In late December 2019, cases of pneumonia of unknown aetiology were reported by Chinese authorities in the city of Wuhan, China [1]. In January 2020, the World Health Organization declared an international public health emergency. The pathogen was identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the disease was named coronavirus disease 2019 (COVID-19) [2]. Africa was last to report SARS-CoV-2 infections [3]. Egypt was the first African country to record a case of COVID-19, 7 weeks after the start of the pandemic [4,5]. As of this writing (2 March 2021), Africa has recorded 2 650 948 cases and 24 464 deaths [6]. In Senegal, the first case of COVID-19 was detected on 2 March 2020. As of 5 February 2021, a total of 23 073 confirmed cases and 666 deaths have been reported (http://www.sante.gouv.sn/). Africa remains less affected by the global pandemic compared to the rest of the world. However, new variants of SARS-CoV-2, such as British variant B.1.1.7, South African variant B.1.351 and Brazilian variant P.1, have been reported [7]. This emergence of new variants appears to be proportional to the increase in the number of new cases worldwide [8]. In West Africa, particularly Senegal and the Gambia, a new variant of SARS-CoV-2 called Marseille-1 has also been reported [9]. Recently, vaccines such as the Pfizer/BioNTech and Moderna have shown good efficacy against the B.1.1.7 variant but less effectiveness against the B.1.351 variant [10].
This study was approved by the National Ethics Committee for Health Research of Senegal (approval 000159/MSAS/CNERS/Sec, 21 August 2020). Free and informed consent was provided by each adult individual who participated in this study.
In Senegal, the Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formations (IRESSEF), collaborated with the Medical Research Council Unit The Gambia at London School of Hygiene and Tropical Medicine (MRCG at LSHTM) to sequence SARS-CoV-2 virus samples and characterize circulating variants within genomes. One hundred nasopharyngeal samples were collected in the Thiès region and at Blaise Diagne International Airport in Senegal. SARS-CoV-2 was confirmed using PCR at the IRESSEF laboratory. RNA was extracted and transported to the MRCG at LSHTM for sequencing. Twenty-four barcode libraries were sequenced on a flow cell using GridION (Nanopore technology) with expected coverage of >100× following version 3 of the ARTIC protocol [11]. The sequences were analysed using the bioinformatic pipeline from the ARTIC Network (https://artic.network/ncov-2019). The Pangolin software package (https://github.com/cov-lineages/pangolin) was used to assign the lineage to each genomic sequence of studied samples. Mutation analysis was performed using the GISAID mutation analysis tool. All the consensus genome sequences have been deposited in the GISAID database (https://www.gisaid.org/).
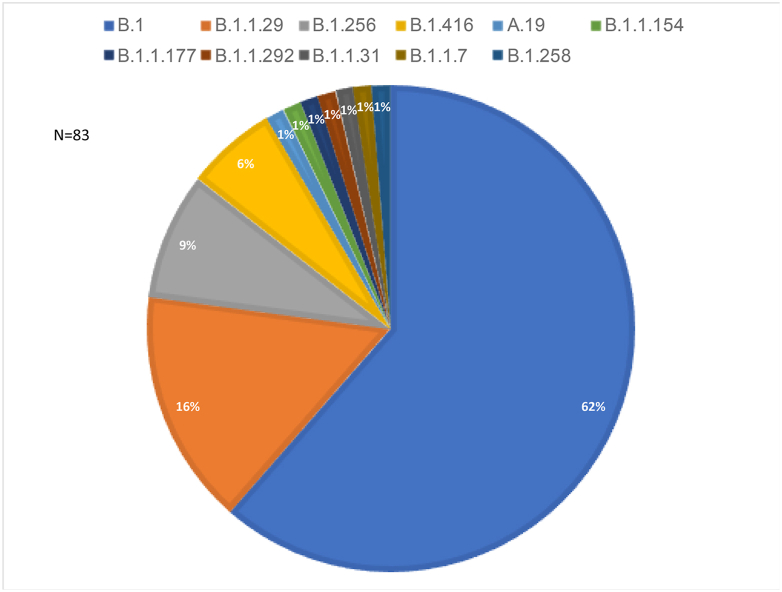
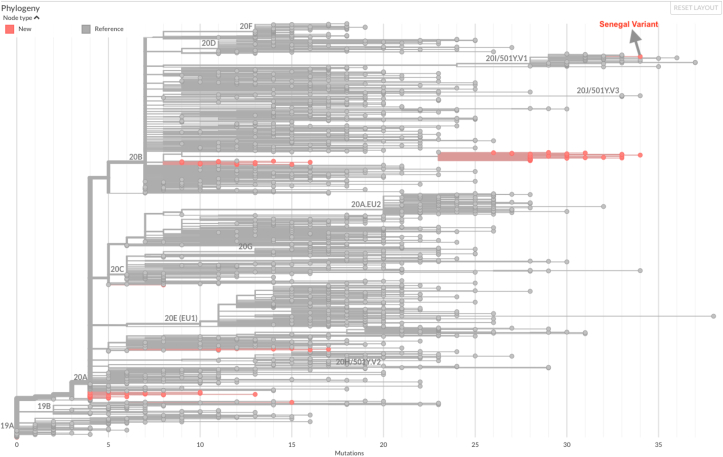
Eighty-three of 100 samples were sequenced successfully. There were 51 samples (61%) that belonged to the B.1 lineage, of which only one corresponded to the B.1.1.7 variant (e.g. the UK variant). This B.1.1.7 variant was isolated from a sample collected from a 23-year-old woman. The patient tested positive on 30 December 2020. She arrived in Senegal from India on 14 August 2020 with a negative PCR test for SARS-CoV-2. She had not traveled since her arrival in Senegal and was only in contact with her husband and her maid. The rest of the samples were distributed as follows: B.1.129 (n = 13), B.1.256 (n = 7), B.1.416 (n = 5), A.19 (n = 1), B.1.1.154 (n = 1), B.1.1.177 (n = 1), B.1.1.292 (n = 1), B.1.1.31 (n = 1), B.1.1.7 (n = 1) and B.1.258 (n = 1) (Fig. 1). Mutation analysis of the B.1.1.7 variant revealed the presence of five mutations (N501Y, D614G, P681H, S982A and D1118H) on the spike glycoprotein. Our genomic sequences were compared to those of the GISAID database, and we constructed a phylogenetic tree to highlight the relationship of Senegal's strains with other strains in the world (Fig. 2).
Fig. 1.
Distribution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages in Senegal.
Fig. 2.
Phylogenetic tree showing Senegal's new variant clustering with 201/50Y.V1 variant.
In conclusion, variant B.1.1.7, termed the ‘UK variant’, ‘British variant’ or ‘English variant’, was detected for the first time in Senegal in an Indian woman. The emergence of new SARS-CoV-2 variants throughout the world should lead health authorities to rely on next-generation sequencing tools to monitor and control the strains circulating in Senegal.
Conflict of interest
None declared.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haider N., Yavlinsky A., Simons D., Osman A.Y., Ntoumi F., Zumla A. Passengers’ destinations from China: low risk of novel coronavirus (2019-nCoV) transmission into Africa and South America. Epidemiol Infect. 2020 doi: 10.1017/S0950268820000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalaoui R. What could explain the late emergence of COVID-19 in Africa? New Microb New Infect. 2020 doi: 10.1016/j.nmni.2020.100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO) WHO Director-General’s remarks at the media briefing on 2019-nCoV on 11 February 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 Available at:
- 7.Voloch C.M., da Silva R., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv. 26 December 2020 doi: 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med. 2021;9:e20. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson P., Levasseur A., Gautret P., Fenollar F., Thuan Hoang V., Delerce J. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: an investigational study. Travel Med Infect Dis. 2021;40:101980. doi: 10.1016/j.tmaid.2021.101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A. Neutralizing activity of BNT162b2-elicited serum – preliminary report. N Engl J Med. 2021 doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quick J. nCoV-2019 sequencing protocol. Protocols.io. 2020;1 doi: 10.17504/protocols.io.bbmuik6w. [DOI] [Google Scholar]