Graphical abstract
Keywords: Recombination map, Chromosome-level genome, Fruiting body development, Candidate genes, Shiitake
Highlights
-
•
We constructed a reference genetic map of Lentinula edodes.
-
•
We re-assembled a chromosome-level genome of L. edodes.
-
•
We disclosed three hotspots regions for fruiting body-related traits in shiitake.
-
•
We scanned candidate genes for fruiting body-related traits.
Abstract
Fruiting body development (FBD) of mushroom-forming fungi has attracted tremendous interest. However, the genetic and molecular basis of FBD is poorly known. Here, using Lentinula edodes (shiitake) as a model, we deciphered the genetic architecture underlying fruiting body-related traits (FBRTs) by combined genomic, genetic and phenotypic data. Using RNA-Seq of fruiting bodies from 110 dikaryons in a bi-parental mapping population, we constructed an ultra-high-density genetic map of L. edodes (Lemap2.0) with a total length of 810.14 cM, which covered 81.7% of the shiitake genome. A total of 94 scaffolds of the shiitake genome were aligned to Lemap2.0 and re-anchored into nine pseudo-chromosomes. Then via quantitative trait locus (QTL) analysis, we disclosed an outline of the genetic architecture of FBD in shiitake. Twenty-nine QTLs and three main genomic regions associated with FBD of shiitake were identified. Using meta-QTL analysis, seven pleiotropic QTLs for multiple traits were detected, which contributed to the correlations of FBRTs. In the mapped QTLs, the expression of 246 genes were found to significantly correlate with the phenotypic traits. Thirty-three of them were involved in FBD and could represent candidate genes controlling the shape and size of fruiting bodies. Collectively, our findings have advanced our understanding of the genetic regulation of FBD in shiitake and mushroom-forming fungi at large.
1. Introduction
Mushroom-forming fungi (Agaricomycetes) are of great importance in ecology, agriculture, industry and medicine. They represent an important and sustainable food source, with favorable medicinal properties [1]. The global production of mushroom-forming fungi has increased rapidly over the last decades [2]. Mushroom-forming fungi have a high morphological diversity, arousing curiosity to understand the molecular mechanisms underlying fruiting body initiation and development [3], [4]. In addition, the morphogenesis and development of fruiting body represent a key innovation in the evolution of mushroom-forming fungi [4].
Lentinula edodes, also known as the shiitake mushroom, is one of the most widely cultivated edible mushrooms, contributing to about 22% of the world’s total production [5]. L. edodes has attracted tremendous interest since it could convert a wide variety of lignocellulosic wastes into high-protein foods with pharmacological effects [6]. L. edodes has a similar life cycle to other basidiomycetes, and produces the typical pileate-stipitate fruiting bodies (sporophores). Because of its ease of growth and fruiting under laboratory conditions, L. edodes has also been used as a model basidiomycete in studies of mushroom genetics and physiology [7].
Triggered by a changing environment (e.g. light, temperature and nutrient factors, etc.), the induction and development of fruiting bodies of mushroom-forming fungi involve a transition from simple multicellular hyphae to a complex multicellular fruiting body initial, a process which is controlled by complicated networks of transcriptional and translational regulation [1], [3], [8], [9]. A number of structural and regulatory genes, including those coding for hydrophobins, blue light receptors, cyclopropane fatty acyl phospholipid synthase, cell wall-modifying enzymes, and transcriptional regulators, have been characterized to be involved in the fruiting of model mushroom-forming basidiomycetes such as Coprinopsis cinerea and Schizophyllum commune [3], [8], [10]. In L. edodes, great efforts were also made to elucidate its fruiting processes, and a few differentially expressed genes at various developmental stages have been characterized [11], including priA [12], lac1 and lac2 [13], Le.hyd1 [14], and Le.flp1 [15]. Gene expression in the fruiting bodies of shiitake was also profiled by using a whole-genome approach [7]. Despite advances in this field, most studies to date have focused on the gene regulation of fruiting body initiation. The development of fruiting body, such as gene regulation of the elongation of pileus and stipe, is poorly known. The shape and size of fruiting bodies are genetically determined, and understanding the molecular basis of fruiting body development (FBD) is important both in biological studies and in commercial production [3].
The pileus and stipe are the two major parts of the basidiocarp in most umbrella-shaped mushrooms, including L. edodes. Traits related to pileus and stipe could reflect the shape of fruiting bodies from different aspects, and are thus known as fruiting body-related traits (FBRTs). FBRTs of shiitake are quantitative traits controlled by multiple genes or quantitative trait loci (QTLs), and the genetic architectures of which are complicated and largely unknown [16]. QTL mapping is a powerful tool for detecting the genetic architectures of quantitative traits, and could greatly facilitate marker-assisted selection and accelerate the breeding progress [17], [18]. In mushroom-forming basidiomycetes, QTL mapping of phenotypic traits was scarce and limited to several cultivated edible mushrooms, such as Pleurotus ostreatus [19], Agaricus bisporus [20], [21], and L. edodes [16]. These QTL detections were mainly of low-resolution and rarely involved in FBRTs. There is an increasing need to decipher the regulatory network of FBRTs with a higher resolution in mushroom-forming basidiomycetes. For accurate identification and characterization of QTLs, high-density genetic maps are required in linkage mapping [22]. RNA sequencing (RNA-Seq) that makes use of next-generation sequencing, is the powerful tool for discovering large-scale single-nucleotide polymorphisms (SNPs), and for generation of dense linkage maps for scaffolding of genomes. This method has been widely applied in plants, and enabled a major technological leap from low-resolution to high-resolution QTL mapping [17], [23], [24].
Fruiting body morphogenesis of mushroom-forming basidiomycetes is a complicated biological process. Systems genetic studies have provided a global view of the molecular architecture of complex phenotypes by combining genomic and genetic data [25]. QTL mapping could be useful to elucidate the genetic program of complex phenotypes such as FBRTs. Despite the fact that an increasing number of mushroom-forming basidiomycetes have been sequenced or re-sequenced [26], the application of high-throughput sequencing in genetic studies of mushrooms is still rare. Herein, the genetic repertoire of the shape of fruiting bodies was deciphered using L. edodes as a model. An ultra-high-density genetic linkage map of L. edodes was constructed via high-throughput RNA-Seq analysis of 110 dikaryons. QTL analysis was then performed using phenotypic data and the linkage map. FBD-related genes were disclosed to be candidate genes of the phenotypic QTLs. Findings of this study have not only provided insights into the genetic and molecular basis of FBRTs in shiitake, but also advanced our understanding of FBD in mushroom-forming fungi at large.
2. Materials and methods
2.1. L. edodes strains and population
All the tested L. edodes strains and mapping population were generated and used previously [27]. Briefly, a total of 146 F1 single-spore isolates (SSIs) were randomly sampled from spores derived from crossing two parental monokaryon strains L205-6 and W1-26. Then, all the 146 SSIs were paired with the monokaryon strain 741–15 to produce the population LQ-15 [27]. All the tested strains were preserved in the Institute of Applied Mycology, Huazhong Agricultural University.
2.2. Phenotype evaluation
Followed as the cultivation procedures described by Gong et al [28], fruiting trials of all the 146 dikaryotic strains in LQ-15 were carried out at Huazhong Agricultural University (Wuhan, China, 114.35°E, 30.48°N). In this study, seven FBRTs (pileus diameter, PD; pileus thickness, PT; pileus weight, PW; stipe length, SL; stipe diameter, SD; stipe weight, SW; and the weight of a single fruiting body, WF), as well as NF (number of fruiting bodies of a single bag), Y (yield of a single bag), and FP (fruiting bodies precocity) were evaluated in cultivation trials both in 2012 and 2016. All strains were allocated in a mushroom house in accordance with the randomized-block design with two blocks, and six culture bags of each strain were included in each replication. During the cultivation trial, only 110 strains could produce >20 fruiting bodies in each block. Thereby, only these 110 strains in LQ-15 were used for phenotypic investigation and subsequently transcriptome sequencing. All the fruiting bodies were harvested while the pileus was still slightly curled in or just as the partial veil breaks away. For each strain, each block, phenotypic evaluation of seven FBRTs were obtained from 20 fruiting bodies from August 2016 to May 2017. From different harvest batches, different bags, these 20 non-malformed fruiting bodies were randomly selected for traits assessment. A brief description of all the ten surveyed traits was summarized in Table 1. Based on the same strains, an additional phenotypic dataset of the ten traits surveyed from September 2012 to May 2013 [28] were also utilized in this study. For simplicity, the phenotypic data of agronomic traits for 2016–2017 were annotated with the suffix “2016” and those for 2012–2013 were annotated with the suffix “2012”.
Table 1.
Descriptive statistics of the ten investigated phenotypic traits.
| Trait | Description | Year | Max value | Min value | Mean ± standard error | Standard deviation | CV a | rb |
|---|---|---|---|---|---|---|---|---|
| PD (mm) | Average diameter of pileus, determined as | 2016 | 55.03 | 25.22 | 36.689 ± 0.48 | 5.03 | 13.84% | 0.404** |
| the mean of two perpendicular diameters | 2012 | 75.95 | 35.88 | 57.368 ± 0.81 | 8.11 | 14.14% | ||
| PT (mm) | Average thickness of pileus | 2016 | 14.44 | 4.74 | 8.032 ± 0.16 | 1.6 | 20.33% | 0.249* |
| 2012 | 24.36 | 11.39 | 17.222 ± 0.25 | 2.49 | 14.43% | |||
| PW (g) | Average weight of pileus | 2016 | 18.16 | 2.37 | 6.917 ± 0.25 | 5.7 | 38.63% | 0.215* |
| 2012 | 35.29 | 5.89 | 19.892 ± 0.63 | 6.34 | 31.89% | |||
| SL (mm) | Average length of stipe | 2016 | 58.4 | 21.82 | 32.806 ± 0.59 | 5.85 | 18.87% | 0.389** |
| 2012 | 54.93 | 18.63 | 34.414 ± 0.68 | 6.84 | 19.87% | |||
| SD (mm) | Average diameter of stipe | 2016 | 17.11 | 6.78 | 10.650 ± 0.15 | 1.67 | 14.91% | 0.373** |
| 2012 | 22.46 | 9.83 | 15.135 ± 0.24 | 2.4 | 15.88% | |||
| SW (mm) | Average weight of stipe | 2016 | 9.52 | 0.86 | 2.564 ± 0.12 | 1.23 | 47.55% | 0.328** |
| 2012 | 13.51 | 1.13 | 4.240 ± 0.21 | 2.07 | 48.82% | |||
| WF (g) | Average weight of the single fruiting body | 2016 | 23.43 | 1.76 | 7.191 ± 0.30 | 3.06 | 43.26% | 0.324** |
| 2012 | 41.67 | 5.41 | 18.795 ± 0.75 | 7.51 | 39.97% | |||
| NF | Average number of total fruiting bodies | 2016 | 73.75 | 3 | 28.133 ± 1.23 | 12.61 | 45.69% | 0.487** |
| (per/bag) | per bag during the whole harvest time | 2012 | 27.83 | 1 | 8.289 ± 0.57 | 5.73 | 69.19% | |
| Y (g/bag) | Total yield per bag | 2016 | 248.28 | 55.42 | 171.205 ± 3.75 | 37.71 | 23.00% | 0.386** |
| during the whole harvest time | 2012 | 244.21 | 12.93 | 127.159 ± 5.98 | 59.83 | 47.05% | ||
| FP (d) | Time interval (in days) from incubation to | 2016 | 233 | 106 | 121.318 ± 1.62 | 15.07 | 13.97% | 0.411** |
| harvest of the first fruiting body | 2012 | 211 | 95 | 132.730 ± 3.00 | 29.96 | 22.57% |
Note: PD: pileus diameter; PT: pileus thickness; PW: pileus weight; SL: stipe length; SD: stipe diameter; SW: stipe weight; WF: weight of a single fruiting body; NF: number of fruiting bodies of single bag; Y: yield of single bag; FP: fruiting bodies precocity.
Coefficient of variation, calculated by dividing the standard deviation by the mean of each trait.
Correlation coefficient of a particular trait between year 2016 and 2012. *p < 0.05; **p < 0.01
SPSS version 22 (SPSS Inc., Chicago, IL, USA) was utilized to perform descriptive statistical analysis, and analysis of variance (ANOVA). For each trait, the phenotypic data were subjected to normality test (Kolmogorov-Smirnov test). Broad-sense heritability (H2) was assessed with the formula: H2 = σG2/[σG2 + σG×E2/nr + σe2/n], where σG2 is the genotypic variance, σe2 is the error variance, σG×E2 is the variance of genotype and year interaction, n is the number of replicates in the experiment, and r is the number of years.
2.3. High-throughput sequencing
Using Illumina HiSeq 2000 platform, the genomes of two monokaryotic strains L205-6 and W1-26 had been re-sequenced previously [16]. In the cultivation trial in 2016, only 110 strains producing >20 fruiting bodies were selected for transcriptome sequencing. Three mature fruiting bodies from each block of each strain were collected separately, longitudinal cut into 1–2 mm thick slices and freezed immediately with liquid nitrogen. For each strain, the collected slices from six fruiting bodies from two blocks were then mixed and ground. Total RNA of each strain was extracted using the Invitrogen™ Trizol Plus RNA Purification kit according to its protocol. mRNA was then enriched by combining Oligo (dT) magnetic beads with polyA tail of mRNA. cDNA was synthesized using NEBNext Ultra II RNA Library Prep Kit for Illumina. Chain-specific transcriptome sequencing was carried out on an Illumina X-Ten sequencing platform. No less than 4 Gb clean data were obtained for each strain. RNA extraction, library construction and sequencing were performed by Wuhan Genoseq Technology Co., Ltd. All DNA and RNA sequencing data were deposited to GenBank Sequence Read Archive (SRA) under the accession PRJNA649389.
2.4. Genotype identification
The raw paired-end reads of 110 dikaryotic strains were trimmed to remove the adaptors using cutadapt (version 1.13, -e 0.1 -O 5 -m 100) [29]. Trimmomatic (version 0.33, SLIDINGWINDOW:4:15 MINLEN:50) was used to remove low-quality bases to obtain high-quality clean data [30]. The trimmed clean reads of L205-6 and W1-26 were aligned to the 0899ss11 genome v1.0 of L. edodes (hereafter as 0899ss11 genome) (https://genome.jgi.doe.gov/Lenedo1/Lenedo1.home.html) via BWA-MEM (version 0.7.15-r1140, -c 200 -M) [31]. Filtered clean reads of the 110 dikaryotic strains were aligned to the 0899ss11 genome by using HISAT2 (version 2.1.0, --score-min L,0,-0.2 --pen-noncansplice 1000000) [32]. SAM files were converted to BAM files by SAMtools (version 1.3.1, with default parameters) [33], and the alignment results were sorted using Picard (version 1.91, with default parameters). SAMtools rmdup was then utilized to remove PCR duplicates. For variants calling, using GATK (version 3.7), SNPs and insertion-deletions (InDels) were excavated for the 110 dikaryotic strains and the parental strains by HaplotypeCaller with default parameters (SNP: QUAL < 40, QD < 2.0, MQ < 40.0, FS > 60.0, SOR > 3.0, MQRankSum < − 12.5, ReadPosRankSum < − 8.0, and InDel: QUAL < 40, QD < 2.0, MQ < 40.0, FS > 200.0, SOR > 3.0, MQRankSum < − 12.5, ReadPosRankSum < − 20.0) [34]. To get high-quality variants, SNPs and InDels were screened using the following criteria: missing data < 20%, minor allele frequency > 20%, and polymorphic between L205-6 and W1-26. Since the parental strains and the tester strain (741–15) were monokaryons, only bi-allelic SNPs and InDels were reserved in further analysis.
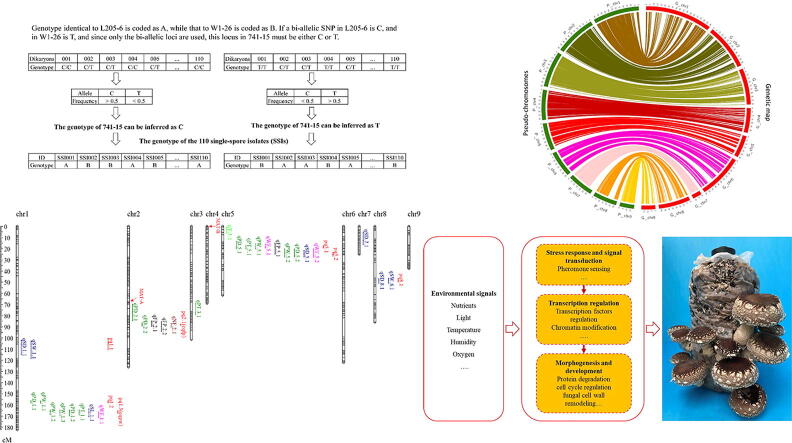
The deduction of genotypes of the SSIs were shown in Fig. 1. At each locus, the genotype identical to L205-6 is coded as A, while that to W1-26 is coded as B. Since only bi-allelic loci were reserved, if a bi-allelic SNP locus is C (cytosine) in L205-6 and T (thymine) in W1-26, this locus in 741–15 must be either C or T. If its genotype in 741–15 is C, only C/C or C/T would be detected in the dikaryotic strains in LQ-15, and vice versa. In this case, C/C in the dikaryotic strain means that the genotype of the corresponding SSI is the same as L205-6 with a C (coded as A), while C/T means that it is the same as W1-26 with a T (coded as B). In the case of the genotype of 741–15 is T, only C/T or T/T would be detected in LQ-15. The dikaryotic strain with C/T means that the genotype of the corresponding SSI is C (same as L205-6, coded as A), while T/T means that it is T (same as W1-26, coded as B). The same deduction process was used for the other types of bi-allelic variants, and the independent deduction of genotypes for each locus was performed. In our previous study, 86 genomic sequence-based InDel markers were used for electrophoresis-based genotyping and linkage mapping in shiitake [16]. To verify the deduction of SSIs genotypes, we compared these 86 InDel genotypes of the SSIs obtained by this deduction procedure with the results of electrophoresis-based genotyping [16].
Fig. 1.
Workflow of genotyping of 110 F1 SSIs. Genotypes of the 110 F1 SSIs were inferred from the genotypes of the 110 dikaryotic strains in LQ-15.
2.5. Construction of linkage map and anchored genomic scaffolds
After filtering, a total of 69,681 markers including 67,138 SNPs and 2,543 InDels were retained. To increase the mapping efficiency, we first merged the adjacent SNPs and InDels within 1 kb into consolidated markers, and only kept one marker for each 1 kb. Therefore, only 826 markers were reserved. To generate a set of non-redundant markers for genetic map construction, the exact co-segregated markers in this population were combined into a recombination bin, and resulted in 488 bin markers. Then, the LOD (logarithm of odds) score between bin markers was calculated using the formula:
where R = number of recombinants, NR = number of non-recombinants. The 488 bin markers were assigned into nine linkage groups (LGs) with LOD > 4. To obtain a preliminary genetic linkage map, MSTMap [35] was used to sort the markers within each LG and to calculate the genetic distance among the bin markers. Then, scaffolds of 0899ss11 genome were anchored to the linkage map by ALLMAPS [36] to construct pseudo-chromosomes corresponding to the nine LGs. The screened SNPs and InDels were sorted according to their physical location on the pseudo-chromosomes. To generate a high-quality genetic linkage map, missing genotypes were filled according to the procedures described by Xie et al [37]. Finally, MSTMap was used to calculate the genetic distances among markers within the pseudo-chromosomes, resulting in a final high-quality genetic linkage map (Lemap2.0). All co-segregated markers located on the same genetic position were combined into a recombination bin. Using the 86 InDel markers, Lemap2.0 and the previously constructed genetic map [15] was also compared. Syntenic relationship between the genetic map and the pseudo-chromosomes was graphically displayed using Circos [38].
2.6. QTL mapping and meta-QTL analysis
Based on Lemap2.0, genome-wide QTL scanning for phenotypic traits was performed by composite interval mapping in WinQTLcart 2.5 [39]. The parameters of QTL mapping were set to model 6 with a scanning step of 1 cM. For all traits, permutation-based LOD score thresholds were calculated (p < 0.05). The QTL confidence interval (CI) was defined as the interval between both sides of the position of LOD peak minus one. QTLs were named in the format of “qSD.1.1”, in which “SD” is the abbreviation of traits and “1.1” means the first SD-QTL on chr1. QTL hotspots were identified manually by searching in a sliding window of 20 cM, in which at least three adjacent or overlapping QTLs were included [40]. To narrow the CIs of QTLs, especially for those co-located QTLs, meta-QTL analysis was performed using BioMercator v2.1 [41]. According to the procedures described by Gong et al [16], using Lemap2.0 as the consensus map, the most probable number of pleiotropic QTLs and their position and the new CIs were determined by the model with the lowest Akaike information criterion score. Genes in the CIs of QTLs were then searched to screen for candidate genes.
2.7. Transcriptome analysis and candidate genes screening
BAM files of RNA-Seq in the 110 dikaryotic strains were used to calculate the expression level of each gene by StringTie [42]. Raw counts of each gene were normalized to FPKM (fragments per kilobase per million reads). Correlation coefficients (r values, ranged between −1 and 1) between FPKM values of all genes and phenotypic values were calculated using the pearsonr function in the SciPy package of python (https://www.scipy.org), and r values < 5% quantile or > 95% quantile indicate significant correlations (p < 0.05) [43]. A correlation coefficient < 0 indicates a negative correlation, whereas those > 0 indicate a positive correlation.
Candidate genes controlling targeted traits could be verified by the correlation between gene expression and phenotypic values [44]. Therefore, genes in the CIs of meta-QTLs, whose expressions were significantly correlated with phenotypic values of the trait (p < 0.05), were identified as putative candidate genes for FBD. Candidate genes were functionally annotated by using Blast2GO [45]. GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis were carried out using WEGO [46] and KAAS [47]. GO and KEGG enrichment were analyzed by clusterProfiler [48]. Candidate genes were considered reliable if they were identified in both phenotypic datasets of 2012 and 2016.
3. Results
3.1. Phenotypic variation of FBRTs in LQ-15
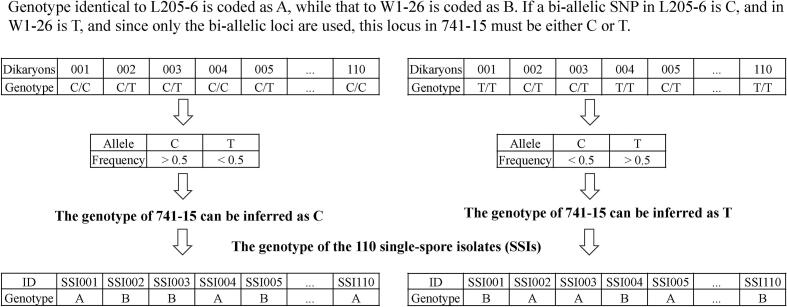
The seven FBRTs and the other three traits in LQ-15 examined in 2016 had a continuous variation (Fig. 2). The phenotypic variations of surveyed traits in 2012 were also shown in Fig. S1. Except WF, NF, Y, and FP, the other six FBRTs traits were distributed normally. The phenotypic performances of all the ten traits investigated in 2012 and 2016 were presented in Table 1. Among the seven FBRTs, three weight-related traits (SW, PW and WF) had a higher coefficient of variation than the other four shape-related traits (SD, SL, PD and PT) in both years. NF also had a relatively high coefficient of variation. Since the same strains in LQ-15 were cultivated in 2012 and 2016, the performances of the same traits were analyzed. For all the surveyed traits, significant correlations were found between the same traits in both years (Table 1). The average yield of dikaryotic strains in LQ-15 had little difference between 2012 and 2016. The dikaryotic strains were earlier-fruiting, and produced more but smaller fruiting bodies in 2016 than in 2012. The phenotypic differences of dikaryotic strains across years was likely caused by the environmental effect, such as temperature and humidity.
Fig. 2.
Frequency distribution of the surveyed traits in LQ-15 of L. edodes in cultivation trial of 2016. For each trait, coefficient of variation was shown as a percentage in the upper right corner of the histogram.
Correlation analysis and ANOVA results showed that genotype, year, and their interaction had significant impact on seven FBRTs (Table S1). The H2 values were able to measure the extent of phenotypic variation affected by genetic factors, which ranged from 0.11 for PT to 0.70 for SL in the seven FBRTs. The three pileus-related traits had lower H2 values as compared to those for stipe-related traits, indicating that environmental factors influenced greatly on these traits.
Correlations among different traits in the same year were also analyzed (Table S2). Significant positive correlations were identified among the seven FBRTs. In addition, significant negative correlations between NF and the seven FBRTs were observed in both years. Most of the FBRTs showed the positive correlations with FP, indicating that strains fruiting late may tend to produce larger fruiting bodies (Table S2).
3.2. Bin map construction and alignment of genomic scaffolds
The reference genome 0899ss11 of L. edodes had a size of 45.59 Mb, consisting of 128 scaffolds with a N50 of 690 kb and L50 of 22 scaffolds. In LQ-15, only 110 strains producing >20 fruiting bodies were selected for transcriptome sequencing. Clean data from RNA-Seq among these 110 dikaryons varied from 4.00 Gb to 7.59 Gb, with an average of 5.47 Gb. The mapping rate of the clean data of the 110 dikaryons to the 0899ss11 genome ranged from 74.6% to 86.7%, with a mean of 83.7%. The percentage of coding sequence covered by the RNA reads, ranged from 69.8% to 74.6%. For the two parental monokaryons L205-6 and W1-26, 74.1% and 65.6% of clean reads were aligned to the 0899ss11 genome, respectively (Table S3). A total of 174, 332 variants were identified between L205-6 and W1-26.
In this study, the 110 F1 SSIs were used for linkage map construction, and their genotypes were inferred from the genotypes of the 110 dikaryotic strains in LQ-15 (Fig. 1). As shown in Dataset 1 (Supplementary material), among 86 InDel markers, 86.5% of SSIs genotypes obtained from deduction procedure were consistent with the results of electrophoresis-based genotyping. After variants calling and filtering, 69,681 markers with allele frequencies of 0.3 to 0.8, were obtained (Fig. S2). After bin calling and correction, 488 bins were obtained for linkage analysis.
The total length of the constructed bin map (Lemap2.0) was 810.1 cM, with an average distance of 1.65 cM between adjacent bin markers (Table 2). The distribution of bins on the LGs was shown in Fig. 3. The size of LGs ranged from 24.8 cM (chr7) to 182.1 cM (chr1), with an average of 90.0 cM (Table 2). Previously, the mating types of all the SSIs were determined by mating tests [25]. Here, by genome annotation and homology search, the MAT-A and MAT-B genes were found to be located on scaffold_1 and scaffold_56, respectively, in accordance with the results of linkage mapping of mating-type loci A and B (Fig. 3). We also compared the genetic position of the 86 InDel markers in Lemap2.0 and the previously constructed shiitake genetic map [16]. A good syntenic relationship was found, and only four InDel markers were discrepant (data not shown).
Table 2.
Summary of Lemap2.0 of L. edodes.
| Chromosome | Physical length (Kb) | Map length (cM) | No. of scaffolds | Recombination rate (cM/Mb) | No. of markers | No. of bins | Marker density (per Mb) | Bin interval (cM) | Max interval (cM) | Mean crossover frequency |
|---|---|---|---|---|---|---|---|---|---|---|
| chr1 | 6297.8 | 182.1 | 15 | 28.9 | 11,676 | 92 | 1854.0 | 2.00 | 16.13 | 1.80 |
| chr2 | 5626.9 | 126.0 | 8 | 22.4 | 14,213 | 86 | 2525.9 | 1.48 | 10.23 | 1.26 |
| chr3 | 5000.0 | 101.2 | 11 | 20.2 | 10,738 | 64 | 2147.6 | 1.61 | 8.33 | 1.01 |
| chr4 | 4198.8 | 68.9 | 17 | 16.4 | 7784 | 47 | 1853.9 | 1.50 | 3.68 | 0.68 |
| chr5 | 3809.0 | 61.7 | 7 | 16.2 | 6633 | 37 | 1741.4 | 1.71 | 9.28 | 0.61 |
| chr6 | 3574.5 | 122.0 | 12 | 34.1 | 5280 | 72 | 1477.1 | 1.72 | 17.16 | 1.21 |
| chr7 | 3489.0 | 24.8 | 6 | 7.1 | 6243 | 24 | 1789.3 | 1.08 | 1.84 | 0.25 |
| chr8 | 3269.4 | 85.8 | 14 | 26.3 | 3112 | 43 | 951.9 | 2.04 | 14.13 | 1.45 |
| chr9 | 1962.5 | 37.7 | 4 | 19.2 | 3955 | 23 | 2015.3 | 1.72 | 9.28 | 0.37 |
| Whole | 37228.0 | 810.1 | 94 | – | 69,634 | 488 | – | – | – | – |
| Average | 4136.4 | 90.0 | 10.4 | 21.8 | 7737.1 | 54.2 | 1870.5 | 1.65 | 10.01 | 0.96 |
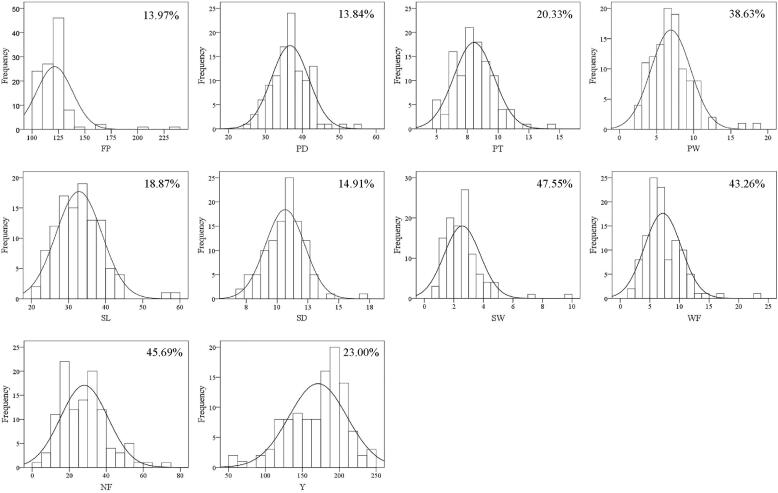
Fig. 3.
Genetic linkage map of L. edodes and distribution of QTLs. The genetic position of bins is shown with a genetic ruler on the left. Bands on the map indicate the bin markers. The bin markers in red on chr2 and chr4 indicate MAT-A (68.05 cM) and MAT-B (0 cM), respectively. LOD-1 confidence interval is indicated by the length of the QTL bar, and the position of the LOD peak is represented by the short line in the middle of the QTL bar. QTLs for PW, PT and PD are marked in green; QTLs for SW, SD and SL are in blue; QTLs for FP are in black; QTLs for WF are in pink; QTLs for NF are in brown; QTLs for Y are in light green; and pleiotropic QTLs identified by meta-QTL analysis are in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A total of 94 scaffolds covering 81.7% (37.23 Mb) of 0899ss11 genome were aligned into nine LGs of Lemap2.0, and were re-anchored into nine pseudo-chromosomes by using ALLMAPS (Table 2, Table S4, Supplementary material Dataset 2). Each pseudo-chromosome contained four to 17 scaffolds. Accordingly, 83.7% (31.16 Mb) of re-anchored scaffolds were oriented. A total of 17 scaffolds of the 0899ss11 genome were dissected into two to four parts (Table S5). The distribution of SNP and InDel markers in the physical map was shown in Fig. S3, and some gaps were found on all nine pseudo-chromosomes. A high level of collinearity between genome scaffolds and Lemap2.0 was found (Fig. S4). Overwhelming majority of the marker orders on LGs were consistent with their position on the genome. The nine pseudo-chromosomes included 11,871 annotated genes accounting for 84.4% of the total L. edodes genes. Recombination rates of the nine pseudo-chromosomes varied from 7.1 cM/Mb to 34.1 cM/Mb, with an average of 21.8 cM/Mb. Limited crossovers occurred in LQ-15 and large continuous segments derived from the same parent were observed in the SSIs (Fig. S5). The crossover frequency per LG per individual varied from 0.2 to 1.8, with an average of 0.96 (Table 2).
3.3. QTL mapping of FBRTs
Based on Lemap2.0, the phenotypic data of the ten traits surveyed in 2012 and 2016 were used to detect seven and 22 QTLs, respectively, and most of them were year-specific (Table 3). One consensus PW-QTL cqpw was identified in both years, contributing to 11.2% and 6.7% of the phenotypic variation of PW, respectively. The other consensus QTL cqfp for FP was also identified in both years. The 29 QTLs were distributed on six LGs, explaining 4.5% to 17.0% of the phenotypic variation. Twenty-four of these QTLs were FBRT-QTLs, while five of them were for the other three traits. Fourteen QTLs for pileus-related traits were identified and seven QTLs for stipe-related traits were detected. Only part of phenotypic variation of investigated traits could be explained by the mapped QTLs (Table S6), which suggests that the existence of other uncharacterized genomic loci associated with FBD.
Table 3.
Meta-analysis of QTLs for fruiting body-related traits of L. edodes.
| Meta-QTL a | Position (cM) | Refined CI (cM) | No. genes | Genes with traits b | Trait | QTL ID | Chr | Position (cM) | LOD | Additive | Parental allele | CI (cM) | R2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pq1.1 | 105.5 | 100.5–110.5 | 11 | 0 | SD-2016 | qSD.1.1 | 1 | 105.5 | 3.02 | −0.49 | L205-6 | 102.1–115 | 9.0 |
| SW-2016 | qSW.1.1 | 1 | 105.5 | 2.95 | −0.38 | L205-6 | 102.4–118.3 | 9.7 | |||||
| pq1.2 | 157.3 | 156.6–158.0 | 1 | 0 | PD-2016 | qPD.1.1 | 1 | 157.3 | 3.02 | −1.55 | L205-6 | 156.1–158.2 | 8.5 |
| PW-2016 | qPW.1.1 | 1 | 157.3 | 4.33 | −0.97 | L205-6 | 156.2–158.2 | 11.9 | |||||
| pq1.3 | 166.3 | 165.3–167.3 | 139 | 33 | PW-2012 | qPW.1.2 | 1 | 165.7 | 2.97 | −3.01 | L205-6 | 164.7–168.4 | 11.2 |
| (cqpw) | PW-2016 | qPW.1.3 | 1 | 166.5 | 4.69 | −0.98 | L205-6 | 164.7–169.8 | 12.9 | ||||
| PD-2016 | qPD.1.2 | 1 | 166.5 | 3.63 | −1.65 | L205-6 | 164.7–169.8 | 10.1 | |||||
| PT-2016 | qPT.1.1 | 1 | 166.5 | 7.30 | −0.68 | L205-6 | 164.7–168.9 | 16.8 | |||||
| SL-2016 | qSL.1.1 | 1 | 166.5 | 3.69 | −2.00 | L205-6 | 163.7–170.6 | 10.2 | |||||
| WF-2016 | qWF.1.1 | 1 | 166.5 | 3.27 | −0.99 | L205-6 | 166.4–171.4 | 9.8 | |||||
| qPD.2.1 | 76.6 | 71.3–81.9 | 5 | 1 | PD-2012 | qPD.2.1 | 2 | 76.6 | 3.09 | 2.71 | W1-26 | 69.7–84.6 | 11.0 |
| pq2.1 | 88.0 | 87.3–88.6 | 211 | 67 | FP-2016 | qFP.2.1* | 2 | 85.5 | 2.12 | 5.71 | W1-26 | 84.6–89.6 | 4.5 |
| (cqfp) | FP-2012 | qFP.2.2 | 2 | 89.3 | 3.05 | 9.72 | W1-26 | 85.5–94.1 | 10.2 | ||||
| PD-2012 | qPD.2.2 | 2 | 88.3 | 3.00 | 2.65 | W1-26 | 86.5–89.3 | 10.5 | |||||
| NF-2012 | qNF.2.1 | 2 | 88.3 | 4.31 | −2.14 | L205-6 | 87.0–90.1 | 13.5 | |||||
| qPT.3.1 | 77.3 | 69.3–79.1 | 53 | 8 | PT-2016 | qPT.3.1 | 3 | 77.3 | 4.10 | 0.49 | W1-26 | 69.3–79.1 | 8.6 |
| qY.5.1 | 2.8 | 0.9–6.1 | 49 | 4 | Y-2016 | qY.5.1 | 5 | 2.8 | 2.86 | 12.79 | W1-26 | 0.9–6.1 | 9.6 |
| pq5.1 | 18.4 | 17.5–19.3 | 15 | 4 | PD-2016 | qPD.5.1 | 5 | 18.4 | 3.58 | −1.65 | L205-6 | 13.3–20.2 | 9.9 |
| PT-2016 | qPT.5.1 | 5 | 18.4 | 7.43 | −0.69 | L205-6 | 17.2–20.2 | 17.0 | |||||
| PW-2016 | qPW.5.1 | 5 | 18.4 | 4.20 | −0.93 | L205-6 | 15.8–20.2 | 11.4 | |||||
| WF-2016 | qWF.5.1 | 5 | 18.4 | 3.30 | −1.03 | L205-6 | 15.9–19.3 | 10.4 | |||||
| FP-2016 | qFP.5.1 | 5 | 18.4 | 2.20 | −4.86 | L205-6 | 15.1–24.9 | 7.7 | |||||
| pq5.2 | 25.0 | 23.3–26.8 | 411 | 74 | PW-2016 | qPW.5.2 | 5 | 23.9 | 3.95 | −0.90 | L205-6 | 20.2–27.7 | 10.8 |
| PD-2016 | qPD.5.2 | 5 | 25.8 | 3.48 | −1.61 | L205-6 | 20.2–27.6 | 9.7 | |||||
| SD-2016 | qSD.5.1 | 5 | 24.8 | 2.99 | −0.49 | L205-6 | 21.5–27.6 | 9.0 | |||||
| WF-2016 | qWF.5.2 | 5 | 25.8 | 2.69 | −0.93 | L205-6 | 20.2–27.9 | 8.6 | |||||
| qSD.7.1 | 12.9 | 5.5–16.5 | 419 | 55 | SD-2016 | qSD.7.1 | 7 | 12.9 | 3.17 | 0.53 | W1-26 | 5.5–16.5 | 9.5 |
| pq8.1 | 48.1 | 46.6–49.7 | 1 | 0 | SD-2012 | qSD.8.1 | 8 | 48.1 | 3.08 | −1.50 | L205-6 | 46.7–51.0 | 11.4 |
| SW-2012 | qSW.8.1 | 8 | 48.1 | 3.75 | −1.43 | L205-6 | 46.5–51 | 13.9 |
* qFP.2.1 was a suggestive locus, since the LOD of this QTL was over 2.0, but lower than permutation-based thresholds (2.15).
Chr was chromosome, LOD was the logarithm of odds, and CI was the QTL confidence interval.
Pleiotropic QTLs were abbreviated as pq.
Number of genes which expression levels were identified to be significantly correlated with the phenotypic traits.
On chr1, chr2 and chr5, three QTL hotspots were identified, on which most FBRT-QTLs were located (Fig. 3, Table 3). The QTL hotspot on chr1 spanned 15.3 cM (156.1–171.4 cM) and mainly contained QTLs for pileus-related traits (Fig. 3). All the eight QTLs showed negative effects, indicating that their favorable alleles were derived from the parental strain L205-6. The stable QTL cqpw was included in this hotspot region. The QTL hotspot on chr2 was located in the region of 69.7 cM to 94.1 cM and contained five QTLs for three traits. The consensus FP-QTL cqfp was located in this hotspot region. On chr5, the third QTL hotspot was mapped on the region of 13.3 cM to 27.9 cM (Fig. 3). A total of nine QTLs with negative effects were included in this hotspot region. This QTL hotspot was found to be associated with multiple traits, such as pileus- and stipe-related traits.
To further illustrate the results of QTL mapping, QTLs for different traits with overlapping CIs were considered to be one pleiotropic QTL. In addition, meta-QTL analysis was performed to clarify the CIs of the pleiotropic QTLs. The 29 QTLs were refined into 11 meta-QTLs, seven of which were pleiotropic loci (Table 3). Each pleiotropic QTL included two to six QTLs. The CIs of the seven pleiotropic QTLs were also narrowed down, varied from 1.4 cM to 10.0 cM, with an average of 3.3 cM. Six pleiotropic QTLs (pq1.1, pq1.2, pq1.3, pq5.1, pq5.2 and pq8.1) were integrated from QTLs with all favorable alleles derived from L205-6.
3.4. Screening of QTL candidate genes
A total of 1,315 annotated genes were identified in the CIs of the 11 meta-QTLs (Table 3). Correlation analysis between FPKM and phenotypic values of these 1,315 genes identified significant correlations between the expression of 246 genes and the phenotypic values of the target traits (Table S7). Among the 246 expression-phenotype-associated genes, 106 genes were associated with multiple traits (Table S7). The 246 genes were then functionally annotated by Blast2GO. GO terms of molecular function mainly covered catalytic activity, binding, molecular function regulator, transporter activity and transcription regulator activity, whereas those of biological process mainly contained metabolic process, cellular process, regulation of biological process, biological regulation, response to stimulus and signaling (Fig. S6). GO and KEGG enrichment analysis were employed for functional annotation for the 246 phenotype-specific expressed genes. Results demonstrated that GO terms of molecular function were enriched in 31 categories and those of biological process were 43 (p < 0.05) (Table S8). Five enriched KEGG pathways were detected in the 246 genes (p < 0.05) (Table S9). As for the reliable candidate genes of QTLs, 47 genes and 36 genes were respectively detected to be associated with PW and FP in both the 2012 and 2016 datasets, with gene expressions significantly correlated with the corresponding phenotypic values (Table S10 and S11). Seven out of the 83 genes (jgi.p|Lenedo1|1157672, jgi.p|Lenedo1|1040279, jgi.p|Lenedo1|1206258, jgi.p|Lenedo1|1164760, jgi.p|Lenedo1|1174308, jgi.p|Lenedo1|1031698 and jgi.p|Lenedo1|1077466), deserved further study, since their functions were relevant to FBD (Table 4). For instance, the gene encoding the cyclase-associated protein was found to be an important candidate gene for PW. In addition, several important candidate genes (such as those encoding fasciclin-like protein and PCI-domain-containing protein), which were reported to be involved in FBD [1], [15], were also identified (Table 4).
Table 4.
Important QTL candidate genes and their functions in fruiting body development.
| Functional category | QTL | Gene name | Functional description | KEGG description | GO description |
|---|---|---|---|---|---|
| Environmental response | pq5.2 | jgi.p|Lenedo1|1114176 | hexose transporter | NA | NA |
| pq2.1 | jgi.p|Lenedo1|1157672a | opt oligopeptide transporter | NA | NA | |
| Signal transduction | pq1.3 | jgi.p|Lenedo1|1040279b | cyclase-associated protein | Membrane trafficking; Exosome | NA |
| qPT.3.1 | jgi.p|Lenedo1|88319 | calcium activated cation channel | NA | Ion channel activity, ion transport | |
| qSD.7.1 | jgi.p|Lenedo1|1194336 | WD40 repeat-like protein | NA | NA | |
| pq1.3 | jgi.p|Lenedo1|1206258b | WD40 repeat-like protein | NA | NA | |
| pq5.2 | jgi.p|Lenedo1|1114625 | WD40 repeat-like protein | RNA transport | Cellular response to osmotic stress | |
| Transcription regulation | pq1.3 | jgi.p|Lenedo1|538809 | transcription factor | NA | Regulation of transcription, DNA-templated |
| qPD.2.1 | jgi.p|Lenedo1|1032489 | HD1 homeodomain mating-type protein Le.a1-2 | NA | Regulation of transcription, DNA-templated | |
| pq2.1 | jgi.p|Lenedo1|1164760a | Transcription and mRNA export factor | NA | Transcription coactivator activity | |
| pq2.1 | jgi.p|Lenedo1|26471 | myb3r transcription factor | Transcription factors | DNA binding | |
| pq2.1 | jgi.p|Lenedo1|30321 | ace1 transcription factor | NA | Regulation of transcription, DNA-templated | |
| pq5.2 | jgi.p|Lenedo1|1067568 | Chromatin modification-related protein EAF7 | NA | Regulation of transcription, DNA-templated | |
| qSD.7.1 | jgi.p|Lenedo1|305037 | predicted protein | NA | Regulation of transcription, DNA-templated | |
| Protein degradation | pq5.2 | jgi.p|Lenedo1|1043151 | e3 ubiquitin-protein ligase rnf13-like | Ubiquitin system | NA |
| pq2.1 | jgi.p|Lenedo1|1076755 | PCI-domain-containing protein | Ubiquitin system | NA | |
| Cell cycle regulation | pq1.3 | jgi.p|Lenedo1|1174308b | cell cycle control protein | NA | NA |
| pq1.3 | jgi.p|Lenedo1|1093770 | TPR-like protein | Cell cycle, Cell cycle - yeast, Meiosis - yeast | Regulation of mitotic cell cycle | |
| Cell adhesion | qSD.7.1 | jgi.p|Lenedo1|1160703 | fasciclin-like protein | NA | NA |
| qSD.7.1 | jgi.p|Lenedo1|1044962 | cell adhesion protein byn-1 | NA | NA | |
| Cell wall remodeling (CAZymes) | qY.5.1 | jgi.p|Lenedo1|1057255 | glycoside hydrolase family 3 protein | Starch and sucrose metabolism | NA |
| pq5.2 | jgi.p|Lenedo1|1101195 | glycoside hydrolase family 17 protein | NA | NA | |
| pq5.2 | jgi.p|Lenedo1|1101211 | glycoside hydrolase family 79 protein | NA | NA | |
| pq5.2 | jgi.p|Lenedo1|881996 | glycoside hydrolase family 61 protein | NA | Hydrolase activity | |
| pq2.1 | jgi.p|Lenedo1|1031698a | glycoside hydrolase family 79 protein | NA | NA | |
| qSD.7.1 | jgi.p|Lenedo1|1205574 | glycoside hydrolase family 47 protein | Various types of N-glycan biosynthesis | NA | |
| Others | pq2.1 | jgi.p|Lenedo1|1164647 | cytochrome p450 | NA | NA |
| pq2.1 | jgi.p|Lenedo1|1032363 | cytochrome P450 | Steroid biosynthesis;Cytochrome P450 | NA | |
| pq2.1 | jgi.p|Lenedo1|1164679 | cytochrome p450 | NA | NA | |
| qSD.7.1 | jgi.p|Lenedo1|1119574 | cytochrome P450 | NA | NA | |
| qSD.7.1 | jgi.p|Lenedo1|1045197 | cytochrome p450 | NA | NA | |
| pq2.1 | jgi.p|Lenedo1|1077466a | BAR-domain-containing protein | Endocytosis; Fc gamma R-mediated phagocytosis | Establishment or maintenance of cell polarity regulating cell shape | |
| pq5.2 | jgi.p|Lenedo1|1168070 | septin family protein | NA | GTP binding |
Genes detected as FP candidate genes in both years.
Genes detected as PW candidate genes in both years.
4. Discussion
4.1. High-density linkage map and chromosome-level genome
In view of its importance in research and the industry of mushroom-forming fungi, the genetics, genomics and breeding of L. edodes have been extensively studied. Mainly based on anonymous and poorly informative PCR-based markers, several genetic maps of L. edodes have been constructed over the past decade [16], [49], [50]. Several genome sequences of L. edodes have also been released [51], [52]. However, until now, there is no L. edodes genome that can correspond to the chromosomal level, and integrative studies combining physical mapping and genetic mapping are unavailable in L. edodes. As a result, the practical use of these genetic and genomic resources is very limited, and the genetic program of complex phenotypes, such as traits related to FBD, remains to be elucidated. In this study, we constructed a second-generation genetic linkage map of L. edodes (Lemap2.0) via high-throughput genotyping by sequencing-based approaches. We then aligned 94 scaffolds of the genome published by JGI to Lemap2.0 and re-assembled nine pseudo-chromosomes in shiitake for the first time.
In most mushroom-forming fungi, heterokaryotic or dikaryotic strains are indispensable for forming fruiting bodies from which phenotypic data of traits (such as FBRTs) are surveyed [16]. F1 progenies such as monokaryotic SSIs are widely used for genotyping and construction of genetic maps. The population inconsistency between the mapping population and trait segregation population causes a large amount of workload in genetic mapping of mushroom-forming fungi. Here, we propose an efficient method to infer the genotype of SSIs from transcriptome sequencing of dikaryotic strains. To verify this genotypes deduction, we compared 86 InDel genotypes of the SSIs obtained by this deduction procedure with our previous data [16]. The results of 86.5% of InDel genotypes of SSIs obtained from deduction procedure were consistent with electrophoresis-based genotyping, suggestive of the robustness of this deduction of SSIs genotypes. Its validity was further verified by the high syntenic relationship between the genetic position of the 86 InDel markers in Lemap2.0 with that in previously constructed shiitake genetic map [16]. Errors in genotype calling and alignment, as well as the location of some InDel markers in intergenic regions, which may account for the inconsistency between these two genotyping strategies. To increase the mapping efficiency, the recombination bin strategy was used in linkage map construction. Since the cultivars of L. edodes in China are closely related [53], the polymorphism between the two parental strains L205-6 and W1-26 was low. Limited recombination events have occurred in the artificial bi-parental population LQ-15. Thereby, a total of 69,634 markers were grouped into only 488 recombination bins, with highly redundant markers in each bin. Similarly in Hericium erinaceus, a relatively small number of recombination events were occurred in a biparental population, and each recombination bin contained massive SNPs [54]. A high collinearity between the genome assemblies and Lemap2.0 indicates that both this genetic map and genome assembly are of high fidelity. The accuracy of Lemap2.0 was also verified by mapping of the mating type loci and InDel markers. Compared with the constructed shiitake genetic maps, Lemap2.0 was highly saturated and could be used as a powerful tool for QTL mapping. More importantly, the integration of the genome assembly and Lemap2.0 could accelerate the identification of compelling candidate genes. Although the molecular marker dataset was from transcriptome sequencing of the mature fruiting bodies, 81.7% of the L. edodes genome was assembled into nine pseudo-chromosomes, which contained 84.4% of all annotated genes. To our knowledge, this is the first chromosome-level genome assembly of shiitake. A high-quality reference genome is beneficial for research of the functional genome of L. edodes. Overall, the newly constructed Lemap2.0 and improved assembly could be used as a reference in future shiitake genetic, genomic, and breeding studies.
4.2. QTLs for fruiting body development
FBD of mushroom-forming fungi has been attracting a lot of attention [3], [4]. However, the genetic underpinnings for FBD of mushroom-forming fungi are poorly understood due to the complicated biological process involved [1]. In the present study, using shiitake as a model, quantitative genetics was employed to reveal the genetic repertoire for fruiting body-related traits at the genome-wide level.
Obtained via genetic mapping, information of the distribution of QTLs provides a basic outline for the genetic architecture of FBD in L. edodes. It is common for the presence of QTL clusters for closely related traits, such as FBRTs surveyed here. As expected, most FBRT-QTLs were clustered in three hotspots on chr1, chr2 and chr5. The shape and size of shiitake fruiting body are regulated by a few main regions in the genome. The QTLs co-localization for FBRTs contributed to the genetic basis for phenotypic correlations between these traits. This result is consistent to that reported in P. ostreatus [19], A. bisporus [20] and our previous study in L. edodes[16]. Pleiotropy and close linkage between QTLs controlling different traits are the major reasons for trait correlations [55]. In most typical pileate-stipitate fruiting bodies of mushroom-forming fungi, the stipe structurally supports the pileus, like in the basidiocarp of shiitake. And the hierarchical structure among FBRTs may represent one possible cause of gene pleiotropy, where a gene is responsible for a trait which leads to, or partly contributes to, another trait [55].
In this study, the segregating population LQ-15 was generated by mated the F1 SSIs with a tester monokaryon. Thereby, the mapped QTLs in LQ-15 reflect the allelic substitution effect of the segregating allele with their interactions with the constant allele from the tester nucleus [16]. Actually, the significant effects of genetic background of tester monokaryon on the expression of traits and QTLs detection had been reported in L. edodes and other mushrooms [16], [19], [20], [21]. This presents the challenge for accurate identification of QTLs.
In mushroom-forming fungi, the complex quantitative traits are sensitive to environmental factors. Thereby, the identification of stable QTLs underlying phenotypic traits across multiple environments is quite challenging. In contrast to previous studies in shiitake [16], where traits were mainly phenotyped in a single environment, we performed QTL analysis based on phenotypic data from two years of cultivation trials in this study. A total of 29 QTLs for ten traits were disclosed, but only two (6.9%) consensus QTLs (cqfp and cqpw) were identified in both years, indicating strong effects of environment on FBD. Moreover, this percentage of stable loci is lower than that in A. bisporus [20] and could be attributed to the fact that the fruiting trials of A. bisporus were carried out at growing rooms with well-controlled environments [20], whereas L. edodes was usually cultivated in simple mushroom houses under varied environments [28]. The identification of stable QTLs is challenging because each locus has only minor phenotypic effects as well. In this study, no QTL could attribute to over 20% of the phenotypic variation, making it difficult to identify these loci simultaneously in different years.
For breeding purposes, findings of QTL mapping have potential to facilitate the genetic improvement of shiitake cultivars. In particular, the stable QTL pq1.3 is a pleiotropic locus for pileus weight, pileus diameter and the weight of a single fruiting body, making it an important locus for genetic improvement of shiitake pileus. For these three traits, the additive values of pq1.3 had the same sign, and only one recombination bin (c01b078) was included in the confidence interval. It is expected that this bin marker can be used in marker-assisted selection to increase the genetic improvement efficiency of the shiitake pileus.
4.3. Candidate genes involved in fruiting body development
Fruiting body is the most complicated multi-cellular structure in fungi, and many genes are involved in FBD, including those function in environmental response, signal transduction, transcription regulation, cell wall remodeling, protein degradation, cell cycle regulation and cell adhesion [1], [3], [8], [56]. In our previous study, some QTLs associated with FBRTs in shiitake had been identified based on the previously constructed genetic map [16]. However, due to a shortage of genome-anchored markers, the identification of candidate genes in the CIs of QTLs was greatly limited. Here, taking advantage of the alignment of genome assembly and Lemap2.0, genes within these identified FBRT-QTLs could be scanned directly. Ten fruiting body-related traits were surveyed here, and there could be connections between these genes and their traits. Correlation analysis between gene expression and phenotypic traits was employed to refine the candidate genes.
FBD is triggered by environmental factors [3] and sensed by fungi, which transduce the signals into cell (Table 4). The gene jgi.p|Lenedo1|1114176 within the CI of meta-QTL pq5.2 encodes a hexose transporter that is induced by different levels of glucose in yeast [57]. The gene jgi.p|Lenedo1|1157672 encoding opt oligopeptide transporter. In fungi, oligopeptide transport plays important roles in sexual differentiation, mating and pheromone sensing [58]. Ca2+ signaling plays regulatory role in primordium differentiation and stipe development of P. ostreatus [59]. The gene jgi.p|Lenedo1|88319 encoding calcium activated cation channel may function in Ca2+ signaling of L. edodes.
Seven QTL candidate genes may be involved in signal transduction in the course of FBD (Table 4). The gene jgi.p|Lenedo1|1040279 encodes a cyclase-associated protein, which is involved in signal transduction of actin polymerization and related to vesicle trafficking and cytoskeletal formation [60], and is also important in fungal morphogenesis [61], [62], [63]. Thus, jgi.p|Lenedo1|1040279 deserves further functional analysis. WD40 repeat protein provides a platform for the interaction and assembly of several proteins into a signalosome, and has diverse functions in fungi, mainly involved in growth, cell differentiation and development [64]. Three genes (jgi.p|Lenedo1|1194336, jgi.p|Lenedo1|1206258 and jgi.p|Lenedo1|1114625) encoding WD40 repeat-like proteins were identified as QTL candidate genes here (Table 4).
Transcriptional regulation is essential for FBD in fungi [56], and transcription factors govern changes in gene expression during FBD. The gene jgi.p|Lenedo1|1032489 encoding HD1 homeodomain mating-type protein of L. edodes, which serves as transcription factor regulating various aspects of FBD in Basidiomycota [65]. Three other genes also code for transcription factors (jgi.p|Lenedo1|538809, jgi.p|Lenedo1|26471 and jgi.p|Lenedo1|30321), and three others (jgi.p|Lenedo1|1164760, jgi.p|Lenedo1|1067568 and jgi.p|Lenedo1|305037) encode proteins whose function are involved in regulation of transcription (Table 4).
Cell-to-cell adhesion in fungal multicellularity is important in 3-dimensional structure of fruiting body [66]. The gene jgi.p|Lenedo1|1044962 encodes cell adhesion protein byn-1. The gene jgi.p|Lenedo1|1160703 encodes a fasciclin-like protein that is capable of promoting cell adhesion and participates in cellular differentiation and development during fruiting body formation in L. edodes [15]. Two genes (jgi.p|Lenedo1|1174308 and jgi.p|Lenedo1|1093770) playing role in cell cycle regulation are also detected as QTL candidate genes.
Protein degradation is required for FBD in filamentous ascomycetes [56]. Proteins encoded by two genes (jgi.p|Lenedo1|1043151 and jgi.p|Lenedo1|1076755) are involved in the ubiquitin system that takes part in protein degradation (Table 4). Carbohydrate-active enzymes (CAZymes) play vital roles in fungal cell wall remodeling during FBD [1]. Six CAZymes were found to be QTL candidate genes here (Table 4).
Five genes encoding cytochrome P450 were found within pq2.1 and qSD.7.1 (Table 4). Cytochrome P450 is related to FBD in L. edodes [67], [68]. In particular, eln2 encodes a novel type of cytochrome P450 enzyme in C. cinerea, which affects stipe elongation during FBD [69]. The gene jgi.p|Lenedo1|1077466 encodes a BAR-domain-containing protein, which regulates fungal morphogenesis [70], [71]. The gene jgi.p|Lenedo1|1168070 encodes a septin family protein, which has been shown to be involved in stipe cell elongation in C. cinerea [72].
Many enriched GO terms were related to FBD. GO:0071470 (cellular response to osmotic stress), GO:0016620 (oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor), and GO:0006457 (protein folding) were probably involved in environmental response. GO:0000281 (mitotic cytokinesis), GO:0007346 (regulation of mitotic cell cycle), GO:0045144 (meiotic sister chromatid segregation), and GO:0044774 (mitotic DNA integrity checkpoint) were concerned with cell cycle regulation. GO:0045893 (positive regulation of transcription, DNA-templated) was related to transcription regulation (Table S8). Three of the five enriched KEGG pathways might be relevant with protein metabolism, i.e., ko00290 (Valine, leucine and isoleucine biosynthesis), ko00260 (Glycine, serine and threonine metabolism), and ko00270 (Cysteine and methionine metabolism) (Table S9).
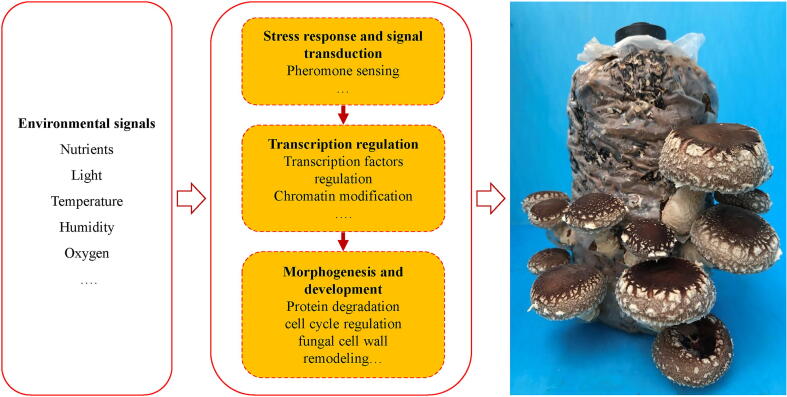
Collectively, findings of candidate genes provided the more detailed view on FBD in shiitake (Table 4, Fig. 4). The genes involved in stress response and signal transduction sense the environmental signals, and play an important role in fruiting body initiation. In the stage of transcription regulation, a number of transcription factors play critical role in transcription regulation of fruiting body formation. Finally, the genes involved in protein degradation, fungal cell wall remodeling, cell adhesion and the critical cell cycle regulation serve the functions in fruiting body morphogenesis and development in shiitake.
Fig. 4.
A process of fruiting body development in L. edodes. Shiitake senses changing environmental conditions and then triggers fruiting body development. Many QTL candidate genes are relevant to fruiting body development, including those involved in environmental response, signal transduction, transcription regulation, and fruiting body morphogenesis and development.
5. Conclusions
In summary, we constructed an ultra-high-density genetic map and re-assembled a chromosome-level genome of L. edodes via high-throughput genotyping. Using systems genetic analysis, we then disclosed the genetic loci and candidate genes for fruiting body-related traits, and provided a global view of the genetic and molecular basis for fruiting body development of shiitake. The newly generated reference genome and genetic map would greatly expand the toolbox for biological studies in L. edodes. Findings of this study could also advance our understanding of fruiting body development in mushroom-forming fungi at large.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was financially supported by the Major Project for Technological Innovation Plans of Hubei Province (2018ABA095), the National Natural Science Foundation of China (Grant No. 32070015), and the National Key Research and Development Program of China (2019YFD1001905-35). The L. edodes 0899ss11 v1.0 genome was produced by the U.S. Department of Energy Joint Genome Institute Community Science Program (Proposal ID 1911, “Comparative and functional genomics of shiitake mushrooms”, D. Hibbett, PI).
Author contributions
Yang Xiao and Yinbing Bian conceived and designed the study. Lin Zhang and Wenbing Gong performed the experiments. Nan Shen, Yang Xiao and Chuang Li performed the analyses. Yang Xiao, Wenbing Gong, Lin Zhang and Ying Gui wrote the manuscript. Hoi Shan Kwan and Man Kit Cheung contributed to finalize the MS. All authors have read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Krizsán K., Almási É., Merényi Z., Sahu N., Virágh M., Kószó T. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc Natl Acad Sci U S A. 2019;116(15):7409–7418. doi: 10.1073/pnas.1817822116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimm D., Wösten H.A.B. Mushroom cultivation in the circular economy. Appl Microbiol Biot. 2018;102(18):7795–7803. doi: 10.1007/s00253-018-9226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakamoto Y. Influences of environmental factors on fruiting body induction, development and maturation in mushroom-forming fungi. Fungal Biol Rev. 2018;32(4):236–248. [Google Scholar]
- 4.Varga T., Krizsán K., Földi C., Dima B., Sánchez-García M., Sánchez-Ramírez S. Megaphylogeny resolves global patterns of mushroom evolution. Nat Ecol Evol. 2019;3(4):668–678. doi: 10.1038/s41559-019-0834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Royse D.J., Baars J., Tan Q. Current overview of mushroom production in the world. In: Diego C.Z., Pardo-Giménez A., editors. Edible and medicinal mushrooms: technology and applications. Wiley-Blackwell; England: 2017. pp. 5–13. [Google Scholar]
- 6.Chang S.T., Wasser S.P. The cultivation and environmental impact of mushrooms. Oxford Res Encyl. 2017 [Google Scholar]
- 7.Chum W.W.Y., Kwan H.S., Au C.H., Kwok I.S.W., Fung Y.W. Cataloging and profiling genes expressed in Lentinula edodes fruiting body by massive cDNA pyrosequencing and LongSAGE. Fungal Genet Biol. 2011;48(4):359–369. doi: 10.1016/j.fgb.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Almási É., Sahu N., Krizsán K., Bálint B., Kovács G.M., Kiss B. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 2019;224(2):902–915. doi: 10.1111/nph.16032. [DOI] [PubMed] [Google Scholar]
- 9.Yoo S.I., Lee H.Y., Markkandan K., Moon S., Ahn Y.J., Ji S. Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes. BMC Genomics. 2019;20(1):121. doi: 10.1186/s12864-019-5509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelkmans J.F., Patil M.B., Gehrmann T., Reinders M.J.T., Wösten H.A.B., Lugones L.G. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci Rep. 2017;7(1):310. doi: 10.1038/s41598-017-00483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chum W.W.Y., Ng K.T.P., Shih R.S.M., Au C.H., Kwan H.S. Gene expression studies of the dikaryotic mycelium and primordium of Lentinula edodes by serial analysis of gene expression. Mycol Res. 2008;112(8):950–964. doi: 10.1016/j.mycres.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 12.Kajiwara S., Yamaoka K., Hori K., Miyazawa H., Saito T., Kanno T. Isolation and sequence of a developmentally regulated putative novel gene, priA, from the basidiomycete Lentinus edodes. Gene. 1992;114(2):173–178. doi: 10.1016/0378-1119(92)90571-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J., Kwan H.S. Characterization, molecular cloning, and differential expression analysis of laccase genes from the edible mushroom Lentinula edodes. Appl Environ Microb. 1999;65(11):4908–4913. doi: 10.1128/aem.65.11.4908-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng W.L., Ng T.P., Kwan H.S. Cloning and characterization of two hydrophobin genes differentially expressed during fruit body development in Lentinula edodes. FEMS Microbiol Lett. 2000;185(2):139–145. doi: 10.1111/j.1574-6968.2000.tb09052.x. [DOI] [PubMed] [Google Scholar]
- 15.Miyazaki Y., Kaneko S., Sunagawa M., Shishido K., Yamazaki T., Nakamura M. The fruiting-specific Le.flp1 gene, encoding a novel fungal fasciclin-like protein, of the basidiomycetous mushroom Lentinula edodes. Curr Genet. 2007;51(6):367–375. doi: 10.1007/s00294-007-0133-2. [DOI] [PubMed] [Google Scholar]
- 16.Gong W.B., Li L., Zhou Y., Bian Y.B., Kwan H.S., Cheung M.K. Genetic dissection of fruiting body-related traits using quantitative trait loci mapping in Lentinula edodes. Appl Microbiol Biot. 2016;100(12):5437–5452. doi: 10.1007/s00253-016-7347-5. [DOI] [PubMed] [Google Scholar]
- 17.Galpaz N., Gonda I., Shem-Tov D., Barad O., Tzuri G. Deciphering genetic factors that determine melon fruit-quality traits using RNA-Seq-based high-resolution QTL and eQTL mapping. Plant J. 2018;94(1):169–191. doi: 10.1111/tpj.13838. [DOI] [PubMed] [Google Scholar]
- 18.Gao W., Qu J., Zhang J., Sonnenberg A., Chen Q., Zhang Y. A genetic linkage map of Pleurotus tuoliensis integrated with physical mapping of the de novo sequenced genome and the mating type loci. BMC Genomics. 2018;19(1):18. doi: 10.1186/s12864-017-4421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larraya L.M., Alfonso M., Pisabarro A.G., Ramírez L. Mapping of genomic regions (quantitative trait loci) controlling production and quality in industrial cultures of the edible basidiomycete Pleurotus ostreatus. Appl Environ Microb. 2003;69(6):3617–3625. doi: 10.1128/AEM.69.6.3617-3625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foulongne-Oriol M., Rodier A., Rousseau T., Savoie J.M. Quantitative trait locus mapping of yield-related components and oligogenic control of the cap color of the button mushroom, Agaricus bisporus. Appl Environ Microb. 2012;78(7):2422–2434. doi: 10.1128/AEM.07516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao W., Weijn A., Baars J.J.P., Mes J.J., Visser R.G.F., Sonnenberg A.S.M. Quantitative trait locus mapping for bruising sensitivity and cap color of Agaricus bisporus (button mushrooms) Fungal Genet Biol. 2015;77:69–81. doi: 10.1016/j.fgb.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Han K., Jeong H.J., Yang H.B., Kang S.M., Kwon J.K., Kim S. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum) DNA Res. 2016;23(2):81–91. doi: 10.1093/dnares/dsv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos C., Almeida N.F., Alves M.L., Horres R., Krezdorn N., Leitão S.T. First genetic linkage map of Lathyrus cicera based on RNA sequencing-derived markers: Key tool for genetic mapping of disease resistance. Hortic Res. 2018;5:4. doi: 10.1038/s41438-018-0047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Xu Y.C., Wang Z.H. Construction of a high-density genetic map by RNA sequencing and eQTL analysis for stem length and diameter in Dendrobium (Dendrobium nobile × Dendrobium wardianum) Ind Crop Prod. 2019;128:48–54. [Google Scholar]
- 25.Civelek M., Lusis A.J. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15(1):34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(D1):D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong W.B., Liu W., Lu Y.Y., Bian Y.B., Zhou Y., Kwan H.S. Constructing a new integrated genetic linkage map and mapping quantitative trait loci for vegetative mycelium growth rate in Lentinula edodes. Fungal Bio-UK. 2014;118(3):295–308. doi: 10.1016/j.funbio.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Gong W.B., Xu R., Xiao Y., Zhou Y., Bian Y.B. Phenotypic evaluation and analysis of important agronomic traits in the hybrid and natural populations of Lentinula edodes. Sci Hortic-Amsterdam. 2014;179:271–276. [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Meth. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., Bhat P.R., Close T.J., Lonardi S., Kruglyak L. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008;4(10):e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H., Zhang X., Miao C., Zhang J., Ming R., Schnable J.C. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015;16(1):3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W., Feng Q., Yu H., Huang X., Zhao Q., Xing Y. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci U S A. 2010;107(23):10578–10583. doi: 10.1073/pnas.1005931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., Bastern C.J., Zeng Z.B. North Carolina State University; Raleigh, NC: 2012. Windows QTL Cartographer 2.5. Department of Statistics. [Google Scholar]
- 40.Marathi B., Guleria S., Mohapatra T., Parsad R., Mariappan N., Kurungara V. QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.) BMC Plant Biol. 2012;12(1):137. doi: 10.1186/1471-2229-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arcade A., Labourdette A., Falque M., Mangin B., Chardon F., Charcosset A. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics. 2004;20(14):2324–2326. doi: 10.1093/bioinformatics/bth230. [DOI] [PubMed] [Google Scholar]
- 42.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J. Huazhong Agricultural University; Wuhan: 2014. Rice gene and sRNA expression quantitative trait loci mapping and regulatory network construction. [Google Scholar]
- 44.Fu J., Cheng Y., Linghu J., Yang X., Kang L., Zhang Z. RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun. 2013;4(1):2832. doi: 10.1038/ncomms3832. [DOI] [PubMed] [Google Scholar]
- 45.Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 46.Ye J., Zhang Y., Cui H.H., Liu J.W., Wu Y.Q. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018;46(W1):W71–W75. doi: 10.1093/nar/gky400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;(35(Web Server issue)):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyazaki K., Huang F., Zhang B., Shiraishi S., Sakai M., Shimaya C. Genetic map of a basidiomycete fungus, Lentinula edodes (shiitake mushroom), constructed by tetrad analysis. Breed Sci. 2008;58(1):23–30. [Google Scholar]
- 50.Terashima K., Matsumoto T., Hayashi E., Fukumasa-Nakai Y. A genetic linkage map of Lentinula edodes (shiitake) based on AFLP markers. Mycol Res. 2002;106(8):911–917. [Google Scholar]
- 51.Chen L.F., Gong Y.H., Cai Y.L., Liu W., Zhou Y., Xiao Y. Genome sequence of the edible cultivated mushroom Lentinula edodes (shiitake) reveals insights into lignocellulose degradation. PLoS ONE. 2016;11(8):e0160336. doi: 10.1371/journal.pone.0160336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shim D., Park S.G., Kim K., Bae W., Lee G.W., Ha B.S. Whole genome de novo sequencing and genome annotation of the world popular cultivated edible mushroom, Lentinula edodes. J Biotechnol. 2016;223:24–25. doi: 10.1016/j.jbiotec.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 53.Chiu S.W., Ma A.M., Lin F.C., Moore D. Genetic homogeneity of cultivated strains of shiitake (Lentinula edodes) used in China as revealed by the polymerase chain reaction. Mycol Res. 1996;100(11):1393–1399. [Google Scholar]
- 54.Gong W.B., Xie C.L., Zhou Y.J., Zhu Z.H., Wang Y.H., Peng Y.D. A resequencing-based ultradense genetic map of Hericium erinaceus for anchoring genome sequences and identifying genetic loci associated with monokaryon growth. Front Microbiol. 2020;10 doi: 10.3389/fmicb.2019.03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y., Lübberstedt T. Molecular basis of trait correlations. Trends Plant Sci. 2010;15(8):454–461. doi: 10.1016/j.tplants.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Pöggeler S., Nowrousian M., Teichert I., Beier A., Kück U. Physiology and Genetics. Springer International Publishing; Cham: 2018. Fruiting-Body Development in Ascomycetes; pp. 1–56. [Google Scholar]
- 57.Ozcan S., Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15(3):1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomolplitinant K.M., Saier M.H. Evolution of the oligopeptide transporter family. J Membrane Biol. 2011;240(2):89–110. doi: 10.1007/s00232-011-9347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu W., Hu J., Li Y., Yang B., Guan Y., Xu C. Comparative proteomic analysis of Pleurotus ostreatus reveals great metabolic differences in the cap and stipe development and the potential role of Ca2+ in the primordium differentiation. Int J Mol Sci. 2019;20(24):6317. doi: 10.3390/ijms20246317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hubberstey A.V., Mottillo E.P. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 2002;16(6):487–499. doi: 10.1096/fj.01-0659rev. [DOI] [PubMed] [Google Scholar]
- 61.Bahn Y.S., Sundstrom P. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J Bacteriol. 2001;183(10):3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takach J.E., Gold S.E. Identification and characterization of Cap1, the adenylate cyclase-associated protein (CAP) ortholog in Ustilago maydis. Physiol Mol Plant Pathol. 2010;75(1-2):30–37. [Google Scholar]
- 63.Zhou X.Y., Zhang H.F., Li G.T., Shaw B., Xu J.R. The cyclase-associated protein Cap1 is important for proper regulation of infection-related morphogenesis in Magnaporthe oryzae. PLoS Pathog. 2012;8(9):e100291. doi: 10.1371/journal.ppat.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain B.P., Pandey S. WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 2018;37(5):391–406. doi: 10.1007/s10930-018-9785-7. [DOI] [PubMed] [Google Scholar]
- 65.Vonk P.J., Ohm R.A. The role of homeodomain transcription factors in fungal development. Fungal Biol Rev. 2018;32(4):219–230. [Google Scholar]
- 66.Nagy L.G., Varga T., Csernetics Á., Virágh M. Fungi took a unique evolutionary route to multicellularity: Seven key challenges for fungal multicellular life. Fungal Biol Rev. 2020;34(4):151–169. [Google Scholar]
- 67.Akiyama R., Sato Y., Kajiwara S., Shishido K. Cloning and expression of cytochrome P450 genes, belonging to a new P450 family, of the basidiomycete Lentinula edodes. Biosci Biotech Bioch. 2002;66(10):2183–2188. doi: 10.1271/bbb.66.2183. [DOI] [PubMed] [Google Scholar]
- 68.Miyazaki Y., Nakamura M., Babasaki K. Molecular cloning of developmentally specific genes by representational difference analysis during the fruiting body formation in the basidiomycete Lentinula edodes. Fungal Genet Biol. 2005;42(6):493–505. doi: 10.1016/j.fgb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 69.Muraguchi H., Kamada T. A Mutation in the eln2 gene encoding a cytochrome P450 of Coprinus cinereus affects mushroom morphogenesis. Fungal Genet Biol. 2000;29(1):49–59. doi: 10.1006/fgbi.2000.1184. [DOI] [PubMed] [Google Scholar]
- 70.Douglas L.M., Martin S.W., Konopka J.B. BAR domain proteins rvs161 and rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect Immun. 2009;77(9):4150–4160. doi: 10.1128/IAI.00683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woraratanadharm T., Kmosek S., Banuett F. UmTea1, a Kelch and BAR domain-containing protein, acts at the cell cortex to regulate cell morphogenesis in the dimorphic fungus Ustilago maydis. Fungal Genet Biol. 2018;121:10–28. doi: 10.1016/j.fgb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Shioya T., Nakamura H., Ishii N., Takahashi N., Sakamoto Y., Ozaki N. The Coprinopsis cinerea septin Cc.Cdc3 is involved in stipe cell elongation. Fungal Genet Biol. 2013;58-59:80–90. doi: 10.1016/j.fgb.2013.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.