Abstract
Mechanics are intrinsic properties which appears throughout the formation, development, and aging processes of biological systems. Mechanics have been shown to play important roles in regulating the development and metastasis of tumors, and understanding tumor mechanics has emerged as a promising way to reveal the underlying mechanisms guiding tumor behaviors. In particular, tumors are highly complex diseases associated with multifaceted factors, including alterations in cancerous cells, tissues, and organs as well as microenvironmental cues, indicating that investigating tumor mechanics on multiple levels is significantly helpful for comprehensively understanding the effects of mechanics on tumor progression. Recently, diverse techniques have been developed for probing the mechanics of tumors, among which atomic force microscopy (AFM) has appeared as an excellent platform enabling simultaneously characterizing the structures and mechanical properties of living biological systems ranging from individual molecules and cells to tissue samples with unprecedented spatiotemporal resolution, offering novel possibilities for understanding tumor physics and contributing much to the studies of cancer. In this review, we survey the recent progress that has been achieved with the use of AFM for revealing micro/nanoscale mechanics in tumor development and metastasis. Challenges and future progress are also discussed.
Keywords: atomic force microscopy, tumor mechanics, cancerous cell, tumor microenvironment, exosome, extracellular matrix
Introduction
Knowledge of tumor metastasis is crucial for understanding the biology of cancers. It has been widely reported that cancer is a major threat to human health worldwide. According to the estimates from the World Health Organization, in 2015, cancer was the first or second leading cause of death for people younger than 70 years old in 91 of 172 countries, and it ranked third or fourth in an additional 22 countries [1]. Recently released statistics by the American Cancer Society has shown that the cancer mortality rate in the United States fell continuously from 1991 to 2017, resulting in an overall decline of 29% [2]. Statistics from the European Union published in recent years also showed the steady downward trend of the cancer death rate, including lung cancer [3], colorectal cancer [4], and breast cancer [5]. For China, researchers have estimated 4,292,000 new cancer cases and 2,814,000 cancer deaths in 2015, and the statistics have shown that the mortality rates since 2006 have decreased significantly for both males (−1.4% per year) and females (−1.1% per year) [6]. Nevertheless, cancer incidence and mortality are rapidly growing worldwide [1]. Notably, metastasis, the process of cancer cells spreading from the primary tumor to surrounding tissues and to distant organs, is responsible for ~90% of cancer deaths [7], a finding supported by a recent statistical study [8]. In cancer patients, the metastasis of tumors to tissues such as sentinel lymph nodes predicts disease progression and often guides treatment decisions [9]. Scientists have identified many processes and factors that contribute to tumor metastasis, including the epithelial–mesenchymal transition (EMT) [10], tumor microenvironment [11], inflammation [12], proteoglycan remodeling [13], and exosomes [14]. However, the biological underpinnings of tumor metastasis remain the least understood aspect of cancer biology, and elucidating the underlying mechanisms driving metastasis is essential to the development of new approaches and therapies for metastatic cancers [15].
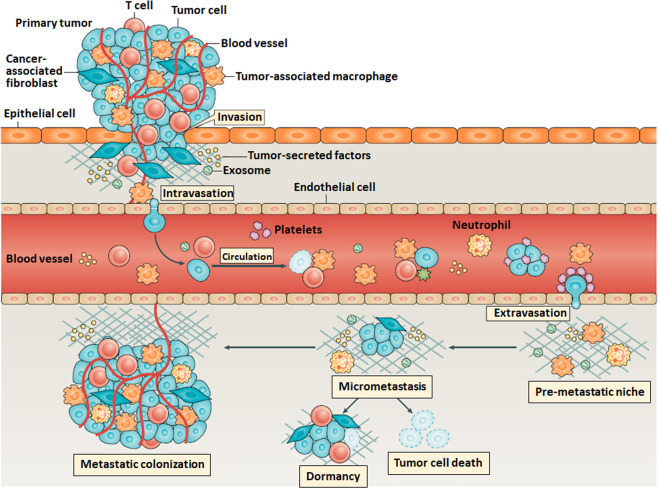
Investigations into the mechanics of tumors have emerged as promising ways to discover the mysteries of tumor metastasis. Mechanics are intrinsic properties that appear throughout the formation, development, and aging processes of biological systems [16–18]. In fact, mechanical force is significantly involved in life activities at nearly all levels, from molecules and cells to tissues and organs [19]. Studies have shown that during biological processes such as embryogenesis and morphogenesis, chemical signaling is insufficient to fully describe how systems grow, and mechanics have been shown to influence spatiotemporal control of the transcriptional activities essential for biological development [20]. Alterations in the biomechanical and biophysical properties of cells and subcellular structures influence and are influenced by the onset and progression of human diseases such as cancer [21]. A cell undergoes many genetic and epigenetic changes as it transitions to malignancy, and cancer biologists have recognized that a critical component of this transformational journey involves marked alterations in the mechanical phenotype of the cell and its surrounding microenvironment [22]. The physical characteristics of tumors are intricately linked to the tumor phenotype and difficulties during treatment [23]. The successful metastasis of primary cancerous cells is the result of comprehensive effects, including the cancer cells themselves and microenvironmental cues, which significantly exhibit mechanical alterations in cancerous cells and tumor microenvironments [24], as shown in Fig. 1. For example, the physical forces generated by growing tumors compress blood and lymphatic vessels, thereby reducing perfusion rates and generating hypoxia [25]. Many factors contribute to the increased stiffness of tumors (e.g., increased matrix deposition, matrix remodeling by forces from cancer cells and stomal fibroblasts, matrix cross-linking, increased cellularity, and the buildup of both solid and interstitial pressure [23]), which recirculate as feedback to promote the migration and invasion of primary cancerous cells [26] and eventually increase tumor invasiveness and reduce therapy efficacy. Consequently, investigating the mechanical cues involved in tumor metastasis contributes to a better understanding of tumor behaviors and probably offers novel therapeutic avenues.
Fig. 1. Schematic illustration of the detailed process of tumor metastasis showing that there are significant physical processes in the process of tumor metastasis and multiple types of microenvironmental cues are involved for promoting the successful metastasis of primary cancerous cells.
Reprinted with permission from Ref. [24]. Copyright 2019 Springer Nature. Tumor metastasis is a complex multistep process. Tumor cell production of angiogenetic factors and TGFβ can activate endothelial cells and fibroblasts to remodel tissues and promote tumor cell invasion of stromal-modified spaces. Intravasation of tumor cells is promoted by binding to macrophages that cause transient permeability in the vasculature. In the circulation, platelets can bind to the circulating tumor cells (CTCs) and protect CTCs from cytotoxic immune cell recognition, escorting tumor cells to the site of extravasation [33]. Preferred colonization sites, termed premetastatic niches, can be prepared in advance of the arrival of tumor cells through the actions of extracellular vesicles such as exosomes. When tumor cells arrive the new sites, only a small subset of tumor cells initiate cell division to form micrometastases, and only a small proportion of these micrometastases persist to become vascularized metastases [34].
In this review, we summarize the applications of atomic force microscopy (AFM), a powerful multifunctional toolbox invented for characterizing living biological systems in their native states with unprecedented spatiotemporal resolution (nanometer spatial resolution and millisecond temporal resolution) under aqueous conditions [27], to investigations of multiple types of micro/nanoscale mechanics, ranging from single cells to microenvironmental cues involved in tumor metastasis. Compared with other high-resolution imaging techniques (as shown in Table 1), such as super-resolution microscopy (stimulated emission depletion (STED), photoactivated localization microscopy (PALM), and stochastic optical reconstruction microscopy (STORM)), scanning electron microscopy (EM), and transmission EM, AFM has unique advantages for simultaneously visualizing the structures and measuring the mechanical properties of living biological specimens under aqueous conditions, making AFM particularly suited for the studies of biointerfaces and mechanobiology [28–32]. AFM has become an important technology for life science research and has been widely used to investigate biological issues in metastasis to reveal novel insights into tumor biology. In the following sections, we survey the recent advances regarding the applications of AFM used to probe the mechanical issues associated with the process of tumor metastasis [24, 33, 34] and illustrate with examples highlighting the exciting capabilities of AFM in addressing fundamental sciences in tumor biology.
Table 1.
Comparison of high-resolution imaging techniques for applications in molecular and cell biology.
| Characteristics | Atomic force microscopy (AFM) | Super-resolution optical microscopy (STED, PALM, and STORM) | Scanning electron microscopy (SEM) | Transmission electron microscopy (TEM) |
|---|---|---|---|---|
| Spatial resolution | 1–50 nm | 20–50 nm | 2–10 nm | 0.2–10 nm |
| Specimen preparations | Living samples immobilized on support | Fluorescent labeling | Dehydrated and gilded samples on metal stubs | Dehydrated or vitrified samples on grids |
| Advantages | Topographical imaging and mechanical measurements under native conditions without pretreatments (e.g., fixation, lysis, labeling, or staining) | Observing three-dimensional cellular structures and molecular activities in life processes with high spatiotemporal resolution | Imaging surfaces of tissues, cells, and interfaces at nanometer resolution | Visualizing atomic structures of proteins and molecular-resolution structures within cells |
| Limitations | Restricted to the surface of samples and cannot access subsurface structures | Fluorescent labeling possibly affects the cellular and molecular behaviors | No life processes | No life processes |
Principles of atomic force microscopy
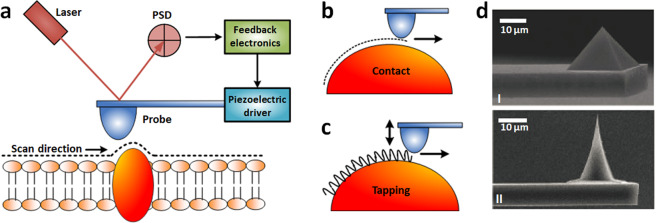
The imaging principle of AFM is illustrated in Fig. 2. AFM uses a sharp tip mounted at the end of a microcantilever (Fig. 2d) to raster scan the specimens [35]. A four-quadrant position sensitive detector (PSD) is used to sense the laser beam reflected from the backside of the cantilever to detect the deformation of the cantilever [36]. The changes in PSD signals are analyzed by feedback electronics, which then control the piezoelectric tube driver to move the AFM probe vertically to sense the interaction forces between the AFM tip and specimen surface (Fig. 2a). Contact mode and tapping mode are two classic AFM imaging modes [28]. In contact mode, the AFM tip is scanned across the surface, and the Coulomb repulsive forces between the AFM tip atoms and specimen surface atoms cause deflections of the AFM cantilever. The piezoelectric tube drives the AFM probe to move vertically to keep the deflections of the cantilever constant and thus maintain the invariable interaction forces (Fig. 2b). Contact mode imaging is based on the repulsive forces that are localized (the repulsive forces between AFM tip atoms and target atoms are not susceptible to forces from the neighboring atoms on the specimen), allowing high spatial resolution imaging [36]. However, the drawback of the contact mode is the lateral mechanical scratch exerted on the sample, which makes it difficult to observe the samples adsorbed loosely onto the substrate. In tapping mode, the cantilever oscillates near its resonant frequency and intermittently touches the sample (Fig. 2c), at which time attractive van der Waals forces between the AFM tip and specimen surface cause changes in the vibration of the cantilever (e.g., amplitude and frequency) [37]. In practice, using the amplitude as the feedback is technically simpler [31], and in this mode, the piezoelectric tube drives the AFM probe to move vertically to keep the amplitude of the vibrating cantilever constant. In recent years, peak force tapping (PFT) [38] has emerged as a novel AFM imaging mode for resolving the ultra-microstructures and mechanical properties of biological systems. For more descriptions about PFT, readers are referred to the literature [39, 40]. Briefly, PFT is based on the AFM force spectroscopy technique, during which the AFM tip is controlled to vertically indent the specimens for recording force curves. The mechanical properties of the specimens are then extracted from the force curves, which are described in detail in the following sections in the context of the mechanics of tumor development and metastasis.
Fig. 2. Principles of AFM imaging.
a Schematic of AFM imaging of biological specimens (an example of cell membrane is shown) attached on the support. b Contact mode AFM imaging. In contact mode, AFM tip is scanned over the specimen surface, while the deflection of the cantilever is maintained constant. c Tapping mode AFM imaging. In tapping mode, commonly the amplitude of the oscillating cantilever is maintained constant. d SEM images showing the different shapes of AFM tips. I Pyramid tip. II Conical tip. d is reprinted with permission from Ref. [35]. Copyright 2016 Springer Nature.
Mechanical alterations of tumor cells during metastasis
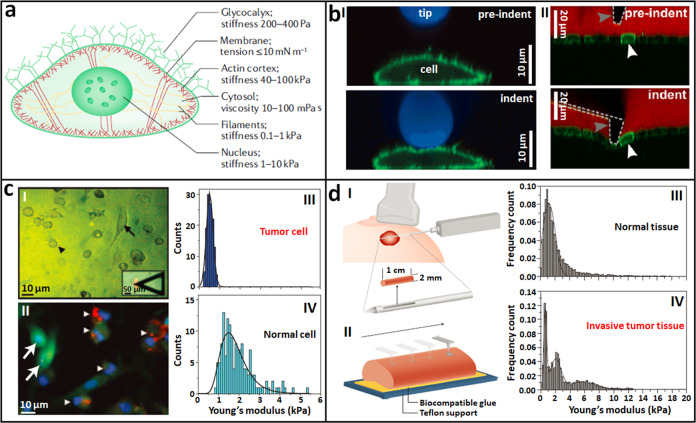
AFM is able to probe the mechanical properties of single living cells with high spatial resolution in indentation force spectroscopy mode. For AFM indentation experiments, the AFM tip is controlled to perform approach–retract movements in the vertical direction on the cell surface, during which both the vertical displacements of the probe and the deflections of the cantilever are recorded, yielding the so-called force curves [32]. By analyzing the force curves with adequate theoretical models, the Young’s modulus of the cells can be determined. Diverse models have been developed for analyzing the force curves (including Hertz–Sneddon, Johnson–Kendall–Roberts, Derjaguin–Muller–Toporov, and Oliver–Pharr [36]), and improvements to these theoretical models have been realized, such as the bottom effect cone correction module for eliminating the influence of substrates [41]. Notably, each theoretical model has its advantages and limitations, and generally, the Hertz–Sneddon model is the most widely used model in practice [42]. For more descriptions about the detailed procedures of AFM indenting experiments used for measuring cellular mechanics and the related data analysis, readers are referred to Refs. [32, 36, 43, 44]. During its approach, the AFM tip encounters different compartments and structures of the cells, including the glycocalyx, membrane, actin cortex, cytosol, filaments, and nucleus, and each of these subcellular structures has unique mechanical properties [29], which are summarized in Fig. 3a. The glycocalyx and the membrane are very soft and can be neglected in AFM indenting experiments [43]. Studies have shown that the cytoskeletons of cells, particularly the actin networks, are important determinants of cellular Young’s modulus as measured by AFM [45, 46]. By utilizing different types of AFM tips (Fig. 3b), the mechanical properties of various subcellular structures can be probed based on the AFM indentation technique. For example, a spherical tip is able to probe the mechanics of the cytoplasm (I in Fig. 3b) [47]; a conical tip is able to access the mechanics of the actin cortex of cells (II in Fig. 3b) [48]; and the use of a needle tip enables probing the mechanics of the cell nucleus [49], allowing the correlation of the mechanics of cellular compartments with their structures and cell functions.
Fig. 3. Stiffness changes of tumor cells during metastasis revealed by AFM indentation assays.
a Schematic diagram of an adherent mammalian cell with a summary of the mechanical properties of the cellular structures and compartments. Reprinted with permission from Ref. [29]. Copyright 2018 Springer Nature. b Different types of AFM tips for indentation assays. I Spherical tip. Zx confocal images of a cell (membrane protein is labeled with green fluorescein) indented by AFM cantilever conjugated with a fluorescent bead (blue). Reprinted with permission from Ref. [47]. Copyright 2013 Springer Nature. II Conical tip. Zx confocal images of a cell monolayer (green) grown on a soft collagen gel (black) during the indentation by an AFM cantilever with conical tip (dotted line). White arrowhead indicates an individual cell and gray arrowhead indicates the tip of the cantilever. A fluorescent dye was added to the cell culture medium (red). Reprinted with permission from Ref. [48]. Copyright 2014 The Company of Biologists Ltd. c AFM probing metastatic tumor cells and normal mesothelial cells prepared from clinical cancer patients. I Optical image of tumor cells (denoted by arrowhead) and normal mesothelial cells (denoted by arrow). The inset shows the alignment of AFM tip over the central region of a cell. II Immunofluorescence of the specimens confirming tumor cells (arrowheads) and normal cells (arrows). Stiffness histogram of tumor cells (III) and normal cells (IV). Reprinted with permission from Ref. [50]. Copyright 2007 Springer Nature. d AFM probing the stiffness changes of tumor tissues during metastasis. I Schematic of an ultrasound-guided biopsy from a patient with a suspicious lesion. II Schematic of utilizing AFM to record multiple stiffness maps (20 × 20 μm) across the biopsy specimen immobilized on the substrate. Stiffness histogram of normal breast tissues (III) and invasive breast tissues (IV). Reprinted with permission from Ref. [51]. Copyright 2012 Springer Nature.
Based on the AFM indentation technique, changes in the stiffness of tumor cells during metastasis have been remarkably revealed. Cross et al. [50] applied AFM to directly probe the mechanical properties of tumor cells and normal cells prepared from the biopsy specimens of clinical metastatic cancer patients, as shown in Fig. 3c. Metastatic cancerous cells and benign mesothelial cells in the pleural effusions of body cavity fluid samples were collected from patients suspected to have metastatic adenocarcinoma. The prepared cell pellets were resuspended in culture medium for 12 h, which promoted the adherence of the cells to the substrate and facilitated the AFM mechanical analysis. During 12 h of incubation, optical microscopy was performed to discriminate the tumor cells and normal cells (I in Fig. 3c). The tumor cells were small and round (indicated by the arrowhead in Fig. 3c (I)), whereas the normal cells were large and flat (indicate by the arrow in Fig. 3c (I)), observations that were confirmed by immunofluorescence assays (II in Fig. 3c). AFM mechanical measurements performed on the living cells showed that the metastatic tumor cells (III in Fig. 3c) were significantly softer than normal mesothelial cells (IV in Fig. 3c). Plodinec et al. [51] used AFM to directly measure tumor tissues prepared from clinical breast cancer patients and identified mechanical changes in tumor tissues during the development and invasion processes of tumors, as shown in Fig. 3d. Cylindrical specimens with a diameter of ~0.2 cm and length of 0.2–1 cm were extracted from suspicious lesions under ultrasound guidance (I in Fig. 3d). The biopsy specimens were immobilized on a plastic dish with a thin layer of epoxy glue (II in Fig. 3d), and then, AFM indentation experiments were performed on the specimens in Ringer’s solution, showing that normal breast tissues (III in Fig. 3d) exhibited a unimodal distribution of Young’s modulus (1.13 ± 0.78 kPa), while the invasive breast tumor tissues (IV in Fig. 3d) exhibited multimodal distributions of Young’s modulus. Further analysis based on hematoxylin and eosin staining of the biopsy specimens showed that the soft peak (0.57 ± 0.16 kPa) corresponded to cancer cells that were surrounded by stiffer stroma (1.99 ± 0.73 kPa). It is widely known that tumors are generally stiffer than comparable healthy tissues (breast tumor tissues can be ten times stiffer than normal breast tissues [52]) due to increased extracellular matrix (ECM) deposition [53]. Studies on tumor tissues based on AFM clearly show the detailed mechanical phenotypes of tumor tissues (combining soft tumor cells and rigid stroma). Combining AFM indentation measurements with confocal microscopy of cytoskeletons has revealed significant differences in the cytoskeletons of tumor cells and healthy cells (normal cells have well-aligned actin filamentous structures, while tumor cells have disorganized actin networks [54]), allowing the establishment of a direct correlation between cell mechanics and cell structures [55]. These studies, based on the utilization of AFM [50, 51, 54, 55], provide novel insights into tumor mechanics and tumor physics, which have significance in translational medicine for developing novel diagnostic or prognostic markers of cancers at the single-cell level [56, 57].
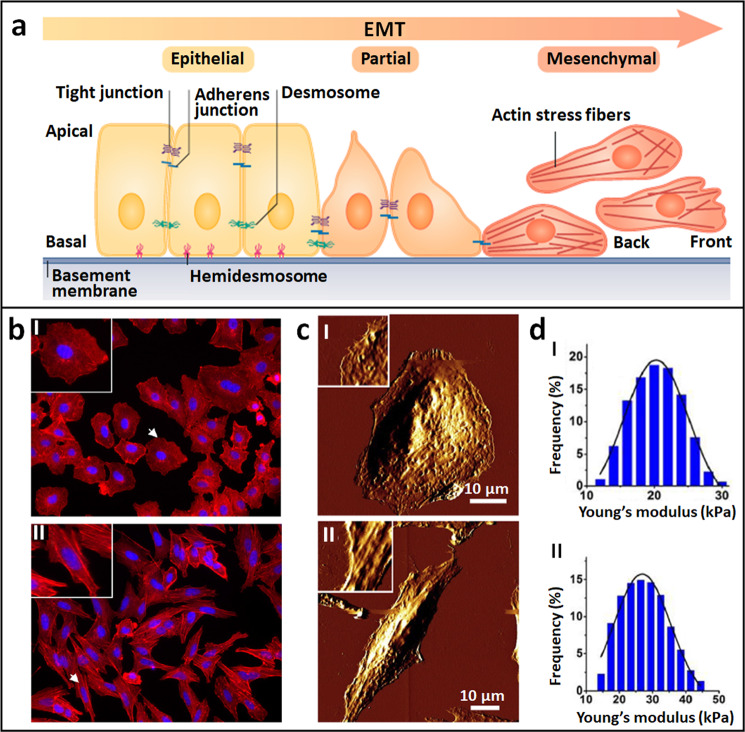
Cellular mechanical alterations during the process of the EMT that promote tumor metastasis have been revealed by AFM. The EMT is a cellular program crucial for malignancy progression, during which epithelial cells lose their polarized organization and acquire migratory and invasive capabilities [58, 59], as shown in Fig. 4a. Studies have shown that the EMT mechanism enhances the detachment of cancer cells from primary tumors preceding metastasis [60]. With the use of AFM, the mechanical cues during the EMT process were revealed. Buckley et al. [61] combined AFM with confocal fluorescence microscopy to investigate cell changes during the EMT process. Transforming growth factor-β1 (TGF-β1), a cytokine that can promote the EMT, was used to induce the EMT process. Both the confocal fluorescence images (Fig. 4b) and AFM images (Fig. 4c) showed that the shape of alveolar epithelial cells changed from being similar to cobblestones to being similar to spindles after TGF-β1 stimulation. Fluorescence staining of the cytoskeletons showed that TGF-β1 treatment resulted in increased numbers of F-actin stress fibers assembled parallel to the long axes of cell bodies (II in Fig. 4b), a finding that was confirmed by AFM imaging of the cytoskeletons (II in Fig. 4c). By obtaining force curves on cells before and after TGF-β1 treatment, the stiffness changes in the cells were determined. Wang et al. [62] used AFM to visualize and quantify the mechanical changes of cells after TGF-β1 stimulation. AFM multiparametric imaging based on PFT qualitatively showed the increased stiffness of cells after TGF-β1 stimulation, while the calculation of Young’s modulus from the obtained force curves quantitatively showed the much higher Young’s modulus values of the cells after treatment with TGF-β1 (Fig. 4d), significantly revealing the stiffening of the cells during the EMT process. Kanlaya et al. [63] used AFM to investigate the inhibitory effects of epigallocatechin-3-gallate (EGCG) on cellular mechanical changes during the TGF-β1-induced EMT process. They used AFM to probe the mechanical properties of regular cells (without treatment), cells treated with TGF-β1, and cells treated with TGF-β1 and EGCG. The AFM measurements showed remarkably increased cell stiffness in the TGF-β1-treated cells, whereas the stiffness of the cells treated with TGF-β1 and EGCG was very similar to that of regular cells, indicating that EGCG efficiently prevented the mechanical effects induced by TGF-β1. These AFM results were confirmed by the immunofluorescence staining of the cytoskeletons. These studies [61–63] revealed the mechanical phenotypes of single cells during the EMT process, which is meaningful for understanding tumor metastasis, providing a novel way to evaluate drug actions from the perspective of cell mechanics.
Fig. 4. Mechanical changes of cells in the process of EMT revealed by AFM.
a Schematic of EMT process. Epithelial cells displaying apical–basal polarity are held together by tight junction, adherens junctions, and desmosomes. Epithelial cells are tethered to the underlying basement membrane by hemidesmosomes. Induction of EMT results in cellular changes that include the disassembly of epithelial cell–cell junctions and the dissolution of apical–basal cell polarity. The loss of epithelial features is accompanied by acquisition of mesenchymal features. Mesenchymal cells display front-to-back polarity and have a reorganized cytoskeleton. After EMT, cells become motile and acquire invasive capabilities. Reprinted with permission from Ref. [58]. Copyright 2019 Springer Nature. b–d Cellular changes during EMT induced by TGF-β1. Cells before TGF-β1 treatment (I) and after TGF-β1 treatment (II). b Confocal fluorescent images. F-actins were stained with red fluorescein and nuclei were stained with blue fluorescein. The insets are the enlarged view of individual cells denoted by the arrows. c AFM morphological changes of cells. The insets are the enlarged view of local structures of the cells. d Statistical histograms of cellular Young’s modulus. b, c are reprinted with permission from Ref. [61]. Copyright 2012 Elsevier Inc. d is reprinted with permission from Ref. [62]. Copyright 2018 Elsevier B.V.
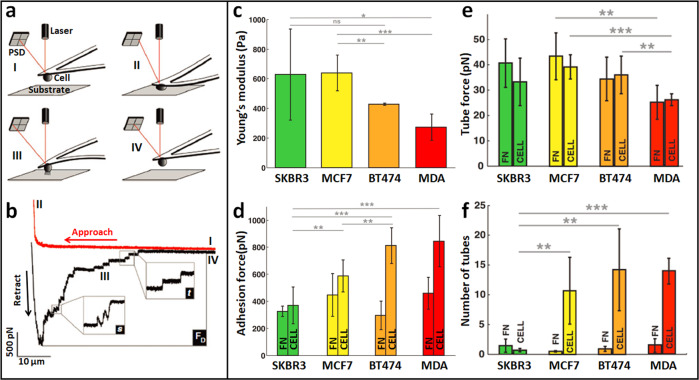
Applications of AFM single-cell force spectroscopy (SCFS) revealed significant changes in the adhesive properties of tumor cells during the process of metastasis. In SCFS, a living cell is attached to the end of the AFM cantilever to prepare a cell probe, and then the cell probe is controlled vertically to perform the approach–retract cycle on the substrate, which can be another cell, a surface or an organic ECM [64], as shown in Fig. 5a. Force curves recorded during the approach–retract cycle reflect the dynamic adhesive interaction process between the cell probe and the substrate, and the molecular binding events (e.g., receptor anchoring and membrane tether [65]) taking place during cell–substrate adhesion are clearly discerned from the force curves, as shown in Fig. 5b. By coating substrates with specific biomolecules, the special molecular interactions and mechanical forces involved in cell adhesion can be investigated by SCFS [66]. For more descriptions about the technical details of SCFS, readers are referred to Refs. [67, 68]. SCFS offers new possibilities for the study of tumor cells. Adhesive forces prevent animal tissues from dissociating into their component cells, and these forces change dynamically to establish new cell contacts during developmental cell movements, tissue renewal and wound repair [69], and they are critically involved in cancer spreading and invasion [70]. Smolyakov et al. [71] used AFM-based SCFS technique to investigate the changes in the adhesive properties of tumor cells during tumor invasion, as shown in Fig. 5c–f. Four types of breast cancer cell lines (SKBR3, MCF7, BT474, and MDA-MB231) with different invasive potentials were used. A living cell (SKBR3, MCF7, BT474, or MDA cell) was attached to a tipless AFM cantilever that was previously coated with concanavalin A, and then, the cell-modified cantilever was used to perform indentation assays either on fibronectin (FN)-coated substrates or on other cells adhered to the FN. Different mechanical parameters were determined from the force curves obtained, including Young’s modulus (obtained from the approach curve) (Fig. 5c), cell adhesion force (maximal force peak in the retraction curve) (Fig. 5d), tube force (force steps corresponding to a single membrane tether in the retraction curve) (Fig. 5e), and number of tubes (number of membrane tethers in a single detachment in the retraction curve) (Fig. 5f), showing that invasive breast cancer cells are softer and more adhesive with membrane tethers that can be readily extruded from invasive cells. The adhesion interactions of the cells were closely related to the adhesive receptors on the cell surface [72], and studies have shown a correlation between cell adhesion molecules (E-cadherin and N-cadherin) and cell adhesion forces [73]. Azadi et al. [74] used SCFS to show that the inhibition of epidermal growth factor receptor (EGFR) activity by monoclonal antibodies and small molecules led to an increase in the cellular adherence of tumor cells. With the use of SCFS, the interactions between cancer cells and endothelial cells have also been studied [75], showing that the glycocalyx strongly modulates the adhesion between cancer cells and endothelial cells. These studies [71–75] significantly demonstrated the feasibility of directly and quantitatively correlating adhesion molecules on the cell surface with cytoadherence and cell functions by AFM-based SCFS, which contributes a better understanding of tumor behaviors.
Fig. 5. AFM-based single-cell force spectroscopy (SCFS) revealing the changes of adhesive properties of tumor cells during metastasis.
a, b Principle of SCFS. a Schematic of performing approach–retract movement on substrate with the use of cell probe in SCFS assays. The cell-conjugated AFM cantilever is first lowered toward the substrate (I) until a preset force is reached (II). After a given contact time, the cantilever retracts from the substrate (III) until cell and substrate are completely separated (IV). b A representative force curve recorded during SCFS showing steps (I–IV) corresponding to those outlined in (a). Several unbinding events can be observed in the retraction curve (s denotes force steps, t denotes unbinding of membrane tethers, Fd denotes maximal detachment force). Reprinted with permission from Ref. [65]. Copyright 2010 Springer Nature. c–f Adhesion properties of tumor cells with different invasion potentials. In the following, breast cell lines are ranked from left to right in ascending order of their invasive character. c Cell Young’s modulus. d Cell adhesion force. e Individual membrane tether force. f Number of membrane tethers. Reprinted with permission from Ref. [71]. Copyright 2016 American Chemical Society.
The utilization of AFM single-molecule force spectroscopy (SMFS) allows probing the mechanics of biomolecules on the surface of tumor cells. In SMFS, ligand molecules are attached to the surface of the AFM tip via flexible cross-linker molecules (often polyethylene glycol (PEG) [76]), and then, the ligand-modified tip is used to vertically perform approach–retract cycles at different areas on the cell surface for the recognition and location of specific receptors on the cell surface [77], enabling single-molecule imaging of cell surfaces with high spatial resolution [78], as shown in Fig. 6a (I). The specific receptor-ligand dissociation events are discriminable from the obtained force curves [79], as shown in Fig. 6a (II, III). The magnitude of the force peak corresponds to the receptor-ligand dissociation force. By obtaining force curves for different retraction velocities of the cantilever, the energetic and kinetic parameters of the receptor-ligand interactions can be obtained [80]. With the use of SMFS, single proteins can be mechanically unfolded, which yields the mechanical dynamics of the unfolding pathway of the proteins [81]. For more details of SMFS and SMFS-based techniques, readers are referred to Refs. [82–84]. The applications of SMFS provide novel insights into the cell-surface activities involved in tumor behaviors at the single-molecule level. When a normal cell becomes cancerous, the organization of cell-surface molecules significantly changes; for example, the expression or activation of EGFR is altered in many epithelial tumors [85], epithelial cell adhesion molecule (EpCAM) is a dominant antigen on tumor cells [86], and tumor cells have a large glycocalyx on their surface for the clustering of integrins and for membrane bending [87]. Using the AFM SMFS-based simultaneous topography and recognition imaging technique, local receptor maps on the surface of tumor cells can be obtained at nanometer spatial resolution [88], as shown in Fig. 6b. Topography images (I in Fig. 6b) of the cell surface and recognition images (II in Fig. 6b) of specific cell-surface receptors can be obtained simultaneously. The clustering behaviors of receptors on the cell surface can be quantitatively characterized from the recognition image [89]. The overly image (III in Fig. 6b) clearly shows the distribution of individual receptors on the complex heterogeneous surface of the tumor cells with high precision, facilitating the establishment of links between tumor cell molecule organizations and cell functions and benefiting the studies of molecular activity on the tumor cell surface. The SMFS-based multiparametric imaging technique, particularly combined with fluorescence microscopy [90, 91], provides a novel way to quantify and map the molecular interactions on the cell surface, contributing to investigations in the mechanics of single molecules on tumor cells. Under optical (I in Fig. 6c) and fluorescent (II in Fig. 6c) guidance, the functionalized AFM tip is moved to target cells for simultaneously imaging cell topography (III in Fig. 6c) and detecting specific molecular interactions on the cell surface (IV in Fig. 6c), and molecular dissociation forces can be obtained from the recorded force curves (V and VI in Fig. 6c). Combining AFM multiparametric imaging with biochemical assays, Hsu et al. [92] revealed the intricate relationship between EpCAM-regulated transcription and altered nanomechanical properties of cells that promoted the epithelial-to-mesenchymal transition. AFM imaging of tumor cells with antibody-conjugated tips clearly showed that the EpCAM molecules were largely dispersed on the surface of endometrial cancer cell lines and that the EpCAM molecular recognition sites vanished after the stimulation with epidermal growth factor, which can bind to EpCAM, and AFM indentation assays on the cells showed altered nanomechanical properties of EpCAM-edited cells, demonstrating a nanomechanical phenotype in advanced cancer progression and illustrating that AFM is an invaluable tool complementing traditional biochemical assays for addressing the mechanical cues in tumor development.
Fig. 6. AFM-based single-molecule force spectroscopy (SMFS) revealing the mechanics of molecules on the surface of tumor cells.
a Principle of SMFS. I Schematic of probing specific receptors on cell surface by SMFS with the use of ligand-conjugated tip. Reprinted with permission from Ref. [77]. Copyright 2019 Springer Nature. II Schematic and III practical force curves recorded during SMFS assays. Reprinted with permission from Ref. [79]. Copyright 2018 Elsevier Ltd. b Mapping nanoscale organizations of receptors on heterogenous surface of tumor cells by AFM. Reprinted with permission from Ref. [88]. Copyright 2018 Elsevier Ltd. c Combining AFM with confocal fluorescence microscopy to reveal specific molecular interactions on cell surface. I Differential interference contrast (DIC) image and II mCherry channel superimposed with DIC channel. The functionalized probe can be seen above the cells. III AFM height image and IV adhesion image of cells recorded in the scan region denoted by the dashed square in (II). Reprinted with permission from Ref. [90]. Copyright 2017 Springer Nature. Distribution of molecular adhesion forces measured on target cells (V) and control cells (VI). Reprinted with permission from Ref. [80]. Copyright 2017 Springer Nature.
Mechanics of tumor cell spheroids
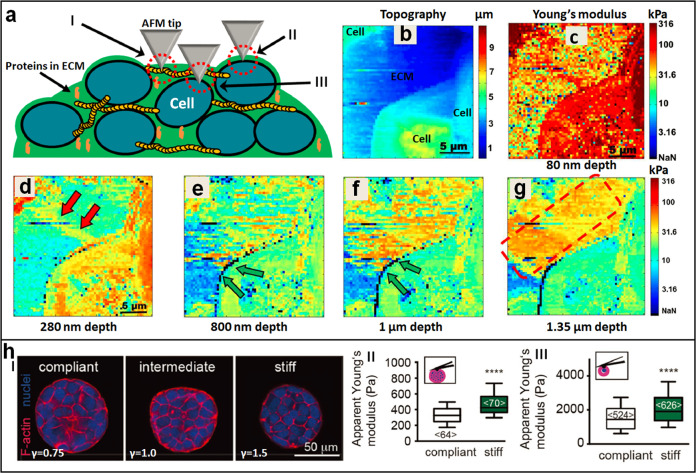
AFM can be used to probe the mechanics of tumor cell spheroids. In recent years, 3D tumor cell spheroids have emerged as promising in vitro models that replicate many features of solid tumors in vivo and are useful for investigating tumor behaviors and drug effects [93]. Currently, 2D monolayer cellular assays serve as the gold standard for studies in the life sciences, such as for investigation of cell activity and drug discovery [94], and monolayer cell cultures are unable to mimic the structure and drug resistance conferred by elements of the tumor microenvironment and its 3D organization, which are often the cause of inaccurate assessments of the biological performance of tumor therapeutics [95]. Diverse fabrication methods have been developed for preparing 3D tumor cell spheroids, including growing cells on nonadherent surfaces or in suspension, seeding cells within polymer scaffolds or within hydrogels, liquid-overlay techniques, and microfluidic devices [96, 97], offering new opportunities for tumor drug testing [98, 99]. With the use of AFM, the detailed nanomechanical properties of tumor cell spheroids can be visualized, significantly contributing to the understanding of tumor behaviors. Recently, Vyas et al. [100] reported utilizing AFM to investigate the mechanics of ECM and cells in tumor spheroids, as shown in Fig. 7a–g. Lung cancer cell spheroids were prepared using a liquid-overlay technique on agar surfaces. After the formation of the spheroids, the lower half of the spheroids were immobilized on the glass coverslip covered in agarose gel (the immobilization prevented lateral motion of the spheroids during AFM measurements), and then, the upper half of the spheroids were exposed to aqueous medium for probing by the AFM tip. Arrays of 128 × 128 force curves were recorded over 20 × 20 μm areas on the spheroids. Each of the indentation curves was split into small fragments, and each fragment was fitted by the Hertz–Sneddon model, yielding data for generating stiffness maps versus indentation depths. With this method, the nanomechanical properties of tumor spheroid structures at various depths were visualized. From the stiffness maps, different components of the spheroids were identified (Fig. 7a), including collagen fibers, the interface of cell membrane and ECM, and cells embedded deep inside the ECM. The results showed that at small depths, cells had a very high modulus compared with the adjacent ECM (Fig. 7c), and there was a gradual decrease in stiffness as the probe indented deeper into the cell (Fig. 7d–g), remarkably visualizing the mechanical maps of the tumor spheroids at different depths. Andolfi et al. [101] designed and fabricated planar AFM macroprobes to measure the whole viscoelasticity of tumor spheroids. The macro AFM probes were fabricated on a silicon wafer using photolithography techniques, and the spring constant of the fabricated probe was calibrated using a commercial AFM cantilever with a known spring constant (the spring constant of the commercial cantilever can be calibrated with the thermal noise method). The results showed that the planar AFM cantilever was well suited to characterize the mechanical properties of large tumor spheroids (with a diameter above 100 μm). These studies [100, 101] showed that AFM can be used not only to probe the heterogeneous mechanics of different structures in tumor spheroids with high precision but also to characterize the whole mechanics of the tumor spheroids, data which are particularly meaningful for understanding the mechanical phenotypes of tumors.
Fig. 7. Probing the mechanics of tumor cell spheroids by AFM.
a–g Visualizing the heterogeneous stiffness signature of tumor cell spheroids at different depths by analyzing the force curves obtained on spheroids. a Schematic showing that three types of nanomechanical topographies are identified during AFM indentation measurements, including collagen type I stress fibers (I), the interface of cell membrane and ECM (II) with high stiffness, and cells embedded deep inside the ECM (III). b Topography image and c–g corresponding stiffness images of a tumor spheroid at different depths. The double red arrows denote the collagen type I stress fibers, the double green arrows denote the interface of cell membrane and ECM, and the dotted red square denotes the cells embedded deep inside the ECM. Reprinted with permission from Ref. [100]. Copyright 2019 Springer Nature. h Mechanical dynamics of tumor spheroids and tumor cells regulated by the rigidity of microenvironment. I Confocal fluorescent images of the spheroids grown in hydrogels of varying stiffness. II AFM indentation assays on the whole tumor spheroids. III AFM measurements on individual tumor cells isolated from the tumor spheroids. Reprinted with permission from Ref. [102]. The insets in (II, III) show the schematic of AFM measurements on tumor spheroids or single tumor cells, respectively. Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
The applications of AFM in measuring the mechanics of tumor spheroids promote understanding of tumor growth in confined environments. Taubenberger et al. [102] combined AFM with biochemical assays to find the mechanical signature of tumor spheroid growth as regulated by microenvironment stiffness (Fig. 7h). MCF7 breast cancer cells were embedded in PEG-heparin hydrogels with various stiffness features. The PEG-heparin ratio (γ) was adjusted to values of 0.75 (compliant), 1.0 (intermediate), and 1.5 (stiff), yielding hydrogels with different Young’s moduli (I in Fig. 7h). In 14 days, multicellular spheroids formed and reached diameters of up to 150 μm. Optical and fluorescence microscopy experiments showed that the tumor spheroids were smaller and more compact when grown in stiff hydrogels (I in Fig. 7h). AFM indentation assays were utilized to characterize the mechanics of tumor spheroids on different scales. The spheroids were first harvested by gel degradation, and then, the collected spheroids were pipetted into glass bottom dishes. After the spheroids were stably attached to the dish, the AFM probe was controlled to indent the spheroids, showing that tumor spheroids grown in stiff hydrogels were significantly stiffer than those grown in compliant hydrogels (II in Fig. 7h). AFM measurements on individual cells after tumor spheroid dissociation also showed that tumor cells grown in stiff hydrogels were significantly stiffer than those grown in compliant hydrogels (III in Fig. 7h). Further experiments performed on cells treated with various drugs showed that Rho-associated kinase was critical for the increased cell stiffness in stiff hydrogels by altering the F-actin cytoskeleton. A study based on AFM [102] revealed that the rigid features of the microenvironment have direct impacts on the growth behaviors of tumor spheroids, which exhibit distinct mechanical phenotypes at different levels, such as altered cancer cell stiffness and spheroid stiffness, providing novel insights into oncology.
Mechanics of exosomes in tumor progression
Exosomes have been shown to play important roles in the metastasis of tumors. All cells, including those in prokaryotes and eukaryotes, release extracellular vesicles (EVs) under normal physiological conditions and during acquired abnormalities [103]. EVs constitute a heterogeneous population of membrane vesicles of various origins and can be classified into two distinct classes: exosomes (with a size typically between 50 and 150 nm) and microvesicles (with a size is typically between 50 nm and 1 μm) [104]. Exosomes are released into the extracellular environment upon exocytic fusion of multivesicular endosomes with the cell surface [105]. These small vesicles are called intraluminal vesicles, and they are contained within multivesicular endosomes and are called exosomes when they are released into the extracellular environment [106], as shown Fig. 8a (I). Exosomes contain bioactive molecules, such as nucleic acids (DNA, mRNA, microRNA, and other noncoding RNAs), proteins (receptors, transcription factors, enzymes, and ECM proteins), and lipids that can redirect the function of a recipient cell [107], as shown in Fig. 8a (II). Once secreted, exosomes bind to neighboring cells or to the ECM or passively move in the bloodstream or other bodily fluids [108]. Exosomes presumably act as important vehicles of intercellular communication between cells locally and distally [109]. Exosomes have been shown to crucially participate in cancer development, and cancer cells are known to secrete more exosomes than their nonmalignant counterparts [110]. Exosome-mediated effects not only influence the tumor microenvironment (e.g., exosomes can enhance cancer cell motility by stabilizing cellular protrusions; proteins such as metalloproteinases contained in exosomes participate in ECM remodeling to promote cell motility; tumor cells can acquire drug resistance via exosomes; and exosomes can promote the differentiation or recruitment of tumor-promoting stromal cells) but also impact distant sites (e.g., exosomes can enter the circulation and travel to sites distant from the primary tumor; various exosome cargoes promote vascular permeability; and exosomes can generate a premetastatic niche by inducing ECM remodeling and promote the recruitment of cancer-associated stromal cells), promoting tumor growth, invasiveness, metastasis, and antineoplastic resistance [111–113]. Exosomes and their components therefore constitute a novel class of emerging therapeutic targets (e.g., exosomes can be blocked from supporting tumor progression and induce the response to therapy [114]), which will have implications for the development of cancer diagnostics and therapeutics [115].
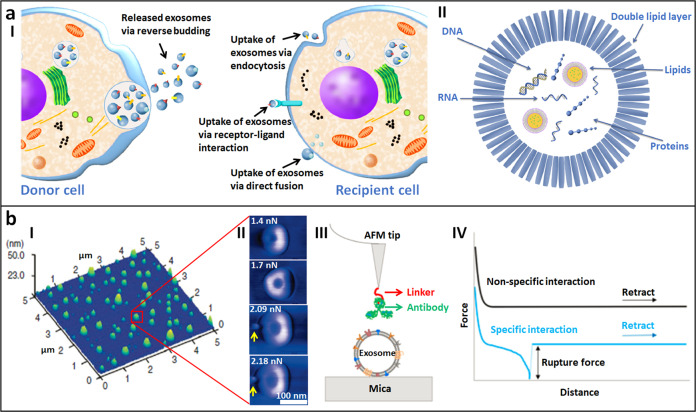
Fig. 8. Imaging and force spectroscopy measurements of individual exosomes by AFM revealing the mechanical phenotypes of exosomes in promoting tumor progression.
a Schematic of exosomes in the communication of cells. I Release of exosomes by donor cells can be induced by diverse signaling pathways and occurs in a reversed budding event. The uptake of exosome is accomplished via endocytosis, receptor-ligand interaction or by direct fusion. II Exosome content. Reprinted with permission from Ref. [106]. Copyright 2017 Elsevier Ltd. b AFM is able to probe the structure and properties of single exosomes. I Imaging the topography of single exosomes. Reprinted with permission from Ref. [116]. Copyright 2014 Elsevier Inc. II Indenting single exosomes. Reprinted with permission from Ref. [118]. Copyright 2010 American Chemical Society. III, IV Probing single receptors on exosomes. III Schematic of using antibody-modified tip to recognize specific receptors on exosomes. IV Force curves obtained during AFM force spectroscopy assays discriminating the specific molecular interactions and nonspecific molecular interactions. Reprinted with permission from Ref. [120]. Copyright 2018 IOP Publishing Ltd.
Applications of AFM to the studies of exosomes offer novel possibilities for understanding the mechanics of exosomes. By attaching exosomes to the substrate, AFM imaging can clearly reveal the detailed morphology of individual exosomes [116], which is useful for analyzing the geometric features of exosomes, such as their sizes, as shown in Fig. 8b (I). Exosomes are commonly characterized by EM or cryo-EM [104, 109], which requires complex pretreatments of the specimens. The pretreatments can inevitably cause changes in exosomes, and thus, the results obtained by EM may not reflect the true structures of living exosomes. AFM can directly obtain the morphology of single exosomes in liquids [117], which is particularly meaningful for investigating the structures and properties of living exosomes in their native states. Controlling the AFM tip to indent individual exosomes enables the characterization of the mechanical properties of single exosomes. The morphological deformation of a single exosome in response to the loading force of the AFM probe can be distinctly observed [118], as shown in Fig. 8b (II). In addition, the blebbing of exosomes can be observed (indicated by the yellow arrows in Fig. 8b (II)) when applying high force to the exosomes, indicating force-induced structural perturbations of the exosomes. By analyzing the force curves obtained for exosomes immobilized on substrates, the mechanical properties of single living exosomes are quantified, such as Young’s modulus, the adhesion force and deformation capability [119]. By using an antibody-conjugated tip, the specific biomolecules on single exosomes can be recognized and located with molecular resolution [120], as shown in Fig. 8b (III). We know various receptors on the surface of exosomes play important roles in regulating the physiological processes of exosomes, such as exosome binding to cells and subsequent internalization [121, 122], and thus, investigating the individual receptors on exosomes may reveal the underlying mechanisms guiding the behaviors of exosomes. From the force curves obtained for exosomes with the use of a functionalized AFM tip, the specific molecular interactions between receptors on exosomes and antibodies on the AFM tip are clearly distinguished (Fig. 8b (IV)), which allows quantifying the binding affinities and the spatial distribution of single receptors on exosomes, providing novel insights into the surface mechanics of exosomes at the single-molecule level.
Mechanics of the ECM for promoting tumor metastasis
It is increasingly evident that the ECM plays an important role in the development and progression of cancers. On the one hand, cells within tissues establish the ECM during development, maintain it under physiological conditions, remodel it during adaptations, and repair it in response to disease and injury [123]. On the other hand, the ECM provides not only a physical scaffold for maintaining the structural integrity of multicellular organisms but also serves as a reservoir for biochemical and biophysical signals to support cell survival, organization, and differentiation [124]. These reciprocal interactions between cells and the ECM have long been recognized to have key roles in cell fate determination [125], and dysregulation of the ECM contributes to several pathological conditions, such as fibrosis and cancer [126]. The ECM is composed of water, proteins (e.g., collagen, elastin, FN, and laminin) and polysaccharides, and each tissue has an ECM with a unique composition and topology that is generated during tissue development [127]. In particular, the microenvironment of tumors is much stiffer than that of normal tissues because of tumor-associated ECM remodeling characterized by increased ECM deposition, fiber alignment, and cross-linking [53]. During the past 10 years, it has become clear that the stiffened ECM and the elevated solid stresses that develop in a tumor are not merely passive byproducts of malignancy; in contrast, these physical features actively participate in tumor progression and modify the tumoral response to therapy [128]. Studies have shown that tumor-derived ECM promotes angiogenesis and tumor cell growth [129], while suppressing ECM expression contributes to drug penetration and successful tumor therapy [130]. It has also been reported that altered gene expression in cancer cells regulates ECM components to promote tumor progression [131]. Consequently, investigating the physical cues (e.g., stiffness, viscoelasticity, pore size, fiber alignment, and molecular composition) of the ECM is important for advancing our understanding of the mechanisms critical for cellular sensing of these properties [53], providing significant novel insights into oncology.
AFM can reveal the mechanical dynamics of the basement membrane during the invasion process of tumor cells. Basement membranes are thin, dense sheets of specialized, self-assembled ECM that surround most animal tissues [132], including epithelial (I in Fig. 9a), endothelial, muscle, and adipose tissues. The basement membrane mainly includes two independent polymeric networks (including type IV collagen and laminin), which are linked by ECM proteins (including nidogen and perlecan), as shown in Fig. 9a (II). Epithelium-derived tumors, also known as carcinomas, represent ~90% of all cancers, and the first barrier against the metastatic spread of these cancers is the basement membrane underlying the epithelium [133]. Studies have shown that in addition to protease-mediated degradation and chemotaxis-stimulated migration, basement membrane invasion by malignant cells is significantly influenced by the stiffness of the associated interstitial ECM and the contractility of the tumor cells [134]. A recent study has shown that preexisting small holes (weak spots) in the basement membrane significantly contribute to recruiting immune cells for attacking cancer cells [135], indicating that the mechanics of the basement membrane play important roles in tumor invasion. Glentis et al. [136], with the use of AFM, revealed the dynamics of the basement membrane during tumor invasion, as shown in Fig. 9b. Mesentery basement membranes were isolated from mice, and the cells (including cancer cells and cancer-associated fibroblasts) were grown on the basement membranes. Morphological imaging by AFM showed that the basement membrane growing cancer cells and fibroblasts had more holes (II in Fig. 9b) than the basement membranes not growing cells (I in Fig. 9b), indicated by the roughened basement membranes upon cell growth (III in Fig. 9b). AFM indentation measurements showed that, after upon cancer cells and fibroblasts growth, the basement membrane became significantly softer (IV in Fig. 9b). The results [136] clearly showed the dynamic mechanics (roughening and softening) of the basement membranes during the tumor cell breaching process, providing novel insights into cancer invasion and metastasis [137]. Notably, in each tissue and in different regions of the same tissue, the matrix structure and composition of the basement membrane can vary [138, 139], indicating that the mechanics of the basement membrane are heterogeneous and are likely associated with the structural organization of the basement membrane. AFM can simultaneously obtain data on the ultra-microstructures and mechanics of biological systems such as the ECM [140] and hydrogel scaffolds for tissue engineering purposes [141], and thus, the utilization of the AFM comprehensively contributes to the characterization of the structures and mechanics of basement membranes, which is meaningful for developing new strategies for blocking cancer progression and metastasis [142].
Fig. 9. Imaging the morphology and measuring the mechanics of basement membranes by AFM.
a Basement membrane localization and composition. I Basement membrane underlies or surround most tissues such as epithelial. II The self-assembling polymeric networks of type IV collagen and laminin provide basement membranes with their core structure and these networks associate with each other through interactions (denoted by arrows) with bridging adapter proteins, such as perlecan and nidogen. The laminin network is closely associated with cell surface through interactions with integrins and dystroglycan receptors as well as sulfated glycolipids. Reprinted with permission from Ref. [132]. Copyright 2017 Elsevier Inc. b AFM revealing the dynamic mechanics of basement membranes during the invasion process of tumor cells. AFM height images of the basement membranes without growing cells (I) and basement membranes treated by cancer cells (CCs) and cancer-associated fibroblasts (CAFs) (II). Statistical histograms of the changes in roughness (III) and stiffness (IV) of basement membranes after growing cells. Reprinted with permission from Ref. [136]. Copyright 2017 Springer Nature.
Applications of AFM for probing decellularized ECM offer novel possibilities for understanding the mechanics of the ECM in oncology. Decellularization is an attractive technique for scaffold preparation in tissue engineering, as the resulting material can maximally recapitulate all the features of the natural ECM of the original tissue [143, 144], which is inaccessible for use with natural or man-made materials. The sources of decellularized matrices can be categorized into two types: ECM from tissue/organs and ECM formed by the cells in culture in vitro [145], as shown in Fig. 10a. Tissue-derived decellularized ECM is similar to the native ECM in terms of composition, mechanics, and structures, but the scarcity of ECM sources limits broader applications. Cultured cell-derived decellularized ECM is easier to obtain on a large scale and is able to reconstitute ECM in a limited region, but cannot completely mimic the composition, mechanics, and structures of native ECM [146]. For more descriptions of the decellularized matrix, readers are referred to the literature [145, 146]. Recently, researchers have applied AFM to characterize the mechanics of a decellularized matrix to garner understanding of the microenvironmental cues in the pathological alterations of cells. Organ-derived decellularized ECM is obtained by perfusing the decellularizing and washing media through a main artery of the organ (I and II in Fig. 10b) [147]. After immobilizing onto the substrate thin slices cut from the decellularized organ, the structures and mechanics of the decellularized ECM can be probed by AFM (III in Fig. 10b). Utilization of AFM on decellularized ECM prepared from healthy organs or diseased organs has revealed that pathological changes significantly cause alterations in ECM mechanics [148, 149], providing important quantitative insights into the mechanical aspects of a variety of physiopathological conditions [150]. Researchers have also used AFM to investigate the mechanics of cultured cell-derived decellularized ECM and established standard procedures (Fig. 10c) [151, 152], allowing the characterization of the stiffness and topography of the matrix deposited by the cells, which is important for understanding how cell behaviors and fates are influenced by these cues in the ECM. Notably, thus far, studies based on AFM assays of decellularized ECM derived from tumor tissues or tumor cells are still scarce, and the established methods (Fig. 10) can be directly applied to the issues associated with tumors. Undoubtedly, these studies (e.g., AFM characterizations on the decellularized matrix prepared from tumors or their corresponding normal tissues) will significantly contribute to the precise understanding of the role of ECM mechanics in tumor invasion and metastasis and may potentially promote the design of reliable biomimetic materials with therapeutic potential.
Fig. 10. AFM assays on decellularized matrix.
a Types of decellularized matrices. I Tissue-/organ-derived decellularized matrices. II Cultured cell-derived decellularized matrices. III Various forms of decellularized matrices. Reprinted with permission from Ref. [145]. Copyright 2017 Royal Society of Chemistry. b AFM assays on decellularized matrix prepared from organ. Photographic image of a mouse lung before (I) and after (II) decellularization by simultaneous tracheal instillation and arterial perfusion of a solution of triton X-100 (0.1%) and sodium dodecyl sulfate (1%). III A slice (12 μm thick) of decellularized mouse lung probed with an AFM placed on the stage of an inverted optical microscope. Reprinted with permission from Ref. [147]. Copyright 2017 Wiley Periodicals, Inc. c Diagram showing the typical steps of AFM force spectroscopy experiments on cultured cell-derived decellularized matrix for measuring matrix stiffness. Reprinted with permission from Ref. [152]. Copyright 2016 Elsevier Inc.
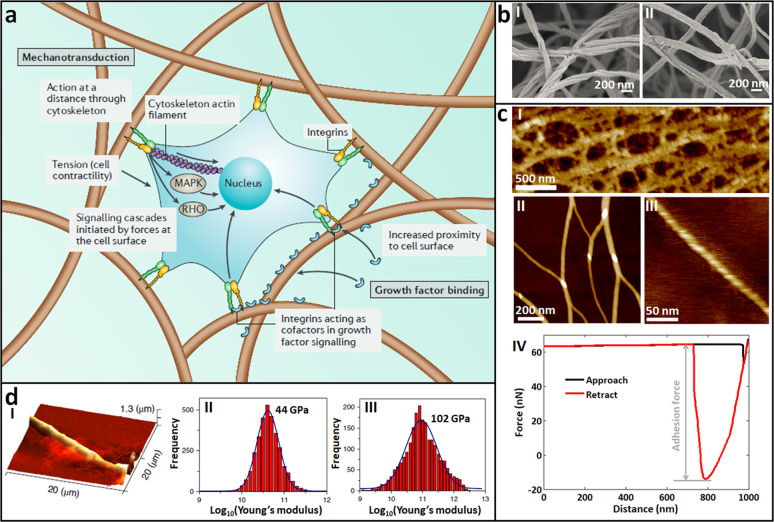
AFM has been used with great success in characterizing the mechanics of the fine structures of the ECM in its native state with unprecedented spatial resolution. On the cell surface, various receptors (such as integrin) can bind to ECM proteins [153], allowing the adhesion of cells onto the ECM. Adhesive interactions between cells and the ECM play important roles in regulating cell behaviors (e.g., survival, migration, polarity, and differentiation), and dysregulated integrin-mediated adhesion is a key precursor in the pathogenesis of many diseases, including cancer [154, 155]. In fact, integrins are implicated in nearly every step of cancer progression from primary tumor development to metastasis [155]. Most ECM proteins are fibrous proteins (e.g., collagen, FN, and elastin), and the filamentous nature of the ECM (Fig. 11a) strongly influences the physical properties of the ECM and its response to mechanical stress [156]. Cells respond to nanoscale surface features (such as nano-topographical surfaces) of the ECM via integrin-mediated cell adhesion, critically affecting cell fate, and cell function [157, 158]. Hence, investigating the structures and properties of the ECM at the nanoscale significantly contributes to the understanding of underlying mechanisms guiding tumor behaviors. However, traditional characterizations based on EM require pretreatments of the ECM (e.g., fixation and drying) (Fig. 11b) [159], which can inevitably damage the native structures of the ECM; thus, the results cannot fully reflect the true ECM in a natural context. The ECM can be clearly visualized by AFM in its native states at high resolution (Fig. 11c) [160, 161]. In addition to the porous polymeric reticulated structures of the ECM (I in Fig. 11c), the individual nanofibrils in the ECM networks (II in Fig. 11c) and even the fine structures in single nanofibrils (e.g., the helical structures of nanofibrils, as shown in III in Fig. 11c) can be visualized by AFM imaging. Based on the AFM high-resolution morphological imaging of ECM nanofibrils, the diverse assembly behaviors of nanofibrils, such as aggregation, can be assessed [162], and the properties that contribute to the rigidity of nanofibrils, such as length persistence, can be quantified [163]. By controlling the AFM tip to perform approach–retract movements in the vertical direction on single nanofibrils to obtain force curves (IV in Fig. 11c), the adhesion forces of nanofibrils can be measured, which are useful for evaluating the adhesion capabilities of ECM and for promoting cell growth. From the force curves obtained on single nanofibrils, the stiffness of different types of nanofibrils can also be ascertained [164, 165], as shown in Fig. 11d. These results [160–165] provide novel insights into the structures and properties of native nanostructures of the ECM. In the future, combining the data on ECM nanostructures obtained by AFM with assays on tumor cell functions will precisely contribute to the analysis of the role of ECM mechanics in tumor invasion and metastasis.
Fig. 11. AFM for visualizing the fine structures and measuring the mechanics of single ECM nanofibrils.
a Schematic illustrating the effect of fibrillar structures on mechanotransduction. Cells can exert forces on the fibers through cell-surface proteins and the cytoskeleton, which are both mechanically coupled to the ECM, initiating signaling pathways via mitogen-activated protein kinase (MAPK) and the RHO family of GTPases (RHO). Reprinted with permission from Ref. [156]. Copyright 2019 Springer Nature. b SEM images of collagen (I) and fibrin (II) gels, respectively. Reprinted with permission from Ref. [159]. Copyright 2015 Authors. c Imaging the fine structures of ECM and measuring the adhesion force of ECM by AFM [160, 161]. I Porous network structures of ECM visualized by large-size AFM scanning. II, III Single nanofibrils visualized by small-size AFM scanning. IV A typical force curve obtained on ECM for evaluating the adhesive capabilities of ECM. d Measuring the stiffness of single nanofibrils by AFM. I AFM image of tripeptide fibers. Statistical histogram of the Young’s modulus of the nanofibrils formed by Pro-Phe-Phe (II) or Hyp-Phe-Phe (III). Reprinted with permission from Ref. [164]. Copyright 2019 Springer Nature.
Conclusion and future perspectives
In this paper, the applications of AFM to investigations into the mechanical phenotypes involved in tumor development and progression are summarized, illustrating that AFM is able to probe the mechanical properties of diverse biological samples associated with tumor behaviors at multiple levels, including single cancerous cells [47, 50], tumor tissue slices [51], biomolecules on the cell surface [77, 88], tumor cell spheroids [100, 102], exosomes secreted by cells [116, 118], and the ECM in tumor microenvironments (e.g., basement membrane [136], decellularized matrix [147, 152], and ECM nanofibrils [160, 164]). The use of AFM reveals the dynamic mechanics of both the cancer cells themselves [50, 71] and microenvironmental cues [51, 136] in the process of tumor invasion and metastasis, providing remarkable novel insights into the regulatory role of mechanics in oncology. The evidence significantly demonstrates that AFM is a powerful multifunctional tool for the studies of biomechanics and biophysics [21], considerably complementing traditional biochemical assays and offering novel possibilities for uncovering the underlying mechanisms guiding tumor processes. Nevertheless, several issues need to be addressed to further advance physical oncology based on AFM.
Utilizing AFM to probe the mechanics of cells involved in the process of tumor metastasis requires overcoming significant challenges. First, the methodology of characterizing cell mechanics by AFM needs to be improved. It is widely known that cell stiffness measured by AFM indentation assays is dependent on many factors [32], including environmental conditions (e.g., temperature, the substrate on which cells are attached, and the cell culture medium) [166], instrumental parameters (e.g., tip shape, approach rate of the probe, and indentation depth) [36, 41], cells (e.g., the cellular areas being probed and cell status such as isolated or connected [167]), and data processing (e.g., theoretical models used for analyzing force curves and contact point determination of the force curves [168]). Hence, strictly speaking, the results (such as cell Young’s modulus) obtained via AFM indentation experiments from different groups can be compared only when the conditions are identical, which requires substantial standardization of the measurements. Particularly, for some applications (e.g., measuring cellular adhesion forces [65] and specific molecular forces by AFM techniques [90]), the tip needs to be functionalized (e.g., attaching cells to the AFM cantilever and linking antibodies to AFM tip), which often involves a complex process and requires specific expertise, adding to the complication of AFM assays. Defining simple standardized protocols will contribute to making AFM appealing to more researchers. Second, there are still shortcomings for current studies about measuring cell mechanics by AFM. Currently, AFM studies are commonly performed on cells grown on 2D flat substrates in vitro [50, 71, 102]; however, significant differences (e.g., phenotype and shape) [169] between cells grown in a 2D environment in vitro and cells grown in a 3D environment in vivo are acknowledged. The results obtained by AFM in a 2D environment cannot directly reflect cell behaviors in vivo. In recent years, researchers have used molecular sensors to successfully sense the mechanics of cells grown in 3D biopolymer networks [170, 171]. Hence, combining AFM-based 2D mechanical measurement data with molecular sensor-based 3D mechanical measurement data to study tumor cells will remarkably contribute to a more thorough understanding of the role of cell mechanics in the process of tumor invasion and metastasis. With an alternative method, researchers have used AFM to directly probe the mechanics of tissue samples [51, 172] to investigate mechanics in vivo, but notably, the tissue samples prepared from tumor patients contain many components, including tumor cells and healthy cells and their respective ECM. Hence, it is challenging to identify the exact contributions of these components to the mechanics measurements.
It is evident that, for utilizing AFM to investigate the mechanics originating from tumor microenvironments, large gaps separate the current studies and practical applications. In addition to cancerous cells, healthy cells secrete exosomes during cellular life activities [103]. During tumor metastasis, primary cancerous cells secrete exosomes to establish premetastatic niches to establish a microenvironment suitable for the growth of cancerous cells [173]. To fully reveal the roles of exosome mechanics in promoting the formation of premetastatic niches, exosomes secreted by primary cancerous cells need to be isolated and analyzed. Researchers have cultured cancerous cells in vitro to produce exosomes [174], but notably, these exosomes cannot completely reflect the true situations of exosomes secreted by primary cancerous cells in vivo. Researchers have developed methods based on combining acoustics with microfluidics for isolating exosomes from whole blood in a label-free and contact-free manner [175], providing novel possibilities for harvesting exosomes secreted by cancerous cells in vivo. However, to date, studies applying AFM to investigate the mechanics of exosomes secreted by primary cancerous cells are rare, and the roles of exosome mechanics on tumor metastasis remain elusive. In the future, utilizing AFM to perform assays on exosomes secreted by cancerous cells from clinical patients will contribute to the understanding of how exosome mechanics influence the metastatic tumor microenvironment. In addition, current studies have demonstrated that AFM can be used to clearly visualize the structures and measure the mechanics of single nanofibrils in ECM [160–164], but how the mechanics of ECM nanofibrils influence the behaviors of tumor cells remains to be determined. Further studies (e.g., combining AFM studies on ECM nanofibrils and biochemical assays on tumor cells grown on an ECM), particularly AFM studies performed on the ECM prepared from the biopsy samples of clinical cancer patients (e.g., surgically excised tissues), are required to establish the correlation between nanoscopic ECM mechanics and tumor cell behaviors.
In general, AFM is very powerful for revealing the multiscale mechanics in tumor invasion and metastasis, providing significant novel insights into physical oncology and offering novel possibilities for cancer therapy. Notably, there is considerable room for the applications of AFM to tumor physics. In the future, more tumor-related studies performed with the use of AFM will lead to further revelations on the regulatory role of mechanics in tumor progression.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (61922081, 61873258, U1613220, and 91748212), the Key Research Program of Frontier Sciences CAS (ZDBS-LY-JSC043), the Youth Innovation Promotion Association CAS (2017243), and the LiaoNing Revitalization Talents Program (XLYC1907072).
Competing interests
The authors declare no competing interests.
Contributor Information
Mi Li, Email: limi@sia.cn.
Lian-qing Liu, Email: lqliu@sia.cn.
References
- 1.Bray F, Ferly J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, Vecchia CL, et al. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol. 2017;28:1117–23.. doi: 10.1093/annonc/mdx033. [DOI] [PubMed] [Google Scholar]
- 4.Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, Vecchia CL, et al. European cancer mortality predictions for the year 2018 with focus on colorectal cancer. Ann Oncol. 2018;29:1016–22.. doi: 10.1093/annonc/mdy033. [DOI] [PubMed] [Google Scholar]
- 5.Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, Vecchia CL, et al. European cancer mortality predictions for the year 2019 with focus on breast cancer. Ann Oncol. 2019;30:781–7. doi: 10.1093/annonc/mdz051. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 7.Lee WC, Kopetz S, Wistuba II, Zhang J. Metastasis of cancer: when and how? Ann Oncol. 2017;28:2045–7. doi: 10.1093/annonc/mdx327. [DOI] [PubMed] [Google Scholar]
- 8.Dillekas H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8:5574–6. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CK, Jeong SH, Jang C, Bae H, Kim YH, Park I, et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science. 2019;363:644–9. doi: 10.1126/science.aav0173. [DOI] [PubMed] [Google Scholar]
- 10.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50.. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theocharis AD, Karamanos NK. Proteoglycans remodeling in cancer: underlying molecular mechanisms. Matrix Biol. 2019;75:220–59.. doi: 10.1016/j.matbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Lobb RJ, Lima LG, Moller A. Exosomes: key mediators of metastasis and pre-metastatic niche formation. Semin Cell Dev Biol. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91.. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard RH, Huang YYS, Terentjev EM. Mechanics of biological networks: from the cell cytoskeleton to connective tissue. Soft Matter. 2014;10:1864–84. doi: 10.1039/c3sm52769g. [DOI] [PubMed] [Google Scholar]
- 17.Phillip JM, Aifuwa I, Walston J, Wirtz D. The mechanobiology of aging. Ann Rev Biomed Eng. 2015;17:113–41.. doi: 10.1146/annurev-bioeng-071114-040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladoux B, Mege RM. Mechanobiology of collective cell behaviors. Nat Rev Mol Cell Biol. 2017;18:743–57.. doi: 10.1038/nrm.2017.98. [DOI] [PubMed] [Google Scholar]
- 19.Dumont S, Prakash M. Emergent mechanics of biological structures. Mol Biol Cell. 2014;25:3461–65. doi: 10.1091/mbc.E14-03-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan P, Sinko R, LeDuc PR, Keten S. The role of mechanics in biological and bio-inspired systems. Nat Commun. 2015;6:7418. doi: 10.1038/ncomms8418. [DOI] [PubMed] [Google Scholar]
- 21.Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007;3:413–38. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–27. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammadi H, Sahai E. Mechanisms and impact of altered tumor mechanics. Nat Cell Biol. 2018;20:766–74.. doi: 10.1038/s41556-018-0131-2. [DOI] [PubMed] [Google Scholar]
- 24.Anderson RL, Balasas T, Callaghan J, Coombes RC, Evans J, Hall JA, et al. A framework for the development of effective anti-metastatic agents. Nat Rev Clin Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell MJ, Jain RK, Langer R. Engineering and physical sciences in oncology: challenges and opportunities. Nat Rev Cancer. 2017;17:659–75.. doi: 10.1038/nrc.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11:512–22. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Xi N, Wang Y, Liu L. Advances in atomic force microscopy for single-cell analysis. Nano Res. 2019;12:703–18.. [Google Scholar]
- 28.Alsteens D, Gaub HE, Newton R, Pfreundschuh M, Gerber C, Muller DJ. Atomic force microscopy-based characterization and design of bioninterfaces. Nat Rev Mater. 2017;2:17008. [Google Scholar]
- 29.Krieg M, Flaschner G, Alsteens D, Gaub BM, Roos WH, Suite GJL, et al. Atomic force microscopy-based mechanobiology. Nat Rev Phys. 2019;1:41–57. [Google Scholar]
- 30.Li M, Dang D, Xi N, Wang Y, Liu L. Nanoscale imaging and force probing of biomolecular systems using atomic force microscopy: from single molecules to living cells. Nanoscale. 2017;9:17643–66. doi: 10.1039/c7nr07023c. [DOI] [PubMed] [Google Scholar]
- 31.Dufrene YF, Ando T, Garcia R, Alsteens D, Martinez-Martin D, Engel A, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12:295–307. doi: 10.1038/nnano.2017.45. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Dang D, Liu L, Xi N, Wang Y. Atomic force microscopy in characterizing cell mechanics for biomedical applications: a review. IEEE Trans Nanobiosci. 2017;16:523–40.. doi: 10.1109/TNB.2017.2714462. [DOI] [PubMed] [Google Scholar]
- 33.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37.. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 35.Lee JS, Song J, Kim SO, Kim S, Lee W, Jackman JA, et al. Multifunctional hydrogel nano-probes for atomic force microscopy. Nat Commun. 2016;7:11566. doi: 10.1038/ncomms11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Xi N, Wang Y, Liu L. Atomic force microscopy in probing tumor physics for nanomedicine. IEEE Trans Nanotechnol. 2019;18:83–113. [Google Scholar]
- 37.Li M, Dang D, Liu L, Xi N, Wang Y. Imaging and force recognition of single molecular behaviors using atomic force microscopy. Sensors. 2017;17:200. doi: 10.3390/s17010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schillers H, Medalsy I, Hu S, Slade AL, Shaw JE. Peakforce tapping resolves individual microvilli on living cells. J Mol Recognit. 2016;29:95–101. doi: 10.1002/jmr.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dufrene YF, Martinez-Martin D, Medalsy I, Alsteens D, Muller DJ. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat Methods. 2013;10:847–54. doi: 10.1038/nmeth.2602. [DOI] [PubMed] [Google Scholar]
- 40.Alsteens D, Muller DJ, Dufrene YF. Multiparametric atomic force microscopy imaging of biomolecular and cellular systems. Acc Chem Res. 2017;50:924–31.. doi: 10.1021/acs.accounts.6b00638. [DOI] [PubMed] [Google Scholar]
- 41.Gavara N, Chadwick RS. Determination of the elastic moduli of thin samples and adherent cells using conical atomic force microscope tips. Nat Nanotechnol. 2012;7:733–6. doi: 10.1038/nnano.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lekka M. Discrimination between normal and cancerous cells using AFM. Bionanoscience. 2016;6:65–80. doi: 10.1007/s12668-016-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radmacher M. Measuring the elastic properties of living cells by the atomic force microscope. Methods Cell Biol. 2002;68:67–90. doi: 10.1016/s0091-679x(02)68005-7. [DOI] [PubMed] [Google Scholar]
- 44.Gavara N. A beginner’s guide to atomic force microscopy probing for cell mechanics. Microsc Res Tech. 2017;80:75–84. doi: 10.1002/jemt.22776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rotsch C, Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys J. 2000;78:520–35. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–92. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moeendarbary E, Valon L, Fritzsche M, Harris AR, Moulding DA, Thrasher AJ, et al. The cytoplasm of living cells behaves as a poroelastic material. Nat Mater. 2013;12:253–61. doi: 10.1038/nmat3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris AR, Daeden A, Charras GT. Formation of adherens junctions leads to the emergence of a tissue-level tension in epithelial monolayers. J Cell Sci. 2014;127:2507–17. doi: 10.1242/jcs.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Wen J, Xiao Y, Liu J, Hopyan S, Radisic M, et al. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano. 2014;8:3821–8. doi: 10.1021/nn500553z. [DOI] [PubMed] [Google Scholar]
- 50.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–3. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 51.Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, et al. The nanomechanical signature of breast cancer. Nat Nanotechnol. 2012;7:757–65. doi: 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- 52.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spill F, Reynolds DS, Kamm RD, Zaman MH. Impact of the physical microenvironment on tumor progression and metastasis. Curr Opin Biotechnol. 2016;40:41–8. doi: 10.1016/j.copbio.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li QS, Lee GYH, Ong CN, Lim CT. AFM indentation study of breast cancer cells. Biochem Biophys Res Commun. 2008;374:609–13. doi: 10.1016/j.bbrc.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 55.Calzado-Martin A, Encinar M, Tamayo J, Calleja M, San Paulo A. Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy. ACS Nano. 2016;10:3365–74. doi: 10.1021/acsnano.5b07162. [DOI] [PubMed] [Google Scholar]
- 56.Alibert C, Goud B, Manneville JB. Are cancer cells really softer than normal cells? Biol Cell. 2017;109:167–89.. doi: 10.1111/boc.201600078. [DOI] [PubMed] [Google Scholar]
- 57.Yu W, Sharma S, Gimzewski JK, Rao J. Nanocytology as a potential biomarker for cancer. Biomark Med. 2017;11:213–6. doi: 10.2217/bmm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 58.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 59.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Tsuji T, Ibaragi S, Hu G. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135–9. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buckley ST, Medina C, Davies AM, Ehrhardt C. Cytoskeletal re-arrangement in TGF-β1-induced alveolar epithelial-mesenchymal transition studied by atomic force microscopy and high-content analysis. Nanomedicine. 2012;8:355–64. doi: 10.1016/j.nano.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 62.Wang N, Zhang M, Chang Y, Niu N, Guan Y, Ye M, et al. Directly observing alterations of morphology and mechanical properties of living cancer cells with atomic force microscopy. Talanta. 2019;191:461–8. doi: 10.1016/j.talanta.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Kanlaya R, Peerapen P, Nilnumkhum A, Plumworasawat S, Sueksakit K, Thongboonkerd V. Epigallocatechin-3-gallate prevents TGF-β1-induced epithelial-mesenchymal transition and fibrotic changes of renal cells via GSK-3β/β-catenin/Snail1 and Nrf2 pathways. J Nutr Biochem. 2020;70:108266. doi: 10.1016/j.jnutbio.2019.108266. [DOI] [PubMed] [Google Scholar]
- 64.Helenius J, Heisenberg CP, Gaub HE, Muller DJ. Single-cell force spectroscopy. J Cell Sci. 2008;121:1785–91. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 65.Friedrichs J, Helenius J, Muller DJ. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nat Protoc. 2010;5:1353–61. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 66.Prystopiuk V, Feuillie C, Herman-Bausier P, Viela F, Alsteens D, Pietrocola G, et al. Mechanical forces guiding staphylococcus aureus cellular invasion. ACS Nano. 2018;12:3609–22.. doi: 10.1021/acsnano.8b00716. [DOI] [PubMed] [Google Scholar]
- 67.Friedrichs J, Legate KR, Schubert R, Bharadwaj M, Werner C, Muller DJ, et al. A practical guide to quantify cell adhesion using single-cell force spectroscopy. Methods. 2013;60:169–78. doi: 10.1016/j.ymeth.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 68.Beaussart A, El-Kirat-Chatel S, Sullan RMA, Alsteens D, Herman P, Derclaye S, et al. Quantifying the forces guiding microbial cell adhesion using single-cell force spectroscopy. Nat Protoc. 2014;9:1049–55. [Google Scholar]
- 69.Harris TJC, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11:502–14. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 70.Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nat Rev Mol Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 71.Smolyakov G, Thiebot B, Campillo C, Labdi S, Severac C, Pelta J, et al. Elasticity, adhesion, and tether extrusion on breast cancer cells provide a signature of their invasive potential. ACS Appl Mater Interfaces. 2016;8:27426–31.. doi: 10.1021/acsami.6b07698. [DOI] [PubMed] [Google Scholar]
- 72.Moreno-Cencerrado A, Iturri J, Pecorari I, Vivanco MDM, Sbaizero O, Toca-Herrera JL. Investigating cell-substrate and cell-cell interactions by means of single-cell-probe force spectroscopy. Microsc Res Tech. 2017;80:124–30.. doi: 10.1002/jemt.22706. [DOI] [PubMed] [Google Scholar]
- 73.Omidvar R, Tafazzoli-Shadpour M, Shokrgozar MA, Rostami M. Atomic force microscope-based single cell force spectroscopy of breast cancer cell lines: an approach for evaluating cellular invasion. J Biomech. 2014;47:3373–9. doi: 10.1016/j.jbiomech.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Azadi S, Tafazzoli-Shadpour M, Omidvar R, Moradi L, Habibi-Anbouhi M. Epidermal growth factor receptor targeting alters gene expression and restores the adhesion function of cancerous cells as measured by single cell force spectroscopy. Mol Cell Biochem. 2016;423:129–39.. doi: 10.1007/s11010-016-2831-x. [DOI] [PubMed] [Google Scholar]
- 75.Malek-Zietek KE, Targosz-Korecka M, Szymonski M. The impact of hyperglycemia on adhesion between endothelial and cancer cells revealed by single-cell force spectroscopy. J Mol Recognit. 2017;30:e2628. doi: 10.1002/jmr.2628. [DOI] [PubMed] [Google Scholar]
- 76.Hinterdorfer P, Dufrene YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3:347–55. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 77.Lo Giudice C, Dumitru AC, Alsteens D. Probing ligand-receptor bonds in physiologically relevant conditions using AFM. Anal Bioanal Chem. 2019;411:6549–59.. doi: 10.1007/s00216-019-02077-6. [DOI] [PubMed] [Google Scholar]
- 78.Hinterdorfer P, Garcia-Parajo MF, Dufrene YF. Single-molecule imaging of cell surfaces using near-field nanoscopy. Acc Chem Res. 2012;45:327–36. doi: 10.1021/ar2001167. [DOI] [PubMed] [Google Scholar]
- 79.Knoops B, Becker S, Poncin MA, Glibert J, Derclaye S, Clippe A, et al. Specific interactions measured by AFM on living cells between peroxiredoxin-5 and TLR4: relevance for mechanisms of innate immunity. Cell Chem Biol. 2018;25:550–9. doi: 10.1016/j.chembiol.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 80.Alsteens D, Newton R, Schubert R, Martinez-Martin D, Delguste M, Roska B, et al. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat Nanotechnol. 2017;12:177–83.. doi: 10.1038/nnano.2016.228. [DOI] [PubMed] [Google Scholar]
- 81.Zocher M, Bippes CA, Zhang C, Muller DJ. Single-molecule force spectroscopy of G-protein-coupled receptors. Chem Soc Rev. 2013;42:7801–15. doi: 10.1039/c3cs60085h. [DOI] [PubMed] [Google Scholar]
- 82.Puntheeranurak T, Newndlinger I, Kinne RKH, Hinterdorfer P. Single-molecule recognition force spectroscopy of transmembrane transporters on living cells. Nat Protoc. 2011;6:1443–52. doi: 10.1038/nprot.2011.370. [DOI] [PubMed] [Google Scholar]
- 83.Thoma J, Sapra KT, Muller DJ. Single-molecule force spectroscopy of transmembrane β-barrel proteins. Ann Rev Anal Chem. 2018;11:375–95.. doi: 10.1146/annurev-anchem-061417-010055. [DOI] [PubMed] [Google Scholar]
- 84.Li M, Xi N, Wang Y, Liu L. Atomic force microscopy as a powerful multifunctional tool for probing the behaviors of single proteins. IEEE Trans Nanobiosci. 2020;19:78–99. doi: 10.1109/TNB.2019.2954099. [DOI] [PubMed] [Google Scholar]
- 85.Hynes NE, Lane HA. Erbb receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 86.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–9. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 87.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–25. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chtcheglova LA, Hinterdorfer P. Simultaneous AFM topography and recognition imaging at the plasma membrane of mammalian cells. Semin Cell Dev Biol. 2018;73:45–56. doi: 10.1016/j.semcdb.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J, Chtcheglova LA, Zhu R, Hinterdorfer P, Zhang B, Tang J. Nanoscale organization of human GnRH-R on human bladder cancer cells. Anal Chem. 2014;86:2458–64. doi: 10.1021/ac403304g. [DOI] [PubMed] [Google Scholar]
- 90.Newton R, Delguste M, Koehler M, Dumitru AC, Laskowski PR, Muller DJ, et al. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat Protoc. 2017;12:2275–92.. doi: 10.1038/nprot.2017.112. [DOI] [PubMed] [Google Scholar]
- 91.Cascione M, De Matteis V, Rinaldi R, Leporatti S. Atomic force microscopy combined with optical microscopy for cells investigation. Microsc Res Tech. 2017;80:109–23. doi: 10.1002/jemt.22696. [DOI] [PubMed] [Google Scholar]
- 92.Hsu YT, Osmulski P, Wang Y, Huang YW, Liu L, Ruan J, et al. EpCAM-regulated transcription exerts influences on nanomechanical properties of endometrial cancer cells that promote epithelial-to-mesenchymal transition. Cancer Res. 2016;76:6171–82.. doi: 10.1158/0008-5472.CAN-16-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y, Kumacheva E. Hydrogel microenvironments for cancer spheroid growth and drug screening. Sci Adv. 2018;4:eaas8998. doi: 10.1126/sciadv.aas8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dyson PJ. The rise of 3D cellular spheroids: efficient culture via upward growth from a superamphiphobic surface. Natl Sci Rev. 2019;6:1068–9. doi: 10.1093/nsr/nwz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Costa EC, Moreira AF, de Melo-Diogo D, Gaspar VM, Carvalho MP, Correia IJ. 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnol Adv. 2016;34:1427–41.. doi: 10.1016/j.biotechadv.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 96.Charoen KM, Fallica B, Colson YL, Zaman MH, Grinstaff MW. Embedded multicellular spheroids as a biomimetic 3D cancer model for evaluating drug and drug-device combinations. Biomaterials. 2014;35:2264–71. doi: 10.1016/j.biomaterials.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Laschke MW, Menger MD. Spheroids as vascularization units: from angiogenesis research to tissue engineering applications. Biotechnol Adv. 2017;35:782–91.. doi: 10.1016/j.biotechadv.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 98.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J Control Release. 2012;164:192–204. doi: 10.1016/j.jconrel.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatzinikolaidou M. Cell spheroids: the new frontiers in in vitro models for cancer drug validation. Drug Discov Today. 2016;21:1553–60.. doi: 10.1016/j.drudis.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 100.Vyas V, Solomon M, D’Souza GGM, Huey BD. Nanomechanical analysis of extracellular matrix and cells in multicellular spheroids. Cell Mol Bioeng. 2019;12:203–14.. doi: 10.1007/s12195-019-00577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andolfi L, Greco SLM, Tierno D, Chignola R, Martinelli M, Giolo E, et al. Planar AFM macro-probes to study the biomechanical properties of large cells and 3D cell spheroids. Acta Biomater. 2019;94:505–13.. doi: 10.1016/j.actbio.2019.05.072. [DOI] [PubMed] [Google Scholar]
- 102.Taubenberger AV, Girardo S, Traber N, Fischer-Friedrich E, Krater M, Wagner K, et al. 3D microenvironment stiffness regulates tumor spheroid growth and mechanics via p21 and ROCK. Adv Biosyst. 2019;3:1900128. doi: 10.1002/adbi.201900128. [DOI] [PubMed] [Google Scholar]
- 103.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28.. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 105.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 106.Steinbichler TB, Dudas J, Riechelmann H, Skvortsova II. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–81.. doi: 10.1016/j.semcancer.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 107.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–48.. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer-implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–38.. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 111.Alderton GK. Metastasis: exosomes drive premetastatic niche formation. Nat Rev Cancer. 2012;12:447. doi: 10.1038/nrc3304. [DOI] [PubMed] [Google Scholar]
- 112.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–42. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–32.. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 114.Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: use them or target them? Semin Cell Dev Biol. 2018;78:13–21. doi: 10.1016/j.semcdb.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 115.Andaloussi SE, Mager I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–57.. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 116.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woo J, Sharma S, Gimzewski J. The role of isolation methods on a nanoscale surface structure and its effect on the size of exosomes. J Circ Biomark. 2016;5:11. doi: 10.5772/64148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, et al. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–6. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sharma S, Das K, Woo J, Gimzewski JK. Nanofilaments on glioblastoma exosomes revealed by peak force microscopy. J R Soc Interface. 2014;11:20131150. doi: 10.1098/rsif.2013.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sharma S, LeClaire M, Gimzewski JK. Ascent of atomic force microscopy as a nanoanalytical tool for exosomes and other extracellular vesicles. Nanotechnology. 2018;29:132001. doi: 10.1088/1361-6528/aaab06. [DOI] [PubMed] [Google Scholar]
- 121.Nakase I, Ueno N, Katayama M, Noguchi K, Takatani-Nakase T, Kobayashi NB, et al. Receptor clustering and activation by multivalent interaction through recognition peptides presented on exosomes. Chem Commun. 2017;53:317–20.. doi: 10.1039/c6cc06719k. [DOI] [PubMed] [Google Scholar]
- 122.Gonda A, Kabagwira J, Senthil GN, Wall NR. Internalization of exosomes through receptor-mediated endocytosis. Mol Cancer Res. 2019;17:337–47.. doi: 10.1158/1541-7786.MCR-18-0891. [DOI] [PubMed] [Google Scholar]
- 123.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–12. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hussey GS, Dziki JL, Badylak SF. Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater. 2018;3:159–73.. [Google Scholar]
- 125.Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–73.. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 126.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leight JL, Drain AP, Weaver VM. Extracellular matrix remodeling and stiffening modulate tumor phenotype and treatment response. Annu Rev Cancer Biol. 2017;1:313–34.. [Google Scholar]
- 129.Romero-Lopez M, Trinh AL, Sobrino A, Hatch MMS, Keating MT, Fimbres C, et al. Recapitulating the human tumor microenvironment: colon tumor-derived extracellular matrix promotes angiogenesis and tumor cell growth. Biomaterials. 2017;116:118–29.. doi: 10.1016/j.biomaterials.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ji T, Lang J, Wang J, Cai R, Zhang Y, Qi F, et al. Designing liposomes to suppress extracellular matrix expression to enhance drug penetration and pancreatic tumor therapy. ACS Nano. 2017;11:8668–78.. doi: 10.1021/acsnano.7b01026. [DOI] [PubMed] [Google Scholar]
- 131.Amelio I, Mancini M, Petrova V, Cairns RA, Vikhreva P, Nicolai S, et al. p53 mutants cooperate with HIF-1 transcriptional regulation of extracellular matrix components to promote tumor progression. Proc Natl Acad Sci USA. 2018;115:E10869–78. doi: 10.1073/pnas.1808314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jayadev R, Sherwood DR. Basement membrane. Curr Biol. 2017;27:R207–11. doi: 10.1016/j.cub.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 133.Sherwood DR. Cell invasion through basement membranes: an anchor of understanding. Trends Cell Biol. 2006;16:250–6. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 134.Chang TT, Thakar D, Weaver VM. Force-dependent breaching of the basement membrane. Matrix Biol. 2017;57:178–89.. doi: 10.1016/j.matbio.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van den Berg MCW, MacCarthy-Morrogh L, Carter D, Morris J, Bravo IR, Feng Y, et al. Proteolytic and opportunistic breaching of the basement membrane zone by immune cells during tumor initiation. Cell Rep. 2019;27:2837–46.. doi: 10.1016/j.celrep.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Glentis A, Oertle P, Mariani P, Chikina A, Marjou FE, Attieh Y, et al. Cancer-associated fibroblasts induce metalloprotease-independent cancer cell invasion of the basement membrane. Nat Commun. 2017;8:924. doi: 10.1038/s41467-017-00985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat Rev Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miller RT. Mechanical properties of basement membrane in health and disease. Matrix Biol. 2017;57:366–73.. doi: 10.1016/j.matbio.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 139.Chaudhuri O, Koshy ST, da Cunha CB, Shin JW, Verbeke CS, Allison KH, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13:970–8. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 140.Achterger VF, Buscemi L, Diekmann H, Smith-Clerc J, Schwengler H, Meister JJ, et al. The nano-scale mechanical properties of the extracellular matrix regulate dermal fibroblast function. J Invest Dermatol. 2014;134:1862–72.. doi: 10.1038/jid.2014.90. [DOI] [PubMed] [Google Scholar]
- 141.Li M, Xi N, Wang Y, Liu L. Tunable hybrid biopolymeric hydrogel scaffolds based on atomic force microscopy characterizations for tissue engineering. IEEE Trans Nanobiosci. 2019;18:597–610. doi: 10.1109/TNB.2019.2922968. [DOI] [PubMed] [Google Scholar]
- 142.Chang J, Chaudhuri O. Beyond proteases: basement membrane mechanics and cancer invasion. J Cell Biol. 2019;218:2456–69.. doi: 10.1083/jcb.201903066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pati F, Jang J, Ha DH, Kim SW, Rhie JW, Shim JH, et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hoshiba T. Cultured cell-derived decellularized matrices: a review towards the next decade. J Mater Chem B. 2017;5:4322–31.. doi: 10.1039/c7tb00074j. [DOI] [PubMed] [Google Scholar]
- 146.Hoshiba T, Lu H, Kawazoe N, Chen G. Decellularized matrices for tissue engineering. Expert Opin Biol Ther. 2010;10:1717–28. doi: 10.1517/14712598.2010.534079. [DOI] [PubMed] [Google Scholar]
- 147.Jorba I, Uriarte JJ, Campillo N, Farre R, Navajas D. Probing micromechanical properties of the extracellular matrix of soft tissues by atomic force microscopy. J Cell Physiol. 2017;232:19–26. doi: 10.1002/jcp.25420. [DOI] [PubMed] [Google Scholar]
- 148.Andreu I, Luque T, Sancho A, Pelacho B, Iglesias-Garcia O, Melo E, et al. Heterogeneous micromechanical properties of the extracellular matrix in healthy and infarcted hearts. Acta Biomater. 2014;10:3235–42. doi: 10.1016/j.actbio.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 149.Gimenez A, Uriarte JJ, Vieyra J, Navajas D, Alcaraz J. Elastic properties of hydrogels and decellularized tissue sections used in mechanobiology studies probed by atomic force microscopy. Microsc Res Tech. 2017;80:85–96. doi: 10.1002/jemt.22740. [DOI] [PubMed] [Google Scholar]
- 150.Alcaraz J, Otero J, Jorba I, Navajas D. Bidirectional mechanobiology between cells and their local extracellular matrix probed by atomic force microscopy. Semin Cell Dev Biol. 2018;73:71–81. doi: 10.1016/j.semcdb.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 151.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–63. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tello M, Spenle C, Hemmerle J, Mercier L, Fabre R, Allio G, et al. Generating and characterizing the mechanical properties of cell-derived matrices using atomic force microscopy. Methods. 2016;94:85–100. doi: 10.1016/j.ymeth.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 153.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–32. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 154.Felding-Habermann B, O’Toole TE, Smith JW, Fransvea E, Ruggeri ZM, Ginsberg MH, et al. Integrin activation controls metastasis in human breast cancer. Proc Natl Acad Sci USA. 2001;98:1853–8. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533–48.. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prince E, Kumacheva E. Design and applications of man-made biomimetic fibrillar hydrogels. Nat Rev Mater. 2019;4:99–115. [Google Scholar]
- 157.Dalby MJ, Gadegaard N, Oreffo ROC. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater. 2014;13:558–69. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 158.Bertocchi C, Wang Y, Ravasio A, Hara Y, Wu Y, Sailov T, et al. Nanoscale architecture of cadherin-based cell adhesion. Nat Cell Biol. 2017;19:28–37. doi: 10.1038/ncb3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moreno-Arotzena O, Meier JG, Del Amo C, Garcia-Aznar JM. Characterization of fibrin and collagen gels for engineering wound healing models. Materials. 2015;8:1636–51.. doi: 10.3390/ma8041636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li M, Xi N, Wang Y, Liu L. Composite nanostructures and adhesion analysis of natural plant hydrogels investigated by atomic force microscopy. IEEE Trans Nanobiosci. 2019;18:448–55.. doi: 10.1109/TNB.2019.2911044. [DOI] [PubMed] [Google Scholar]
- 161.Li M, Xi N, Wang Y, Liu L. Nanoscale multiparametric imaging of peptide-assembled nanofibrillar hydrogels by atomic force microscopy. IEEE Trans Nanotechnol. 2019;18:315–28.. [Google Scholar]
- 162.Adamcik J, Jung JM, Flakowski J, De Los Rios P, Dietler G, Mezzenga R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat Nanotechnol. 2010;5:423–8. doi: 10.1038/nnano.2010.59. [DOI] [PubMed] [Google Scholar]
- 163.Usov I, Nystrom G, Adamcik J, Handschin S, Schutz C, Fall A, et al. Understanding nanocellulose chirality and structure-properties relationship at the single fibril level. Nat Commun. 2015;6:7564. doi: 10.1038/ncomms8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bera S, Mondal S, Xue B, Shimon LJW, Cao Y, Gazit E. Rigid helical-like assemblies from a self-aggregating tripeptide. Nat Mater. 2019;18:503–9. doi: 10.1038/s41563-019-0343-2. [DOI] [PubMed] [Google Scholar]
- 165.Cao Y, Bolisetty S, Adamcik J, Mezzenga R. Elasticity in physically cross-linked amyloid fibril networks. Phys Rev Lett. 2018;120:158103. doi: 10.1103/PhysRevLett.120.158103. [DOI] [PubMed] [Google Scholar]
- 166.Li M, Liu L, Xi N, Wang Y. Atomic force microscopy studies on cellular elastic and viscoelastic properties. Sci China Life Sci. 2018;61:57–67. doi: 10.1007/s11427-016-9041-9. [DOI] [PubMed] [Google Scholar]
- 167.Guz NV, Patel SJ, Dokukin ME, Clarkson B, Sokolov I. AFM study shows prominent physical changes in elasticity and pericellular layer in human acute leukemia cells due to inadequate cell-cell communication. Nanotechnology. 2016;27:494005. doi: 10.1088/0957-4484/27/49/494005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li M, Liu L, Xi N, Wang Y. Nanoscale monitoring of drug actions on cell membrane using atomic force microscopy. Acta Pharmacol Sin. 2015;36:769–82. doi: 10.1038/aps.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Polacheck WJ, Chen CS. Measuring cell-generated forces: a guide to the available tools. Nat Methods. 2016;13:415–23. doi: 10.1038/nmeth.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, et al. Three-dimensional force microscopy of cells in biopolymer networks. Nat Methods. 2016;13:171–6. doi: 10.1038/nmeth.3685. [DOI] [PubMed] [Google Scholar]
- 171.Roca-Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat Cell Biol. 2017;19:742–51.. doi: 10.1038/ncb3564. [DOI] [PubMed] [Google Scholar]
- 172.Ciasca G, Sassun TE, Minelli E, Antonelli M, Papi M, Santoro A, et al. Nano-mechanical signature of brain tumors. Nanoscale. 2016;8:19629–43.. doi: 10.1039/c6nr06840e. [DOI] [PubMed] [Google Scholar]
- 173.Kiberstis PA. Metastasis: an evolving story. Science. 2016;352:162–3. doi: 10.1126/science.352.6282.162. [DOI] [PubMed] [Google Scholar]
- 174.Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018;37:6105–18.. doi: 10.1038/s41388-018-0391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA. 2017;114:10584–9. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]