Abstract
Systemic lupus erythematosus (SLE, lupus) is a chronic autoimmune disease characterized by loss of peripheral tolerance to nuclear self-antigens. It is increasingly recognized that aberrant T cell metabolism is a critical mediator of SLE immunopathology. Hypoxia inducible factor 1α (HIF-lα) is a key transcription factor that regulates T cell metabolism in response to immune stimuli. T cell activation induces HIF-1α expression and transcriptional activation of HIF-responsive genes. HypoxamiRs are a group of microRNAs sensitive to HIF-1α transcriptional regulation that function to fine-tune the HIF-driven transcriptional program. The ‘master’ hypoxamiR, miR-210 is transcriptionally regulated by HIF-1α and negatively regulates HIF-1α activity. Although a key role for HIF-1α in has been described in a number of autoimmune and inflammatory diseases and abnormal microRNA expression profiles correlate with poor clinical outcome in a number of rheumatologic diseases, the expression and function of HIF-1α and miR-210 in lupus remains largely uncharacterized. Here we report HIF-1α and miR-210 differential and lineage-specific expression in systemic lupus erythematosus. We show that HIF-1α mRNA and protein is overexpressed in human lupus CD4+ cells but not in CD8+ or CD19+ cells. RORγt, was upregulated in human lupus lymphocytes while FoxP3 expression remained unchanged. We show that miR-210 expression in lupus-prone mice correlates with disease activity and is robustly and selectively upregulated in CD4+ cells from both human lupus patients and lupus-prone mice. Our results suggest that abnormal HIF-1α and miR-210 expression contributes to SLE immune pathology and that HIF-1α/miR-210 may represent a novel and important regulatory axis in SLE.
Keywords: HIF-1α, microRNA, miR-210, SLE, Lupus, Lymphocytes, PBMC, T cell, TCR, CD4, Autoimmune disease, Th17, IL-17
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by loss of tolerance to nuclear components leading to accumulation of autoantibodies, immune complex formation and possible end-organ damage (Tsokos, 2011). It is increasingly recognized that abnormal T cells are critical mediators of SLE immunopathology (Moulton and Tsokos, 2015). Autoreactive T cells from both human lupus patients and lupus-prone mice present multiple abnormalities including a number of metabolic disturbances (Morel, 2017; Perl, 2016). The importance of T cell metabolism in the etiology of SLE immunopathology is illustrated by studies in lupus-prone mice demonstrating that normalization of aberrant metabolic programs reverses autoimmune phenotypes (Yin et al., 2015; Yin et al., 2016).
Hypoxia inducible factor la (HIF-1α) is a key transcription factor that regulates cellular metabolism in response to oxygen stress (Semenza, 2009; G. L. Wang et al., 1995). Studies in T cells have shown that while the primary function of HIF-1α is detection and response to oxygen tension, a number of other mechanisms can induce HIF-1α activity under normoxic conditions, allowing T cells to utilize the HIF-1α axis in response to immune stimuli (Palazon et al., 2014). T cell receptor (TCR) ligation induces HIF-1α expression and subsequent transcriptional activation of HIF-responsive genes influence T cell function and lineage differentiation (McNamee et al., 2013). HIF-1α regulates development of Tregulatory (Treg) and Thelper17 (Th17) cells by direct transcriptional activation of RORγt expression and suppression of FoxP3 transcriptional activity (Dang et al., 2011; Shi et al., 2011).
MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs that exert a regulatory influence on almost every aspect of cellular biology (Bartel, 2018). Several miRNAs sensitive to HIF-1α transcriptional regulation, collectively termed ‘hypoxamiRs’, have been identified (Kulshreshtha et al., 2007). MiR-210 has emerged as the ‘master hypoxamir’ due to its unique status as the only miRNA whose expression is consistently and robustly induced by HIF-1α activity (Ivan and Huang, 2014). Although aberrant miRNA expression profiles correlate with poor clinical outcome in a number of rheumatologic diseases (Esteller, 2011), the expression and function of miR-210 in lupus remains largely uncharacterized.
Here we report HIF-1α and miR-210 differential and lineage-specific expression in systemic lupus erythematosus. We purified peripheral lymphocyte subsets from lupus-prone mice and showed that miR-210 is overexpressed specifically in the CD4+T cell lineage and positively correlates with disease activity. We purified peripheral lymphocyte subsets from SLE patients and detected overexpression of both HIF-1α and miR-210 in CD4+ T cells but not in CD8+ or CD19+ cells. We showed that RORγt is up-regulated in human lupus peripheral lymphocytes and positively correlates with HIF-1α overexpression. Taken together, our results suggest that abnormal HIF-1α and miR-210 expression may contribute to SLE immune dysregulation and that HIF-1α/miR-210 may represent a novel and important regulatory pathway in SLE.
Methods
Human subjects
Subject peripheral blood samples were collected with prior written informed consent and in accordance with the ethical standards of the Institutional Review Boards (IRB) at Thomas Jefferson University, Philadelphia, PA (TJU IRB Control #15D.243). Candidate subjects were identified by evaluation of patient medical records by a qualified Rheumatologist in full compliance with all Federal and institutional HIPAA regulations.
Animals
Breeder pairs of B6.Sle123 (Sle123) (Morel et al., 2000) mice were a kind gift from Dr. Laurence Morel at the University of Florida (Gainesville, FL). The lupus-prone Sle123 strain is a spontaneous lupus model that develops an autoimmune phenotype beginning at approximately two months of age (no/mild disease activity) progressing to advanced disease activity by 8 months of age (Morel et al., 2000). C57BL/6 (B6, WT) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Colonies were bred and maintained in an Institutional Animal Care and Use Committee (IACUC) approved facility at the University of Pennsylvania (Philadelphia, PA). All animal care and experimental procedures were conducted with prior approval by the IACUC at the University of Pennsylvania.
Purification of murine splenic lymphocytes
Speen tissue was harvested from euthanized animals and placed immediately into dishes containing ice-cold cell separation buffer (1X DPBS, 0.5% BSA, 1 mM EDTA). Single-cell lymphocyte suspensions were prepared by mechanical tissue disruption through a 70 υm nylon filter followed by red blood cell lysis with RBC Lysis Buffer (eBioscience, San Diego, CA) according to the manufacturer's instructions. Cells were collected by centrifugation at 300 RCF for 10 minutes and washed 3X with an appropriate volume of FACS buffer.
Purification of murine lymphocyte subsets
Murine single-cell lymphocyte suspensions were prepared as described above. Cells were suspended at 1 x 107 cells/mL in cell separation buffer (1X DPBS, 0.1% BSA, 2 mM EDTA) and incubated with 2.5 μg/mL Mouse Fc Block (BD Pharmingen) for 10 minutes at room temperature. Cells were then stained with the following fluorophore-conjugated antibodies against cell surface markers CD3ε, CD4, CD8, CD19 and CD45R. Antibody details provided in Supplementary Methods. Lymphocyte populations were separated on a BD FACS Aria cytometer (Becton Dickinson, Franklin Lakes, NJ) on the basis of cell surface marker expression.
Purification of human peripheral blood mononuclear cells
Human subject peripheral blood samples were drawn into BD Vacutainer lithium heparin blood collection tubes (Becton Dickinson, Franklin Lakes, NJ) and stored upright at room temperature until processed not more than 2 hours post-collection. Peripheral blood mononuclear cells (PBMCs) were isolated under sterile conditions using Leucosep cell separation tubes (Greiner Bio-One, Monroe, NC) prepared with Ficoll-Paque Plus (GE Healthcare, Piscataway, NJ) density gradient solution according to the manufacturer's instructions. Briefly, peripheral blood was diluted 2X with room tempearture HBSS (Corning, Manassas, VA), decanted into prepared Leucosep cell separation tubes and centrifuged at 800 RCF for 20 minutes at room temperature. Buffy coats were removed and washed 3X with ice-cold HBSS. PBMC’s were either used directly or further separated into distinct subsets. If used directly, purity was evaluated by flow cytometry on the basis of forward/side light scatter. Representative plots are provided in Supplementary Methods.
Purification of human lymphocyte subsets
PBMC’s were isolated from human subject peripheral blood as described above. Cells were suspended at 1 x 107 cells/mL in separation buffer (1X DPBS, 0.1% BSA, 2 mM EDTA) and incubated with 2.5 μg/mL Human Fc Block (BD Pharmingen) for 10 minutes at room temperature. Cells were incubated with 10 ug/mL biotinylated primary antibody for 10 minutes at 4 °C, washed with 2 volumes of separation buffer then incubated with Dynabeads Biotin Binder magnetic beads (Life Technologies, Carlsbad, CA) following the manufacturer's instructions. Bead-bound cells were captured with a magnet, washed twice with ice-cold PBS and aliquoted for downstream applications. Primary antibody details provided in Supplementary Methods. Purity of the separations was evaluated by flow cytometry. Antibodies used and representative plots are provided in Supplementary Methods.
Total RNA purification
Cells were collected by centrifugation and lysed in an appropriate volume of Trizol tri-reagent (Invitrogen, Carlsbad, CA). RNA was isolated using Direct-zol Trizol RNA MiniPrep Plus kit components following the manufacturer's instructions (Zymo Research, Irvine, CA). Genomic DNA contamination was reduced by on-column DNase I digestion (Thermo Fisher Scientific, USA). RNA purity was determined by evaluating 260/280 nm and 260/230 nm absorbtion ratios. RNA preparations with 260/280 nm ratios > 1.9 and 260/230 nm ratios > 2.0 were considered pure.
Total protein purification
Cells were collected by centrifugation and resuspended in an appropriate volume of RIPA buffer (Cell Signaling Technology, Danvers, MA) supplemented with 1 mM PMSF (Millipore Sigma, USA). Cells were lysed by repetitive pipetting and incubated in ice for at least 5 minutes. Crude extracts were centrifuged at 14,000 RCF for 10 minutes at 4 °C and the supernatants used for subsequent immunoblotting experiments.
Reverse transcription PCR
For gene expression assays, 100 ng total RNA was reverse-transcribed into cDNA using Superscript III reverse transcription kit components following the manufacturer's instructions (Invitrogen, Carlsbad, CA) except that reactions were primed with a cocktail of oligo(dT)20 (50 μM) and random hexamers (50 ng/μL). cDNA was recovered using DNA Clean & Concentrator kit components in a final elution volume of 10 μL (Zymo Research, Irvine, CA). 2 μL eluent was used as input DNA in subsequent quantitative real-time PCR experiments. For miR-210 expression, RNA templates were reverse-transcribed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). Reactions were primed using Taqman miR-210 or U6 snRNA reverse-transcription primers (Applied Biosystems, Foster City, CA). Primer details provided in Supplemental Methods. Reverse-transcription products were diluted 1:20 and 5 μL used as input DNA in subsequent quantitative real-time PCR experiments. For both gene and miRNA expression experiments, no-reverse-transcriptase control (NRT) reactions were included for each RNA template.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) reactions were ran in triplicate on either Applied Biosystems 7500 or StepOnePlus real-time PCR systems (Applied Biosystems, Foster City, CA). Each 20 μL gene expression qPCR reaction contained the following components: 1X PowerUp SYBR Green master mix (Applied Biosystems, Foster City, CA), 400 nM KiCqStart SYBR Green gene assay primers (Sigma-Aldrich, St. Louis, MO) and 2 μL reverse-transcription product prepared as described above. Primer sequences provided in Supplemental Methods. Each 20 μL microRNA qPCR reaction contained the following components: 1X TaqMan Fast Advanced Master Mix, 1X Taqman real-time assay primers (Applied Biosystems, Foster City, CA) and 5 μL 1:20-diluted reverse-transcription product. Assay details provided in Supplemental Methods. Baseline-adjusted quantification cycle (Cq) values were determined using instrument software with automatic threshold enabled.
Immunoblotting
Total protein was resolved on NuPAGE 4-12% bis-tris polyacrylamide mini gels (Invitrogen, Carlsbad, CA) with MOPS-SDS running buffer. Proteins were transferred to nitrocellulose membranes and stained with either HIF-1α or β-actin primary antibodies (Cell Signaling Technology, Danvers, MA). Antibody details provided in Supplemental Methods. Immunostained membranes were probed with horseradish peroxidase-conjugated secondary antibody and detected by incubation with enhanced chemiluminescent reagent (Pierce Biotechnology, Rockford, IL). Membranes were imaged on an iBright CL750 imaging system (Invitrogen, Carlsbad, CA).
Northern blotting
Total RNA was resolved on 15% urea-polyacrylamide gels, transferred to nitrocellulose membranes and hybridized with radiolabelled DNA probes complementary to the mature miR-210-3p sequence. Small-nucleolar RNA 429 (sno429), previously verified to be not regulated in the Sle123 model, served as a loading control. Probe sequences provided in Supplemental Methods.
Determination of differential expression
For quantitative PCR, differential gene expression was determined using the 2(-ΔΔCT) method as described by Livak and Schmittgen (Livak and Schmittgen, 2001). For Northern blot analysis, exposed blots were imaged on a Storm 840 phosphorimager and the resulting bands quantified using the ImageJ image processing software program (National Institutes of Health and the Laboratory for Optical and Computational Instrumentation). Briefly, regions of equal area were drawn around bands of interest. The measured image integrated density within each region was used to evaluate differential expression. For Western blot analyses, membranes were imaged on an iBright CL750 imaging system (Invitrogen, Carlsbad, CA) and the resulting bands quantified as described for Northern blot analysis.
Statistical analysis
Statistical analyses were performed using the R Foundation for Statistical Computing software package. Mean, error and significance values were determined using an unpaired Student's t test assuming equal variation. p-values < 0.05 were considered significant.
Results
MiR-210 expression in lupus-prone Sle123 mice is disease-dependent and lineage-specific
Splenic lymphocytes were harvested from lupus-prone female Sle123 mice with mild or advanced lupus-like disease and assayed for miR-210 expression by Northern blotting. As shown in Figure 1A, miR-210 expression in lymphocytes from mice with advanced disease was 2.3 ± 0.2-fold higher than in lymphocytes from mice with mild disease suggesting that miR-210 expression in Sle123 lymphocytes positively associates with disease activity. To determine if miR-210 expression in these mice may be lineage-specific, we purified CD4+, CD8+ and CD19+ lymphocyte subsets from lupus-prone female Sle123 mice with mild or advanced lupus-like disease and from age- and gender-matched C57BL/6 (WT) mice by flow cytometry and quantified miR-210 expression by quantitative real-time PCR (qPCR). As shown in Figure 1B, miR-210 was only moderately up-regulated in CD4+ and CD8+ cells from Sle123 mice with mild disease compared to WT controls (2.2 ± 0.01-fold and 1.5 ± 0.03-fold respectively). MiR-210 expression in CD19+ cells from Slel23 mice with mild disease was similar to WT controls. In contrast, miR-210 was robustly up-regulated in CD4+ cells from Sle123 mice with advanced disease (359 ± 36.5-fold) but only moderately up-regulated in CD8+ (2.5 ± 0.08-fold) and CD19+ cells (2.3 ± 0.06-fold) suggesting that miR-210 is predominantly expressed by the CD4+ T cell lineage. Our Northern blot experiments (Figure 1A) suggest that miR-210 expression in Sle123 lymphocytes associates with disease activity. As shown in Figure 1B, miR-210 expression in CD4+ cells from mice with advanced lupus-like disease was 165.2 ± 16.8-fold greater than miR-210 expression in CD4+ cells from mice with mild disease. In contrast, miR-210 was only moderately up-regulated in CD8+ and CD19+ cells from mice with advanced disease (1.6 ± 0.04-fold and 2.4 ± 0.08-fold respectively) compared to miR-210 expression in CD8+ and CD19+ cells from mice with mild disease. Our results show that miR-210 is overexpressed in lupus-prone Sle123 lymphocytes and suggests that miR-210 overexpression in these mice is disease-dependent and specific to the CD4+ T cell lineage.
Figure 1.
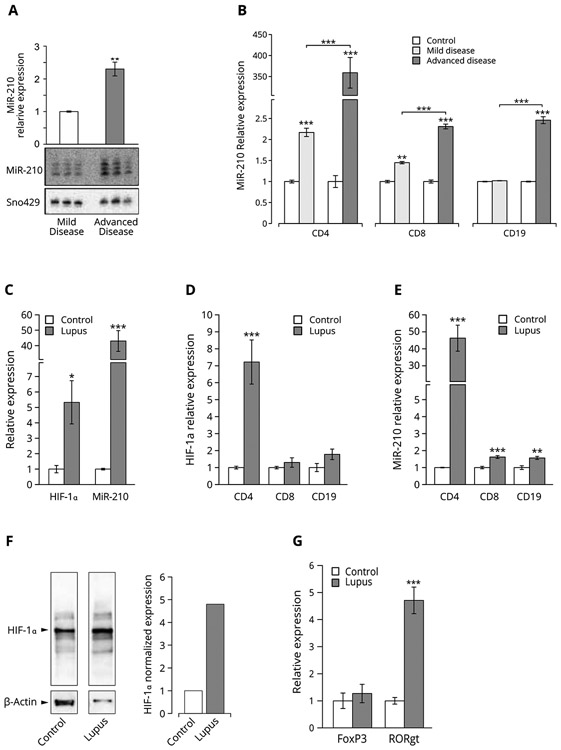
(A) MiR-210 expression in lymphocytes from lupus-prone mice is disease-dependent. Total RNA was purified from splenic lymphocytes isolated from lupus-prone female Sle123 mice with no/mild or advanced disease and subjected to urea PAGE Northern blot analysis for miR-210 expression. MiR-210 expression was detected with radio-labeled anti-sense DNA probes specific for the mature mmu-miR-210-3p species. Differential loading was detected using radio-labeled anti-sense DNA probes specific for small-nucleolar RNA 429 (mmu-sno429), previously verified to be not regulated in the Sle123 model. Exposed blots were imaged on a Storm 840 phosphorimager and the resulting bands quantified using the ImageJ software program as described in Materials and Methods. Bar heights represent mean relative expression and error bars represent mean relative expression ± SEM of three independent biological replicates. *p < 0.05, n = 3. (B) MiR-210 expression in lymphocytes from lupus-prone mice is lineage-specific. Splenic lymphocytes were harvested from lupus-prone female Sle123 mice with no/mild or advanced disease activity and purified into CD4+, CD8+ and CD19+ subsets by flow cytometry. Total RNA was purified from each subset and assayed for miR-210 expression by qPCR as described in Materials and Methods. Expression of small-nucleolar RNA 429 (mmu-sno429), previously verified to be not regulated in the Sle123 model, served as the loading control. Bar heights represent mean relative expression and error bars represent mean relative expression ± SEM of three independent biological replicates. **p < 0.01 and ***p < 0.001, n = 3. (C) HIF-1α is up-regulated in human lupus and expressed mainly by CD4+ T cells. Peripheral lymphocytes were isolated from lupus patients with active disease and from healthy control subjects and separated into CD4+, CD8+ and CD19+ subsets as described in Materials and Methods. Total RNA was purified from each subset and assayed for HIF-1α expression by qPCR. Bar heights represent mean relative HIF-1α expression. For control subjects, error bars represent mean relative expression ± SEM of four independent biological replicates (n = 4). For lupus patients, error bars represent mean relative expression ± SEM of six independent biological replicates (n = 6). *** p < 0.001. (D) MiR-210 is up-regulated in human lupus and expressed mainly by CD4+ T cells. Peripheral lymphocytes were isolated from lupus patients with active disease and from healthy control subjects and separated into CD4+, CD8+ and CD19+ subsets as described in Materials and Methods. Total RNA was purified from each subset and assayed for miR-210 expression by qPCR. Bar heights represent mean relative miR-210 expression. For control subjects, error bars represent mean relative expression ± SEM of four independent biological replicates (n = 4). For lupus patients, error bars represent mean relative expression ± SEM of six independent biological replicates (n = 6). ** p < 0.01 and *** p < 0.001. (E) FoxP3 and RORγt expression associate with HIF-1α and miR-210 overexpression in human lupus peripheral lymphocytes. Peripheral lymphocytes were isolated from lupus patients with active disease and healthy subjects and assayed for HIF-1α, miR-210, FoxP3 and RORγt expression by qPCR as described in Materials and Methods. Bar heights represent mean relative expression and error bars represent mean relative expression ± SEM of four independent biological replicates. * p < 0.05 and *** p < 0.001, n = 4. (F) HIF-1α protein is up-regulated in human lupus CD4+ T cells. Total protein was purified from CD4+ T cells isolated from lupus patients with active disease and healthy subjects and subjected to Western blot analysis for HIF-1α protein expression as described in Materials and Methods.
HIF-1α and miR-210 are up-regulated in human lupus and expressed mainly by CD4+ T cells
We purified peripheral blood mononuclear cells (PBMC) from lupus patients and quantified HIF-1α and miR-210 expression levels by qPCR. As shown in Figure 1C, we measured overexpression of both HIF-1α (5.3 ± 0.56-fold ) and miR-210 (43.4 ± 1.3-fold) in human lupus PBMCs compared to healthy controls. We then asked if HIF-1α or miR-210 may be expressed by distinct lymphocyte lineages in human lupus. We purified CD4+, CD8+ and CD19+ lymphocytes from lupus patients and healthy control subjects and quantified HIF-1α and miR-210 expression by qPCR. We measured robust and statistically significant up-regulation of HIF-1α (Figure 1D) and miR-210 (Figure 1E) in the CD4+ lineage (7.2 ± 1.6-fold and 46.2 ± 1.8-fold respectively) compared to HIF-1α and miR-210 expression in control subject CD4+ cells. In contrast, HIF-1α and miR-210 expression in human lupus CD8+ and CD19+ cells was not significantly different from controls. HIF-1α and miR-210 expression in CD4+ cells was similar to HIF-1α and miR-210 expression in lupus patient PBMC’s suggesting that HIF-1α and miR-210 are expressed mainly by the CD4+ T cell lineage in human lupus. To confirm HIF-1α differential expression at the protein level, we purified CD4+ cells from lupus patients with active disease and healthy control subjects and subjected protein lysates to Western blot analysis for HIF-1α expression. As shown in Figure 1F, HIF-1α protein levels were significantly elevated in CD4+ cells from lupus patients compared to control subjects. Taken together, our results show that HIF-1α and miR-210 are overexpressed in human lupus and that the majority of HIF-1α and miR-210 activity is contained in the CD4+ T cell compartment.
RORγt is up-regulated in human lupus and associates with HIF-1γ activity
RORγt is a direct transcriptional target for HIF-1α. Having shown HIF-1α overexpression in lupus peripheral lymphocytes (Figure 1C), we asked if RORγt is also overexpressed. Peripheral lymphocytes were purified from lupus patients and healthy controls and assayed for FoxP3 and RORγt expression by qPCR. As shown in Figure 1G, we detected a significant 4.6 ± 0.49-fold up-regulation of RORγt mRNA levels that positively correlated with HIF-1α expression in these cells. In contrast, FoxP3 expression did not significantly differ between lupus and control cells. These data support the hypothesis that HIF-1γ overexpression in human lupus CD4+ cells may influence the Treg/Th17 cell ratio by transcriptional up-regulation of RORγt.
Discussion
It is increasingly appreciated that cellular metabolism profoundly influences effector T cell differentiation and function. T cell activation and clonal expansion triggers extensive cellular metabolic reprogramming characterized by a transition from oxidative to glycolytic metabolism (Pearce, 2010). In SLE, autoreactive T cells display a sustained activated phenotype due to a lowered threshold of activation and persistent stimulation by autoantigens (La Cava, 2009). T cells from both lupus-prone mice and human lupus patients present a hyperactive metabolic profile characterized by increased rates of both glycolysis and oxidative phosphorylation (Vukelic et al., 2020). The importance of T cell metabolism in the etiology of SLE immunopathology is illustrated by metabolic studies in lupus-prone mice. Inhibition of Glut1 in MRL/lpr T cells ameliorates disease activity (Jacobs et al., 2008) and normalization of the hyperactive metabolic phenotype in the Sle123 model reverses autoimmune phenotypes (Yin et al., 2015; Yin et al., 2016).
Regulation of T cell metabolism is the primary function of the transcription factor HIF-1α under both oxygen-dependent and -independent conditions (Del Rey et al., 2017). Elevated HIF-1α levels strongly associate with increased disease activity in a variety of autoimmune and inflammatory diseases with diverse pathologies, including rheumatoid arthritis (Ivan and Huang, 2014), type 1 diabetes mellitus (Loukovaara et al., 2014), multiple sclerosis (Graumann et al., 2003), and inflammatory bowel disease (Mimouna et al., 2011). Dang et. al. showed that HIF-1α-deficient mice are strongly resistant to induction of experimental autoimmune encephalitis (Dang et al., 2011), Kojima et. al. report development of an autoimmune phenotype in Hif1α–/– → Rag2–/– chimeric mice (Kojima et al., 2002) and recently, Zhao et.al reported that RNAi silencing of HIF-1α reduced development of an autoimmune phenotype in MRL/lpr mice (W. Zhao et al., 2018). Taken together, these reports suggest that inappropriate HIF-1α expression contributes to the development of autoimmune immunopathology.
In the current study, we measured a 7-fold HIF-1α up-regulation in human lupus CD4+ T cells (Figure 1D) that was statistically different from controls and paralleled HIF-1α up-regulation in lupus PBMCs (Figure 1C). In contrast, HIF-1α expression in CD8+ and CD19+ cells was similar to controls (Figures 1B and 1E). These data suggest that the majority of HIF-1α activity is contained in the CD4 T cell lineage. None of the lupus subjects had active infection, hypoxia due to chronic lung disease or was an active smoker, suggesting that HIF-1α overexpression in these subjects may be ascribed to chronic activation of autoreactive T cells. Lupus patients with moderate-to-severe disease or those unresponsive to conventional therapeutics are often prescribed one or more non-specific immunosuppressive medications (Tsokos, 2011). Although effective in many cases, these medications can cause undesirable side-effects and increased risk of infection or other complications. In the context of the above discussion, our results suggest that achieving or maintaining “normal” HIF-1α expression in lupus patients may be a novel therapeutic strategy.
MicroRNAs exert a regulatory influence on numerous aspects of cellular biology (Bartel, 2009, 2018). MiR-210 is the only miRNA consistently and robustly up-regulated by HIF-1α (Ivan and Huang, 2014). HIF-1α binds a highly conserved hypoxia-responsive element (HRE) on the proximal miR-210 promoter and mutation of this site completely abolishes the responsiveness of the miR-210 promoter to HIF-1α (X. Huang et al., 2009; H. Wang et al., 2014). HIF-1α is also a direct regulatory target of miR-210. MiR-210 regulates HIF-1α expression at both the protein and transcript levels via a non-canonical miR-210 target site in the HIF-1α mRNA 3’ UTR (H. Wang et al., 2014). The miR-210 host gene coding sequence is highly conserved across species suggesting an important functional role for miR-210 and its regulatory targets (Ivan and Huang, 2014).
Inappropriate miRNA expression profiles characterize many human diseases (Esteller, 2011) including SLE (de Yebenes et al., 2008; Garchow et al., 2011; Gururajan et al., 2010; Zan et al., 2014). MiR-210 expression has been well characterized in diseases with an hypoxic signature (Bavelloni et al., 2017) and in a number of autoimmune and inflammatory diseases. Zhao, et al. reports miR-210 overexpression in CD4+ T cells from psoriasis vulgaris patients and showed that miR-210 influences the immunosuppressive functions of Treg cells by targeting FOXP3 (M. Zhao et al., 2014). Wu et.al show that miR-210 induces Th17 and Th1 cell differentiation and inhibits Th2 differentiation in psoriasis by repression of STAT6 and LYN activity (Wu et al., 2018). Zheng, et al. detected elevated serum miR-210 levels in Graves’ disease patients (Zheng et al., 2018) and Chen, et al. reports significantly increased miR- 210 expression in placenta from women with preeclampsia (Chen et al., 2019). In contrast, the expression and function of miR-210 in lupus has been largely unexplored.
In the current study, we show that miR-210 is overexpressed in lupus-prone mouse lymphocytes (Figure 1B) and positively associates with increased disease activity (Figure 1A). We detected robust miR-210 overexpression in lupus patient PBMC’s (Figure 1C) consistent with Huang et al., who report miR-210 overexpression in lupus patient PBMC’s (Q. Huang et al., 2018). In a large-scale miRNA-Seq experiment, Kuchen, et. al. profiled miRNA expression patterns during lymphopoiesis and in several non-lymphoid tissues (Kuchen et al., 2010). The authors found that miR-210 was one of 49 miRNAs preferentially upregulated in lymphocytes and that miR-210 up-regulation was exclusive to CD4+ T cells. Consistent with this report, we measured robust miR-210 up-regulation in CD4+ T cells from both lupus-prone mice (Figure 1B) and in human lupus (Figure 1E) but only modest miR-210 up-regulation in CD8+ and CD19+ cells. We detected similar miR-210 expression levels in human lupus CD4+ T cells and PBMC’s suggesting that the majority of miR-210 activity is contained in the CD4 T cell compartment. MiR-210 expression human lupus correlated with HIF-1α expression at both the mRNA (Figure 1D) and protein levels (Figure 1F) supporting the hypothesis of HIF-1α transcriptional regulation of miR-210 expression.
FoxP3 and RORγt are the master transcription factors that define the Tregulatory (Treg) and Thelper17 (Th17) cell lineages respectively. RORgt regulates the development of Th17 cells which secrete IL-17, an inflammatory cytokine that promotes development of autoimmunity (McGeachy et al., 2019; Park et al., 2005; Rutz et al., 2013). Absolute numbers of IL-17+ cells and IL-17 levels are elevated in several autoimmune diseases including SLE (Wong et al., 2008) although a positive correlation between increased IL-17 levels and SLE disease activity has not been firmly established. In contrast, Tregs help maintain self-tolerance by suppressing the activity of autoreactive lymphocytes, however, the function of Tregs in SLE remains controversial (Li et al., 2019).
HIF-1α promotes Th17 differentiation in vitro and in experimental autoimmune encephalomyelitis (EAE) by direct transcriptional activation of RORγt expression and suppression of FoxP3 transcriptional activity (Dang et al., 2011; Shi et al., 2011; H. Wang et al., 2014). We consistently detected robust RORγt up-regulation in human lupus PBMCs that positively correlated with HIF-1α overexpression (Figure 1F). In contrast, FoxP3 expression was not significantly different from controls (Figure 1F). These data suggest that increased HIF-1α activity in lupus T cells may promote Th17 differentiation and inhibit Treg development in lupus.
In summary, our results suggest that a HIF-1α/miR-210 regulatory pathway may play an important and previously unrecognized role in lupus immunopathology. Dissection of HIF-1α/miR-210 and HIF-1α/miR-210-regulated pathways may represent a novel and productive direction for future etiological studies in lupus.
Supplementary Material
Highlights.
A key role for HIF-1α in has been described in a number of autoimmune and inflammatory diseases with diverse pathologies but remains unexplored in SLE.
MiR-210 expression in lupus-prone mice associates with disease activity and is robustly and selectively upregulated in the CD4+ T cell lineage.
HIF-1α and miR-210 are over-expressed in human SLE patient peripheral lymphocytes and positively associate with RORγt and IL-17 expression.
Results suggest that HIF-1α/miR-210 may represent a novel and and previously unrecognized regulatory axis in lupus.
Acknowledgments
Funding
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases [R03AR060988 and R21AR063416] and Thomas Jefferson University [080-18040-913251],
Abbreviations
- SLE:
Systemic Lupus Erythematosus
- HIF-1α
Hypoxia-inducible factor 1-alpha
- MiR-210
Micro-RNA-210 (hsa-miR-210-3p)
- RORγt
Retineic-acid-receptor-related orphan nuclear receptor gamma
- IL-17
Interleukin 17A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no competing financial interest.
Ethics and Consent
Human subject samples were collected with prior written informed consent and in accordance with the ethical standards of the Institutional Review Boards at Thomas Jefferson University. All animal care and experimental procedures were conducted with prior approval by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
References
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2018. Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, Faenza I, 2017. MiRNA-210: A Current Overview. Anticancer Res. 37, 6511–6521. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhao L, Wang D, Xu Y, Gao H, Tan W, Wang C, 2019. Contribution of regulatory T cells to immune tolerance and association of microRNA210 and Foxp3 in preeclampsia. Mol Med Rep 19, 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F, 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR, 2008. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med 205, 2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rey MJ, Valin A, Usategui A, Garcia-Herrero CM, Sanchez-Arago M, Cuezva JM, Galindo M, Bravo B, Canete JD, Blanco FJ, Criado G, Pablos JL, 2017. Hif-1alpha Knockdown Reduces Glycolytic Metabolism and Induces Cell Death of Human Synovial Fibroblasts Under Normoxic Conditions. Sci Rep 7, 3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, 2011. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, Obad S, Petri A, Kauppinen S, Kiriakidou M, 2011. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol. Med 3, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann U, Reynolds R, Steck AJ, Schaeren-Wiemers N, 2003. Molecular changes in normal appearing white matter in multiple sclerosis are characteristic of neuroprotective mechanisms against hypoxic insult. Brain Pathol. 13, 554–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD, 2010. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int.Immunol 22, 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Chen SS, Li J, Tao SS, Wang M, Leng RX, Pan HF, Ye DQ, 2018. miR-210 expression in PBMCs from patients with systemic lupus erythematosus and rheumatoid arthritis. Ir. J. Med. Sci 187, 243–249. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ, 2009. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 35, 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Huang X, 2014. miR-210: fine-tuning the hypoxic response. Adv. Exp. Med. Biol 772, 205–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC, 2008. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol 180, 4476–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Gu H, Nomura S, Caldwell CC, Kobata T, Carmeliet P, Semenza GL, Sitkovsky MV, 2002. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1alpha -deficient chimeric mice. Proc. Natl. Acad. Sci. U. S. A 99, 2170–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O'Shea J, Rajewsky K, Casellas R, 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M, 2007. A microRNA signature of hypoxia. Mol. Cell. Biol 27, 1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cava A, 2009. Lupus and T cells. Lupus 18, 196–201. [DOI] [PubMed] [Google Scholar]
- Li W, Deng C, Yang H, Wang G, 2019. The Regulatory T Cell in Active Systemic Lupus Erythematosus Patients: A Systemic Review and Meta-Analysis. Front Immunol 10, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loukovaara S, Koivunen P, Ingles M, Escobar J, Vento M, Andersson S, 2014. Elevated protein carbonyl and HIF-1alpha levels in eyes with proliferative diabetic retinopathy. Acta Ophthalmol 92, 323–327. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Cua DJ, Gaffen SL, 2019. The IL-17 Family of Cytokines in Health and Disease. Immunity 50, 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee EN, Korns Johnson D, Homann D, Clambey ET, 2013. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol. Res 55, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimouna S, Goncalves D, Barnich N, Darfeuille-Michaud A, Hofman P, Vouret-Craviari V, 2011. Crohn disease-associated Escherichia coli promote gastrointestinal inflammatory disorders by activation of HIF-dependent responses. Gut Microbes 2, 335–346. [DOI] [PubMed] [Google Scholar]
- Morel L, 2017. Immunometabolism in systemic lupus erythematosus. Nat. Rev. Rheumatol 13, 280–290. [DOI] [PubMed] [Google Scholar]
- Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, Wakeland EK, 2000. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. U. S. A 97, 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton VR, Tsokos GC, 2015. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J. Clin. Invest 125, 2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, Johnson RS, 2014. HIF transcription factors, inflammation, and immunity. Immunity 41, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C, 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol 6, 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, 2010. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol 22, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A, 2016. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat. Rev. Rheumatol 12, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Eidenschenk C, Ouyang W, 2013. IL-22, not simply a Th17 cytokine. Immunol. Rev 252, 116–132. [DOI] [PubMed] [Google Scholar]
- Semenza GL, 2009. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiol. (Bethesda) 24, 97–106. [DOI] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H, 2011. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med 208, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC, 2011. Systemic lupus erythematosus. N. Engl. J. Med 365, 2110–2121. [DOI] [PubMed] [Google Scholar]
- Vukelic M, Kono M, Tsokos GC, 2020. T cell Metabolism in Lupus. Immunometabolism 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL, 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A 92, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A, 2014. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat. Immunol 15, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Lit LC, Tam LS, Li EK, Wong PT, Lam CW, 2008. Hyperproduction of IL-23 and IL-17 in patients with systemic lupus erythematosus: implications for Th17-mediated inflammation in auto-immunity. Clin. Immunol 127, 385–393. [DOI] [PubMed] [Google Scholar]
- Wu R, Zeng J, Yuan J, Deng X, Huang Y, Chen L, Zhang P, Feng H, Liu Z, Wang Z, Gao X, Wu H, Wang H, Su Y, Zhao M, Lu Q, 2018. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J. Clin. Invest 128, 2551–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L, 2015. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med 7, 274ra218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Choi SC, Xu Z, Zeumer L, Kanda N, Croker BP, Morel L, 2016. Glucose Oxidation Is Critical for CD4+ T Cell Activation in a Mouse Model of Systemic Lupus Erythematosus. J. Immunol 196, 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Tat C, Casali P, 2014. MicroRNAs in lupus. Autoimmunity 47, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wang LT, Liang GP, Zhang P, Deng XJ, Tang Q, Zhai HY, Chang CC, Su YW, Lu QJ, 2014. Up-regulation of microRNA-210 induces immune dysfunction via targeting FOXP3 in CD4(+) T cells of psoriasis vulgaris. Clin. Immunol 150, 22–30. [DOI] [PubMed] [Google Scholar]
- Zhao W, Wu C, Li LJ, Fan YG, Pan HF, Tao JH, Leng RX, Ye DQ, 2018. RNAi Silencing of HIF-1alpha Ameliorates Lupus Development in MRL/lpr Mice. Inflammation 41, 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhuang C, Wang X, Ming L, 2018. Serum miR-146a, miR-155, and miR-210 as potential markers of Graves' disease. J. Clin. Lab. Anal 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


