Abstract
The BAF (mSWI/SNF) chromatin remodeling complex is of importance in development and has been linked to prostate oncogenesis. The oncogenic MUC1-C protein promotes lineage plasticity in the progression of neuroendocrine prostate cancer (NEPC); however, there is no known association between MUC1-C and BAF. We report here that MUC1-C binds directly to the E2F1 transcription factor and that the MUC1-C→E2F1 pathway induces expression of embryonic stem cell esBAF components BRG1, ARID1A, BAF60a, BAF155, and BAF170 in castrate-resistant (CRPC) and NEPC cells. In concert with this previously unrecognized pathway, MUC1 was associated with increased expression of E2F1 and esBAF components in NEPC tumors as compared to CRPC, supporting involvement of MUC1-C in activating the E2F1→esBAF pathway with progression to NEPC. MUC1-C formed a nuclear complex with BAF and activated cancer stem cell (CSC) gene signatures and the core pluripotency factor gene network. The MUC1-C→E2F1→BAF pathway was necessary for induction of both the NOTCH1 effector of CSC function and the NANOG pluripotency factor, and collectively, this network drove CSC self-renewal. These findings indicate that MUC1-C promotes NEPC progression by integrating activation of E2F1 and esBAF with induction of NOTCH1, NANOG, and stemness.
Keywords: MUC1-C, E2F1, BAF, NOTCH1, NANOG, stemness
Introduction
The mammalian Brg/Brahama-associated factor (BAF, mSWI/SNF) complex functions in the remodeling of chromatin in development and oncogenesis (1,2). BAF contributes to the regulation of gene transcription and DNA repair and acts in concert with Polycomb Repressive Complexes (PRCs) for regulating gene expression and cellular identity (3). The canonical BAF (cBAF) complex consists of core subunits that include (i) mutually exclusive SMARCA4/BRG1 or SMARCA2/BRM ATPase subunits that catalyze energy dependent repositioning of nucleosomes (4), (ii) SMARCB1, which is essential for complex formation and enhancer targeting (5), and (iii) the SMARCC1 and SMARCC2 leucine zipper containing proteins. Lineage-specific subunits include the AT-rich interactive domain-containing proteins ARID1A and ARID1B, which maintain BAF complexes, particularly those at enhancers, and regulate pausing of RNA polymerase II (6,7). The functional specificity of the BAF complex is determined by distinct subunit compositions. In this respect, an embryonic stem cell (ESC) specific BAF (esBAF) complex that regulates the ESC transcriptome and pluripotency consists of BRG1, ARID1A, SMARCD1/BAF60a, SMARCC1/BAF155 and SMARCC2/BAF170 (8,9).
The oncogenic MUC1-C protein promotes lineage plasticity in the progression to neuroendocrine prostate cancer (NEPC) (10). MUC1-C drives multiple hallmarks of the cancer cell that include reprogramming of the epigenome (11). MUC1-C activates DNA methyltransferase (DNMT) 1 and DNMT3b, and induces promoter-specific and global increases in DNA methylation patterns (11). MUC1-C also activates PRC1, binds directly to BMI1 and promotes H2A ubiquitylation (12). Moreover, MUC1-C induces expression of PRC2 components, interacts with EZH2 and regulates H3K27 trimethylation at promoters of tumor suppressor genes (TSGs) (13). Other work has shown that MUC1-C activates the nucleosome remodeling and deacetylation complex (NuRD) complex in driving dedifferentiation and induction of the Yamanaka reprogramming factors (10,14,15), supporting a role for MUC1-C in integrating pluripotency and stemness. This involvement of MUC1-C in lineage plasticity is associated with induction (i) the epithelial-mesenchymal transition (EMT), (ii) the cancer stem cell (CSC) state, and (iii) self-renewal capacity and tumorigenicity (11,16).
BAF functions in concert with PRCs in regulating cell fate (3) and has been associated with promoting prostate oncogenesis (17). MUC1-C activates PRC1/2 (11) and drives PC lineage plasticity and progression (10), invoking the possibility that MUC1-C might contribute to BAF regulation. The present results demonstrate that MUC1-C binds directly to the E2F1 transcription factor and that MUC1→E2F1 signaling induces expression of the BRG1, ARID1A, BAF60a, BAF155 and BAF170 embryonic stem cell esBAF components in PC cells. MUC1-C also associates with BAF and drives BAF-dependent induction of the NOTCH1 effector of CSC self-renewal (18), the NANOG pluripotency factor, which is upregulated in CSCs (19), and thereby self-renewal capacity. These findings provide support for involvement of MUC1-C in integrating the activation of E2F1 and esBAF with CSC function.
Materials and Methods
Cell culture.
Human LNCaP-AI cells were grown in phenol red-free RPMI1640 medium (ThermoFisher Scientific, Waltham, MA, USA) containing 10% charcoal-stripped FBS (Millipore Sigma, Burlington, MA, USA) (10). Human DU-145 (ATCC) cells were cultured in RPMI1640 medium (Corning Life Sciences, Corning, NY, USA) containing 10% heat-inactivated FBS (GEMINI Bio-Products, West Sacramento, CA, USA). Human NCI-H660 NEPC cells (ATCC) were cultured in RPMI1640 medium with 5% FBS, 10 nM β-estradiol (Millipore Sigma), 10 nM hydrocortisone, 1% insulin-transferrin-selenium (ThermoFisher Scientific) and 2 mM L-glutamine (ThermoFisher Scientific). Cells were treated with the MUC1-C inhibitor GO-203 (20). Authentication of the cells was performed by short tandem repeat (STR) analysis. Cells were monitored for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Rockland, ME, USA). Studies were performed on cells within 3–4 months of culture.
Tetracycline-inducible and stable vector expression.
MUC1shRNA (MISSION shRNA TRCN0000122938), E2F1shRNA (MISSION shRNA TRCN0000010328), E2F1shRNA#2 (MISSION shRNA TRCN0000039658), BRG1shRNA (MISSION shRNA TRCN0000231102), BRG2shRNA#2 (MISSION shRNA TRCN0000379829), ARID1AshRNA (MISSION shRNA TRCN0000059092), ARID1AshRNA#2 (MISSION shRNA TRCN0000059090), STAT3shRNA (MISSION shRNA TRCN0000329811), NOTCH1shRNA (MISSION shRNA TRCN0000003359), NOTCH2shRNA#2 (MISSION shRNA TRCN0000003362), NANOGshRNA (MISSION shRNA TRCN0000004888), NANOGshRNA#2 (MISSION shRNA TRCN0000004884), a control scrambled shRNA (CshRNA)(Millipore Sigma), and a MUC1-C vector (14) was inserted into pLKO-puro or pLKO-tet-puro (Plasmid #21915; Addgene, Cambridge, MA, USA). Guide RNA (CATCGTCAGGTTATATCGAG) targeting MUC1-C was cloned into the lentiCRISPRv2 vector (Addgene #52961). The viral vectors were produced in 293T cells (10). Cells transduced with the vectors were selected for growth in 1–3 μg/ml puromycin. For tet-inducible vectors, cells were treated with 0.1% DMSO as the vehicle control or 500 ng/ml doxycycline (DOX; Millipore Sigma).
Quantitative reverse-transcription PCR (qRT-PCR).
Total cellular RNA was isolated using Trizol reagent (ThermoFisher Scientific). cDNAs were synthesized and amplified as described (10). Primers used for qRT-PCR are listed in Supplementary Table 1.
Immunoblotting.
Total lysates prepared from subconfluent cells were immunoblotted with anti-MUC1-C (HM-1630-P1ABX, 1:400 dilution; ThermoFisher Scientific, Waltham, MA, USA), anti-E2F1 (3742, 1:1000; Cell Signaling Technology (CST), Danvers, MA, USA), anti-BRG1 (ab110641, 1:10000; abcam, Cambridge, MA, USA), anti-ARID1A (12354, 1:500; CST), anti-BAF60a (ab81621, 1:1000; abcam), anti-BAF155 (11956, 1:5000; CST), anti-BAF170 (12760, 1:5000; CST), anti-BRD9 (71232, 1:1000; CST), anti-NOTCH1 (3608, 1:1000; CST), anti-cleaved NOTCH1 (N1ICD) (4147, 1:500, CST), anti-E-cadherin (3195, 1:10000, CST), anti-vimentin (5741, 1:1000; CST), anti-NANOG (4903, 1:1000; CST), anti-STAT3 (9139, 1:5000; CST), anti-pSTAT3 (9145, 1:5000; CST), anti-β-actin (A5441, 1:100,000; Sigma) and anti-GAPDH (5174, 1:5000, CST).
Protein binding assays.
pGEX-E2F1 (Plasmid #21668; Addgene) was used to purify full-length (FL) GST-E2F1(aa 1–437) and to generate GST-E2F1 fragments containing the (i) DNA binding domain GST-E2F1(DBD; aa 1–191), (ii) dimerization domain GST-E2F1(DD; aa 192–284) and (iii) transactivation domain GST-E2F1(TAD; aa 285–437). GST-MUC1-CD (FL; aa 1–72), GST-MUC1-CD(aa 1–45), GST-MUC1-CD(aa 46–72) and GST-MUC1-CD(CQC→AQA) were prepared as described (14). GST-E2F1 and GST-MUC1-CD proteins were cleaved with thrombin to remove GST (14). Purified E2F1 was incubated with GST or GST-MUC1-CD proteins bound to glutathione beads and the adsorbates were analyzed by immunoblotting with anti-E2F1. Purified MUC1-CD was incubated with GST or GST-E2F1 proteins bound to glutathione beads and the adsorbates were analyzed by immunoblotting with anti-MUC1-CD CD1 antibody (21).
Chromatin immunoprecipitation (ChIP) assays.
Formaldehyde cross-linked and sheered soluble chromatin was precipitated with pre-cleared magnetic dynabeads (ThermoFisher Scientific) and 2 μg/ml of anti-MUC1-C (HM-1630-P1ABX; ThermoFisher Scientific), anti-E2F1 (3742; CST), anti-STAT3 (9139; CST), anti-BRG1 (ab110641; abcam), anti-ARID1A (12354, 2 μg/ml; CST) or a control non-immune IgG (Santa Cruz Biotechnology). The DNA-antibody precipitates were reverse cross-linked at 65°C for 18 h. DNAs were purified using gel extraction columns (QIAGEN, Germantown, MD, USA) and analyzed by qPCR using the Power SYBR Green PCR Master Mix and the ABI Prism 7300 sequence detector (Applied Biosystems). Data are reported as relative fold enrichment (10). Primers used for ChIP qPCR are listed in Supplementary Table 2.
RNA-seq analysis.
Total RNA from cells cultured in triplicates was isolated using Trizol reagent (Invitrogen). TruSeq Stranded mRNA (Illumina, San Diego, CA, USA) was used for library preparation. Raw sequencing reads were aligned to the human genome (GRCh38.74) using STAR. Raw feature counts were normalized and differential expression analysis using DESeq2. Differential expression rank order was performed using Gene Set Enrichment Analysis (GSEA) and the fgsea (v1.8.0) package in R. Gene sets queried included those from the Hallmark Gene Sets available through the Molecular Signatures Database (MSigDB).
Tumorsphere formation assays.
Cells (5 × 103) were seeded per well in 6-well ultra-low attachment culture plates (Corning Life Sciences) in DMEM/F12 50/50 medium (Corning Life Sciences) with 20 ng/ml EGF (Millipore Sigma), 20 ng/ml bFGF (Millipore Sigma) and 1% B27 supplement. Cells were treated with vehicle or DOX for 10–14 days. Tumorspheres were counted under an inverted microscope in triplicate wells.
Statistical analysis.
Each experiment was performed at least three times. Data are expressed as the mean±SD. The unpaired Mann-Whitney U test or Student’s t-test was used to determine differences between means of groups. A p-value of <0.05 denoted by an asterisk (*) was considered statistically significant.
Data Availability.
The accession number for the RNA-seq data is GEO Submission GSE139335.
Results
MUC1-C drives BRG1 and ARID1A expression.
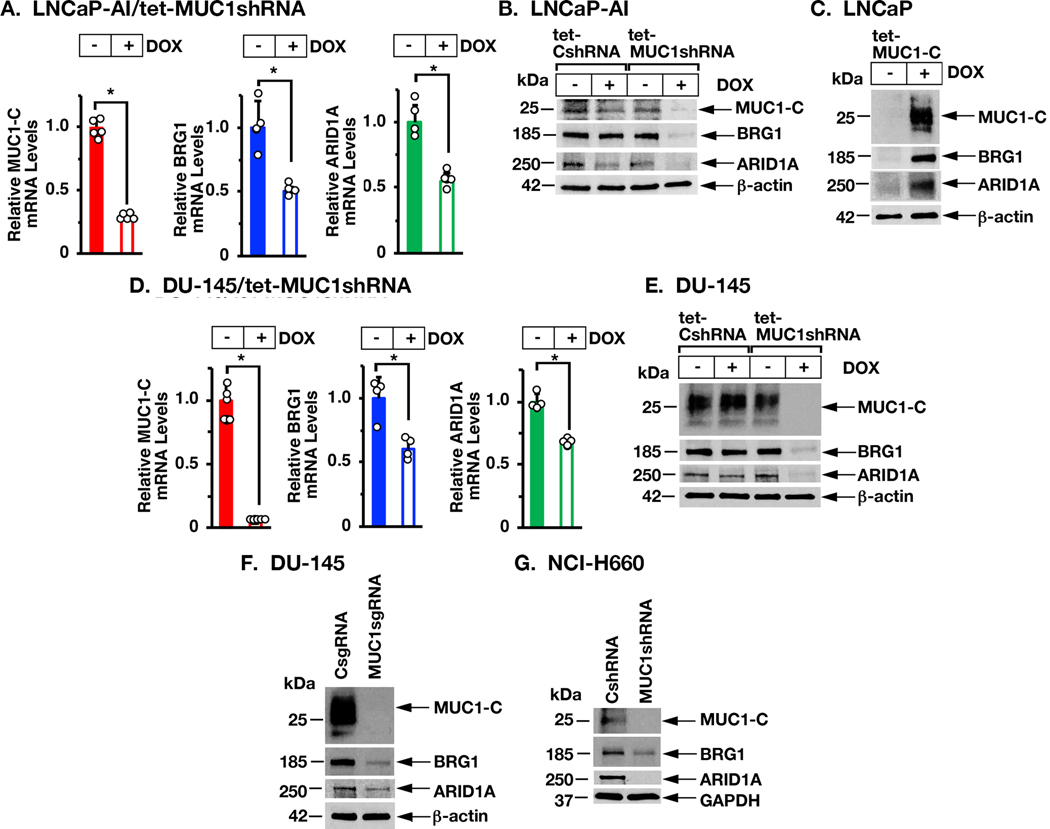
In addressing the contention that MUC1-C may interact with BAF, we found that DOX-inducible silencing of MUC1-C in androgen-independent LNCaP-AI PC cells is associated with downregulation of BRG1 and ARID1A expression at the mRNA (Fig. 1A) and protein (Fig. 1B) levels. As support for these observations, inducible expression of MUC1-C in MUC1-null LNCaP PC cells resulted in upregulation of BRG1 and ARID1A (Fig. 1C). DU-145 CRPC cells similarly responded to MUC1-C silencing with decreases in BRG1 and ARID1A expression (Figs. 1D and 1E). Stable MUC1-C silencing with a MUC1sgRNA (Fig. 1F) or a MUC1shRNA (Supplemental Fig. S1A) also resulted in suppression of BRG1 and ARID1A. Additionally, we found that MUC1-C induces BRG1 and ARID1A in NCI-H660 NEPC cells (Fig. 1G). Similar results obtained in BT-549 triple-negative breast cancer (TNBC) cell (Supplemental Fig. S1B) and SW620 colon cancer cell (Supplemental Fig. S1C) models of MUC1-C-driven plasticity (14,15) further indicated that MUC1-C activates BRG1 and ARID1A expression in the progression of different types of carcinoma cells.
Figure 1. MUC1-C induces BRG1 and ARID1A expression.
A. LNCaP-AI cells stably transduced with a tet-inducible MUC1shRNA were treated with control vehicle or DOX for 7 days. The indicated mRNA levels were analyzed by qRT-PCR. The results (mean±SD of 4–5 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). B. LNCaP-AI cells expressing a control tet-CshRNA or tet-MUC1shRNA were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. C. LNCaP cells expressing a tet-inducible MUC1-C (tet-MUC1-C) vector were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. D. DU-145/tet-MUC1shRNA cells were treated with control vehicle or DOX for 7 days. The indicated mRNA levels were analyzed by qRT-PCR. The results (mean±SD of 4–5 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). E. DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. F. Lysates from DU-145/CsgRNA and DU-145/MUC1sgRNA cells were immunoblotted with antibodies against the indicated proteins. G. Lysates from NCI-H660 NEPC cells expressing a CshRNA or MUC1shRNA were immunoblotted with antibodies against the indicated proteins.
MUC1-C induces BRG1 and ARID1A expression by an E2F1-mediated pathway.
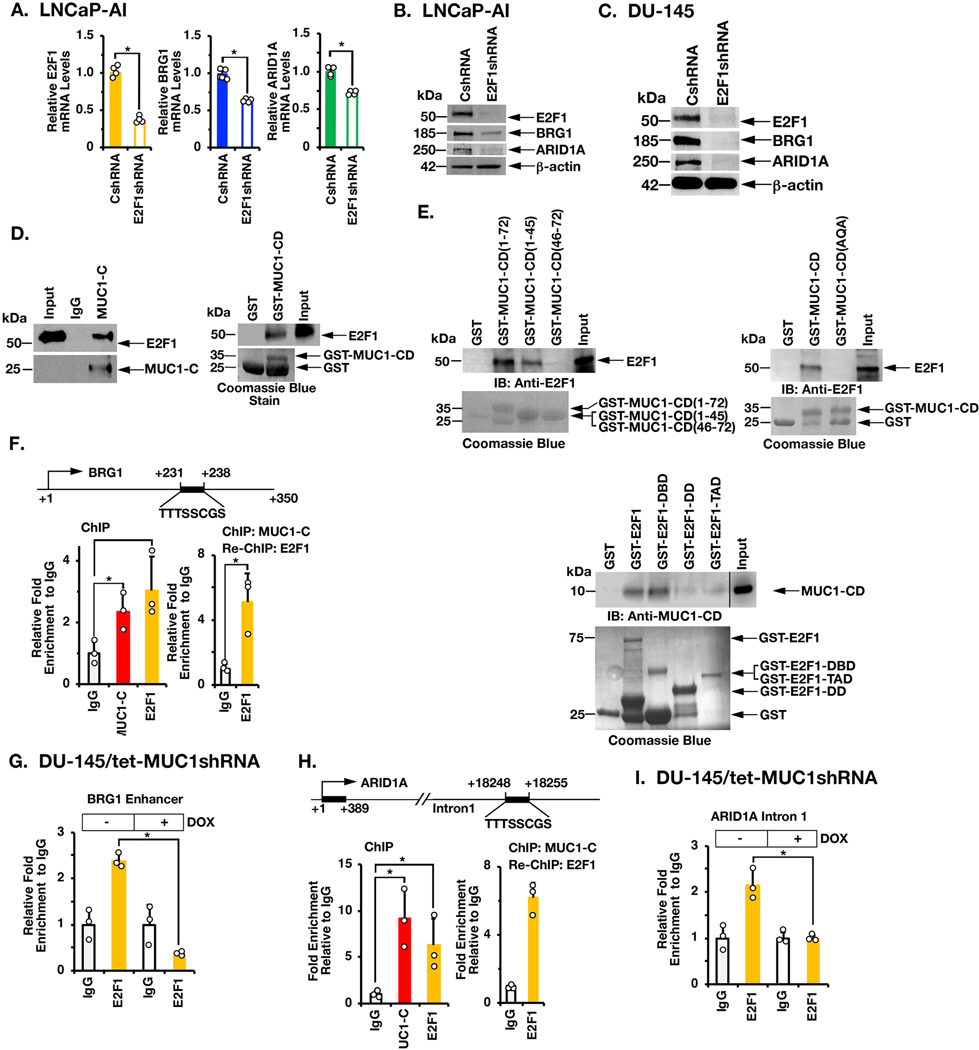
MUC1-C activates the PRC2 complex by an E2F1-dependent mechanism (11). In the present studies, we found that (i) expression of MUC1-C in LNCaP cells significantly associates with the activation of E2F target genes (pvalue=0.002; Supplemental Fig. S2A), and (ii) silencing E2F1 in LNCaP-AI cells decreases BRG1 and ARID1A expression (Figs. 2A and 2B). These results were extended in DU-145 (Fig. 2C; Supplemental Fig. S2B) and BT-549 (Supplemental Figs. S2C and S2D) cells, supporting involvement of a MUC1-C→E2F1 pathway. Little is known regarding interactions between MUC1-C and E2F1. Here, nuclear co-immunoprecipitation studies demonstrated that MUC1-C associates with E2F1 (Fig. 2D, left). Moreover, in vitro binding experiments showed that the MUC1-C cytoplasmic domain (CD; aa 1–72) interacts with E2F1 (Fig. 2D, right). Further analysis demonstrated that MUC1-CD(1–45) and not MUC1-CD(46–72) confers the interaction (Fig. 2E, left). Consistent with this result, we found that mutating the MUC1-C cytoplasmic domain CQC (aa 1–3) motif to AQA abrogated the interaction with E2F1 (Fig. 2E, right). We also found that MUC1-CD binds to the region of E2F1 that contains the DNA binding domain (DBD) and not the transactivation domain (TAD) or dimerization domain (DD)(Fig. 2E, lower), confirming that the MUC1-C cytoplasmic domain binds directly to the E2F1 DBD. Specificity of the MUC1-C→E2F1 interaction for BAF activation was further supported by the demonstration that expression of MUC1-C(AQA) with mutation of the CQC motif to AQA abrogates the induction of BRG1 and ARID1A (Supplemental Fig. S2E). Based on these results, we identified a putative E2F binding motif in the BRG1 gene downstream of the transcription start site (Fig. 2F). ChIP studies of that BRG1 region demonstrated occupancy of MUC1-C and E2F1 (Fig. 2F, left). Re-ChIP analysis further demonstrated the detection of MUC1-C/E2F1 complexes (Fig. 2F, right). Additionally, silencing MUC1-C decreased E2F1 occupancy on the BRG1 enhancer (Fig. 2G). We also identified a putative E2F1 binding site in the ARID1A intron 1 region and found that MUC1-C/E2F1 complexes occupy that region (Figs. 2H, left and right). As shown for the BRG1 enhancer, silencing MUC1-C was associated with downregulation of E2F1 occupancy on the ARID1A intron 1 region (Fig. 2I), providing evidence for a MUC1-C→E2F1 pathway that induces BRG1 and ARID1A.
Figure 2. MUC1-C interacts with E2F1 in driving BRG1 and ARID1A expression.
A. LNCaP-AI/CshRNA and LNCaP-AI/E2F1shRNA cells were analyzed for the indicated mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). B. Lysates from LNCaP-AI/CshRNA and LNCaP-AI/E2F1shRNA cells were immunoblotted with antibodies against the indicated proteins. C. Lysates from DU-145/CshRNA and DU-145/E2F1shRNA cells were immunoblotted with antibodies against the indicated proteins. D. Nuclear lysates from LNCaP-AI cells were incubated with anti-MUC1-C or a control IgG. The input and precipitates were analyzed by immunoblotting with anti-E2F1 and anti-MUC1-C (left). Purified E2F1 was incubated with GST or GST-MUC1-CD(FL; 1–72) bound to glutathione beads. Adsorbates were immunoblotted with anti-E2F1 (right). Input of the GST proteins was assessed by Coomassie blue staining. E. Purified E2F1 was incubated with GST, GST-MUC1-CD(FL; 1–72) or the indicated GST-MUC1-CD fragments bound to glutathione beads (left). Purified E2F1 was incubated with GST, GST-MUC1-CD(FL; 1–72) or the GST-MUC1-CD(CQC→AQA) mutant bound to glutathione beads (right). Adsorbates were immunoblotted with anti-E2F1. Purified MUC1-CD was incubated with GST, GST-E2F1(FL, 1–437), GST-E2F1(DBD; aa 1–191), GST-E2F1(DD; aa 192–284) and or GST-E2F1(TAD; aa 285–437) bound to glutathione beads (lower). Adsorbates were immunoblotted with anti-MUC1-CD. Input of the GST proteins was assessed by Coomassie blue staining. F. Schema of the BRG1 enhancer region with positioning of the putative E2F1 binding motif. Soluble chromatin from DU-145 cells was precipitated with anti-MUC1-C, anti-E2F1 or a control IgG (left). Soluble chromatin was precipitated with anti-MUC1-C (ChIP) and then reprecipitated with anti-E2F1 or a control IgG (re-ChIP) (right). G. DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Soluble chromatin was precipitated with anti-E2F1 or a control IgG. The DNA samples were amplified by qPCR with primers for the BRG1 enhancer region. The results (mean±SD of 3 determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). H. Schema of the ARID1A intron 1 region with positioning of the putative E2F1 binding motif. Soluble chromatin from DU-145 cells was precipitated with anti-MUC1-C, anti-E2F1 or a control IgG (left). Soluble chromatin was precipitated with anti-MUC1-C (ChIP) and then reprecipitated with anti-E2F1 or a control IgG (re-ChIP) (right). I. DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Soluble chromatin was precipitated with anti-E2F1 or a control IgG. The DNA samples were amplified by qPCR with primers for the ARID1A intron 1 region. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1).
MUC1-C→E2F1 pathway activates expression of esBAF subunits.
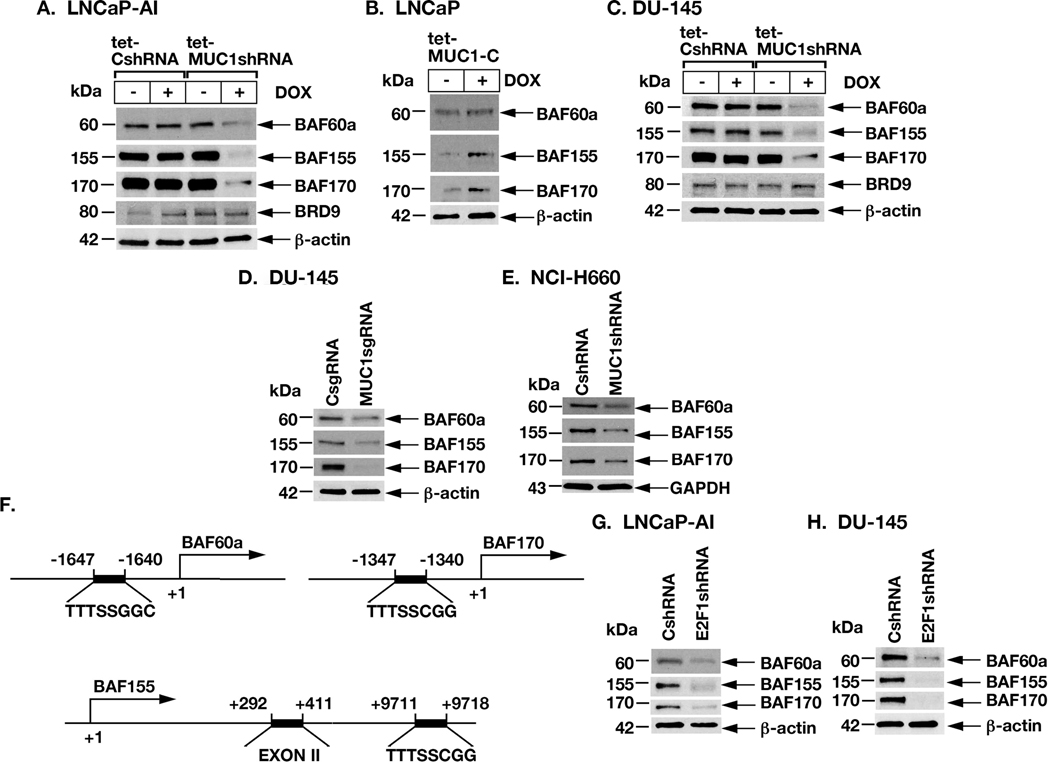
BRG1 and ARID1A are components of canonical cBAF and certain other BAF complexes (8,9). In addition to BRG1 and ARID1A, we found that silencing MUC1-C in LNCaP-AI cells decreases expression of BAF60a and BAF155, which are components of the esBAF complex (Fig. 3A) (8,9). Silencing MUC1-C was also associated with suppression of BAF170, another esBAF component that maintains pluripotency of human ESCs (Fig. 3A) (9,22). In contrast, MUC1-C silencing had little effect on BRD9, a subunit of the non-canonical ncBAF/GBAF complex (Fig. 3A) (23). As found for BRG1 and ARID1A, inducible expression of MUC1-C in LNCaP cells was associated with upregulation of BAF60a, BAF155 and BAF170 (Fig. 3B). Moreover, in studies of DU-145 cells, we found that MUC1-C is necessary for induction of BAF60a, BAF155 and BAF170, but not BRD9 (Fig. 3C). These findings were confirmed by MUC1-C silencing in DU-145/MUC1sgRNA (Fig. 3D) and NCI-H660/MUC1shRNA (Fig. 3E) cells. Putative E2F binding motifs were also identified in the BAF60a and BAF170 promoter regions and in BAF155 between exons II and III (Fig. 3F), invoking their possible activation by MUC1-C→E2F1 signaling. Indeed, silencing E2F1 in LNCaP-AI (Fig. 3G) and DU-145 (Fig. 3H) cells resulted in downregulation of BAF60a, BAF155 and BAF170 expression.
Figure 3. MUC1-C and E2F1 drive expression of BAF60a, BAF155 and BAF170.
A. LNCaP-AI/tet-CshRNA and LNCaP-AI/tet-MUC1shRNA were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. B. LNCaP/tet-MUC1-C cells were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. C. DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA were treated with vehicle or DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. D. Lysates from DU-145/CsgRNA and DU-145/MUC1sgRNA cells were immunoblotted with antibodies against the indicated proteins. E. Lysates from NCI-H660/CshRNA and NCI-H660/MUC1shRNA cells were immunoblotted with antibodies against the indicated proteins. F. Schema of the BAF60a, BAF170 and BAF155 genes with positioning of putative E2F1 binding motifs. G. Lysates from LNCaP-AI/CshRNA and LNCaP-AI/E2F1shRNA cells were immunoblotted with antibodies against the indicated proteins. H. Lysates from DU-145/CshRNA and DU-145/E2F1shRNA were immunoblotted with antibodies against the indicated proteins.
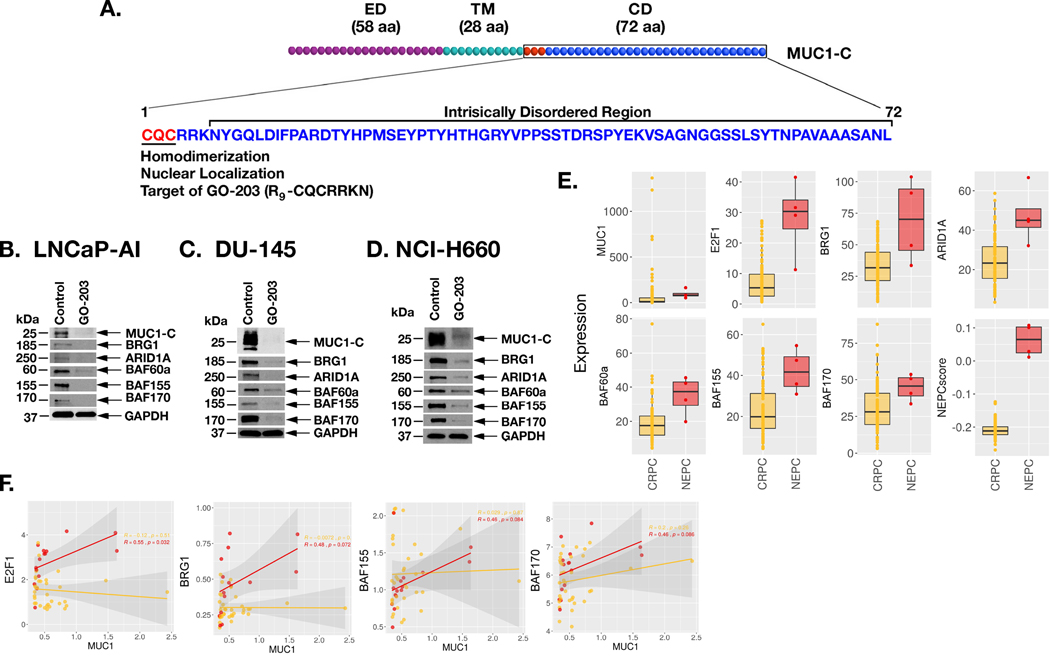
MUC1-C consists of 58-aa extracellular, 28-aa transmembrane and 72-aa cytoplasmic domains (Fig. 4A). The MUC1-C cytoplasmic domain includes a CQC motif, which is necessary for homodimerization and nuclear import (11,24). The cell-penetrating GO-203 inhibitor (R9-CQCRRKN) binds to the CQC motif and blocks MUC1-C homodimerization and function (20,25,26), phenocopies the effects of MUC1-C silencing, including downregulation of (i) MYC→BRN2 signaling, (ii) NE markers, and (iii) the OSKM pluripotency factors (10). Treatment with GO-203 was also associated with inhibition of self-renewal and tumorigenicity, indicating that this agent is active in targeting MUC1-C-induced stemness (10). Treatment of LNCaP-AI cells with GO-203 was associated with downregulation of MUC1-C, BRG1 and ARID1A (Fig. 4B). Moreover, we found that targeting MUC1-C with GO-203 decreases expression of BAF60a, BAF155 and BAF170 (Fig. 4B). Similar results were obtained in studies of GO-203-treated DU-145 (Fig. 4C) and NCI-H660 (Fig. 4D) cells, confirming that MUC1-C drives these esBAF components. In regard to the effects of targeting more than one BAF component, we found that silencing BRG1 is associated with downregulation of ARID1A, BAF60a, and BAF170 (Supplemental Fig. S3A). Moreover, silencing ARID1A decreased BAF60a, BAF155 and BAF170 (Supplemental Fig. S3B). These findings are consistent with interconnectivity of BAF component expression and indicate that the effects of silencing BRG1 or ARID1A are reflective of downregulating multiple components of the BAF complex. In extending these results to clinical samples, we found that E2F1, BRG1, ARID1A, BAF60a, BAF155 and BAF170 are significantly increased in NEPC as compared to CRPC tumors (Fig. 4E). Further analysis demonstrated that MUC1 associates with E2F1, BRG1, BAF155 and BAF170 in NEPC and not CRPC tumors (Fig. 4F), supporting involvement of MUC1-C in activating the E2F1→BAF pathway with progression to NEPC.
Figure 4. Targeting MUC1-C with the GO-203 inhibitor suppresses expression of BAF subunits.
A. Schema of MUC1-C depicting the 58 aa extracellular domain (ED), 28 aa transmembrane domain (TM) and 72 aa cytoplasmic domain (CD). MUC1-C homodimerization and thereby nuclear import is dependent on the CQC motif, which is targeted by the cell penetrating GO-203 inhibitor (R9-CQCRRKN). Downstream to the CQC motif is an intrinsically disordered region as found in multiple oncogenic proteins that function as nodes in integrating multiple signaling pathways linked to transformation. B-D. LNCaP-AI (B), DU-145 (C) and NCI-H660 (D) cells were left untreated or treated with 5 μM GO-203 for 48 h. Lysates were immunoblotted with antibodies against the indicated proteins. E. Analysis of the SU2C PC dataset comparing expression of the indicated genes and the NEPC score in CRPC and NEPC tumor samples. The asterisk (*) denotes a pvalue<0.05 (Wilcox-test). F. Analysis of the Beltran PC dataset assessing the correlation of MUC1 with expression of the indicated genes in CRPC (yellow) and NEPC (red) tumor samples.
MUC1-C interacts with BAF in driving NOTCH1.
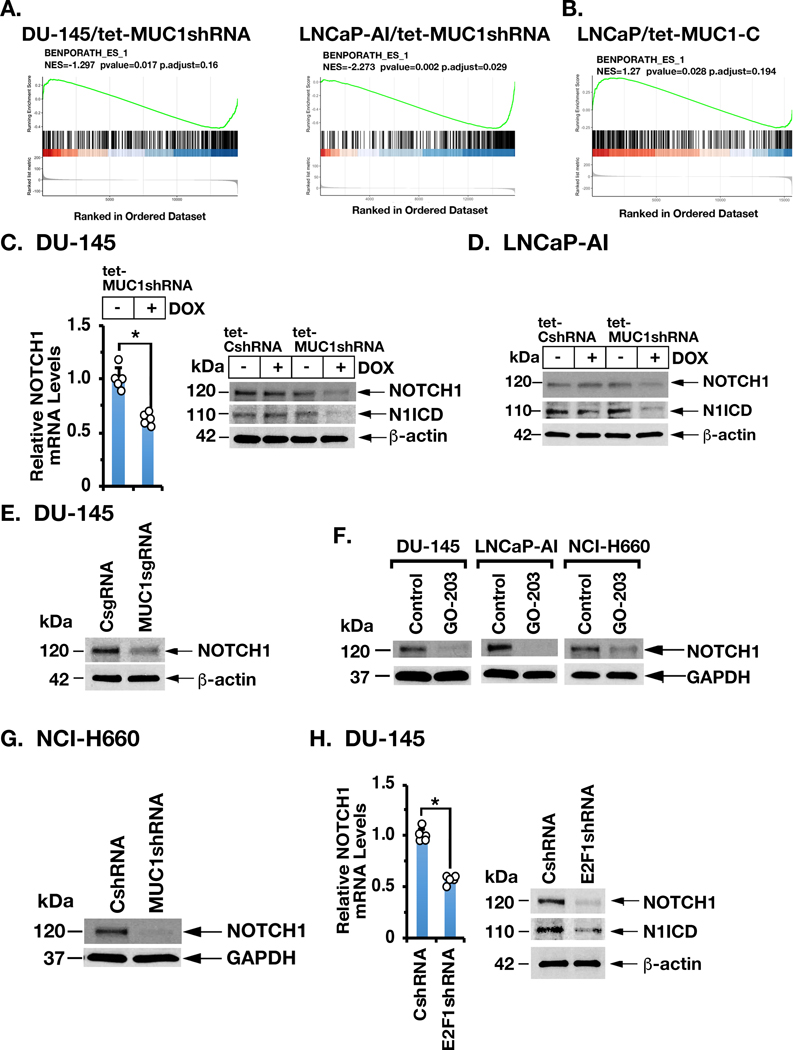
esBAF is necessary for ESC self-renewal and differentiation by regulating ESC gene expression (8,9,27,28). We found by RNA-seq analysis that MUC1-C significantly associates with regulation of (i) the BENPORATH_ES1 signature (Figs. 5A and 5B), which is derived from genes enriched in ESCs and CSC-like phenotypes (29), and (ii) other ESC and NOTCH gene signatures (Supplemental Table S3). As found for MUC1-C, NOTCH1 drives androgen-independent PC and is upregulated, albeit by unclear mechanisms, in progression to CRPC (30). Along these lines, we found that silencing MUC1-C in DU-145 (Fig. 5C) and LNCaP-AI (Fig. 5D) cells decreases expression of NOTCH1 (120 kDa) and the activated form of cleaved NOTCH1 (N1ICD) (110 kDa). These results were extended by targeting MUC1-C with a MUC1sgRNA (Fig. 5E) and with the GO-203 inhibitor (Fig. 5F). Additional studies in NCI-H660 (Fig. 5G) and other types of carcinoma cells (Supplemental Figs. S4A and S4B) confirmed that MUC1-C drives NOTCH1 expression. Moreover, as found for MUC1-C, silencing E2F1 resulted in the suppression of NOTCH1 (Fig. 5H; Supplemental Figs. S4C and S4D), supporting a previously unrecognized MUC1-C→E2F1→NOTCH1 pathway.
Figure 5. MUC1-C→E2F signaling induces NOTCH1 expression.
A and B. RNA-seq was performed in triplicate on LNCaP-AI/tet-MUC1shRNA (A, left), DU-145/tet-MUC1shRNA (A, right) and LNCaP/tet-MUC1-C cells (B) treated with vehicle or DOX for 7 days. The datasets were analyzed using the BENPORATH_ES_1 gene signature. C. DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days were analyzed for NOTCH1 mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1) (left). Lysates from DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days were immunoblotted with antibodies against the indicated proteins (right). D. Lysates from LNCaP-AI/tet-CshRNA and LNCaP-AI/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days were immunoblotted with antibodies against the indicated proteins. E. Lysates from DU-145/CsgRNA and DU-145/MUC1sgRNA cells were immunoblotted with antibodies against the indicated proteins. F. The indicated cells were left untreated or treated with 5 μM GO-203 for 48 h. Lysates were immunoblotted with antibodies against the indicated proteins. G. Lysates from NCI-H660/CshRNA and NCI-H660/MUC1shRNA cells were immunoblotted with antibodies against the indicated proteins. H. DU-145/CshRNA and DU-145/E2F1shRNA cells were analyzed for NOTCH1 mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1) (left). Lysates were immunoblotted with antibodies against the indicated proteins (right).
We also found that, in addition to inducing BAF expression, MUC1-C forms a nuclear complex with BRG1 and ARID1A (Supplemental Fig. S5A), extending the potential involvement of MUC1-C in regulating BAF function. We therefore studied cells with BRG1 silencing and found that, like MUC1-C and E2F1, BRG1 is necessary for driving NOTCH1 expression (Supplemental Figs. S5B–S5D). Similar results were obtained by silencing ARID1A (Supplemental Figs. S5E–S5G), supporting a MUC1-C→E2F1→BAF→NOTCH1 pathway (Supplemental Figs. S5H and S5I). As confirmation of these findings, silencing MUC1-C (Supplemental Fig. S5J), E2F1 (Supplemental Fig. S5K), BRG1 (Supplemental Fig. S5L) or ARID1A (Supplemental S5M) was associated with (i) downregulation of the NOTCH1 target genes, HES1 and HEY1, which like NOTCH1 contribute to EMT and stemness (31,32), and (ii) suppression of the EMT phenotype, as evidenced by induction of E-cadherin and suppression of vimentin (Supplemental Figs. S6A–S6D).
MUC1-C→E2F1→BAF→NOTCH1 pathway induces NANOG expression.
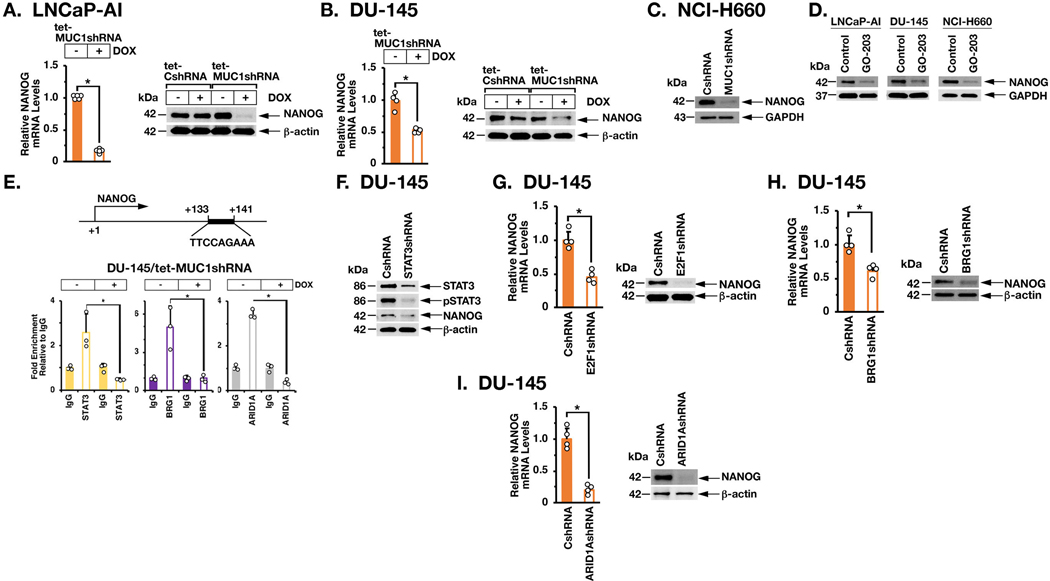
Silencing MUC1-C results in downregulation of the Yamanaka OSKM (OCT4, SOX2, KLF4 and MYC) pluripotency factors in the DU-145 and LNCaP-AI cell models (10). Here, we found that E2F1, BRG1 and ARID1A have distinct effects on OSKM regulation (Supplemental Fig. S7A). For example, in contrast to MUC1-C, silencing E2F1 and ARID1A was associated with increases in SOX2, whereas silencing BRG1 had no apparent effect on SOX2 expression (Supplemental Fig. S7A), indicating that MUC1-C-driven pathways other than MUC1-C→E2F1→BAF regulate the OSKM pluripotency factors. We also found that MUC1 significantly associates with activation of the BENPORATH_NANOG_TARGETS gene signature (Supplemental Figs. S7B and S7C). Accordingly, we focused on the core NANOG pluripotency factor and found that silencing MUC1-C results in downregulation of NANOG expression (Figs. 6A, left and right). Similar results were obtained in DU-145 (Fig. 6B), NCI-H660 (Fig. 6C) and other carcinoma cells (Supplemental Figs. S8A and S8B). Targeting MUC1-C with GO-203 also suppressed NANOG (Fig. 6D), confirming involvement of MUC1-C in driving NANOG expression. In addition, and like NOTCH1, we found that enforced expression of MUC1-C in LNCaP cells induces NANOG (Supplemental Fig. S8C). The NANOG gene contains a STAT3 binding motif downstream of the TSS (Fig. 6E), which is activated by STAT3 when complexed with BAF (33,34). In cancer cells, nuclear MUC1-C binds directly to STAT3, promotes JAK1-mediated phosphorylation of pSTAT3 and forms an auto-inductive MUC1-C/STAT3 circuit (35). ChIP studies of NANOG in that region demonstrated the detection of STAT3, BRG1 and ARID1A (Fig. 6E). Silencing MUC1-C was associated with marked decreases in STAT3, BRG1 and ARID1A occupancy (Fig. 6E). Moreover, downregulation of STAT3 (Fig. 6F), E2F1 (Fig. 6G; Supplemental Fig. S8D), BRG1 (Fig. 6H; Supplemental Fig. S8E) and ARID1A (Fig. 6I; Supplemental Fig. S8F) decreased NANOG expression, indicating that MUC1-C-induced activation of NANOG is dependent on STAT3 and BAF.
Figure 6. MUC1-C→E2F→BAF signaling activates NANOG expression.
A. LNCaP-AI/tet-MUC1shRNA cells were treated with control vehicle or DOX for 7 days. NANOG mRNA levels were analyzed by qRT-PCR (left). The results (mean±SD of 5 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). LNCaP-AI/tet-CshRNA and LNCaP-AI/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days (right). Lysates were immunoblotted with antibodies against the indicated proteins. B. DU-145/tet-MUC1shRNA cells were treated with control vehicle or DOX for 7 days. NANOG mRNA levels were analyzed by qRT-PCR (left). The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days (right). Lysates were immunoblotted with antibodies against the indicated proteins. C. Lysates from NCI-H660/CshRNA and NCI-H660/MUC1shRNA cells were immunoblotted with antibodies against the indicated proteins. D. The indicated cells were left untreated or treated with 5 μM GO-203 for 48 h. Lysates were immunoblotted with antibodies against the indicated proteins. E. Schema of the NANOG enhancer region with positioning of the STAT3 binding site. DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Soluble chromatin was precipitated with anti-STAT3, anti-BRG1, anti-ARID1A or a control IgG. The DNA samples were amplified by qPCR with primers for the NANOG enhancer region. The results (mean±SD of 3 determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). F. Lysates from DU-145/CshRNA and DU-145/STAT3shRNA cells were immunoblotted with antibodies against the indicated proteins. G-I. DU-145/CshRNA, DU-145/E2F1shRNA (G), DU-145/BRG1shRNA (H) and DU-145/ARID1AshRNA (I) cells were analyzed for NANOG mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1) (left). Lysates were immunoblotted with antibodies against the indicated proteins (right).
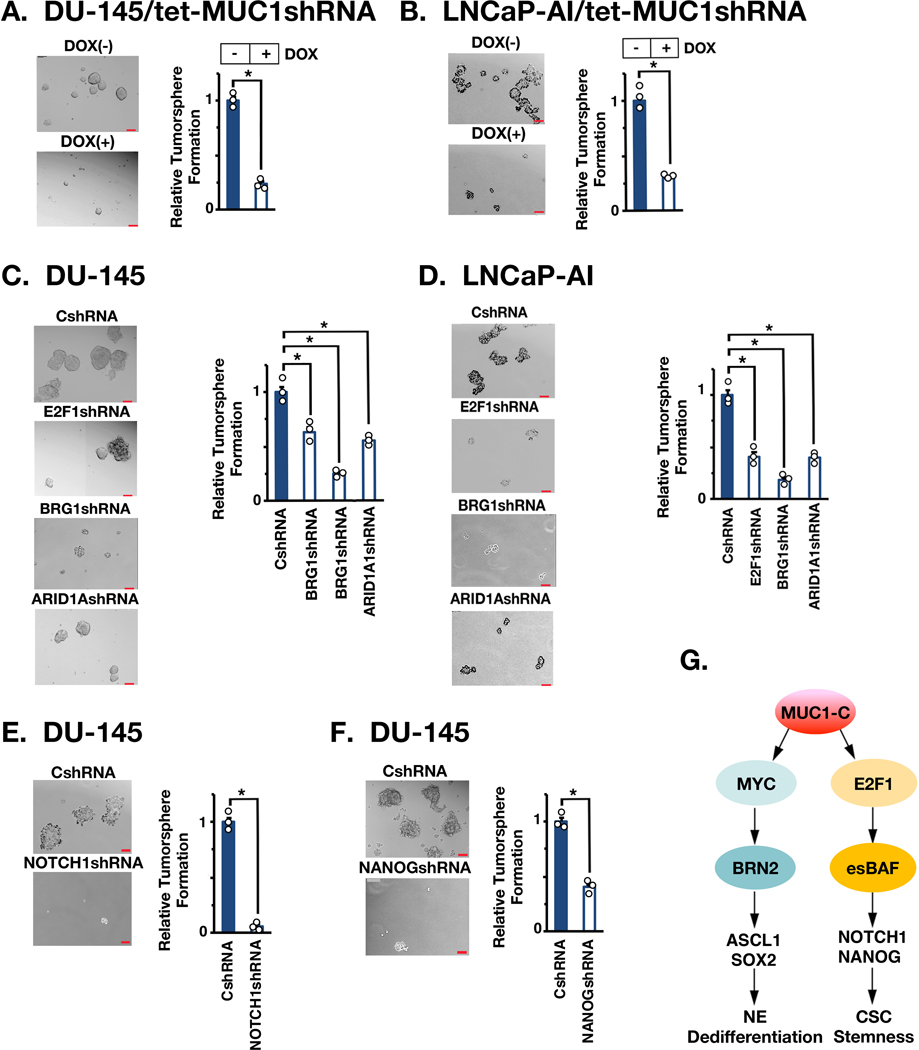
MUC1-C→E2F1→BAF→NOTCH1→NANOG pathway drives self-renewal capacity.
MUC1-C→MYC signaling promotes progression to NEPC as evidenced by induction of BRN2 and NE differentiation markers (AURKA and SYP) (10). Silencing E2F1, BRG1 or ARID1A had little if any effect on these markers (Supplemental Fig. S9A), indicating that the MUC1-C→MYC and MUC1-C→BAF pathways promote integrated, but separate, downstream networks. Therefore, to assess involvement of MUC1-C→E2F1→BAF signaling in the CSC state, we investigated the involvement of this pathway in PC self-renewal capacity. Silencing MUC1-C significantly decreased self-renewal capacity in DU-145 (Fig. 7A) and LNCaP-AI (Fig. 7B) cells as assessed by tumorsphere formation. We also found that silencing E2F1, BRG1 and ARID1A in DU-145 cells suppresses self-renewal (Fig. 7C). Similar responses were observed in LNCaP-AI cells (Fig. 7D). In addition, silencing NOTCH1 (Supplemental Fig. S9B) and NANOG (Supplemental Fig. S9C) suppressed self-renewal capacity (Figs. 7E and 7F; Supplemental Figs. S9D and S9E). As additional evidence for effects on CSCs, we found that silencing MUC1-C (Supplemental Fig. S9F), E2F1 (Supplemental Fig. S9G, left), BRG1 (Supplemental Fig. S9G, middle) and ARID1A (Supplemental Fig. S9G, right) suppresses CD44, supporting a MUC1-C→E2F1→BAF pathway in integrating NOTCH1 and NANOG expression with stemness (Fig. 7G).
Figure 7. MUC1-C→E2F→BAF→NOTCH1→NANOG pathway is necessary for self-renewal.
A and B. DU-145/tet-MUC1shRNA (A) and LNCaP-AI/tet-MUC1shRNA (B) cells treated with vehicle or DOX for 10 days were assayed for tumorsphere formation (left). Scale bar: 100 mm. The results (mean±SD of 3 biologic replicates) are expressed as relative tumorsphere number per field compared to the CshRNA control (right). C. The indicated DU-145 cells were assayed for tumorsphere formation (left). The results (mean±SD of 3 biologic replicates) are expressed as relative tumorsphere number per field compared to the CshRNA control (right). D. The indicated LNCaP-AI cells were assayed for tumorsphere formation (left). The results (mean±SD of 3 biologic replicates) are expressed as relative tumorsphere number per field compared to the CshRNA control (right). E and F. DU-145 cells expressing a CshRNA, NOTCH1shRNA (E) or NANOGshRNA (F) were assayed for tumorsphere formation (left). The results (mean±SD of 3 biologic replicates) are expressed as relative tumorsphere number per field compared to the CshRNA control (right). G. Proposed model for involvement of MUC1-C in integrating MYC and E2F1 signaling pathways in NEPC progression. MUC1-C binds directly to MYC and drives activation of BRN2, the Yamanaka OSK pluripotency factors and NE dedifferentiation (10). The present results demonstrate that MUC1-C binds directly to E2F1 and activates the esBAF complex (BRG1, ARID1A, BAF60a, BAF155 and BAF170). The MUC1-C→E2F1→esBAF pathway drives the induction of NOTCH1, NANOG and CSC self-renewal.
Discussion
MUC1-C is aberrantly expressed in CRPCs and NEPCs, suppresses AR axis signaling and by unclear mechanisms induces lineage plasticity and stemness associated with NEPC progression (10). The present work extends those findings by identifying a previously unrecognized role for MUC1-C in activating the BAF complex in cancer cells. BRG1, SMARCB1 and ARID1A are subject to mutations, translocations and deletions in ~20% of human cancers, supporting their functions as tumor suppressors (4,36). The BRG1 and ARID1A gene mutations contribute to dysregulation of BAF in controlling enhancer functions and cell fate (4). Our findings that MUC1-C induces expression of BRG1 and ARID1A was therefore somewhat unexpected for an oncogenic protein that promotes cancer progression (10,14,16). Nonetheless, BAF has been linked to PC by an ERG-mediated mechanism (17). In addition, BRG1 expression is increased in multiple cancer types and is associated with aggressive disease (37,38), raising the possibility that BRG1 may play distinct roles depending on cell context. In this respect and of importance, little is known about the activation of BRG1, as well as ARID1A, expression in cancer cells. Our findings that MUC1-C is involved in inducing BRG1 and ARID1A thus uncover a common pathway responsible for integrating their expression (Fig. 7G).
BAF interacts with PRCs in regulating gene expression and cellular identity, and the balance of these complexes is of importance in development and oncogenesis (3,39). MUC1-C activates the repressive PRC1/2 complexes in cancer cells (11–13). Moreover and of potential relevance for the present work, MUC1-C induces expression of the PRC2 subunits, EZH2, SUZ12 and EED, by E2F-dependent activation of their promoter regions (11,13). The E2F proteins play a role in cell proliferation, genomic stability and pluripotent stem cell fate (40,41). BAF controls pluripotency and self-renewal of ESCs (8,42); however, there has been no known link between E2Fs and BAF in ESCs or CSCs. We found that MUC1-C interacts directly with E2F1 and that MUC1-C/E2F1 complexes occupy the BRG1 and ARID1A genes and contribute to their activation. BRG1 and ARID1A are components of cBAF, ncBAF and neuronal BAF (nBAF) and esBAF, the latter of which includes BAF60a, BAF155 and BAF170 (8,9). The interchange of BAF155 and BAF170 as esBAF components may contribute to regulation of ESC functions in self-renewal and lineage specification (8,9). We identified putative E2F binding motifs in BAF60a, BAF155 and BAF170, and found that, like BRG1 and ARID1A, silencing MUC1-C or E2F1 decreases their expression. Whereas subsequent studies will be needed to investigate MUC1-C/E2F1 occupancy and activation of those genes, our findings reveal a MUC1-C→E2F1 pathway that activates the esBAF complex in cancer cells (Fig. 7G).
MUC1-C regulates lineage plasticity in the progression to NEPC by activating in part a MYC→BRN2 pathway with induction of NE differentiation markers (10). The present results indicate that MUC1-C integrates MYC and E2F1 signaling and that, in contrast to MUC1-C→MYC→BRN2, the MUC1-C→E2F1→BAF network functions in driving a distinct pathway in parallel with NE dedifferentiation (Fig. 7G). Indeed, identification of the murine ESC specific esBAF complex established dependence on these components for the maintenance of ESC pluripotency and self-renewal (27,28). Our results demonstrate that an esBAF-like complex plays a role in CSC self-renewal. In this respect, we identified a previously unrecognized MUC1-C→E2F1→BAF pathway that drives NOTCH1 expression in PC cells. NOTCH1 signaling has been linked to PC progression (30) and PC CSC function (18,43). Additionally, the NOTCH1 downstream effectors, HES1 and HEY1, promote integration of EMT and CSC self-renewal (31,32). Other work has shown that NOTCH1 interacts with NANOG in expanding CSCs (44). Along these lines, we found that the MUC1-C→E2F1→BAF pathway is necessary for expression of NOTCH1 and NANOG in PC cells. BAF promotes binding of NANOG to its genomic targets (8,45,46). Moreover, NANOG is essential for CSC self-renewal (47,48) and promotes treatment resistance and poor clinical outcomes (19,49). Consistent with inducing NOTCH1 and NANOG, our results demonstrate that, in contrast to NE differentiation driven by MUC1-C→MYC signaling (10), the MUC1-C→E2F1→BAF pathway is necessary for CSC self-renewal capacity (Fig. 7G).
MUC1-C promotes lineage plasticity and stemness in NEPC, TNBC and colon cancer cells (10,14,15). The present results extend those findings by demonstrating that MUC1-C (i) activates the BAF complex in cancer cells, (ii) links E2F1 to the induction of BAF, and (iii) drives a E2F1→BAF pathway that induces NOTCH1, NANOG and stemness (Fig. 7G). Lineage plasticity in cancer is increasingly being associated with treatment resistance, immune evasion and poor patient outcomes (50). Our findings that MUC1-C confers CSC lineage plasticity in CRPC/NEPC progression lends support for this oncogenic protein as a potential prognostic factor and therapeutic target in advanced recalcitrant cancers.
Supplementary Material
Statement of Significance.
Findings show that MUC1-C, which promotes prostate cancer (PC) progression, activates a novel pathway that drives the BAF remodeling complex, induces NOTCH1 and NANOG, and promotes self-renewal of PC CSC.
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant numbers CA97098, CA166480 and CA233084 awarded to D. Kufe and CA232979 awarded to S. Liu.
Abbreviations:
- BAF
Brg/Brahama-associated factors
- esBAF
embryonic stem cell BAF
- NEPC
neuroendocrine prostate cancer
- CRPC
castration-resistant prostate cancer
- MUC1-C
MUC1 C-terminal subunit
- CSC
cancer stem cell
- NOTCH1
NOTCH homolog 1
- EMT
epithelial-mesenchymal transition
- DNMT
DNA methyltransferase
- PRC
polycomb repressive complex
- NuRD
nucleosome remodeling and deacetylation complex
- MUC1-CD
MUC1-C cytoplasmic domain
- DBD
DNA binding domain
- TAD
transactivation domain
- DD
dimerization domain
- N1ICD
cleaved NOTCH1 intracellular domain
- OSKM factors
OCT4, SOX2, KLF4 and MYC
- TNBC
triple-negative breast cancer
Footnotes
DK has equity interests in Genus Oncology, Reata Pharmaceuticals and Hillstream Biopharma and is a paid consultant to Reata and CanBas. The other authors declared no potential conflicts of interest.
References
- 1.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med 2016;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 2017;171:34–57. [DOI] [PubMed] [Google Scholar]
- 3.Bracken AP, Brien GL, Verrijzer CP. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev 2019;33:936–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev 2017;42:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 2017;49:289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 2017;49:296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trizzino M, Barbieri E, Petracovici A, Wu S, Welsh SA, Owens TA, et al. The tumor suppressor ARID1A controls global transcription via pausing of RNA polymerase II. Cell Rep 2018;23:3933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol 2010;26:503–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenetics Chromatin 2019;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasumizu Y, Rajabi H, Jin C, Hata T, Pitroda S, Long MD, et al. MUC1-C drives lineage plasticity in progression to neuroendocrine prostate cancer. Nat Commun 2020;11:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajabi H, Hiraki M, Kufe D. MUC1-C activates polycomb repressive complexes and downregulates tumor suppressor genes in human cancer cells. Oncogene 2018;37:2079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, et al. MUC1-C activates BMI1 in human cancer cells. Oncogene 2017;36:2791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajabi H, Hiraki M, Tagde A, Alam M, Bouillez A, Christensen CL, et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep 2017;7:7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hata T, Rajabi H, Takahashi H, Yasumizu Y, Li W, Jin C, et al. MUC1-C activates the NuRD complex to drive dedifferentiation of triple-negative breast cancer cells. Cancer Res 2019;79:5711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Zhang N, Jin C, Long MD, Rajabi H, Yasumizu Y, et al. MUC1-C drives stemness in progression of colitis to colorectal cancer. JCI Insight 2020;5:137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kufe D. MUC1-C in chronic inflammation and carcinogenesis; emergence as a target for cancer treatment. Carcinogenesis 2020;41:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandoval GJ, Pulice JL, Pakula H, Schenone M, Takeda DY, Pop M, et al. Binding of TMPRSS2-ERG to BAF chromatin remodeling complexes mediates prostate oncogenesis. Mol Cell 2018;71:554–66 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther 2020;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeter CR, Yang T, Wang J, Chao HP, Tang DG. Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells 2015;33:2381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One 2015;10:e0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panchamoorthy G, Rehan H, Kharbanda A, Ahmad R, Kufe D. A monoclonal antibody against the oncogenic mucin 1 cytoplasmic domain. Hybridoma 2011;30:531–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, et al. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports 2014;3:460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatchalian J, Malik S, Ho J, Lee DS, Kelso TWR, Shokhirev MN, et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat Comms 2018;9:5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene 2013;32:1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther 2011;10:806–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol 2012;40:1643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA 2009;106:5181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA 2009;106:5187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008;40:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoyanova T, Riedinger M, Lin S, Faltermeier CM, Smith BA, Zhang KX, et al. Activation of Notch1 synergizes with multiple pathways in promoting castration-resistant prostate cancer. PNAS USA 2016;113:E6457–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu ZH, Dai XM, Du B. Hes1: a key role in stemness, metastasis and multidrug resistance. Cancer Biol Ther 2015;16:353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Sanders AJ, Liang G, Song E, Jiang WG, Gong C. HEY factors at the crossroad of tumorigenesis and clinical therapeutic modulation of HEY for anticancer treatment. Mol Cancer Ther 2017;16:775–86. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 2010;7:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 2011;13:903–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmad R, Rajabi H, Kosugi M, Joshi M, Alam M, Vasir B, et al. MUC1-C oncoprotein promotes STAT3 activation in an auto-inductive regulatory loop. Sci Signal 2011;4:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C, Allis CD. SWI/SNF complex in cancer. Nat Genet 2017;49:178–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Q, Lian JB, Stein JL, Stein GS, Nickerson JA, Imbalzano AN. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017;9:919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerrero-Martinez JA, Reyes JC. High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Sci Rep 2018;8:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010;18:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julian LM, Blais A. Transcriptional control of stem cell fate by E2Fs and pocket proteins. Front Genet 2015;6:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer 2019;19:326–38. [DOI] [PubMed] [Google Scholar]
- 42.Panamarova M, Cox A, Wicher KB, Butler R, Bulgakova N, Jeon S, et al. The BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development 2016;143:1271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien R, Marignol L. The Notch-1 receptor in prostate tumorigenesis. Cancer Treat Rev 2017;56:36–46. [DOI] [PubMed] [Google Scholar]
- 44.Choi HY, Siddique HR, Zheng M, Kou Y, Yeh DW, Machida T, et al. p53 destabilizing protein skews asymmetric division and enhances NOTCH activation to direct self-renewal of TICs. Nat Commun 2020;11:3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hainer SJ, Gu W, Carone BR, Landry BD, Rando OJ, Mello CC, et al. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev 2015;29:362–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iv Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. Int J Cancer 2014;135:2741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Sui Y, Ni J, Yang T. Insights into the Nanog gene: a propeller for stemness in primitive stem cells. Int J Biol Sci 2016;12:1372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene 2014;33:2655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quintanal-Villalonga A, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol 2020;17:360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data is GEO Submission GSE139335.