Abstract
Addiction is a chronic and relapsing brain disorder characterized by compulsive seeking despite adverse consequences. There are both heritable and epigenetic mechanisms underlying drug addiction. Emerging evidence suggests that non-coding RNAs (ncRNAs) such as microRNAs (miRNAs), long non-coding RNAs, and circular RNAs regulate synaptic plasticity and related behaviors caused by substances of abuse. These ncRNAs modify gene expression and may contribute to the behavioral phenotypes of addiction. Among the ncRNAs, the most widely researched and impactful are miRNAs. The goal in this systematic review is to provide a detailed account of recent research involving the role of miRNAs in addiction.
This article is categorized under:
RNA Interactions with Proteins and Other Molecules > Small Molecule-RNA Interactions
RNA in Disease and Development > RNA in Disease
Keywords: addiction, epigenetic, microRNA, non-coding RNA
1 | INTRODUCTION
Addiction is a chronic and relapsing brain disorder characterized by compulsive seeking. Both sporadic and genetic factors contribute to the negative symptoms associated with addiction, such as drug-seeking and relapse. The consequences of repeated exposure to drugs of abuse often lead to difference in feeling, preoccupant behavior, temporary satiation, loss of control, and negative consequences, all of which are symptoms of addiction (Sussman & Sussman, 2011). The easiest measure of addiction is a user’s behavioral preoccupancy with obtaining more drugs; in humans, this behavior is complex and can be challenging to measure, however, in animal models, addiction can be measured through Pavlovian-style classical conditioning such as conditioned placement preference (CPP) or operant conditioned lever-pressing self-administration (SA). Animal models have consistently shown that addiction will cause compulsive-like SA even in the presence of aversive consequences (Hopf & Lesscher, 2014; Hyman & Malenka, 2001; Vanderschuren & Everitt, 2004). The change from a positive reward, commonly called a “high,” to negative stimuli during abstinence is one of the hallmarks of addiction (Koob & Le Moal, 2001). These uncontrolled changes often contribute to the user’s inability to cease drug taking regardless of negative symptoms. Treating addiction disorders is a substantial challenge given the far-reaching consequences, which include medical, financial, and civil costs. In the United States alone, the cost of addiction to legal drugs surpasses tens of billions of dollars (Florence, Zhou, Luo, & Xu, 2016; Sacks, Gonzales, Bouchery, Tomedi, & Brewer, 2015; Xu, Bishop, Kennedy, Simpson, & Pechacek, 2015). Furthermore, according to the Centers for Disease Control and Prevention, the death toll from illicit substance poisoning is above 70,000 per year (Kochanek, 2019).
Understanding the molecular mechanisms underlying addiction, such as the regulation of gene expression, is critical. It is well established that altered gene expression affects function of individual neurons and their neural circuits, ultimately leading to the behavioral abnormalities observed in drug addicts (McClung & Nestler, 2008; Nestler, Hope, & Widnell, 1993). Alterations of gene expression can occur through regulation of transcription factors, chromatin structure, and small non-coding RNA (ncRNA). In this review, we will focus on the ability of ncRNA to transcriptionally regulate gene expression in the context of drug addiction. Although there are many classes of ncRNA—microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), piwi-interfering RNA, small nucleolar RNA, and small interfering RNA—the most abundant and widely studied are miRNAs, which will be the primary focus of this review article.
1.1 | Drugs of abuse and microRNA
miRNAs are small (~21–23 nucleotides) ncRNA transcripts that are capable of regulating gene expression at the post-transcriptional level (Lim et al., 2005). These were first discovered in Caenorhabditis elegans in 1993 (R. C. Lee, Feinbaum, & Ambros, 1993), and to date 1,917 miRNAs have been identified in humans (http://www.mirbase.org). However, less than a third of these miRNAs are robustly supported as miRNA genes (Fromm et al., 2015). miRNAs impact posttranscriptional regulation via destabilizing or cleaving mRNA, making them ideal tools for regulating gene expression (Fabian, Sonenberg, & Filipowicz, 2010). Furthermore, the silencing effect of miRNAs on their target genes is highly specific and allows them to precisely regulate physiological responses. These abilities have caused miRNAs to be the subject of research in a variety of neurological diseases, including addiction (Hammond, 2015). In humans, the brain expresses a greater variety of unique miRNAs than any other tissues, thus denoting their prominence in regulating development and plasticity of the brain (Adlakha & Saini, 2014). As of today, a PubMed search (“miRNA” and “addiction”) has yielded 5,602 manuscripts, of which 739 have been published within the last year.
Drugs of abuse can have a wide array of impacts and varying pharmacological mechanisms of action in the brain. However, a common connection associated with drugs of abuse is the ability to induce plasticity that impacts overall brain function, thus leading to addiction (Kauer & Malenka, 2007). Importantly, the number of uniquely expressed miRNAs in the brains of higher organisms is key in the development of the diverse cellular mechanisms behind cognitive function, namely plasticity (Z. Hu & Li, 2017; McCreight, Schneider, Wilburn, & Swanson, 2017). miRNAs have been heavily investigated to help understand how drugs of abuse affect the biological machinery to cause the learned behavioral and biochemical dependence on a drug. Some miRNAs are conserved across drugs of addiction and modalities, whereas others can be seen in specific drugs or drug classes. The most commonly researched drugs of abuse fall into three main classes depending on their physiologic effects: psychostimulants, opiates, and depressants. We present below the key miRNAs impacted by these different classes of drugs across different model systems.
2 | PSYCHOSTIMULANTS
Psychostimulants, whether illicit or legal, are the most widely abused class of drugs. The canonical reward pathway, or dopaminergic pathway, is conserved throughout most drugs of abuse, including psychostimulants, and is comprised of the activation of the dopaminergic neurons in the ventral tegmental area (VTA) and substantia nigra, which affect GABAergic neurons in the striatum (N. D. Volkow & Morales, 2015). The complex transition from reward to addiction also involves the activation of higher brain structures including the limbic system and pre-frontal cortex (PFC) (N. D. Volkow & Morales, 2015). Psychostimulants stimulate the central nervous system by enhancing or mimicking various catecholamine neurotransmitters such as dopamine (Treatment for Stimulant Use Disorders, 1999; Wise, 1978). Some psychostimulants are also capable of acting on serotonin receptors to induce hallucinogenic and psychogenic effects (Müller & Homberg, 2015); both of these pathways have been investigated for clinical applications of therapeutics. This review will focus on methamphetamine (METH), cocaine, and nicotine, the three most widely abused and highly addictive psychostimulants.
2.1 | Methamphetamine
METH is a potent psychostimulant known for its significant negative health effects such as hallucinations, neurological dysfunction, and accelerated aging. According to self-reported data from human studies as well as from studies utilizing rodent SA addiction models, METH is highly addictive and can cause drastic negative withdrawal symptoms even after short dosing periods (Kitamura, Wee, Specio, Koob, & Pulvirenti, 2006; Marshall, Belcher, Feinstein, & O’Dell, 2007). It has recently been shown that even a single dose of METH in rats drastically increased SA of METH after the fifth day of exposure, and this rate of increase persisted for 18 days (Jayanthi, Torres, Ladenheim, & Cadet, 2020). In humans, similarly increased drug craving can persist up to 6 weeks (Zorick et al., 2010). Given METH’s potential to induce dependence and relapse, elucidating the different miRNAs involved in these processes could provide a greater understanding of the mechanisms that make METH such a potently addictive drug and provide further insight into addiction to other drugs of abuse.
2.1.1 | miRNAs in METH abuse
One of the characteristic features of METH is that it can disrupt the blood brain barrier (BBB) with both chronic and acute use (Turowski & Kenny, 2015). Indeed, miR-125a-5p, miR-29b, and miR-143, which are associated with METH abuse, have been shown to increase BBB degradation (Bai et al., 2016). A significant clinical challenge when addressing METH addiction in humans is accurately characterizing the status of abuse and addiction via readily accessible biomarkers in plasma or serum. In humans, several circulating plasma miRNAs are differentially regulated in METH abusers. miR-4799, miR-4776, miR-550b, and miR-9 were increased in active METH users while miR-181a, miR-15b, miR-let-7e, and miR-let-7d were decreased significantly in recovering METH users (Gu et al., 2020; Y. Zhao et al., 2016). However, as observed in chronic models, different miRNAs were associated with chronic versus acute METH usage; therefore, future studies of plasma or serum samples should seek to identify potential biomarkers of addiction regarding the duration of drug use (i.e., chronic versus acute), not just drug-specific biomarkers.
In chronic versus acute usage of METH, distinct miRNA regulatory differences exist. One study revealed 132 miRNAs were differentially regulated in the nucleus accumbens (NAc) between a single acute dose and chronic escalating doses of METH as compared to control rats (Sim et al., 2017). Specifically, 32 miRNAs were uniquely regulated in acute groups, and 38 miRNAs were uniquely regulated in chronic groups. Intriguingly, the final dose prior to sacrifice was identical in both conditions, suggesting that METH dosage can affect the expression of miRNAs in NAc (Sim et al., 2017). Specifically, the study identified one miRNA, miR-496-3p, to be 13-fold increase in the chronic group (Sim et al., 2017). Though the exact role of miR-496-3p in addiction has not been explored, its role in learning and memory via activity-dependent regulation of dendritogenesis has been documented (Rago, Beattie, Taylor, & Winter, 2014; W. Wang, Kwon, & Tsai, 2012), indicating that miR-496-3p differential expression could affect synaptic plasticity and, therefore, behavior. Interestingly, both miR-496-3p and miR-496-5p were upregulated in a study of nicotine, further suggesting it may play a regulatory role in psychostimulant addiction (S. Lee, Woo, Kim, & Im, 2015).
In another METH SA rat study, Bosch, Benton, Macartney-Coxson, and Kivell (2015) revealed 78 miRNAs and 150 mRNA transcripts were differentially expressed in the VTA. Of significance is the identification of the miR-29 family, which was downregulated in the VTA. Intriguingly, a recent study from Su et al. showed similar downregulation of the miR-29 family in the NAc in a chronic rat model. Specifically, they showed that miR-29c inhibition attenuated METH-induced locomotor sensitization (Su et al., 2019). Furthermore, miRNA-29c has been shown to be an inhibitor of BACE1, a key enzyme in myelination that is also upregulated with METH exposure (Shukla, Maitra, Hernandez, Govitrapong, & Vincent, 2019; Zong et al., 2011). Inhibition of miR-29c during chronic exposure is likely the condition through which METH-induced increase of BACE1 occurs. Together, miR-29c’s differential regulation in chronic METH groups and its impact on locomotor sensitization indicate that it plays a role in reward addiction. Ultimately, behavioral studies using miRNA regulation in animal models, such as knockouts or miRNA-inhibition, are necessary to validate miRNAs as regulators of addiction. Such behaviorally validated miRNAs, like miR-29c, could elucidate other potential miRNAs for investigation and validation based on their mRNA targets.
Another interesting longitudinal study performed in METH SA rats found unique miRNA differences in the PFC related to duration of drug access (Du et al., 2016). During longer SA time periods of 6 hr, miR-127, miR-186, miR-222, and miR-24 were increased significantly while miR-329 was decreased (Du et al., 2016). During much shorter exposure times such as 1 hr, only miR-186 increased while miR-195 and miR-329 were decreased (Du et al., 2016). miR-24, miR-195, and miR-222 have been previously shown to target the apoptosis regulating gene Bcl2 (Singh & Saini, 2012). It is suggested by Du et al. that Bcl2-mediated apoptosis is promoted in the short access group via decreased miR-195 expression whereas Bcl2 is decreased in long access groups via increased miR-24, miR-195, and miR-222. Additionally, a KEGG pathway analysis by Du et al. (2016) showed several of the 6-hr specific miRNAs’ putative targets to be axon guidance, MAPK signaling, and Wnt signaling. This differential apoptotic regulation alongside an increase in synaptic plasticity-associated genes suggests that extended use of METH contributes to the learned behavior of addiction whereas the shorter exposure times do not. This study therefore provides insight on the role of miRNAs at a regulatory level. One of the most widely researched miRNAs from the Du et al. paper is miR-24, which is a poly-drug-linked miRNA associated in METH, oxycodone, and alcohol use. miR-24 targets p38 MAPK, a MAPK family related to the serotonergic sensitization that contributes heavily to addiction-associated behavior such as CPP, stress, and anxiety (Bruchas et al., 2011; El Rawas, Amaral, & Hofer, 2020).
Other studies of METH-associated miRNAs have focused on miR-128, which has key roles in learning and behavior. miR-128’s differential expression has been repeatedly noted in chronic cocaine and METH abuse (Hollander et al., 2010; J. Li et al., 2020; Lin et al., 2011; Sim et al., 2017). Recently, it was validated that only chronically exposed METH rats showed a significant difference in miR-128 expression in the NAc (J. Li et al., 2020). miR-128’s overexpression and under-expression were shown to exacerbate or mitigate METH-seeking, respectively (J. Li et al., 2020). Interestingly, Sim et al. (2017) reported dysregulation of miR-128 in the NAc with both acute and chronic METH treatments. Importantly, miR-128 targets three major proteins: Arf6, Cpeb3, and Nlgn1 (J. Li et al., 2020). These proteins are important for dendritic spine morphology, synaptic transmission, and neurogenesis, the three key modalities for synaptic plasticity (S. Choi et al., 2006; Y. S. Huang, Kan, Lin, & Richter, 2006; Südhof, 2008).
In summary, understanding the role of miRNAs in METH addiction will help in understanding how METH abuse dysregulates specific brain regions. Due to the severe neurotoxicity of METH, several biological responses from neurons and glia are elicited in all brain regions, however much of this may not directly contribute to addiction. METH studies have produced an astounding array of data regarding miRNA dysregulation, but for the purpose of studying addiction it is critical to understand which miRNAs play a role in the acquisition of METH-dependent behaviors such as drug seeking and withdrawal. The miRNAs addressed here that have the greatest promise for further investigation are miR-128 and miR-29c for their ability to affect METH-seeking behavior (J. Li et al., 2020; Su et al., 2019). However, there are other miRNAs that are noted in SA studies that deserve further investigation due to their targeted pathways, particularly miR-24 and miR-496-3p, both of which have roles in synaptic plasticity and learning behavior.
2.2 | Cocaine
Cocaine, like METH, pharmacologically operates through presynaptic monoamine receptors and is a highly addictive drug. The mechanism of addiction likely lies in synaptic plasticity and memory pathways (Vanderschuren & Everitt, 2004). The reward pathway of addiction is highly conserved through different drugs, as illustrated in Figures 1 and 2, and a few miRNAs appear to be conserved across multiple types of drugs, including cocaine. In addition to these conserved miRNAs, cocaine dependency (CD) has shown a variety of uniquely differentially regulated miRNAs.
FIGURE 1.
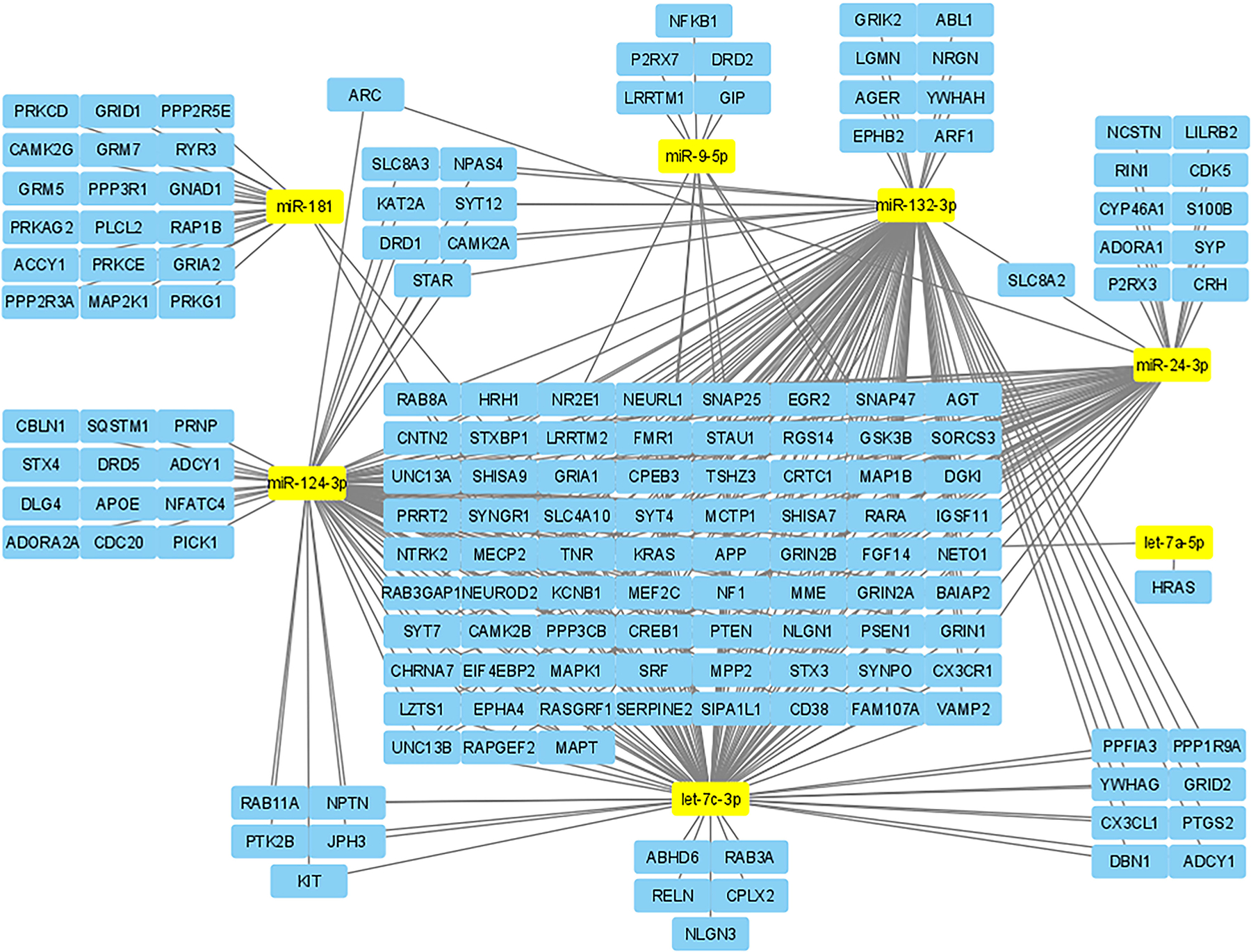
Using miRPathDB and Cytoscape, we mapped the seven miRNAs with the most poly-drug associations in this paper to putative target genes related to synaptic plasticity. This allows for a visual representation of the strength that some miRNAs have in synaptic plasticity and likely is also representative of their strength in regulating addiction
FIGURE 2.
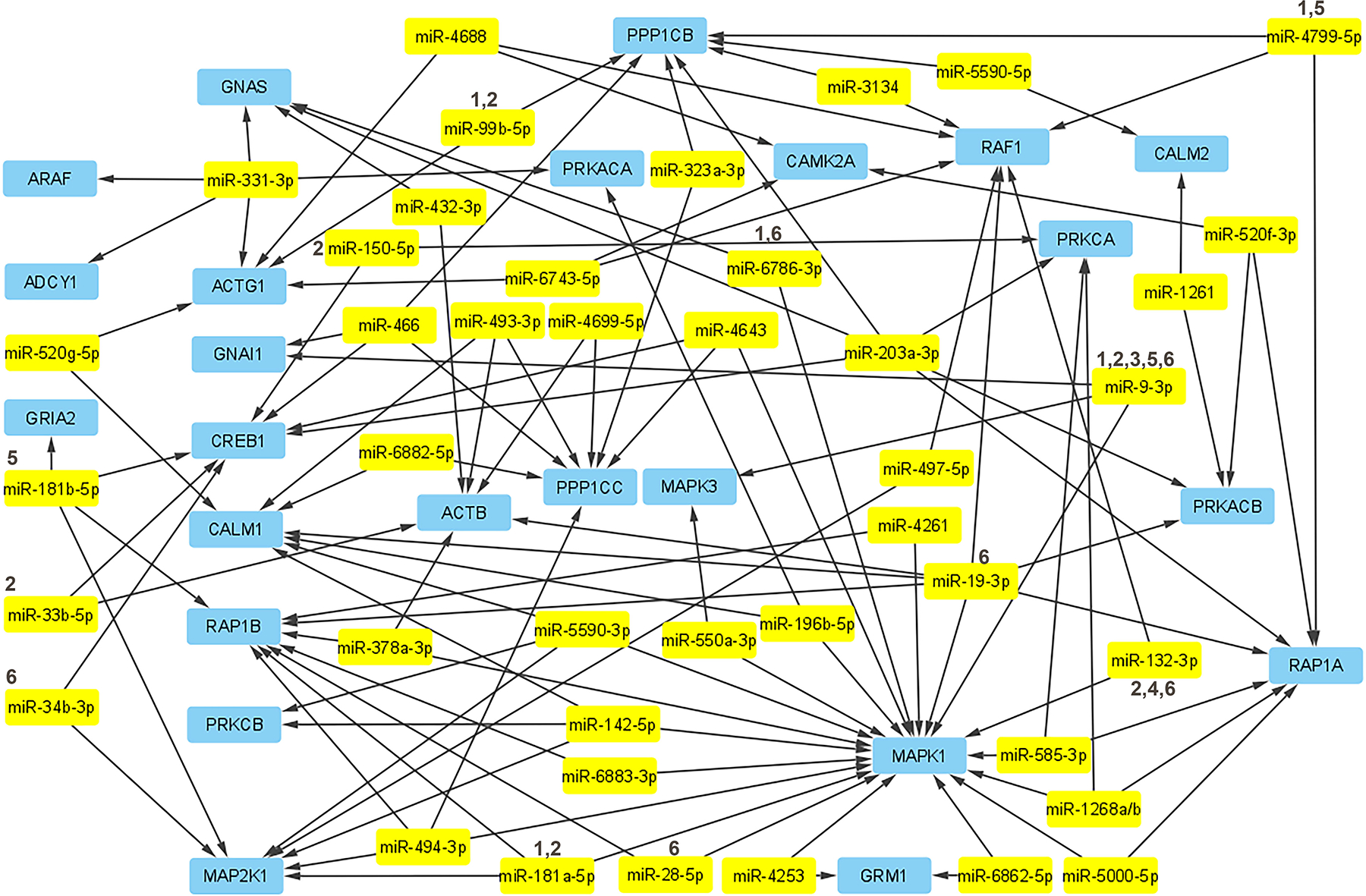
The underlying genes in the drug addiction pathway in relation to miRNAs experimentally linked to these genes have been mapped using Cytoscape. There are many more non-confirmed miRNAs that are putatively targeting these genes, but for the sake of validity and simplicity the map has been limited to those that were confirmed by miRPathDB to have experimental validity linking them to impacting these genes. The miRNAs which have experimental data linking them to a drug of abuse mentioned in this article have been denoted with a number according to the key above: (1) METH, (2) cocaine, (3) nicotine, (4) oxycodone, (5) morphine/heroin, (6) alcohol
2.2.1 | miRNAs in cocaine abuse
A study performed using the neuroblastoma SH-SY5Y cell line found that 30-min and 6-hr cocaine treatments revealed a total of 90 differentially expressed miRNAs (Cabana-Domínguez, Arenas, Cormand, & Fernàndez-Castillo, 2018). The most significantly downregulated of these were miR-9-5p, miR-101-3p, miR-124-3p, miR-124-5p, miR-137, miR-153-3p, and miR-369-3p. Following the identification of these miRNAs, the authors further investigated a human dataset comprised of 2,085 individuals with CD and 4,293 controls. Three of these miRNAs were confirmed to be associated with CD in humans; miR-124-3p, miR-153, and miR-9 were all significantly downregulated (Cabana-Domínguez et al., 2018). Interestingly, of these three miRNAs, miR-124-3p was shown to be a key regulator in cocaine-induced synaptic plasticity (Chandrasekar & Dreyer, 2009, 2011). Interestingly, miR-153 is widely known for playing a role in tumorigenesis, but more investigation is needed to establish its relevance in addiction (W. J. Chen et al., 2015; Ghasemi, Fallah, & Ansari, 2016; Xia et al., 2015). Although miR-9 has not been shown to directly regulate CD, it has been shown to directly target several key transcriptional regulators that control neurogenesis (Bonev, Stanley, & Papalopulu, 2012; Shibata, Nakao, Kiyonari, Abe, & Aizawa, 2011; Yao et al., 2014; C. Zhao, Sun, Li, & Shi, 2009). The role of miR-9 in activation of the NF-κB pathway and RE1-silencing transcription factor (REST), both of which have been shown to be directly involved in alcohol and METH addiction, further endorses it as a target of interest for CD (Ruffle, 2014; Sadakierska-Chudy et al., 2017; H. Tian et al., 2016).
Other studies using the SH-SY5Y cell line investigated an enzyme called poly[ADP-ribose] polymerase 1 (PARP-1) for its role in plasticity, memory, and CD (Cohen-Armon et al., 2004; Scobie et al., 2014). Furthering this line of research, an in vivo study of PARP-1 found it was modulated by miR-125b, a miRNA significantly downregulated with cocaine exposure (Dash et al., 2017). Increasing the dose of cocaine was shown to gradually decrease the expression of miR-125b, demonstrating the dose-dependent nature of addiction (Dash et al., 2017). Dash et al. (2020) further investigated PARP-1 and found that it also has significant binding overlap with miR-124. Although PARP-1 is regulated by both miR-125b and miR-124, miR-124 appears to be the primary regulator. It is also interesting to note that while CD downregulated both miR-125b and miR-124 in the NAc, only miR-124 appeared to be impacted in the hippocampus (HIP) and VTA (Chandrasekar & Dreyer, 2009; Dash et al., 2017; Dash et al., 2020).
Another in vivo experiment using mice found that miR-495, let-7, and miR-212/132 were significantly down-regulated in the NAc shell by SA of cocaine and showed that these miRNAs could regulate several canonical reward pathways (Bastle et al., 2018). Using cocaine infused with green fluorescent protein-tagged lentivirus for miR-495, Bastle et al. further showed that miR-495 was two-fold more enriched in the NAc in chronic SA mice. Furthermore, the same study showed that overexpression of miR-495 in the NAc shell recovered lever-pressing conditioning in CD rats (Bastle et al., 2018). It was also shown that miR-495 targeted the key genes BDNF and ARC as well as CAMK2A, a gene associated with long-term memory potentiation known to regulate CD in humans (Easton et al., 2014). The role of miR-495 in addiction-related behavior and its regulation of three key synaptogenesis-related genes mark it as a significant potential therapeutic target.
In a rat SA model of cocaine, it was shown that miR-212/132 family was differentially expressed in the striatum (Im, Hollander, Bali, & Kenny, 2010; Sadakierska-Chudy et al., 2017). miR-212 and miR-132 are transcribed from the same pre-miRNA (Aten, Hansen, Hoyt, & Obrietan, 2016). It has been shown that miR-212/132 is critical in CREB cycle-mediated synaptic plasticity and memory (Hollander et al., 2010). Hollander et al. (2010) showed that controlling striatal miR-212/132 levels in a SA rat model directly affected the development of cocaine addiction. A recently published study took this analysis a step further by illustrating two key points using an active and yoked SA model; the active subject could press a lever that would result in cocaine delivery to both the active and yoked subjects. If the yoked or saline cagemates pressed the lever, however, no cocaine would be administered. The first novel discovery was that altered expression of miR-212/132 extended through 10 days of cocaine abstinence for active group SA rats, but yoked condition cagemates did not show extended differential miR-212/132 expression despite receiving the same cocaine doses (Sadakierska-Chudy et al., 2017). This suggests that the duration of miR-212/132 dysregulation is dependent on the learned behavior of lever pressing not just cocaine exposure, further establishing miR-212/132 as a behavioral regulator of addiction. The second significant finding was that miR-212/132 impacted REST expression differentially in the active condition versus the yoked condition, suggesting that REST may be a key protein in the motivational formation of addiction (Sadakierska-Chudy et al., 2017). REST has also been implicated in CREB cycle-mediated synaptic plasticity associated with multiple drugs of abuse (Hollander et al., 2010; Jayanthi et al., 2014; Mavrikaki et al., 2019). These points highlight miR-212/132 as likely being key to the development of learned addiction as well as potentially contributing to relapse. These factors make miR-212/132 one of the most attractive targets for translationally relevant therapeutics regarding cocaine addiction and its negative outcomes.
Together these studies reveal that miRNAs play an important regulatory role in CD. The miRNAs responsible for mediating the behavior of addiction such as miR-212/132, miR-495, and miR-124 are the most relevant for future study. Many of these miRNAs, such as miR-124 are not only impactful in CD, but they are also associated with multiple drugs of abuse and several brain diseases. This widespread influence may suggest that novel miRNAs which impact similar disorders and pathways could merit further investigation. Particularly, miRNAs that are seen to be active across multiple drug modalities could be key in understanding addiction. Some of the miRNAs not fully established in addiction behavior paradigms are seen across several drugs of abuse.
2.3 | Nicotine
Nicotine is the major stimulant present in tobacco with a strong addictive potential. Typically taken through inhalation, nicotine accesses the brain in one of most direct drug delivery methods outside of injection. While commonly abused and less stigmatic than METH or cocaine, it is still a highly addictive drug of abuse (McLaughlin, Dani, & De Biasi, 2015). Nicotine acts as an agonist to the nicotinic acetylcholine receptors on dopaminergic neurons and, like cocaine and METH, primarily affects the mesocortolimbic dopamine system (Nisell, Nomikos, & Svensson, 1994). Despite the similarities in neural activation, the reinforcement of nicotine addiction may rely on different neural substrates as compared to other psychostimulants. One particular brain region involved in the reward-addiction pathway, known as the habenular nuclei, has been shown to be highly significant in mediating nicotine addiction (Baldwin, Alanis, & Salas, 2011). The habenular nuclei has a high density of nicotinic acetylcholine receptors that, when bound by nicotine, are responsible for mediating the release of various neurotransmitters, including dopamine (Grady et al., 2007; Jackson, Toma, Contreras, Alkhlaif, & Damaj, 2019). Although nicotine has some miRNA overlap with psychostimulants, it also presents a unique miRNA profile due to the differences in how it affects the brain.
2.3.1 | miRNAs in nicotine abuse
Recently, a mouse model of nicotine SA found that a total of 44 miRNAs were differentially regulated in the habenular nuclei (S. Lee et al., 2015). miR-496, which was also identified in a METH model of addiction, was found to be upregulated (S. Lee et al., 2015; Sim et al., 2017). Lee et al. also noted 6 other miRNAs that had a direct effect on 10 mRNA targets associated with addiction: miR-223-3p, miR-467c-3p, miR-669c-3p, miR-721, miR-3474, and miR-5621-3p. These miRNAs have not been experimentally shown to impact the behavior of addiction, however many of their target mRNAs, such as NF-κB, BDNF, and Mapk1, have been strongly implicated in addiction (Robison & Nestler, 2011). Another study considered miRNA expression in nicotine addiction as related to environment enrichment and found that nicotine-dosed rats in stimulus-enriched arenas had significantly upregulated levels of miR-221, miR-150, miR-202, miR-330, miR-380, miR-221, and miR-483 in the PFC (Gomez et al., 2015). Of these, the two most significantly upregulated were miR-483 and miR-221, both of which play a role in pathways heavily associated with depression and neurological disorders (Ausió, 2016; Chouri et al., 2018; N. Lian et al., 2018). Gomez et al. (2015) further validated that miR-221 was a key regulator of nicotine-associated locomotor sensitization. The findings by Gomez et al. identified miRNAs specifically in the PFC and suggested that, due to their putative targets, miR-483 and miR-221 might be related to the negative emotional effects of nicotine-induced depression, which impacts drug relapse (Kendler et al., 1993).
Furthermore, sex specific miRNAs have been identified with nicotine dependency. Using RNA-Seq analysis on the prefrontal cortex, Pittenger et al. (2018) identified miR-199a and miR-214 in response to nicotine dependency. The authors also noted a total of 23 differentially regulated miRNAs in female rats and only 21 in males, regardless of similar addictive tendencies during nicotine SA training. Whether or not this indicates that nicotine’s impact on miRNA expression is regulated differently in females versus males requires further investigation into these two miRNAs. Interestingly, miR-140-5p was highlighted in this study and in a study in perinatal nicotine exposure, where it was upregulated in males but not in females (Keller, Dragomir, Yantao, Akay, & Akay, 2018; Pittenger et al., 2018). Interestingly, several miRNA expression differences have also been noted in spermatozoa and seminal plasma: 21 upregulated and 7 downregulated miRNAs (Marczylo, Amoako, Konje, Gant, & Marczylo, 2012). Specifically, miR-146 was differentially regulated in sperm cells; the 5p strand was downregulated while the 3p strand was upregulated. Furthermore, many of the miRNAs noted by Marczyla et al. are associated with reproductive disorders and apoptosis signaling (J. Lian et al., 2009; C. Wang et al., 2011). Additionally, 18 of the identified miRNAs may play a role in the strongly observed epigenetic effects of nicotine exposure that may cause subsequent generations to be more susceptible to nicotine addiction (Marczylo et al., 2012; Taki, Pan, Lee, & Zhang, 2014).
Another interesting area of research in nicotine addiction is the effect of prenatal nicotine exposure and the potential for abuse in later stages of life (Agrawal et al., 2010). This prenatal exposure can cause affected infants to have an increased likelihood of addiction in later life (Levin et al., 2006). An in vivo rat model of perinatal nicotine exposure showed an upregulation of 58 miRNAs and downregulation of 16 miRNAs in the VTA neurons (Keller et al., 2018). Keller et al. post-validated miR-1224 and miR-6216 as significantly upregulated and miR-130a-3p and miR-9a-3p as significantly downregulated. While none of these miRNAs has been behaviorally confirmed to be relevant in nicotine addiction, miR-9a-3p has also been identified in cocaine and METH addiction, perhaps suggesting it for further nicotine addiction studies (Cabana-Domínguez et al., 2018; Gu et al., 2020). Among the miRNAs reported by Keller et al. (2018), it is important to note that, despite not being post-validated, miR-140-3p, miR-140-5p, and miR-125a-5p were also seen to be significantly upregulated in their microarray data. Another study on a fetal stem cell line found that miR-140-3p was significantly upregulated with nicotine exposure (Balaraman, Winzer-Serhan, & Miranda, 2012). In cell culture models, nicotine has been shown to affect Dnm1, a canonical protein coding gene responsible for endocytosis of synaptic terminals via upregulation of miR-140-3p (W. Huang & Li, 2009b; Mears et al., 2011). Upregulation of Dnm1 is heavily correlated with morphologically significant synaptic changes (Hwang & Li, 2006). This not only further validates miR-140-3p as impactful in nicotine addiction but also emphasizes that the dopaminergic pathway is critically impacted by nicotine via miRNA dysregulation (W. Huang & Li, 2009b). Interestingly, miR-125a-5p was previously associated with METH-related BBB degradation, and it may play a similar role in nicotine exposure (Bai et al., 2016; Hawkins et al., 2004). A study of placentas from nicotine-exposed mothers noted three significantly downregulated miRNAs: miR-21, miR-146a, and miR-182 (Maccani et al., 2010). Maccani et al. further validated these findings by exposing TCL-1 cells to nicotine and benzo(a)pyrene, another common chemical found in cigarettes. This revealed that miR-146a was significantly downregulated by both nicotine and benzo(a)pyrene, while miR-21 and miR-182 were affected only by nicotine exposure. The inclusion of this chemical is important as the plethora of chemicals in cigarettes may cause confounding results in human studies of nicotine abuse. Overall, nicotine exposure during development and how it can affect addiction in later life will likely be an increasingly active area of study.
miRNAs associated with nicotine addiction are often less researched in animal models when compared to other psychostimulants. This is likely due to nicotine being seen as less addictive than psychostimulants such as cocaine or METH. However, nicotine provides a perfect opportunity for psychostimulant studies of addiction since its use is prevalent among humans. Though the mechanism of addiction may not be entirely conserved, future nicotine studies may provide a deeper translational understanding of psychostimulant addiction.
3 | OPIOIDS
Narcotics such as oxycodone and other opioids are typically prescribed for pain management, but they are also frequently abused and known for their addictive potential (Kolodny et al., 2015; Silverstein, Silva, & Iberti, 1993). Unlike legal drugs of abuse such as nicotine and alcohol, these narcotics are prescribed and used in surgeries. In addition to pharmaceuticals in use today, several illegal opioids such as heroin and opium are also widely abused (Karbakhsh & Zandi, 2007; Rudd et al., 2014; Seth, Rudd, Noonan, & Haegerich, 2018). The unifying factor all of these opioids possess is that they act on opioid receptors that are widely dispersed in the brain and spinal cord, allowing for complex addiction pharmacodynamics. There are three principal opioid receptors (μ, κ, δ) that act on GABAergic neurons (Kosten & George, 2002). This activation occurs throughout the brain, but it is primarily studied in the canonical mesolimbic pathway (Kosten & George, 2002). On the surface, opioid addiction occurs in a very similar network to psychostimulants; however, the targets and substrates that mediate opioid addiction impact significantly different receptors. As such, the miRNA expression profile of addiction is highly varied from other classes of drugs. The most commonly abused opiates are morphine, heroin, and oxycodone.
3.1 | Morphine/heroin
Morphine and its semi-synthetic relative heroin are very closely related. While morphine is a naturally occurring and widely used painkiller, heroin is synthesized from morphine and used illicitly as a narcotic. Both are pharmacokinetically similar and will be discussed here together. Morphine and heroin operate predominantly as agonists to the μ-δ opioid receptor heteromer (Yekkirala, Kalyuzhny, & Portoghese, 2010). These receptors are spread throughout the brain, but most are concentrated in the posterior amygdala, hypothalamus, thalamus, and cortices (Pilapil, Welner, Magnan, Zamir, & Quirion, 1986).
3.1.1 | miRNAs in morphine/heroin abuse
In an in vivo study using a mouse model of morphine SA, 33 upregulated and 29 downregulated miRNAs were identified in the NAc (J. Kim, Im, & Moon, 2018). Many of these miRNAs operate in a network to collaboratively affect mRNA targets. The upregulated miR-32 and miR-451a were putatively found to co-regulate Nsmal as well as interact with miR-202 to co-regulate B3galt2 (J. Kim et al., 2018). The function of these genes in addiction requires further validation as not much is known at this time. The downregulated miR-20b and miR-193 shared four common targets: Arid4b, Vldlr, Fbxw11, and Ddhd1. Each of these possesses diverse functions and is not in the same canonical pathways despite being co-targeted by miR-20b and miR-193 in addiction. Kim et al. also identified a total of 7,752 mRNA involved in 43 different pathways that were putatively suggested to be impacted by the 62 identified miRNA. Of these, the most important and commonly suggested targets were involved in the Wnt and MAPK signaling pathways, which are ubiquitously altered in addiction, and long-term depression (J. Kim et al., 2018).
In a rat model of heroin addiction, miR-218 was found to be the most significantly differentially regulated miRNA between chronic saline and chronic heroin groups (Yan et al., 2017). This study focused on the NAc and showed that chronic doses of heroin significantly lowered expression of miR-218. It was also shown that lentiviral-mediated overexpression of miR-218 caused heroin-seeking behavior extinction (Yan et al., 2017). Putative targets of miR-218 include Gabrb3, GluR2, Dnmt3a, Ube3a, Nrxn1, Sema6b, Gng3, and Mecp2, all of which are highly relevant to synaptic plasticity. One study of psychostimulants in Mecp2 knockout and enhanced mice found that Mecp2-enhanced mice were resistant to CPP training while Mecp2 knockout mice learned CPP more readily (Deng et al., 2010). This suggests that miR-218 could be a primary regulator of heroin addiction in the NAc via downregulation of the transcription factor Mecp2 (Deng et al., 2010). Another previously discussed miRNA cluster, miRNA-132/212, is also a key regulator of opioid receptor expression via Mecp2 regulation (Garcia-Concejo, Jimenez-Gonzalez, & Rodríguez, 2016). With regard to Mecp2, the interactions among brain region specificity, miRNA modality, and drug of abuse may provide critical insight into the mechanisms of morphine addiction.
Opioids such as morphine are considered cofactors in HIV infection because they act synergistically with HIV proteins to enhance infection (Reddy, Pilakka-Kanthikeel, Saxena, Saiyed, & Nair, 2012). One study of the miRNAs upregulated in HIV-infected monocyte-derived macrophages treated with morphine found 14 downregulated and 12 upregulated miRNAs within a selected group of miRNAs that were predicted to target growth factors (Dave & Khalili, 2010). Of these, the most downregulated was miR-181b, which was previously addressed in both cocaine- and METH-exposed conditions (Chandrasekar & Dreyer, 2009; Y. Zhao et al., 2016). As in those examples, the putative targets of miR-181 (NFAT5, ITGA2, and CAMK2D) are implicated in inflammation via MCP-2 and IL-6 as well as Tbl1x, which targets the Wnt signaling pathway (Dave & Khalili, 2010). The most upregulated miRNA was miR-15b, which is putatively suggested to act on the Wnt signaling pathway via Btrc regulation (Dave & Khalili, 2010). Further investigation into these miRNAs as they impact the key Wnt signaling pathway, which is strongly associated with both memory and addiction (Dave & Khalili, 2010; Maguschak & Ressler, 2011).
miRNAs can also be shuttled to different areas of the body through extracellular vesicles (EV), which serve as important cell–cell communicators by transporting proteins, lipids, and small nucleotides (J. Tian, Casella, Zhang, Rostami, & Li, 2020). Though widely researched in the fields of oncology and pathology, EV research in the context of addiction is still in its infancy. A study of primary astrocyte cultures found that astrocyte-derived EVs treated with morphine had differentially regulated miRNAs. Most notably, miR-23, miR-98, and miR-138 were significantly upregulated while miR-221 was significantly downregulated (G. Hu et al., 2018). The majority of the upregulated miRNAs found in morphine-treated astrocyte EVs were thought to activate the TLR7/8 gene to cause NF-κB-induced overexpression of lincRNA-Cox2. Hu et al. showed that morphine-treated astrocyte-derived EVs could induce upregulation of lincRNA-Cox2 expression in microglia culture. Previous work on morphine’s ability to activate NF-κB signaling suggests that astrocytes, which possess a high quantity of mu opioid receptor (OPRM1), utilize lncRNA-Cox2 to facilitate the deleterious effects seen in morphine addiction (Welters et al., 2000). EVs provide a promising potential avenue for investigation for all drugs of abuse, but opioid studies in particular have made recent strides investigating miRNAs shuttled by EVs (Odegaard et al., 2020; Shahjin, Guda, et al., 2019).
3.2 | Oxycodone
Structurally similar to morphine, oxycodone has been established as a potent and widely abused prescription opioid. Designed for pain management, oxycodone operates as a full agonist to OPRM1, though it also has a low binding affinity to both the κ and δ receptor (Chahl, 1996). The binding of oxycodone to μ opioid receptors causes the subsequent inhibition of neurotransmitter release by decreasing cAMP production (Chahl, 1996). Importantly, oral doses of extended release of oxycodone are thought to be twice as potent as a similar dose of morphine (Freye, 2008). Oxycodone addiction is a clinically relevant and rapidly increasing field of research, particularly with regard to miRNA.
3.2.1 | miRNAs in oxycodone abuse
One study assessing oxycodone and hydromorphone, two opiates primarily used in a clinical setting, found several differentially regulated miRNAs (Toyama, Kiyosawa, Watanabe, & Ishizuka, 2017). This study found 22 upregulated and 32 downregulated miRNAs, of which 13 were increased and 14 were decreased in only oxycodone exposure. Of the increased miRNAs in oxycodone exposure, let-7, miR-146a, miR-24, and miR-221 stood out. Each of these miRNAs is differentially regulated in previously mentioned psychostimulant paradigms, but not always in a similar manner. miR-146a was upregulated in oxycodone exposure but downregulated significantly in placental nicotine exposure; miR-221 was also increased in in vivo experiments for both oxycodone and nicotine (Gomez et al., 2015; Maccani et al., 2010). Furthermore, miR-24 was increased in both METH SA and oxycodone exposure (Du et al., 2016). let-7 is also a reoccurring miRNA that is affected by SA in METH, cocaine, alcohol, and now opiates; for a full account of let-7’s interaction with drugs of abuse see Table 1. Additionally, another miRNA noted across several substances of abuse, miR-9, was found to increase SA of oxycodone in rats when overexpressed in the NAc (Mavrikaki et al., 2019). This is believed to be caused by miR-9’s inhibition of REST. REST is a transcriptional repressor shown to be relevant in opioid abuse via inhibition of the expression of dopamine receptor D2 (DRD2) and the OPRM1 (Chidambaran et al., 2017; Y.-M. Sun et al., 2005). Overexpression of miR-9 caused a significant increase in DRD2, which is suggested to be the cause of increased oxycodone SA (Mavrikaki et al., 2019). miR-9 should also be further investigated in heroin models of addiction, as it has been confirmed that DRD2 expression is increased in rats treated with increasing doses of heroin (Y. Li, Xia, Li, Yin, & Liang, 2017).
TABLE 1.
The miRNAs affected by the drugs of focus in this paper
| miRNA | Drug | Paradigm | Effect | Sample Source | Citation |
|---|---|---|---|---|---|
| 7a-3p | Alcohol | ↑ | PFC | Nunez et al. (2013) | |
| 9-3p | Nicotine | ↓ | VTA | Keller et al. (2018) | |
| 9-5p | Cocaine | ↓ | NAc | Cabana-Domínguez et al. (2018) | |
| Methamphetamine | ↓ | VTA | Bosch et al. (2015) | ||
| Morphine | ↓ | PFC | Tapocik et al. (2014) | ||
| Cocaine | ↑ | NAc | Eipper Mains et al. (2011) | ||
| Cocaine | ↓ | Striatum | |||
| Alcohol | ↑ | Pituitary, striatum, supraoptic nucleus | Pietrzykowski (2010) | ||
| Alcohol | ↓ | Fetal NSC | Balaraman et al. (2012) | ||
| 10b-5p | Alcohol | ↑ | Midbrain | Gorini, et al. (2013) | |
| 15b | Methamphetamine | ↓ | Plasma | Y. Zhao et al. (2016) | |
| Morphine | ↑ | Monocyte-derived macrophages | Dave and Khalili (2010) | ||
| 15b-3p | Alcohol | ↑ | PFC | Lewohl et al. (2011) | |
| 16 | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 19a-3p | Alcohol | ↓ | Cortex | Gorini, et al. (2013) | |
| 20b-5p | Morphine | ↓ | NAc | J. Kim et al. (2018) | |
| 23b-3p | Morphine | ↑ | Plasma | Wu et al. (2009) | |
| 24 | Methamphetamine | ↑ | PFC | Du et al. (2016) | |
| 24-3p | Oxycodone | ↑ | Plasma | Toyama et al. (2017) | |
| 24-3p/1-5p/2-5p | Alcohol | ↑ | Frontal Cortex | Nunez et al. (2013) | |
| 28-3p | Alcohol | ↓ | Midbrain | Gorini, et al. (2013) | |
| 29a/b | Cocaine | ↑ | Limbic forebrain, PFC | Lippi et al. (2011) | |
| Nicotine | ↑ | PFC, HIP | |||
| Methamphetamine | ↑ | Limbic Forebrain, HIP | |||
| 29a/b/c | Methamphetamine | ↓ | VTA | Bosch et al. (2015) | |
| 29c | Methamphetamine | ↓ | NAc | Su et al. (2019) | |
| 29a-3p/c-3p | Alcohol | ↓ | Ventral HIP | Asimes, Kim, Rao, Bartelt, and Pak (2019) | |
| 29b-3p/c-3p/a-5p | Methamphetamine | ↓ | NAc | Zhu et al. (2016) | |
| 32 | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 32-5p | Morphine | ↑ | NAc | J. Kim et al. (2018) | |
| 33 | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 34a-3p | Alcohol | ↓ | Ventral HIP | Asimes et al. (2019) | |
| 34b-3p/b-5p/c-5p | Alcohol | ↑ | NAc | Mamdani et al. (2015) | |
| 34c-5p | Alcohol | ↑ | Frontal cortex, PFC | Nunez et al. (2013) | |
| 54 | Nicotine | ↑ | C. elegans | Taki et al. (2014) | |
| 55 | Nicotine | ↓ | C. elegans | Taki et al. (2014) | |
| 58 | Nicotine | ↓ | C. elegans | Taki et al. (2014) | |
| 80 | Nicotine | ↑ | C. elegans | Taki et al. (2014) | |
| 99b | THC | ↑ | Intestine | Chandra et al. (2014) | |
| Cocaine | ↓ | HIP | Chen et al. (2013) | ||
| 101-3p | Cocaine | ↓ | NAc | Cabana-Domínguez et al. (2018) | |
| 124 | Alcohol | ↑ | Whole brain | Mizuo, Okazaki, Watanabe, Inoue, and Matsumoto (2016) | |
| Alcohol | ↑ | mPFC | Darcq et al. (2015) | ||
| 124a | Alcohol | ↓ | DLS | Bahi and Dreyer (2013) | |
| 124-3p/5p | Cocaine | ↓ | NAc, SH-SY5Y neuroblastoma cells | Cabana-Domínguez et al. (2018) | |
| 125a-5p | Methamphetamine | ↓ | VTA | Bosch et al. (2015) | |
| Alcohol | ↓ | PFC | H. Tian et al. (2016) | ||
| Nicotine | ↑ | VTA | Keller et al. (2018) | ||
| 125b | Cocaine | ↓ | NAc | Dash et al. (2017) | |
| 127 | Methamphetamine | ↑ | PFC | Du et al. (2016) | |
| 127-5p | Alcohol | ↑ | Cortex | Gorini, et al. (2013) | |
| 128 | Methamphetamine | ↑ | NAc | J. Li et al. (2020), Lin et al. (2011) | |
| 128b | Cocaine | ↑ | Striatum | Hollander et al. (2010) | |
| 130a-3p | Nicotine | ↓ | VTA | Keller et al. (2018) | |
| 130b-3p | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 132 | Cocaine | ↑ | Striatum | Sadakierska-Chudy et al. (2017) | |
| Alcohol | ↑ | Whole brain | Mizuo et al. (2016) | ||
| 132-3p | Opioids | ↑ | Zebrafish embryos | Chiarlone et al. (2016) | |
| Alcohol | ↑ | NAc | Mamdani et al. (2015) | ||
| 134-5p | Alcohol | ↑ | NAc | Mamdani et al. (2015) | |
| 137 | Cocaine | ↓ | NAc | Cabana-Domínguez et al. (2018) | |
| 140-3p | Nicotine | ↑ | PC12 cells | W. Huang and Li (2009b) | |
| Nicotine | ↑ | Fetal NSC | Balaraman et al. (2012) | ||
| Alcohol | ↓ | Fetal NSC | |||
| Alcohol | ↓ | Cortex, midbrain | Gorini, et al. (2013) | ||
| 140-5p | Nicotine | ↑ | PFC | Pittenger et al. (2018) | |
| 145 | Methamphetamine | ↓ | VTA | Bosch et al. (2015) | |
| 150-5p | Cocaine | ↓ | Striatum | Eipper Mains et al. (2011) | |
| 152 | Alcohol | ↑ | PFC | Lewohl et al. (2011) | |
| 153-3p | Cocaine | ↓ | NAc | Cabana-Domínguez et al. (2018) | |
| 181a | Methamphetamine | ↓ | Plasma | Y. Zhao et al. (2016) | |
| Cocaine | ↑ | NAc, CPU, HIP | Chandrasekar and Dreyer (2009, 2011) | ||
| 181b | Morphine | ↓ | Monocyte-derived macrophages | Dave and Khalil (2010) | |
| 186 | Methamphetamine | ↑ | PFC | Du et al. (2016) | |
| 190 | Ketamine/Morphine | ↓ | Primary HIP rat culture | Zheng et al. (2010) | |
| 190-5p | Alcohol | ↑ | Cortex | Gorini, et al. (2013) | |
| 193-3p | Morphine | ↓ | NAc | J. Kim et al. (2018) | |
| 195 | Methamphetamine | ↓ | PFC | Du et al. (2016) | |
| Alcohol | ↑ | PFC | Darcq et al. (2015) | ||
| 199a-5p | Nicotine | ↑ | PFC | Pittenger et al. (2018) | |
| 200a/b/c | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 202 | Nicotine | ↑ | PFC | Gomez et al. (2015) | |
| 202-5p | Morphine | ↑ | NAc | J. Kim et al. (2018) | |
| 204 | Nicotine | ↑ | Choroid plexus | Lallai et al. (2019) | |
| 204-5p | Nicotine | ↓ | VTA | Keller et al. (2018) | |
| 206 | Alcohol | ↑ | mPFC | Tapocik et al. (2014) | |
| 210 | Nicotine | ↑ | Whole brain | L. Wang et al. (2017) | |
| 212 | Cocaine | ↑ | Striatum | Sadakierska-Chudy et al. (2017) | |
| 212-3p | Alcohol | ↑ | NAc | Mamdani et al. (2015) | |
| 214-3p | Nicotine | ↑ | PFC | Pittenger et al. (2018) | |
| 218 | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 221 | Nicotine | ↑ | PFC | Gomez et al. (2015) | |
| 221-3p | Oxycodone | ↑ | Plasma | Toyama et al. (2017) | |
| 222 | Methamphetamine | ↑ | PFC | Du et al. (2016) | |
| 223-3p | Nicotine | ↓ | LHb | S. Lee et al. (2015) | |
| Nicotine | ↑ | MHb | |||
| 329 | Methamphetamine | ↓ | PFC | Du et al. (2016) | |
| 330 | Nicotine | ↑ | PFC | Gomez et al. (2015) | |
| 335 | Alcohol | ↓ | Fetal NSC | Balaraman et al. (2012) | |
| 346-3p | Alcohol | ↓ | Midbrain | Gorini, et al. (2013) | |
| 361-3p | Alcohol | ↑ | Midbrain | Gorini, et al. (2013) | |
| 369-3p | Cocaine | ↓ | NAc | Cabana-Domínguez et al. (2018) | |
| 370 | Cocaine | ↓ | Striatal PSD | Eipper Mains et al. (2011) | |
| 380 | Nicotine | ↑ | PFC | Gomez et al. (2015) | |
| 382 | Alcohol | ↓ | Nac | J. Li et al. (2013) | |
| 429 | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 451a | Morphine | ↑ | NAc | J. Kim et al. (2018) | |
| 483 | Nicotine | ↑ | PFC | Gomez et al. (2015) | |
| 488-3p | Alcohol | ↑ | Cortex, midbrain | Gorini, et al. (2013) | |
| 495 | Cocaine | ↓ | NAc | Bastle et al. (2018) | |
| 496-3p | Methamphetamine | ↑ | NAc | Sim et al. (2017) | |
| 496a-5p | Nicotine | ↑ | Habenular nuclei | S. Lee et al. (2015) | |
| 582-5p | Cocaine | ↑ | Striatal PSD | Eipper Mains et al. (2011) | |
| 669c-3p | Nicotine | ↓ | LHb | S. Lee et al. (2015) | |
| Nicotine | ↑ | MHb | |||
| 721 | Nicotine | ↓ | MHb | S. Lee et al. (2015) | |
| Nicotine | ↑ | LHb | |||
| 770-5p | Cocaine | ↓ | Striatal PSD | Eipper Mains et al. (2011) | |
| 1224 | Nicotine | ↑ | VTA | Keller et al. (2018) | |
| 1224-3p | Alcohol | ↓ | Midbrain | Gorini, et al. (2013) | |
| 3474 | Nicotine | ↑ | LHb | S. Lee et al. (2015) | |
| Nicotine | ↓ | MHb | |||
| 4799-5p | Methamphetamine | ↑ | Serum | Gu et al. (2020) | |
| 5621-3p | Nicotine | ↑ | LHb | S. Lee et al. (2015) | |
| Nicotine | ↓ | MHb | |||
| 6216 | Nicotine | ↑ | VTA | Keller et al. (2018) | |
| Let-7c | Cocaine | ↓ | NAc, CPU, HIP | Chandrasekar and Dreyer (2009, 2011) | |
| Methanandamide | ↑ | SH-SY5Y neuroblastoma cells | Chiarlone et al. (2016) | ||
| Alcohol | ↑ | Frontal cortex, PFC | Nunez et al. (2013) | ||
| Hydromorphone | ↑ | Plasma | Toyama et al. (2017) | ||
| Let-7d/7e | Methamphetamine | ↓ | Plasma | Y. Zhao et al. (2016) | |
| Let-7b-3p | Hydromorphone | ↓ | Plasma | Toyama et al. (2017) | |
| Let-7a-5p | Oxycodone | ↑ | Plasma | Toyama et al. (2017) | |
| Let-7d-5p/7e-5p/7f-5p | Hydromorphone | ↑ | Plasma | Toyama et al. (2017) | |
| Let-7g-5p | Oxycodone | ↓ | Plasma | Toyama et al. (2017) |
Oxycodone, like morphine, has also been shown to alter the miRNA cargo of EVs. In a rat study, in utero and postnatal exposure to oxycodone resulted in unique, differentially expressed miRNA signatures in brain-derived EVs (BDEs) (Shahjin, Guda, et al., 2019). Among these, miR-504, which is expressed in the cortical and hippocampal regions of the human brain, was upregulated in rats treated in utero. Interestingly, miR-504 is involved in the regulation of hippocampal dendritic spine density (S.-Y. Choi et al., 2015), downregulation of DRD2 in the NAc in response to maternal deprivation (Zhang et al., 2013), and modulation of DRD1 expression by a DRD1 polymorphism (W. Huang & Li, 2009a; Jiménez, Pereira-Morales, & Forero, 2018). Indeed, when primary neuron cultures were treated with BDEs from the in utero oxycodone group, there was a significant reduction in spine density, an effect that may possibly be attributed to the upregulation of miR-504 in the in utero exposure group BDEs.
Oxycodone abuse is a growing focus for research largely due to the current ongoing opioid epidemic (Manchikanti et al., 2012). miRNAs may be critical regulators behind the transition of intermittent use to abuse of oxycodone. The continued study of these differentially regulated miRNAs is clinically important; a miRNA therapy could potentially halt or mitigate the transition from use to abuse. One such therapy could lie in the recent research regarding EV-mediated miRNA transport. This emerging field could have drastic implications across several drugs of abuse (Rao, O’Connell, & Finnerty, 2018).
4 | DEPRESSANTS
Depressants are named for their ability to impair and depress both behavioral and mechanical functions. Alcohol, which is similar to nicotine in legality, is one of the most abused drugs in the world. Δ9-tetrahydrocannabinol (THC), the primary psychoactive component in marijuana, has also received significant attention recently due to its increasing social and legal acceptance worldwide. THC is sometimes considered a psychostimulant drug, but functionally it has been observed to depress reaction to stimuli and will be considered as a depressant for this review. While both of these are described canonically as depressants, their methods of action in the brain are very different. Interestingly, we see some small conserved effects in miRNAs between alcohol and THC that are in contrast to those identified in the stimulant studies.
4.1 | Alcohol
Alcohol exerts its psychoactive effects on the brain via complex interactions with the lipid bilayer (Ingólfsson & Andersen, 2011) and by interacting with membrane proteins directly (Treistman & Martin, 2009). Unlike most of the other drugs mentioned in this paper, alcohol requires a relatively high concentration to take effect. However, alcohol’s effects on the brain are widespread, affecting GABA receptors, glutamate receptors, NMDA receptors, nicotinic acetylcholine receptors, and serotonin receptors (Narahashi et al., 2001; Wu, Gao, & Taylor, 2014). Acute alcohol use increases extracellular dopamine release and alters synaptic plasticity (Grace, 2000). Interestingly, acute and chronic alcohol paradigms have identified many miRNAs linked to alcohol exposure that coincide with other drugs of abuse. This is likely due to alcohol’s widespread mechanism of action. Despite alcohol’s relatively low potency, alcohol addiction has a robust effect on miRNA regulation. Analysis of miRNA expression in alcohol addiction in specific brain regions may provide insight into alcohol’s mechanism of action.
4.1.1 | miRNAs in alcohol abuse
In studies of fetal and adult alcohol exposure, miR-9 was upregulated in the pituitary, supraoptic nucleus, and striatum. (Omkaram, Shaima, Olivia, & Dipak, 2017; Pietrzykowski et al., 2008). miR-9 is a brain-enriched miRNA that plays a role in both neurogenesis and synaptic plasticity (Krichevsky, King, Donahue, Khrapko, & Kosik, 2003; Pietrzykowski, 2010). Targets of miR-9 that are relevant to alcohol addiction include the large-conductance calcium-and-voltage-activated potassium (BK) channel and DRD2. The BK channel, which is itself a target for alcohol (Treistman & Martin, 2009), is expressed throughout the body. The α subunit of the BK channel is ubiquitously expressed in the brain, and the channel plays a key role in neurotransmitter release and dendritic excitability (Martin et al., 2004). Several mechanisms involving tolerance levels and durations of exposure have been proposed to explain how the BK channel contributes to molecular alcohol tolerance (Treistman & Martin, 2009). One possible mechanism involves miR-9 selectively inhibiting isoforms of the BK channel that have lower tolerance. miR-9 allows more tolerant isoforms of the BK channel to remain, thus contributing to overall alcohol tolerance, which increases the amount of alcohol required to achieve similar levels of sensation.
As discussed in the opioid section, DRD2 is thought to play a role in the development of addiction for several substances (Noble, 2000). One study found that chronic alcohol use in rats led to reduced mRNA levels of the long DRD2 isoform in the NAc (Feltmann et al., 2018). As miR-9 has previously been shown to regulate DRD2 expression (Shi et al., 2014), the upregulation of miR-9 with alcohol exposure may result in this reduced expression of DRD2 mRNA.
miR-124, another miRNA that has been linked to alcohol exposure in several studies, is involved in neuronal plasticity (Smith & Kenny, 2018). In a mouse model of acute ethanol exposure, miR-124 and miR-132 were found to be significantly upregulated across the whole brain 1 hr after ethanol administration and remained elevated for a full 12 hr (Mizuo et al., 2016). The researchers observed that the increase of miR-124 and miR-132 was caused by histone H3 acetylation mediated by the decrease of HDAC4 (Mizuo et al., 2016). A similar pattern of alcohol-mediated H3 acetylation and HDAC4 inhibition was also previously observed in the rat amygdala (Pandey, Ugale, Zhang, Tang, & Prakash, 2008). Upon further investigation of miR-124, researchers found miR-124 downregulation, as well as upregulation of its target BDNF, in the dorsolateral striatum (DLS) of rats after 2 weeks of ethanol exposure (Bahi & Dreyer, 2013). This study also observed that lentiviral-mediated upregulation of miR-124 enhanced CPP and alcohol intake; further, overexpression of BDNF caused reduced CPP and alcohol intake (Bahi & Dreyer, 2013). The authors proposed that continuous exposure to alcohol attenuated miR-124 expression and subsequently amplified BDNF expression. This hypothesis is given further weight by an additional study that showed a difference in alcohol consumption linked to BDNF infusions in the DLS (Jeanblanc et al., 2009). This difference in consumption is believed to be caused by the binding of BDNF to its receptor TrkB, which attenuates alcohol SA (Jeanblanc et al., 2006). Alcohol administration significantly dysregulates miR-124 expression across several brain regions, and miR-124 plays a direct role in alcohol addiction-related behavior. These data, along with miR-124’s role in other drugs of abuse, suggest that it is a significant global mediator of drug addiction.
Another study implicated miR-124, along with miR-30a-5p, miR-195, and miR-1, in the modulation of BDNF in the medial PFC (mPFC) during binge drinking and abstinence (Darcq et al., 2015); miR-195 and miR-30a-5p were upregulated after binge drinking and after a 24-hr abstinence period. Furthermore, miR-124 expression was increased after abstinence while miR-1 was only upregulated during binge drinking. This is interesting as miR-124’s upregulation in abstinence means that it could play a role in alcohol relapse. Furthermore, moderate levels of alcohol consumption led to no significant change in the expression of BDNF or any of the previously mentioned miRNAs studied, suggesting that binge drinking may trigger their expression. In addition to miR-124, miR-206 has also been shown to regulate BDNF. Furthermore, after 3 weeks of alcohol exposure, rats going through withdrawal had upregulated miR-206 and downregulated BDNF in the mPFC, but not in the NAc, VTA, or amygdala (Tapocik et al., 2014). The same study found that BDNF attenuation was replicated in rats that were treated with viral-mediated overexpressed miR-206 in the mPFC; alcohol SA increased in these animals after treatment. Together, these data provide evidence for BDNF’s role in controlling the consumption of alcohol in the mPFC and the DLS.
Bioinformatics studies have also linked the miR-34 family to chronic alcohol exposure; following alcohol exposure, miR-34 was upregulated in the NAc of humans (Mamdani et al., 2015) and in the PFC of mice (Nunez et al., 2013). miR-34 was downregulated in the ventral HIP in rats (Asimes et al., 2019). miR-134, miR-212, and miR-132 were also found to be differentially expressed in the NAc (Mamdani et al., 2015). miR-34 and miR-134 are both capable of targeting SIRT1, which influences the learning and memory process (Ferguson et al., 2013; Gao et al., 2010; Zovoilis et al., 2011). Although highlighted here in alcohol studies, miR-132 and miR-212 also play a role in CD (Smith & Kenny, 2018).
As with opioids, EVs appear to play a role in alcohol addiction. A study by Tseng et al. (2019) found that, after 3 days of alcohol exposure, EVs released from fetal neural stem cells and the neural stem cells themselves contained miRNAs that were differentially expressed. Among their findings, miR-140, which is also sensitive to nicotine exposure, was the most overexpressed, resulting in increased proliferation of neural stem cells. Much like in the Darcq et al. study, miR-195, miR-30a-5p, and miR-30c were upregulated. In addition to this study of fetal samples, several differentially regulated miRNAs have also been observed in the serum of alcohol-abusing human mothers: miR-122, miR-126, miR-216b, miR-221, miR-3119, miR-3942, miR-4704, miR-4743, miR-514, and miR-602 (Gardiner et al., 2016). Studies into maternal alcohol use and fetal alcohol exposure are critical, as many miRNAs have effects in neuronal differentiation and may impact addiction and development in utero (Lau & Hudson, 2010).
Studies of miRNAs in alcohol abuse are growing in the field. Additionally, both miR-206 and miR-124 have promise for further investigation as they can directly regulate alcohol-seeking behavior (Bahi & Dreyer, 2013; Tapocik et al., 2014). miR-206 and miR-124 both act to regulate BDNF, and, as seen in both psychostimulants and opioids, BDNF-regulating miRNAs play a significant role in addiction behavior. Alcohol’s wide range of targets in the brain, including similar targets to both psychostimulants and opioids, indicates that some of these miRNA regulatory pathways could be conserved across several drugs of abuse. Further alcohol addiction studies into miR-9, miR-let7, and miR-212/132, miRNAs seen across psychostimulants, opioids, and depressants, may identify them as conserved regulators of addiction.
4.2 | Cannabis
Currently, literature in the context of cannabis addiction and miRNA expression is very scarce. THC is the primary active component in cannabis, the other being cannabidiol. THC mainly binds to the cannabinoid 1 (CB1) receptor. Evidence suggests that THC-mediated dopamine release occurs in the striatum, and THC alters neural components related to salience processing and the mesolimbic pathway (Zehra et al., 2018). Rodent models show that chronic THC exposure leads to downregulation of CB1 receptors; receptor levels in all regions of the brain except the HIP returned to baseline after 4 weeks of abstinence, however (Hirvonen et al., 2012).
4.2.1 | miRNAs in cannabis abuse
The results of a study by Egervari et al. suggest that chronic cannabis usage could interfere with the expression of miR-365. This study provided evidence that the single nucleotide polymorphism (SNP) rs2235749, a genetic variant that codes for the hormone prodynorphin (PDYN), alters PDYN expression by impairing the binding of miR-365 with the 3′-UTR of PDYN in the striatum (Egervari et al., 2016). This study found that the Pdyn-miR365 interaction in the NAcSh directly influenced novelty-seeking exploratory behavior and facilitated SA. Specifically, cannabis users with rs2235749 had higher novelty-seeking traits. While there appeared to be a relationship between miR-365 and PDYN in novelty-seeking and reinforcement decision-making, the authors noted that it was unclear how cannabis exposure contributed to these effects; they proposed follow-up studies of cannabis exposure and PDYN expression in translational animal models. This proposed future study has merit, as striatal PDYN expression changes have been linked to alcoholism (Guitart-Masip et al., 2006), opioid dependence (Clarke, Krause, Li, & Schumann, 2009), and CD (Yuferov et al., 2009). With the overlap of miRNAs and differential expression of their targets across different substances of abuse, there may be a common addiction pathway in the brain that is vulnerable to substance-specific alterations in miRNAs.
5 | DISCUSSION
This article summarized known miRNAs involved in drug addiction that may have potential in diagnosing and treating addition in the future. Although this review found some miRNAs that were specific to a single substance of abuse, other miRNAs, like the miR-132/212 family, miR-124, and miR-9, were differentially expressed in multiple drugs of abuse. These miRNAs appear to be pleiotropic in nature, and their expression can differ depending on the type of tissue, anatomical region, stage of development, and the presence or absence of other factors/molecules in their environment. In the context of addiction, their expression can be further altered by the use of addictive substances, the type of substance, and the paradigm (e.g., acute vs. chronic) through which those substances are ingested.
miRNA levels have been shown to differ with acute and chronic use, indicating that miRNAs behave in a manner almost comparable to that of a protein–protein interaction network (Zou, Li, Song, Zeng, & Wang, 2015), meaning that miRNA isoforms implicated in multiple substances of abuse and brain regions (e.g., let-7, miR-212/132) potentially act as regulatory hubs for addictive behavior. Evidence further suggests that miRNA strands act competitively, behaving in a hierarchical manner, and some miRNA targets may have stronger binding affinities than others (Clevers, 2006; Negi & Chan, 2017; Rzepiela et al., 2018).
Future research in the field of addiction biology should continue to investigate miRNAs with a greater focus on how miRNA expression changes over time. In particular, emphasis should be placed on understanding the difference in miRNA expression during acute and chronic drug usage, as well as how expression changes during drug abstinence and relapse. Another avenue of study for future research concerns polydrug usage. Known polydrug studies explore the usage of alcohol, nicotine, and occasionally cannabis in various combinations. However, of particular interest is whether miRNAs are implicated in multiple drugs of abuse and whether they are further up- or down-regulated in the presence of more than one substance. Because miRNAs are associated with alleged gateway drugs such as nicotine, alcohol, marijuana, and opiates, understanding their role in poly-drug abuse is essential. To highlight prospective poly-drug links, we have created a map featuring the miRNAs affected by drugs of abuse in the context of synaptic plasticity pathway genes (Figure 2). Identifying these miRNAs and their associated pathways allows for the potential development of therapeutics for the behavior of addiction.
Presently, several pre-clinical studies are being performed using miRNAs as biomarkers or therapeutics for diseases, but no clinical trials, and few pre-clinical trials, exist specifically for addiction (Creed, 2018; Dennis et al., 2020; Edemann-Callesen, Barak, Hadar, & Winter, 2020; Whyte, Torregrossa, Barker, & Gourley, 2018). Many drug abuse studies rely on self-reported data, and reliable diagnostics are difficult and expensive (Nora D. Volkow, Koob, & Baler, 2015). Biomarkers would remedy these issues and are therefore one of the most investigated subjects at the moment. Despite many challenges, the pre-clinical studies utilizing small RNA therapies show promise for the use of miRNAs in addiction therapeutics. One of the biggest challenges in utilizing miRNAs is that they often exert a wide range of downstream effects, as shown in Table 1. Indeed, promising work with circRNAs has shown their ability to act as a miRNA sponge and potentially block negative effects associated with an increase in a specific miRNA (J. Li et al., 2019). However, as with miRNAs, circRNAs can have unintended effects. miRNA effects are widely varied depending on the system and tissues involved, so absorption of miRNAs by circRNAs may pose issues in therapy development. Aside from potential unintended effects, miRNA delivery in humans has been challenging due to extant nucleases and passage through phospholipid bilayers (Fu, Chen, & Huang, 2019). One popular method for overcoming the delivery hurdle is using inorganic carriers such as gold, mesoporous silicon, graphene oxide, and iron oxide nanoparticles to pass phospholipid bilayers (Y. Chen, Xianyu, & Jiang, 2017; H. Y. Kim et al., 2017; Y. Li et al., 2018; S. Sun et al., 2017). Other successful vectors for miRNAs include naturally occurring and synthetic lipid nanocarriers. Naturally occurring EVs, such as microvesicles and exosomes, are also very promising candidates for future research as they can be both bio-labeled and receptor-targeted while having low toxicity and antigenicity (Shahjin, Chand, & Yelamanchili, 2019). Another key benefit that prompts EV research for drug delivery is their ability to pass the blood brain barrier, which is incredibly pertinent for addiction therapeutics (Fu et al., 2019; Shahjin, Chand, et al., 2019).
A large amount of data regarding miRNAs associated with drugs of addiction has been generated over the years. Recent research listed here has focused on addiction-specific scenarios such as SA, withdrawal, and relapse. Future research should address specific elements of addiction or amelioration of the negative effects of addiction. Another important consideration for research are the time-specific miRNA changes in addiction models and the subsequent miRNA activity as drug-taking transitions into drug-seeking behavior, dependency, and withdrawal. From a therapeutic standpoint, promising research uses miRNA vectors for disease amelioration, and these vectors have been successfully applied in some models of addiction mentioned above. Overall, pre-clinical work regarding miRNA treatment of drug addiction is still in its infancy and rapid progress is being made.
ACKNOWLEDGMENTS
The authorship of this review was supported by National Institutes of Health grants R01DA042379, R21DA046855 awarded to Sowmya V. Yelamanchili and R01DA046852 awarded to Gurudutt Pendyala and Sowmya V. Yelamanchili.
Funding information
Center for Scientific Review, Grant/Award Numbers: R01DA042379, R01DA046852, R21DA046855
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
REFERENCES
- Adlakha YK, & Saini N (2014). Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Molecular Cancer, 13, 33–33. 10.1186/1476-4598-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Scherrer JF, Grant JD, Sartor CE, Pergadia ML, Duncan AE, … Bucholz KK (2010). The effects of maternal smoking during pregnancy on offspring outcomes. Preventive Medicine, 50(1–2), 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimes A, Kim CK, Rao YS, Bartelt K, & Pak TR (2019). microRNA expression profiles in the ventral hippocampus during pubertal development and the impact of peri-pubertal binge alcohol exposure. Non-coding RNA, 5(1), 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten S, Hansen KF, Hoyt KR, & Obrietan K (2016). The miR-132/212 locus: A complex regulator of neuronal plasticity, gene expression and cognition. RNA and Disease (Houston, Texas), 3(2), e1375. [PMC free article] [PubMed] [Google Scholar]
- Ausió J (2016). MeCP2 and the enigmatic organization of brain chromatin. Implications for depression and cocaine addiction. Clinical Epigenetics, 8(1), 58. 10.1186/s13148-016-0214-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi A, & Dreyer J-L (2013). Striatal modulation of BDNF expression using microRNA124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. European Journal of Neuroscience, 38(2), 2328–2337. 10.1111/ejn.12228 [DOI] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M, … Yao H (2016). Silencing microRNA-143 protects the integrity of the blood-brain barrier: Implications for methamphetamine abuse. Scientific Reports, 6(1), 35642. 10.1038/srep35642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Winzer-Serhan UH, & Miranda RC (2012). Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical-derived neural progenitor cells. Alcoholism, Clinical and Experimental Research, 36(10), 1669–1677. 10.1111/j.1530-0277.2012.01793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin PR, Alanis R, & Salas R (2011). The role of the habenula in nicotine addiction. Journal of Addiction Research & Therapy, S1(2), 002. 10.4172/2155-6105.S1-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastle RM, Oliver RJ, Gardiner AS, Pentkowski NS, Bolognani F, Allan AM, … Perrone-Bizzozero NI (2018). In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Molecular Psychiatry, 23(2), 434–443. 10.1038/mp.2016.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Stanley P, & Papalopulu N (2012). MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Reports, 2(1), 10–18. 10.1016/j.celrep.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch PJ, Benton MC, Macartney-Coxson D, & Kivell BM (2015). mRNA and microRNA analysis reveals modulation of biochemical pathways related to addiction in the ventral tegmental area of methamphetamine self-administering rats. BMC Neuroscience, 16(1), 43. 10.1186/s12868-015-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas MR, Schindler AG, Shankar H, Messinger DI, Miyatake M, Land BB, … Chavkin C (2011). Selective p38α MAPK deletion in serotonergic neurons produces stress resilience in models of depression and addiction. Neuron, 71(3), 498–511. 10.1016/j.neuron.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana-Domínguez J, Arenas C, Cormand B, & Fernàndez-Castillo N (2018). miR-9, miR-153 and miR-124 are down-regulated by acute exposure to cocaine in a dopaminergic cell model and may contribute to cocaine dependence. Translational Psychiatry, 8(1), 173–173. 10.1038/s41398-018-0224-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahl LA (1996). Opioids – Mechanisms of action. Australian Prescribers, 19, 52–54. [Google Scholar]
- Chandra LC, Kumar V, Torben W, Stouwe CV, Winsauer P, Amedee A, … Mohana M (2014). Chronic administration of Δ9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute SIV infection of rhesus macaques. Journal of Virology: JVI. 01754–01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar V, & Dreyer J-L (2009). microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Molecular and Cellular Neuroscience, 42(4), 350–362. [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, & Dreyer J-L (2011). Regulation of miR-124, let-7d, and miR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology, 36(6), 1149–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Liu H, & Guan X (2013). Changes in microRNA expression profile in hippocampus during the acquisition and extinction of cocaine-induced conditioned place preference in rats. Journal of biomedical science, 20(1), 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Zhang EN, Zhong ZK, Jiang MZ, Yang XF, Zhou DM, & Wang XW (2015). MicroRNA-153 expression and prognosis in non-small cell lung cancer. International Journal of Clinical and Experimental Pathology, 8(7), 8671–8675. [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xianyu Y, & Jiang X (2017). Surface modification of gold nanoparticles with small molecules for biochemical analysis. Accounts of Chemical Research, 50(2), 310–319. [DOI] [PubMed] [Google Scholar]
- Chiarlone A, Börner C, Martín-Gómez L, Jiménez-González A, García-Concejo A, García-Bermejo ML, … Martella E (2016). Micro-RNA let-7d is a target of cannabinoid CB1 receptor and controls cannabinoid signaling. Neuropharmacology, 108, 345–352. [DOI] [PubMed] [Google Scholar]
- Chidambaran V, Zhang X, Martin LJ, Ding L, Weirauch MT, Geisler K, … Ji H (2017). DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmacogenomics and Personalized Medicine, 10, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee J-R, Lee HW, Kim K, Chung HS, … Kim E (2006). ARF6 and EFA6A regulate the development and maintenance of dendritic spines. Journal of Neuroscience, 26(18), 4811–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S-Y, Pang K, Kim JY, Ryu JR, Kang H, Liu Z, … Han K (2015). Post-transcriptional regulation of SHANK3 expression by microRNAs related to multiple neuropsychiatric disorders. Molecular Brain, 8(1), 74–74. 10.1186/s13041-015-0165-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouri E, Servaas NH, Bekker CPJ, Affandi AJ, Cossu M, Hillen MR, … Radstake T (2018). Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. Journal of Autoimmunity, 89, 162–170. 10.1016/j.jaut.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Clarke T-K, Krause K, Li T, & Schumann G (2009). Genetic study: An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addiction Biology, 14(3), 366–370. 10.1111/j.1369-1600.2009.00151.x [DOI] [PubMed] [Google Scholar]
- Clevers H (2006). Wnt/β-catenin signaling in development and disease. Cell, 127(3), 469–480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Katzoff A, Levitan D, Susswein AJ, Klein R, … Schwartz JH (2004). Long-term memory requires polyADP-ribosylation. Science, 304(5678), 1820–1822. [DOI] [PubMed] [Google Scholar]
- Creed M (2018). Current and emerging neuromodulation therapies for addiction: Insight from pre-clinical studies. Current Opinion in Neurobiology, 49, 168–174. [DOI] [PubMed] [Google Scholar]
- Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, & Ron D (2015). MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Molecular Psychiatry, 20(10), 1240–1250. 10.1038/mp.2014.120 [DOI] [PubMed] [Google Scholar]
- Dash S, Balasubramaniam M, Martínez-Rivera FJ, Godino A, Peck EG, Patnaik S, … Pandhare J (2020). Cocaine-regulated micro-RNA miR-124 controls poly (ADP-ribose) polymerase-1 expression in neuronal cells. Scientific Reports, 10(1), 11197. 10.1038/s41598-020-68144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Balasubramaniam M, Rana T, Godino A, Peck EG, Goodwin JS, … Pandhare J (2017). Poly (ADP-ribose) polymerase-1 (PARP-1) induction by cocaine is post-transcriptionally regulated by miR-125b. eNeuro, 4(4). 10.1523/ENEURO.0089-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave RS, & Khalili K (2010). Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: Impact on inflammation and oxidative stress in the central nervous system. Journal of Cellular Biochemistry, 110(4), 834–845. 10.1002/jcb.22592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim I-H, Wetsel WC, & West AE (2010). MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nature Neuroscience, 13(9), 1128–1136. 10.1038/nn.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis B, Sanger N, Bawor M, Naji L, Worster A, Baptist-Mohseni N, … Samaan Z (2020). A call for consensus in defining efficacy in clinical trials for opioid addiction: Combined results from a systematic review and qualitative study in patients receiving pharmacological assisted therapy for opioid use disorder. Trials, 21(1), 30. 10.1186/s13063-019-3995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H-Y, Cao D-N, Chen Y, Wang L, Wu N, & Li J (2016). Alterations of prefrontal cortical microRNAs in methamphetamine self-administering rats: From controlled drug intake to escalated drug intake. Neuroscience Letters, 611, 21–27. 10.1016/j.neulet.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Easton AC, Lourdusamy A, Havranek M, Mizuno K, Solati J, Golub Y, … Müller CP (2014). αCaMKII controls the establishment of cocaine’s reinforcing effects in mice and humans. Translational Psychiatry, 4(10), e457–e457. 10.1038/tp.2014.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edemann-Callesen H, Barak S, Hadar R, & Winter C (2020). Choosing the optimal brain target for neuromodulation therapies as alcohol addiction progresses—Insights from pre-clinical studies. Current Addiction Reports, 7(3), 237–244. [Google Scholar]
- Egervari G, Jutras-Aswad D, Landry J, Miller ML, Anderson SA, Michaelides M, … Hurd YL (2016). A functional 3′UTR polymorphism (rs2235749) of prodynorphin alters microRNA-365 binding in ventral striatonigral neurons to influence novelty seeking and positive reward traits. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 41(10), 2512–2520. 10.1038/npp.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, & Graveley BR (2011). microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. Rna, 17(8), 1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Rawas R, Amaral IM, & Hofer A (2020). Is p38 MAPK associated to drugs of abuse-induced abnormal behaviors? International Journal of Molecular Sciences, 21(14), 4833. 10.3390/ijms21144833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, & Filipowicz W (2010). Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry, 79(1), 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Feltmann K, Borroto-Escuela DO, Rüegg J, Pinton L, de Oliveira Sergio T, Narváez M, … Steensland P (2018). Effects of long-term alcohol drinking on the dopamine D2 receptor: Gene expression and heteroreceptor complexes in the striatum in rats. Alcoholism: Clinical and Experimental Research, 42(2), 338–351. 10.1111/acer.13568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, … Nestler EJ (2013). Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(41), 16088–16098. 10.1523/JNEUROSCI.1284-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence CS, Zhou C, Luo F, & Xu L (2016). The economic burden of prescription opioid overdose, abuse, and dependence in the United States, 2013. Medical Care, 54(10), 901–906. 10.1097/MLR.0000000000000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freye E (2008). Opioids in medicine: A comprehensive review on the mode of action and the use of analgesics in different clinical pain states, New York, NY: Springer Science & Business Media. [Google Scholar]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, … Peterson KJ (2015). A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annual Review of Genetics, 49, 213–242. 10.1146/annurev-genet-120213-092023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Chen J, & Huang Z (2019). Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA, 1(1), 24. 10.1186/s41544-019-0024-y [DOI] [Google Scholar]
- Gao J, Wang W-Y, Mao Y-W, Gräff J, Guan J-S, Pan L, … Tsai L-H (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature, 466(7310), 1105–1109. 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Concejo A, Jimenez-Gonzalez A, & Rodríguez RE (2016). μ opioid receptor expression after morphine administration is regulated by miR-212/132 cluster. PLoS One, 11(7), e0157806–e0157806. 10.1371/journal.pone.0157806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner AS, Gutierrez HL, Luo L, Davies S, Savage DD, Bakhireva LN, & Perrone-Bizzozero NI (2016). Alcohol use during pregnancy is associated with specific alterations in microRNA levels in maternal serum. Alcoholism: Clinical and Experimental Research, 40(4), 826–837. 10.1111/acer.13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi A, Fallah S, & Ansari M (2016). miR-153 as a tumor suppressor in glioblastoma multiforme is downregulated by DNA methylation. Clinical Laboratory, 62(4), 573–580. 10.7754/clin.lab.2015.150738 [DOI] [PubMed] [Google Scholar]
- Gomez AM, Altomare D, Sun W-L, Midde NM, Ji H, Shtutman M, … Zhu J (2015). Prefrontal microRNA-221 mediates environmental enrichment-induced increase of locomotor sensitivity to nicotine. The International Journal of Neuropsychopharmacology, 19(1), pyv090. 10.1093/ijnp/pyv090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini G, et al. (2013). Integration of miRNA and Protein Profiling Reveals Coordinated Neuroadaptations in the Alcohol-Dependent Mouse Brain. PLOS ONE, 8(12), e82565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA (2000). The tonic/phasic model of dopamine system regulation and its implications for understanding alcohol and psychostimulant craving. Addiction, 95(8s2), 119–128. 10.1046/j.1360-0443.95.8s2.1.x [DOI] [PubMed] [Google Scholar]
- Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, & Marks MJ (2007). The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochemical Pharmacology, 74(8), 1235–1246. 10.1016/j.bcp.2007.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W-J, Zhang C, Zhong Y, Luo J, Zhang C-Y, Zhang C, & Wang C (2020). Altered serum microRNA expression profile in subjects with heroin and methamphetamine use disorder. Biomedicine & Pharmacotherapy, 125, 109918. 10.1016/j.biopha.2020.109918 [DOI] [PubMed] [Google Scholar]
- Guitart-Masip M, Giménez-Llort L, Fernández-Teruel A, Cañete T, Tobeña A, Ögren SO, … Johansson B (2006). Reduced ethanol response in the alcohol-preferring RHA rats and neuropeptide mRNAs in relevant structures. European Journal of Neuroscience, 23(2), 531–540. 10.1111/j.1460-9568.2005.04556.x [DOI] [PubMed] [Google Scholar]
- Hammond SM (2015). An overview of microRNAs. Advanced Drug Delivery Reviews, 87, 3–14. 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins BT, Abbruscato TJ, Egleton RD, Brown RC, Huber JD, Campos CR, & Davis TP (2004). Nicotine increases in vivo blood–brain barrier permeability and alters cerebral microvascular tight junction protein distribution. Brain Research, 1027(1–2), 48–58. [DOI] [PubMed] [Google Scholar]
- Hirvonen J, Goodwin RS, Li CT, Terry GE, Zoghbi SS, Morse C, … Innis RB (2012). Reversible and regionally selective down-regulation of brain cannabinoid CB1 receptors in chronic daily cannabis smokers. Molecular Psychiatry, 17(6), 642–649. 10.1038/mp.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, & Kenny PJ (2010). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nature Neuroscience, 13(9), 1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im H-I, Amelio AL, Kocerha J, Bali P, Lu Q, … Kenny PJ (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature, 466(7303), 197–202. 10.1038/nature09202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf FW, & Lesscher HM (2014). Rodent models for compulsive alcohol intake. Alcohol (Fayetteville, NY), 48(3), 253–264. 10.1016/j.alcohol.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Liao K, Niu F, Yang L, Dallon BW, Callen S, … Buch S (2018). Astrocyte EV-induced lincRNA-Cox2 regulates microglial phagocytosis: Implications for morphine-mediated neurodegeneration. Molecular Therapy – Nucleic Acids, 13, 450–463. 10.1016/j.omtn.2018.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, & Li Z (2017). miRNAs in synapse development and synaptic plasticity. Current Opinion in Neurobiology, 45, 24–31. 10.1016/j.conb.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, & Li MD (2009a). Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biological Psychiatry, 65(8), 702–705. 10.1016/j.biopsych.2008.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, & Li MD (2009b). Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1). International Journal of Neuropsychopharmacology, 12(4), 537–546. 10.1017/s1461145708009528 [DOI] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, & Richter JD (2006). CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. The EMBO Journal, 25(20), 4865–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YY, & Li MD (2006). Proteins differentially expressed in response to nicotine in five rat brain regions: Identification using a 2-DE/MS-based proteomics approach. Proteomics, 6(10), 3138–3153. [DOI] [PubMed] [Google Scholar]
- Hyman SE, & Malenka RC (2001). Addiction and the brain: The neurobiology of compulsion and its persistence. Nature Reviews Neuroscience, 2(10), 695–703. [DOI] [PubMed] [Google Scholar]
- Ingólfsson HI, & Andersen OS (2011). Alcohol’s effects on lipid bilayer properties. Biophysical Journal, 101(4), 847–855. 10.1016/j.bpj.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AB, Toma W, Contreras KM, Alkhlaif Y, & Damaj MI (2019). The β3 subunit of the nicotinic acetylcholine receptor is required for nicotine withdrawal-induced affective but not physical signs or nicotine reward in mice. Pharmacology, Biochemistry, and Behavior, 183, 1–5. 10.1016/j.pbb.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi S, McCoy MT, Chen B, Britt JP, Kourrich S, Yau H-J, … Cadet JL (2014). Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biological Psychiatry, 76(1), 47–56. 10.1016/j.biopsych.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi S, Torres OV, Ladenheim B, & Cadet JL (2020). A single prior injection of methamphetamine enhances methamphetamine self-administration (SA) and blocks SA-induced changes in DNA methylation and mRNA expression of potassium channels in the rat nucleus accumbens. Molecular Neurobiology, 57(3), 1459–1472. 10.1007/s12035-019-01830-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He D-Y, Carnicella S, Kharazia V, Janak PH, & Ron D (2009). Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. Journal of Neuroscience, 29(43), 13494–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He D-Y, McGough NNH, Logrip ML, Phamluong K, Janak PH, & Ron D (2006). The dopamine D3 receptor is part of a homeostatic pathway regulating ethanol consumption. The Journal of Neuroscience, 26(5), 1457–1464. 10.1523/jneurosci.3786-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez KM, Pereira-Morales AJ, & Forero DA (2018). A functional polymorphism in the DRD1 gene, that modulates its regulation by miR-504, is associated with depressive symptoms. Psychiatry Investigation, 15(4), 402–406. 10.30773/pi.2017.10.16.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbakhsh M, & Zandi NS (2007). Acute opiate overdose in Tehran: The forgotten role of opium. Addictive Behaviors, 32(9), 1835–1842. [DOI] [PubMed] [Google Scholar]
- Kauer JA, & Malenka RC (2007). Synaptic plasticity and addiction. Nature Reviews Neuroscience, 8(11), 844–858. [DOI] [PubMed] [Google Scholar]
- Keller RF, Dragomir A, Yantao F, Akay YM, & Akay M (2018). Investigating the genetic profile of dopaminergic neurons in the VTA in response to perinatal nicotine exposure using mRNA-miRNA analyses. Scientific Reports, 8(1), 13769–13769. 10.1038/s41598-018-31882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, MacLean CJ, Heath AC, Eaves LJ, & Kessler RC (1993). Smoking and major depression: A causal analysis. Archives of General Psychiatry, 50(1), 36–43. [DOI] [PubMed] [Google Scholar]
- Kim HY, Li F, Park JY, Kim D, Park JH, Han HS, … Char K (2017). In vivo visualization of endogenous miR-21 using hyaluronic acid-coated graphene oxide for targeted cancer therapy. Biomaterials, 121, 144–154. [DOI] [PubMed] [Google Scholar]
- Kim J, Im H-I, & Moon C (2018). Intravenous morphine self-administration alters accumbal microRNA profiles in the mouse brain. Neural Regeneration Research, 13(1), 77–85. 10.4103/1673-5374.224374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura O, Wee S, Specio SE, Koob GF, & Pulvirenti L (2006). Escalation of methamphetamine self-administration in rats: A dose–effect function. Psychopharmacology, 186(1), 48–53. 10.1007/s00213-006-0353-z [DOI] [PubMed] [Google Scholar]
- Kochanek KD (2019). Deaths: Final data for 2017. National Vital Statistics Reports: Centers for Disease Control and Prevention. [PubMed] [Google Scholar]
- Kolodny A, Courtwright DT, Hwang CS, Kreiner P, Eadie JL, Clark TW, & Alexander GC (2015). The prescription opioid and heroin crisis: A public health approach to an epidemic of addiction. Annual Review of Public Health, 36, 559–574. [DOI] [PubMed] [Google Scholar]
- Koob GF, & Le Moal M (2001). Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology, 24(2), 97–129. 10.1016/s0893-133x(00)00195-0 [DOI] [PubMed] [Google Scholar]
- Kosten TR, & George TP (2002). The neurobiology of opioid dependence: Implications for treatment. Science & Practice Perspectives, 1 (1), 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, & Kosik KS (2003). A microRNA array reveals extensive regulation of micro-RNAs during brain development. RNA, 9(10), 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallai V, Grimes N, Fowler JP, Sequeira PA, Cartagena P, Limon A, … Fowler CD (2019). Nicotine Acts on Cholinergic Signaling Mechanisms to Directly Modulate Choroid Plexus Function. Eneuro, 6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, & Hudson LD (2010). MicroRNAs in neural cell differentiation. Brain Research, 1338, 14–19. 10.1016/j.brainres.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, & Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75(5), 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- Lee S, Woo J, Kim YS, & Im H-I (2015). Integrated miRNA-mRNA analysis in the habenula nuclei of mice intravenously self-administering nicotine. Scientific Reports, 5, 12909–12909. 10.1038/srep12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ED, Lawrence S, Petro A, Horton K, Seidler FJ, & Slotkin TA (2006). Increased nicotine self-administration following prenatal exposure in female rats. Pharmacology Biochemistry and Behavior, 85(3), 669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, & Mayfield RD (2011). Up-Regulation of MicroRNAs in Brain of Human Alcoholics. Alcoholism: Clinical and Experimental Research, 35(11), 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li J, Liu X, Qin S, Guan Y, Liu Y, … Xiong M (2013). Micro RNA expression profile and functional analysis reveal that mi R-382 is a critical novel gene of alcohol addiction. EMBO molecular medicine, 5(9), 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi Q, Wang Q, Tan X, Pang K, Liu X, … Sun J (2019). Profiling circular RNA in methamphetamine-treated primary cortical neurons identified novel circRNAs related to methamphetamine addiction. Neuroscience Letters, 701, 146–153. 10.1016/j.neulet.2019.02.032 [DOI] [PubMed] [Google Scholar]
- Li J, Zhu L, Su H, Liu D, Yan Z, Ni T, … Chen T (2020). Regulation of miR-128 in the nucleus accumbens affects methamphetamine-induced behavioral sensitization by modulating proteins involved in neuroplasticity. Addiction Biology, e12881. 10.1111/adb.12881 [DOI] [PubMed] [Google Scholar]
- Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S, … Yu X (2018). Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. International Journal of Nanomedicine, 13, 1241–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xia B, Li R, Yin D, & Liang W (2017). Changes in expression of dopamine, its receptor, and transporter in nucleus accumbens of heroin-addicted rats with brain-derived neurotrophic factor (BDNF) overexpression. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 23, 2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, … Sun F (2009). Altered microRNA expression in patients with non-obstructive azoospermia. Reproductive Biology and Endocrinology, 7(1), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian N, Niu Q, Lei Y, Li X, Li Y, & Song X (2018). miR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell cycle (Georgetown, Texas), 17(24), 2745–2755. 10.1080/15384101.2018.1556060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, … Johnson JM (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature, 433(7027), 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, … Bredy TW (2011). The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nature Neuroscience, 14(9), 1115–1117. 10.1038/nn.2891 [DOI] [PubMed] [Google Scholar]
- Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, … Young KW (2011). Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. Journal of Cell Biology, 194(6), 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, & Marsit CJ (2010). Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics, 5(7), 583–589. 10.4161/epi.5.7.12762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguschak KA, & Ressler KJ (2011). Wnt signaling in amygdala-dependent learning and memory. Journal of Neuroscience, 31(37), 13057–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, … Vladimirov VI (2015). Integrating mRNA and miRNA weighted gene co-expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PLoS One, 10(9), e0137671–e0137671. 10.1371/journal.pone.0137671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchikanti L, Helm Ii S, Fellows B, Janata JW, Pampati V, Grider JS, & Boswell MV (2012). Opioid epidemic in the United States. Pain Physician, 15(3 Suppl), ES9–ES38. [PubMed] [Google Scholar]
- Marczylo EL, Amoako AA, Konje JC, Gant TW, & Marczylo TH (2012). Smoking induces differential miRNA expression in human spermatozoa: A potential transgenerational epigenetic concern? Epigenetics, 7(5), 432–439. [DOI] [PubMed] [Google Scholar]
- Marshall JF, Belcher AM, Feinstein EM, & O’Dell SJ (2007). Methamphetamine-induced neural and cognitive changes in rodents. Addiction, 102(Suppl. 1), 61–69. 10.1111/j.1360-0443.2006.01780.x [DOI] [PubMed] [Google Scholar]
- Martin G, Puig S, Pietrzykowski A, Zadek P, Emery P, & Treistman S (2004). Somatic localization of a specific large-conductance calcium-activated potassium channel subtype controls compartmentalized ethanol sensitivity in the nucleus accumbens. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 24(29), 6563–6572. 10.1523/JNEUROSCI.0684-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikaki M, Anastasiadou E, Ozdemir RA, Potter D, Helmholz C, Slack FJ, & Chartoff EH (2019). Overexpression of miR-9 in the nucleus accumbens increases oxycodone self-administration. International Journal of Neuropsychopharmacology, 22(6), 383–393. 10.1093/ijnp/pyz015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, & Nestler EJ (2008). Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology, 33(1), 3–17. 10.1038/sj.npp.1301544 [DOI] [PubMed] [Google Scholar]
- McCreight JC, Schneider SE, Wilburn DB, & Swanson WJ (2017). Evolution of microRNA in primates. PLoS One, 12(6), e0176596–e0176596. 10.1371/journal.pone.0176596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin I, Dani JA, & De Biasi M (2015). Nicotine withdrawal. Current Topics in Behavioral Neurosciences, 24, 99–123. 10.1007/978-3-319-13482-6_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, & Hinshaw JE (2011). Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature Structural & Molecular Biology, 18(1), 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuo K, Okazaki S, Watanabe S, Inoue H, & Matsumoto H (2016). Acute ethanol administration increases miR-124 expression via histone acetylation in the brain. Journal of Alcoholism and Drug Dependence, 4(232), 2. [Google Scholar]
- Müller CP, & Homberg JR (2015). The role of serotonin in drug use and addiction. Behavioural Brain Research, 277, 146–192. 10.1016/j.bbr.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Narahashi T, Kuriyama K, Illes P, Wirkner K, Fischer W, Mühlberg K, … Sato N (2001). Neuroreceptors and ion channels as targets of alcohol. Alcoholism: Clinical and Experimental Research, 25(Suppl. 1), 182S–188S. 10.1111/j.1530-0277.2001.tb02394.x [DOI] [PubMed] [Google Scholar]
- Negi V, & Chan SY (2017). Discerning functional hierarchies of microRNAs in pulmonary hypertension. JCI Insight, 2(5), e91327–e91327. 10.1172/jci.insight.91327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hope BT, & Widnell KL (1993). Drug addiction: A model for the molecular basis of neural plasticity. Neuron, 11(6), 995–1006. [DOI] [PubMed] [Google Scholar]
- Nisell M, Nomikos GG, & Svensson TH (1994). Systemic nicotine-induced dopamine release in the rat nucleus accumbens is regulated by nicotinic receptors in the ventral tegmental area. Synapse, 16(1), 36–44. [DOI] [PubMed] [Google Scholar]
- Noble EP (2000). Addiction and its reward process through polymorphisms of the D2 dopamine receptor gene: A review. European Psychiatry, 15(2), 79–89. 10.1016/S0924-9338(00)00208-X [DOI] [PubMed] [Google Scholar]
- Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, & Mayfield RD (2013). Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics, 14(1), 725. 10.1186/1471-2164-14-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard KE, Chand S, Wheeler S, Tiwari S, Flores A, Hernandez J, … Yelamanchili SV (2020). Role of extracellular vesicles in substance abuse and HIV-related neurological pathologies. International Journal of Molecular Sciences, 21(18), 6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omkaram G, Shaima J, Olivia W, & Dipak KS (2017). MicroRNA-9 regulates fetal alcohol-induced changes in D2 receptor to promote prolactin production. Journal of Endocrinology, 235(1), 1–14. 10.1530/JOE-17-0135 [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, & Prakash A (2008). Brain chromatin remodeling: A novel mechanism of alcoholism. The Journal of Neuroscience, 28(14), 3729–3737. 10.1523/jneurosci.5731-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ (2010). The role of microRNAs in drug addiction: A big lesson from tiny molecules. In Reilly MT & Lovinger DM (Eds.), International review of neurobiology (Vol. 91, pp. 1–24). Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, … Treistman SN (2008). Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron, 59(2), 274–287. 10.1016/j.neuron.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilapil C, Welner S, Magnan J, Zamir N, & Quirion R (1986). Mu opioid receptor binding sites in human brain. NIDA Research Monograph, 75, 319–322. [PubMed] [Google Scholar]
- Pittenger ST, Schaal VL, Moore D, Guda RS, Koul S, Yelamanchili SV, … Pendyala G (2018). MicroRNA cluster miR199a/214 are differentially expressed in female and male rats following nicotine self-administration. Scientific Reports, 8(1), 17464–17464. 10.1038/s41598-018-35747-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago L, Beattie R, Taylor V, & Winter J (2014). miR 379–410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. The EMBO Journal, 33(8), 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P, O’Connell K, & Finnerty TK (2018). Potential role of extracellular vesicles in the pathophysiology of drug addiction. Molecular Neurobiology, 55(8), 6906–6913. [DOI] [PubMed] [Google Scholar]
- Reddy PVB, Pilakka-Kanthikeel S, Saxena SK, Saiyed Z, & Nair MPN (2012). Interactive effects of morphine on HIV infection: Role in HIV-associated neurocognitive disorder. AIDS Research and Treatment, 2012, 953678. 10.1155/2012/953678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, & Nestler EJ (2011). Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience, 12(11), 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd RA, Paulozzi LJ, Bauer MJ, Burleson RW, Carlson RE, Dao D, … Fernandes JC (2014). Increases in heroin overdose deaths—28 states, 2010 to 2012. MMWR. Morbidity and Mortality Weekly Report, 63(39), 849. [PMC free article] [PubMed] [Google Scholar]
- Ruffle JK (2014). Molecular neurobiology of addiction: What’s all the (Δ)FosB about? The American Journal of Drug and Alcohol Abuse, 40 (6), 428–437. 10.3109/00952990.2014.933840 [DOI] [PubMed] [Google Scholar]
- Rzepiela AJ, Ghosh S, Breda J, Vina-Vilaseca A, Syed AP, Gruber AJ, … Zavolan M (2018). Single-cell mRNA profiling reveals the hierarchical response of miRNA targets to miRNA induction. Molecular Systems Biology, 14(8), e8266–e8266. 10.15252/msb.20188266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, & Brewer RD (2015). 2010 national and state costs of excessive alcohol consumption. American Journal of Preventive Medicine, 49(5), e73–e79. 10.1016/j.amepre.2015.05.031 [DOI] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Frankowska M, Miszkiel J, Wydra K, Jastrzębska J, & Filip M (2017). Prolonged induction of miR-212/132 and REST expression in rat striatum following cocaine self-administration. Molecular Neurobiology, 54(3), 2241–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie KN, Damez-Werno D, Sun H, Shao N, Gancarz A, Panganiban CH, … Kaufman L (2014). Essential role of poly (ADP-ribosyl) ation in cocaine action. Proceedings of the National Academy of Sciences of the United States of America, 111(5), 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Rudd RA, Noonan RK, & Haegerich TM (2018). Quantifying the epidemic of prescription opioid overdose deaths. American Journal of Public Health, 108(4), 500–502. 10.2105/AJPH.2017.304265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahjin F, Chand S, & Yelamanchili SV (2019). Extracellular vesicles as drug delivery vehicles to the central nervous system. Journal of Neuroimmune Pharmacology, 15, 443–458. 10.1007/s11481-019-09875-w [DOI] [PubMed] [Google Scholar]
- Shahjin F, Guda RS, Schaal VL, Odegaard K, Clark A, Gowen A, … Yelamanchili SV (2019). Brain-derived extracellular vesicle microRNA signatures associated with in utero and postnatal oxycodone exposure. Cell, 9(1), 21. 10.3390/cells9010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Leites C, He D, Schwartz D, Moy W, Shi J, & Duan J (2014). MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression, and the microRNA-mediated expression regulation is altered by a genetic variant. The Journal of Biological Chemistry, 289(19), 13434–13444. 10.1074/jbc.M113.535203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, & Aizawa S (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. The Journal of Neuroscience, 31(9), 3407–3422. 10.1523/JNEUROSCI.5085-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla M, Maitra S, Hernandez J-F, Govitrapong P, & Vincent B (2019). Methamphetamine regulates βAPP processing in human neuroblastoma cells. Neuroscience Letters, 701, 20–25. 10.1016/j.neulet.2019.02.023 [DOI] [PubMed] [Google Scholar]
- Silverstein JH, Silva DA, & Iberti TJ (1993). Opioid addiction in anesthesiology. Anesthesiology: The Journal of the American Society of Anesthesiologists, 79(2), 354–375. [DOI] [PubMed] [Google Scholar]
- Sim MS, Soga T, Pandy V, Wu YS, Parhar IS, & Mohamed Z (2017). MicroRNA expression signature of methamphetamine use and addiction in the rat nucleus accumbens. Metabolic Brain Disease, 32(6), 1767–1783. 10.1007/s11011-017-0061-x [DOI] [PubMed] [Google Scholar]
- Singh R, & Saini N (2012). Downregulation of BCL2 by miRNAs augments drug-induced apoptosis – A combined computational and experimental approach. Journal of Cell Science, 125(6), 1568–1578. [DOI] [PubMed] [Google Scholar]
- Smith ACW, & Kenny PJ (2018). MicroRNAs regulate synaptic plasticity underlying drug addiction. Genes, Brain and Behavior, 17(3), e12424. 10.1111/gbb.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Zhu L, Li J, Wang R, Liu D, Han W, … Chen T (2019). Regulation of microRNA-29c in the nucleus accumbens modulates methamphetamine -induced locomotor sensitization in mice. Neuropharmacology, 148, 160–168. 10.1016/j.neuropharm.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Südhof TC (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature, 455(7215), 903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wang Y, Zhou R, Deng Z, Han Y, Han X, … Hong D (2017). Targeting and regulating of an oncogene via nanovector delivery of MicroRNA using patient-derived xenografts. Theranostics, 7(3), 677–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-M, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, … Buckley NJ (2005). Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Molecular Biology of the Cell, 16(12), 5630–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman S, & Sussman AN (2011). Considering the definition of addiction. International Journal of Environmental Research and Public Health, 8(10), 4025–4038. 10.3390/ijerph8104025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki FA, Pan X, Lee M-H, & Zhang B (2014). Nicotine exposure and transgenerational impact: A prospective study on small regulatory microRNAs. Scientific Reports, 4, 7513–7513. 10.1038/srep07513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, … Heilig M (2014). microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. The Journal of Neuroscience, 34(13), 4581–4588. 10.1523/jneurosci.0445-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ye X, Hou X, Yang X, Yang J, & Wu C (2016). SVCT2, a potential therapeutic target, protects against oxidative stress during ethanol-induced neurotoxicity via JNK/p38 MAPKs, NF-κB and miRNA125a-5p. Free Radical Biology and Medicine, 96, 362–373. 10.1016/j.freeradbiomed.2016.03.039 [DOI] [PubMed] [Google Scholar]
- Tian J, Casella G, Zhang Y, Rostami A, & Li X (2020). Potential roles of extracellular vesicles in the pathophysiology, diagnosis, and treatment of autoimmune diseases. International Journal of Biological Sciences, 16(4), 620–632. 10.7150/ijbs.39629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama K, Kiyosawa N, Watanabe K, & Ishizuka H (2017). Identification of circulating miRNAs differentially regulated by opioid treatment. International Journal of Molecular Sciences, 18(9), 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treatment for Stimulant Use Disorders. (1999). How stimulants affect the brain and behavior. Substance Abuse and Mental Health Services Administration (US). [PubMed] [Google Scholar]
- Treistman SN, & Martin GE (2009). BK channels: Mediators and models for alcohol tolerance. Trends in Neurosciences, 32(12), 629–637. 10.1016/j.tins.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AM, Chung DD, Pinson MR, Salem NA, Eaves SE, & Miranda RC (2019). Ethanol exposure increases miR-140 in extracellular vesicles: Implications for fetal neural stem cell proliferation and maturation. Alcoholism: Clinical and Experimental Research, 43 (7), 1414–1426. 10.1111/acer.14066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski P, & Kenny B-A (2015). The blood–brain barrier and methamphetamine: Open sesame? Frontiers in Neuroscience, 9, 156–156. 10.3389/fnins.2015.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren LJMJ, & Everitt BJ (2004). Drug seeking becomes compulsive after prolonged cocaine self-administration. Science, 305 (5686), 1017–1019. 10.1126/science.1098975 [DOI] [PubMed] [Google Scholar]
- Volkow ND, Koob G, & Baler R (2015). Biomarkers in substance use disorders. ACS Chemical Neuroscience, 6(4), 522–525. 10.1021/acschemneuro.5b00067 [DOI] [PubMed] [Google Scholar]
- Volkow ND, & Morales M (2015). The brain on drugs: From reward to addiction. Cell, 162(4), 712–725. 10.1016/j.cell.2015.07.046 [DOI] [PubMed] [Google Scholar]
- Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, … Shao Y (2011). Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clinical Chemistry, 57(12), 1722–1731. [DOI] [PubMed] [Google Scholar]
- Wang L, Ke J, Li Y, Ma Q, Dasgupta C, Huang X, … Xiao D (2017). Inhibition of miRNA-210 reverses nicotine-induced brain hypoxic-ischemic injury in neonatal rats. International journal of biological sciences, 13(1), 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kwon EJ, & Tsai L-H (2012). MicroRNAs in learning, memory, and neurological diseases. Learning & Memory, 19(9), 359–368. [DOI] [PubMed] [Google Scholar]
- Welters ID, Menzebach A, Goumon Y, Cadet P, Menges T, Hughes TK, … Stefano GB (2000). Morphine inhibits NF-κB nuclear binding in human neutrophils and monocytes by a nitric oxide-dependent mechanism. Anesthesiology: The Journal of the American Society of Anesthesiologists, 92(6), 1677–1684. [DOI] [PubMed] [Google Scholar]
- Whyte AJ, Torregrossa MM, Barker JM, & Gourley SL (2018). Long-term consequences of adolescent drug use: Evidence from pre-clinical and clinical models. Frontiers in Behavioral Neuroscience, 12, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA (1978). Catecholamine theories of reward: A critical review. Brain Research, 152(2), 215–247. [DOI] [PubMed] [Google Scholar]
- Wu J, Gao M, & Taylor DH (2014). Neuronal nicotinic acetylcholine receptors are important targets for alcohol reward and dependence. Acta Pharmacologica Sinica, 35(3), 311–315. 10.1038/aps.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang L, Law PY, Wei LN, & Loh HH (2009). Long-Term Morphine Treatment Decreases the Association of μ-Opioid Receptor (MOR1) mRNA with Polysomes through miRNA23b. Molecular Pharmacology, 75(4), 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Ma X, Li X, Dong H, Yi J, Zeng W, & Yang Z (2015). miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting Snail. Oncology Reports, 34(2), 655–662. 10.3892/or.2015.4008 [DOI] [PubMed] [Google Scholar]
- Xu X, Bishop EE, Kennedy SM, Simpson SA, & Pechacek TF (2015). Annual healthcare spending attributable to cigarette smoking: An update. American Journal of Preventive Medicine, 48(3), 326–333. 10.1016/j.amepre.2014.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Hu Z, Yao W, Le Q, Xu B, Liu X, & Ma L (2017). miR-218 targets MeCP2 and inhibits heroin seeking behavior. Scientific Reports, 7(1), 40413. 10.1038/srep40413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, … Buch S (2014). miR-9 promotes microglial activation by targeting MCPIP1. Nature Communications, 5(1), 4386. 10.1038/ncomms5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekkirala AS, Kalyuzhny AE, & Portoghese PS (2010). Standard opioid agonists activate heteromeric opioid receptors: Evidence for morphine and [d-Ala2-MePhe4-Glyol5]Enkephalin as selective μ–δ agonists. ACS Chemical Neuroscience, 1(2), 146–154. 10.1021/cn9000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V, Ji F, Nielsen DA, Levran O, Ho A, Morgello S, … Kreek MJ (2009). A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology, 34(5), 1185–1197. 10.1038/npp.2008.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehra A, Burns J, Liu CK, Manza P, Wiers CE, Volkow ND, & Wang G-J (2018). Cannabis addiction and the brain: A review. Journal of Neuroimmune Pharmacology: The Official Journal of the Society on NeuroImmune Pharmacology, 13(4), 438–452. 10.1007/s11481-018-9782-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu X, Bai M, Zhang L, Xue L, & Yi J (2013). Maternal deprivation enhances behavioral vulnerability to stress associated with miR-504 expression in nucleus accumbens of rats. PLoS One, 8(7), e69934–e69934. 10.1371/journal.pone.0069934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, & Shi Y (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Structural & Molecular Biology, 16(4), 365–371. 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang K, Jiang H, Du J, Na Z, Hao W, … Zhao M (2016). Decreased expression of plasma microRNA in patients with methamphetamine (MA) use disorder. Journal of Neuroimmune Pharmacology, 11(3), 542–548. 10.1007/s11481-016-9671-z [DOI] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zeng Y, Loh HH, & Law PY (2010). Yin Yang 1 phosphorylation contributes to the differential effects of μ-opioid receptor agonists on microRNA-190 expression. Journal of Biological Chemistry, 285(29), 21994–22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Li J, Dong N, Guan F, Liu Y, Ma D, … Chen T (2016). mRNA changes in nucleus accumbens related to methamphetamine addiction in mice. Scientific reports, 6, 36993–36993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Wang H, Dong W, Quan X, Zhu H, Xu Y, … Qin C (2011). miR-29c regulates BACE1 protein expression. Brain Research, 1395, 108–115. 10.1016/j.brainres.2011.04.035 [DOI] [PubMed] [Google Scholar]
- Zorick T, Nestor L, Miotto K, Sugar C, Hellemann G, Scanlon G, … London ED (2010). Withdrawal symptoms in abstinent methamphetamine-dependent subjects. Addiction, 105(10), 1809–1818. 10.1111/j.1360-0443.2010.03066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Li J, Song L, Zeng X, & Wang G (2015). Similarity computation strategies in the microRNA-disease network: A survey. Briefings in Functional Genomics, 15(1), 55–64. 10.1093/bfgp/elv024 [DOI] [PubMed] [Google Scholar]
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, … Fischer A (2011). microRNA-34c is a novel target to treat dementias. The EMBO Journal, 30(20), 4299–4308. 10.1038/emboj.2011.327 [DOI] [PMC free article] [PubMed] [Google Scholar]


