Abstract
Patients with myelodysplastic syndromes (MDS) display severe anemia but the mechanisms underlying this phenotype are incompletely understood. Right open-reading-frame kinase 2 (RIOK2) encodes a protein kinase located at 5q15, a region frequently lost in MDS del(5q) patients. Here, we show that hematopoietic cell-specific haploinsufficient deletion of Riok2 (Riok2f/+Vav1cre) led to reduced erythroid precursor frequency leading to anemia. Proteomic analysis of Riok2f/+Vav1cre erythroid precursors suggested immune system activation and transcriptomic analysis revealed an increase in p53-dependent interleukin-22 (IL-22) in Riok2f/+Vav1cre CD4+ T cells (TH22). Further, we discovered that the IL-22 receptor, IL-22RA1, was unexpectedly present on erythroid precursors. Blockade of IL-22 signaling alleviated anemia not only in Riok2f/+Vav1cre mice but also in wild-type mice. Serum concentrations of IL-22 were increased in the subset of del(5q) MDS patients as well as patients with anemia secondary to chronic kidney disease (CKD). This work reveals a possible therapeutic opportunity for reversing many stress-induced anemias by targeting IL-22 signaling.
Keywords: IL-22, del(5q) MDS, anemia, myeloproliferation, Riok2, p53
Introduction
Myelodysplastic syndromes (MDS) are a group of cancers characterized by failure of blood cells in the bone marrow to mature. About 7 out of 100,000 people are affected and the typical survival time following diagnosis is less than three years. While a sizable percentage of MDS cases progress to acute myelogenous leukemia (AML), most of the morbidity and mortality associated with MDS results not from transformation to AML but rather from hematological cytopenias.
Anemia is the most common hematologic manifestation of MDS, particularly in the subset of patients with del(5q) MDS. Del(5q), either isolated or accompanied by additional cytogenetic abnormalities, is the most commonly detected chromosomal abnormality in MDS, reported in 10–15% of patients1-4. The severe anemia in del(5q) MDS patients has been linked to haploinsufficiency of ribosomal proteins such as RPS14 and RPS195; 6. Previous studies using mice with haploinsufficient 5q gene deletions revealed diminished erythroid progenitor frequency7-10 but the mechanisms underlying this phenotype are incompletely understood. Right open-reading-frame kinase 2 (RIOK2) encodes an atypical serine-threonine protein kinase with an indispensable function as a component of the pre-40S ribosome subunit16.
There is growing evidence for the role of activated innate immunity and inflammation as well as immune dysregulation in the pathogenesis of MDS17-20. Abnormal expression of numerous cytokines has been reported in MDS21-24. Chronic immune stimulation in both hematopoietic stem and progenitor cells (HSPCs) and the bone marrow (BM) microenvironment25 was suggested to be central to the pathogenesis of MDS. In patients with chronic inflammation, cytokines in the BM have been associated with inhibition of erythropoiesis26. Despite growing evidence for a link between the immune system and MDS pathogenesis, no study has identified the mechanism by which the immune microenvironment may initiate or contribute to the MDS phenotype. Further it remains unclear how ribosomal protein haploinsufficiency is connected with the immune system in MDS.
Results
Riok2 haploinsufficiency leads to anemia
We previously noted that Riok2 expression was reduced in T cells lacking the endoplasmic reticulum stress transcription factor Xbp127. RIOK2 is a little-studied atypical serine-threonine protein kinase11 encoded by RIOK2 at 5q15 in the human genome (Extended Data Fig. 1a), adjacent to the 5q commonly deleted regions in MDS and frequently lost in MDS and acute myeloid leukemia12-15.Gene expression commons (GEXC)28 analysis revealed that in mouse BM, Riok2 expression is highest in primitive colony-forming-unit erythroid (pCFU-E) cells, suggesting that RIOK2 may be involved in maintaining red blood cell (RBC) output (Extended Data Fig. 1d). To further study the role of RIOK2 in hematopoiesis, we generated Vav1-Cre transgenic floxed Riok2 (Riok2f/+Vav1cre) mice in which the Cre recombinase is under the control of the hematopoietic cell–specific Vav1 promoter. Riok2 floxed mice were generated with exons 5 and 6 flanked by loxP sites (Extended Data Fig. 1b, c). Interestingly, no Vav1-Cre floxed Riok2 homozygous-deficient mice (Riok2f/fVav1cre) were recovered (Extended Data Fig. 1f), indicating embryonic lethality from complete hematopoietic deletion of Riok2. However, heterozygous Riok2f/+Vav1cre mice were viable with approximately 50% Riok2 mRNA expression in hematopoietic cells compared to that of Vav1Cre controls (Extended Data Fig. 1e). As seen with other ribosomal protein haploinsufficiency mouse models10, BM cells from Riok2f/+Vav1cre mice showed reduced nascent protein synthesis in vivo compared to Vav1Cre controls (Extended Data Fig. 1g), consistent with a RIOK2 role in maturation of the pre-40S ribosome. A recent study29 showed that ribosomal protein deficiency-mediated reduced protein synthesis predominantly affects erythropoiesis over myelopoiesis.
Consistent with the high expression of Riok2 in pCFU-e cells in the BM, aged (>60 wks) mice with heterozygous deletion of Riok2 in hematopoietic cells (Riok2f/+Vav1cre) displayed anemia with reduced peripheral blood (PB) RBC numbers, hemoglobin (Hb), and hematocrit (HCT) (Fig. 1a). We next determined whether Riok2 haploinsufficiency-mediated anemia was secondary to a defect in erythroid development in the BM, the major site of erythropoiesis. We characterized the stages (referred to here as RI, RII, RIII and RIV) of erythropoiesis by flow cytometry using the expression of Ter119 and CD71 (Extended Data Fig. 2a). Riok2f/+Vav1cre mice had impaired erythropoiesis in the BM (Fig. 1b, Extended Data Fig. 2b). Moreover, Riok2 haploinsufficiency led to increased apoptosis in erythroid precursors compared to controls (Fig. 1c). Additionally, Riok2f/+Vav1cre erythroid precursors showed a decrease in cell quiescence with cell cycle block at the G1 phase (Extended Data Fig. 2c). A block in cell cycle is driven by a group of proteins known as cyclin-dependent kinase inhibitors (CKI). The expression of p21 (a CKI encoded by Cdkn1a) was increased in erythroid precursors from Riok2f/+Vav1cre mice compared to Riok2+/+Vav1cre controls (Extended Data Fig. 2d).
Figure 1. Riok2 haploinsufficient (Riok2f/+Vav1cre) mice display anemia and myeloproliferation.
(a) Peripheral blood (PB) RBC numbers, hemoglobin (Hb), and hematocrit (HCT) in Riok2f/+Vav1cre mice in comparison to Riok2+/+Vav1cre controls (n=5/group). (b) Frequency of erythroid progenitor/precursor populations among viable bone marrow (BM) cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=5/group). (c) Frequency of apoptotic erythroid precursors among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=5/group). (d) PB RBC numbers, Hb, and HCT in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls undergoing phenylhydrazine (PhZ)-induced stress erythropoiesis (n=7/group). (e) Frequency of RIII and RIV erythroid precursor populations among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls day 6 after PhZ treatment (n=4/group). (f) Number of CFU-e colonies in Epo-containing MethoCult assay using Lin−c-kit+CD71+ cells from Riok2f/+Vav1cre mice (n=4) in comparison to Riok2+/+Vav1cre controls (n=6). (g) Percentage of monocytes (CD11b+Ly6G−Ly6Chi) and neutrophils (CD11b+Ly6G+) in the PB of Riok2f/+Vav1cre mice (n=6) in comparison to Riok2+/+Vav1cre controls (n=5). (h) Percentage of Ki-67+ granulocyte-macrophage progenitors (GMPs) in the BM of Riok2f/+Vav1cre mice (n=3) and Riok2+/+Vav1cre controls (n=4). (i) Percentage of CD11b+ cells obtained from Lin−Sca-1+c-kit+ BM cells from Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls cultured in MethoCult for 7 days (n=5/group). Unpaired two-tailed t-test (a to c, e to i) and 1-way ANOVA with Tukey’s correction for multiple comparisons (d) used to calculate statistical significance. * p < 0.05, ** p < 0.01, *** p < 0.001. Data are shown as mean ± s.e.m and are representative of two (c, d, f-h) or three (a, b, e, i) independent experiments.
We next examined the effect of Riok2 haploinsufficiency on stress-induced erythropoiesis using 8-12 wk old mice in which hemolysis was induced by non-lethal phenylhydrazine treatment (25 mg/kg on days 0 and 1). After acute hemolytic stress, Riok2f/+Vav1cre mice developed more severe anemia and had a delayed RBC recovery response, as compared to Riok2+/+Vav1cre control mice (Fig. 1d) and succumbed faster to a lethal dose of phenylhydrazine (35 mg/kg on days 0 and 1) compared to Vav1cre controls (Extended Data Fig. 2e). The anemia in young phenylhydrazine-administered Riok2f/+Vav1cre mice seen on day 7 was preceded by a reduction in BM RIII and RIV erythroid precursor frequency on day 6, highlighting an erythroid differentiation defect in Riok2 haploinsufficient mice (Fig. 1e, Extended Data Fig. 2f). In line with a role for RIOK2 in driving erythroid differentiation, fewer CFU-e colonies were observed in erythropoietin-containing MethoCult cultures from Riok2 haploinsufficient Lin−c-kit+CD71+ cells compared to Riok2 sufficient cells (Fig. 1f). To determine whether Riok2 haploinsufficiency in BM cells drives anemia, we generated BM chimeras. Wild-type (WT) mice transplanted with Riok2f/+Vav1cre whole BM developed anemia compared to Riok2+/+Vav1cre BM transplanted wt mice (Extended Data Fig. 2g). In addition, tamoxifen-inducible deletion of Riok2 in Riok2f/+Ert2cre mice led to reduction in PB RBCs, Hb, and HCT compared to Riok2+/+Ert2cre controls (Extended Data Fig. 2h). Taken together, these data show that Riok2 haploinsufficiency leads to anemia owing to defective bone marrow erythroid differentiation.
Riok2 haploinsufficiency increases myelopoiesis
In addition to the reduction in RBC numbers in PB from aged Riok2f/+Vav1cre mice, we also observed an increased percentage of monocytes (monocytosis) and decreased percentage of neutrophils (neutropenia) compared to controls (Fig. 1g, Extended Data Fig. 2i). Granulocyte macrophage progenitors (GMPs) in the BM give rise to PB myeloid cells. The percentage of proliferating (Ki67+) GMPs in the BM was increased in Riok2f/+Vav1cre mice compared to Riok2+/+Vav1cre controls (Fig. 1h, Extended Data Fig. 2j). To analyze the effect of Riok2 haploinsufficiency on myelopoiesis in the absence of in vivo compensatory mechanisms, we cultured LSK (lineage−Sca-1+Kit+) cells from the BM of Riok2f/+Vav1cre and Riok2+/+Vav1cre controls in a MethoCult assay supplemented with growth factors (interleukin 6 (IL-6), IL-3, stem cell factor (SCF) but devoid of erythropoietin). LSKs from Riok2f/+Vav1cre mice gave rise to an increased percentage of CD11b+ myeloid cells (Fig. 1i, Extended Data Fig. 2k), suggesting a cell-intrinsic myeloproliferative effect due to Riok2 haploinsufficiency consistent with a myelodysplasia phenotype.
We also evaluated whether Riok2 haploinsufficiency affects early hematopoietic progenitors. Frequency and numbers of early hematopoietic progenitors were comparable between young Riok2f/+Vav1cre and Riok2+/+Vav1cre mice (Extended Data Fig. 3a), however, long-term hematopoietic stem cells (LT-HSCs) were increased in the BM of aged Riok2f/+Vav1cre mice (Extended Data Fig. 3a). To further corroborate this data, we analyzed the capacity of Riok2 haploinsufficient cells in a competitive transplantation assay. Starting at 8 weeks after tamoxifen treatment to induce Riok2 deletion, Riok2 haploinsufficient cells out-competed CD45.1+ competitor cells, while Ert2cre control cells had no competitive advantage (Extended Data Fig. 3b). Similar to non-transplanted mice (Extended Data Fig. 3a), in competitive transplant experiments the frequency of Riok2-haploinsufficient LT-HSCs was significantly higher than Riok2-sufficient LT-HSCs in relation to competitor CD45.1+ cells (Extended Data Fig. 3c). Thus, in addition to its effect on erythroid differentiation, Riok2 haploinsufficiency increases myelopoiesis and affects early hematopoietic progenitor differentiation.
Reduced Riok2 induces alarmins in erythroid precursors
To elucidate a mechanism for the erythroid differentiation defect observed in Riok2f/+Vav1cre mice, we performed quantitative proteomic analysis of purified erythroid precursors using mass spectrometry30. Riok2 haploinsufficiency led to upregulation of 564 distinct proteins (adjusted p-value < 0.05) in erythroid precursors compared to those from Vav1cre controls (Fig. 2a). Interestingly, Riok2 haploinsufficiency resulted in down-regulation of other ribosomal proteins, loss of some of which (RPS5, PRL11) has been implicated in driving anemias (Extended Data Fig. 4a). The alarmins including S100A8, S100A9, CAMP, NGP, and others were the most highly upregulated proteins in our dataset and interestingly, correlated significantly with those observed upon haploinsufficiency of Rps1410, another component of the 40S ribosomal complex (Fig. 2b). Using the 26 upregulated proteins in the Rps14 haploinsufficient dataset as an ‘Rps14 signature’ (Supplementary Table 1), gene set enrichment analysis (GSEA) revealed a marked enrichment for the Rps14 signature in the Riok2 haploinsufficient dataset, suggesting a shared proteomic signature upon deletion of distinct ribosomal proteins (Fig. 2c). The increased expression of S100A8 and S100A9 in Riok2f/+Vav1cre mice was confirmed by flow cytometry and qRT-PCR (Extended Data Fig. 4b-e).
Figure 2. Quantitative proteomics of Riok2 haploinsufficient erythroid precursors reveals immune activation signatures.
(a) Proteomic analysis of changes in protein expression in erythroid progenitors from Riok2 haploinsufficient mice and Vav1cre controls. n = 4-5 mice/group. (b) Comparison of upregulated proteins with their respective p-values and log fold-change values in erythroid precursors from Riok2 haploinsufficient mice and Rps14 haploinsufficient mice with their respective controls. (c to d, f) GSEA performed on proteomics data shown in (a) to reveal similarity with Rps14 haploinsufficient data (c), activation of immune response (d) and enrichment of IL-22 signature genes (f). NES = Normalized enrichment score, FDR = False discovery rate. (e) MetaCore analysis of the Riok2 proteomics dataset shown in (a). Two sample moderated t-test with multiple hypothesis corrections used to calculate the statistical significance in (b).
In Riok2f/+Vav1cre erythroid precursors cells, the upregulated proteins with the highest fold-change (S100A8, S100A9, CAMP, NGP) are proteins with known immune functions such as antimicrobial defense. GSEA analysis of the proteomics data indicated a possible role for the immune system in driving the proteomic changes seen in Riok2 haploinsufficient erythroid precursors (Fig. 2d). An independent analysis of the Riok2 proteomics dataset using MetaCore pathway analysis software showed immune response as the top differentially regulated pathway in Riok2f/+Vav1cre mice (Fig. 2e). To assess if Riok2 haploinsufficiency leads to changes in immune cell function, we subjected naïve T cells from Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls to in vitro polarization towards known CD4+ T helper cell lineages (TH1, TH2, TH17, TH22) and regulatory T cells (Treg). Secretion of interferon-γ (IFN-γ), IL-2, IL-4, IL-5, IL-13, IL-17A, and the frequency of Foxp3+ Treg cells was similar between Riok2f/+Vav1cre and Riok2+/+Vav1cre T cells (Extended Data Fig. 5a - g). Strikingly, however, we observed an exclusive increase in IL-22 secretion from Riok2f/+Vav1cre naïve T cells polarized towards the TH22 lineage (Fig. 3a). The frequency of IL-22+CD4+ T cells was higher in Riok2f/+Vav1cre TH22 cultures compared to Vav1cre control TH22 cultures (Fig. 3b). The concentration of IL-22 in the serum and BM fluid (BMF) of aged Riok2f/+Vav1cre mice was also significantly higher as compared to age-matched Vav1cre controls (Fig. 3c). Using known IL-22 target genes from the literature, we curated an ‘IL-22 signature’ (Supplementary Table 1) gene set, which showed a statistically significant enrichment in the Riok2-haploinsufficient proteomics dataset using GSEA (Fig. 2f), further suggesting that IL-22-induced inflammation is a contributing factor for Riok2 haploinsufficiency-mediated ineffective erythropoiesis and anemia. Increased numbers of splenic IL-22+ CD4+ T, natural killer T (NKT), and innate lymphoid cells (ILCs) were observed in aged Riok2f/+Vav1cre mice compared to Riok2+/+Vav1cre mice (Fig. 3d, Extended Data Fig. 5h, i). Interestingly, mild anemia was observed in mice lacking Riok2 only in T cells (Extended Data Fig. 5k). Expression of IL-23, required for IL-22 production, was enhanced in Riok2-haploinsufficient dendritic cells (Extended Data Fig. 5j). Rps14 haploinsufficient TH22 cells also secreted elevated concentrations of IL-22 compared to Vav1cre control TH22 cells (Extended Data Fig. 5l). Mutation(s) in the gene adenomatosis polyposis coli (Apc), also found on human chromosome 5q, lead to anemia in addition to adenomas31. In vitro generated TH22 cells from ApcMin mice secreted elevated IL-22 compared to littermate controls (Extended Data Fig. 5m). In total, our analysis of three distinct heterozygous deletions of genes found on human chromosome 5q suggests that increased IL-22 is a generalized phenomenon observed upon heterozygous loss of genes found on chromosome 5q leading to anemia.
Figure 3. Riok2 haploinsufficiency-driven p53 upregulation drives increased IL-22.
(a) Secreted IL-22 and (b) Percentage of IL-22+CD4+ T cells from in vitro polarized TH22 cells from Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=5/group). (c) IL-22 levels in the serum (left) and bone marrow fluid (BMF) (right) in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=5/group). (d) Number of IL-22+CD4+ cells in the spleens of Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=5/group). (e) Volcano plot showing transcriptomic changes in purified IL-22+ cells from Riok2f/+Vav1cre mice in comparison to Riok2+/+Vav1cre controls. n=5/group. (f) GSEA analysis showing activation of the p53 pathway in IL-22+ cells from Riok2f/+Vav1cre mice in comparison to Riok2+/+Vav1cre controls. (g) Snapshot of differentially expressed genes in the p53 pathway shown in (F) in Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. (h) Flow cytometry plot showing p53 expression in in vitro polarized TH22 cells from Riok2f/+Vav1cre and Riok2+/+Vav1cre controls. (i) Graphical representation of data shown in (H). n=5/group. (j) Predicted p53 binding site in the Il22 promoter region. (k) Chromatin immunoprecipitation showing p53 occupancy at the Il22 promoter in T cells. n=2 independent experiments. (l) Secreted IL-22 from wt TH22 cells cultured in the presence or absence of p53 inhibitor, pifithrin-α, p-nitro (1 μM). n=5 mice/group. (m) Secreted IL-22 from WT TH22 cells cultured in the presence or absence of p53 activator, Nutlin-3 (100 nM). n=4 mice/group. (n) Secreted IL-22 from in vitro polarized TH22 cells from the indicated strains. n=5/group. Unpaired two-tailed t-test (a to d, i, k-m) and 1-way ANOVA with Tukey’s correction for multiple comparison (n) used to calculate statistical significance. * p < 0.05, ** p < 0.01, *** p < 0.001. Data are shown as mean ± s.e.m and are representative of two (c - d, h, i, k - n) or three (a,b) independent experiments. Data in (k) is represented as mean ± s.d. and is pooled from two independent experiments.
p53 upregulation drives increased IL-22 secretion upon Riok2 loss
To identify cell-intrinsic molecular mechanism(s) driving the increase in IL-22 secretion upon Riok2 haploinsufficiency, we performed RNA-sequencing (RNA-Seq) on in vitro polarized IL-22+ (TH22) cells purified by flow cytometry from Riok2+/+Il22tdtomato/+Vav1cre and Riok2f/+Il22tdtomato/+Vav1cre mice (Fig. 3e). GSEA analysis of the RNA-Seq dataset identified activation of the p53 pathway in Riok2f/+Vav1cre mice (Fig. 3f, g). p53 increase in TH22 cells from Riok2f/+Vav1cre mice was confirmed by flow cytometry (Fig. 3h, i). p53 upregulation was also observed in Riok2f/+Vav1cre erythroid precursors (Extended Data Fig. 3f, g). The p53 pathway is activated by decreased expression of ribosomal protein genes5; 10, however, its involvement in IL-22 regulation has not been known.
p53 is a transcription factor with well-defined consensus binding sites. To assess whether p53 drives Il22 transcription, we analyzed the Il22 promoter for potential p53 binding sites using LASAGNA algorithm and found putative p53 consensus binding sequences in the Il22 promoter (Fig. 3j). Chromatin immunoprecipitation (ChIP) confirmed the presence of p53 on the Il22 promoter (Fig. 3k). In line with the ChIP data, p53 inhibition by pifithrin-α, p-nitro decreased IL-22 concentrations while p53 activation by nutlin-3 increased IL-22 from in vitro polarized wild-type TH22 cells (Fig. 3l, m). Treatment with either pifithrin-α, p-nitro or nutlin-3 did not decrease cell viability (Extended Data Fig. 5n). Accordingly, genetic deletion of Trp53 blunted the increase in IL-22 secretion observed upon Riok2 haploinsufficiency (Fig. 3n). A significant decrease in IL-22 secretion was also observed upon Trp53 deletion in Riok2-sufficient cells further suggesting a homeostatic role for p53 in controlling IL-22 production (Fig. 3n). Taken together, these data show that Riok2 haploinsufficiency-mediated p53 upregulation drives increased IL-22 secretion in Riok2f/+Vav1cre mice.
IL-22 neutralization alleviates stress-induced anemia
Mice with compound genetic deletion of Il22 on the Riok2 haploinsufficient background (Riok2f/+Il22+/−Vav1cre) exhibited increased numbers of PB RBCs compared to IL-22 sufficient Riok2 haploinsufficient mice on day 7 after two treatments with 25 mg/kg phenylhydrazine treatment (Fig. 4a). Interestingly, we also saw an increase in PB RBCs in Riok2 sufficient mice heterozygous for Il22 deletion (Riok2+/+Il22+/−Vav1cre) compared to Riok2 sufficient IL-22 sufficient mice (Riok2+/+Il22+/+Vav1cre). PB Hb and HCT also were increased in Il22 haploinsufficient mice, regardless of Riok2 background, however, this difference did not reach statistical significance (Fig. 4a). Next, we assessed whether the increase in PB RBCs in Riok2f/+Il22+/−Vav1cre mice was due to increased erythropoiesis in the BM of these mice. We observed an increase in RII and RIV erythroid precursors in Riok2f/+Il22+/−Vav1cre compared to Riok2f/+Il22+/+Vav1cre mice (Fig. 4b, Extended Data Fig. 6a). Treatment of mice with a neutralizing IL-22 antibody in vivo, also reversed phenylhydrazine-induced anemia as evidenced by increase in PB RBCs and HCT in Riok2f/+Vav1cre mice as well as Riok2 sufficient (Riok2+/+Vav1cre) mice (Fig. 4c). Unexpectedly, IL-22 neutralization also reduced the frequency of apoptotic erythroid precursors in both Riok2 sufficient as well as Riok2f/+Vav1cre mice (Fig. 4d). These data indicate that IL-22 neutralization reverses anemia, at least in part, by reducing apoptosis of erythroid precursors. Recently, dampening IL-22 signaling in intestinal epithelial stem cells was shown to reduce apoptosis32. The effect of IL-22 deficiency (genetic as well as antibody-mediated) in alleviating anemia in genetically wild-type mice indicated a role for IL-22 in reversing anemia regardless of ribosomal haploinsufficiency. Accordingly, treatment of C57BL/6J mice undergoing phenylhydrazine-induced anemia with anti-IL-22 significantly increased PB RBCs, Hb, and HCT compared to isotype antibody-treated mice (Extended Data Fig. 7b). This increase in PB RBCs could be attributed to the increased frequency of RIII and RIV erythroid precursors in the BM of mice treated with anti-IL-22 compared to isotype-administered controls (Extended Data Fig. 7c). Of note, PB RBCs, Hb, and HCT did not differ in healthy, non-anemic wild-type mice injected with anti-IL-22 versus isotype-matched antibody (Extended Data Fig. 7a). Thus, we show here that IL-22 neutralization, either by genetic deletion or antibody blockade, alleviates stress-induced anemia in Riok2f/+Vav1cre as well as wild-type mice.
Figure 4. IL-22 neutralization alleviates stress-induced anemia in Riok2 sufficient and haploinsufficient mice.
(a) PB RBC numbers, Hb, and HCT in the indicated strains undergoing PhZ-induced stress erythropoiesis. n=6,5,5, and 5 mice for Riok2+/+Il22+/+Vav1cre, Riok2+/+Il22+/−Vav1cre, Riok2f/+Il22+/+Vav1cre, Riok2f/+Il22+/−Vav1cre, respectively. (b) Frequency of erythroid progenitor/precursor populations among viable BM cells in the indicated strains undergoing PhZ-induced stress erythropoiesis (n=4-5/group). (c) PB RBC numbers, Hb, and HCT in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls undergoing PhZ-induced stress erythropoiesis treated with either an isotype control or anti-IL-22 antibody (n=4-5/group). (d) Frequency of apoptotic erythroid precursors among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls undergoing PhZ-induced stress erythropoiesis treated with either an isotype control or anti-IL-22 antibody. n=4,5,4, and 5 mice for isotype-treated Riok2+/+Vav1cre, anti-IL-22-treated Riok2+/+Vav1cre, isotype-treated Riok2f/+Vav1cre, and anti-IL-22-treated Riok2f/+Vav1cre mice, respectively. 1-way ANOVA with Tukey’s correction for multiple comparison (a to d) used to calculate statistical significance. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data are shown as mean ± s.e.m and are representative of two (c, d) or three (a, b) independent experiments.
IL-22 worsens stress-induced anemia in wild-type mice
Phenylhydrazine administration to wild-type C57BL/6J mice treated intraperitoneally with recombinant IL-22 (rIL-22) led to decreased PB RBCs, Hb, and HCT owing to decreased BM erythroid precursor cell frequency and number (Fig. 5a, c, Extended Data Fig. 6b). rIL-22 treatment led to increased apoptosis of erythroid precursors (Fig. 5d). This treatment also led to an increase in PB reticulocytes, an indication of increased erythropoiesis under stress (Fig. 5b). Recombinant IL-22 also dose-dependently decreased terminal erythropoiesis in an in vitro erythropoiesis assay (Fig. 5e, f). Importantly, IL-22-mediated inhibition of in vitro erythropoiesis led to induction of p53 suggesting a feedback loop between IL-22 and p53 in driving dyserythropoiesis (Fig. 5g). Overall, these data show that exogenous recombinant IL-22 exacerbates stress-induced anemia in wild-type mice.
Figure 5. Recombinant IL-22 exacerbates PhZ-induced anemia in wt mice.
(a) PB RBC numbers, Hb, and HCT in wt C57BL/6J mice administered PBS (n=5) or rIL-22 (n=4) and subsequently treated with PhZ. n=4-5 mice/group. (b) PB reticulocytes in mice treated as in (a). n=4 mice/group. (c) Percentage of RII-RIV erythroid precursors in the BM of PBS- or rIL-22-treated C57BL/6J mice 7 days after PhZ administration. n=4 mice/group. (d) Percentage of apoptotic RII erythroid precursors in mice treated as in (C). n=4 mice/group. (e) Effect of recombinant IL-22 (500 ng/mL) on the frequency (left) and cell number (right) in an in vitro erythropoiesis assay and (f) dose dependent effect of recombinant IL-22. n=5 and 4 for PBS and IL-22 groups, respectively. (g) p53 expression in in in vitro erythropoiesis culture treated with rIL-22 or PBS. n=5 and 4 mice for PBS and IL-22 groups, respectively. Data are shown as mean ± s.e.m and are representative of three (a, b) or two (c to g) independent experiments. Unpaired two-tailed t-test (A to D, G), multiple unpaired two-tailed t-tests with Holm-Sidak method (e) and 1-way ANOVA with Tukey’s correction for multiple comparisons (f) used to calculate statistical significance. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Erythroid precursors express IL-22RA1 receptors
IL-22 signals through a cell surface heterodimeric receptor composed of IL-10Rβ and IL-22RA1 (encoded by Il22ra1)33. IL-22RA1 expression has been reported to be restricted to cells of non-hematopoietic origin (e.g., epithelial cells and mesenchymal cells). We discovered, however, that erythroid precursors in the BM also express IL-22RA1 (Fig. 6a, Extended Data Fig. 8a). Moreover, we observed that among BM hematopoietic progenitors, IL-22RA1-expressing cells were exclusively of the erythroid lineage (Extended Data Fig. 8b). Using a second IL-22RA1-specific antibody (targeting a different epitope than the antibody used in Fig. 4A), we confirmed the presence of IL-22RA1 on erythroid precursors (Extended Data Fig. 8c). Il22ra1 mRNA expression was detected exclusively in erythroid precursors among all lineage-negative cells in the BM (Extended Data Fig. 8d).
Figure 6. Genetic deletion of Il22ra1 alleviates anemia in Riok2 haploinsufficient mice.
(a) IL-22RA1 expression on BM erythroid precursors in wild-type (WT) mice assessed by flow cytometry using antibody from Novus Biologicals targeting the extracellular domain of IL-22RA1. (b) PB RBC numbers, Hb, and HCT in the indicated strains undergoing PhZ-induced stress erythropoiesis. n=6,5,6, and 4 mice for Riok2+/+Il22r1a+/+Vav1cre, Riok2+/+Il22ra1f/fVav1cre, Riok2f/+Il22ra1+/+Vav1cre, Riok2f/+Il22ra1f/fVav1cre, respectively. (c) Frequency of erythroid progenitor/ precursor populations among viable BM cells in the indicated strains undergoing PhZ-induced stress erythropoiesis (n=5/group). (d) Flow cytometry plots showing p53 expression in IL-22RA1+ and IL-22RA1− erythroid precursors in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls. n=5/group. (e) Graphical representation of data shown in (D). (f) Gene expression of Trp53 (p53) and listed p53 target genes in IL-22RA1+ and IL-22RA1− erythroid precursors from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice assessed by qRT-PCR. n=3/group. (g) Frequency of apoptotic cells (assessed by flow cytometry) with (n=4) or without (n=5) p53 inhibitor, pifithrin-α, p-nitro (1 μM), in an in vitro erythropoiesis assay using Lin− BM cells from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. 2-way ANOVA with Tukey’s correction for multiple comparisons (e, g) and 1-way ANOVA with Tukey’s correction for multiple comparison (b, c) used to calculate statistical significance. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data are shown as mean ± s.e.m and are representative of two (D-G) or three (a to c) independent experiments.
Deletion of Il22ra1 in Riok2 haploinsufficient mice (Riok2f/+Il22ra1f/fVav1cre) led to improvement in PB RBCs and HCT as compared to IL-22RA1 sufficient Riok2 haploinsufficient mice (Riok2f/+Il22ra1+/+Vav1cre) (Fig. 6b). This improvement could be attributed to the increase in RIII and RIV erythroid precursors in the BM of Riok2f/+Il22ra1f/fVav1cre mice (Fig. 6c, Extended Data Fig. 6c).
In accordance with the upregulation of p53 upon in vitro IL-22 stimulation in an in vitro erythropoiesis assay (Fig. 5g) and p53 upregulation in erythroid precursors upon Riok2 haploinsufficiency (Extended Data Fig. 4f, g), we observed a synergistic effect of Riok2 haploinsufficiency in IL-22-responsive (IL-22RA1+) erythroid precursors in Riok2f/+Vav1cre mice (Fig. 6d, e). p53 target genes such as Gadd45a and Cdkn1a1 were also increased in IL-22RA1+ Riok2 haploinsufficient erythroid precursors compared to IL-22RA1+ Riok2 sufficient erythroid precursors (Fig. 6f). Given that rIL-22 induced apoptosis in erythroid precursors in vivo (Fig. 5d), we sought to determine if Riok2 haploinsufficiency-mediated p53 upregulation played an independent role in apoptosis induction. In an IL-22-free in vitro erythropoiesis assay, p53 inhibition by pifithrin-α, p-nitro inhibited the apoptosis induced by Riok2 haploinsufficiency (Fig. 6g).
Erythroid cell-specific deletion of IL-22RA1 (using cre recombinase driven by the erythropoietin receptor (Epor) promoter) also increased numbers of PB RBCs and HCT (Fig. 7a) due to the increase in RIII and RIV erythroid precursors in the BM (Fig. 7b, Extended Data Fig. 6d). Additionally, rIL-22 failed to exacerbate phenylhydrazine-induced anemia in Il22ra1f/fEporcre mice lacking the IL-22 receptor only on erythroid cells (Fig. 7c). These data reinforce our view that IL-22 signaling plays an important role in regulating erythroid development regardless of ribosomal haploinsufficiency. Thus, using three different approaches to neutralize IL-22 signaling, we demonstrate that IL-22 plays a critical role in controlling RBC production by directly regulating early stages of erythropoiesis.
Figure 7. Erythroid-specific deletion of IL-22RA1 alleviates stress-induced anemia.
(a) PB RBC numbers, Hb, and HCT in the indicated strains undergoing PhZ-induced stress erythropoiesis (n=6/group). (b) Frequency of erythroid progenitor/precursor populations among viable BM cells in the indicated strains undergoing PhZ-induced stress erythropoiesis (n=5/group). (c) PB RBC numbers, Hb, and HCT in Il22ra1+/+Eporcre (n=5) and Il22ra1f/fEporcre (n=4) mice administered rIL-22 and subsequently treated with PhZ. n=4-5 mice/group. Unpaired two-tailed t-test (a-c) used to calculate statistical significance. * p < 0.05, *** p < 0.001. Data are shown as mean ± s.e.m and are representative of two (a-c) independent experiments.
IL-22 is increased in del(5q) MDS patients
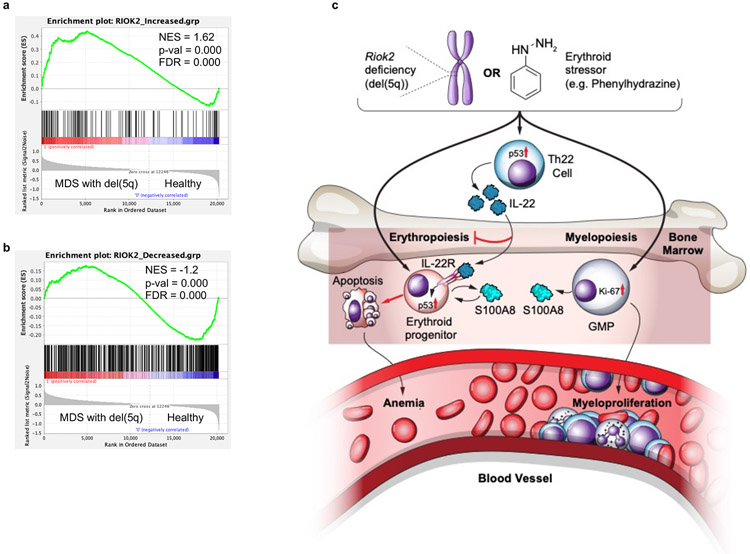
Next, we assessed whether IL-22 expression is increased in human disorders that display dyserythropoiesis due to ribosomal protein haploinsufficiencies. Given the localization of RIOK2 on human chromosome 5, we focused on MDS with 5q deletion and compared it to MDS without 5q deletion and healthy controls. We observed a significant increase in IL-22 levels in the BM fluid (BMF) of del(5q) MDS patients compared to BMF from healthy controls and non-del(5q) MDS patients (Fig. 8a). Interestingly, a strong negative correlation between cellular RIOK2 mRNA expression and BMF IL-22 concentration was evident in the del(5q) MDS cohort (Fig. 8b) indicating that a decrease in RIOK2 expression is associated with increased IL-22 expression. In our MDS cohort, S100A8 concentrations were found to be higher than healthy controls regardless of del(5q) status (Fig. 8c). However, IL-22 positively correlated with S100A8 concentrations only in the del(5q) MDS group (Fig. 8d). Of note, S100A8 concentrations were higher in BMF from non-del(5q) MDS patients compared to MDS patients with del(5q) (Fig. 8c). These data suggest that the regulation of S100A8 expression may be IL-22-mediated in del(5q) MDS patients, but IL-22-independent in other subtypes of MDS. In a second cohort of MDS patients, the frequency of CD4+ T cells producing IL-22 (TH22 cells) among freshly isolated peripheral blood mononuclear cells (PBMCs) was significantly higher in MDS patients with 5q deletion compared to healthy controls (Fig. 8e - cumulative data of representative flow plots shown in Extended Data Fig. 9a). Our independent analysis of a large-scale microarray sequencing dataset of CD34+ cells from normal, del(5q) MDS, and non-del(5q) MDS subjects showed that RIOK2 mRNA was significantly decreased in the del(5q) MDS cohort (78% (37/47)). Additionally, expression of known IL-22 target genes such as S100A10, S100A11, PTGS2, RAB7A, and LCN2 was specifically increased in the del(5q) MDS cohort compared to both the healthy control and non-del(5q) groups (Extended Data Fig. 9b). Using differentially expressed proteins (adjusted p-value < 0.01) from the Riok2 haploinsufficient proteomics dataset as a reference set, GSEA analyses of the CD34+ microarray dataset revealed significant enrichment scores (Extended Data Fig. 10a, b) further suggesting that the mouse model of Riok2 haploinsufficiency faithfully recapitulates the molecular changes seen in patients with del(5q) MDS.
Figure 8. MDS patients exhibit increased IL-22 levels and IL-22 associated signature.
(a) IL-22 concentration in the BM fluid of healthy controls (n=12), del(5q) MDS (n=11), and non-del(5q) MDS (n=22) patients. (b) Correlation between RIOK2 mRNA and BM IL-22 concentration in the del(5q) cohort shown in (a), n=10. (c) S100A8 concentration in the samples shown in (a). n = 11, 10, and 19 for healthy, del(5q) MDS, and non-del(5q) MDS, respectively. (d) Correlation between IL-22 concentration and S100A8 concentration in BM fluid of del(5q) (left, n=10) and non-del(5q) (right, n=19) samples. (e) Frequency of IL-22 producing CD4+ T cells in the PB of healthy controls (n=11), del(5q) (n=3) and non-del(5q) (n=24) MDS patients. (f) Plasma IL-22 concentration in healthy subjects (n=10) and chronic kidney disease (CKD) patients with (n=13) or without (n=13) secondary anemia. (g) Correlation between plasma IL-22 concentration and hemoglobin (HGB) in CKD patients with (n=13) or without (n=13) anemia. Kruskal-Wallis test with Dunn’s correction for multiple comparisons (a to c, e), 1-way ANOVA with Tukey’s correction for multiple comparisons (f) used to calculate statistical significance. Pearson correlation co-efficient (b, d, g), used to calculate statistical significance and correlation coefficient. ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data are shown as mean ± s.e.m (c). Solid lines represent median and dashed lines represent quartiles (a, e, f).
High IL-22 in anemic chronic kidney disease patients
Anemia is frequently observed in CKD patients and is associated with poor outcomes. Anemia of CKD is resistant to erythropoiesis-stimulating agents (ESAs) in 10-20% of patients34, suggesting that pathogenic mechanisms other than erythropoietin deficiency are at play. We found a significant increase in IL-22 concentration in the plasma of CKD patients with secondary anemia compared to healthy controls and to CKD patients without anemia (Fig. 8f). Plasma IL-22 concentration negatively correlated with hemoglobin in CKD patients (Fig. 8g), suggesting a function for IL-22 in driving anemia in some patients with CKD.
Discussion
The overall conclusion of this study is that IL-22 signaling directly controls bone marrow erythroid differentiation and that its neutralization is a potential therapeutic approach for anemias and MDS. By exploring the function of a little-studied atypical kinase Riok2 in mammalian biology, we identified the erythroid precursors as a novel target for IL-22 action via the IL-22RA1. We further identify IL-22 as a disease biomarker for the del(5q) subtype of MDS and lastly, identify IL-22 signaling blockade as a potential therapeutic for stress-induced anemias irrespective of the genetic background. Interestingly, we also detect elevated levels of IL-22 in patients with anemia secondary to chronic kidney disease (CKD) suggesting that IL-22 signaling blockade may be therapeutic in reversing anemia in a much wider patient population.
Del(5q), either isolated or accompanied by additional cytogenetic abnormalities, is the most commonly detected chromosomal abnormality in MDS, reported in 10-15% of patients and enriched in therapy-related MDS. The severe anemia in MDS patients with isolated del(5q) has been linked to haploinsufficiency of ribosomal proteins such as RPS14 10 and RPS19 5. While much research has focused on the effect of such gene deletions or mutations in hematopoietic stem cells and lineage-committed progenitors, the immunobiology underlying this MDS subtype has remained largely unexplored, thus impeding the development of immune-targeted therapies. With the exception of the TNF-α inhibitor etanercept35 which proved to be ineffective, the only other therapy against immune cell-derived cytokines, is luspatercept36, a recombinant fusion protein derived from human activin receptor type IIb37 which has just been approved for use in anemia in lower risk MDS patients. Here, we identify two critical and independent functions of an understudied atypical kinase, RIOK2, that synergize to induce dyserythropoiesis and anemia. One effect of Riok2 loss in erythroid precursors is an intrinsic block in erythroid differentiation owing to its indispensable role in the maturation of the pre-40S ribosomal complex, leading to increased apoptosis and cell cycle arrest. The second effect of Riok2 loss is the induction of the erythropoiesis-suppressive cytokine IL-22 in T cells which then directly acts on the IL-22RA1 on erythroid precursors (Extended Data Fig. 10c). While IL-22RA1 is known to be widely expressed on epithelial cells and hepatocytes, its expression has also been recently reported on specialized cells such as retinal Müller glial cells38 and now described here, on erythroid precursors. Our data reveal a novel molecular link between haploinsufficiency of a ribosomal protein and induction of erythropoiesis-suppressive cytokine IL-22. While IL-22 has been shown to modulate RBC production by controlling the expression of iron-chelating proteins such as hepcidin39 and haptoglobin40, we have uncovered a novel role for IL-22 in directly binding to the previously unknown IL-22R on erythroid precursors leading to their apoptosis. Diminished expression of ribosomal proteins has been shown to increase p53 levels. We show here that Riok2 haploinsufficiency leads to p53 upregulation in T cells which drives the increase in IL-22 secretion. Additionally, we also show that IL-22-responsive erythroid precursors express elevated p53 further suggesting a role for p53 downstream of IL-22 signaling in driving dyserythropoiesis. Using banked and fresh del(5q) MDS and CKD patient samples, we also show that IL-22 is elevated in these human diseases.
The role of inflammatory cytokines in directly regulating various aspects of BM hematopoiesis in steady-state and diseased conditions is increasingly being recognized41-43. IL-22 is known to play a pathogenic role in some autoimmune diseases44; 45. Interestingly, autoimmune diseases such as colitis, Behçet's disease, and arthritis are common in MDS patients, with features of autoimmunity observed in up to 10% of patients46. It is intriguing to hypothesize that IL-22 may account both for the onset of MDS and autoimmunity in this subset of patients. Studies have reported that in patients with co-existence of MDS and autoimmunity, treatment for one can alleviate the symptoms of the other47; 48. Low-level exposure to benzene, a hydrocarbon, has been associated with an increased risk of MDS49. Hydrocarbons are known ligands for aryl hydrocarbon receptor (AHR), the transcription factor that controls IL-22 production in T cells. Stemregenin 1, an AHR antagonist, was shown to promote the ex vivo expansion of human HSCs, with the highest fold expansion seen in the erythroid lineage50. Overall, these observations suggest that inhibition of the AHR-IL-22 axis may be an attractive approach for treating red blood cell disorders that arise from dyserythropoiesis.
Further, we provide evidence that neutralization of IL-22 signaling may be effective not only in the treatment of MDS and other stress-induced anemias, but also in the anemia of chronic diseases such as CKD, which are very much in need of new therapeutic approaches. With currently approved MDS therapies (lenalidomide and other hypomethylating agents, erythropoiesis-stimulating agents), the survival time of MDS patients after diagnosis is only 2.5-3 yrs. Patients also develop resistance to these therapies thus intensifying the need for additional therapeutic modalities. IL-22-based therapies could be used in conjunction with already existing therapeutics or after first-line therapies have failed due to acquisition of resistance.
METHODS
Human samples and processing
MDS and CKD patient samples were collected under IRB-approved protocols at Dana-Farber Cancer Institute (DFCI) and Brigham and Women’s Hospital (BWH), respectively. All samples were de-identified at the time of inclusion in the study. All patients provided informed consent and the data collection was performed in accordance with the Declaration of Helsinki.
Peripheral blood mononuclear cells (PBMCs) from EDTA-treated whole blood were isolated using density gradient centrifugation. PBMCs were then incubated in RPMI with 10% FBS and Cell Activation Cocktail (Tonbo Biosciences) for 4 h and then processed for flow cytometry as described below. Relevant clinical information of MDS samples is provided in Supplemental Table Adult CKD plasma samples were stored at −80 °C until further use. Relevant clinical information of CKD samples is provided in Supplemental Table 3.
Generation of Riok2 floxed mice
Riok2f/f mice were generated using frozen sperm obtained from Mutant Mouse Resource and Research Centers (MMRRC) (Riok2tm1a(KOMP)Wtsi)51. In brief, a floxed Riok2 allele was created by inserting an FRT-flanked IRES-LacZ-neor cassette into intron 4 of the Riok2 gene. LoxP sites were inserted to flank exons 5 and 6. After germline transmission, the FRT cassette was removed by crossing to FLPe deleter mice52 and resulting floxed mice were bred with individual cre driver strains to create conditional Riok2-deleted mice (Extended Data Fig. 1a). Genotyping (Extended Data Fig. 1c) was carried out using the following primers:
Forward primer: 5′ GCATCAGTGATTTACAGACTAAAATGCC 3′
Reverse primer 1: 5′ GCTCTTACCCACTGAGTCATCTCACC 3′
Reverse primer 2: 5′ CCCAGACTCCTTCTTGAAGTTCTGC 3′
Mice
Wild-type C57BL/6J mice (Stock no. 000664), Vav-icre mice (Stock no. 008610), R26-CreErt2 mice (Stock no. 008463), Il22ra1-floxed (Stock no. 031003), CD45.1 C57BL/6J mice (Stock no. 002014), Trp53−/− (Stock no. 002101), Cd4-cre (Stock no. 022071), and ApcMin (Stock no. 002020) mice were purchased from The Jackson Laboratory. Il22−/− mice were provided by R. Caspi (National Institutes of Health, Bethesda, MD) with permission from Genentech (San Francisco, CA). Epor-cre53 mice were a gift from U. Klingmüller (Deutsches Krebsforschungszentrum (DFKZ), Germany). Il22-tdtomato (Catch-22)54 mice were a gift from R. Locksley (University of California at San Francisco, CA). Rps14-floxed mice55 were a gift from B. Ebert (Dana-Farber Cancer Institute, Boston, MA). Mice were housed in the Animal Research Facility (ARF) at DFCI under ambient temperature and humidity with 12 h light/12 h dark cycle. Animal procedures and treatments were in compliance with the guidelines set forth by the Institutional Animal Care and Use Committee (IACUC) at DFCI. Age- and gender- matched mice were used within experiments.
Competitive bone marrow (BM) transplantation
2 × 106 freshly isolated BM cells from CD45.2+ Riok2f/+Ert2cre or Riok+/+Ert2cre mice were transplanted in competition with 2 × 106 freshly isolated CD45.1+ wild-type (WT) BM cells via retro-orbital injection into lethally irradiated 8-10 week old CD45.1+ WT recipient mice. The donor cell chimerism was determined in the peripheral blood four weeks after transplantation before the excision of Riok2 was induced by tamoxifen injection as well as every four to eight weeks as indicated. Tamoxifen (75 mg/kg) (Cayman Chemical, Cat # 13258) was administered for five consecutive days.
Flow cytometry and cell isolation
Whole bone marrow (BM) cells were isolated by crushing hind leg bones (femur and tibia) with mortar and pestle in staining buffer (PBS (Corning) supplemented with 2% heat-inactivated fetal bovine serum (FBS, Atlanta Biologicals) and EDTA (GIBCO). Whole BM was lysed with 1X PharmLyse (BD Biosciences) for 90 s, and the reaction was terminated by adding an excess of staining buffer. Cells were labeled with fluorochrome-conjugated antibodies in staining buffer for 30 min at 4 °C. For flow cytometric analysis, cells were incubated with combinations of fluorochrome-conjugated antibodies to the following cell surface markers: CD3 (17A2, 1:500), CD5 (53–7.3, 1:500), CD11b (M1/70, 1:500), Gr1 (RB6–8C5, 1:500), B220 (RA3–6B2, 1:500), Ter119 (TER119, 1:500), CD71 (C2, 1:500), c-kit (2B8, 1:500), Sca-1 (D7, 1:500), CD16/32 (93, 1:500), CD150 (TC15–12F12.2, 1:150), CD48 (HM48-1, 1:500). For sorting of lineage-negative cells, lineage markers included CD3, CD5, CD11b, Gr1 and Ter119. For sorting erythroid progenitor cells, the lineage cocktail did not include Ter119. All reagents were acquired from BD Biosciences, Thermo Fisher Scientific, Novus Biologicals, Tonbo Biosciences, or BioLegend. Identification of apoptotic cells was carried out using the Annexin V Apoptosis Detection Kit (BioLegend). Intracytoplasmic and intranuclear staining was performed using Foxp3/Transcription Factor Staining Kit (Thermo Fisher Scientific) or 0.1% saponin in PBS supplemented with 3% FBS. For staining performed with AF647 p53 antibody (Cell Signaling Technology, 1:50), cells were permeabilized with 90% ice-cold methanol. To increase the sorting efficiency, whole BM samples were lineage-depleted using magnetic microbeads (Miltenyi Biotec) and autoMACS Pro magnetic separator (Miltenyi Biotec). Cell sorting was performed on a FACS Aria flow cytometer (BD Biosciences), data acquisition was performed on a BD Fortessa X-20 instrument equipped with 5 lasers (BD Biosciences) employing FACSDiva software. Data were analyzed by FlowJo (Tree Star) version 9 software. Flow analyses were performed on viable cells by exclusion of dead cells using either DAPI or a fixable viability dye (Tonbo Biosciences). Gating for early and committed hematopoietic progenitors was performed as described elsewhere56. ILCs and NKT cells were identified as Lin−CD45+CD90+CD12+ and CD3ε+ NK1.1+, respectively.
Complete blood count
Mice were bled via the submandibular facial vein to collect blood in EDTA-coated tubes (BD Microtainer ™ Capillary Blood Collector, BD 365974). Complete blood counts were obtained using the HemaVet CBC Analyzer (Drew Scientific) or Advia 120 (Siemens Inc.,) instruments.
In vivo measurement of protein synthesis
100 μL of a 20mM solution of O-Propargyl-Puromycin (OP-Puro; BioMol) was injected intraperitoneally in mice and mice were then rested for 1 h. Mice injected with PBS were used as controls. BM was harvested after 1 h and stained with antibodies against cell surface markers, washed to remove excess unbound antibodies, fixed in 1% paraformaldehyde, and permeabilized in PBS with 3% FBS and 0.1% saponin. The azide-alkyne cyclo-addition was performed using the Click-iT Cell Reaction Buffer Kit (Thermo Fisher Scientific) and azide conjugated to Alexa Fluor 647 (Thermo Fisher Scientific) at 5 μM final concentration for 30 min. Cells were washed twice and analyzed by flow cytometry. ‘Relative rate of protein synthesis’ was calculated by normalizing OP-Puro signals to whole bone marrow after subtracting autofluorescence.
Methylcellulose assay
250 – 500 BM Lin−c-kit+ Sca-1+ cells were flow sorted and plated in semi-solid methylcellulose culture medium (M3534, StemCell Technologies) and incubated at 37 °C in a humidified atmosphere for 7-10 days. At the end of the incubation period, each well was triturated with staining buffer to collect cells and then processed for flow cytometry as described above. Enumeration of colonies in MethoCult media was performed with StemVision instrument StemCell Technologies).
Phenylhydrazine treatment
Phenylhydrazine (PhZ) was purchased from Sigma and injected intraperitoneally on 2 consecutive days (days 0 and 1) at the dose of 25 mg/kg (sublethal model) or 35 mg/kg (lethal model). Peripheral blood was collected 3-4 days before the start of treatment and at day 4, 7, and 11. PhZ treatment experiments were carried out in 8-12 week old mice.
T cell polarization
Single-cell suspensions of mouse spleens were prepared by pressing tissue through a 70-μm cell strainer followed by red blood cell lysis using PharmLyse. Total splenic CD4+ T cells were isolated using CD4 T cell isolation kit (Miltenyi Biotec). Enriched CD4+ T cells were then incubated with fluorochrome-conjugated antibodies to CD4, CD8, CD25, CD62L, and CD44 to purify naïve CD4+ T cells using fluorescence activated cell sorting (FACS). In the presence of 1 μg/mL plate-bound anti-CD3ε and soluble 1 μg/mL anti-CD28, T cell polarizations were carried out in IMDM supplemented with 10% FBS, 2mM L-glutamine, 100 mg/mL penicillin-streptomycin, HEPES (pH 7.2-7.6) , non-essential amino acids, 100 μM β-mercaptoethanol (BME), and sodium pyruvate as follows – TH1 (20 ng/mL IL-12, 10 μg/mL anti-IL-4), TH2 (20 ng/mL IL-4, 10 μg/mL anti-IFN-γ), TH17 (30 ng/mL IL-6, 20 ng/mL IL-23, 20 ng/mL IL-1β, 10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ), Treg cells (1 ng/mL TGF-β, 10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ), and TH22 (30 ng/mL IL-6, 20 ng/mL IL-23, 20 ng/mL IL-1β, 10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ, 400 nM FICZ). Pifithrin-α, p-Nitro and Nutlin-3a were purchased from Santa Cruz Biotechnology and Tocris Biosciences, respectively.
IL-22 neutralization and reconstitution
Monoclonal anti-IL-22 (Clone IL22JOP) blocking antibody and isotype control IgG2aκ (Clone eBR2a) were purchased from Thermo Fisher Scientific. Mice were administered anti-IL-22 (50 μg/mouse) or isotype intraperitoneally every 48 h until the conclusion of the experiment. For \recombinant IL-22 treatment, mice were injected with recombinant IL-22 (500 ng/mouse; PeproTech) intraperitoneally every 24 h until the conclusion of the experiment. Mice were administered these reagents at least five times before inducing PhZ-mediated anemia.
Cytokine quantitation
IL-22 in human samples was quantified using either Human IL-22 Quantikine ELISA Kit (D2200, R&D Systems) or SMCTM Human IL-22 High Sensitivity Immunoassay Kit (EMD Millipore, 03-0162-00) according to manufacturers’ instructions. The SMC assay was read on a SMC Pro (EMD Millipore) instrument. The lower limit of quantification (LLOQ) of this immunoassay is 0.1 pg/mL. IL-22 in mouse samples was quantified using ELISA MAXTM Deluxe Set Mouse IL-22 (BioLegend, 436304). The LLOQ of this immunoassay is 3.9 pg/mL. S100A8 in human samples was quantified using Human S100A8 DuoSet ELISA (DY4570, R&D Systems).
Concentration of lineage-associated cytokines in cell culture supernatants of polarized T cells were quantified using a custom-made ProCarta Plex assay (Thermo Fisher Scientific) acquired on a Luminex platform. Hepcidin in mouse serum was quantified using a colorimetric assay from Hepcidin Murine-Compete ™ ELISA Kit from Intrinsic LifeSciences (HMC-001).
mRNA quantitation
Cells were flow sorted directly into the lysis buffer provided with the Cells-to-CT™ 1-Step TaqMan™ Kit (A25605, Thermo Fisher Scientific) and processed according to the manufacturer’s instructions. Pre-designed TaqMan gene expression assays were used to quantify mRNA expression by qPCR using QuantStudio 6 (Thermo Fisher Scientific). Hprt was used as housekeeping control. Relative expression was calculated using the ΔCt method. Please refer to Supplementary Table 4 for primers details.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using EZ-ChIP Kit (EMD Millipore) according to manufacturer’s instructions. Briefly, cells were fixed and cross-linked with 1% formaldehyde at 25 °C for 10 min and quenched with 125 mM glycine for an additional 10 min. Cells pellet was resuspended in lysis buffer and shearing was carried out Diagenode Bioruptor sonication system for a total of 40 cycles beads. Pre-cleared lysates were incubated with control Mouse IgG or anti-p53 (Santa Cruz Biotechnology) antibodies. Il22 promoter-specific primer pair was designed using Primer 3.0 Input and were as follows:
Forward Primer: 5′ CCAAACTTAACTTGACCTTGGC 3′
Reverse Primer: 5′ TTCTTCACAGCTCCCA TTGC 3′
In vitro erythroid differentiation
Whole BM cells were labeled with biotin-conjugated lineage antibodies (cocktail of anti-CD3ε, anti-CD11b, anti-CD45R/B220, anti-Gr1, anti-CD5, and anti-TER-119) (BD Pharmingen) and purified using anti-biotin beads and negative selection on the AutoMACS Pro (Miltenyi). Purified cells were then seeded in fibronectin-coated (2 μg/cm2) tissue-culture treated polystyrene wells (Corning® BioCoat™ Cellware) at a cell density of 105/mL. Erythroid differentiation was carried out according to modified published protocols57. The erythropoietic medium was IMDM supplemented with erythropoietin at 10 U/mL, 10 ng/mL stem cell factor SCF, PeproTech), 10 μM Dexamethasone (Sigma-Aldrich), 15% FBS, 1% detoxified BSA (StemCell Technologies), 200 μg/mL holotransferrin (Sigma-Aldrich), 10 mg/mL human insulin (Sigma-Aldrich), 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, and penicillin-streptomycin. After 48 h, the medium was replaced by IMDM medium containing 20% FBS, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, and penicillin-streptomycin. 50% of the culture media was replaced after 48 h and cell density was maintained at 0.5 × 106/mL. Total culture period for the assay was 6 days. Recombinant mouse IL-22 (PeproTech/Cell Signaling) was used where indicated. RII-RIV populations were gated as shown in Extended Data Fig. 2a.
Proteomic profiling
Proteomic profiling of sort-purified erythroid progenitors was performed as described elsewhere58. Briefly, cells were captured in collection microreactors and stored at −80 °C. Cell lysis was performed by adding 10 μL of 8 M urea, 10 mM TCEP and 10 mM iodoacetamide in 50 mM ammonium bicarbonate (ABC) to the cell pellet of 1 × 106 erythroid progenitors and incubatedat room temperature for 30 min, shaking in the dark. 50 mM ABC was used to dilute the urea to less than 2 M and the appropriate amount of trypsin for a 1:100 enzyme to substrate ratio was added and allowed to incubate at 37 °C overnight. Once digestion is completed, the lysate is spun through the glass mesh directly onto a C18 Stage tip (Empore59) at 3500 x g until the entire digest passes through the C18 resin. 75 μL 0.1% formic acid (FA) is then used to ensure transfer of peptides to the C18 resin from the mesh while washing away the lysis buffer components. C18-bound peptides were immediately subjected to on-column TMT labeling.
On-column TMT Labeling -
Resin was conditioned with 50 μL methanol (MeOH), followed by 50 μL 50% acetonitrile (ACN)/0.1% FA, and equilibrated with 75 μL 0.1% FA twice. The digest was loaded by spinning at 3500 × g until the entire digest passed through. One μL of TMT reagent in 100% ACN was added to 100 μL freshly made HEPES, pH 8, and passed over the C18 resin at 350 × g until the entire solution passed through. The HEPES and residual TMT was washed away with two applications of 75 μL 0.1% FA and peptides were eluted with 50 μL 50% ACN/0.1% FA followed by a second elution with 50% ACN/20 mM ammonium formate (NH4HCO2), pH 10. Peptide concentrations were estimated using an absorbance reading at 280 nm and checking of label efficiency was performed on 1/20th of the elution. After using 1/20th of the elution to test for labeling efficiency, the samples are mixed before fractionation and analysis.
Stage tip bSDB Fractionation -
200 μL pipette tips were packed with two punches of sulfonated divinylbenzene (SDB-RPS, Empore) with a 16-gauge needle. After loading ~20 μg peptides total, a pH switch was performed using 25 μL 20 mM NH4HCO2, pH 10, and was considered part of fraction one. Then, step fractionation was performed using ACN concentrations of 5, 7.5, 10, 12.5, 15, 17.5, 20, 25, 42, and 50%. Each fraction was transferred to autosampler vials and dried via vacuum centrifugation and stored at 80 °C until analysis.
Data Acquisition -
Chromatography was performed using a Proxeon UHPLC at a flow rate of 200 nl/min. Peptides were separated at 50 °C using a 75 μm i.d. PicoFrit (New Objective) column packed with 1.9 μm AQ-C18 material (Dr. Maisch, Germany) to 20 cm in length over a 110 min run. The on-ine LC gradient went from 6% B at 1 min to 30% B in 85 mins, followed by an increase to 60% B by minute 94, then to 90% by min 95, and finally to 50% B until the end of the run. Mass spectrometry was performed on a Thermo Scientific Lumos Tribrid mass spectrometer. After a precursor scan from 350 to 1800 m/z at 60,000 resolution, the topmost intense multiply charged precursors in a 2 second window were selected for higher energy collisional dissociation (HCD) at a resolution of 50,000. Precursor isolation width was set to 0.7 m/z and the maximum MS2 injection time was 110 msecs for an automatic gain control of 6e4. Dynamic exclusion was set to 45 s and only charge states two to six were selected for MS2. Half of each fraction was injected for each data acquisition run.
Data Processing -
Data were searched all together with Spectrum Mill (Agilent) using the Uniprot Mouse database (28 Dec. 2017), containing common laboratory contaminants and 553 smORFs. A fixed modification of carbamidomethylation of cysteine and variable modifications of N-terminal protein acetylation, oxidation of methionine, and TMT-11plex labels were searched. The enzyme specificity was set to trypsin and a maximum of three missed cleavages was used for searching. The maximum precursor-ion charge state was set to six. The MS1 and MS2 mass tolerance were set to 20 ppm. Peptide and protein FDRs were calculated to be less than 1% using a reverse, decoy database. Proteins were only reported if they were identified with at least two distinct peptides and a Spectrum Mill score protein level score ~ 20.
TMT11 reporter ion intensities in each MS/MS spectrum were corrected for isotopic impurities by the Spectrum Mill protein/peptide summary module using the afRICA correction method which implements determinant calculations according to Cramer’s Rule and general correction factors obtained from the reagent manufacturer’s certificate of analysis.
Differential Protein Abundance Analysis -
The median normalized, median absolute deviation- scale data set was subjected to a moderated F-test, followed by Benjamini-Hochberg Procedure correcting for multiple hypothesis testing. We drew an arbitrary cutoff at adj. p val < 0.05.
RNA Sequencing (RNA-Seq)
5000 IL-22 +(CD4+IL-22(tdtomato)+) cells were FACS sorted directly into TLC Buffer (Qiagen) with 1% β-mercaptoethanol. For the preparation of libraries, cell lysates were thawed and RNA was purified with 2.2x RNAClean SPRI beads (Beckman Coulter Genomics) without final elution60. The RNA captured beads were air-dried and processed immediately for RNA secondary structure denaturation (72 °C for 3 min) and cDNA synthesis. SMART-seq2 was performed on the resultant samples following the published protocol61 with minor modifications in the reverse transcription (RT) step. A 15 μL reaction mix was used for subsequent PCR and performed 10 cycles for cDNA amplification. The amplified cDNA from this reaction was purified with 0.8× Ampure SPRI beads (Beckman Coulter Genomics) and eluted in 21 μL TE buffer. We used 0.2 ng cDNA and one-eighth of the standard Illumina NexteraXT (Illumina FC-131-1096) reaction volume to perform both the tagmentation and PCR indexing steps. Uniquely indexed libraries were pooled and sequenced with NextSeq 500 high output V2 75 cycle kits (Illumina FC-404-2005) and 38 × 38 paired-end reads on the NextSeq 500 instrument. Reads were aligned to the mouse mm10 transcriptome using Bowtie62, and expression abundance TPM estimates were obtained using RSEM63.
Pathway analysis
Gene set enrichment analysis (GSEA) was performed with Broad Institute’s GSEA Software64. The ‘IL-22_Signature’ and ‘Rps14_Increased’ gene sets were created from the literature55,65 (Fig. 2c,f). Full lists of genes in the individual gene sets can be found in Supplementary Table 1. Other reference gene sets are available from MSigDB66. For GSEA analyses, mouse UniProt IDs were converted to their orthologous human gene symbols using MSigDB 7.1 CHIP file mappings. Pathway enrichment (Fig. 2e) was performed using Clarivate Analytics’ MetaCore™ software67.
Microarray data analysis
Microarray data of CD34+ cells from healthy, del(5q), and non-del(5q) MDS subjects was obtained from a previously published study68 submitted in Gene Expression Omnibus69,70 accessible under GSE19429.
Statistical tests
Data are presented as mean ± s.e.m unless otherwise indicated. Comparison of two groups was performed using paired or unpaired two-tailed t-test. For multiple group comparisons, analysis of variance (ANOVA) with Tukey’s correction or Kruskal-Wallis test with Dunn’s correction was depending on data requirements. Statistical analyses were performed using GraphPad Prism v8.0 (GraphPad Software Inc.). A P-value of less than 0.05 was considered significant.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Localization and expression of Riok2.
(a) Location of RIOK2 gene on human chromosome 5. (b) Schematic representation of the Riok2tm1a(KOMP)Wtsi allele and generation of Riok2 floxed mice. (c) Agarose gel showing genotyping of Riok2 floxed mice. Riok2Δ indicates deletion of Riok2. No band expected in the Riok2wt lane. (d) Expression of Riok2 mRNA in mouse BM cells. Modified from Gene Expression Commons. Numbers next to bars indicate expression level. (e) Riok2 mRNA expression by qRT-PCR in BM cells from Riok2 haploinsufficient mice and Vav1cre controls. n=5 mice/group. (f) Frequency of the genotypes indicated on the X-axis among 4 litters from 4 different breeding crosses of the genotypes mentioned. (g) In vivo protein synthesis rates in the indicated cell types from Riok2 haploinsufficient mice (n=2) and Vav1cre controls (n=8). Unpaired two-tailed t-test (e), multiple unpaired two-tailed t-tests with Holm-Sidak method (g) used to calculate statistical significance. Data are shown as mean ± s.e.m (e,f) or mean ± s.d. (g) and are representative of two (e, g) or four (c, f) independent experiments. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Extended Data Fig. 2. Riok2 haploinsufficient mice display anemia and myeloproliferation.
(a) Gating strategy used for the identification of erythroid progenitor/ precursor cells in the BM. (b) Number of erythroid progenitor populations among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls. n=5/group. (c) Cell cycle analysis of erythroid progenitor/ precursor cells from Riok2 haploinsufficient mice in comparison to Vav1cre controls. n=5 mice/group. (d) Cdkn1a mRNA expression by qRT-PCR in erythroid progenitors from Riok2 haploinsufficient mice and Vav1cre controls. n=3 mice/group. (e) Kaplan-Meier survival curve for Riok2 haploinsufficient mice and Vav1cre controls subjected to lethal dose of PhZ. (f) Number of RIII and RIV erythroid precursor populations among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls day 6 after PhZ treatment. n=4/group. (g) PB RBC numbers, Hb, and HCT in mice transplanted with either Riok2 haploinsufficient mice or Vav1cre BM cells. n=5 mice/group. (h) PB RBC numbers, Hb, and HCT in mice with tamoxifen-inducible deletion of Riok2. Tamoxifen administered on days 3 – 7. n=8 and 7 for Riok2+/+Ert2cre and Riok2f/+Ert2cre mice, respectively. (i) Representative flow cytometry plots showing frequency of monocytes (CD11b+Ly6G−Ly6Chi) and neutrophils (CD11b+Ly6G+) in the PB of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. (j) Representative flow cytometry plots showing Ki-67+ GMPs in the BM of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. (k) Number of CFU-GM colonies in MethoCult from Lin−Sca-1+c-kit+ BM cells from Riok2f/+Vav1cre mice (n=5) and Riok2+/+Vav1cre controls (n=4) after a 7-day culture period. n=4-5/group. Multiple unpaired two-tailed t-tests with Holm-Sidak method (b, c), unpaired two-tailed t-test (d, f, g, k), log-rank test (e), and 2-way ANOVA with Sidak’s correction for multiple comparisons (h) used to calculate statistical significance. Data are shown as mean ± s.e.m and are representative of two (b to k) independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
Extended Data Fig. 3. Riok2 haploinsufficiency alters early hematopoietic progenitors in an age-dependent fashion.
(a) Frequency and number of indicated cell types in the bone marrow of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n = 4/group. LT-HSC = long term hematopoietic stem cells, ST-HSC = short term hematopoietic stem cells, MPP = multipotent progenitors, CLP = common lymphoid progenitors. (b) % CD45.2 (donor) chimerism in PB from competitive BM transplant with CD45.1 recipient cells. Time point ‘−1’ reflects first bleeding 4 weeks after transplantation and one day before tamoxifen induced deletion of Riok2. Donor (CD45.2) chimerism of the HSC compartment in the BM of competitive transplantation experiments. n = 5/group. (c) Frequency of donor (CD45.2+) early hematopoietic progenitors 24 weeks after tamoxifen treatment in a competitive transplantation assay as described in (b). n=5 and 4 for Riok2+/+Ert2cre and Riok2f/+Ert2cre mice, respectively. Unpaired two-tailed t-test (a, c) and 2-way ANOVA with Sidak’s multiple comparison test (b) used to calculate statistical significance. Data are shown as mean ± s.e.m and are representative of two (a-c) independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Extended Data Fig. 4. Riok2 haploinsufficient erythroid precursors express increased S100 proteins.
(a) Expression of ribosomal proteins quantified by proteomics in Riok2f/+Vav1cre and Riok2+/+Vav1cre erythroid precursors. S100A8 (b) and S100A9 (c) expression assessed by flow cytometry in BM erythroid precursors from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n=4mice/group. (d, e) S100a8 and S100a9 mRNA expression in erythroid precursors isolated from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n=4 mice/group. (f) p53 expression assessed by flow cytometry in BM erythroid precursors from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. (g) Graphical representation of data shown in (e). n=5 mice/group. Data are shown as mean ± s.e.m and are representative of two (b to g) independent experiments. Unpaired two-tailed t-test (b to g) used to calculate statistical significance. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Extended Data Fig. 5. Expression of lineage-associated T cell cytokines is comparable between Riok2 haploinsufficient and sufficient T cells.
a-g, Concentration of IL-2 (a), IFN-γ (b), IL-4 (c), IL-5 (d), IL-13 (e), IL-17A (f) and frequency of Foxp3+ cells (g) from in vitro polarized T cells of the indicated genotypes. n=3 mice/group. (h-i) Number of IL-22+ NKT cells (H) and ILCs (I) in the spleens of Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n=4/group). (j) Frequency of IL_23p19+ DCs in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls. n=4 mice/group. (k) PB RBC numbers, Hb, and HCT in Riok2f/fCd4cre mice (n=3) in comparison to Riok2+/+Cd4cre controls (n=5) . (l) Secreted IL-22 from in vitro polarized TH22 cells from Rps14 haploinsufficient mice and Vav1cre controls. n=4 mice/group. (m) Secreted IL-22 from in vitro polarized TH22 cells from ApcMin mice and littermate controls. n=4 mice/group. (n) Viable cells (expressed as percentage of total cells in culture) for the indicated treatments assessed by flow cytometry. n=5 mice/group. Data are shown as mean ± s.e.m and are representative of two (a to n) independent experiments. Unpaired two-tailed t-test (a to n) used to calculate statistical significance. * p < 0.05, ** p < 0.01.
Extended Data Fig. 6. Neutralization of IL-22 signaling increases number of erythroid precursors.
a-d, Number of RI-RIV erythroid populations among viable BM cells in the indicated strains undergoing PhZ-induced stress erythropoiesis. For (a), n=5,5,4, and 4 for Riok2+/+Il22+/+Vav1cre, Riok2+/+Il22+/−Vav1cre, Riok2f/+Il22+/+Vav1cre, Riok2f/+Il22+/−Vav1cre, respectively. For (d), n=5/group. Data are shown as mean ± s.e.m and are representative of three (a, c) or two (b, d) independent experiments. 1-way ANOVA with Tukey’s correction (a, c) or unpaired two-tailed t-test (b, d) used to calculate statistical significance. * p < 0.05, ** p < 0.01.
Extended Data Fig. 7. IL-22 neutralization alleviates anemia in wt mice undergoing PhZ-induced stress erythropoiesis.
(a) PB RBC numbers, Hb, and HCT in naïve wt C57BL/6J mice treated with isotype control (Rat IgG2aκ, 50 mg/mouse) or anti-IL-22 antibody (50 mg/mouse). n=5 mice/group. (b) PB RBC numbers, Hb, and HCT in wt C57BL/6J mice undergoing PhZ-induced stress erythropoiesis treated with isotype control or anti-IL-22 antibody. n=5 mice/group. (c) Percentage of RI-RIV erythroid precursors in the BM of mice treated as in (b). n=4 and 5 for Il22ra1+/+Eporcre and Il22ra1f/fEporcre mice, respectively. Data are shown as mean ± s.e.m and are representative of three (a, b) or two (c) independent experiments. Unpaired two-tailed t-test (a to c) used to calculate statistical significance. * p < 0.05, *** p < 0.001.
Extended Data Fig. 8. Erythroid precursors express IL-22RA1.
(a) Gating strategy employed for assessing IL-22RA1 expression on erythroid precursors. (b) Gating strategy to show that majority of IL-22RA1+ cells in the mouse BM are erythroid precursors. (c) IL-22RA1 expression on erythroid precursors assessed using flow cytometry and a second antibody targeting a different epitope of IL-22RA1. (d) Il22ra1 mRNA expression in the indicated cell types assessed by qRT-PCR. T cells and liver represent negative and positive controls, respectively. n=4 mice/group. Data are shown as mean ± s.e.m (d) and are representative of three (a to c) or two (d) independent experiments.
Extended Data Fig. 9. Increased IL-22 and its signature genes in del(5q) MDS subjects.
(a) Representative flow cytometry plots showing frequency of CD4+IL-22+ cells among total PBMCs in the peripheral blood of MDS patients and healthy subjects. Pre-gated on viable CD3ε+CD4+ cells. Cumulative data shown in Figure 4e. (b) Expression of indicated IL-22 signature genes in CD34+ cells from healthy controls and del(5q) and non-del(5q) MDS patients. n = 17, 47, and 136 for healthy, del(5q) MDS, and non-del(5q) MDS, respectively. Kruskal-Wallis test with Dunn’s correction for multiple comparisons (b) used to calculate statistical significance * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Solid lines represent median and dashed lines represent quartiles (b).
Extended Data Fig. 10. Riok2 haploinsufficiency recapitulates del(5q) MDS transcriptional changes.
(a to b) GSEA enrichment plots comparing proteins up-regulated (a) and down-regulated (b) upon Riok2 haploinsufficiency to the transcriptional changes seen in del(5q) MDS. (c) Schematic of mechanism underlying Riok2 haploinsufficiency-induced, IL-22 –induced anemia.
Supplementary Material
Acknowledgements
We thank R. Caspi (National Eye Institute, US National Institutes of Health (NIH)) and Genentech for providing Il22 knockout mice; B. L. Ebert (Dana-Farber Cancer Institute) for providing Rps14 floxed mice; R. Locksley (University of California, San Francisco) for providing Catch22 mice; and U. Klingmüller (Deutsches Krebsforschungszentrum (DFKZ), Germany) for providing Epor-cre mice. We also thank the Animal Resources Facility (ARF), The Ted and Eileen Pasquarello Tissue Bank in Hematologic Malignancies and flow cytometry core facility at DFCI for their valuable technical assistance. We thank E. Smith for assistance with creating the graphical illustration. We thank P. Májek (Ústav hematologie a krevní transfuse, ÚHKT) for providing plasma proteomic profiling data of healthy subjects and MDS patients.
Funding: This work was supported by a Discovery Research Grant from the Edward P. Evans Foundation (L.H.G.) and institutional funding from Dana-Farber Cancer Institute (L.H.G). D.P.S. is supported by the Edward P. Evans Foundation and by NIH Leukemia SPORE 1P50CA206963 and 2P01CA066996. This work was supported in part by grants from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (to S.A.C.).
Footnotes
Competing Interests statement
An invention disclosure has been filed based on the data generated in this study.
From August 4th, 2020, Meromit Singer is an employee of Guardant Health.
S.S.W. has served on the steering committee of GSK trial on an oral hypoxia-inducible factor prolyl hydroxylase inhibitor, as a potential treatment for anemia associated with chronic kidney disease; S.S.W. has also received consulting fees from Public Health Advocacy Institute, CVS, Roth Capital Partners, Kantum Pharma, Mallinckrodt, Wolters Kluewer, GE Health Care, Allena Pharmaceuticals, Mass Medical International, JNJ, Venbio, Strataka, Takeda, Cerus, and Pfizer. D.P.S. has served on independent data safety monitoring committees for clinical trials supported by Takeda, Astex, Janssen and Onconova; has consulted for Celgene and Daiichi Sankyo; and has received research support (to the institution) for clinical trials sponsored by Aprea, H3 Biosciences, Syros and Astra Zeneca. J.R. reports research funding from Amgen, Equillium, and Kite Pharma; and consulting income from Aleta Biotherapeutics, Avrobio, Celgene, Falcon Therapeutics, LifeVault Bio, Rheos Medicines, Talaris Therapeutics and TScan Therapeutics.
R.M.S. has served on independent data safety monitoring committees for trials supported by Celgene, Takeda and Argenix; has consulted for AbbVie, Actinium, Agios, Amgen, Arog, Astellas, AstraZeneca, Biolinerx, Celgene, Daiichi Sankyo, Fujifilm, Janssen, Juno, Macrogenics, Novartis, Ono, Orsenix, Pfizer, Roche, Stemline, Sumitomo, Takeda, and Trovagene; and has received research support (to the institution) for clinical trials sponsored by AbbVie, Agios, Arog, and Novartis. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and a scientific advisor to Pfizer and Biogen.
A.R. is a SAB member of ThermoFisher Scientific, Neogene Therapeutics, Asimov and Syros Pharmaceuticals. A.R. is a cofounder of and equity holder in Celsius Therapeutics and an equity holder in Immunitas. From August 1, 2020, AR is an employee of Genentech.
L.H.G. is a former Director of Bristol-Myers Squibb and the Waters Corporation and is currently on the board of directors of and holds equity in GlaxoSmithKline Pharmaceuticals and Analog Devices, Inc. She also serves on the scientific advisory boards of Repare Therapeutics, Abpro Therapeutics and Kaleido Therapeutics. All other authors declare no relevant competing interest.
Data availability
The original mass spectra may be downloaded from MassIVE (http:\\massive.ucsd.edu), MSV000085287. The data are directly accessible via ftp://massive.ucsd.edu/MSV000085287/. Raw RNA-Seq data is accessible via Gene Expression Omnibus (GEO) under the accession code GSE165467.
Source data for all applicable figures (Main and Extended) will be provided with the paper. The remaining data supporting the findings of this study are available from the corresponding authors upon reasonable request. Materials will be provided with material transfer agreements (MTA) as appropriate.
References
- 1.Giagounidis AA, Germing U & Aul C Biological and prognostic significance of chromosome 5q deletions in myeloid malignancies. Clin. Cancer Res 12, 5–10 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Haase D et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood 110, 4385–4395 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Hofmann WK, Lubbert M, Hoelzer D & Phillip Koeffler H Myelodysplastic syndromes. Hematol. J 5, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Sole F et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo Cooperativo Espanol de Citogenetica Hematologica. Br. J. Haematol 108, 346–356 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Dutt S et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebert BL et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar MS et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood 118, 4666–4673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribezzo F et al. Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia, (2019). [DOI] [PubMed] [Google Scholar]
- 9.Schneider RK et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell 26, 509–520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider RK et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med 22, 288–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira-Cerca S et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol 19, 1316–1323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royer-Pokora B et al. Delineation by molecular cytogenetics of 5q deletion breakpoints in myelodyplastic syndromes and acute myeloid leukemia. Cancer Genet. Cytogenet 167, 66–69 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Tang G et al. Isolated del(5q) in Patients Following Therapies for Various Malignancies May Not All Be Clinically Significant. Am. J. Clin. Pathol 144, 78–86 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Boultwood J et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood 99, 4638–4641 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Lai F et al. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q). Genomics 71, 235–245 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Zemp I et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol 185, 1167–1180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muto T et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol 21, 535–545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MA et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol 21, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starczynowski DT & Karsan A Innate immune signaling in the myelodysplastic syndromes. Hematol. Oncol. Clin. North Am 24, 343–359 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Qian Y, Eksioglu E, Epling-Burnette PK & Wei S The inflammatory microenvironment in MDS. Cell. Mol. Life Sci 72, 1959–1966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allampallam K et al. Measurement of mRNA expression for a variety of cytokines and its receptors in bone marrows of patients with myelodysplastic syndromes. Anticancer Res. 19, 5323–5328 (1999). [PubMed] [Google Scholar]
- 22.Schipperus MR et al. Interleukin-6 and interleukin-1 enhancement of GM-CSF-dependent proliferation of haematopoietic progenitor cells in myelodysplastic syndromes. Br. J. Haematol 77, 515–522 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Shao LL et al. Th22 cells as well as Th17 cells expand differentially in patients with early-stage and late-stage myelodysplastic syndrome. PLoS One 7, e51339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhoef GE et al. Measurement of serum cytokine levels in patients with myelodysplastic syndromes. Leukemia 6, 1268–1272 (1992). [PubMed] [Google Scholar]
- 25.Starczynowski DT & Karsan A Deregulation of innate immune signaling in myelodysplastic syndromes is associated with deletion of chromosome arm 5q. Cell Cycle 9, 855–856 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Means RT Jr. Pathogenesis of the anemia of chronic disease: a cytokine-mediated anemia. Stem Cells 13, 32–37 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Song M et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seita J et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One 7, e40321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khajuria RK et al. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103 e119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers SA et al. Streamlined Protocol for Deep Proteomic Profiling of FAC-sorted Cells and Its Application to Freshly Isolated Murine Immune Cells. Mol. Cell. Proteomics 18, 995–1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su LK et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Gronke K et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotenko SV et al. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta ) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J. Biol. Chem 276, 2725–2732 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Kdoqi & National Kidney, F. KDOQI Clinical Practice Guidelines and Clinical Practice Recommendations for Anemia in Chronic Kidney Disease. Am. J. Kidney Dis 47, S11–145 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Maciejewski JP et al. A pilot study of the recombinant soluble human tumour necrosis factor receptor (p75)-Fc fusion protein in patients with myelodysplastic syndrome. Br. J. Haematol 117, 119–126 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Fenaux P et al. Luspatercept in Patients with Lower-Risk Myelodysplastic Syndromes. N. Engl. J. Med 382, 140–151 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Suragani RN et al. Transforming growth factor-beta superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med 20, 408–414 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Mattapallil MJ et al. Interleukin 22 ameliorates neuropathology and protects from central nervous system autoimmunity. J. Autoimmun 102, 65–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CL et al. IL-22 regulates iron availability in vivo through the induction of hepcidin. J. Immunol 191, 1845–1855 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto K et al. IL-22 Controls Iron-Dependent Nutritional Immunity Against Systemic Bacterial Infections. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietras EM et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol 18, 607–618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita M & Passegue E TNF-alpha Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell 25, 357–372 e357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang TY et al. IL-6 blockade reverses bone marrow failure induced by human acute myeloid leukemia. Sci. Transl. Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai T et al. Increased expression of IL-22 is associated with disease activity in Behcet's disease. PLoS One 8, e59009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto-Furusho JK et al. Colonic epithelial upregulation of interleukin 22 (IL-22) in patients with ulcerative colitis. Inflamm. Bowel Dis 16, 1823 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Lee SJ et al. Certain Autoimmune Manifestations Are Associated With Distinctive Karyotypes and Outcomes in Patients With Myelodysplastic Syndrome: A Retrospective Cohort Study. Medicine (Baltimore) 95, e3091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fozza C, La Nasa G & Caocci G The Yin and Yang of myelodysplastic syndromes and autoimmunity: The paradox of autoimmune disorders responding to therapies specific for MDS. Crit. Rev. Oncol. Hematol 142, 51–57 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Wolach O & Stone R Autoimmunity and Inflammation in Myelodysplastic Syndromes. Acta Haematol. 136, 108–117 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Schnatter AR, Glass DC, Tang G, Irons RD & Rushton L Myelodysplastic syndrome and benzene exposure among petroleum workers: an international pooled analysis. J. Natl. Cancer Inst 104, 1724–1737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boitano AE et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods Only):
- 51.Skarnes WC et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farley FW, Soriano P, Steffen LS & Dymecki SM Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 (2000). [PubMed] [Google Scholar]
- 53.Heinrich AC, Pelanda R & Klingmuller U A mouse model for visualization and conditional mutations in the erythroid lineage. Blood 104, 659–666 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Savage AK, Liang HE & Locksley RM The Development of Steady-State Activation Hubs between Adult LTi ILC3s and Primed Macrophages in Small Intestine. J. Immunol 199, 1912–1922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider RK et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med 22, 288–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pronk CJ et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1, 428–442 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Shuga J, Zhang J, Samson LD, Lodish HF & Griffith LG In vitro erythropoiesis from bone marrow-derived progenitors provides a physiological assay for toxic and mutagenic compounds. Proc. Natl. Acad. Sci. U. S. A 104, 8737–8742 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myers SA et al. Streamlined Protocol for Deep Proteomic Profiling of FAC-sorted Cells and Its Application to Freshly Isolated Murine Immune Cells. Mol. Cell. Proteomics 18, 995–1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rappsilber J, Ishihama Y & Mann M Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663–670 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Shalek AK et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Picelli S et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudakov JA, Hanash AM & van den Brink MR Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol 33, 747–785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikolsky Y, Ekins S, Nikolskaya T & Bugrim A A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett 158, 20–29 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Pellagatti A et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Barrett T et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra may be downloaded from MassIVE (http:\\massive.ucsd.edu), MSV000085287. The data are directly accessible via ftp://massive.ucsd.edu/MSV000085287/. Raw RNA-Seq data is accessible via Gene Expression Omnibus (GEO) under the accession code GSE165467.
Source data for all applicable figures (Main and Extended) will be provided with the paper. The remaining data supporting the findings of this study are available from the corresponding authors upon reasonable request. Materials will be provided with material transfer agreements (MTA) as appropriate.