Abstract
Erythroferrone (ERFE) is the main erythroid regulator of hepcidin, the homeostatic hormone controlling plasma iron levels and total body iron. When the release of erythropoietin from the kidney stimulates the production of new red blood cells, it also increases the synthesis of ERFE in bone marrow erythroblasts. Increased ERFE then suppresses hepcidin synthesis, thereby mobilizing cellular iron stores for use in heme and hemoglobin synthesis. Recent mechanistic studies have shown that ERFE suppresses hepcidin transcription by inhibiting bone morphogenetic protein signaling in hepatocytes. In ineffective erythropoiesis, pathological overproduction of ERFE by an expanded population of erythroblasts suppresses hepcidin and causes iron overload, even in non-transfused patients. ERFE may be a useful biomarker of ineffective erythropoiesis and an attractive target for treating its systemic effects.
Keywords: iron homeostasis, hepcidin, ineffective erythropoiesis, β-thalassemia, erythroferrone, bone morphogenetic proteins
1. Erythropoiesis and iron homeostasis—an overview
1.1. Iron homeostasis
Iron is an essential micronutrient for nearly all living organisms because of its important role in catalyzing redox reactions and the transport and storage of oxygen. Many organisms with circulatory systems use myoglobin to store oxygen reserves in skeletal muscle and hemoglobin to deliver oxygen to all body tissues. In both myoglobin and hemoglobin, oxygen binds to an iron atom at the center of heme groups. Total iron content in healthy human adults is 3–4 g, of which more than half is found in hemoglobin of erythrocytes. Macrophages continuously recycle iron from old erythrocytes and other senescent cells, returning it to a dynamic plasma iron pool. Conservation effectively limits iron losses to a tiny fraction of the total content (~1 mg/day) in children, men, and post-menopausal women. Pregnant, lactating, and menstruating women experience increased iron loss due to the iron demands of these physiological processes. Non-heme iron is transported through plasma bound to transferrin protein which constrains the propensity of iron to catalyze the generation of reactive oxygen species. Once imported into cells through transferrin receptors, iron is stored within ferritin. Most cellular iron stores are located in the liver and spleen.
Tight regulation of iron flows ensures that iron is available to developing red blood cells while avoiding the potential toxicity of excess extracellular iron, and limiting its availability to infectious microorganisms. The body’s iron traffic is controlled by hepcidin, the sole homeostatic hormone responsible for regulating both plasma iron levels and the total iron content of the body. The 25 amino acid peptide is produced in the liver in proportion to plasma iron concentrations and hepatic iron stores. When iron is abundant, high concentrations of hepcidin bind the cellular iron exporter ferroportin, causing its occlusion, internalization (Nemeth 2004), and degradation. Intestinal absorption of dietary iron and iron release from cellular stores are thereby inhibited. When plasma iron is scarce, hepatic hepcidin secretion is low which allows ferroportin to export iron to plasma, thereby stimulating intestinal iron absorption and mobilization of cellular stores.
1.2. Erythropoiesis and its interaction with iron homeostasis
The intensely energy-consuming renal medulla effectively monitors oxygen delivery by blood, determines its hemoglobin content, the amount of oxygen bound to hemoglobin, and the dissociation of oxygen from hemoglobin. When oxygen delivery to this tissue does not meet demand, interstitial fibroblasts sense hypoxia and begin producing and excreting erythropoietin (EPO) whose synthesis is transcriptionally regulated by hypoxia-inducible factor 2 (HIF-2). Erythroid precursors in the bone marrow respond to EPO signaling by cell division and differentiation towards mature erythrocytes. Hemoglobin synthesis in the developing erythrocytes requires iron-containing heme. Iron-deficiency inhibits the production of heme and hemoglobin, as well as the production and maturation of erythrocytes.
Erythropoiesis even at baseline consumes most of the iron flowing into the plasma compartment. Stimulation of erythropoiesis, by blood loss, hypoxia, or injection of exogenous erythropoietin strongly suppresses the production of hepatic hepcidin in mice and humans, allowing more dietary iron and iron from stores to enter blood plasma for heme and hemoglobin synthesis by developing erythrocytes in the marrow (Figure 1, A-B). Arguing from clinical observations and experimental models, investigators in the last century proposed that an erythroid regulator strongly influences iron homeostasis. After the discovery of hepcidin as an iron-regulatory hormone, it became clear that the erythroid regulator does not directly affect iron transport but rather acts by suppressing hepcidin. Although erythropoietin itself was a reasonable candidate for a hepcidin suppressor, in vitro studies showed that EPO does not directly suppress hepcidin in isolated liver cells (Gammella et al. 2015) implying the existence of an intermediary EPO-responsive suppressor of hepcidin.
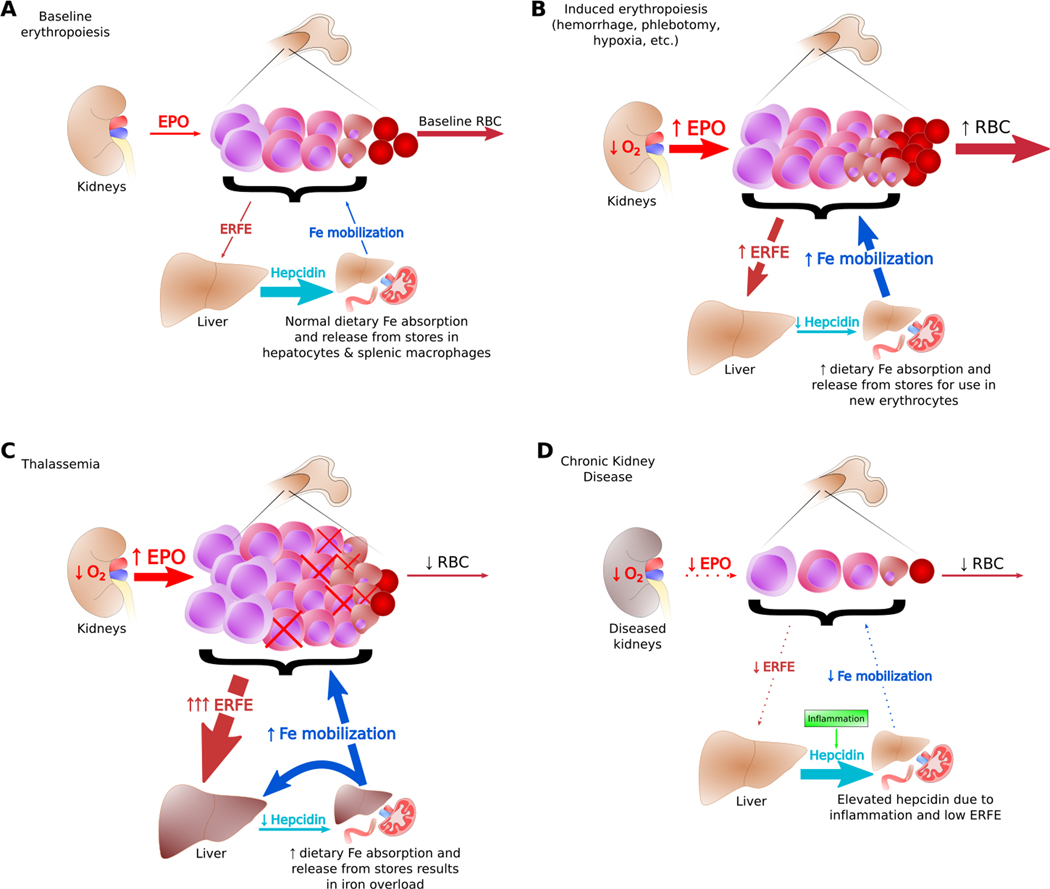
Figure 1. Effective and Ineffective Erythropoiesis.
(A) Baseline erythropoiesis generates red blood cells to replace old and damaged cells. Physiological levels of ERFE and hepcidin at baseline provide sufficient iron for steady state production of erythrocytes. (B) When the kidneys sense cellular hypoxia, they secrete EPO which stimulates erythropoiesis and the production of ERFE. As ERFE suppresses hepcidin, iron is mobilized from stores for use by the expanded population of maturing red blood cells. (C) In β-thalassemia, most erythroblasts do not generate mature erythrocytes, causing anemia and tissue hypoxia. This results in high levels of EPO and ERFE, chronically low hepcidin, and iron overload. (D) In chronic kidney disease, low EPO production, low clearance of hepcidin by the kidney, and inflammation can lead to low iron availability in the erythroid system.
2. The Erythroferrone Molecule
2.1. Discovery of Erythroferrone
Motivated by evidence that hepcidin expression is influenced by erythropoietic activity (Pak et al. 2006; Vokurka et al. 2006), a search was performed for mRNAs encoding secreted proteins that were induced in mouse bone marrow within hours after blood loss (Kautz et al. 2014). A transcript identified at the time as Fam132b and since renamed Erfe was induced shortly before hepcidin suppression and sustained its high expression for 24–48 hours, mirroring serum EPO concentrations. EPO injections resulted in a similar but faster increase in Erfe expression indicating that its gene product may be directly EPO-inducible. qPCR analysis revealed EPO-stimulated bone marrow, and specifically erythroblasts, to be the highest ERFE-expressing cell type. Though the erythroferrone protein had been catalogued previously under the names CTRP15 and myonectin (Seldin, Tan, and Wong 2014), its prominent role in the regulation of hepcidin and iron homeostasis was a novel finding. ERFE met the criteria for the putative erythroid regulator of iron homeostasis that could explain the observed iron-regulatory responses to anemia, hemorrhage, hypoxia, and disorders of ineffective erythropoiesis including β-thalassemia and myelodysplastic syndromes (Figure 1, C-D). The development of an immunoassay for serum ERFE (Ganz et al. 2017) allowed for the analysis of ERFE in patients with these diseases, as discussed below. In a short time, the measurement of ERFE levels has become a common research tool for studies of the pathophysiology of anemias and other erythroid disorders.
2.1.1. Human Erythroferrone Immunoassay
In 2017, a rabbit monoclonal antibody-based sandwich immunoassay test was developed for the detection of serum ERFE from human clinical samples (Ganz et al. 2017). The immunoassay was validated using blood samples taken from three sets of human volunteers: healthy adult males, EPO-treated geriatric patients with anemia, and β-thalassemic patients (both non-transfused and transfusion-dependent). ERFE levels detected by the assay rose in the healthy volunteers after phlebotomy with a matching decline in serum hepcidin levels as confirmation. The EPO-treated patients likewise manifested the expected changes in their ERFE and hepcidin values. ERFE concentrations in non-transfused and pre-transfused β-thalassemia patients were elevated relative to normal samples, as expected, and levels were much lower post-transfusion. These data recapitulated the expected trends for ERFE concentrations in each population and were validated by inverse correlations with serum hepcidin. A similar validated human ERFE immunoassay using mouse monoclonal antibodies targeted to the same antigen is available commercially (Intrinsic LifeSciences, La Jolla, CA).
2.1.2. Mouse Erythroferrone Immunoassay
Another ELISA using mouse monoclonal antibodies was developed by Silarus Therapeutics for the detection of murine ERFE. We modified the assay to increase its sensitivity by employing the DELFIA reporter system instead of the conventional horse-radish peroxidase-based colorimetric readout. Another ELISA for murine ERFE, making use of polyclonal antibodies against mouse ERFE, is commercially available (Intrinsic LifeSciences). Using this assay, ERFE levels are below the threshold of detection in healthy male C57BL/6 mice, but mice with the Hbbth3/+ model of β-thalassemia have elevated and detectable ERFE levels (Gutschow et al. 2019). A number of other ERFE ELISA kits are also marketed but should be viewed with caution unless supported by peer-reviewed validation data.
2.2. The C1q/TNF-Related Protein (CTRP) Family
Erythroferrone belongs to the C1q/TNF-Related Protein (CTRP) family of structurally related, secreted hormones found in all vertebrate organisms. The 16 identified CTRP proteins share structural similarities and 4 distinct domains: an N-terminal signal peptide directing the protein for secretion, a variable region that differentiates each member, a collagenous linker with Gly-X-Y repeats, and finally a C-terminal globular head with homology to complement factor C1q and the TNFα family of cytokines (Wong et al. 2004) (Figure 2A). Gene expression profiling shows that some CTRPs are preferentially produced in certain tissue types. Adiponectin, the most extensively studied member of this family, is expressed highly and nearly exclusively in adipose tissues. CTRP6, CTRP12, and ERFE likewise have similarly restricted expression in the placenta, small intestine, and erythropoietic bone marrow, respectively. Early attempts to characterize the proteins of this large family have focused on their metabolic effects and potential to function as inter-organ communicators (Schäffler and Buechler 2012; Seldin et al. 2014). Adiponectin has held special interest for decades, with numerous studies that continue to elaborate its roles in insulin resistance, dyslipidemia, and atherosclerosis (Yadav et al. 2013). Some of adiponectin’s metabolic functions are believed to be mediated by its various oligomerized forms. The protein can exist as a trimer, hexamer, and the high molecular weight 18-mer and 30-mer species in some organisms. Functional data show differing and sometimes opposing biological effects of adiponectin depending on the oligomer type, with the higher molecular weight species thought to be the most metabolically active (Wang et al. 2008). The ability of adiponectin to multimerize is dependent on interactions in the collagenous domain, specifically between conserved lysine residues, and their post-translational modifications (PTMs). Bacterially-derived adiponectin can form trimeric and hexameric structures, but the lack of PTMs prevents higher order oligomerization. The formation of these high molecular weight species from mammalian cells can be disrupted by de-glycosylation of the key lysine residues or mutation of these lysines to other amino acids. The presence and conservation of the collagenous domain in all CTRPs suggests that multimerization is likely relevant for the native structures and biological functions of all family members. The varied expression patterns of CTRP members suggest that they mediate communication among distinct organ systems. Mechanisms for CTRP action are currently a topic of active research, and there are as of yet no known cell surface receptors that mediate their physiological functions, with the exception of adiponectin receptors AdipoR1 and AdipoR2 (Yamauchi et al. 2003).
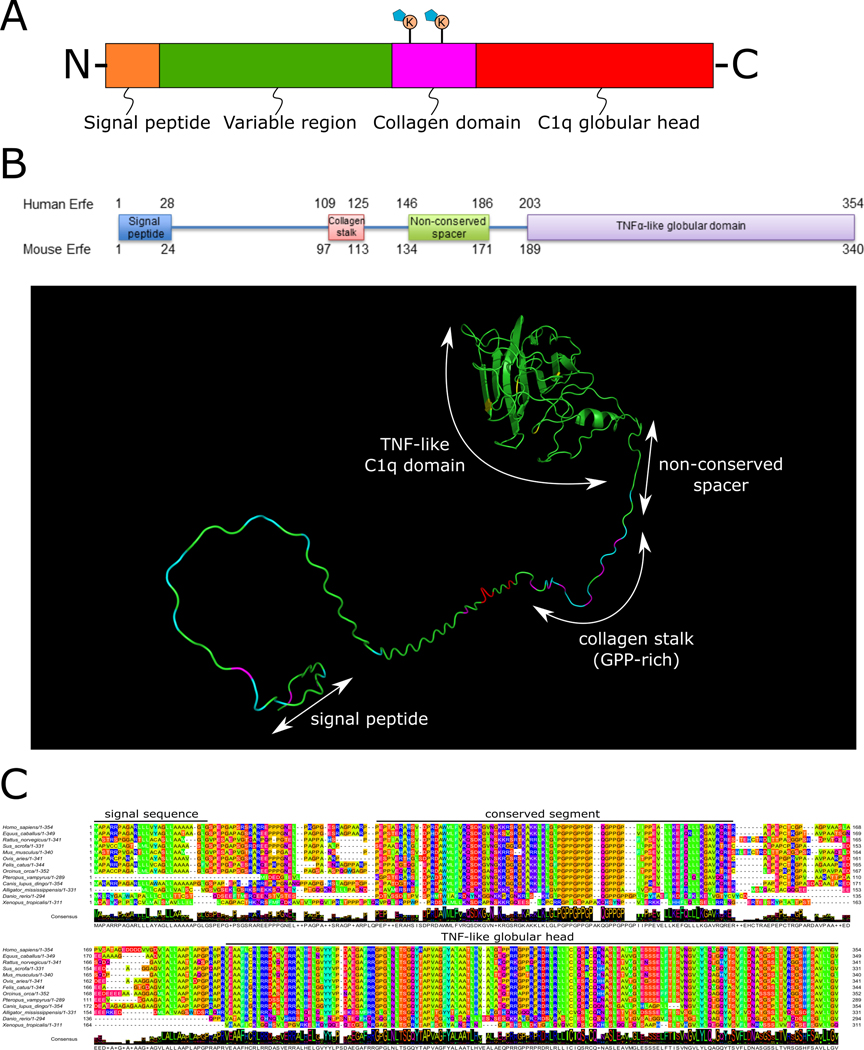
Figure 2. The C1q/TNF-related protein (CTRP) and ERFE Structure.
(A) The generalized domain structure of CTRPs. The signal peptide directs the protein for secretion and is removed in the final form. The variable region is the least conserved domain between CTRP family members. The collagen domain is glycine- and proline-rich and may contain glycosylated lysine residues known to promote multimerization (e.g. adiponectin). The C1q globular head is predicted to be the most structured part of the protein and is highly conserved between CTRP family members. (B) Human and murine erythroferrone domains and prediction of folded structure. (C) Erythroferrone sequence alignment across vertebrate species highlighting the conserved segments.
2.3. Erythroferrone Structure and Properties
Like other CTRPs, ERFE has a 4-domain structure with a unique N-terminus and significant sequence homology to other family members at the C-terminus. The full-length human and mouse proteins are composed of 354 and 340 amino acids, respectively, and have predicted masses of 37.3 kDa and 36.3 kDa. The glycosylation of one (human) or two (mouse) asparagine residue(s) (Blom et al. 2004) coupled with co-translational removal of the signal peptides all contribute to an apparent weight of 35–40 kDa as determined by reducing SDS-PAGE electrophoresis. The globular C-terminal domain is predicted to fold into the highly structured TNF/C1q head while the N-terminus of the molecule is expected to have an extended and flexible secondary structure (Figure 2B). In addition to the globular head, the short proline-rich collagenous linker connecting the two larger domains is thought to promote multimerization of the protein as is seen in the native, oligomerized forms of adiponectin (Wang et al. 2008), but lacks the lysines that are thought to facilitate higher order multimer assembly. Because it contains two predicted PCSK3/furin recognition sites, ERFE may exist as multiple cleaved isoforms after protein processing. Little is currently understood about multimerization or the regulation and differential function of the cleaved forms.
2.4. Evolutionary Analysis of Erythroferrone
ERFE is encoded in the genomes of all vertebrates, indicating its importance to the physiology of so many forms of life. Amino acid sequence alignment reveals a striking conservation across species, down to and including Xenopus tropicalis (Figure 2C). Beyond the N-terminal signal sequence that directs each ERFE ortholog for secretion, there is a stretch of nearly 100 amino acid residues with high levels of conservation in both charge and polarity. These conserved regions include hydrophobic stretches, positively charged patches, and the remarkably preserved glycine/proline-rich collagen linker which could all be integral to ERFE’s cellular function. The C-terminal TNF domain is the most highly conserved region across species but unexpectedly, functional evidence rules this region out as the primary mediator of ERFE’s effect on hepcidin.
2.5. Erythroferrone Mechanism of Action
When plasma iron concentrations and liver iron stores are high, hepcidin synthesis is induced by bone morphogenetic protein activation of the Smad1/5/8 pathway, predominantly through binding of the BMP2/6 heterodimer to the heterotetrameric BMP receptor (Wang et al. 2020). Although technical limitations may interfere with the detection of the active phosphorylated forms of Smad1/5/8 in vivo, the weight of evidence indicates that erythroferrone potently suppresses this induction both in vivo and in Hep3B in vitro cell systems (Wang et al. 2017:5). Various molecular mechanisms for this suppression have been postulated including signaling by dedicated ERFE receptors and interaction with the negative BMP regulator matripase-2 (Arezes et al. 2018; Aschemeyer et al. 2017). Matripase-2, expressed from the TMPRSS6 gene, is a serine protease that cleaves the membrane-bound form of the positive hepcidin regulator hemojuvelin (and likely also other proteins in the iron-regulatory complex) and thereby suppresses hepcidin transcription. Although matripase-2 was a reasonable candidate to consider as part of the ERFE mechanism of action, experiments with Tmprss6 KO mice have demonstrated that erythroferrone and matripase-2 regulate hepcidin independently of one another (Aschemeyer et al. 2017). These results showed that while ablation of matripase-2 increases hepcidin expression overall, ERFE still suppresses transcription by the same relative amount as in WT mice.
Recent data have provided unexpected but well-founded evidence for ERFE’s action as a novel BMP trap that diminishes hepcidin expression by sequestering BMP ligands away from their cell surface receptors (Arezes et al. 2018), specifically ALK3 (Wang et al. 2020:3) (Figure 3). Surface plasmon resonance (SPR) studies confirm the ability of ERFE to bind BMPs. The strongest interactions take place between ERFE and the homodimers of BMP2, BMP6, and the BMP2/6 heterodimer (Arezes et al. 2020; Wang et al. 2020). Other previously proposed erythroid regulators of hepcidin, such as the TGFβ-family ligand GDF15 (Kim and Nemeth 2015), lack any binding interaction (Arezes et al. 2020). Our unpublished data on the binding of other CTRPs to BMPs also show varied preference for BMP species between family members, suggesting a general model for how their physiological effects may be carried out. In studies of ERFE truncation mutants, the N-terminal variable region is sufficient to suppress hepcidin expression nearly to the same extent as the WT protein, and the C1q globular domain carries no suppressive activity (Arezes et al. 2020). Moreover, antibodies specifically targeted toward the N-terminus of the protein compete for binding with BMP6, unlike antibodies that target the C-terminus. SPR analysis of ERFE truncation mutants has confirmed the specific binding of BMPs to the N-terminus only. Nevertheless, it is likely that the globular head facilitates the function of ERFE in vivo, perhaps by promoting multimerization, enhancing stability, or in some other manner yet to be elucidated.
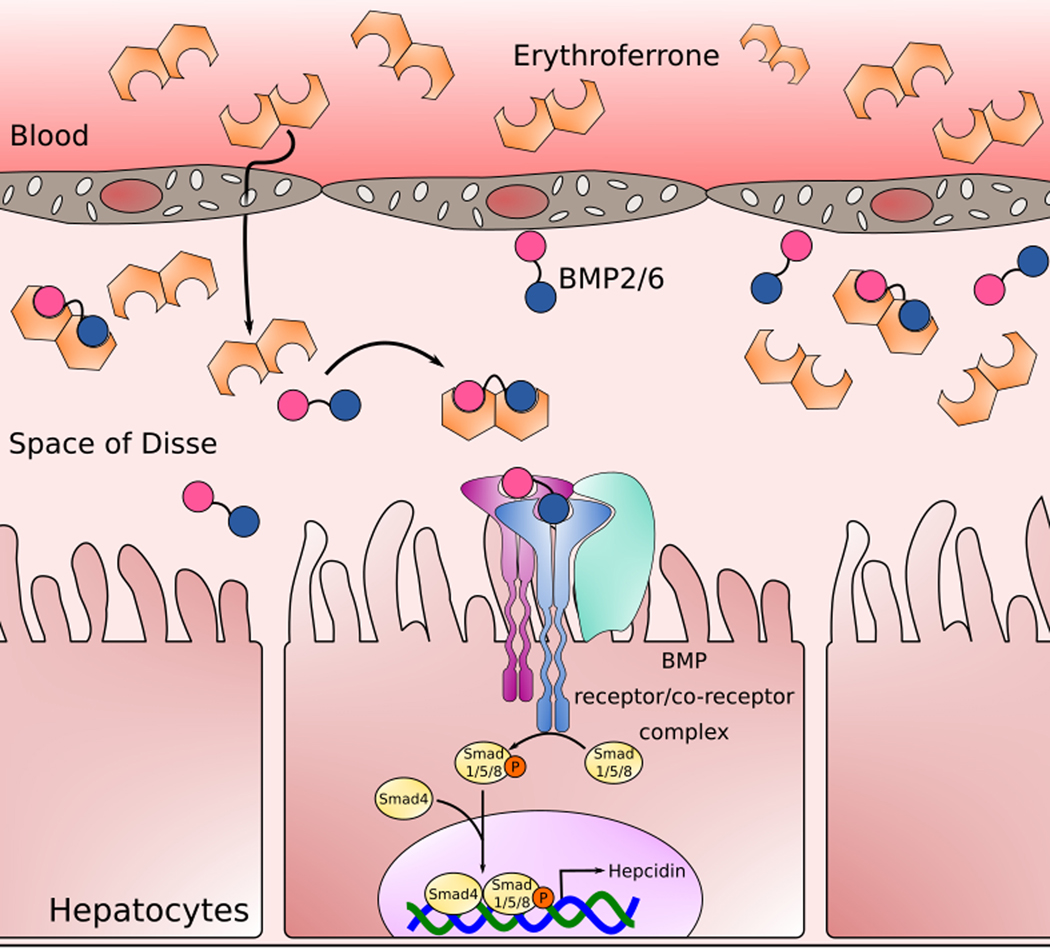
Figure 3. Erythroferrone Mechanism of Action.
Erythroferrone molecules secreted into blood by erythroblasts in the marrow reach the liver where it permeates through fenestrations in sinusoidal endothelial cells into the Space of Disse, a perisinusoidal space separating hepatocytes from sinusoidal endothelial cells. Sinusoidal endothelial cells secrete BMP2/6 heterodimers, but ERFE sequesters these before they engage the BMP receptor complex on hepatocytes, lowering activation of the Smad1/5/8 signal transduction pathway, and reducing hepcidin transcription.
3. Erythroferrone Pathophysiology
3.1. Erythroferrone in Baseline and Stress Erythropoiesis
Basal erythropoiesis in the marrow produces red cells at a relatively constant rate to replace old and damaged ones whose iron content is recycled mostly by splenic and hepatic macrophages (Figure 1A). Anemia or hypoxia decrease oxygen delivery to tissues, a change sensed as cellular hypoxia by interstitial fibroblasts in the intensely energy-consuming regions of the kidney, and these cells respond by HIF-2 mediated increases in the production of EPO (Haase 2013; Hodges et al. 2007). Elevated plasma EPO concentrations enhance the survival of erythroid precursors, increase the number of erythroblasts, and stimulate those erythroblasts to produce and secrete ERFE, thereby suppressing hepcidin synthesis in the liver. With less hepcidin available to inhibit ferroportin function, iron-exporting cells deliver more cellular iron into plasma. Dietary iron absorption increases, and cellular iron stores are mobilized from macrophages and hepatocytes for heme and hemoglobin synthesis in newly produced red blood cells (Figure 1B). Erythroferrone is most important early in the response to erythropoietic stimuli, as ERFE knockout mice fail to rapidly suppress hepcidin after acute blood loss or EPO injection. This leads to a delay of several days in their recovery from anemia compared to WT animals (Kautz et al. 2014).
The increased ERFE production in response to anemia has two components: when erythropoietic tissues are stimulated by EPO, the population of erythroid precursor cells expands. Secondly, within this population, each individual cell produces more ERFE. In anemias with ineffective erythropoiesis, the erythroid precursor population is greatly expanded and stimulated by EPO, but most of these cells do not generate mature erythrocytes. These dead-end erythroid precursors secrete high levels of ERFE that chronically suppress hepcidin and thereby cause iron overload. High concentrations of non-transferrin bound iron in these iron-loading anemias are known to cause tissue injury by catalyzing the formation of reactive oxygen species (Figure 1C). Iron toxicity is manifested as cellular damage in hepatocytes, cardiomyocytes, and endocrine glands as well as an increased risk of infection (Vento, Cainelli, and Cesario 2006). To prevent these potentially lethal complications of iron overload, treatment with iron chelators is required to trap and eliminate excess iron through the urinary and gastrointestinal routes.
3.2. Effects of Erythroferrone Loss-of-Function and Over-Expression
ERFE knockout mice that are allowed to develop without external erythropoietic stresses have normal iron and blood parameter phenotypes with the exception of a transient iron-restrictive anemia at around 6 weeks of age (Kautz et al. 2014). This is a period of rapid growth in mice, with expansion of erythrocyte mass and increased iron demand. Lower hemoglobin levels in ERFE knockout mice catch up to their WT counterparts by 12 weeks. While ERFE ablation prevents knockout animals from quickly suppressing hepcidin after an acute erythropoietic stimulus as previously mentioned, an appropriate, if delayed, hepcidin response is seen after a prolonged erythropoietic stimulus such as multiple EPO injections (Coffey et al. 2018). The delayed but complete compensation for the lack of ERFE may be due, at least in part, to the depletion of iron stores which itself is a strong negative regulator of hepcidin expression (Figure 4). Because differic transferrin is a strong inducer of hepcidin transcription, the transient decrease of diferric transferrin concentrations caused by EPO-inducible expression of transferrin receptor-1 on erythroblasts may contribute to the small initial drop in hepcidin (Mirciov et al. 2018) that is seen even in ERFE KO mice, and could synergize with the suppressive effect of ERFE on hepcidin.
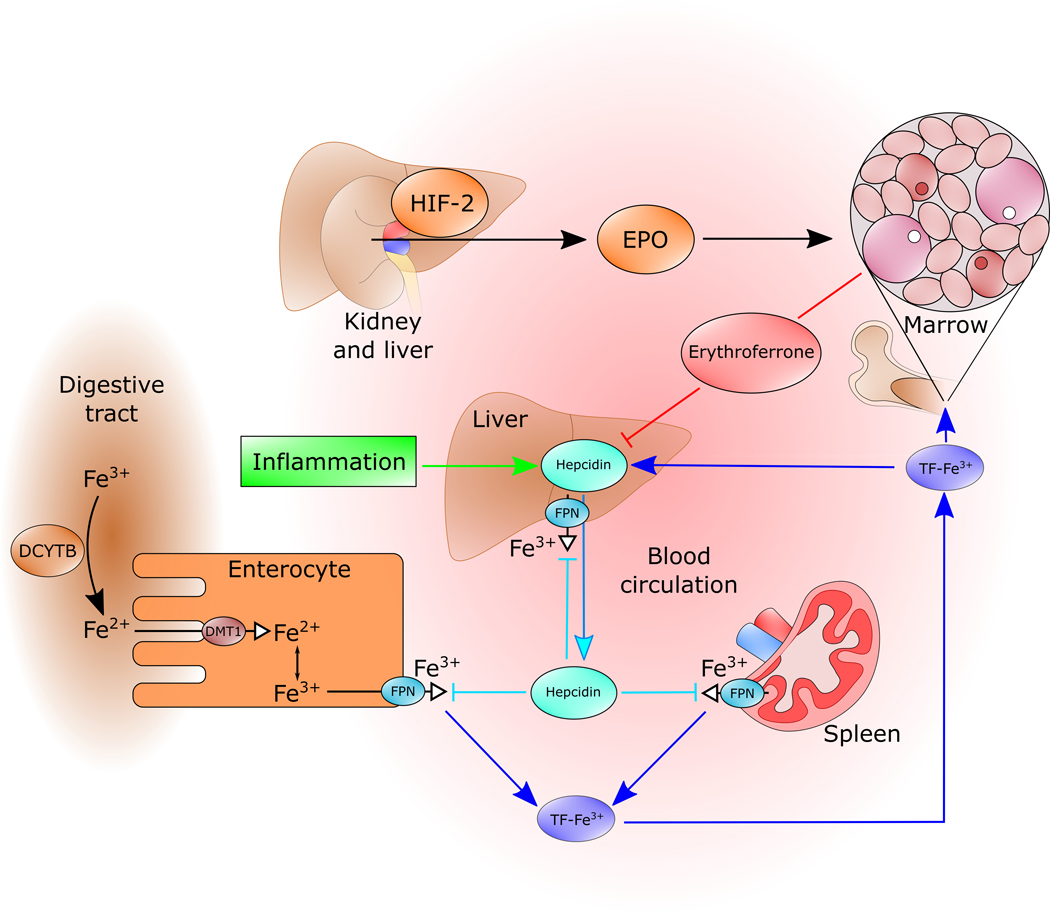
Figure 4. ERFE-Hepcidin-Ferroportin Axis.
Interstitial fibroblasts in the kidney and hepatocytes in the liver respond to cellular hypoxia by HIF-2 induced transcription and production of EPO. EPO stimulates the expansion of erythroblasts as well as increasing production of ERFE. ERFE inhibits the transcription of hepatic hepcidin thereby stabilizing the cellular iron exporter ferroportin on the surfaces of hepatocytes and macrophages. Cellular iron stores are then mobilized into plasma to be used for hemoglobin synthesis by the expanded population of maturing erythrocytes. EPO, erythropoietin; ERFE, erythroferrone; HIF-2, hypoxia-inducible factor 2.
In unpublished data, our lab has developed mice that over-express ERFE under control of the β-globin promoter, restricting the secretion of ERFE to erythroid cells. Highly over-expressing strains—with ERFE mRNA levels that are 10-fold higher than WT—have suppressed hepcidin levels and thereby develop pathological iron overload. Interestingly, these animals manifest lower body weights and a number of other systemic abnormalities, some of which are also seen in human anemias with ineffective erythropoiesis. We hypothesize that these effects are mediated by the interference of ERFE with BMP signaling during development and growth.
3. 3. The Pathological Role of ERFE in Diseases of Altered Erythropoiesis
The inherited blood disorder β-thalassemia results from mutations in β-globin genes that limit the production of β-globin relative to α-globin, causing an imbalance that decreases erythroblast survival in the marrow and erythrocyte lifespan in blood circulation. Ineffective erythropoiesis is characteristic of β-thalassemia because the erythroblast population becomes greatly expanded but generates relatively few mature erythrocytes. In the most severe cases known as β-thalassemia major, patients require regular blood transfusions in order to survive or to prevent severe systemic complications. Each transfused unit of packed erythrocytes delivers the equivalent of 200–250 mg of iron. With transfusions needed every two to five weeks and physiological iron losses limited to a few mg/day, total body iron will quickly exceed the normal adult endowment of 3–4 g by many-fold. Iron chelation is therefore necessary to prevent the overload that results from transfusion. In the less severe form termed β-thalassemia intermedia, red blood cell production is usually sufficient to avoid regular transfusions, but ineffective erythropoiesis may be as or more severe. This is the case because transfusion therapy for thalassemia major increases oxygenation by increasing the number of functional red blood cells. This lowers erythropoietin production by the kidneys and relieves the stimulus for erythropoiesis (Figure 1C).
Several mouse models of anemias—induced genetically, by administration of a hemolytic agent or through dietary iron deprivation—show increased ERFE expression relative to control littermates (Mirciov et al. 2017). Mouse models of anemias with ineffective erythropoiesis would be expected to show particularly high ERFE levels, because in these anemias the erythroblast population, which secretes ERFE, is expanded out of proportion to the severity of anemia. One such model, the Hbbth3/+ mouse model recapitulates a non-transfusion dependent β-thalassemia with moderately decreased hemoglobin concentrations, markedly lower hepcidin during growth compared to WT, and increased plasma and hepatic iron concentrations. The model is haploinsufficient for β-globin leading to the precipitation of excess unpaired α-globin chains. ERFE production in the bone marrow and spleens of these mice is increased relative to WT consistently throughout their lives, between 8-fold and 32-fold depending on age (Kautz et al. 2015). The beneficial effect of anti-ERFE-neutralizing antibodies on anemia in this model suggests that excess ERFE contributes not only to iron overload (Kautz et al. 2015) but also to the pathogenesis of anemia in this model (Arezes et al. 2020). Importantly, reticulocyte percentage is decreased with antibody treatment, indicative of increased quality and lifespan of erythrocytes. It remains to be seen if ERFE inhibition for the treatment of human β-thalassemia can decrease iron absorption and iron deposition in organs sufficiently to avoid chelation therapies—the current standard of care.
Myelodysplastic syndromes (MDS) represent another class of hematopoietic disorders where ERFE pathology could be therapeutically targeted. MDS is a group of malignancies characterized by impairment in the differentiation of blood cell precursors. Many of these undifferentiated cells undergo apoptosis in the bone marrow, but mutation and clonal expansion of surviving abnormal cells eventually selects for more malignant clones that progress to leukemia. Patients with the ring sideroblasts subtype (MDS-RS) develop nucleated erythroblasts containing excess iron that accumulates in mitochondria. These cells fail to mature or enter circulation. MDS is accompanied by mutations in splicing factor genes in 40–80% of patients, particularly in those with MDS-RS (Rozovski, Keating, and Estrov 2013). Mutations in the splicing factor SF3B1 are especially relevant, as patients with MDS-RS and SF3B1 mutations present with systemic iron overload even in the absence of blood transfusions. Bone marrow transcriptome analysis in a cohort of human MDS subjects revealed a mutated ERFE transcript in SF3B1MUT cells attributable to dysregulated splicing. The mutation adds four amino acids to the translated protein, and the final product is still able to suppress hepcidin with efficacy comparable to the WT variant. Furthermore, ERFE mRNA abundance was significantly higher in these mutated cells relative to the SF3B1WT cells that were profiled (Bondu et al. 2019). Importantly, increased expression of this mutant ERFE was associated with longer survival of SF3B1MUT MDS patients. Since ERFE expression is induced in erythroblasts, it may be useful for these patients as an indicator that the abnormal clones are capable of differentiation and therefore less likely to become frankly malignant.
Chronic kidney disease (CKD) is associated with a characteristic anemia caused by a combination of relative erythropoietin deficiency and iron-restriction (Hanudel et al. 2017) (Figure 1D). The pathogenesis of anemia in CKD is multifactorial, including systemic inflammation, relative deficiency of EPO production, and low clearance of hepcidin by the diseased kidney. Current treatment guidelines suggest the monitoring of erythrocyte and iron parameters and the administration of iron and EPO to patients who become iron-deficient and anemic (Anemia Work Group 2012). A study of hepcidin knockout mice with an adenine diet-induced CKD showed that ablation of hepcidin improved the associated anemias (Hanudel et al. 2017). For those CKD patients whose anemia is mainly caused by increased hepcidin, future ERFE mimetics and activators could prove useful for the treatment of anemia. While the EPO administration currently used as therapy does induce endogenous ERFE, a mouse study revealed that serum ERFE levels have a delayed response in CKD mice relative to WT (Hanudel et al. 2018). The resulting delay in iron mobilization could make EPO less effective as an erythropoiesis-stimulating agent, raising the possibility that administration of bioengineered ERFE may have therapeutic utility in helping to mobilize iron in anemia of CKD.
3.4. Erythroferrone Variants
Another consideration of ERFE pathophysiology is the effect of excessive concentrations of the protein on iron-loading anemias. ERFE variants could change the bioactivity of the protein or modify the stability of mRNA transcripts or proteins, or a combination of all three. One example is the A260S substitution caused by a point mutation in the C1q domain. Healthy people with this mutation express higher levels of ERFE at the RNA and protein levels compared to their WT equivalents. The same increased expression is seen in patients with congenital dyserythropoietic anemia type II (CDAII) who also have the ERFE mutation (Andolfo et al. 2019). These patients have very high circulating ERFE protein levels at baseline because of ineffective erythropoiesis. The ERFEA260S mutation found in only a subset of CDAII patients drives RNA and protein expression up even higher. Remarkably, ERFEA260S is associated with severe anemia (Andolfo et al. 2019), possibly by making more iron available for erythropoiesis and increasing oxidative stress in the already impaired erythroblasts.
4. Erythroferrone as a Biomarker in Competitive Sports
4.1. Blood Doping
Monitoring the use of performance-enhancing drugs is a cat-and-mouse game between hypercompetitive athletes who try to skirt the rules and the organizations responsible for upholding the integrity of their sports competitions. Historically, anabolic steroids were commonly used to boost performance, but these drugs can be readily detected by the presence their metabolites in urine. Blood doping is the use of techniques or substances that increases the number of circulating red blood cells. The boosted oxygen carrying capacity from blood doping brings increased stamina and performance, and the artificial enhancement is more difficult to detect than steroids. Two common blood doping methods are erythrocyte transfusions and the administration of erythropoiesis-stimulating agents (ESAs) such as EPO. Unlike steroids, enhanced red cell mass may be unsurprising—especially in athletes—and challenging to detect. In particular, the transfusion of blood shortly before a competition, especially if it is the athlete’s own blood, is difficult to detect. The World Anti-Doping Agency has therefore recognized the need to monitor athletes’ biological parameters over time to reveal the time-dependent alterations caused by doping rather than testing for the agents, substances, or methods themselves. They refer to these profiles as Athlete Biological Passports, and they are currently the most effective deterrent to the use of blood doping (Schumacher et al. 2012).
Hematological parameters that have been proposed for the detection of blood doping include reticulocyte percentage, hemoglobin concentration, plasma iron, ferritin, and hepcidin. A study in healthy, male athletes showed a positive correlation between hepcidin, hemoglobin, iron, and ferritin (Leuenberger et al. 2017). The same study went on to measure the direct effects of multiple recombinant EPO injections on these hematological values in healthy, male volunteers finding the expected increase in reticulocyte percentage and decrease in hepcidin. The drop in hepcidin came more quickly than the rise in reticulocytes for most participants: 2 days versus 4 days after the beginning of treatment. Hepcidin is a useful biomarker in this context because of its rapid and dramatic response to erythropoietic stimuli. It is important to note that hepcidin is also negatively regulated by plasma iron concentrations and positively regulated by inflammation (Nemeth et al. 2003) (Figure 4). Since intense exercise has the potential to induce both inflammation and erythropoiesis, this could lead to the generation of both positive and negative signals for hepcidin transcription. A review of the hepcidin response to exercise found an observed increase in serum levels in a vast majority of studies, largely regardless of exercise duration or intensity (Domínguez et al. 2018). This indicates that elevation of hepcidin by inflammation could mask the expected suppression of hepcidin by erythropoietic stimulants.
While hepcidin is regulated by multiple distinct pathways, marrow-derived ERFE is only known to be responsive to erythropoietic stimulation by EPO in the context of iron regulation. (For contexts where ERFE is produced in other tissues in response to non-erythropoietic stimuli, see the sections on myonectin and Xenopus.) Strict EPO regulation creates the potential for ERFE to serve as a blood doping biomarker with as much sensitivity as hepcidin and fewer confounders. As discussed in the myonectin section below, the stimuli for ERFE in non-erythropoietic contexts are not fully understood, but the contributions of nonerythroid sources to the systemic ERFE concentrations appear to be small. The following studies are part of a new and important effort to characterize the usefulness of ERFE in the detection of blood doping.
A small study in healthy males compared the effects of several EPO micro-doses (20 IU/kg) and small doses (50 IU/kg) on hepcidin and ERFE (Robach et al. 2020). As expected, serum hepcidin levels were quickly reduced in both dosing groups. Serum ERFE levels rose in response to the EPO injections, and unlike hepcidin, the response was dose-dependent rather than all-or-nothing. Another study compared the effects of EPO and two other common ESAs on serum ERFE levels. All three agents significantly increased ERFE levels starting from one day after administration (Ramirez Cuevas et al. 2020). The associated increase from a single dose was sustained for 6–13 days, with the duration depending on the ESA used. Iron supplementation and exercise-induced muscle damage did not affect the ERFE readings. If these findings hold true in larger studies, a sensitive and standardized human ERFE assay could be a useful addition to an athlete’s hematological profile.
4.2. Altitude Training
Altitude training is a method of inducing erythropoiesis without the use of exogenous substances. Living at high altitude exposes the body to relative hypoxia which induces EPO production and increases erythropoiesis. The increased red cell mass and oxygen carrying capacity become advantageous when the athlete returns to sea level for training and competitions. Studies in athletes and healthy adults have shown that repetitive altitude training, or the use of normobaric hypoxia chambers to simulate the low-oxygen effects of high altitude, results in elevated EPO production (Frese and Friedmann-Bette 2010; Mackenzie, Watt, and Maxwell 2008), and an estimated 3.3% increase in hemoglobin mass after 20 days of altitude training compared to baseline (Gore et al. 2013). Normobaric hypoxia caused a decrease in plasma hepcidin within 2 days that lasted 2 weeks (Govus et al. 2017), and 15 hours of high altitude exposure was enough to increase serum ERFE concentrations (Robach et al. 2020). Altitude training is considered a legal form of blood doping, and as such, the practice is widely used in preparation for competitions. It remains to be determined whether ERFE measurement would enhance existing methodologies that distinguish between legal and illegal blood doping.
5. Erythroferrone beyond erythropoiesis and iron homeostasis
5.1. Myonectin
In 2012, a novel skeletal muscle-derived member of the CTRP family was characterized in mice and named myonectin (Seldin et al. 2012:15). Circulating levels of this new myokine were increased after exercise, and transcription of its RNA in cultured myotubes was induced by a rise in cellular cAMP or calcium levels. Based on these results, the authors proposed that the protein may be a marker or regulator of metabolism in muscle. It was later found to be expressed from the same gene and transcript as ERFE, but for the purposes of its metabolic roles and expression in muscle tissues, the name myonectin will continue to be used here. As part of their characterization of myonectin, the authors described various ways the protein could be induced in mice including fasting followed by refeeding with normal chow, glucose, or emulsified lipids. C2C12 myotubes cultured in vitro with glucose or palmitate also increased expression of myonectin 4-fold compared to serum-starved conditions. Injection of recombinant myonectin in mice caused a reduction in non-esterified fatty acid levels that was believed to be the result of increased fatty acid uptake into cells. Isolated adipocytes and hepatocytes cultured with recombinant myonectin showed increased expression of a number of fatty acid binding and transport proteins as well as dose-dependent augmented palmitate uptake. After the initial characterization, several groups investigated the new myokine in various contexts.
5.1.1. Myonectin in glucose and fat metabolism
Whole body myonectin deletion in mice fed a high-fat diet led to increased fat storage in adipose lipid droplets (Little et al. 2019). This suggests the possibility that myonectin may be protective against insulin resistance. In order to test for associations in relevant populations, a study of circulating myonectin levels was performed in humans with type 2 diabetes and healthy control subjects (Li et al. 2019). Both groups were further stratified into lean, overweight, and obese subcategories. Serum myonectin was found to be significantly decreased in diabetes patients. Within both healthy and diabetic groups, increasing BMI was associated with lower levels of myonectin. Moreover, patients with higher BMI had lower circulating levels regardless of disease state, but the effect sizes in both analyses were small. A multivariate stepwise regression analysis of multiple parameters confirmed that high BMI was a main independent predictor of low myonectin in their study (Li et al. 2019). A similar study comparing type 2 diabetics to people with impaired and normal glucose tolerance found the exact opposite: metabolic disease parameters and obesity associated with higher circulating myonectin levels (Li et al. 2018). It should be noted that measurements of circulating myonectin in these studies differed by a factor of 10 despite using the same ELISA assay system for detection. Authors of both studies agreed on myonectin’s potential as a biomarker in predicting the development of obesity and type 2 diabetes, but the questions of cause, effect, and directionality remain open. Adding to the uncertainty, a calorie restriction experiment in rats showed no change in plasma myonectin levels between animals fed ad libitum and those restricted to 60% of standard. This is despite a drop in plasma levels of insulin and a rise in adiponectin in the calorie restricted group (Sharma, Castorena, and Cartee 2012). It is possible that myonectin is affected only during extreme stresses, whereas insulin sensitivity and other modes of fat regulation respond in a more sensitive manner, and possibly more consistently. As with all ERFE/myonectin detection systems, it is important to validate the assays using appropriate controls before the results can be reliably interpreted.
Experiments in vitro and in vivo were reported as showing that myonectin suppressed starvation-induced autophagy. Administration of recombinant myonectin to fasted animals and serum-starved hepatocytes reduced the expression of autophagy genes and prevented formation of autophagosomes (Seldin et al. 2013). Mechanistic studies using various pathway inhibitors revealed that this activity is accomplished through mTOR/Akt/PI3K. In vitro time course experiments in rat cardiomyocytes confirmed these findings by demonstrating an induction of pAkt, pCREB, and pGSK-3β within 15 minutes of treatment with recombinant myonectin (Otaka et al. 2018), though no cell surface receptors have been described to mediate this signaling. It was not reported if or how the recombinant myonectin protein was validated for their experiments.
In addition to refeeding, myonectin can be induced by exercise. A study in rats confirmed the original reports that myonectin protein levels increase in muscle after chronic exercise. Confusingly, RNA abundance of the myonectin transcript in diaphragm muscle was lower in the exercised group than in the control. This suppression of RNA but not protein was observed in both lean rats and in a genetic model of obesity where caloric intake from a normal diet is increased due to knockout of the leptin receptor (Peterson, Mart, and Bond 2014). A study in overweight human subjects found that chronic exercise significantly increased serum myonectin levels (Pourranjbar et al. 2018), but these results can be difficult to interpret. Since exercise can induce erythropoiesis, the change they observed may have been circulating ERFE that originated in the bone marrow. Pairing this with the contradictory RNA data highlights the need for a more complete analysis of the metabolic landscape. This analysis must include the effects on renal EPO production and marrow ERFE secretion in order to document the predominant tissue sources of ERFE/myonectin and the specific pathways that stimulate the increase in ERFE/myonectin plasma concentrations. More extensive assay validation is also needed to assure that the measurements are sufficiently sensitive and specific, and the consistent use of knockout animals as controls is required for appropriate attribution of myonectin effects.
Just upstream of ERFE/myonectin induction, erythropoietin has also been implicated in the regulation of cellular metabolism including glucose tolerance and fatty acid metabolism. Knockout of the mouse EPO receptor in all non-hematopoietic tissues resulted in increased body mass compared to WT animals with differences appearing as early as one week after birth (Teng et al. 2011). Transgenic mice that over-express EPO, or WT mice that are administered exogenous EPO, show a reduction in body mass and increase in glucose tolerance compared to untreated WT controls. Notably, these changes are not mediated by an EPO-related increase in erythropoiesis (Foskett et al. 2011; Teng et al. 2011). Indeed, experiments in erythroferrone knockout mice demonstrated that a lack of the ERFE protein did not impair either glucose tolerance or clearance of non-esterified fatty acids after chronic or acute EPO treatment (Coffey et al. 2018). Taken together, these results demonstrate that the effects of erythropoietin on cellular metabolism are independent of its role in stimulating erythropoiesis.
5.1.2. Myonectin as a cardioprotective hormone
A comprehensive study on cardiac ischemia-reperfusion injury (IRI) in myonectin knockout and over-expressing mice found that myonectin was protective of heart muscle subjected to IRI, with knockouts experiencing more apoptosis in the heart than WT, and over-expressers suffering less cardiac damage. Specifically, pro-inflammatory gene expression levels and myocardial infarct size were both increased in myonectin KO mice subjected to IRI. Over-expression of myonectin in skeletal muscle resulted in reduced cardiac injury after IRI via an attenuation of cardiomyocyte apoptosis (Otaka et al. 2018). The mechanisms of cardio-protection seen in these experiments are unclear. It is also unknown how the various reported effects of myonectin—ranging from metabolic modulation in the contexts of feeding and exercise to this report of apoptosis mitigation in the heart—fit together in the big picture. Further complicating the matter are the known effects of ERFE in the erythroid system and its mechanism of action. While a BMP trapping mechanism is proving very important for hepcidin suppression by ERFE, its relevance is still unclear in the context of myonectin. Little progress has been made regarding research into the mechanism(s) of action of myonectin.
5.2. Erythroferrone in Xenopus development
Erythroferrone ortholog genes are present in all vertebrates. In a Xenopus tropicalis cDNA library developmental gain-of-function screen, transgenic erfe over-expression caused axis duplication in the embryo (Melchert et al. 2020). This effect is known in other settings to be induced by either stimulation of Wnt/β-Catenin or inhibition of BMP signaling. Consistent with mammalian data, RT-qPCR analysis showed Xenopus erfe to be a potent inhibitor of BMP target genes. As in mammals, the active domain was found to be the N-terminal region and not the C1q globular head. Importantly, Xenopus erfe inhibited BMP4 signaling while murine ERFE did not. Consistent with this finding, injection of murine ERFE RNA did not cause secondary axis duplication unlike the Xenopus version, however over-expression of the murine variant did cause a mild dorsalization phenotype. The difference may be attributable to murine ERFE’s relatively weaker binding to BMP4 than to BMP2 or BMP6 (Arezes et al. 2020), as primary axis formation during Xenopus development is mainly controlled by BMP4 (De Robertis 2009; De Robertis and Kuroda 2004). These data raise two possibilities for why our ERFE transgenic mice are less affected during development than the Xenopus embryos. One explanation is that divergent evolution in erythroferrone proteins leading to differential BMP4 binding may explain the difference in developmental abnormalities. Alternatively, overexpression of ERFE in our transgenic ERFE is driven by the β-globin promoter-enhancer, so Erfe synthesis in the mouse model begins in mid-gestation when β-globin synthesis starts in the mouse, perhaps too late for more dramatic developmental defects to appear. Xenopus erfe loss-of-function resulted in severely impaired blood circulation and edema at the tadpole stage. Together, these data demonstrate that erfe activity is required for normal morphological development and vascularization in Xenopus, raising the possibility that Erfe may have additional as of yet undescribed roles in mammals.
Conclusion
Since its discovery in 2014, erythroferrone has become recognized as a key erythroid regulator of iron homeostasis. In response to erythropoietic stimuli, increased ERFE levels suppress hepcidin production, and the consequent stabilization of ferroportin mobilizes cellular iron stores into plasma for use by maturing erythroblasts. This important regulatory axis allows vertebrate species to respond to and rapidly recover from blood loss. It also allows for quick adaptation to low oxygen concentrations found at high altitudes—a physiological feature that some athletes use to gain advantages when competing at sea level. The potential of ERFE to serve as a biomarker of blood doping could have a beneficial impact on the fairness and safety of sports competitions. In hematological diseases with ineffective erythropoiesis like β-thalassemia and some subclasses of myelodysplastic syndrome, dysregulated ERFE leads to iron overload. Iron chelation is currently used as the standard-of-care therapy, but recent studies of anti-ERFE antibodies show potential utility in preventing iron overload in these disorders. Patients with chronic kidney disease have the opposite problem: high circulating levels of hepcidin and iron-restricted anemia. In these cases, the therapeutic administration of ERFE or ERFE mimetics could become a useful modality in the management of anemia. Despite the rapid progress made in understanding the role of erythroferrone in baseline, stress, and disease-state erythropoiesis, there is much about the hormone that remains to be explained. We are only beginning to understand how ERFE suppresses hepcidin, with the best-supported mechanism dependent on the ability of ERFE to trap BMPs and prevent their interactions with the BMP receptor complex that regulates hepcidin transcription. Furthermore, the effects of multimerization on the regulation and potency of ERFE remain unknown. Much remains to be discovered about the role of erythroferrone in embryonic development and its roles in muscle biology and systemic metabolism. Discoveries in these areas may have important implications for understanding the entire CTRP family whose biological functions remain largely unknown.
Acknowledgments
The authors thank Elizabeta Nemeth for reviewing the manuscript and acknowledge Richard Coffey for sharing pre-publication data on ERFE transgenic mice. Funding for work related to this article was provided by NIH R01DK126680 (TG).
Footnotes
Conflict of Interest Statement
TG is a scientific co-founder of Intrinsic LifeSciences and Silarus Pharma and has consulted for ADARx, Akebia, Pharmacosmos, Ionis, Gossamer Bio, Global Blood Therapeutics, American Regent, Disc Medicine, and Rockwell Scientific. TG is also listed as an inventor on patents related to erythroferrone. DNS declares no conflicts.
References
- Andolfo Immacolata, Barbara Eleni Rosato Roberta Marra, Gianluca De Rosa Francesco Manna, Gambale Antonella, Iolascon Achille, and Russo Roberta. 2019. “The BMP-SMAD Pathway Mediates the Impaired Hepatic Iron Metabolism Associated with the ERFE-A260S Variant.” American Journal of Hematology 94(11):1227–35. doi: 10.1002/ajh.25613. [DOI] [PubMed] [Google Scholar]
- Anemia Work Group. 2012. “KDIGO Clinical Practice Guideline for Anemia in Chronic Kidney Disease.” Kidney International Supplements 2(4):279–335. doi: 10.1038/kisup.2012.40. [DOI] [Google Scholar]
- Arezes João, Foy Niall, Kirsty McHugh Doris Quinkert, Benard Susan, Sawant Anagha, Frost Joe N., Armitage Andrew E., Pasricha Sant-Rayn, Pei Jin Lim May S. Tam, Lavallie Edward, Pittman Debra D., Cunningham Orla, Lambert Matthew, Murphy John E., Draper Simon J., Jasuja Reema, and Drakesmith Hal. 2020. “Antibodies against the Erythroferrone N-Terminal Domain Prevent Hepcidin Suppression and Ameliorate Murine Thalassemia.” Blood 135(8):547–57. doi: 10.1182/blood.2019003140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezes João, Foy Niall, Kirsty McHugh Anagha Sawant, Quinkert Doris, Terraube Virginie, Brinth Alette, Tam May, LaVallie Edward R., Taylor Stephen, Armitage Andrew E., Sant-Rayn Pasricha, Cunningham Orla, Lambert Matthew, Draper Simon J., Jasuja Reema, and Drakesmith Hal. 2018. “Erythroferrone Inhibits the Induction of Hepcidin by BMP6.” Blood 132(14):1473–77. doi: 10.1182/blood-2018-06-857995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschemeyer Sharraya, Gabayan Victoria, Ganz Tomas, Nemeth Elizabeta, and Kautz Léon. 2017. “Erythroferrone and Matriptase-2 Independently Regulate Hepcidin Expression.” American Journal of Hematology 92(5):E61–63. doi: 10.1002/ajh.24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom Nikolaj, Thomas Sicheritz-Pontén Ramneek Gupta, Gammeltoft Steen, and Brunak Søren. 2004. “Prediction of Post-Translational Glycosylation and Phosphorylation of Proteins from the Amino Acid Sequence.” PROTEOMICS 4(6):1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bondu Sabrina, Alary Anne-Sophie, Carine Lefèvre Alexandre Houy, Jung Grace, Lefebvre Thibaud, Rombaut David, Boussaid Ismael, Bousta Abderrahmane, Guillonneau François, Perrier Prunelle, Alsafadi Samar, Wassef Michel, Margueron Raphaël, Rousseau Alice, Droin Nathalie, Cagnard Nicolas, Kaltenbach Sophie, Winter Susann, Kubasch Anne-Sophie, Bouscary Didier, Santini Valeria, Toma Andrea, Hunault Mathilde, Stamatoullas Aspasia, Gyan Emmanuel, Cluzeau Thomas, Platzbecker Uwe, Lionel Adès Hervé Puy, Stern Marc-Henri, Karim Zoubida, Mayeux Patrick, Nemeth Elizabeta, Park Sophie, Ganz Tomas, Kautz Léon, Kosmider Olivier, and Fontenay Michaëla. 2019. “A Variant Erythroferrone Disrupts Iron Homeostasis in SF3B1 -Mutated Myelodysplastic Syndrome.” Science Translational Medicine 11(500):eaav5467. doi: 10.1126/scitranslmed.aav5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey Richard, Sardo Ugo, Kautz Léon, Gabayan Victoria, Nemeth Elizabeta, and Ganz Tomas. 2018. “Erythroferrone Is Not Required for the Glucoregulatory and Hematologic Effects of Chronic Erythropoietin Treatment in Mice.” Physiological Reports 6(19):e13890. doi: 10.14814/phy2.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM 2009. “Spemann’s Organizer and the Self-Regulation of Embryonic Fields.” Mechanisms of Development 126(11–12):925–41. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertis De, Edward M, and Hiroki Kuroda. 2004. “DORSAL-VENTRAL PATTERNING AND NEURAL INDUCTION IN XENOPUS EMBRYOS.” Annual Review of Cell and Developmental Biology 20(1):285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez Raúl, Antonio Sánchez-Oliver Fernando Mata-Ordoñez, Adrián Feria-Madueño Moisés Grimaldi-Puyana, López-Samanes Álvaro, and Pérez-López Alberto. 2018. “Effects of an Acute Exercise Bout on Serum Hepcidin Levels.” Nutrients 10(2):209. doi: 10.3390/nu10020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foskett Amanda, Alnaeeli Mawadda, Wang Li, Teng Ruifeng, and Noguchi Constance T.. 2011. “The Effects of Erythropoietin Dose Titration during High-Fat Diet-Induced Obesity.” Journal of Biomedicine and Biotechnology 2011:1–8. doi: 10.1155/2011/373781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese F, and Friedmann-Bette B. 2010. “Effects of Repetitive Training at Low Altitude on Erythropoiesis in 400 and 800 m Runners.” International Journal of Sports Medicine 31(06):382–88. doi: 10.1055/s-0030-1248328. [DOI] [PubMed] [Google Scholar]
- Gammella Elena, Diaz Victor, Recalcati Stefania, Buratti Paolo, Samaja Michele, Dey Soumyadeep, Constance Tom Noguchi Max Gassmann, and Cairo Gaetano. 2015. “Erythropoietin’s Inhibiting Impact on Hepcidin Expression Occurs Indirectly.” American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 308(4):R330–35. doi: 10.1152/ajpregu.00410.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz Tomas, Jung Grace, Naeim Arash, Ginzburg Yelena, Pakbaz Zahra, Walter Patrick B., Kautz Léon, and Nemeth Elizabeta. 2017. “Immunoassay for Human Serum Erythroferrone.” Blood 130(10):1243–46. doi: 10.1182/blood-2017-04-777987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore Christopher J., Sharpe Ken, Garvican-Lewis Laura A., Saunders Philo U., Humberstone Clare E., Robertson Eileen Y., Wachsmuth Nadine B., Clark Sally A., McLean Blake D., Friedmann-Bette Birgit, Neya Mitsuo, Pottgiesser Torben, Schumacher Yorck O., and Schmidt Walter F.. 2013. “Altitude Training and Haemoglobin Mass from the Optimised Carbon Monoxide Rebreathing Method Determined by a Meta-Analysis.” British Journal of Sports Medicine 47(Suppl 1):i31–39. doi: 10.1136/bjsports-2013-092840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govus AD, Peeling P, Abbiss CR, Lawler NG, Swinkels DW, Laarakkers CM, Thompson KG, Peiffer JJ, Gore CJ, and Garvican-Lewis LA. 2017. “Live High, Train Low - Influence on Resting and Post-Exercise Hepcidin Levels.” Scandinavian Journal of Medicine & Science in Sports 27(7):704–13. doi: 10.1111/sms.12685. [DOI] [PubMed] [Google Scholar]
- Gutschow Patrick, Han Huiling, Olbina Gordana, Westerman Keith, Westerman Eileen, Marc Ruiz Martinez Yelena Ginzburg, Nemeth Elizabeta, Ganz Tomas, and Ostland Vaughn. 2019. “A Novel Sandwich ELISA to Quantify Erythroferrone in Mouse Serum.” Blood 134(Supplement_1):2237–2237. doi: 10.1182/blood-2019-130947. [DOI] [Google Scholar]
- Haase Volker H. 2013. “Regulation of Erythropoiesis by Hypoxia-Inducible Factors.” Blood Reviews 27(1):41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanudel Mark R., Rappaport Maxime, Chua Kristine, Gabayan Victoria, Qiao Bo, Jung Grace, Salusky Isidro B., Ganz Tomas, and Nemeth Elizabeta. 2018. “Levels of the Erythropoietin-Responsive Hormone Erythroferrone in Mice and Humans with Chronic Kidney Disease.” Haematologica 103(4):e141–42. doi: 10.3324/haematol.2017.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanudel Mark R., Rappaport Maxime, Gabayan Victoria, Jung Grace, Salusky Isidro B., Nemeth Elizabeta, Ganz Tomas, and Zaritsky Joshua. 2017. “Increased Serum Hepcidin Contributes to the Anemia of Chronic Kidney Disease in a Murine Model.” Haematologica 102(3):e85–88. doi: 10.3324/haematol.2016.150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges Vivien M., Rainey Susan, Lappin Terence R., and Peter Maxwell A. 2007. “Pathophysiology of Anemia and Erythrocytosis.” Critical Reviews in Oncology/Hematology 64(2):139–58. doi: 10.1016/j.critrevonc.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Kautz Léon, Jung Grace, Du Xin, Gabayan Victoria, Chapman Justin, Nasoff Marc, Nemeth Elizabeta, and Ganz Tomas. 2015. “Erythroferrone Contributes to Hepcidin Suppression and Iron Overload in a Mouse Model of β-Thalassemia.” Blood 126(17):2031–37. doi: 10.1182/blood-2015-07-658419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautz Léon, Jung Grace, Valore Erika V., Rivella Stefano, Nemeth Elizabeta, and Ganz Tomas. 2014. “Identification of Erythroferrone as an Erythroid Regulator of Iron Metabolism.” Nature Genetics 46(7):678–84. doi: 10.1038/ng.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Airie, and Nemeth Elizabeta. 2015. “New Insights into Iron Regulation and Erythropoiesis:” Current Opinion in Hematology 22(3):199–205. doi: 10.1097/MOH.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger Nicolas, Bulla Emanuele, Salamin Olivier, Nicoli Raul, Robinson Neil, Baume Norbert, and Saugy Martial. 2017. “Hepcidin as a Potential Biomarker for Blood Doping: Hepcidin and Blood Doping.” Drug Testing and Analysis 9(7):1093–97. doi: 10.1002/dta.2122. [DOI] [PubMed] [Google Scholar]
- Li Kejia, Liao Xin, Wang Kuan, Mi Qiao, Zhang Tingran, Jia Yanjun, Xu Xiaohuei, Luo Xiaoheu, Zhang Cheng, Liu Hua, Zhen Hongting, Li Ling, and Yang Gangyi. 2018. “Myonectin Predicts the Development of Type 2 Diabetes.” The Journal of Clinical Endocrinology & Metabolism 103(1):139–47. doi: 10.1210/jc.2017-01604. [DOI] [PubMed] [Google Scholar]
- Li Zhu, Yang Yan-Ling, Zhu Yan-Juan, Li Chen-Guang, Tang Yun-Zhao, Ni Chang-Lin, Chen Li-Ming, and Niu Wen-Yan. 2019. “Circulating Serum Myonectin Levels in Obesity and Type 2 Diabetes Mellitus.” Experimental and Clinical Endocrinology & Diabetes a-0896–8548. doi: 10.1055/a-0896-8548. [DOI] [PubMed] [Google Scholar]
- Little Hannah C., Rodriguez Susana, Lei Xia, Tan Stefanie Y., Stewart Ashley N., Sahagun Ageline, Sarver Dylan C., and G. William Wong. 2019. “Myonectin Deletion Promotes Adipose Fat Storage and Reduces Liver Steatosis.” The FASEB Journal 33(7):8666–87. doi: 10.1096/fj.201900520R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie Richard W. A., Watt Peter W., and Maxwell Neil S.. 2008. “Acute Normobaric Hypoxia Stimulates Erythropoietin Release.” High Altitude Medicine & Biology 9(1):28–37. doi: 10.1089/ham.2007.1043. [DOI] [PubMed] [Google Scholar]
- Melchert Juliane, Henningfeld Kristine A., Richts Sven, Lingner Thomas, Jonigk Danny, and Pieler Tomas. 2020. “The Secreted BMP Antagonist ERFE Is Required for the Development of a Functional Circulatory System in Xenopus.” Developmental Biology 459(2):138–48. doi: 10.1016/j.ydbio.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Mirciov Cornel S. G., Wilkins Sarah J., Dunn Linda A., Anderson Gregory J., and Frazer David M.. 2017. “Characterization of Putative Erythroid Regulators of Hepcidin in Mouse Models of Anemia” edited by K. Pantopoulos. PLOS ONE 12(1):e0171054. doi: 10.1371/journal.pone.0171054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirciov Cornel S. G., Wilkins Sarah J., Hung Grace C. C., Helman Sheridan L., Anderson Gregory J., and Frazer David M.. 2018. “Circulating Iron Levels Influence the Regulation of Hepcidin Following Stimulated Erythropoiesis.” Haematologica 103(10):1616–26. doi: 10.3324/haematol.2017.187245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E. 2004. “Hepcidin Regulates Cellular Iron Efflux by Binding to Ferroportin and Inducing Its Internalization.” Science 306(5704):2090–93. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Nemeth Elizabeta, Valore Erika V., Territo Mary, Schiller Gary, Lichtenstein Alan, and Ganz Tomas. 2003. “Hepcidin, a Putative Mediator of Anemia of Inflammation, Is a Type II Acute-Phase Protein.” Blood 101(7):2461–63. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- Otaka Naoya, Shibata Rei, Ohashi Koji, Uemura Yusuke, Kambara Takahiro, Enomoto Takashi, Ogawa Hayato, Ito Masanori, Kawanishi Hiroshi, Maruyama Sonomi, Joki Yusuke, Fujikawa Yusuke, Narita Shingo, Unno Kazumasa, Kawamoto Yoshiyuki, Murate Takashi, Murohara Toyoaki, and Ouchi Noriyuki. 2018. “Myonectin Is an Exercise-Induced Myokine That Protects the Heart From Ischemia-Reperfusion Injury.” Circulation Research 123(12):1326–38. doi: 10.1161/CIRCRESAHA.118.313777. [DOI] [PubMed] [Google Scholar]
- Pak Mihwa, Lopez Miguel A., Gabayan Victroia, Ganz Tomas, and Rivera Seth. 2006. “Suppression of Hepcidin during Anemia Requires Erythropoietic Activity.” Blood 108(12):3730–35. doi: 10.1182/blood-2006-06-028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson Jonathan M., Mart Ryan, and Bond Cherie E.. 2014. “Effect of Obesity and Exercise on the Expression of the Novel Myokines, Myonectin and Fibronectin Type III Domain Containing 5.” PeerJ 2:e605. doi: 10.7717/peerj.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourranjbar Mohammad, Arabnejad Najmeh, Naderipour Khatereh, and Rafie Forouzan. 2018. “Effects of Aerobic Exercises on Serum Levels of Myonectin and Insulin Resistance in Obese and Overweight Women.” Journal of Medicine and Life 11(4):381–86. doi: 10.25122/jml-2018-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas Ramirez, Kelvin Céline Schobinger, Gottardo Emeric, Sven Christian Voss Tiia Kuuranne, Tissot Jean-Daniel, Favrat Bernard, Townsend Nathan, and Leuenberger Nicolas. 2020. “Erythroferrone as a Sensitive Biomarker to Detect Stimulation of Erythropoiesis.” Drug Testing and Analysis 12(2):261–67. doi: 10.1002/dta.2720. [DOI] [PubMed] [Google Scholar]
- Robach Paul, Gammella Elena, Recalcati Stefania, Girelli Domenico, Castagna Annalisa, Roustit Matthieu, Lundby Carsten, Lundby Anne-Kristine, Bouzat Pierre, Samuel Vergès Guillaume Séchaud, Banco Pierluigi, Uhr Mario, Cornu Catherine, Sallet Pierre, and Cairo Gaetano. 2020. “Induction of Erythroferrone in Healthy Humans by Micro-Dose Recombinant Erythropoietin or High-Altitude Exposure.” Haematologica haematol.2019.233874. doi: 10.3324/haematol.2019.233874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovski Uri, Keating Michael, and Estrov Zeev. 2013. “The Significance of Spliceosome Mutations in Chronic Lymphocytic Leukemia.” Leukemia & Lymphoma 54(7):1364–66. doi: 10.3109/10428194.2012.742528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffler Andreas, and Buechler Christa. 2012. “CTRP Family: Linking Immunity to Metabolism.” Trends in Endocrinology & Metabolism 23(4):194–204. doi: 10.1016/j.tem.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Schumacher Yorck Olaf, Saugy Martial, Pottgiesser Torben, and Robinson Neil. 2012. “Detection of EPO Doping and Blood Doping: The Haematological Module of the Athlete Biological Passport: The Haematological Module of the Athlete Biological Passport.” Drug Testing and Analysis 4(11):846–53. doi: 10.1002/dta.406. [DOI] [PubMed] [Google Scholar]
- Seldin Marcus M., Lei Xia, Tan Stefanie Y., Stanson Kevin P., Wei Zhikui, and William Wong G. 2013. “Skeletal Muscle-Derived Myonectin Activates the Mammalian Target of Rapamycin (MTOR) Pathway to Suppress Autophagy in Liver.” Journal of Biological Chemistry 288(50):36073–82. doi: 10.1074/jbc.M113.500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin Marcus M., Peterson Jonathan M., Byerly Mardi S., Wei Zhikui, and William Wong G. 2012. “Myonectin (CTRP15), a Novel Myokine That Links Skeletal Muscle to Systemic Lipid Homeostasis.” Journal of Biological Chemistry 287(15):11968–80. doi: 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin Marcus M., Tan Stefanie Y., and William Wong G. 2014. “Metabolic Function of the CTRP Family of Hormones.” Reviews in Endocrine and Metabolic Disorders 15(2):111–23. doi: 10.1007/s11154-013-9255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Naveen, Castorena Carlos M., and Cartee Gregory D.. 2012. “Greater Insulin Sensitivity in Calorie Restricted Rats Occurs with Unaltered Circulating Levels of Several Important Myokines and Cytokines.” Nutrition & Metabolism 9(1):90. doi: 10.1186/1743-7075-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Ruifeng, Gavrilova Oksana, Suzuki Norio, Chanturiya Tatyana, Schimel Daniel, Hugendubler Lynne, Mammen Selin, Yver Dena R., Cushman Samuel W., Mueller Elisabetta, Yamamoto Masayuki, Hsu Lewis L., and Noguchi Constance Tom. 2011. “Disrupted Erythropoietin Signalling Promotes Obesity and Alters Hypothalamus Proopiomelanocortin Production.” Nature Communications 2(1):520. doi: 10.1038/ncomms1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento Sandro, Cainelli Francesca, and Cesario Francesco. 2006. “Infections and Thalassaemia.” The Lancet Infectious Diseases 6(4):226–33. doi: 10.1016/S1473-3099(06)70437-6. [DOI] [PubMed] [Google Scholar]
- Vokurka M, Krijt J, Sulc K, and Necas E. 2006. “Hepcidin MRNA Levels in Mouse Liver Respond to Inhibition of Erythropoiesis.” Physiological Research 55(6):667–74. [DOI] [PubMed] [Google Scholar]
- Wang Chia-Yu, Core Amanda B., Canali Susanna, Zumbrennen-Bullough Kimberly B., Ozer Sinan, Umans Lieve, Zwijsen An, and Babitt Jodie L.. 2017. “Smad1/5 Is Required for Erythropoietin-Mediated Suppression of Hepcidin in Mice.” Blood 130(1):73–83. doi: 10.1182/blood-2016-12-759423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Chia-Yu, Xu Yang, Traeger Lisa, Dogan Deniz Y., Xiao Xia, Steinbicker Andrea U., and Babitt Jodie L.. 2020. “Erythroferrone Lowers Hepcidin by Sequestering BMP2/6 Heterodimer from Binding to the BMP Type I Receptor ALK3.” Blood 135(6):453–56. doi: 10.1182/blood.2019002620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Yu, Lam Karen S. L., Yau Ming-hon, and Xu Aimin. 2008. “Post-Translational Modifications of Adiponectin: Mechanisms and Functional Implications.” Biochemical Journal 409(3):623–33. doi: 10.1042/BJ20071492. [DOI] [PubMed] [Google Scholar]
- Wong GW, Wang J, Hug C, Tsao TS, and Lodish HF. 2004. “A Family of Acrp30/Adiponectin Structural and Functional Paralogs.” Proceedings of the National Academy of Sciences 101(28):10302–7. doi: 10.1073/pnas.0403760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav Amita, Kataria Megha A., Saini Vandana, and Yadav Anil. 2013. “Role of Leptin and Adiponectin in Insulin Resistance.” Clinica Chimica Acta 417:80–84. doi: 10.1016/j.cca.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Yamauchi Toshimasa, Kamon Junji, Ito Yusuke, Tsuchida Atsushi, Yokomizo Takehiko, Kita Shunbun, Sugiyama Takuya, Miyagishi Makoto, Hara Kazuo, Tsunoda Masaki, Murakami Koji, Ohteki Toshiaki, Uchida Shoko, Takekawa Sato, Waki Hironori, Tsuno Nelson H., Shibata Yoichi, Terauchi Yasuo, Froguel Philippe, Tobe Kazuyuki, Koyasu Shigeo, Taira Kazunari, Kitamura Toshio, Shimizu Takao, Nagai Ryozo, and Kadowaki Takashi. 2003. “Cloning of Adiponectin Receptors That Mediate Antidiabetic Metabolic Effects.” Nature 423(6941):762–69. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]