Summary
Thermogenic beige fat found in white adipose tissue is a potential therapeutic target to curb the global obesity and diabetes epidemic. However, these inducible thermogenic beige adipocytes have been thought to be short-lived and rapidly convert to “white-like” adipocytes after discontinuing stimuli. In this study, using effective labelling techniques and genetic mouse tools, we demonstrated that a subset of UCP1+ cells that exist within white adipose tissue are able to self-divide and contribute to new beige adipocyte recruitment in response to β3 stimuli. When these cells were depleted or their adipogenic capability was blocked, β3-induced beige adipocyte formation was impaired. We also identified a cell cycle machinery of p21 and CDKN2A as a molecular basis of beige adipocyte regulation. Collectively, our findings provide new insights into the cellular and molecular mechanisms of beige adipocyte regulation and potentially therapeutic opportunities to induce beige phenotype and treat metabolic disease.
Keywords: Beige adipocytes, UCP1, Cell cycle regulators, p21, CDKN2A, Obesity
eTOC blurb
Park et al. demonstrate that new UCP1+ beige fat cells in the inguinal white adipose tissue arise from pre-existing UCP1+ cells in response to b3-agonist activation in a region selective manner in mice. Moreover, cell cycle regulators play an important role in beige fat cell expansion in this process.
Introduction
The epidemic of obesity and diabetes has prominently increased and is becoming one of the leading health problems in western countries (Friedman, 2009; Spiegelman and Flier, 2001; WHO, 2013); thus, mitigating obesity is a major medical need. Uncontrolled expansion of white adipose tissue (WAT) results in compromised metabolic health related to diminished lifespan and poor life quality (Gesta et al., 2007; Jiang et al., 2012; Shin et al., 2020a). Within WAT, there exist two distinct types of adipocytes: white and beige, which play contrasting physiological roles in regulating energy homeostasis. Whereas white adipocytes store excessive energy as triacylglycerol, beige adipocytes act as cellular furnaces to burn blood glucose and fatty acids to generate heat.
Recent studies indicate that adult humans possess beiging capacity, which confers metabolic benefits such as reduced blood glucose and increased energy expenditure (Chondronikola et al., 2014; Finlin et al., 2018; Finlin et al., 2020; van der Lans et al., 2013; Weir et al., 2018; Yoneshiro et al., 2013). Beige adipocytes can be recruited by multiple physiological and pharmacological stimuli such as cold exposure or β3-adrenergic agonists. As described in several recent reviews, these beige adipocytes within WAT are developmentally, molecularly, and functionally distinct from classical brown adipocytes (Cohen and Spiegelman, 2015; Ikeda et al., 2018; Seale et al., 2008; Timmons et al., 2007; Wu et al., 2012; Wu et al., 2013). Beige adipocytes, rather than classical brown adipocytes, are increasingly recognized as a cellular target for therapeutic approaches against obesity partly because: (1) beige adipocytes can be largely recruited throughout WAT and convert glucose and fatty acids into heat, whereas brown adipocytes are limited to small deposits; (2) adult humans have beiging capacity, while brown adipocytes are restricted to infants; (3) beige adipocytes can be induced by various mechanisms that exert no effects on brown adipocytes.
Despite the progress made over the past 10 years, fundamental knowledge pertaining to beige adipocyte regulation is lacking in several aspects. First, the origin of beige adipocytes remains unclear although multiple studies have suggested that beige adipocytes can arise from both progenitors and differentiated unilocular white adipocytes (Acharya et al., 2019; Berry et al., 2016; Jiang et al., 2017; Lee et al., 2015; Long et al., 2014; McDonald et al., 2015; Min et al., 2016; Oguri et al., 2020; Shao et al., 2019; Vishvanath et al., 2016; Wang et al., 2014). Second, the function of beige adipocytes as a metabolic sink for fat and glucose appears to be temporary. For instance, in both rodents and humans, beige adipocytes express high levels of uncoupling protein 1 (UCP1) and other thermogenic genes in response to cold exposure or β3-agonists; however, upon removal of external stimuli such as transferring mice from cold environments (6°C) to thermoneutral conditions (30°C), the UCP1+ multilocular beige adipocytes convert into dormant unilocular adipocytes with expression of white adipocyte-enriched genes (Altshuler-Keylin et al., 2016; Lu et al., 2018; Paulo and Wang, 2019; Roh et al., 2018; Rosenwald et al., 2013). This presents a key challenge of using beige fat as a sustained long-term therapy to diminish adiposity and to improve glucose metabolism and insulin sensitivity.
In the present study, we sought to investigate the function and behavior of beige adipocytes and their effects on metabolic health. We identified a class of unique UCP1+ cells within the peri-lymph node region of inguinal WAT (IGW) that can self-divide and give rise to new beige adipocytes in response to β3 stimulation. Transient depletion of UCP1+ cells or blocking UCP1+ adipogenic potential impairs beige adipocyte formation in response to β3 stimuli. To dissect the underlying mechanism, we found either overexpression of p21 (a cyclin-dependent kinase inhibitor) or deleting cyclin-dependent kinase inhibitor 2A (Cdkn2a) conditionally and specifically in UCP1+ cells results in the alteration of the beige lineage in animal models. Taken together, our results strongly suggest that a subgroup of UCP1+ cells display progenitor-like characteristics which are regulated by cell cycle genes in beige adipocyte development.
Results
UCP1+ cells generate new beige adipocytes in response to β3-adrenergic stimulation
UCP1 was previously thought to be expressed only in mature brown and beige adipocytes that do not proliferate. However, it was recently reported that UCP1+ brown adipocytes display proliferative ability in response to cold stimuli (Fukano et al., 2016; Lee et al., 2015; Okamatsu-Ogura et al., 2017). To test whether UCP1+ pre-existing beige adipocytes within IGW could proliferate and give rise to new UCP1+ beige adipocytes, we revisited a previously developed Ucp1-CreERT2; Rosa26RRFP (UCP1-RFP) mouse model (Figure 1A) (Jiang et al., 2017; Rosenwald et al., 2013). In this fate mapping model, Rosa26RRFP reporter allele expresses RFP in UCP1 expressing cells and potential descendants (Soriano, 1999). During the 2–3 days pulse-labeling period, we observed that RFP specifically and efficiently labeled BAT and inguinal WAT (IGW) around peri-lymph node region: without Tamoxifen (TM) administration, there was no RFP reporter activity; upon TM injection, RFP displayed robust labeling of UCP1+ cells in the BAT and IGW by whole mount imaging and histological sections (Figure S1A–C). Critically, other tissues, including heart, kidney, liver, muscle, and spleen, were not labeled with RFP reporter in both male and female mice, except the epididymis in males (Figure S1A). Consistent with the Cre expression in beige adipocytes, we observed the strong RFP signal in adipocyte fraction but not in fresh isolated stromal vascular fraction (SVF) cells or the SVF cells cultured for 7 days (data not shown). In addition to the Rosa26RRFP reporter, we alternatively employed the Rosa26RmT/mG reporter. Ucp1-CreERT2; Rosa26RmT/mG pulse data showed the same result as Rosa26RRFP (Figure S1B). To further examine whether Ucp1-CreERT2 was truly specific to adipocytes, we isolated non-Tamoxifen IGW SVF from Ucp1-CreERT2; Rosa26RRFP mice at P60 and then treated these SVFs with 4-hydroxy-Tamoxifen (4-OHT). The data showed that upon 4-OHT treatment, there remained no RFP expression, indicating that UCP1-Cre does not turn on in other cell populations within SVF (Figure S1D). Therefore, we conclude that Ucp1-CreERT2 faithfully labels tissue resident UCP1+ multilocular brown and beige adipocytes, consistent with the previous results (Rosenwald et al., 2013).
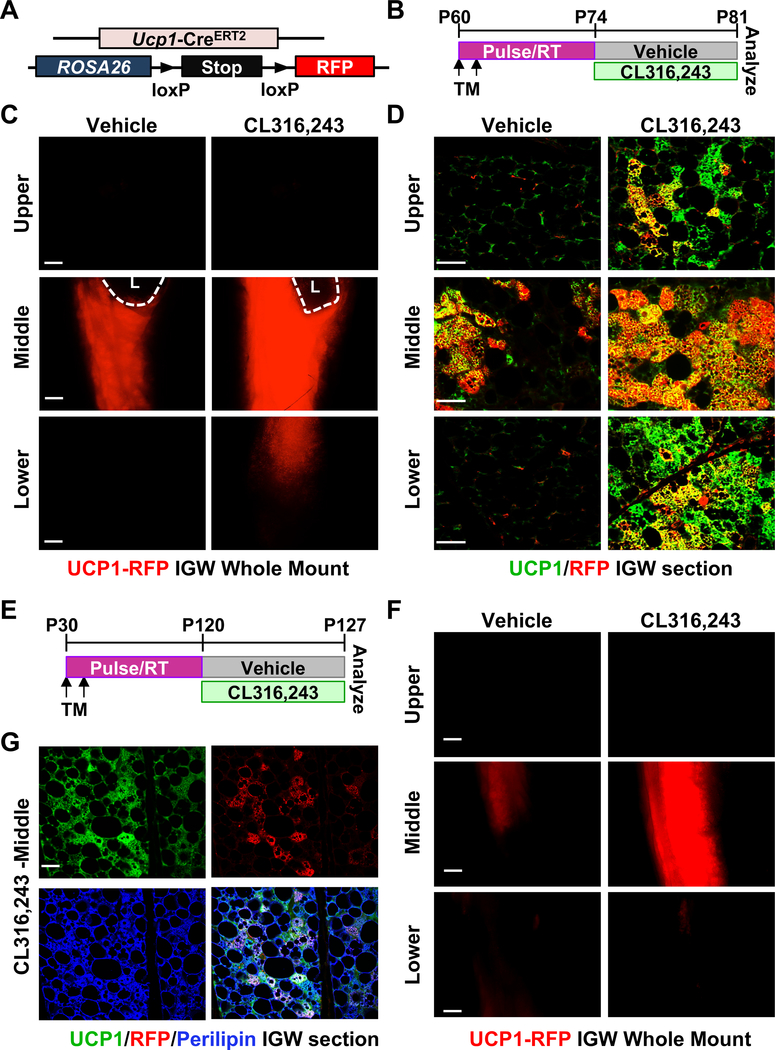
Figure 1. UCP1+ cells generate new beige adipocytes in response to β3-adrenergic stimulation.
(A) UCP1-RFP (Ucp1-CreERT2; Rosa26RRFP) mouse model.
(B) Experimental procedure. TM-induced 2-month-old UCP1-RFP male mice were administered with vehicle or CL316,243 (CL; β3 agonist) for 7 consecutive days before analyses.
(C) Whole IGW depots of mice described in (A) and (B) were imaged for direct RFP (n=5 mice per group). L: lymph node. Scale = 100 μm.
(D) Representative immunostaining of RFP (red) and UCP1 (green) on IGW sections of mice described in (B) (n=5 mice per group). Scale = 200 μm.
(E) Experimental procedure. TM-induced one-month-old UCP1-RFP male mice were administered vehicle or CL316,243 after 3 months to wash-out TM, and terminal analyses were undertaken after 7 days of treatment.
(F) Whole IGW depots of mice described in (E) were imaged for direct UCP1-RFP (n=4 mice per group). Scale = 100 μm.
(G) Representative immunostaining of UCP1 (green), RFP (red), and Perilipin (blue, also known as lipid droplet-associated protein which labels adipocytes) on IGW sections of mice described in (A) and (E) (n=4 mice per group). Scale = 100 μm.
We then administered CL316,243 (CL; a β3-agonist) to TM-induced 2-month-old UCP1-RFP mice (Figure 1A and 1B) and subsequently observed an increase in the number of multilocular beige adipocytes within IGW depot based on hematoxylin and eosin (H&E) staining (Figure S2A and S2B). Whole mount RFP imaging revealed an increase in overall RFP intensity around the lymph node and lower area (Figure 1C and S2C). To further confirm the results from whole mount IGW imaging, we conducted immunostaining of the IGW sections and found that more than 90% of the UCP1+ beige adipocytes around the lymph node were RFP positive, suggesting that pre-labeled UCP1+ RFP beige adipocytes give rise to CL-induced new beige adipocytes. In contrast, the upper and lower IGW regions, where little to no preexisting beige adipocytes were found without the stimulus, showed comparably low labeling (~20–25%) of the RFP+ UCP1+ beige adipocytes with CL treatment (Figure 1D), indicating that these CL-induced beige adipocytes are generated from non-UCP1+ sources. Given the potential TM residue within body, we did CL treatment into UCP1-RFP animals after 3 months of TM injection, when TM activity was completely washed out in mice (Figure 1E). Compared to vehicle, CL treatment significantly increased UCP1-RFP intensity in the peri-lymph node region (Figure 1F). Our histological examination revealed that ~70–80% UCP1+ beige adipocytes were RFP+ (Figure 1G), suggesting that a large majority of the long-lived UCP1+ cells have maintained their beige potential and are poised to expand in response to CL stimuli in adult life. Together, these results indicate that a subset of UCP1+ beige adipocytes are capable of generating new beige adipocytes in response to β3-adrenergic stimulation.
UCP1+ cells proliferate in response to β3-agonist in vivo
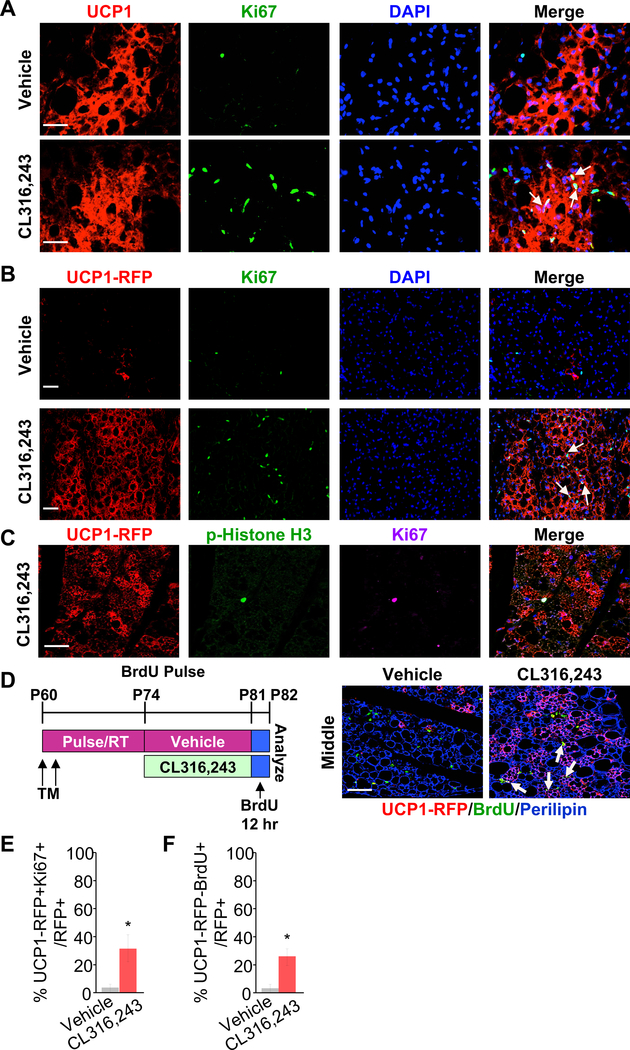
The fate mapping data above suggest that UCP1+ cells self-divide and give rise to new beige adipocytes in response to β3 stimulation, especially in the area of IGW around the lymph node. To investigate if beige adipocytes were capable of proliferation, we performed immunostaining analysis on 2-month-old TM induced UCP1-RFP IGW depots after CL treatment. The immunostaining on the IGW sections around the lymph node showed that CL treatment increased the number of cells expressing Ki67, a cellular proliferation marker, in UCP1+ beige adipocytes (Figure 2A). UCP1-RFP+ recapitulated the activity of UCP1+ cells: increased number of Ki67+ cells in UCP1-RFP+ cells (Figure 2B, quantified in Figure 2E). In agreement, a number of beige adipocytes were also positive for the proliferative marker phosphorylation of Histone 3, indicating active division (Figure 2C). To further confirm that these Ki67+ cells were adipocytes, we included caveolin-1 to visualize adipocyte plasma membranes, co-stained with Ki67 and p-Histone H3, as well as DAPI to visualize nuclei. We observed the overlapping among DAPI, Ki67 and caveolin-1, indicating that the Ki67 signal is from adipocyte nuclei (Figure S3A). In addition, we found similar results using BrdU pulse labeling after CL treatment with co-staining of RFP, BrdU and Perilipin (Figure 2D, quantified in Figure 2F). In contrast, cold exposed beige adipocytes within the IGW peri-lymph node region were Ki67 negative analyzed right after cold exposure (Figure S3B). As a control, we also examined BAT and observed that the number of Ki67+ cells within BAT was comparable between vehicle and CL treatment (Figure S3C). Therefore, these results suggest that a subset of UCP1+ beige adipocytes display progenitor-like characteristics to proliferate and give rise new beige adipocytes in response to β3 stimulation.
Figure 2. UCP1+ cells proliferate in response to β3 agonist in vivo.
(A-G) TM-induced 2-month-old UCP1-RFP male mice were administered with vehicle or CL316,243 (β3 agonist) for 7 consecutive days (n=3–5 mice per group).
(A) Representative immunostaining of UCP1 (red) and Ki67 (green) on IGW sections. DAPI (blue) stained the nuclei. Arrows showed Ki67+ cells that are UCP1+ as well. Scale = 200 μm.
(B) Representative fluorescence imaging of UCP1-RFP (red) and Ki67 (green) immunostaining on IGW sections. DAPI (blue) stained the nuclei. Scale = 100 μm.
(C) Representative fluorescence imaging of UCP1-RFP (red) and p-Histone H3 (green) immunostaining on IGW sections. DAPI (blue) stained the nuclei. Scale = 100 μm.
(D) Experimental procedure. TM-induced 2-month-old UCP1-RFP male mice were BrdU pulsed after vehicle or CL316,243 treatment (n=4 mice per group). Representative fluorescence imaging of UCP1-RFP (red), BrdU (green), and Perilipin (Blue) on IGW sections. Arrows showed BrdU+ cells that were UCP1+ as well. Scale = 200 μm.
(E-F) Quantification of Ki67 and Brdu in total RFP+ cells from IGW depots of vehicle or CL (CL316,243) treated mice (n=3–5 mice per group). *, P<0.05 between CL and vehicle-treated mice. Data are presented as means ± SEM.
(E) Percentage of Ki67+ beige adipocytes among all UCP1-RFP+ cell population.
(F) Percentage of BrdU+ beige adipocytes among all UCP1-RFP+ cell population, after BrdU pulse-labelling in Fig. 2D.
To further test this proliferative feature of the UCP1+ beige adipocytes in response to β3 stimulation, we isolated SV cells from un-induced Ucp1-CreERT2; Rosa26RRFP 2-month-old male mice, and SV cells were then induced with beige adipogenic media for 7 days. We then labeled these UCP1+ beige adipocytes with RFP reporter by a pulse treatment of 4-OHT. After 3 days of 4-OHT washout, we treated induced beige adipocytes with either vehicle or CL for 4 days and then quantified the RFP signal. Compared to vehicle condition, we observed a significantly increased number of RFP+ cells in the CL condition (Figure S3D). Altogether, these data are consistent with our in vivo fate mapping data and imply that beige adipocytes can turn on the proliferative program and generate new beige adipocytes in response to β3 stimulation.
UCP1+ cells are an important source of newly generated beige adipocytes in response to β3 stimulation
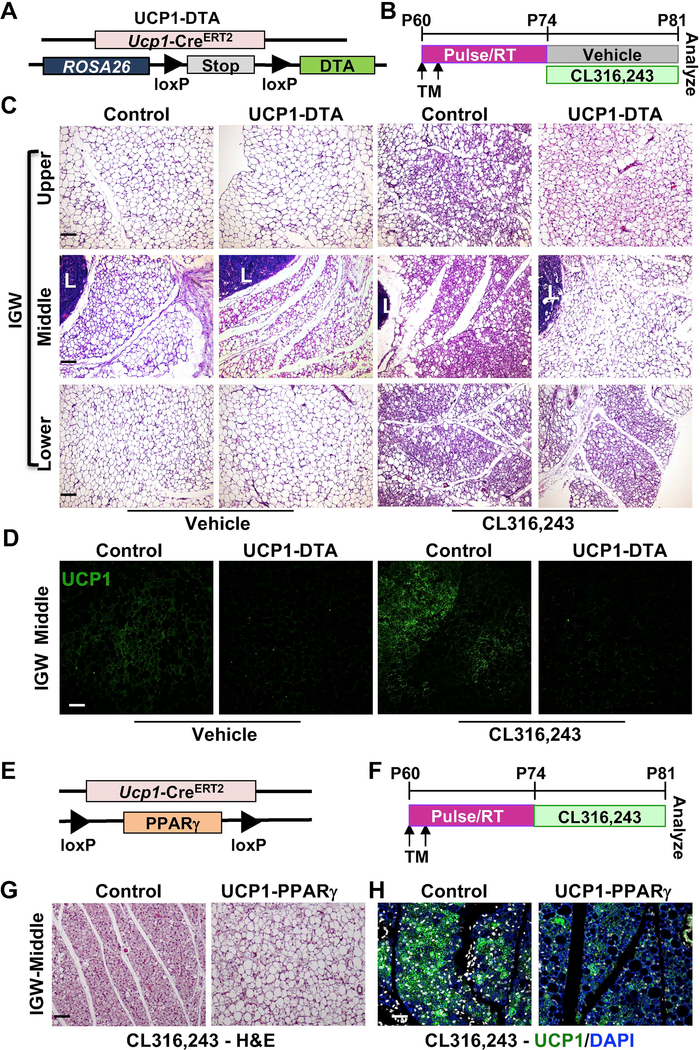
To test if UCP1+ cells were necessary for new beige adipocyte formation induced by β3 stimulation, we generated an inducible cell ablation model mediated by diphtheria toxin fragment A (DTA) to delete UCP1+ cells: Ucp1-CreERT2; DTAfl/fl (UCP1-DTA, Figure 3A). We injected vehicle or CL to TM-induced 2-month-old UCP1-DTA mice to temporally ablate UCP1+ cells, and then examined the beige adipocyte formation (Figure 3B). Both H&E staining and UCP1-immunostaining showed that CL-induced beige adipocyte formation was substantially reduced, especially near the lymph node in IGW of UCP1-DTA mice (Figure 3C,3D, S4A and S4B). There were rare RFP+ cells in UCP1-DTA IGW compared to controls, indicating efficient ablation of the UCP1+ cells in IGW depots (Figure S4B). In contrast, UCP1-DTA mice did not significantly impact the morphology of BAT based on H&E staining under either vehicle or CL-induced conditions (Figure S4C). This was likely due to the insufficient ablation in the BAT because the majority of brown adipocytes in UCP1-DTA still maintained the RFP+ brown adipocytes (Figure S4D). However, we could not rule out another possibility that there could be new brown adipocytes quickly regenerated from UCP1+ cells during our experimental time window.
Figure 3. UCP1+ cells are an important source of newly generated beige adipocytes in response to β3 agonist.
(A) UCP1-DTA (Ucp1-CreERT2; Rosa26RRFP; DTAfl/fl) mouse model.
(B) Experimental procedure. TM-induced 2-month-old UCP1-DTA male mice were treated with vehicle or CL316,243 (β3 agonist) for 7 days before terminal analyses (n=5 mice per group).
(C) Representative H&E staining on IGW sections of mice described in (A). IGW depots were shown at 3 locations: upper, middle, and lower. L: lymph node. Scale = 100 μm.
(D) Representative fluorescence imaging of UCP1 (green) on IGW sections. Scale = 100 μm.
(E) UCP1-PPARγ (Ucp1-CreERT2; PPARγfl/fl) mouse model.
(F) Experimental procedure. TM-induced 2-month-old UCP1-PPARγ male mice were treated with CL316,243 (β3 agonist) for 7 days before terminal analyses (n=5 mice per group).
(G) Representative H&E staining on IGW sections of mice described in (E). Scale = 100 μm.
(H) Representative fluorescence imaging of UCP1 (green) immunostaining on IGW sections. DAPI (blue) stained the nuclei. Scale = 100 μm.
To further support the necessity of the unique subset of UCP1+ cells for new beige adipocyte formation in response to external stimuli, we employed a functional deletion model to impair the adipogenic capability of UCP1+ cells by deleting peroxisome proliferator-activated receptor γ (PPARγ), a master regulator of adipose tissue development (Farmer, 2006; Lehrke and Lazar, 2005) (UCP1-PPARγ=UCP1-CreERT2; PPARγfl/fl) (Figure 3E and 3F). We then randomized TM-induced UCP1-PPARγ mice to vehicle or CL for 7 days. In this model, we observed similar results as the UCP1-DTA model, where beige adipocyte formation was reduced substantially near the lymph node area based on H&E and immunostaining of UCP1 (Figure 3G and 3H). Together, our UCP1+ cell ablation and functional deletion model studies indicate an essential role of UCP1+ cells in new beige adipocyte formation in response to stimuli.
Overexpressing cell cycle inhibitor p21 in UCP1+ cells inhibits beige adipocyte formation
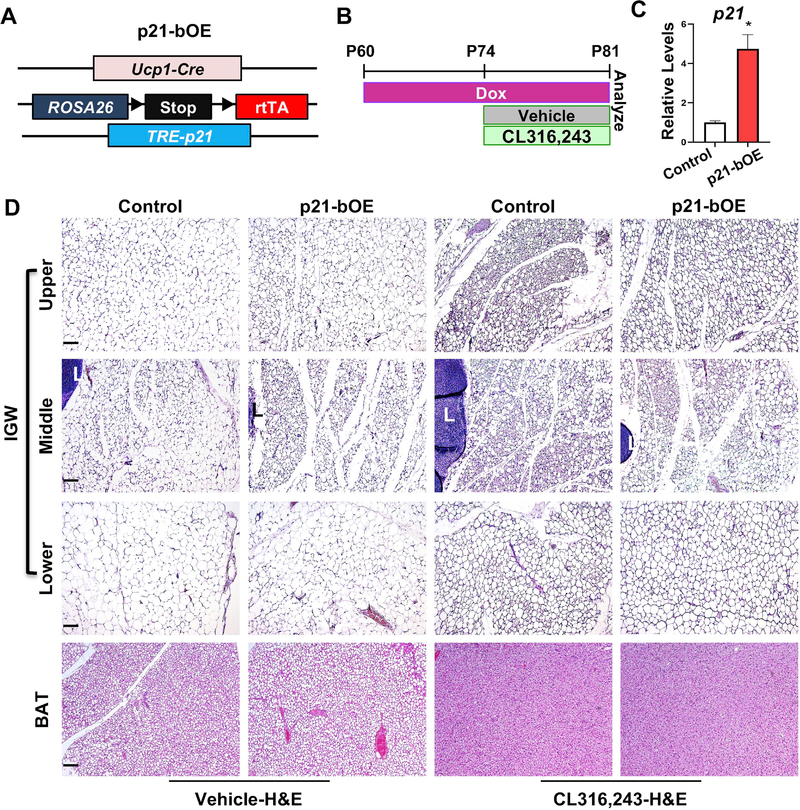
The above models could not differentiate the role of fully differentiated mature UCP1+ cells from that of dividing UCP1+ cells since all the UCP1+ cells were deleted or disrupted in these two models regardless of their cell division potential. Therefore, to investigate the specific contribution of the dividing UCP1+ cells, we induced conditional overexpression of p21, a cell cycle inhibitor encoded by cyclin-dependent kinase inhibitor 1A, in UCP1+ cells: Ucp1-Cre, Rosa26R-rtTA; TRE-p21 (p21-bOE, Figure 4A). p21 has been shown to trigger cell cycle arrest in other stem cell lineages (Cheng et al., 2000). After providing doxycycline (Dox) in drinking water for two weeks, we injected vehicle or CL to Dox-induced 2-month-old control and p21-bOE mice (Figure 4B). Real-time qPCR analysis verified that there was a ~4-fold induction of p21 expression in the p21-bOE model (Figure 4C). Consistent with the role in cell cycle, we observed that the p21 overexpression model had reduced Ki67 positive signals compared to controls (Figure S4E). H&E staining of IGW and BAT sections indicates that increased p21 expression did not significantly alter adipocyte size and morphology in vehicle conditions (Figure 4D). As expected, multilocular beige adipocytes widely accumulated in IGW of the control mice upon CL treatment (Figure 4D). However, this CL-induced beige adipocyte formation was largely blocked in p21-bOE mice (Figure 4D). Interestingly, we observed that the beige fat induction appears to be equally compromised in the upper and lower region of the depot, which suggests a non-cell-autonomous effect from the middle area of the IGW depots. In contrast, p21-bOE mice did not significantly change the morphology of BAT based on H&E staining under CL treatment (Figure 4D). These data suggest that overexpressing cell cycle inhibitor p21 in UCP1+ cells disrupts beiging potential.
Figure 4. Overexpression of p21 in UCP1+ cells inhibits beige adipocyte formation in response to β3 agonist.
(A) Mouse model used to overexpress p21 in UCP1+ cells.
(B) Experimental procedure. TM-induced 2-month-old male control or p21-bOE mice were administered vehicle or CL316,243 for 7 consecutive days (n=4 mice per group).
(C) Real-time qPCR analysis of p21 in control or p21-bOE IGW depots before CL316,243 treatment (n=4 mice per group). Data are expressed as means ± SEM. *P<0.01.
(D) Representative H&E images of IGW and BAT sections from control and p21-bOE. IGW depots were separated into 3 portions: upper, middle, and lower. L: lymph node. Scale = 100 μm.
Deleting cell cycle inhibitor Cdkn2a in UCP1+ cells promotes beige adipocyte formation
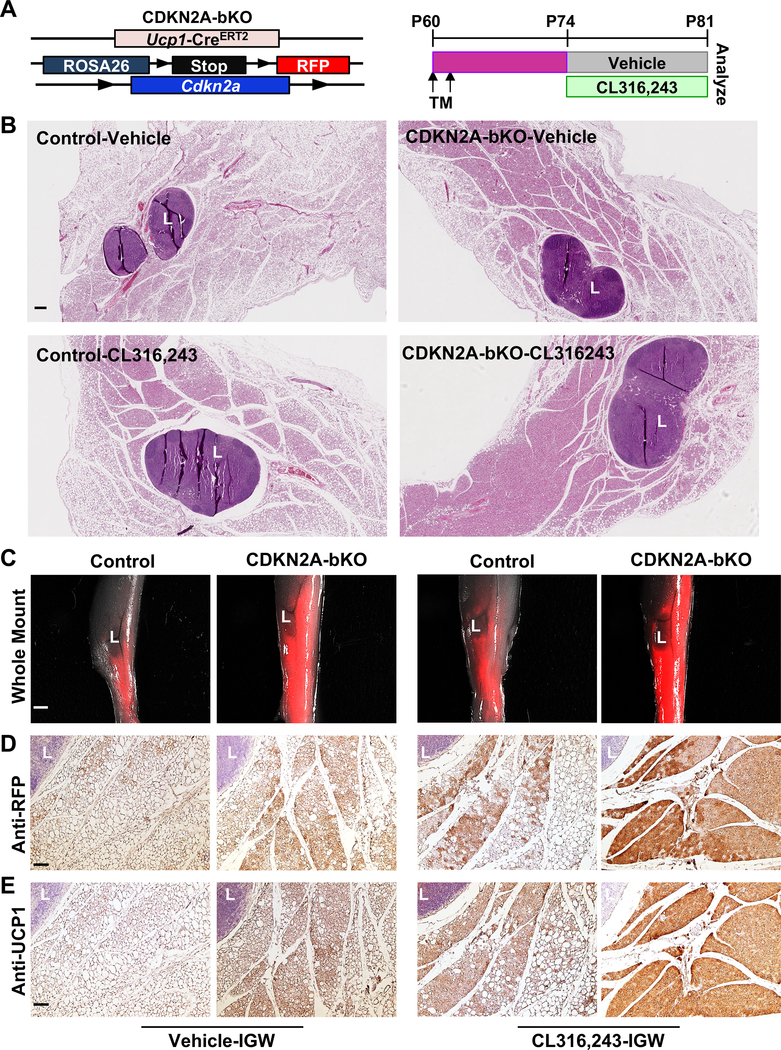
To further assess the importance of UCP1+ cell proliferative events in CL-induced beige adipocyte formation, we investigated whether we could specifically increase the cell division of UCP1+ cells and test the subsequent effects. Cdkn2a, a gene encoding for p16Ink4a and p14Arf, is associated with the risk of obesity and type 2 diabetes (Annicotte et al., 2009; Hannou et al., 2015; Morris et al., 2012; Pal et al., 2016). A recent CDKN2A whole-body knockout study suggested the potential significance of CDKN2A in whole-body energy metabolism through promoting adipose tissue browning (Rabhi et al., 2018). We have previously reported a role of CDKN2A in SMA+ beige progenitor cells isolated from aged mice and humans (Berry et al., 2017), but the function of CDKN2A in the UCP1+ beige adipocytes remains undetermined. Therefore, we crossed our inducible Ucp1-CreERT2 mouse model with a Rosa26RRFP indelible labeling reporter and Cdkn2a floxed mice: Ucp1-CreERT2; Rosa26RRFP; Cdkn2afl/fl (CDKN2A-bKO, Figure 5A). This model allowed us to delete Cdkn2a conditionally and specifically in the UCP1+ beige adipocytes and to simultaneously mark and track the beige adipocytes. Fourteen days after administering TM to 2-month-old control and CDKN2A-bKO mutant mice, we induced beige adipocyte formation by injecting CL for 7 consecutive days (Figure 5B). With CL treatment, control mice induced beige adipocyte formation around the lymph node areas in IGWs. Interestingly, CDKN2A-bKO IGW sections had notably increased beige adipocyte formation compared to controls (Figure 5A, 5B, S5A, and S5B). Accordingly, we observed that the mRNA levels of key components of the thermogenic genes, such as Ucp1, Cox8b, and Pgc1a, were significantly higher in CDKN2A-bKO than controls (Figure S5C).
Figure 5. Deleting cell cycle inhibitor Cdkn2a in UCP1+ cells promotes beige adipocyte formation.
(A) Scheme of CDKN2A-bKO mouse model used to conditionally knockout Cdkn2a in UCP1+ cells (Ucp1-CreERT2; Rosa26RRFP; Cdkn2afl/fl) and experimental procedure. TM-induced 2-month-old male UCP1-RFP (control) or CDKN2A-bKO mice were administered CL316,243 for 7 consecutive days before they are sacrificed for terminal analyses (n=6 mice per group).
(B) Representative H&E staining on IGW sections from control and CDKN2A-bKO described in (A). L: lymph node. Scale = 0.5 mm
(C) Whole IGW depots of mice described in (A) were imaged for direct RFP (n=6 mice per group). L: lymph node. Scale = 100 μm.
(D) Representative IHC of UCP1-RFP staining on IGW sections. Scale = 100 μm.
(E) Representative IHC of UCP1 staining on IGW sections. Scale = 100 μm.
We also assessed beige adipocyte formation in CDKN2A-bKO mice without CL-treatment. Remarkably, the histology sections revealed that CDKN2A-bKO had a higher number of beige adipocytes compared to controls under both vehicle and CL-treatment conditions (Figure 5B). Whole mount imaging of IGWs confirmed that even without CL, the CDKN2A-bKO mice had higher RFP+ intensity compared to controls (Figure 5C). Notably, we observed baseline beige adipocyte around the lymph node area of control IGWs both in histology section and whole mount imaging, suggesting a stimulus for beige adipocytes even at room temperature (22 °C). Consistent with the whole mount imaging, IHCs of RFP and UCP1 revealed that CDKN2A-bKO IGWs had more beige adipocytes than controls under both vehicle and CL treated conditions (Figure 5D and 5E). Unlike IGWs, the histology sections of CDKN2A-bKO BATs were similar to those of control BAT depots under CL-treatment based on both H&E staining and anti-UCP1 staining (Figure S5D). Of note, vehicle-treated CDKN2A-KO BATs appeared to have lower UCP1 expression compared to control BATs. This suggests that there may be a thermogenic defect in CDKN2A-bKO BATs under the vehicle condition, which may drive a compensatory increase of beiging in IGW. This speculation also lines up with the unchanged body weight and adiposity in CDKN2A-bKO mice. Meanwhile, a shorter treatment of CL (3 consecutive days, Figure S5E) was also effective to induce multilocular UCP1-RFP+ beige adipocytes in CDKN2A-bKO IGWs compared to controls (Figure S5F). Together, these data suggest that deleting cell cycle inhibitor Cdkn2a in UCP1+ cells promotes beige adipocyte formation with or without CL stimuli.
Since thermogenic adipocytes can increase energy expenditure and reduce body fat, we expected that TM-induced 2-month-old CDKN2A-bKO mice would have less body weight and reduced body fat after vehicle or CL treatment. However, we did not observe significant difference in body weights among vehicle or CL-treated control and CDKN2A-bKO mice (Figure S6A and S6B). Neither did we observe a difference in serum analysis of non-esterified fatty acids (NEFA) (Figure S6C), cholesterol (Figure S6D), glucose (Figure S6E), and triglyceride levels (Figure S6F) of vehicle or CL-treated control and CDKN2A-bKO mice. The adipose tissue weights were similar in both control and CDKN2A-bKO mice regardless of vehicle or CL treatment (Figure S6G). CDKN2A deletion was confirmed by qPCR in IGW depots (Figure S6H). These results suggest that CDKN2A-bKO mice do not display immediate metabolic phenotypes even with increased number of beige adipocytes.
An inverse correlation between cell cycle gene expression and beige activity
To address the physiological role of CDKN2A in beige adipocyte development, we assessed the expression level of cell cycle genes in response to beige adipocyte expansion. We observed that levels of cell cycle inhibitors including both p16Ink4a and p14ARF within IGW were reduced in response to both cold stimulus and CL treatment. In contrast, cell cycle activator CCND1 (Cyclin D1) was increased as assessed by qPCR (Figure S7A). We also tested the effect of Rosiglitazone (Rosi) which is a thiazolidinedione (TZD) that acts as a PPARγ agonist and is also known as a beiging inducer. Similar to cold exposure, we observed reduction of p16Ink4a and p14ARF after Rosi treatment (Figure S7B). Furthermore, our previous finding showed that 6-month-old mice had significantly reduced beiging potential compared to 2-month-old mice (Berry et al., 2017). Consistent with the potential role of cell cycle gene expression in beige activity, these older mice had increased levels of p16Ink4a, p14ARF and reduced level of cyclinD1 in IGW depots (Figure S7C). Together, our expression data suggest that there is an inverse correlation between the level of cell cycle genes and beige adipocyte activity, further supporting the notion that cell cycle genes are involved in controlling beige adipocyte expansion and their activity.
A dual role of Cdkn2a in beige adipocyte expansion
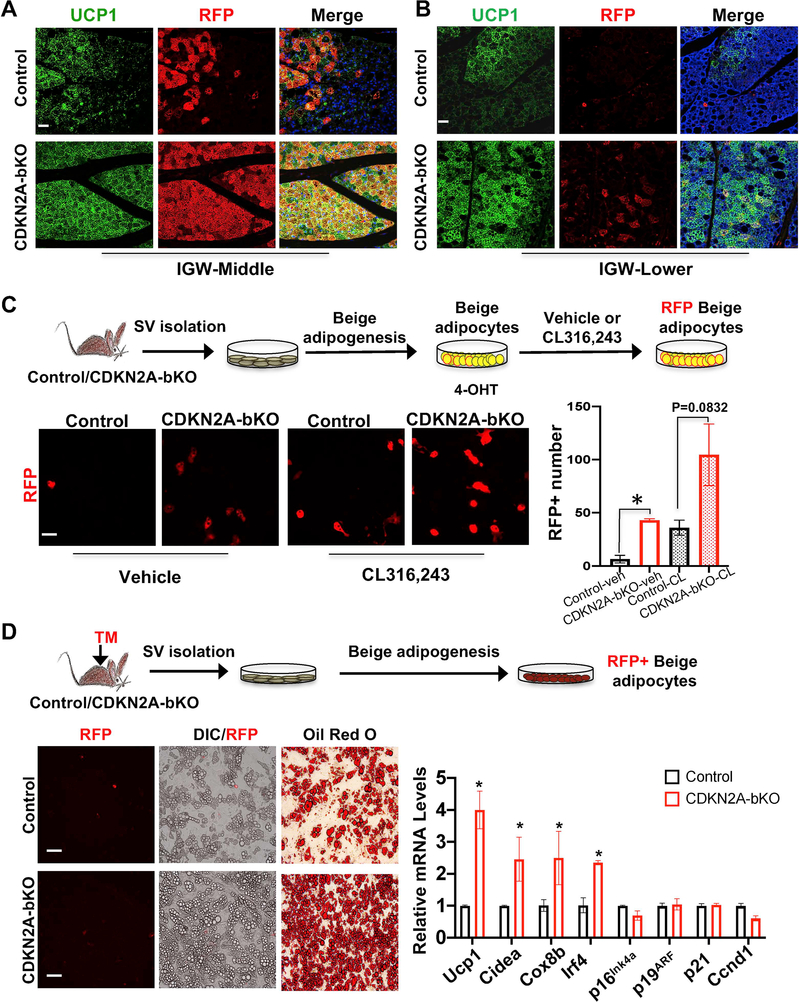
To investigate how Cdkn2a impacted the beige adipocyte formation in UCP1+ cells, we first tested if CL-induced beige adipocyte expansion in CDKN2A-bKO mutant mice derived exclusively from a UCP1+ lineage or not. Our whole mount imaging suggests that loss of CDKN2A generated more RFP+ beige adipocytes around peri-lymph node area of IGW compared to control depots (Figure 5C). This whole mount data was supported by our IHC staining of RFP and UCP1 on tissue sections from IGW middle (Figure 5D and 5E). Through UCP1 and RFP co-staining quantification of IGW sections, we found that the majority (70–80%) of the UCP1+ beige adipocytes in control mice were RFP+, and a similar labeling rate was observed in CDKN2A-bKO IGW depots around the lymph nodes (Figure 6A). This indicates that the majority of new beige adipocytes, which were formed around the lymph nodes in response to CL treatment in the CDKN2A-bKO mice, arise from the existing CDKN2A-deficient UCP1+ cells. In addition to peri-lymph node area of IGW, we also observed a large number of beige adipocytes formed in the upper and lower part of IGW (Figure S5B), which was not close to the lymph node. Interestingly, the majority of these beige adipocytes were RFP-negative in both control and mutants (Figure 6B). These results suggest that CDKN2A in UCP1+ cells play a dual role: a cell-autonomous role in expanding beige fat, and a non-cell-autonomous role in stimulating new beige fat formation.
Figure 6. Both cell-autonomous and non-cell-autonomous roles of CDKN2A in beige expansion in response to CL treatment.
(A-B) Representative fluorescence imaging of UCP1 (green) and UCP1-RFP (red) immunostaining on IGW sections. Scale = 100 μm. (n=3–4 mice per group).
(C-D) Beige adipogenesis in vitro. (n=3–4 mice per group).
(C) SV cells isolated from IGW of non-Tamoxifen treated UCP1-RFP (control) or mutant CDKN2A-bKO mice were differentiated into beige adipocytes in vitro first and then treated with 4-OHT to induce deletion, then treated with vehicle or CL316,243; RFP imaging; Scale = 100 μm. Data are expressed as means ± SEM; *P<0.05.
(D) Scheme to isolate SV cells after 2 days of TM injection to UCP1-RFP (control) or mutant CDKN2A-bKO mice, and then induce beige fat differentiation. DIC, RFP and Oil Red O imaging; Scale = 100 μm. Real-time qPCR analysis of thermogenic and cell cycle genes; Data are expressed as means ± SEM; *P<0.05.
To probe how loss of Cdkn2a in UCP1+ cells promoted beige adipocyte expansion in a cell-autonomous manner, we directly tested whether the expansion of beige fat by CDKN2a depletion was mediated by its effect on cell proliferation. We first assessed RFP+ beige cell proliferation in vivo by providing BrdU drinking water to the mice right after Tamoxifen administration until the end of the 7-day CL treatment (Figure S7D). BrdU incorporation into the RFP+ compartment was analyzed by flow cytometry (BrdU and RFP). CDKN2A-bKO mutants had increased BrdU+ in RFP+ cells compared to controls, and this phenotype appeared to be specific to IGW but not BAT (Figure S7E). Next, we further tested this proliferative effect using our established in vitro system. Briefly, SV cells from uninduced control and CDKN2A-bKO mice were first differentiated into beige adipocytes. We then induced CDKN2A deletion in these beige adipocytes in vitro by a pulse treatment of 4-OHT. After 3 days of 4-OHT washout, we treated control and mutant beige adipocytes with either vehicle or CL for 4 days and then quantified the RFP signal. Consistent with our in vivo data, we observed increased number of the RFP+ cells in CDKN2A-bKO in both vehicle and CL conditions (Figure 6C). Together, these data suggest that CDKN2A inhibits beige adipocyte proliferation and expansion in a cell-autonomous manner.
To further examine the non-cell-autonomous role of CDKN2A in beige expansion in response to CL stimuli, we isolated the SV cells from the Tamoxifen pulse control and CDKN2A-bKO mutant mice at P60 and subsequently cultured them in beige adipogenic media. Our qPCR data showed there were no changes in the levels in CDKN2A and other cell cycle genes (Figure 6D), consistent with the notion that CDKN2A was not affected in SV cells. In contrast, the beige adipogenic potential of SV cells from CDKN2A-bKO mice appeared to increase compared to control SV cells as assessed by lipid-containing adipocytes, Oil Red O staining and expression of thermogenic genes (Figure 6D). To further support the notion that the primary effects of CDKN2A-bKO mice were not from SV compartments, we isolated SV cells from un-induced control and CDKN2A-bKO mice. We then cultured the cells in adipogenic media containing 4-OHT to delete Cdkn2a. In contrast to Tamoxifen injection in vivo, SV cells from control and CDKN2A-bKO mice that received 4-OHT underwent beige adipogenesis similarly as indicated by Oil Red O staining (Figure S7F).
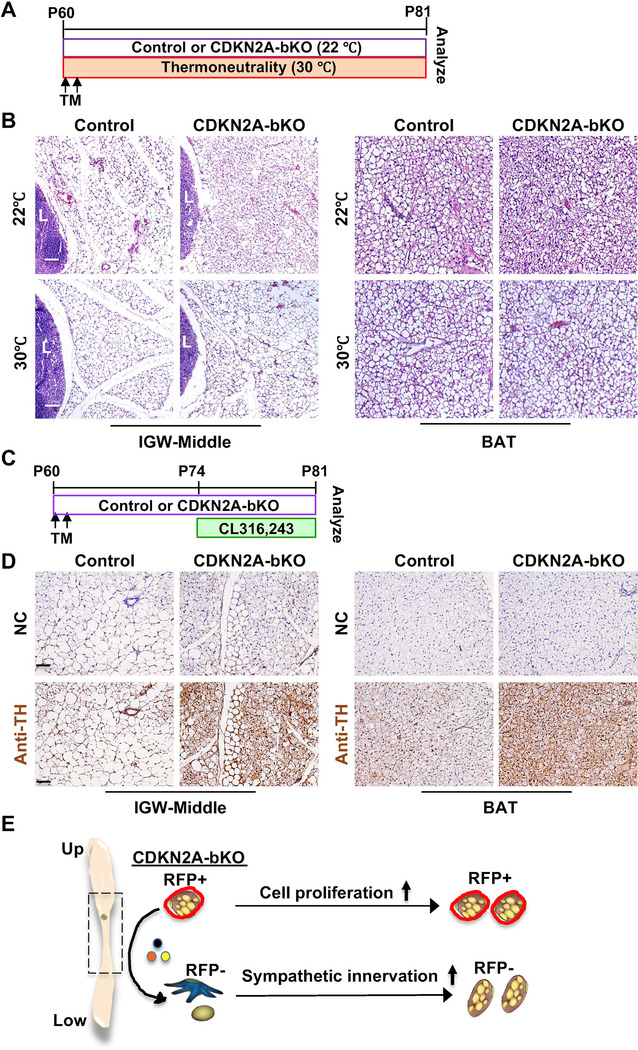
Sympathetic nerve innervation is required for WAT beiging (Cao et al., 2019). The observation that CDKN2A-bKO mutant mice have more beige adipocytes in the absence of CL treatment is interesting. To remove the low-level thermal stress and sympathetic flow at standard housing temperature (22°C), we kept the control and CDKN2A-bKO mutant mice at thermoneutrality (30°C) right after TM induction (Figure 7A). Unlike the mice kept at 22°C, we found that both control and CDKN2A-bKO mutant mice had blunted ability to induce multilocular beige adipocyte formation even in the peri-lymph node area of IGW based on H&E staining (Figure 7B). Of note, we did not observe apparent morphological difference in BAT H&E staining (Figure 7B). These data suggest that the beige adipocyte expansion observed in CDKN2A-bKO mutant mice is largely dependent on sympathetic innervation.
Figure 7. Deleting Cdkn2a in UCP1+ cells increases sympathetic innervation.
(A) Experimental procedure. TM-induced 2-month-old male control or CDKN2A-bKO mice were kept at room temperature (22°C) or under thermoneutrality (30°C). (n=3–4 mice per group).
(B) Representative H&E staining on IGW and BAT sections from control and CDKN2A-bKO described in (A). L: lymph node. Scale = 100 μm.
(C) Experimental procedure. TM-induced 2-month-old male control or CDKN2A-bKO mice were administered vehicle or CL316,243 for 7 consecutive days before terminal analyses (n=5–6 mice per group).
(D) Representative IHC of TH staining on IGW and BAT sections of mice described in (C). Scale = 100 μm.
(E) Working model of CDKN2A-bKO on beige formation. Deletion of Cdkn2a in UCP1+ cells stimulates new beige fat formation through both cell proliferation (a cell-autonomous role; RFP+; mainly in the IGW upper part) and enhanced sympathetic innervation (a non-cell-autonomous role; RFP-; mainly in the IGW lower part).
To evaluate sympathetic nerve activity, we performed tyrosine hydroxylase (TH, a marker of norepinephrine turnover) immunostaining. Notably, TH immunoreactivity of the CDKN2A-bKO mutant IGW and BAT were substantially enhanced relative to controls (Figure 7C and 7D). Interestingly, we did not observe any significant difference in lipolysis and lipogenesis activities in IGW and BAT of control and mutant mice, suggested by qPCR analyses of Atgl, Lpl, Acly (Figure S7G). These data suggest that CDKN2A-bKO mutants have enhanced sympathetic innervation to induce WAT beiging. All together, these data suggest that loss of CDKN2A in UCP1+ cells stimulates new beige fat formation through both cell proliferation (a cell-autonomous role) and enhanced sympathetic innervation (a non-cell-autonomous role) (Figure 7E).
Discussion
Tremendous efforts have been made to find signals to induce beige adipocyte formation in WAT and identify the cellular origins. It is thought that beige adipocytes are terminally differentiated and non-proliferative, and they are converted into white adipocytes after discontinued cues including β3 agonist and cold stimuli (Altshuler-Keylin et al., 2016; Lu et al., 2018; Paulo and Wang, 2019; Roh et al., 2018; Rosenwald et al., 2013). In contrast, relatively little work has been done to investigate the fates of beige adipocytes and their regulations. In this study, we report a new finding that a class of UCP1+ cells within the peri-lymph node region of inguinal WAT can proliferate and generate new beige adipocytes in response to β3 stimulation, indicating that this class of UCP1+ cells possess progenitor-like characteristics. Further, we provide evidence that this beige adipogenesis is modulated by cell cycle proteins including p21 and CDKN2A. Our work provides mechanistic insights on beige adipocyte development and has implications in potential therapeutics against obesity and aging.
We and others have previously shown that cold exposure and β3 agonists stimulate beige adipocyte recruitment through distinct cellular sources (Acharya et al., 2019; Berry et al., 2016; Jiang et al., 2017; Lee et al., 2015; Long et al., 2014; McDonald et al., 2015; Min et al., 2016; Oguri et al., 2020; Rosenwald et al., 2013; Shao et al., 2019; Vishvanath et al., 2016; Wang et al., 2014). Particularly we have demonstrated that vascular smooth muscle cells (SMA) can be converted into beige adipocytes by cold treatments, whereas β3 agonists interconvert white adipocytes to form beige adipocytes (Jiang et al., 2017). In this study, using UCP1-RFP fate mapping tools, we followed the fate of the UCP1+ beige adipocytes and demonstrated that a subset of the UCP1+ beige adipocytes around the peri-lymph node of WAT were capable of generating new beige adipocytes in response to β3 stimulation. The fact that the UCP1+ cells close to the lymph node area of the IGWs can proliferate and produce new beige adipocytes in response to β3 stimulation may seem surprising, considering differentiated beige adipocytes are post-mitotic. However, the findings from a recent report suggest the induction of UCP1 expression in nonadipocytes (Shamsi et al., 2020). Several recent studies have established the remarkable plasticity within mature adipocyte populations (Giordano et al., 2014; Wang et al., 2018; Zhang et al., 2019). Wang et al. showed that pre-existing mammary adipocytes could act as preadipocyte-like precursors and undergo de-differentiation and re-differentiation during pregnancy and lactation (Wang et al., 2018). Zhang et al. reported that in dermal adipocytes dedifferentiated cells could proliferate and re-differentiate into adipocytes upon various challenges (Zhang et al., 2019). Therefore, the finding that a subset of UCP1+ cells, which are thought to be post-mitotic, can proliferate and generate new beige adipocytes in response to β3 stimulation, representing a new source of beige adipogenesis.
Mechanistically, we demonstrated that overexpression of p21, a cell cycle inhibitor, in UCP1+ cells inhibits beige adipocyte formation. In contrast, deletion of Cdkn2a, another cell cycle inhibitor, promotes beige adipocyte formation. Our results suggest that CDKN2A in UCP1+ cells can negatively impact new beige fat formation through two distinctive mechanisms: cell proliferation (a cell-autonomous role) and enhanced sympathetic innervation (a non-cell-autonomous role). Of note, it remains to be determined if sympathetic activation and cell cycle regulated by CDKN2A are dependent events or not, and if yes, which one occurs first. In addition, we demonstrated that the levels of expression of cell cycle inhibitors including both p16Ink4a and p14ARF were reduced in response to both cold stimulus and CL treatment. In contrast, cell cycle activator CCND1, also known as cyclinD1, was increased as assessed by qPCR. These results provide strong evidence that cell cycle genes are involved in modulating beige adipogenesis. Interestingly, although our results suggest that p21 and Cdkn2a play an essential role in beige expansion, they appear to play no significant role in BAT expansion. Since proliferation of mature brown adipocytes has been reported to contribute to cold-induced BAT hyperplasia (Fukano et al., 2016), we hypothesize that other cell cycle genes may play a role in BAT expansion. Indeed, overexpression of Cdkn1b (Cyclin-dependent kinase inhibitor 1B, which encodes a cell cycle inhibitory protein p27Kip1) in mature adipocytes was shown to impair development of BAT, but not that of WAT (Okamatsu-Ogura et al., 2017). Together, these data indicate that cell cycle regulatory genes may have divergent effects on brown and beige adipocytes.
Increased number of thermogenic adipocytes is often accompanied by a profound improvement in metabolic health, including reduced adiposity and improved lipid profile. Interestingly, our CDKN2A-bKO mice had no significant difference in body weight and adiposity, even with increased number of beige adipocytes. We reasoned this could be due to the timing of our analysis, and CDKN2A-bKO would have long-term metabolic improvement over an extended period of time. Importantly, our data suggest that the formation of beige adipocytes in CDKN2A-bKO mice can be triggered at standard housing room temperature, which has modest thermal stress but without external stimuli such as cold and β3 agonists. This could be significant because both prolonged cold exposure and β3 agonists have side effects, especially for those with heart problems. Because both obesity and aging have significantly diminished ability to form beige adipocytes (Berry et al., 2017; Khanh et al., 2018; Shin et al., 2017), it makes thermogenic brown and beige adipocytes a potential target for anti-obesity and even anti-aging by reducing fats and sugars. We and others have reported that obesity and aging have increased level of CDKN2A. Therefore, it is also conceivable that this high level of CDKN2A may impair the proliferation of UCP1+ beige adipocytes, leading to beiging failure in obese and old mice. The possibility that beiging potential can be retained in old CDKN2A-bKO mice is noteworthy because many of the previous beige studies were performed on lean healthy rodents and humans, resulting in increased energy expenditure, weight loss, and improved metabolic health. It is not yet clear if the same program will work on older, obese individuals.
In summary, this study shows that a unique subset of UCP1+ cells around the lymph node of white adipose tissues is capable of self-dividing in response to β3 stimulus and generating new beige adipocytes, which is a previously unappreciated cellular mechanism of beige adipocyte formation. We believe that more research should be conducted exploring the use of beiging potential of beige adipocytes as a new therapeutic approach against the global obesity epidemic.
Limitations of the Study
One limitation of our study is that the nature of these proliferative UCP1+ beige adipocytes in response to β3 stimulation remains unknown. Although UCP1+ expression is restricted to mature beige and brown adipocytes at pulse in our studies, it is conceivable that UCP1+ beige adipocytes may lose mature adipocyte features and gain progenitor-like characteristics in response to β3 agonists. With future single-cell RNA-seq and spatial transcriptomics analysis (Burl et al., 2018; Hepler et al., 2018; Liu et al., 2019; Min et al., 2019; Raajendiran et al., 2019; Schwalie et al., 2018; Weinstock et al., 2019), we expect it will shed insight into the molecular heterogeneity and trajectories within UCP1+ beige adipocytes in response to beige stimuli. Further, expansion of the pre-existing UCP1+ cells is most prominent around lymph nodes of IGW. This local specific effect correlates well with the observation that many more UCP1+ adipocytes are found here in the mice housed at room temperature, suggesting that the lymph nodes may act as a signal producing center for beige adipocyte formation in response to CL stimuli. Although multiple studies support the link between lymphatic function and obesity (Escobedo and Oliver, 2017; Greene and Maclellan, 2013; Harvey et al., 2005; Hespe et al., 2016), the potential role of lymphatic systems in beige adipocyte formation remains unclear. Further studies into this relationship could help elucidate molecular and cellular mechanisms on how beige adipocytes develop in response to sympathetic activation.
STAR METHODS
Resource Availability
Lead Contact
Requests for further information, reagent, and resource should be directed to and will be fulfilled by the Lead Contact, Yuwei Jiang (yuweij@uic.edu).
Materials Availability
This study did not generate any new unique reagents.
Data and Code Availability
N/A
Experimental Models and Subject Details
Mouse Models
All the studies were performed according to procedures reviewed and approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago. Mice were housed in a 14:10 light:dark cycle with a standard rodent chow diet and water unless otherwise indicated. Ucp1-Cre (Stock No. 24670), Rosa26R-M2rtTA (Stock No. 6965), Rosa26RDTA (Stock No. 006331), Pparγl/fl (JAX Stock No: 004584), Rosa26RmT/mG (Stock No. 007676) and Rosa26RRFP (Stock No. 007914) mice were obtained from the Jackson Laboratory (Bar harbor, ME, USA). TRE-p21 was previously generated in Dr. Jonathan M. Graff lab (Berry et al., 2017). Ucp1-CreERT2 and Cdkn2afl/fl mice were generously provided by Dr. Eric N. Olson (University of Texas Southwestern Medical Center).
Cell Culture
The isolated SV cells were cultured in DMEM/F12 media (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO) and 1% Penicillin/Streptomycin (Gibco, Waltham, MA). Beige adipogenesis was induced by treating confluent cells with DMEM/F12 containing 10% FBS, 1 μg/mL insulin (Sigma-Aldrich, St. Louis, MO), 1 μM dexamethasone (Cayman, Ann Arbor, MI), 0.5 mM isobutylmethylxanthine (Sigma-Aldrich, St. Louis, MO), 60 μM indomethacin (Sigma-Aldrich, St. Louis, MO), 1 nM triiodo-L-thyronine (Sigma-Aldrich, St. Louis, MO), and 1 μM rosiglitazone (Sigma-Aldrich, St. Louis, MO) for the first 3 days and with DMEM/F12 containing 10% FBS and 1 μg/mL insulin, 1 nM triiodo-L-Thyronine, and 1 μM rosiglitazone for every 3 days. To induce thermogenic genes, cells were treated with 5 μM CL316,243 for 4 days.
Method Details Tamoxifen treatment
Cre recombination was induced by administering 1.5mg kg−1 body weight of Tamoxifen (TM; Cayman Chemical, Ann Arbor, MI, USA) dissolved in sunflower oil (Sigma-Aldrich, St. Louis, MO, USA) through intraperitoneal (IP) injection for 2 consecutive days. For Cre induction in cell culture system, we used 2 μM/mL 4-hydroxy-Tamoxifen (4-OHT, Sigma, Sigma-Aldrich, St. Louis, MO).
Beige adipocyte induction in mice
CL316,243 (CL; Tocris, Minneapolis, MN, USA) was dissolved in water and injected at one dose (1 mg kg−1day−1) for 7 consecutive days through IP injection. For cold exposure experiments, mice were placed in a 6°C environmental chamber (Environmental & Temperature Solutions, Hoffman Estates, IL, USA) for 7 days while the control group was maintained at room temperature (22°C).
Serum analysis
Blood was collected into heparinized tubes from the heart and then centrifuged at 6,000 × g for 8 min, the plasma was collected, and aliquots were stored at −80°C until analysis. Plasma glucose concentration was measured spectrophotometrically using the Trinder Assay (Sigma-Aldrich, St. Louis, MO, USA). Non-esterified fatty acids (NEFA), triglyceride (TG), and cholesterol concentration in serum were measured with NEFA-C (Wako), Triglyceride E tests (Thermo Fisher Scientific) and cholesterol liquicolor kits (Stanbio), respectively. Assays were performed according to manufacturer instructions.
Isolation of stromal vascular fraction (SVF)
The SVF cells were isolated as previously described (Shin et al., 2020b). Briefly, subcutaneous WAT was pooled and digested at 37°C for 1 hour in isolation buffer (100mM HEPES, 0.12 M NaCl, 50 mM KCl, 5mM D-glucose, 1.5% BSA, 1mM CaCl2, pH 7.3) containing 1 mg ml−1 collagenase. After removing the floating layer containing mature adipocytes, the aqueous phase was centrifuged at 1000 Χ g for 10 minutes. The pellet, including a crude SVF, was resuspended in red blood cell (RBC) lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 minutes and centrifuged at 1000 g for 5 minutes. The pellet was washed once 1X PBS, resuspended, and strained through 70 μm mesh.
Histological staining
Hematoxylin and eosin (H&E) staining was conducted on paraffin sections using standard methods. Briefly, adipose tissues were fixed in formalin overnight, dehydrated, embedded in paraffin, and sectioned with a microtome at 4–8 μm thicknesses. For immunohistochemistry (IHC), sections were deparaffinized, boiled in antigen-retrieval solution, treated with rabbit anti-UCP1 antibody (1:500, ab23841, Abcam), mouse anti-RFP (1:200, 632392, Takara), or rabbit anti-TH (tyrosine hydroxylase, 1:500, ab6211, Abcam) in blocking buffer (5% normal donkey serum in 1X PBS) at 4°C overnight, treated with secondary antibody for 2 hours at room temperature in blocking buffer, and stained with Vectastain ABC KIT (PK-6100, Vector Laboratories, Burlingame, CA) and DAB KIT (SK-4100, Vector Laboratories, Burlingame, CA). For immunofluorescence staining, paraffin sections were incubated with permeabilization buffer (0.3% Triton X-100 in PBS) for 30 minutes at room temperature, with primary antibody at 4°C overnight, and with secondary antibody for 2 hours at room temperature, all in blocking buffer (5% normal donkey serum in 1X PBS). Antibodies used for immunostaining are rabbit anti-UCP1 (1:500, ab23841, Abcam), mouse anti-RFP (1:200, 632392, Takara), goat anti-Perilipin (1:500, ab61682, Abcam), rabbit anti-Ki67 (1:200, ab15580, Abcam), rat anti-Ki67 antibody (eBioscience, 14–5698-80), mouse anti-phospho-Histone H3 (Ser10) (Mitosis Marker, 1:200, Cell Signaling Technology), mouse anti-BrdU (1:200, G3G4, DSHB), rabbit anti-Caveolin-1 antibody (1:200, ab2910), mouse anti-SMA antibody (1:500, ab7817, Abcam). Secondary antibodies including cy3 donkey anti-mouse, Alexa donkey anti-rabbit, cy5 donkey anti-rat, and cy5 donkey anti-goat were from Jackson ImmunoResearch. All secondary antibodies were used at a 1:500 dilution. Immunostaining images were taken using a Leica DMi8 microscope (Leica, Wetzlar, Germany). To stain lipid, cryostat sections were formaldehyde-fixed and incubated in LipidTox Green (Invitrogen, H34475) at 1:200 in PBS for 30 min before washing in PBS and mounting for imaging. Whole-mount images were taken on a Leica M205 FA microscope (Leica, Wetzlar, Germany).
Oil Red O Staining
In vitro differentiated cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature. After washing with 1X PBS twice, cells were rinsed with 60% isopropanol (Sigma-Aldrich, St. Louis, MO). Cells stained with 60% filtered Oil Red O working solution (vol/vol in distilled water) of Oil red O stock solution (Sigma-Aldrich, St. Louis, MO) at room temperature for 15 minutes. After Oil Red O solution was removed, cells were washed with distilled water before imaging.
Flow Cytometry
SV cells were isolated, washed, centrifuged at 1000 Χ g for 10 minutes, and sorted with a MoFlo Astrios Cell Sorter (Beckman Coulter, Brea, CA). BrdU detection of SV cells was performed with the APC BrdU Flow Cytometry Kit, according to the manufacturer’s instructions. For gating strategies of both RFP sorting and flow analysis, live cells were selected by size on the basis of FSC and SSC. Single cells were then gated on both SSC and FSC Width singlet’s. SVF cells isolated from RFP-negative mice, along with primary-minus-one controls, were used as a negative control to determine background fluorescence levels.
Quantitative real-time PCR (qPCR)
Total RNA was extracted using Tripure Isolation Reagent (Roche, Basel, Switzerland) from adipose tissues using Bullet Blender Homogenizer (Next Advance, Troy, NY, USA) according to the manufacturer’s protocol. cDNA was generated from 1 μg of total RNA using iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA), and qPCR was performed using ViiA7 system (Applied Biosystems, Forster City, CA, USA) according to the manufacturer’s protocol. Briefly, appropriate amount of the reverse transcription reaction mixture was amplified with specific primers using the iTaq universal SYBR Green Supermix (Bio-Rad Laboratories) in total volume of 10 μL. Data were analyzed using the comparative Ct method and the qPCR value were normalized by 18s rRNA expression. mRNA levels were expressed as the fold-increase relative to basal transcription levels. Primer sequences are available in Table S1.
Quantification and Statistical analysis
All labeling quantifications were performed in at least 3 animals, with a minimal of 3 distinct sections being imaged and counted per animal. Two-tailed unpaired Student’s t-test was conducted using Graphpad Prism software (Graphpad, La Jolla, CA, USA). Data was presented as mean ± standard errors of the means (Chen et al.). Studies were performed on two or three independent cohorts and were performed 3 to 5 mice per group unless specified. Asterisks indicate statistically significant difference between groups. *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-UCP1 antibody | Abcam | Cat# ab23841 |
| mouse anti-RFP | Takara | Cat# 632392 |
| rabbit anti-TH (tyrosine hydroxylase) | Abcam | Cat# ab6211 |
| goat anti-Perilipin | Abcam | Cat#: ab61682 |
| rabbit anti-Ki67 | Abcam | Cat#: ab15580 |
| rat anti-Ki67 | eBioscience | Cat#:14-5698-80 |
| mouse anti-SMA | Abcam | Cat#: ab7817 |
| mouse anti-phospho-Histone H3 (Ser10) | Cell Signaling Technology | Cat#: #9701 |
| mouse anti-BrdU | DSHB | Cat#: G3G4 |
| rabbit anti-Caveolin-1 antibody | Abcam | Cat#: ab2910 |
| cy3 donkey anti-mouse | Jackson ImmunoResearch | Cat#: 715-165-150 |
| Alexa donkey anti-rabbit | Jackson ImmunoResearch | Cat#: 711-545-152 |
| cy5 donkey anti-rat | Jackson ImmunoResearch | Cat#: 712-175-153 |
| cy5 donkey anti-goat | Jackson ImmunoResearch | Cat#: 705-175-147 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5-Bromo-2’-deoxyuridine | Sigma-Aldrich | Cat#: B5002 |
| Collagenase, Type 1 | Worthington Biochemical | Cat#: LS004197 |
| Dexamethasone | Sigma-Aldrich | Cat#: D4902 |
| Forskolin | Sigma-Aldrich | Cat#: F6886 |
| Fetal Bovine Serum | Corning | Cat#: 35-015-CV |
| Glucose | Sigma-Aldrich | Cat#: G7021-100G |
| Isobutylmethylxanthine (IBMX) | Sigma-Aldrich | Cat#: I5879 |
| triiodo-L-thyronine (T3) | Sigma-Aldrich | Cat#: T2877 |
| Insulin | Sigma-Aldrich | Cat#: I6634 |
| CL316243 disodium salt | Tocris | Cat#: 151126-84-0 |
| 4-hydroxy-Tamoxifen | Sigma-Aldrich | Cat#: H7904 |
| DAPI | Sigma-Aldrich | Cat#: D9564 |
| Doxycycline (hyclate) | Cayman Chemical | Cat#: 14422 |
| Tamoxifen | Sigma-Aldrich | Cat#: T5648 |
| Rosiglitazone | Sigma-Aldrich | Cat#: R-2408 |
| Reagent or resource | ||
| Vectastain ABC KIT | Vector Laboratories | Cat#: PK-6100 |
| DAB KIT | Vector Laboratories | Cat#: SK-4100 |
| FITC BrdU Flow kit | BD PharMingen | Cat#: 557891 |
| iScript cDNA Synthesis Kit | BIORAD | Cat#: 1708890 |
| oil red O | Sigma-Aldrich | Cat#: O1391-500ML |
| hydrogen peroxide solution | Sigma-Aldrich | Cat#: H1009 |
| Nefa Linearity Set Solution | Wako Chemicals | Cat#: 991-34891 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| Cholesterol LiquiColor® | Stanbio Laboratory | Cat#: 1010-430 |
| Triglycerides - Liquid Reagent Set | Thermo Fisher Scientific | Cat#: 23-666-410 |
| 0.05% Trypsin 0.53mM EDTA | Corning | Cat#: MT25052CI |
| LipidTox Green | Invitrogen | Cat#: H34475 |
| Critical Commercial Assays | ||
| Vectastain ABC KIT | Vector Laboratories | Cat#: PK-6100 |
| DAB KIT | Vector Laboratories | Cat#: SK-4100 |
| APC BrdU Flow kit | BD Biosciences | Cat#: 552598 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#: 554714 |
| iScript cDNA Synthesis Kit | BIORAD | Cat#: 1708891 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat#: 4368813 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Ucp1-Cre | Jackson laboratory | 24670 |
| Mouse: Rosa26R-M2rtTA | Jackson laboratory | 6965 |
| Mouse: Rosa26RDTA | Jackson laboratory | 006331 |
| Mouse: Pparγfl/fl | Jackson laboratory | 004584 |
| Mouse: Ucp1-CreERT2 | Rosenwald et al., 2013 | N/A |
| Mouse: Cdkn2afl/fl | Berry et al., 2017 | N/A |
| Mouse: TRE-p21 | Berry et al., 2017 | N/A |
| Mouse: Rosa26RmT/mG | Jackson laboratory | 007676 |
| Mouse: Rosa26RRFP | Jackson laboratory | 007914 |
| Oligonucleotides | ||
| A full list of qRT-PCR primers, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Word | Microsoft | N/A |
| Excel | Microsoft | N/A |
| Fiji (ImageJ) | Fiji | https://fiji.sc/ |
| Flowjo software | Flowjo | https://www.flowjo.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism | GraphPad Software | Graphpad Software |
| Other | ||
| FACS Aria Fusion | BDBioscience | N/A |
| Zeiss LSM8800 Confocal Microscope | Zeiss | N/A |
| MoFlo Astrios Cell Sorter | Beckman Coulter | N/A |
| Leica DMi8 microscope | Leica | N/A |
| Leica M205 FA microscope | Leica | N/A |
Highlights.
A subset of UCP1+ cells present in white adipose tissue generate new beige fat cells
UCP1+ cell ablation or functional disruption impairs beige adipocyte formation
Blocking division of UCP1+ cells inhibits beige adipocyte formation
Promoting division of UCP1+ cells enhances beige adipocyte formation
Acknowledgements
We thank Dr. Jonathan M Graff for supporting this work and giving fruitful advice. We thank Drs. Cynthia Rose Adams and Jeanette Purcell for assistance with mouse husbandry, Dr. Stefan J. Green and the Research Resources Center for real-time qPCR analysis, Dr. Brian Layden and Metabolic Phenotyping Core for analytical and phenotypical mouse measurements, Dr. Balaji Ganesh, Dr. Kiwook Kim, and Flow Cytometry Core facility for FACS and flow analysis, and members of the Jiang laboratory for helpful comments on the manuscript. We thank the reviewers for their insights and suggestions. We thank Professor Christian Wolfrum for useful discussion. This work was supported by the National Institute of Diabetes and Digestive and Kidney Disease grant K01 DK111771 and Pilot & Feasibility Diabetes Research & Training Center (DRTC) Award (P30DK020595) to Y.J.
Footnotes
Declaration of Interests
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acharya A, Berry DC, Zhang H, Jiang Y, Jones BT, Hammer RE, Graff JM, and Mendell JT (2019). miR-26 suppresses adipocyte progenitor differentiation and fat production by targeting Fbxl19. Genes Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, and Kajimura S (2016). Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metab 24, 402–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annicotte JS, Blanchet E, Chavey C, Iankova I, Costes S, Assou S, Teyssier J, Dalle S, Sardet C, and Fajas L (2009). The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat Cell Biol 11, 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Jiang Y, Arpke RW, Close EL, Uchida A, Reading D, Berglund ED, Kyba M, and Graff JM (2017). Cellular Aging Contributes to Failure of Cold-Induced Beige Adipocyte Formation in Old Mice and Humans. Cell Metab 25, 166–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Jiang Y, and Graff JM (2016). Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nat Commun 7, 10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee YH, and Granneman JG (2018). Deconstructing Adipogenesis Induced by beta3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell Metab 28, 300–309 e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Jing J, Cui X, Shi H, and Xue B (2019). Sympathetic nerve innervation is required for beigeing in white fat. Physiol Rep 7, e14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Goh KJ, Rocha N, Groeneveld MP, Minic M, Barrett TG, Savage D, and Semple RK (2017). Evaluation of human dermal fibroblasts directly reprogrammed to adipocyte-like cells as a metabolic disease model. Dis Model Mech. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, and Scadden DT (2000). Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808. [DOI] [PubMed] [Google Scholar]
- Chondronikola M, Volpi E, Borsheim E, Porter C, Annamalai P, Enerback S, Lidell ME, Saraf MK, Labbe SM, Hurren NM, et al. (2014). Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 63, 4089–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, and Spiegelman BM (2015). Brown and Beige Fat: Molecular Parts of a Thermogenic Machine. Diabetes 64, 2346–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo N, and Oliver G (2017). The Lymphatic Vasculature: Its Role in Adipose Metabolism and Obesity. Cell Metab 26, 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR (2006). Transcriptional control of adipocyte formation. Cell Metab 4, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Memetimin H, Confides AL, Kasza I, Zhu B, Vekaria HJ, Harfmann B, Jones KA, Johnson ZR, Westgate PM, et al. (2018). Human adipose beiging in response to cold and mirabegron. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Memetimin H, Zhu B, Confides AL, Vekaria HJ, El Khouli RH, Johnson ZR, Westgate PM, Chen J, Morris AJ, et al. (2020). The beta3-adrenergic receptor agonist mirabegron improves glucose homeostasis in obese humans. J Clin Invest 130, 2319–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM (2009). Obesity: Causes and control of excess body fat. Nature 459, 340–342. [DOI] [PubMed] [Google Scholar]
- Fukano K, Okamatsu-Ogura Y, Tsubota A, Nio-Kobayashi J, and Kimura K (2016). Cold Exposure Induces Proliferation of Mature Brown Adipocyte in a ss3-Adrenergic Receptor-Mediated Pathway. PLoS One 11, e0166579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, and Kahn CR (2007). Developmental origin of fat: tracking obesity to its source. Cell 131, 242–256. [DOI] [PubMed] [Google Scholar]
- Giordano A, Smorlesi A, Frontini A, Barbatelli G, and Cinti S (2014). White, brown and pink adipocytes: the extraordinary plasticity of the adipose organ. Eur J Endocrinol 170, R159–171. [DOI] [PubMed] [Google Scholar]
- Greene AK, and Maclellan RA (2013). Obesity-induced Upper Extremity Lymphedema. Plast Reconstr Surg Glob Open 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannou SA, Wouters K, Paumelle R, and Staels B (2015). Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab 26, 176–184. [DOI] [PubMed] [Google Scholar]
- Harvey NL, Srinivasan RS, Dillard ME, Johnson NC, Witte MH, Boyd K, Sleeman MW, and Oliver G (2005). Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat Genet 37, 1072–1081. [DOI] [PubMed] [Google Scholar]
- Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, et al. (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespe GE, Kataru RP, Savetsky IL, Garcia Nores GD, Torrisi JS, Nitti MD, Gardenier JC, Zhou J, Yu JZ, Jones LW, et al. (2016). Exercise training improves obesity-related lymphatic dysfunction. J Physiol 594, 4267–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Maretich P, and Kajimura S (2018). The Common and Distinct Features of Brown and Beige Adipocytes. Trends Endocrinol Metab 29, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Berry DC, and Graff J (2017). Distinct cellular and molecular mechanisms for beta3 adrenergic receptor induced beige adipocyte formation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jo AY, and Graff JM (2012). SnapShot: adipocyte life cycle. Cell 150, 234–234 e232. [DOI] [PubMed] [Google Scholar]
- Khanh VC, Zulkifli AF, Tokunaga C, Yamashita T, Hiramatsu Y, and Ohneda O (2018). Aging impairs beige adipocyte differentiation of mesenchymal stem cells via the reduced expression of Sirtuin 1. Biochem Biophys Res Commun 500, 682–690. [DOI] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Konkar AA, and Granneman JG (2015). Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J 29, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrke M, and Lazar MA (2005). The many faces of PPARgamma. Cell 123, 993–999. [DOI] [PubMed] [Google Scholar]
- Liu X, Xiang Q, Xu F, Huang J, Yu N, Zhang Q, Long X, and Zhou Z (2019). Single-cell RNA-seq of cultured human adipose-derived mesenchymal stem cells. Sci Data 6, 190031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. (2014). A Smooth Muscle-Like Origin for Beige Adipocytes. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Altshuler-Keylin S, Wang Q, Chen Y, Henrique Sponton C, Ikeda K, Maretich P, Yoneshiro T, and Kajimura S (2018). Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald ME, Li C, Bian H, Smith BD, Layne MD, and Farmer SR (2015). Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell 160, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SY, Desai A, Yang Z, Sharma A, DeSouza T, Genga RMJ, Kucukural A, Lifshitz LM, Nielsen S, Scheele C, et al. (2019). Diverse repertoire of human adipocyte subtypes develops from transcriptionally distinct mesenchymal progenitor cells. Proc Natl Acad Sci U S A 116, 17970–17979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SY, Kady J, Nam M, Rojas-Rodriguez R, Berkenwald A, Kim JH, Noh HL, Kim JK, Cooper MP, Fitzgibbons T, et al. (2016). Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nat Med 22, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. (2012). Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguri Y, Shinoda K, Kim H, Alba DL, Bolus WR, Wang Q, Brown Z, Pradhan RN, Tajima K, Yoneshiro T, et al. (2020). CD81 Controls Beige Fat Progenitor Cell Growth and Energy Balance via FAK Signaling. Cell 182, 563–577 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamatsu-Ogura Y, Fukano K, Tsubota A, Nio-Kobayashi J, Nakamura K, Morimatsu M, Sakaue H, Saito M, and Kimura K (2017). Cell-cycle arrest in mature adipocytes impairs BAT development but not WAT browning, and reduces adaptive thermogenesis in mice. Sci Rep 7, 6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Potjer TP, Thomsen SK, Ng HJ, Barrett A, Scharfmann R, James TJ, Bishop DT, Karpe F, Godsland IF, et al. (2016). Loss-of-Function Mutations in the Cell-Cycle Control Gene CDKN2A Impact on Glucose Homeostasis in Humans. Diabetes 65, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo E, and Wang B (2019). Towards a Better Understanding of Beige Adipocyte Plasticity. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raajendiran A, Ooi G, Bayliss J, O’Brien PE, Schittenhelm RB, Clark AK, Taylor RA, Rodeheffer MS, Burton PR, and Watt MJ (2019). Identification of Metabolically Distinct Adipocyte Progenitor Cells in Human Adipose Tissues. Cell Rep 27, 1528–1540 e1527. [DOI] [PubMed] [Google Scholar]
- Rabhi N, Hannou SA, Gromada X, Salas E, Yao X, Oger F, Carney C, Lopez-Mejia IC, Durand E, Rabearivelo I, et al. (2018). Cdkn2a deficiency promotes adipose tissue browning. Mol Metab 8, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh HC, Tsai LTY, Shao M, Tenen D, Shen Y, Kumari M, Lyubetskaya A, Jacobs C, Dawes B, Gupta RK, et al. (2018). Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab 27, 1121–1137 e1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rulicke T, and Wolfrum C (2013). Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol 15, 659–667. [DOI] [PubMed] [Google Scholar]
- Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, et al. (2018). A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103–108. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi F, Xue R, Huang TL, Lundh M, Liu Y, Leiria LO, Lynes MD, Kempf E, Wang CH, Sugimoto S, et al. (2020). FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nat Commun 11, 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Wang QA, Song A, Vishvanath L, Busbuso NC, Scherer PE, and Gupta RK (2019). Cellular Origins of Beige Fat Cells Revisited. Diabetes 68, 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, El-Sabbagh AS, Lukas BE, Tanneberger SJ, and Jiang Y (2020a). Adipose stem cells in obesity: challenges and opportunities. Biosci Rep 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Pang Y, Park J, Liu L, Lukas BE, Kim SH, Kim KW, Xu P, Berry DC, and Jiang Y (2020b). Dynamic control of adipose tissue development and adult tissue homeostasis by platelet-derived growth factor receptor alpha. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin W, Okamatsu-Ogura Y, Machida K, Tsubota A, Nio-Kobayashi J, and Kimura K (2017). Impaired adrenergic agonist-dependent beige adipocyte induction in aged mice. Obesity (Silver Spring) 25, 417–423. [DOI] [PubMed] [Google Scholar]
- Soriano P (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, and Flier JS (2001). Obesity and the regulation of energy balance. Cell 104, 531–543. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. (2007). Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A 104, 4401–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lans AA, Hoeks J, Brans B, Vijgen GH, Visser MG, Vosselman MJ, Hansen J, Jorgensen JA, Wu J, Mottaghy FM, et al. (2013). Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J Clin Invest 123, 3395–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvanath L, MacPherson KA, Hepler C, Wang QA, Shao M, Spurgin SB, Wang MY, Kusminski CM, Morley TS, and Gupta RK (2016). Pdgfrbeta+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab 23, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Song A, Chen W, Schwalie PC, Zhang F, Vishvanath L, Jiang L, Ye R, Shao M, Tao C, et al. (2018). Reversible De-differentiation of Mature White Adipocytes into Preadipocyte-like Precursors during Lactation. Cell Metab 28, 282–288 e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, and Seale P (2014). Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A 111, 14466–14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock A, Brown EJ, Garabedian ML, Pena S, Sharma M, Lafaille J, Moore KJ, and Fisher EA (2019). Single-Cell RNA Sequencing of Visceral Adipose Tissue Leukocytes Reveals that Caloric Restriction Following Obesity Promotes the Accumulation of a Distinct Macrophage Population with Features of Phagocytic Cells. Immunometabolism 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir G, Ramage LE, Akyol M, Rhodes JK, Kyle CJ, Fletcher AM, Craven TH, Wakelin SJ, Drake AJ, Gregoriades ML, et al. (2018). Substantial Metabolic Activity of Human Brown Adipose Tissue during Warm Conditions and Cold-Induced Lipolysis of Local Triglycerides. Cell Metab 27, 1348–1355 e1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2013). Obesity and overweight. World Health Organization. [Google Scholar]
- Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, and Schaart G (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, and Spiegelman BM (2013). Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev 27, 234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, and Saito M (2013). Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest 123, 3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shao M, Hepler C, Zi Z, Zhao S, An YA, Zhu Y, Ghaben AL, Wang MY, Li N, et al. (2019). Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice. J Clin Invest 129, 5327–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
N/A