Abstract
Forsythin extracted from Forsythiae Fructus is widely used to treat fever caused by the common cold or influenza in China, Japan and Korea. The present study aimed to analyze the pharmacokinetics, metabolism and excretion routes of forsythin in humans and determine the major enzymes and transporters involved in these processes. After a single oral administration, forsythin underwent extensive metabolism via hydrolysis and further sulfation. In total, 3 of the 13 metabolites were confirmed by comparison to reference substances, i.e., aglycone M1, M1 sulfate (M2), and M1 glucuronide (M7). Hydrolysis was the initial and main metabolic pathway of the parent compound, followed by extensive sulfation to form M2 and a reduced level of glucuronidation to form M7. In addition, the plasma exposure of M2 and M7 were 86- and 4.2-fold higher than that of forsythin. Within 48 h, ~75.1% of the administered dose was found in urine, with M2 accounting for 71.6%. Further phenotyping experiments revealed that sulfotransferase 1A1 and UDP-glucuronosyltransferase 1A8 were the most active hepatic enzymes involved in the formation of M2 and M7, respectively. The in vitro kinetic study provided direct evidence that M1 showed a preference for sulfation. Sulfated conjugate M2 was identified as a specific substrate of organic anion transporter 3, which could facilitate the renal excretion of M2. Altogether, our study demonstrated that sulfation dominated the metabolism and pharmacokinetics of forsythin, while the sulfate conjugate was excreted mainly in the urine.
Keywords: forsythin, pharmacokinetics, drug metabolism, drug excretion, sulfation, glucuronidation, organic anion transporter 3
Introduction
Forsythin is a compound extracted from Forsythiae Fructus (FF, Lianqiao in Chinese) [1]. FF is widely used as an antipyretic agent in China, Japan and Korea [2]. According to the Pharmacopoeia of China 2015 Edition, there are at least 114 Chinese herbal preparations with FF as an active ingredient, including Shuanghuanglian oral solution, Niuhuang Shangqing tablet, and Yinqiao Jiedu tablet etc [3]. Forsythin belongs to a class of natural glycosidic lignan compounds [4], and can be used to treat fevers caused by the common cold or influenza. Phase II clinical trials of forsythin are currently under way in China (http://www.chinadrugtrials.org.cn/, registration number: CTR20192482), and the proposed dosing regimen is 100 or 200 mg TID.
Sulfation and glucuronidation are considered to be common and important phase II metabolic reactions mediated by sulfotransferases (SULTs) and UDP glucuronic acid transferases (UGTs). The majority of phenolic compounds are sensitive to extensive sulfation and glucuronidation. Typically, sulfation displays high affinity and low capacity for xenobiotic conjugation, while glucuronidation acts in the opposite way. For instance, with increasing dose, the percent acetaminophen that is conjugated with sulfate decreases, while that conjugated with glucuronic acid increases. In addition, UGT-mediated glucuronidation is generally considered to be the main pathway of polyphenol metabolism [5], whereas SULT-mediated sulfation is often regarded as the secondary pathway [6].
Therefore, this study aimed to (1) determine the metabolism of forsythin in humans; (2) analyze the pharmacokinetic profiles and excretion routes of forsythin and its metabolites in humans; (3) evaluate the roles of metabolizing enzymes in forsythin biotransformation; and (4) assess the roles of transporters in forsythin excretion.
Materials and methods
Chemicals
The forsythin reference substance (purity 94.9%) was supplied by the National Institute for Food and Drug Control (Beijing, China). The hydrolyzed metabolite aglycone (M1, purity 99.5%), M1 sulfate (M2, purity 99.5%), and M1 glucuronide (M7, purity 96.6%) were synthesized by Dalian Fusheng Pharmaceutical Co., Ltd (Dalian, China). The structures of these four reference substances were confirmed based on nuclear magnetic resonance (NMR) and mass spectrometry (MS) data (Supplementary file).
The internal standard teniposide (TNP) was supplied by Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). The other internal standard, C52241 (used for the quantitative analyses of urine and fecal samples), was kindly provided by Dr. Jing-shan Shen’s laboratory, Shanghai Institute of Materia Medica, Chinese Academy of Sciences. Probenecid, aclacinomycin, Hanks’ balanced salt solution (HBSS), uridine-5′-diphospho-glucuronic acid (UGDPA), and 3′-phosphoadenosine-5′-phosphosulfate (PAPS) were supplied by Sigma-Aldrich Co. (St. Louis, MO, USA). Metformin, para-aminohippurate (PAH) and estrone-3-sulfate (E3S) were supplied by the National Institute for Food and Drug Control, MCE (Monmouth Junction, NJ, USA) and TCL Pharma (Toronto, Canada). Penicillin, streptomycin and 0.05% trypsin-EDTA were obtained from Gibco (Carlsbad, CA, USA). The bicinchoninic acid (BCA) protein assay kit was purchased from Beyotime (Shanghai, China). Fetal bovine serum (FBS), poly-D-lysine-coated plates, Dulbecco’s modified Eagle’s medium (DMEM) and recombinant human UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17 isoenzymes were purchased from Corning Gentest (Woburn, MA, USA). Recombinant human SULT1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1E1, and 2A1 were supplied by Cypex (Dundee, Scotland, UK). Purified and deionized water was prepared using a Milli-Q gradient water purification system (Millipore, Molsheim, France). All other solvents and reagents were of either high-performance liquid chromatography (HPLC) or analytic grade.
Research protocol and sample collection
This was a single-center, randomized, double-blind, single ascending dose phase I study to evaluate the pharmacokinetics of metabolism of forsythin in healthy subjects. The clinical study was conducted in the Phase I Clinical Trial Unit, First Hospital of Jilin University. Ethical approval for this trial protocol was obtained from the Ethics Committee of the First Hospital of Jilin University (Changchun, China). The design of clinical research monitoring was in compliance with the Declaration of Helsinki and in accordance with the guidelines of Good Clinical Practice. Written informed consent was obtained from all the study participants before the initiation of the study procedures. Sixty-two healthy volunteers aged 18–45 were recruited. A total of six dose groups were set up, which were 50, 100, 200, 400, 600, and 800 mg of forsythin. Two additional subjects as a pilot group were enrolled before the trial in the 50 mg dose group. The blood sample collection time was adjusted based on the pharmacokinetic results of these two subjects. There were 10 subjects in each group, of which eight received the test drug and the other two received placebo; the ratio of males to females was close to 1. A summary of the demographics of the study subjects is shown in Supplementary Table S1.
After overnight fasting, eight healthy volunteers from each group were orally administered forsythin capsules in the morning (50 mg/capsule). Different groups received different doses with 240–480 mL of water (e.g., the 800 mg group received 16 capsules). Blood sample collection was performed at 0 (predose), 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 6, 8, 10, 12, 14, 16, 24, and 36 h postdose (hpd). The design of the study is shown in Supplementary Fig. S1. After 72 h of hospitalization, if forsythin is safe for the subjects, then we can increase the dose. For the excretion study of forsythin and its metabolites, urine and feces were collected in the 100 mg dose group. Urine sample collection was performed at 0 (predose), 0–4, 4–8, 8–12, 12–24, 24–48, and 48–72 hpd. Fecal sample collection was performed at 0 (predose) and 0–72 hpd. All samples were preserved at −70 °C until further analysis.
Metabolite profiling and identification
Plasma and urine sample preparation
All plasma samples (predose and 4 hpd) from three subjects of the same sex were pooled at equal volumes. Urine samples (predose and 0–48 hpd) were pooled by using the volumes in proportion to the overall volume across the time intervals. Approximately 600 µL of acetonitrile was added to 200 µL of the pooled plasma and urine samples, followed by vortexing for 1 min. Then, the mixture was centrifuged at 14,000 rpm for 5 min. Subsequently, the supernatant was transferred into a 10 mL tube and evaporated to dryness at 40 °C under a stream of nitrogen. The residue was reconstituted in 120 µL of acetonitrile/water (5:95, v/v), and the resulting solution (7 µL) was subjected to ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF MS) analysis.
Fecal sample preparation
The fecal samples were thawed and then weighed. Approximately 10 mL of methanol/water (1:1, v/v) was added to 2 g of a fecal sample, followed by blending for 10 min and sonication for 20 min. The fecal samples (predose and 0–24 hpd) from three subjects of the same sex were pooled. Then, 200 µL of the fecal homogenate was mixed with 600 µL of acetonitrile for 1 min. After centrifugation at 14,000 rpm for 5 min, the supernatant was evaporated to dryness at 40 °C under nitrogen. Finally, the residue was reconstituted in 120 μL of acetonitrile/water (5:95, v/v), and the resulting solution was subjected to UPLC/Q-TOF MS analysis.
UPLC/Q-TOF MS analysis
Chromatographic separation was performed on an Acquity UPLC HSS T3 column (1.8 μm; 100 mm × 2.1 mm) using an Acquity UPLC system (Waters, Milford, MA, USA). The mobile phase consisted of 5 mM ammonium acetate [0.01% ammonia] (A) and acetonitrile (B). The gradient elution was maintained at 5% B for 2 min, increased linearly to 60% B over 10 min and held for 2 min, further increased to 95% B over the next 1 min, and ultimately reverted back to 5% B for 2 min. The column temperature and flow rate were fixed at 40 °C and 0.4 mL/min, respectively. Fractions were collected during elution and detected by UV at 280 nm.
MS analysis was conducted with a Synapt G2-Si Q-TOF high-resolution mass spectrometer (Waters, Milford, MA, USA) in negative electrospray ionization (ESI) mode. The desolvation gas flow was optimized to 800 L/h at 450 °C, with the capillary voltage set at 3.0 kV and source temperature fixed at 120 °C. At the time of acquisition, data were centroided within the range of 50–1200 Da using 50 ng/mL leucine enkephalin infused at 10 µL/min in order to produce a reference ion at m/z 554.2615. The MSE scan function was used to simultaneously analyze both precursor and fragment ions in a single run by rapidly alternating between two independent collision energy (CE) parameters. At low CE background, the transfer and trap CEs were 2 and 5 V, respectively. At high CE background, the transfer CE was 15 V, while the trap CE ramped from 10 to 20 V. Data collection and analysis were conducted by MassLynx software version 4.1 and UNIFI 1.8.2 software (Waters, Milford, MA, USA), respectively.
Pharmacokinetics of forsythin and its metabolites in human plasma
Analytical method
The plasma samples were kept frozen at −70 °C and then analyzed during storage. The plasma concentrations of forsythin and its main metabolites M1, M2, and M7 were assessed using an LC-MS/MS method validated based on the Chinese Pharmacopoeia guidelines [7]. After removing proteins through precipitation with methanol, the analytes and internal standard teniposide were eluted using a Gemini C18 column (110 Å, 2.0 mm × 50 mm, 5 µm; Phenomenex). The mobile phase consisted of methanol-acetonitrile (2:1, v/v) and 0.01% ammonia in water with gradient elution. The total run time was 5.5 min. MS detection was performed in ESI (–) multiple reaction monitoring mode. The calibration curves were linear over the plasma concentration ranges of 1.00–1000, 0.500–500, 5.00–5000, and 2.50–2500 ng/mL for forsythin, M1, M2, and M7, respectively. The lower limits of quantification for forsythin, M1, M2, and M7 were 1.00, 0.500, 5.00, and 2.50 ng/mL, respectively.
Calculation of pharmacokinetic parameters
WinNonlin (V7.0, Pharsight, Mountain View, CA, USA) with noncompartmental analysis was employed to calculate the pharmacokinetic parameters. The maximum plasma concentration (Cmax) and the time to achieve Cmax (Tmax) were obtained directly from the pharmacokinetic data. The area under the plasma concentration–time curve (AUC) was calculated according to the linear trapezoidal with linear interpolation method to the last measurable concentration (AUC0–t). The AUC from zero to infinity (AUC0–∞) was calculated by AUC0–∞ = AUC0–t + Clast/λz (Clast, the last measurable drug concentration, and λz, the terminal elimination rate estimated by log-linear regression of plasma concentrations observed during the terminal phase of elimination). The apparent elimination half-life (t1/2) was calculated as t1/2 = ln2/λz. Other pharmacokinetic parameters included the apparent volume of distribution (VZ/F) and apparent clearance (CL/F), which was calculated by CL/F = dose/AUC0–∞. Tmax was expressed as the median with interquartile range; all other parameters are expressed as the mean ± standard deviation (SD).
Excretion of forsythin and its metabolites in human urine and feces
The urine and fecal concentrations of forsythin, M1, M2, and M7 were assessed using the validated LC-MS/MS method. C52241 was chosen as the internal standard at a concentration of 500 ng/mL. Briefly, the analytes and internal standard were separated using a BEH C18 column (1.7 µm, 50 × 2.1 mm, Waters) after methanol-based protein precipitation. The mobile phase was composed of methanol and 2 mM ammonium acetate with a gradient elution program. A total run time of 2.6 min was used for chromatographic separation at a 0.6 mL/min flow rate. The standard curves ranged from 10.0 to 5000, 10.0 to 5000, 20.0 to 10,000, and 10.0 to 5000 ng/mL for forsythin, M1, M2, and M7, respectively, in human urine, while the standard curves ranged from 25.0 to 12500, 50.0 to 25,000, 150 to 75,000, and 25.0 to 12,500 ng/g for forsythin, M1, M2, and M7, respectively, in human feces. Finally, the cumulative urinary and fecal excretion rates of the drug were calculated.
Human SULT isoenzyme phenotyping
The total volume for the incubation system was 200 µL and consisted of 10 mM PBS (pH 7.4), 250 µM PAPS, 50 µM M1, and 0.25 mg/mL human recombinant SULT isoform (SULT1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1E1, or 2A1). Before adding the recombinant SULT isoform, preincubation was conducted at 37 °C for 3 min. After 1 h of reaction, an equal amount of ice-cold acetonitrile was added to terminate the reaction. All assays were carried out in triplicate. The final concentration of organic solvent in all reactions was <0.1%. All phenotyping samples were kept at −70 °C prior to analysis.
Inhibition of the sulfation reaction of M1 in human liver cytosols (HLCs) by the SULT1A1- and SULT1E1-sensitive inhibitor quercetin and SULT inhibitor 2,6-dichloro-4-nitrophenol (DCNP) was performed. The other conditions were similar to those described above. Sulfation of M1 in inhibited samples was compared with that of the control group to determine the remaining enzyme activity.
Human UGT isoenzyme phenotyping
The total volume for the incubation system was 100 µL, containing 50 mM Tris-HCl buffer (pH 7.5), 50 µM M1, 25 µg/mL aclacinomycin, 2 mM UDPGA, and 0.5 mg/mL human recombinant UGT isoform (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, or 2B17). Before adding the recombinant UGT isoform, preincubation was conducted at 37 °C for 3 min. After 1 h of reaction, an equal amount of ice-cold acetonitrile was added to terminate the reaction. All assays were carried out in duplicate. The final concentration of organic solvent in all reactions was <0.1%. All phenotyping samples were kept at −70 °C prior to analysis.
The effect of the general UGT inhibitor flurbiprofen on the formation of M7 was also evaluated using human liver microsomes (HLMs). The other conditions were similar to those described above. Glucuronidation of M1 in inhibited samples was also compared with that of the control group.
Transporter phenotype of sulfation conjugate M2
Cell culture
Human embryonic kidney 293 (HEK293) cells transfected with human organic anion transporter 1 (OAT1), OAT3 and organic cation transporter 2 (OCT2) as well as empty-vector (mock)-transfected cells were constructed by HD Biosciences Co., Ltd. (Shanghai, China). The functional roles of these three transporters were assessed using the corresponding substrate or inhibitor. The cells were cultured in DMEM containing 10% FBS, 100 µg/mL hygromycin B, 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C, 5% CO2, and 95% humidity. Cells were dispensed at a density of 2 × 105 cells/well and grown in a 24-well plate coated with BD Biocoat poly-D-lysine. Upon reaching confluency (2 days after seeding), cell uptake experiments were carried out.
In vitro uptake experiments
Prior to the in vitro uptake experiments, the cells were washed three times with prewarmed HBSS (pH 7.4, 37 °C) and then incubated with 300 µL of HBSS at 37 °C for 10 min. After removing the pre-equilibrated HBSS solution, the uptake reaction was initiated by adding 300 µL of 50 µM M2-containing HBSS solution in the presence or absence of inhibitors. The uptake reaction was terminated by removing the solution and rinsing three times with ice-cold HBSS. Cell lysis was performed with 200 µL of deionized water after 3 freeze-thaw cycles. To minimize the potential impacts of nonspecific binding and passive uptake, mock-transfected HEK293 cells were included as controls.
Following cell lysis, the solubilized protein samples (50 µL), water/methanol (1:1, v/v; 50 µL), C52241 internal standard (500 ng/mL; 100 µL), and methanol (200 µL) were mixed and vortexed. After centrifugation at 14,000 rpm for 5 min, a 100 μL aliquot of the supernatant was diluted with 200 µL of water, and 15 µL of the resulting solution was subjected to LC-MS/MS analysis. The protein content of the cell lysate was evaluated with a BCA protein assay kit.
The uptake rates of HEK293-OAT1, HEK293-OAT3, HEK293-OCT2, and mock cells were calculated. We considered that active transport occurs when the substrate ratio of the transporter is >2 [8]. The formula used for calculation is as follows:
Among these, the unit of the uptake rate (Vuptake) is pmol·min−1·mg−1 protein; Ccell lysate is the level of drug in the cell lysate with the unit of nM; the unit of uptake time (tuptke) is minute; and the unit of protein concentration (Cprotein) is µg/mL.
Kinetic analysis of sulfation in SULT1A1*1 and glucuronidation in UGT1A8
The pre-experiment optimized the reaction times and enzyme concentrations so that metabolite formation was within the linear range and the consumption of substrate M1 did not exceed 20%. The incubation systems were the same as mentioned above (see Human SULT isoenzyme phenotyping and Human UGT isoenzyme phenotyping parts). Nonlinear regression analysis was used to calculate the kinetic parameters (Km, the Michaelis constant and Vmax, the maximum velocity) according to the Michaelis–Menten equation in GraphPad Prism software 5.0 (GraphPad Software Inc., San Diego, CA, USA).
Results
UPLC/Q-TOF MS analysis of forsythin
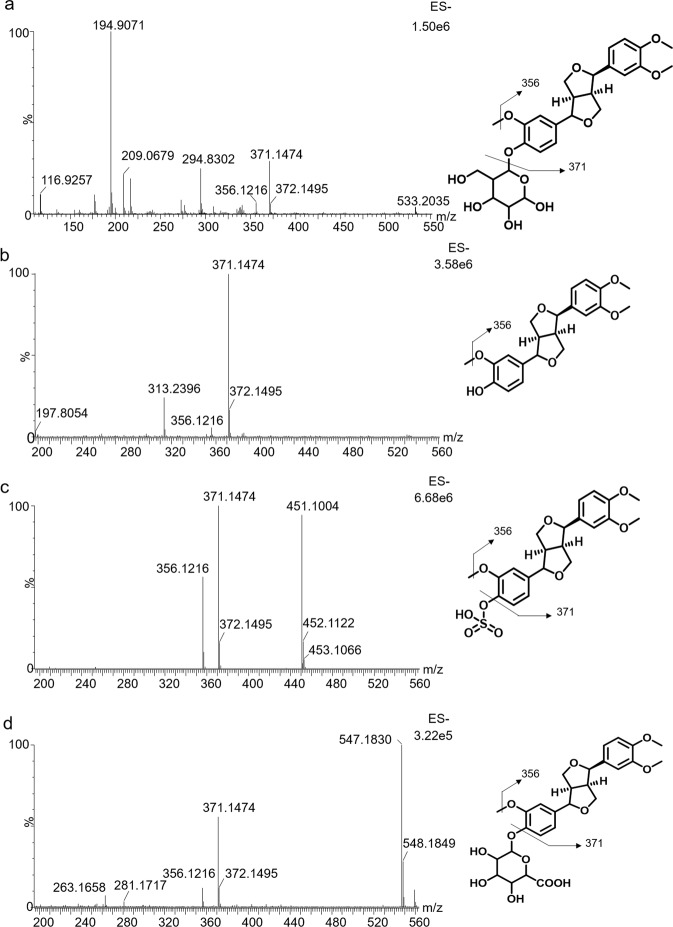
First, the MS/MS fragmentation patterns and chromatographic behaviors of forsythin were studied. The retention time of forsythin was 7.54 min under the specific chromatographic conditions. In the negative scan mode, forsythin generated a deprotonated molecular ion [M – H]– at m/z 533.2035. The product ion spectrum of forsythin at high collision energy is demonstrated in Fig. 1a. The fragment ions were detected at m/z 371.1474 (–C6H10O5) and 356.1216 (–C6H10O5, –CH3). Several tentative fragmentation patterns were speculated according to the high-resolution mass spectral data (Fig. 1a).
Fig. 1. MS2 spectrum of four substances and their proposed fragmentation pathways.
a Forsythin; b M1; c M2 and d M7.
Metabolite profiling and characterization of forsythin
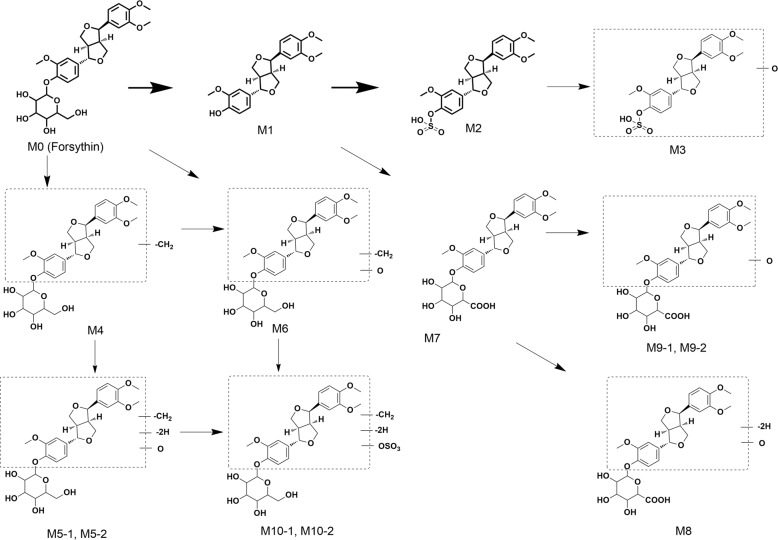
After MS data processing with MassLynx V4.1 and UNIFI V1.8.2, the plasma, urine and fecal metabolic profiles of forsythin and its metabolites from healthy volunteers were obtained (Fig. 2). A total of 4, 12, and 3 metabolites were detected from the plasma, urine and fecal samples, respectively. The identities of the three proposed metabolites were verified with reference standards (details are summarized in Table 1). The postulated metabolic pathways of forsythin are illustrated in Fig. 3. Characterization of the metabolites are as follows.
Fig. 2. Metabolic profiles of forsythin in humans (100 mg dose group).
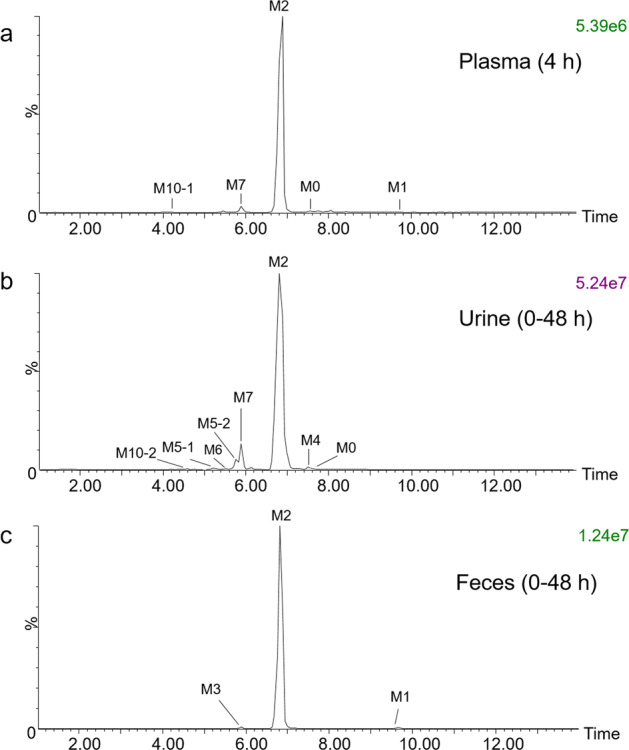
a Pooled plasma at 4 h; b pooled urine at 0–48 h; c pooled feces at 0–48 h.
Table 1.
Characterization of forsythin metabolites in humans by UPLC/Q-TOF MS.
| Metabolite | Retention time (min) | Observed mass (m/z) | Mass error (ppm) | Elemental composition | Mass area (*103) | Metabolic pathway | ||
|---|---|---|---|---|---|---|---|---|
| Plasma | Urine | Feces | ||||||
| M0 | 7.54 | 533.2031 | 0.4 | C27H34O11 | 0.390 | 6.65 | – | Parent |
| M1a | 9.66 | 371.1484 | −4.3 | C21H24O6 | 0.344 | 0.611 | 6.00 | Hydrolysis |
| M2a | 6.83 | 451.1072 | 0.8 | C21H24O9S | 543 | 3390 | 543 | M1 sulfation |
| M3 | 5.89 | 467.1012 | −1.0 | C21H24O10S | – | 34.3 | 2.09 | M1 sulfation + oxidation |
| M4 | 7.28 | 519.1891 | 3.7 | C26H32O11 | – | 0.754 | – | Demethylation |
| M5-1 | 5.23 | 533.1664 | 0.1 | C26H30O12 | – | 26.2 | – | M4 oxidation + dehydrogenation |
| M5-2 | 5.77 | 533.1666 | 0.4 | C26H30O12 | – | 109 | – | M4 oxidation + dehydrogenation |
| M6 | 5.35 | 535.1819 | −0.4 | C26H32O12 | – | 50.7 | – | M4 oxidation |
| M7a | 5.91 | 547.1819 | −0.4 | C27H32O12 | 12.7 | 154 | – | M1 glucuronidation |
| M8 | 5.68 | 561.1619 | 1.1 | C27H30O13 | – | 2.04 | – | M7 oxidation + dehydrogenation |
| M9-1 | 5.29 | 563.1769 | −0.1 | C27H32O13 | – | 2.88 | – | M7 oxidation |
| M9-2 | 5.58 | 563.1756 | −2.3 | C27H32O13 | – | 5.17 | – | M7 oxidation |
| M10-1 | 4.19 | 613.1229 | −0.5 | C26H30O15S | 4.55 | – | – | M5 sulfation |
| M10-2 | 4.57 | 613.1236 | 0.6 | C26H30O15S | – | 18.4 | – | M5 sulfation |
aConfirmed using reference standards.
Fig. 3.
Proposed metabolic pathways of forsythin in humans.
Metabolite M1
M1 eluted at 9.66 min and displayed an [M – H]– ion at m/z 371.1484. Its elemental composition was deduced to be C21H24O6. Similar to M0, M1 exhibited a fragment ion at m/z 356.1216 (Fig. 1b). The chromatographic and mass spectrometric behaviors of M1 were similar to the reference standard, indicating that M1 is an aglycone metabolite produced from the parent drug via the hydrolysis process.
Metabolite M2
The elemental composition of M2 was inferred to be C21H24O9S, suggesting the addition of SO3 to M1. Metabolite M2 exhibited an [M – H]– ion at m/z 451.1072, with an elution time of 6.83 min. The product ion spectrum of M2 at high collision energy is demonstrated in Fig. 1c. Fragment ions at m/z 371.1473 and 356.1216 were detected, which were similar to those of the parent drug. There were high similarities in the chromatographic and mass spectral behaviors of M2 with the reference aglycone sulfate conjugate, confirming that M2 is a sulfated metabolite of M1.
Metabolite M3
Metabolite M3 displayed an elemental composition of C21H24O10S, suggesting that an oxygen atom was introduced into M1. The retention time of the deprotonated ion [M – H]– at m/z 467.1012 was 5.89 min. The high-energy MS spectrum of M3 demonstrated fragment ions at m/z 387.1808 and 327.1235. The deprotonated ion [M – H]– of M3 at m/z 467.1012 exhibited a mass that was 15.9940 Da greater than that of M2 at m/z 451.1072, signifying the inclusion of an oxygen atom. Therefore, M3 was designated to be a monooxygenated metabolite of M2 with an uncertain oxidation site.
Metabolite M4
Metabolite M4 (formula: C26H32O11) exhibited a retention time of 7.28 min and was generated by the loss of CH2 from M0. The product ion scan spectrum was not collected. Thus, it was presumed that M4 was a demethylated metabolite of M0, and its demethylation site could not be determined.
Metabolite M5
Metabolite M5 displayed an elemental composition of C26H30O12. This composition was suggestive of the occurrence of +O–2H from the M4 moiety. M5-1 and M5-2 displayed highly similar ions at m/z 357.1280 and 357.1360 in the mass spectrum with high collision energy. Hence, M5 could be formed through the demethylation, monooxidation and dehydrogenation of forsythin. However, its metabolic sites remain uncertain.
Metabolite M6
The chromatographic retention time of M6 was 5.35 min, with an [M – H]– ion at m/z 535.1819. According to the accurate mass information, the formula of M6 was inferred to be C26H32O12, suggesting the addition of O onto the M4 moiety. It yielded a main fragment ion at m/z 359.1530. It could be presumed that M6 was formed through the demethylation and monooxidation of M0.
Metabolite M7
Metabolite M7 eluted at 5.91 min, which was earlier than its parent drug, suggesting increased polarity. M7 showed an [M – H]– ion at 547.1819, with an elemental composition of C27H32O12. In ESI (–), M7 displayed main fragment ions at m/z 371.1474 with the neural loss of glucuronic acid (176.0335 Da) and m/z 356.1216. The product ions of M7 were the same as those of M0. The chromatographic and mass spectrometric behaviors of M7 were similar to the reference standard, indicating that M7 is a glucuronide conjugate of M1.
Metabolite M8
Metabolite M8 displayed an elemental composition of C27H30O13 and eluted at 5.68 min. This composition was suggestive of a +O–2H modification to M7. The product ion scan spectra of M8 were not collected. Hence, M8 was postulated to be a monooxidated and dehydrogenated metabolite of M7.
Metabolite M9
Metabolite M9 with an elemental composition of C27H32O13 was indicative of a monooxygenation product of M7. The characteristic fragment ion of M9 at m/z 387.1390 was 15.9916 Da greater than that of M7 at m/z 371.1474, further supporting the incorporation of an oxygen atom. Taken together, M9-1 and M9-2 could be generated from the parent drug through the hydroxylation process.
Metabolite M10
The retention times of metabolites M10-1 (found in plasma) and M10-2 (observed in urine) were 4.19 and 4.57 min, respectively. They represented the sulfated conjugates of M5, as revealed by their elemental composition (C26H30O15S), with increased SO3 compared with M5. M10-1 exerted a main fragment ion at m/z 437.0830, while M10-2 produced a major fragment ion at m/z 533.1650 under high collision energy. The product ion of M10-1 at m/z 437.0830 was 79.955 Da (SO3) higher than that of M5-1 at m/z 371.1474. In addition, the fragment ion of M10-2 at m/z 533.1650 was produced by the loss of 79.9586 (SO3) from the precursor ion at m/z 612.1236. These fragments reveal that M10-1 and M10-2 are the sulfated conjugates of M5.
Pharmacokinetics of forsythin
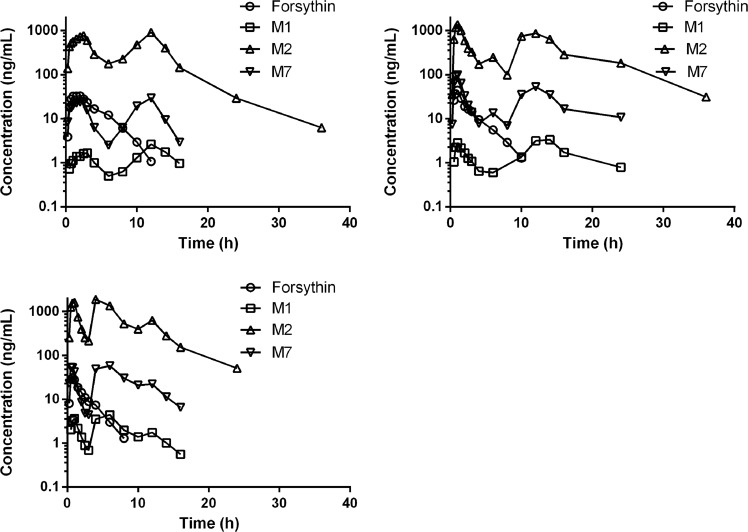
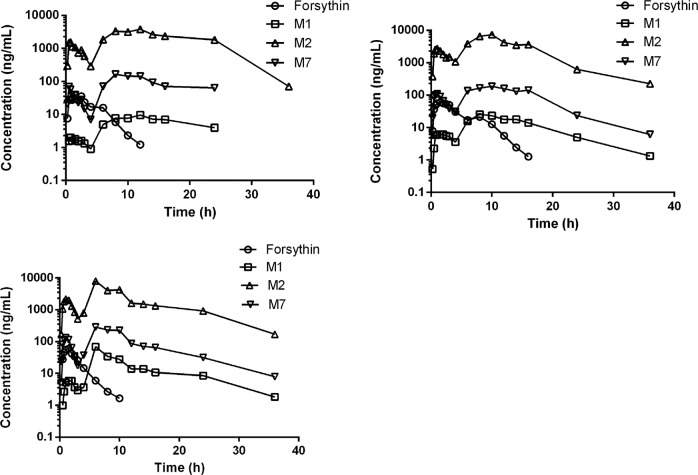
For the pharmacokinetic analysis, the plasma concentrations of forsythin, M1, M2, and M7 were detected using the abovementioned LC-MS/MS technique. Following a single oral dose administration of 100 mg of forsythin capsules, the absorption and elimination of the drug was rapid. The plasma concentration of the parent drug reached its peak at 1.25 h, with a half-life value of 1.75 h. The plasma concentration–time profiles of forsythin, M1, M2, and M7 following the administration of 100 and 800 mg forsythin capsules are shown in Figs. 4 and 5, and their respective pharmacokinetic parameters are summarized in Tables 2 and 3, respectively. The plasma concentration–time profiles of the other doses of forsythin, M1, M2, and M7 are presented in Supplementary Figs. S2–S5, and the respective plasma concentration–time profiles are presented in Supplementary Tables S2–S5. At the dose of 100 mg, the plasma concentration and exposure of M2 were the highest. Notably, the peak concentration and AUC0–∞ of M2 were 46- and 86-fold greater than those of the parent drug, respectively. In contrast, the concentration of M1 was lower, and its peak concentration was ~1/10 that of the parent drug. Nevertheless, the concentration of M7 was higher, and its peak concentration and AUC0–∞ were 2.3- and 4.2-fold higher than those of the parent drug, respectively. The mean (SD) plasma concentration–time profiles did not show the characteristics of each individual, so three typical concentration–time profiles of healthy volunteers are presented. The clearance values of forsythin were 942 and 3070 L/h at doses of 100 and 800 mg, respectively. As the dose increased, the AUC0–∞ of forsythin increased, but the increase in the AUC0–∞ of forsythin was significantly lower than the dose increase rate. The apparent clearance was calculated by CL/F = dose/AUC0–∞. Thus, the clearance values increased nonlinearly with dose. Overall, no serious adverse events were reported throughout the study. No volunteers withdrew because of adverse events.
Fig. 4.
Plasma concentration–time profiles of forsythin, M1 (aglycone), M2 (M1 sulfate), and M7 (M1 glucuronide) following administration of 100 mg forsythin capsules in three healthy volunteers.
Fig. 5.
Plasma concentration–time profiles of forsythin, M1 (aglycone), M2 (M1 sulfate), and M7 (M1 glucuronide) following administration of 800 mg forsythin capsules in three healthy volunteers.
Table 2.
Pharmacokinetic parameters of forsythin, M1 (aglycone), M2 (M1 sulfate), and M7 (M1 glucuronide) following administration of 100 mg forsythin capsules in 8 healthy subjects.
| Parameters | Forsythin | M1 | M2 | M7 |
|---|---|---|---|---|
| Cmax (ng/mL) | 31.8 ± 6.5 | 3.81 ± 1.24 | 1450 ± 792 | 72.2 ± 55.1 |
| Tmax (h) | 1.25 (0.75–3.00) | 11.0 (4.00–14.0) | 5.00 (1.00–12.0) | 6.00 (1.00–12.0) |
| Cmax1 (ng/mL) | 29.7 ± 8.23 | 1.88 ± 0.97 | 893 ± 462 | 35.7 ± 29.6 |
| Tmax1 (h) | 1.25 (0.72–2.00) | 0.75 (0.50–3.00) | 0.875 (0.25–2) | 1.25 (0.72–2.50) |
| Cmax2 (ng/mL) | 22.0 ± 13.6 | 3.30 ± 1.79 | 1203 ± 946 | 59.8 ± 58.3 |
| Tmax2 (h) | 2.50 (3.00–6.00) | 6.00 (2.50–12.0) | 6.00 (3.00–12.0) | 6.00 (2.50–12.0) |
| AUC0–t (h·ng/mL) | 111 ± 34 | 31.3 ± 11.8 | 9700 ± 1890 | 417 ± 132 |
| AUC0–∞ (h·ng/mL) | 114 ± 34 | 45.3 ± 7.39 | 9860 ± 1960 | 477 ± 127 |
| t1/2 (h) | 1.75 ± 0.26 | 4.57 ± 2.88 | 4.36 ± 1.61 | 3.32 ± 1.47 |
| Vz/F (L) | 2370 ± 731 | – | – | – |
| CL/F (L/h) | 942 ± 265 | – | – | – |
Cmax maximum observed plasma concentration, Tmax time of the maximum concentration, AUC0–t area under the concentration–time curve from time zero to the last measurable concentration, AUC0–∞ area under the concentration–time curve from 0 h to the infinite time, tmax time to the Cmax, t1/2 apparent elimination half-life, Vz/F apparent volume of distribution, CL/F apparent clearance.
Table 3.
Pharmacokinetic parameters of forsythin, M1 (aglycone), M2 (M1 sulfate), and M7 (M1 glucuronide) following administration of 800 mg forsythin capsules in 8 healthy subjects.
| Parameters | Forsythin | M1 | M2 | M7 |
|---|---|---|---|---|
| Cmax (ng/mL) | 65.9 ± 15.5 | 22.7 ± 19.8 | 5590 ± 2290 | 241 ± 89.5 |
| Tmax (h) | 1.50 (1.00–2.50) | 11.0 (6.00–12.0) | 11.0 (1.50–12.0) | 10.0 (1.50–12.0) |
| Cmax1 (ng/mL) | 59.9 ± 18.2 | 3.91 ± 1.41 | 1890 ± 562 | 102 ± 35.0 |
| Tmax1 (h) | 1.25 (0.50–2.50) | 0.875 (0.50–1.50) | 0.875 (0.50–1.50) | 0.75 (0.5–1.5) |
| Cmax2 (ng/mL) | 56.3 ± 22.7 | 19.8 ± 20.8 | 4275 ± 2965 | 235 ± 95.8 |
| Tmax2 (h) | 2.5 (1.5–8) | 7.00 (6.00–12.0) | 6.00 (2.00–12.0) | 9.00 (6.00–12.0) |
| AUC0–t (h·ng/mL) | 279 ± 80 | 222 ± 137 | 57,700 ± 19200 | 2820 ± 1140 |
| AUC0–∞ (h·ng/mL) | 283 ± 80 | 244 ± 134 | 59,400 ± 19700 | 3050 ± 1060 |
| t1/2 (h) | 1.83 ± 0.24 | 7.00 ± 2.29 | 5.74 ± 2.31 | 6.6 ± 3.55 |
| Vz/F (L) | 7920 ± 2110 | – | – | – |
| CL/F (L/h) | 3070 ± 1020 | – | – | – |
Urinary and fecal excretion of forsythin
A wide variety of metabolites, including sulfate conjugate, were detected in both urine and fecal samples, and the total urinary excretion accounted for 75.1% of the administered dose. The cumulative urinary excretion percentages of forsythin, M1, M2, and M7 in 8 healthy volunteers administered 100 mg of forsythin are shown in Fig. 6. The cumulative excretion percentages of forsythin, M1, M2, and M7 in urine samples were 0.752%, 0.00416%, 71.6%, and 2.79%, respectively, while those of forsythin, M1, M2, and M7 in fecal samples were 0, 0.161%, 0.651%, and 0.00502%, respectively. Therefore, the sulfated conjugate M2 was excreted mainly through the kidneys, accounting for 71.6% of the administered dose, while the other related substances accounted for ~3%.
Fig. 6.
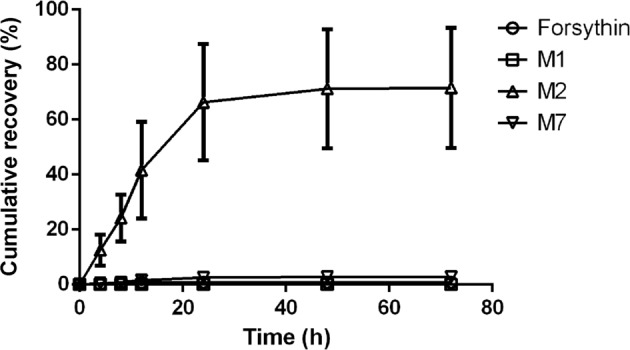
Percentage of cumulative excretion of forsythin, M1 (aglycone), M2 (M1 sulfate), and M7 (M1 glucuronide) in urine after oral administration of 100 mg of forsythin capsules in 8 healthy subjects.
Identification of the SULT isoenzyme responsible for the sulfation of M1
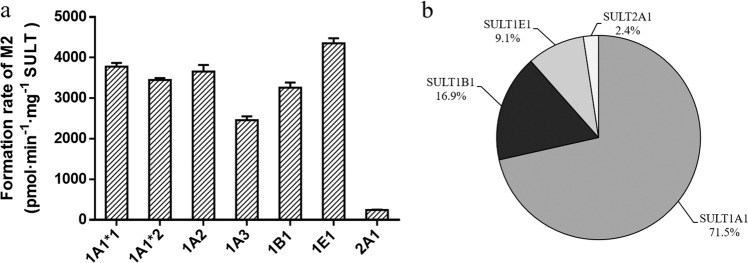
To characterize the SULT isoforms associated with M2 formation, the activities of SULT1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1E1, and 2A1 were assessed. It was found that all 7 SULT isoforms could affect the yield of M2 (Fig. 7a). The formation rates of M2 ranged from 244 to 4351 pmol·min−1·mg−1 protein, in descending order as follows: SULT1E1 > 1A1*1 ≈ 1A2 ≈ 1A1*2 > 1B1 > 1A3 > 2A1 (Fig. 7a). After normalizing the rates of M2 formation to the native contents of the SULT isoforms [9], sulfation of M2 was found to be primarily catalyzed by SULT1A1 (Fig. 7b), followed by 1B1, 1E1, and 2A1. The SULT inhibitors DCNP (1 µM), DCNP (10 µM), and quercetin (25 µM) suppressed M2 formation in HLCs by 72.1%, 87.6%, and 94.9%, respectively (Supplementary Fig. S6).
Fig. 7. Identification of the SULT isoenzyme responsible for the sulfation of M1.
a Formation rate of M2 (M1 sulfate) in the incubation of M1 (50 μM) with 7 human recombinant SULTs (0.25 mg protein/mL) supplemented with PAPS (250 μM). b Contribution of each SULT isoform to the sulfation of M1 after normalization with regard to their native abundance in human liver cytosolic fraction.
Identification of the UGT isoenzyme involved in glucuronidation of M1
To determine the main UGT isoenzymes involved in the glucuronidation of M1 to M7, human UGT phenotyping experiments were carried out. We investigated the formation of M7 by 12 recombinant human UGT isozymes, namely, UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17. As shown in Fig. 8, all 12 UGT recombinant enzymes could catalyze the transformation of M1 to M7. In the incubation systems of UGT1A4, 1A6, 2B4, 2B7, and 2B17, the formation rates of M1 glucuronide conjugates were all <1.0 pmol·min−1·mg−1 protein. In the other UGT isoenzyme incubation systems, the formation rates ranged from 3.71 to 149 pmol·min−1·mg−1 protein, in descending order as follows: UGT1A8 > 1A10 > 1A3 ≈ 2B15 > 1A9 ≈ 1A7 ≈ 1A1. As a result, the principal enzyme that contributed to the formation of glucuronide conjugate M7 was identified to be UGT1A8. Inhibition of the glucuronidation of M1 in HLMs by flurbiprofen was performed. The UGT inhibitor flurbiprofen (1 mM) suppressed M7 formation in HLMs by 75.4% (Supplementary Fig. S7).
Fig. 8.
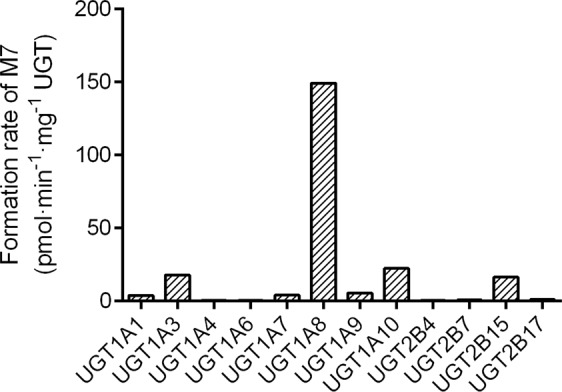
Formation rate of M7 (M1 glucuronide) in the incubation of M1 (50 μM) with 12 human recombinant UGTs (0.5 mg protein/mL) supplemented with UDPGA (2 mM).
Uptake of sulfated conjugate M2 by transfected HEK293 cells
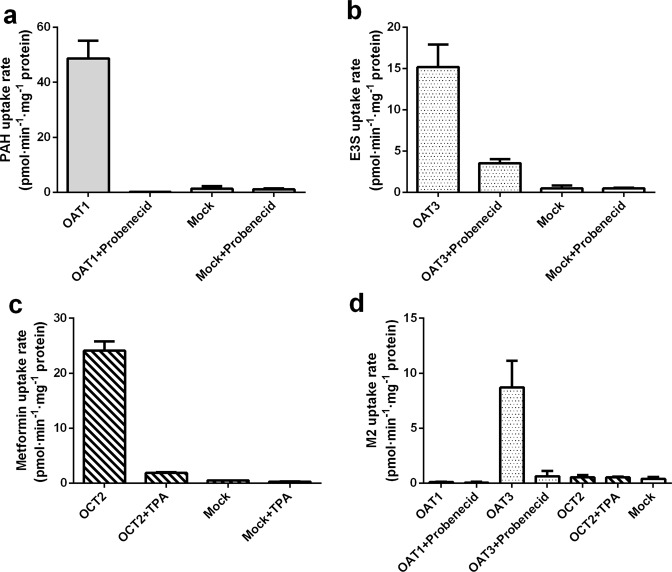
To evaluate the functions of OAT1, OAT3, and OCT2, an initial study was performed to compare the differences in positive substrates (PAH for OAT1, E3S for OAT3, and metformin for OCT2) in OAT1-, OAT3-, OCT2-, or mock-transfected HEK293 cells. Notably, the uptake rates of positive substrates in transporter-transfected HEK293 cells were all greater than those in mock-transfected HEK293 cells (Fig. 9). Moreover, OAT1-, OAT3-, and OCT2-mediated uptake of PAH, E3S, and metformin, respectively, was remarkably inhibited by 200 µM probenecid (OAT inhibitor) or 200 µM TPA (OCT2 inhibitor).
Fig. 9. Uptake of standard substrates and test compound in the absence or presence of specific transporter inhibitors in transporter-transfected and mock HEK293 cells.
Substrates used. a PAH; b E3S; c metformin; d aglycone sulfate conjugate M2 (M1 sulfate).
To elucidate the roles of renal uptake transporters during sulfate conjugate M2 elimination, the activities of the three transporters (i.e., OAT1, OAT3, and OCT2) were determined. As presented in Fig. 9, the uptake rate of M2 by OAT1 or OCT2 was relatively close to that of the mock cells (the ratio was <2), suggesting that M2 is not a substrate of OAT1 or OCT2. The uptake rate of M2 by OAT3 was 22.1 times that of the mock-transfected cells, and this difference was statistically significant. In addition, probenecid decreased the uptake rate of M2 by OAT3 by 90%, implying that M2 is an active substrate for OAT3. Nevertheless, the uptake rates of M2 were not markedly altered by other inhibitors, further showing that OAT1 and OCT2 are not responsible for M2 uptake.
Kinetic analysis of sulfation and glucuronidation
The kinetic analysis of sulfation and glucuronidation of M1 both met with the Michaelis–Menten kinetics (Supplementary Fig. S8). The Km and Vmax values are shown in Table 4. Sulfation of M1 by SULT1A1*1 showed a markedly higher Vmax value with a lower Km value than M1 glucuronidation. The Vmax/Km value of sulfation of M1 was 1400-fold higher than that of M1 glucuronidation. The in vitro kinetic study provided direct evidence that M1 showed a preference for sulfation in vitro.
Table 4.
Kinetic parameters of M1 sulaftion in human recombinant SULT1A1*1 and M1 glucuronidation in human recombinant UGT1A8.
| Incubation system | SULT1A1*1 | UGT1A8 |
|---|---|---|
| Protein concentration (mg protein/mL) | 0.0250 | 0.100 |
| Incubation time (min) | 15.0 | 60.0 |
| Concentration range (µM) | 0.200–20.0 | 2.00–250 |
| Km (µM) | 3.68 | 63.6 |
| Vmax (pmol·min−1·mg−1 protein) | 44,391 | 548 |
| Vmax/Km (µL·min−1·mg−1 protein) | 12,062 | 8.61 |
Discussion
In this study, we evaluated the pharmacokinetics, metabolism, and excretion routes of forsythin. We also evaluated the roles of metabolizing enzymes involved in the metabolism of aglycone M1. Our results demonstrated that forsythin could undergo extensive metabolism in humans and generated a total of 13 metabolites in plasma, urine and feces. However, compared with M2, the contents of the other metabolites were all lower than 10%, further suggesting that M2 was the principal metabolite of forsythin. Based on available pharmacodynamic data, both M1 and M2 were found to be active. The activity of M2 is higher than that of M1 but lower than that of forsythin. It is worth noting that enterohepatic circulation can occur through intestinal reabsorption and biliary excretion, often accompanied by intestinal deconjugation and hepatic conjugation. In general, it may prolong the pharmacologic actions of specific drugs and drug metabolites [10]. In this study, as shown in Figs. 4 and 5, enterohepatic recycling was observed in the plasma concentration–time profiles of M1, M2, and M7. At the other doses, enterohepatic recycling was also demonstrated (Supplementary Figs. S2–S5). We inferred that after M1 was reabsorbed by the small intestine, it was quickly sulfated to form M2 since the small intestine contains a large amount of SULTs, with SULT1B1 (36%) and SULT1A3 (31%) being the major enzymes present [9]. The liver is not the only organ containing UGTs, and the distribution of UGTs in each tissue is different. Thus, a second peak on the metabolite curves was observed. In addition, the half-life values of M2 and M7 were ~4–6 h, which were longer than those of the parent drug.
Glucuronidation is a pivotal metabolic pathway of some drugs and endogenous substances in humans. Glucuronidation of phenols usually occurs with nucleophilic groups such as hydroxyl groups [11]. In addition, numerous drugs (e.g., morphine) and flavonoid glycosides (e.g., puerarin) directly undergo glucuronidation [12–14].
Similar to glucuronidation, sulfation is also a common phase II reaction that occurs during the activation and detoxification of various exogenous and endogenous compounds in humans [15, 16]. Typically, sulfation displays a high affinity and low capacity for xenobiotic conjugation, while glucuronidation acts in the opposite way. Acetaminophen is a xenobiotic substrate for both SULTs and UGTs. The sulfation and glucuronidation abilities of acetaminophen are dose-dependent. Acetaminophen sulfate is the principal conjugate metabolized by SULTs at low doses. With an increasing dose of acetaminophen, the percent conjugated with sulfate decreased, while that conjugated with glucuronic acid increased. However, considering the substrate inhibition of SULT, the absolute amount of the sulfate conjugate can decrease at high doses. This phenomenon has also been observed in the Caco-2 model. The excretion of apigenin glucuronide at high concentrations (>25 µM) has been reported, while that of the sulfate conjugate is more significant at low concentrations [17, 18]. However, in our study, the sulfate conjugate M2 constituted the principal circulating metabolite rather than glucuronide conjugate M7, and M2 dominated the pharmacokinetics, metabolism, and excretion of forsythin within the 50–800 mg dosing range.
According to the pharmacokinetic results, M2 was more easily generated than M7. To verify this conjecture and characterize the phenotypes of the major metabolizing enzymes, further experiments were carried out. In vitro phenotyping experiments revealed that SULT1A1*1, 1A1*2, 1A2, 1A3, 1B1, 1E1, and 2A1 could all mediate the transformation of M1 to M2, of which SULT1A1 represented the main subtype for M1 sulfation. In addition, we assessed the roles of UGT during M7 formation at the same substrate concentration (50 µM) because the exposure of M7 was greater than that of the parent drug. Our findings indicated that the glucuronidation of M1 was catalyzed mainly by UGT1A8, and its formation rate was 149 pmol·min−1·mg−1 protein. Although all 12 UGT subtypes could mediate the transformation of M1 to M7, the formation rate of M1 to M7 mediated by UGT was much lower than that of M1 to M2 mediated by SULT. This indicated that M1 preferred sulfation to glucuronidation, which also explained to some extent the large amounts of M2 that were detected instead of M7. In addition, the in vitro kinetic study showed that M1 had greater affinity for SULT1A1 than UGT1A8, providing direct evidence that M1 more easily generated M2 than M7 in vitro. One volunteer was orally administered 100 mg of aglycon M1. The main drug-related component in his plasma was sulfate-conjugated M2 instead of M7, further confirming that whether the concentration of aglycon M1 was low or high, sulfation played a dominant role in the phase II reactions of M1. Thus, after oral administration or injection of Shuanghuanglian, the main component in the human body will be totally different. After oral administration of Shuanghuanglian, forsythin undergoes first-pass metabolism mainly via SULT1A1, leading to M2 as the main drug-related component in systemic circulation. Our previous work has shown that the human plasma protein binding rates of forsythin, M2, and M7 are 69.2%, 79.4%, and 94.7%, respectively.
Human SULT isoenzyme phenotyping experiments revealed that SULT1A1 significantly contributed to the formation of M2. There are three common genetic variants in the SULT1A1 gene: SULT1A1*1, 1A1*2, and 1A1*3 [19, 20]. Although the activity of SULT1A1*2 is 40% lower than that of SULT1A1*1 for the sulfation of troglitazone [21], relatively similar activity levels of SULT1A1*1 and 1A1*2 were noted during M2 formation (Fig. 7a). Nonetheless, the activity of SULT1A1*3 was not examined due to its commercial unavailability.
Considering that the sulfate conjugate M2 is a hydrophilic compound with a clogD value of −2.41, which was calculated using ACD software version 14.0 (ACD/Labs, Advanced Chemistry Development, Inc., Toronto, ON, Canada), it does not readily pass through biological membranes via passive diffusion. In addition, ~75.1% of the dose was recovered in urine, with M2 accounting for 71.6% of the dose. Therefore, we further examined the uptake of M2 by the transporters OAT1, OAT3, and OCT2. Members of the OAT family, particularly OAT1 and OAT3, are responsible for the renal transport of organic anions. Members of the OCT family are involved in the transport of various xenobiotics as well as endogenous organic cations [8]. In this study, the uptake rate of M2 by OAT1 and OCT2 was less than 2-fold, suggesting that M2 may not be transported by OAT1 and OCT2. Notably, the uptake rate of M2 was markedly higher in HEK293 cells transfected with OAT3 than in mock-transfected cells. The findings of OAT inhibition with probenecid further verified that OAT3 was the principal transporter of M7. We also carried out uptake experiments of M7 by transfected HEK293 cells. The results showed that M7 is also a substrate of OAT3. Secretory transporters are also expressed on the apical membrane, such as multidrug and toxin extrusion transporter 1 (MATE1), breast cancer resistance proteins (BCRP), and multidrug resistance associated proteins 2 and 4 (MRP2 and MRP4) [22]. Further studies should therefore be conducted.
Clinically, many drugs are substrates for OAT1 and/or OAT3. In patients with chronic renal insufficiency, these drugs can exhibit certain pharmacokinetic changes, such as decreased renal clearance and increased plasma exposure, due to the obstruction of renal excretion. In a chronic renal insufficiency state, the expression or function of transporters may be altered [23]. For instance, the sulfated conjugate of morinidazole is a sensitive substrate of OAT1 and OAT3, whereas the glucuronides are limited to OAT3 only. In patients with chronic kidney failure, the three conjugates resulted in a 15-fold increase in total plasma exposure to the drug. Moreover, the conjugates may exert pharmacologic or adverse effects [24]. For tenofovir, its plasma exposure increased by 8.5-fold in patients with end-stage renal disease [25].
In conclusion, our study reveals that forsythin is well absorbed and extensively metabolized following oral administration, and the sulfate conjugate M2 is the main metabolite in plasma. Moreover, renal excretion is the principal excretion pathway for M2, and M2 is a substrate for OAT3. In addition, SULT1A1 predominantly mediates the sulfation of M1, while UGT1A8 is the primary enzyme involved in glucuronidation. The rate of M1 sulfation was much higher than that of glucuronidation. The significantly greater exposure and plasma concentration of M2 compared with the parent drug indicate that severe renal impairment patients may be subjected to a remarkable increase in the plasma concentration of M2 compared with healthy subjects. Therefore, the safety of forsythin in renally impaired patients should be placed under close monitoring.
Supplementary information
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 81521005 and 81903701) and the National Key Research Project of the Chinese Academy of Sciences (grant no. XDA12050306).
Author contributions
LLP, XXD, LF, and DFZ participated in research design. LLP and YY conducted experiments. MH, SW, and LF contributed new reagents or analytic tools. LLP and YY performed data analysis. YHD, CYL, and HZ participated in clinical trials. LLP, XXD, and DFZ contributed to the writing of the paper.
Competing interests
The authors declare no competing interests. The authors alone are responsible for the content and writing of this article.
Contributor Information
Yan-hua Ding, Email: dingyanhua2003@126.com.
Li Fu, Email: 2729677386@qq.com.
Xing-xing Diao, Email: xxdiao@simm.ac.cn.
Da-fang Zhong, Email: dfzhong@simm.ac.cn.
Supplementary information
The online version of this article (10.1038/s41401-020-0481-8) contains supplementary material, which is available to authorized users.
References
- 1.Wang Z, Xia Q, Liu X, Liu W, Huang W, Mei X, et al. Phytochemistry, pharmacology, quality control and future research of forsythia suspensa (Thunb.) Vahl: a review. J Ethnopharmacol. 2018;210:318–39.. doi: 10.1016/j.jep.2017.08.040. [DOI] [PubMed] [Google Scholar]
- 2.Ozaki Y, Rui J, Tang Y, Satake M. Antiinflammatory effect of forsythia suspensa vahl and its active fraction. Biol Pharm Bull. 1997;20:861–64. doi: 10.1248/bpb.20.861. [DOI] [PubMed] [Google Scholar]
- 3.Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China. China Medical Science Press; 2015.
- 4.Han Z, Lei XL, Zhang H, Liu L, Chen ZS, Yang W, et al. Evaluating the safety of forsythin from forsythia suspensa leaves by acute and sub-chronic oral administration in rodent models. Asian Pac J Trop Med. 2017;10:47–51. doi: 10.1016/j.apjtm.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45:1121–32. doi: 10.1016/j.biocel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Suiko M, Kurogi K, Hashiguchi T, Sakakibara Y, Liu MC. Updated perspectives on the cytosolic sulfotransferases (SULTs) and SULT-mediated sulfation. Biosci Biotechnol Biochem. 2017;81:63–72. doi: 10.1080/09168451.2016.1222266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinese Pharmacopoeia Commission. Guidance for bioanalytical method validation, Pharmacopoeia of the People’s Republic of China. China Med Sci Press. 2015;4:363–8. [Google Scholar]
- 8.Zhong K, Li XL, Xie C, Zhang YF, Zhong DF, Chen XY. Effects of renal impairment on the pharmacokinetics of morinidazole: uptake transporter-mediated renal clearance of the conjugated metabolites. Antimicrob Agents Chemother. 2014;58:4153–61. doi: 10.1128/AAC.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos. 2009;37:2255–61. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohpatic circulation. Clin Pharmacokinet. 2002;41:751–90. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 11.Wu B, Kulkarni K, Basu S, Zhang S, Hu M. First-pass metabolism via UDP-glucuronosyltransferase: a barrier to oral bioavailability of phenolics. J Pharm Sci. 2011;100:3655–81. doi: 10.1002/jps.22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang G, Ge S, Singh R, Basu S, Shatzer K, Zen M, et al. Glucuronidation: driving factors and their impact on glucuronide disposition. Drug Metab Rev. 2017;49:105–38.. doi: 10.1080/03602532.2017.1293682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen G, Christrup L, Sjøgren P. Among morphine metabolism, pain and side effects during long-term treatment: an update. J Pain Symptom Manag. 2003;25:74–91. doi: 10.1016/S0885-3924(02)00531-6. [DOI] [PubMed] [Google Scholar]
- 14.Luo CF, Cai B, Hou N, Yuan M, Liu SM, Ji H, et al. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for puerarin metabolism in human liver microsomes. Arch Toxicol. 2012;86:1681–90. doi: 10.1007/s00204-012-0874-7. [DOI] [PubMed] [Google Scholar]
- 15.Tian X, Huo X, Dong P, Wu B, Wang X, Wang C, et al. Sulfation of melatonin: enzymatic characterization, differences of organs, species and genders, and bioactivity variation. Biochem Pharmacol. 2015;94:282–96. doi: 10.1016/j.bcp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Barbosa ACS, Feng Y, Yu C, Huang M, Xie W. Estrogen sulfotransferase in the metabolism of estrogenic drugs and in the pathogenesis of diseases. Expert Opin Drug Metab Toxicol. 2019;15:329–39.. doi: 10.1080/17425255.2019.1588884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Hu M. Absorption and metabolism of flavonoids in the Caco-2 cell culture model and a perused rat intestinal model. Drug Metab Dispos. 2002;30:370. doi: 10.1124/dmd.30.4.370. [DOI] [PubMed] [Google Scholar]
- 18.Hu M, Chen J, Lin H. Metabolism of flavonoids via enteric recycling: mechanistic studies of disposition of apigenin in the Caco-2 cell culture model. J Pharmacol Exp Ther. 2003;307:314–21. doi: 10.1124/jpet.103.053496. [DOI] [PubMed] [Google Scholar]
- 19.J Carlini E, B Raftogianis R, Wood T, Jin F, Zheng W, Rebbeck T, et al. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics. 2001;11:57–68. doi: 10.1097/00008571-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Rasool MI, Bairam AF, Gohal SA, El Daibani AA, Alherz FA, Abunnaja MS, et al. Effects of the human SULT1A1 polymorphisms on the sulfation of acetaminophen, O-desmethylnaproxen, and tapentadol. Pharmacol Rep. 2019;71:257–65.. doi: 10.1016/j.pharep.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honma W, Shimada M, Sasano H, Ozawa S, Miyata M, Nagata K, et al. Phenol sulfotransferase, ST1A3, as the main enzyme catalyzing sulfation of troglitazone in human liver. Drug Metab Dispos. 2002;30:944–49. doi: 10.1124/dmd.30.8.944. [DOI] [PubMed] [Google Scholar]
- 22.Morrissey KM, Stocker SL, Wittwer MB, Xu L, Giacomini KM. Renal transporters in drug development. Annu Rev Pharmacol Toxicol. 2013;53:503–29. doi: 10.1146/annurev-pharmtox-011112-140317. [DOI] [PubMed] [Google Scholar]
- 23.Schwenk MH, Pai AB. Drug transporter function-implications in CKD. Adv Chronic Kidney Dis. 2016;23:76–81. doi: 10.1053/j.ackd.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Kong FD, Pang XY, Zhong K, Guo ZT, Li XL, Zhong DF, et al. Increased plasma exposures of conjugated metabolites of morinidazole in renal failure patients: a critical role of uremic toxins. Drug Metab Dispos. 2017;45:593–603. doi: 10.1124/dmd.116.074492. [DOI] [PubMed] [Google Scholar]
- 25.Kearney BP, Yale K, Shah J, Zhong L, Flaherty JF. Pharmacokinetics and dosing recommendations of tenofovir disoproxil fumarate in hepatic or renal impairment. Clin Pharmacokinet. 2006;45:1115–24. doi: 10.2165/00003088-200645110-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.