Abstract
Vision begins when light is captured by the outer segment organelle of photoreceptor cells in the retina. Outer segments are modified cilia filled with hundreds of flattened disk-shaped membranes. Disk membranes are separated from the surrounding plasma membrane, and each membrane type has unique protein components. The mechanisms underlying this protein sorting remain entirely unknown. In this study, we investigated the outer segment delivery of the rod cyclic nucleotide-gated (CNG) channel, which is located in the outer segment plasma membrane, where it mediates the electrical response to light. Using Xenopus and mouse models of both sexes, we now show that the targeted delivery of the CNG channel to the outer segment uses the conventional secretory pathway, including protein processing in both ER and Golgi, and requires preassembly of its constituent α1 and β1 subunits. We further demonstrate that the N-terminal glutamic acid-rich protein (GARP) domain of CNGβ1 contains two distinct functional regions. The glutamic acid-rich region encodes specific information targeting the channel to rod outer segments. The adjacent proline-enriched region connects the CNG channel to photoreceptor disk rims, likely through an interaction with peripherin-2. These data reveal fine functional specializations within the structural domains of the CNG channel and suggest that its sequestration to the outer segment plasma membrane requires an interaction with peripherin-2.
SIGNIFICANCE STATEMENT Neurons and other differentiated cells have a remarkable ability to deliver and organize signaling proteins at precise subcellular locations. We now report that the CNG channel, mediating the electrical response to light in rod photoreceptors, contains two specialized regions within the N terminus of its β-subunit: one responsible for delivery of this channel to the ciliary outer segment organelle and another for subsequent channel sequestration into the outer segment plasma membrane. These findings expand our understanding of the molecular specializations used by neurons to populate their critical functional compartments.
Keywords: cilium, CNG channel, membrane trafficking, outer segment, photoreceptor
Introduction
Photoreceptor cells of the retina are responsible for light detection. They contain a modified primary cilium, called the outer segment, which houses all the molecular components necessary for capturing light and eliciting an electrical response. To increase light sensitivity, outer segments contain a stack of flattened membrane disks, so that incoming photons pass through hundreds of membrane surfaces packed with the visual pigment, rhodopsin. Another unique outer segment feature is that it undergoes constant regeneration, which places a heavy demand on the trafficking of signaling and structural proteins to this compartment (Pearring et al., 2013; Goldberg et al., 2016; Spencer et al., 2020). Defects in these trafficking mechanisms underlie many forms of inherited retinal degeneration, ultimately leading to blindness (Rattner et al., 1999).
Within the outer segment, there is functional specialization between the membranes of photoreceptor disks and the surrounding plasma membrane. Whereas disks primarily harbor components of the visual signaling pathway (Molday and Molday, 1987; Skiba et al., 2013), the cyclic nucleotide-gated (CNG) channel required to elicit an electrical response to light and the Na/Ca-K exchanger regulating Ca2+ dynamics are confined to the plasma membrane (Bauer, 1988; Reid et al., 1990). Previous studies, focused primarily on disk-resident transmembrane proteins, showed that their outer segment delivery relies on specific sequences for targeting. These proteins include rhodopsin (Sung et al., 1994), peripherin-2 (Tam et al., 2004; Salinas et al., 2013), RDH8 (Luo et al., 2004), and R9AP (Pearring et al., 2014). Two other proteins, guanylyl cyclase-1 (GC-1) and PRCD, have been shown to be delivered to the outer segment in a complex with rhodopsin (Pearring et al., 2015; Spencer et al., 2016). Less is known about the outer segment delivery of plasma membrane-resident proteins and even less about their segregation specifically into this membrane.
In this study, we investigated the outer segment targeting and plasma membrane sorting of the CNG channel. This channel is composed of four subunits: three α1 and one β1 (Weitz et al., 2002; Zheng et al., 2002; Zhong et al., 2002). Early studies of the CNG channel, performed by heterologously expressing its subunits in Xenopus oocytes, showed that both CNGα1 and the CNGα1/CNGβ1 complex, but not CNGβ1 alone, are effectively delivered to the oocyte plasma membrane and form functional channels. Yet, only the channel formed by both subunits recapitulates electrical and gating properties of the native channel (Kaupp et al., 1989; Chen et al., 1993; Colville and Molday, 1996; Trudeau and Zagotta, 2002; Zheng et al., 2002). Transgenic expression of the channel in Xenopus rods showed that removing the N-terminal glutamic acid-rich protein (GARP) domain from CNGβ1 results in accumulation of the mutant subunit in the internal inner segment membranes and aberrant delivery to the plasma membrane of the inner segment (Nemet et al., 2014), suggesting an important role for the GARP domain in CNGβ1 transport.
The CNG channel was reported to associate with the Na/Ca-K exchanger within the outer segment plasma membrane (Molday and Molday, 1998). In addition, the channel is connected to the photoreceptor disk rim through binding to a rim-specific protein, peripherin-2 (Poetsch et al., 2001; Conley et al., 2010). The latter interaction is believed to link the disks with the plasma membrane, likely stabilizing the disk stack within the outer segment.
We now demonstrate that the CNG channel travels through both the ER and Golgi on its route to the rod outer segment and that its specific outer segment delivery requires the channel's preassembly. This targeting is mediated by the glutamic acid-rich region of CNGβ1's GARP domain. Whereas this targeting region confers outer segment localization, it does not define specific plasma membrane localization. We observed that the adjacent R1-4 region of the GARP domain accumulates in disk incisures likely through peripherin-2 binding. Together, our results show that outer segment targeting and peripherin-2 interaction are performed by two distinct sites within the CNGβ1 subunit's GARP domain and suggest that the plasma membrane sequestration of the channel relies on peripherin-2 interaction.
Materials and Methods
Animals
Mice were handled following the protocols approved by the Institutional Animal Care and Use Committees at the University of Michigan (registry number A3114–01) and Duke University (registry number A011-14-01). Albino WT CD1 mice used in electroporation experiments were obtained from Charles River Laboratories. Cngb1-X1−/− mice (Zhang et al., 2009) used in Western blotting and electroporation experiments were kindly provided by Steven Pittler (University of Alabama at Birmingham). Pigmented wild-type (WT) C57BL/6J mice used for Western blotting were from The Jackson Laboratory. All mice were housed under a 12/12-h light cycle. The experimenters were not blinded to genotype.
In vivo electroporation
Retinal transfection of neonatal mice by the in vivo electroporation technique (Matsuda and Cepko, 2007) was used with modifications described in Pearring et al. (2015) and Salinas et al. (2017) to express exogenous constructs in mouse rods. DNA constructs were electroporated into the retinas of WT CD1 or Cngb1-X1−/− neonatal mice. Following anesthetization of neonatal mice (P0–P2) on ice, the eyelid and sclera were punctured at the periphery of the eye using a 30-gauge needle. A blunt-end 32-gauge needle was advanced through the puncture wound until reaching the subretinal space, and 0.3–0.5 µl of concentrated plasmid DNA (2 µg/µl construct of interest and 1 µg/µl soluble mCherry to visualize transfected cells) was deposited. A tweezer-type electrode (BTX) was placed over the mouse's head with the positive electrode overlying the injected eye. Five 100V pulses of 50 ms duration were applied using an ECM830 square pulse generator (BTX). Neonates were returned to their mother and allowed to develop until postnatal day 21 when mice were sacrificed by CO2 inhalation followed by decapitation, and retinal tissue was collected for analysis.
Generation of transgenic tadpoles
Transgenic Xenopus tadpoles were produced using restriction enzyme-mediated integration developed previously (Kroll and Amaya, 1996; Amaya and Kroll, 1999), with modifications described in Batni et al. (2000) and Whitaker and Knox (2004).
Primary antibodies
The following antibodies were generously provided by Robert Molday, University of British Columbia (mAb 1D1, anti-CNGa1; and mAb 8G8, anti-GARP); Steven Pittler, University of Alabama at Birmingham (pAb, anti-CNGβ1, C-terminal peptide QEPPEPKDPPKPPGC); Gabriel Travis, University of California, Los Angeles (pAb, anti-peripherin-2). The polyclonal antibody against Rom-1 was generated in the Arshavsky Laboratory (Gospe et al., 2011). Rabbit antibody against R9AP sequence 144–223 is described in Keresztes et al. (2003). Commercial antibodies were mAb 1D4, antirhodopsin (ab5417, Abcam); pAb M-18, anti-ABCA4 (sc-21 460, Santa Cruz Biotechnology); mAb anti-Na/K-ATPase (sc-58 628, Santa Cruz Biotechnology); mAb M2, anti-FLAG (F1804, Sigma-Aldrich); pAb, anti-FLAG (F7425, Sigma-Aldrich); pAb, anti-GFP conjugated to Alexa Fluor 488 (Invitrogen); and pAb, anti-MYC (2278S, Cell Signaling Technology).
DNA constructs
DNA constructs were generated using standard PCR-based subcloning methods. Point mutations were generated using the QuikChange II XL Kit (Stratagene). All DNA constructs were cloned between 5′ AgeI and 3′ NotI sites and sequence confirmed. For mouse in vivo electroporation, the pRho plasmid driving gene expression under the 2.2 kb bovine rhodopsin promoter was used (plasmid #11 156, Addgene). For Xenopus transgenics, the XOP1.5 vector was used for constructs containing GFP, whereas untagged and Myc-tagged CNGβ1 constructs were expressed using a dual promoter strategy described in Baker et al., (2008). In brief, along with the Xenopus opsin promoter driving expression of the various CNGβ1 constructs in rods, a cassette containing the γ-crystalline promoter was used to drive GFP expression in the lens. DNA templates were obtained as follows: human Cngb1 was a gift from Van Bennett (Duke University), mouse Cnga1 and Cngb1 were amplified from mouse retina cDNA [Stratagene; the Myc epitope was added using overlap extension PCR (Zhu et al., 2007)]. All chimeric constructs were produced using overlap extension PCR. All forward and reverse primers were designed to introduce 5′-AgeI and 3′-NotI sites respectively. A complete list of primers can be found in Table 1.
Table 1.
Oligonucleotide primers to generate and sequence confirm constructs used in this study
| Primer Name | Sequence (from 5′ to 3′) | Usage |
|---|---|---|
| Mcngb1_fwd | ATGTTGGGCTGGGTCCAAAGGG | PCR mouse CNGβ1 from cDNA |
| mCNGb1_rev | TCACTCCGCCCCCTCTTTCTTC | PCR mouse CNGβ1 from cDNA |
| mCNGB1 ssF1 | GCACAGATCCCAGAACCTTG | Sequencing mouse CNGβ1 |
| mCNGB1 ssF2 | GAGCAGGAAGAGGATGAGGA | Sequencing mouse CNGβ1 |
| mCNGB1 ssF3 | TGCAGCAGGAAGAAGCTACC | Sequencing mouse CNGβ1 |
| mCNGB1 ssF4 | GCGGGGACATCATTACAGAC | Sequencing mouse CNGβ1 |
| mCNGB1 ssF5 | GCAGAACCGTGTCAAGACCT | Sequencing mouse CNGβ1 |
| mCNGB1 ssF6 | CCGAAGCTCTTCAATGCTG | Sequencing mouse CNGβ1 |
| mCNGB1 ss7 | GCATCTTGTCCGGAAGTTGT | Sequencing mouse CNGβ1 |
| mCNGB1 ss8 | GAGGGTTGATGCCAAGTTTG | Sequencing mouse CNGβ1 |
| mCNGB1 ss9 | GGGACTCAGCTGGTGACAG | Sequencing mouse CNGβ1 |
| mCNGB1 ss10 | CACCAAACAGCTATCCAGCA | Sequencing mouse CNGβ1 |
| mCNGB1 ss11 | ATTCTGCTCAAGCCACCTGA | Sequencing mouse CNGβ1 |
| AgeI_hCNGB_F1 | CACATAACCGGTATGTTGGGCTGGGTCCAGAGG | Clone untagged human CNGβ1 |
| hCNGB1ssF1 | TCACAGCATCACGGAGGACC | Sequencing human CNGβ1 |
| hCNGB1ssF2 | GTGCAGACCATCAGCATCCT | Sequencing human CNGβ1 |
| hCNGB1ssF3 | TCAGGAGTGCCTGCCACGAA | Sequencing human CNGβ1 |
| hCNGB1ssF4 | GAGGAGCACTATTGCGACAT | Sequencing human CNGβ1 |
| hCNGB1ssF5 | GTGTACAGGGTCATCAGGAC | Sequencing human CNGβ1 |
| hCNGB1ssF6 | GTTAGCAAAGTCGCACTCTT | Sequencing human CNGβ1 |
| hCNGB1ssF7 | GGCAAAAGGCGGCAAACTTG | Sequencing human CNGβ1 |
| hCNGBtmd-F1 | GGATCCATGCTGACCAACCTGATGTATG | Sequencing human CNGβ1 |
| hCNGBtmd-R1 | ACCGGTATCACATCTCTCATCTGTCCGA | Sequencing human CNGβ1 |
| hCNGBntd-F1 | GGATCCATGTTGGGCTGGGTCCAGAG | Sequencing human CNGβ1 |
| hCNGBntd-R1 | GACCGGTATCAGGTTGGTCAGCGGGTCA | Sequencing human CNGβ1 |
| hCNGB1-ssR1 | GTGGAGAGCTCGGTGGAGAC | Sequencing human CNGβ1 |
| hCNGB1-ssR2 | CCAACAGCCAGCAAGCTTAT | Sequencing human CNGβ1 |
| AgeI_EGFP-Fwd | CACATAACCGGTATGGTGAGCAAGGGCGAGGAGCT | Add N-terminal eGFP tag |
| Age_myc-F | CACATAACCGGTATGGAACAAAAACTCATCTCAGA | Add N-terminal myc tag |
| NotI_myc_Rev | GCATTGGCGGCCGCTCACAGATCCTCTTCTGAGATAAGCTT | Add C-terminal myc tag |
| hCNGBtmd-NotI-R1 | AGAGTCGCGGCCGCTTACACATCTCTCATCTGTCCGA | Truncate human CNGβ1 after TMD domain |
| NotI_GARP_Rev | AGAGTCGCGGCCGCTTTATCCTGAACTGGCAGCCTCGG | Truncate human CNGβ1 after GARP domain |
| NotI-hCNGBntd-R | AGAGTCGCGGCCGCTTACAGGTTGGTCAGCGGGTCAATG | Truncate human CNGβ1 after NTD domain |
| hB1343-NotI-R New | ATAGAGTCGCGGCCGCTTTACTTCTCCACAGCTTTGTTCTCTTC | Truncate human CNGβ1 after R domain |
| AgeI_mCNGA1_Fwd | CAATGCACCGGTGCCGCCACCATGAAGACAAATATTATCAATA | PCR mouse CNGα1 from cDNA |
| NotI_mCNGA1_Rev | GCATTGGCGGCCGCTCAGTCCTGTAGAGACTCCGT | PCR mouse CNGα1 from cDNA |
| mCNGA1-ssF1 | TTCCAGAGGACGGAAACAAG | Sequencing mouse CNGα1 |
| mCNGA1-ssR1 | GGTGGCGAAAATTAAAACTCC | Sequencing mouse CNGα1 |
| xR9APtm_Rev | TTACAGAATAGAATAGAGTGTAATAGCTACAAGGGCTGTTCCACATAAAAGCAAGGACACAATTAACTGTCCGTCCAGGTTGGTCAGCGGGTCAATG | Add C-terminal single-pass TM from R9AP |
| hCNGB-AAAA-Fwd | GCCCTCGGTGAGGATCGCTGCGGCT | Mutagenesis primers for 4A mutant |
| hCNGB-AAAA-Rev | CCGGGATCAGGGCCTGCAGCCGCAG | Mutagenesis primers for 4A mutant |
| mB1-535-Not-R | AGAGTCGCGGCCGCTTCAGGGGCCACTATCAGCATCCG | Truncate mouse CNGβ1 after GLU domain |
| mB1-358-Not-R | AGAGTCGCGGCCGCTTCATATCTCCACTATGGCCTCAC | Truncate mouse CNGβ1 after R domain |
| myc-mCNGB1-F | CAAAAACTCATCTCAGAAGAGGATCTGATGTTGGGCTGGGTCCAAAG | Add myc tag to mouse CNGβ1 N terminal |
| myc-hCNGB1-F | CAAAAACTCATCTCAGAAGAGGATCTGTTGGGCTGGGTCCAGAGGGT | Add myc tag to human CNGβ1 N terminal |
| myc-mB1-359-F | CAAAAACTCATCTCAGAAGAGGATCTGCCCAGGGAGCTGACGAAGAT | Add myc tag to mouse CNGβ1 GLU domain |
| myc-hCNGB1-344-F | CAAAAACTCATCTCAGAAGAGGATCTGATGCCCAGAGAGCTGTCC | Add myc tag to human CNGβ1 GLU domain |
Immunofluorescence
Xenopus retinal cross-sections: GFP expressing transgenic tadpoles at developmental stages 43–45 were anesthetized in 0.2% Tricaine, fixed in 4% paraformaldehyde in PBS, cryoprotected in 30% sucrose, and frozen in OCT tissue-freezing compound (Fisher). 12-µm-thick sections were cryosectioned for immunostaining. To detect human CNGβ1 or Myc-tagged constructs, tissue sections were permeabilized for 10 min with 0.1% Triton X-100 in PBS (PBX), blocked with 5% goat serum and 0.5% Triton X-100 in PBS for 1 h at 22°C. Specific primary antibodies were diluted 1:2000 in blocking and incubated overnight at 4°C. The next day, sections were rinsed in 0.5% PBX and incubated with antimouse IgG secondary antibodies conjugated to Alexa Fluor 594 (Jackson ImmunoResearch) and 10 µg/ml Hoechst 33 342 (Thermo Fisher Scientific, H3569) in blocking solution for 2 h at 22°C. Slides were coverslipped using Immu-Mount (Thermo Fisher Scientific).
Mouse retinal cross-sections: Posterior eyecups were fixed for 1 h in 4% paraformaldehyde in PBS, rinsed 3 times in PBS, and embedded in 7.0% low-melt agarose (A3038, Sigma). 100 µm cross-sections through the central retina were collected using a vibratome (Leica VT1200S), placed in a 24-well plate, and blocked in 5% goat serum and 0.5% Triton X-100 in PBS for 1 h at 22°C. Sections were incubated in primary antibody diluted in blocking solution overnight at 4°C, rinsed 3 times in PBS, and incubated with 10 µg/ml Hoechst 33 342 and goat or donkey secondary antibodies conjugated with Alexa Fluor 488, 568, or 647 (Jackson ImmunoResearch) in blocking solution for 2 h at 22°C. Sections were mounted with Immu-Mount and coverslipped.
At Duke University, images were acquired using a Nikon Eclipse 90i upright microscope equipped with a 100x oil-immersion objective (1.40 NA, Plan Apo VC) and an A1 confocal scanner controlled by NIS-Elements AR software (Nikon). At the University of Michigan, images were acquired using a Zeiss Observer 7 inverted microscope equipped with a 63x oil-immersion objective (1.40 NA) and a LSM 800 confocal scanhead controlled by Zen 5.0 software (Zeiss). Manipulation of images was limited to adjusting the brightness level, image size, rotation, and cropping using FIJI (ImageJ) and Illustrator (Adobe).
Protein deglycosylation assay
Eyecups from C57BL/6J mice were collected at P21 and homogenized by pestle, followed by sonication in 250 ml of 1% sodium dodecyl sulfate and 1x cOmplete protease inhibitor mixture (Roche) in PBS. Lysates were cleared at 15,000 rpm for 10 min at 22°C. Total protein concentration was measured using the DC Protein Assay Kit (Bio-Rad), and 20 µg of protein were used for each condition. For PNGase F treatment, lysate is combined with 2 µL each Glycoprotein Denaturing Buffer, GlycoBuffer 2 and 10% NP-40, 1 µL PNGase F (P0708L, New England Biolabs), and H2O as necessary to a final volume of 20 µL. The reaction is then incubated for 1 h at 37°C. For Endo H (P0702L, New England Biolabs) treatment, the same procedure was followed, except that GlycoBuffer 3 was used instead of GlycoBuffer 2. For control experiments, reactions used GlycoBuffer 2 without enzymes. The resulting reactions were terminated by cooling in ice or freezing at −20°C. 5x sample buffer with 100 mm dithiothreitol (DTT) was added to each sample. For rhodopsin and peripherin-2 detection, 5 µg total protein were loaded on SDS-PAGE for Western blot. For all other proteins, 10 µg total protein were loaded on SDS-PAGE for Western blot.
Experimental design and statistical analyses
All in vivo electroporated mouse retinas were analyzed at postnatal day 21. All transgenic Xenopus tadpoles were collected at 14 d postinjection (stage 43–45). For both mice and Xenopus, a minimum of three expressing eyes or tadpoles were analyzed for every DNA construct. Animals of both sexes were used for all experimental models. The immunofluorescence signal in tangential sections through the outer segment was analyzed by comparing the mean intensity of a 5-pixel-wide perimeter line at the edge of the outer segment (selected from corresponding Wheat Germ Agglutinin (WGA) staining of each outer segment) divided by the mean intensity of a matching contour line reduced to 80% of the original size, which crosses the incisures inside the outer segment (see Fig. 6). From this analysis, constructs that are present within the incisures yield a value <1, whereas those residing in the plasma membrane yield a value >1. This analysis was performed for 11–14 expressing rods in a minimum of three transgenic tadpoles for each construct. Values were plotted using Prism 9.0 (GraphPad Software), and an unpaired Student's t test was performed between full-length CNGβ1 and the other three constructs.
Figure 6.
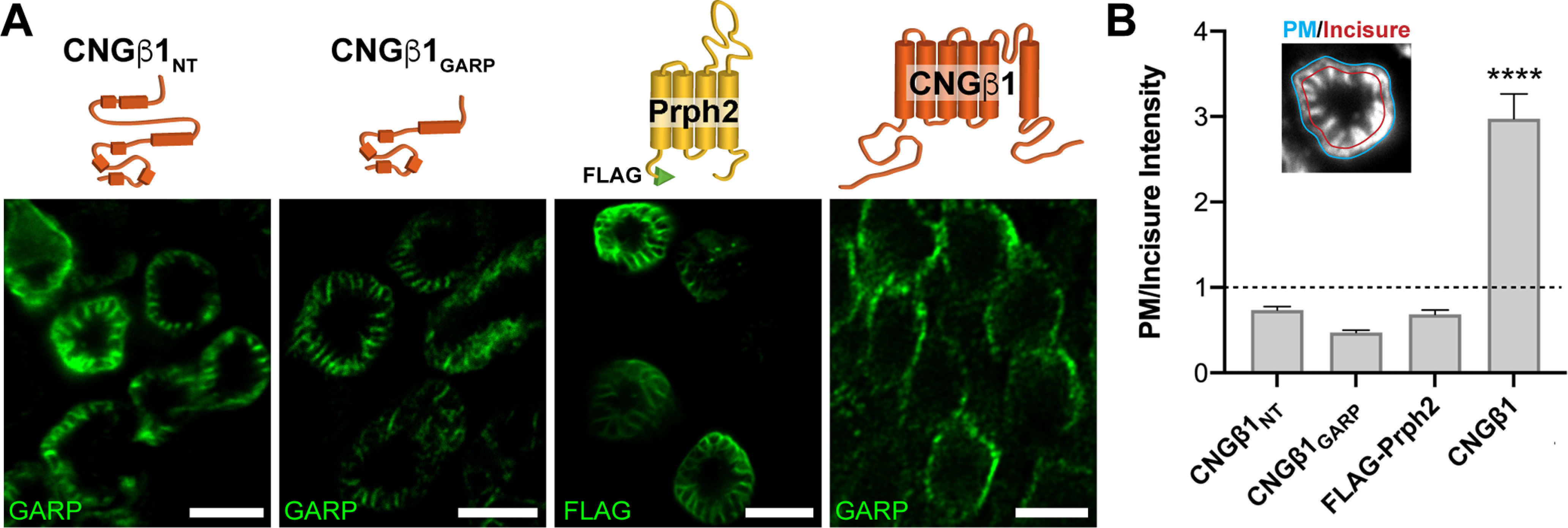
CNGβ1's N-terminal GARP domain localizes to disk incisures within rod outer segments. A, Tangential sections through Xenopus rod outer segments expressing soluble CNGβ1NT and CNGβ1GARP; FLAG-tagged full-length bovine peripherin-2 or untagged full-length CNGβ1. Diagram of transfected constructs are shown above their corresponding panel. Constructs that include N terminal of human CNGβ1 were detected using an anti-GARP antibody. Scale bar: 5 µm in all panels. B, Quantification of plasma membrane intensity (blue line, inset) normalized to incisure intensity (red line, inset) was performed for all the constructs shown in panel A. Protein residing in the plasma membrane yield a value >1, whereas proteins residing in disk incisures yield a value <1. Unpaired Student's t test was performed for full-length CNGβ1 compared with the other three constructs, p < 0.0001.
Results
Domain structure of the rod CNG channel subunits
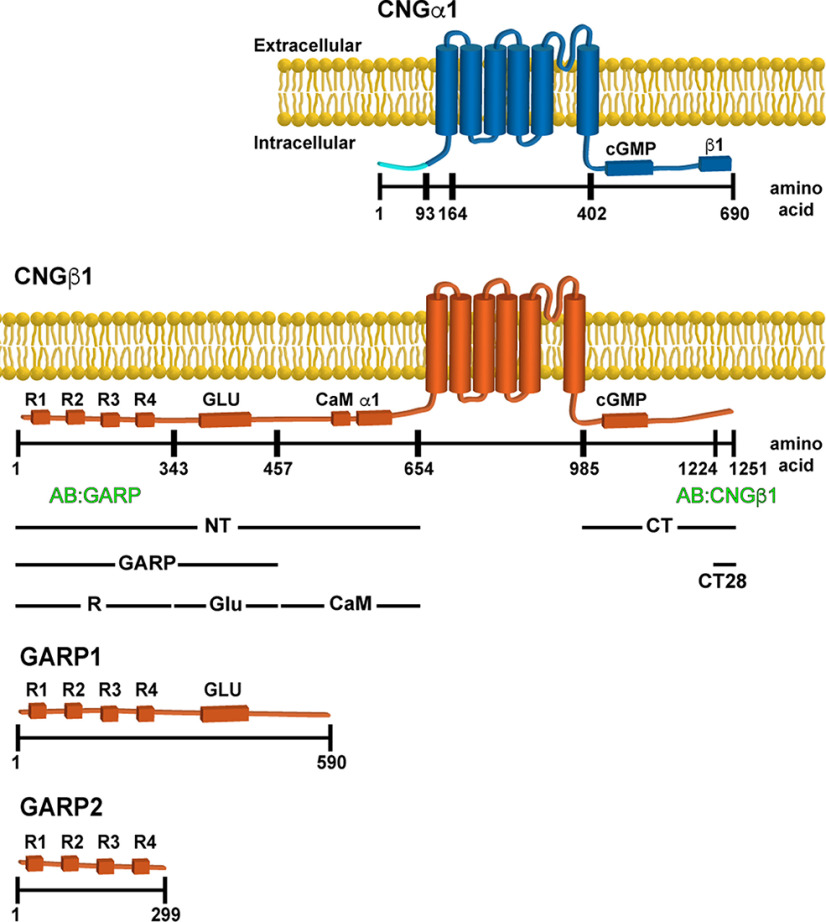
The domain composition of CNG subunits is shown in Figure 1. Both subunits contain a cytosolic N terminus, a hexahelical transmembrane core, and a cytosolic C-terminus bearing the cGMP-binding domain (Molday and Molday, 1998). The channel's subunits interact through sites located at the C-terminus of CNGα1 and N-terminus of CNGβ1 (Trudeau and Zagotta, 2002). The N terminus of CNGβ1 is also responsible for binding calmodulin (Grunwald et al., 1998; Weitz et al., 1998) and peripherin-2 (Ritter et al., 2011; Milstein et al., 2017). The latter is conveyed through a relatively large, intrinsically disordered GARP domain (Sugimoto et al., 1991; Colville and Molday, 1996; Batra-Safferling et al., 2006; Haber-Pohlmeier et al., 2007), which can be subdivided into two regions. The first contains four short proline-enriched repeats and is responsible for peripherin-2 binding (Ritter et al., 2011; Milstein et al., 2017). We refer to this region as R. The second contains nearly threefold higher density of glutamic acid residues, which we refer to as Glu, than the R region (40% vs 15%). Notably, rods also express two soluble proteins, GARP1 and GARP2, formed through alternative splicing of the Cngb1 gene and include various fragments of the CNGβ1 N terminus (Ardell et al., 1995, 1996, 2000).
Figure 1.
Domain composition of CNGα1, CNGβ1, GARP1 and GARP2. Cytosolic domains include cGMP binding, calmodulin (CaM) binding, CNGα1 (α1) and CNGβ1 (β1) binding, Glu, and four proline-rich repeats (R1–R4). Notably, the first 92 amino acids of CNGα1 (marked in light blue) are cleaved during intracellular CNG channel processing (Molday et al., 1991). The epitope location for anti-GARP and anti-CNGβ1 antibodies as well as boundaries of CNGβ1-derived constructs expressed in this study are shown below the diagram. The numbering of amino acids corresponds to human protein sequences. For mouse variants, the corresponding amino acids for CNGα1 would be 1, 93, 156, 394, and 684, and for CNGβ1 would be 1, 358, 512, 731, 935, 1278, and 1305.
The CNG channel uses a conventional trafficking pathway to reach the outer segment
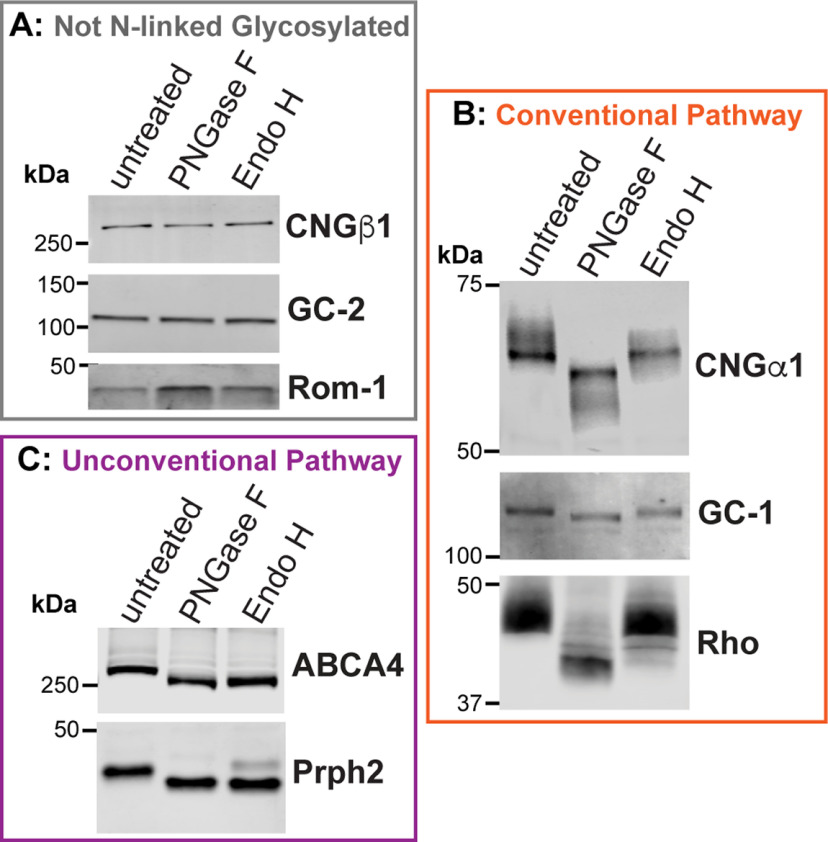
Membrane proteins can use two secretory pathways to reach their final destinations: the conventional pathway through both the ER and Golgi and the unconventional pathway directly from the ER. One way to determine which pathway is used by a particular protein is to assess whether its glycosylation is sensitive to cleavage by Endoglycosidase H (Endo H). N-glycosylation takes place in ER, where high-mannose oligosaccharides are transferred to proteins. This high-mannose N-glycan is sensitive to Endo H cleavage, so proteins exiting from the ER can be fully deglycosylated by this enzyme. If a glycoprotein is transferred and is processed through the Golgi, its oligosaccharide moieties are modified to produce hybrid oligosaccharide chains that are resistant to Endo H cleavage. Accordingly, proteins trafficked through the unconventional secretory pathway are Endo H sensitive, and those using the conventional pathway are Endo H resistant. In contrast, all N-linked glycoproteins are sensitive to general amidases, like PNGase F.
To understand how outer segment proteins use these two secretory pathways, we treated mouse retinal lysates with PNGase F and Endo H to examine the glycosylation status of eight outer segment-resident membrane proteins (Fig. 2). We found that the electrophoretic mobility of three proteins, CNGβ1, guanylyl cyclase-2 (GC-2) and Rom-1, did not change on PNGaseF or Endo H treatment (Fig. 2A), suggesting that they are not N-linked glycoproteins. The other five proteins, CNGα1, rhodopsin, peripherin-2, GC-1, and ABCA4, had increased mobility on PNGaseF treatment demonstrating that they are N-linked glycoproteins. We also treated retinal lysates with Endo H and found that three proteins, CNGα1, GC-1, and rhodopsin, are resistant to this treatment (Fig. 2B), indicating that they are trafficked via the conventional pathway. Whereas this route of outer segment delivery was previously established for rhodopsin (Deretic and Papermaster, 1991), the findings for CNGα1 and GC-1 represent original observations. Notably, the behavior of GC-1 is consistent with our previous result that it is delivered to the outer segment in a complex with rhodopsin (Pearring et al., 2015). Consistent with previous observations (Connell and Molday, 1990; Illing et al., 1997), ABCA4 was completely deglycosylated by Endo H, whereas peripherin-2 displayed a mixed pattern whereby the majority was deglycosylated, and a smaller fraction, shown to be in complex with Rom-1 (Conley et al., 2019), was not (Fig. 2C).
Figure 2.
The rod CNG channel is trafficked using the conventional secretory pathway. Mouse retinal lysates (untreated) were incubated with PNGase F or Endo H and analyzed by Western blotting. A, Electrophoretic mobility of nonglycosylated proteins CNGβ1, guanylate cyclase-2 (GC-2) and Rom-1 is unaffected by enzymatic treatments. B, CNGα1, GC-1, and rhodopsin are sensitive to PNGase F and resistant to Endo H treatment, indicating their processing through the conventional secretory pathway. C, ABCA4 and peripherin-2 are sensitive to both PNGase F and Endo H treatments, indicating their processing through the unconventional secretory pathway.
These results reveal an interesting analogy between the outer segment delivery of the peripherin-2/Rom-1 and CNGα1/CNGβ1 complexes. Both complexes use the conventional secretory pathway for outer segment delivery, and only one subunit in the complex becomes N-link glycosylated en route.
Outer segment localization of the CNG channel requires subunit preassembly and is targeted by the CNGβ1-subunit
To investigate the relationship between the CNG channel subunits' delivery to the rod outer segment, we first expressed epitope-tagged constructs representing each subunit in both WT and CNGβ1 knock-out (Cngb1-X1−/−) mice. The Cngb1-X1−/− mouse has a targeted deletion of exons 1 and 2 from the Cngb1 gene resulting in the CNGβ1 knockout (Zhang et al., 2009). It is also characterized by an ∼30-fold reduction in the expression level of CNGα1, suggesting that the cellular stability of CNGα1 relies on its association with CNGβ1. Expression of CNGα1-MYC in WT mice resulted in its predominantly normal outer segment localization (Fig. 3A). However, when expressed in Cngb1-X1−/− mice, CNGα1-MYC was localized in both outer segments and other cellular compartments in a pattern typical for untargeted membrane proteins expressed in mouse rods (Pearring et al., 2014). These observations suggest that CNGβ1 contains targeting information required for the outer segment delivery of the entire channel. Consistently, coexpression of CNGα1-MYC with CNGβ1-FLAG in Cngb1-X1−/− rods redirected CNGα1-MYC to the outer segment, along with expressed CNGβ1 (Fig. 3B).
Figure 3.
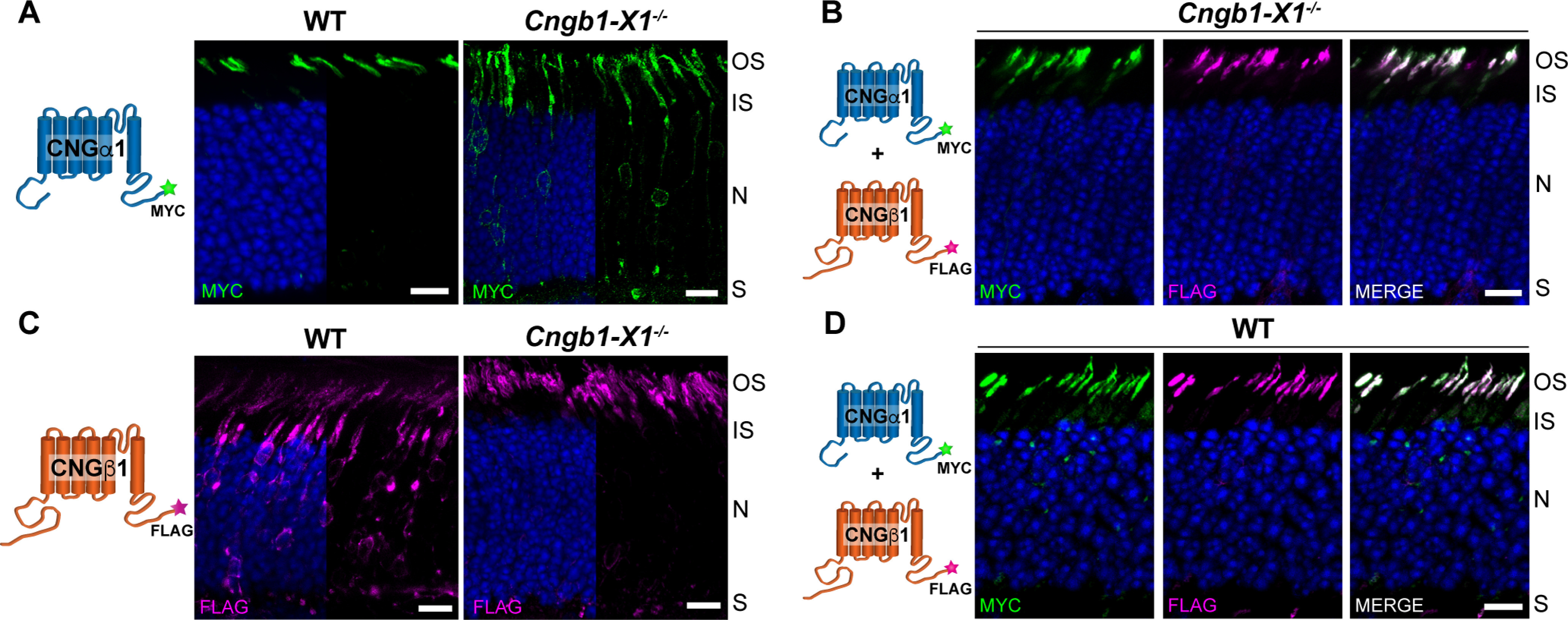
Outer segment localization of the CNG channel requires expression of the CNGβ1 subunit. A, Retinal cross sections from WT and Cngb1-X1−/− rods transfected with MYC-tagged, full-length mouse CNGα1. B, Cngb1-X1−/− rods were cotransfected with CNGα1-MYC and CNGβ1-FLAG. The merged image is shown on the far right. C, WT or Cngb1-X1−/− rods transfected with FLAG-tagged, full-length mouse CNGβ1. D, WT rods were cotransfected with CNGα1-MYC and CNGβ1-FLAG. In all panels, retinal sections were stained for each construct using an anti-MYC (green) and/or anti-FLAG (magenta) antibodies as indicated in the panel, and nuclei are stained by Hoechst (blue). Cartoon of transfected constructs are depicted on the left. Scale bar, 10 µm. Here and in the following figures, photoreceptor cell layer abbreviations are outer segment (OS), inner segment (IS), nucleus (N), and synapse (S). All mice are analyzed at P21.
There is another important aspect of the data shown in Figure 3A. The fact that all overexpressed CNGα1 is normally targeted in WT rods suggests that these cells naturally express more CNGβ1 than is incorporated in the mature channel. In this case, the extra CNGβ1 is normally cleared by the cell but can become a part of a mature channel when CNGα1 is overexpressed. Further evidence that the cellular level of the mature CNG channel is determined by the amount of expressed CNGα1 comes from the behavior of CNGβ1-FLAG expressed in WT and Cngb1-X1−/− rods. Whereas CNGβ1-FLAG was normally localized to outer segments of Cngb1-X1−/− rods, a large fraction was retained in the soma of WT rods in a pattern consistent with retention in the internal membranes of the inner segment following their biosynthesis (Fig. 3C). This suggests that knock-out rods contain enough CNGα1 to associate with expressed CNGβ1-FLAG and form a mature channel, but WT rods do not have enough CNGα1 to assemble with both endogenous CNGβ1 and overexpressed CNGβ1-FLAG. Consistent with this interpretation, coexpression of CNGα1-MYC with CNGβ1-FLAG in WT rods resulted in normal localization of both subunits in the outer segment (Fig. 3D), showing that increased CNGα1 levels are sufficient to assemble with both endogenous and overexpressed CNGβ1.
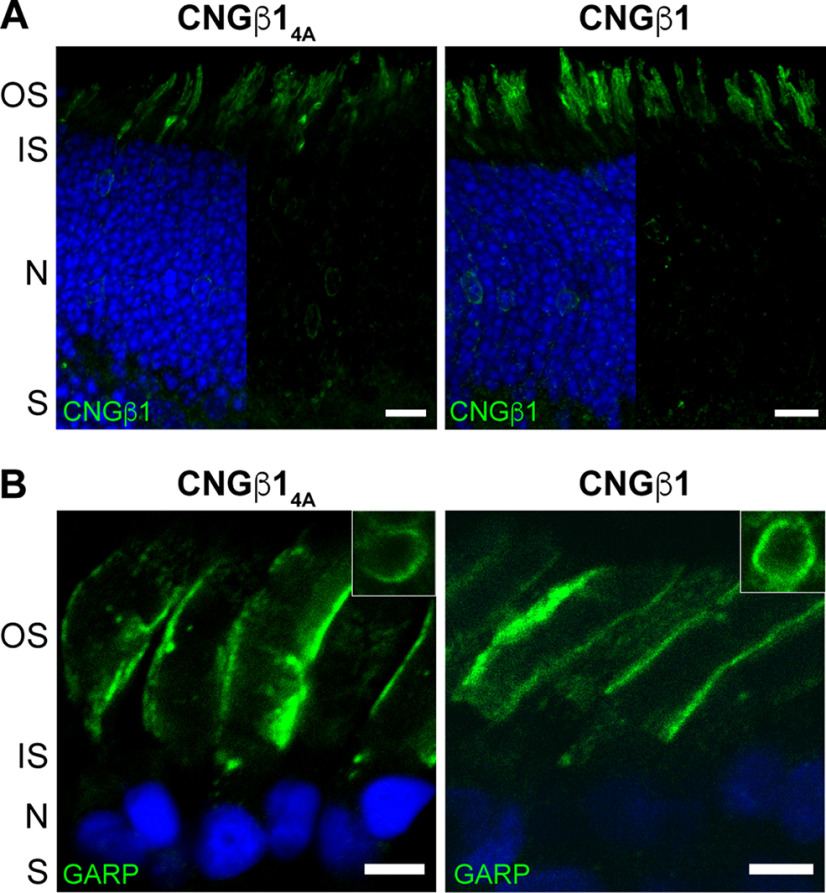
C-terminal RVxP motif of CNGβ1 is not required for outer segment localization
Previous studies of the olfactory CNG channel localization in cell culture showed that its ciliary targeting requires an RVxP motif on the C-terminus of the CNGβ1b subunit, which is spliced from the same Cngb1 gene as CNGβ1 expressed in rods (Jenkins et al., 2006; 2009). Therefore, we examined whether this sequence in CNGβ1 (RVSP in both mouse and Xenopus) is required for the outer segment CNG channel targeting in rods. We generated a mutant construct in which these four residues were replaced with alanines (CNGβ14A) and expressed it in Cngb1-X1−/− rods. The outer segment localization of this mutant was indistinguishable from the WT CNGβ1 control (Fig. 4A), demonstrating that this motif is not necessary for outer segment targeting. A similar result was obtained on transgenic expression of the corresponding mutant of human CNGβ1 subunit in Xenopus rods. Both outer segment targeting and subcellular localization of the mutant construct to the outer segment plasma membrane were normal (Fig. 4B). Notably, and unlike in WT mice, we did not observe CNGβ1 retention in the internal inner segment membranes on overexpression, which may be explained by differences in relative overexpression levels and/or cross-species differences in the clearance efficiency of unprocessed proteins.
Figure 4.
The C-terminus RVxP motif is not involved in CNG channel localization to the plasma membrane of the outer segment. A, Cngb1-X1−/− rods were transfected with untagged mouse CNGβ14A mutant or CNGβ1 control constructs. Retinal cross-sections were stained for each construct using an anti-CNGβ1 (green) antibody. Scale bar: 10 µm. B, Transgenic Xenopus rods expressing full-length human CNGβ14A mutant or CNGβ1 control. Retinal cross-sections were stained for each construct using an anti-GARP (green) antibody. Scale bar, 5 µm. Nuclei are counterstained with Hoechst (blue).
Outer segment targeting of CNGβ1 is mediated by its N terminus
In the next set of experiments, we undertook a search for the outer segment targeting and sorting signal(s) within CNGβ1. Of note, we expressed many constructs that included either all of or part of the transmembrane domain region of CNGβ1; however, all of these constructs were retained in the membranes inside the inner segment, so their contribution to the channels' targeting could not be experimentally assessed. We therefore focused on the cytosolic regions of the channel. We first expressed the cytosolic C-terminus of human CNGβ1 in transgenic Xenopus (Fig. 1, CT construct). To ensure membrane association while preserving its relative membrane topology, this construct was fused with an N-terminal single-pass transmembrane domain, subcloned from the activin receptor. However, the resulting TM-CNGβ1CT construct was retained in the internal inner segment membranes (Fig. 5A) precluding us from analyzing its targeting capacity. To circumvent this problem, we appended the CT sequence to a construct containing YFP and the untargeted lipidation sequence of rhodopsin (YFP-RhoCTΔ5), a strategy previously applied to achieve membrane attachment of C-terminal protein fragments (Tam et al., 2004; Salinas et al., 2013; YFP-RhoCTΔ5-CNGβ1CT; Fig. 5B). However, expression of the YFP-RhoCTΔ5-CNGβ1CT construct resulted in a pattern indicative of a soluble protein, suggesting that the long full-length CT construct prevented posttranslational lipidation of the rhodopsin C-terminus. Therefore, we resorted to testing a short C-terminal sequence of 28 amino acids previously suggested to contain outer segment targeting information (Kizhatil et al., 2009). Somewhat unexpectedly, when we expressed the YFP-RhoCTΔ5-CNGβ1CT28 its subcellular distribution was essentially identical to that of the base YFP-RhoCTΔ5 construct (Fig. 5C,D). Both constructs followed a default distribution pattern for untargeted membrane-associated proteins, in which the majority is localized in the outer segment, but appreciable fractions are also present in the plasma membrane of the cell body and the synaptic region (Baker et al., 2008).
Figure 5.
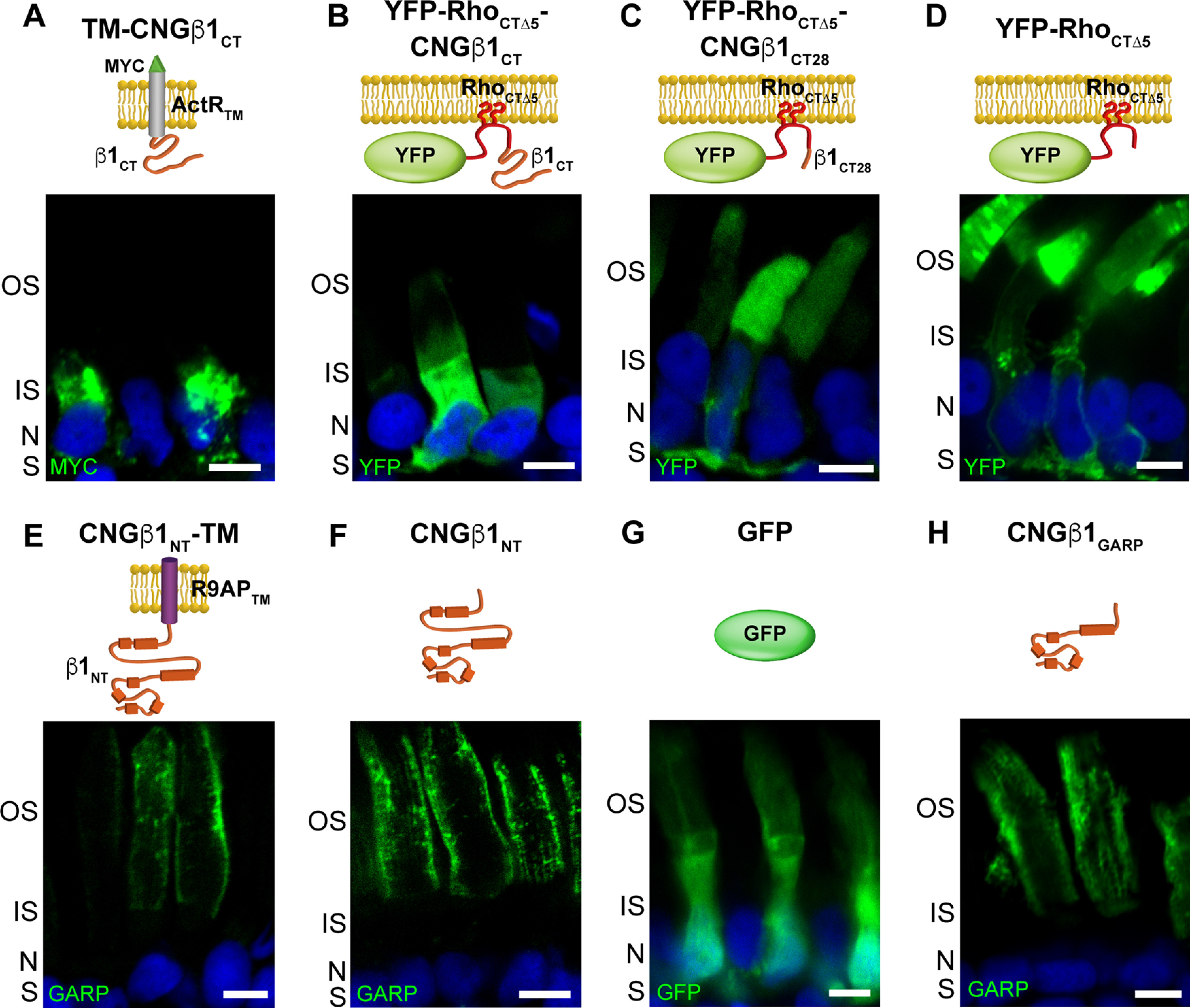
CNGβ1 N-terminal GARP domain is confined to the rod outer segment. A–H, Retinal cross-sections from transgenic Xenopus rods expressing membrane-anchored TM-CNGβ1CT (A), lipidated YFP-RhoCTΔ5-CNGβ1CT (B), lipidated YFP-RhoCTΔ5-CNGβ1CT28 (C), lipidated YFP-RhoCTΔ5 (D), membrane-anchored CNGβ1NT-TM (E), soluble CNGβ1NT (F), soluble GFP (G), and soluble CNGβ1GARP constructs (H). Diagram of transfected constructs are shown above their corresponding panel. Constructs that include N terminal of human CNGβ1 were detected using an anti-GARP antibody. Nuclei are counterstained with Hoechst (blue). Scale bar: 5 µm in all panels. All Xenopus tadpoles were analyzed at stage 43–45.
Next, we focused on the targeting properties of the cytosolic CNGβ1 N terminus (NT construct in Fig. 1). Its proper membrane attachment and topology were established by fusing with the single-pass transmembrane domain from R9AP lacking specific targeting information (Baker et al., 2008; Pearring et al., 2014). The resulting CNGβ1NT-TM construct was localized exclusively to the outer segment, suggesting that it bears sufficient targeting information (Fig. 5E). However, when we expressed the same construct as a soluble protein (CNGβ1NT), it was also localized to the outer segments with the same staining pattern (Fig. 5F). The latter is inconsistent with the well-described distribution of soluble proteins, such as GFP, located predominantly in the cytosol-rich inner segment (Tam et al., 2000; Najafi et al., 2012; Fig. 5G). The distribution of CNGβ1NT resembles that of soluble GARP1/2 proteins composed predominantly from the parts of the CNGβ1 N terminus (Hüttl et al., 2005; DeRamus et al., 2017) and suggests that its outer segment localization is conveyed through an interaction with another outer segment-resident protein, such as peripherin-2 located at the disk rims or CNGα1 located in the outer segment plasma membrane (Poetsch et al., 2001; Ritter et al., 2011). We then expressed a soluble construct encoding CNGβ1's GARP domain (CNGβ1GARP; Fig. 5H) and found that it is distributed like the entire N terminus. Because the interaction site with CNGα1 resides outside the GARP domain (Fig. 1), this result favors peripherin-2 as the interacting partner for these constructs. This was further supported by the analysis of tangential sections cut across rod outer segments expressing soluble N-terminal constructs (Fig. 6). Both CNGβ1NT and CNGβ1GARP displayed a flower petal-like pattern, which corresponds to invaginating incisures at the disk rims (Papermaster et al., 1978). This pattern is similar to that of FLAG-tagged peripherin-2 (FLAG-Prhp2) but is different from that of CNGβ1, which lacks immunostaining of the disk incisures (Fig. 6A). To formalize this observation for each construct in quantitative terms, plasma membrane intensity was normalized to incisure intensity, as described in Materials and Methods (Fig. 6B). In this analysis, constructs that are present within the incisures (CNGβ1NT and CNGβ1GARP and FLAG-Prph2) yield a value <1, whereas the construct residing in the plasma membrane (full-length CNGβ1) yields a value >1.
CNGβ1's GARP domain encodes both outer segment targeting and peripherin-2 interaction site
Demonstrating that the GARP domain of CNGβ1 binds to the disk rims might suggest that the CNG channel uses an interaction with peripherin-2 for outer segment delivery rather than using its own targeting signal. To explore this possibility, we expressed smaller fragments of the CNGβ1 N terminus—CNGβ1CaM, CNGβ1R, and CNGβ1Glu (Fig. 1)—as both soluble and membrane-anchored proteins. Soluble CNGβ1CaM was localized predominantly in the inner segment, suggesting that it is not engaged in any outer segment interactions, whereas the corresponding membrane-anchored construct (CNGβ1CaM-TM) was retained in the internal inner segment membranes (Fig. 7A). Soluble CNGβ1R was biased to the outer segment where it tended to accumulate at the periphery (Fig. 7B). However, a portion of this construct was also found in the cell body and synaptic termini, suggesting that its affinity for periphern-2 is not as high as that of longer N-terminal constructs (Fig. 5E,G). Membrane-anchored CNGβ1R-TM was successfully processed and delivered to both inner and outer segments (Fig. 7B), indicating that it lacks specific outer segment targeting information. Within the outer segment, CNGβ1R-TM was located at disk incisures similar to longer N-terminal constructs, as evidenced from the flower petal-like pattern seen in tangential sections (Fig. 7B). Together, these data suggest that the CNGβ1R region is sufficient for peripherin-2 interaction, but not for specific outer segment targeting.
Figure 7.
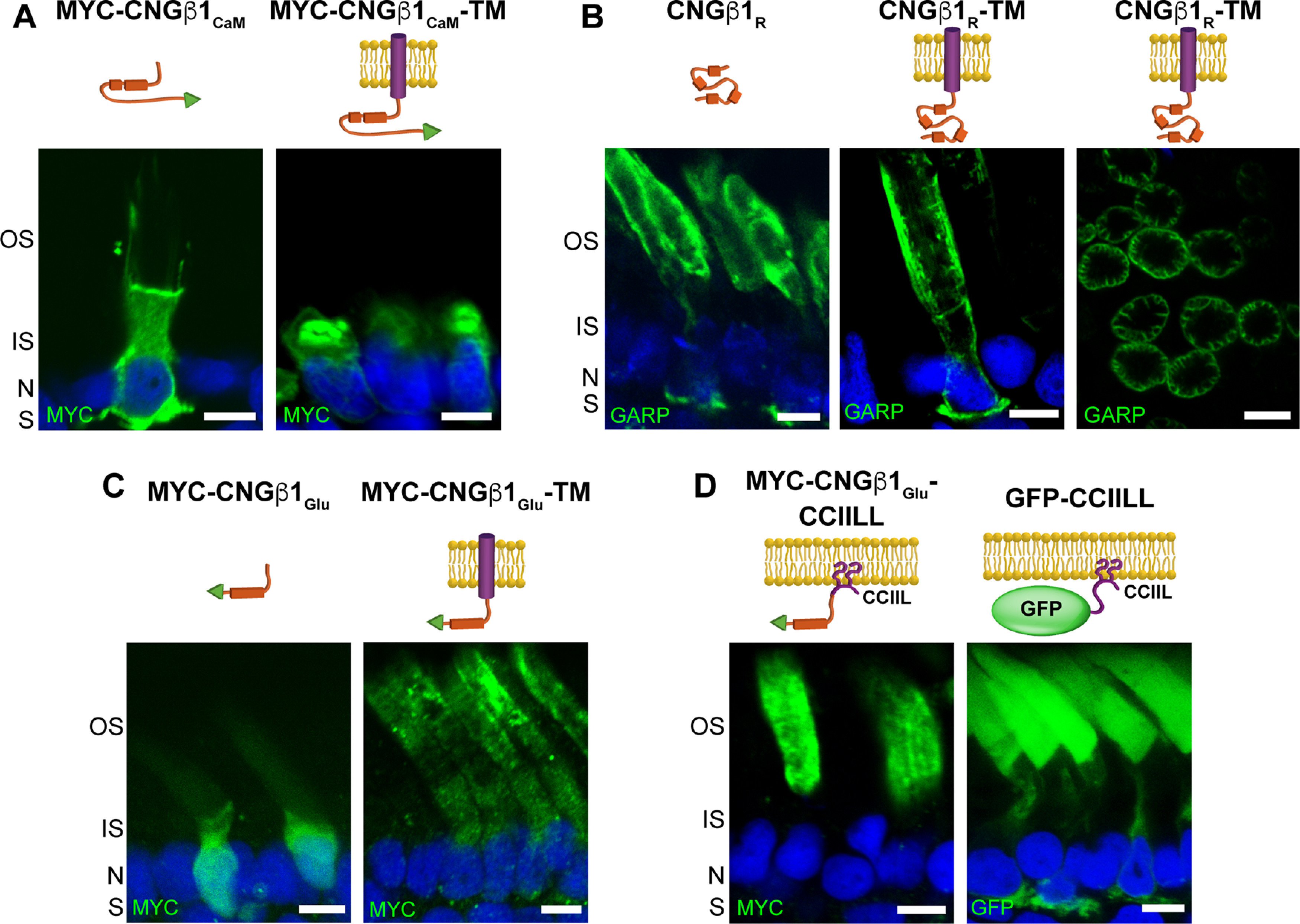
CNGβ1's GARP domain contains separate sites for peripherin-2 binding and outer segment targeting. A–D, Retinal cross-sections from transgenic Xenopus rods comparing localization of soluble and membrane-anchored CNGβ1CaM (A), soluble and membrane-anchored CNGβ1R and a tangential image of outer segment's expressing CNGβ1R-TM (B), soluble and membrane-anchored CNGβ1Glu (C), and CNGβ1Glu or GFP fused to a C-terminal CCIIL double lipidation anchor (D). Cartoon of transgenic constructs are shown above their corresponding panel. Constructs that include the N terminal of human CNGβ1 were detected using an anti-GARP antibody. All other constructs were N-terminally tagged with the MYC epitope and detected using an anti-MYC antibody. Scale bar, 5 µm. Nuclei are counterstained with Hoechst (blue).
We next analyzed CNGβ1Glu constructs (Fig. 7C). Soluble CNGβ1Glu behaved as a typical cytosolic protein with diffuse staining within the photoreceptor cytoplasm, suggesting the lack of specific outer segment interactions. The membrane-anchored CNGβ1Glu-TM was found in both inner and outer segments. However, its inner segment fraction was located inside the cell and not within the surrounding plasma membrane. This suggests that only a portion of this construct is properly processed following its biosynthesis, precluding us from determining whether CNGβ1Glu may contain an outer segment targeting signal. To avoid protein misprocessing, we expressed CNGβ1Glu as a lipidated construct by adding a C-terminal CCIIL sequence for double lipidation (palmitoylation and geranylgeranylation), previously introduced in similar studies (Tam et al., 2004; Baker et al., 2008; Pearring et al., 2014; Fig. 7D). We have found that CNGβ1Glu-CCIIL was localized exclusively in the outer segments, whereas the control untargeted GFP-CCIIL construct displayed a typical default pattern with an appreciable amount localized in other parts of the rod cell. This result shows that CNGβ1Glu contains a specific outer segment targeting signal. Within the outer segment, CNGβ1Glu-CCIIL was evenly distributed across all membranes without showing any bias toward periphery. Combined with the behavior of the soluble CNGβ1Glu construct, this suggests that this region of CNGβ1 is not sufficient for binding to peripherin-2.
Discussion
Protein targeting to the light-sensitive photoreceptor outer segment is still far from being understood and remains a subject of active investigation. Except for a few proteins shown to be delivered within larger complexes, such as GC-1 and PRCD associated with rhodopsin (Pearring et al., 2015; Spencer et al., 2016) or RGS9 and Gβ5 associated with R9AP (Hu and Wensel, 2002; Keresztes et al., 2004; Krispel et al., 2006; Gospe et al., 2011), most membrane proteins have targeting information encoded in their sequences. Among them, the CNG channel is particularly interesting because it needs to be delivered to the outer segment and then sequestered to the plasma membrane surrounding the disks. In this study, we demonstrated that the rod outer segment delivery of this channel requires preassembly of its constituent subunits on their biosynthesis and that the targeting signal is encoded within the glutamic acid-rich region of CNGβ1's GARP domain. Whereas acidic clusters have previously been associated with packaging membrane proteins into clathrin-coated vesicles (Navarro Negredo et al., 2017), to our knowledge, this is the first time an acidic cluster has been implicated in ciliary targeting of a membrane protein. This targeting signal is sufficient for outer segment localization but not subsequent outer segment plasma membrane sequestration, suggesting that the final sorting of the CNG channel to the outer segment plasma membrane occurs independently of its initial targeting.
We further showed that the cellular level of the mature CNG channel in rods is determined by the expression level of CNGα1. This follows the pattern in which the total amount of a mature multisubunit protein complex is determined by the expression level of one of its subunits. The best studied example in photoreceptors is the RGS9-Gβ5-R9AP GTPase activating complex for transducin, whose cellular content is set by the expression of R9AP, whereas the fractions of other subunits expressed in excess of R9AP are efficiently cleared by the cell (Keresztes et al., 2004; Krispel et al., 2006).
Another aspect of outer segment protein delivery explored in this study is that some proteins, such as rhodopsin, use the conventional secretory pathway through the ER and Golgi (Deretic and Papermaster, 1991; Deretic et al., 1998; Murray et al., 2009), whereas others, such as ABCA4, use the unconventional pathway bypassing the Golgi (Illing et al., 1997; Tsybovsky et al., 2011). Peripherin-2, known to interact with the CNG channel in outer segments, is also delivered through the unconventional pathway (Connell and Molday, 1990). The exception is a fraction of peripherin-2 engaged in the hetero-tetrameric complex with Rom-1, which is diverted into the conventional pathway (Zulliger et al., 2015; Conley et al., 2019). The latter is species specific: it was documented for mice, yet the entire pool of peripherin-2 in bovine and Xenopus is delivered via the unconventional pathway (Connell and Molday, 1990; Tian et al., 2014).
Our new data show that the CNG channel is delivered to the outer segment via the conventional pathway, which may suggest that the CNG channel and peripherin-2 are trafficked to this compartment independently of one another, particularly because the entire pool of peripherin-2 was shown to use the unconventional pathway in Xenopus (Tian et al., 2014). However, another Xenopus study using fluorescence complementation assays concluded that the interaction between the CNG channel and peripherin-2 first takes place in the inner segment (Ritter et al., 2011). Furthermore, CNGβ1 is completely absent from the outer segment membrane material of peripherin-2 knock-out mice despite all other known outer segment proteins, including CNGα1 and Rom-1, being found in this preparation (Spencer et al., 2019). This argues that the CNGβ1 cannot be delivered to the outer segment without peripherin-2, and they ought to interact before reaching this destination. A possible trafficking mechanism reconciling all of these observations was proposed in a recent study suggesting that, at least in cones, peripherin-2 is trafficked through the late endosome before entering the outer segment (Otsu et al., 2019). Likewise, the CNG channel might pass through endosomes, based on its mislocalization from the outer segment in conditional double knockout mice lacking the endocytic adaptor proteins Numb and Numb-like in rods (Ramamurthy et al., 2014). The authors further showed that, in cell culture, Numb binds and redirects CNGα1 to endosomes. Therefore, it is conceivable that the CNG channel and peripherin-2 are first trafficked to the inner segment endosomes using the conventional and unconventional secretory pathways, respectively, and then are delivered to the outer segment as a complex.
If the CNG channel and peripherin-2 are in fact delivered to the outer segment as a complex, then how do they next segregate into two different membrane subcompartments—the disks and the plasma membrane? It is logical to assume that they segregate on the remodeling of outer segment membranes during the process of disk enclosure (Spencer et al., 2020), after which they remain connected in-trans forming a link between the disk rim and the plasma membrane. However, no direct evidence for this mechanism have been obtained so far. It also remains unknown whether any additional proteins, such as the Na/Ca-K exchanger associated with the CNG channel, are involved in the channel's plasma membrane segregation. It has been proposed that this process involves a cytoskeletal protein, ankyrin-G (Kizhatil et al., 2009). However, subsequent proteomic studies have not identified any ankyrin-G peptides in the outer segment (Skiba et al., 2013; Spencer et al., 2019), challenging the existence of ankyrin-G in this subcellular compartment. Thus, elucidating the exact outer segment trafficking route of the CNG channel and the mechanism responsible for its subsequent sorting into the outer segment plasma membrane remain the challenges for future studies.
Footnotes
This work was supported by the National Institutes of Health (Grants EY025732 to J.N.P., EY012859 to V.Y.A., EY030451 to V.Y.A., EY007003 to University of Michigan, and EY005722 to Duke University). This work was also supported by the Matilda E. Ziegler Research Award to J.N.P. and Research to Prevent Blindness.
The authors declare no competing financial interests.
References
- Amaya E, Kroll KL (1999) A method for generating transgenic frog embryos. Methods Mol Biol 97:393–414. 10.1385/1-59259-270-8:393 [DOI] [PubMed] [Google Scholar]
- Ardell MD, Makhija AK, Oliveira L, Miniou P, Viegas-Péquignot E, Pittler SJ (1995) cDNA, gene structure, and chromosomal localization of human GAR1 (CNCG3L), a homolog of the third subunit of bovine photoreceptor cGMP-gated channel. Genomics 28:32–38. 10.1006/geno.1995.1102 [DOI] [PubMed] [Google Scholar]
- Ardell MD, Aragon I, Oliveira L, Porche GE, Burke E, Pittler SJ (1996) The β subunit of human rod photoreceptor cGMP-gated cation channel is generated from a complex transcription unit. FEBS Lett 389:213–218. 10.1016/0014-5793(96)00588-1 [DOI] [PubMed] [Google Scholar]
- Ardell MD, Bedsole DL, Schoborg RV, Pittler SJ (2000) Genomic organization of the human rod photoreceptor cGMP-gated cation channel β-subunit gene. Gene 245:311–318. 10.1016/S0378-1119(00)00023-8 [DOI] [PubMed] [Google Scholar]
- Baker SA, Haeri M, Yoo P, Gospe SM, Skiba NP, Knox BE, Arshavsky VY (2008) The outer segment serves as a default destination for the trafficking of membrane proteins in photoreceptors. J Cell Biol 183:485–498. 10.1083/jcb.200806009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batni S, Mani SS, Schlueter C, Ji M, Knox BE (2000) Xenopus rod photoreceptor: model for expression of retinal genes. Methods Enzymol 316:50–64. 10.1016/s0076-6879(00)16716-3 [DOI] [PubMed] [Google Scholar]
- Batra-Safferling R, Abarca-Heidemann K, Körschen HG, Tziatzios C, Stoldt M, Budyak I, Willbold D, Schwalbe H, Klein-Seetharaman J, Kaupp UB (2006) Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J Bio Chem 281:1449–1460. [DOI] [PubMed] [Google Scholar]
- Bauer PJ (1988) Evidence for two functionally different membrane fractions in bovine retinal rod outer segments. J Physiol 401:309–327. 10.1113/jphysiol.1988.sp017164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TY, Peng YW, Dhallan RS, Ahamed B, Reed RR, Yau KW (1993) A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature 362:764–767. 10.1038/362764a0 [DOI] [PubMed] [Google Scholar]
- Colville CA, Molday RS (1996) Primary structure and expression of the human beta-subunit and related proteins of the rod photoreceptor cGMP-gated channel. J Biol Chem 271:32968–32974. 10.1074/jbc.271.51.32968 [DOI] [PubMed] [Google Scholar]
- Conley SM, Ding XQ, Naash MI (2010) RDS in cones does not interact with the beta subunit of the cyclic nucleotide gated channel. Adv Exp Med Biol 664:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley SM, Stuck MW, Watson JN, Zulliger R, Burnett JL, Naash MI (2019) Prph2 initiates outer segment morphogenesis but maturation requires Prph2/Rom1 oligomerization. Hum Mol Genet 28:459–475. 10.1093/hmg/ddy359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell GJ, Molday RS (1990) Molecular cloning, primary structure, and orientation of the vertebrate photoreceptor cell protein peripherin in the rod outer segment disk membrane. Biochemistry 29:4691–4698. 10.1021/bi00471a025 [DOI] [PubMed] [Google Scholar]
- DeRamus ML, Stacks DA, Zhang Y, Huisingh CE, McGwin G, Pittler SJ (2017) GARP2 accelerates retinal degeneration in rod cGMP-gated cation channel β-subunit knockout mice. Sci Rep 7:42545. 10.1038/srep42545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Papermaster DS (1991) Polarized sorting of rhodopsin on post-Golgi membranes in frog retinal photoreceptor cells. J Cell Biol 113:1281–1293. 10.1083/jcb.113.6.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH (1998) Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc Natl Acad Sci U S A 95:10620–10625. 10.1073/pnas.95.18.10620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AF, Moritz OL, Williams DS (2016) Molecular basis for photoreceptor outer segment architecture. Prog Retin Eye Res 55:52–81. 10.1016/j.preteyeres.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospe SM, Baker SA, Kessler C, Brucato MF, Winter JR, Burns ME, Arshavsky VY (2011) Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors. J Neurosci 31:14660–14668. 10.1523/JNEUROSCI.3516-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald ME, Yu WP, Yu HH, Yau KW (1998) Identification of a domain on the beta-subunit of the rod cGMP-gated cation channel that mediates inhibition by calcium-calmodulin. J Biol Chem 273:9148–9157. 10.1074/jbc.273.15.9148 [DOI] [PubMed] [Google Scholar]
- Haber-Polmeier S, Abarca-Heidemann K, Korschen HG, Kaur Dhiman H, Heberle J, Schwalbe H, Klein-Seetharaman J, Kaupp UB, Pohlmeier A (2007) Binding of Ca2+ to glutamic acid-rich polypeptides from the rod outer segment. Biophys J 92:3207–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wensel TG (2002) R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A 99:9755–9760. 10.1073/pnas.152094799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttl S, Michalakis S, Seeliger M, Luo DG, Acar N, Geiger H, Hudl K, Mader R, Haverkamp S, Moser M, Pfeifer A, Gerstner A, Yau KW, Biel M (2005) Impaired channel targeting and retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGB1. J Neurosci 25:130–138. 10.1523/JNEUROSCI.3764-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing M, Molday LL, Molday RS (1997) The 220-kDa rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J Biol Chem 272:10303–10310. 10.1074/jbc.272.15.10303 [DOI] [PubMed] [Google Scholar]
- Jenkins PM, Hurd TW, Zhang L, McEwen DP, Brown RL, Margolis B, Verhey KJ, Martens JR (2006) Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol 16:1211–1216. 10.1016/j.cub.2006.04.034 [DOI] [PubMed] [Google Scholar]
- Jenkins PM, Zhang L, Thomas G, Martens JR (2009) PACS-1 mediates phosphorylation-dependent ciliary trafficking of the cyclic-nucleotide-gated channel in olfactory sensory neurons. J Neurosci 29:10541–10551. 10.1523/JNEUROSCI.1590-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB, Niidome T, Tanabe T, Terada S, Bönigk W, Stühmer W, Cook NJ, Kangawa K, Matsuo H, Hirose T (1989) Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature 342:762–766. 10.1038/342762a0 [DOI] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Hibino H, Hudspeth AJ, Heller S (2003) Expression patterns of the RGS9-1 anchoring protein R9AP in the chicken and mouse suggest multiple roles in the nervous system. Mol Cell Neurosci 24:687–695. 10.1016/S1044-7431(03)00231-8 [DOI] [PubMed] [Google Scholar]
- Keresztes G, Martemyanov KA, Krispel CM, Mutai H, Yoo PJ, Maison SF, Burns ME, Arshavsky VY, Heller S (2004) Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem 279:1581–1584. 10.1074/jbc.C300456200 [DOI] [PubMed] [Google Scholar]
- Kizhatil K, Baker SA, Arshavsky VY, Bennett V (2009) Ankyrin-G promotes cyclic nucleotide-gated channel transport to rod photoreceptor sensory cilia. Science 323:1614–1617. 10.1126/science.1169789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME (2006) RGS expression rate-limits recovery of rod photoresponses. Neuron 51:409–416. 10.1016/j.neuron.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E (1996) Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122:3173–3183. [DOI] [PubMed] [Google Scholar]
- Luo W, Marsh-Armstrong N, Rattner A, Nathans J (2004) An outer segment localization signal at the C terminus of the photoreceptor-specific retinol dehydrogenase. J Neurosci 24:2623–2632. 10.1523/JNEUROSCI.5302-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL (2007) Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA 104:1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein ML, Kimler VA, Ghatak C, Ladokhin AS, Goldberg AFX (2017) An inducible amphipathic helix within the intrinsically disordered C terminus can participate in membrane curvature generation by peripherin-2/rds. J Biol Chem 292:7850–7865. 10.1074/jbc.M116.768143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday RS, Molday LL (1987) Differences in the protein composition of bovine retinal rod outer segment disk and plasma membranes isolated by a ricin-gold-dextran density perturbation method. J Cell Biol 105:2589–2601. 10.1083/jcb.105.6.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday RS, Molday LL (1998) Molecular properties of the cGMP-gated channel of rod photoreceptors. Vision Res 38:1315–1323. 10.1016/s0042-6989(97)00409-4 [DOI] [PubMed] [Google Scholar]
- Molday RS, Molday LL, Dosé A, Clark-Lewis I, Illing M, Cook NJ, Eismann E, Kaupp UB (1991) The cGMP-gated channel of the rod photoreceptor cell characterization and orientation of the amino terminus. J Biol Chem 266:21917–21922. [PubMed] [Google Scholar]
- Murray AR, Fliesler SJ, Al-Ubaidi MR (2009) Rhodopsin: the functional significance of asn-linked glycosylation and other post-translational modifications. Ophthalmic Genet 30:109–120. 10.1080/13816810902962405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M, Maza NA, Calvert PD (2012) Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci U S A 109:203–208. 10.1073/pnas.1115109109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Negredo P, Edgar JR, Wrobel AG, Zaccai NR, Antrobus R, Owen DJ, Robinson MS (2017) Contribution of the clathrin adaptor AP-1 subunit µ1 to acidic cluster protein sorting. J Cell Biol 216:2927–2943. 10.1083/jcb.201602058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I, Tian G, Imanishi Y (2014) Organization of cGMP sensing structures on the rod photoreceptor outer segment plasma membrane. Channels (Austin) 8:528–535. 10.4161/19336950.2014.973776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu W, Hsu YC, Chuang JZ, Sung CH (2019) The late endosomal pathway regulates the Ciliary Targeting of Tetraspanin Protein Peripherin 2. J Neurosci 39:3376–3393. 10.1523/JNEUROSCI.2811-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papermaster DS, Schneider BG, Zorn MA, Kraehenbuhl JP (1978) Immunocytochemical localization of a large intrinsic membrane protein to the incisures and margins of frog rod outer segment disks. J Cell Biol 78:415–425. 10.1083/jcb.78.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Salinas RY, Baker SA, Arshavsky VY (2013) Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retin Eye Res 36:24–51. 10.1016/j.preteyeres.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Lieu EC, Winter JR, Baker SA, Arshavsky VY (2014) R9AP targeting to rod outer segments is independent of rhodopsin and is guided by the SNARE homology domain. Mol Biol Cell 25:2644–2649. 10.1091/mbc.E14-02-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Spencer WJ, Lieu EC, Arshavsky VY (2015) Guanylate cyclase 1 relies on rhodopsin for intracellular stability and ciliary trafficking. Elife 4:12058. 10.7554/eLife.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poetsch A, Molday LL, Molday RS (2001) The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J Biol Chem 276:48009–48016. 10.1074/jbc.M108941200 [DOI] [PubMed] [Google Scholar]
- Ramamurthy V, Jolicoeur C, Koutroumbas D, Mühlhans J, Le YZ, Hauswirth WW, Giessl A, Cayouette M (2014) Numb regulates the polarized delivery of cyclic nucleotide-gated ion channels in rod photoreceptor cilia. J Neurosci 34:13976–13987. 10.1523/JNEUROSCI.1938-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner A, Sun H, Nathans J (1999) Molecular genetics of human retinal disease. Annu Rev Genet 33:89–131. 10.1146/annurev.genet.33.1.89 [DOI] [PubMed] [Google Scholar]
- Reid DM, Friedel U, Molday RS, Cook NJ (1990) Identification of the sodium-calcium exchanger as the major ricin-binding glycoprotein of bovine rod outer segments and its localization to the plasma membrane. Biochemistry 29:1601–1607. 10.1021/bi00458a035 [DOI] [PubMed] [Google Scholar]
- Ritter LM, Khattree N, Tam B, Moritz OL, Schmitz F, Goldberg AF (2011) In situ visualization of protein interactions in sensory neurons: glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J Neurosci 31:11231–11243. 10.1523/JNEUROSCI.2875-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas RY, Baker SA, Gospe SM, Arshavsky VY (2013) A single valine residue plays an essential role in peripherin/rds targeting to photoreceptor outer segments. PLoS One 8:e54292. 10.1371/journal.pone.0054292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas RY, Pearring JN, Ding JD, Spencer WJ, Hao Y, Arshavsky VY (2017) Photoreceptor discs form through peripherin-dependent suppression of ciliary ectosome release. J Cell Biol 216:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiba NP, Spencer WJ, Salinas RY, Lieu EC, Thompson JW, Arshavsky VY (2013) Proteomic identification of unique photoreceptor disc components reveals the presence of PRCD, a protein linked to retinal degeneration. J Proteome Res 12:3010–3018. 10.1021/pr4003678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WJ, Pearring JN, Salinas RY, Loiselle DR, Skiba NP, Arshavsky VY (2016) Progressive rod-cone degeneration (PRCD) protein requires N-terminal S-acylation and rhodopsin binding for photoreceptor outer segment localization and maintaining intracellular stability. Biochemistry 55:5028–5037. 10.1021/acs.biochem.6b00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WJ, Lewis TR, Phan S, Cady MA, Serebrovskaya EO, Schneider NF, Kim KY, Cameron LA, Skiba NP, Ellisman MH, Arshavsky VY (2019) Photoreceptor disc membranes are formed through an Arp2/3-dependent lamellipodium-like mechanism. Proc Natl Acad Sci U S A 116:27043–27052. 10.1073/pnas.1913518117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WJ, Lewis TR, Pearring JN, Arshavsky VY (2020) Photoreceptor discs: built like ectosomes. Trends Cell Biol 11:904–915. 10.1016/j.tcb.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Yatsunami K, Tsujimoto M, Khorana HG, Ichikawa A (1991) The amino acid sequence of a glutamic acid-rich protein from bovine retina as deduced from the cDNA sequence. Proc Natl Acad Sci U S A 88:3116–3119. 10.1073/pnas.88.8.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Makino C, Baylor D, Nathans J (1994) A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J Neurosci 14:5818–5833. 10.1523/JNEUROSCI.14-10-05818.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL, Hurd LB, Papermaster DS (2000) Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J Cell Biol 151:1369–1380. 10.1083/jcb.151.7.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL, Papermaster DS (2004) The C terminus of peripherin/rds participates in rod outer segment targeting and alignment of disk incisures. Mol Biol Cell 15:2027–2037. 10.1091/mbc.e03-09-0650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Ropelewski P, Nemet I, Lee R, Lodowski KH, Imanishi Y (2014) An unconventional secretory pathway mediates the cilia targeting of peripherin/rds. J Neurosci 34:992–1006. 10.1523/JNEUROSCI.3437-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau MC, Zagotta WN (2002) An intersubunit interaction regulates trafficking of rod cyclic nucleotide-gated channels and is disrupted in an inherited form of blindness. Neuron 34:197–207. 10.1016/S0896-6273(02)00647-5 [DOI] [PubMed] [Google Scholar]
- Tsybovsky Y, Wang B, Quazi F, Molday RS, Palczewski K (2011) Posttranslational modifications of the photoreceptor-specific ABC transporter ABCA4. Biochemistry 50:6855–6866. 10.1021/bi200774w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz D, Zoche M, Müller F, Beyermann M, Körschen HG, Kaupp UB, Koch KW (1998) Calmodulin controls the rod photoreceptor CNG channel through an unconventional binding site in the N-terminus of the beta-subunit. EMBO J 17:2273–2284. 10.1093/emboj/17.8.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz D, Ficek N, Kremmer E, Bauer PJ, Kaupp UB (2002) Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36:881–889. 10.1016/s0896-6273(02)01098-x [DOI] [PubMed] [Google Scholar]
- Whitaker SL, Knox BE (2004) Conserved transcriptional activators of the Xenopus rhodopsin gene. J Biol Chem 279:49010–49018. 10.1074/jbc.M406080200 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Molday LL, Molday RS, Sarfare SS, Woodruff ML, Fain GL, Kraft TW, Pittler SJ (2009) Knockout of GARPs and the β-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J Cell Sci 122:1192–1200. 10.1242/jcs.042531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Trudeau MC, Zagotta WN (2002) Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 36:891–896. 10.1016/s0896-6273(02)01099-1 [DOI] [PubMed] [Google Scholar]
- Zhong H, Molday LL, Molday RS, Yau KW (2002) The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 420:193–198. 10.1038/nature01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Cai G, Hall EO, Freeman GJ (2007) In-fusion Assembly: seamless Engineering of Multidomain Fusion Proteins, Modular Vectors, and Mutations. Biotechniques 43:354–359. 10.2144/000112536 [DOI] [PubMed] [Google Scholar]
- Zulliger R, Conley SM, Mwoyosvi ML, Stuck MW, Azadi S, Naash MI (2015) SNAREs interact with retinal degeneration slow and rod outer segment membrane protein-1 during conventional and unconventional outer segment targeting. PLoS One 10:e0138508. 10.1371/journal.pone.0138508 [DOI] [PMC free article] [PubMed] [Google Scholar]