Summary
The Worldwide Protein Data Bank (wwPDB) has provided validation reports based on recommendations from community Validation Task Forces for structures in the PDB since 2013. To further enhance validation of small molecules as recommended from the 2016 Ligand Validation Workshop, wwPDB, Global Phasing Ltd., and The Noguchi Institute, recently formed a public/private partnership to incorporate some of their software tools into the wwPDB validation package. Augmented wwPDB validation report features include: two-dimensional (2D) diagrams of small-molecule ligands and carbohydrates, highlighting geometric validation outcomes; 2D topological diagrams of oligosaccharides present in branched entities generated using 2D Symbol Nomenclature For Glycan representation; and views of 3D electron density maps for ligands and carbohydrates, illustrating the goodness-of-fit between the atomic structure and experimental data (X-ray crystallographic structures only). These improvements will impact confidence in ligand conformation and ligand-macromolecular interactions that will aid in understanding biochemical function and contribute to small-molecule drug discovery.
eTOC Blurb
The Worldwide Protein Data Bank (wwPDB) has enhanced validation reports generated for small molecule ligands and carbohydrates to help ensure PDB data quality. Feng et al. describe improvements which include two-dimensional (2D) diagrams of small-molecule ligands and carbohydrates and 3D electron density maps and difference maps for X-ray structures.
Graphical Abstract
Introduction
Since 1971, the Protein Data Bank archive (PDB) (Protein Data Bank, 1971) has served as the global repository of information regarding 3D structures of proteins, nucleic acids, and complex assemblies. The PDB archive has grown from just seven X-ray crystal structures in 1971 to >170,000 structures as of early 2021. The Worldwide PDB (wwPDB) (Berman, et al., 2003, wwPDB consortium, 2019) organization manages the singular PDB archive and ensures that PDB structure data are freely and publicly available to the global community according to the FACT principles of Fairness-Accuracy-Confidentiality-Transparency (van der Aalst, et al., 2017) and the FAIR principles of Findability-Accessibility-Interoperability-Reusability (Wilkinson, et al., 2016). Current wwPDB members include the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB, US), Protein Data Bank in Europe (PDBe, UK), Protein Data Bank Japan (PDBj, Japan), and Biological Magnetic Resonance Data Bank (BMRB, US). Open access to PDB structure data plays critical roles in research and education across the natural, biomedical, and engineering sciences by enabling insight into biological function on the basis of form or 3D structure. A recent review reported that >420 fully-operational biodata resources enumerated in the Nucleic Acids Research Molecular Biology Database Collection utilize PDB data across major categories and subcategories (Markosian, et al., 2018), and corresponding resources in the collection’s “golden set” (Galperin, et al., 2017). The PDB archive is revered as a pioneer of open access and the wwPDB has been certified by CoreTrustSeal (CoreTrustSeal.org) as trustworthy since 2017.
The wwPDB is committed to providing the highest data quality in the global PDB archive. Method-specific wwPDB Validation Task Forces (VTFs, wwpdb.org/task/validation-task-forces) for macromolecular crystallography (MX), nuclear magnetic resonance (NMR) spectroscopy, electron microscopy (3DEM), and small-angle scattering were established to develop consensus community recommendations for structure validation and identify software applications to perform validation tasks. Each of the VTFs has published white papers (Read, et al., 2011, Montelione, et al., 2013, Henderson, et al., 2012, Trewhella, et al., 2013). VTF recommendations have been implemented within the wwPDB validation software (beginning for MX in 2013 and subsequently for NMR and 3DEM in 2016 (Gore, et al., 2017)). Initially, the wwPDB focused on protein/nucleic acid 3D structure quality assessments of polymer substituent bond lengths, bond angles, torsion angles, and sidechain rotamers for MX, NMR, and 3DEM methods, and fit of atomic coordinates to electron density maps for X-ray structures. Since launch of the wwPDB validation system, various improvements have been made, encompassing updates on percentile statistics, third-party software upgrades (e.g., Mogul (Bruno, et al., 2004), CCP4/Refmac (Winn, et al., 2011), and Phenix (Adams, et al., 2010)), and identification of ligands with poor fit of electron density (replacing LLDF (Smart, et al., 2018a) with RSR (Jones, et al., 1991) and RSCC (Smart, et al., 2018a) calculations). wwPDB validation reports are available in both PDF and XML formats (Gore, et al., 2017). Assessments of the impact of wwPDB validation reports have been published (Shao, et al., 2017, Smart, et al., 2018b, Shao, et al., 2018, Horsky, et al., 2019).
Since the launch of the unified wwPDB OneDep system for structure deposition (Young, et al., 2017), validation (Gore, et al., 2017), and biocuration (Young, et al., 2018) in 2014, wwPDB validation reports have been made available at various stages, including standalone anonymous validation server and application programming interface (API) generated reports intended for use pre-deposition; preliminary validation reports issued at the time of deposition; “official” wwPDB validation reports issued following completion of biocuration process; and public wwPDB validation reports issued at the time of data release. Upon public release of the deposited structure, the wwPDB validation report becomes part of the open access PDB archive. The standalone anonymous wwPDB validation server (validate.wwpdb.org) and corresponding API (wwpdb.org/validation/onedep-validation-web-service-interface) are used by structural biologists and computational modelers of macromolecular structures. In 2019, wwPDB recorded 27,571 validation API calls and 62,426 standalone validation server sessions supporting ~12,600 unique users. A growing number of scientific journals (e.g., Journal of Biological Chemistry, eLife, and International Union of Crystallography (IUCr) journals) require that “official” wwPDB validation reports accompany manuscripts describing new macromolecular structures.
wwPDB validation undergoes continuous improvement with the benefit of feedback from wwPDB VTFs, PDB data depositors, and PDB data consumers. Small molecules (ligands and carbohydrates) appearing in PDB structures have been prioritized for enhanced validation. These small chemicals play key biological and biochemical roles in energy transduction, cell signaling, and enzyme inhibition when bound either covalently or non-covalently to macromolecules. Understanding precisely how small molecules interact with biological macromolecules is central to understanding their roles in fundamental biology and in human health and disease.
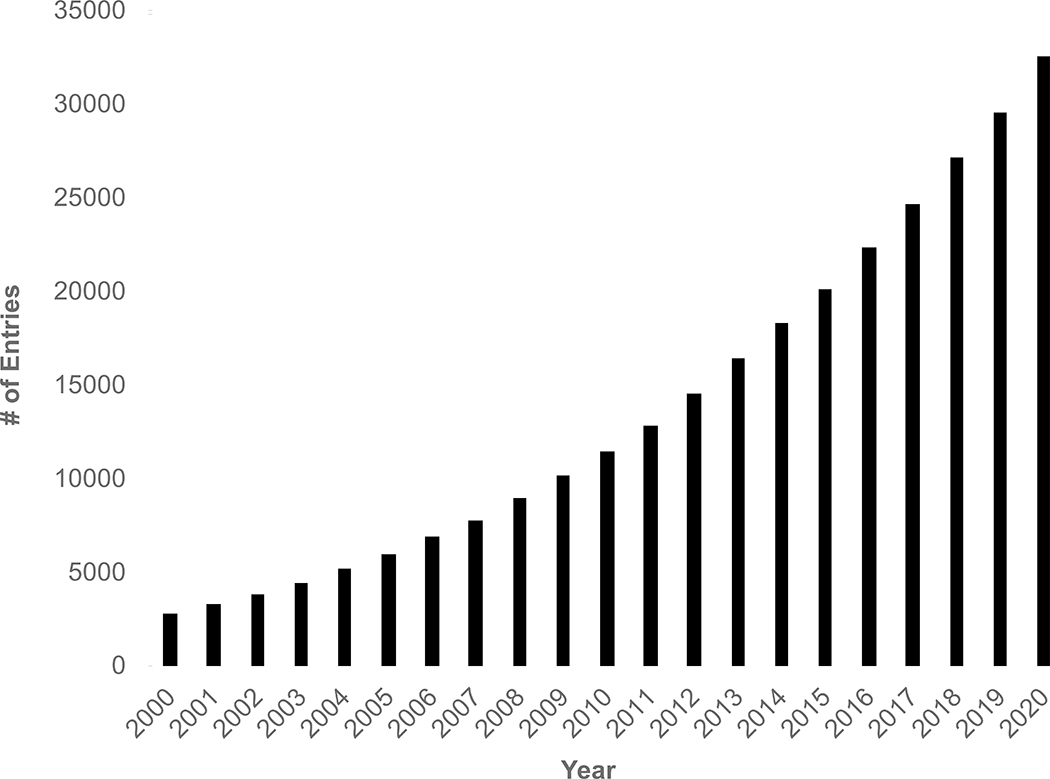
The PDB archive provides a wealth of information on small molecule (or drug)-protein interactions in co-crystal structures that are central to structure-based drug discovery and essential for understanding biochemical function at the atomic level. Every ligand (i.e., Chemical Component) represented in the PDB archive (including amino acids, nucleotides, organic compounds, and ions) is defined in the wwPDB Chemical Component Dictionary (CCD) (Westbrook, et al., 2015). The wwPDB CCD currently houses >32,500 unique ligands (Figure 1.) It is used to standardize atom nomenclature and chemical naming using 2D graphic matching during OneDep biocuration (Young, et al., 2018). In addition, ligand geometry and chirality checking is performed using Mogul (Bruno, et al., 2004) (courtesy of the Cambridge Crystallographic Data Centre (CCDC) (Groom, et al., 2016), as recommended by wwPDB X-ray VTF (Read, et al., 2011)). The initial implementation of wwPDB validation software supported only limited validation of ligands.
Figure 1.
Cumulative growth of unique Chemical Components (ligands) in the PDB archive from 2000–2020.
Highlighting and validating protein- or nucleic acid-bound small molecules that play important biological and biochemical roles is critical for making PDB data more valuable to millions of users globally. As of early 2021, more than 900 SARS-CoV-2 protein structures, some with bound drugs and small molecules, had been deposited to the PDB. Inhibition of the SARS-CoV-2 main protease blocks polyprotein processing, which is essential for viral infection. PDB IDs 7K6D and 7K6E (PDB doi:10.2210/pdb7K6D/pdb, doi:10.2210/pdb7K6E/pdb) revealed how the SARS-CoV-2 main proteinase active site is covalently modified by the US Food and Drug Administration antiviral drug telaprevir (CCD ID SV6 is a bound form of Telaprevir with a reduced C to O double bond). Telaprevir was previously visualized in another co-crystal structure revealing Hepatitis C virus protease inhibition (PDB ID 3SV6 (Romano, et al., 2012)). There are many more examples of functional studies on the interaction of small molecules with proteins in the PDB, including studies of Fentanyl (CCD ID 7V7; PDB ID 5TZO)(Bick, et al., 2017), morphine (CCD ID MOI; PDB ID 1Q0Y) (Pozharski, et al., 2004), hyoscyamine (CCD ID HYO; PDB ID 6TTM) (Kluza, et al., 2020), cholesterol (CCD ID Y01; found in tens of integral membrane protein structures), atropine (CCD ID OIN; PDB ID 6WJC) (Maeda, et al., 2020), and galanthamine (CCD ID GNT, a drug treatment for Alzheimer disease in present in many PDB structures). Hence, recent improvements to the wwPDB validation system have been focused on small molecules.
In 2015, a wwPDB/Cambridge Crystallographic Data Centre/Drug Design Data Resource Ligand Validation Workshop (hereafter LVW) assembled co-crystal structure determination experts from academe and industry together with X-ray crystallography and computational chemistry software developers to discuss, develop, and recommend best practices for validation of co-crystal structures; editorial/refereeing standards for publishing co-crystal structures; and ligand representation across the PDB archive (Adams, et al., 2016).
Results
Workshop Recommendations:
The LVW addressed challenges that structure depositors, PDB users, and journal editors/manuscript reviewers face when trying to assess the quality and accuracy of 3D structures of macromolecule-ligand complexes (Adams, et al., 2016). It has been noted repeatedly by some that spurious electron density difference map features have been mistakenly interpreted in a small number of cases as indicating the presence of particular bound small molecules. More commonly, wwPDB biocurators encounter chemical transformations upon protein or nucleic acid binding that are not reflected in the atomic-level PDB structure; usage of incorrect restraint value targets for ligands during structure refinement; and inaccurate or incomplete chemical descriptors supplied by structure depositors. LVW recommendations included provision of unambiguous chemical definitions for ligands, identification of Ligand(s) of Interest (LOI(s), research focus and/or biologically important), and provision of informative images of ligand pose(s) with electron density maps using a presentation style comparable to the Global Phasing Ltd. buster-report tool (SmartBricogne, 2015).
wwPDB partners first addressed identification of the Ligand of Interest (LOI) by modifying the OneDep software system (Young, et al., 2017). Since 2017, depositors have been required to flag the ligand(s) in the structure is their research focus and/or of biological importance. This LOI information is captured by the OneDep system and used by the augmented wwPDB validation software. All LOIs are highlighted in the wwPDB validation report with 2D depictions of geometric quality and, for MX structures, with 3D depictions of (1) the fit of the atomic model to the electron density map, and (2) the significant features (if any) of the difference map indicating imperfection in that fit, both maps being computed from appropriate Fourier coefficients produced by the final refinement step.
Community Software Re-use by the wwPDB Validation System:
To implement LVW recommendations concerning use of buster-report presentation style, wwPDB collaborated with Global Phasing Ltd. under a formal agreement to re-use their software for generating 3D graphic depictions of the goodness-of-fit of ligands to electron density maps, plus 2D graphic depictions of atomic stick-figures of ligands labeled with geometric, stereochemical, and unassessed annotations.
As summarized in Table 1, Maxit (Feng) extracts LOI and oligosaccharide chemical descriptors from atomic coordinate files. The buster-report software (SmartBricogne, 2015) creates 3D views of the atomic-level structure fit to experimental data and 2D views of annotated chemical diagrams. Fast Fourier transform and mapmask programs from CCP4 (Winn, et al., 2011) are used to generate bias-corrected 2m|Fo|-D|Fc| and m|Fo|-D|Fc| electron density maps, where m denotes the figure of merit and D denotes the Sigma-A weighting factor. Pymol (DeLano, 2002) is used for graphical display of the electron density features in these two maps surrounding the bound ligand. Mogul (Bruno, et al., 2004) analyses of ligand geometric properties (bond lengths, bond angles, dihedral angles, and a ring planarity measure) are performed to identify geometrical outliers. OpenBabel (O’Boyle, et al., 2011) is used to generate a 2D renditions of the atomic coordinates. Quality assessment from the Mogul output file is then extracted and converted to quality color coding (Green: good; pink: outlier; and gray: not assessed by Mogul) for each bond length, bond angle, torsion angle, and ring planarity measure. A Pymol script is used to project the Mogul quality assessments onto 2D chemical stick-figure drawings.
Table 1.
Summary of wwPDB Validation Software Integration SNFG: standard monosaccharide Symbol Nomenclature For Glycans
| Feature | Details | Software Package(s) |
|---|---|---|
| Atomic coordinates and bond order information for all ligands | Extracts LOI (Ligand Of Interest) ligands and WURCS descriptor of branched entities for carbohydrates from the atomic coordinate file | Maxit (Feng) PDB2Glycan (gitlab.com/glyconavi/pdb2glycan) |
| 2D SNFG images | Generates 2D SNFG images using wurcs2pic software that uses GlycanBuilder2 as a library, if WURCS descriptors are present in the branched entities | wurcs2Pic (gitlab.com/glycoinfo/wurcs2pic) GlycanBuilder2 (Tsuchiya, et al., 2017) |
| Geometric quality assessment | Mogul is used to assess geometric quality for all ligands. | Mogul (Bruno, et al., 2004) |
| 2D image of quality | Creates 2D graphical depiction of Mogul quality analyses for each LOI and/or each branched oligosaccharide | buster-report (SmartBricogne, 2015) |
| 3D graphical views | Creates 3D graphical views of electron density difference maps for each LOI and/or each branched oligosaccharide | buster-report (SmartBricogne, 2015) |
Oligosaccharides in the PDB are treated as special cases (i.e., branched entity representation) with a Web3 Unique Representation of Carbohydrate Structures (WURCS) descriptor (Matsubara, et al., 2017) generated by PDB2Gycan software (gitlab.com/glyconavi/pdb2glycan) in collaboration with The Noguchi Institute (Japan). Java code-based wurcs2pic using GlycanBuilder2 (Tsuchiya, et al., 2017) as a library is used to generate 2D Symbol Nomenclature For Glycan (SNFG) (Varki, et al., 2015, Neelamegham, et al., 2019) images in both .png and .svg file formats for each WURCS descriptor (Matsubara, et al., 2017). The protocols used for ligands are also used for oligosaccharides when generating 2D graphical depiction and 3D views of the goodness-of-fit with electron density maps for MX structures (Table 1).
Enhanced wwPDB Validation of Ligands:
The two-dimensional graphical depiction (SmartBricogne, 2015) of Mogul quality analysis of bond lengths, bond angles, torsion angles, and ring geometry is provided for all ligands that have been designated as LOI by the structure depositor and for any ligands with molecular weight >250 Daltons that have outliers flagged in the wwPDB validation report (Gore, et al., 2017). Individual bond lengths or angles with a Z-score (the difference between an observed value and expected or average value, divided by the standard deviations of the latter) less than −2 or greater than 2 are flagged as outliers. This scoring system represents a simplification of the four-color scheme (with three Z-score thresholds: 1.5, 2.5, and 4.0) used in the buster-report. It was adopted by wwPDB to conform to the classification scheme recommended by the wwPDB VTFs (Read, et al., 2011, Montelione, et al., 2013, Henderson, et al., 2012, Trewhella, et al., 2013). For torsion angles, Mogul (Bruno, et al., 2004) provides so-called local density measurements from the Cambridge Structural Database (CSD) (Groom, et al., 2016). Ring conformations are considered as unusual if less than 5% of the experimental values extracted from the CSD archive fall within 10 degrees of the conformation in the PDB ligand.
2D depictions of ligands are color-coded according to validation results with green indicating commonly observed values, magenta indicating “unusual” values, and gray indicating that there was insufficient data to derive a validation score. Unusual values can reflect issues with model quality and/or atomic model fit to the electron density map. For atomic model quality, individual bond lengths or bond angles with a Z-score less than −2 or greater than +2, torsion angles with less than 5% of local density measure from Mogul calculation, or root-mean-square deviation (RMSD) >60 degrees are considered unusual and colored coded with magenta.
3D graphical representation of atomic model fit to the electron density map is also provided in the new wwPDB validation report. Poor fits of atomic models to electron density maps are flagged as outliers whenever the calculated real space correlation coefficient (RSCC) is less than 0.8 or the calculated Real-space R (RSR) is greater than 0.4. Electron density maps are color-coded as follows: 2m|Fo|-D|Fc| map-gray; and m|Fo|-D|Fc| map-green for positive values and magenta for negative values.
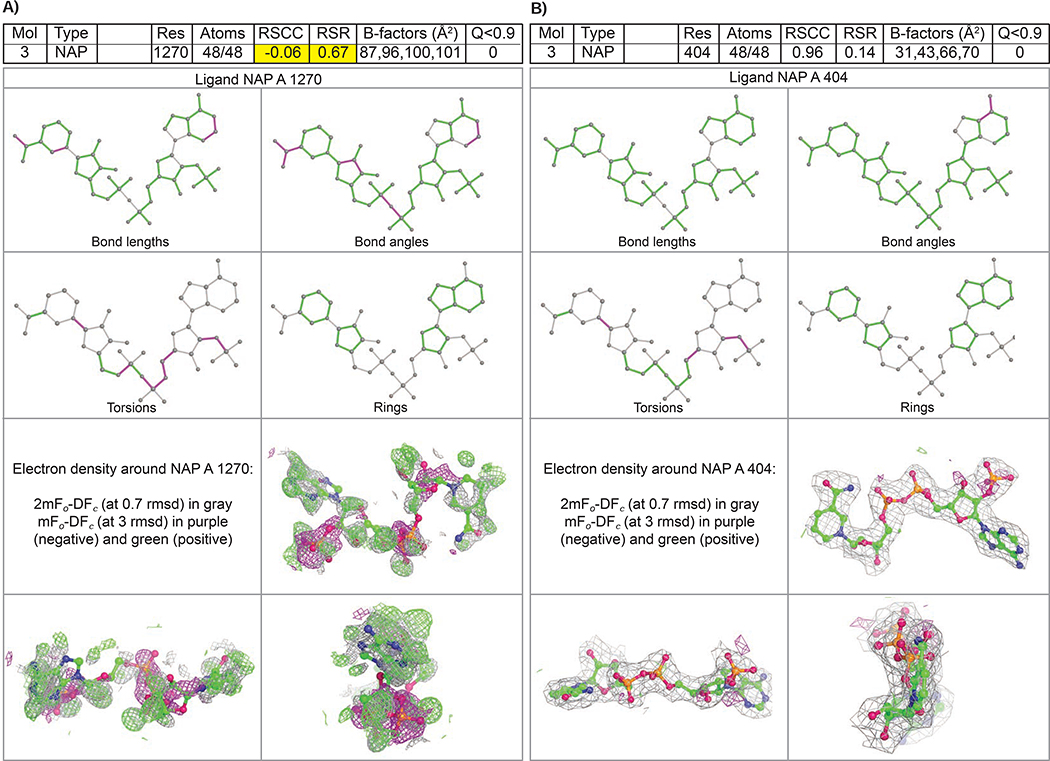
Figure 2 illustrates ligand quality metrics for two examples of the Nicotinamide Adenine Dinucleotide Phosphate (NADP) ligand (CCD ID NAP). PDB ID 1ZK4 (Khanppnavar, et al., 2019) represents a case of lower atomic model and/or data quality (Figure 2A). PDB ID 5ZIX (Khanppnavar, et al., 2019) represents a case of higher atomic model and/or data quality (Figure 2B). In PDB ID 1ZK4, a considerable number of outliers for bond lengths, bond angles, and torsion angles in ligand NAP 1270 (associated with polypeptide chain A) were highlighted in the augmented wwPDB validation report ligand geometry table and colored as magenta in the 2D depiction. The RSR value for NAP 1270 in PDB ID 1ZK4 is unusually high (RSR~0.67) indicating poor fit of the atomic model to the experimental data. The 3D representation of ligand fit shows significant positive (green) and negative (magenta) features in the m|Fo|-D|Fc| electron density map, which are diagnostic of poor fit of atomic model to the experimental data and/or data quality. No such issues were identified for the NADP ligand in PDB ID 5ZIX.
Figure 2.
2D depictions of Mogul quality analysis of bond lengths, bond angles, torsion angles, and ring geometry, and 3D depictions of the atomic model fit to experimental electron density map (drawn in gray) with the surrounding difference map features (drawn in green and magenta) for NADP. Each fit is shown from a different orientation to approximate a three-dimensional view. (Left) PDB ID 1ZK4 represents a lower structure and/or data quality, with multiple features in the difference map. (Right) PDB ID 5ZIX represents higher structure and/or data quality, with a good fit of the model to the experimental electron density map and an essentially featureless difference map.
Improved Carbohydrate Representation in PDB Structures:
Wholesale improvements in the representation of carbohydrates in the PDB archive (Shao, et al.) were recently incorporated into the augmented wwPDB validation reports. Each oligosaccharide present in a branched entity is identified with a systematic name and illustrated schematically with a 2D SNFG image generated using wurcs2pic software (Tsuchiya, et al., 2017). Residue-property plots for quality assessment at the chain level are provided for each oligosaccharide instance. Geometric quality assessments at the residue level are described in a new carbohydrates section of the new wwPDB validation report, wherein monosaccharide atomic structure outliers are listed for each component of the branched entity representation oligosaccharide chain. Augmented wwPDB validation reports also incorporate visualization of oligosaccharide validation outcomes as above for small-molecule ligands. These enhancements include 2D diagrams of oligosaccharides, highlighting geometric validation criteria, and 3D views of the atomic structure superimposed on displays of the electron density maps for MX structures.
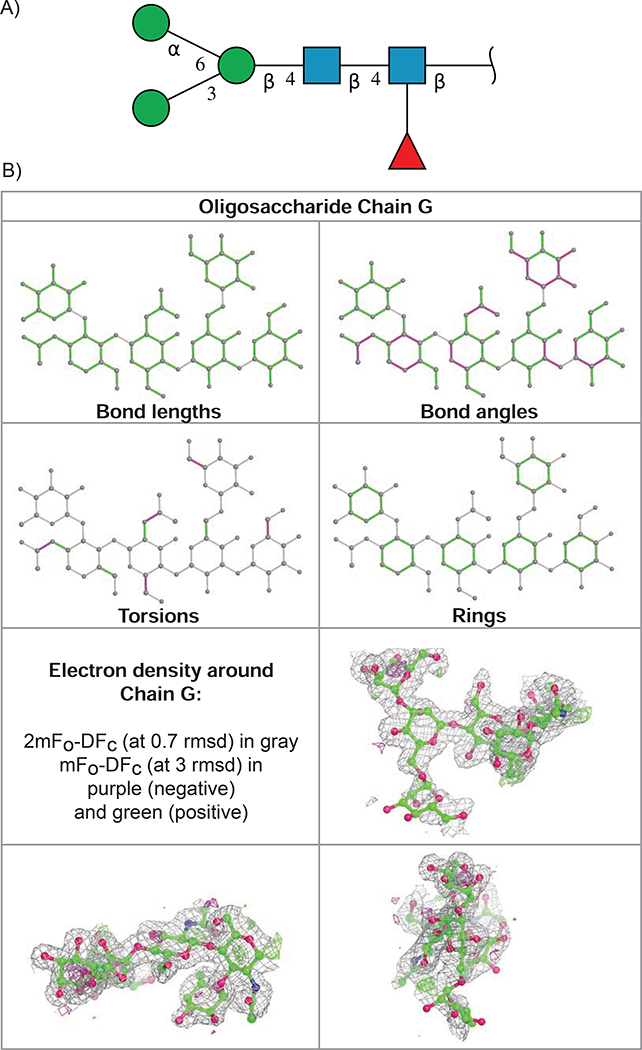
For example, the 2D SNFG image for oligosaccharide chain G, alpha-D-mannopyranose-(1–3)-[alpha-D-mannopyranose-(1–6)]beta-D-mannopyranose-(1–4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1–4)-[alpha-L-fucopyranose-(1–3)]2-acetamido-2-deoxy-beta-D-glucopyranose (PDB ID 1B5F) (Frazao, et al., 1999) is shown in Figure 3A. Validation results for this branched carbohydrate entity are illustrated in Figure 3B. The ability to represent an oligosaccharide as a branched entity has enabled users to easily identify and review an oligosaccharide with unambiguous topology for oligosaccharide sequence and connectivity.
Figure 3.
Carbohydrate validation for PDB ID 1B5F. (A) 2D SNFG symbol image that indicates carbohydrate anomer (α versus β) and connectivity positions (1–3, 1–4 or 1–6 glycosidic connection) for oligosaccharide chain G. (B) 2D depiction of Mogul quality analysis of bond lengths, bond angles, torsion angles, and ring geometry (top), and three orthogonal projection views of the superposition of the atomic coordinates for oligosaccharide chain G onto the electron density map (in gray), with the very few significant features in the difference map in green and magenta (bottom).
Discussion
Enhanced validation of small molecules present in the PDB archive is now reflected in the augmented wwPDB validation report. All ligands that are the focus of the structure determination (LOI) and/or some ligands with higher molecular weight (>250 Daltons) identified as outliers are now provided with 2D geometrical quality assessments and 3D views of electron density maps (X-ray crystallographic structures only). Each oligosaccharide present in the PDB archive is now represented as a branched entity and depicted using 2D SNFG images. Graphical representations of oligosaccharide geometry quality and goodness-of-fit of the atomic model to electron density maps (MX structures only) are also provided.
The wwPDB is committed to improving the quality of data archived in the PDB by making validation information available to PDB data depositors and data consumers. Relevant wwPDB services include anonymous, pre-deposition validation through web user interface or a programmatic API; validation reports at the time of deposition; validation reports furnished for use during publication peer-review; and validation reports and visualization capabilities for every publicly one for the more than 170,000 released PDB IDs. To the same end, the wwPDB provides a coordinate versioning mechanism that allows a depositor to retrospectively improve or correct the structure for an earlier deposition while retaining the original PDB ID.
New depictions of electron density fit and geometrical outliers from the CCDC Mogul analysis (Bruno, et al., 2004) are provided to enable identification of incorrect restraint values used by depositors. The wwPDB OneDep system has been modified recently to capture depositor-provided restraints for ligands. Such input restraint values are used to search for a matching ligand in the wwPDB CCD and aiding in validating the structure of the ligand(s). As a next step, wwPDB intends to include depositor-provided restraint values in a further updated version of the wwPDB validation report, wherein they may be compared directly with CCDC Mogul target values.
As wwPDB partners receive additional recommendations from wwPDB VTFs and feedback from our diverse community of users, we will continue to improve validation tools and reporting. Current efforts are underway to incorporate NMR restraint validation into wwPDB validation software tools. In addition, a 3DEM Data Management Workshop was held at the European Bioinformatics Institute in early 2020 to consider how PDB should collect and validate 3DEM data. Recommendations from this exercise are being prepared for publication and will be implemented in due course. One outstanding recommendation pertaining to ligands dating from the LVW concerns provision of percentile scores for the overall ligand quality (a so-called ligand ranking slider). This feature will be introduced after new enhancements are implemented in NMR restraints and 3DEM data validation.
A new method known as PanDDA (Pearce, et al., 2017) performs multi-dataset crystallographic analysis for identification of weakly bound ligands in co-crystal structures deposited to the PDB. This method compares structure factors measured from a crystal containing a weakly bound ligand to structure factors measured from a large number apo protein crystals. Aggregation of the apo protein structure factor data enables more sensitive detection of electron density map features corresponding to weakly bound ligands. Currently, wwPDB requires depositors to provide all the information in the structure factor file necessary for other researchers to repeat the depositor’s PanDDA analysis. Recommended additional information includes structure factor data used the final refinement cycle; map coefficients for apo protein crystal data sets; and native structure factor data for apo protein crystals. PanDDA structures are validated with structure factors from the final round of refinement cycle, as is the case for all MX structures. wwPDB is aware that this universal approach is not entirely suitable for structures determined using the PanDDA method. wwPDB will seek X-ray VTF and community recommendations regarding best practices for future validation of PanDDA structures.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jasmine Young (jasmine.young@rcsb.org)
Materials Availability
This study did not generate new unique reagents.
Data and Code availability
wwPDB validation tools are publicly accessible. The wwPDB validation server is provided at https://validate.wwpdb.org and the wwPDB validation API is accessible at http://www.wwpdb.org/validation/onedep-validation-web-service-interface. wwPDB validation report for each PDB entry is provided for users to download at PDB archive, https://ftp.wwpdb.org/pub/pdb/validation_reports/. These validation reports are also accessible at wwPDB website via PDB DOI links, e.g., https://www.wwpdb.org/pdb?id=pdb_0000{PDB 4-letter ID} (DOI: 10.2210/pdb{PDB ID}/pdb). Maxit program is available at https://sw-tools.rcsb.org/apps/MAXIT/index.html. Community tools are available at https://www.globalphasing.com/buster/wiki/index.cgi?BusterReport for buster-report and gitlab.com/glyconavi/pdb2glycan for WURCS descriptor.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
There was no model used.
METHOD DETAILS
Java code-based wurcs2pic (Matsubara, et al., 2017) is called to generate 2D SNFG images in both png and svg formats for each WURCS descriptor. Maxit (Feng) extracts atomic coordinates and bond order information on all ligands and WURCS descriptors on branched entities of oligosaccharides. Mogul analysis (Bruno, et al., 2004) including all small molecule ligands and monosaccharides in branched oligosaccharide entities is performed in a multi-threading mode. Buster-report (SmartBricogne, 2015) script is called to create 2D graphical depiction of Mogul quality analyses for each LOI and/or each branched oligosaccharide. A Pymol (DeLano, 2002) script was developed that takes projected 2D coordinates and Mogul quality assessments as inputs and renders 2D chemical images and 3D graphical views.
QUANTIFICATION AND STATISTICAL ANALYSIS
No analysis was performed.
Highlights.
2D geometrical depiction of small molecules now in wwPDB ligand validation
3D views of electron density fits for X-ray small molecules are also included
Depositor-defined research focus (Ligand of Interest, LOI) highlighted in report
Carbohydrate branched representations and 2D SNFG images are introduced in the report
Acknowledgments
We thank the tens of thousands of structural biologists who have contributed structures to the PDB and the many millions of PDB data consumers around the world, members of the wwPDB Validation Task Forces, and participants in the wwPDB/CCDC/D3R Ligand Validation Workshop. We also thank all the staff members of the wwPDB partners for their support and feedback, and wwPDB biocurators for testing and feedback of the wwPDB validation software. RCSB PDB is funded by the National Science Foundation (DBI-1832184), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM133198. Protein Data Bank in Europe is supported by the European Molecular Biology Laboratory-European Bioinformatics Institute and Wellcome Trust [104948]. Protein Data Bank Japan is supported by the Database Integration Coordination Program from the National Bioscience Database Center (NBDC)-JST (Japan Science and Technology Agency), the Platform Project for Supporting in Drug Discovery and Life Science Research from AMED, and the joint usage program of Institute for Protein Research, Osaka University. BMRB is supported by the US National Institute of General Medical Sciences under grant R01GM109046. The GlyCosmos project has been supported by the Integration Promotion Program of the Japan Science and Technology Agency and the National Bioscience Database Center (NBDC).
Author contributions
The wwPDB validation software package is maintained by wwPDB partners headed by S.K.B., S.V., G.K., and J.C.H. The work reported herein was carried out by RCSB PDB at Rutgers, the State University of New Jersey. The buster-report software was contributed free of charge by Global Phasing Ltd. The PDB2Glycan software was contributed through the GlyCosmos project by the Noguchi Institute and Soka University (Japan). Integration of buster-report and PDB2Glycan software into the wwPDB validation software and development of branched representation of carbohydrates were carried out by Z.F. The technical plan was provided by J.D.W. The collection of authors’ LOI was implemented by R.S. Project management was provided by J.Y.Y. The article was written by J.Y.Y. and reviewed by all co-authors. S.K.B. provided overall leadership to the RCSB PDB team.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Aertgeerts K, Bauer C, Bell JA, Berman HM, Bhat TN, Blaney JM, Bolton E, Bricogne G, Brown D, et al. 2016. Outcome of the first wwpdb/ccdc/d3r ligand validation workshop. Structure. 24:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallographica Section D. 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Henrick K, Nakamura H 2003. Announcing the worldwide protein data bank. Nature Structure Biology. 10:980. [DOI] [PubMed] [Google Scholar]
- Bick MJ, Greisen PJ, Morey KJ, Antunes MS, La D, Sankaran B, Reymond L, Johnsson K, Medford JI, Baker D. 2017. Computational design of environmental sensors for the potent opioid fentanyl. eLife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IJ, Cole JC, Kessler M, Luo J, Motherwell WD, Purkis LH, Smith BR, Taylor R, Cooper RI, Harris SE, et al. 2004. Retrieval of crystallographically-derived molecular geometry information. J Chem Inf Comput Sci. 44:2133–2144. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002). The pymol molecular graphics system.
- Feng Z Maxit: Macromolecular exchange and input tool, https://sw-tools.Rcsb.Org/apps/maxit/index.Html, https://sw-tools.rcsb.org/apps/MAXIT/index.html. [Google Scholar]
- Frazao C, Bento I, Costa J, Soares CM, Verissimo P, Faro C, Pires E, Cooper J, Carrondo MA 1999. Crystal structure of cardosin a, a glycosylated and arg-gly-asp-containing aspartic proteinase from the flowers of cynara cardunculus l. J Biol Chem. 274:27694–27701. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Fernandez-Suarez XM, Rigden DJ 2017. The 24th annual nucleic acids research database issue: A look back and upcoming changes. Nucleic Acids Research. 45:D1–D11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore S, Sanz Garcia E, Hendrickx PMS, Gutmanas A, Westbrook JD, Yang H, Feng Z, Baskaran K, Berrisford JM, Hudson BP, et al. 2017. Validation of structures in the protein data bank. Structure. 25:1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom CR, Bruno IJ, Lightfoot MP, Ward SC 2016. The cambridge structural database. Acta Crystallogr B. 72:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, et al. 2012. Outcome of the first electron microscopy validation task force meeting. Structure. 20:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsky V, Bendova V, Tousek D, Koca J, Svobodova R 2019. Valtrendsdb: Bringing protein data bank validation information closer to the user. Bioinformatics. 35:5389–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 47:110–119. [DOI] [PubMed] [Google Scholar]
- Khanppnavar B, Chatterjee R, Choudhury GB, Datta S 2019. Genome-wide survey and crystallographic analysis suggests a role for both horizontal gene transfer and duplication in pantothenate biosynthesis pathways. Biochim Biophys Acta Gen Subj. 1863:1547–1559. [DOI] [PubMed] [Google Scholar]
- Kluza A, Wojdyla Z, Mrugala B, Kurpiewska K, Porebski PJ, Niedzialkowska E, Minor W, Weiss MS, Borowski T 2020. Regioselectivity of hyoscyamine 6β-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and qm/mm calculations Dalton Trans. 49:4454–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Xu J, FM NK, Clark MJ, Zhao J, Tsutsumi N, Aoki J, Sunahara RK, Inoue A, Garcia KC, et al. 2020. Structure and selectivity engineering of the m1 muscarinic receptor toxin complex. Science. 369:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosian C, Di Costanzo L, Sekharan M, Shao C, Burley SK, Zardecki C 2018. Analysis of impact metrics for the protein data bank. Sci Data. 5:180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M, Aoki-Kinoshita KF, Aoki NP, Yamada I, Narimatsu H 2017. Wurcs 2.0 update to encapsulate ambiguous carbohydrate structures. J Chem Inf Model. 57:632–637. [DOI] [PubMed] [Google Scholar]
- Montelione GT, Nilges M, Bax A, Guntert P, Herrmann T, Richardson JS, Schwieters CD, Vranken WF, Vuister GW, Wishart DS, et al. 2013. Recommendations of the wwpdb nmr validation task force. Structure. 21:1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lutteke T, O’Boyle N, Packer NH, Stanley P, Toukach P, et al. 2019. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 29:620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR 2011. Open babel: An open chemical toolbox. J Cheminform. 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce NM, Krojer T, Bradley AR, Collins P, Nowak RP, Talon R, Marsden BD, Kelm S, Shi J, Deane CM, et al. 2017. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nature communications. 8:15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozharski E, Wilson MA, Hewagama A, Shanafelt AB, Petsko G, Ringe D 2004. Anchoring a cationic ligand: The structure of the fab fragment of the anti-morphine antibody 9b1 and its complex with morphine. J Mol Biol. 337:691–697. [DOI] [PubMed] [Google Scholar]
- Protein Data Bank. 1971. Crystallography: Protein data bank. Nature (London), New Biol. 233:223–223.16063295 [Google Scholar]
- Read RJ, Adams PD, Arendall WB 3rd, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, et al. 2011. A new generation of crystallographic validation tools for the protein data bank. Structure. 19:1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, et al. 2012. The molecular basis of drug resistance against hepatitis c virus ns3/4a protease inhibitors. PLoS Pathog. 8:e1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Feng Z, Westbrook J, Peisach E, Berrisford JM, Ikegawa Y, Kurisu G, Velankar S, Burley SK, Young JY Modernized uniform representation of carbohydrate molecules in the protein data bank. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Liu Z, Yang H, Wang S, Burley SK 2018. Outlier analyses of the protein data bank archive using a probability-density-ranking approach. Sci Data. 5:180293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang H, Westbrook JD, Young JY, Zardecki C, Burley SK 2017. Multivariate analyses of quality metrics for crystal structures in the protein data bank archive. Structure. 25:458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O, Bricogne G (2015). Multifaceted roles of crystallography in modern drug discovery, edited by Scapin G PD, Arnold E, pp. 165–181. Netherlands: Springer. [Google Scholar]
- Smart OS, Horsky V, Gore S, Svobodova Varekova R, Bendova V, Kleywegt GJ, Velankar S 2018a. Validation of ligands in macromolecular structures determined by x-ray crystallography. Acta crystallographica. Section D, Structural biology. 74:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Horsky V, Gore S, Svobodova Varekova R, Bendova V, Kleywegt GJ, Velankar S 2018b. Worldwide protein data bank validation information: Usage and trends. Acta crystallographica. Section D, Structural biology. 74:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM 2013. Report of the wwpdb small-angle scattering task force: Data requirements for biomolecular modeling and the pdb. Structure. 21:875–881. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Aoki NP, Shinmachi D, Matsubara M, Yamada I, Aoki-Kinoshita KF, Narimatsu H 2017. Implementation of glycanbuilder to draw a wide variety of ambiguous glycans. Carbohydr Res. 445:104–116. [DOI] [PubMed] [Google Scholar]
- van der Aalst WMP, Bichler M, Heinzl A 2017. Responsible data science. Business & Information Systems Engineering. 59:311–313. [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook JD, Shao C, Feng Z, Zhuravleva M, Velankar S, Young J 2015. The chemical component dictionary: Complete descriptions of constituent molecules in experimentally determined 3d macromolecules in the protein data bank. Bioinformatics. 31:1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. 2016. The fair guiding principles for scientific data management and stewardship. Sci Data. 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. 2011. Overview of the ccp4 suite and current developments. Acta Crystallographica. Series D. 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wwPDB consortium. 2019. Protein data bank: The single global archive for 3d macromolecular structure data. Nucleic Acids Res. 47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JY, Westbrook JD, Feng Z, Peisach E, Persikova I, Sala R, Sen S, Berrisford JM, Swaminathan GJ, Oldfield TJ, et al. 2018. Worldwide protein data bank biocuration supporting open access to high-quality 3d structural biology data. Database (Oxford). 2018:bay002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JY, Westbrook JD, Feng Z, Sala R, Peisach E, Oldfield TJ, Sen S, Gutmanas A, Armstrong DR, Berrisford JM, et al. 2017. Onedep: Unified wwpdb system for deposition, biocuration, and validation of macromolecular structures in the pdb archive. Structure. 25:536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
wwPDB validation tools are publicly accessible. The wwPDB validation server is provided at https://validate.wwpdb.org and the wwPDB validation API is accessible at http://www.wwpdb.org/validation/onedep-validation-web-service-interface. wwPDB validation report for each PDB entry is provided for users to download at PDB archive, https://ftp.wwpdb.org/pub/pdb/validation_reports/. These validation reports are also accessible at wwPDB website via PDB DOI links, e.g., https://www.wwpdb.org/pdb?id=pdb_0000{PDB 4-letter ID} (DOI: 10.2210/pdb{PDB ID}/pdb). Maxit program is available at https://sw-tools.rcsb.org/apps/MAXIT/index.html. Community tools are available at https://www.globalphasing.com/buster/wiki/index.cgi?BusterReport for buster-report and gitlab.com/glyconavi/pdb2glycan for WURCS descriptor.