Abstract
Organoid technology has rapidly transformed basic biomedical research and contributed to significant discoveries in the last decade. With the application of protocols to generate organoids from cancer tissue, organoid technology has opened up new opportunities for cancer research and therapy. Using organoid cultures derived from healthy tissues, different aspects of tumour initiation and progression are widely studied including the role of pathogens or specific cancer genes. Cancer organoid cultures, on the other hand, are applied to generate biobanks, perform drug screens, and study mutational signatures. With the incorporation of cellular components of the tumour microenvironment such as immune cells into the organoid cultures, the technology is now also exploited in the rapidly advancing field of immuno-oncology. In this review, I discuss how organoid technology is currently being utilised in cancer research and what obstacles are still to be overcome for its broader use in anti-cancer therapy.
Keywords: Cancer, Organoids, 3D culture, Pre-clinical models, Personalised medicine, Drug screening, Immuno-oncology
Introduction
Cancer remains a major threat to quality of life with significant risks of morbidity and mortality worldwide [1], despite extraordinary progress made in cancer research, prevention, detection, and therapy in the past decades. For example in the USA, incidences of the prominent cancer types of lung or colorectal cancer are decreasing partially due to increased knowledge on cancer biology and, hence, improved prevention, while incidence rates of other cancer types such as liver or oral cancers are increasing [2]. However, cancer is a heterogenous disease with a broad range of types and subtypes, which can be defined based on their anatomical location, histological appearance, and genetical makeup. In order to guide the way to improved targeted therapy, pre-clinical model systems are essential to better capture the inter- and intra-tumour heterogeneity. For instance, animal cancer models, in particular genetically engineered mouse models (GEMMs), have provided significant insights into the cellular and genetic basis of cancer [3]. However, their application is rather costly, time-consuming, and often cannot be translated into therapy, due to major differences to human pathology and tumourigenesis (reviewed in [4, 5]). Using human cancer models such as cancer cell lines and patient-derived xenografts (PDTXs), some of these limitations were overcome: in principle, these models can be generated from a larger cohort of patients better presenting the inter-tumour heterogeneity [6]. Yet, these models also have significant drawbacks. Cancer cell lines often do not sufficiently retain the intra-tumour cellular and genetic heterogeneity in vitro, as only robust and colony-forming cells (i.e. clones) can be maintained in culture long-term, which are frequently genetically instable [6, 7]. Furthermore, cancer cell lines are devoid of the cellular microenvironment of the tumour in vivo, including the tumour stroma as well as immune infiltrate [6]. In most cases, cancer cell lines also lack matched cell lines established from normal tissue as reference control [6]. PDTXs are generated by transplanting primary patient tumour material into immunocompromised mice [8]. As such, PDTX models allow for the spontaneous development of a tumour stroma of murine origin and the investigation of metastasis formation [6, 8]. PDTX-based approaches therefore do model some critical aspects of the tumour and its microenvironment. Yet, PDTXs still lack the human-specific immune components, require the use of animals (potentially causing mouse-specific features not found in human cancer), and are both expensive and time-consuming (reviewed in [9, 10]).
A promising alternative to these conventional cancer models is based on the discovery that adult stem cells (ASCs) proliferate and spontaneously self-organise into three-dimensional (3D) organotypic cellular structures—so-called organoids—in culture, when they are embedded into a hydrogel rich in extracellular matrix (ECM) proteins such as Matrigel or Basement Membrane Extract (BME) [11]. A key to organoid technology is the tissue-specific growth factor cocktail provided to the culture. In the case of the first organoid model system—murine small intestinal organoids generated from Lgr5+ intestinal epithelial stem cells—the growth medium contained the Wnt pathway agonist and ligand of LGR5 R-spondin-1, epidermal growth factor, and the bone morphogenetic protein (BMP) pathway inhibitor Noggin [11]. By designing a growth factor cocktail specific to each stem cell type, tissue, and species, the protocol was adapted to allow for the generation of organoid cultures from other murine and human epithelial tissues (Fig. 1; reviewed in [12, 13]) such as the bladder [14], breast [15], colon (and rectum) [16, 17], endometrium [18], fallopian tubes [19], kidney [20], liver [21, 22], lung [23], oesophagus [24], oral mucosa [25], pancreas [26], prostate [27, 28], salivary gland [29], skin epidermis [30], stomach [31], and taste buds [32], and, most recently, even non-mammalian tissue such as snake venom glands [33]. As such, it has also been possible to generate ASC-enriched epithelial organoids from tissue pieces [11] or tissue pieces containing both epithelial and stromal cells [34]. Organoids allow for the long-term expansion of stem cells and their spontaneous differentiation into specialised tissue cells, most prominently seen in the crypt–villus differentiation observed in murine small intestinal organoids [11, 35] or stratification and cornification described for murine skin epidermal organoids [30] as well as murine and human oral epithelial organoids [25]. Long-term analyses further suggest that ASC-derived organoids remain largely phenotypically and genetically stable, reflecting their tissue of origin [36]. Comparative mutational analysis of organoid cultures generated from different murine and human tissues further demonstrated that tissue-specific mutational signatures can be defined using organoids [36–38]. In addition, organoid cultures are amenable to a wide range of experimental tools, including single-cell transcriptomics [39], gene editing and tagging [40], (whole-mount) imaging [41], xenotransplantation [42], and co-culture with other cells such as immune cells (reviewed in [43]). Another complementary approach is to generate organoids using pluripotent stem cells (PSCs), namely embryonic stem cells or induced pluripotent stem cells (iPSCs) (reviewed in [12, 13]). PSC-derived organoids reflecting various types of tissues and organs including the brain [44], intestine [45], kidney [46, 47], and retina [48, 49] have been described. As PSC-derived organoids typically remain phenotypically and transcriptionally immature (reviewed in [50]) and, therefore, are more similar to embryonic-like tissue, their use for cancer research has been limited so far (reviewed in [51]). Another important disadvantage of PSC-derived organoids in cancer research is the lack of acquired cancer gene mutations in individual tumour organoid lines. These models are therefore less attractive for biobanking or drug screening, in contrast to organoids directly derived from tumour biopsies that preserve cancer gene mutations in culture. However, improved methods may allow for broader utilisation of PSC-derived organoid models in cancer research in the future.
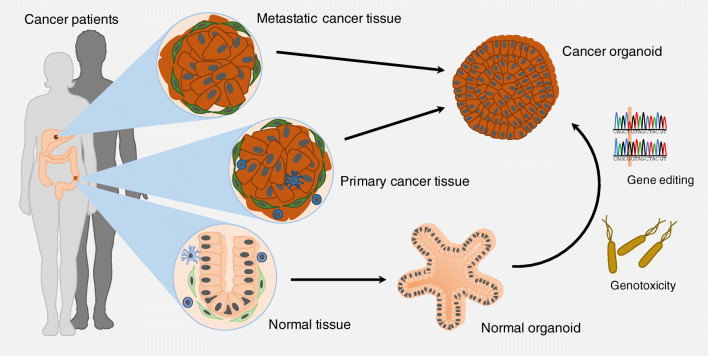
Fig. 1.
Generation of patient-derived normal and cancer organoids. Patient-derived cancer organoids can be established from primary and metastatic cancer tissue. Matched normal organoids can be generated from normal tissue. Through gene editing, normal organoids may be transformed into cancer organoids. By exposure to genotoxic factors, it may also allow for malignant transformation of normal organoids in vitro, as a recent study showed that normal organoids incubated with genotoxic bacteria acquired mutational signature characteristic of cancer subsets [63]. Some images were modified from the medical art database at https://smart.servier.com/
In this review, I discuss the use of human ASC-based organoid technology in basic and translational cancer research. I explore how epithelial organoid cultures serve as a base to study the processes of tumourigenesis and metastasis, including the role of the tissue microenvironment. I further highlight the use of cancer organoids generated from primary patient material in biobanks for drug discovery, personalised anti-cancer therapy, and immuno-oncology. I close by discussing challenges of organoid technology to be overcome to allow its wider use in the clinic and future prospects of its further exploitation in cancer research.
Genotoxic factors and cancer initiation
Cancer develops in a multistep process from normal tissue by acquisitions of somatic mutations in so-called cancer driver genes [52]. Cells throughout our body are continuously challenged by different endogenous or exogenous genotoxic factors that can damage their DNA [53, 54]. While most of the DNA damage induced is being continuously repaired by one of the DNA repair pathways, errors in the DNA repair process may result in the gain of mutations at a low frequency [52, 55]. However, prolonged exposure to such genotoxic factors may further increase the risk of developing cancer. The International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) has published a list of exogenous cancer hazards, including physical agents such as ionising radiation (i.e. gamma rays, X-rays, and higher spectrum ultraviolet light), carcinogenic chemicals, and certain types of pathogenic infections (‘IARC monographs’, https://monographs.iarc.fr/). While animal models are widely used for research into agents associated with an increased cancer risk, it is often rather difficult to define a mechanism of action. In many cases, it is therefore not possible to determine whether a suspected carcinogenic agent can be classified as carcinogen to humans [54]. As epithelial organoid cultures remain essentially phenotypically and genetically stable long-term, they are an excellent model system to study the genotoxic potential of different agents. Attempting this strategy, a recent study described the testing of different genotoxic chemicals, namely ethyl methanesulfonate (EMS), acrylamide (AA), and 7,12-dimethylbutylamine (DMBA), on mouse-derived epithelial organoids for their potential to generated tumours upon transplantation into nude mice [56]. Murine mammary gland organoids with partial loss of Trp53 treated with DMBA displayed tumourigenicity upon transplantation, while wild-type Trp53 counterparts did not develop tumours in vivo, in line with earlier work done in mouse models. Murine lung organoids, irrespective of their Trp53 status, developed tumours in vivo following exposure to EMS and AA [56], overall demonstrating that organoids may be used as chemical carcinogenicity studies. Further improvements should aim to allow for the assessment of genotoxic potential of chemicals in human tissue–derived organoids with a setup that is entirely in vitro.
Apart from ionising radiation and carcinogenic chemicals, pathogenic infections have been linked to tumourigenesis. Again, organoids may allow for the investigation of pathogenic infections and their contribution to cancer development. Different organoid–pathogen co-culture protocols have been described (reviewed in [57, 58]). For instance, gastric organoids have been cultured with Helicobacter pylori [31], a known pathogen populating the stomach that has long been linked to gastric cancer [59]. Other studies demonstrated, for example, that human noroviruses can replicate in human small intestinal organoid cultures [60]; that human papilloma viruses (HPV), described as oncogenic factors in subtypes of head and neck cancer [61], can infect human oral epithelial organoids [25]; and that the parasite Cryptosporidium can complete its life cycle in human small intestinal and lung organoids [62]. A recent study addressed whether the microbiome directly contributes to tumourigenesis using organoid technology [63], as it has long been suggested that gut microbiota may have been involved in colorectal cancer (CRC) development [64]. The authors repeatedly injected a strain of genotoxic Escherichia coli (pks E. coli) expressing an enzyme that synthesises colibactin into the lumen of cystic human intestinal organoids for a period of up to 5 months. In line with a previous report demonstrating that colibactin induces double-strand breaks in cultured cells [65], the authors found that organoids co-cultured with pks E. coli had increased levels of DNA damage [63]. Subsequently, whole-genome sequencing (WGS) was performed on the DNA collected from organoids before and after exposure to pks E. coli. Data were then compared with those obtained from organoid lines exposed to an isogenic control strain of E. coli incapable of producing active colibactin (pks-mutant E. coli) [63]. Organoids exposed to pks E. coli acquired distinct mutational signatures, which were present neither in organoids before bacteria co-culture nor in those co-cultured with pks-mutant E. coli. Assessment of more than 5000 genomes of human cancer metastases revealed that the same mutational signatures were present in 11% of CRC genomes as well as genomes of other cancers including head and neck cancers and urinary tract cancers [63]. In a second CRC-specific patient cohort, the pks E. coli-induced mutational signatures were found in more than 20% of all CRC genomes analysed [63]. This suggests that pathogenic bacteria may directly contribute to malignant transformation (Fig. 1). Organoids that grow as cystic spheroids such as human intestinal organoids are well suited for studying host–pathogen interactions using intra-luminal injections [62, 63]. However, future progress may expand possibilities to study organoid–pathogen interactions in basic cancer research. For instance, modelling more complex communities of the gut microbiota in organoid–bacteria co-cultures may allow for the functional validation of the microbiome alternations linked to CRC formation and progression [66, 67].
Cancer organoids and ‘living’ biobanks of cancer
The adaption of protocols to generate organoids from human ASCs allowed the derivation of cancer organoids from patient material, typically surgical specimen or needle biopsies [68]. Establishment of cancer organoid cultures has been described for primary and/or metastatic tumour tissue sampled from the bladder [14, 69], brain [70, 71], breast [15], colon [17, 72–74], endometrium [18, 75], head and neck [25, 68], kidney [76, 77], liver [78], lung [23], oesophagus [17], ovaries [79, 80], pancreas [26, 81, 82], prostate [83], rectum [72, 84], and stomach [85, 86] (Fig. 1). Overgrowth or contamination by normal (epithelial) cells is a major drawback for the generation of organoid cultures containing only epithelial tumour cells [68, 87]. For instance, based on the observation that cultures lacked copy number alterations and were free of any mutations in fifty common cancer-associated genes, Gao et al. reported that several of their organoid cultures generated from prostate cancer metastases into lung and liver had likely been overgrown by normal epithelial cells [83]. This observation highlights two considerations when establishing cancer organoid cultures. Firstly, due to higher rates of cell death by mitotic catastrophes and other aberrations [40, 88], cancer organoids often grow slower than normal organoids, and, secondly, many normal organoid cultures thrive under surprisingly simple growth conditions or require similar conditions as their cancerous counterparts. To avoid this, different strategies have been developed. On the one hand, cancer organoid cultures may be established from metastatic tissue, ideally, taken from sites devoid of normal epithelial cells such as lymph node biopsies, bone biopsies, or ascites fluid. On the other hand, pure cancer organoid cultures may be generated by providing a minimal or selective medium inhibiting the growth of normal epithelial cells. Activating mutations in the Wnt/β-catenin signalling pathway are found in about 95% of all cases of CRC. Therefore, pure CRC organoid cultures have been achieved by removal of Wnt pathway stimulants such as Wnt ligands and R-spondins from the growth medium [17, 72, 88]. Withdrawal of other growth factors may be used to select for tumour cells when mutations in other specific signalling pathway have been identified. For instance, epidermal growth factor (EGF) withdrawal allows for the selection of rat sarcoma viral oncogene homologue (RAS) mutants, hence EGF receptor (EGFR)–signalling independent tumour cells [88]. In accordance, combinatory withdrawal of growth medium components has been used to select for CRC subtypes with specific growth factor independencies [73, 88]. Furthermore, cancer organoid cultures have been robustly purified from non-small cell lung carcinomas, which are frequently mutant for TP53 by selection with the MDM2 agonist Nutlin-3 [23]. In line with observations reported for organoids established from normal (i.e. healthy) primary material, cancer organoids histologically, transcriptionally, and genetically largely retain the bulk characteristics of the tumour epithelium of origin. Based on cancer organoid protocols, large efforts have been made to generate ‘living’ biobanks of patient-derived cancer organoids (Fig. 2; Table 1 and references therein), often with their matched normal counterparts. An important consideration here is the use of patient-matched pairs of normal and cancer organoid lines for subsequent analyses such as drug screenings (see “Personalised anti-cancer therapy”). Such matched pairing of organoid lines may allow accounting for genetic and phenotypic variation among normal human epithelial organoids derived from different patients [36–38, 89], which may even exceed the effects of single cancer gene mutations. As the complexity of cancer results in cancer types being further broken down into subtypes, it is important to note that the majority of these biobanks provide cancer organoid cultures representing different cancer subtypes. Overall, more and more cancer biobanks are being described. However, as most of the existing cancer organoid protocols were developed for epithelial carcinomas, future research should aim at generating more organoid cultures from non-epithelial cancers as those recently described for glioblastoma [70] and rhabdoid tumours of the kidney [76].
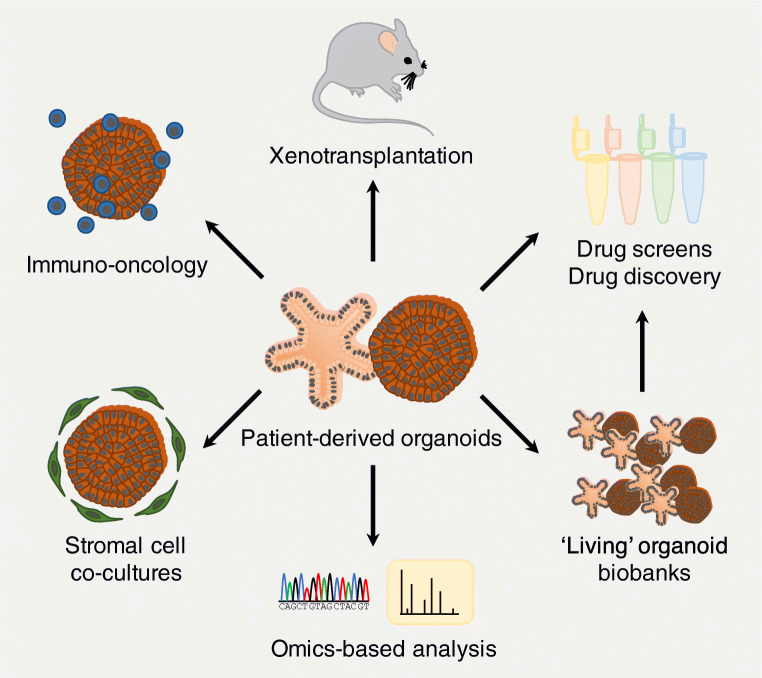
Fig. 2.
Utilisation of patient-derived organoids in cancer research. Patient-derived (cancer) organoids have already been used to generate ‘living’ organoid biobanks that can be exposed to different drugs for efficacy screenings and drug discovery validations. Organoids have further been used to study inter- and intra-tumour heterogeneity by analysis of mutational signatures, gene expression patterns, or proteomics. By transplanting cancer organoids into mice, tumour cell invasiveness and potential to metastasise can be tested. Finally, approaches to incorporate cells of the tumour microenvironment such as stromal cells (such as cancer-associated fibroblasts) or immune cells (i.e. immuno-oncology) are being developed. Some images were modified from the medical art database at https://smart.servier.com/
Table 1.
Overview of published patient-derived cancer organoid biobanks. Listed are only studies describing cancer organoid collections with cancer organoid lines established from more than three patients
| Cancer type | Source | Validation and analysis | Therapy testing | Year of publication | References | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary cancer tissue | Metastatic cancer tissue | Other tumour tissue | Matched normal tissue | Tumour-specific medium | Histology | DNAa | RNAb | Xenografting | Other analysisc | Drug screen | Radiotherapy | Immunotherapy | Clinical response | |||
| Biliary tract cancer | ● | ● | ● | E | M/P | ● | ● | 2019 | [139] | |||||||
| Bladder cancer | ● | ● | T/E | R | ● | ● | 2018 | [69] | ||||||||
| ● | ● | ● | R | ● | 2019 | [14] | ||||||||||
| Breast cancer | ● | ● | ● | G | R | ● | ● | 2018 | [15] | |||||||
| Colorectal cancer | ● | ● | ● | ● | E | M | ● | Pro | ● | 2015 | [72, 124, 125] | |||||
| ● | E | 2015 | [74] | |||||||||||||
| ● | ● | ● | ● | ● | T/E | R | ● | 2016 | [73] | |||||||
| ● | ● | ● | ● | 2016 | [126] | |||||||||||
| ● | ● | ● | E/G | R | ● | 2017 | [127] | |||||||||
| ● | G | ● | ● | 2019 | [104] | |||||||||||
| Endometrial cancer | ● | ● | ● | ● | ● | E | R | ● | EM | ● | 2019 | [75] | ||||
| Gastric cancer | ● | ● | ● | G | ● | 2018 | [128] | |||||||||
| ● | ● | ● | ● | ● | ● | E | R | ● | ● | 2018 | [85] | |||||
| ● | ● | G | R | ● | ● | 2019 | [129] | |||||||||
| Gastrointestinal cancerd | ● | ● | G | R | ● | ● | ● | 2018 | [87] | |||||||
| Glioblastoma | ● | ● | E | R/scR | ● | ● | ● | ● | 2020 | [70] | ||||||
| Head and neck cancer | ● | ● | ● | T/E | R | ● | EM | ● | ● | ● | 2019 | [25] | ||||
| Kidney cancer | ● | ● | ● | E | R | ● | ● | 2019 | [77] | |||||||
| ● | ● | ● | ● | G | scR | Me | ● | 2020 | [76] | |||||||
| Liver cancer | ● | ● | ● | E | R | ● | ● | 2017 | [78] | |||||||
| ● | ● | E | ● | 2019 | [130] | |||||||||||
| Lung cancer | ● | ● | ● | ● | E | ● | ● | 2019 | [131] | |||||||
| ● | ● | ● | G | ● | ● | 2019 | [23] | |||||||||
| ● | ● | ● | 2020 | [84] | ||||||||||||
| Multiple cancerse | ● | ● | E | ● | ● | 2017 | [132] | |||||||||
| Multiple cancersf | ● | ● | ● | M | ● | ● | 2018 | [133, 134] | ||||||||
| Ovarian cancer | ● | ● | G | ● | 2017 | [135] | ||||||||||
| ● | ● | ● | G/scG | R | ● | EM/Me | ● | ● | 2019 | [79, 136] | ||||||
| Pancreatic cancer | ● | ● | ● | ● | ● | T/E | P | ● | 2015 | [26] | ||||||
| ● | ● | ● | ● | ● | E | M/P | ● | Me | 2018 | [81] | ||||||
| ● | ● | ● | ● | E/G | R | ● | ● | ● | 2018 | [82] | ||||||
| ● | ● | ● | ● | G | R | ● | ● | 2019 | [137] | |||||||
| Prostate cancer | ● | ● | E | R | ● | A | ● | 2014 | [83] | |||||||
| Rectal cancer | ● | ● | ● | ● | ● | E | P | ● | ● | ● | ● | 2019 | [138] | |||
| ● | ● | E | ● | ● | ● | 2020 | [84] | |||||||||
aE, whole-exome sequencing; G, whole-genome sequencing; scG, single-cell genome sequencing; T, targeted (cancer gene) sequencing
bM, microarray; P, qRT-PCR; scR, single-cell mRNA sequencing; R, mRNA sequencing
cA, array CGH; EM, electron microscopy; Me, DNA methylation analysis; Pro, proteomics
dColorectal cancer, gastro-oesophageal cancer, liver cancer
eBladder cancer, brain cancer, breast cancer, colorectal cancer, gastro-oesophageal cancer, kidney cancer, lung cancer, ovarian cancer, prostate cancer, small intestinal cancer, soft tissue cancer, and uterus cancer
fBreast cancer, digestive organ cancer, lung cancer, ovarian cancer, peritoneal cancer, and uterus cancer
Molecular genetics meets cancer organoid technology
To better understand the molecular genetics of cancer, organoid cultures have been utilised for two complementary approaches. The first strategy is the mutational analysis of patient-derived cancer organoids via WGS, whole-exome sequencing (WES), or targeted sequencing for cancer gene mutations (Fig. 2). On the other hand, the second strategy aims at probing the consequences of specific mutations on tumourigenesis by introducing putative or validated cancer gene mutations into either normal organoids or cancer organoids using gene editing technology.
Mutational analysis of patient-derived cancer organoids
As discussed above, cancer organoid biobanks were previously sequenced to confirm that cancer organoids robustly retain the genetics of the tumour epithelium on the bulk level (Table 1 and references therein). Mutational analysis of these biobanks revealed that the individual cancer organoid lines could be assigned to different molecular subtypes of cancer [72], which demonstrates that cancer organoids can represent inter-tumour heterogeneity in vitro. As normal organoid cultures remain largely genetically stable [36] and cancer organoids reflect the bulk genetic makeup of the cancer of origin [72], cancer organoid cultures allow for the investigation of clonal dynamics within cancer, a critical feature of intra-tumour heterogeneity and cancer responses to therapy. A recent study, for example, generated organoids from high-grade serous ovarian carcinoma tissue obtained from a single patient at different time points [79]. About 800 single-cell DNA sequencing profiles were generated from the primary tumour samples as well as corresponding tumour organoid lines from two different passages. For each single sequenced cell copy number variation (CNV), profiles were calculated and cluster analysis was performed. The analysis revealed that the primary tumour cells clustered into five different clusters [79]. All organoid cells were also assigned to one of the five clusters, suggesting that intra-tumour heterogeneity was maintained in vitro. Interestingly, one of the clusters contained diploid cells, and cells obtained from late passaged organoids were less abundant in this cluster. The authors suggested that these cells were likely normal cells that were outgrown by aneuploid tumour cells (present in the remaining four clusters) upon extended organoid culture [79]. Another study generated clonal organoid lines from several locations within CRC tissue as well as from adjacent normal colorectal epithelium from three different patients and analysed genome, epigenome, and transcriptome of each line [89]. WES analysis revealed a dramatically higher mutational load and a robust diversification of the mutational signatures in the CRC cells in comparison with the normal intestinal cells. Reconstruction of phylogenetic trees demonstrated that most mutations present in the cancer were absent from normal colorectal cells and were acquired in the late stages of clonal expansion [89]. Importantly, the mutational alterations likely formed the basis of the inter- and intra-tumour diversification, as epigenetic and transcriptional changes were aligned with the mutations present within each clonal CRC organoid line [89]. As this study highlights clonal (genetic) diversity within one cancer, it remains important for many applications of organoid technology in cancer research to consider and, if possible, prevent clonal drift in tumour organoids [90]. Here, clonal drift describes the process during which genetically diverse and polyclonal (cancer) organoid cultures reach a bottleneck with only one or a few dominant genetic clones surviving, as also seen in the homoeostatic small intestinal epithelium in vivo [91]. Different ways to minimise the risk of clonal drift in organoid cultures may be passaging the entire culture plate (instead of only a smaller fraction of it), returning to cryopreserved early passages of the cultures, or keeping the culture period as short as possible [38].
Probing the role of (cancer) gene mutations in organoids
Epithelial organoid cultures are amendable to gene editing via different genetical tools, including CRISPR–Cas9 and RNAi [30, 40, 90, 92]. This versatility allows for the probing of specific gene functions in cancer formation or the modelling of cancer progression using organoids (Fig. 1). For instance, several studies have been published on the stepwise introduction of classical CRC driver mutations—APC−/−, TP53−/−, SMAD4−/−, and KRASG12D/+—following the so-called Vogelgram cancer progression model [93]. Starting from patient-derived normal colon organoid cultures, the groups replicated key features of CRC progression, including independence of niche factors [88, 94], chromosome instability, aneuploidy, invasiveness [88], and ability to metastasise when the gene-edited quadruple mutated organoids were orthotopically transplanted into the caecum of NSG mice [95]. CRISPR–Cas9 gene editing of human colon organoids has further been applied to investigate the role of mutations in DNA mismatch repair genes such as MLH1 [96] or BRAFV600E mutations [97] in CRC. In another study, human liver ductal organoids were modified to harbour loss-of-function mutations for the tumour suppressor BAP1 [98]. To allow for selection of mutant organoid clones, the authors co-injected targeting plasmids for both the genes encoding BAP1 and TP53. As described above, Nutlin-3 could then be used to select for TP53 mutant organoids, which very likely were also BAP1 mutant due to the high efficiency of plasmid co-transfection observed in the system [98]. TP53−/− BAP1−/− double-mutant organoids showed abnormal morphology with loss of cell polarity, perturbation of the epithelial layer, and increased cell motility. Profiling of transcriptome and proteome provided further evidence for alterations in the expression of cytoskeletal and cell–cell junctional components, which are essential for the proper functioning of epithelia. In elegant rescue assays, the authors demonstrated that only catalytically active BAP1 localised to the nucleus can recover the homeostatic phenotype in the organoids [98]. When introducing other known liver cancer (cholangiocarcinoma) gene mutations in the organoids in combination with the BAP1 mutation, the authors could show that BAP1 loss-of-function is required for tumourigenesis when mutant organoids are transplanted into mice [98]. The authors highlighted their study as an example to probe cancer gene function mechanistically in human tissue by combining organoid technology with gene editing tools such as CRISPR–Cas9 [98, 99]. An essential advantage of using organoid technology to probe the role of (cancer) gene mutations is the possibility to introduce the gene mutations of interest into a normal organoid line, which—in its unmodified form—serves as an isogenic control. This strategy helps to avoid artefacts introduced by the individual variability due to the genetic background of donors and should be carefully considered for such studies.
Personalised anti-cancer therapy
Organoid technology is already exploited for personalised therapy of cystic fibrosis (CF) patients. CF is a monogenic disease caused by a wide spectrum of mutations in the cystic fibrosis transmembrane conductor regulator (CFTR) gene encoding a chloride ion transport channel [100]. An in vitro assay was developed using normal intestinal organoids generated from rectal biopsies of patients that allows for the prediction of patient response to CF drugs [101]. Following the proof-of-concept, several further studies validated the suitability of the drug testing platform utilising organoid cultures [102, 103]. Using different proxies to show inhibition of cancer cell growth or induction of cytotoxicity, limited drug screens have also been performed on cancer organoid biobanks (Fig. 2 and Table 1; [68]). However, robustness, reproducibility, and applicability of the assays to different cancer types still require further assessment before cancer organoid-based drug (or small molecule) screenings may be more broadly used for personalised medicine approaches [68, 87, 104, 105]. Another critical issue is whether patient-derived cancer organoids at all have the potential to predict patient response to anti-cancer therapy. Several studies aimed at resolving this concern comparing cancer organoid responses with chemo- or chemoradiotherapy with clinical outcomes. In one study, the authors generated fifty cancer organoid lines from metastatic tissues of different gastrointestinal cancers (i.e. CRC, gastro-oesophageal cancer, and cholangiocarcinoma) [87]. Following genotypical and phenotypical characterisation and validation of the cancer organoids as well as transcriptional profiling, the authors tested a variety of drugs directly in vitro on the organoids and in vivo using organoid-based murine xenograft models. Strikingly, when comparing their results with the clinical data, the authors found an 88% positive predictive value and 100% negative predictive value of organoid-based targeted therapy or chemotherapy [87]. Another study using organoids derived from metastatic CRC patients demonstrated almost similar predictive values for the drug irinotecan alone or in combination with 5-fluorouracil, while treatment with only oxaliplatin or oxaliplatin combined with 5-fluorouracil could not be validated [104]. A third study tested irradiation, 5-fluorouracil, and irinotecan on a set of rectal cancer-derived organoid lines and found a diagnostic accuracy of almost 85% using the organoids [84]. Lastly, a study described that cohorts of pancreatic cancer patients could be stratified based on transcriptional signatures and chemosensitivity profiles obtained from cancer organoid cultures of these patients [82]. Collectively, these efforts make a strong case that cancer organoids may have a significant predictive value for patient response to anti-cancer treatments. However, many variables, including the tumour microenvironment and drug metabolism or toxicity by peripheral organs, remain to be addressed.
Immuno-oncology and the tumour microenvironment in a dish
The tumour microenvironment plays a critical role in tumour formation and progression (reviewed in [6]). Hence, the interaction of the tumour with its microenvironment is a heavily studied hallmark of cancer [106]. Bidirectional communication between tumour cells and cellular components of the tumour microenvironment such as fibroblasts, the vasculature, and immune cells plays a critical role in tumour promotion [106]. For instance, cancer cells may stimulate endothelial cells to induce angiogenesis or chronic inflammation mediated by tissue-infiltrating immune cells may provide survival factor or mitogens promoting tumour growth [106]. On the other hand, an active immune response may suppress tumour growth and, therefore, cancer cells may develop means to avoid immune destruction [106]. A better understanding of the influence of the tumour microenvironment on tumour growth dynamics is essential to guide anti-cancer therapy and minimise resistance against treatment [6]. However, the tumour microenvironment is composed of a heterogeneous pool of cells with a variety of features that may promote or prevent tumour growth, making it a highly complex biological system to be studied [6].
While cancer organoid cultures lack cellular components of the tumour microenvironment, they may serve as very good reductionist in vitro model systems to study the influence of tumour microenvironment on cancer growth. Recently, several immuno-oncological protocols have been developed using organoid technology (Fig. 2; reviewed in [43]). One such approach used cancer organoid co-cultures with peripheral blood mononuclear cells (PBMCs) to generate patient-specific tumour-reactive cytotoxic T cells [107, 108]. Since high degree of neoantigen presentation is critical to elicit a robust anti-tumour immune response by antigen-specific T cells [109, 110], the authors chose organoids generated from specific subtypes of CRC and non-small cell lung cancer with a high mutational burden. Through serial co-cultivation of cancer organoids and PBMCs in the presence of a T cell-stimulating growth factor cocktail, it was possible to select for and expand antigen-specific cytotoxic T cells in about half of the samples. Importantly, co-cultures of expanded T cells with organoid generated from adjacent healthy epithelial tissue resulted in undisturbed organoid expansion without significant levels of organoid cytotoxicity [107]. In another approach, tumour-infiltrating T cells and CRC organoids were separately expanded using their respective gold standard methods and then combined to assess T cell-mediated organoid killing [111]. Interestingly, the extent of cell death in the in vitro co-culture assay correlated well with the patient’s response to chemotherapy and immune checkpoint blockade [111]. Furthermore, the authors tested immune checkpoint blockade using PD1 antibodies in their co-culture model system and demonstrated that organoid killing by PD1high T cells was improved upon antibody treatment. Two other groups described organoid-based immuno-oncology assays that utilised cytotoxic lymphocytes engineered to recognise defined antigens and kill organoids presenting such antigens. The first group generated chimeric antigen receptor-engineered natural killer cells recognising the antigens of choice [112], while the second group generated T cell receptor transgenic cytotoxic T cells [113]. In both cases, robust antigen-specific cytotoxicity was observed against cancer organoids presenting the antigen of choice [112, 113]. Co-cultures with matched normal epithelial organoids or cancer organoids presenting control antigens did not show significant levels of antigen-specific cytotoxicity. Apart from these reductionist approaches, other protocols to test lymphocyte-mediated tumour organoid killing have been established. One such approach utilises the air–liquid interface (ALI) cultures of patient-derived cancer organoids that preserved not only the tumour epithelium but also significant cellular components of the tumour microenvironment including fibroblasts, macrophages, and lymphocytes for about 1 month [114]. These ALI-based cultures could be established from various cancers such as CRC, lung cancer, head and neck cancer, and melanoma and also allowed for modelling of immune checkpoint blockade [114].
Apart from the immune infiltrate, fibroblasts may also critically contribute to tumour initiation and progression. A recent study in mice, for instance, described the presence of a rare population of Ptgs2-expressing fibroblasts that reside under the crypt epithelium and constitute the intestinal stem cell niche in mice [115]. The authors then showed that Ptgs2-expressing fibroblasts secreted prostaglandin E2 (PGE2), which promoted adenoma formation in the classical ApcMin/+ tumour mouse model in vivo [116]. As there is growing evidence for the tumour-promoting features of PGE2 [117, 118], but its cellular source remained elusive, the authors investigated further. To decipher the bidirectional signalling between stem cells and their mesenchymal niche, the authors then developed a co-culture method of mouse small intestinal organoids and wild-type primary mouse intestinal fibroblasts (Fig. 2). In the absence of fibroblasts, organoids started budding and displayed the typical crypt–villus architecture [11]. However, when co-cultured with fibroblasts, organoids formed cystic spheroids. This is usually only seen for a very short culture period or under high Wnt conditions with increased stemness and inhibited differentiation of the cultures [119]. By generating organoids lacking the main PGE2 receptor in the (murine and human) intestinal epithelium (prostaglandin E2 receptor 4, EP4; encoded by Ptger4 in mice and PTGER4 in humans), the authors then elegantly demonstrated that fibroblast-secreted PGE2 directly acts on the epithelial stem cells, as spheroids did not form when culturing Ptger4-depleted organoids [115], aphenotype that was readily reproduced by applying an EP4 inhibitor to the co-cultures [115]. Interestingly, PGE2 is a critical component of the growth factor cocktail for human intestinal organoid culture promoting stem cell proliferation and formation of the very characteristic cystic spheroids [16, 17]. In line with the results on murine intestinal organoids, the effect of PGE2 on human colon organoid cultures could also be blocked by PTGER4 inhibition [115], suggesting a conservation of the mechanism of action between mice and humans. Further along the line of tumour progression, cancer-associated fibroblasts (CAFs) play a major role in the tumour microenvironment, for example by providing mitogenic factors to the growing cancer as well as mediating resistance against anti-cancer therapy [6]. CAFs are typically studied following xenotransplantation of human cancer cells or organoids into mice; however, the tumour stroma generated in this setting is entirely composed of murine cells. In order to better understand the interaction between cancer cells and their mesenchymal niche, a few co-culture methods have been published over the last couple of years. One study, for example, described the development of a co-culture protocol of organoids and CAFs derived from pancreatic ductal adenocarcinoma (PDAC) to investigate stem cell niche factor dependency during tumour progression [81]. In their PDAC organoid biobank, the authors found that the organoids showed different levels of dependency on Wnt/R-spondin supplementation. Some organoids survived in the absence of both ligands, while others were fully dependent on exogenous Wnt ligands or both exogenous Wnt and R-spondin. The cellular source of Wnt ligands remained unknown. However, as PDACs are characterised by robust stromal cell infiltration, it was hypothesised that these may be the Wnt source promoting survival of Wnt-dependent PDAC subtypes in vivo. To test this, the authors went on to generate stroma-attached organoids by letting PDAC cells and CAFs (derived from the same patient) aggregate together [81]. In this co-culture system, Wnt-dependent PDAC organoids formed in the absence of Wnt supplementation [81], as Wnt3A was provided by the CAFs in short range to PDAC cells, in line with earlier studies describing a short-range Wnt gradient in intestinal organoids [120]. A similar effect was not observed when CAF-conditioned medium was provided or when Wnt-dependent PDAC organoids and CAFs were co-cultured without direct physical contact [81]. PDACs self-producing Wnt ligands have been shown to be more aggressive [81], which suggests that PDACs dependent on Wnt supplementation by CAFs may represent an initial stage of tumour initiation that is lost during tumour progression into an aggressive and metastatic cancer.
Challenges and outlook
In order to use organoids for cancer therapy, several challenges remain to be overcome. ASC-derived organoid models have mostly been established from epithelial tissues, and, in line, cancer organoid cultures are in most cases only available for epithelial cancer such as different types of carcinomas with the exception of the recently described culture protocols for glioblastoma organoids [70] and rhabdoid tumour organoids [76]. Another critical issue is the efficiency at which cancer organoid cultures can be established [87], as well as the culture purity, as contamination with normal epithelial cells remains a problem, making organoid culture from some primary cancers such as prostate cancer very difficult [83]. Furthermore, to allow high-throughput assays, improved methods are required to decrease the time and costs of organoid generation as well as the input material needed to establish cultures. At the same time, other prerequisites for personalised (precision) medicine using cancer organoids are a better understanding of the clonal dynamics of cancers as well as the role of cellular components of the tumour microenvironment, which are too often still poorly understood. While some in vitro approaches have been developed to incorporate cells of the tumour microenvironment such as immune cells and fibroblasts into the cancer organoid culture [43, 107, 112, 114, 115], existing methods need further improvement and incorporation of more (non-epithelial) cell types. In addition, advancements to co-culture organoids with bacteria still need broader implementation and explorations. A major challenge remains the use of (non-human) animal products for organoid cultures such as murine-derived extracellular matrix (ECM) hydrogels (such as Matrigel, BME, Geltrex) or bovine-derived foetal calf serum or bovine serum in growth factor-conditioned media [68]. Bioengineering approaches such as the development of hydrogels using artificial matrices [121] or the use of alternatives to conditioned media may help to overcome some of these limitations in the future [122]. However, the need to test invasiveness or metastatic potential of cancer organoids using mouse xenograft models is still without robust alternatives [95]. Research into finding suitable replacements for such models should be fostered in the future. Lastly, ethical implications of cancer organoid biobanks preserving viable patient material require further considerations and may result in a stronger legislative regulation in the future [123].
Organoid technology has been developed just over 10 years ago. Its rapid implementation by numerous research groups worldwide led to many breakthroughs in the field of cell and developmental biology, but also in pre-clinical (cancer) research. The application of organoid technology in basic cancer research has provided many new experimental models and led to a variety of new discoveries. With the establishment of living biobanks of cancer organoids, new possibilities arise for the broader testing and development of anti-cancer drugs as well as the better stratification of cancer patient cohorts. With its versatility, robust ability to model in vivo situations, and fast-evolving set of applications, organoid technology is expected to keep making a significant impact on basic cancer research and clinical cancer therapy in the future.
Acknowledgements
The author thanks Kim E. Boonekamp and Jarno Drost for comments and critical reading of the manuscript. K.K. is funded by the Deutsche Krebshilfe (MSNZ Würzburg).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Competing interests
KK is named inventor on a patent pending related to immune cell–organoid cultures.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.GBD 2015 Mortality and Causes of Death Collaborators, Global, regional, and national life expectancy (2016) all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388(10053):1459–1544 [DOI] [PMC free article] [PubMed]
- 2.Ward EM, Sherman RL, Henley SJ, Jemal A, Siegel DA, Feuer EJ, Firth AU, Kohler BA, Scott S, Ma J, Anderson RN, Benard V, Cronin KA. Annual report to the nation on the status of cancer, featuring cancer in men and women age 20-49 years. J Natl Cancer Inst. 2019;111(12):1279–1297. doi: 10.1093/jnci/djz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheon DJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119. doi: 10.1146/annurev.pathol.3.121806.154244. [DOI] [PubMed] [Google Scholar]
- 4.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364(6444):952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 5.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 6.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 7.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman RM. Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat Rev Cancer. 2015;15(8):451–452. doi: 10.1038/nrc3972. [DOI] [PubMed] [Google Scholar]
- 9.Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and organoid model systems in cancer research. EMBO J. 2019;38(15):e101654. doi: 10.15252/embj.2019101654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 12.Kretzschmar K, Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev Cell. 2016;38(6):590–600. doi: 10.1016/j.devcel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 14.Mullenders J, de Jongh E, Brousali A, Roosen M, Blom JPA, Begthel H, Korving J, Jonges T, Kranenburg O, Meijer R, Clevers HC. Mouse and human urothelial cancer organoids: a tool for bladder cancer research. Proc Natl Acad Sci U S A. 2019;116(10):4567–4574. doi: 10.1073/pnas.1803595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sachs, N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A.A., Lelieveld D., van Hoeck A., Ernst R.F., Blokzijl F., Nijman I.J., Hoogstraat M., van de Ven M., Egan D.A., Zinzalla V., Moll J., Boj S.F., Voest E.E., Wessels L., van Diest P.J., Rottenberg S., Vries R.G.J., Cuppen E., Clevers H., A living biobank of breast cancer organoids captures disease heterogeneity. Cell, 2018. 172(1–2): p. 373–386 e10 [DOI] [PubMed]
- 16.Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17(10):1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 17.Sato T, Stange DE, Ferrante M, Vries RGJ, van Es JH, van den Brink S, van Houdt WJ, Pronk A, van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 18.Turco MY, Gardner L, Hughes J, Cindrova-Davies T, Gomez MJ, Farrell L, Hollinshead M, Marsh SGE, Brosens JJ, Critchley HO, Simons BD, Hemberger M, Koo BK, Moffett A, Burton GJ. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. 2017;19(5):568–577. doi: 10.1038/ncb3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M, Sehouli J, Fotopoulou C, Meyer TF. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, Yengej FY, Derakhshan S, de Winter-de Groot KM, Artegiani B, van Boxtel R, Cuppen E, Hendrickx APA, van den Heuvel-Eibrink MM, Heitzer E, Lanz H, Beekman J, Murk JL, Masereeuw R, Holstege F, Drost J, Verhaar MC, Clevers H. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol. 2019;37(3):303–313. doi: 10.1038/s41587-019-0048-8. [DOI] [PubMed] [Google Scholar]
- 21.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MMA, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JNM, Nieuwenhuis EES, Hoekstra R, Strom S, Vries RRG, van der Laan LJW, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160(1–2):299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, H., Gehart H., Artegiani B., LÖpez-Iglesias C., Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J., van den Born M., Zou C., Quirk C., Chiriboga L., Rice C.M., Ma S., Rios A., Peters P.J., de Jong Y.P., Clevers H., Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell, 2018. 175(6): p. 1591–1606 e19 [DOI] [PubMed]
- 23.Sachs N et al (2019) Long-term expanding human airway organoids for disease modeling. EMBO J 38(4):e100300 [DOI] [PMC free article] [PubMed]
- 24.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9(2):701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driehuis E, Kolders S, Spelier S, Lõhmussaar K, Willems SM, Devriese LA, de Bree R, de Ruiter EJ, Korving J, Begthel H, van Es JH, Geurts V, He GW, van Jaarsveld RH, Oka R, Muraro MJ, Vivié J, Zandvliet MMJM, Hendrickx APA, Iakobachvili N, Sridevi P, Kranenburg O, van Boxtel R, Kops GJPL, Tuveson DA, Peters PJ, van Oudenaarden A, Clevers H. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 2019;9(7):852–871. doi: 10.1158/2159-8290.CD-18-1522. [DOI] [PubMed] [Google Scholar]
- 26.Boj SF, Hwang CI, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Öhlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FHM, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RGJ, Clevers H, Tuveson DA. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RGJ, Cuppen E, Chen Y, Sawyers CL, Clevers HC. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chua CW, Shibata M, Lei M, Toivanen R, Barlow LMJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, Shen MM. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16(10):951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, Giepmans BNG, Vries RGJ, Clevers H, de Haan G, van Os R, Coppes RP. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Reports. 2016;6(1):150–162. doi: 10.1016/j.stemcr.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boonekamp KE, Kretzschmar K, Wiener DJ, Asra P, Derakhshan S, Puschhof J, López-Iglesias C, Peters PJ, Basak O, Clevers H. Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures. Proc Natl Acad Sci U S A. 2019;116(29):14630–14638. doi: 10.1073/pnas.1715272116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartfeld, S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H., In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology, 2015. 148(1): p. 126–136 e6 [DOI] [PMC free article] [PubMed]
- 32.Ren W, Lewandowski BC, Watson J, Aihara E, Iwatsuki K, Bachmanov AA, Margolskee RF, Jiang P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc Natl Acad Sci U S A. 2014;111(46):16401–16406. doi: 10.1073/pnas.1409064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Post, Y., Puschhof J., Beumer J., Kerkkamp H.M., de Bakker M.A.G., Slagboom J., de Barbanson B., Wevers N.R., Spijkers X.M., Olivier T., Kazandjian T.D., Ainsworth S., Iglesias C.L., van de Wetering W.J., Heinz M.C., van Ineveld R.L., van Kleef R.G.D.M., Begthel H., Korving J., Bar-Ephraim Y.E., Getreuer W., Rios A.C., Westerink R.H.S., Snippert H.J.G., van Oudenaarden A., Peters P.J., Vonk F.J., Kool J., Richardson M.K., Casewell N.R., Clevers H., Snake venom gland organoids. Cell, 2020. 180(2): p. 233–247 e21 [DOI] [PubMed]
- 34.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ (2009) Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 15(6):701–706 [DOI] [PMC free article] [PubMed]
- 35.Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Challet Meylan L, Stadler MB, Strnad P, Papasaikas P, Vischi D, Waldt A, Roma G, Liberali P. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569(7754):66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, Nijman IJ, Martincorena I, Mokry M, Wiegerinck CL, Middendorp S, Sato T, Schwank G, Nieuwenhuis EES, Verstegen MMA, van der Laan LJW, de Jonge J, IJzermans JNM, Vries RG, van de Wetering M, Stratton MR, Clevers H, Cuppen E, van Boxtel R. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538(7624):260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, Tarpey PS, Roerink S, Blokker J, Maddison M, Mudie L, Robinson B, Nik-Zainal S, Campbell P, Goldman N, van de Wetering M, Cuppen E, Clevers H, Stratton MR. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513(7518):422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuijk E, Jager M, van der Roest B, Locati MD, van Hoeck A, Korzelius J, Janssen R, Besselink N, Boymans S, van Boxtel R, Cuppen E. The mutational impact of culturing human pluripotent and adult stem cells. Nat Commun. 2020;11(1):2493. doi: 10.1038/s41467-020-16323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 40.Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, Chuva de Sousa Lopes S, van Zon J, Tans S, Clevers H. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol. 2020;22(3):321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 41.Dekkers JF, Alieva M, Wellens LM, Ariese HCR, Jamieson PR, Vonk AM, Amatngalim GD, Hu H, Oost KC, Snippert HJG, Beekman JM, Wehrens EJ, Visvader JE, Clevers H, Rios AC. High-resolution 3D imaging of fixed and cleared organoids. Nat Protoc. 2019;14(6):1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 42.Fumagalli A, Suijkerbuijk SJE, Begthel H, Beerling E, Oost KC, Snippert HJ, van Rheenen J, Drost J. A surgical orthotopic organoid transplantation approach in mice to visualize and study colorectal cancer progression. Nat Protoc. 2018;13(2):235–247. doi: 10.1038/nprot.2017.137. [DOI] [PubMed] [Google Scholar]
- 43.Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol. 2020;20(5):279–293. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 44.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14(1):53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, Little MH. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16(1):118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 48.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 49.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Holloway EM, Capeling MM, Spence JR (2019) Biologically inspired approaches to enhance human organoid complexity. Development 146(8):dev166173 [DOI] [PMC free article] [PubMed]
- 51.Papapetrou EP. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat Med. 2016;22(12):1392–1401. doi: 10.1038/nm.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat Rev Cancer. 2011;11(3):218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luch A. Nature and nurture-lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5(2):113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 55.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16(1):20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 56.Naruse M, Masui R, Ochiai M, Maru Y, Hippo Y, Imai T (2020) An organoid-based carcinogenesis model induced by in vitro chemical treatment. Carcinogenesis bgaa011 [DOI] [PubMed]
- 57.Fujii M, Sato T (2020) Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat Mater [DOI] [PubMed]
- 58.Bartfeld S. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev Biol. 2016;420(2):262–270. doi: 10.1016/j.ydbio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Amieva M, Peek RM., Jr Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353(6306):1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363(9420):1488–1489. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 62.Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, Peters PJ, Riggs MW, O’Connor R, Clevers H. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. 2018;3(7):814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pleguezuelos-Manzano C, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580(7802):269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 2019;11(1):11. doi: 10.1186/s13073-019-0621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 66.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, Sunagawa S, Coelho LP, Schrotz-King P, Vogtmann E, Habermann N, Niméus E, Thomas AM, Manghi P, Gandini S, Serrano D, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Waldron L, Naccarati A, Segata N, Sinha R, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Driehuis E, Kretzschmar K, Clevers H (2020) Establishment of patient-derived cancer organoids for drug screening applications. Nat Protoc 15(10):3380–3409 [DOI] [PubMed]
- 69.Lee, S.H., Hu W., Matulay J.T., Silva M.V., Owczarek T.B., Kim K., Chua C.W., Barlow L.M.J., Kandoth C., Williams A.B., Bergren S.K., Pietzak E.J., Anderson C.B., Benson M.C., Coleman J.A., Taylor B.S., Abate-Shen C., McKiernan J.M., al-Ahmadie H., Solit D.B., Shen M.M., Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell, 2018. 173(2): p. 515–528 e17 [DOI] [PMC free article] [PubMed]
- 70.Jacob, F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., Liu D.A., Qian X., Petrov D., Lucas T., Chen H.I., Dorsey J.F., Christian K.M., Binder Z.A., Nasrallah M.L., Brem S., O’Rourke D.M., Ming G.L., Song H., A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell, 2020. 180(1): p. 188–204 e22 [DOI] [PMC free article] [PubMed]
- 71.Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, Sato T. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 74.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CGM, van der Velden DL, Peeper DS, Cuppen EPJG, Vries RG, Clevers H, Voest EE. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112(43):13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boretto M, Maenhoudt N, Luo X, Hennes A, Boeckx B, Bui B, Heremans R, Perneel L, Kobayashi H, van Zundert I, Brems H, Cox B, Ferrante M, Uji-i H, Koh KP, D’Hooghe T, Vanhie A, Vergote I, Meuleman C, Tomassetti C, Lambrechts D, Vriens J, Timmerman D, Vankelecom H. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 2019;21(8):1041–1051. doi: 10.1038/s41556-019-0360-z. [DOI] [PubMed] [Google Scholar]
- 76.Calandrini C, Schutgens F, Oka R, Margaritis T, Candelli T, Mathijsen L, Ammerlaan C, van Ineveld RL, Derakhshan S, de Haan S, Dolman E, Lijnzaad P, Custers L, Begthel H, Kerstens HHD, Visser LL, Rookmaaker M, Verhaar M, Tytgat GAM, Kemmeren P, de Krijger RR, al-Saadi R, Pritchard-Jones K, Kool M, Rios AC, van den Heuvel-Eibrink MM, Molenaar JJ, van Boxtel R, Holstege FCP, Clevers H, Drost J. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat Commun. 2020;11(1):1310. doi: 10.1038/s41467-020-15155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grassi L, Alfonsi R, Francescangeli F, Signore M, de Angelis ML, Addario A, Costantini M, Flex E, Ciolfi A, Pizzi S, Bruselles A, Pallocca M, Simone G, Haoui M, Falchi M, Milella M, Sentinelli S, di Matteo P, Stellacci E, Gallucci M, Muto G, Tartaglia M, de Maria R, Bonci D. Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death Dis. 2019;10(3):201. doi: 10.1038/s41419-019-1453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broutier L, Mastrogiovanni G, Verstegen MMA, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, IJzermans JNM, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJW, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H, van Wijk LM, Revilla SA, Theeuwsen R, van de Ven M, van Roosmalen MJ, Ponsioen B, Ho VWH, Neel BG, Bosse T, Gaarenstroom KN, Vrieling H, Vreeswijk MPG, van Diest PJ, Witteveen PO, Jonges T, Bos JL, van Oudenaarden A, Zweemer RP, Snippert HJG, Kloosterman WP, Clevers H. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25(5):838–849. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 80.Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ, Jr, Feltmate CM, Nucci MR, Swisher EM, Nguyen H, Yang C, Morizane R, Kochupurakkal BS, Do KT, Konstantinopoulos PA, Liu JF, Bonventre JV, Matulonis UA, Shapiro GI, Berkowitz RS, Crum CP, D’Andrea AD. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 2018;8(11):1404–1421. doi: 10.1158/2159-8290.CD-18-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seino, T., Kawasaki S., Shimokawa M., Tamagawa H., Toshimitsu K., Fujii M., Ohta Y., Matano M., Nanki K., Kawasaki K., Takahashi S., Sugimoto S., Iwasaki E., Takagi J., Itoi T., Kitago M., Kitagawa Y., Kanai T., Sato T., Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell, 2018. 22(3): p. 454–467 e6 [DOI] [PubMed]
- 82.Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche RE, Jang GH, Miyabayashi K, Young CM, Patel H, Ma M, LaComb JF, Palmaira RLD, Javed AA, Huynh JC, Johnson M, Arora K, Robine N, Shah M, Sanghvi R, Goetz AB, Lowder CY, Martello L, Driehuis E, LeComte N, Askan G, Iacobuzio-Donahue CA, Clevers H, Wood LD, Hruban RH, Thompson E, Aguirre AJ, Wolpin BM, Sasson A, Kim J, Wu M, Bucobo JC, Allen P, Sejpal DV, Nealon W, Sullivan JD, Winter JM, Gimotty PA, Grem JL, DiMaio DJ, Buscaglia JM, Grandgenett PM, Brody JR, Hollingsworth MA, O’Kane GM, Notta F, Kim E, Crawford JM, Devoe C, Ocean A, Wolfgang CL, Yu KH, Li E, Vakoc CR, Hubert B, Fischer SE, Wilson JM, Moffitt R, Knox J, Krasnitz A, Gallinger S, Tuveson DA. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8(9):1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao, Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M., Guo Q., Gao X., Li Y., Rao X., Zhou Y., Liang L., Wang Y., Zhang J., Zhang H., Li G., Zhang L., Peng J., Cai S., Hu C., Gao J., Clevers H., Zhang Z., Hua G., Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell, 2020. 26(1): p. 17–26 e6 [DOI] [PubMed]
- 85.Yan, H.H.N., Siu H.C., Law S., Ho S.L., Yue S.S.K., Tsui W.Y., Chan D., Chan A.S., Ma S., Lam K.O., Bartfeld S., Man A.H.Y., Lee B.C.H., Chan A.S.Y., Wong J.W.H., Cheng P.S.W., Chan A.K.W., Zhang J., Shi J., Fan X., Kwong D.L.W., Mak T.W., Yuen S.T., Clevers H., Leung S.Y., A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell, 2018. 23(6): p. 882–897 e11 [DOI] [PubMed]
- 86.Nanki, K., Toshimitsu K., Takano A., Fujii M., Shimokawa M., Ohta Y., Matano M., Seino T., Nishikori S., Ishikawa K., Kawasaki K., Togasaki K., Takahashi S., Sukawa Y., Ishida H., Sugimoto S., Kawakubo H., Kim J., Kitagawa Y., Sekine S., Koo B.K., Kanai T., Sato T., Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell, 2018. 174(4): p. 856–869 e17 [DOI] [PubMed]
- 87.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJPL, Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 89.Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA, Pronk A, Smakman N, van Gorp J, Anderson E, Gamble SJ, Alder C, van de Wetering M, Campbell PJ, Stratton MR, Clevers H. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556(7702):457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 90.Michels, B.E., Mosa M.H., Streibl B.I., Zhan T., Menche C., Abou-el-Ardat K., Darvishi T., Członka E., Wagner S., Winter J., Medyouf H., Boutros M., Farin H.F., Pooled in vitro and in vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell, 2020. 26(5): p. 782–792 e7 [DOI] [PubMed]
- 91.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143(1):134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 92.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, Cantrell MA, Rack PG, Neal JT, Chan CWM, Yeung T, Gong X, Yuan J, Wilhelmy J, Robine S, Attardi LD, Plevritis SK, Hung KE, Chen CZ, Ji HP, Kuo CJ. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20(7):769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 94.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21(3):256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 95.Fumagalli A, Drost J, Suijkerbuijk SJE, van Boxtel R, de Ligt J, Offerhaus GJ, Begthel H, Beerling E, Tan EH, Sansom OJ, Cuppen E, Clevers H, van Rheenen J. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A. 2017;114(12):E2357–E2364. doi: 10.1073/pnas.1701219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, Nik-Zainal S, Kuiper RP, Cuppen E, Clevers H. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358(6360):234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fessler E, Drost J, Hooff SR, Linnekamp JF, Wang X, Jansen M, de Sousa E Melo F, Prasetyanti PR, IJspeert JEG, Franitza M, Nürnberg P, Noesel CJM, Dekker E, Vermeulen L, Clevers H, Medema JP. TGFbeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8(7):745–760. doi: 10.15252/emmm.201606184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Artegiani, B., van Voorthuijsen L., Lindeboom R.G.H., Seinstra D., Heo I., Tapia P., López-Iglesias C., Postrach D., Dayton T., Oka R., Hu H., van Boxtel R., van Es J.H., Offerhaus J., Peters P.J., van Rheenen J., Vermeulen M., Clevers H., Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell, 2019. 24(6): p. 927–943 e6 [DOI] [PubMed]
- 99.Doudna JA, Charpentier E, Genome editing, The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 100.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16(1):45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NWM, Bijvelds MJC, Scholte BJ, Nieuwenhuis EES, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19(7):939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 102.Dekkers JF et al (2016) Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med 8(344):344ra84 [DOI] [PubMed]
- 103.Boj SF et al (2017) Forskolin-induced swelling in intestinal organoids: an in vitro assay for assessing drug response in cystic fibrosis patients. J Vis Exp 120:55159 [DOI] [PMC free article] [PubMed]
- 104.Ooft SN et al (2019) Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 11(513):eaay2574 [DOI] [PubMed]
- 105.Betge J et al. (2019), Multiparametric phenotyping of compound effects on patient derived organoids. bioRxiv:660993
- 106.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 107.Dijkstra, K.K., Cattaneo C.M., Weeber F., Chalabi M., van de Haar J., Fanchi L.F., Slagter M., van der Velden D.L., Kaing S., Kelderman S., van Rooij N., van Leerdam M.E., Depla A., Smit E.F., Hartemink K.J., de Groot R., Wolkers M.C., Sachs N., Snaebjornsson P., Monkhorst K., Haanen J., Clevers H., Schumacher T.N., Voest E.E., Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell, 2018. 174(6): p. 1586–1598 e12 [DOI] [PMC free article] [PubMed]
- 108.Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, van den Brink S, Schumacher TN, Voest EE. Tumor organoid-T-cell coculture systems. Nat Protoc. 2020;15(1):15–39. doi: 10.1038/s41596-019-0232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, Mulder GE, Toebes M, Vesely MD, Lam SSK, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee HG, Melief CJM, Mardis ER, Gillanders WE, Artyomov MN, Schreiber RD. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 111.Kong JCH, et al. Tumor-infiltrating lymphocyte function predicts response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. JCO Precision Oncology. 2018;2:1–15. doi: 10.1200/PO.18.00075. [DOI] [PubMed] [Google Scholar]
- 112.Schnalzger TE et al (2019) 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J 38(12):e100928 [DOI] [PMC free article] [PubMed]
- 113.Bar-Ephraim YE et al (2018) Modelling cancer immunomodulation using epithelial organoid cultures. bioRxiv:377655
- 114.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, Deutsch BC, Liao L, Zemek AJ, Zhao F, Karlsson K, Schultz LM, Metzner TJ, Nadauld LD, Tseng YY, Alkhairy S, Oh C, Keskula P, Mendoza-Villanueva D, de la Vega FM, Kunz PL, Liao JC, Leppert JT, Sunwoo JB, Sabatti C, Boehm JS, Hahn WC, Zheng GXY, Davis MM, Kuo CJ. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175(7):1972–1988. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E, Frommelt LS, Qu R, Knapp MS, Henriques A, Chalkidi N, Koliaraki V, Jiao J, Brewer JR, Bacher M, Blackburn HN, Zhao X, Breyer RM, Aidinis V, Jain D, Su B, Herschman HR, Kluger Y, Kollias G, Flavell RA. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 2020;580(7804):524–529. doi: 10.1038/s41586-020-2166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 117.Xia D, Wang D, Kim SH, Katoh H, DuBois RN. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat Med. 2012;18(2):224–226. doi: 10.1038/nm.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang, D., Fu L., Sun H., Guo L., DuBois R.N., Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology, 2015. 149(7): p. 1884–1895 e4 [DOI] [PMC free article] [PubMed]
- 119.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11(1):106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, de Punder K, Angers S, Peters PJ, Maurice MM, Clevers H. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 121.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539(7630):560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 122.Janda CY, Dang LT, You C, Chang J, de Lau W, Zhong ZA, Yan KS, Marecic O, Siepe D, Li X, Moody JD, Williams BO, Clevers H, Piehler J, Baker D, Kuo CJ, Garcia KC. Surrogate Wnt agonists that phenocopy canonical Wnt and beta-catenin signalling. Nature. 2017;545(7653):234–237. doi: 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bredenoord AL, Clevers H, Knoblich JA (2017) Human tissues in a dish: the research and ethical implications of organoid technology. Science 355(6322):eaaf9414 [DOI] [PubMed]
- 124.Cristobal A, van den Toorn HWP, van de Wetering M, Clevers H, Heck AJR, Mohammed S. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep. 2017;18(1):263–274. doi: 10.1016/j.celrep.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 125.Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, Gerwen B, van der Ven M, Wetering M, Egan DA, Bernards R, Clevers H, Bos JL, Snippert HJ (2016) Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. Elife 5:e18489 [DOI] [PMC free article] [PubMed]
- 126.Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, Haybaeck J, Liebs S, Lange M, Schäfer R, Regenbrecht CRA, Reinhard C, Velasco JA. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J Biomol Screen. 2016;21(9):931–941. doi: 10.1177/1087057116650965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schutte M, et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun. 2017;8:14262. doi: 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gao M, Lin M, Rao M, Thompson H, Hirai K, Choi M, Georgakis GV, Sasson AR, Bucobo JC, Tzimas D, D’Souza LS, Buscaglia JM, Davis J, Shroyer KR, Li J, Powers S, Kim J. Development of patient-derived gastric cancer organoids from endoscopic biopsies and surgical tissues. Ann Surg Oncol. 2018;25(9):2767–2775. doi: 10.1245/s10434-018-6662-8. [DOI] [PubMed] [Google Scholar]
- 129.Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grützmann K, Sommer U, Schweitzer C, Schölch S, Uhlemann H, Gaebler AM, Werner K, Krause M, Baretton GB, Welsch T, Koo BK, Aust DE, Klink B, Weitz J, Stange DE. Human gastric cancer modelling using organoids. Gut. 2019;68(2):207–217. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li L et al (2019) Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 4(2):e121490 [DOI] [PMC free article] [PubMed]
- 131.Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, Jung DJ, Shin TH, Jeong GS, Kim DK, Choi EK, Jeong SY, Taylor AM, Jain S, Meyerson M, Jang SJ. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10(1):3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tamura H, Higa A, Hoshi H, Hiyama G, Takahashi N, Ryufuku M, Morisawa G, Yanagisawa Y, Ito E, Imai JI, Dobashi Y, Katahira K, Soeda S, Watanabe T, Fujimori K, Watanabe S, Takagi M. Evaluation of anticancer agents using patient-derived tumor organoids characteristically similar to source tissues. Oncol Rep. 2018;40(2):635–646. doi: 10.3892/or.2018.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takahashi N et al (2019) An in vitro system for evaluating molecular targeted drugs using lung patient-derived tumor organoids. Cells 8(5):481 [DOI] [PMC free article] [PubMed]
- 135.Jabs J, Zickgraf FM, Park J, Wagner S, Jiang X, Jechow K, Kleinheinz K, Toprak UH, Schneider MA, Meister M, Spaich S, Sütterlin M, Schlesner M, Trumpp A, Sprick M, Eils R, Conrad C. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol Syst Biol. 2017;13(11):955. doi: 10.15252/msb.20177697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de Witte CJ, Espejo Valle-Inclan J, Hami N, Lõhmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P, Nguyen L, Clevers H, Kloosterman WP, Cuppen E, Snippert HJG, Zweemer RP, Witteveen PO, Stelloo E. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 2020;31(11):107762. doi: 10.1016/j.celrep.2020.107762. [DOI] [PubMed] [Google Scholar]
- 137.Driehuis E, van Hoeck A, Moore K, Kolders S, Francies HE, Gulersonmez MC, Stigter ECA, Burgering B, Geurts V, Gracanin A, Bounova G, Morsink FH, Vries R, Boj S, van Es J, Offerhaus GJA, Kranenburg O, Garnett MJ, Wessels L, Cuppen E, Brosens LAA, Clevers H. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé CEG, Adileh M, Wasserman I, Marco MR, Kim AS, Shady M, Sanchez-Vega F, Karthaus WR, Won HH, Choi SH, Pelossof R, Barlas A, Ntiamoah P, Pappou E, Elghouayel A, Strong JS, Chen CT, Harris JW, Weiser MR, Nash GM, Guillem JG, Wei IH, Kolesnick RN, Veeraraghavan H, Ortiz EJ, Petkovska I, Cercek A, Manova-Todorova KO, Saltz LB, Lavery JA, DeMatteo RP, Massagué J, Paty PB, Yaeger R, Chen X, Patil S, Clevers H, Berger MF, Lowe SW, Shia J, Romesser PB, Dow LE, Garcia-Aguilar J, Sawyers CL, Smith JJ (2019) A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med 25(10):1607–1614 [DOI] [PMC free article] [PubMed]
- 139.Yoshimasa Saito, Toshihide Muramatsu, Yae Kanai, Hidenori Ojima, Aoi Sukeda, Nobuyoshi Hiraoka, Eri Arai, Yuko Sugiyama, Juntaro Matsuzaki, Ryoei Uchida, Nao Yoshikawa, Ryo Furukawa, Hidetsugu Saito, (2019) Establishment of Patient-Derived Organoids and Drug Screening for Biliary Tract Carcinoma. Cell Reports 27 (4):1265–1276.e4 [DOI] [PubMed]