Abstract
Severe asthma exacerbations are a major cause of school absences and healthcare costs in children, particularly those in high-risk racial/ethnic groups.
To identify susceptibility genes for severe asthma exacerbations in Latino children and adolescents, we conducted a meta-analysis of genome-wide association studies (GWAS) in 4,010 Latino youth with asthma in four independent cohorts, including 1,693 Puerto Ricans, 1,019 Costa Ricans, 640 Mexicans, 256 Brazilians, and 402 members of other Latino subgroups. We then conducted methylation quantitative trait locus (mQTL), expression quantitative trait locus (eQTL), and expression quantitative trait methylation (eQTM) analyses to assess whether the top SNP in the meta-analysis is linked to DNA methylation and gene expression in nasal (airway) epithelium in separate cohorts of Puerto Rican and Dutch children and adolescents.
In the meta-analysis of GWAS, a SNP in FLJ22447 (rs2253681) was significantly associated with 1.55 increased odds of severe asthma exacerbations (95% confidence interval= 1.34 to 1.79, P=6.3×10−9). This SNP was significantly associated with DNA methylation of a CpG site (cg25024579) at the FLJ22447 locus, which was in turn associated with increased expression of KCNJ2-AS1 in nasal airway epithelium from Puerto Rican children and adolescents (β=0.10, P=2.18 x 10−7).
SNP rs2253681 was significantly associated with both DNA methylation of a cis-CpG in FLJ22447 and severe asthma exacerbations in Latino youth. This may be partly explained by changes in airway epithelial expression of a gene recently implicated in atopic asthma in Puerto Rican children and adolescents (KCNJ2-AS1).
INTRODUCTION
Asthma is the most common chronic respiratory disease among children [1]. In the USA, total costs related to asthma exceed $81 billion per year [2]. Severe asthma exacerbations (SAEs), defined as episodes of disease worsening that require a change in treatment to prevent a serious outcome [3], are a major cause of school or work absences and healthcare costs. Of the ~6.1 million children with asthma in the U.S., 2.1% report ≥1 asthma-related hospitalization and 10.7% report ≥1 asthma-related visit to the emergency department (ED) in the previous year [4]. Despite recent advances, the best predictor of SAEs remains having had one in the previous year [5].
Although genome-wide association studies (GWASs) have identified susceptibility loci for asthma [6-8], little is known about genetic determinants of asthma exacerbations, which may be distinct from those for asthma per se. A GWAS in Danish children with asthma (ages 2–6 years) identified cadherin-related family member 3 (CDHR3), a gene not previously associated with asthma, as a susceptibility locus for recurrent SAEs [9]. In a combined GWAS of two cohorts of non-Hispanic white children with asthma, four intronic single nucleotide polymorphisms (SNPs) in cadherin-associated protein alpha 3 (CTNNA3) were significantly associated with SAEs [10], but such association was not replicated in an independent cohort including 786 children and adults with asthma. Moreover, a GWAS of SAEs among 806 non-Hispanic white children and adults with asthma who were being treated with inhaled corticosteroids found no genome-wide significant results [11].
The burden of asthma varies across racial or ethnic groups in the U.S. and Latin America. For example, Puerto Rican children have a greater prevalence of morbidity and mortality from asthma than non-Hispanic white children in the USA [12], and Costa Rican adolescents have a greater burden of asthma than those in other Latin American countries [13]. Moreover, recent evidence suggests that some susceptibility variants for asthma-related outcomes are ethnic-specific [14, 15]. We hypothesized that there would be susceptibility variants for SAEs that would be more common or exert a greater effect in Latino subgroups at risk for morbidity from asthma. To test this hypothesis, we conducted a meta-analysis of GWAS of SAEs among Latino youth with asthma in four independent studies.
METHODS
Please see the Online Supplement for more details.
Study populations included in the meta-analysis of GWAS of severe exacerbations
Hartford-Puerto Rico study
Hartford-Puerto Rico (HPR) study is a case-control study of childhood asthma in Puerto Ricans [6]. SAEs were defined as ≥1 hospitalization for asthma or ≥1 ED/urgent care visit for asthma requiring treatment with systemic corticosteroids in the previous year, or ≥1 course of systemic corticosteroids for asthma in the previous year. After quality control (QC) measures, 554 independent children with asthma (236 of whom had ≥1 SAE in the previous year) were included in the genome-wide association analysis, which was conducted using logistic regression under an additive genetic model, adjusting for age, sex, inhaled steroid use, and the first two principal components (PCs), calculated using smartPCA [16].
The Genetics of Asthma in Latino Americans study
The Genetics of Asthma in Latino Americans (GALA II) study is a case-control study of asthma in Latino children and youth [17]. An SAE was defined as having ≥1 of the following events in the previous year: hospitalizations for asthma, ED visits or unscheduled and urgent doctor's visits because of asthma, or treatment with systemic corticosteroids for asthma. The current analysis focused on 2181 children with asthma (1283 of whom had ≥1 SAE) and self-reported Latino ethnicity (1139 Puerto Rican, 640 Mexican, and 402 from other groups). Association testing was conducted using logistic regression under an additive genetic model, adjusting for age, sex, inhaled steroid use, ethnicity, and two PCs.
The Genetics of Asthma in Costa Rica Study
The Genetics of Asthma in Costa Rica Study (GACRS) is a genetic study of nuclear families of children with asthma in Costa Rica [18, 19]. An SAE was defined as ≥1 ED visit for asthma or ≥1 hospitalization for asthma in the previous year. After QC, 1019 independent children with asthma (851 of whom had ≥1 SAE) were included in the analysis, which was also conducted using logistic regression under an additive genetic model, adjusting for age, sex, inhaled steroid use, and the first two PCs.
The Social Changes, Asthma and Allergy in Latin America study, Bahia, Brazil
The Social Changes, Asthma and Allergy in Latin America (SCAALA) study is a longitudinal study of asthma and allergic diseases of Brazilian children [20]. Children who reported asthma symptoms and had data on SAEs (n=256) were included in this analysis. An SAE was defined as ≥1 ED visit or ≥1 hospitalization due to wheeze in the previous year. The analysis was conducted using logistic regression under an additive genetic model, with adjustment for age, sex, and the first two PCs from genotypic data.
Study populations included in molecular quantitative trait analyses in nasal epithelium
The Epigenetic Variation and childhood Asthma in Puerto Rico study
Nasal (airway) epithelium can serve as a surrogate marker for DNA methylation and gene expression in bronchial (airway) epithelium [21]. In the Epigenetic Variation and childhood Asthma in Puerto Rico (EVA-PR) study, whole-genome methylation assays were performed using HumanMethylation450 BeadChips (Illumina, San Diego, CA). After QC, 227 901 CpG probes remained for the analysis of nasal epithelium, and M-values were used in all downstream analyses. RNA sequencing was performed with the Illumina NextSeq 500 platform, paired-end reads at 75 cycles, and 80 million reads per sample. After QC, 16 737 genes were retained for the analysis. The R function sva was used to estimate latent factors that capture unknown data heterogeneity [22]. Of the 543 study participants, 457 had complete genome-wide data for genotypes, methylation, and transcriptomics in nasal epithelium (see Supplementary Figure 1).
Prevention and Incidence of Asthma and Mite Allergy study
The Prevention and Incidence of Asthma and Mite Allergy (PIAMA) study is a birth cohort study of children born in the Netherlands in 1996 and 1997 [23, 24]. A total of 479 nasal epithelial samples were hybridized to the Infinium HumanMethylation450 BeadChip arrays. After QC, 455 samples and 436 824 probes remained, and 432 samples had matched genotype data. RNA-sequencing was performed with the Illumina HiSeq 2500 platform, paired-end sequencing. After stringent QC, 17 156 genes and 326 samples remained, with 233 samples having matched genotype data [25].
Meta-analysis of GWAS of SAEs
METAL [26] software was used to perform the meta-analysis of GWAS of SAEs, using data from HPR, GALA II, GACRS, and SCAALA.
Molecular quantitative trait locus analyses
To estimate the effects of our top SNP on DNA methylation and gene expression, we conducted a methylation quantitative trait locus (mQTL) analysis to test for association between our top SNP and genome-wide DNA methylation in nasal epithelium, and an expression quantitative trait locus (eQTL) analysis to test for an association between our top SNP and genome-wide gene expression in nasal epithelium. We then conducted an expression quantitative trait methylation (eQTM) analysis to test whether the top CpG site identified in the mQTL analysis was associated with genome-wide gene expression in nasal epithelium. All analyses were adjusted for age, sex, asthma status, atopy status, the top five PCs from genotypic data, and latent factors estimated from sva [22]. In addition, RNA-sequencing batch and RNA sample sorting protocol (i.e., whole cells or CD326-positive nasal epithelial cells) [21] were adjusted for in the eQTM analyses, and methylation batch was adjusted for in the mQTL and eQTM analyses; both cis- and trans- effects were considered.
Pathway analysis
A pathway analysis was performed with MAGMA [27], which conducts SNP-wise gene analysis of summary statistics with correction for linkage disequilibrium between variants and genes, to test whether sets of genes are jointly associated with a phenotype (i.e. asthma exacerbation) compared to other genes across the genome. Adaptive permutation was used to produce an empirical p-value and false discovery rate (FDR). Gene sets used in the analyses were from GO [28, 29], Kyoto Encyclopedia of Genes and Genomes (KEGG) [30, 31], Reactome [32, 33], and BioCarta pathways.
RESULTS
The characteristics of the 4010 participants in the four studies included in the meta-analysis of GWAS of SAEs are shown in Table 1. In total, there are 2509 children with asthma and ≥1 SAE (cases) and 1501 children with asthma but no SAEs (controls). Compared to subjects who participated in the HPR or SCAALA studies, those in GALA II were older and those in the GACRS were younger. Compared to subjects in the other studies, those in the GACRS were more likely to be male and to have had ≥1 severe asthma exacerbation in the previous year.
Table 1:
Summary of main characteristics of study participants
| HPR (n=554) |
GALA II (n=2181) |
GACRS (n=1019) |
SCAALA (n=256) |
|
|---|---|---|---|---|
| Age in years (mean ± SD) | 10.0±2.7 | 12.7±3.3 | 9.2±1.9 | 7.2±1.9 |
| Male sex (n, %) | 300 (54.2) | 1196 (54.8) | 598 (58.7) | 139 (54.3) |
| Asthma exacerbation (n, %) | 236 (42.6) | 1283 (58.8) | 851 (83.5) | 139 (54.3) |
| Inhaled steroid use (n, %) | 187 (33.8) | 996 (45.7) | 521 (51.1) | Not available |
| FEV1 % pred#(mean ± SD) | 86.8±16.0 | 90.6±16.3 | 99.1±17.3 | Not available |
| FEV1/FVC % pred#(mean ± SD) | 91.8±9.8 | 96.3± 8.8 | 94.5±8.7 | Not available |
| Study sites | Hartford (CT) and San Juan (Puerto Rico) | Chicago (IL), Bronx (NY), Houston (TX), San Francisco (CA) and Puerto Rico | Costa Rica | Salvador (Bahia), Brazil |
| Genotyping platform | Illumina 2.5M | Affymetrix Axiom® LAT1 | Illumina Human Omni Express-12v1_A | Illumina Human Omni 2.5-8v1 |
HPR: Hartford-Puerto Rico study. GALA II: Genetics of Asthma in Latino Americans II study. GACRS: Genetics of Asthma in Costa Rica Study. SCAALA: Social Changes, Asthma and Allergy in Latin America study; FEV1: forced expiratory volume in 1s; FVC: forced vital capacity.
: for comparability, all percent predicted values across the three studies were calculated using reference values for Mexican American youth [45].
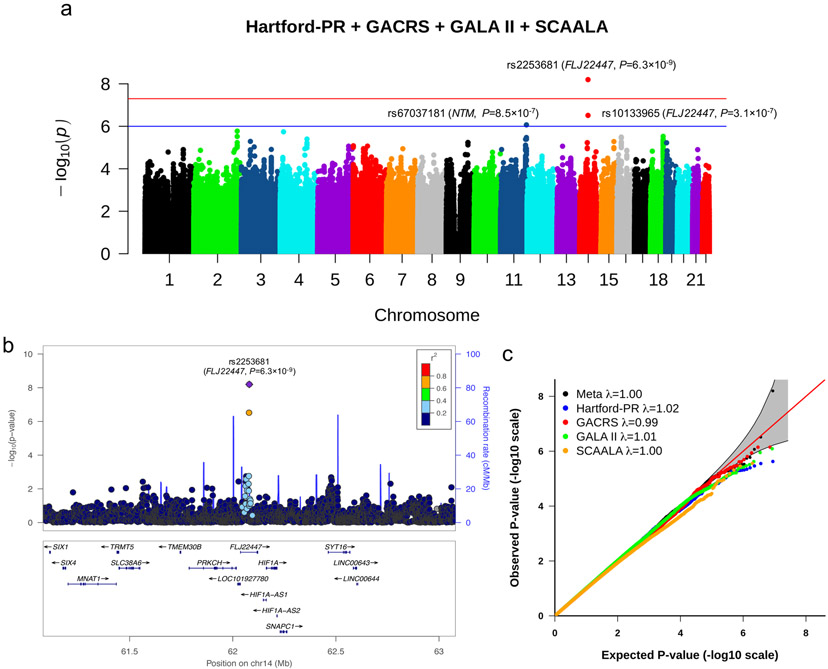
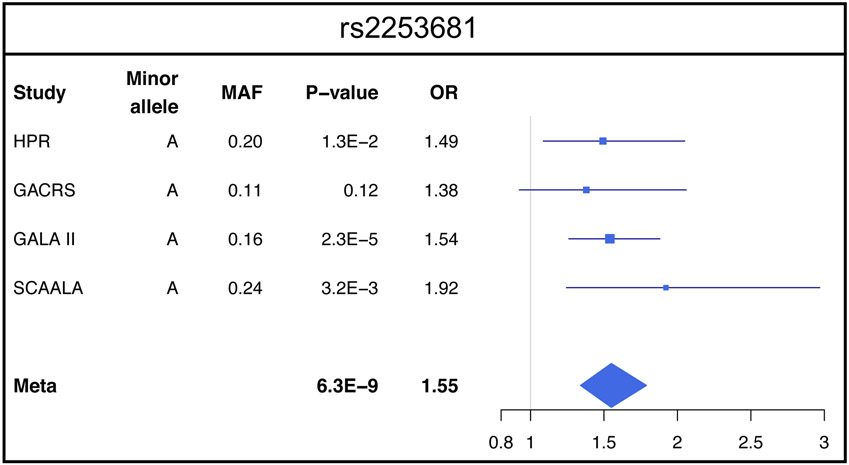
Approximately 6 million genotyped and imputed SNPs with a minor allele frequency (MAF) ≥0.05 were included in the meta-analysis of GWAS of SAEs. In this meta-analysis, one SNP (rs2253681, in FLJ22447 on chromosome 14q23.2) was significantly associated with SAEs at P < 5×10−8 (Figure 1a). This SNP was genotyped in HPR, GALA II and SCAALA, and imputed in GACRS with imputation quality r2=0.96. Each copy of the minor allele (A) of SNP rs2253681 was associated with 1.55 times increased odds of SAEs (95% CI 1.34 –1.79, P=6.3×10−9, Figure 1b). The Q-Q plot (Figure 1c) showed no inflation for the results for each of the four individual cohorts or for the pooled analysis (Figure 2). There was no significant interaction between SNP rs2253681 and inhaled steroid use on SAEs in the GWAS conducted in the HPR, GALA II, and GACRS cohorts (P for interaction ≥0.15 in all instances).
Figure 1.
1a) Manhattan plot of meta-analysis results: Manhattan plot showing the summary meta-analysis results of HPR, GALA II, GACRS, and SCAALA. The chromosomal position of each SNP is displayed along the X-axis and the negative logarithm of the association P-value is displayed on the Y-axis. The blue line represents the suggestive significance line (P < 1×10−6). The red line represents the genome-wide significance line (P < 5×10−8). HPR: Hartford-Puerto Rico cohort. GALA II: Genetics of Asthma in Latino Americans II. GACRS: Genetics of Asthma in Costa Rica Study, and the SCAALA (Social Changes, Asthma and Allergy in Latin America) Study. 1b) Results of the meta-analysis on the chromosome 14 region: The relative location of genes and the direction of transcription are shown in the lower portion of the figure, and the chromosomal position is shown on the x axis. The blue line shows the recombination rate across the region (right y axis), and the left y axis shows the significance of the associations. The purple diamond shows the P-value for rs2253681 that is the most significant SNP in the meta-analysis. The circles show the P-values for all other SNPs and are color coded according to the level of LD with rs2253681 in the 1000 Genome Project Admixed American (AMR) population. This plot was generated at www.locuszoom.org/. 1c) QQ plots for the meta-analysis. λ is the genomic control value.
Figure 2. Forest plots of odds ratio and 95% CI for the association with asthma for rs2253681, the most significant SNP in the meta-analysis.
The heterogeneity measure I2=0 that implies no heterogeneity among the ORs from the four studies (see supplementary text for details). HPR: Hartford-Puerto Rico cohort. GALA II: Genetics of Asthma in Latino Americans II. GACRS: Genetics of Asthma in Costa Rica Study, SCAALA: Social Changes, Asthma and Allergy in Latin America; MAF: minor allele frequency.
We then examined whether SNPs previously associated with asthma in either a multi-ancestry meta-analysis [7] or a meta-analysis from UK Biobank [34] were associated with SAEs in our analysis (Supplementary Table 1). None of the previously reported asthma-susceptibility SNPs was significantly associated with SAEs in our meta-analysis of Latino youth. Moreover, no SNPs associated with SAEs or asthma hospitalizations in previous candidate-gene studies (e.g., IL13, IL4RA) or GWAS (e.g., CDHR3, CTNNA3) [9, 10, 18, 35-38] were significantly associated with SAEs in our meta-analysis (Meta-P≥0.05 in all instances, see Supplementary Table 2). Although the previously reported SNP rs7216389 in ORMDL3 on chromosome 17q21 was nominally associated with SAEs in HPR (P=1.6×10−3), it did not reach statistical significance in our meta-analysis (Meta-P=0.06, see Supplementary Table 2).
To examine whether SNP rs2253681 affects methylation of the FLJ22447 locus in nasal epithelium, we first conducted a mQTL analysis. In this analysis, rs2253681 was a significant cis-acting mQTL in nasal epithelium for cg25024579 in FLJ22447 (Beta=0.55, P=3.6×10−16 and FDR-P=8.1×10−11 in EVA-PR; Beta=0.34, P=1.2×10−11 in PIAMA; and Meta-P=9.5×10−25 for a combined analysis of EVA-PR and PIAMA; Table 2). In addition, among the top 20 mQTL, cg05223396 in SH3PXD2B, cg13127574 in ADSSL1 and cg21115391 in CCDC67 were associated with rs2253681 at P<0.05 in PIAMA, in the same direction as in EVA-PR (Table 2). Next, we conducted an eQTL analysis of SNP rs2253681 in the FLJ22447 locus and gene expression in nasal epithelium, and found no cis- or trans- genome-wide significant results (Supplementary Table 3). Among the top 20 eQTL, SRF was associated with rs2253681 at P<0.05 in PIAMA, in the same direction as in EVA-PR (Supplementary Table 3).
Table 2:
Top 20 mQTLs for rs2253681 in EVA-PR nasal epithelial cells and replication results from PIAMA
| CpG | Chr | Position | Nearest gene |
Distance to gene |
EVA-PR | PIAMA | Meta | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | FDR* | Effect | P-value | Effect | P-value | |||||
| cg25024579 | 14 | 62 076 297 | FLJ22447 | 0 | 0.5520 | 3.55×10−16 | 8.08×10−11 | 0.3365 | 1.16×10−11 | 0.4114 | 9.45×10−25 |
| cg02787615 | 2 | 99 434 688 | KIAA1211L | 0 | −0.1080 | 9.68×10−7 | 1.10×10−1 | 0.0415 | 1.73×10−2 | −0.0161 | 2.41×10−1 |
| cg05223396 | 5 | 171 830 155 | SH3PXD2B | 0 | −0.2175 | 1.89×10−6 | 1.44×10−1 | −0.0641 | 2.52×10−2 | −0.1074 | 9.56×10−6 |
| cg24718015 | 17 | 40 489 721 | STAT3 | 0 | −0.1581 | 5.01×10−6 | 2.80×10−1 | 0.0215 | 5.00×10−1 | −0.0609 | 9.48×10−3 |
| cg21785067 | 1 | 54 587 182 | TCEANC2 | 8989 | 0.2305 | 7.39×10−6 | 2.80×10−1 | 0.0824 | 6.42×10−2 | 0.1458 | 1.48×10−5 |
| cg21917349 | 15 | 29 213 860 | APBA2 | 0 | −0.2427 | 7.89×10−6 | 2.80×10−1 | −0.0441 | 2.22×10−1 | −0.1050 | 4.82×10−4 |
| cg15904664 | 14 | 104 569 653 | ASPG | 0 | −0.2093 | 9.96×10−6 | 2.80×10−1 | −0.0574 | 1.24×10−1 | −0.1156 | 8.11×10−5 |
| cg11374933 | 8 | 9 766 172 | MIR124-1 | 5189 | −0.1660 | 1.18×10−5 | 2.80×10−1 | 0.0357 | 2.55×10−1 | −0.0464 | 5.49×10−2 |
| cg15375424 | 5 | 131 823 451 | IRF1 | 0 | −0.2272 | 1.37×10−5 | 2.80×10−1 | −0.0623 | 9.38×10−2 | −0.1177 | 1.02×10−4 |
| cg07975634 | 1 | 147 232 608 | GJA5 | 0 | −0.2236 | 1.49×10−5 | 2.80×10−1 | 0.0396 | 6.77×10−2 | 0.0002 | 9.92×10−1 |
| cg08487909 | 6 | 30 904 981 | DPCR1 | 3794 | −0.1356 | 1.59×10−5 | 2.80×10−1 | −0.0364 | 8.15×10−2 | −0.0668 | 1.23×10−4 |
| cg00203736 | 7 | 55 022 567 | EGFR | 64156 | −0.2440 | 1.86×10−5 | 2.80×10−1 | 0.0034 | 9.20×10−1 | −0.0614 | 3.54×10−2 |
| cg23696248 | 19 | 45 260 501 | BCL3 | 0 | −0.3099 | 2.01×10−5 | 2.80×10−1 | 0.0230 | 7.09×10−1 | −0.1164 | 1.33×10−2 |
| cg13127574 | 14 | 105 196 523 | ADSSL1 | 0 | −0.2116 | 2.11×10−5 | 2.80×10−1 | −0.0445 | 9.01×10−3 | −0.0621 | 1.18×10−4 |
| cg08403064 | 11 | 44 068 709 | ACCSL | 820 | −0.2011 | 2.23×10−5 | 2.80×10−1 | 0.0472 | 1.78×10−1 | −0.0406 | 1.50×10−1 |
| cg21115391 | 11 | 93 143 810 | CCDC67 | 0 | −0.2452 | 2.23×10−5 | 2.80×10−1 | −0.0945 | 9.51×10−3 | −0.1374 | 8.40×10−6 |
| cg01220257 | 19 | 45 260 935 | BCL3 | 0 | −0.1474 | 2.30×10−5 | 2.80×10−1 | 0.0269 | 5.78×10−1 | −0.0879 | 1.87×10−3 |
| cg12765028 | 4 | 13 526 659 | LINC01097 | 1282 | 0.1566 | 2.34×10−5 | 2.80×10−1 | −0.0063 | 7.70×10−1 | 0.0349 | 6.07×10−2 |
| cg11136041 | 1 | 158 111 350 | LOC646268 | 919 | −0.2204 | 2.39×10−5 | 2.80×10−1 | 0.0249 | 4.28×10−1 | −0.0404 | 1.33×10−1 |
| cg01735277 | 15 | 75 077 691 | CSK | 0 | −0.2291 | 2.46×10−5 | 2.80×10−1 | −0.0460 | 1.30×10−1 | −0.0897 | 7.22×10−4 |
FDR is adjusted for the whole genome in EVA-PR
Given the association between SNP rs2253681 and methylation of cg25024579, we then examined the effect of that CpG on gene expression in nasal epithelium, by conducting an eQTM analysis (Table 3). In this analysis, methylation of cg25024579 was significantly associated in trans with expression of the gene KCNJ2 antisense RNA 1 (KCNJ2-AS1) on chromosome 17q24.3 (Beta=0.10, P=2.2×10−7 and FDR-P=3.2×10−3) in EVA-PR. Although this was not replicated in PIAMA, the observed association remained significant in the combined analysis of the two cohorts (Beta=0.09, P=1.4×10−6). The top cis-eQTM gene in the current analysis in EVA-PR was protein kinase C-η (PKCη), encoded by PRKCH (Beta=0.07, P=4.1×10−5 and FDR-P=1.5×10−1). PRKCH expression was also associated with cg2504579 methylation in PIAMA, in the same direction of association as in EVA-PR (Beta=0.11, P=1.4×10−2; and P=2.4×10−6 in the combined analysis of EVA-PR and PIAMA). Among the top 20 eQTM in EVA-PR, SLC27A2 expression was associated with cg25024579 methylation in PIAMA, in the same direction of association as in EVA-PR (Beta=0.22, P=4.2×10−5 in PIAMA; and Beta=0.09, P=4.9×10−7 in the combined analysis of the two cohorts). In addition, expression of FOCAD, PIGF and ERLEC1 was associated with cg25024579 methylation in PIAMA at P<0.05, in the same direction as in EVA-PR.
Table 3:
Top 20 eQTMs for cg25024579 in EVA-PR nasal epithelial cells and replication results from PIAMA
| Gene | Chr | Start | End | EVA-PR | PIAMA | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | FDR* | Effect | P-value | Effect | P-value | ||||
| KCNJ2-AS1 | 17 | 68 163 101 | 68 165 543 | 0.1019 | 2.18×10−7 | 3.18×10−3 | −0.1265 | 1.95×10−1 | 0.0930 | 1.40×10−6 |
| CCDC125 | 5 | 68 576 518 | 68 616 410 | 0.0669 | 2.11×10−5 | 1.22×10−1 | 0.0439 | 4.03×10−1 | 0.0650 | 1.60×10−5 |
| FARS2 | 6 | 5 261 583 | 5 771 816 | 0.0563 | 2.50×10−5 | 1.22×10−1 | 0.0262 | 6.10×10−1 | 0.0544 | 2.58×10−5 |
| PRKCH | 14 | 61 788 514 | 62 017 698 | 0.0705 | 4.08×10−5 | 1.49×10−1 | 0.1119 | 1.35×10−2 | 0.0757 | 2.44×10−6 |
| TBL2 | 7 | 72 983 276 | 72 993 013 | 0.0567 | 5.81×10−5 | 1.53×10−1 | −0.0624 | 1.06×10−1 | 0.0427 | 1.27×10−3 |
| FOCAD | 9 | 20 658 307 | 20 995 954 | 0.0764 | 7.38×10−5 | 1.53×10−1 | 0.1647 | 2.70×10−2 | 0.0819 | 1.13×10−5 |
| ERLIN1 | 10 | 101 909 846 | 101 945 734 | 0.0596 | 9.62×10−5 | 1.53×10−1 | 0.0530 | 2.01×10−1 | 0.0588 | 4.10×10−5 |
| LOC401052 | 3 | 10 048 101 | 10 052 779 | 0.0001 | 9.75×10−5 | 1.53×10−1 | NA | NA | NA | NA |
| SMIM14 | 4 | 39 552 545 | 39 640 481 | 0.0752 | 9.97×10−5 | 1.53×10−1 | −0.0787 | 8.53×10−2 | 0.0519 | 3.55×10−3 |
| SLC27A2 | 15 | 50 474 392 | 50 528 589 | 0.0772 | 1.19×10−4 | 1.53×10−1 | 0.2221 | 4.23×10−5 | 0.0947 | 4.92×10−7 |
| BLOC1S6 | 15 | 45 879 416 | 45 901 909 | 0.0445 | 1.43×10−4 | 1.53×10−1 | 0.0104 | 7.89×10−1 | 0.0417 | 1.99×10−4 |
| TRMT12 | 8 | 125 463 047 | 125 465 266 | 0.0526 | 1.46×10−4 | 1.53×10−1 | 0.0887 | 9.43×10−2 | 0.0549 | 4.17×10−5 |
| CDK19 | 6 | 110 931 180 | 111 136 412 | 0.0728 | 1.48×10−4 | 1.53×10−1 | −0.1092 | 7.73×10−2 | 0.0568 | 1.93×10−3 |
| ACSS1 | 20 | 24 986 865 | 25 013 342 | 0.0720 | 1.48×10−4 | 1.53×10−1 | 0.0173 | 6.87×10−1 | 0.0630 | 2.81×10−4 |
| SEC22A | 3 | 122 920 773 | 122 992 982 | 0.0485 | 1.74×10−4 | 1.53×10−1 | −0.0870 | 1.15×10−1 | 0.0415 | 9.79×10−4 |
| PIGF | 2 | 46 808 412 | 46 844 251 | 0.0624 | 1.76×10−4 | 1.53×10−1 | 0.1262 | 4.12×10−2 | 0.0667 | 3.29×10−5 |
| NME6 | 3 | 48 335 588 | 48 342 848 | 0.0474 | 1.89×10−4 | 1.53×10−1 | 0.0335 | 3.94×10−1 | 0.0461 | 1.36×10−4 |
| ERLEC1 | 2 | 54 014 067 | 54 045 956 | 0.0532 | 2.07×10−4 | 1.53×10−1 | 0.0675 | 4.21×10−2 | 0.0554 | 2.53×10−5 |
| KATNAL2 | 18 | 44 526 786 | 44 628 614 | 0.0783 | 2.27×10−4 | 1.53×10−1 | 0.0687 | 3.38×10−1 | 0.0775 | 1.41×10−4 |
| PKHD1L1 | 8 | 110 374 705 | 110 543 500 | 0.1190 | 2.43×10−4 | 1.53×10−1 | −0.3652 | 1.87×10−1 | 0.1125 | 4.82×10−4 |
FDR is adjusted for the whole genome in EVA-PR
To further assess the biological relevance of the FLJ22447 locus, we conducted a SNP-wise pathway analysis of SAEs using the meta-analysis results and evaluated public repositories and databases. Although there were no pathways associated with SAEs after adjusting for multiple testing, there were a number of nominally significant pathways that included genes on chromosome 14q23.2, near FLJ22447 (Supplementary Table 4).
In a sensitivity analysis, we repeated the GWAS of SAEs after imputing genotypes using the 1,000 Genomes Ad Mixed American (AMR) reference panel instead of the Haplotype Reference Consortium (HRC) r1.1 2016 reference panel (see Online Supplement), obtaining very similar results (see Supplementary Figure 2 and Supplementary Table 5).
DISCUSSION
Our combined analysis including 4010 youth in four study cohorts showed that a novel SNP in FLJ22447 (rs2253681) is significantly associated with SAEs (P=6.3×10−9) among children and adolescents in Latino subgroups affected with asthma (Puerto Ricans, Costa Ricans, Mexicans and Brazilians) [12]. In an mQTL analysis in nasal epithelium, this SNP was significantly associated with DNA methylation of a CpG site at the FLJ22447 locus (cg25024579), which was in turn significantly associated with the expression of KCNJ2-AS1 in nasal epithelium.
Gene FLJ22447 codes for a long non-coding RNA, a type of RNA that does not get translated to protein but may serve pre- and post-transcriptional regulatory functions. FLJ22447 partially overlaps with AL355916.3, which encodes a protein kinase C paralog expressed in epithelial tissues. Of note, a SNP in AL355916.3 has been associated with forced expiratory volume in 1 s/forced vital capacity and bronchodilator response in adults [34]. FLJ22447 mediates interleukin (IL)-33 upregulation in activated fibroblasts; together, FLI22447 and IL-33 can increase fibroblast expression of α smooth muscle actin (α-SMA), vimentin, and N-cadherin [35], which are associated with myofibroblast differentiation and airway remodeling [36, 37].
KCNJ2-AS1 also encodes a long non-coding RNA. Expression of KCNJ2-AS1 (log2(fold-change)=0.34; P=3.9×10−6; FDR-P=3.1×10−5) was significantly associated with atopic asthma in our recent transcriptome-wide association study (TWAS) of atopic asthma in Puerto Rican children and adolescents [38]. SNPs rs8066985 and rs312750 near KCNJ2-AS1 are associated with body mass index and waist-to-hip ratio [39, 40], which are associated with worse asthma outcomes in children and adults. To our knowledge, this is the first study linking FLJ22447 or KCNJ2-AS1 to SAEs in children and adolescents.
While not significant genome wide, the top cis-eQTM gene in EVA-PR was PRKCH, which is adjacent to FLJ22447 and most highly expressed in the lungs [41]. Both methylation and transcription of this gene in nasal (airway) epithelium were recently associated with atopic asthma in Puerto Rican children and adolescents [21]. PKCη, encoded by PRKCH, plays a key role in the assembly and maintenance of epithelial tight junctions. PKCη phosphorylates occludin on threonine residues (T403 and T404), and such phosphorylation is required for the assembly and/or maintenance of occludin in epithelial tight junctions that are key to the integrity and function of the airway epithelial barrier [42].
Recent GWAS of exacerbations in non-Hispanic white children identified SNPs in two genes, CDHR3 and CTNNA3 [9, 10], neither of which replicated in our analysis, despite having similar or larger sample sizes than those in the original studies. Similarly, a previously reported association between a SNP on chromosome 17q21 (rs7216389) and SAEs was not statistically significant in our meta-analysis (P=0.06). While an association between the 17q21 locus and asthma is widely recognized, whether such locus is linked to severe disease exacerbations among children with asthma is less clear. The initial report in pediatric asthma analyzed a cohort of 376 children and reported 80 with recurrent wheezing and 66 with asthma [43]. The authors then analyzed the rate of severe exacerbations within the full cohort (57 among the 376 participants), rather than comparing children with asthma and an exacerbation to those with asthma but no exacerbation. A recent meta-analysis reported a significant association between rs7216389 and asthma hospitalizations and ED visits in children, but, while the pooled estimates were significant, only four of the 13 cohorts showed significant results [44]. Although lack of an association between CDHR3 and SAEs in the current study may be due to differences in the age of participants and outcome definitions across studies, our results for FLJ22447 are novel and further highlight the importance of studying asthma outcomes in children and adolescents of diverse races and ethnicities.
We recognize several study limitations. First, we lack data on viral infections or air pollution, which may interact with genetic or epigenetic mechanisms on causing SAEs in children. Second, we cannot assess temporal relationships between DNA methylation or gene expression and SAEs in our cross-sectional analysis. Third, we had insufficient statistical power to detect either uncommon risk alleles or alleles with modest genetic effects on SAEs. Moreover, we had limited statistical power to detect an interaction between our top SNP and inhaled steroid use on SAEs or for our molecular quantitative trait analyses, and lacked a replication cohort for analyses of DNA methylation or gene expression in nasal epithelium and SAEs (because PIAMA lacks adequate data on asthma exacerbations). Fourth, the definitions of SAEs varied across the study cohorts. However, we obtained similar results in a sensitivity analysis for the HPR and GALA II cohorts, in which we re-ran the GWAS after excluding subjects who received systemic corticosteroids but did not report an unscheduled and acute visit for asthma (data not shown).
In summary, we identified a novel SNP in FLJ22447 that is significantly associated with both SAEs in Latino children and adolescents and DNA methylation of a cis-CpG in FLJ22447 in nasal epithelium. This CpG is, in turn, linked to nasal epithelial expression of a gene recently implicated in atopic asthma in children and adolescents (KCNJ2-AS1). Future longitudinal studies of asthma omics should assess whether and how genetic variants affect airway epithelial function and SAEs in childhood.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants HL079966, HL117191, and MD011764 from the U.S. National Institutes of Health (NIH) to J.C.C. Dr. Yan’s contribution was supported by grant HL138098 from the U.S. NIH. Dr. Forno’s contribution was supported by grant HL149693 from the U.S. NIH. The GALA II study was supported by U.S. NIH grants to E.G.B: HL088133, HL004464, HL117004, ES015794, ES24844, TRDRP 24RT 0025, MD006902, and GM007546. M.P.Y. was supported by the Ramón y Cajal Program (RYC-2015-17205) and by grant SAF2017-83417R from the Spanish Ministry of Economy, Industry and Competitiveness. The PIAMA study was supported by The Netherlands Organization for Health Research and Development; The Netherlands Organization for Scientific Research; the Lung Foundation of the Netherlands (with methylation and gene expression studies supported by AF 4.1.14.001); The Netherlands Ministry of Spatial Planning, Housing, and the Environment; and The Netherlands Ministry of Health, Welfare, and Sport. C.Q. was supported by a grant from the China Scholarship Council.
REFERENCES
- 1.von Schacky C. Prophylaxis of atherosclerosis with marine omega-3 fatty acids. A comprehensive strategy. [DOI] [PubMed] [Google Scholar]
- 2.Nurmagambetov T, Kuwahara R, Garbe P. The Economic Burden of Asthma in the United States, 2008-2013. Ann Am Thorac Soc 2018: 15(3): 348–356. [DOI] [PubMed] [Google Scholar]
- 3.Fuhlbrigge A, Peden D, Apter AJ, et al. Asthma outcomes: exacerbations. J Allergy Clin Immunol 2012: 129(3 Suppl): S34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005-2009. Natl Health Stat Report 2011(32): 1–14. [PubMed] [Google Scholar]
- 5.Puranik S, Forno E, Bush A, et al. Predicting Severe Asthma Exacerbations in Children. Am J Respir Crit Care Med 2017: 195(7): 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Q, Brehm J, Pino-Yanes M, et al. A meta-analysis of genome-wide association studies of asthma in Puerto Ricans. Eur Respir J 2017: 49(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demenais F, Margaritte-Jeannin P, Barnes KC, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 2018: 50(1): 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 2011: 43(9): 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnelykke K, Sleiman P, Nielsen K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet 2014: 46(1): 51–55. [DOI] [PubMed] [Google Scholar]
- 10.McGeachie MJ, Wu AC, Tse SM, et al. CTNNA3 and SEMA3D: Promising loci for asthma exacerbation identified through multiple genome-wide association studies. J Allergy Clin Immunol 2015: 136(6): 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlin A, Denny J, Roden DM, et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immunity, inflammation and disease 2015: 3(4): 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosser FJ, Forno E, Cooper PJ, et al. Asthma in Hispanics. An 8-year update. Am J Respir Crit Care Med 2014: 189(11): 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forno E, Gogna M, Cepeda A, et al. Asthma in Latin America. Thorax 2015: 70(9): 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkart KM, Sofer T, London SJ, et al. A Genome-Wide Association Study in Hispanics/Latinos Identifies Novel Signals for Lung Function. The Hispanic Community Health Study/Study of Latinos. Am J Respir Crit Care Med 2018: 198(2): 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak ACY, White MJ, Eckalbar WL, et al. Whole-Genome Sequencing of Pharmacogenetic Drug Response in Racially Diverse Children with Asthma. Am J Respir Crit Care Med 2018: 197(12): 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006: 38(8): 904–909. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. American journal of respiratory and critical care medicine 2013: 188(3): 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunninghake GM, Soto-Quiros ME, Avila L, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol 2007: 120(1): 84–90. [DOI] [PubMed] [Google Scholar]
- 19.Hunninghake GM, Soto-Quiros ME, Avila L, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007: 119(3): 654–661. [DOI] [PubMed] [Google Scholar]
- 20.Barreto ML, Cunha SS, Alcantara-Neves N, et al. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study). BMC Pulm Med 2006: 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forno E, Wang T, Qi C, et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med 2019: 7(4): 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012: 28(6): 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunekreef B, Smit J, de Jongste J, et al. The prevention and incidence of asthma and mite allergy (PIAMA) birth cohort study: design and first results. Pediatr Allergy Immunol 2002: 13(s15): 55–60. [DOI] [PubMed] [Google Scholar]
- 24.Wijga A, Smit HA, Brunekreef B, et al. Are children at high familial risk of developing allergy born into a low risk environment? The PIAMA Birth Cohort Study. Prevention and Incidence of Asthma and Mite Allergy. Clin Exp Allergy 2001: 31(4): 576–581. [DOI] [PubMed] [Google Scholar]
- 25.Qi C, Jiang Y, Yang IV, et al. Nasal DNA methylation profiling of asthma and rhinitis. J Allergy Clin Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010: 26(17): 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw CA, Mooij JM, Heskes T, et al. MAGMA: generalized gene-set analysis of GWAS data. PLoS computational biology 2015: 11(4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000: 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gene Ontology C Gene Ontology Consortium: going forward. Nucleic Acids Res 2015: 43(Database issue): D1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic acids research 2016: 44(D1): D457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogata H, Goto S, Sato K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research 1999: 27(1): 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabregat A, Sidiropoulos K, Garapati P, et al. The Reactome pathway Knowledgebase. Nucleic acids research 2016: 44(D1): D481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croft D, O'Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic acids research 2011: 39(Database issue): D691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Lee PH, Chaffin MD, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet 2018: 50(6): 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding L, Ren J, Zhang D, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis 2018: 39(3): 397–406. [DOI] [PubMed] [Google Scholar]
- 36.Hackett TL, Warner SM, Stefanowicz D, et al. Induction of epithelial-mesenchymal transition in primary airway epithelial cells from patients with asthma by transforming growth factor-beta1. Am J Respir Crit Care Med 2009: 180(2): 122–133. [DOI] [PubMed] [Google Scholar]
- 37.Liu T, Liu Y, Miller M, et al. Autophagy plays a role in FSTL1-induced epithelial mesenchymal transition and airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol 2017: 313(1): L27–L40. [DOI] [PubMed] [Google Scholar]
- 38.Forno E, Zhang R, Jiang Y, et al. Transcriptome-wide and differential expression network analyses of childhood asthma in nasal epithelium. J Allergy Clin Immunol 2020: 146(3): 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015: 518(7538): 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler TW, Justice AE, Graff M, et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet 2015: 11(10): e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013: 45(6): 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T, Elias BC, Seth A, et al. PKC eta regulates occludin phosphorylation and epithelial tight junction integrity. Proc Natl Acad Sci U S A 2009: 106(1): 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bisgaard H, Bonnelykke K, Sleiman PM, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med 2009: 179(3): 179–185. [DOI] [PubMed] [Google Scholar]
- 44.Farzan N, Vijverberg SJ, Hernandez-Pacheco N, et al. 17q21 variant increases the risk of exacerbations in asthmatic children despite inhaled corticosteroids use. Allergy 2018: 73(10): 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 1999: 159(1): 179–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.