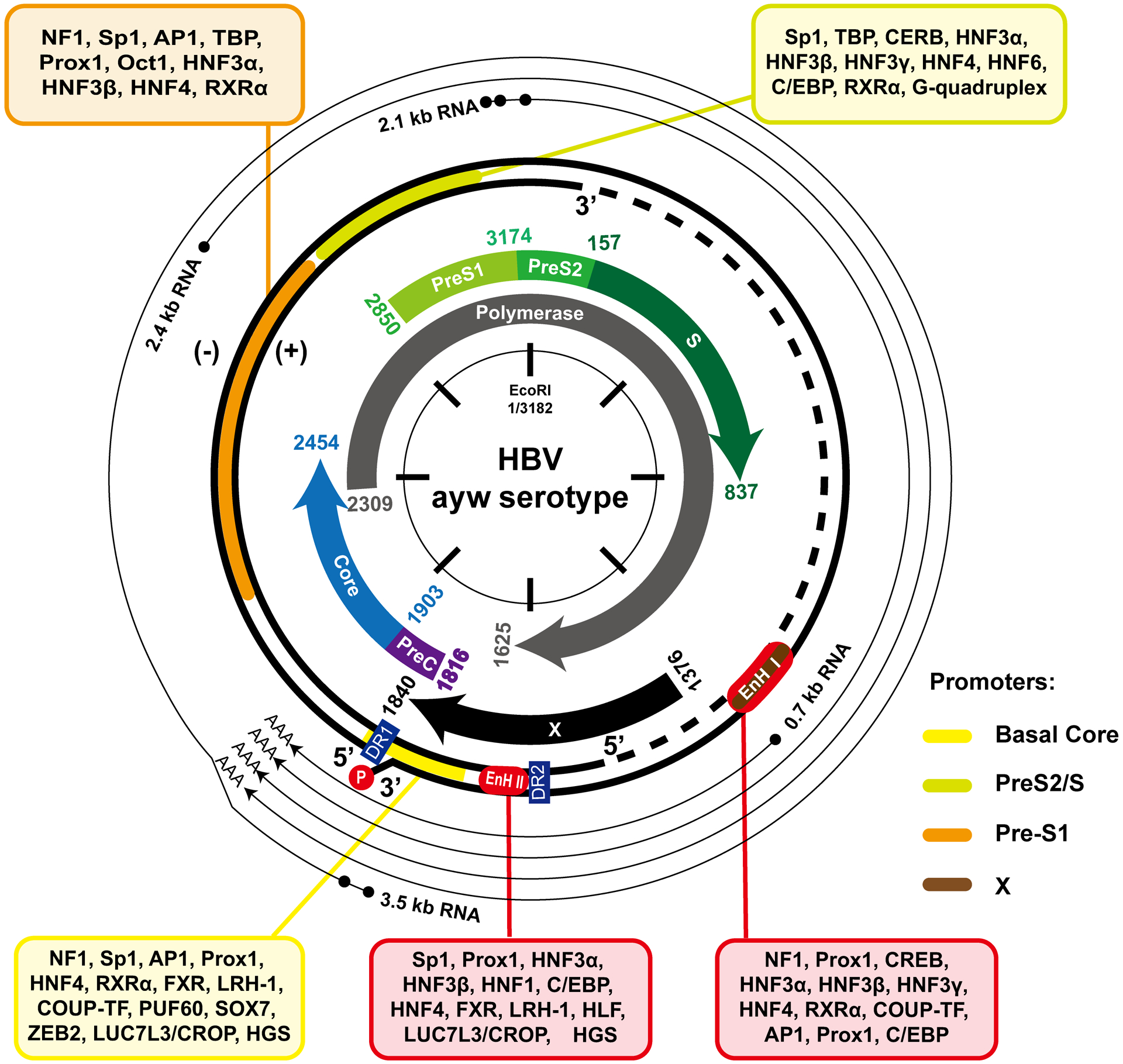
The relaxed circular (rc) DNA genome structure, ORFs and cis-transcriptional regulation elements are depicted. The transcription factors bound to each of the transcriptional elements are presented. The nucleotide numbering is based on genotype D HBV (ayw).
Landscape of cccDNA-associated histone modification
Histone modifications play important roles in every aspect of chromatin functions, including DNA replication, transcription and repair as well as chromatin assembly and remodeling, by primarily serving as platforms for recruitment of other cellular proteins to participate and regulate chromatin metabolism. Using ChIP-Seq technology, cccDNA minichromosome genome-wide histone post-translational modifications (PTMs) had been examined. This study revealed that high levels of histone PTMs associated with active transcription, such as H3K4me3, H3K27ac and H3K122ac are enriched at positions overlapping the four viral promoters34. Interestingly, IFN-α silence of cccDNA transcription is associated with the reduction of histone modifications specifying active transcription, but not the increase of histone modifications related to the silence of gene expression, such as H3K9me3 and H3K27me3 34–35. These findings suggest that while activation of cccDNA transcription is regulated by similar histone PTMs reported for cellular chromatin, silence of cccDNA transcription by IFN-α apparently works via a mechanism distinct from silencing of host chromatin transcription.
As illustrated in Fig. 3, the active HBV cccDNA transcription state is attributed to the presence of HBx, a small viral regulatory protein36. In the absence of HBx, histone methyltransferase SETDB1 as well as histone deacetylases HDAC1 and Sirt1 are recruited to cccDNA minichromosomes, resulting in deposition of transcriptionally suppressive histone makers and erasing of active histone markers37. On the contrary, the presence of HBx can counteract SETDB1 mediated suppression, as well as bring in acetyltransferases p300/CBP, histone lysine demethylase-1 (LSD1) and other factors to establish transcriptionally permissive cccDNA state37–38. In addition to directly participating in cccDNA epigenetic coding, HBx was demonstrated to facilitate cccDNA transcription by counteracting the recruitment of a tudor domain protein Spindlin1 onto cccDNA which otherwise serves as a transcription suppressor39, inhibiting protein arginine methyltransferase1 (PRMT1) activity to relieve PRMT1-mediated repression on cccDNA transcription40, and stabilizing transcription factor p-CREB41. Most importantly, recent studies indicated that cccDNA minichromosome is transcriptionally silenced by the binding of cellular structural maintenance of chromosomes 5/6 (SMC5/6) complex, including Smc5, Smc6, Nse1, Nse2, Nse3, and Nse4 proteins42. Moreover, it appears that SMC5/6 complex suppression of cccDNA transcription requires its co-localization with nuclear domain 10 bodies (ND10), a nuclear protein aggregates containing PML, Sp100 and other proteins42c. By interacting with DNA-damage binding protein 1 (DDB1), HBx protein recruits DDB1-Cullin4 E3 ubiquitin ligase complex to degrade SMC5/6 complex and relieves its restriction on cccDNA transcription42. As a key regulator of viral infection, the stability of HBx is regulated by poly-ubiquitination mediated proteasomal degradation and E3 ligase HDM2 mediated Neddylation that stabilizes HBx43.
Figure 3. Epigenetic regulation of HBV cccDNA transcription by HBx protein.
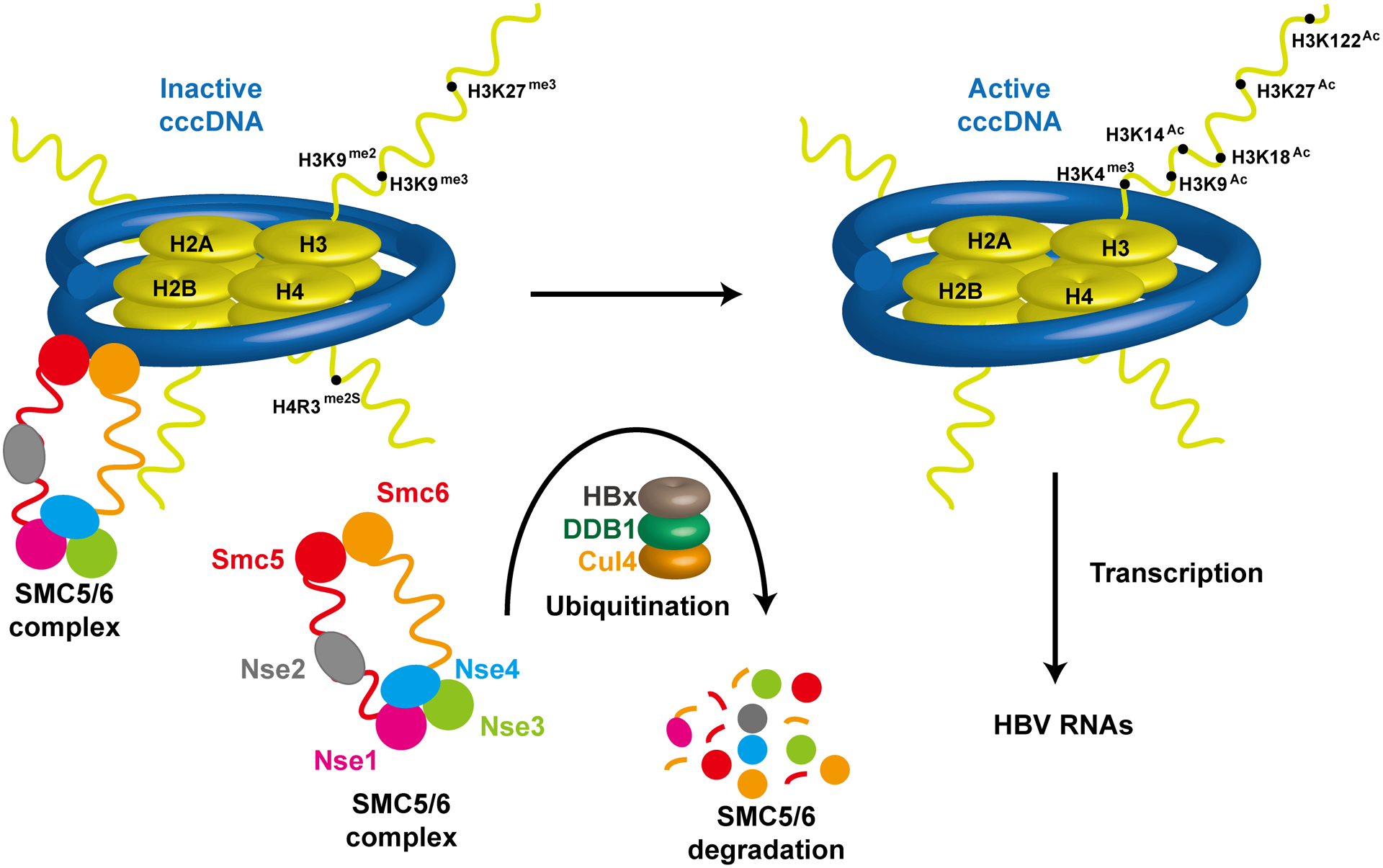
Binding of SMC5/6 complex to cccDNA maintains the cccDNA in a transcriptionally silencing state, characterized by H3K9me3, H3K29me3 and H4R3me2s. Through interaction with DDB1, HBx recruits E3 ubquitin ligase Cullin 4 to degrade SMC5/6 complex and induces the establishment of a transcriptionally permissive cccDNA state.
Recently, a suppressive histone modification, symmetric dimethylation of H4R3, H4R3me2s, catalyzed by protein arginine methyltransferase PRMT5 was found on cccDNA minichromosome even in the presence of HBx, leading to epigenetic silencing of cccDNA44. Interestingly, the recruitment of PRMT5 to cccDNA was dependent on HBV core protein, suggesting a role of core protein in regulating cccDNA transcription. Furthermore, binding of core protein to cccDNA CpG islands is associated with an epigenetic permissive state of cccDNA45. It is conceivable that selectively disrupting distinct interactions between HBx and/or core protein with their host binding partners might provide a chance to dismantle their roles in supporting cccDNA transcription and consequentially inactivate cccDNA, which may result in a functional cure of chronic hepatitis B9.
In addition to histone modifications, cccDNA methylation also regulates its transcriptional activity. There are three CpG islands in HBV genome. Studies have shown that patients with high methylation frequency in CpG islands 2 and 3 are associated with lower viremia and serum HBV surface antigen (HBsAg) levels, respectively, probably due to the reduced transcription activity of HBV X and S genes46. In addition, hypermethylation in CpG islands 1 and 3 is associated with increased risk of HCC47. However, the cellular DNMT(s) that catalyze cccDNA CpG methylation remains to be identified.
Transcription factors and cccDNA transcription regulation
As illustrated in Fig. 2 and reviewed recently48, the transcription activity of every HBV promoter or enhancer is regulated by multiple ubiquitous and hepatocyte enriched transcription factors. Particularly, the hepatocyte nuclear factors (HNF), including HNF1α, HNF3β, HNF4α, HNF6 and C/EBP, play the most important roles in cccDNA transcription and, at least in part, specify the hepatotropism of HBV48a. However, the regulatory activities of some transcriptional factors vary among the different experimental systems utilized. For instance, it has been demonstrated that HNF1α stimulates transcriptional activity of pre-S1 promoter and Enh II in HBV replicon plasmid-transfected hepatoma cells49. However, HBV transcription was not affected, whereas HBV DNA replication was modestly increased with appearance of cccDNA in HNF1α-null HBV transgenic mice50. Because the studies on HBV transcription have been done mostly in transiently transfected human hepatoma cells or HBV transgenic mice that cccDNA is not the transcription template, the biological function of those transcription factors in HBV cccDNA transcription should be further evaluated in HBV infected human hepatocytes.
HBV transcription is regulated by many cellular factors, such as cytokines and hormones, by modulating the function and abundance of transcription factors and epigenetic landscape of cccDNA minichromosome. For examples, interferon (IFN)-γ regulates HNF4 and C/EBP levels to affect HBV transcription and replication51. IL-6 controls HBV transcription by reducing the levels of HNF4α and HNF1α52. Interleukin-4 (IL-4) down-regulates C/EBPα to inhibit the activity of HBV core and preS2 promoter activity and pgRNA synthesis53. NF-ƙB represses the HBV gene expression through repression of Sp1 and HNF4-mediated transcriptional activation54. Hepatocystin down-regulates HNF4 through the Ras/MAPK pathway to impair HBV replication, and is important for determining the susceptibility of HBV to IFN-γ55. The activation of ERK1/2, ASPK/JNK or PI3K-AKT-mTOR pathway suppresses HBV transcription by negative regulation of HNF4α in both Huh7 and HepG2 cells56,57. Estrogen represses transcription of HBV genes by up-regulating ER-α, which interacts with and alters binding of HNF-4α to the HBV Enh I58. On the contrary, androgen increases the transcription of HBV through direct binding to the androgen-responsive element sites in viral Enh I59. This may explain a higher HBV titer in male carriers and an increased risk of HCC.
Stability and maintenance of cccDNA
The cccDNA minichromosomes are stable in non-dividing hepatocytes and possibly persist throughout the lifespan of host cells60. The high stability of cccDNA is further highlighted by the fact that CRISPR/Cas9 targeted cleavage of cccDNA rarely results in the loss of cccDNA, because the cleaved products are efficiently re-circularized by cellular NHEJ DNA repair pathway61. As illustrated in Fig. 4, while an important role of cytolytic T lymphocyte killing of infected hepatocytes is clearly evident in the resolution of acute HBV infection, the non-cytolytic cure of infected hepatocytes also occurs and is essential for the clearance of viral infection62. However, how the host antiviral immune response non-cytolytically eliminates cccDNA from infected hepatocytes remains controversial. Interestingly, Lucifera and Xia et al reported that treatment of HBV-infected primary human hepatocytes with IFN-α, IFN-γ, TNF-α or lymphotoxin-β receptor (LTβR) agonists non-cytolytically reduces the amount of cccDNA63. Mechanistically, it was shown that these inflammatory cytokines induce the expression of APOBEC3A and APOBEC3B, which are recruited to cccDNA via interaction with HBV core protein and deaminate cytosines in the negative strand of cccDNA. Deamination of cccDNA results in uracil-DNA glycosylase cleavage of uracils to form apurinic/apyrimidinic sites, which are subsequently recognized and cleaved by apurinic/apyrimidinic (AP) endonucleases63a. Interestingly, while a recent study demonstarted that while incubation of HBV replicating human hepatoma cells (Hep G2.1.15) with HBV-specific T-cell receptor mRNA-transfecetd resting human T lymphocytes non-cytolytically reduced HBV cccDNA by activation of APOBEC3 expression64, IFN-α-induced reduction of cccDNA in HBV-infected HepaRG cells and/or PHHs was not obviously noted in other studies65.
Figure 4. Mechanisms to eliminate cccDNA.
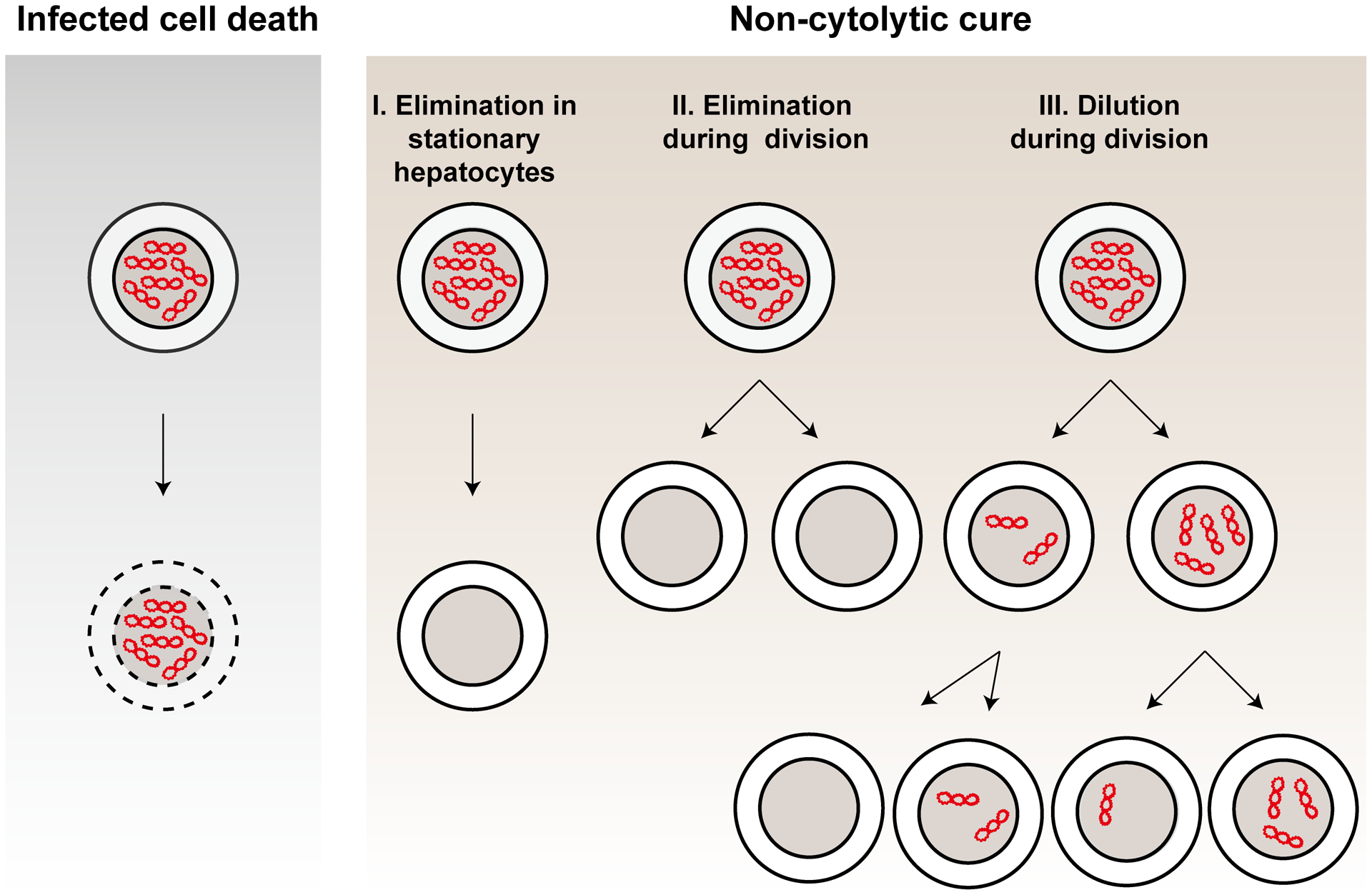
Pre-existing cccDNA can be eliminated by killing of infected hepatocytes (A) or non-cytolytically cure of infected hepatocytes via three distinct mechanisms (B).
Another potential mechanism of cccDNA elimination is loss or dilution through cell division. Due to the lack of kinetochore structure, episomal DNAs are not equally partitioned into daughter cells during cell division and may be left in the cytoplasm and degraded after the cell division. Other DNA viruses, such as Kaposi’s sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV) and bovine papillomavirus (BPV), tether their episomal genomes onto host chromosomes via interacting with a host bromodomain containing protein BRD4, to survive cell division66. A previous study in HBV infected chimeric mice suggested that both dilution of the cccDNA pool among daughter cells and significant loss of cccDNA occur during hepatocyte proliferation67. By tracking cccDNA fate with fluorescence in situ hybridization (FISH) technology, a recent study obtained evidence suggesting that cccDNA are symmetrically distributed into daughter cells68. However, there is no evidence suggesting that cccDNA are attached to metaphase chromosomes68. It is conceivable that therapeutic disruption of cccDNA distribution into daughter cells during hepatocyte proliferation should accelerate cccDNA decay and facilitate resolution of chronic HBV infection.
pgRNA splicing
Unlike classic retroviruses, RNA splicing is not required for HBV to produce viral envelope or regulatory proteins. However, pgRNA splicing products are readily detectable in the livers infected with HBV, woodchuck hepatitis virus (WHV) or DHBV69. The first HBV splicing product SP1RNA, a 2.2 kb RNA that shared the 5’ cap and 3’ poly(A) tail with pgRNA but with deletion of sequence between the last codon of core and a portion of Pol and S ORFs, was discovered in 1989 (Fig. 5, Example A). Up to now, more than 18 single or multiple spliced HBV RNA species have been identified. In cells actively transcribing HBV pgRNA, spliced (sp) RNA consists 10–30% of total pgRNA70. Splicing activity of pgRNA is regulated by the HBV post transcriptional regulatory element (PRE)70b, 71 and host splicing related factors such as La and PSF72.
Figure 5. Examples of pgRNA splicing.
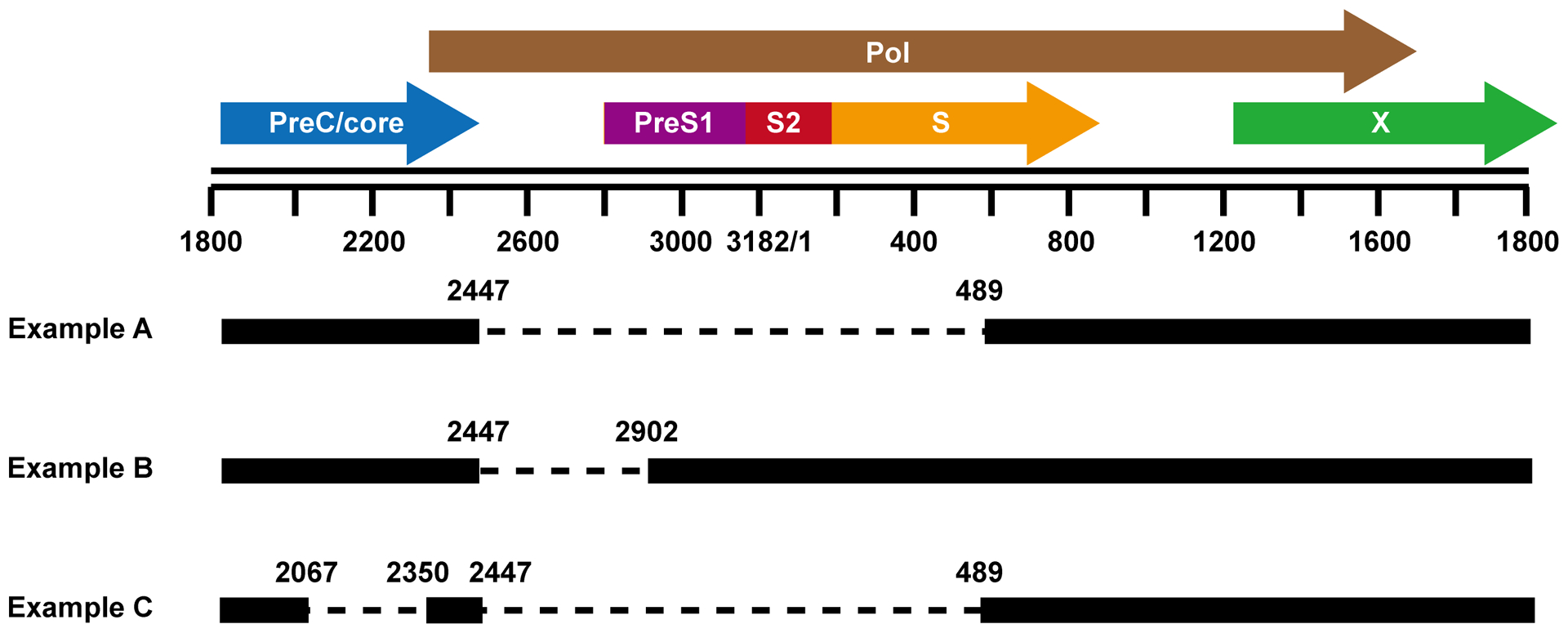
In pgRNA, the nucleotides 2447 and 2007 are two most common splicing donor sites. The nucleotides 489, 2902 and 2355 are common splicing acceptor sites. Three examples of single and double spliced pgRNA are depicted. Examples A and B produce fusion proteins that modulate virion morphogenesis and cell death pathway, respectively.
Although Core and Pol ORFs are disrupted, many spRNA species have intact 5’ epsilon (ɛ) structure and can thus be packaged into capsids and reverse transcribed into DNA. Because polymerase provided in trans seems in favor of in situ priming of positive strand DNA synthesis, the spRNA-derived DNA are largely dslDNAs, which are preferred substrates for integration into host chromosomal DNA73. In PLC/PRF/5 hepatoma cells, integrated HBV DNA had a deletion between splice donor (nt 2447) and acceptor (nt 2902) sites, indicating that the integrant is probably originated from spHBV DNA74. Moreover, levels of defective HBV particles derived from spRNA have been recently shown to be correlated with disease status. Particularly, presence of mutant HBV with preS1 deletion between aa 57 and 119, a consequence of alternative splicing, is significantly associated with development of chronic hepatitis, cirrhosis and HCC75.
Although it is not absolutely required, the role for spRNA and their derived DNA or proteins in modulating HBV infection and pathogenesis cannot be ruled out. For instance, co-transfection of a spliced 2.2 kb RNA with wild-type HBV in HepG2 cells significantly enhanced HBV replication, in an HBx-dependent manner76. It was also reported that transfection of SP1 RNA into Huh7 cells up-regulated the expression of HBcAg and HBeAg from the cotransfected HBV replicon77. Interestingly, DHBV large surface protein can be translated from either preS1 promoter-directed authentic mRNA or a transcript derived from pgRNA splicing. Ironically, the presence of spliced preS1 mRNA was crucial for virion production in both animals and primary duck hepatocytes, but not in immortalized cell lines69c.
Some of the spRNAs can support the synthesis of novel viral proteins which are collectively named as HBV splicing-generated proteins (HBSPs). To date, there are only two spliced RNAs that have been reported to be translated into HBSPs. The first one was found to form a 43 kD fusion protein that contained the N-terminal 47 aa of pol and the C-terminal 19 aa of preS1, plus the following preS2 and S domains (Fig. 5, Example A). This fusion protein lost the preS1 myristylation site but retained the core interacting domain and surface protein transmembrane domains. The fusion protein was incorporated into both Dane and subviral particles78. Another HBSP is a 10 kD fusion protein derived from SP1 RNA. It contains 46 amino acid (aa) residues from the N terminal of Pol and 65 aa residues from a frame shift after nt 489 (Fig. 5, Example B)79. In the 1.3.32 HBV transgenic mice, treatment with carbon tetrachloride (CCL4) or lipopolysaccharide (LPS) induced a significant increase of SP1 RNA in the liver. The elevation of SP1 RNA inhibited the recruitment of monocytes and macrophages and subsequently down-modulated liver damage72.
In summary, pgRNA splicing is regulated by host pathobiological factors. The advent of biologically relevant HBV infection cell culture and animal models should facilitate the investigation toward uncovering the roles of spRNA as well as derived proteins and DNA products in HBV infection and pathogenesis.
Viral RNA nuclear export and decay
Similar to retroviruses, to avoid aberrant RNA splicing, nuclear export of intron-less HBV RNA is facilitated by host cellular factors through interaction with post-transcriptional regulatory element (PRE) at the 3′ region of HBV transcripts. HBV PRE contains approximately 450 nucleotides spanning nt 1151 to 1582 and is divided into 3 sub-elements HPREα (nt 1151–1346), HPREβ1 (nt1347–1457) and HPREβ2 (nt 1458–1582)80. Unlike HIV PRE that interaction with viral protein Rev is required for full-length viral RNA nuclear export, HBV PRE mediated viral RNA nuclear export does not require the involvement of any viral protein. Instead, cellular lupus-associated (La) protein and polypyrimidine tract binding protein (PTB1) have been found to bind the HPREα and HPREβ2 region and consequentially stabilize and facilitate HBV mRNA nuclear export, respectively81. Intriguingly, it was reported recently that a small molecule dihydroquinolizinone, 6-R2-10-methoxy-9-R1-2-oxo-2,3,4,6,7,11b-hexahydro-1H-pyrido[2,1-a]-isoquinoline-3-carboxylicacid (DHQ-1), selectively induced HBV RNA degradation in a manner depending on a 109-nucleotide RNA element in HPREα region82. It was further demonstrated that DHQ-1 specifically interacts with host cellular proteins non-canonical poly(A) polymerases D5 (PAPD5) and D7 (PAPD7), and knocking down the expression of these cellular proteins reduces HBV RNA and HBsAg expression83. It will be extremely interesting to know how the cellular 3’-end polymerases stabilize HBV RNA and how their interaction with DHQ-1 results in HPREα sequence-dependent HBV RNA degradation.
HBV mRNAs are also targeted by host innate and adaptive immune response to control viral infection. For example, adoptive transfer of HBV-specific CTLs into HBV transgenic mice induces HBV RNA decay, which is tightly associated with the cytokine-induced proteolytic cleavage of La, a protein that binds HPREα element and stabilize HBV RNA81a, 84. In addition, investigation of IFN antiviral mechanism against HBV reveals two IFN-induced cellular proteins, zinc finger antiviral protein (ZAP) and ISG20, that selectively degrade HBV RNA through interaction with HBV RNA terminal redundant region containing ε stem-loop structure85.
pgRNA packaging and reverse transcription
HBV DNA synthesis occurs in cytoplasmic nucleocapsids by viral DNA polymerase self-primed reverse transcription of pgRNA. The ε signal located on the 5’ end of pgRNA is recognized by the viral polymerase to initiate pgRNA packaging. The roles of HBV core protein and polymerase as well as related host factors in pgRNA packaging and reverse transcription have been extensively investigated86.
Cellular chaperone proteins play critical role in pgRNA packaging
Early studies indicate that the stem-loop structure ε located at the 5’ end of pgRNA is its packaging signal and viral DNA polymerase is required for pgRNA package by core protein86. It was soon discovered that Hsp90 is required for viral polymerase to bind ε RNA element, an essential step for the packaging of viral polymerase-pgRNA complex into nucleocapsids and the priming of negative strand DNA synthesis87. In vitro reconstitution studies elegantly demonstrated that Hsp90 and its co-chaperones, Hsp70, Hsp40, Hop/p60, and p23, are all required for viral polymerase binding of pgRNA and priming of DNA synthesis88. In addition, through mediating interaction between viral polymerase and Hsp90, co-chaperone p50 increases pgRNA packaging and DNA replication89. While p23 has been shown to be packaged with viral polymerase and pgRNA into nucleocapsids, whether other chaperones are co-assembled has not been experimentally determined90.
Other host factors regulate pgRNA encapsidation
In addition to chaperones, protein arginine methyltransferase 5 (PRMT5) has been reported to inhibit pgRNA encapsidation. One study showed that PRMT5 binds to the RNase H domain of viral DNA polymerase and prevents its interaction with pgRNA, which results in inhibition of pgRNA packaging and DNA replication in a methyltransferase activity-independent manner44. Another study showed that PRMT5 interacts with the HBV core protein and di-methylates arginine residues R150 and R156 in the CTD of core protein to modulate its subcellular distribution. While symmetric di-methylation favors core protein nuclear accumulation, monomethylation of core protein results in cytoplasmic accumulation91. The altered intracellular trafficking of core protein should not only interfere with capsid assembly, but also affect core protein-mediated PRMT5 recruitment and the formation of H4R3me2s repressive mark on cccDNA minichromosome44. Moreover, it has been known for a long time that IFN-α treatment inhibited pgRNA encapsidation92, but the detailed mechanism involved in the cytokine-induced cellular response leading to disruption of pgRNA packaging remains elusive.
Core protein dynamic phosphorylation regulates pgRNA packaging
HBV core protein consists of N-terminal assembly domain and arginine-rich CTD with seven conserved serines or threonine that are dynamically phosphorylated/dephosphorylated during the viral replication cycle (Fig. 6). Phosphomimetic mutagenesis and metabolic labeling studies indicated that CTD phosphorylation, particularly at S162 and S170, is required for pgRNA packaging. Our recent studies showed that pgRNA encapsidation is associated with core protein dephosphorylation93. Interestingly, while studies with HBV and DHBV demonstrated that CDK2 is assembled into nucleocapsids and phosphorylates core protein94, several other cellular kinases, such as SRPK1, SRPK295, PKA, CK2-aPKAIα/CK2-aPKAIIα, GAPD-PK, PKC, and PLK1, have also been reported to phosphorylate one or multiple phosphoaceptor sites in core protein CTD96. As depicted in Fig. 6, it has been reported that S155, S162 and S170 can be phosphorylated by CDK294, 97, S168, S176, and S178 can be phosphorylated by PLK1 and PKA, S155, S162, S168, S170, S176, S178 and S181 can be phosphorylated by SRPK98. Interestingly, it appeared that phosphorylation of S168, S176 and S178 is dependent on prior phosphorylation of S155, S162 and S17099. Cellular phosphatases that dephosphorylate core protein have not been identified.
Figure 6. HBV core protein and CTD phosphorylation.
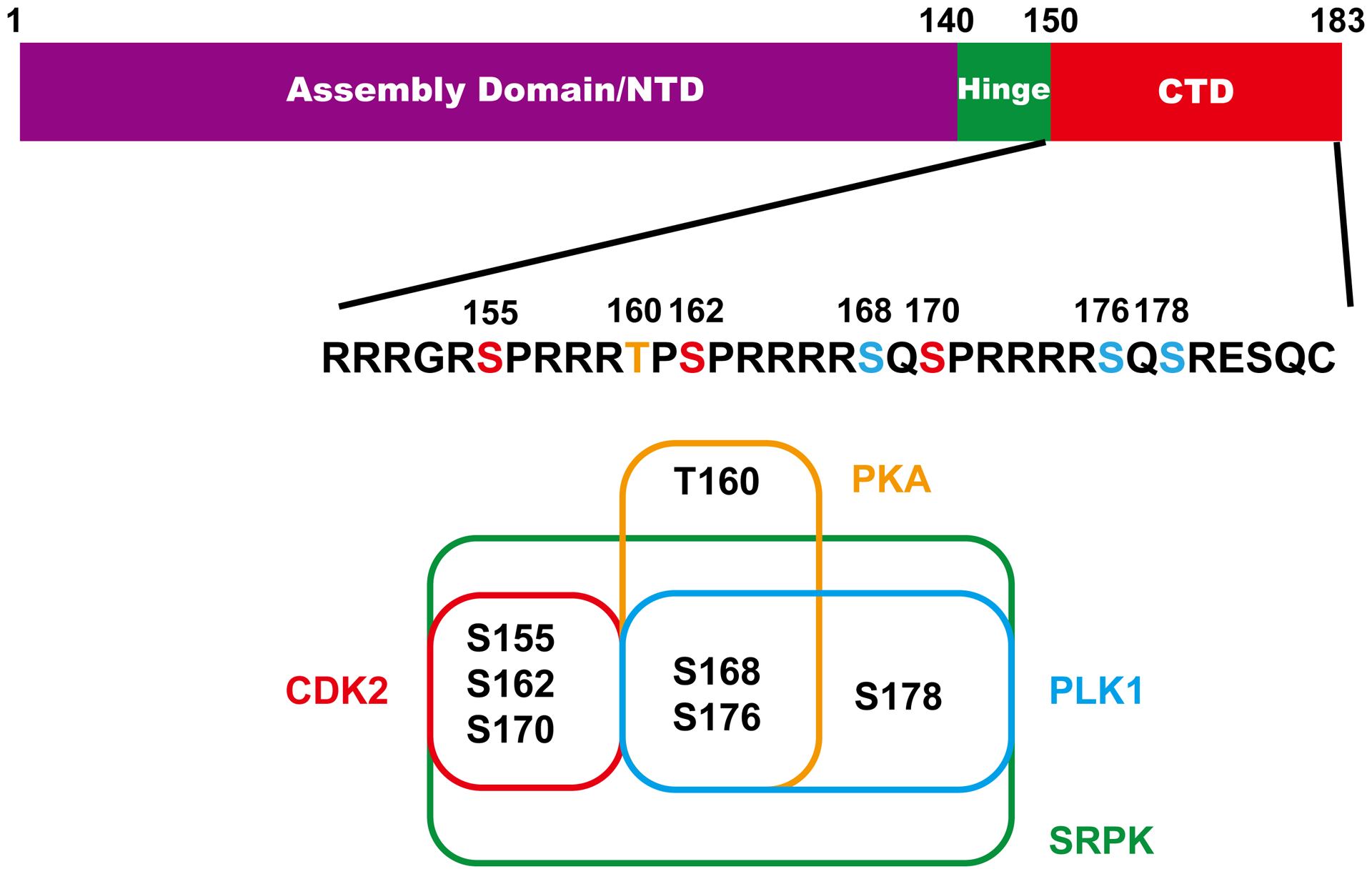
The domain structure of core protein and sequence of CTD domain are presented. The three major (red) and four minor (blue) phosphoacceptor sites are highlighted. Kinases and their ability to phosphorylate the specific serines/thorenine are also depicted.
Host functions regulate reverse transcription
HBV DNA synthesis in nucleocapsids is also regulated by host cellular factors. For example, although only 5 to 10% of DHBV genome in DHBV replicating cells are dslDNA, in vitro synthesis of DHBV DNA from purified pgRNA-containing capsids yields predominantly dslDNA22a, suggesting that the primer translocation/genome circularization is facilitated by host cellular factor(s) that are lost during the purification of pgRNA-containing capsids. In agreement with this observation, a recent study showed that the increased proportion of dslDNA in circulating HBV virions is associated with liver inflammation100. Furthermore, antiviral protein APOBEC3G can be packaged in nucleocapsids to inhibit reverse transcription and deaminate viral negative strand DNA, the later activity results in G to A hypermutation of HBV DNA101. However, although translation initiation factor eIF4E was demonstrated to be encapsidated with viral polymerase and pgRNA, its biological function has not been determined102.
Cellular functions are required for secretion of viral and subviral particles
As illustrated in Fig. 1, in addition to double-stranded DNA-containing mature nucleocapsids, viral RNA-containing capsids and empty capsids can also be enveloped and secreted out as virion-like particles, or RNA-containing virions and genome-free (GF) virions, respectively103. Interestingly, core proteins are hyperphosphorylated in GF-virions, but dephosphorylated in DNA-containing virions, or complete virions. However, the dephosphorylation is not required for mature nucleocapsid envelopementation104. In the blood of HBV-infected individuals, GF-virions are usually more abundant than that of complete virions, but RNA-containing virions are less abundant than that of complete virions105. The RNA species in RNA-containing virions can be either pgRNA or spliced viral RNA106. How those different cytoplasmic capsids are selectively enveloped and secreted as virions or virion-like particles is poorly understood103. Accumulating evidence suggests that assembly and secretion of complete HBV virions depend on intraluminal vesicles of maturing endosomes, i.e., the multivesicular bodies (MVB), and endosomal sorting complex required for transport (ESCRT) systems. Specifically, mature nucleocapsids are recognized by Nedd4. Ubiquitinated Nedd4 interacts with the ubiquitin-interacting adaptor γ2-adaptin and bridges membrane studded with envelope proteins and nucleocapsids to ESCRT machinery for virion assembly107. However, a recent study suggested that α-taxilin interacts with LHBs and the ESCRT I component tsg101 to recruit mature nucleocapsids to ESCRT machinery108. Nevertheless, the requirement of HBV morphogenesis on Vps4A/B and Alix/AIP1 implies that the pre-assembly complex utilizes the scission function of ESCRT-III/VPS4 to bud into the late endosomes or MVBs and exits cells by the exosome pathway109.
Besides the virions and virion-like particles, HBV infected hepatocytes also secrete two types of subviral particles (SVPs) that contain only viral envelope proteins, but not capsids, the small spheres and filaments of 20 nm in diameter. SVPs exist in the blood of infected individuals at 1,000 to 100,000-fold in excess over virions. While the small spheres, that contain SHBs and MHBs, bud into the lumen of the ER/ERGIC and exit cells via the constitutive secretion pathway, the filements, that share similar viral envelope protein contents with virions110, assembled and released via the ESCRT/MVB pathway like the infectious virions111.
2. Cellular non-coding and viral RNAs regulate HBV replication and pathogenesis
Virus-host interactions can also be mediated by RNA molecules encoded by viral and cellular genomes, such as non-coding (nc) RNA and microRNA. Many cellular microRNAs have been reported to modulate HBV replication by either directly targeting viral RNA or indirectly targeting cellular mRNA encoding proteins regulating HBV infection, which have been reviewed recently112. Herein, we only discuss the regulatory effects of recently identified non-coding RNA transcribed from cccDNA or integrated HBV DNA on viral replication and pathogenesis.
Using cap analysis of gene expression technology to quantitatively map the global transcriptional start site (TSS) distribution over the entire HBV genome in liver tissues, Altinel and colleagues identified 17 robust TSSs, including all the classically identified transcripts and some minor transcripts. Importantly, two minor viral transcripts in antisense polarity were identified, which might be involved in the regulation of the neighboring promoter activity113. Moreover, Yang and colleagues identified a HBV encoded microRNA, designated as HBV-miR-3, with its sequence mapped to the highly conserved HBsAg coding region of various HBV genotypes. HBV-miR-3 can be derived from 2.1kb, 2.4kb or 3.5kb mRNA, and targets the 3.5 kb mRNA at the ORF region of core gene to specifically reduce core protein expression, levels of pgRNA, and consequential viral replication114. The biological significance of HBV-miR-3 in HBV infection and pathogenesis in humans remains to be determined.
As discussed in detail below, integrated HBV DNA can transcribe viral-human chimeric RNAs. For instance, integration of HBV DNA into a long interspersed nuclear elements (LINE) in a normally silent intergenic region, chr.8p11.21, results in the production of an oncogenic HBx-LINE1 chimeric transcript. HBx-LINE1 can be detected in 23.3% of HBV-associated HCC tumors and correlates with poorer patient survival. Functioning as a long non-coding RNA, HBx-LINE1 activates Wnt/β-Catenin signaling and promotes cell motility through epithelial-to-mesenchymal transition, which contributes to the ultimate development of liver cancer115. Interestingly, HBx-LINE1 RNA has six binding sites for miR-122, a major microRNA in hepatocytes with important functions in cellular metabolism, development and differentiation116. It is thus possible that the epithelial-to-mesenchymal transition promoted by HBx-LINE1 works through a miR-122-sequestering mechanism117.
3. Genomic response of HBV infection and its biological consequences
Despite not a requirement for replication, HBV DNA occasionally integrates into chromosomal DNA of infected hepatocytes. Integration can be detected as early as a few days post infection and the frequency of integration increases in the infected livers with the duration of viral infection118. The dslDNA, a minor species of HBV genomic DNA derived from in situ priming of positive-stranded DNA synthesis, is the preferential substrate for integration119. Results obtained from genetic studies and sequence analyses of viral-host DNA junctions support the hypothesis that hepadnaviral DNA integration occurred at sites of host cell DNA damage, preferentially double strand DNA break, through non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ) DNA repair pathway120. Not surprisingly, it is well documented that DNA damage promotes viral DNA integration121. Moreover, a recent study revealed that the abundance of dslDNA in the circulating virions increases under the condition of liver inflammation100. Hence, the increased levels of dslDNA and host cellular DNA damage in inflammatory livers may facilitate HBV DNA integration and promote oncogenesis of infected hepatocytes via activation of oncogenes, inactivation of tumor suppressor genes, inducing genome instability and other mechanisms. As illustrated in Fig. 7, due to the disruption and loss of HBV DNA sequence corresponding to the 5’ and 3’ terminus of dslDNA, the integrated HBV DNAs are not possible to produce an authentic pgRNA to support viral DNA replication. However, at least some of transcripts derived from integrated DNA contain complete open reading frame of viral envelope proteins and can thus translate HBsAg. In fact, the significant contribution of integrated HBV DNA to circulating HBsAg in humans, particularly in patients who are HBeAg-negative or receive long-term therapy with NUCs, is only appreciated recently in a clinical trial of HBV mRNA-targeting small interference RNA (siRNA) therapy122. In addition to full-length dslDNA, as mentioned above, dslDNA derived from spliced pgRNA can also integrated into host cellular chromosomes to produce viral RNA as well as truncated viral proteins, viral-viral and/or viral-host fusion proteins and RNAs to modulate host cellular functions and play important role in HBV carcinogenesis123.
Figure 7. Structure and function of integrated HBV DNA.
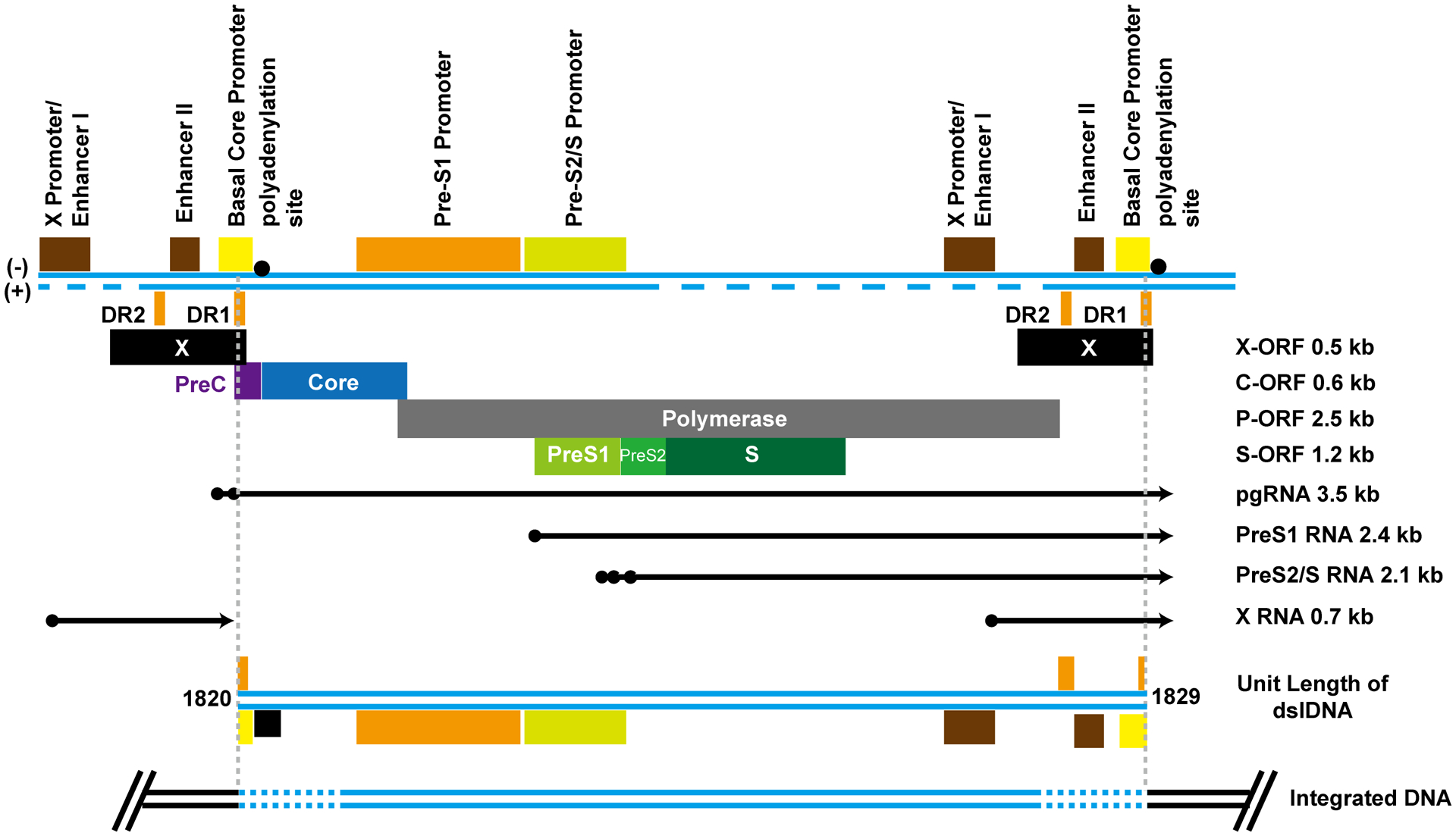
Top panel, HBV transcriptional cis regulatory elements, ORFs, and transcripts are presented in reference to linear DNA longer than unit-length of HBV genome. Middle panel, the structure of HBV DNA double stranded linear (dsl) DNA genome is presented. Low panel, the structure of integrated HBV DNA is presented. Due to the processing of the dslDNA ends during integration, variable length of DNA sequences may be lost (dashed lines).
4. Virological basis for the cure of chronic HBV infection
Complete (or sterilized) cure of HBV infection means the eradication of all free viruses as well as killing and/or cure of all HBV-infected cells from an infected individual. Due to the fast clearance of cell-free viruses from circulation124, cure of a HBV-infected hepatocyte or individual can be achieved by complete inhibition of HBV DNA replication for a period of time (Tcure) that allows the complete decay of the most stable HBV DNA replication intermediate, cccDNA. Obviously, the time required to cure an infected individual is determined by the fraction of infected cells with longest Tcure, such as the cells with very low rate of turnover and long life span. Selective killing of this fraction of HBV infected cells will accelerate the cure. Because complete inhibition of HBV DNA replication has not been achieved by current available antiviral agents125, the Tcure for an infected individual is difficult to estimate. It is our hope that combination therapy of multiple HBV DNA replication inhibitors, such as NUCs and CpAMs, will more efficiently inhibit HBV replication and cure the HBV infection with a finite, ideally a reasonably short Tcure. However, accumulating evidence suggests that immunological resolution of acute HBV infection is not a complete cure, but a tight immune control of a very low level of residual HBV infection (replication)126. Therefore, the complete cure of HBV infection is desirable, but may not be realistic. Practically, the therapeutic goal of chronic hepatitis B is to achieve a sustained loss of HBsAg or more ideally, anti-HBs seroconversion, an indication of stable immune control of HBV infection or a functional cure127. In addition to complete inhibition of viral replication by combination antiviral therapy24a and selective killing of HBV-infected cells by immunotherapies128, elimination or transcriptional silencing of pre-existing cccDNA in the nuclei of infected hepatocytes will cure HBV infected cells or reduce viral antigen load, which may be essential for restoration of functional host antiviral adaptive immune response and ultimately achieve a functional cure5, 9. Although it has been demonstrated that CRISPR/Cas9 or other DNA editing technologies can efficiently mutate and functionally inactivate cccDNA in HBV infected hepatocytes61, it is a challenge to deliver the DNA editing molecules into every HBV-infected hepatocytes to inactivate all the cccDNA molecules.
To therapeutically silence cccDNA transcription, it is essential to specifically target the transcription factors or epigenetic modifications that are unique to cccDNA minichromosomes. Unfortunately, such unique cccDNA transcription regulation properties have not yet been identified. However, recent findings about the critical role of HBx protein in cccDNA transcription regulation suggest that HBx itself is an ideal target for the future discovery of small molecules towards permanent silencing of cccDNA transcription. For instance, small molecules that selectively bind HBx to disrupt its interaction with Cullin4-DDB1-E3 ligase complex would inhibit degradation of Smc5/6 in infected hepatocytes and restore the heterochromatin status of cccDNA minichromosomes (Fig. 3). As discussed in previous sections, several lines of circumstantial evidence indicate that HBV core protein may also bind cccDNA minichromosome and regulate its transcriptional activity. If this is the case, disruption of core protein interaction with cccDNA is also an ideal therapeutic target to silence cccDNA function.
As a stealthy virus, HBV escapes cell intrinsic antiviral mechanisms through avoiding recognition by pattern recognition receptors (PRRs) rather than blocking its effecter functions. In line with this notion, HBV replication can be inhibited by agonists of several endosomal and cytoplasmic PRRs129. Due to the difficulty to eliminate residual amounts of HBV cccDNA from infected individuals, a functional cure requires activation of host adaptive antiviral immune response, particularly, HBV-specific cytolytic T cell response, to maintain a sustained surveillance of HBV reactivation.
Finally, recent findings that integrated HBV DNA serves as the significant contributor of HBsAg in circulation as well as HBx-LINE1 RNA plays an important role in HCC oncogenesis highlight the importance to therapeutically target integrated DNA and their transcripts for the functional cure of chronic HBV infection and prevention of HCC development. While CRISPR/Cas9-targeted cleavage and inactivation of integrated DNA may result in chromatin DNA break, RNA interference and small molecules specifically inhibiting cellular PAPD5/7 have been demonstrated to selectively induce degradation of viral RNA transcribed from cccDNA and integrated HBV DNA. It is anticipated that further investigation of HBV-host interaction may reveal novel molecular targets for safer and more efficient elimination or inactivation of cccDNA, restore host antiviral immune response and ultimately cure chronic HBV infection.
Acknowledgement
Writing this review article is supported by a NIH grant R01 AI113267 and the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award No. W81XWH-17-1-0600. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
Conflict interest statement
The authors declared the following competing financial interest(s): J.-T. G receives research fund from and holds stock of Arbutus Biopharma, Inc. J.C. receives research fund from Arbutus Biopharma, Inc..
BRBREVIATIONS
- cccDNA
covalently closed circular DNA
- CpAM
core protein allosteric modulator
- DDB1
DNA-damage binding protein 1
- DHBV
duck hepatitis B virus
- DHQ-1
dihydroquinolizinone
- dslDNA
double-stranded linear DNA
- EBV
Epstein-Barr virus
- ESCRT
endosomal sorting complex required for transport
- HBsAg
hepatitis B virus surface antigen
- HBV
hepatitis B virus
- HNF
hepatocyte nuclear factor
- HSPG
heparan sulfate proteoglycan
- ISG20
interferon stimulated gene 20
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- MMEJ
microhomology-mediated end joining
- MVB
multivesicular bodies
- NHEJ
non-homologous end joining
- NLS
nuclear localization signal
- NPCs
nuclear pore complexes
- NTCP
sodium taurocholate cotransporting polypeptide
- PAPD5
poly(A) polymerases D5
- pgRNA
pregenomic RNA
- PRE
post-transcriptional regulatory element
- PRMT
protein arginine methyltransferase
- rcDNA
relaxed circular DNA
- SMC5/6
structural maintenance of chromosomes 5/6
- TDP2
Tyrosol-DNA phosphodiesterase 2
- WHV
woodchuck hepatitis virus
- ZAP
zinc finger antiviral protein
References
- 1.Kaufmann SHE; Dorhoi A; Hotchkiss RS; Bartenschlager R, Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov 2018, 17 (1), 35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puschnik AS; Majzoub K; Ooi YS; Carette JE, A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol 2017, 15 (6), 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe T; Kawaoka Y, Influenza virus-host interactomes as a basis for antiviral drug development. Curr Opin Virol 2015, 14, 71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Block TM; Guo H; Guo JT, Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis 2007, 11 (4), 685–706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo JT; Guo H, Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 2015, 122, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott JJ; Stevens GA; Groeger J; Wiersma ST, Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30 (12), 2212–9. [DOI] [PubMed] [Google Scholar]
- 7.Mortalit GBD Causes of Death, C., Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385 (9963), 117–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Dienstag JL, Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology 2009, 49 (5 Suppl), S112–21; [DOI] [PubMed] [Google Scholar]; (b) Perrillo R, Benefits and risks of interferon therapy for hepatitis B. Hepatology 2009, 49 (5 Suppl), S103–11. [DOI] [PubMed] [Google Scholar]
- 9.Tang L; Zhao Q; Wu S; Cheng J; Chang J; Guo JT, The current status and future directions of hepatitis B antiviral drug discovery. Expert opinion on drug discovery 2017, 12 (1), 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Wieland S; Thimme R; Purcell RH; Chisari FV, Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 2004, 101 (17), 6669–74; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Guo F; Tang L; Shu S; Sehgal M; Sheraz M; Liu B; Zhao Q; Cheng J; Zhao X; Zhou T; Chang J; Guo JT, Activation of STING in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cheng X; Xia Y; Serti E; Block PD; Chung M; Chayama K; Rehermann B; Liang TJ, Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mutz P; Metz P; Lempp FA; Bender S; Qu B; Schoneweis K; Seitz S; Tu T; Restuccia A; Frankish J; Dachert C; Schusser B; Koschny R; Polychronidis G; Schemmer P; Hoffmann K; Baumert TF; Binder M; Urban S; Bartenschlager R, HBV Bypasses the Innate Immune Response and Does not Protect HCV From Antiviral Activity of Interferon. Gastroenterology 2018. [DOI] [PubMed] [Google Scholar]
- 11.Schulze A; Gripon P; Urban S, Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46 (6), 1759–68. [DOI] [PubMed] [Google Scholar]
- 12.(a) Yan H; Zhong G; Xu G; He W; Jing Z; Gao Z; Huang Y; Qi Y; Peng B; Wang H; Fu L; Song M; Chen P; Gao W; Ren B; Sun Y; Cai T; Feng X; Sui J; Li W, Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, e00049; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ni Y; Lempp FA; Mehrle S; Nkongolo S; Kaufman C; Falth M; Stindt J; Koniger C; Nassal M; Kubitz R; Sultmann H; Urban S, Hepatitis B and D viruses exploit sodium taurocholate cotransporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146 (4), 1070–83. [DOI] [PubMed] [Google Scholar]
- 13.Petersen J; Dandri M; Mier W; Lutgehetmann M; Volz T; von Weizsacker F; Haberkorn U; Fischer L; Pollok JM; Erbes B; Seitz S; Urban S, Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol 2008, 26 (3), 335–41. [DOI] [PubMed] [Google Scholar]
- 14.Verrier ER; Colpitts CC; Bach C; Heydmann L; Weiss A; Renaud M; Durand SC; Habersetzer F; Durantel D; Abou-Jaoude G; Lopez Ledesma MM; Felmlee DJ; Soumillon M; Croonenborghs T; Pochet N; Nassal M; Schuster C; Brino L; Sureau C; Zeisel MB; Baumert TF, A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 2016, 63 (1), 35–48. [DOI] [PubMed] [Google Scholar]
- 15.(a) Macovei A; Radulescu C; Lazar C; Petrescu S; Durantel D; Dwek RA; Zitzmann N; Nichita NB, Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J Virol 2010, 84 (1), 243–53; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huang HC; Chen CC; Chang WC; Tao MH; Huang C, Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J Virol 2012, 86 (17), 9443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macovei A; Petrareanu C; Lazar C; Florian P; Branza-Nichita N, Regulation of hepatitis B virus infection by Rab5, Rab7, and the endolysosomal compartment. J Virol 2013, 87 (11), 6415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucifora J; Protzer U, Attacking hepatitis B virus cccDNA--The holy grail to hepatitis B cure. J Hepatol 2016, 64 (1 Suppl), S41–S48. [DOI] [PubMed] [Google Scholar]
- 18.(a) Mason WS; Halpern MS; England JM; Seal G; Egan J; Coates L; Aldrich C; Summers J, Experimental transmission of duck hepatitis B virus. Virology 1983, 131 (2), 375–84; [DOI] [PubMed] [Google Scholar]; (b) Tuttleman JS; Pourcel C; Summers J, Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 1986, 47 (3), 451–60. [DOI] [PubMed] [Google Scholar]
- 19.(a) Cui X; Guo JT; Hu J, Hepatitis B Virus Covalently Closed Circular DNA Formation in Immortalized Mouse Hepatocytes Associated with Nucleocapsid Destabilization. J Virol 2015, 89 (17), 9021–8; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Guo H; Jiang D; Zhou T; Cuconati A; Block TM; Guo JT, Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 2007, 81 (22), 12472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y; Gao Z; Xu G; Peng B; Liu C; Yan H; Yao Q; Sun G; Liu Y; Tang D; Song Z; He W; Sun Y; Guo JT; Li W, DNA Polymerase kappa Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog 2016, 12 (10), e1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osseman Q; Gallucci L; Au S; Cazenave C; Berdance E; Blondot ML; Cassany A; Begu D; Ragues J; Aknin C; Sominskaya I; Dishlers A; Rabe B; Anderson F; Pante N; Kann M, The chaperone dynein LL1 mediates cytoplasmic transport of empty and mature hepatitis B virus capsids. J Hepatol 2017. [DOI] [PubMed] [Google Scholar]
- 22.(a) Guo H; Mao R; Block TM; Guo JT, Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol 2010, 84 (1), 387–96; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gallucci L; Kann M, Nuclear Import of Hepatitis B Virus Capsids and Genome. Viruses 2017, 9 (1); [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Roseman AM; Berriman JA; Wynne SA; Butler PJ; Crowther RA, A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci U S A 2005, 102 (44), 15821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C; Wang JC; Pierson EE; Keifer DZ; Delaleau M; Gallucci L; Cazenave C; Kann M; Jarrold MF; Zlotnick A, Importin beta Can Bind Hepatitis B Virus Core Protein and Empty Core-Like Particles and Induce Structural Changes. PLoS Pathog 2016, 12 (8), e1005802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Guo F; Zhao Q; Sheraz M; Cheng J; Qi Y; Su Q; Cuconati A; Wei L; Du Y; Li W; Chang J; Guo JT, HBV core protein allosteric modulators differentially alter cccDNA biosynthesis from de novo infection and intracellular amplification pathways. PLoS Pathog 2017, 13 (9), e1006658; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cui X; Luckenbaugh L; Bruss V; Hu J, Alteration of Mature Nucleocapsid and Enhancement of Covalently Closed Circular DNA Formation by Hepatitis B Virus Core Mutants Defective in Complete-Virion Formation. J Virol 2015, 89 (19), 10064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alber F; Dokudovskaya S; Veenhoff LM; Zhang W; Kipper J; Devos D; Suprapto A; Karni-Schmidt O; Williams R; Chait BT; Sali A; Rout MP, The molecular architecture of the nuclear pore complex. Nature 2007, 450 (7170), 695–701. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz A; Schwarz A; Foss M; Zhou L; Rabe B; Hoellenriegel J; Stoeber M; Pante N; Kann M, Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog 2010, 6 (1), e1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Guo H; Xu C; Zhou T; Block TM; Guo JT, Characterization of the Host Factors Required for Hepadnavirus Covalently Closed Circular (ccc) DNA Formation. PLoS One 2012, 7 (8), e43270; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Long Q; Yan R; Hu J; Cai D; Mitra B; Kim ES; Marchetti A; Zhang H; Wang S; Liu Y; Huang A; Guo H, The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog 2017, 13 (12), e1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schellenberg MJ; Appel CD; Adhikari S; Robertson PD; Ramsden DA; Williams RS, Mechanism of repair of 5’-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol 2012, 19 (12), 1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koniger C; Wingert I; Marsmann M; Rosler C; Beck J; Nassal M, Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci U S A 2014, 111 (40), E4244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui X; McAllister R; Boregowda R; Sohn JA; Cortes Ledesma F; Caldecott KW; Seeger C; Hu J, Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One 2015, 10 (6), e0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J; Cui X; Gao L; Hu J, Identification of Intermediate in Hepatitis B Virus CCC DNA Formation and Sensitive and Selective CCC DNA Detection. J Virol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song M; Sun Y; Tian J; He W; Xu G; Jing Z; Li W, Silencing Retinoid X Receptor Alpha Expression Enhances Early-stage Hepatitis B Virus Infection In Cell Cultures. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bock CT; Schranz P; Schroder CH; Zentgraf H, Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes 1994, 8 (3), 215–29. [DOI] [PubMed] [Google Scholar]
- 34.Tropberger P; Mercier A; Robinson M; Zhong W; Ganem DE; Holdorf M, Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci U S A 2015, 112 (42), E5715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F; Campagna M; Qi Y; Zhao X; Guo F; Xu C; Li S; Li W; Block TM; Chang J; Guo JT, Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog 2013, 9 (9), e1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Breugel PC; Robert EI; Mueller H; Decorsiere A; Zoulim F; Hantz O; Strubin M, Hepatitis B virus X protein stimulates gene expression selectively from extrachromosomal DNA templates. Hepatology 2012, 56 (6), 2116–24. [DOI] [PubMed] [Google Scholar]
- 37.(a) Belloni L; Pollicino T; De Nicola F; Guerrieri F; Raffa G; Fanciulli M; Raimondo G; Levrero M, Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 2009, 106 (47), 19975–9; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Riviere L; Gerossier L; Ducroux A; Dion S; Deng Q; Michel ML; Buendia MA; Hantz O; Neuveut C, HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol 2015, 63 (5), 1093–102. [DOI] [PubMed] [Google Scholar]
- 38.Alarcon V; Hernandez S; Rubio L; Alvarez F; Flores Y; Varas-Godoy M; De Ferrari GV; Kann M; Villanueva RA; Loyola A, The enzymes LSD1 and Set1A cooperate with the viral protein HBx to establish an active hepatitis B viral chromatin state. Sci Rep 2016, 6, 25901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ducroux A; Benhenda S; Riviere L; Semmes OJ; Benkirane M; Neuveut C, The Tudor domain protein Spindlin1 is involved in intrinsic antiviral defense against incoming hepatitis B Virus and herpes simplex virus type 1. PLoS Pathog 2014, 10 (9), e1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benhenda S; Ducroux A; Riviere L; Sobhian B; Ward MD; Dion S; Hantz O; Protzer U; Michel ML; Benkirane M; Semmes OJ; Buendia MA; Neuveut C, Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. J Virol 2013, 87 (8), 4360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cougot D; Allemand E; Riviere L; Benhenda S; Duroure K; Levillayer F; Muchardt C; Buendia MA; Neuveut C, Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci Signal 2012, 5 (205), ra1. [DOI] [PubMed] [Google Scholar]
- 42.(a) Decorsiere A; Mueller H; van Breugel PC; Abdul F; Gerossier L; Beran RK; Livingston CM; Niu C; Fletcher SP; Hantz O; Strubin M, Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016, 531 (7594), 386–9; [DOI] [PubMed] [Google Scholar]; (b) Murphy CM; Xu Y; Li F; Nio K; Reszka-Blanco N; Li X; Wu Y; Yu Y; Xiong Y; Su L, Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep 2016, 16 (11), 2846–2854; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Niu C; Livingston CM; Li L; Beran RK; Daffis S; Ramakrishnan D; Burdette D; Peiser L; Salas E; Ramos H; Yu M; Cheng G; Strubin M; Delaney WI; Fletcher SP, The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS One 2017, 12 (1), e0169648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.(a) Hu Z; Zhang Z; Doo E; Coux O; Goldberg AL; Liang TJ, Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol 1999, 73 (9), 7231–40; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu N; Zhang J; Yang X; Jiao T; Zhao X; Li W; Zhu J; Yang P; Jin J; Peng J; Li Z; Ye X, HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function. J Virol 2017, 91 (16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W; Chen J; Wu M; Zhang X; Zhang M; Yue L; Li Y; Liu J; Li B; Shen F; Wang Y; Bai L; Protzer U; Levrero M; Yuan Z, PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology 2017, 66 (2), 398–415. [DOI] [PubMed] [Google Scholar]
- 45.Guo YH; Li YN; Zhao JR; Zhang J; Yan Z, HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 2011, 6 (6), 720–6. [DOI] [PubMed] [Google Scholar]
- 46.(a) Zhang Y; Mao R; Yan R; Cai D; Zhang Y; Zhu H; Kang Y; Liu H; Wang J; Qin Y; Huang Y; Guo H; Zhang J, Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 2014, 9 (10), e110442; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhong C; Lu H; Han T; Tan X; Li P; Huang J; Xie Q; Hou Z; Qu T; Jiang Y; Wang S; Xu L; Zhong Y; Huang T, CpG methylation participates in regulation of hepatitis B virus gene expression in host sperm and sperm-derived embryos. Epigenomics 2017, 9 (2), 123–125. [DOI] [PubMed] [Google Scholar]
- 47.(a) Jain S; Chang TT; Chen S; Boldbaatar B; Clemens A; Lin SY; Yan R; Hu CT; Guo H; Block TM; Song W; Su YH, Comprehensive DNA methylation analysis of hepatitis B virus genome in infected liver tissues. Sci Rep 2015, 5, 10478; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kaur P; Paliwal A; Durantel D; Hainaut P; Scoazec JY; Zoulim F; Chemin I; Herceg Z, DNA methylation of hepatitis B virus (HBV) genome associated with the development of hepatocellular carcinoma and occult HBV infection. J Infect Dis 2010, 202 (5), 700–4. [DOI] [PubMed] [Google Scholar]
- 48.(a) Kim DH; Kang HS; Kim KH, Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J Gastroenterol 2016, 22 (31), 7017–29; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Quasdorff M; Protzer U, Control of hepatitis B virus at the level of transcription. J Viral Hepat 2010, 17 (8), 527–36. [DOI] [PubMed] [Google Scholar]
- 49.Raney AK; Easton AJ; Milich DR; McLachlan A, Promoter-specific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. Journal of Virology 1991, 65 (11), 5774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.(a) Raney AK; Eggers CM; Kline EF; Guidotti LG; Pontoglio M; Yaniv M; McLachlan A, Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol 2001, 75 (6), 2900–11; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cai YN; Zhou Q; Kong YY; Li M; Viollet B; Xie YH; Wang Y, LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell research 2003, 13 (6), 451–8. [DOI] [PubMed] [Google Scholar]
- 51.Pasquetto V; Wieland SF; Uprichard SL; Tripodi M; Chisari FV, Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J Virol 2002, 76 (11), 5646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.(a) Xiang WQ; Feng WF; Ke W; Sun Z; Chen Z; Liu W, Hepatitis B virus X protein stimulates IL-6 expression in hepatocytes via a MyD88-dependent pathway. Journal of hepatology 2011, 54 (1), 26–33; [DOI] [PubMed] [Google Scholar]; (b) Hosel M; Quasdorff M; Wiegmann K; Webb D; Zedler U; Broxtermann M; Tedjokusumo R; Esser K; Arzberger S; Kirschning CJ; Langenkamp A; Falk C; Buning H; Rose-John S; Protzer U, Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology 2009, 50 (6), 1773–82. [DOI] [PubMed] [Google Scholar]
- 53.Lin SJ; Shu PY; Chang C; Ng AK; Hu CP, IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. Journal of immunology 2003, 171 (9), 4708–16. [DOI] [PubMed] [Google Scholar]
- 54.Lin YC; Hsu EC; Ting LP, Repression of hepatitis B viral gene expression by transcription factor nuclear factor-kappaB. Cellular microbiology 2009, 11 (4), 645–60. [DOI] [PubMed] [Google Scholar]
- 55.Shin GC; Ahn SH; Choi HS; Kim J; Park ES; Kim DH; Kim KH, Hepatocystin contributes to interferon-mediated antiviral response to hepatitis B virus by regulating hepatocyte nuclear factor 4alpha. Biochimica et biophysica acta 2014, 1842 (9), 1648–57. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Y; Li J; Johnson DL; Ou JH, Regulation of hepatitis B virus replication by the ras-mitogen-activated protein kinase signaling pathway. J Virol 2003, 77 (14), 7707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo H; Zhou T; Jiang D; Cuconati A; Xiao GH; Block TM; Guo JT, Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J Virol 2007, 81 (18), 10072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang SH; Yeh SH; Lin WH; Yeh KH; Yuan Q; Xia NS; Chen DS; Chen PJ, Estrogen receptor alpha represses transcription of HBV genes via interaction with hepatocyte nuclear factor 4alpha. Gastroenterology 2012, 142 (4), 989–998 e4. [DOI] [PubMed] [Google Scholar]
- 59.Wang SH; Yeh SH; Lin WH; Wang HY; Chen DS; Chen PJ, Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology 2009, 50 (5), 1392–402. [DOI] [PubMed] [Google Scholar]
- 60.Moraleda G; Saputelli J; Aldrich CE; Averett D; Condreay L; Mason WS, Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol 1997, 71 (12), 9392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seeger C; Sohn JA, Complete Spectrum of CRISPR/Cas9-induced Mutations on HBV cccDNA. Mol Ther 2016, 24 (7), 1258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Guidotti LG; Rochford R; Chung J; Shapiro M; Purcell R; Chisari FV, Viral clearance without destruction of infected cells during acute HBV infection. Science 1999, 284 (5415), 825–9; [DOI] [PubMed] [Google Scholar]; (b) Guo JT; Zhou H; Liu C; Aldrich C; Saputelli J; Whitaker T; Barrasa MI; Mason WS; Seeger C, Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J Virol 2000, 74 (3), 1495–505; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang YY; Zhang BH; Theele D; Litwin S; Toll E; Summers J, Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc Natl Acad Sci U S A 2003, 100 (21), 12372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.(a) Lucifora J; Xia Y; Reisinger F; Zhang K; Stadler D; Cheng X; Sprinzl MF; Koppensteiner H; Makowska Z; Volz T; Remouchamps C; Chou WM; Thasler WE; Huser N; Durantel D; Liang TJ; Munk C; Heim MH; Browning JL; Dejardin E; Dandri M; Schindler M; Heikenwalder M; Protzer U, Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343 (6176), 1221–8; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xia Y; Stadler D; Lucifora J; Reisinger F; Webb D; Hosel M; Michler T; Wisskirchen K; Cheng X; Zhang K; Chou WM; Wettengel JM; Malo A; Bohne F; Hoffmann D; Eyer F; Thimme R; Falk CS; Thasler WE; Heikenwalder M; Protzer U, Interferon-gamma and Tumor Necrosis Factor-alpha Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis. Gastroenterology 2016, 150 (1), 194–205. [DOI] [PubMed] [Google Scholar]
- 64.Koh S; Kah J; Tham CYL; Yang N; Ceccarello E; Chia A; Chen M; Khakpoor A; Pavesi A; Tan AT; Dandri M; Bertoletti A, Non-lytic Lymphocytes Engineered to Express Virus-specific T-cell Receptors Limit HBV Infection by Activating APOBEC3. Gastroenterology 2018. [DOI] [PubMed] [Google Scholar]
- 65.(a) Shen F; Li Y; Yang W; Sozzi V; Revill PA; Liu J; Gao L; Yang G; Lu M; Sutter K; Dittmer U; Chen J; Yuan Z, Hepatitis B Virus Sensitivity to Interferon-alpha in Hepatocytes is More Associated with Cellular Interferon Response than with Viral Genotype. Hepatology 2017; [DOI] [PubMed] [Google Scholar]; (b) Niu C; Li L; Daffis S; Lucifora J; Bonnin M; Maadadi S; Salas E; Chu R; Ramos H; Livingston CM; Beran RK; Garg AV; Balsitis S; Durantel D; Zoulim F; Delaney W. E. t.; Fletcher SP, Toll-Like Receptor 7 Agonist GS-9620 Induces Prolonged Inhibition of HBV via a Type I Interferon-Dependent Mechanism. J Hepatol 2017. [DOI] [PubMed] [Google Scholar]
- 66.(a) Ballestas ME; Chatis PA; Kaye KM, Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 1999, 284 (5414), 641–4; [DOI] [PubMed] [Google Scholar]; (b) Hung SC; Kang MS; Kieff E, Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci U S A 2001, 98 (4), 1865–70; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) You J; Croyle JL; Nishimura A; Ozato K; Howley PM, Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 2004, 117 (3), 349–60. [DOI] [PubMed] [Google Scholar]
- 67.Allweiss L; Volz T; Giersch K; Kah J; Raffa G; Petersen J; Lohse AW; Beninati C; Pollicino T; Urban S; Lutgehetmann M; Dandri M, Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 2018, 67 (3), 542–552. [DOI] [PubMed] [Google Scholar]
- 68.Li M; Sohn JA; Seeger C, Distribution of Hepatitis B Virus Nuclear DNA. J Virol 2018, 92 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.(a) Su TS; Lai CJ; Huang JL; Lin LH; Yauk YK; Chang CM; Lo SJ; Han SH, Hepatitis B virus transcript produced by RNA splicing. J Virol 1989, 63 (9), 4011–8; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ogston CW; Razman DG, Spliced RNA of woodchuck hepatitis virus. Virology 1992, 189 (1), 245–52; [DOI] [PubMed] [Google Scholar]; (c) Obert S; Zachmann-Brand B; Deindl E; Tucker W; Bartenschlager R; Schaller H, A splice hepadnavirus RNA that is essential for virus replication. EMBO J 1996, 15 (10), 2565–74. [PMC free article] [PubMed] [Google Scholar]
- 70.(a) Terre S; Petit MA; Brechot C, Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J Virol 1991, 65 (10), 5539–43; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Heise T; Sommer G; Reumann K; Meyer I; Will H; Schaal H, The hepatitis B virus PRE contains a splicing regulatory element. Nucleic Acids Res 2006, 34 (1), 353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chowdhury JB; Roy D; Ghosh S, Identification of a unique splicing regulatory cluster in hepatitis B virus pregenomic RNA. FEBS Lett 2011, 585 (20), 3348–53. [DOI] [PubMed] [Google Scholar]
- 72.Duriez M; Mandouri Y; Lekbaby B; Wang H; Schnuriger A; Redelsperger F; Guerrera CI; Lefevre M; Fauveau V; Ahodantin J; Quetier I; Chhuon C; Gourari S; Boissonnas A; Gill U; Kennedy P; Debzi N; Sitterlin D; Maini MK; Kremsdorf D; Soussan P, Alternative splicing of hepatitis B virus: A novel virus/host interaction altering liver immunity. J Hepatol 2017, 67 (4), 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abraham TM; Lewellyn EB; Haines KM; Loeb DD, Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology 2008, 379 (1), 30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziemer M; Garcia P; Shaul Y; Rutter WJ, Sequence of hepatitis B virus DNA incorporated into the genome of a human hepatoma cell line. J Virol 1985, 53 (3), 885–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin YM; Chen BF, A putative hepatitis B virus splice variant associated with chronic hepatitis and liver cirrhosis. Virology 2017, 510, 224–233. [DOI] [PubMed] [Google Scholar]
- 76.Ma ZM; Lin X; Wang YX; Tian XC; Xie YH; Wen YM, A double-spliced defective hepatitis B virus genome derived from hepatocellular carcinoma tissue enhanced replication of full-length virus. J Med Virol 2009, 81 (2), 230–7. [DOI] [PubMed] [Google Scholar]
- 77.Rosmorduc O; Petit MA; Pol S; Capel F; Bortolotti F; Berthelot P; Brechot C; Kremsdorf D, In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology 1995, 22 (1), 10–9. [PubMed] [Google Scholar]
- 78.Huang HL; Jeng KS; Hu CP; Tsai CH; Lo SJ; Chang C, Identification and characterization of a structural protein of hepatitis B virus: a polymerase and surface fusion protein encoded by a spliced RNA. Virology 2000, 275 (2), 398–410. [DOI] [PubMed] [Google Scholar]
- 79.Soussan P; Garreau F; Zylberberg H; Ferray C; Brechot C; Kremsdorf D, In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Invest 2000, 105 (1), 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.(a) Smith GJ 3rd; Donello JE; Luck R; Steger G; Hope TJ, The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res 1998, 26 (21), 4818–27; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schwalbe M; Ohlenschlager O; Marchanka A; Ramachandran R; Hafner S; Heise T; Gorlach M, Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element. Nucleic Acids Res 2008, 36 (5), 1681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.(a) Heise T; Guidotti LG; Chisari FV, La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J Virol 1999, 73 (7), 5767–76; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zang WQ; Li B; Huang PY; Lai MM; Yen TS, Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J Virol 2001, 75 (22), 10779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.(a) Zhou T; Block T; Liu F; Kondratowicz AS; Sun L; Rawat S; Branson J; Guo F; Steuer HM; Liang H; Bailey L; Moore C; Wang X; Cuconatti A; Gao M; Lee ACH; Harasym T; Chiu T; Gotchev D; Dorsey B; Rijnbrand R; Sofia MJ, HBsAg mRNA degradation induced by a dihydroquinolizinone compound depends on the HBV posttranscriptional regulatory element. Antiviral Res 2018, 149, 191–201; [DOI] [PubMed] [Google Scholar]; (b) Mueller H; Wildum S; Luangsay S; Walther J; Lopez A; Tropberger P; Ottaviani G; Lu W; Parrott NJ; Zhang JD; Schmucki R; Racek T; Hoflack JC; Kueng E; Point F; Zhou X; Steiner G; Lutgehetmann M; Rapp G; Volz T; Dandri M; Yang S; Young JAT; Javanbakht H, A novel orally available small molecule that inhibits hepatitis B virus expression. J Hepatol 2018, 68 (3), 412–420. [DOI] [PubMed] [Google Scholar]
- 83.Han X; Hassan J; Mueller H; Wang Y; Yang S, PAPD5 and PAPD7 inhibitors for treating a hepatitis infection. International Patent 2017, WO 2017/216391 A1. [Google Scholar]
- 84.Heise T; Guidotti LG; Cavanaugh VJ; Chisari FV, Hepatitis B virus RNA-binding proteins associated with cytokine-induced clearance of viral RNA from the liver of transgenic mice. J Virol 1999, 73 (1), 474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.(a) Liu Y; Nie H; Mao R; Mitra B; Cai D; Yan R; Guo JT; Block TM; Mechti N; Guo H, Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog 2017, 13 (4), e1006296; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mao R; Nie H; Cai D; Zhang J; Liu H; Yan R; Cuconati A; Block TM; Guo JT; Guo H, Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 2013, 9 (7), e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nassal M, Hepatitis B viruses: reverse transcription a different way. Virus Res 2008, 134 (1–2), 235–49. [DOI] [PubMed] [Google Scholar]
- 87.Hu J; Seeger C, Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci U S A 1996, 93 (3), 1060–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu J; Toft D; Anselmo D; Wang X, In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J Virol 2002, 76 (1), 269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X; Grammatikakis N; Hu J, Role of p50/CDC37 in hepadnavirus assembly and replication. J Biol Chem 2002, 277 (27), 24361–7. [DOI] [PubMed] [Google Scholar]
- 90.Hu J; Toft DO; Seeger C, Hepadnavirus assembly and reverse transcription require a multi- component chaperone complex which is incorporated into nucleocapsids. Embo J 1997, 16 (1), 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lubyova B; Hodek J; Zabransky A; Prouzova H; Hubalek M; Hirsch I; Weber J, PRMT5: A novel regulator of Hepatitis B virus replication and an arginine methylase of HBV core. PLoS One 2017, 12 (10), e0186982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schultz U; Summers J; Staeheli P; Chisari FV, Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J Virol 1999, 73 (7), 5459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Q; Hu Z; Cheng J; Wu S; Luo Y; Chang J; Hu J; Guo JT, Hepatitis B virus core protein dephosphorylation occurs during pregenomic RNA encapsidation. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ludgate L; Ning X; Nguyen DH; Adams C; Mentzer L; Hu J, Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol 2012, 86 (22), 12237–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.(a) Daub H; Blencke S; Habenberger P; Kurtenbach A; Dennenmoser J; Wissing J; Ullrich A; Cotten M, Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. Journal of virology 2002, 76 (16), 8124–37; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zheng Y; Fu XD; Ou JH, Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 2005, 342 (1), 150–8. [DOI] [PubMed] [Google Scholar]
- 96.(a) Okabe M; Enomoto M; Maeda H; Kuroki K; Ohtsuki K, Biochemical characterization of suramin as a selective inhibitor for the PKA-mediated phosphorylation of HBV core protein in vitro. Biological & pharmaceutical bulletin 2006, 29 (9), 1810–4; [DOI] [PubMed] [Google Scholar]; (b) Enomoto M; Sawano Y; Kosuge S; Yamano Y; Kuroki K; Ohtsuki K, High phosphorylation of HBV core protein by two alpha-type CK2-activated cAMP-dependent protein kinases in vitro. FEBS letters 2006, 580 (3), 894–9; [DOI] [PubMed] [Google Scholar]; (c) Duclos-Vallee JC; Capel F; Mabit H; Petit MA, Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. The Journal of general virology 1998, 79 ( Pt 7), 1665–70; [DOI] [PubMed] [Google Scholar]; (d) Kang H; Yu J; Jung G, Phosphorylation of hepatitis B virus core C-terminally truncated protein (Cp149) by PKC increases capsid assembly and stability. The Biochemical journal 2008, 416 (1), 47–54. [DOI] [PubMed] [Google Scholar]
- 97.Liu K; Ludgate L; Yuan Z; Hu J, Regulation of multiple stages of hepadnavirus replication by the carboxyl-terminal domain of viral core protein in trans. Journal of virology 2015, 89 (5), 2918–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen C; Wang JC; Zlotnick A, A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS pathogens 2011, 7 (11), e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.(a) Jung J; Hwang SG; Chwae YJ; Park S; Shin HJ; Kim K, Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication. J Virol 2014, 88 (16), 8754–67; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Diab A; Foca A; Fusil F; Lahlali T; Jalaguier P; Amirache F; N’Guyen L; Isorce N; Cosset FL; Zoulim F; Andrisani O; Durantel D, Polo-like-kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology 2017, 66 (6), 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao XL; Yang JR; Lin SZ; Ma H; Guo F; Yang RF; Zhang HH; Han JC; Wei L; Pan XB, Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 2016, 65 (3), 502–11. [DOI] [PubMed] [Google Scholar]
- 101.(a) Nguyen DH; Hu J, Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol 2008, 82 (14), 6852–61; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Baumert TF; Rosler C; Malim MH; von Weizsacker F, Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology 2007, 46 (3), 682–9. [DOI] [PubMed] [Google Scholar]
- 102.Kim S; Wang H; Ryu WS, Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J Virol 2010, 84 (1), 52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu J; Liu K, Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 2017, 9 (3), E56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ning X; Basagoudanavar SH; Liu K; Luckenbaugh L; Wei D; Wang C; Wei B; Zhao Y; Yan T; Delaney W; Hu J, Capsid Phosphorylation State and Hepadnavirus Virion Secretion. J Virol 2017, 91 (9), e00092–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luckenbaugh L; Kitrinos KM; Delaney W. E. t.; Hu J, Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J Viral Hepat 2015, 22 (6), 561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.(a) Wang J; Shen T; Huang X; Kumar GR; Chen X; Zeng Z; Zhang R; Chen R; Li T; Zhang T; Yuan Q; Li PC; Huang Q; Colonno R; Jia J; Hou J; McCrae MA; Gao Z; Ren H; Xia N; Zhuang H; Lu F, Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016; [DOI] [PubMed] [Google Scholar]; (b) Lam AM; Ren S; Espiritu C; Kelly M; Lau V; Zheng L; Hartman GD; Flores OA; Klumpp K, Hepatitis B Virus Capsid Assembly Modulators, but Not Nucleoside Analogs, Inhibit the Production of Extracellular Pregenomic RNA and Spliced RNA Variants. Antimicrob Agents Chemother 2017, 61 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rost M; Mann S; Lambert C; Doring T; Thome N; Prange R, Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation. J Biol Chem 2006, 281 (39), 29297–308. [DOI] [PubMed] [Google Scholar]
- 108.Hoffmann J; Boehm C; Himmelsbach K; Donnerhak C; Roettger H; Weiss TS; Ploen D; Hildt E, Identification of alpha-taxilin as an essential factor for the life cycle of hepatitis B virus. J Hepatol 2013, 59 (5), 934–41. [DOI] [PubMed] [Google Scholar]
- 109.Lambert C; Doring T; Prange R, Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol 2007, 81 (17), 9050–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Patient R; Hourioux C; Roingeard P, Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol 2009, 11 (11), 1561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang B; Himmelsbach K; Ren H; Boller K; Hildt E, Subviral Hepatitis B Virus Filaments, like Infectious Viral Particles, Are Released via Multivesicular Bodies. J Virol 2015, 90 (7), 3330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mizuguchi Y; Takizawa T; Uchida E, Host cellular microRNA involvement in the control of hepatitis B virus gene expression and replication. World journal of hepatology 2015, 7 (4), 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Altinel K; Hashimoto K; Wei Y; Neuveut C; Gupta I; Suzuki AM; Dos Santos A; Moreau P; Xia T; Kojima S; Kato S; Takikawa Y; Hidaka I; Shimizu M; Matsuura T; Tsubota A; Ikeda H; Nagoshi S; Suzuki H; Michel ML; Samuel D; Buendia MA; Faivre J; Carninci P, Single-Nucleotide Resolution Mapping of Hepatitis B Virus Promoters in Infected Human Livers and Hepatocellular Carcinoma. J Virol 2016, 90 (23), 10811–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang X; Li H; Sun H; Fan H; Hu Y; Liu M; Li X; Tang H, Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol 2017, 91 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lau CC; Sun T; Ching AK; He M; Li JW; Wong AM; Co NN; Chan AW; Li PS; Lung RW; Tong JH; Lai PB; Chan HL; To KF; Chan TF; Wong N, Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014, 25 (3), 335–49. [DOI] [PubMed] [Google Scholar]
- 116.Chang J; Nicolas E; Marks D; Sander C; Lerro A; Buendia MA; Xu C; Mason WS; Moloshok T; Bort R; Zaret KS; Taylor JM, miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 2004, 1 (2), 106–13. [DOI] [PubMed] [Google Scholar]
- 117.Liang HW; Wang N; Wang Y; Wang F; Fu Z; Yan X; Zhu H; Diao W; Ding Y; Chen X; Zhang CY; Zen K, Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol 2016, 64 (2), 278–291. [DOI] [PubMed] [Google Scholar]
- 118.(a) Summers J; Mason WS, Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc Natl Acad Sci U S A 2004, 101 (2), 638–40; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mason WS; Gill US; Litwin S; Zhou Y; Peri S; Pop O; Hong ML; Naik S; Quaglia A; Bertoletti A; Kennedy PT, HBV DNA Integration and Clonal Hepatocyte Expansion in Chronic Hepatitis B Patients Considered Immune Tolerant. Gastroenterology 2016, 151 (5), 986–998 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang W; Summers J, Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol 1999, 73 (12), 9710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bill CA; Summers J, Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A 2004, 101 (30), 11135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomez-Moreno A; Garaigorta U, Hepatitis B Virus and DNA Damage Response: Interactions and Consequences for the Infection. Viruses 2017, 9 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wooddell CI; Yuen MF; Chan HL; Gish RG; Locarnini SA; Chavez D; Ferrari C; Given BD; Hamilton J; Kanner SB; Lai CL; Lau JYN; Schluep T; Xu Z; Lanford RE; Lewis DL, RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017, 9 (409). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levrero M; Zucman-Rossi J, Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol 2016, 64 (1 Suppl), S84–S101. [DOI] [PubMed] [Google Scholar]
- 124.Nowak MA; Bonhoeffer S; Hill AM; Boehme R; Thomas HC; McDade H, Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A 1996, 93 (9), 4398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang P; Liu F; Guo F; Zhao Q; Chang J; Guo JT, Characterization of novel hepadnaviral RNA species accumulated in hepatoma cells treated with viral DNA polymerase inhibitors. Antiviral Res 2016, 131, 40–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rehermann B; Ferrari C; Pasquinelli C; Chisari FV, The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med 1996, 2 (10), 1104–8. [DOI] [PubMed] [Google Scholar]
- 127.Block TM; Gish R; Guo H; Mehta A; Cuconati A; Thomas London W; Guo JT, Chronic hepatitis B: what should be the goal for new therapies? Antiviral Res 2013, 98 (1), 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kah J; Koh S; Volz T; Ceccarello E; Allweiss L; Lutgehetmann M; Bertoletti A; Dandri M, Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J Clin Invest 2017, 127 (8), 3177–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chang J; Guo JT, Treatment of chronic hepatitis B with pattern recognition receptor agonists: Current status and potential for a cure. Antiviral Res 2015, 121, 152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


