Abstract
Purpose
To investigate the role of consumption phenotypes as genetic proxies for alcohol misuse and nicotine dependence.
Methods
We leveraged GWAS data from well-powered studies of consumption, alcohol misuse, and nicotine dependence phenotypes measured in individuals of European ancestry from the UK Biobank (UKB) and other population-based cohorts (largest total N=263,954), and performed genetic correlations within a medical-center cohort, BioVU (N=66,915). For alcohol, we used quantitative measures of consumption and misuse via AUDIT from UKB. For smoking, we used cigarettes per day from UKB and non-UKB cohorts comprising the GSCAN consortium, and nicotine dependence via ICD codes from UKB and Fagerström Test for Nicotine Dependence from non-UKB cohorts.
Results
In a large phenome-wide association study, we show that smoking consumption and dependence phenotypes show similar strongly negatively associations with a plethora of diseases, whereas alcohol consumption shows patterns of genetic association that diverge from those of alcohol misuse.
Conclusions
Our study suggests that cigarette smoking consumption, which can be easily measured in the general population, may be good a genetic proxy for nicotine dependence, whereas alcohol consumption is not a direct genetic proxy of alcohol misuse.
Keywords: Alcohol, nicotine, consumption, dependence, genetics, polygenic analysis, PheWAS
1. INTRODUCTION
Recent large genome-wide association studies (GWAS) of substance use disorder phenotypes consistently show that there is only modest overlap [rg=0.38–0.77 (Kranzler et al., 2019; Sanchez-Roige et al., 2018)] between genetic factors that influence alcohol consumption and alcohol use disorder [AUD, (Johnson et al., 2020; Kranzler et al., 2019; Liu et al., 2019; Sanchez-Roige et al., 2018; Walters et al., 2018)], whereas smoking (cigarettes per day [CPD]) and nicotine dependence (ND) are almost genetically identical [rg=0.95 (Quach et al., 2020)].
Importantly, the alcohol consumption and use disorder studies show divergent patterns of genetic association with other diseases (Sanchez-Roige et al., 2020). Whereas alcohol consumption is genetically correlated with higher educational attainment, lower body mass index and lower risk of coronary heart disease and type 2 diabetes; AUD and aspects of alcohol misuse share genetic associations with psychiatric disorders (Kranzler et al., 2019; Sanchez-Roige et al., 2018). In contrast, genetic correlation analyses show consistent associations of both CPD and ND with higher risks of psychiatric disorders, lower educational attainment, and higher risks of coronary heart disease or its predisposing factors (Liu et al., 2019; Quach et al., 2020).
Sample sizes for GWAS of consumption phenotypes range from thousands to more than a million subjects from healthy volunteer collections, primarily from the GSCAN consortium, including UK Biobank (UKB) and 23andMe, and the Million Veterans Program (Kranzler et al., 2019; Liu et al., 2019; Sanchez-Roige et al., 2020, 2018; Zhou et al., 2019; Xu et al., 2020); however, past GWAS of dependence phenotypes and genetic correlation analyses included smaller samples of some high-risk populations (Hancock et al., 2018; Quach et al., 2020; Walters et al., 2018). Both ascertainment strategies can introduce bias. In this short communication, we are leveraging GWAS data from well-powered studies of consumption and misuse/dependence phenotypes. Unlike previous studies, we focus largely on the UKB to control for potential selection biases, but also comparing results from non-UKB cohorts, and perform genetic correlations within a medical-center cohort from Vanderbilt University Medical Center (BioVU). These analyses provide an evaluation of the degree to which the more easily and broadly obtained consumption phenotypes are good proxies for alcohol misuse and nicotine dependence.
2. MATERIAL AND METHODS
2.1. Participants and measurements
We used GWAS summary statistics largely from the UKB to control for potential selection biases that may differ across different cohorts. For alcohol phenotypes, we used GWAS summary statistics for alcohol consumption and misuse via the AUDIT (AUDIT-C and AUDIT-P, respectively) from our previous work [UKB, N=121, 604 (Sanchez-Roige et al., 2018)]. For smoking phenotypes, we used GWAS summary statistics for CPD from GSCAN, for which 45.7% were UKB participants (total N =263,954 across 25 independent cohorts, excluding 23andMe (Liu et al., 2019)]. For ND, we used GWAS summary statistics from 244,890 UKB participants with an International Classification of Disease (ICD version 10) code for ND (Watanabe et al., 2019). Because our measure of ND was binary, unlike all of our other quantitative variables, we also included data from a quantitative measure (the Fagerström Test of ND), available only from non-UKB cohorts in the Nicotine Dependence GenOmics (iNDiGO) consortium (total N=46,213 across 20 cohorts (Quach et al., 2020)].
2.2. Phenome-wide association analysis
We computed polygenic risk scores (PRSs) for the four alcohol and nicotine phenotypes using the PRS-CS (Ge et al., 2019) “auto” version (i.e., the global shrinkage parameter phi was learned from the data in a Bayesian approach) for each of the 66,915 genotyped individuals of European descent in BioVU. BioVU is one of the largest biobanks in the United States, consisting of electronic health record data from the Vanderbilt University Medical Center on ∼250,000 patients captured from 1990 to 2017. Genotyping and QC of this sample have been described elsewhere (Dennis et al., 2019). In the genotyped BioVU sample, we fitted a logistic regression model to each of 1,335 case/control disease phenotypes to estimate the odds of each diagnosis given the PRS, after adjustment for sex, median age of the longitudinal electronic health record measurements, and first ten principal components of ancestry. Phenome-wide association study (pheWAS) analyses were run using the PheWAS R package v0.12 (Carroll et al., 2014). We required the presence of 100 cases with at least two or more ICD codes that mapped to a PheWAS disease category (otherwise known as “phecode”, aka the hierarchical groupings of ICD codes curated for pheWAS analyses; Phecode Map 1.2, https://phewascatalog.org/phecodes) to assign “case” status. We used the standard Benjamini—Hochberg False Discovery Rate (FDR 5%) to correct for multiple testing. This threshold, however, is likely conservative because it incorrectly assumes independence between phecodes. To explore whether pleiotropic effects of the PRS were mediated by the diagnosis of tobacco use disorder (TUD), we also conducted PheWAS analyses using TUD (phecode 318) as an additional covariate for each PRS. In addition, we repeated the PheWAS analyses using AUD diagnoses (phecodes 317, 317.1) as additional covariates for each PRS.
3. RESULTS
PRSs for both alcohol consumption and misuse were associated with AUD in BioVU (Figure 1, Supplementary Tables 1–2). Of 1,335 phenotypes, PRSs for alcohol misuse were positively associated (after 5% FDR corrections) with other mental disorders, including mood disorders, major depressive disorder, bipolar disorder, and suicidal ideation or attempt, replicating previous findings using a PRS of a clinical alcohol dependence (AD)/AUD measure (Zhou et al., 2019). In contrast, and replicating previous associations, alcohol consumption was negatively genetically correlated with metabolic conditions, such as type 2 diabetes and obesity. Adjusting the associations between alcohol consumption and metabolic disorders for AUD or TUD did not meaningfully change the magnitude of these associations, although the magnitude of the p-values increased slightly for some associations (Supplementary Tables 3–4). Similarly, adjusting the associations with alcohol misuse use for AUD or TUD increased the magnitude of the p-value (likely due to reductions in sample size) but the effect sizes remained largely unchanged (Supplementary Tables 5–6).
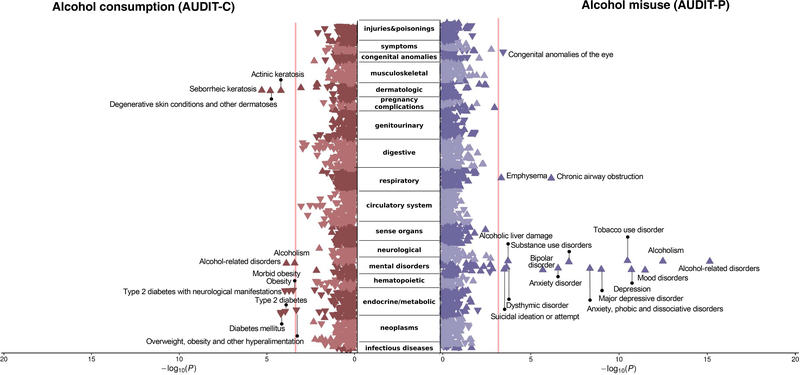
Figure 1. Phenome-wide association study of polygenic risk scores for alcohol consumption (AUDIT-C, first 3 items of the Alcohol Use Disorder Identification Test, AUDIT; left panel) and alcohol misuse (AUDIT-P, final 7 items; right panel), both traits measured in a subset of the UK Biobank cohort, against 1,335 phenotypes available in the biobank from Vanderbilt University Medical Center, BioVU.
The orientation of the triangles represents the direction of the effect sizes (beta coefficients). Of the 1,3355 phenotypes, 11 and 16 were significant after correction for multiple testing for AUDIT-C and AUDIT-P, respectively. The range of effect sizes, significant after FDR correction, were 0.913–1.119, 99% CI [0.909–1.143] for AUDIT-C, and 0.700–1.262, 99% CI [0.669–1.293] for AUDIT-P. The PheWAS of both alcohol consumption and misuse revealed positive genetic associations with alcohol use disorders, as well as identified divergent patterns of genetic association between consumption and misuse phenotypes. Only alcohol misuse, but not consumption, was positively genetically associated with psychopathology. Most of the associations became non-significant when corrected for tobacco or alcohol use disorder, although the magnitude and direction of the associations remained unchanged.
Conversely, both CPD and ND were negatively genetically associated with hundreds of other diseases in BioVU (Figure 2), including those known to be associated with smoking, such as chronic airway obstruction, lung cancer, and metabolic diseases (Supplementary Tables 7–8). Most of the associations between CPD or ND and psychiatric disorders were attenuated, and no longer significant, when we adjusted for TUD or AUD (Supplementary Tables 9–12). We repeated our analyses using a quantitative measure of ND and obtained very similar findings (Supplementary Tables 13–15).
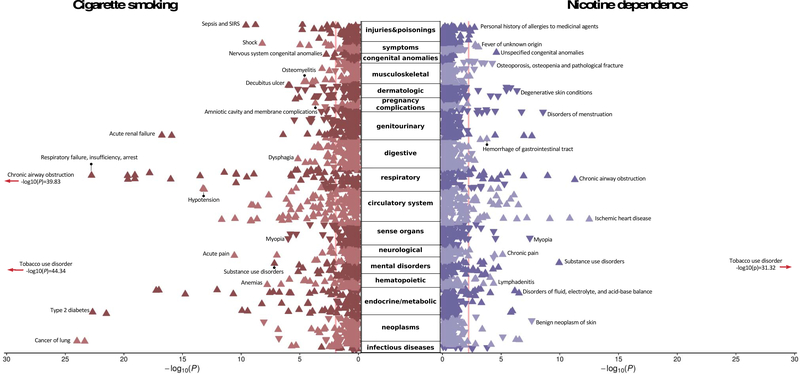
Figure 2. Phenome-wide association study of polygenic risk scores for cigarette smoking (cigarettes per day; left panel) and nicotine dependence (right panel), both traits measured primarily in the UK Biobank cohort, against 1,335 phenotypes available in the biobank from Vanderbilt University Medical Center, BioVU.
Of the 1,335 phenotypes, 321 and 50 were significant after correction for multiple testing for cigarettes per day and nicotine dependence, respectively. The range of effect sizes, significant after FDR correction, were 0.784–1.383, 99% CI [0.779–1.388] for cigarettes per day, and 0.766–1.214, 99% CI [0.758–1.221] for nicotine dependence. PheWAS of both smoking consumption and dependence revealed positive genetic associations with numerous medical conditions from nearly every bodily system. Only the strongest associations in each category are depicted. Most of the associations became non-significant when corrected for tobacco or alcohol use disorder.
All pairs of PRSs showed significant correlations, except for AUDIT-C PRS and ND PRS (Supplementary Table 16). All r coefficients were positive, the strongest association being between CPD and FTND (quantitative measure of ND, r=0.347), except for AUDIT-C’s associations with and CPD and with FTND, which showed a negative association (r=−0.043 and r=−0.020 respectively).
4. DISCUSSION
The current study examines smoking and alcohol consumption phenotypes as genetic surrogates for nicotine dependence and alcohol misuse, respectively, using PRSs constructed from well-powered GWAS in the UKB and other population-based non-UKB cohorts. In applying the PRSs to a large pheWAS, we found that smoking consumption was a good proxy for dependence, but alcohol consumption (as measured via AUDIT-C) was not a good proxy for alcohol misuse (as measured via AUDIT-P).
Ascertainment bias may explain some of the inverse genetic correlations between alcohol consumption and, for example, obesity and type 2 diabetes (Kranzler et al., 2019; Sanchez-Roige et al., 2018; Xue et al., 2020). UKB and other similar collections based on voluntary participation, are only available to individuals who are relatively healthy and who have both the means and opportunity to participate, resulting in an overrepresentation of data from individuals with higher education levels and socioeconomic status and alcohol consumption than the general population but, crucially, lower levels of metabolic disorders and problem drinking.
Importantly, both alcohol consumption (AUDIT-C) and alcohol misuse (AUDIT-P) were measured in UKB. Thus, the difference between alcohol consumption and misuse could indicate that the genetic overlap between alcohol consumption and AUD is dependent on the specific patterns of drinking (e.g. frequency of use vs. binge drinking or pathological use). For example, Polimanti et al (2019) identified a positive genetic correlation between alcohol dependence and alcohol drinking quantity (rg=0.75), but not frequency. Similarly, Marees et al (2020) showed that high alcohol consumption frequency was associated with high socioeconomic status and low risk of substance use disorders and other psychiatric disorders, whereas the opposite applied for high alcohol consumption quantity. Furthermore, these genetic correlations may be dissimilar to those observed when analyzing alcohol consumption in alcohol dependent individuals; such studies have yet to be performed.
Notably, even though studying alcohol consumption has shown some utility, it is apparent that this phenotype measured in volunteer collections is not an optimal proxy for AUD. Similar observations have recently been described for cannabis use (as measured primarily in UKB) versus disorder with regards to proxy measures of psychosocial and anthropometric indices (Johnson, 2020). Initial stages of recreational use may be etiologically distinct from later stages of pathological use for commonly used substances such as alcohol and cannabis, with only the latter stages of dependence and abuse indexing vulnerability to psychiatric impairment. Whereas use of nicotine may be a more addictive, alcohol, particularly measured in population-based cohorts such as the UKB, may represent a social habit.
In contrast with alcohol, the genetic correlation between smoking consumption (measured as CPD) and ND was almost identical, and both scores showed similar patterns of genetic association with psychiatric and smoking-related comorbid diseases. We speculate that consumption phenotypes (i.e., drinks/week and CPD) represent distinct indices of use depending on the drug: cigarette smoking may be a more accurate phenotype than drinks consumed (which may vary in sizes and patterns of drinking), in addition to being a better index outcome of problematic use, such that quantity of cigarettes smoked may reflect ND. Indeed, CPD is a major component of standard measures that are used to define ND, most notably the Fagerström Test for ND. Although studying the genetics of ND alongside other smoking traits (e.g., initiation and cessation) is key to gaining a better understanding of the neurobiological processes that influence the trajectory of smoking behaviors and their treatment implications, our findings suggest that smoking consumption phenotypes measured in volunteer cohorts can capture relevant sources of genetic information applicable to later stages of dependence and abuse.
Our analyses are not without limitations. We lack information regarding the potencies of cigarette and alcohol products used by individuals in the discovery (UKB) and target (BioVU) samples. High-potency substance use is associated with increased severity of dependence, especially at younger ages. Relatedly, our results may be restricted to PRSs calculated in populations with low levels of alcohol-related problems, like UKB. Furthermore, although we used a proxy measure of alcohol misuse (AUDIT-P), instead of a clinical diagnosis of AD, the pheWAS findings using problematic alcohol use (a combination of AUDIT-P and AD/AUD) in BioVU suggest that the results of AUDIT-P PRS are similar to AD PRS (Zhou et al., 2019). In addition, results from our sensitivity analyses revealed that the associations were slightly attenuated after diagnosis of AUD or TUD were included as covariates. It is plausible that many of the relationships between alcohol misuse and ND and psychopathology, detected in BioVU, may be consequences of an AUD or TUD diagnosis rather than due to shared genetic risk. Alternatively, these associations (or lack of thereof) could reflect pleiotropy (e.g., with substance use disorders and other mental disorders) or be a causal peripheral effect of alcohol/nicotine persistent/pathological use. The reduction in the number of statistically significant associations after adjustment for AUD or TUD may imply shared genetic liability between these disorders and comorbid psychopathology, or that, simply, our correction for AUD or TUD was too stringent considering that most of the effect sizes were essentially unchanged and that we expected some degree of collinearity between AUD/TUD diagnosis and the PRSs that we calculated. Future studies should aim at exploring causal mechanisms. Lastly, our estimates of genetic overlap may be sensitive to environmental factors, for example when comparing results from UKB (generally older) to younger cohorts.
In summary, we performed a pheWAS of consumption and dependence/misuse polygenic scores. We conclude that smoking consumption (but not alcohol consumption) measured in healthy volunteer cohorts is a powerful proxy for genetic studies of ND (but not AUD). For alcohol consumption, by using multivariate approaches that give statistically-derived weights to alcohol phenotypes (e.g. AUDIT-C items) or by including further restrictions to the study cohort (Xue et al., 2020), we may be able to mitigate some of the inverse associations between alcohol consumption and poor health and, in doing so, we may realize the full potential of alcohol consumption phenotypes as proxies for AUD. Moreover, and as a collateral finding, we identified very robust associations between well-characterized measures of alcohol and nicotine consumption, misuse, and clinical diagnoses from a real-world medical-center setting (BioVU). These series of analyses demonstrate the value of using broad electronic health record measures (i.e. presence of ICD codes) for genetic studies of substance use disorders.
Supplementary Material
HIGHLIGHTS.
Cigarettes smoked per day is a good genetic proxy for nicotine dependence.
Dissimilar genetic association patterns occur between alcohol use and misuse.
Alcohol phenotypes may be more influenced by measurement and ascertainment biases.
ACKNOWLEDGEMENTS AND DISCLOSURES
The authors declare no conflict of interest. We thank Dr. Mallard for sharing the code to construct the graphics, and Mariela V Jennings for her assistance in the preparation of this manuscript.
FUNDING
SSR was supported by the Families for Borderline Personality Disorder Research (Beth and Rob Elliott) 2018 NARSAD Young Investigator Grant (#27676) and funds from the Tobacco-Related Disease Research Program of the University of California (Grant #T29KT0526).
The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/. DBH and EOJ were supported by NIDA and NIAAA grant numbers R01DA042090 and R01AA027049, respectively. NJC and LKD obtained support from 1R01MH113362, 1R01MH118233, and 1R56MH120736.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
REFERENCES
- Carroll RJ, Bastarache L, Denny JC, 2014. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 30, 2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, Sealock J, Levinson RT, Farber-Eger E, Franco J, Fong S, Straub P, Hucks D, Linton MF, Song W-L, Fontanillas P, Elson SL, Ruderfer D, Abdellaoui A, Sanchez-Roige S, Palmer AA, Boomsma DI, Cox N, Chen G, Mosley JD, Wells QS, Davis L, 2019. Genetic risk for major depressive disorder and loneliness in gender-specific associations with coronary artery disease. Mol. Psychiatry. 10.1038/s41380-019-0614-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW, 2019. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Guo Y, Reginsson GW, Gaddis NC, Lutz SM, Sherva R, Loukola A, Minica C, Markunas CA, Han Y, Young KA, Gudbjartsson DF, Gu F, McNeil DW, Qaiser B, Glasheen C, Olson S, Landi MT, Madden PAF, Farrer LA, Vink J, Saccone NL, Neale MC, Kranzler HR, McKay J, et al. , 2018. Genome-wide association study across European and African American ancestries identifies a SNP in DNMT3B contributing to nicotine dependence. Mol. Psychiatry 23, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, Sanchez-Roige S, Paul SE, Wendt FR, Clarke T-K, Lai D, Reginsson GW, Zhou H, He J, Baranger DAA, Gudbjartsson DF, Wedow R, Adkins DE, Adkins AE, Alexander J, Bacanu S-A, Bigdeli TB, Boden J, Brown SA, Bucholz KK, et al. , 2020. Genome-wide association study meta-analysis of cannabis use disorder in over 380,000 individuals reveals distinct genetic architectures of cannabis use and use disorder. Lancet Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Sanchez-Roige S, Acion L, Adams MJ, Bucholz KK, Chan G, Chao MJ, Chorlian DB, Dick DM, Edenberg HJ, Foroud T, Hayward C, Heron J, Hesselbrock V, Hickman M, Kendler KS, Kinreich S, Kramer J, Kuo SI-C, Kuperman S, Lai D, McIntosh AM, Meyers JL, Plawecki MH, Porjesz B, et al. , 2020. Polygenic contributions to alcohol use and alcohol use disorders across population-based and clinically ascertained samples. Psychol. Med. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J, 2019. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, 23andMe Research Team, HUNT All-In Psychiatry, Choquet H., Docherty AR., Faul JD., Foerster JR., Fritsche LG., Gabrielsen ME., Gordon SD., Haessler J., Hottenga JJ., Huang H., Jang SK., Jansen PR., et al. , 2019. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJA, Ong J-S, MacGregor S, An J, Denys D, Vorspan F, van den Brink W, Derks EM, 2020. Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychol. Med. 50, 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Peterson RE, Ong J-S, MacGregor S, Edwards AC, Clarke T-K, Frank J, Gerring Z, Gillespie NA, Lind PA, Maes HH, Martin NG, Mbarek H, Medland SE, Streit F, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Agrawal A., Edenberg HJ., Kendler KS., Lewis CM., Sullivan PF., Wray NR., Gelernter J., Derks EM., 2019. Evidence of causal effect of major depression on alcohol dependence: findings from the psychiatric genomics consortium. Psychol. Med. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, Zellers S, Sherva R, Aliev F, Nothnagel M, Young KA, Marks J, Young H, Guo Y, Waldrop A, Sey N, Landi MT, McNeil DW, Farrer LA, Markunas CA, Vink J, Hottenga J-J, Iacono WG, Kranzler HR, Saccone NL, et al. , 2020. Expanding the Genetic Architecture of Nicotine Dependence and its Shared Genetics with Multiple Traits: Findings from the Nicotine Dependence GenOmics (iNDiGO) Consortium. Nat. Commun. 11, 5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Clarke T-K, 2020. Recent Efforts to Dissect the Genetic Basis of Alcohol Use and Abuse. Biol. Psychiatry 87, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, McIntosh AM, Clarke T-K, 2018. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. AJP 176, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million Veteran Program Xu K, Li B., McGinnis KA., Vickers-Smith R., Dao C., Sun N., Kember RL., Zhou H., Becker WC., Gelernter J., Kranzler HR., Zhao H., Justice AC., 2020. Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nat. Commun. 11, 5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen L-S, Clarke T-K, Chou Y-L, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, et al. , 2018. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, Posthuma D, 2019. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348. [DOI] [PubMed] [Google Scholar]
- Xue A, Jiang L, Zhu Z, Wray NR, Visscher PM, Zeng J, Yang J, 2020. Genome-wide analyses of behavioural traits biased by misreports and longitudinal changes. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey D, Cheng Z, Li B, Polimanti R, Kember RL, Smith RV, Thygesen JH, Morgan MY, Atkinson SR, Thursz MR, Nyegaard M, Mattheisen M, Børglum AD, Johnson EC, Program VMV., Justice AC., Palmer AA., McQuillin A., Davis LK., Edenberg HJ., Agrawal A., et al. , 2019. Meta-analysis of problematic alcohol use in 435,563 individuals identifies 29 risk variants and yields insights into biology, pleiotropy and causality. Nat. Neurosci. 23, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.