Abstract
Background:
Phenotypic changes in vesicular compartments are an early pathological hallmark of many peripheral and central diseases. For example, accurate assessment of early endosome pathology is crucial to the study of Down syndrome (DS) and Alzheimer’s disease (AD), as well as other neurological disorders with endosomal-lysosomal pathology.
New Method:
We describe a method for quantification of immunolabeled early endosomes within transmitter-identified basal forebrain cholinergic neurons (BFCNs) using 3-dimensional (3D) reconstructed confocal z-stacks employing Imaris software.
Results:
Quantification of 3D reconstructed z-stacks was performed using two different image analysis programs: ImageJ and Imaris. We found ImageJ consistently overcounted the number of early endosomes present within individual BFCNs. Difficulty separating densely packed early endosomes within defined BFCNs was observed in ImageJ compared to Imaris.
Comparison with Existing Methods:
Previous methods quantifying endosomal-lysosomal pathology relied on confocal microscopy images taken in a single plane of focus. Since early endosomes are distributed throughout the soma and neuronal processes of BFCNs, critical insight into the abnormal early endosome phenotype may be lost as a result of analyzing only a single image of the perikaryon. Rather than relying on a representative sampling, this protocol enables precise, direct quantification of all immunolabeled vesicles within a defined cell of interest.
Conclusions:
Imaris is an ideal program for accurately counting punctate vesicles in the context of dual label confocal microscopy. Superior image resolution and detailed algorithms offered by Imaris make precise and rigorous quantification of individual early endosomes dispersed throughout a BFCN in 3D space readily achievable.
Keywords: Early endosome, basal forebrain cholinergic neuron, 3D reconstruction, Alzheimer’s disease, Down syndrome, Imaris, ImageJ
1. Introduction
Proper functionality of the endosomal-lysosomal system and autophagy pathway is essential to the maintenance of cellular homeostasis. This is particularly true for neurons, which rely on a tightly regulated endocytic pathway for intracellular trafficking and synaptic transmission (Perez et al., 2015). Endosomes are vesicular organelles involved in the internalization, modification, and recycling of extracellular nutrients and signaling molecules (Nixon, 2005). Endocytic trafficking defects have been described in many neurodegenerative disorders, including Alzheimer’s disease (AD) (Perez et al., 2015; Nixon, 2017), Huntington’s disease (Kegel et al., 2000; Pal et al., 2006; Malik et al., 2019), amyotrophic lateral sclerosis (Mitra et al., 2019), Niemann-Pick disease (Jin et al., 2004; Cabeza et al., 2012), and Down syndrome (DS) (Salehi et al., 2006; Colacurcio et al., 2018), among others.
Extracellular molecules internalized via endocytosis are first sequestered into protease-rich vesicles called early endosomes (Nixon, 2005). Early endosomes shuttle their contents throughout the neuron, delivering cargo to late endosomes and lysosomes for processing and degradation, to the Golgi for further utilization, or to recycling endosomes for return to the plasma membrane (Nixon and Cataldo, 2006). Morphological abnormalities in early endosomes are an early pathological hallmark of both AD and DS, oftentimes appearing unusually enlarged and in increased abundance within the cell soma (Cataldo et al., 1997, 2008; Ginsberg et al., 2010a, 2010b, 2011). This aberrant endosomal phenotype has been documented within vulnerable neuronal populations in human AD and DS patients (Perez et al., 2015; Colacurcio et al., 2018), in human DS fibroblasts (Cataldo et al., 2008), and in trisomic Ts2 and Ts65Dn mice (Cataldo et al., 2003; Jiang et al., 2016), two well-characterized murine models of DS. Importantly, the appearance of enlarged early endosomes often precedes the deposition of amyloid-β plaques and neurofibrillary tangles in the brains of AD patients by decades, and is observable in individuals with DS before birth (Cataldo et al., 2000). Given their connection to key components of AD pathology, the premature manifestation of endosomal defects provides a unique avenue by which to study prodromal AD, and underscores the importance of being able to accurately visualize and quantify vesicular abnormalities.
Many studies perform endosomal quantitation on confocal microscopy images using the software program ImageJ (Rasband, 1997; Schneider et al., 2012). These images are typically taken on a single plane of focus at high magnification to enable counting of fluorescently labeled puncta. (Cataldo et al., 2008; Jiang et al., 2010; Choi et al., 2013; Kaur et al., 2017, 2018). While this methodology is sound, technological improvements related to image acquisition and processing have led to an increase in the number of analytical programs that offer high resolution visualization, precise quantification, and smooth integration of statistical analysis within a single software package (Zinchuk and Grossenbacher-Zinchuk, 2009; Eliceiri et al., 2012).
One such advancement is the availability of quantitative software suites that enable the construction of 3-dimensional (3D) models from confocal microscopy images taken at multiple depths along the z-axis. This series of images can then be combined to render a representation of what an object of interest looks like in 3D space (Halbhuber and König, 2003; Bankhead, 2013). A number of multidimensional image analysis programs exist within the current quantitative landscape. Some, like BioImageXD and Icy, are similar to ImageJ, with a modular design and focus on coding allowing for a high degree of pipeline customization (Eliceiri et al., 2012; Schneider et al., 2012; Miura et al., 2016). Others are highly specialized for use within a certain system or to perform a particular task. Programs such as Cell Profiler and Velocity focus on object tracking over time, while Vaa3D and NeuroStudio are specifically geared towards neuroscientists interested in measuring filamentous structures such as dendritic arborization (Eliceiri et al., 2012; Miura et al., 2016).
Use of 3D reconstructions helps facilitate reproduceable quantitative morphometric analyses of tissue (Fricker et al., 2006). Acquiring a series of images taken at consistently spaced depth intervals enables a high degree of histological precision (Taylor and Levinson, 2006). This is particularly useful when attempting to visualize complex structures, like the brain or vasculature, and densely packed small structures, including immunofluorescently labeled vesicles (Pham et al., 2000; Tischer and Tosi, 2016). Early endosomes are distributed throughout both the cell body and neuronal processes, which often project in multiple directions. Important aspects of the enlarged early endosome phenotype may therefore be missed by analyzing only a single image of the perikaryon. Utilizing 3D methods can also alleviate some of the drawbacks associated with 2-dimensional (2D) microscopy, including shallow depth of field when working on high objectives (McGavin, 2014). We demonstrate quantifying neuronal puncta from 3D reconstructions, rather than 2D images, allows for a more complete assessment of endosomal pathology.
In the present study, we quantified Rab5-immunolabelled early endosomes within 3D reconstructions of individual basal forebrain cholinergic neurons (BFCNs), a neuronal population that is selectively vulnerable to degeneration in AD and DS. 3D reconstructed z-stacks were generated and analyzed using two different software packages: ImageJ, an open access image analysis program developed by the NIH, and Imaris, a proprietary software package offered by Bitplane that specializes in 3D quantification of fluorescence microscopy data (Miura et al., 2016). Image processing, analytical workflow, and final data output were extensively compared between the two programs. Our findings indicate Imaris provided accurate and reproducible quantification. Herein, we provide a method for quantifying Rab5-immunoreactive early endosomes within choline acetyltransferase (ChAT)-immunoreactive BFCNs using tissue sections obtained from the mouse basal forebrain via a double label confocal microscopy approach.
2. Materials and Methods
2.1. Mice
Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Nathan Kline Institute/NYU Grossman School of Medicine and were in accordance with NIH guidelines. Breeder pairs of mixed background were purchased from Jackson Laboratories (Bar Harbor, ME) and mated at the Nathan Kline Institute. Upon arrival, mice were bred and fed a normal chow diet and ad libitum water. Standard cages contained paper bedding and several objects for enrichment (e.g., plastic igloo, t-tube, and cotton square nestlet). Mice were maintained on a 12-hour light-dark cycle under temperature- and humidity-controlled conditions. Mice were aged to ~11 months of age (MO).
2.2. Tissue preparation
Mice were anesthetized via an intraperitoneal injection of ketamine (83 mg/kg) and xylazine (13 mg/kg) and perfused transcardially with ice-cold 0.15 M phosphate buffer (PB). Brains were removed from the skull and drop-fixed in 4% paraformaldehyde buffered in PB for 48 h on an orbital shaker at 4 °C. Brains were transferred through a series of sucrose solutions in PB (12%, 18%, 30%) for post-fixation. Brains were kept in each sucrose solution for a minimum of 24 h at 4 °C. Sectioning was performed in the coronal plane at 40 μm on a cryostat (CM1860; Leica Biosystems, Buffalo Grove, IL) set to −25 °C. Tissue sections were stored in a cryoprotectant solution (30% glycerol, 30% ethylene glycol, 40% PB) (Kelley et al., 2014) at −20 °C until immunolabeling.
2.3. Immunolabeling
Free-floating sections were rinsed in phosphate buffered saline (PBS; pH 7.4) and blocked in a PBS solution containing 2% bovine serum albumin, 3% fetal bovine serum, and 0.8% Triton X-100 (Choi et al., 2013; Kaur et al., 2018) for 1 h on an orbital shaker at room temperature. The blocking solution was used as the diluent in all subsequent antibody dilutions. Tissue sections were incubated overnight in a rabbit polyclonal antibody directed against Rab5c (1:500; ab18211, Abcam, Cambridge, MA) on a shaker at 4 °C. Following overnight incubation, sections were washed in dilution buffer for 30 min and incubated in Alexa-Fluor donkey anti-rabbit 568 secondary antibody (1:500; Invitrogen, Eugene, OR) for 2 h on an orbital shaker at room temperature in a darkened room. All ensuing steps after initial secondary antibody incubation were performed out of direct light. Tissue sections were washed in PBS at room temperature and incubated in a goat polyclonal primary antibody against ChAT (1:250; AB144P, Millipore Sigma, Temecula, CA) for 36 h on an orbital shaker at 4 °C. Tissue sections were washed in dilution buffer for 30 min and incubated in Alexa-Fluor donkey anti-sheep 488 secondary antibody (1:500; Invitrogen) (Buchwalow et al., 2011; Kaur et al., 2017, 2018) for 2 h on an orbital shaker at room temperature. Sections were rinsed in PBS, mounted on gelatin-coated glass slides, and coverslipped with VectaShield mounting medium (Vector Labs, Burlingame, CA). Coverslips were sealed using clear nail polish (Electron Microscopy Sciences, Ft. Washington, PA). Slides were stored in an opaque slide box at room temperature until imaging.
2.4. Image acquisition
Z-stacks of individual BFCNs within the medial septal nucleus (MSN) were imaged on a confocal microscope (LSM510, Zeiss, White Plains, NY) with the accompanying LSM5 software (Zeiss). Multi-track acquisition was performed with excitation lines set at 488 nm at 5% and 543 nm at 50%. Emission filters utilized were BP 505–530 nm and LP 560 nm, and beam splitters were HFT 488/543 and NFT 545. Z-stack images of BFCNs were acquired using a Plan-Apochromat 100X/1.4 oil objective. Stacks were collected at a 0.5 μm slice interval, stepping through the entire soma of the neuron. Frame size was set to 1024 × 1024 pixels and pixel time to 3.20 μsec. Scanning was performed unidirectionally at a 12-bit pixel depth. Scan averaging was set to 4 to cut down on background noise. All z-stack files were saved in (.lsm) format prior to export and quantitative analysis.
2.5. BFCN Selection
A total of (n=6) ChAT-immunoreactive BFCNs were selected for quantification from a larger pool of (n=29) BFCN z-stacks imaged from the brain of a single 11 MO mouse. Primary selection criteria included the size and shape of the neuron, as well as their proximity to non-cholinergic cells within the MSN.
2.6. Endosome quantification in ImageJ v. 1.52
Z-stacks were opened in ImageJ in their native format. The split red and green channels were merged to form a composite color image (Image > Color > Make Composite), which was subsequently converted from a 16-bit stack to a RGB image (Image > Color > Stack to RGB). Individual slices comprising the entire z-stack were compiled onto a single plane via the “3D project” function (Image > Stacks > 3D Project). The brightest point projection method was employed to produce the 3D image, with the y-axis set as the axis of rotation. The generated projection was then converted to a 16-bit grayscale image prior to image segmentation (Image > Type > 8-bit > 16-bit).
A minimum threshold value of 50 was used across all images to identify Rab5 immunolabeled endosomes and differentiate them from the image background (Image > Adjust > Threshold). To enable accurate quantification, the paintbrush tool was used to break up endosomes in close proximity that became fused during binarization. The original z-stack images were used as a reference guide when segmenting endosomes visually, and the brush width was adjusted as needed to achieve the greatest degree of precision. The watershed algorithm, an image processing function commonly used to automatically split touching objects, was generally not applied as it was often insufficient to fully and accurately partition endosomes in 3D.
To restrict vesicle measurements to individual BFCNs of interest, a region of interest (ROI) was drawn by tracing the perimeter of the cell with the freehand selection tool. The area of the ROI was calculated using the command Analyze > Measure. Early endosome quantification was performed using the Analyze > Analyze Particles command. The lower bound for particle size was set to 0.2 μm2, to match the limit of detection of the Imaris Spots analysis.
2.7. Endosome quantification in Imaris v. 9.1.2
Z-stacks were opened in Imaris in their native format. Z-stacks are automatically reconstructed into a multi-channel 3D model during input into Imaris, requiring no further image pre-processing. To designate individual BFCNs of interest, the Surface creation tool was used to generate a ROI. In the Surface creation wizard, the source channel was set to “Green Channel-Ch2-T1”, corresponding to the Alexa Fluor 488 secondary antibody. To create a clean border around a desired cell, smoothing surface detail was enabled with the surface grain size set between 0.5–1.0 μm, dependent upon the shape of the individual neuron and the distribution of early endosomes within the cytoplasm. Background subtraction (local contrast) was used to separate the cell in the foreground from the background environment. The auto-threshold value was utilized during background subtraction, without user adjustments. The size and shape of the generated surface were a direct map of the intensity distribution of ChAT immunolabeling within the cytoplasm as detected by Imaris. Any adjustments to the Surface shape were made by altering the Surface grain size to reduce extraneous noise around the periphery during detection, rather than manually shrink or expand the 3D model. Cell width was measured at its widest point in Slice Mode. This measured value was rounded down to the nearest whole number and entered as the “diameter of the largest sphere which fits into the object” in the Surface creation wizard.
To isolate Rab5-positive early endosomes within individual BFCNs of interest and exclude vesicles within neighboring cells, a new source channel was created (Scene > Surface > Edit > Mask Properties > Mask All). “Red Channel-Ch3-T2”, corresponding to the Alexa Fluor 568 secondary antibody, was hidden outside the Surface by creating a “Mask Channel” wherein all red voxels outside the ROI were set to “0.000”. The newly created source channel was designated “Red Channel-Masked Ch3-T2”.
To quantify early endosomes within the BFCN, the Spots creation tool was used. In the Spots creation wizard, “different spot sizes (region growing)” was selected to enable the measurement of endosomes across a range of diameters. The source channel for detection was set to “Red Channel-Masked Ch3-T2”. Estimated XY diameter for Spot detection was 0.45 μm. The background subtraction (local contrast) method was used to threshold endosomes, with the threshold value held at 65 across all images. During Spot detection, the region growing diameter was measured from the region volume.
2.8. Comparison of image processing functions
To determine whether 3D reconstructions generated in ImageJ could be improved by utilizing Imaris methodology within the ImageJ framework, several different image processing functions were applied to a reconstructed stack after “3D Project” and conversion to greyscale. These included: i) application of a Gaussian filter (Process > Filters > Gaussian Blur) followed by background subtraction (Process > Filters > Subtract Background), ii) local thresholding (Image > Adjust > Auto Local Threshold > Otsu’s Method) followed by watershed (Process > Binary > Watershed), and iii) all four processes in combination (Gaussian Blur > Subtract Background > Otsu’s Method > Watershed) (Ferreira and Rasband, 2012).
2.9. Statistical Analysis
Data are presented as total number of Rab5-immunoreactive early endosomes identified within the ROI. A Wilcoxon-Mann-Whitney test was used to evaluate differences between analysis programs in terms of the number of Rab5-immunoreactive early endosomes quantified. Statistical significance was set at (p < 0.05).
3. Results
ImageJ 1.52s, the most up-to-date version at the time of data quantification, was downloaded from imagej.nih.gov (imagej.nih.gov/ij/download.html) as a zip file bundled with Java. No additional plugins, macros, or scripts were used to extend capabilities beyond the core ImageJ program. Imaris Start (Bitplane, Concord, MA) was downloaded through the Imaris customer portal. Imaris version 9.1.2 was utilized for all image processing and quantification.
Both ImageJ and Imaris allowed for easy maneuverability around an image. Imaris was highly interactive, allowing for user-controlled, 360° rotation of the entire reconstructed z-stack along both the x- and y-axis, as well as zoom capability along the z-axis. The graphical user interface (GUI) in ImageJ was largely menu based, which offered a wide variety of image processing functions (Miura et al., 2016). Most functions existed as individual commands, allowing for a high degree of protocol customization during analysis. In contrast, Imaris image analysis tools were organized into guided workflows, or wizards. The user was walked through a pre-programmed sequence and prompted to enter necessary parameters at appropriate stages during quantification.
Similar strategies were employed in the ImageJ and Imaris 3D endosome quantification protocols to enable comparison. A side-by-side workflow detailing the quantification of Rab5-immunoreactive endosomes within a representative ChAT-immunoreactive BFCN is presented in Figure 1. The analyzed BFCN was imaged from the rostral MSN within the basal forebrain of a female mouse at 11 MO. In this protocol, the BFCN of interest was designated as the sole region within a z-stack of confocal images in which individual Rab5-positive early endosomes were quantified.
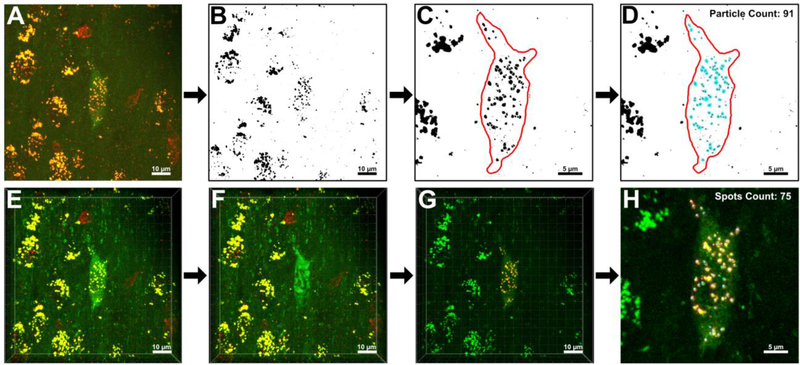
Figure 1.
Image analysis protocol for the quantification of Rab5-positive early endosomes (yellow) within individual ChAT-positive BFCNs (green) in both ImageJ (A-D) and Imaris (E-H). Representative images are presented side-by-side to enable comparison of the general workflow between analysis programs. A. In Step 1 of the ImageJ protocol, a color composite, 3D reconstruction of an individual BFCN of interest is constructed from 27 consecutive z-slices using the “3D Project” function in ImageJ. B. A binarized version of the image presented in (A). In Step 2 of the ImageJ protocol, thresholding is applied to the image to segment foreground objects (early endosomes: black) from the image background (white). C. In Step 3 of the ImageJ protocol, the ROI within the 3D reconstruction is designated by tracing the outline of the BFCN using ImageJ’s “freehand selection” tool. Discrete early endosomes that fused together during thresholding are separated using the ImageJ “paintbrush” tool (brush width set to 1–3 pixels, as necessary for precision). Individual slices from the original z-stack were used as a point of reference while separating endosomes to accurately determine where boundaries should be drawn. D. In Step 4 of the ImageJ protocol, the “Analyze Particle” function is performed within the ROI. ImageJ detected 91 Rab5-immunoreactive early endosomes (blue) within the representative BFCN. E. In Step 1 of the Imaris protocol, a z-stack file imported into Imaris is automatically reconstructed into a color composite, 3D model. F. In Step 2 of the Imaris protocol, the “Surface” creation tool is utilized to encase the BFCN of interest and designate it as the ROI. In this representative image, the generated “Surface” maps to the cytoplasmic ChAT antibody staining, creating a green shell around the BFCN. G. In Step 3 of the Imaris protocol, Rab5-immunoreactive early endosomes in surrounding non-cholinergic cells are hidden from the program via creation of a mask overlay that resets all red voxels external to the ROI to “0.000”. H. In Step 4 of the Imaris protocol, the “Spots” creation tool is utilized to quantify only early endosomes within the BFCN of interest. Imaris detected 75 Rab5-immunoreactive early endosomes (blue spheres) within the representative BFCN. Scale bar A-B, E-G: 10 μm. Scale bar C-D, H: 5 μm.
Multi-channel, color composite images were generated in both ImageJ and Imaris of the ChAT-positive BFCN and Rab5-immunolabeled early endosomes from 27 consecutive z-slices taken at a 0.5 μm slice interval. ImageJ required the user to manually merge red and green source channels to form a composite image as part of Step 1 (Fig. 1A), while Imaris did not require this rendering step (Fig. 1E) (Ferreira and Rasband, 2012; Bitplane, 2017). As a result, Imaris was able to run commands on either the red or the green channel independently while still presenting the BFCN as a composite color model. The ImageJ “3D Project” function noticeably reduced resolution of the generated image. Compression of the z-stack into a single plane resulted in the reconstructed BFCN appearing flattened and static (Fig. 1A). In contrast, Imaris rendered a z-stack as an interactive 3D “Scene”, preserving dimensionality of the BFCN, endosomes, and surrounding neuropil (Fig. 1E).
In Step 2 of the ImageJ protocol, a reconstructed z-stack was segmented to denote foreground and background regions. A histogram of pixel intensity was used to determine an appropriate threshold value to classify pixels as either foreground or background objects (Ferreira and Rasband, 2012; Bankhead, 2013). The thresholding operation enabled endosomes in the foreground of the image to appear dark against a white background (Fig. 1B). Thresholding routinely resulted in multiple endosomes clustered in close proximity to appear as if they were a single large object. In Step 3, merged early endosomes were separated using the paintbrush tool. A freehand outline was traced around the BFCN, designating it as the region of interest (ROI) (Fig. 1C). In ImageJ, the ROI is applied as an overlay on top of the image, not integrated into the 3D reconstruction itself (Bankhead, 2013). This overlay can be drawn around the BFCN of interest at any stage post-reconstruction and does not affect image processing.
In the final step of the ImageJ protocol, the “Analyze Particle” command was run. ImageJ used edge-detection, a boundary extraction method, to identify individual particles and classify each as a discrete object (Shapiro and Stockman, 2001). ImageJ scanned within the ROI for the edges of endosomes, defined as the point where a black pixel abruptly met a white pixel (Fig. 1D). This boundary was then traced for each endosome until ImageJ returned to the initial border pixel (Ferreira and Rasband, 2012). The ImageJ “Analyze Particle” function outlined, counted, and measured 91 Rab5-positive early endosomes in the representative BFCN shown in Figure 1. The process took between 30–60 minutes to perform on a single image in ImageJ, with the majority of the time spent separating endosomes with the paintbrush tool.
The Imaris protocol diverged from the ImageJ method in its order of operations. In Step 2 of the Imaris workflow, a ROI was constructed around the BFCN of interest using the “Surface” creation tool (Fig. 1F). Unlike ImageJ, it was imperative that the ROI be designated before any subsequent steps were taken in Imaris. The ROI in Step 2 did not need to be manually traced around the BFCN, as in ImageJ. Instead, the “Surface” creation wizard first prompted the user to select the source channel the program would utilize while building the ROI (in this case Green Channel-Ch2-T1). Imaris automatically detected boundaries of the labeled cell by employing a two-step background subtraction protocol (Bitplane, 2017). Using the intensity of the ChAT immunolabeling, this function first applied a Gaussian smoothing filter to reduce noise and estimate the background intensity of each identifiable voxel on the green source channel (Oberholzer et al., 1996; Sezgin and Sankur, 2004; Bitplane, 2017). Baseline subtraction was then performed by subtracting this variable background from every voxel in the image (Bitplane, 2017). The finalized “Surface” encased the BFCN of interest, demarcating the ROI. This generated “Surface” was fully integrated into the 3D “Scene” as a discrete, manipulatable object, rather than a superimposed overlay as in ImageJ.
In Step 3 of the Imaris protocol, early endosomes within the BFCN of interest were partitioned from the rest of the 3D environment (Fig. 1G). To isolate endosomes, a mask was applied over “Red Channel-Ch3-T2” external to the generated “Surface”. Upon application, this mask reset the intensity of all red voxels not enclosed by the “Surface” to 0.000. By converting the intensity of all exterior voxels to 0, voxels on the red source channel not within the ROI became invisible to the program and were subsequently ignored by the algorithm as quantification progressed (Oberholzer et al., 1996; Bitplane, 2017).
In the final step of the Imaris protocol, the “Spots” creation wizard was run (Fig. 1H). Unlike ImageJ, which used edge-detection to identify individual endosomes, Imaris used a combination of background subtraction and seeded region growing (Bitplane, 2017). Imaris first identified all red voxels not hidden by the previously constructed mask, i.e. only those red voxels that were located within the BFCN of interest. Background subtraction was again applied, this time on the masked red source channel. For each distinct collection of red voxels within the ROI, a seed-point was placed at the voxel of highest intensity (Bitplane, 2017). From this central seed voxel, each neighboring voxel was examined to determine its similarity to the seed point (Jamil et al., 2011). Each region was iteratively grown from this seed point until the endosomal borders detected during background subtraction were reached (Khalifa, 2010). In total, the protocol took 5–10 minutes to perform on a single z-stack image. Imaris’ “Spots” analysis identified, counted, and measured 75 Rab5-positive early endosomes in the representative BFCN shown in Figure 1.
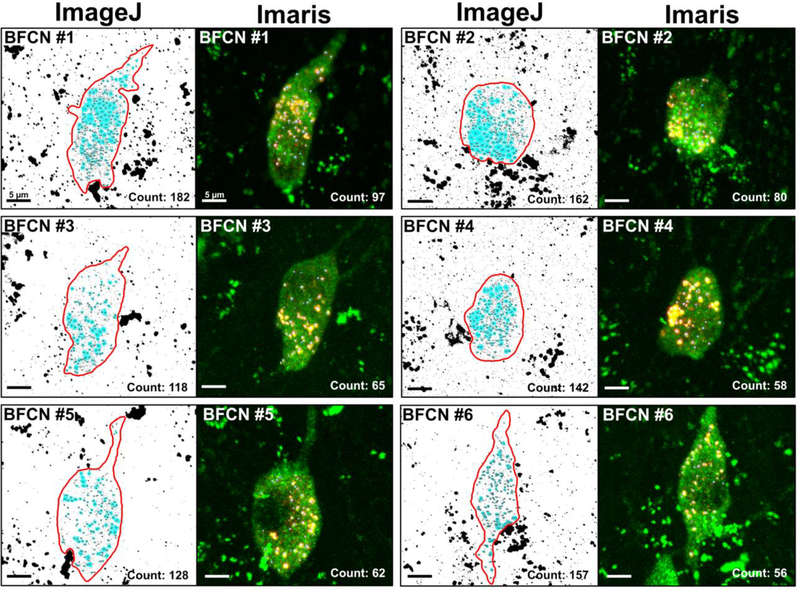
To compare ImageJ and Imaris in terms of accuracy when quantifying Rab5-immuolabeled early endosomes within transmitter-identified BFCNs, 6 individual BFCNs were analyzed in parallel on both platforms (Fig. 2). All BFCNs analyzed were imaged from within the MSN of the same ~11 MO female mouse. The 6 neurons chosen for quantification were selected from a larger pool of 29 BFCNs based upon the clarity of the z-stack post-3D reconstruction and the absence of any staining artifacts or obfuscating structures such as blood vessels or adjacent cells. BFCNs of different shapes and sizes, as well as ones with variations in the pattern of early endosome distribution within the cell soma, were prioritized. Additionally, to get a representative sampling of different cholinergic neurons throughout the basal forebrain, all 6 BFCNs selected were from different positions along the rostrocaudal axis of the MSN. The total numbers of Rab5-positive early endosomes quantified per BFCN analyzed in both ImageJ and Imaris are presented in Table 1.
Figure 2.
Representative images comparing image analysis and data output of both early endosome quantification protocols within 3D reconstructions of 6 BFCNs in ImageJ and Imaris. All images displayed are processed models, not the original confocal microscopy images. Tissue sections were double labeled with a primary antibody against Rab5c (yellow) for early endosomes and ChAT (green) for BFCNs. Z-stacks of individual BFCNs were taken at 100x magnification with a 0.5 μm slice interval prior to 3D reconstruction. All BFCNs were from the MSN of the same ~11 MO female mouse. The ImageJ “Analyze Particle” protocol classified more small pixels as early endosomes (highlighted in blue) when compared to Imaris. ImageJ also underestimated the number of early endosomes present within endosomal clusters in the soma of the BFCN due to object occlusion. The Imaris “Spots” protocol (blue spheres superimposed over yellow early endosomes) was more adept at filtering out noise present within the z-stack and splitting touching objects through the combined usage of local thresholding and seeded region growing. Scale bar in each panel: 5 μm.
Table 1.
Comparison of the total number of Rab5-immunoreactive early endosomes quantified in each ChAT-immunoreactive BFCN in ImageJ and Imaris. A Wilcoxon-Mann-Whitney test showed statistically a significant difference in the number of early endosomes identified by each program (z = 2.3434, p = 0.028), with approximately twice the number of early endosomes counted per BFCN in ImageJ compared to Imaris.
| BFCN number | Number of z-slices in original stack | Total Number of Rab5-immunoreactive early endosomes per neuron |
||
|---|---|---|---|---|
| ImageJ | Imaris | |||
| 1 | 33 | 182 | 97 | |
| 2 | 33 | 162 | 80 | |
| 3 | 27 | 118 | 65 | |
| 4 | 36 | 142 | 58 | |
| 5 | 29 | 128 | 62 | |
| 6 | 33 | 157 | 56 | |
| Mean (± St Dev): | 148.17 (± 23.53) | 69.67 (±15.85) | p=0.0281 | |
Significantly more Rab5-immunoreactive early endosomes were quantified in ImageJ in each of the ChAT-immunoreactive BFCNs compared to Imaris (Wilcoxon-Mann-Whitney z = 2.3434, p = 0.028). On average (mean ± standard deviation), the ImageJ “Particle Analysis” algorithm counted 148.17 (± 23.53) Rab5-positive early endosomes per BFCN compared to the Imaris “Spots” analysis which quantified an average of 69.67 (± 15.86) early endosomes per BFCN. Though the lower bound for particle size in ImageJ was set to match the lower bound identified by Imaris (0.2 μm2), ImageJ counted noticeably more small particles than Imaris in each of the cells quantified (Fig. 2). Upon closer examination in Imaris, most of these small particles fell into 3 different categories: i) endosomes or other small objects that, upon rotation of the frame, were not within the BFCN of interest, but were instead located either in front of or behind it, ii) small objects on the exterior surface of the neuron that were filtered out during “Surface” creation, and iii) low intensity staining artifacts around the nucleus that were removed via background subtraction (Fig. 3).
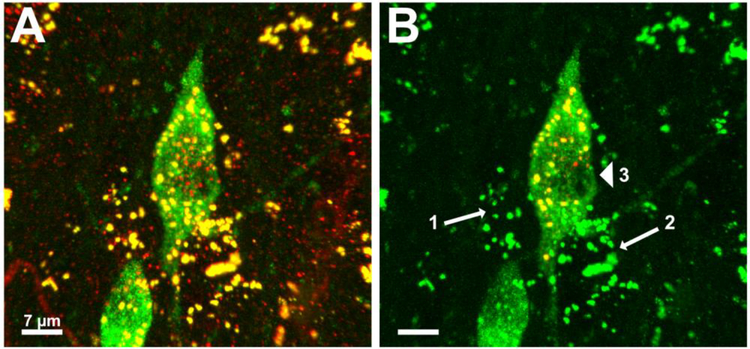
Figure 3.
Representative images demonstrating Imaris’ ability to accurately quantify Rab5-immunlabeled early endosomes only within a specified BFCN of interest. A. 3D reconstruction of Rab5-immumolabeled early endosomes and 2 ChAT-immunolabeled BFCNs, prior to image analysis. B. The same 3D reconstruction after creation of the ROI. The centered BFCN has been selected as the neuron for analysis. Early endosomes in the neighboring BFCN and in surrounding non-cholinergic cells appear green after application of a mask overlay resetting all red voxels outside of the BFCN of interest to “0.000”. Imaris was able to recognize and exclude early endosomes in multiple adjacent non-cholinergic cells from quantification (arrows 1 and 2). Small staining artifacts around the nucleus (arrowhead) were filtered out by Imaris during the background subtraction step of “Surface” creation. Scale bar A-B: 7 μm.
In addition to overcounting small particles, ImageJ undercounted the number of endosomes present within large clusters. Clusters of endosomes often appeared as black pixelated masses after thresholding in ImageJ. While a non-segmented image was used as a visual guide when manually breaking up endosomes that appeared fused, it was not always possible to accurately replicate the original curvature of individual endosomes using the paintbrush tool. Even prior to thresholding, conversion of the z-stack into a 3D projection in ImageJ resulted in a loss of definition of discrete vesicles. It was therefore predictable that some endosomes in the background would be concealed by those in the foreground. Due to the inherent 3D nature of the program, Imaris was much more adept at parsing and quantifying distinct endosomes that were clustered in proximity within the soma.
To determine whether ImageJ could generate 3D reconstructions comparable to those in Imaris when similar methodology was applied, several different ImageJ functions were performed on a BFCN of interest (Fig. 4). These tests included: i) application of a Gaussian filter followed by background subtraction, ii) local thresholding followed by watershed, and iii) all four processes in combination (Ferreira and Rasband, 2012).
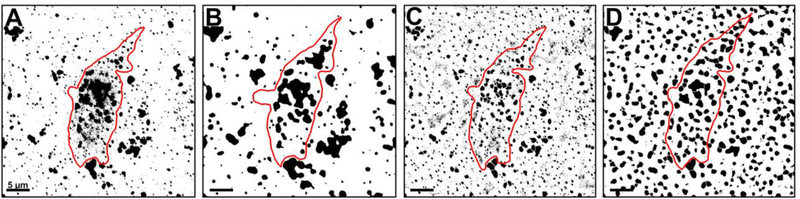
Figure 4.
ImageJ was unable to replicate the image analysis performed in Imaris, despite application of similar methodology. A. Representative image of a 3D reconstructed BFCN (outlined in red) after thresholding in ImageJ. Early endosomes (black) have merged to form large masses making analysis equivocal. B. Application of a Gaussian filter and background subtraction akin to the Imaris “Surface” creation step reduced the number of staining artifacts and salt-and-pepper noise classified as foreground pixels during thresholding. Clusters of endosomes were still difficult to resolve. C. Application of Otsu’s local thresholding, similar to the Imaris “Spots” creation step, reduced excessive noise around the periphery of endosomal clusters and enabled more precise application of watershed. Early endosomes were more reminiscent of their original shape prior to the global thresholding originally performed in (A), however image noise was still high. D. Application of all 4 techniques produced an over processed image that was still difficult to analyze. Scale bar A-D: 5 μm.
Background subtraction and application of the Gaussian filter prior to thresholding noticeably reduced the number of low intensity staining artifacts that were erroneously recognized as foreground objects during image segmentation (Fig. 4B). Salt and pepper noise, the presence of a single light pixel in a dark region or a single dark pixel in a light region (Shapiro and Stockman, 2001), was also greatly diminished when compared to the original thresholded image (Fig. 4A). Application of these filters did not, however, help resolve clusters of early endosomes.
Performing Otsu’s automatic thresholding on the image effectively rendered clear endosomal clusters (Fig. 4C). Otsu’s method automatically selects a threshold value based on minimization of within-group variance between groups of pixels (Otsu, 1979; Shapiro and Stockman, 2001). This technique minimized the number of extraneous background pixels adjacent to endosomal clusters that were misclassified as foreground pixels due to their grey-value (Chaubey, 2016; https://imagej.net/Principles, 2020). Resulting clusters were well defined and reminiscent of their original shape prior to image binarization. While excessive noise around the periphery of endosomal clusters was eliminated, individual endosomes within a given cluster were still not well delineated. Global noise was reduced with application of Otsu’s thresholding method, though it was not as proficient at filtering out small particulate as the application of background subtraction. The watershed protocol, which is designed to separate overlapping objects, was more successful at identifying contours when combined with Otsu’s thresholding (Schulze and Tosi, 2016; http://imagej.net/Particle_Analysis, 2020). Watershed separation operates by equating pixel intensity to height on a topographical map, with the deepest intensities considered basins (Beucher and Lantuéjoul, 1979; Meyer, 1994). Watershed lines are drawn by ImageJ where water would theoretically overflow between basins, akin to a dam between adjacent regions (Vincent and Soille, 1991; Bankhead, 2013). Removal of extraneous pixels via Otsu’s method enabled a greater degree of accuracy when calculating watershed lines (Fig. 4C). However, this was not sufficient to replace the use of the paintbrush tool.
Combining a Gaussian filter, background subtraction, auto local thresholding, and watershed in a manner similar to the way Imaris processes images did not render a 3D reconstruction of comparable quality (Fig. 4D). The application of all four functions produced a BFCN that was more complicated to analyze rather than less complicated, likely due to over-processing of the image. Despite intensive efforts, quantification in ImageJ could not reproduce accuracy levels attained with Imaris in terms of quantifying Rab5-immunoreactive early endosomes on identified ChAT-immunoreactive BFCNs in 3D reconstructions.
4. Discussion
ImageJ and Imaris are popular software packages in image analysis fields. Both platforms offer users a host of tools to assist with in-depth analysis of biological systems. Despite many core similarities, ease of use, image processing, and data output differ substantially between the two programs when quantifying 3D reconstructed z-stacks acquired via confocal microscopy in the present study. Our data indicates Imaris is a more suitable software choice in terms of performing quantitative analysis involving dual immunolabeling of endosomes within defined neurons.
Discrepancies in total endosome counts between ImageJ and Imaris can be largely attributed to two major differences in the way ImageJ and Imaris process 3D reconstructions: i) the thresholding technique employed to segment images and ii) the preservation and utilization of spatial information to identify and measure individual Rab5-positive endosomes.
Image segmentation, the partitioning of an image into non-overlapping, constituent regions based on pre-defined similarities, is a key component of image analysis (Pham et al., 2000). ImageJ’s “Particle Analysis” function is most accurate when performed on binary black and white images, necessitating segmentation (Bankhead, 2013; http://imagej.net/Particle_Analysis, 2020). ImageJ accomplishes the conversion from greyscale to binary via a global thresholding technique (Fig. 5A). Pixel intensity is measured throughout a reconstructed image. A single threshold value is subsequently applied to the entire image to partition pixels into foreground or background regions (Sahoo et al., 1988; Oberholzer et al., 1996; Bankhead, 2013). Variables including image resolution, object size, staining intensity variation, and the signal-to-noise ratio of the z-stack impact the ability to discriminate endosomes in the foreground from the image background (Lee et al., 1990). Excess noise within low resolution 3D reconstructions can therefore result in pixel misclassification once binarized (Lin et al., 2003). In the present study, thresholded endosomes localized within a BFCN of interest are difficult to differentiate from high intensity extracellular particulate on a flat white background with no visible landmarks. Endosomal clusters often appear as black pixelated masses, necessitating extensive use of ImageJ’s paintbrush tool to separate objects, which can introduce user bias.
Figure 5.
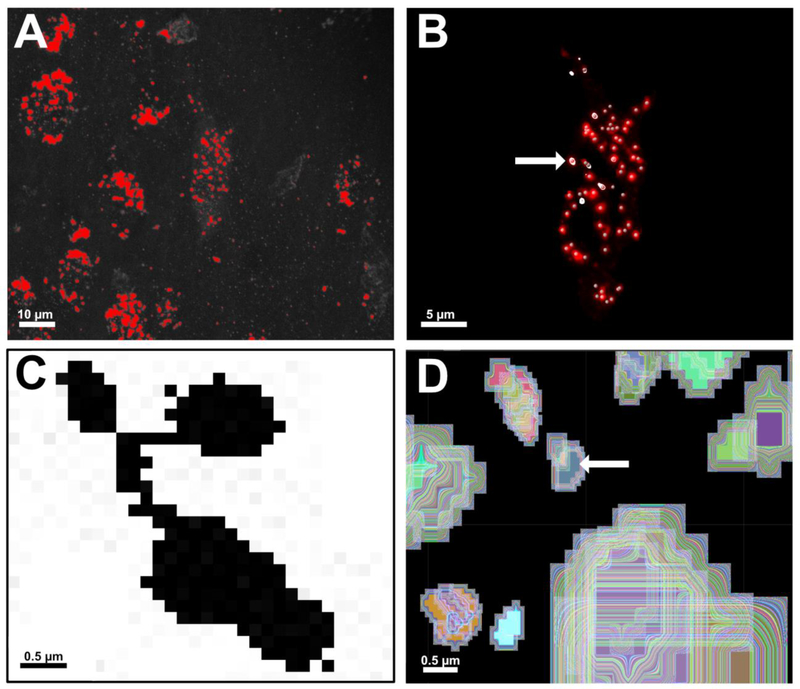
A comparison of global thresholding versus local thresholding of early endosomes (A-B) and edge detection versus seeded region growing (C-D) in ImageJ and Imaris, respectively. A. Global thresholding being performed on a 3D reconstruction in ImageJ to convert the image from greyscale to binary black and white. Early endosomes are colored red, signifying they will be classified as foreground pixels in the subsequent binary image. Global thresholding measures pixel intensity across the entire image, making it vulnerable to inconsistencies in image resolution, staining intensity, and signal-to-noise ratios. Scale bar: 10 μm. B. Local thresholding being performed in Imaris using “Spots”. Local thresholding measures voxel intensities within a locally defined neighborhood. Creation of the mask channel forces “Spots” to measure red voxels only within the ROI, leading to more accurate detection of Rab5-immunolabeled early endosomes (central point denoted by grey spheres). Early endosomes highlighted in white (arrow) are all located on the same XY plane of focus within the 3D reconstruction. Use of the XY Orthoslicing tool enables precise assessment of a chosen threshold value for each endosome detected within the BFCN. Scale bar: 5 μm. C. A zoomed-in image of a cluster of ~4–5 early endosomes in ImageJ after thresholding. The “Analyze Particle” command in ImageJ uses 8-neighbor pixel connectivity during edge detection. All black pixels in the image would be counted as a single endosome unless manually separated with white pixels using the “Paintbrush” tool. D. Imaris employs seeded region growing to count and measure individual Rab5-positive early endosomes. A seed point is placed on the voxels of highest intensity at the center of an identified object (arrow). The region is then grown outward in 3 dimensions until borders detected during prior background subtraction steps are reached. In this representative image, each trace around the seed point has been color coded to illustrate this principle. Scale bar C-D: 0.5 μm.
In contrast to the global thresholding approach implemented by ImageJ, Imaris employs local thresholding during the region growing phase of its “Spots” analysis (Fig. 5B). Local thresholding subdivides an image and determines an appropriate threshold value based solely on local characteristics (Sahoo et al., 1988; Lee et al., 1990). Utilizing local thresholding techniques help circumvent many issues experienced with global methods involving uneven background illumination (Otsu, 1979; Chaubey, 2016). This is especially important when attempting to detect numerous small objects scattered throughout an image. Generating the ROI in Step 2 of the Imaris protocol, prior to image thresholding, is integral to our quantitative endosomal analysis. Partitioning an identified neuron into its own insulated neighborhood ensures only local red voxels are used as a source of comparison when performing “Spots”. A caveat of employing local thresholding methods is they require significant free memory to perform optimally (Bitplane, 2017).
Another critical factor contributing to quantification discrepancies between Imaris and ImageJ involves utilization of spatial information to identify Rab5-positive early endosomes. Some segmentation methods, such as the global thresholding applied in ImageJ, rely only on pixel intensities, and therefore function independently of the image domain. In contrast, region growing algorithms, like those employed by Imaris, operate differently depending on the dimensionality of the image (Rivest et al., 1992; Pham et al., 2000). When working with 3D reconstructions, utilizing analytical methods that discount spatial information is likely to introduce error into the quantification. Ignoring spatial relationships between objects and using only image features to model a 3D reconstruction leads to excessively noisy images (Saxena et al., 2008), as evidenced in Fig. 1A.
Interposition is the concept that objects closer to the viewer in a visual field occlude parts of objects farther away (Shapiro and Stockman, 2001). Subtle changes in texture and shading, as well as rotation of a scene or light source around a fixed point, can help elucidate whether two objects are in-line or located on different planes (Jense and Huusmans, 1989). In Imaris this is easily achieved, as the program allows for full rotation of a dataset during analysis. This type of rendering is not possible in ImageJ without the “ImageJ 3D Viewer” plugin, an add-on often bundled with Fiji that assists with qualitative visualization of 3D data (Schmid et al., 2010; Schindelin et al., 2012, 2015). In our quantitative analysis, projection of the 3D z-stack onto a single plane of focus actively degrades available depth cues. As a result, occlusion of Rab5-positive endosomes in the background by those in the foreground was observed in every BFCN analyzed in ImageJ (Fig. 2). This becomes particularly problematic after binarization. Without the ability to discern edges and contours, it becomes increasingly difficult to determine whether neighboring regions or objects within an image are physically connected (Hoiem et al., 2007).
Proper boundary refinement is one of the most important components of accurate image segmentation (Taneja et al., 2015). ImageJ relies on boundary detection as part of the “Analyze Particles” algorithm. This technique defines an edge as a region where there is an abrupt discontinuity in the intensity of adjacent pixels (Khalifa, 2010). Edge detection methods work well for small, uncomplicated images with well separated objects (Shapiro and Stockman, 2001). However, these methods are very sensitive to image noise and prone to error when boundaries are not clearly defined (Lin et al., 2003; Jamil et al., 2011). Therefore, noise levels in 3D reconstructions pose a problem for ImageJ users. Interposition, coupled with noise, leads to the creation of the large, pixelated masses of endosomes seen after thresholding.
ImageJ’s use of 8-neighbor pixel connectivity further complicates quantitative analysis (Bankhead, 2013). “Analyze Particles” considers pixels to be part of the same object when diagonally adjacent to each other (Fig. 5C). Meticulous use of the paintbrush tool by the user is necessary to ensure edges cannot be traced along diagonal lines that inadvertently link two unrelated regions together. Ideally the watershed algorithm would help automatically disconnect these regions. However, as seen in Fig. 4C, watershed did not perform well when used in this analysis. Reasons for this failure are twofold. First, watershed assumes touching objects exhibit a narrow “neck” at the point where adjacent catchment basins connect (Lin et al., 2003). If this narrowing isn’t present, particularly if there is a large amount of object occlusion, watershed will not draw dams. Second, watershed depends on the Euclidean distance map (EDM) to identify intensity minima and maxima (Roerdink and Meijster, 2001; Ferreira and Rasband, 2012; Paul-Gilloteaux and Tosi, 2016). Euclidean distance refers to the straight-line distance between two points in geometric space. In ImageJ, this translates to the distance between two pixels within an image (Oberholzer et al., 1996; Ferreira and Rasband, 2012; Burger and Burge, 2016). Since depth is not well preserved and noise is exacerbated in ImageJ after 3D reconstruction, adjacent pixels within the flattened reconstruction cannot be assumed to have been adjacent in the original z-stack. The EDM is therefore not as accurate as one generated from a 2D image, as calculations are based on potentially erroneous information.
In contrast to ImageJ, the seeded region growing Imaris employs as part of “Spots” retains and incorporates spatial information. Region growing is generally preferred over other extraction methods, like edge detection, when both the image background and the objects being detected have inconsistent staining intensity (Jamil et al., 2011; Bitplane, 2017). Additionally, region growing in “Spots” is particularly adept at splitting small touching objects, though its accuracy can decrease as endosome density within a cluster increases (Lin et al., 2003; Bitplane, 2017).
The “Spots” algorithm in Imaris combines seeded region growing with a boundary detection mechanism akin to a 3D watershed (Fig. 5D). During “Spots” creation, a user defined estimation of the diameter of an endosome is utilized to determine the width of the Gaussian filter and locate intensity maxima that serve as seed points (Bitplane, 2017). Rather than simply tracing border pixels, Imaris grows a spot in 3 dimensions until it fills the volume occupied by red voxels. In our analysis, seeded region growing is more effective at accurately counting early endosomes because it can overcome the problems ImageJ’s watershed experienced with respect to object occlusion and 2D Euclidean distance calculations.
A significant drawback of seeded region growing is the amount of manual interaction necessary to “plant” seeds (Pham et al., 2000). A general understanding of the composition of the image is required to ensure seeds are placed at appropriate locations, which can introduce bias and steer the analysis in the direction the user desires (Jamil et al., 2011). However, the less automated nature of region growing is also what makes it more accurate than the edge detection used in ImageJ.
In conclusion, both ImageJ and Imaris software packages offer significant contributions to confocal image analysis. Our quantitative assessment illustrates Imaris outperforms ImageJ in terms of accuracy when it comes to counting Rab5-immunoreactive early endosomes within transmitter-identified BFCNs in 3D reconstructed z-stacks. The high degree of precision demonstrated by Imaris makes it a valuable tool in the evaluation of cellular and molecular pathology of vesicular structures within defined cells.
Highlights.
A comparison of ImageJ and Imaris early endosome quantification was performed
Vesicle quantification using Imaris’ 3D reconstructions increases accuracy
Seeded region growing in Imaris circumvents object occlusion in 3D reconstructions
Choice of analysis software is crucial when quantifying vesicles like endosomes
Acknowledgements
This study was supported by grants AG014449, AG043375, and AG017617 from the National Institutes of Health and the Alzheimer’s Association. We thank Dr. Melissa J. Alldred, Ph.D., and Arthur Saltzman, M.S., for expert technical assistance with colony breeding and maintenance, Philip Stavrides, M.S., for technical assistance with confocal microscopy, and Dr. Sang H. Lee, Ph.D., for statistical analysis.
Footnotes
Financial Disclosure
Megan K. Gautier and Stephen D. Ginsberg have no financial disclosures to declare.
Megan K. Gautier and Stephen D. Ginsberg have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bankhead P Analyzing fluorescence microscopy images with ImageJ. Heidelberg University, Queen’s University Belfast: Nikon Imaging Center, 2013. [Google Scholar]
- Beucher S, Lantuéjoul C. Use of watersheds in contour detection. International Workshop on Image Processing: Real-Time Edge and Motion Detection/Estimation: Rennes, France, 1979. [Google Scholar]
- Bitplane. IMARIS 9.1 Reference Manual. Bitplane: Concord, MA, 2017. [Google Scholar]
- Buchwalow I, Samoilova V, Boecker W, Tiemann M. Non-specific binding of antibodies in immunohistochemistry: fallacies and facts. Sci Rep, 2011; 1: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger W, Burge MJ. Digital Image Processing. Texts in Computer Science. Springer-Verlag, London, 2016. [Google Scholar]
- Cabeza C, Figueroa A, Lazo OM, Galleguillos C, Pissani C, Klein A, Gonzalez-Billault C, Inestrosa NC, Alvarez AR, Zanlungo S, Bronfman FC. Cholinergic abnormalities, endosomal alterations and up-regulation of nerve growth factor signaling in Niemann-Pick type C disease. Mol Neurodegener, 2012; 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci, 1997; 17: 6142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, Mullaney K, Neve RL, Gruenberg J, Nixon RA. Down syndrome fibroblast model of Alzheimer-related endosome pathology: accelerated endocytosis promotes late endocytic defects. Am J Pathol, 2008; 173: 370–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, Villar A, Carlson EJ, Staufenbiel M, Nixon RA. App gene dosage modulates endosomal abnormalities of Alzheimer’s disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci, 2003; 23: 6788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol, 2000; 157: 277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey AK. Comparison of The Local and Global Thresholding Methods in Image Segmentation. World Journal of Research and Review, 2016; 2. [Google Scholar]
- Choi JH, Kaur G, Mazzella MJ, Morales-Corraliza J, Levy E, Mathews PM. Early endosomal abnormalities and cholinergic neuron degeneration in amyloid-β protein precursor transgenic mice. J Alzheimers Dis, 2013; 34: 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacurcio DJ, Pensalfini A, Jiang Y, Nixon RA. Dysfunction of autophagy and endosomal-lysosomal pathways: Roles in pathogenesis of Down syndrome and Alzheimer’s Disease. Free Radic Biol Med, 2018; 114: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri KW, Berthold MR, Goldberg IG, Ibáñez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, Stuurman N, Swedlow JR, Tomancak P, Carpenter AE. Biological imaging software tools. Nat Methods, 2012; 9: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Rasband WS. ImageJ User Guide, IJ 1.46 ed: imagej.nih.gov/ij/docs/guide/, 2010–2012.
- Fricker M, Runions J, Moore I. Quantitative fluorescence microscopy: from art to science. Annu Rev Plant Biol, 2006; 57: 79–107. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, Wuu J, Chao MV, Mufson EJ, Nixon RA, Che S. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry, 2010; 68: 885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, Che S. Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Chem Neuroanat, 2011; 42: 102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Counts SE, Wuu J, Alldred MJ, Nixon RA, Che S. Regional selectivity of rab5 and rab7 protein upregulation in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis, 2010; 22: 631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbhuber KJ, König K. Modern laser scanning microscopy in biology, biotechnology and medicine. Ann Anat, 2003; 185: 1–20. [DOI] [PubMed] [Google Scholar]
- Hoiem D, Efros AA, Hebert M. Recovering Surface Layout from an Image. International Journal of Computer Vision, 2007; 75: 151–72. [Google Scholar]
- ImageJ.net, 2020. Particle Analysis. https://imagej.net/Particle_Analysis (accessed February 2020)
- ImageJ.net, 2020. Principles. https://imagej.net/Principles (accessed April 2020)
- Jamil N, Soh HC, Tengku Sembok TM, Bakar ZA. A Modified Edge-Based Region Growing Segmentation of Geometric Objects. Springer Berlin; Heidelberg: Berlin, Heidelberg, 2011: 99–112. [Google Scholar]
- Jense GJ, Huijsmans DP. Interactive voxel-based graphics for 3D reconstruction of biological structures. Computers & Graphics, 1989; 13: 145–50. [Google Scholar]
- Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A, Ginsberg SD, Cataldo AM, Mathews PM, Nixon RA. Alzheimer’s-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci U S A, 2010; 107: 1630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Rigoglioso A, Peterhoff CM, Pawlik M, Sato Y, Bleiwas C, Stavrides P, Smiley JF, Ginsberg SD, Mathews PM, Levy E, Nixon RA. Partial BACE1 reduction in a Down syndrome mouse model blocks Alzheimer-related endosomal anomalies and cholinergic neurodegeneration: role of APP-CTF. Neurobiol Aging, 2016; 39: 90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LW, Shie FS, Maezawa I, Vincent I, Bird T. Intracellular accumulation of amyloidogenic fragments of amyloid-beta precursor protein in neurons with Niemann-Pick type C defects is associated with endosomal abnormalities. Am J Pathol, 2004; 164: 975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Gauthier SA, Perez-Gonzalez R, Pawlik M, Singh AB, Cosby B, Mohan PS, Smiley JF, Levy E. Cystatin C prevents neuronal loss and behavioral deficits via the endosomal pathway in a mouse model of down syndrome. Neurobiol Dis, 2018; 120: 165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Pawlik M, Gandy SE, Ehrlich ME, Smiley JF, Levy E. Lysosomal dysfunction in the brain of a mouse model with intraneuronal accumulation of carboxyl terminal fragments of the amyloid precursor protein. Mol Psychiatry, 2017; 22: 981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci, 2000; 20: 7268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CM, Powers BE, Velazquez R, Ash JA, Ginsberg SD, Strupp BJ, Mufson EJ. Sex differences in the cholinergic basal forebrain in the Ts65Dn mouse model of Down syndrome and Alzheimer’s disease. Brain Pathol, 2014; 24: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa A Evaluating the effectiveness of region growing and edge detection segmentation algorithms. Journal of American Science, 2010; 6. [Google Scholar]
- Lee SU, Yoon Chung S, Park RH. A comparative performance study of several global thresholding techniques for segmentation. Computer Vision, Graphics, and Image Processing, 1990; 52: 171–90. [Google Scholar]
- Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry A, 2003; 56: 23–36. [DOI] [PubMed] [Google Scholar]
- Malik BR, Maddison DC, Smith GA, Peters OM. Autophagic and endo-lysosomal dysfunction in neurodegenerative disease. Mol Brain, 2019; 12: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin MD. Factors affecting visibility of a target tissue in histologic sections. Vet Pathol, 2014; 51: 9–27. [DOI] [PubMed] [Google Scholar]
- Meyer F Topographic distance and watershed lines. Signal Processing, 1994; 38: 113–25. [Google Scholar]
- Mitra J, Hegde PM, Hegde ML. Loss of endosomal recycling factor RAB11 coupled with complex regulation of MAPK/ERK/AKT signaling in postmortem spinal cord specimens of sporadic amyotrophic lateral sclerosis patients. Mol Brain, 2019; 12: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Tosi S, Möhl C, Zhang C, Paul-Gilloteaux P, Schulze U, Nørrelykke SF, Tischer C, Pengo T. Bioimage Analysis Tools. In Miura K, editor. Bioimage Data Analysis. Wiley-VCH Verlag GmbH & Co. KGaA: Boschstr. 12, 69469 Weinheim, Germany, 2016: 4–18. [Google Scholar]
- Nixon RA. Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: inseparable partners in a multifactorial disease. FASEB J, 2017; 31: 2729–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging, 2005; 26: 373–82. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Cataldo AM. Lysosomal system pathways: genes to neurodegeneration in Alzheimer’s disease. J Alzheimers Dis, 2006; 9: 277–89. [DOI] [PubMed] [Google Scholar]
- Oberholzer M, Östreicher M, Christen H, Brühlmann M. Methods in quantitative image analysis. Histochemistry and Cell Biology, 1996; 105: 333–55. [DOI] [PubMed] [Google Scholar]
- Otsu N A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics, 1979; 9: 62–6. [Google Scholar]
- Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington’s disease. J Cell Biol, 2006; 172: 605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Gilloteaux P, Tosi S. Quantitative Evaluation of Multicellular Movements in Drosophila Embryo. In Miura K, editor. Bioimage Data Analysis. Wiley-VCH Verlag GmbH & Co. KGaA: Boschstr. 12, 69469 Weinheim, Germany, 2016: 170–96. [Google Scholar]
- Perez SE, He B, Nadeem M, Wuu J, Ginsberg SD, Ikonomovic MD, Mufson EJ. Hippocampal endosomal, lysosomal, and autophagic dysregulation in mild cognitive impairment: correlation with aβ and tau pathology. J Neuropathol Exp Neurol, 2015; 74: 345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annu Rev Biomed Eng, 2000; 2: 315–37. [DOI] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health: Bethesda, Maryland, USA, 1997–2018. [Google Scholar]
- Rivest J-F, Serra J, Soille P. Dimensionality in image analysis. Journal of Visual Communication and Image Representation, 1992; 3: 137–46. [Google Scholar]
- Roerdink J, Meijster A. The Watershed Transform: Definitions, Algorithms and Parallelization Strategies. Fundam Inf, 2001; 41: 187–228. [Google Scholar]
- Sahoo PK, Soltani S, Wong AKC. A survey of thresholding techniques. Computer Vision, Graphics, and Image Processing, 1988; 41: 233–60. [Google Scholar]
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, Takimoto-Kimura R, Kleschevnikov AM, Sambamurti K, Chung PP, Xia W, Villar A, Campbell WA, Kulnane LS, Nixon RA, Lamb BT, Epstein CJ, Stokin GB, Goldstein LS, Mobley WC. Increased App expression in a mouse model of Down’s syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron, 2006; 51: 29–42. [DOI] [PubMed] [Google Scholar]
- Saxena A, Chung SH, Ng AY. 3-D Depth Reconstruction from a Single Still Image. International Journal of Computer Vision, 2008; 76: 53–69. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods, 2012; 9: 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev, 2015; 82: 518–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics, 2010; 11: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods, 2012; 9: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze U, Tosi S. FISH Spot Detection in Human Spermatozoids. In Miura K, editor. Bioimage Data Analysis. Wiley-VCH Verlag GmbH & Co. KGaA: Boschstr. 12, 69469 Weinheim, Germany, 2016: 98–118. [Google Scholar]
- Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. Journal of Electronic Imaging, 2004; 13. [Google Scholar]
- Stockman GC, Shapiro LG. Computer Vision, 1st Edition ed. Prentice Hall PTR, 2001. [Google Scholar]
- Taneja A, Ranjan P, Ujjlayan A. A performance study of image segmentation techniques. 2015 4th International Conference on Reliability, Infocom Technologies and Optimization (ICRITO) (Trends and Future Directions), 2015: 1–6. [Google Scholar]
- Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology, 2006; 49: 411–24. [DOI] [PubMed] [Google Scholar]
- Tischer C, Tosi S. Tumor Blood Vessels: 3D Tubular Network Analysis. In Miura K, editor. Bioimage Data Analysis. Wiley-VCH Verlag GmbH & Co.KGaA: Boschstr. 12, 69469 Weinheim, Germany, 2016: 219–35. [Google Scholar]
- Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Transactions on Pattern Analysis and Machine Intelligence, 1991; 13: 583–98. [Google Scholar]
- Zinchuk V, Grossenbacher-Zinchuk O. Recent advances in quantitative colocalization analysis: focus on neuroscience. Prog Histochem Cytochem, 2009; 44: 125–72. [DOI] [PubMed] [Google Scholar]