Abstract
Coronavirus disease, first detected in late 2019 (COVID-19), has spread fast throughout the world, leading to high mortality. This condition can be diagnosed using RT-PCR technique on nasopharyngeal and throat swabs with sensitivity values ranging from 30 to 70%. However, chest CT scans and X-ray images have been reported to have sensitivity values of 98 and 69%, respectively. The application of machine learning methods on CT and X-ray images has facilitated the accurate diagnosis of COVID-19. In this study, we reviewed studies which used machine and deep learning methods on chest X-ray images and CT scans for COVID-19 diagnosis and compared their performance. The accuracy of these methods ranged from 76% to more than 99%, indicating the applicability of machine and deep learning methods in the clinical diagnosis of COVID-19.
Keywords: COVID-19, machine learning, detection, biomarker, X-ray image
Introduction
First identified in Wuhan, China, severe pneumonia caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) quickly spread all over the world. The resultant disorder was named coronavirus disease (COVID-19) (1, 2). COVID-19 has various clinical symptoms, including fever, cough, dyspnea, fatigue, myalgia, headache, and gastrointestinal complications (3–5). Diagnosis of COVID-19 infection through RT-PCR on nasopharyngeal and throat swab samples has been reported to yield positive results in 30–70% of cases (6, 7). On the other hand, chest CT scans and X-ray images have been reported to have sensitivity values of 98 and 69%, respectively (7–9). The most typical radiological signs in these patients include multifocal and bilateral ground-glass opacities and consolidations, particularly in the peripheral and basal sites (10). However, interpretation of the results of these imaging techniques by expert radiologists might encounter some problems leading to reduced sensitivity (11). Artificial intelligence has recently gained the attention of both clinicians and researchers for the appropriate management of the COVID-19 pandemic (12). As an accurate method, artificial intelligence is able to identify abnormal patterns of CT and X-ray images. Using this method, it is possible to assess certain segment regions and take precise structures in chest CT images facilitating diagnostic purposes. Artificial intelligence methods have been shown to detect COVID-19 and distinguish this condition from other pulmonary disorders and community-acquired pneumonia (13). Both deep learning and machine learning approaches have been used to predict different aspects of the COVID-19 outbreak. Support vector and random forest are among the most applied machine learning methods, while Convolutional Neural Network (CNN), Long Short-Term Memory (LSTM), Generative Adversarial Networks (GAN), and Residual Neural network are among the deep learning methods used in this regard (14). In this study, we reviewed studies which used machine and deep learning methods on chest X-ray images and CT scans for the purpose of COVID-19 diagnosis and compared their performance.
Methods
Search Strategy
The research question was: “What are the applications of machine learning techniques and their performances in COVID-19 diagnosis using X-ray images?”. The search of the present review was based on the PICO elements, which were as follows:
P (Problem/Patient/Population): Patients' CT scans and Chest X-rays.
I (Intervention/Indicator): Machine/deep learning models for diagnosis of Covid-19 patients
C (Comparison): Ground truth or reference standards
O (Outcome): Performance measurements including accuracy, AUC score, sensitivity, and specificity.
In other words, we were looking for publications that evaluated the performance of any machine learning or deep learning approaches based on inclusion and exclusion criteria. Studies that used other types of medical image modalities (e.g., ultrasound images) were excluded. An electronic search was conducted on PubMed, Google Scholar, Scopus, Embase, arXiv, and medRxiv for finding the relevant literature. Duplicate studies were removed. Studies that were cited within the retrieved papers were reviewed for finding missing studies. For identifying proper journal papers and conference proceedings, investigators screened the title and abstracts based on inclusion and exclusion criteria independently. Finally, considering the inclusion and exclusion criteria, investigators identified the eligible publications in this stage independently.
Inclusion Criteria
The following inclusion criteria were used in the selection of the articles: (1) Studies that applied machine learning or deep learning algorithms, (2) Studies that evaluated the measurement of model outcomes in comparison with ground truth or gold standards, and (3) Studies that used algorithms to analyze radiographic images (CT scan, Chest X-ray, etc.).
Exclusion Criteria
The following studies were excluded: (1) Studies that used any machine learning or deep learning approaches for problems not directly related to the COVID-19 imaging, (2) Studies that used other artificial intelligence techniques or classic computer vision approaches, (3) Studies that did not provide a clear explanation of the machine learning or deep learning model that was used to solve their problem, and (4) Review studies. The latter were excluded as we did not aim to review the data on an original level without any second-hand interpretations (summation, inferences, etc.).
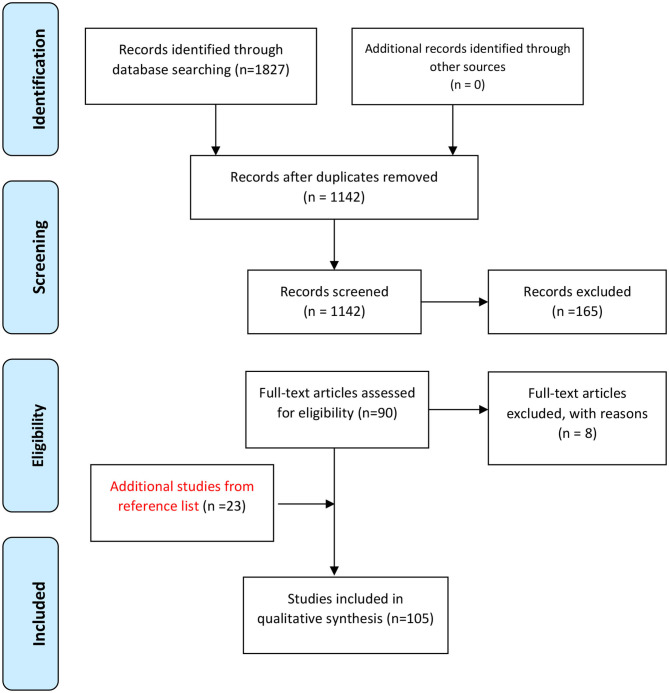
Figure 1 shows the flowchart of the study design.
Figure 1.
PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) chart showing the process of systematic identification, screening, and selection of articles.
Results
We obtained 105 studies that used machine or deep learning methods to assess chest images of COVID-19 patients. These studies have used different analytical methods. For instance, Ardakani et al. (15) have assessed radiological features of CT images obtained from patients with COVID-19 and non-COVID-19 pneumonia. They used decision tree, K-nearest neighbor, naïve Bayes, support vector machine, and ensemble classifiers to find the computer-aided diagnosis system with the best performance in distinguishing COVID-19 patients from non-COVID-19 pneumonia. They reported that site and distribution of pulmonary involvement, the quantity of the pulmonary lesions, ground-glass opacity, and crazy-paving as the most important characteristics for differentiation of these two sets of patients. Their computer-aided diagnosis method yielded the accuracy of 91.94%, using an ensemble (COVIDiag) classifier. Alazab et al. (16) have developed an artificial-intelligence method based on a deep CNN to evaluate chest X-ray images and detection of COVID-19 patients. Their method yielded an F-measure ranging from 95 to 99%. Notably, three predicting strategies could forecast the numbers of COVID-19 confirmations, recoveries, and mortalities over the upcoming week. The average accuracy of the prediction models were 94.80 and 88.43% in two different countries. Albahli has applied deep learning-based models on CT images of COVID-19 patients. He has demonstrated a high performance of a Deep Neural Network model in detecting COVID-19 patients and has offered an efficient assessment of chest-related disorders according to age and sex. His proposed model has yielded 89% accuracy in terms of GAN-based synthetic data (17). Automatic detection of COVID-19 based on X-ray images has been executed through the application of three deep learning models, including Inception ResNetV2, InceptionNetV3, and NASNetLarge. The best results have been obtained from InceptionNetV3, which yielded the accuracy levels of 98.63 and 99.02% with and without application of data augmentation in model training, respectively (18). Alsharman et al. (19) have used the CNN method to detect COVID-19 based on chest CT images in the early stages of disease course. Authors have reported high accuracy of GoogleNet CNN architecture for diagnosis of COVID-19. Altan et al. (20) have used a hybrid model comprising two-dimensional curvelet transformation, chaotic salp swarm algorithm, and deep learning methods for distinguishing COVID-19 from other pneumonia cases. Application of their proposed model on chest X-ray images has led to accurate diagnosis of COVID-19 patients (Accuracy = 99.69%, Sensitivity = 99.44% and Specificity = 99.81%). Apostolopoulos et al. (21) have used a certain CNN strategy, namely MobileNet on X-Ray images of COVID-19 patients. This method has yielded more than 99% accuracy in the diagnosis of COVID-19. In another study, Ardakani et al. (22) used 10 CNN strategies, namely AlexNet, VGG-16, VGG-19, SqueezeNet, GoogleNet, MobileNet-V2, ResNet-18, ResNet-50, ResNet-101, and Xception, to differentiate COVID-19 cases from non-COVID-19 patients. They have demonstrated the best diagnostic values for ResNet-101 and Xception, both of them having area under curve (AUC) values higher than 0.99 which is superior to the performance of the radiologist. Das et al. (23) have used the CNN model Truncated InceptionNet to diagnose COVID-19 from other non-COVID and/or healthy cases based on chest X-ray. Their suggested model yielded AUC of 1.0 in distinguishing COVID-19 patients from combined Pneumonia and healthy subjects. Tables 1, 2 summarize the features of studies which adopted machine learning methods in CT images and chest X-ray of COVID-19 patients, respectively.
Table 1.
Characteristics of papers that used CT images or a combination of X-ray and CT images.
| Author, year | Data source | Data structure and size | Data preprocessing | Best model structure(s) | Performance measurements (on the best model) | References | |||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | AUC score | Sensitivity | Specificity | ||||||
| Abbasian et al. (2020) | Iran University of Medical Sciences (IUMS) | 306 COVID-19 patients; 306 COVID-19 pneumonia (CT images) |
Extracting 20 features of CT images | Ensemble | 91.94% | 0.965 | 93.54% | 90.32% | (15) |
| Alsharman et al. (2020) | “COVID-CT-dataset” | CT images | Binarization (the separation of the object and background is known as Binarization); Converting input image from 2D Grayscale to 3D Color |
GoogleNet CNN | 82.14% | (19) | |||
| Ardakani et al. (2020) | Private dataset | 108 COVID-19 patients; 86 viral pneumonia diseases (CT images) |
Converted to the gray-scale Cropped and resized to 60 * 60 pixels | ResNet-101 Xception |
Resnet: 99.51% Xception: 99.02% (compared to 86.7% in human) |
Resnet: 0.994 Xception: 0.994% (compared to 0.873 in human) |
Resnet: 100% Xception: 98.04% (compared to 89.21% in human) |
Resnet: 99.02% Xception: 100% (compared to 83.33% in human) |
(22) |
| Aswathy et al. (2020) | “National Cancer Institute and the Cancer Image Archive” | 1,763 normal patients; 63 pneumonia patients |
Thresholding; Texture-based feature extractionwith a wrapper |
CNN | 99% | – | – | – | (24) |
| Bai et al. (2020) | Private dataset |
521 COVID-19 patients; 665 other pulmonary diseases (CT images) |
Lung segmentation; Generate an 8-bit image for each axial slice by applying Lung windowing to the Hounsfield units |
EfficientNet B4 | 96% (compared to 85% in human) | 0.95 | 95% (compared to 79% in human) | 96% (compared to 88% in human) | (11) |
| Bridge et al. (2020) |
“Toy dataset;” “Italian Society of Radiology;” “Shenzhen Hospital X-Ray dataset;” “ChestX-Ray8;” “COVID-CT-Dataset” |
129 COVID-19 patients; 62,267 normal patients; 5,689 pneumonia patients (X-ray images) 30 COVID-19 patients; 1,919 normal patients (CT images) |
Using the GEV activation function for unbalanced data | Inception V3 | 100% | – | 100% | 100% | (25) |
| Butt et al. (2020) | Not mentioned |
219 images from 110 COVID-19 patients; 399 normal patients (CT images) |
Image processing method base on HU values | 3D CNN | – | 0.996 | 98.2% | 92.2% | (26) |
| Dey et al. (2020) | “COVID-19 CT segmentation dataset;” “Chest X-rays (Radiopaedia)” |
200 COVID-19 patients; 200 normal patients (grayscale lung CTI images) |
Segmenting lung area related to pneumonia infection; Extracting CWT, DWT, EWT features from original image and Haralick, Hu moments from binary segmented area Feature selection based on statistical tests |
KNN | 87.75% | – | 89.00% | 86.50% | (27) |
| El Asnaoui et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Kermany et al. (28) |
2,780 Bacterial pneumonia patients; 1,493 Coronavirus patients; 231 COVID-19 patients; 1,583 normal patients (X-ray and CT images) |
Intensity Normalization; Contrast Limited Adaptive Histogram Equalization |
Inception ResNetV2; Densnet201 | Inception-ResNetV2: 92.18% Densnet201: 88.09% |
Inception-ResNetV2: 0.920 Densnet201: 0.879 |
Inception-ResNetV2: 92.11% Densnet201: 87.99% |
Inception-ResNetV2: 96.6% Densnet201: 94.00% |
(29) |
| Han et al. (2020) | “COVID-19 hospitals in Shandong Province” | 79 COVID-19 patients; 100 pneumonia patients; 130 normal patients (CT images) |
Data augmentation | AD3D-MIL | 97.9% | 0.99 | 97.9% | 97.9% | (30) |
| Harmon et al. (2020) | Private dataset |
386 COVID-19 patients; 1,011 negative COVID-19 patients (CT images) |
Lung segmentation; clipping images to HU range (−1,000, 500); Data augmentation (flipping, rotation, image intensity and contrast adjustment, adding random Gaussian noise); |
Hybrid 3D based on Densnet-121 | 90.8% | – | 84% | 93% | (31) |
| Hasan et al. (2020) | “Radiopaedia and the cancer imaging archive websites” | 118 COVID-19 patients; 96 pneumonia patients; 107 normal patients (CT images) |
Histogram Thresholding; Dilation; Hole Filling |
LSTM | 99.68% | – | 100% | – | (32) |
| Hu et al. (2020) | “Hospital of Wuhan Red Cross Society;” “Shenzhen Hospital;” “TCIA dataset;” “Cancer Centre Archive (TCIA) Public Access;” “MD Anderson Cancer Centre;” “Memorial Sloan-Kettering Cancer Center;” “MAASTRO clinic” |
150 COVID-19 patients; 150 pneumonia patients; 150 normal patients (CT images) |
Data augmentation | CNN | 96.2% | 0.970 | 94.5% | 95.3% | (33) |
| Jaiswal et al. (2020) | “The SARS-CoV-2 CT scan dataset” | 1,262 COVID-19 patients; 1,230 non-COVID-19 patients (CT images) | Data augmentation (rotation up to 15, slant-angle of 0.2, horizontal flipping, filling new pixels as “nearest” for better robustness) | DenseNet201 | 96.25% | 0.97 | 96.29% | 96.21% | (34) |
| Kang et al. (2020) | “Tongji Hospital of Huazhong University of Science and Technology;” “China-Japan Union Hospital of Jilin University;” “Ruijin Hospital ofShanghai Jiao Tong University” |
1,495 COVID-19 patients; 1,027 community-acquired pneumonia (CAP) patients (CT images) |
Normalization; Standardization |
NN | 93.90% | – | 94.60% | 91.70% | (35) |
| Lessmann et al. (2020) | “Emergency wards of an Academic center and teaching hospital in the Netherlands in March and April 2020” | 237 COVID-19 patients; 606 normal patients (CT images) |
Resampling; Normalization |
CORADS-AI | – | 0.95 | 85.7% | 89.8% | (36) |
| Li et al. (2020) | Private | 1,296 COVID-19 patients; 1,325—patients; 1,735 community-acquired (CT images) |
Segmenting lung area with U-net | COVNet (ResNet-50) | – | 0.96 | 90% | 96% | (13) |
| Li et al. (2020) | More than 10 medical centers between Nov. 11th, 2010 and Feb. 9th, 2020 | 305 images from 251 COVID-19 patients; 872 images from 869 pneumonia patients; 1,498 images from 1,475 non-pneumonia patients (CT images) |
DL-based algorithm Image processing method base on HU values; Data augmentation |
3D ResNet-18 | Recall = 88% Precision = 89.6% F1 score = 87.8% |
(37) | |||
| Liu et al. (2020) | Private | 73 COVID-19 patients; 27 general pneumonia patients (CT images) |
ROI delineation based on ground-glass opacities (GGOs); 13 gray level co-occurrence matrix (GLCM) features, 15 gray level-gradient co-occurrence matrix (GLGCM) features, and six histogram features were extracted; Feature selection by ReliefF; |
An ensemble of bagged tree (EBT) | 94.16% | 0.99 | 88.62% | 100% | (38) |
| Mei et al. (2020) | Private | 419 COVID-19 patients 486 non-COVID-19 patients (CT images) |
Selecting pertinent slices by image segmentation to detect parenchymal tissue; Segmenting lung in CT images; |
ResNet-18 | 79.6% | 0.86 | 83.6% | 75.9% | (39) |
| Panwar et al. (2020) | “COVID-chest X-ray;” “SARS-COV-2 CT-scan;” “Chest X-Ray Images (Pneumonia);” |
206 COVID-19 patients; 364 Pneumonia patients (X-ray and CT images) |
– | VGG-19 | 95.61% (COVID-19 vs. Pneumonia) | – | 96.55% (COVID-19 vs. Pneumonia) | 95.29% (COVID-19 vs. Pneumonia) | (40) |
| Pathak et al. (2020) | 2 different COVID-19 datasets of chest-CT images | CT images | – | Deep bidirectional long short-term memory network with mixture density network (DBM) | 96.19% (multi-class) | 0.96 (multi-class) | 96.22% (multi-class) | 96.16% (multi-class) | (41) |
| Pathak et al. (2020) | “COVID-19 open datasets of chest CT images” | 413 COVID-19 patients; 439 normal or pneumonia infected patients (CT images) |
– | ResNet-50 | 93.01% | – | 91.45% | 94.77% | (41) |
| Peng et al. (2020) | Collected from PMC | 606 COVID-19 patients; 222 Influenza; 397 Normal or other disease patients (CT images) |
– | DenseNet121 | – | 0.87 | 72.3% | 85.2% | (42) |
| Pu et al. (2020) | Private | 498 COVID-19 patients; 497 community-acquired pneumonia (CAP) (CT images) |
Data augmentation [rotation, translation, vertical/horizontal flips, Hounsfield Unit (HU) shift, smoothing (blurring) operation, Gaussian noise] | 3D CNNs | 99% | 0.7 | – | – | (43) |
| Raajan et al. (2020) | X-ray images on public medical Github repositories; Kaggle chest X-ray database |
349 images from 216 COVID-19 patients; 1,341 Normal patients (CT images) |
Normalization | ResNet-16 | 95.09% | – | 100% | 81.89% | (44) |
| Rajaraman et al. (2020) | “Pediatric CXR dataset;” “RSNA CXR dataset;” “Twitter COVID-19 CXR dataset;” “Montreal COVID-19 CXR dataset” |
313 COVID-19 patients; 7,595 pneumonia of unknown type patients; 2,780 bacterial pneumonia; 7,595 Normal patients (X-ray images) |
Median filtering; Normalization; Standardization |
Inception-V3 | 99.01% | 0.997 | 98.4% | – | (45) |
| Sakagianni et al. (2020) | COVID-19 articles on medRxiv and bioRxiv | 349 COVID-19 patients; 397 non-COVID-19 patients (CT images) |
– | AutoML Cloud Vision | – | 0.94 | 88.31% | – | (46) |
| Sharma (2020) | Dataset from Italian Society of Medical and Interventional Radiology; COVID-CT available in GitHub; Dataset from hospitals in Moscow, Russia; Dataset from SAL Hospital, Ahmedabad, India; |
800 COVID-19 patients; 600 Viral Pneumonia; 800 normal patients (CT images) |
Ground-glass opacities (GGO), consolidation and pleural effusion are the features | ResNet | 91% | – | 92.1% | 90.29% | (47) |
| Singh et al. (2020) | Not mentioned | CT images | – | Multi-objective differential evolution (MODE) based CNN | 90.22% | – | 91.17% | 89.23% | (48) |
| Song et al. (2020) | Private (two hospitals in China); | 98 COVID-19 patients; 103 non-COVID-19 pneumonia (CT images) |
– | BigBiGAN | – | 0.972 | 92% | 91% | (49) |
| Wang et al. (2020) | Private | 1,315 COVID-19 patients; 2,406 ILD patients; 936 Normal patients (CT images) |
Lobe Segmentation by 3D-Unet; Converting CT numbers to grayscale |
PA-66 model | 93.3% | 0.973 | 97.6% | – | (50) |
| Wang et al. (2020) | COVID-19 dataset (private); CT-epidermal growth factor receptor (CT-EGFR) dataset (private); |
754 COVID-19 patients; 271 bacterial pneumonia 29 viral pneumonia; 42 Other pneumonia (CT images) *The CT-EGFR dataset was used for auxiliary training of the DL system |
Lung segmentation; Using a fully automatic DL model (DenseNet121-FPN); suppress the intensities of non-lung areas inside the lung ROI; |
COVID-19Net (DenseNet-like architecture) | Test-set1: 78.32% Test-set2: 80.12% |
Test-set1: 0.87 Test-set2: 0.88 |
Test-set1: 80.39% Test-set2: 79.35% |
Test-set1: 76.61% Test-set2: 81.16% |
(51) |
| Warman et al. (2020) | “Public sources” | 606 COVID-19 patients; 224 viral pneumonias patients; 74 Normal patients (CT images) |
Data augmentation | YOLOv3 model | 96.80% | 0.966 | 98.33% | 94.95% | (52) |
| Wu et al. (2020) | Private | 368 COVID-19 patients; 127 other pneumonia (CT images) |
Lung region in each axial, coronal and sagittal CT slices were segmented using threshold segmentation and morphological optimization algorithms; The slice with the most pixels in the segmented lung area from each of the axial, coronal and sagittal views was selected as the inputs of the deep learning network; |
Multi-view fusion ResNet50 architecture | 76% | 0.819 | 81.1% | 61.5% | (53) |
| Xu et al. (2020) | Private “Hospitals in Zhejiang Province, China.” | 219 images from 110 COVID-19 patients; 224 Influenza-A viral pneumonia patients; 175 Normal patients (CT images) |
Image processing method base on HU values | 3D CNN segmentation model | 86.7% | – | 86.7% | – | (54) |
| Xu et al. (2020) | Private | 432 COVID-19 patients; 76 other viral pneumonia; 350 bacterial pneumonia; 418 normal patients (CT images) |
Sampling 5 subsets of CT slices from all sequential images of one CT case to picture the infected lung regions. | 3D-Densenet | – | 0.98 | 97.5% (differentiating COVID-19 from three types of non-COVID-19 cases) (compared to 79% in human) | 89.4% (differentiating COVID-19 from three types of non-COVID-19 cases) (compared to 90% in human) | (55) |
| Yan et al. (2020) | Private | 416 images from 206 COVID-19 patients; 412 common pneumonia patients (CT images) |
Transferring image slices to JPG; Normalization |
MSCNN | 97.7% | 0.962 | 99.5% | 95.6% | (56) |
| Yang et al. (2020) | Private | 146 COVID-19 patients; 149 normal patients (CT images) |
For patients, images containing round-glasses opacity (GGO), GGO with consolidation was selected; for healthy control, every 3 slices containing pulmonary parenchyma were selected; Lung windowing is performed over all image slices; |
DenseNet | 92% (compared to 95% in human) | 0.98 | 97% (compared to 94% in human) | 87% (compared to 96% in human) | (57) |
| Yu et al. (2020) | Private | 202 COVID-19 patients (CT images) | – | DenseNet-201 with the cubic SVM model | 95.2% | 0.99 | 91.87% | 96.87% | (58) |
| Al-Karawi et al. (2020) | “COVID-CT-Dataset” | 275 COVID-19 patients; 195 normal patients (CT images) |
Adaptive winner filter followed by inversion; Feature extraction by the FFT-spectrum |
SVM | 95.37% | – | 95.99% | 94.76% | (59) |
| Alom et al. (2020) | Publicly available datasets; “Kaggle repository” |
3,875 pneumonia patients; 1,341 normal patients (X-Ray images) 178 COVID-19 patients; 247 normal patients (CT images) |
Data augmentation; Adaptive Thresholding Approach |
IRRCNN model; NABLA-3 network model |
X-ray images: 84.67% CT images: 98.78% |
0.93 | – | – | (60) |
| Barstugan et al. (2020) | From the Italian Society of Medical and Interventional Radiology | 150 COVID-19 patients (CT images) | 13 features were extracted by Gray Level Size Zone Matrix (GLSZM) | SVM | 98.77% | – | 97.72% | 99.67% | (61) |
| Chen et al. (2020) | Private dataset | 25,989 images from 51 COVID-19 patients; 20,107 images from 55 normal patients (retrospective dataset); 13,911 images from 27 consecutive patients (prospective dataset) (CT images) |
Filtering | Deep learning model | Retrospective dataset: 95.24%; Prospective dataset: 92.59% (per patient) |
– | Retrospective dataset: 100%; Prospective dataset: 100% (per patient) |
Retrospective dataset:93.55%; Prospective dataset: 81.82% (per patient) |
(62) |
| Farid et al. (2020) | Kaggle database | 51 COVID-19 patients (CT images) | Feature extraction (MPEG7 Histogram Filter, Gabor Image Filter, Pyramid of Rotation-Invariant Local Binary Pattern, Fuzzy 64-bin Histogram Image Filter); Feature selection by composite hybrid feature selection |
CHFS-Stacked (jrip, RF) with Naïve Bayes classifier | 96.07% | – | – | – | (63) |
| Gozes et al. (2020) | Dataset1:ChainZ; Dataset2: Private; Dataset3: ChainZ; |
50 suspicious COVID-19 patients from dataset1 used for training; 56 COVID-19 patients; 51 normal patients (CT images) used for testing |
Data augmentation (rotation, horizontal flips and cropping) | Resnet-50-2D | – | 0.996 | 98.2% | 92.2% | (64) |
| Jin et al. (2020) | Three centers in China; “LIDC-IDRI;” “Tianchi-Alibaba;” “CC-CCII” |
2,529 images from 1,502 COVID-19 patients; 1,338 images from 1,334 CAP patients; 135 images from 83 influenza-A/B patients; 258 images from 258 normal patients (CT images) |
– | CNN | – | 0.977 | 90.19% | 95.76% | (65) |
| Jin et al. (2020) | Data from three different centers in Wuhan; Data from three publicly available databases, LIDC-IDRI26, Tianchi-Alibaba27, and CC-CCII18; |
1,502 COVID-19 patients; 83 influenza-A/B patients; 1,334 CAP patients except for influenza; 258 healthy subjects (CT images) |
Segmenting lung area with U-net | ResNet152 | – | 0.971 | 90.19% | 95.76% | (66) |
| Hosseinzadeh Kassani et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “Kaggle chest X-ray database;” “Kaggle RSNA Pneumonia Detection dataset” |
117 COVID-19 patients; 117 normal patients (X-Ray images); 20 COVID-19 patients; 20 normal patients (CT images) |
Normalization | DenseNet121 with Bagging tree classifier | 99% | – | 96% | – | (67) |
| Ozkaya et al. (2020) | From the Italian Society of Medical and Interventional Radiology | 53 COVID-19 patients (CT images) | Feature vectors obtained from Pre-trained VGG-16, GoogleNet and ResNet-50 networks and fusion method; Feature ranking by t-test method |
SVM | 98.27% | – | 98.93% | 97.60% | (68) |
| Shi et al. (2020) | From Tongji Hospital, Shanghai Public Health Clinical Center, and China-Japan Union Hospital (all in China) | 183 COVID-19 patients; 5,521 Pneumonia patients (CT images) | Segmentation by a deep learning network (VB-Net) | Infection size-aware random forest | 87.9% | 0.942 | 90.7% | 83.3% | (69) |
| Song et al. (2020) | From the Renmin Hospital of Wuhan University | 88 COVID-19 patients (CT images) | We extracted the main regions of lungs and filled the blank of lung segmentation with the lung itself | Details Relation Extraction neural network | 86% | 0.96 | 96% | – | (3) |
| Wang et al. (2020) | Private dataset | 44 COVID-19 patients; 55 Pneumonia patients (CT images) | Random selection of ROI; Feature extraction using Transfer Learning | Fully connected network and combination of Decision tree and Adaboost | 82.9% | 0.90 | 81% | 84% | (6) |
| Zheng et al. (2020) | Private dataset | 313 COVID-19 patients; 229 non-COVID-19 patients (CT images) | Data augmentation; Producing lung masks by a trained UNet | 3D deep convolutional neural network | 90.8% | 0.959 | – | – | (70) |
Data Source: The source(s) that images were acquired from, Data Structure and Size: Number of images, image modalities, sample groups, Data Preprocessing: cleaning, Instance selection, normalization, transformation, feature extraction, selection, etc. The product of data preprocessing is the final training set, Best Model Structure(s): Best machine algorithm or deep learning model reported in the selected paper based on its performance, Performance Measurements (on the best model): The measurement of the model's output performance based on accuracy, sensitivity, specificity, and AUC score.
Table 2.
Characteristics of papers that used X-ray images.
| Author, year | Data source | Data structure and size | Data preprocessing | Best model structure(s) | Performance measurements (on the best model) | References | |||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | AUC score | Sensitivity | Specificity | ||||||
| Alazab et al. (2020) | Kaggle database | 70 COVID-19 patients 28 normal patients (X-ray images) |
Augmented to 1,000 images | VGG-16 | F1 Score: 0.99 | (16) | |||
| Albahli et al. (2020) | “ChestX-ray8” combined with the few samples of rare classes from the Kaggle challenge | 108,948 X-ray images of 32,717 unique patients. Including 15 kinds of chest disease | Data augmentation (rotation, height shift, zoom, horizontal flip) | ResNet | 89% | – | – | – | (17) |
| Albahli et al. (2020) | Open source COVIDx dataset | 850 COVID-19 patients; 500 non-COVID-19 pneumonia cases; 915 normal patients (X-ray images) |
Data augmentation | InceptionNetV3 | 99.02% | – | – | – | (18) |
| Altan et al. (2020) | Not mentioned | 7,980 chest X-ray image (2,905 real raw 5,075 synthetic chests X-ray images) | Data augmentation; The feature matrix is formed by 2D Curvelet transformation Coefficients; Optimizing the coefficients in the feature matrix with the CSSA |
Hybrid model | 99.69% | – | 99.44% | 99.81% | (20) |
| Apostolopoulos et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Common Bacterial and Viral Pneumonia X-ray Images by Kermany et al.; Public datasets (Radiological Society of North America, Radiopaedia, and the Italian Society of Medical and Interventional Radiology); “NIH Chest X-ray Dataset” |
455 COVID-19 patients; 910 viral pneumonia; 2,540 other pulmonary diseases (X-ray images) |
Data augmentation (randomly rotated by a maximum of 10° and randomly shifted horizontally or vertically by a maximum of 20 pixels toward any direction) | MobileNet v2 | 99.18% | – | 97.36% | 99.42% | (21) |
| Apostolopoulos et al. (2020) | X-ray images on public medical Github repositories; “Radiological Society of North America;” “Radiopaedia, and Italian Society of Medicine and Interventional Radiology” |
Dataset 1: 224 COVID-19 patients; 700 bacterial pneumonia patients; 504 normal patients (X-ray images) Dataset 2: 224 Covid-19 patients; 714 bacterial and viral pneumonia patients; 504 normal patients (X-ray images) |
- | MobileNet v2 | 96.78% | – | 98.66% | 96.46% | (71) |
| Brunese et al. (2020) | COVID-19 image data collection; COVID-19 X-ray image database developed by Cohen JP; “ChestX-ray8;” “NIH Chest X-ray Dataset” |
250 COVID-19 patients; 2,753 other pulmonary diseases; 3,520 normal patients (X-Ray images) |
Data augmentation (15 degrees rotation clockwise or counterclockwise) | VGG-16 | 96% (comparison between COVID-19 and other pulmonary diseases) | – | 87% 96% |
94% 98% |
(72) |
| Chowdhury et al. (2020) | Kaggle chest X-ray database; “Italian Society of Medical and Interventional Radiology COVID-19 database;” “Novel Corona Virus 2019 Dataset;” GitHub database; “COVID-19 Chest imaging at thread reader;” “RSNA-Pneumonia-Detection-Challenge” |
423 COVID-19 patients; 1,485 viral pneumonia patients; 1,579 normal patients (X-ray images) |
Data augmentation | CNN | 99.7% | – | 99.7% | 99.55% | (73) |
| Civit-Masot et al. (2020) | COVID-19 and Pneumonia Scans Dataset | 132 COVID-19 patients; 132 normal patients; 132 Pneumonia patients (X-ray images) |
Histogram equalization | VGG16 | 85% | – | 85% | 92% | (74) |
| Das et al. (2020) | COVID-19 collection; “Kaggle CXR collection;” “Tuberculosis collections;” “U.S. National Library of Medicine;” “National Institutes of Health;” Pneumonia collections |
162 COVID-19 patients; 1,583 normal patients |
Histogram matching | Truncated Inception Net | 100% (Pneumonia collections) | 1.0 | 100% | 100% | (23) |
| Elaziz et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “Chest X-Ray Images Pneumonia;” Italian Society of Medical and Interventional Radiology COVID-19 DATABASE; |
219 COVID-19 patients; 1,341 negative COVID-19 patients (X-ray images) |
Feature extraction by Fractional Multichannel Exponent Moments (FrMEMs); Feature selection by modified Manta-Ray Foraging Optimization based on differential evolution |
KNN | 98.09 | – | 98.91 | – | (75) |
| Hassantabar et al. (2020) | “COVID-CT-Dataset” | 315 COVID-19 patients; 367 non-COVID-19 patients (X-ray images) | – | CNN | 93.2% | – | 96.1% | 99.71% | (76) |
| Islam et al. (2020) | “GitHub;” “Radiopaedia;” “Cancer Imaging Archive;” “Italian Society of Radiology;” “Kaggle repository;” NIH dataset |
1,525 COVID-19 patients; 1,525 pneumonia patients; 1,525 normal patients (X-ray images) |
Normalization | CNN-LSTM | 99.4% | 0.999 | 99.3% | 99.2% | (77) |
| Khan et al. (2020) | “Covid-chestxray-dataset” “Chest X-Ray Images (Pneumonia)” |
284 COVID-19 patients; 330 Pneumonia Bacterial 327 Pneumonia Viral; 310 normal patients (X-ray images) |
Random under-sampling (to overcome the unbalanced data problem) | CoroNet (based on Xception) | 89.6% | – | 89.92% | 96.4% | (78) |
| Khuzani et al. (2020) | “GitHub” | 140 COVID-19 patients; 140 non-COVID-19 pneumonia patients; 140 normal patients (X-ray images) |
PCA method; Min-Max Normalization; Adaptive Histogram Equalization |
ML | 94% | 0.91 | 100% | – | (79) |
| Ko et al. (2020) | Private; Italian Society of Medical and Interventional Radiology COVID-19 DATABASE; |
1,194 COVID-19 patients; 1,442 non-pneumonia patients; 1,357 Pneumonia patients (X-ray images) |
Data augmentation (rotation, zoom) | FCONet (ResNet-50) | 99.58% | – | 99.58% | 100% | (80) |
| Loey et al. (2020) | COVID-19 X-ray image database developed by Cohen JP | 69 COVID-19 patients; 79 pneumonia bacterial patients; 79 |
Data augmentation | Googlenet | 80.56% (Four classes) | – | 80.56% | – | (81) |
| Mahmud et al. (2020) | Private | 1,583 normal patients; 1,493 non-COVID viral pneumonia; 2,780 bacterial pneumonia; 305 COVID-19 patients (X-ray images) |
– | CovXNet (CNN based architecture) | 90.2% (multi-class) | 0.911 (multi-class) | 89.9% (multi-class) | 89.1% (multi-class) | (82) |
| Martínez et al. (2020) | COVID-19 X-ray image database developed by Cohen JP | 120 COVID-19 patients; 120 normal patients (X-ray images) |
Data augmentation; Normalization |
NASNet-type convolutional | 97% | – | 97% | 97% | (83) |
| Minaee et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “ChexPert dataset” |
40 COVID-19 patients; 3,000 normal patients (X-ray images) |
Regularization | SqueezeNet | 97% | – | 97.5% | 97.8% | (84) |
| Narayan Das et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “ChestX-ray8” |
125 COVID-19 patients; 500 pneumonia patients; 500 normal patients (X-ray images) |
– | Xception | 97.4% | 0.986 | 97.09% | 97.29% | (85) |
| Nour et al. (2020) | “Public COVID-19 radiology database;” “Italian Society of Medical and Interventional Radiology;” “COVID-19 Database;” “Novel Corona Virus 2019 Dataset;” “COVID-19 positive chest X-ray images from different articles;” |
219 COVID-19 patients; 1,345 Viral Pneumonia patients; 1,341 Normal patients (X-ray images) |
Data augmentation | CNN | 97.14% | 0.995 | 94.61% | 98.29% | (86) |
| Novitasari et al. (2020) | GitHub and Kaggle | 102 COVID-19 patients; 204 Pneumonia and Normal patients (X-ray images) |
Feature extraction by Googlenet, Resnet18, Resnet50, Resnet101; Feature selection by PCA, Relief; |
SVM | 97.33% (multi class) | – | 96% | 98% | (87) |
| Oh et al. (2020) | “Japanese Society of Radiological Technology;” “SCR database;” “U.S. National Library of Medicine” |
180 COVID-19 patients; 20 Viral Pneumonia patients; 54 pneumonia bacterial patients; 57 Tuberculosis patients; 191 Normal patients (X-ray images) |
Data normalization; Data type casting; Histogram equalization; Gamma correction |
(FC)-DenseNet103 | 88.9% | – | 85.9% | 96.4% | (88) |
| Ozturk et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “ChestX-ray8;” |
(X-ray images) | DarkCovidNet inspired by the DarkNet architecture | 87.02% | – | 85.35% | 92.18% | (89) | |
| Pandit et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Kaggle chest X-ray database |
224 COVID-19 patients; 700 pneumonia bacterial patients; 504 Normal patients (X-ray images) |
Data augmentation | VGG-16 | 92.53% (Three class output) | – | 86.7% | 95.1% | (90) |
| Panwar et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Radiopedia.org website; Kaggle chest X-ray database |
142 COVID-19 patients; 142 other (“Normal” “Bacterial Pneumonia” and “Viral Pneumonia”) (X-ray images) |
Data augmentation | nCOVnet | 88.10% | 0.880 | 97.62% | 78.57% | (40) |
| Pereira et al. (2020) | “RYDLS-20;” Radiopedia Encyclopedia “Chest X-ray14” |
90 COVID-19 patients; 1,000 Normal patients; 10 MERS patients; 11 SARS patients; 10 Varicella patients; 12 Streptococcus patients; 11 Pneumocystis patients (X-ray images) |
Resampling algorithms; Fusion techniques; |
Pre-trained CNN | F1 score = 89% | (91) | |||
| Rahaman et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “Chest X-Ray Images (pneumonia)” | 260 COVID-19 patients; 300 Pneumonia; 300 Normal patients (X-ray images) |
Data augmentation (rotate, shift, shear, zoom, horizontal and vertical flip) | VGG19 | 89.3% | – | 89% | – | (92) |
| Rahimzadeh et al. (2020) | “Covid chestxray dataset;” “RSNA pneumonia detection challenge” |
180 COVID-19 patients; 6,054 Pneumocystis patients; 8,851 Normal patients (X-ray images) |
Data augmentation | Xception ResNet50V2 concatenated |
91.4% | – | 80.53% | 99.56% | (93) |
| Rajaraman et al. (2020) | Pediatric CXR dataset; RSNA CXR dataset; CheXpert CXR dataset; NIH CXR-14 dataset; Twitter COVID-19 CXR dataset; Montreal COVID-19 CXR dataset; |
4,683 Bacterial Pneumonia; 3,883 Viral Pneumonia (X-Ray images) |
Segmenting lung area with dilated dropout U-Net; Image thresholding to remove very bright pixels; In-painting missing pixels using the surrounding pixel values; Using median-filter to remove noise and preserve edges; |
VGG-16 | 94.05% | 0.96 | 98.77% | 86.24% | (45) |
| Rajaraman et al. (2020) | “Pediatric CXR dataset;” “RSNA CXR dataset;” “Twitter COVID-19 CXR dataset;” “Montreal COVID-19 CXR dataset” |
313 COVID-19 patients; 7,595 pneumonia of unknown type patients; 2,780 bacterial pneumonia; 7,595 Normal patients (X-ray images) |
Median Filtering; Normalization; Standardization |
Inception-V3 | 99.01% | 0.997 | 98.4% | – | (45) |
| Sethy et al. (2020) | X-ray images on public medical Github repositories; Kaggle chest X-ray database |
127 COVID-19 patients; 127 Pneumonia patients; 127 Normal patients (X-ray images) |
– | ResNet50 plus SVM | 98.66% | – | 95.33% | – | (94) |
| Shibly et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “RSNA pneumonia detection challenge dataset;” Kaggle chest X-ray database; “COVIDx” |
183 COVID-19 patients; 5,551 Pneumonia patients; 8,066 Normal patients (X-ray images) |
– | Faster R-CNN | 97.36% | – | 97.65% | – | (95) |
| Togaçar et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Kaggle COVID-19 dataset created by a team of researchers from Qatar University, medical doctors from Bangladesh, and collaborators from Pakistan and Malaysia. |
295 COVID-19 patients; 98 Pneumonia; 65 normal patients (X-ray images) |
Restructuring images using the Fuzzy Color technique and stacking them with the original images; Feature extracting using deep learning models (MobileNetV2, SqueezeNet) using the Social Mimic optimization method; |
SVM | 100% | – | 100% | 100% | (96) |
| Toraman et al. (2020) | COVID-19 X-ray image database developed by Cohen JP | 231 COVID-19 patients; 1,050 Pneumonia patients; 1,050 Normal patients (X-ray images) |
Data augmentation; | Convolutional capsnet | 97.24% (Binary class) | – | 97.42% | 97.04% | (97) |
| Tsiknakis et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; Dataset originated from the QUIBIM imagingcovid19 platform database and various public repositories, including RSNA, IEEE, RadioGyan and the British Society of Thoracic Imaging; Publicly available X-ray dataset of patients with pneumonia; |
137 COVID-19 patients; 150 Virus Pneumonia; 150 Bacteria Pneumonia; 150 normal patients (X-ray images) |
Data augmentation (rotation, shear, zoom) | Inception V3 | 76% (multi-class) | 0.93 (multi-class) | 93% (multi-class) | 91.8% (multi-class) | (98) |
| Tuncer et al. (2020) | GitHub website; Kaggle chest X-ray database |
87 COVID-19 patients; 234 Normal patients (X-ray images) |
Converting X-ray image to grayscale; ResExLBP and IRF based method |
SVM | 100% | – | 98.29% | 100% | (99) |
| Ucar et al. (2020) | “COVID chest X-ray dataset;” “Kaggle chest X-ray pneumonia dataset;” | 403 COVID-19 patients; 721 normal patients (X-ray images) |
Data augmentation (noise, shear, brightness increase, brightness decrease) | Bayes-SqueezeNet | 98.26% (multi-class) | – | – | 99.13% (multi-class) | (100) |
| Vaid et al. (2020) | Set of lately published articles; NIH dataset |
181 COVID-19 patients; 364 Normal patients (X-ray images) |
Normalization | VGG-19 | 96.3% | – | 97.1% | – | (101) |
| Waheed et al. (2020) | “IEEE Covid Chest X-ray dataset;” “COVID-19 Radiography Database” “COVID-19 Chest X-ray Dataset;” |
403 COVID-19 patients; 721 normal patients (X-ray images) |
Data augmentation using CovidGAN | VGG16 | 95% | – | 90% | 97% | (102) |
| Yildirim et al. (2020) | “COVID-19 Chest X-Ray dataset;” Kaggle chest X-ray database |
136 COVID-19 patients; 162 Pneumonia patients; 245 Normal patients (X-ray images) |
– | Hybrid model | 96.30% | – | 96.30% | 98.73% | (103) |
| Yoo et al. (2020) | “COVID-Chest XrayDataset;” Eastern Asian Hospital; Shenzen data; |
162 COVID-19 Patients; 162 TB patients; 162 Non-TB patients (X-ray images) |
Data augmentation (rotated, translated, and horizontally flipped) | ResNet18 | 95% Average of (COVID-19/TB) and (COVID-19/non-TB) | 0.95 Average of (COVID-19/TB) and (COVID-19/non-TB) | 97% Average of (COVID-19/TB) and (COVID-19/non-TB) | 93% Average of (COVID-19/TB) and (COVID-19/non-TB) | (104) |
| Ghoshal et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; “Kaggle chest X-ray database” |
68 COVID-19 patients; 2,786 Bacterial Pneumonia patients; 1,504 Viral Pneumonia patients; 1,583 normal patients (X-Ray images) |
Standardization; Data augmentation |
Bayesian ResNet50V2 model | 89.82% | – | – | – | (105) |
| Hall et al. (2020) | “X-ray images on public medical Github repositories;” “Radiopaedia;” “Italian Society of Medical and Interventional Radiology (SIRM)” |
135 COVID-19 patients; 320 Viral and Bacterial Pneumonia patients (X-Ray images) |
Data augmentation | Resnet50 and VGG16 plus CNN |
91.24% | 0.94 | – | – | (106) |
| Hammoudi et al. (2020) | “Chest XRay Images (Pneumonia) dataset;” COVID-19 X-ray image database developed by Cohen JP; |
148 Bacterial pneumonia; 148 Viral pneumonia; 148 Normal patients (X-Ray Images) |
– | DenseNet169 | 95.72% | – | – | – | (107) |
| El-Din Hemdan et al. (2020) | COVID-19 X-ray image database developed by Cohen JP; COVID-19 X-ray image database by Dr. Adrian Rosebrock |
25 COVID-19 patients; 25 normal patients (X-Ray images) |
Scaling to 224*224 pixels; One-hot encoding |
COVIDX-Net (VGG19 and DenseNet201 models) | VGG19 = 90%; DenseNet201 = 90% |
VGG19 = 0.90; DenseNet201 = 0.90 |
VGG19 = 100%; DenseNet201 = 100% |
– | (108) |
| Jain et al. (2020) | “Chest XRay Images (Pneumonia) dataset;” COVID-19 X-ray image database developed by Cohen JP; |
250 COVID-19 patients; 300 Bacterial pneumonia; 350 Viral pneumonia; 315 Normal patients (X-Ray Images) |
Normalize images according to the images in the ImageNet database; Data augmentation (rotation and Gaussian blur); |
ResNet50 | 97.77% | – | 97.14% | – | (109) |
| Luz et al. (2020) | “COVIDx dataset;” “RSNA Pneumonia Detection Challenge dataset;” “COVID-19 image data collection” |
183 COVID-19 patients; 5,521 Pneumonia patients; 8,066 normal patients (X-Ray images) |
Intensity normalization; Data augmentation |
EfficientNet B3 | 93.9% | – | 96.8% | – | (110) |
| Ozkaya et al. (2020) | From the Italian Society of Medical and Interventional Radiology | 53 COVID-19 patients (CT images) |
Feature vectors obtained from Pre-trained VGG-16, GoogleNet and ResNet-50 networks and fusion method; Feature ranking by t-test method |
SVM | 98.27% | – | 98.93% | 97.60% | (68) |
| Ozturk et al. (2020) | “covid-chestxray-dataset available at: https://github.com/ieee8023/covid-chestxray-dataset” | 4 ARds images, 101 COVID images, 2 No finding images, 2 pneumocystis-pneumonia images, 11 Sars images, and 6 streptococcus (X-Ray images) | Data augmentation; SMOTE oversampling; creating feature vectors with sAE and PCA; feature extraction by feature vectors, Gray Level Co-occurrence Matrix, Local Binary Gray Level Co-occurrence Matrix, Gray Level Run Length Matrix, and Segmentation-based Fractal Texture Analysis | SVM | 94.23% | 0.99 | 91.88% | 98.54% | (111) |
| Wang et al. (2020) | COVIDx dataset | 266 COVID-19 patients; 5,536 Pneumonia patients; 8,066 normal patients (X-Ray images) | – | COVID-Net Network Architecture using a “lightweight residual projection-expansion- projection-extension design pattern” (Customized CNN) | 93.3% | 91.0% | – | (1) | |
| Zhang et al. (2020) | X-COVID, OpenCOVID | 599 COVID-19 patients; 2,107 non-COVID-19 patients (non-viral pneumonia and healthy) (X-Ray images) | Data augmentation; Feature extraction using EfficientNet | Confidence-aware anomaly detection | 78.57% | 0.844 | 77.13% | 78.97% | (112) |
Data Source: The source(s) that images were acquired from, Data Structure and Size: Number of images, image modalities, sample groups, Data Preprocessing: cleaning, Instance selection, normalization, transformation, feature extraction, selection, etc. The product of data preprocessing is the final training set, Best Model Structure(s): Best machine algorithm or deep learning model reported in the selected paper based on its performance, Performance Measurements (on the best model): The measurement of the model's output performance based on accuracy, sensitivity, specificity, and AUC score.
Discussion
Machine and deep learning methods have been proven as valuable strategies to assess massive high-dimensional characteristics of medical images. CT or X-Ray findings of COVID-19 patients have similarities with other atypical and viral pneumonia diseases. Therefore, machine and deep learning methods might facilitate automatic discrimination of COVID-19 from other pneumonia conditions. The differential diagnosis of COVID also includes drug-induced diseases or immune pneumonitis. However, most of the studies reviewed here lack these kinds of samples. This point is the limitation of these studies. Different methods, such as Ensemble, VGG-16, ResNet, InceptionNetV3, MobileNet v2, Xception, CNN, VGG16, Truncated Inception Net, and KNN, have been used for the purpose of assessment of chest images of COVID-19 patients. Notably, the application of these methods on X-rays has offered promising results. Such a finding is particularly important since X-rays are easily accessible and low cost. These methods not only can diagnose COVID-19 patients from non-COVID pneumonia cases, but can also predict the severity of COVID-19 pneumonia and the risk of short-term mortality. In spite of the low expense of X-ray compared with CT images, the numbers of studies that assessed these two types of imaging using machine/deep learning methods are not meaningfully different. However, few studies have used these methods on both types of imaging (25, 29, 40). CNN-based methods have achieved accuracy values above 99% in classifying COVID-19 patients from other cases of pneumonia or related disorders, as reported by several independent studies, suggesting these strategies as screening methods for initial evaluation of COVID-19 cases.
Although both deep learning and machine learning strategies can be used for the mentioned purpose, they differ in some respects. For instance, deep learning methods usually need a large amount of labeled training data to make a concise conclusion. However, machine learning can apply a small amount of data delivered by users. Moreover, deep learning methods need high-performance hardware. Machine learning, on the other hand, needs features to be precisely branded by users, deep learning generates novel features by itself, thus requires more time to train. Machine learning classifies tasks into small fragments and subsequently combines obtained results into one conclusion, whereas deep learning resolves the problems using end-to-end principles.
Several studies have diagnosed COVID-19 patients through the application of machine learning methods rather than using deep learning methods by retrieving the features from the images. These studies have yielded high recognition outcomes and have the advantage of high learning speed (12). Pre-processing is an essential step for reducing the impacts of intensity variations in CT slices and getting rid of noise. Subsequent thresholding and morphological operations have also enhanced the analytical performance. Data augmentation and histogram equalization are among the most applied preprocessing methods.
One of the most promising approaches used in the included studies was transfer learning. Transfer learning is defined as using model knowledge on a huge dataset (which is referred to as the “pre-trained model”) and transferring it to use on a new problem. This is very useful in settings like medical imaging, where there is a limited number of labeled data (113). Previous studies showed favorable outcomes of the transfer learning approaches in medical imaging tasks (114, 115). Among the included studies, Bridge et al. (25) even reached 100% classification accuracy on COVID-19 using the pre-trained InceptionV3.
The availability of public databases of CT and X-ray images of patients with COVID-19 has facilitated the application of machine learning methods on large quantities of clinical images and execution of training and verification steps. However, since these images have come from various institutes using different scanners, preprocessing of the obtained data is necessary to make them uniform and facilitate further analysis (12). Appraisal of demographic and clinical data of COVID-19 patients and their association with CT/ X-ray images features as well as the accuracy of machine learning prediction methods would provide more valuable information in the stratification of COVID-19 patients. Moreover, one of the major challenges of deep learning models in medical applications is its unexplainable features due to its black-box nature, which should be solved (116). Future studies can focus on approaches that provide interpretation besides black-box predictions.
Conclusion
Deep and machine learning methods have high accuracy in the differentiation of COVID-19 from non-COVID-19 pneumonia based on chest images. These techniques have facilitated the automatic evaluation of these images. However, deep learning methods suffer from the absence of transparency and interpretability, as it is not possible to identify the exact imaging feature that has been applied to define the output (13). As no single strategy has the capacity to distinguish all pulmonary disorders based merely on the imaging presentation on chest CT scans, the application of multidisciplinary approaches is suggested for overcoming diagnostic problems (13).
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
HM-R, MN, and AG-L collected the data and designed the tables. MT and SG-F designed the study, wrote the draft, and revised it. All the authors read the draft and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Wang L, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. arXiv. (2020) Preprint arXiv:200309871. 10.1038/s41598-020-76550-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghafouri-Fard S, Noroozi R, Vafaee R, Branicki W, Poṡpiech E, Pyrc K, et al. Effects of host genetic variations on response to, susceptibility and severity of respiratory infections. Biomed Pharmacother. (2020) 128:110296. 10.1016/j.biopha.2020.110296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samsami M, Mehravaran E, Tabarsi P, Javadi A, Arsang-Jang S, Komaki A, et al. Clinical and demographic characteristics of patients with COVID-19 infection: statistics from a single hospital in Iran. Human Antibodies. (2020) 1–6. 10.3233/HAB-200428 [DOI] [PubMed] [Google Scholar]
- 5.Ghafouri-Fard S, Noroozi R, Omrani MD, Branicki W, Pośpiech E, Sayad A, et al. Angiotensin converting enzyme: a review on expression profile and its association with human disorders with special focus on SARS-CoV-2 infection. Vascular Pharmacol. (2020) 130:106680. 10.1016/j.vph.2020.106680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv. (2020) 14:1–9. 10.1101/2020.02.14.20023028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. (2020) 296:1–2. 10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Tian S, Lou J, Chen Y. Familial cluster of COVID-19 infection from an asymptomatic. Crit Care. (2020) 24:1–3. 10.1186/s13054-020-2817-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei Y, Zhang H-W, Yu J, Patlas MN. COVID-19 Infection: Early Lessons. Los Angeles, CA: Sage; (2020). [DOI] [PubMed] [Google Scholar]
- 10.Rousan LA, Elobeid E, Karrar M, Khader Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulmonary Med. (2020) 20:1–9. 10.1186/s12890-020-01286-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. (2020) 296:1–8. 10.1148/radiol.2020200823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozsahin I, Sekeroglu B, Musa MS, Mustapha MT, Uzun Ozsahin D. Review on diagnosis of COVID-19 from chest CT images using artificial intelligence. Comput Math Methods Med. (2020) 2020:1–10. 10.1155/2020/9756518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. (2020) 296:E65–71. 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.rekha Hanumanthu S. Role of intelligent computing in COVID-19 prognosis: a state-of-the-art review. Chaos Solitons Fractals. (2020) 138:109947. 10.1016/j.chaos.2020.109947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasian Ardakani A, Acharya UR, Habibollahi S, Mohammadi A. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur Radiol. (2020) 31:1–10. 10.1007/s00330-020-07087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alazab M, Awajan A, Mesleh A, Abraham A, Jatana V, Alhyari S. COVID-19 prediction and detection using deep learning. Int J Comput Information Syst Indus Manage Appl. (2020) 12:168–81. 10.1016/j.chaos.2020.110338 [DOI] [Google Scholar]
- 17.Albahli S. Efficient GAN-based Chest Radiographs (CXR) augmentation to diagnose coronavirus disease pneumonia. Int J Med Sci. (2020) 17:1439–48. 10.7150/ijms.46684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albahli S, Albattah W. Detection of coronavirus disease from X-ray images using deep learning and transfer learning algorithms. J Xray Sci Technol. (2020) 28:841–50. 10.3233/XST-200720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsharman N, Jawarneh I. GoogleNet CNN neural network towards chest CT-coronavirus medical image classification. J Comput Sci. (2020) 16:620–5 10.3844/jcssp.2020.620.625 [DOI] [Google Scholar]
- 20.Altan A, Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals. (2020) 140:110071. 10.1016/j.chaos.2020.110071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apostolopoulos ID, Aznaouridis SI, Tzani MA. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. (2020) 40:1–8. 10.1007/s40846-020-00529-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. (2020) 121:103795. 10.1016/j.compbiomed.2020.103795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das D, Santosh KC, Pal U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys Eng Sci Med. (2020) 43:1–11. 10.21203/rs.3.rs-20795/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aswathy SU, Jarin T, Mathews R, Nair LM, Rroan M. CAD systems for automatic detection and classification of COVID-19 in nano CT lung image by using machine learning technique. Int J Pharm Res. (2020) 12:1865–70. 10.31838/ijpr/2020.12.02.247 [DOI] [Google Scholar]
- 25.Bridge J, Meng Y, Zhao Y, Du Y, Zhao M, Sun R, et al. Introducing the GEV activation function for highly unbalanced data to develop COVID-19 diagnostic models. IEEE J Biomed Health Inform. (2020) 24:1–10. 10.1109/JBHI.2020.3012383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butt C, Gill J, Chun D, Babu BA. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. (2020) 6:1–7. 10.1007/s10489-020-01714-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey N, Rajinikanth V, Fong SJ, Kaiser MS, Mahmud M. Social group optimization-assisted Kapur's entropy and morphological segmentation for automated detection of COVID-19 infection from computed tomography images. Cognit Comput. (2020) 12:1–13. 10.20944/preprints202005.0052.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kermany D, Zhang K, Goldbaum M. Labeled optical coherence tomography (OCT) and Chest X-Ray images for classification. Mendeley Data. (2018) 2. 10.17632/RSCBJBR9SJ.2 [DOI] [Google Scholar]
- 29.El Asnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. (2020) 1–12. 10.1080/07391102.2020.1767212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Z, Wei B, Hong Y, Li T, Cong J, Zhu X, et al. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans Med Imaging. (2020) 39:2584–94. 10.1109/TMI.2020.2996256 [DOI] [PubMed] [Google Scholar]
- 31.Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun. (2020) 11:4080. 10.1038/s41467-020-17971-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasan AM, Al-Jawad MM, Jalab HA, Shaiba H, Ibrahim RW, Al-Shamasneh AR. Classification of Covid-19 coronavirus, pneumonia and healthy lungs in CT scans using Q-deformed entropy and deep learning features. Entropy. (2020) 22:517. 10.3390/e22050517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu S, Gao Y, Niu Z, Jiang Y, Li L, Xiao X, et al. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. (2020) 8:118869–83. 10.1109/ACCESS.2020.3005510 [DOI] [Google Scholar]
- 34.Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J Biomol Struct Dyn. (2020) 1–8. 10.1080/07391102.2020.1788642 [DOI] [PubMed] [Google Scholar]
- 35.Kang H, Xia L, Yan F, Wan Z, Shi F, Yuan H, et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans Med Imaging. (2020) 39:2606–14. 10.1109/TMI.2020.2992546 [DOI] [PubMed] [Google Scholar]
- 36.Lessmann N, Sánchez CI, Beenen L, Boulogne LH, Brink M, Calli E, et al. Automated assessment of CO-RADS and chest CT severity scores in patients with suspected COVID-19 using artificial intelligence. Radiology. (2020) 202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Dong W, Chen J, Cao S, Zhou H, Zhu Y, et al. Efficient and effective training of COVID-19 classification networks with self-supervised dual-track learning to rank. IEEE J Biomed Health Inform. (2020) 24:1–10. 10.1109/JBHI.2020.3018181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Wang X, Liu C, Sun Q, Peng W. Differentiating novel coronavirus pneumonia from general pneumonia based on machine learning. Biomed Eng Online. (2020) 19:66. 10.1186/s12938-020-00809-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. (2020) 26:1224–8. 10.1038/s41591-020-0931-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals. (2020) 138:109944. 10.1016/j.chaos.2020.109944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK. Deep transfer learning based classification model for COVID-19 disease. Ing Rech Biomed. (2020) 1–6. 10.1016/j.irbm.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng Y, Tang YX, Lee S, Zhu Y, Summers RM, Lu Z. COVID-19-CT-CXR: a freely accessible and weakly labeled chest X-ray and CT image collection on COVID-19 from biomedical literature. ArXiv. (2020). 10.1109/TBDATA.2020.3035935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pu J, Leader J, Bandos A, Shi J, Du P, Yu J, et al. Any unique image biomarkers associated with COVID-19? Eur Radiol. (2020) 30:1–7. 10.1007/s00330-020-06956-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raajan NR, Lakshmi VSR, Prabaharan N. Non-invasive technique-based novel corona (COVID-19) virus detection using CNN. Natl Acad Sci Lett. (2020) 1–4. 10.1007/s40009-020-01009-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. (2020) 8:115041–50. 10.1109/ACCESS.2020.3003810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakagianni A, Feretzakis G, Kalles D, Koufopoulou C, Kaldis V. Setting up an easy-to-use machine learning pipeline for medical decision support: a case study for COVID-19 diagnosis based on deep learning with CT scans. Stud Health Technol Inform. (2020) 272:13–6. 10.3233/SHTI200481 [DOI] [PubMed] [Google Scholar]
- 47.Sharma S. Drawing insights from COVID-19-infected patients using CT scan images and machine learning techniques: a study on 200 patients. Environ Sci Pollut Res Int. (2020) 27:1–9. 10.21203/rs.3.rs-23863/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh D, Kumar V, Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur J Clin Microbiol Infect Dis. (2020) 39:1379–89. 10.1007/s10096-020-03901-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song J, Wang H, Liu Y, Wu W, Dai G, Wu Z, et al. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur J Nucl Med Mol Imaging. (2020) 47:1–9. 10.1007/s00259-020-04929-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Bao Y, Wen Y, Lu H, Luo H, Xiang Y, et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans Med Imaging. (2020) 39:2572–83. 10.1109/TMI.2020.2994908 [DOI] [PubMed] [Google Scholar]
- 51.Wang S, Zha Y, Li W, Wu Q, Li X, Niu M, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. (2020) 56:2000775. 10.1183/13993003.00775-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warman A, Warman P, Sharma A, Parikh P, Warman R, Viswanadhan N, et al. Interpretable artificial intelligence for COVID-19 diagnosis from chest CT reveals specificity of ground-glass opacities. medRxiv. (2020) 1–13. 10.1101/2020.05.16.20103408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Hui H, Niu M, Li L, Wang L, He B, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiol. (2020) 128:109041. 10.1016/j.ejrad.2020.109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. (2020) 6:1–7. 10.1016/j.eng.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Ma L, Yang F, Chen Y, Ma K, Yang J, et al. A collaborative online AI engine for CT-based COVID-19 diagnosis. medRxiv. (2020). 10.1101/2020.05.10.20096073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan T, Wong PK, Ren H, Wang H, Wang J, Li Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals. (2020) 140:110153. 10.1016/j.chaos.2020.110153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang S, Jiang L, Cao Z, Wang L, Cao J, Feng R, et al. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med. (2020) 8:450. 10.21037/atm.2020.03.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Z, Li X, Sun H, Wang J, Zhao T, Chen H, et al. Rapid identification of COVID-19 severity in CT scans through classification of deep features. Biomed Eng Online. (2020) 19:63. 10.1186/s12938-020-00807-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Karawi D, Al-Zaidi S, Polus N, Jassim S. Machine learning analysis of chest CT scan images as a complementary digital test of coronavirus (COVID-19) patients. medRxiv. (2020) 1–8. 10.1101/2020.04.13.20063479 [DOI] [Google Scholar]
- 60.Alom MZ, Rahman M, Nasrin MS, Taha TM, Asari VK. COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. arXiv. (2020) Preprint arXiv:200403747. [Google Scholar]
- 61.Barstugan M, Ozkaya U, Ozturk S. Coronavirus (covid-19) classification using ct images by machine learning methods. arXiv. (2020) Preprint arXiv:200309424. [Google Scholar]
- 62.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci Rep. (2020) 10:1–11. 10.1101/2020.02.25.20021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farid AA, Selim GI, Awad H, Khater A. A novel approach of CT images feature analysis and prediction to screen for corona virus disease (COVID-19). Int J Sci Eng Res. (2020) 11:1–9. 10.14299/ijser.2020.03.02 [DOI] [Google Scholar]
- 64.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, et al. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv. (2020) Preprint arXiv:200305037. [Google Scholar]
- 65.Jin C, Chen W, Cao Y, Xu Z, Zhang X, Deng L, et al. Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv. (2020) 11:1–14. 10.1101/2020.03.20.20039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. (2020). 10.1101/2020.03.19.20039354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kassani SH, Kassasni PH, Wesolowski MJ, Schneider KA, Deters R. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning-based approach. arXiv. (2020) Preprint arXiv:200410641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ozkaya U, Ozturk S, Barstugan M. Coronavirus (COVID-19) classification using deep features fusion and ranking technique. arXiv. (2020) Preprint arXiv:200403698. 10.1007/978-3-030-55258-9_17 [DOI] [Google Scholar]
- 69.Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, et al. Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv. (2020) Preprint arXiv:200309860. 10.1088/1361-6560/abe838 [DOI] [PubMed] [Google Scholar]
- 70.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, et al. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. (2020) 1–13. 10.1101/2020.03.12.20027185 [DOI] [Google Scholar]
- 71.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. (2020) 43:635–40. 10.1007/s13246-020-00865-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Programs Biomed. (2020) 196:105608. 10.1016/j.cmpb.2020.105608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. (2020) 8:132665–76. 10.1109/ACCESS.2020.3010287 [DOI] [Google Scholar]
- 74.Civit-Masot J, Luna-Perejón F, Morales MD, Civit A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci. (2020) 10:4640. 10.3390/app10134640 [DOI] [Google Scholar]
- 75.Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE. (2020) 15:e0235187. 10.1371/journal.pone.0235187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hassantabar S, Ahmadi M, Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung x-ray image using convolutional neural network approaches. Chaos Solitons Fractals. (2020) 140:110170. 10.1016/j.chaos.2020.110170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. (2020) 20:100412. 10.1016/j.imu.2020.100412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. (2020) 196:105581. 10.1016/j.cmpb.2020.105581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khuzani AZ, Heidari M, Shariati SA. COVID-Classifier: an automated machine learning model to assist in the diagnosis of COVID-19 infection in chest x-ray images. medRxiv. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ko H, Chung H, Kang WS, Kim KW, Shin Y, Kang SJ, et al. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J Med Internet Res. (2020) 22:e19569. 10.2196/19569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. (2020) 12:651. 10.3390/sym12040651 [DOI] [Google Scholar]
- 82.Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. (2020) 122:103869. 10.1016/j.compbiomed.2020.103869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez F, Martínez F, Jacinto E. Performance evaluation of the NASnet convolutional network in the automatic identification of COVID-19. Int J Adv Sci Engin Information Technol. (2020) 10:662–7. 10.18517/ijaseit.10.2.11446 [DOI] [Google Scholar]
- 84.Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. (2020) 65:101794. 10.1016/j.media.2020.101794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narayan Das N, Kumar N, Kaur M, Kumar V, Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Ing Rech Biomed. (2020) 1–7. 10.1016/j.irbm.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl Soft Comput. (2020) 97:1–14. 10.1016/j.asoc.2020.106580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Novitasari DCR, Hendradi R, Caraka RE, Rachmawati Y, Fanani NZ, Syarifudin A, et al. Detection of COVID-19 chest x-ray using support vector machine and convolutional neural network. Commun Math Biol Neurosci. (2020) 2020:1–19. 10.28919/cmbn/476533363252 [DOI] [Google Scholar]
- 88.Oh Y, Park S, Ye JC. Deep Learning COVID-19 Features on CXR using limited training data sets. IEEE Trans Med Imaging. (2020) 39:2688–700. 10.1109/TMI.2020.2993291 [DOI] [PubMed] [Google Scholar]
- 89.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. (2020) 121:103792. 10.1016/j.compbiomed.2020.103792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pandit MK, Banday SA. SARS n-CoV2-19 detection from chest x-ray images using deep neural networks. Int J Pervasive Comput Commun. (2020) 16:1–9. 10.1108/IJPCC-06-2020-006032568675 [DOI] [Google Scholar]
- 91.Pereira RM, Bertolini D, Teixeira LO, Silla CN, Jr., Costa YMG. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed. (2020) 194:105532. 10.1016/j.cmpb.2020.105532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rahaman MM, Li C, Yao Y, Kulwa F, Rahman MA, Wang Q, et al. Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J Xray Sci Technol. (2020) 28:1–19. 10.3233/XST-200715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform Med Unlocked. (2020) 19:100360. 10.1016/j.imu.2020.100360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manage Sci. (2020) 5:643–51. 10.33889/IJMEMS.2020.5.4.05233738639 [DOI] [Google Scholar]
- 95.Shibly KH, Dey SK, Islam MT, Rahman MM. COVID faster R-CNN: a novel framework to diagnose novel coronavirus disease (COVID-19) in X-ray images. Inform Med Unlocked. (2020) 20:100405. 10.1016/j.imu.2020.100405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Togaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. (2020) 121:103805. 10.1016/j.compbiomed.2020.103805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. (2020) 140:110122. 10.1016/j.chaos.2020.110122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsiknakis N, Trivizakis E, Vassalou EE, Papadakis GZ, Spandidos DA, Tsatsakis A, et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp Ther Med. (2020) 20:727–35. 10.3892/etm.2020.8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tuncer T, Dogan S, Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image. Chemometr Intell Lab Syst. (2020) 203:104054. 10.1016/j.chemolab.2020.104054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ucar F, Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. (2020) 140:109761. 10.1016/j.mehy.2020.109761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vaid S, Kalantar R, Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int Orthop. (2020) 44:1539–42. 10.1007/s00264-020-04609-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. CovidGAN: data augmentation using auxiliary classifier GAN for improved Covid-19 detection. IEEE Access. (2020) 8:91916–23. 10.1109/ACCESS.2020.2994762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yildirim M, Cinar A. A deep learning based hybrid approach for covid-19 disease detections. Traitement Signal. (2020) 37:461–8. 10.18280/ts.370313 [DOI] [Google Scholar]
- 104.Yoo SH, Geng H, Chiu TL, Yu SK, Cho DC, Heo J, et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front Med. (2020) 7:427. 10.3389/fmed.2020.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv. (2020) Preprint arXiv:200310769. [Google Scholar]
- 106.Hall LO, Paul R, Goldgof DB, Goldgof GM. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv. (2020) 40:1–14. 10.36227/techrxiv.12083964 [DOI] [Google Scholar]
- 107.Hammoudi K, Benhabiles H, Melkemi M, Dornaika F, Arganda-Carreras I, Collard D, et al. Deep learning on chest X-ray images to detect and evaluate pneumonia cases at the Era of COVID-19. arXiv. (2020) Preprint arXiv:200403399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hemdan EE-D, Shouman MA, Karar ME. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv. (2020) Preprint arXiv:200311055. [Google Scholar]
- 109.Jain G, Mittal D, Thakur D, Mittal MK. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybernet Biomed Eng. (2020). 10.1016/j.bbe.2020.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luz E, Silva PL, Silva R, Moreira G. Towards an efficient deep learning model for covid-19 patterns detection in x-ray images. arXiv. (2020) 31:1–10. [Google Scholar]
- 111.Ozturk S, Ozkaya U, Barstugan M. Classification of coronavirus images using shrunken features. medRxiv. (2020). 10.1101/2020.04.03.20048868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, Xie Y, Li Y, Shen C, Xia Y. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv. (2020) Preprint arXiv:200312338. [Google Scholar]
- 113.Ravishankar H, Sudhakar P, Venkataramani R, Thiruvenkadam S, Annangi P, Babu N, et al. Understanding the mechanisms of deep transfer learning for medical images. In: Deep Learning and Data Labeling for Medical Applications: Springer; (2016). p. 188–96. [Google Scholar]
- 114.Hosny KM, Kassem MA, Foaud MM. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE. (2019) 14:e0217293. 10.1371/journal.pone.0217293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khan S, Islam N, Jan Z, Din IU, Rodrigues JJC. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett. (2019) 125:1–6. 10.1016/j.patrec.2019.03.022 [DOI] [Google Scholar]
- 116.Singh A, Sengupta S, Lakshminarayanan V. Explainable deep learning models in medical image analysis. J Imaging. (2020) 6:52. 10.3390/jimaging6060052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.