SUMMARY
Human testis development in prenatal life involves complex changes in germline and somatic cell identity. To better understand, we profiled and analyzed ~32,500 single-cell transcriptomes of testicular cells from embryonic, fetal and infant stages. Our data shows that at 6-7 weeks post-fertilization as the testicular cords are established, the Sertoli and interstitial cells originate from a common heterogeneous progenitor pool, which then resolves into fetal Sertoli cells (expressing tube-forming genes) or interstitial cells (including Leydig-lineage cells expressing steroidogenesis genes). Almost ten weeks later, beginning at 14-16 weeks post-fertilization, the male primordial germ cells exit mitosis, downregulate pluripotent transcription factors and transition into cells that strongly resemble the ‘State 0’ spermatogonia originally defined in the infant and adult testes. Therefore, we termed these fetal spermatogonia ‘State f0’. Overall, we reveal multiple insights into the coordinated and temporal development of the embryonic, fetal and postnatal male germline together with the somatic niche.
Graphical Abstract

eTOC Blurb:
Guo et al. provide a transcriptional cell atlas of the fetal and postnatal human testes. Remarkably, starting from ~14 weeks post-fertilization, fetal primordial germ cells transition to a cell state highly similar to postnatal spermatogonial stem cells. Furthermore, somatic niche specification precedes this transition, consistent with guiding fetal germline development.
INTRODUCTION
As the germline stem cells of the adult testis, spermatogonial stem cells (SSCs) must properly balance self-renewal and differentiation to maintain lifelong spermatogenesis and fertility (Kanatsu-Shinohara and Shinohara, 2013). Adult SSCs are the culmination of a complex developmental process that begins in the embryo, and continues through distinct fetal, juvenile, pubertal, and adult stages. The human germline is specified through the formation of primordial germ cells (PGCs), which occurs in the peri-implantation human embryo around the time of gastrulation (Chen et al., 2019; Tang et al., 2016; Witchi, 1948). Here, studies in the cynomolgus macaque and the porcine embryo (Kobayashi et al., 2017; Sasaki et al., 2016), as well as through differentiation of human embryonic stem cells, suggest that primate and human PGCs originate during amnion specification and also from the posterior end of the nascent primitive streak (Chen et al., 2019; Zheng et al., 2019). Following specification, PGCs migrate through the hindgut, dorsal mesentery and ultimately into the genital ridges at around the 4-5 weeks post-fertilization (Witchi, 1948). At around 6 weeks post-fertilization, the genital ridges differentiate into either the male or female gonads, with SRY being essential for testicular development in males (Hanley et al., 2000; Mamsen et al., 2017; Yang et al., 2019). One of the earliest morphological changes in the male gonad at 6 weeks is the formation of nascent ‘cord-like’ structures comprised of PGCs and Sertoli-lineage cells surrounded by fetal Leydig and interstitial cells. In humans, this basic niche structure persists through fetal and postnatal stages – as the formation of an organized seminiferous tubule does not occur until pubertal stages in humans (Guo et al., 2020; Paniagua and Nistal, 1984).
Within the developing fetal testicular niche, recent genomics profiling and immunofluorescence (IF) imaging approaches have revealed that male germline cells undergo major developmental changes (Gkountela et al., 2013, 2015; Guo et al., 2015; Li et al., 2017; Tang et al., 2015). Notably, the germline transitions from pluripotent-like PGCs migrating to and into the developing gonad, to pluripotent-like and mitotically active PGCs in the gonad [termed fetal germ cells (FGCs) or gonocytes] followed by the transition to ‘mitotically-arrested’ germ cells that repress the pluripotency-like program at/after week 14-18 (Li et al., 2017). Here, a key unanswered question during this stage of germline development involves the relationship between the ‘mitotically-arrested’ germ cells that arise during week 14-18, and postnatal spermatogonial stem cells (SSCs); are prenatal germ cells nearly identical to postnatal SSCs, or are there major additional developmental stages that occur during prenatal stages? Notably, our prior work on the adult testis identified five distinct spermatogonial states (termed States 0-4) accompanying human spermatogonial differentiation, with State 0 identified as the most naïve and undifferentiated state (Guo et al., 2017, 2018, 2020), a result supported by single cell RNA-seq (scRNA-seq) profiling from other groups (Hermann et al., 2018; Li et al., 2017; Shami et al., 2020; Sohni et al., 2019; Wang et al., 2018). Consistent with this notion, State 0 is the predominant SSC state present in the infant testis, and State 0 SSCs express hundreds of state-specific markers including PIWIL4, TSPAN33, MSL3 and EGR4 (Guo et al., 2018). The key markers identified in State 0 SSCs are also expressed in the undifferentiated spermatogonial states identified by others in recent studies, such as the SSC1-B (Sohni et al., 2018) or SPG-1 adult spermatogonia population (Shami et al., 2020), as well as in spermatogonia profiled from human neonates (Sohni et al., 2019) and in undifferentiated spermatogonia from macaques (Shami et al., 2020). Here, we explore whether the previously identified ‘mitotically-arrested’ prenatal germ cells transcriptionally resemble State 0 postnatal spermatogonia, or instead represents a unique precursor that undergoes additional prenatal changes prior to birth.
The testis niche plays an important role in guiding the survival and differentiation of the male germline. In the adult testis, somatic niche cells, including Sertoli, Leydig and myoid cells, provide physical and hormonal support for the successful execution of spermatogenesis from SSCs (Guo et al., 2018). The development of the functional adult testis and its organized tubule-like structure is completed at puberty, during which the final specification and maturation of all somatic niche cells takes place. Our prior work, which utilized single cell RNA-seq (scRNA-seq) to study human testis development during puberty, revealed a common progenitor for Leydig and myoid cells that exists prior to puberty in humans, which is analogous to the somatic progenitor observed in fetal mice (Guo et al., 2020). However, during prenatal life several key issues still remain elusive – such as how the human testicular niche cell lineages are initially specified, whether they have a common progenitor, how the nascent gonad initially forms cords, and how niche cells differentiate further during subsequent fetal developmental stages to arrive at their postnatal states.
To address these questions, we profiled a total of approximately 32,500 unsorted single testicular cells from embryonic, fetal and postnatal samples through 10x Genomics Chromium platform. This unbiased profiling allowed us to examine the specification process in the somatic cell niche, and the development of both the germline and niche cells – which enabled a detailed comparison to the cell types and developmental processes in infant, pubertal and adult testis.
RESULTS
Single-Cell Transcriptomes of Human Embryonic, Fetal and Postnatal Testes
We obtained human testis tissues from three embryonic stages (6-, 7- and 8-weeks post fertilization), three fetal stages (12-, 15- and 16-weeks post fertilization) and one young infant stage (5 months post birth) – for comparisons to prior datasets from older infants, juveniles and adults. To systematically investigate both germ cell and somatic cell development across embryonic and fetal stages, we prepared single cell suspensions from these testicular tissues and performed scRNA-seq using the 10x Genomics platform. For embryonic and fetal samples, we profiled ~5,000 single cells per sample; for the young infant sample, we performed two replicates, and profiled ~2,500 single cells. From a total of approximately 32,500 cells, 30,045 passed standard quality control dataset filters and were retained for downstream analysis (see Methods for details). We obtained approximately 80,000-120,000 reads/cell, which enabled the analysis of approximately 1,800-2,500 genes/cell.
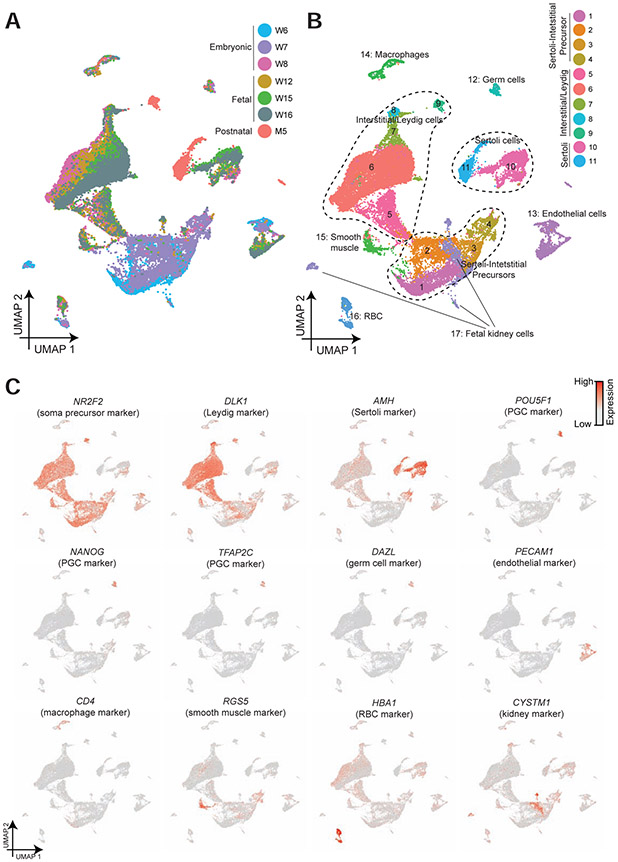
To analyze the dataset, we first performed UMAP (dimension reduction analysis) on the combined datasets using the Seurat package (Fig 1A and Fig S1A) (Butler et al., 2018). Interestingly, we observed a trend in which cells from 6- and 7-weeks cluster closely, and likewise cells from 8-, 12-, 15- and 16-weeks cluster closely (Fig 1A and Fig S1A), while also displaying temporal changes in particular cell types (Fig S1B-C). Further clustering analysis yielded seventeen major clusters or cell types (Fig 1B) that were subsequently annotated using known gene markers (Fig 1C and Fig S2). Clusters 1-4 are testicular niche cells from 6- and 7-week embryos, which uniquely express NR2F2 and TCF21. Clusters 5-9 correspond to somatic cells from the interstitial and Leydig lineage from 8-week and older samples, which express DLK1. Clusters 10-11 are Sertoli lineage cells from 8-week and older samples, which express AMH and SOX9. Cluster 12 includes germ cells from all samples, which express known germ cell markers (e.g. TFAP2C and DAZL) with a subset expressing markers of pluripotency (e.g POU5F1 and NANOG). Clusters 13-17 correspond to endothelial cells (Cluster 13, PECAM1+), macrophages (Cluster 14, CD4+), smooth muscle cells (Cluster 15, RGS5+), red blood cells (Cluster 16, HBA1+) and fetal kidney cells (Cluster 17, CYSTM1+), respectively. We also provide examples of the many additional markers that were used to define these cell types (Fig S2).
Figure 1. Single cell transcriptome profiling and analysis of the human fetal and postnatal testis.
(A) Dimension reduction presentation (via UMAP) of combined single-cell transcriptome data from embryonic, fetal and infant human testes (n=30,045). Each dot represents a single cell and is colored according to its age/donor of origin.
(B) Dimension reduction presentation of combined single-cell transcriptome data from (Fig 1A) labeled with corresponding cell categories and colored according to its cell type identity.
(C) Expression patterns of selected markers projected on the UMAP plot (Fig 1A). For each cell cluster, one cell marker is shown in the main figure accompanied by a gallery of additional markers in Figure S2.
See also Figures S1 and S2.
Emergence of State 0 SSCs as PGCs Exit Mitosis and Repress Pluripotency
Development of the male germline was first examined by parsing out and analyzing the germ cells separately from the somatic cells of the prenatal and postnatal (5 months) testes (Cluster 12 from Fig 1B). To place the embryonic, fetal and postnatal germ cells in a more complete developmental timeline, and enable comparisons, we combined this data with data from infant germ cells (1-year old) and adult spermatogonial states (States 0-4) from our prior published work (Guo et al., 2018), which was also profiled on the 10x Genomics platform. A combination of dimension reduction (via t-SNE) and pseudotime analysis revealed seven defined clusters, and a single pseudo-developmental trajectory that ordered and linked germ cells from the different stages (Fig 2A). Following the order of pseudotime, we observed that the first cluster of germ cells was largely composed of cells from 6- to 12-weeks, as well as a portion of germ cells from week 15 (Fig 2A and Fig S3A). This cluster was termed the ‘embryonic-fetal group’ and their transcriptional identity is consistent with that of PGCs, including expression of TFAP2C, KIT, NANOG, POUF51, SOX17 and others (Fig 2B), consistent with prior scRNA-seq results (Li et al., 2017). The next developmental stage along pseudotime consists of cells from 15- and 16-week fetal samples which group together with cells from the 5-month and 1-year old postnatal samples, and was thus termed the ‘fetal-infant group’ (Fig 2A and Fig S3B). Interestingly, cells from the fetal-infant group lacked expression of PGC markers mentioned above, and instead initiated expression of multiple key State 0-specific markers (PIWIL4, EGR4, MSL3, TSPAN33, others) which were previously defined in the adult, infant and neonatal testis. The subsequent clusters correspond to States 0-4 spermatogonia from adult, which display sequential expression of markers associated with the subsequent developmental states: quiescent/undifferentiated (State 1; GFRA1+), proliferative (States 2-3; MKI67+, TOP2A+) and differentiating (State 4; SYCP3+) (Fig 2A-B and Fig S3C), consistent with our prior work (Guo et al., 2017, 2018). This pseudotime order was further supported by orthogonal Monocle-based pseudotime analysis (Fig S3D-E). A more systematic analysis via heatmap and clustering yielded 2,448 dynamic genes, and provided a format to explore and display the identity, gene ontology (GO) terms, and magnitude of genes that show dynamic expression along this germ cell differentiation timeline (Fig 2C and Table S1). The embryo-fetal group (PGCs) displayed high expression of genes (Cluster 1) associated with signaling, gonad and stem cell development, which were then abruptly repressed between week 15-16, coinciding with the transition to the subsequent fetal-infant group. Here, we also observe upregulation of many transcription and homeobox related genes (Cluster 2) in the fetal-infant group, and the clear upregulation of markers of State 0 spermatogonia. Interestingly, the transition from the fetal-infant group to State 0 spermatogonia is characterized by a deepening and reinforcement of the State 0 gene expression signature, rather than a large number of new genes displaying upregulation. For example, differential gene expression analysis comparing fetal germ cells to adult State 0 spermatogonia identified only 2 genes (ID3 and GAGE12H; 2-fold, p-value <0.05) that display fetal-specific expression (Fig S4G). Consistent with prenatal-postnatal similarity, we observe germ cells from both younger and older infants located in both the fetal-infant and adult State 0 clusters. Taken together, our results revealed that the spermatogonia present in young and older infants (termed State 0) are highly similar to the fetal germline cells that emerge directly after PGCs exit the pluripotent-like state. Given this similarity, we will term these fetal (f) cells ‘State f0’.
Figure 2. Gene expression dynamic during the development of human PGCs to adult spermatogonia.

(A) Focused analysis (t-SNE and pseudotime) of the profiled germ cells (Cluster 12 from Figure 1B) combined with infant germ cells and adult spermatogonia States (from Guo et al., 2018) revealed a single pseudo-developmental trajectory for germ cell development from embryo to adult. Cells are colored based on the ages of the donors.
(B) Expression patterns of known PGC and germ cell markers projected onto the tSNE plot from Figure 2A.
(C) K-means clustering of genes exhibiting differential expression (n=2448) along the germ cell pseudo-developmental trajectory. Each row represents a gene, and each column represents a single cell, with columns/cells placed in pseudotime order defined in Figure 2A. Differential gene expression levels utilize a Z-score as defined by the color key; associated GO terms (using DAVID v6.7) are given on the right of the corresponding gene clusters.
D) Protein co-immunofluorescence for markers of proliferation (MKI67, yellow), pluripotency (NANOG, magenta) and germ cells (DDX4, cyan) in samples from 5 to 19 weeks, and their corresponding quantification.
(E) Protein co-immunofluorescence for germ cell (DDX4) and State 0 (PIWIL4) markers in samples from 8 to 17 weeks.
(F) Quantification of the proportion of PIWIL4+ germ cells (DDX4+) in W12-W16 fetal testis samples. At least 100 cells per replicate and 3 replicates were counted. Each replicate was from a unique donor. Data show the mean ± SEM (One-way ANOVA followed by a Tukey's post-test). Adjusted P-values *=0.0136;**=0.0048;***≤0.0008.
See also Figures S3, S4 and Table S1.
To validate our scRNA-seq profiles at the protein level, we performed IF staining for key markers. First, the proportion of NANOG+ (PGC marker) and also MKI67+ (proliferation marker), decreased from 5- to 19-weeks (Fig 2D and Fig S3G), supporting the notion that the exit from the pluripotent-like state and entry into G0 are temporally linked. We note that for NANOG, the loss of RNA signal based on transcription profiling appears more abrupt than the loss of protein, suggesting heterogeneity in rates of protein loss. Regarding the acquisition of State 0 markers, no PIWIL4 positivity was detected in the 8- and 10-week samples; however, from week 14 onwards, PIWIL4+ cells were clearly detected, specifically in DDX4+ germ cells (Fig 2E-F and Fig S3H). Thus, for the key pluripotency, proliferation and State 0 markers tested, our IF staining results validate our scRNA-seq results.
Network Expression Dynamics during Embryonic, Fetal and Postnatal Germ Cell Development
To define candidate key genes and networks linked to germline developmental stages and transitions, we conducted network analysis. Utilizing Weighted Correlation Network Analysis (WGCNA) (Langfelder and Horvath, 2008), we identified gene-gene interactions that display dynamic expression patterns during PGC differentiation to State f0 spermatogonia. Here, for the PGC upregulated network (termed “PGC network”, Fig S4A and S4D), we identified 2,126 genes and 122,360 interactions, and present the top 11 hub genes (and their interactions). As expected, several genes with known expression in PGCs were present, including POU5F1, NANOG, NANOS3, SOX15 and TFAP2C (Gkountela et al., 2015; Guo et al., 2015; Tang et al., 2015), confirming the robustness of our analysis. In addition, this analysis revealed PHLDA3, PDPN, ITM2C, RNPEP, THY1, and ETV4 as prominent markers in mitotic PGCs, providing candidates for future analysis. For example, PDPN, ITM2C and THY1 all encode cell surface proteins, and PDPN has successfully been used to isolate PGCs differentiated from human pluripotent stem cells (Sasaki et al., 2016). Regarding networks that accompany the differentiation of PGCs into State f0 spermatogonia, a large fraction of the identified genes show relatively broad expression within all subsequent spermatogonia stages, and thus term this network as “Spermatogonia network” (Fig S4B and S4E). We identified 771 genes and 31,557 interactions, and present the top 10 hub genes. Here, roles for EGR4, DDX4, TCF3 and MORC1 in mammalian germ cells are well known. Interestingly, our analysis also indicates several additional factors (e.g. RHOXF1, STK31, CSRP2, ASZ1, SIX1, THRA) worthy of further exploration. For example, RHOXF1 mutations in humans confer male infertility (Borgmann et al., 2016), and MORC1 and ASZ1 both play important roles in protecting the germline genome by repressing transposable element activity (Ma et al., 2009; Pastor et al., 2014) – raising the possibility that they may coordinate with the PIWIL4 factor described below. We also examined the networks that were exclusively expressed in State 0 SSCs (“State 0 network”, Fig S4C and S4F). We identified 190 genes and 8,841 interactions, and present the top 9 hub genes. Among them, EGR4, CAMK2B, MSL3, PLPPR5, APBB1 and PIWIL4 were already identified in prior work (Guo et al., 2018; Sohni et al., 2019), whereas here NRG2, RGS14 and DUSP5 emerge as additional factors. Thus, our analysis confirms the roles of many known factors, and provides a list of key candidate genes with less studied functions in germ cell development, providing multiple avenues for future studies.
Embryonic Specification and Fetal Development of Interstitial and Sertoli Lineages
Our cell type analyses revealed that the human embryonic and fetal testis stages consist primarily of somatic niche cells, including Sertoli cells and interstitial cells (including Leydig cells) (Fig S1B-C). Notably, we did not observe cells that resemble fetal myoid cells by examining myoid markers including ACTA2 and MYH11, which contrasts with observations in mice (Wen et al., 2016). Here, our profiling of early embryonic (week 6-7) testes provided the opportunity to examine Sertoli and interstitial/Leydig cell specification. To this end, we parsed out the fetal somatic niche cells that belong to the interstitial/Leydig and Sertoli lineages, along with the early cells of indeterminate cell type (Clusters 1-8 & 10 from Fig 1B), and performed further analysis. Interestingly, reclustering and subsequent pseudotime analysis revealed one cell cluster at early pseudotime, which transcriptionally bifurcates into two distinct lineages later in pseudotime (Fig 3A). Notably, the early cluster was composed exclusively of cells from week 6-7, whereas cells from week 7 onwards align along two distinct paths (Fig 3A-B and Fig S5A). Examination of known markers suggested the two developmental paths represent Sertoli (left trajectory) or interstitial/Leydig (right trajectory) lineages, respectively (Fig 3C-D), and the existence of a heterogeneous pool of cells at week 6-7 from which both these trajectories originate, raising the possibility of a common somatic progenitor population. Based on our clustering analysis, we then classified the embryonic-fetal interstitial and Sertoli development into seven stages (A-G), beginning with: candidate common somatic progenitors (A) that differentiate into embryonic interstitial/Leydig progenitors (B) which undergo active proliferation (expressing high MKI67). The mostly quiescent embryonic Sertoli progenitors emerge around week 7 (F). The embryonic interstitial progenitors (A) appear to differentiate into fetal interstitial progenitors (C & D) and also fetal Leydig cells (E), and embryonic Sertoli progenitors will differentiate into fetal Sertoli cells (G). Thus, our computational analysis suggests a heterogeneous multipotential progenitor for interstitial cells and Sertoli cells at 6-7 weeks, which then differentiates into Sertoli and interstitial (including Leydig) lineages between week 7-8.
Figure 3. The specification of interstitial and Sertoli lineages.

(A) Focused analysis (UMAP and pseudotime) of the testicular niche cells (Clusters 1-11 from Figure 1B), with cells colored according to ages of the donors.
(B) Deconvolution of the plot in Figure 3A according to the ages of donors.
(C) Focused analysis (in Figure 3A) of the testicular niche cells (Clusters 1-11 from Figure 1B), with cells colored according to ages/donors of origin.
(D) Expression patterns of known progenitor, interstitial/Leydig and Sertoli markers projected onto the plot from Figure 3A.
See also Figure S5.
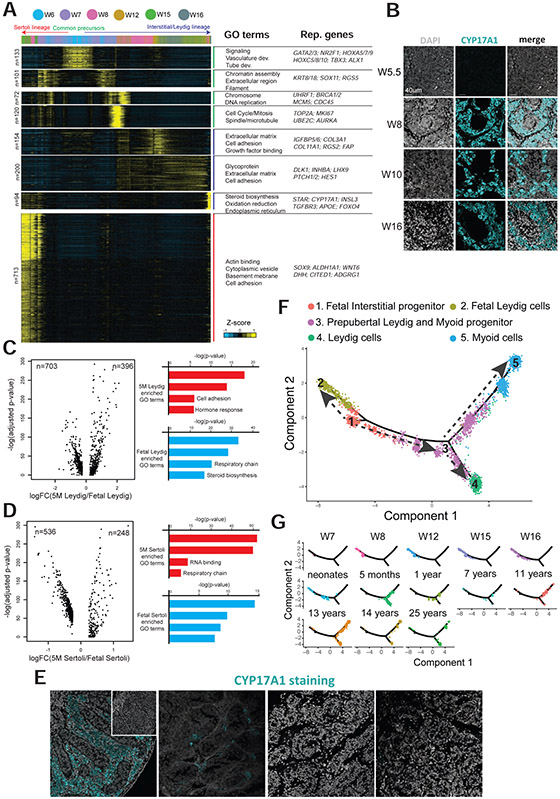
To further define the gene expression programs that accompany male sex determination, we performed gene expression clustering analysis (k-means), to identify the gene groups that display dynamic expression patterns along the pseudotime developmental trajectories (Fig 4A and Table S1). Notably, the candidate progenitors (at weeks 6-7) express multiple notable transcription factors including GATA2, GATA3, NR2F1, HOXA, HOXC factors and others, with enriched gene ontology (GO) terms that include signaling and vasculature development. Particularly, several genes involved in tube development (e.g. TBX3, ALX1 and HOXA5) are specifically expressed in these candidate progenitors, consistent with the initiation of tubule formation to create the testis cords at week 6 (Fig 4A and Fig S5B).
Figure 4. Gene expression dynamic during specification of interstitial and Sertoli lineages.
(A) K-means clustering of genes exhibiting differential expression (n=1578) along interstitial/Leydig and Sertoli specification. Each row represents a gene, and each column represents a single cell, with columns/cells placed in pseudotime order defined in Figure 3A. Differential gene expression levels utilize a Z score as defined by the color key; associated GO terms (using DAVID v6.7) are given on the right of the corresponding gene clusters.
(B) Immunostaining of Leydig marker, CYP17A1 (cyan), in samples from 5 to 16 weeks.
(C-D) Analysis to reveal differentially expressed genes during Leydig (Fig 4C) or Sertoli (Fig 4D) cell differentiation from fetal to infant stages. Violin plot on the left of each panel display the fold change (x-axis) and adjusted p value (y-axis). Right part of each panel represents the enriched gene ontology terms and the associated p-value.
(E) Immunostaining of Leydig marker, CYP17A1 (cyan), in fetal and postnatal testis samples.
(F) Pseudotime trajectory (combined Monocle analysis) of fetal interstitial cells, prepubertal Leydig/myoid cells and the adult Leydig and myoid cells. Cells are colored according to their predicted locations along pseudotime. Neonatal data were from Sohni et al., 2019, 1 year and 25 year old data were from Guo et al., 2018, and 7-14 year old data were from Guo et al., 2020.
(G) Deconvolution of the Monocle pseudotime plot according to ages/donors of origin.
This population of cells then bifurcates into distinct transcriptional programs consistent with embryonic Leydig or Sertoli cell progenitors. Along the Sertoli lineage, expressed genes are associated with chromatin assembly, extracellular region and filament formation. Along the Leydig lineage, cells first express genes related to DNA replication, proliferation and cell cycle, indicating a phase of Leydig lineage amplification, consistent with a much higher number of cells present on the Leydig lineage trajectory at and after 8 weeks compared to the Sertoli lineage (Fig 3B, 4A and Fig S5A). This is followed in the Leydig lineage by upregulation of terms linked to extracellular matrix, cell adhesion and glycoproteins, and components and gene targets associated with both Notch and hedgehog signaling. Consistent with known roles for fetal Leydig cells in androgen production in mice (Shima et al., 2013, 2015), fetal Leydig cells placed at the end of pseudotime express high levels of genes related to steroid biosynthesis (e.g. HSD3B2, Fig 3D) and secretion. Interestingly these cells emerge very early, by week 7, and persist for the remainder of the stages examined – suggesting both an early and persistent role. For the Sertoli lineage, the fetal Sertoli cells express high level of genes associated with structural functions. To validate the temporal features of steroidogenesis genes, we performed IF staining of CYP17A1, a marker for steroidogenesis highly expressed in fetal Leydig cells (Shima et al., 2013) (Fig 4B and Fig S5D). Notably, we found that CYP17A1 is absent in the genital ridge epithelium at 5.5 weeks, whereas robust staining is observed in the interstitial (non-cord) areas in all samples at 7 weeks and older, strongly suggesting that Leydig cell specification occurs around week 7, consistent with our scRNA-seq findings. Furthermore, we observed that at week 8 not all interstitial cells are positive for CYP17A1. Here, we speculate that the fetal CYP17A1-negative interstitial cells may be the interstitial cell population that gives rise to postnatal Leydig and peritubular cells.
Relationship Between Fetal and Infant Leydig and Sertoli Cells
Our datasets provided an opportunity to compare and contrast fetal versus postnatal human Leydig and Sertoli cells. First, we found 396 or 703 genes differentially expressed (upregulated or downregulated, respectively) when comparing fetal to infant Leydig cells, respectively (bimodal test; adjust p-value <0.01; ∣logFC∣>0.25) (Fig 4C). As Leydig cells transition from fetal to infant, genes associated with extracellular matrix, secretion, cell adhesion and hormonal response are upregulated, while those with mitochondrial function and steroid biosynthesis (e.g. CYP17A1, HSD3B2, STAR etc) are downregulated (Fig 4C). Likewise, we found 536 or 248 genes differentially expressed in the infant or fetal Sertoli cells, respectively (Fig 4D). As Sertoli cells transition from fetal to infant, genes associated with translation and respiratory chain are upregulated, and these with endoplasmic reticulum and steroid biosynthesis are downregulated (Fig 4D). To confirm, we performed IF staining of CYP17A1 (Shima et al., 2013), and found that its expression is undetected in the postnatal samples (Fig 4E), suggesting that fetal Leydig cells disappear or differentiate after birth in humans, consistent with discoveries in mice (Svingen and Koopman, 2013). Taken together, our results suggest that human fetal Leydig and Sertoli cells both exhibit expression of steroid biosynthetic genes, whereas this property is downregulated in the postnatal samples tested.
Our prior work based on juvenile human testes showed that Leydig and myoid cells share a common progenitor at pre-pubertal stage (Guo et al., 2020). To gain a deeper understanding of the relationship between the fetal interstitial progenitors and prepubertal Leydig/myoid progenitors, as well as insight into how the common progenitor for the Leydig and myoid lineage is specified from fetal and postnatal precursor cells, we performed additional analysis. Here, we combined in silico the scRNA-seq datasets from fetal interstitial cells (Clusters C,D&E from Fig 3C), neonatal Leydig cells (Sohni et al., 2019), as well as the postnatal and adult Leydig/myoid cells (Guo et al., 2020). Following cell combination, we performed Monocle pseudotime analysis, which aims to provide the developmental order of the analyzed cells through computational prediction (Fig 4F-G). Here, the pseudotime trajectories (depicted by the dashed arrows in Fig 4F) nicely agree with developmental order based on age (Fig 4G), suggesting that fetal interstitial progenitor cells give rise to the postnatal and prepubertal Leydig/myoid progenitor cells. In addition, the analysis suggests that the fetal Leydig cells, which originate from the fetal interstitial progenitors, are absent in the postnatal and infant stages – a result confirmed by our immunostaining data (Fig 4E).
Key Factors Correlated with Embryonic Specification of Interstitial and Sertoli Lineages
Whereas testicular niche cells from 8-16 weeks expressed transcription factors characteristic of advanced interstitial or Sertoli cell lineages, the cells from the 6-week gonads lack these late markers, which initially emerge at week 7 (Fig 3A-C). In order to better understand the genes expressed during the time of somatic specification, we parsed out the 6- and 7-week cells (from Fig 3A), and performed more detailed analysis. Here, principal component analysis (PCA) of the 6- and 7-week cells revealed that a large portion of the cells did not display markers distinctive of either interstitial or Sertoli cells (Fig 5A), suggesting a heterogeneous population in which the Sertoli and Leydig/Interstitial precursors are emerging. An orthogonal analysis via Monocle also confirmed similar patterns and properties (Fig S6C-E). Based on the gene expression patterns (Fig 5B), we can assign the cells at the bottom as the embryonic interstitial/Leydig lineage (expressing DLK1 and TCF21), and the cells at the top right as the embryonic Sertoli lineage (expressing SRY, DMRT1, SOX9, AMH, others).
Figure 5. Key transcription factors involving the specification of interstitial and Sertoli cells.
(A) Principal component analysis of testicular niche progenitors from 6- and 7 weeks cells, revealing the existence of interstitial/Leydig and Sertoli lineage bifurcation.
(B) Expression patterns of key factors that show specific patterns during the progenitor differentiation.
(C) Staining of transcription factors GATA3 (cyan) in the 5- and 8-week samples.
(D) Staining of transcription factors GATA4 (cyan) in the 6- and 17-week samples.
(E) Co-staining of Sertoli (DMRT1, magenta) and germ cell (DDX4, cyan) markers in the 5- and 8-week samples.
(F) Co-staining of two Sertoli cell markers, DMRT1 and SOX9, in the 5.5- to 17-week samples.
See also Figure S6.
Next, we sought to identify candidate key transcription factors that might participate in initial somatic lineage specification (Fig 5B). Interestingly, a set of GATA-family factors displayed sequential and largely non-overlapping patterns: GATA3 expression was earliest, at the top and left edge of the PCA plot (mostly 7-week), GATA2 started to express somewhat later, and GATA4 was expressed in a later population that was progressing toward the Sertoli lineage. Many other factors also display sequential expression. For example, NR2F1, MAFB and TCF21 show relatively early expression (similar to GATA2), while TCF21 expression persists through the development of the Leydig lineage, but not the Sertoli lineage. Notably, both ARX and NR0B1 are expressed at the bifurcation stage. For the Sertoli lineage, these early markers cease expression at lineage specification, followed by expression of SRY and DMRT1 as the earliest markers of the lineage, followed by SOX9.
Finally, we performed extensive IF to validate our genomics findings. First, we observed GATA3 throughout the genital ridge epithelium at week 5, which became restricted to a subpopulation of interstitial cells at week 6-7 and by week 8 GATA3 protein becomes undetectable (Fig 5C). In addition, GATA4 expression is evident both inside and outside the cords from week 6 and onward (Fig 5D and Fig S5B). To evaluate Sertoli lineage specification, we stained for DMRT1 alongside either a germ cell marker (DDX4) or an additional Sertoli cell marker (SOX9) (Fig 5E-F). As expected, DMRT1 and SOX9 protein were undetectable in the GATA3/GATA2 positive genital ridge epithelium containing DDX4 positive PGCs at week 5 (Fig 5E). However, by 8 weeks (after cord formation), DMRT1+ and SOX9+ Sertoli cells are identified (Fig 5F). Taken together, our IF staining results confirm key markers discovered through our genomics approaches, and provide additional insights into the physiology of testis cord development in the embryonic and fetal stages.
DISCUSSION
PGCs are specified in the early embryo, followed by migration to the genital ridge (Chen et al., 2019; Tang et al., 2016; Witchi, 1948). The genital ridge then undergoes exquisite developmental programming to form the somatic cells of the testicular niche that support the survival and differentiation of the male germline during fetal life. Although prior studies from mice provide rich knowledge on the formation and lineage specification in the embryonic testis (reviewed in Svingen and Koopman, 2013), our understanding of human embryonic and fetal testis development has been much less studied, particularly with regard to the specification of the somatic lineages. Here, through the application of single cell sequencing of unselected testicular cells, together with IF staining, we provide a detailed molecular overview of human fetal testis development, to help delineate the temporal molecular changes involved in human embryonic and fetal testis development and further differentiation.
One critical question we aimed to address is the transition of PGCs into spermatogonia - specifically the transcriptional relationship of differentiating male human PGCs during fetal life to postnatal State 0 SSCs, which have been identified as the most undifferentiated male germline stem cells in human infants and adults(Guo et al., 2018; Sohni et al., 2019), as well as primates (Shami et al., 2020). Combined with prior work (Guo et al., 2017, 2018, 2020; Sohni et al., 2019), our current work provides an evidence-based and detailed model for human germline development that spans embryonic, fetal, infant, pubertal and adult stages (Fig 6A). During 6-12 weeks post fertilization, as the male somatic cell linages are being specified, human male PGCs express high levels of transcription factors associated with pluripotency (e.g. POU5F1 and NANOG), together with classic well-characterized PGC transcription factors (e.g. SOX17 and TFAP2C) and are proliferative. At 14 weeks, a subpopulation of PGCs initiates repression of the pluripotency-like program, and extinguishes expression of the early PGC genes (Li et al., 2017), while simultaneously turning on the State f0 spermatogonia programs (e.g. PIWIL4, MSL3, EGR4, TSPAN33). These State f0 spermatogonia are transcriptionally highly similar to the State 0 spermatogonia, and are found from fetal stages through infants within the seminiferous cords. Interestingly, when we examine the expression patterns of many key PGC or State f0 markers in a prior FGC dataset (Li et al., 2017) (Fig S4H), we found that the ‘mitotically-arrested’ FGCs exhibit specific and high expression of State 0 genes (e.g. PIWIL4, EGR4, MSL3 and TSPAN33) and low expression of PGC genes (e.g. POU5F1, NANOG, TFAP2C and SOX17). This observation strongly suggests that the previously defined ‘mitotically-arrested’ FGCs (Li et al., 2017), which also emerge at ~14 weeks post fertilization (Fig S4I), are likely the same cells as the State f0 defined in our study. Here, our prior derivation of infant State 0 cellular identity, and their demonstrated similarity to the fetal population in the current study defines a critical linkage: PGCs differentiate and transition into State f0 spermatogonia, and reinforce their State 0-like transcriptome as they transition between fetal germ cells to postnatal germ cells. By 5 months, all germline cells display a State 0 spermatogonial transcriptome, and cells with a PGC transcriptome are below the limit of detection. Consistent with our observations at 5 months and in infants, State 0 markers are also expressed in human neonatal germ cells (Sohni et al., 2019). Taken together, we have revealed that State 0-like spermatogonia originate from PGCs at ~week 14-16 of fetal life, and persist through all prenatal and postnatal developmental stages, to provide a pool of undifferentiated spermatogonia in adults available for niche-guided transitions to more differentiated spermatogonial states, and ultimately gametogenesis (Fig 6A).
Figure 6. Proposed models for human germline development and somatic niche cell specification during prenatal and postnatal stages.
(A) Schematic summarizing the combined gene expression programs and cellular events accompanying human PGCs differentiation into adult SSCs.
(B) The timeline and proposed model for human testicular somatic niche cell development at embryonic, fetal and postnatal stages. Specification of a unique progenitor cell population towards Sertoli and interstitial/Leydig lineages begins at around 7 weeks post fertilization, when the cord formation occurs.
Prior work in mouse models has revealed several factors and pathways that play important roles in lineage specification and progression of testicular somatic cells in mice (Liu et al., 2016; Svingen and Koopman, 2013; Yao et al., 2002). Recently, scRNA-seq has proven to be a powerful tool to study embryonic and neonatal mouse testis development (Stévant et al., 2019; Tan et al., 2020). Here, our work demonstrates that several key factors in early somatic lineages (e.g. WT1, NR2F1, SOX9, SRY, DMRT1) are shared between humans and mice. Furthermore, through our systematic examination of prenatal human testes via single cell profiling and IF staining, we provide many additional candidate factors for future characterization, and reveal multiple human-mouse differences. For example, through IF staining of the genital ridge epithelium we find no evidence of Sertoli cell or Leydig cell identity prior to 6 weeks post-fertilization. Then, starting at week 6, our unbiased/unselected single cell transcriptome profiling identified rare fetal Leydig- and Sertoli-like cells. We also identified in pseudotime a large, closely related population of cells that are heterogeneously positive for developmental transcription factors, notably NR2F1, GATA3, and GATA4 RNA. GATA3 protein analysis demonstrated that GATA3 is uniformly expressed by the genital ridge epithelium at week 5 post-fertilization before specification of Sertoli and Leydig cells. Notably, at week 6 when cord formation initiates, GATA3 expression is restricted to a subpopulation of cells in the interstitium. In counter distinction, GATA4 expression is evident and broad at 6-7 weeks post fertilization, and remains detectable in 17 weeks post fertilization. In the mouse embryo, GATA4 is known to be critical for genital ridge formation, and in the absence of GATA4, the bipotential gonads do not form (Hu et al., 2013). Given that GATA3 is expressed in the genital ridge epithelium prior to GATA4, we speculate that GATA3 may have a role in specifying the genital ridge in humans, whereas GATA4 instead may be involved in maintaining the somatic cell lineages after 6 weeks post fertilization when GATA3 expression is reduced. In the mouse, NR5A1 (also called SF1) is another major transcription factor required for specifying the genital ridge epithelium (Hatano et al., 1996; Luo et al., 1994). However, we do not observe clear expression of NR5A1 in the GATA3 positive human progenitors, providing a second example where formation of the genital ridge epithelium in human embryos appears different from the mouse (Fig S5B). Analysis at the week 6-7 time point suggests that Leydig and Sertoli cell specification occur at or near the same developmental time. Indeed, our IF studies at week 7 show both Sertoli cells in cords and Leydig cells outside of the cords. This result represents a major difference from the mouse, where Sertoli cells are specified first, and then Leydig cells are subsequently specified (Svingen and Koopman, 2013). Considering that the size of the fetal human testis is proportionally much larger than that of mice, the human testis progenitors may commit relatively early in development, followed by waves of proliferation, which may partly explain the developmental differences.
In addition to being specified at an equivalent developmental stage, we also discovered that the 6- and 7- week somatic niche progenitors expressed markers consistent with their ability to differentiate into interstitial/Leydig and Sertoli lineages by transiently expressing (in a small subset of cells) key transcription factors including ARX, NR0B1 or SRY. This identity is further reinforced at 8 weeks, when all cells are distinguishable as interstitial/Leydig or Sertoli lineage cells. Notably, the establishment of the male somatic cell lineages in the embryonic testis occurs almost two months before the PGCs begin differentiating into State f0 (at 14-18 weeks). In contrast, in mice there is only a two-day delay in the timing of the male niche cell differentiation (at day 12) to the initiation of mouse PGC differentiation into prospermatogonia (at embryonic day 14 )(Saitou and Yamaji, 2012; Svingen and Koopman, 2013; Western et al., 2008). The purpose of this two-month delay in which human PGCs are shielded from initiating differentiation into State f0 spermatogonia in the seminiferous cord niche may be related to the need to increase the number of male germ cells through proliferation, given that these cells are MKI67 positive, before initiation of State f0 differentiation and male-specific epigenetic reprogramming (Fig 6B).
The testis produces gametes in adult males through continuous niche-guided differentiation of SSCs, and a deep understanding of this biology is needed to improve male reproductive health. Here, our work provides major insights in defining the timing and strategy of human testis formation and its development prior to and after birth. Notably, the State f0 germ cells that emerge at ~15 weeks during fetal life, display remarkable similarities to the infant and adult State 0 cells, and thus allow us to link and depict the complete developmental progression of PGCs to adult State 0 cells. Furthermore, we provide a detailed molecular characterization of a common somatic progenitor pool and its amplification and transition to testicular niche cells, as well as initial insights into testicular cord formation and possible roles in guiding germ cell development. These results should provide a foundation for future hypothesis-driven research, and could also help guide the reconstruction and study of the human early testis in vitro.
Limitations of the Study
Our current work primarily focuses on transcriptomic profiling with additional protein validation of key markers; however, RNA expression does not always linearly reflect protein abundance. For example, as PGCs transition to State f0 SSCs (at/after 14 weeks), transcript levels for key PGC markers (e.g. DDX4, NANOG and MKI67) fall abruptly, whereas their protein levels reduce gradually, suggesting that complex post transcriptional mechanisms exist to modulate protein levels. Furthermore, given the variations that may exist among different embryos, and the challenges in accurately assessing the embryo ages, future studies with additional samples may refine temporal aspects of our findings, and may also reveal additional details regarding developmental processes and transitions. For example, although we know the transition from PGC to State f0 begins at ~week 14, there could be heterogeneity and individual variation regarding the time at which this conversion is complete. Finally, our identification of a common human fetal somatic cell precursor was based on transcriptional profiling and computational prediction. Here, further studies that utilize lineage tracing approaches in non-human primate models may provide a more definitive test of our model.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Bradley R. Cairns (brad.cairns@hci.utah.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All software tools can be found online (see Key Resources Table). The accession number for all sequencing data reported in this paper is GEO: GSE143356 and GEO:GSE161617.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-PIWIL4 Dilution: 1:200 |
Thermo Fisher Scientific | Cat#: PA5-3144 RRID: AB_2548922 |
| Mouse monoclonal (CloneB56) anti-MKI67 Dilution: 1:200 |
BD Biosciences | Cat#: 556003 RRID: AB_396287 |
| Goat polyclonal anti-DDX4 Dilution: 1:100 |
R&D Systems | Cat#: AF2030 RRID: AB_2277369 |
| Rabbit monoclonal (D73G4) anti-NANOG Dilution: 1:100 |
Cell Signaling Technology | Cat#:4903 RRID: AB_10559205 |
| Mouse monoclonal anti-CYP17A1 Dilution: 1:200 |
Santa Cruz Biotechnology | Cat#: SC-374244 RRID: AB_10988393 |
| Mouse monoclonal (1A12-1D9) anti-GATA3 Dilution: 1:100 |
Thermo Fisher Scientific | Cat#: MA1028 RRID: AB_2536713 |
| Mouse monoclonal (G-4) anti-GATA4 Dilution: 1:100 |
Santa Cruz Biotechnology | Cat#: SC-25310 RRID: AB_627667 |
| Mouse monoclonal anti-DMRT1 Dilution: 1:100 |
Santa Cruz Biotechnology | Cat#: SC-377167 |
| Rabbit polyclonal anti-SOX9 Dilution: 1:200 |
Millipore | Cat#: AB5535 RRID: AB_2239761 |
| AF488 goat-anti mouse IgG2a | Invitrogen | Cat#: A21131 RRID: AB_2535771 |
| AF594 donkey-anti-mouse IgG | Invitrogen | Cat#: A21203 RRID: AB_2535789 |
| AF594 goat-anti-mouse IgG1 | Invitrogen | Cat#: A21125 RRID: AB_2535767 |
| AF594 donkey-anti-rabbit IgG | Jackson ImmunoResearch | Cat#: 711-585-152 RRID: AB_2340621 |
| AF647 donkey-anti-goat IgG | Invitrogen | Cat#: A21447 RRID: AB_2535864 |
| Biological Samples | ||
| Human testis samples from postnatal donors | DonorConnect | NA |
| Human testis samples from embryonic and fetal stages | University of Washington-Birth Defects Research Lab | NA |
| Human testis samples from Jan’s lab | Karolinska Institutet | NA |
| Deposited Data | ||
| Single cell RNA-seq for embryonic and fetal human testes | This paper | GEO: GSE143356 |
| Single cell RNA-seq for postnatal testes | This paper | GEO: GSE161617 |
| Software and Algorithms | ||
| Seurat (2.3.4) | Butler et al., 2018 | https://satijalab.org/seurat/ |
| Monocle (2.10.1) | (Qiu et al., 2017) | http://cole-trapnell-lab.github.io/monocle-release/ |
| GO (David 6.7) | Huang et al., 2009 | https://david-d.ncifcrf.gov |
| Cell Ranger (2.2.0) | NA | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Cluster 3.0 | NA | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| WGCNA (1.68) | (Langfelder and Horvath, 2008) | https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/ |
| Cytoscape (3.7.2) | (Otasek et al., 2019) | https://cytoscape.org |
| Other | ||
| Single cell RNA-seq for infant and adult human testes | Guo et al., 2018 | GEO: GSE120508 |
| Single cell RNA-seq for neonatal human testes | Sohni et al., 2019 | GEO: GSE124263 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Testicular Tissues
Prenatal male gonads from 6 to 16 weeks post-fertilization were obtained from three collaborating laboratories at University of Washington Birth Defects Research Laboratory (BDRL), University of Tubingen and Karolinska Institutet. At BRDL, the prenatal gonads were obtained with regulatory oversight from the University of Washington IRB approved Human Subjects protocol, combined with a Certificate of Confidentiality from the Federal Government. The research project was also approved by the research ethics committee of the University of Tubingen. All consented material was donated anonymously and carried no personal identifiers. Human first trimester tissue was collected after elective surgical terminations with maternal written informed consent. The Regional Human Ethics Committee, Stockholm, Sweden, approved the collection (Dnr 2007/1477-31 with complementary permissions 2011/1101-32 and 2013/564-32. The ethical approval to perform the gonadal studies: Dnr 2013/457-31/4). Developmental age was documented by BDRL and University of Tubingen as days post fertilization using a combination of prenatal intakes and Carnegie staging. Developmental age was documented by Karolinska Institutet as days post fertilization by the examination of anatomical landmarks such as nervous system, limb, eye and gonadal development according to the atlas of England. Formalin fixed and paraffin embedded adult testis from biobank samples without underlaying testicular pathologies was obtained at the Department of Pathology at the Karolinska Institutet, and Karolinska University Hospital (ethical approval: Dnr 2014/267-31/4).
Postnatal human testicular sample (5 months old) was obtained through the University of Utah Andrology laboratory and DonorConnect. This sample was removed from deceased individuals who consented to organ donation for transplantation and research.
METHOD DETAILS
Sample Transportation and Storage
The prenatal samples collected at BDRL used for single cell transcriptome profiling were shipped overnight in HBSS with an ice pack for immediate processing in Los Angeles. From University of Tubingen samples were delivered to UCLA within 24-48 hours after the procedure. The postnatal whole testis was transported to the research laboratory on ice in saline and processed within 1 hour of removal by surgery. Around 90% of each testis was divided into smaller portions (~500 mg – 1g each) using scissors and directly transferred into cryovials (Corning cat # 403659) in DMEM medium (Life Technologies cat # 11995073) containing 10% DMSO (Sigma-Aldrich cat # D8779), 15% fetal bovine serum (FBS) (Gibco cat # 10082147) and cryopreserved in Mr. Frosty container (Thermo Fisher Scientific cat #5100-0001) at a controlled slow rate, and stored at −80°C for overnight. Cryovials were transferred to liquid nitrogen for long-term storage.
Human Testis Sample Preparation for Single Cell RNA Sequencing
Prenatal tissues were processed within 24-48 hours after termination. Upon arrival to UCLA tissues were gently washed with PBS and placed in dissociation buffer containing collagenase IV 10mg/ml (Life Technologies #17104-019), Dispase II 250 ug/ml (Life Technologies #17105041), DNAse I 1:1000 (Sigma 4716728001), 10% FBS (Life Technologies 10099141) in 1x PBS. After every 5 minutes tissues were gently pipetted with P1000 pipette against the bottom of Eppendorf tube. This process was repeated 3 times for a total of 15 minutes. Afterwards, cells were centrifuged for 5 minutes at 500g and pellet was resuspended in 1x PBS with 0.04% BSA and strained through 40μm strainer and counted using automated cell counter (Thermo Fisher, Countess II). The cell concentration was adjusted to 800-1200 cells per microliter and immediately used for scRNA-seq. For postnatal tissues, 1 cryovial of tissue was thawed quickly, which was then washed twice with PBS, and subject to digestion as described previously (Guo et al., 2018). Tissues were washed twice in 1 x PBS and minced into small pieces for better digestion outcome. Tissues were then treated with trypsin/ethylenediaminetetraacetic acid (EDTA; Invitrogen cat # 25300054) for 20-25 min and collagenase type IV (Sigma Aldrich cat # C5138-500MG) at 37°C. Single testicular cells were obtained by filtering through 70 μm (Fisher Scientific cat # 08-771-2) and 40 μm (Fisher Scientific cat # 08-771-1) strainers. The cells were pelleted by centrifugation at 600 g for 15 min and washed with PBS twice. Cell number was counted using a hemocytometer, and the cells were then resuspended in PBS + 0.4% BSA (Thermo Fisher Scientific cat # AM2616) at a concentration of ~1,000 cells/uL ready for single-cell sequencing.
Single Cell RNA-seq Performance, Library Preparation and Sequencing
We targeted to capture ~6,000-7,000 cells. The prenatal sequencing was conducted in UCLA, and the postnatal sequencing was conducted at University of Utah. Briefly, cells were diluted following manufacturer’s instructions, and 33.8 μL of total mixed buffer together with cells were loaded into 10x Chromium Controller using the Chromium Single Cell 3’ v3 reagents. The sequencing libraries were prepared following the manufacturer’s instructions, using 13 cycles of cDNA amplification, followed by an input of ~100 ng of cDNA for library amplification using 12 cycles. The resulting libraries were then sequenced on a 2 X 150 cycle paired-end run on an Illumina Novaseq 6000 instruments.
Processing of Single Cell RNA-seq Data
Raw data were demultiplexed using mkfastq application (Cell Ranger v2.2.0) to make Fastq files. Fastq files were then run with count application (Cell Ranger v2.2.0) using default settings, which performs alignment (using STAR aligner), filtering and UMI counting. The UMI count tables were used for further analysis.
Immunostaining of testicular tissues
Intact testes were fixed in 4% PFA at room temperature for 2 hours on a platform rocker. Tissues were washed 3 times with PBS for 10 minutes each wash then placed into paraffin blocks (Histogel,Thermo Scientific HG4000012) for sectioning onto slides. Sections were deparaffinized and rehydrated in a Xylene then ethanol series (100%, 95%, 70%, 50%, water) respectively. Antigen retrieval was performed in either Tris-EDTA solution (pH 9.0) or Sodium Citrate Solution (pH 6.0) in a hot water bath (95°C) for 40 minutes. Sections were washed in PBS, 0.2% Tween-20 (PBS-T) 3 times, 5 minutes each then permeabilized in PBS, 0.05% Trition X-100 for 20 minutes. Sections were blocked with blocking solution (10% Normal Donkey Serum (NDS), PBS-T) for 30 minutes at room temperature in a humid chamber. Primary Antibodies were diluted in 2.5% NDS, PBS-T at the appropriate dilutions (see key resource table) and incubated overnight at 4°C in a humid chamber. After 3 washes in PBS-T (5 minutes each) secondary antibodies were added and allowed to incubate at room temperature for 1 hour in a humid chamber. After 3 washes in PBS-T, DAPI was added to the sections for approximately 5 minutes, then washed 3 times 5 minutes each in PBS-T. Prolong Gold antifade mountant (Invitrogen P10144) was added to the sections. Coverslips were placed onto slides then sealed with nail polish. Slides were allowed to cure overnight, in the dark, at room temperature then subsequently stored at 4°C until ready to image. For sections stained with PIWIL4 antibody, the blocking buffer used was Superblock blocking buffer (Thermo Scientific 37580). In addition, the SignalBoost Immunoreaction Enhancer Kit (Millipore 407207) was used to dilute primary and secondary antibodies for experiments involving PIWIL4 antibody.
Microscopy
A Zeiss LSM 880 with Airyscan controlled by the Zen Black software, equipped with the Plan-Apochromat 20×/0.8 NA and the Plan-Apochromat 63×/1.4 NA M27 oil immersion objective, was used to acquire confocal images. Saved CZI files were converted to Imaris format files (.ims) using the Imaris File converter (Bitplane), then processed using the image analysis software IMARIS 9.3 (Bitplane). An Olympus BX-61 light microscope was used to examine Hematoxylin and Eosin (H&E) stained slides. Stitched images were built using the ImageJ2(NIH) Grid/Collection Stitching plugin.
QUANTIFICATIONS AND STATISTICAL ANALYSIS
The Seurat program (http://satijalab.org/seurat/, R package, v.2.3.4) was used as a first analytical package. To start with, UMI count tables from both replicates from all four juvenile donors were loaded into R using Read10X function, and Seurat objects were built from each experiment. Each experiment was filtered and normalized with default settings. Specifically, cells were retained only if they contained > 500 expressed genes and had < 25% reads mapped to mitochondrial genome. t-SNE and clustering analysis were first run on each replicate, which resulted in similar t-SNE map. Data matrices from different donors and replicates were then combined with the previously published infant and adult data (Guo et al 2018). Next, cells were normalized to the total UMI read counts, as instructed in the tutorial (http://satijalab.org/seurat/). t-SNE and clustering analyses were performed on the combined data using the top 6,000 highly variable genes and 1-30 PCs, which showed the most significant p-values.
Detailed pseudotime for different cell types were performed using the Monocle package (v2.10.1) following the default settings. After pseudotime coordinates/order were determined, gene clustering analysis was performed to establish the accuracy of pseudotime ordering. Here, cells (in columns) were ordered by their pseudotime, and genes (in rows) were clustered by k-means clustering using Cluster 3.0. Different k-mean numbers were performed to reach the optimal clustering number. Cell cycle analysis was performed using scran program (https://bioconductor.org/packages/3.7/bioc/vignettes/scran/inst/doc/scran.html, R Package; v1.6.5).
Weighted correlation network analysis
Hub genes in PGC, spermatogonia and State 0 were found by WGCNA (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/, R package, v1.68). When finding hub genes in PGC and spermatogonia, gene expression data of 40 cells from PGC and State 0 respectively were randomly extracted from the UMI count tables of scRNA-seq data. Genes were filtered by selecting those genes expressed in more than 20 cells since scRNA-seq data had a high drop-out rate and low expression genes may represent noise. Then the counts were normalized by total reads (x*100000/total reads) and then log-transformed (log2(x+1)). Afterwards, one-step network construction and module detection were performed. In this step, we chose parameters including signed hybrid network type, Pearson correlation method and the default soft-threshold power β to reach the scale-free network topology. To identify the modules that were significantly correlated with PGC or spermatogonia, biweight mid-correlation (robustY = FALSE) was used. The quality of the modules was checked by the strong correlation between module eigengenes and traits of interest as well as the strong correlation between gene module membership and gene-trait correlation. Finally, hub genes inside those modules were selected from the top 40 genes with the highest intramodular connectivity (sum of in-module edge weights). Specifically, in order to find hub genes in State 0 rather than spermatogonia, we added gene expression data of 40 cells from State1 to rule out the genes expressing broadly in States 0-4 and performed the same analysis to determine the modules that were significantly correlated with State 0. Ten hub genes were selected by the same standard. Finally, the networks were visualized by Cytoscape Software 3.7.2.
Supplementary Material
Figure S1. Single cell transcriptome profiling and analysis of the human fetal and postnatal testis. Related to Figure 1.
Figure S2. Expression patterns of additional markers projected on the UMAP plot. Related to Figure 1.
Figure S3. Transition of human PGCs to State f0. Related to Figure 2.
Figure S4. Network expression dynamic during fetal and postnatal germ cell development. Related to Figure 2.
Figure S5. Somatic niche cell specification at embryonic and fetal stages. Related to Figures 3, 4 and 5.
Figure S6. Proposed models for human germline development and somatic niche cell specification during embryonic, fetal and postnatal stages. Related to Figures 4 and 5.
Table S1: Genes that display dynamic expression along germ cell pseudo-developmental trajectory shown in Fig 2C. Related to Figure 2.
Table S2: Genes that display dynamic expression during human testicular somatic cell specification as shown in Fig 4A. Related to Figure 4.
Highlights:
A transcriptional single cell atlas of the fetal and postnatal human testes
Somatic niche cell types derive from a common progenitor ~7 weeks after fertilization
PGCs transition directly into fetal State 0-like cells (State f0) starting at week 14
Fetal somatic niche cell specification precedes the PGC-to-State f0 transition
ACKNOWLEDGEMENTS
We are grateful to the donors and their families, who made this work possible. We thank Brian Dalley and Opal Allen in the HCI High-Throughput Genomics Shared Resource for sequencing expertise, Chris Stubben and Tim Parnell in the HCI Bioinformatics Share Resource for bioinformatics assistance, and DonorConnect staff for family consents and postnatal sample handling. Financial support was from Howard Hughes Medical Institute (HHMI) to B.R.C. and NCI P30CA042014 to Huntsman Cancer Institute core facilities. We thank the Technology Center for Genomics and Bioinformatics at the UCLA Johnson Comprehensive Cancer Center (JCCC) and the Next Generation Sequencing core at BSCRC for help with genomics approaches, and the Translational Pathology Core Laboratory for help with histology. We thank microscopy cores at the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Center (BSCRC) for help with imaging. Financial support was from the NIH to A.T.C. (R01HD079546). K.P. was supported by the BSCRC at UCLA, the David Geffen School of Medicine at UCLA, the UCLA JCCC, the NIH (R01HD098387, P01 GM099134), and a Faculty Scholar grant from HHMI. T.C. was supported by Boehringer Ingelheim PhD fellowship. E.S. was supported by the BSCRC Post-doctoral Fellowship at UCLA. We acknowledge Ian Glass from the University of Washington Birth Defects laboratory for fetal tissue, supported by Human fetal tissue research grant 5R24HD000836-53. J.B.S. was supported by the Swedish Childhood Cancer Foundation (PR2019-0123; TJ2020-0026), Magnus Bergvalls Foundation, Birgitta and Carl-Axel Rydbeck’s Research grant for Pediatric Research and the Swedish Research Council (2018-03094).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Borgmann J, Tüttelmann F, Dworniczak B, Röpke A, Song H-W, Kliesch S, Wilkinson MF, Laurentino S, and Gromoll J (2016). The human RHOX gene cluster: target genes and functional analysis of gene variants in infertile men. Hum Mol Genet 25, 4898–4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, et al. (2019). Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Reports 29, 4568–4582.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Li Z, Vincent JJ, Zhang KX, Chen A, Pellegrini M, and Clark AT (2013). The ontogeny of cKIT+ human primordial germ cells: A resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao W-W, Hargan-Calvopiña J, Chen P-Y, and Clark AT (2015). DNA Demethylation Dynamics in the Human Prenatal Germline. Cell 161, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, et al. (2015). The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 161, 1437–1452. [DOI] [PubMed] [Google Scholar]
- Guo J, Grow EJ, Yi C, Mlcochova H, Maher GJ, Lindskog C, Murphy PJ, Wike CL, Carrell DT, Goriely A, et al. (2017). Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 21, 533–546.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, Guo Y, Takei Y, Yun J, Cai L, et al. (2018). The adult human testis transcriptional cell atlas. Cell Research 28, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Nie X, Giebler M, Mlcochova H, Wang Y, Grow EJ, Kim R, Tharmalingam M, Matilionyte G, Lindskog C, et al. (2020). The Dynamic Transcriptional Cell Atlas of Testis Development during Human Puberty. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley NA, Hagan DM, Clement-Jones M, Ball SG, Strachan T, Salas-Cortés L, McElreavey K, Lindsay S, Robson S, Bullen P, et al. (2000). SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mechanisms of Development 91, 403–407. [DOI] [PubMed] [Google Scholar]
- Hatano O, Takakusu A, Nomura M, and Morohashi K (1996). Identical origin of adrenal cortex and gonad revealed by expression profiles of Ad4BP/SF-1. Genes to Cells 1, 663–671. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen I-C, Gildersleeve H, Lehle JD, Mayo M, Westernströer B, et al. (2018). The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Reports 25, 1650–1667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y-C, Okumura LM, and Page DC (2013). Gata4 Is Required for Formation of the Genital Ridge in Mice. PLOS Genetics 9, e1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, and Shinohara T (2013). Spermatogonial Stem Cell Self-Renewal and Development. Annual Review of Cell and Developmental Biology 29, 163–187. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, Sybirna A, Dietmann S, Contreras DA, Webb R, et al. (2017). Principles of early human development and germ cell program from conserved model systems. Nature 546, 416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, et al. (2017). Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell 20, 858–873.e4. [DOI] [PubMed] [Google Scholar]
- Liu C, Rodriguez K, and Yao HH-C (2016). Mapping lineage progression of somatic progenitor cells in the mouse fetal testis. Development 143, 3700–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, and Parker KL (1994). A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77, 481–490. [DOI] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. (2009). GASZ Is Essential for Male Meiosis and Suppression of Retrotransposon Expression in the Male Germline. PLoS Genet 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamsen LS, Ernst EH, Borup R, Larsen A, Olesen RH, Ernst E, Anderson RA, Kristensen SG, and Andersen CY (2017). Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Scientific Reports 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D, Morris JH, Bouças J, Pico AR, and Demchak B (2019). Cytoscape Automation: empowering workflow-based network analysis. Genome Biol 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, and Nistal M (1984). Morphological and histometric study of human spermatogonia from birth to the onset of puberty. J Anat 139, 535–552. [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc’his D, et al. (2014). MORC1 represses transposable elements in the mouse male germline. Nature Communications 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nature Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, and Yamaji M (2012). Primordial Germ Cells in Mice. Cold Spring Harb Perspect Biol 4, a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. (2016). The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Developmental Cell 39, 169–185. [DOI] [PubMed] [Google Scholar]
- Shami AN, Zheng X, Munyoki SK, Ma Q, Manske GL, Green CD, Sukhwani M, Orwig KE, Li JZ, and Hammoud SS (2020). Single-Cell RNA Sequencing of Human, Macaque, and Mouse Testes Uncovers Conserved and Divergent Features of Mammalian Spermatogenesis. Dev. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Miyabayashi K, Haraguchi S, Arakawa T, Otake H, Baba T, Matsuzaki S, Shishido Y, Akiyama H, Tachibana T, et al. (2013). Contribution of Leydig and Sertoli Cells to Testosterone Production in Mouse Fetal Testes. Mol Endocrinol 27, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Matsuzaki S, Miyabayashi K, Otake H, Baba T, Kato S, Huhtaniemi I, and Morohashi K (2015). Fetal Leydig Cells Persist as an Androgen-Independent Subpopulation in the Postnatal Testis. Mol Endocrinol 29, 1581–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohni A, Tan K, Song H-W, Burow D, de Rooij DG, Laurent L, Hsieh T-C, Rabah R, Hammoud SS, Vicini E, et al. (2019). The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Reports 26, 1501–1517.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stévant I, Kühne F, Greenfield A, Chaboissier M-C, Dermitzakis ET, and Nef S (2019). Dissecting Cell Lineage Specification and Sex Fate Determination in Gonadal Somatic Cells Using Single-Cell Transcriptomics. Cell Rep 26, 3272–3283.e3. [DOI] [PubMed] [Google Scholar]
- Svingen T, and Koopman P (2013). Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev. 27, 2409–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Song H-W, and Wilkinson MF (2020). Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, and Surani MA (2015). A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 161, 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Kobayashi T, Irie N, Dietmann S, and Surani MA (2016). Specification and epigenetic programming of the human germ line. Nat. Rev. Genet 17, 585–600. [DOI] [PubMed] [Google Scholar]
- Wang M, Liu X, Chang G, Chen Y, An G, Yan L, Gao S, Xu Y, Cui Y, Dong J, et al. (2018). Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 23, 599–614.e4. [DOI] [PubMed] [Google Scholar]
- Wen Q, Wang Y, Tang J, Cheng CY, and Liu Y-X (2016). Sertoli Cell Wt1 Regulates Peritubular Myoid Cell and Fetal Leydig Cell Differentiation during Fetal Testis Development. PLOS ONE 11, e0167920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western PS, Miles DC, van den Bergen JA, Burton M, and Sinclair AH (2008). Dynamic Regulation of Mitotic Arrest in Fetal Male Germ Cells. STEM CELLS 26, 339–347. [DOI] [PubMed] [Google Scholar]
- Witchi E (1948). Migration of the germ cells of human embryos from the yolk sac to the primitive gonadal folds. Contrib Embryol Carnegie Inst 32, 67–80. [Google Scholar]
- Yang Y, Workman S, and Wilson MJ (2019). The molecular pathways underlying early gonadal development. Journal of Molecular Endocrinology 62, R47–R64. [DOI] [PubMed] [Google Scholar]
- Yao HH-C, Whoriskey W, and Capel B (2002). Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 16, 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, et al. (2019). Controlled modelling of human epiblast and amnion development using stem cells. Nature 573, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Single cell transcriptome profiling and analysis of the human fetal and postnatal testis. Related to Figure 1.
Figure S2. Expression patterns of additional markers projected on the UMAP plot. Related to Figure 1.
Figure S3. Transition of human PGCs to State f0. Related to Figure 2.
Figure S4. Network expression dynamic during fetal and postnatal germ cell development. Related to Figure 2.
Figure S5. Somatic niche cell specification at embryonic and fetal stages. Related to Figures 3, 4 and 5.
Figure S6. Proposed models for human germline development and somatic niche cell specification during embryonic, fetal and postnatal stages. Related to Figures 4 and 5.
Table S1: Genes that display dynamic expression along germ cell pseudo-developmental trajectory shown in Fig 2C. Related to Figure 2.
Table S2: Genes that display dynamic expression during human testicular somatic cell specification as shown in Fig 4A. Related to Figure 4.
Data Availability Statement
All software tools can be found online (see Key Resources Table). The accession number for all sequencing data reported in this paper is GEO: GSE143356 and GEO:GSE161617.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-PIWIL4 Dilution: 1:200 |
Thermo Fisher Scientific | Cat#: PA5-3144 RRID: AB_2548922 |
| Mouse monoclonal (CloneB56) anti-MKI67 Dilution: 1:200 |
BD Biosciences | Cat#: 556003 RRID: AB_396287 |
| Goat polyclonal anti-DDX4 Dilution: 1:100 |
R&D Systems | Cat#: AF2030 RRID: AB_2277369 |
| Rabbit monoclonal (D73G4) anti-NANOG Dilution: 1:100 |
Cell Signaling Technology | Cat#:4903 RRID: AB_10559205 |
| Mouse monoclonal anti-CYP17A1 Dilution: 1:200 |
Santa Cruz Biotechnology | Cat#: SC-374244 RRID: AB_10988393 |
| Mouse monoclonal (1A12-1D9) anti-GATA3 Dilution: 1:100 |
Thermo Fisher Scientific | Cat#: MA1028 RRID: AB_2536713 |
| Mouse monoclonal (G-4) anti-GATA4 Dilution: 1:100 |
Santa Cruz Biotechnology | Cat#: SC-25310 RRID: AB_627667 |
| Mouse monoclonal anti-DMRT1 Dilution: 1:100 |
Santa Cruz Biotechnology | Cat#: SC-377167 |
| Rabbit polyclonal anti-SOX9 Dilution: 1:200 |
Millipore | Cat#: AB5535 RRID: AB_2239761 |
| AF488 goat-anti mouse IgG2a | Invitrogen | Cat#: A21131 RRID: AB_2535771 |
| AF594 donkey-anti-mouse IgG | Invitrogen | Cat#: A21203 RRID: AB_2535789 |
| AF594 goat-anti-mouse IgG1 | Invitrogen | Cat#: A21125 RRID: AB_2535767 |
| AF594 donkey-anti-rabbit IgG | Jackson ImmunoResearch | Cat#: 711-585-152 RRID: AB_2340621 |
| AF647 donkey-anti-goat IgG | Invitrogen | Cat#: A21447 RRID: AB_2535864 |
| Biological Samples | ||
| Human testis samples from postnatal donors | DonorConnect | NA |
| Human testis samples from embryonic and fetal stages | University of Washington-Birth Defects Research Lab | NA |
| Human testis samples from Jan’s lab | Karolinska Institutet | NA |
| Deposited Data | ||
| Single cell RNA-seq for embryonic and fetal human testes | This paper | GEO: GSE143356 |
| Single cell RNA-seq for postnatal testes | This paper | GEO: GSE161617 |
| Software and Algorithms | ||
| Seurat (2.3.4) | Butler et al., 2018 | https://satijalab.org/seurat/ |
| Monocle (2.10.1) | (Qiu et al., 2017) | http://cole-trapnell-lab.github.io/monocle-release/ |
| GO (David 6.7) | Huang et al., 2009 | https://david-d.ncifcrf.gov |
| Cell Ranger (2.2.0) | NA | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Cluster 3.0 | NA | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| WGCNA (1.68) | (Langfelder and Horvath, 2008) | https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/ |
| Cytoscape (3.7.2) | (Otasek et al., 2019) | https://cytoscape.org |
| Other | ||
| Single cell RNA-seq for infant and adult human testes | Guo et al., 2018 | GEO: GSE120508 |
| Single cell RNA-seq for neonatal human testes | Sohni et al., 2019 | GEO: GSE124263 |