Abstract
MDM2 regulates p53 degradation by functioning as an E3 ubiquitin ligase. The role of MDMX, an MDM2 homolog that lacks E3 ligase activity, in the regulation of p53 degradation remains incompletely understood and sometime controversial. This confusion is due at least in part to studies of p53 degradation mainly carried out in in vitro settings, as elimination of either MDM2 or MDMX from mice results in p53-dependent embryonic lethality, thus obfuscating in vivo studies of the individual roles of MDM2 and MDMX in p53 degradation. To overcome this problem, we generated mice expressing an inducible p53 allele under various MDM2 and MDMX deletion and mutation statuses and studied in vivo p53 degradation. Degradation of p53 in vivo was largely prevented in mice and MEF retaining MDM2 but lacking MDMX. While MDM2 and MDMX interacted with p53 in the absence of each other, they bound p53 more efficiently as a heterodimer. MDMX, but not MDM2, interacted with ubiquitin-conjugating enzyme UbcH5c, an interaction that was essential for MDMX to enable MDM2 E3 ligase activity for p53 degradation. Grafting the C-terminal residues of MDMX to the C-terminus of MDM2 allowed MDM2 to interact with UbcH5c and enhanced MDM2-mediated p53 degradation in the absence of MDMX. Together, these data indicate that MDMX plays an essential role for p53 degradation in vivo by recruiting UbcH5c to facilitate MDM2 E3 ligase function.
INTRODUCTION
TP53 is the most commonly mutated gene in human cancers. As a transcription factor, p53 monitors genomic damage and inhibits tumor development through promoting numerous activities, including cell cycle arrest, apoptosis, and autophagy(1). In unstressed cells, p53 expression and activity is kept at low levels primarily by ubiquitin-mediated proteasomal degradation facilitated by its negative regulator MDM2, which harbors E3 ubiquitin ligase activity in its C-terminal RING (really interesting new gene) finger domain. MDM2 ubiquitinates p53, targeting it for export from the nucleus and degradation in the cytoplasm(2). In response to stress, a rapid increase in p53 abundance occurs through the inhibition of MDM2 mediated p53 degradation and accumulates p53 protein in the nucleus. Moreover, MDM2 can inhibit p53 activity by binding to the p53 N-terminal transactivation domain(3). Deletion of MDM2 in mice causes increase in p53 protein levels and transcriptional activity and induces embryonic lethality; and the lethality can be rescued by co-deletion of p53, demonstrating that a primary function of MDM2, at least during mouse embryogenesis, is to keep p53 at low levels and inactive(4,5). MDMX, also known as MDM4, is a structural homolog of MDM2, but it does not harbor intrinsic E3 ubiquitin ligase activity. MDMX, like MDM2, binds p53 at its N-terminal transactivation domain and contains a C-terminal RING domain through which to interact with the MDM2 RING domain. Deletion of MDMX in mice, also like that of MDM2, leads to embryonic lethality in a p53-dependent manner(6). The p53-dependent embryonic lethality observed in MDM2 and MDMX deletion mice indicates that MDM2 and MDMX play essential yet non-redundant roles in p53 regulation. How MDM2 and MDMX play together to regulate p53, particularly under physiological conditions, remains partially understood.
In particular, the role of MDMX in the regulation of p53 degradation is inadequately understood. In vitro studies have suggested that MDMX primarily inhibits p53 transcriptional activity but not its protein stability, which is regulated principally by MDM2 E3 ligase(7,8). Other studies have shown that overexpression of MDMX can stabilize p53 and reverse MDM2-mediated p53 degradation(9), and that MDMX increases MDM2 levels by reducing MDM2 E3 ligase activity toward itself(10). Conversely, studies also shown that the MDM2-MDMX heterodimer functions as a better E3 ubiquitin ligase than MDM2 alone, suggesting that MDMX facilitates MDM2 E3 activity(11). Consistent with this notion, ectopic overexpression of MDMX enhances MDM2 E3 ligase activity toward p53(12) and rescues the activity of certain MDM2 mutants lacking E3 ligase activity(13).
Our recent study using the Mdm2Y487A mutant mice, in which the MDM2Y487A mutation inactivates MDM2 E3 ligase activity without affecting its ability to bind MDMX, demonstrated that in vivo the MDM2-MDMX heterodimer plays an essential role in suppressing p53 transcriptional activity independent of MDM2 E3 ligase function, while the MDM2 E3 ligase function becomes indispensable under stress conditions such as DNA damage(14). The study recapitulates two recent MDMX mutant mouse models addressing questions about the in vivo functions of MDMX. Deletion of the RING domain of MDMX (MdmxΔRING) or introduction of a C462A mutation in the MDMX RING finger (MdmxC462A) disrupted MDM2-MDMX binding and caused embryonic lethality in mice(15,16). In both models, the lethality can be rescued by p53 deletion, suggesting that disruption of the MDMX RING domain, which will interrupt MDM2-MDMX heterodimerization, results in uncontrolled activation of p53, implicating once again the essential role of MDM2-MDMX heterodimer for p53 suppression. Nevertheless, the contributions of MDMX in MDM2-mediated p53 ubiquitination and degradation remain an important yet unresolved issue in the field.
Because deletion of either MDM2 or MDMX results in p53-dependent embryonic lethality, dissecting the individual contributions of MDM2 and MDMX to p53 regulation under truly physiological conditions is difficult. We took advantage of the p53ERTAM (p53ER hereafter) mouse model, in which the endogenous p53 gene is replaced by one encoding full-length p53 fused with the hormone-binding domain of a modified estrogen receptor at its C-terminus(17). The p53 activity in these mice can be rapidly switched “on” and “off” in the presence and absence of 4-hydroxytamoxifen (4-OHT), allowing generation of mice containing WT p53 under various MDM2 and MDMX deletion and mutation backgrounds. The expression of p53ER is controlled by the native p53 promoter, and the p53ER protein functions normally as the WT p53(17), except an slightly extended half-life of ~50 min(18), as compared to ~20 min for WT p53(19). We generated p53ER mice under various MDM2 and MDMX deletion and mutation backgrounds, and studied the contribution of MDMX to MDM2 E3 ligase function in vivo.
MATERIALS AND METHODS
Mouse Experiments
Mice and were bred and maintained strictly under protocols approved by the Institutional Animal Care and Use Committee at UNC (16–026). Mice were treated with vehicle or 10 mg/mL 4-OHT dissolved in peanut oil as previously described(20). After 6 hours, mice were euthanized and tissue was harvested.
Cell lines
The human osteosarcoma SJSA, osteosarcoma U2OS, and breast cancer MCF7 cells were obtained from the ATCC and came with comprehensive authentication and quality controls. The cells are grown in DMEM with 10% FBS supplemented with 100 U penicillin/streptomycin and maintained at 37°C and 5% CO2. The cells were actively passaged for less than 6 months. All these cell lines were routinely tested for Mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, LT07–118), and the last Mycoplasma test was performed in July 2020. Mycoplasma-free cell lines were used in all of our experiments.
Cloning and Transfections
MDMX and MDM2 mutations were generated using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, cat. no. 200521). Wild-type human MDMX or MDM2 encoded in pcDNA3-Myc3 or pcDNA3.1(–)-GFP vectors were used as templates for reactions. Transfections were performed using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions.
Protein Analysis
MEFs were treated with 4-OHT in the presence or absence of MG132 for the indicated times, after which the cells were harvested for analysis. Tissues were lysed as previously described(14). IP and immunoblotting were performed as described(21).
In vivo Ubiquitination Assay
Early passage MEFs were treated with 4-OHT for 2 hours, then with MG132 for 5 hours. Cells were trypsinized and collected. One-fifth of each sample was lysed as an input control. The remaining cells were boiled in hot Manabu’s buffer. Then, lysates were diluted in 0.1% NP-40 lysis buffer for p53 IP. Western blotting was performed to probe for ubiquitinated forms of p53.
Protein Half-life Assay
Early passage MEFs were treated with 4-OHT for 2 hours prior to treatment with 100 μg/mL cycloheximide. Cells were harvested using hot SDS lysis buffer at the indicated time points. Actin and p53 levels were analyzed by western blot. Protein bands were quantified, normalized to actin, and plotted as the relative amount of protein compared to the 0 minute treatment. Protein bands were quantified using ImageJ.
Cell Fractionation Assay
Early passage MEF cells were treated with 100 nM 4-OHT for 1 hour. The cells were collected by centrifugation at 1000 rpm for 5 min, and the pellets were resuspended in TM-2 buffer (0.01 M Tris-HCl, pH 7.4; 0.002 M MgCl2 and 0.5 mM PMSF) and incubate at RT for 1 minute, followed by incubation on ice for 5 minutes. Triton X-100 was then added into the cell suspension to a final concentration of 0.5% (v/v) and the cells were incubated on ice for an additional 5 minutes. The supernatant cytosol fraction was collected by centrifugation at 1,500 rpm at 4°C for 10 minutes. The pellet containing the nuclei was washed with TM-2 buffer twice and precipitated by centrifugation at 1,500 rpm at 4°C for 10 minutes. The harvested the nuclei were lysed with NP-40 buffer. The levels of p53, MDM2, and MDMX were analyzed by western blotting. Tubulin and PARP were used as controls for cytoplasmic and nuclear fractions, respectively. The bands were compared quantitatively using ImageJ software.
Mouse Breeding, Maintenance, and Genotyping
Mdm2−/− and Mdmx−/− mice were gifts from Guillermina Lozano (University of Texas, M.D. Anderson Cancer Center). p53ERTAM knock-in mice were a gift from Gerard Evan (University of Cambridge, UK). Mdm2−/− and Mdmx−/− mice were crossed with p53ERTAM mice to generate Mdm2−/−;p53ER/- and Mdmx−/−;p53ER/- compound mice. Genotyping of the Mdm2−/−, Mdmx−/−, and p53ERTAM alleles was performed as described previously(5,6,17). Mice were bred and maintained strictly under protocols (16–026.0) approved by the Institutional Animal Care and Use Committee in the UNC Animal Care Facility. Mice 6–8 weeks of age were treated with either peanut oil or 10 mg/mL 4-OHT dissolved in peanut oil as previously described(20). After 6 hours of treatment, mice were euthanized and tissue was harvested.
Cell Culture and Reagents
Primary MEF cells were cultured in a 37°C incubator with 5% CO2, and 3% O2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco) and 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco). For activation of p53ERTAM, 100 nM 4-OHT dissolved in 100% ethanol was added to the culture medium. Cycloheximide was purchased from Sigma (cat. no. C7698). MG132 was purchased from Calbiochem (cat. no. 474790). siRNAs against MDMX or UbcH5c were synthesized by Jima (Jima, Shanghai, China).
Cloning and Plasmids
MDMX and MDM2 mutations were generated using the QuikChange II XL site-directed mutagenesis protocol (Agilent Technologies, cat. no. 200521). Briefly, WT human MDMX or MDM2 encoded in the pcDNA3-Myc3 or pcDNA3.1(–)-GFP vector were used as a template for all site-directed mutagenesis reactions. PCR reactions were performed according to manufacturer’s instruction. The primers used are as follows: MDMXΔC7: Forward, 5’-TGCAAGAAAGAG ATTCAGCTGTAAGGATCCGTTTTTATAGCATAAGCGGGC-3’; Reverse: 5’-CGC TTATGCTATAAAAACGGATCCTTACAGCTGAATCTCTTTCTTGCA-3’; MDM2XC7: Forward: 5’-TGTAGACAACCAATTCAAATGGTTATTAAGGTTTTTATAGCATAAAATTCTGCAGTCGACGGTACC-3’; Reverse: 5’-GGTACCGTCGACTGCAGAATTTTATGC TATAAAAACCTTAATAACCATTTGAATTGGTTGTCTACATACTGG-3’. Mutagenesis primers were used to amplify the intended product using a thermocycler (Applied Biosystems, model 2720). PCR reactions were digested with DpnI (New England Biolabs) for two hours, and then 5 μl of each reaction was transformed into chemically competent XL-1 blue Escherichia coli cells. All clones were submitted to the University of North Carolina Genome Analysis Facility for sequence verification.
Transfections
Cells were plated for overnight and transfections were performed using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions. All transfections included a pcDNA3.1(–)-GFP plasmid to visually confirm transfection efficiency (in all transfections, at least 50% of cells were GFP-positive).
Protein Analysis
Early passage MEFs were treated with 100 nM 4-OHT in the presence or absence of MG-132 for the indicated time points, after which the cells were lysed in 0.1% NP-40 for immunoprecipitation and 0.5% NP-40 buffer for western blot analysis. Tissue was lysed as previously described(14). Procedures and conditions for immunoprecipitation and immunoblotting were also described previously(21). Mouse monoclonal anti-MDM2 (2A-10 and 4B11), mouse monoclonal anti-MDMX (MDMX-82), mouse monoclonal anti-p53 (pAb122), and mouse monoclonal anti-Actin (MAB1501) antibodies were purchased commercially. Mouse monoclonal anti-MDMX (8C6 and 10C2) was gift from Jiandong Chen (Moffitt Cancer Center).
In vivo Ubiquitination Assay
Early passage Mdm2+/+;Mdmx+/+;p53ER/-, Mdm2+/+;Mdmx−/−;p53ER/-, Mdm2−/−;Mdmx+/+;p53ER/- and Mdm2C462A/C462A;Mdmx+/+;p53ER/- MEFs were treated with 100 nM 4-OHT for six hours, after which cells were treated with 10 μM (final concentration) of MG132 proteasome inhibitor for another five hours. U2OS cells were transfected with indicated plasmid DNA for 24 hours followed by treatment with 10 μM MG132 for five hours. Then cells were trypsinized and collected. One-fifth of each sample was lysed with 0.1% NP-40 lysis buffer as an input control. The remaining cells were boiled in hot Manabu’s buffer containing 1X protease inhibitor (Sigma), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium orthovanadate (NaVO4), and 1 mM dithiothreitol (DTT) for 10 minutes. Then, the lysates were diluted into 0.1% NP-40 lysis buffer containing protease inhibitor and subjected to immunoprecipitation. Western blotting was performed to probe for the ubiquitinated forms of p53.
Protein Half-life Assay
Early passage Mdm2+/+;Mdmx+/+;p53ER/-, Mdm2+/+;Mdmx−/−;p53ER/- and Mdm2−/−;Mdmx+/+;p53ER/- MEFs were treated with 100 nM 4-OHT for two hours prior to treatment with 100 μg/mL cycloheximide. Cells were harvested using sodium dodecyl sulfate (SDS) lysis buffer at the indicated time points. The levels of p53 and Actin were analyzed by western blotting. p53 bands were quantified after normalization to Actin, and were plotted as the relative amount of protein remaining compared to the zero minute treatment time. Bands were compared quantitatively using ImageJ software.
GST Pulldown Assay
GST pulldown assay was performed as described (22). Briefly, GST and GST-MDMX were expressed in the Escherichia coli BL21 strain and purified according to manufacturer’s instructions (GE Healthcare). The UbcH5c protein, which was obtained from the whole cell lysates of 293T cells transfected with the pcDNA3.1(+)-ubcH5c plasmid, was incubated with GST and GST-MDMX bound to GST beads in 1 mL of binding buffer containing protease inhibitor cocktail at 4°C for 6 h. GST beads were then washed three times, resuspended in 30 μL of 1× SDS-PAGE loading buffer and detected by immunoblotting.
Immunofluorescence
Immunofluorescence was performed as described(23). In brief, cells growing in glass bottom cell culture dish, were treated with 4-OHT for 1 hour, fixed with 4% paraformaldehyde (VICMED, Xuzhou), and permeabilized with 0.2% Triton X-100 (VICMED) for 5 min. Following primary and secondary antibody incubations in blocking buffer (0.5% bovine serum albumin in 1 phosphate-buffered saline), the stained cells were analyzed using an Olympus IX-81 microscope with SPOT-camera and software.
RESULTS
MDMX deletion in mice causes p53 accumulation
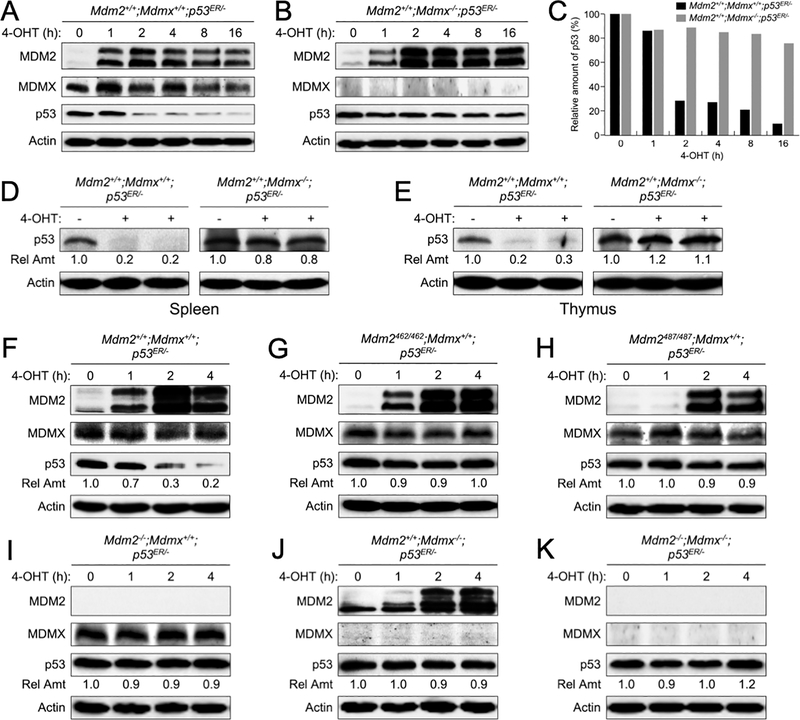
Because previous studies showed that under Mdm2-null background, two copies of p53ER alleles (p53ER/ER) could cause partial embryonic lethality due to a “leakage” of the p53ER/ER alleles(18), we therefore generated mice containing one p53ER allele and one p53 deletion allele, and bred the p53ER/- mice with mice containing WT MDMX or no MDMX to generate Mdm2+/+;Mdmx+/+;p53ER/- or Mdm2+/+;Mdmx−/−;p53ER/- mice, respectively. Mouse embryonic fibroblasts (MEFs) were isolated from these mice and cultured in the presence of 100 nM 4-OHT to restore p53 function. As expected, after exposure to 4-OHT the MDM2 levels increased immediately in both Mdm2+/+;Mdmx+/+;p53ER/- and Mdm2+/+;Mdmx−/−;p53ER/- MEFs, indicative of p53 activation (Figure 1A–B). The p53 protein levels were decreased after 2 hours of 4-OHT exposure in the Mdm2+/+;Mdmx+/+;p53ER/- MEFs, apparently due to MDM2-mediated degradation (Figure 1A, 1C). Surprisingly, p53 protein levels were barely decreased in the Mdm2+/+;Mdmx−/−;p53ER/- MEFs despite significantly elevated the MDM2 protein levels (Figure 1B–C). This observation suggested that MDMX might be necessary for MDM2-mediated p53 degradation in unstressed primary MEF cells.
Figure 1. Deletion of MDMX in mice causes p53 accumulation.
Early passages of (A) Mdm2+/+;Mdmx+/+;p53ER/- and (B) Mdm2+/+;Mdmx−/−;p53ER/- mouse embryonic stem (MEF) cells were treated with 4-hydroxytamoxifen (4-OHT) for the indicated times. The levels of MDM2, MDMX, p53 and actin were analyzed by western blot. (C) The amounts of p53 remained at each time point in A-B were quantified by densitometry, normalized to actin, and plotted. (D) Spleen and (E) thymus tissues were isolated from 8 weeks old Mdm2+/+;Mdmx+/+;p53ER/- and Mdm2+/+;Mdmx−/−;p53ER/- mice after treatment with 4-OHT for 6 hours. Tissue lysates were analyzed by western blot. Relative amounts of p53 were indicated. (F-K) MEFs of varies MDM2 and MDMX statuses were treated with 4-OHT for the indicated times and cell lysates were analyzed by western blot. Relative amounts of p53 were indicated.
To substantiate the observations made in MEFs, we examined p53 protein levels in mouse tissues. We injected Mdm2+/+;Mdmx+/+;p53ER/- and Mdm2+/+;Mdmx−/−;p53ER/- mice intraperitoneally with peanut oil or 4-OHT dissolved in peanut oil. Six hours after injection the mice were sacrificed, spleen and thymus tissues were isolated and examined for p53 protein levels. In the Mdm2+/+;Mdmx+/+;p53ER/- mouse tissues, p53 protein levels reduced to nearly undetectable after 4-OHT injection (Figure 1D–E), indicative p53 degradation. However, p53 protein levels were basically unchanged and remained high in the Mdm2+/+;Mdmx−/−;p53ER/- mouse tissues 6 hours after 4-OHT injection (Figure 1D–E), indicating that elimination of MDMX blocked p53 degradation.
To validate the above observation was indeed due to a loss of MDM2 E3 ligase activity, we generated additional p53ER/− mice containing MDM2C462A or MDM2Y487A mutations as well as containing MDM2 or MDMX deletions. Both MDM2C462A and MDM2Y487A mutations abolish MDM2 E3 ubiquitin ligase function, while MDM2C462A mutation also disrupts MDM2-MDMX dimerization(24) and MDM2Y487A mutation did not affect MDM2-MDMX dimerization(14). We examined p53 levels in MEFs isolated from these mice. Upon treatment with 4-OHT, p53 levels were decreased in Mdm2+/+;Mdmx+/+;p53ER/- MEFs (Figure 1F). In comparison, p53 levels were unchanged in MEFs containing MDM2 E3 ligase mutations (Mdm2C462A/C462A;Mdmx+/+;p53ER/- and Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- ) (Figure 1G–H), or lacking MDM2 or/and MDMX (Mdm2−/−;Mdmx+/+;p53ER/-, Mdm2+/+;Mdmx−/−;p53ER/-, Mdm2−/−;Mdmx−/−;p53ER/-) (Figure 1I–K). Together, these data support that in vivo MDMX plays an essential role in allowing MDM2 E3 function.
Deletion of MDMX impedes p53 polyubiquitination and proteasomal degradation
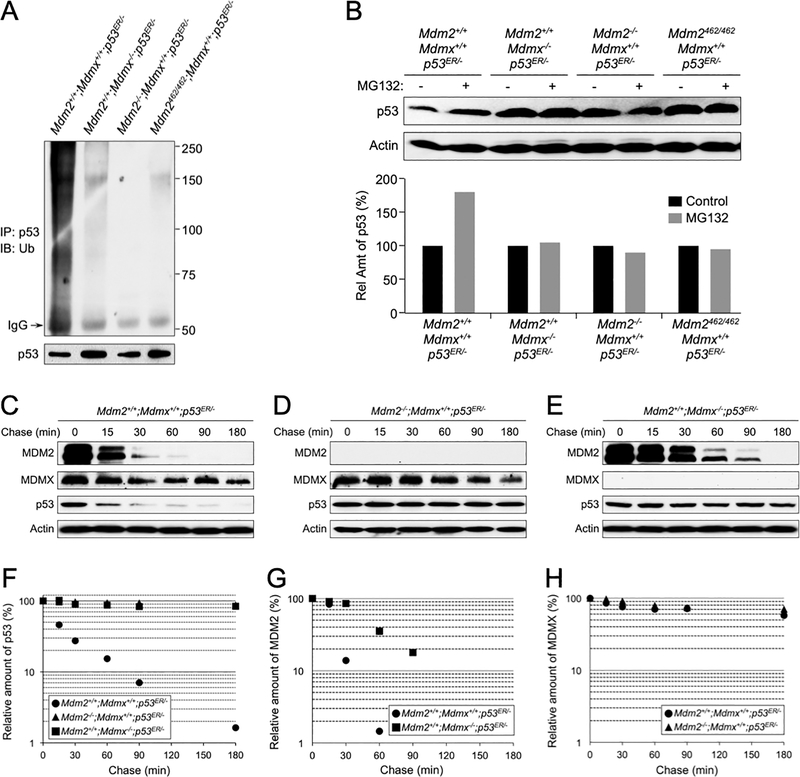
To investigate if the failure of MDM2 to degrade p53 in the absence of MDMX is due to a loss of MDM2 E3 ligase activity, we examined p53 ubiquitination in MEFs of various MDM2 and MDMX statuses, including Mdm2+/+;Mdmx+/+;p53ER/-, Mdm2+/+;Mdmx−/−;p53ER/-, Mdm2−/−;Mdmx+/+;p53ER/-, and Mdm2C462A/C462A;Mdmx+/+;p53ER/- genotypes. We treated the MEFs with 4-OHT for 2 hours to activate p53ER and to induce MDM2 expression followed by treating the cells with proteasome inhibitor MG132 for 5 hours. Cell lysates were then harvested and p53 was immunoprecipitated by a p53 antibody and analyzed by western blot using an anti-ubiquitin antibody. High molecular weight smears, which are indicative of polyubiquitinated protein species, were observed in Mdm2+/+;Mdmx+/+;p53ER/- MEF lysate, but not in MEF lysates lacking either MDM2 or MDMX, or with MDM2C462A mutation (Figure 2A). Furthermore, treating MEFs with MG132 resulted in p53 accumulation only in Mdm2+/+;Mdmx+/+;p53ER/- MEFs, but not in MEFs lacking either MDM2 or MDMX, or with MDM2C462A mutation (Figure 2B). These data indicated that MDMX, like MDM2, plays an essential role for p53 polyubiquitination and proteasomal degradation.
Figure 2. Deletion of MDMX impedes p53 polyubiquitination and proteasomal degradation.
(A) MEFs of indicated genotypes were treated with 4-OHT for 2 hours and then with 10 μM MG132 for 5 hours before harvesting. The cell lysates were isolated and immunoprecipitated with an anti-p53 antibody and analyzed by western blot using an anti-ubiquitin antibody. (B) MEFs of indicated genotypes were treated with 4-OHT for 2 hours and then with MG132 for additional 4 hours. Cell lysates were isolated and the levels of p53 and actin were analyzed by western blot. Relative amounts of p53 were quantified and shown as a bar graph below. MEFs of Mdm2+/+;Mdmx+/+;p53ER/-(C), Mdm2−/−;Mdmx+/+;p53ER/- (D), and Mdm2+/+;Mdmx−/−;p53ER/- (E) genotypes were treated with 4-OHT for 2 hours followed by treatment with 100 μg/mL cycloheximide and harvested at the indicated time points. MDM2, MDMX, p53 and actin were analyzed by western blot. The relative levels of p53 (F), MDM2 (G), and MDMX (H) was quantified, normalized to actin, and plotted.
To further investigate p53 degradation that is affected by MDMX, we performed a protein half-life assay in using the primary MEFs. In the presence of WT MDM2 and WT MDMX (Mdm2+/+;Mdmx+/+;p53ER/-), p53 exhibited a short half-life of approximately 20 min (Figure 2C). By contrast, in the absence of MDM2 (Mdm2−/−;Mdmx+/+;p53ER/-), p53 degradation was largely inhibited showing a half-life of beyond 3 hours (Figure 2D). Importantly, in the absence MDMX (Mdm2+/+;Mdmx−/−;p53ER/-), similar to MEFs lacking MDM2, p53 degradation was also inhibited showing a protein half-life more than 3 hours (Figure 2E). The relative levels of p53 was quantified, normalized to actin, and plotted in Figure 2F). We noticed that in the absence of MDMX, MDM2 degradation was somewhat impeded (compare MDM2 panels in Figure 2C and 2E); and the half-life of MDM2 was extended from less than 30 min in cells containing MDMX to about 60 min in cells lacking MDMX (Figure 2G). On the other hand, MDMX was a stable protein and its stability was not affected by the status of MDM2 (compare MDMX panels in Figure 2C and 2D), and its half-life was well beyond 3 hours (Figure 2H). Together, these data strongly support for an indispensable role of MDMX in MDM2 mediated p53 ubiquitination and proteasomal degradation.
MDM2-MDMX heterodimerization facilitates MDM2-p53 binding
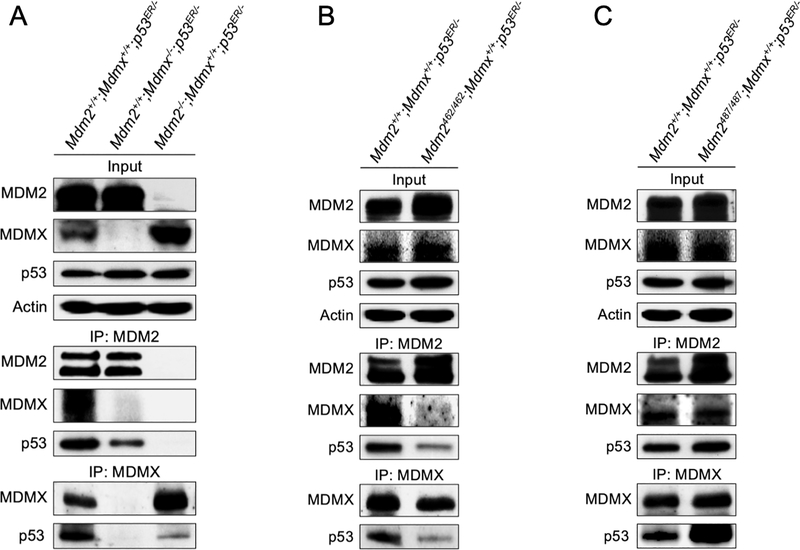
Recent in vivo evidences demonstrated that the MDM2-MDMX heterodimerization is required for p53 suppression(14–16). In search of mechanisms by which MDMX facilitates MDM2-mediated p53 degradation, we hypothesize that the MDM2-MDMX heterodimer might be necessary for or facilitating MDM2-p53 binding so that to support MDM2 mediated p53 degradation. To test this hypothesis, we performed immunoprecipitation-coupled western blot (IP-western) analysis in MEF cell lysates to determine the role of MDMX in MDM2-p53 interaction. IP of MDM2 pulled down a significant amount of MDMX and p53 in the Mdm2+/+;Mdmx+/+;p53ER/- MEF cell lysates (Figure 3A, left lane). In comparison, in the absence of MDMX a similar amount of MDM2 pulled down much less p53 in the Mdm2+/+;Mdmx−/−;p53ER/- cell lysates (Figure 3A, middle lane), suggesting MDMX is important for MDM2-p53 binding. Likewise, IP of MDMX pulled down more p53 in the presence of MDM2 (Mdm2+/+;Mdmx+/+;p53ER/- cell lysates) than in the absence of MDM2 (Mdm2−/−;Mdmx+/+;p53ER/- cell lysates), even though the later expresses a higher level of MDMX (Figure 3A, IP: MDMX, compare left lane and right lane). We noticed that in our co-IP assay the MDMX antibody (mouse monoclonal anti-MDMX antibody clone MDMX-82) was unable to pull down MDM2, and the reason is currently unknown.
Figure 3. MDM2-MDMX heterodimerization facilitates MDM2-p53 binding.
(A) MEFs of indicated genotypes were treated with 4-OHT for 2 hours. The cells were then harvested, lysed, and immunoprecipitated (IP) for MDM2 and MDMX, and blotted with the indicated proteins. Input represents 5% of total cell lysate utilized for IP. (B) MEFs of indicated genotypes were treated and blotted as in A. (C) MEFs of indicated genotypes were treated and blotted as in A.
To determine whether the MDM2-MDMX physical interaction, rather than merely the presence of both proteins in the cell lysates, is necessary for efficient MDM2-p53 binding, we assessed p53 binding capability to MDM2 in Mdm2C462A/C462A;Mdmx+/+;p53ER/- cell lysates. In these MEF cells, both MDM2 and MDMX proteins are expressed, but the MDM2-MDMX heterodimer is disrupted by the MDM2C462A mutation(24). The binding efficacy of p53 by either MDM2C462A or MDMX was significantly lower in the Mdm2C462A/C462A;Mdmx+/+;p53ER/- MEF cell lysates, as compared to the binding efficacy of p53 by MDM2 and MDMX in the Mdm2+/+;Mdmx+/+;p53ER/- MEF cell lysates (Figure 3B). Because the MDM2C462A mutation not only disrupted MDM2-MDMX binding but also eliminated MDM2 E3 ligase function, we want to determine whether the MDM2 E3 ligase function might contribute to MDM2-p53 and MDMX-p53 binding. We took advantage of the Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEF cells, in which the MDM2Y487A mutation inactivates MDM2 E3 ligase activity without affecting its interaction with MDMX(14). The binding of MDM2Y487A or MDMX to p53 appeared unaffected in the Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEFs (Figure 3C), suggesting that the MDM2 E3 ligase activity is not required for MDM2-p53 binding, but rather the presence of the MDM2-MDMX physical interaction is important for p53 binding. We detected somewhat higher levels of p53 present in the IP of MDM2 and IP of MDMX in the Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEF cell lysates than that of the Mdm2+/+;Mdmx+/+;p53ER/- MEF cell lysates. We interpreted this is likely due to the presence of higher levels of p53 in the Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEF cells, and also a possibility of lacking MDM2 E3 activity might facilitate p53 binding. Collectively, these data demonstrated that in vivo the MDM2-MDMX heterodimerization facilitates MDM2-p53 binding, implicating that the mechanism by which MDMX promotes MDM2-mediated p53 degradation is at least in part through promoting MDM2-p53 interaction.
MDM2-MDMX heterodimerization facilitates p53 cytoplasmic localization
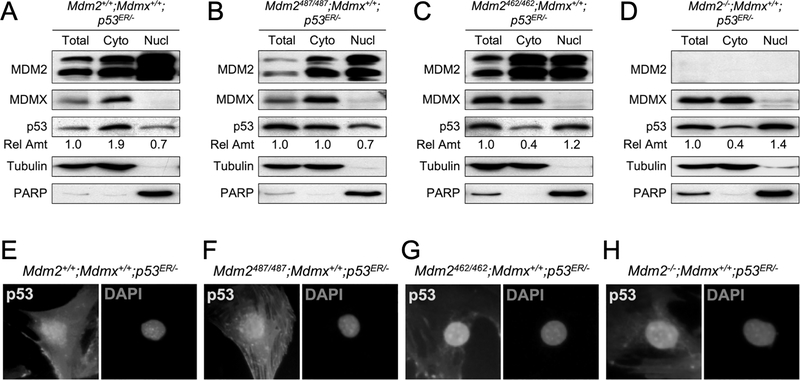
Because proteasomal degradation occurs mainly in the cytoplasm and MDMX is predominantly a cytoplasmic protein, we hypothesize that MDMX might facilitate p53 degradation by promoting its cytoplasmic localization. To test this possibility, we carried out fractionation assays using MEF cell lysates to determine the relative amount of p53 in the cytoplasmic and nuclear fractions under various MDM2 and MDMX statuses. We found that the relative amount of p53 in cytoplasm was higher in cells possessing MDM2-MDMX heterodimers (Mdm2+/+;Mdmx+/+;p53ER/-, Figure 4A; Mdm2Y487A/Y487A;Mdmx+/+;p53ER/-, Figure 4B) than in cells lacking MDM2-MDMX heterodimers (Mdm2C462A/C462A;Mdmx+/+;p53ER/-, Figure 4C; Mdm2−/−;Mdmx+/+;p53ER/- Figure 4D). Note that the levels of p53 in the cytoplasmic as well as the nuclear fractions shown in these western blots are relative levels, and they do not reflect actual amount of p53 in each fractions. We noticed that MDM2 was readily identified in both the nuclear and the cytoplasmic fractions, and the relative levels of MDM2 in the cytoplasm seemed somewhat increased in the Mdm2C462A/C462A;Mdmx+/+;p53ER/- MEF cells (Figure 4C). In contrast, MDMX was detected predominantly in the cytoplasm in ay of the cell types and it was not affected by the status of MDM2. To support observations from the fractionation assay, we also carried out immunofluorescence staining of p53 in these MEFs. We found that in the Mdm2+/+;Mdmx+/+;p53ER/- and Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEFs, where MDM2 and MDMX form heterodimers, p53 was found in both the nucleus and the cytoplasm with relatively more p53 in the cytoplasm (Figure 4E–4F). On the other hand, in the Mdm2C462A/C462A;Mdmx+/+;p53ER/- and Mdm2−/−;Mdmx+/+;p53ER/- MEFs, where MDM2 and MDMX do not form heterodimers, p53 was mainly observed in the nucleus with little cytoplasmic presence (Figure 4G–4H). These immunofluorescence results were consistent with the data from fractionation assays, and together they suggested that the MDM2-MDMX heterodimerization promotes p53 cytoplasmic localization.
Figure 4. MDM2-MDMX heterodimerization facilitates p53 cytoplasmic localization.
Cell lysates were isolated from (A) Mdm2+/+;Mdmx+/+;p53ER/-, (B) Mdm2487/487;Mdmx+/+;p53ER/-, (C) Mdm2462/462;Mdmx+/+;p53ER/-, and (D) Mdm2+/+;Mdmx−/−;p53ER/- MEFs. The lysates were either untreated (Total) or separated into cytoplasmic (Cyto) and nuclear (Nucl) fractions and analyzed for MDM2, MDMX, and p53 by western blot. Tubulin and PARP were used as controls for cytoplasmic fraction and nuclear fraction, respectively. The relative amounts of p53 in each fraction were shown. (E-H) MEFs of indicated genotypes were stained with p53 antibody and counter-stained with DAPI, and cell images were taken by microscope. Representative images were shown.
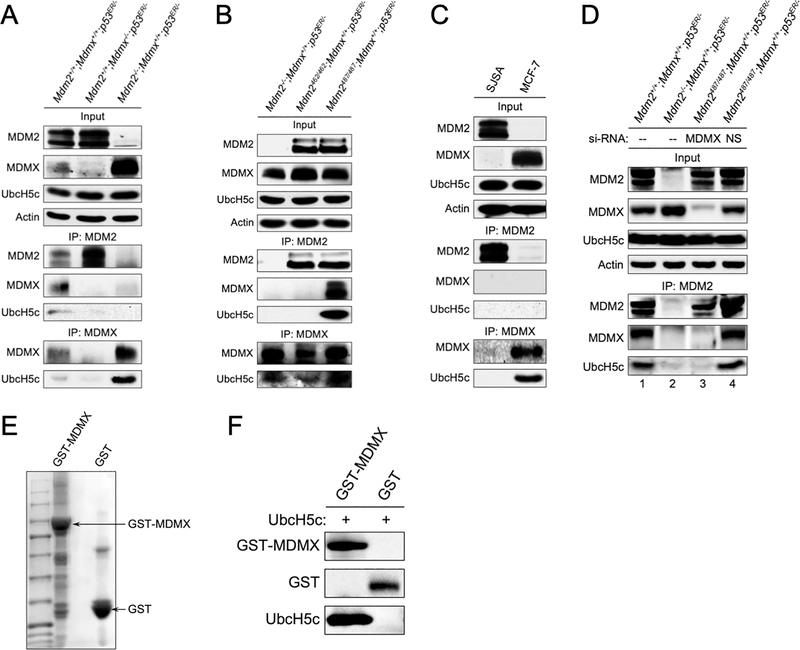
MDMX interacts with UbcH5c
Studies have shown that down-regulation of UbcH5c increases p53 expression(25), suggesting that UbcH5c may be a decisive E2 for MDM2 mediated p53 degradation. Consistent with UbcH5c being critical for MDM2 E3 ligase activity, in vitro study demonstrated that UbcH5c interacts with MDM2(26). Given that MDMX appears to be critical for the function of MDM2 E3 ligase, we sought to examine whether under in vivo conditions MDMX is involved in MDM2-UbcH5c interaction. We performed IP of MDM2 and IP of MDMX using WT (Mdm2+/+;Mdmx+/+;p53ER/-) MEFs and MEFs lacking either MDM2 (Mdm2−/−;Mdmx+/+;p53ER/-) or MDMX (Mdm2+/+;Mdmx−/−;p53ER/-) and probed for UbcH5c binding. Both MDM2 and MDMX were able to pull down UbcH5c from WT MEF lysates (Figure 5A, left lane). Surprisingly, MDM2-UbcH5c binding was barely detectable in Mdm2+/+;Mdmx−/−;p53ER/- cell lysates, despite a high level of MDM2 was immunoprecipitated (Figure 5A, middle lane). In contrast, IP of MDMX was able to pull down substantial amount of UbcH5c in Mdm2−/−;Mdmx+/+;p53ER/- cell lysates (Figure 5A, right lane). The data suggested that in vivo most likely MDMX, but not MDM2, directly interacts with UbcH5c, and the UbcH5c that pulled down by MDM2 IP in the Mdm2+/+;Mdmx+/+;p53ER/- cell lysates could be bridged by MDMX. To test this possibility, we performed MDM2 IP using Mdm2C462A/C462A;Mdmx+/+;p53ER/- MEF cell lysates, where the mutant MDM2C462A does not bind MDMX(24), and Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- MEF cell lysates, where the mutant MDM2Y487A binds MDMX(14). While both MDM2 and MDMX were abundantly expressed in these cell lysates, IP of MDM2 could only pull down UbcH5c in the Mdm2Y487A/Y487A;Mdmx+/+;p53ER/- cell lysates, correlating with the ability of MDM2Y487A to interact with MDMX, suggesting that MDMX bridges MDM2-UbcH5c interaction (Figure 5B). We also tested MDM2 and MDMX binding to UbcH5c in human osteosarcoma SJSA cells, which express high levels of MDM2 but no detectable MDMX, and breast cancer MCF-7 cells, which express high levels of MDMX but no detectable MDM2(27). Consistent with MEF cell data, although MDM2 was abundantly expressed in SJSA cells, no MDM2-UbcH5c binding was detected, whereas MDMX IP identified UbcH5c in MCF-7 cells (Figure 5C). To further confirm the observed MDM2-UbcH5c interaction is bridged by MDMX, we carried out MDM2 IP assays in MEFs in the presence of MDMX knockdown. Consistent with the notion that MDMX is required for MDM2-UbcH5c interaction, knockdown MDMX diminished MDM2 IP pulling down of UbcH5c (Figure 5D, compare lane 3 and lane 4). Moreover, we tested whether the interaction between MDMX and UbcH5c is direct using purified MDMX and UbcH5c proteins. Our data showed that purified MDMX could bind purified UbcH5c in an in vitro assay, indicating that they directly interact (Figure 5E–5F). All together, these data indicated that MDMX, but not MDM2, interacts with UbcH5c, and suggested that MDMX could act as an “E2 carrier” to bring UbcH5c to the proximity of MDM2 to facilitate MDM2-mediated p53 ubiquitination and degradation.
Figure 5. MDMX interacts with UbcH5c.
(A) MEFs of indicated genotypes were treated with 4-OHT for 2 hours, cell lysates were harvested and immunoprecipitated (IP) for MDM2 or MDMX, and blotted for indicated proteins. Input represents 5% of total cell lysate utilized for IP. (B) MEFs of indicated genotypes were treated as in A and immunoprecipitated for MDM2 and MDMX and blotted for indicated proteins. (C) SJSA and MCF-7 cells were immunoprecipitated (IP) for MDM2 or MDMX, and immunoblotted with indicated antibodies. Input represents 5% of total cell lysate utilized for IP. (D) MEFs of indicated genotypes were treated with siRNA against Mdmx (MDMX) or non-specific sequences (NS) for 24 hour followed by treatment with 4-OHT for 2 hours. Cell lysates were harvested and immunoprecipitated for MDM2 and blotted for indicated proteins. (E) MDMX expressed in bacterial was analyzed by SDS-PAGE and Commassie Blue staining. (F) MDMX-UbcH5c complexes were resolved by SDS-PAGE and blotted for MDMX, GST, and UbcH5c.
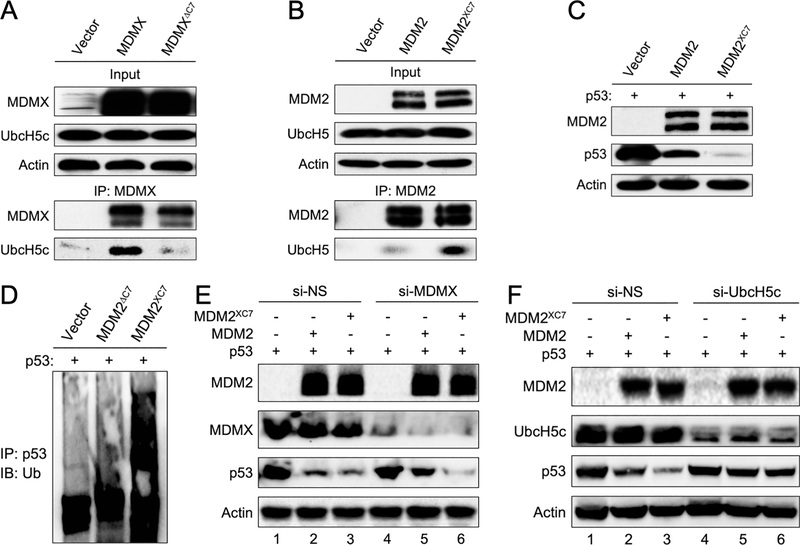
The C-terminus of MDMX is essential for UbcH5c binding and MDM2 E3 activity
Previous studies have shown that deletion or mutation of the C-terminal residues of MDM2 ablate its ability to degrade p53, and the diminished MDM2 E3 ligase activity can be restored by substituting the C-terminus of MDM2 with the corresponding C-terminus of MDMX(13,28). We speculated that the reason for MDMX C-terminus being able to substitute MDM2 C-terminus and restore MDM2 E3 activity is because the MDMX C-terminus is important for UbcH5c binding. We constructed MYC-tagged WT MDMX and MDMX mutant lacking the C-terminal seven residues (deleting residues 484–490, MDMXΔC7). We ectopically expressed these MDMX proteins in U2OS cells and examined their interaction with endogenous UbcH5c by IP-western assays. Ectopic WT MDMX interacted with UbcH5c (Figure 6A, middle lane), however ectopic MDMXΔC7 interacted with UbcH5c very poorly (Figure 6A, right lane), suggesting that the C-terminal seven residues of MDMX are important for UbcH5c binding. Since MDM2 does not detectably interact with UbcH5c (Figure 5), we wondered whether the differences in the extreme C-terminal amino acid sequences between MDM2 and MDMX determine their UbcH5c binding ability. We constructed MDM2 mutant in which the last seven amino acids were replaced by those of MDMX (termed as MDM2XC7) and tested binding of MDM2XC7 with UbcH5c. Remarkably, the binding affinity of MDM2XC7 with UbcH5c significantly increased as compared with WT MDM2 (Figure 6B). The increased UbcH5c binding capability of MDM2XC7 was also reflected in its activity to ubiquitinate and degrade p53, as shown by increased p53 polyubiquitination and degradation by the MDM2XC7 mutant in co-transfected Mdm2−/−;p53−/− MEF cells (Figure 6C–6D). Hence, the C-terminus of MDMX is critical for interacting with UbcH5c and grafting the C-terminal residues to MDM2 enabled MDM2 to interact with UbcH5c and enhanced MDM2 E3 ligase activity for p53 degradation.
Figure 6. MDMX C-terminus is essential for UbcH5c binding and p53 degradation.
(A) U2OS cells were transfected with empty vector DNA, MDMX or MDMXΔC7 plasmid DNA. IP was performed with an anti-MDMX antibody and blotted for the indicated proteins. (B) U2OS cells were transfected with vector, MDM2 or MDM2XC7 plasmid DNA. IP was performed with an anti-MDM2 antibody and blotted for with the indicated proteins. (C) 2KO (Mdm2−/−;p53−/−) MEFs were co-transfected with p53 and MDM2 or MDM2XC7 plasmid DNA. The levels of MDM2, p53 and actin were analyzed by western blot. (D) 2KO MEFs were co-transfected with vector, MDMXΔC7 or MDM2XC7 plasmid DNA along p53 DNA. The cell lysates were isolated and immunoprecipitated with an anti-p53 antibody and analyzed by western blot using an anti-ubiquitin antibody. (E) 2KO MEFs were treated with siRNA against MDMX or non-specific sequences (NS) for 24 hours followed by transfection with indicated plasmid DNA for another 24 hours. Cell lysates were blotted for the indicated proteins. (F) 2KO MEFs were treated with siRNA against UbcH5c or non-specific sequences (NS) for 24 hours followed by transfection with indicated plasmid DNA for another 24 hours. Cell lysates were blotted for the indicated proteins.
We reasoned that if a role for MDMX is to interact with UbcH5c and bring it to MDM2 for its E3 activity, a fusion MDM2XC7 mutant, which can interact UbcH5c, might be able to degrade p53 in the absence of MDMX. We tested this possibility by knockdown MDMX in Mdm2−/−;p53−/− MEFs followed by co-expressing MDM2 and MDM2XC7 mutant with p53. Knockdown MDMX hindered p53 degradation by WT MDM2 (Figure 6E, compare lane 2 and lane 5), but it hardly had any effect on p53 degradation by MDM2XC7 mutant (Figure 6E, compare lane 3 and lane 6). To further demonstrate UbcH5c is involved in MDM2 mediated p53 degradation, we knocked down UbcH5c in Mdm2−/−;p53−/− MEFs and showed that reducing UbcH5c decreased p53 degradation by both MDM2 and MDM2XC7 (Figure 6F).
DISCUSSION
MDM2 E3 ligase activity is thought to be the primary mechanism for MDM2-mediated p53 regulation, whereas MDMX is thought to either enhance or inhibit MDM2 E3 activity. The contribution of MDMX to MDM2-mediated p53 regulation, particularly under physiological conditions, remains controversial, partly because most studies have been performed using ectopically expressed proteins. Due to the fact that deletion of either MDM2 or MDMX causes p53-induced embryonic lethality in mice, it has been challenging to study regulation of p53 by MDM2 or MDMX individually under in vivo conditions. To resolve the confusion of the role of MDMX in p53 degradation and to study p53 regulation by MDM2 and MDMX under physiological conditions, we took advantage of an inducible p53ER system, in which the endogenous p53 gene is replaced by one encoding full-length p53 fused C-terminally with the hormone-binding domain of estrogen receptor(17), and the p53 activity in these mice can be switched “on” and “off” in the presence and absence of 4-OHT, respectively. We generated a series of mice that express p53ER under various MDM2 and MDMX deletion or mutation backgrounds. Contrary to previous studies indicating MDM2 can degrade p53 by itself(29), using inducible p53ER mice and MEF cells our data showed that in the absence of MDMX p53 protein is barely degraded in mice and cells containing WT MDM2 (Figure 1), suggesting that in vivo MDMX plays an essential role in MDM2-induced p53 degradation. Our work is consistent with recent in vivo studies reporting the positive contribution of MDMX to the regulation of p53 protein expression(15,16). Our study revealed that MDMX augments MDM2-mediated p53 degradation most likely through multiple mechanisms including promoting MDM2-p53 binding (Figure 3), facilitating p53 cytoplasmic localization (Figure 4), and interacting with UbcH5c and bringing it to MDM2 proximity (Figure 5). We recognize that the p53ER fusion protein, although is regulated by the native promoter and functions similarly as the endogenous WT p53, is not exactly the same as the WT p53. For example, the p53ER has a longer half-life around 50 min compared to WT p53’s half-life of 15–30 min(18). Nonetheless, the inducible feature of the p53ER makes it a convenient system to study in vivo regulation of p53 without encountering the lethality caused by deletion or mutation of MDM2 and MDMX.
The MDM2-MDMX heterodimer is the preferred form for p53 binding
Several mouse models have demonstrated the importance of the MDM2-MDMX heterodimer in p53 regulation. For example, p53-dependent embryonic lethality is observed in MdmxΔRING and MdmxC462A mice, where MDM2-MDMX heterodimerization is disrupted(15,16). Similar to the MDMXC462A RING finger mutant mice, the MDM2C462A mice, in which the MDM2C462A RING figure mutation disrupts E3 ligase activity and blocks MDMX binding, die in embryogenesis(24). In contrast, the MDM2Y487A mice, in which MDM2 E3 ligase activity is inhibited by the Y487A mutation but the MDM2-MDMX interaction is retained, live a normal lifespan, demonstrating that the MDM2-MDMX heterodimer is critical for p53 suppression, at least under normal unstressed growth conditions(14). Both MDM2 and MDMX bind the p53 transactivation domain through their N terminus(3), but the p53 binding efficiency by either protein alone had not been determined under physiological conditions. We demonstrated that under physiological conditions although both MDM2 and MDMX can bind p53 by themselves, the MDM2-MDMX heterodimer binds p53 more efficiently than either individual protein (Figure 3).
There are several ways in which the MDM2-MDMX heterodimer could facilitate stronger p53 binding than either protein alone. In vitro studies have shown that the acidic domain (AD) of MDM2 weakly interacts with the DNA-binding domain of p53(30). Unlike the RING domain of MDM2 that is required for MDM2 homodimerization with itself and heterodimerization with MDMX, the AD of MDM2 is required for MDM2 homodimerization, but not for MDM2-MDMX heterodimerization(31). Hence, it is possible that when MDM2 or MDMX is present as a homodimer, their two acidic domains interact, competing against p53 binding, while MDM2-MDMX heterodimerization prevents acidic domain interaction and facilitates p53 binding.
A recent study demonstrated that the AD and the RING domain of MDMX interact internally with the p53 binding domain(32), and the AD of MDMX can also form an intramolecular interaction with p53 binding domain and RING domain(33), which could prevent MDMX-p53 binding. Since in vivo MDMX homodimer has not been detected, it is thought that MDMX exists either as a monomer or as a heterodimer with MDM2(34). The RING domain of MDM2 has a higher affinity to bind the RING domain of MDMX than that of other MDM2 molecules(35). It is possible that upon the formation of an MDM2-MDMX heterodimer, the AD of both MDM2 and MDMX may release from their respective intermolecular and intramolecular interactions, and open up for p53 binding.
The MDM2-MDMX heterodimer facilitates p53 cytoplasmic localization
MDMX is a predominantly cytoplasmic protein. The function of MDMX in the cytoplasm remains partially understood. It has been proposed that cytoplasmic MDMX can play both and anti-apoptosis and a pro-apoptosis roles, and the choice of which depends on the types of cellular stress and levels of MDM2(36). Moreover, MDMX can act as a sequestering factor for cytoplasmic p53 and inhibit p53-induced apoptosis(37). However, opposing models suggest that MDMX sequesters p53 in the cytoplasm and protects it from MDM2 mediated degradation(38). We and others have previously shown that blocking p53 nuclear export prevents MDM2 mediated p53 degradation(39,40), suggesting that p53 degradation occurs mainly in the cytoplasm. Given that MDMX plays an essential role for MDM2 mediated p53 degradation, it can be envisioned that one of the mechanisms of MDMX to promote p53 degradation is to facilitate p53 to localize into the cytoplasm. Our data showed that MDMX is indeed plays a role in promoting p53 cytoplasmic accumulation (Figure 4). Interestingly, this function of MDMX seems to require the formation of MDM2-MDMX heterodimer, as mere presence of MDMX without MDM2 (Figure 4D), or in the presence of a mutant MDM2 that cannot form heterodimer with MDMX (Figure 4C), high level of p53 accumulation in the cytoplasm is not observed.
MDMX interacts with UbcH5c
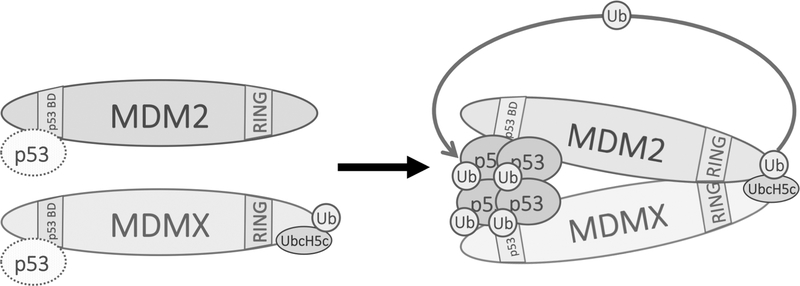
Previous studies have shown that UbcH5b/c functions as an E2 enzyme for MDM2 E3 ligase(25). However, whether UbcH5b/c directly interacts with MDM2 was unknown. Our study demonstrated that under physiological conditions MDMX, but not MDM2, interacts with UbcH5c (Figure 5). Importantly, we found that a segment of seven residues from MDMX C-terminus are critical for UbcH5c binding (Figure 6A). Grafting the seven residues to MDM2 rendered MDM2 ability to bind UbcH5c (Figure 6B), enhanced MDM2 E3 ligase activity (Figure 6C–D), and made MDM2 E3 activity somewhat independent of MDMX (Figure 6E). However, it appears that the seven C-terminal residues of MDMX are necessary but not sufficient for UbcH5c binding since grafting the seven residues to GFP did not give the GFP fusion protein ability to bind UbcH5c. Hence, it seems that sequences/residues in MDMX other than the last seven residues are involved in UbcH5c binding. In case of the MDM2XC7 fusion protein, we believe the C-terminally fused residues from MDMX together with other part of MDM2 gave the fusion protein the UbcH5c binding ability. Collectively, our data suggest a model that in vivo MDMX interacts with UbcH5c via its C-terminus and brings UbcH5c to MDM2 proximity to facilitate transfer of UbcH5c to MDM2 (Figure 7). Previous studies have shown that ectopically expressed MDMX can restore MDM2 E3 ligase activity in C-terminal MDM2 mutants(13). Our study complements these studies by showing evidences suggesting that MDMX enables MDM2 E3 ligase activity through bringing UbcH5c to nearby MDM2 RING domain thereby facilitating MDM2 transferring ubiquitin from UbcH5c to p53. Alternatively, it is also possible that MDM2-MDMX binding induces conformational changes in the MDM2 RING domain, allowing for MDM2 itself to interact with UbcH5c and enable its E3 ligase activity. This type of conformation-changing interaction has been demonstrated in other E3 ligases, such as cIAP1-SMAC or DIABLO(41). Furthermore, previous studies have shown that the AD of MDM2 is not only necessary for MDM2 homodimerization but is also involved in activating MDM2 catalytic activity(31,42). Binding of MDMX to MDM2 could release the AD, allowing it to “turn on” MDM2 catalytic activity and thus, ubiquitin transfer.
Figure 7. A hypothetic model for MDMX facilitating MDM2 E3 ligase function.
Singular MDM2 or MDMX weakly interact with p53 (shown by dashed molecules of p53) through their N-terminus. When MDM2 and MDMX form a heterodimer via their respective C-terminal RING domains, the binding affinity of the MDM2-MDMX heterodimer with p53 is increased (shown by solid molecules of p53). At the same time, MDMX brings UbcH5c to MDM2 proximity, activates MDM2 RING E3 ligase activity to transfer ubiquitin to p53.
STATEMENT OF SIGNIFICANCE.
This study provides the first in vivo evidence of MDMX facilitating MDM2-mediated p53 degradation, clarifying its role in the regulation of this critical tumor suppressor.
ACKNOWLEDGEMENTS
We thank Klaus Hahn, Robert Bagnell, Daniel Marston, Yong Liu, Shijie Liu, Patrick Leslie, Derek Franklin, and Orrin Stone for their helpful advice and technical assistance. We thank Jiandong Chen for sharing reagents. This research was supported by grants from the NIH (CA167637, CA155235, CA212407) to Y. Zhang; the 333 projects of Jiangsu Province (LGY2017094), the Foundation for Key Program of Universities of Jiangsu Province (No. 17KJA320010), the Jiangsu Distinguished Professorship Program and Jiangsu Shuangchuang Program to J. Yang; the Jiangsu Provincial Key Medical Discipline and the Project of Invigorating Health Care through Science, Technology and Education (No. ZDXKA2016014) to J. Zheng.
Footnotes
CONFLICTS OF INTEREST STATEMENT
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009;137(3):413–31. [DOI] [PubMed] [Google Scholar]
- 2.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nature cell biology 2000;2:569–73. [DOI] [PubMed] [Google Scholar]
- 3.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69(7):1237–45. [DOI] [PubMed] [Google Scholar]
- 4.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206–08. [DOI] [PubMed] [Google Scholar]
- 5.Luna RM, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203–06. [DOI] [PubMed] [Google Scholar]
- 6.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29(1):92–5. [DOI] [PubMed] [Google Scholar]
- 7.Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G. Mdm2 and Mdm4 loss regulates distinct p53 activities. Molecular cancer research : MCR 2008;6(6):947–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proceedings of the National Academy of Sciences of the United States of America 2006;103(9):3232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Molecular and cellular biology 2000;20(3):1001–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS letters 2009;583(17):2710–4. [DOI] [PubMed] [Google Scholar]
- 11.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proceedings of the National Academy of Sciences of the United States of America 2003;100(21):12009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. The Journal of biological chemistry 2011;286(27):23725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. The EMBO journal 2007;26(1):102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tollini LA, Jin A, Park J, Zhang Y. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer cell 2014;26(2):235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proceedings of the National Academy of Sciences of the United States of America 2011;108(29):11995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proceedings of the National Academy of Sciences of the United States of America 2011;108(29):12001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet 2005;37(7):718–26. [DOI] [PubMed] [Google Scholar]
- 18.Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer cell 2006;10(6):501–14. [DOI] [PubMed] [Google Scholar]
- 19.Alarcon R, Koumenis C, Geyer RK, Maki CG, Giaccia AJ. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer research 1999;59(24):6046–51. [PubMed] [Google Scholar]
- 20.Garcia D, Warr MR, Martins CP, Brown Swigart L, Passegue E, Evan GI. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev 2011;25(16):1746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein Involved in ribosome biogenesis and cell proliferation. Molecular cell 2003;12(5):1151–64. [DOI] [PubMed] [Google Scholar]
- 22.Wu QY, Zhu YY, Liu Y, Wei F, Tong YX, Cao J, et al. CUEDC2, a novel interacting partner of the SOCS1 protein, plays important roles in the leukaemogenesis of acute myeloid leukaemia. Cell death & disease 2018;9(7):774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstrom MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. The Journal of biological chemistry 2008;283(23):15568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, et al. Targeted Inactivation of Mdm2 RING Finger E3 Ubiquitin Ligase Activity in the Mouse Reveals Mechanistic Insights into p53 Regulation. Cancer cell 2007;12(4):355–66. [DOI] [PubMed] [Google Scholar]
- 25.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. The Journal of biological chemistry 2004;279(40):42169–81. [DOI] [PubMed] [Google Scholar]
- 26.Ranaweera RS, Yang X. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. The Journal of biological chemistry 2013;288(26):18939–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose M, Yamato K, Endo S, Saito R, Ueno T, Hirai S, et al. MDM4 expression as an indicator of TP53 reactivation by combined targeting of MDM2 and MDM4 in cancer cells without TP53 mutation. Oncoscience 2014;1(12):830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. The EMBO journal 2007;26(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299–303. [DOI] [PubMed] [Google Scholar]
- 30.Cross B, Chen L, Cheng Q, Li B, Yuan ZM, Chen J. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. The Journal of biological chemistry 2011;286(18):16018–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leslie PL, Ke H, Zhang Y. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. The Journal of biological chemistry 2015;290(20):12941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Wu S, Song T, Chen L, Gao M, Borcherds W, et al. Secondary interaction between MDMX and p53 core domain inhibits p53 DNA binding. Proceedings of the National Academy of Sciences of the United States of America 2016;113(19):E2558–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Borcherds W, Wu S, Becker A, Schonbrunn E, Daughdrill GW, et al. Autoinhibition of MDMX by intramolecular p53 mimicry. Proceedings of the National Academy of Sciences of the United States of America 2015;112(15):4624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Molecular cancer research : MCR 2009;7(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer research 2007;67(13):6026–30. [DOI] [PubMed] [Google Scholar]
- 36.Mancini F, Moretti F. Mitochondrial MDM4 (MDMX): an unpredicted role in the p53-mediated intrinsic apoptotic pathway. Cell Cycle 2009;8(23):3854–9. [DOI] [PubMed] [Google Scholar]
- 37.Ohtsubo C, Shiokawa D, Kodama M, Gaiddon C, Nakagama H, Jochemsen AG, et al. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Sci 2009;100(7):1291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. MolCell Biol 2000;20:1001–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Xiong Y. Mutation in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Molecular cell 1999;3:579–91. [DOI] [PubMed] [Google Scholar]
- 40.Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. MolCell Biol 1998;18:7288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 2011;334(6054):376–80. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Q, Song T, Chen L, Chen J. Autoactivation of the MDM2 E3 ligase by intramolecular interaction. Molecular and cellular biology 2014;34(15):2800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]