Abstract
The long-known resistance to pathogens provided by host-associated microbiota fostered the notion that adding protective bacteria could prevent or attenuate infection. However, the identification of endogenous or exogenous bacteria conferring such protection is often hindered by the complexity of host microbial communities. Here, we used zebrafish and the fish pathogen Flavobacterium columnare as a model system to study the determinants of microbiota-associated colonization resistance. We compared infection susceptibility in germ-free, conventional and reconventionalized larvae and showed that a consortium of 10 culturable bacterial species are sufficient to protect zebrafish. Whereas survival to F. columnare infection does not rely on host innate immunity, we used antibiotic dysbiosis to alter zebrafish microbiota composition, leading to the identification of two different protection strategies. We first identified that the bacterium Chryseobacterium massiliae individually protects both larvae and adult zebrafish. We also showed that an assembly of 9 endogenous zebrafish species that do not otherwise protect individually confer a community-level resistance to infection. Our study therefore provides a rational approach to identify key endogenous protecting bacteria and promising candidates to engineer resilient microbial communities. It also shows how direct experimental analysis of colonization resistance in low-complexity in vivo models can reveal unsuspected ecological strategies at play in microbiota-based protection against pathogens.
Subject terms: Microbiome, Microbial ecology
Introduction
Animal resident microbial consortia form complex and long-term associations with important community-level functions essential for host development and physiology [1, 2]. Microbial ecosystems also provide protection against exogenous pathogens by inhibition of pathogen settlement and growth and/or stimulation of the host immune system [3–8]. From the perspective of microbial community composition, a shift or reduction in resident microbial diversity, a phenomenon generally referred to as dysbiosis, is often associated with increased susceptibility to infection due to the loss or change in abundance of key microbial community members [3, 9]. These observations early supported the notion that addition or promotion of individually or communally protective bacteria (such as probiotics) could minimize microbiota dysbiosis or directly prevent infection to restore host health [10–12].
Although the efficacy of probiotics has been shown in animals and humans, their mechanisms of action are poorly understood and low throughput experimental models often offer limited information on the individual contribution of probiotic species to community functions [1, 6, 7, 13, 14]. Moreover, characterization of bacterial strains improving colonization resistance is still hindered by the complexity of host-commensal ecosystems. Zebrafish have recently emerged as a powerful tool to study microbe-microbe and host-microbe interactions [15–19]. Zebrafish can be easily reared germ-free or gnotobiotically in association with specific bacterial species [15, 20]. Moreover, zebrafish bacterial communities are increasingly well characterized and a number of phylogenetically distinct zebrafish gut bacteria can be cultured, making this model system directly amenable to microbiota manipulation and assessment of probiotic effect on host infection resistance [21–24]. Several studies have used zebrafish to evaluate the effect of exogenous addition of potential probiotics on host resistance to infection by various pathogens [22–29]. However, the role of the endogenous microbial community in protecting against invasive pathogens was rarely assessed and the reported protections were often partial, illustrating the difficulty in identifying fully protective exogenous probiotics.
One major fish pathogen causing such problematic seasonal outbreaks is Flavobacterium columnare, a ubiquitously distributed freshwater bacterium that is the etiological agent of columnaris disease [30]. This disease affects a broad range of wild and cultured species including carp, channel catfish, goldfish, eel, salmonids and tilapia [30–34]. Different F. columnare strains exhibit different degrees of virulence but relatively similar infection phenotypes [30, 31, 35–37]. The symptoms primarily associated with strains with low virulence are gross tissue damages of gills, skin, fins, and tail, whilst such damages are not observed in highly virulent strains, leading to mortality within hours [31, 38]. Although F. columnare infection causes important losses in aquaculture, there is no consensus on the determinants of its virulence. Recently, however, type IX secretion system (T9SS) was shown to be involved in F. columnare pathogenesis in adult zebrafish, but the nature of the secreted virulence factors remains unclear [39].
Here we used germ-free and conventional zebrafish larvae to mine the indigenous commensal microbiota for bacterial species protecting against F. columnare. We identified two distinct infection resistance strategies preventing mortality caused by F. columnare, mediated either by an individual member of the microbiota, the Bacteroidetes Chryseobacterium massiliae or by an assembly of 9 individually non-protecting bacterial species that formed a protective community. Our results demonstrated that mining host microbiota constitutes a powerful approach to identify key mediators of intrinsic colonization resistance, providing insight into how to engineer ecologically resilient and protective microbial communities.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains isolated from zebrafish microbiota are listed in Table 1. F. columnare strains (Supplementary Table S1) were grown at 28 °C in tryptone yeast extract salts (TYES) broth [0.4% (w/v) tryptone, 0.04% yeast extract, 0.05% (w/v) MgSO4 7H2O, 0.02% (w/v) CaCl2 2H2O, 0.05% (w/v) D-glucose, pH 7.2]. F. columnare strains were assigned into four genomovar groups using 16S rRNA restriction fragment length polymorphism analysis, including genomovar I, I/II, II, and III [40]. All 10 strains of the core zebrafish microbiota species were grown in TYES or LB at 28 °C.
Table 1.
The 10 strains composing a core assembly of zebrafish larvae microbiota.
| Bacterial species of the core zebrafish microbiota | ANI %a | 16S rRNA (%)b | recA (%)c | rplC (%)d |
|---|---|---|---|---|
| Aeromonas veronii 1G | 96.52 | 98.27 | 97.00 | 99.84 |
| Aeromonas veronii 2G | 96.58 | 99.53 | 98.31 | 99.68 |
| Aeromonas caviaeG | 97.97 | 99.94 | 98.78 | 99.84 |
| Chryseobacterium massiliaeF | 95.85 | 99.86 | 96.61 | 99.84 |
| Phyllobacterium myrsinacearumA | 98.58 | 99.86 | 99.72 | 100 |
| Pseudomonas sediminisG | 96.12 | 99.73 | 97.70 | 99.84 |
| Pseudomonas mosselliiG | 99.39 | 98.27 | 100 | 99.84 |
| Pseudomonas nitroreducenseG | 92.14 | 99.80 | 94.95 | 99.06 |
| Pseudomonas pelieG | 88.84 | 99.20 | 91.51 | 95.44 |
| Stenotrophomas maltophiliaeG | 90.94 | 97.85 | 95.38 | 99.08 |
Bacterial strains consistently detected at all time points (6 and 11 dpf) in all experiment runs by clone library generation (16S rRNA %) and by whole genome sequencing of the culture isolates (ANI %, recA %, rplC %).
These 10 strains constitute the core of conventional zebrafish larval microbiota and their taxonomic affiliation.
aAverage nucleotide identity value.
b16S rRNA gene sequence similarity.
crecA gene sequence similarity.
drplC gene sequence similarity.
eSpecies ambiguously identified; GGammaproteobacteria; FFlavobacteria; AAlphaproteobacteria.
General handling of zebrafish
Wild-type AB fish, originally purchased from the Zebrafish International Resource Center (Eugene, OR, USA), or myd88-null mutants (myd88hu3568/hu3568) [41], kindly provided by A.H. Meijer, (Leiden University, the Netherlands), were raised in our facility. A few hours after spawning, eggs were collected, rinsed, and sorted under a dissecting scope to remove faeces and unfertilized eggs. All following procedures were performed in a laminar microbiological cabinet with single-use disposable plasticware. Fish were kept in sterile 25 cm3 vented cap culture flasks containing 20 mL of water (0-6 days post fertilization (dpf), 15 fish per flask) or 24-well microtiter plates (6-15 dpf,1 fish per 2 mL well) in autoclaved mineral water (Volvic) at 28 °C. Fish were fed 3 times a week from 4 dpf with germ-free Tetrahymena thermophila protozoans [22]. Germ-free zebrafish were produced after sterilizing the egg chorion protecting the otherwise sterile egg, with antibiotic and chemical treatments (see below), whereas conventional larvae (with facility-innate microbiota) were directly reared from non-sterilized eggs and then handled exactly as the germ-free larvae.
Sterilization of zebrafish eggs
Egg sterilization was performed as previously described with some modifications [22]. Freshly fertilized zebrafish eggs were first bleached (0.003%) for 5 min, washed 3 times in sterile water under gentle agitation and maintained overnight in groups of 100 eggs per 75 cm3 culture flasks with vented caps containing 100 mL of autoclaved Volvic mineral water supplemented with methylene blue solution (0.3 µg/mL). Afterwards, eggs were transferred into 50 mL Falcon tubes (100 eggs per tube) and treated with a mixture of antibiotics (500 μL of penicillin G: streptomycin, 10,000 U/ml: 10 mg/mL GIBCO #P4333), 200 μL of filtered kanamycin sulfate (100 mg/mL, SERVA Electrophoresis #26899) and antifungal drug (50 μL of amphotericin B solution Sigma-Aldrich (250 μg/mL) #A2942) for 2 h under agitation at 28 °C. Eggs were then washed 3 times in sterile water under gentle agitation and bleached (0.003%) for 15 min, resuspending the eggs every 3 min by inversion. Eggs were washed again 3 times in water and incubated 10 min with 0.01% Romeiod (COFA, Coopérative Française de l’Aquaculture). Finally, eggs were washed 3 times in water and transferred into 25 cm3 culture flasks with vented caps containing 20 mL of water. After sterilization, eggs were transferred with approximately 30 to 35 eggs / flasks and were transferred into new flasks at 4 dpf before reconventionalization with 10 to 15 fish / flask. We monitored sterility at several points during the experiment by spotting 50 μL of water from each flask on LB, TYES and on YPD agar plates, all incubated at 28 °C under aerobic conditions. Plates were left for at least 3 days to allow slow-growing organisms to multiply. Spot checks for bacterial contamination were also carried out by PCR amplification of water samples with the 16S rRNA gene primers and procedure detailed further below. If a particular flask was contaminated, those fish were removed from the experiment.
Procedure for raising germ-free zebrafish
After hatching, fish were fed with germ-free T. thermophila 3 times per week from 4 dpf onwards. (i) T. thermophila stock. A germ-free line of T. thermophila was maintained at 28 °C in 20 mL of PPYE (0.25% proteose peptone BD Bacto #211684, 0.25% yeast extract BD Bacto #212750) supplemented with penicillin G (10 unit/mL) and streptomycin (10 µg/mL). Medium was inoculated with 100 μL of the preceding T. thermophila stock. After one week of growth, samples were taken, tested for sterility on LB, TYES, and YPD plates and restocked again. (ii) Growth. T. thermophila were incubated at 28 °C in MYE broth (1% milk powder, 1% yeast extract) inoculated from stock suspension at a 1:50 ratio. After 24 h of growth, T. thermophila were transferred to Falcon tubes and washed (4400 rpm, 3 min at 25 °C) 3 times in 50 mL of autoclaved Volvic water. Finally, T. thermophila were resuspended in sterile water and added to culture flasks (500 µL in 20 mL) or 24-well plates (50 µL / well). Sterility of T. thermophila was tested by plating and 16S rRNA PCR as described in the section above.
(iii) Fine-powder feeding. When indicated, fish were fed with previously γ-ray-sterilized fine-powdered food suitable for an early first feeding gape size (ZM-000 fish feed, ZM Ltd) every 48 h [42].
Reconventionalization of germ-free zebrafish
At 4 dpf, just after hatching, zebrafish larvae were reconventionalized with a single bacterial population or a mix of several. The 10 bacterial strains constituting the core protective microbiota were grown for 24 h in suitable media (TYES or LB) at 28 °C. Bacteria were then pelleted and washed twice in sterile water, and all adjusted to the same cell density (OD600 = 1 or 5.107 colony forming units (cfu)/mL) (i) Reconventionalization with individual species. Bacteria were resuspended and transferred to culture flasks containing germ-free fish at a final concentration of 5.105 cfu/mL. (ii) Reconventionalization with bacterial mixtures. For the preparation of Mix10, Mix9, Mix8 and all other mixes used, equimolar mixtures were prepared by adding each bacterial species at initial concentration to 5.107 cfu/mL. Each bacterial mixture suspension was added to culture flasks containing germ-free fish at a final concentration of 5.105 cfu/mL.
Infection challenges
F. columnare strains (Supplementary Table S1) were grown overnight in TYES broth at 28 °C. Then, 2 mL of culture were pelleted (10,000 rpm for 5 min) and washed once in sterile water. GF zebrafish were brought in contact with the tested pathogens at 6 dpf for 3 h by immersion in culture flasks with bacterial doses ranging from 5.102 to 5.107 cfu/mL. Fish were then transferred to individual wells of 24-well plates, containing 2 mL of water and 50 μL of freshly prepared GF T. thermophila per well. Mortality was monitored daily as described in [22], and measured in days post infection (dpi), with 0 dpi corresponding to the infection day, i.e. 6 dpf-old larvae. As few as 54 ± 9 cfu/larva of F. columnare were recovered from infected larvae. All zebrafish experiments were stopped at day 9 post-infection and zebrafish were euthanized with tricaine (MS-222) (Sigma-Aldrich #E10521). Each experiment was repeated at least 3 times and between 10 and 15 larvae were used per condition and per experiment.
Collection of eggs from other zebrafish facilities
Conventional zebrafish eggs were collected in 50 mL Falcon tubes from the following facilities: Facility 1 - zebrafish facility in Hospital Robert Debré, Paris; Facility 2 - Jussieu zebrafish facility A2, University Paris 6; Facility 3 - Jussieu zebrafish facility C8 (UMR7622), University Paris 6; Facility 4- AMAGEN commercial facility, Gif sur Yvette; Larvae were treated with the same rearing conditions, sterilization and infection procedures used in the Institut Pasteur facility.
Determination of fish bacterial load using cfu count
Zebrafish were euthanized with tricaine (MS-222) (Sigma-Aldrich #E10521) at 0.3 mg/mL for 10 min. Then they were washed in 3 different baths of sterile PBS-0.1% Tween to remove bacteria loosely attached to the skin. Finally, they were transferred to tubes containing calibrated glass beads (acid-washed, 425 μm to 600 μm, SIGMA-ALDRICH #G8772) and 500 μL of autoclaved PBS. They were homogenized using FastPrep Cell Disrupter (BIO101/FP120 QBioGene) for 45 s at maximum speed (6.5 m/s). Finally, serial dilutions of recovered suspension were spotted on TYES agar and cfu were counted after 48 h of incubation at 28 °C.
Characterization of zebrafish microbial content
Over 3 months, the experiment was run independently 3 times and 3 different batches of eggs were collected from different fish couples in different tanks. Larvae were reared as described above. GF and Conv larvae were collected at 6 dpf and 11 dpf for each batch. Infected Conv larvae were exposed to F. columnareALG for 3 h by immersion as described above. For each experimental group, triplicate pools of 10 larvae (one in each experimental batch) were euthanized, washed and lysed as above. Lysates were split into 3 aliquots, one for culture followed by 16S rRNA gene sequencing (A), one for 16S rRNA gene clone library generation and Sanger sequencing (B), and one for Illumina metabarcoding-based sequencing (C).
Bacterial culture followed by 16S rRNA gene-based identification
Lysates were serially diluted and immediately plated on R2A, TYES, LB, MacConkey, BHI, BCYE, TCBS and TSB agars and incubated at 28 oC for 24-72 h. For each agar, colony morphotypes were documented, and colonies were picked and restreaked on the same agar in duplicate. In order to identify the individual morphotypes, individual colonies were picked for each identified morphotype from each agar, vortexed in 200 μL DNA-free water and boiled for 20 min at 90 oC. Five μL of this bacterial suspension were used as template for colony PCR to amplify the 16S rRNA gene with the universal primer pair for the Domain bacteria 8 f (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1492r (5’-GGT TAC CTT GTT ACG ACT T-3’). Each primer was used at a final concentration of 0.2 μM in 50 μL reactions. PCR cycling conditions were - initial denaturation at 94 °C for 2 min, followed by 32 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 2 min, with a final extension step at 72 °C for 10 min. 16S rRNA gene PCR products were verified on 1% agarose gels, purified with the QIAquick® PCR purification kit and two PCR products for each morphotype were sent for sequencing (Eurofins, Ebersberg, Germany). 16S rRNA sequences were manually proofread, and sequences of low quality were removed from the analysis. Primer sequences were trimmed, and sequences were compared to GenBank (NCBI) with BLAST, and to the Ribosomal Database Project with SeqMatch. For genus determination a 95% similarity cut-off was used, for Operational Taxonomic Unit determination, a 98% cut-off was used.
16S rRNA gene clone library generation
Total DNA was extracted from the lysates with the Mobio PowerLyzer® Ultraclean® kit according to manufacturer’s instructions. Germ-free larvae and DNA-free water were also extracted as control samples. Extracted genomic DNA was verified by Tris-acetate-EDTA-agarose gel electrophoresis (1%) stained with GelRed and quantified by applying 2.5 μL directly to a NanoDrop® ND-1000 Spectrophotometer. The 16S rRNA gene was amplified by PCR with the primers 8 f and 1492r, and products checked and purified as described above. Here, we added 25–50 ng of DNA as template to 50 μL reactions. Clone libraries were generated with the pGEM®-T Easy Vector system (Promega) according to manufacturer’s instructions. Presence of the cloned insert was confirmed by colony PCR with vector primers gemsp6 (5’-GCT GCG ACT TCA CTA GTG AT-3’) and gemt7 (5’-GTG GCA GCG GGA ATT CGA T-3’). Clones with an insert of the correct size were purified as above and sent for sequencing (Eurofins, Ebersberg, Germany). Blanks using DNA-free water as template were run for all procedures as controls. For the three independent runs of the experiment, 10 Conv fish per condition (6 and 11 dpf, exposed or not to F. columnare) and per repeat were pooled. Each pool of 10 fish was sequenced separately, generating 3 replicates for each condition (n = 12), resulting in a total of 857 clones. Clone library coverage was calculated with the following formula [1-(n1/N2)] x 100, where n1 is the number of singletons detected in the clone library, and N2 is the total number of clones generated for this sample. Clone libraries were generated to a minimum coverage of 95%. Sequence analysis and identification was carried out as above.
by 16S rRNA V3V4 amplicon Illumina sequencing
To identify the 16S rRNA gene diversity in our facility and fish collected from 4 other zebrafish facilities, fish were reared as described above. GF fish were sterilized as above, and uninfected germ-free and conventional fish were collected at 6 dpf and 11 dpf. Infection was carried out as above with F. columnareALG for 3 h by bath immersion, followed by transfer to clean water. Infected conventional fish were collected at 6 dpf 6 h after infection with F. columnare and at 11 dpf, the same as uninfected fish. GF infected larvae were only collected at 6 dpf 6 h post infection, as at 11 dpf all larvae had succumbed to infection. Triplicate pools of 10 larvae were euthanized, washed and lysed as above. Total DNA was extracted with the Mobio PowerLyzer® Ultraclean® kit as described above and quantified with a NanoDrop® ND-1000 Spectrophotometer and sent to IMGM Laboratories GmbH (Germany) for Illumina sequencing. Primers Bakt_341F (5’-CCTACGGGNGGCWGCAG-3’) and Bakt_805R (5’-GACTACHVGGGTATCTAATCC-3’), amplifying variable regions 3 and 4 of the 16S gene were used for amplification [43]. Each amplicon was purified with solid phase reversible immobilization (SPRI) paramagnetic bead-based technology (AMPure XP beads, Beckman Coulter) with a Bead:DNA ratio of 0.7:1 (v/v) following manufacturers instructions. Amplicons were normalized with the Sequal-Prep Kit (Life Technologies), so each sample contained approximately 1 ng/μl DNA. Samples, positive and negative controls were generated in one library. The High Sensitivity DNA LabChip Kit was used on the 2100 Bioanalyzer system (both Agilent Technologies) to check the quality of the purified amplicon library. For cluster generation and sequencing, MiSeq® reagents kit 500 cycles Nano v2 (Illumina Inc.) was used. Before sequencing, cluster generation by two-dimensional bridge amplification was performed, followed by bidirectional sequencing, producing 2 × 250 bp paired-end (PE) reads.
MiSeq® Reporter 2.5.1.3 software was used for primary data analysis (signal processing, demultiplexing, trimming of adapter sequences). CLC Genomics Workbench 8.5.1 (Qiagen) was used for read-merging, quality trimming, and QC reports and OTU definition were carried out in the CLC plugin Microbial Genomics module.
Comparison of whole larvae vs intestinal bacterial content
Larvae reconventionalized with Mix10 and infected with F. columnareALG at 6 dpf for 3 h were euthanized and washed. DNA was extracted from pools of 10 whole larvae or of pools of 10 intestinal tubes dissected with sterile surgical tweezer and subjected to Illumina 16S rRNA gene sequencing. GF larvae and dissected GF intestines were sampled as controls. As dissection of the larval guts involved high animal loss and was a potential important contamination source, we proceeded with using entire larvae for the rest of the study.
Whole genome sequencing
Chromosomal DNA of the ten species composing the core of zebrafish larvae microbiota was extracted using the DNeasy Blood & Tissue kit (QIAGEN) including RNase treatment. DNA quality and quantity were assessed on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
DNA sequencing libraries were made using the Nextera DNA Library Preparation Kit (Illumina Inc.) and library quality was checked using the High Sensitivity DNA LabChip Kit on the Bioanalyzer 2100 (Agilent Technologies). Sequencing clusters were generated using the MiSeq reagents kit v2 500 cycles (Illumina Inc.) according to manufacturer’s instructions. DNA was sequenced at the Helmholtz Centre for Infection Research by bidirectional sequencing, producing 2 × 250 bp paired-end reads. Between 1,108,578 and 2,914,480 reads per sample were retrieved with a median of 1,528,402. Reads were quality filtered, trimmed and adapters removed with trimmomatic 0.39 [44] and genomes assembled using SPAdes 3.14 [45].
Bacterial species identification
Whole genome-based bacterial species identification was performed by the TrueBac ID system (v1.92, DB:20190603) [46]. Species-level identification was performed based on the algorithmic cut-off set at 95% ANI when possible or when the 16S rRNA gene sequence similarity was >99 %.
Monitoring of bacterial dynamics
Three independent experiments were run over 6 weeks with eggs collected from different fish couples from different tanks to monitor establishment and recovery. Larvae were reared, sterilized and infected as above with the only difference that 75 cm3 culture flasks with vented caps (filled with 50 mL of sterile Volvic) were used to accommodate the larger number of larvae required, as in each experiment. Larvae for time course Illumina sequencing were removed sequentially from the experiment that monitored the survival of the larvae. Animals were pooled (10 larvae for each time point/condition), euthanized, washed and lysed as described above and stored at −20o C until the end of the survival monitoring, and until all triplicates had been collected.
Community establishment
In order to follow the establishment of the 10 core strains in the larvae, GF larvae were reconventionalized with an equiratio Mix10 as above. Re-convMix10 larvae were sampled at 4 dpf immediately after addition of the 10 core species and then 20 min, 2 h, 4 h and 8 h after. Germ-free, conventional larvae and the inoculum were also sampled as controls.
Induction of dysbiosis
Different doses of kanamycin (dose 1 = 200 µg/mL; dose 2 = 50 µg/mL; dose 3 = 25 µg/mL) and a penicillin/streptomycin antibiotic mix dose 1 = 250 µg/mL; dose 2 = 15.6 µg/mL were tested on re-convMix10 4 dpf zebrafish larvae by adding them to the flask water to identify antibiotic treatments that were non-toxic to larvae but that caused dysbiosis.
After 16 h of treatment, antibiotics were extensively washed off with sterile water and larvae were challenged with F. columnareALG, leading to the death of all larvae – e.g. successful abolition of colonization resistance with best results in all repeats with 250 µg/mL penicillin/streptomycin and 50 µg/mL kanamycin as antibiotic treatment.
Community recovery
As above, after 8 h of incubation, 4 dpf re-convMix10 larvae were treated with 250 µg/mL penicillin/streptomycin and 50 µg/mL kanamycin for 16 h. Antibiotics were extensively washed off and larvae were now left to recover in sterile water for 24 h to assess resilience of the bacterial community. Samples (pools of 10 larvae) were taken at 3 h, 6 h, 12 h, 18 h, and 24 h during recovery and sent for 16S rRNA Illumina sequencing. Larvae were then challenged at 6 dpf with F. columnareALG for 3 h and survival was monitored daily for 9 days post-infection. All time course samples were sequenced by IMGM Laboratories GmbH, as described above.
Statistical analysis of metataxonomic data
16S RNA analysis was performed with SHAMAN [47]. Library adapters, primer sequences, and base pairs occurring at 5’ and 3’ends with a Phred quality score <20 were trimmed off by using Alientrimmer (v0.4.0). Reads with a positive match against zebrafish genome (mm10) were removed. Filtered high-quality reads were merged into amplicons with Flash (v1.2.11). Resulting amplicons were clustered into operational taxonomic units (OTU) with VSEARCH (v2.3.4) [48]. The process includes several steps for de-replication, singletons removal, and chimera detection. The clustering was performed at 97% sequence identity threshold, producing 459 OTUs. The OTU taxonomic annotation was performed against the SILVA SSU (v132) database [49] completed with 16S sequence of 10 bacterial communities using VSEARCH and filtered according to their identity with the reference [50]. Annotations were kept when the identity between the OTU sequence and reference sequence is ≥78.5% for taxonomic Classes, ≥82% for Orders, ≥86.5% for Families, ≥94.5% for Genera and ≥98% for species. Here, 73.2% of the OTUs set was annotated and 91.69% of them were annotated at genus level.
The input amplicons were then aligned against the OTU set to get an OTU contingency table containing the number of amplicon associated with each OTU using VSEARCH global alignment. The matrix of OTU count data was normalized for library size at the OTU level using a weighted non-null count normalization. Normalized counts were then summed within genera. The generalized linear model (GLM) implemented in the DESeq2 R package [51] was then applied to detect differences in abundance of genera between each group. We defined a GLM that included the treatment (condition) and the time (variable) as main effects and an interaction between the treatment and the time. Resulting P values were adjusted according to the Benjamini and Hochberg procedure [45].
The statistical analysis can be reproduced on SHAMAN by loading the count table, the taxonomic results with the target and contrast files that are available on figshare 10.6084/m9.figshare.11417082.v2.
Determination of cytokine levels
Total RNA from individual zebrafish larvae were extracted using RNeasy kit (Qiagen), 18 h post pathogen exposure (12 h post-wash). Oligo(dT17)-primed reverse transcriptions were carried out using M-MLV H- reverse- transcriptase (Promega). Primer specificity was initially tested by sequencing the amplicons from a positive control template. At the end of each real-time qPCR assay, a denaturation step was conducted to determine the melt curve of the amplicon, for comparison with a positive control sample systematically included. Quantitative PCRs were performed using Takyon SYBR Green PCR Mastermix (Eurogentec) on a StepOne thermocycler (Applied Biosystems). Primers for ef1a (housekeeping gene, used for cDNA amount normalization), il1b, il10 and il22 are described in [22]. Data were analyzed using the ∆∆Ct method. Four larvae were analyzed per condition. Zebrafish genes and proteins mentioned in the text: ef1a NM_131263; il1b BC098597; il22 NM_001020792; il10 NM_001020785; myd88 NM_212814.
Histological comparisons of GF, Conv and Re-Conv fish GF infected or not with F. columnare
Fish were collected 24 h after infection (7 dpf) and were fixed for 24 h at 4 °C in Trump fixative (4% methanol-free formaldehyde, 1% glutaraldehyde in 0.1 M PBS, pH 7.2) and sent to the PIBiSA Microscopy facility services (https://microscopies.med.univ-tours.fr/) in the Faculté de Médecine de Tours (France), where whole fixed animals were processed, embedded in Epon. Semi-thin sections (1 µm) were cut using a X ultra-microtome and then either dyed with toluidine blue for observation by light microscopy and imaging or processed for Transmission electron microscopy.
Adult zebrafish pre-treatment with C. massiliae
The zebrafish line AB was used. Fish were reared at 28 °C in dechlorinated recirculated water, then transferred into continuous flow aquaria when aging 3–4 months for infection experiments. C. massiliae was grown in TYES broth at 150 rpm and 28 °C until stationary phase. This bacterial culture was washed twice in sterile water and adjusted to OD600nm = 1. Adult fish reconventionalization was performed by adding C. massiliae bacterial suspension directly into the fish water (1 L) at a final concentration of 2.106 cfu/mL. Bacteria were maintained in contact with fish for 24 h by stopping the water flow then subsequently removed by restoring the water flow. C. massiliae administration was performed twice after water renewal. In the control group, the same volume of sterile water was added.
Adult zebrafish infection challenge
F. columnare infection was performed just after fish reconventionalization with C. massiliae. The infection was performed as previously described by Li and co-workers with few modifications [39]. Briefly, F. columnare strain ALG-00-530 was grown in TYES broth at 150 rpm and 28 °C until late-exponential phase. Then, bacterial cultures were diluted directly into the water of aquaria (200 mL) at a final concentration of 5.106 cfu/mL. Bacteria were maintained in contact with fish for 1 h by stopping the water flow then subsequently removed by restoring the water flow. Sterile TYES broth was used for the control group. Bacterial counts were determined at the beginning of the immersion challenge by plating serial dilutions of water samples on TYES agar. Water was maintained at 28 °C and under continuous oxygenation for the duration of the immersion. Groups were composed of 10 fish. Virulence was evaluated according to fish mortality 10 days post-infection.
Statistical methods
Statistical analyses were performed using unpaired, nonparametric Mann–Whitney test or unpaired t-tests. Analyses were performed using Prism v8.2 (GraphPad Software).
Evenness: The Shannon diversity index was calculated with the formula (HS = −Σ[P(ln(P)])) where P is the relative species abundance. Total evenness was calculated for the Shannon index as E = HS/Hmax. The less evenness in communities between the species (and the presence of a dominant species), the lower this index is.
Results
Flavobacterium columnare kills germ-free but not conventional zebrafish
To investigate microbiota-based resistance to infection in zebrafish, we compared the sensitivity of germ-free (GF) and conventional (Conv) zebrafish larvae to F. columnare, an important fish pathogen affecting carp, channel catfish, goldfish, eel, salmonids and tilapia and previously shown to infect and kill adult zebrafish [12, 30, 33, 39, 52]. We used bath immersion to expose GF and Conv zebrafish larvae at 6 days post-fertilization (dpf), to a collection of 28 F. columnare strains, belonging to four different genomovars for 3 h at the chosen median infection dose of 5.105 colony forming units (cfu)/mL (see Fig. 1). Daily monitoring showed that 16 out of 28 F. columnare strains killed GF larvae in less than 48 h (Supplementary Fig. S1A), whereas Conv larvae survived exposure to all tested virulent F. columnare strains (Supplementary Fig. S1B). Exposure to the highly virulent strain ALG-00–530 (hereafter F. columnareALG) also showed that GF mortality was fast (1 day post-infection -dpi) and dose-dependent and that Conv zebrafish survived all but the highest dose (107 cfu/mL) (Fig. 1). Similar survival of infected Conv larvae was obtained with zebrafish AB strain eggs obtained from 4 different zebrafish facilities (Supplementary Fig. S2), suggesting that conventional zebrafish microbiota could provide protection against F. columnare infection.
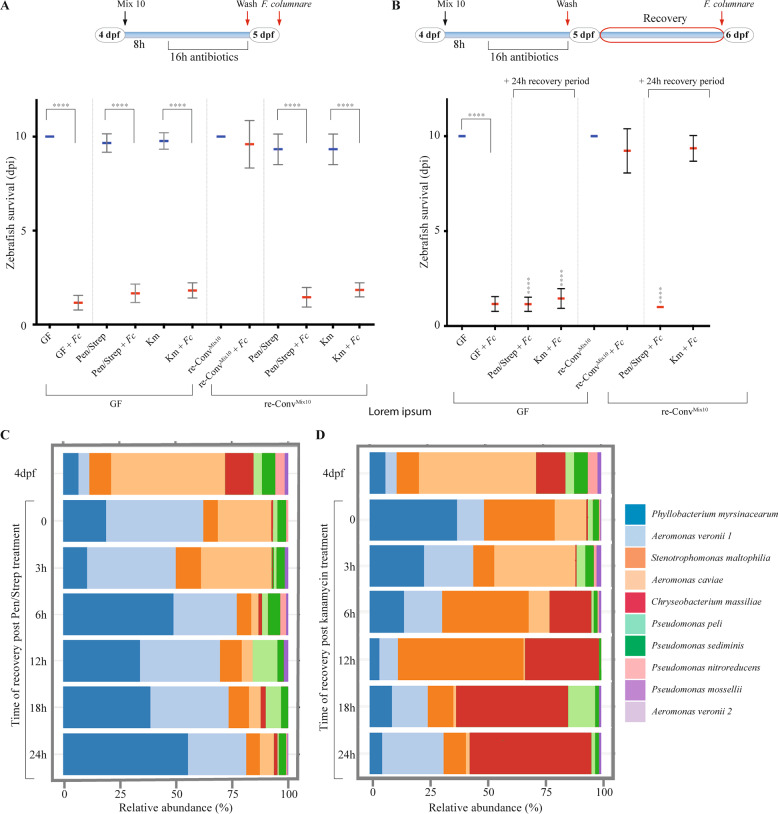
Fig. 1. Flavobacterium columnare kills germ-free but not conventional zebrafish.
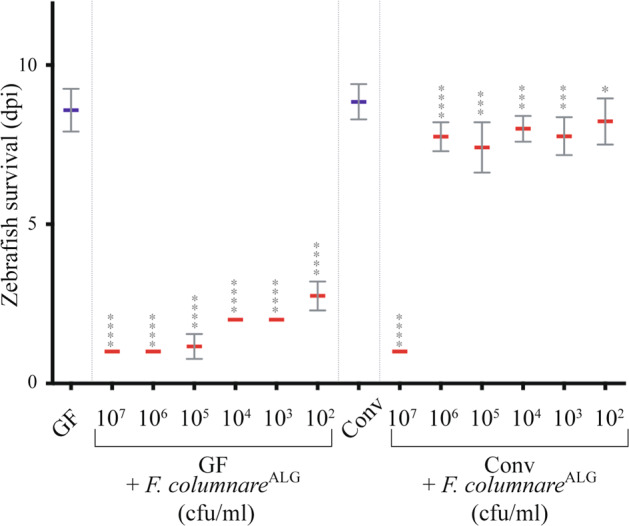
Six dpf fish (corresponding to fish at 0 day post infection- or - dpi) or Conv zebrafish larvae were exposed 3 h to F. columnareALG doses ranging from 5 × 102 to 5 × 107 cfu/mL, using bath immersion before transfer into sterile water. Infection doses of 5 × 104 to 5 × 106 cfu/mL led to a robust read-out of lethality (GF) or survival (Conv). Below 5 × 104 cfu/mL, lethality phenotype was slower and less reproducible while use of more than 5 × 106 cfu/mL, killed GF too fast. 5 × 105 cfu/mL was chosen as a median infection dose in the rest of the study. Mean survival is represented by a thick horizontal bar with standard deviation. For each condition, n = 12 zebrafish larvae. Larvae mortality rate was monitored daily and surviving fish were euthanized at day 9 post infection (9 dpi). Statistics correspond to unpaired, nonparametric Mann–Whitney test comparing all conditions to noninfected GF (left) or Conv (right). ****p < 0.0001; ***p < 0.001; *p < 0.05, absence of *: non-significant. Blue mean bars correspond to nonexposed larvae and red mean bars correspond to larvae exposed to F. columnare.
A community of 10 culturable bacterial strains protects against F. columnare infection
In our rearing conditions, the conventional larval microbiota is acquired after hatching from microorganisms present on the egg chorion and in fish facility water. To test the hypothesis that microorganisms associated with conventional eggs provided protection against F. columnareALG, we exposed sterilized eggs to either fish facility tank water or to non-sterilized conventional eggs at 0 or 4 dpf (before or after hatching, respectively). In both cases, these reconventionalized (re-Conv) zebrafish survived F. columnareALG infection as well as Conv zebrafish (Supplementary Fig. S3). To determine the composition of conventional zebrafish microbiota, we generated 16S rRNA gene clone libraries from homogenate pools of Conv larvae aged 6 and 11 dpf exposed or not to F. columnareALG, sampled over 3 months from 3 different batches of larvae (n = 10). A total of 857 clones were generated for all samples. We identified 15 operational taxonomical units (OTUs), 10 of which were identified in all experiments (Supplementary Table S2, in Table 1 the 16S rRNA gene similarity is shown). Two OTUs (belonging to an Ensifer sp. and a Hydrogenophaga sp.) were only detected once, and a Delftia sp., a Limnobacter sp. and a Novosphingobium sp. were detected more than once (2, 3, and 2 times, respectively), but not consistently in all batches of fish (Supplementary Table S2). Moreover, deep-sequencing of the 16S rRNA V3-V4 region of gDNA retrieved from larvae originating from the other four zebrafish facilities described above, revealed that OTUs for all of these 10 species were also detected in Conv larvae, with the exception of A. veronii 2 that was not detected in all samples (Supplementary Table S3).
To isolate culturable zebrafish microbiota bacteria, we plated dilutions of homogenized 6 dpf and 11 dpf larvae pools on various growth media and we identified 10 different bacterial morphotypes. 16S-based analysis followed by full genome sequencing identified 10 bacteria corresponding to 10 strains of 9 different species that were also consistently detected by culture-free approaches (Table 1 shows the average nucleotide identity value for the culture isolates). To assess the potential protective role of these 10 strains, we reconventionalized GF zebrafish at 4 dpf with a mix of all 10 identified culturable bacterial species (hereafter called Mix10), each at a concentration of 5.105 cfu/mL and we monitored zebrafish survival after exposure to F. columnareALG at 6 dpf. We showed that zebrafish reconventionalized with the Mix10 (Re-ConvMix10) displayed a strong level of protection against all identified highly virulent F. columnare strains (Supplementary Fig. S4). These results demonstrated that the Mix10 constitutes a core protective bacterial community providing full protection of zebrafish larvae against F. columnare infection.
Community dynamics under antibiotic-induced dysbiosis reveal a key contributor to resistance to F. columnare infection
To further analyze the determinants of Mix10 protection against F. columnareALG infection, we inoculated 4 dpf germ-free larvae with an equal-ratio mix of the 10 bacteria (at 5.105 cfu/mL each) and monitored their establishment over 8 h. We first verified that whole larvae bacterial content (OTU abundance) was not significantly different from content of dissected intestinal tubes (p = 0.99, two-tailed t test) (Supplementary Fig. S5) and proceeded to use entire larvae to monitor bacterial establishment and recovery in the rest of the study. We then collected pools of 10 larvae immediately after reconventionalization (t0), and then at 20 min, 2 h, 4 h, and 8 h in three independent experiments. Illumina sequencing of the 16S rRNA gene was used to follow bacterial relative abundance. At t0, all species were present at >4% in the zebrafish, apart from A. veronii strains 1 (0.2%) and 2 (not detected) (Supplementary Fig. S6). Aeromonas caviae was detected as the most abundant species (33%), followed by Stenotrophomonas maltophilia (23%) and Chryseobacterium massiliae (12%), altogether composing 68% of the community (Supplementary Fig. S6). The relative species abundance, possibly reflecting initial colonization ability, was relatively stable for most species during community establishment, with similar species evenness at t0 (E = 0.84) and t8h (E = 0.85). Whereas both Conv and Re-ConvMix10 larvae were protected against F. columnareALG infection, the global structure of the reconstituted Mix10 population was different from the conventional one at 4 dpf (Supplementary Fig. S6).
To test the sensitivity to disturbance and the resilience of the protection provided by Mix10 bacterial community, we determined the minimal inhibitory concentration to penicillin/streptomycin and kanamycin of the strains composing the Mix10 microbiota (Supplementary Fig. S7A) and tested different dose of penicillin/streptomycin and kanamycin treatment on zebrafish survival to F. columnare infection (Supplementary Fig. S7B). We then subjected Re-ConvMix10 zebrafish to identify a non-toxic antibiotic treatment at 4 dpf using either 250 µg/mL penicillin/streptomycin combination (all members of the Mix10 bacteria are sensitive to penicillin/streptomycin) or 50 µg/mL kanamycin (affecting all members of the Mix10 bacteria except C. massiliae, P. myrsinacearum and S. maltophilia) (Supplementary Fig. S7A). At 5 dpf, after 16 h of exposure, antibiotics were washed off and zebrafish were immediately exposed to F. columnareALG. Both antibiotic treatments resulted in complete loss of the protection against F. columnareALG infection observed in Re-ConvMix10 (Fig. 2a). We then used the same antibiotic treatments but followed by a 24 h recovery period after washing off the antibiotics at 5 dpf, therefore only performing the infection at 6 dpf (Fig. 2b). Whilst Re-ConvMix10 larvae treated with penicillin/streptomycin showed similar survival to infected GF larvae, kanamycin-treated Re-ConvMix10 zebrafish displayed restored protection after 24 h recovery and survived similarly to untreated conventionalized fish (Fig. 2b). Sampling and 16S gene analysis during recovery experiments at different time points showed that bacterial community evenness decreased after antibiotic administration for both treatments (E = 0.85 for 4 dpf control, E = 0.72 for t0 kanamycin and E = 0.7 for t0 penicillin/streptomycin), and continued to decrease during recovery (E = 0.6 and 0.64 for kanamycin and penicillin/streptomycin treatment after 24 h recovery, respectively). Although C. massiliae remained detectable immediately after both antibiotic treatments, penicillin/streptomycin treatment led to a significant reduction in its relative abundance (0.21%) (Fig. 2c). By contrast, C. massiliae relative abundance rebounded 6 h after cessation of kanamycin treatment and was the dominant member (52%) of the reconstituted microbiota after 24 h recovery period (Fig. 2d), suggesting that the protective effect observed in the kanamycin-treated larvae might be due to the recovery of C. massiliae.
Fig. 2. Analysis of protection against F. columnare infection after antibiotic dysbiosis.
a Response of zebrafish larvae to exposure to F. columnareALG after antibiotic-induced dysbiosis with a diagram showing timing and treatments of the experiment. b A 24 h period after antibiotic treatment allows the recovery of protection in kanamycin-treated zebrafish larvae with a diagram showing timing and treatments. Mean survival is represented by a thick horizontal bar with standard deviation. For each condition, n = 12 zebrafish larvae. Blue mean bars correspond to larvae not exposed to the pathogen and red mean bars correspond to exposed larvae. Larvae mortality rate was monitored daily and surviving fish were euthanized at day 9 post exposition to the pathogen (9 dpi). Indicated statistics correspond to unpaired, nonparametric Mann–Whitney test. ****p < 0.0001; absence of *: non-significant. c Community recovery profile of re-ConvMix10 larvae with streptomycin/penicillin treatment. d Community recovery profile of re-ConvMix10 larvae with kanamycin treatment. Pools of 10 larvae were collected for 16S rRNA sequencing for both antibiotic treatments.
Resistance to F. columnare infection is provided by both individual and community-level protection
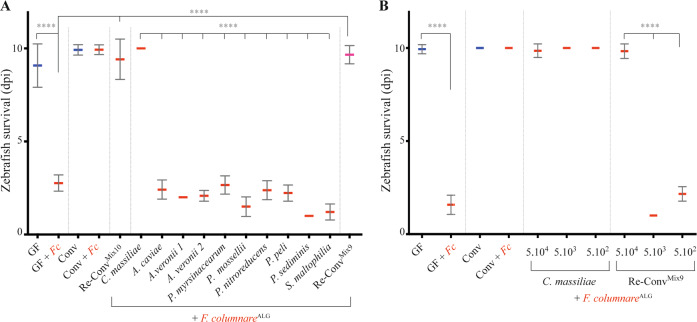
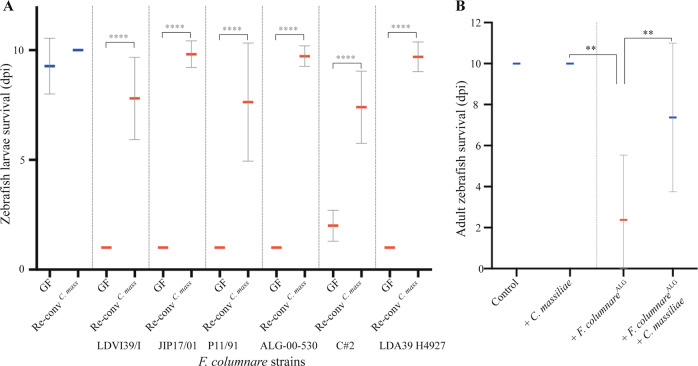
To test the potential key role played by C. massiliae in protection against F. columnareALG infection, we exposed 4 dpf GF zebrafish to C. massiliae only and showed that it conferred individual protection at doses as low as 5.102 cfu/mL (Fig. 3a). Whereas none of the 9 other species composing the Mix10 were protective individually (Fig. 3a), their equiratio combination (designated as Mix9) conferred protection to zebrafish, although not at doses lower than 5.104 cfu/mL (Fig. 3b) and not as reproducibly as with C. massiliae. To identify which association of species protected Re-ConvMix9 zebrafish against F. columnareALG infection, we tested all 9 combinations of 8 species (Mix8), as well as several combinations of 7, 6, 4, or 3 species and showed no protection (Supplementary Fig. S8A and Supplementary Table S4). We then tested whether lack of protection of Mix8 compared to Mix9 could rely on a density effect by doubling the concentration of any of the species within the nonprotective Mix8a (Supplementary Fig. S8B) and showed no protection. Interestingly, monitoring C. massiliae and the bacteria composing the Mix9 in conventional fish challenged by F. columnare only resulted (after 1 day) in an increase of P. sediminis and P. nitroreducens (p = <0.0001) and reduction of Phyllobacterium myrsinacearum, but no change in C. massiliae (Supplementary Fig. S9). Evenness also increased after infection from 0.65 in unchallenged Conv larvae to 0.82 for larvae infected with F. columnare. These results therefore also indicate that microbiota-based protection against F. columnareALG infection can rely on either C. massiliae-dependent membership effect or on a community-structure-dependent effect mediated by the Mix9 consortium.
Fig. 3. Protection against F. columnare in zebrafish reconventionalized with individual or mixed bacterial strains isolated from zebrafish.
a Determination of the level of protection provided by each of the 10 bacterial strains composing the core protective zebrafish microbiota. Bacteria were added individually to the water on hatching day (dose 5.105 cfu/mL). b Level of protection provided by different amount of C. massiliae and Mix9. Mix9 only protected at the highest inoculum doses. Mean survival is represented by a thick horizontal bar with standard deviation. Blue mean bars correspond to larvae not exposed to the pathogen and red mean bars correspond to exposed larvae. Larvae mortality rate was monitored daily and surviving fish were euthanized at day 9 post exposition to the pathogen. Indicated statistics correspond to unpaired, nonparametric Mann–Whitney test. ****p < 0.0001; absence of *: non-significant.
Pro- and anti- inflammatory cytokine production does not contribute to microbiota-mediated protection against F. columnareALG infection
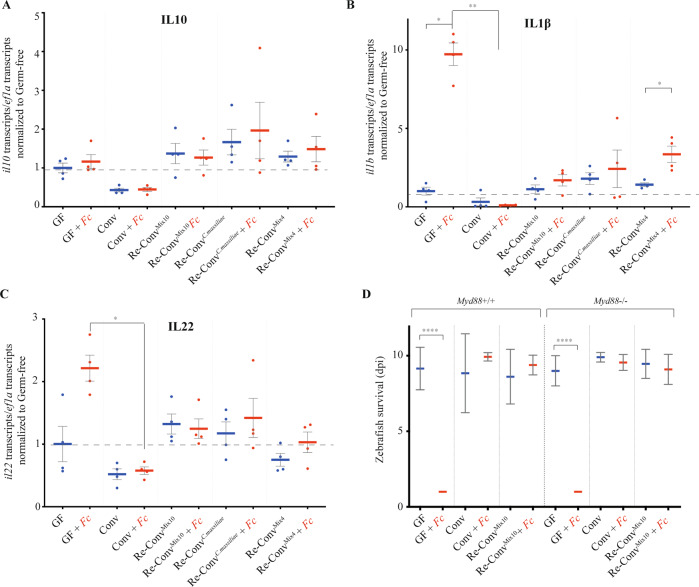
To test the contribution of the immune response of zebrafish larvae to resistance to F. columnare infection, we used qRT-PCR to measure cytokine mRNA expression in GF and Conv zebrafish exposed or not to F. columnareALG. We also tested the impact of reconventionalization with C. massiliae (re-ConvCm), Mix10 (re-ConvMix10) or with Mix4 (A. caviae, both A. veronii spp., P. mossellii) as a nonprotective control (Supplementary Table S4). We tested genes encoding IL1β (pro-inflammatory), IL22 (promoting gut repair), and IL10 (anti-inflammatory) cytokines. While we observed some variation in il10 expression among noninfected reconventionalized larvae, this did not correlate with protection. Furthermore, il10 expression was not modulated by infection in any of the tested conditions (Fig. 4a). By contrast, we observed a strong induction of il1b and il22 in GF zebrafish exposed to F. columnareALG (Fig. 4b, c). However, although this induction was reduced in protected Conv, Re-ConvCm and Re-ConvMix10, it was also observed in nonprotective Re-ConvMix4 larvae, indicating that down-modulation of the inflammatory response induced by F. columnare does not correlate with resistance to infection. Consistently, the use of a myd88 mutant, a key adapter of IL-1 and toll-like receptor signaling deficient in innate immunity [41, 53], showed that Conv or Re-ConvMix10, but not GF myd88 mutants survived F. columnare as well as wild-type zebrafish (Fig. 4d). Moreover, il1b induction by F. columnare infection was observed only in GF larvae and was myd88-independent (Supplementary Fig. S10). These results therefore indicated that the tested cytokine responses do not play a significant role in the microbiota-mediated protection against F. columnare infection.
Fig. 4. Zebrafish immune response to F. columnare infection.
a–c qRT-PCR analysis of host gene expression, 18 h after exposure to F. columnare, in larvae reconventionalized with indicated bacteria or bacterial mixes; each point corresponds to an individual larva. Expression of il10 (a), il1b (b), and il22 (c), by wild-type AB zebrafish; d: Comparison of the survival of myd88−/− and background-matched myd8+/+ zebrafish after reconventionalization and exposure to F. columnareALG. Mean survival is represented by a thick horizontal bar with standard deviation. For each condition, n = 12 zebrafish larvae. Larvae mortality rate was monitored daily and surviving fish were euthanized at day 9 post exposition to the pathogen (9 dpi). a–d Blue bars correspond to larvae not exposed to the pathogen and red mean bars correspond to exposed larvae. Indicated statistics correspond to unpaired, nonparametric Mann–Whitney test. ****p < 0.0001; **p < 0.005*: p < 0.05, absence of *: non-significant.
C. massiliae and Mix9 protect zebrafish from intestinal damages upon F. columnareALG infection
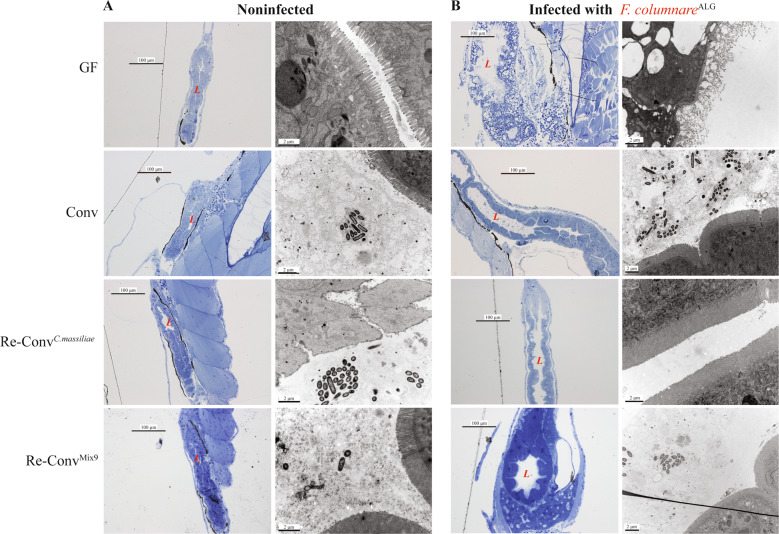
Histological analysis of GF larvae fixed 24 h after exposure to F. columnareALG revealed extensive intestinal damage (Fig. 5a) prior to noticeable signs in other potential target organs such as gills or skin. To test the requirement for gut access in F. columnareALG infection process, we modified our standard rearing protocol of GF fish, which involves feeding with live germ-free T. thermophila. We found that, if left unfed, GF zebrafish did not die after F. columnareALG exposure, while feeding with either T. thermophila or another food source such as sterile fish food powder, restored sensitivity to F. columnareALG infection (Supplementary Fig S11), suggesting that successful infection requires feeding and ingestion.
Fig. 5. Intestine of F. columnare infected germ-free zebrafish displays severe disorganization compared to conventional and reconventionalized larvae.
Germ-free, conventional and reconventionalized zebrafish larvae. Reconventionalized zebrafish were inoculated at 4 dpf with Mix9 or C. massiliae. a Representative picture of intestines of noninfected larvae. Fish were fixed for histology analysis or electron microscopy at 7 dpf. b Representative picture of intestines of infected larvae exposed at 7 dpf to F.columnareALG. In (a and b): Left column: Toluidine blue staining of Epon-embedded zebrafish larvae for Light microscopy. Right column: Transmission electron microscopy at 7 dpf (right). L = intestinal lumen.
Histological sections consistently showed severe disorganization of the intestine with blebbing in the microvilli and vacuole formation in F. columnareALG-infected GF larvae (Fig. 5). In contrast, zebrafish pre-incubated with either C. massiliae or Mix9 consortium at 4 dpf, and then exposed to F. columnareALG at 6 dpf showed no difference compared to noninfected larvae or conventional infected larvae (Fig. 5), confirming full protection against F. columnareALG at the intestinal level.
C. massiliae protects larvae and adult zebrafish against F. columnare
The clear protection provided by C. massiliae against F. columnareALG infection prompted us to test whether exogenous addition of this bacterium could improve microbiota-based protection towards this widespread fish pathogen. We first showed that zebrafish larvae colonized with C. massiliae were fully protected against all virulent F. columnare strains identified in this study (Fig. 6a). To test whether C. massiliae could also protect adult zebrafish from F. columnare infection, we pre-treated conventional 3–4-month-old Conv adult zebrafish with C. massiliae for 48 h before challenging them with a high dose (5.106 cfu/mL) of F. columnareALG. Monitoring of mortality rate showed that pre-treatment with C. massiliae significantly increased the survival rate of adult zebrafish upon F. columnareALG infection compared to non-treated conventional fish (p = 0.0084 Mann–Whitney test, Fig. 6b). Taken together, these results show that C. massiliae is a promising probiotic protecting zebrafish against columnaris disease caused by F. columnare.
Fig. 6. Pre-exposure to C. massiliae protects larval and adult zebrafish against F. columnare infection.
a Zebrafish larvae were inoculated at 4 dpf with 5.105 cfu/mL of C. massiliae for 48 h before infection at 6 dpf with virulent F. columnare strains. b Survival of adult zebrafish with or without pre-exposure to C. massiliae (2.106 cfu/mL for 48 h) followed by exposure to F. columnareALG (5.106 cfu/mL for 1 h) Mean survival is represented by a thick horizontal bar with standard deviation. For each condition, n = 12 zebrafish larvae or 10 adult. Zebrafish mortality rate was monitored daily and surviving fish were euthanized at day 9 post exposition to the pathogens (9 dpi). Blue bars correspond to larvae not exposed to the pathogen and red mean bars correspond to exposed larvae. Indicated statistics correspond to unpaired, nonparametric Mann–Whitney test. ****p < 0.0001; **p < 0.005; absence of *: non-significant.
Discussion
In this study, we used gnotobiotic zebrafish reconventionalized with relevant but relatively simple zebrafish larval microbiota in order to identify communities involved in colonization resistance against the fish pathogen F. columnare. We chose to work on larvae instead of adult fish because zebrafish microbiotas complexity increases when shifting from larval to later developmental stages [15, 54], while avoiding the important husbandry challenges associated with rearing germ-free adult zebrafish [20]. Using reconventionalization of otherwise germ-free zebrafish larvae we showed that conventional-level protection against infection by a broad range of highly virulent F. columnare strains is provided by a set of 10 culturable bacterial strains, belonging to 9 different species, isolated from the indigenous standard laboratory zebrafish microbiota. With the exception of the Bacteroidetes C. massiliae, this protective consortium was dominated by Proteobacteria such as Pseudomonas and Aeromonas spp., bacteria commonly found in aquatic environments [55, 56]. Despite the relative permissiveness of zebrafish larvae microbiota to environmental variations and inherent variability between samples [54], we showed that these ten bacteria were consistently identified in four different zebrafish facilities, suggesting the existence of a core microbiota assemblage with important colonization resistance functionality. Use of controlled combinations of these 10 bacterial species enabled us to show a very robust species-specific protection effect in larvae mono-associated with C. massiliae. We also identified a community-level protection provided by the combination of the 9 other species that were otherwise unable to protect against F. columnare when provided individually. This protection was however less reproducible and required a minimum inoculum of 5.104 cfu/mL, compared to 5.102 cfu/mL with C. massiliae. These results therefore suggest the existence of two distinct microbiota-based protection scenarios: one based on a membership effect provided by C. massiliae, and the other mediated by the higher-order activity of the Mix9 bacterial community.
Although protection against F. columnare infection does not seem to rely on microbiota-based immuno-modulation, we cannot exclude that, individually, some members of the studied core zebrafish microbiota could induce pro- or anti-inflammatory responses masked in presence of the full Mix10 consortium [1]. Whereas the identification of the mechanisms involved in the community-level Mix9 protection will require further studies, reconventionalization and dysbiosis and recovery experiments demonstrated the key role of C. massiliae in resistance against F. columnare. The mechanisms underlying this protection may be multi-factorial. First, these two phylogenetically close Bacteroidetes bacteria could compete for similar resources and directly antagonize each other [6, 13]. For example, we identified a cluster of 11 genes in the genome of C. massiliae (tssB, tssC, tssD, tssE, tssF, tssG, tssH, tssI, tssK, tssN, and tssP) encoding a putative contact-dependent type VI secretion system (T6SS) potentially injecting toxins [57]. Second, we also identified a gene encoding a putative pore-forming toxin of the Membrane Attack Complex/Perforin superfamily, which has been shown to contribute to interbacterial competition that occurs between phylogenetically close Bacteroidetes species [57–60]. Finally, all the genes associated with a functional T9SS involved in gliding motility as well as secretion of carbohydrate-active enzymes and other toxin or virulence factors are also conserved in C. massiliae and could contribute to its protective activity [61]. We cannot, however, exclude other mechanisms of protection such as nutrient depletion or pathogen exclusion upon direct competition for adhesion to host tissues [11, 22, 26], and experiments are currently underway to identify nonprotective C. massiliae mutants to uncover the bases of its activity against F. columnare. Interestingly, infected larvae reconventionalized with either C. massiliae or Mix9 showed no signs of the intestinal damage displayed by germ-free larvae, suggesting that both C. massiliae and Mix9 provide similar intestinal resistance to F. columnare infection. Whereas microbial colonization contributes to gut maturation and stimulates the production of epithelial passive defenses such as mucus [62, 63], lack of intestinal maturation is unlikely to be contributing to F. columnare-induced mortality, as mono-colonized larvae or larvae reconventionalized with nonprotective mixes died as rapidly as germ-free larvae.
Several studies have monitored the long-term assembly and development of the zebrafish microbiota from larvae to sexually mature adults, however little is known about the initial colonization establishment of the larvae after hatching [64, 65]. Neutral (stochastic) and deterministic (host niche-based) processes [66–68] lead to microbial communities that are mostly represented by a limited number of highly abundant species with highly diverse low-abundant populations. In our experiments, the Mix10 species inoculum corresponded to an equiratio bacterial mix, thus starting from an engineered and assumed total evenness (E = 1) [69, 70]. Evenness was still relatively high (0.84) and remained similar up until 8 h in our study, indicating that most of the ten species were able to colonize the larvae. From the perspective of community composition, a loss of diversity is often associated with decreased colonization resistance, but it remains unclear whether this increased susceptibility is due to the loss of certain key member species of the microbial community and/or a change in their prevalence [3, 9].
We further investigated resistance to infection by exposing established bacterial communities to different antibiotic perturbations, followed by direct challenge with F. columnare (to study core microbiota sensitivity to disturbance) or after recovery (to study its resilience) [12, 71]. Antibiotics are known to shift the composition and relative abundances of the microbiota according to their spectrum [13, 72]. We observed that penicillin/streptomycin treatment that would affect most of the core species, reduced the abundance of all but two species (A. veronii 1 and P. myrsinacearum) that became relatively dominant during recovery, but failed to provide protection against F. columnare. With the kanamycin treatment, colonization resistance was fully restored at the end of the 24 h recovery period, indicative of a resilience that could result from species recovering quickly to their pre-perturbation levels due to fast growth rates, physiological flexibility or mutations [73]. Interestingly, even taking into account potential biases associated with the use of the 16S rRNA as a proxy index to determine relative abundance [74, 75], evenness was similarly reduced during recovery for both treatments, but abundance at phylum level changed to 48% for Proteobacteria, and 52% for Bacteroidetes compared to the >98% of Proteobacteria with the penicillin/streptomycin treatment. Furthermore, C. massiliae was detected as rare (<1%) in conventional larvae, suggesting that it could have a disproportionate effect on the community or that community-level protection provided by the nine other bacteria was also responsible for the protection of conventional larvae to F. columnare infection.
We showed that germ-free zebrafish larvae are highly susceptible to a variety of different F. columnare genomovars isolated from different hosts, demonstrating that they are a robust animal model for the study of its pathogenicity. Recently, F. columnare mutants in T9SS were shown to be avirulent in adult zebrafish, suggesting that proteins secreted by the T9SS are likely to be key, but still largely unidentified, F. columnare virulence determinants [39]. Body skin, gills, fins and tail are also frequently damaged in salmonid fish, whereas severe infection cases are associated with septicemia [38]. We could not identify such clear F. columnare infection sites in zebrafish larvae, perhaps due to the very low dose of infection used, with less than 100 cfu recovered from infected moribund larvae. However, several lines of evidence suggest that the gut is the main target of F. columnare infection in our model: (i) unfed germ-free larvae survived exposure, (ii) histology analysis showing severe disruption of the intestinal region just hours after infection in germ-free larvae, and (iii) induction of il22 in germ-free larvae exposed to F. columnare, since a major function of IL-22 is to promote gut repair [76]. This induction appears to be a consequence of the pathogen-mediated damage, as there was no observed induction in conventional or reconventionalized larvae. The very rapid death of larvae likely caused by this severe intestinal damage may explain why other common target organs of columnaris disease showed little damage.
In this study, we showed that C. massiliae is a promising probiotic candidate that could contribute to reduce the use of antibiotics to prevent columnaris diseases in research and aquaculture settings. Whereas C. massiliae provided full and robust protection against all tested virulent F. columnare genomovars and significantly increased survival of exposed adult conventional zebrafish, further studies are needed to elucidate C. massiliae protection potential in other teleost fish. However, the endogenous nature of C. massiliae suggests that it could establish itself as a long-term resident of the zebrafish larval and adult microbiota, an advantageous trait when seeking a stable modulation of the bacterial community over long periods [43].
In conclusion, the use of a simple and tractable zebrafish larval model to mine indigenous host microbial communities allowed us to identify two independent protection strategies against the same pathogen. Whereas further study will determine how these strategies may contribute to protection against a wider range of pathogens, this work also provides insights into how to engineer stable protective microbial communities with controlled colonization resistance functions.
Supplementary information
Acknowledgements
We thank Mark McBride, Pierre Boudinot, and Rebecca Stevick for critical reading of the manuscript. We are grateful to the late Covadonga Arias for the gift of F. columnare ALG 00–530, to Mark McBride for F. columnare C#2 strain and to Jean-François Bernardet for all other F. columnare strains. Prof. Annemarie Meijer (Leiden University) kindly provided the myd88 mutant zebrafish line. We thank Chloé Baron for her help, Julien Burlaud-Gaillard and Rustem Uzbekov from the IBiSA Microscopy facility, Tours University, France and the following zebrafish facility teams for providing eggs: José Perez and Yohann Rolin (Institut Pasteur), Nadia Soussi-Yanicostas (INSERM Robert Debré), Sylvie Schneider-Manoury and Isabelle Anselme (UMR7622, University Paris 6) and Frédéric Sohm (AMAGEN Gif-sur-Yvette).
Funding
This work was supported by the Institut Pasteur, the French Government’s Investissement d’Avenir program: Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant no. ANR-10-LABX-62-IBEID to J-MG.), the Fondation pour la Recherche Médicale (grant no. DEQ20180339185 to J-MG). FS was the recipient of a post-doctoral Marie Curie fellowship from the EU-FP7 program, JBB was the recipient of a long-term post-doctoral fellowship from the Federation of European Biochemical Societies (FEBS) and by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 842629. DP-P was supported by an Institut Carnot MS Postdoctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
FAS, JBB, DP-P, J-PL, and J-MG designed the experiments. OR contributed to the initial experiments. VB and J-PL provided zebrafish material and advice. FAS, JBB, DP-P, BA, VB, and J-PL performed the experiments. SB, SH performed bacterial genome sequencing and analysis, AG, SV, FAS, and DP-P performed the bioinformatic and sequence analyses. FAS, JBB, DP-P, J-PL, and J-MG analysed the data and wrote the paper with significant contribution from OR and ED.
Data availability
The raw sequences generated for the study can be found in the NCBI Short Read Archive under BioProject No. PRJNA649696. Bacterial genome sequences obtained in the present study are available at the European Nucleotide Archive with the project number PRJEB36872, under accession numbers ERS4385993 (Aeromonas veronii 1); ERS4386000 (Aeromonas veronii 2); ERS4385996 (Aeromonas caviae); ERS4385998 (Chryseobacterium massiliae); ERS4385999 (Phyllobacterium myrsinacearum); ERS4406247 (Pseudomonas sediminis); ERS4385994 (Pseudomonas mossellii) ERS4386001 (Pseudomonas nitroreducens); ERS4385997 (Pseudomonas peli); ERS4385995 (Stenotrophomas maltophilia).
Compliance with ethical standards
Conflict of interest
The authors of this manuscript have the following conflict of interest: a provisional patent application has been filed: “bacterial strains for use as probiotics, compositions thereof, deposited strains and method to identify probiotic bacterial strains” by J-MG, FAS, DP-P, and JBB The other authors declare no conflict of interest in relation to the submitted work.
Ethics
All animal experiments described in the present study were conducted at the Institut Pasteur (larvae) or at INRA Jouy-en-Josas (adults) according to European Union guidelines for handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and were approved by the relevant institutional Animal Health and Care Committees.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Franziska A. Stressmann, Joaquín Bernal-Bayard
Supplementary information
The online version of this article (10.1038/s41396-020-00807-8) contains supplementary material, which is available to authorized users.
References
- 1.Rolig AS, Parthasarathy R, Burns AR, Bohannan BJ, Guillemin K. Individual members of the microbiota disproportionately modulate host innate immune responses. Cell Host Microbe. 2015;18:613–20. doi: 10.1016/j.chom.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 3.van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-van der Wees JEC. Colonization resistance of the digestive tract and the spread of bacteria to the lymphatic organs in mice. J Hyg. 1972;70:335–42. doi: 10.1017/s0022172400022385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–58. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 5.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–14. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Falcinelli S, Rodiles A, Unniappan S, Picchietti S, Gioacchini G, Merrifield DL, et al. Probiotic treatment reduces appetite and glucose level in the zebrafish model. Sci Rep. 2016;6:18061. doi: 10.1038/srep18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsan EE, Byndloss MX, Faber F, Rivera-Chavez F, Tsolis RM, Baumler AJ. Colonization resistance: The deconvolution of a complex trait. J Biol Chem. 2017;292:8577–81. doi: 10.1074/jbc.R116.752295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–23. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heselmans M, Reid G, Akkermans LM, Savelkoul H, Timmerman H, Rombouts FM. Gut flora in health and disease: potential role of probiotics. Curr Issues Intest Microbiol. 2005;6:1–7. [PubMed] [Google Scholar]
- 11.Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol. 2007;23:679–92. doi: 10.1097/MOG.0b013e3282f0cffc. [DOI] [PubMed] [Google Scholar]
- 12.Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–76. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–14. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill HS. Probiotics to enhance anti-infective defences in the gastrointestinal tract. Best Pr Res Clin Gastroenterol. 2003;17:755–73. doi: 10.1016/s1521-6918(03)00074-x. [DOI] [PubMed] [Google Scholar]
- 15.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, Guillemin K, et al. Study of host-microbe interactions in zebrafish. Methods cell Biol. 2011;105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns AR, Guillemin K. The scales of the zebrafish: host-microbiota interactions from proteins to populations. Curr Opin Microbiol. 2017;38:137–41. doi: 10.1016/j.mib.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas AE. Simple animal models for microbiome research. Nat Rev Microbiol. 2019;17:764–75. doi: 10.1038/s41579-019-0242-1. [DOI] [PubMed] [Google Scholar]
- 19.Flores EM, Nguyen AT, Odem MA, Eisenhoffer GT, Krachler AM. The zebrafish as a model for gastrointestinal tract-microbe interactions. Cell Microbiol. 2020;22:e13152. doi: 10.1111/cmi.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melancon E, Gomez De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods cell Biol. 2017;138:61–100. doi: 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantas L, Sorby JR, Alestrom P, Sorum H. Culturable gut microbiota diversity in zebrafish. Zebrafish. 2012;9:26–37. doi: 10.1089/zeb.2011.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rendueles O, Ferrieres L, Fretaud M, Begaud E, Herbomel P, Levraud JP, et al. A new zebrafish model of oro-intestinal pathogen colonization reveals a key role for adhesion in protection by probiotic bacteria. PLoS Pathog. 2012;8:e1002815. doi: 10.1371/journal.ppat.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caruffo M, Navarrete NC, Salgado OA, Faundez NB, Gajardo MC, Feijoo CG, et al. Protective Yeasts Control V. anguillarum Pathogenicity and Modulate the Innate Immune Response of Challenged Zebrafish (Danio rerio) Larvae. Front Cell Infect Microbiol. 2016;6:127. doi: 10.3389/fcimb.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Ramos A, Mohedano ML, Pardo MA, Lopez P. Beta-glucan-producing pediococcus parvulus 2.6: test of probiotic and immunomodulatory properties in zebrafish models. Front Microbiol. 2018;9:1684. doi: 10.3389/fmicb.2018.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu W, Zhou S, Zhu W, Zhuang X. Quorum quenching bacteria Bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci Rep. 2014;4:5446. doi: 10.1038/srep05446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Ren Z, Fu L, Su X. Two highly adhesive lactic acid bacteria strains are protective in zebrafish infected with Aeromonas hydrophila by evocation of gut mucosal immunity. J Appl Microbiol. 2016;120:441–51. doi: 10.1111/jam.13002. [DOI] [PubMed] [Google Scholar]
- 27.Qin C, Zhang Z, Wang Y, Li S, Ran C, Hu J, et al. EPSP of L. casei BL23 Protected against the Infection Caused by Aeromonas veronii via Enhancement of Immune Response in Zebrafish. Front Microbiol. 2017;8:2406. doi: 10.3389/fmicb.2017.02406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girija V, Malaikozhundan B, Vaseeharan B, Vijayakumar S, Gobi N, Del Valle Herrera M, et al. In vitro antagonistic activity and the protective effect of probiotic Bacillus licheniformis Dahb1 in zebrafish challenged with GFP tagged Vibrio parahaemolyticus Dahv2. Microb Pathogenesis. 2018;114:274–80. doi: 10.1016/j.micpath.2017.11.058. [DOI] [PubMed] [Google Scholar]
- 29.Lin YS, Saputra F, Chen YC, Hu SY. Dietary administration of Bacillus amyloliquefaciens R8 reduces hepatic oxidative stress and enhances nutrient metabolism and immunity against Aeromonas hydrophila and Streptococcus agalactiae in zebrafish (Danio rerio) Fish Shellfish Immunol. 2019;86:410–9. doi: 10.1016/j.fsi.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 30.Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, Decostere A. Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Vet Res. 2013;44:27. doi: 10.1186/1297-9716-44-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decostere A, Haesebrouck F, Devriese LA. Characterization of four Flavobacterium columnare (Flexibacter columnaris) strains isolated from tropical fish. Vet Microbiol. 1998;62:35–45. doi: 10.1016/s0378-1135(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 32.Figueiredo HC, Klesius PH, Arias CR, Evans J, Shoemaker CA, Pereira DJ, Jr, et al. Isolation and characterization of strains of Flavobacterium columnare from Brazil. J fish Dis. 2005;28:199–204. doi: 10.1111/j.1365-2761.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 33.Soto E, Mauel MJ, Karsi A, Lawrence ML. Genetic and virulence characterization of Flavobacterium columnare from channel catfish (Ictalurus punctatus) J Appl Microbiol. 2008;104:1302–10. doi: 10.1111/j.1365-2672.2007.03632.x. [DOI] [PubMed] [Google Scholar]
- 34.Suomalainen LR, Bandilla M, Valtonen ET. Immunostimulants in prevention of columnaris disease of rainbow trout, Oncorhynchus mykiss (Walbaum) J fish Dis. 2009;32:723–6. doi: 10.1111/j.1365-2761.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- 35.Pacha RE, Ordal EJ Myxobacterial infections of salmonids. American Fisheries Society, Diseases of Fishes and Shellfishes 1970:12.
- 36.Amin NE, Abdallah IS, Faisal M, Easa Me-S, Alaway T, Alyan SA. Columnaris infection among cultured Nile tilapia Oreochromis niloticus. Antonie Van Leeuwenhoek. 1988;54:509–20. doi: 10.1007/BF00588387. [DOI] [PubMed] [Google Scholar]
- 37.Decostere A, Haesebrouck F, Charlier G, Ducatelle R. The association of Flavobacterium columnare strains of high and low virulence with gill tissue of black mollies (Poecilia sphenops) Vet Microbiol. 1999;67:287–98. doi: 10.1016/s0378-1135(99)00050-4. [DOI] [PubMed] [Google Scholar]
- 38.Bernardet J-F, Bowman JP. The genus flavobacterium. Prokaryotes. 2006;7:481–531. [Google Scholar]
- 39.Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, et al. The type IX secretion system is required for virulence of the fish pathogen flavobacterium columnare. Appl Environ Microbiol. 2017;83:e01769–17. doi: 10.1128/AEM.01769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia JC, LaFrentz BR, Waldbieser GC, Wong FS, Chang SF. Characterization of atypical Flavobacterium columnare and identification of a new genomovar. J Fish Dis. 2018;41:1159–64. doi: 10.1111/jfd.12778. [DOI] [PubMed] [Google Scholar]
- 41.van der Vaart M, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Models Mech. 2013;6:841–54. doi: 10.1242/dmm.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–75. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, Ser B,57, 289-300. 1995;57:289–300. [Google Scholar]
- 46.Ha SM, Kim CK, Roh J, Byun JH, Yang SJ, Choi SB, et al. Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann Lab Med. 2019;39:530–6. doi: 10.3343/alm.2019.39.6.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volant S, Lechat P, Woringer P, Motreff L, Campagne P, Malabat C, et al. SHAMAN: a user-friendly website for metataxonomic analysis from raw reads to statistical analysis. BMC Bioinforma. 2020;21:345. doi: 10.1186/s12859-020-03666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–45. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olivares-Fuster O, Bullard SA, McElwain A, Llosa MJ, Arias CR. Adhesion dynamics of Flavobacterium columnare to channel catfish Ictalurus punctatus and zebrafish Danio rerio after immersion challenge. Dis Aquat Org. 2011;96:221–7. doi: 10.3354/dao02371. [DOI] [PubMed] [Google Scholar]
- 53.Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108:4570–7. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016;10:644–54. doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mena KD, Gerba CP. Risk assessment of pseudomonas aeruginosa in water. Rev Environ contamination Toxicol. 2009;201:71–115. doi: 10.1007/978-1-4419-0032-6_3. [DOI] [PubMed] [Google Scholar]
- 56.Goncalves Pessoa RB, de Oliveira WF, Marques DSC, Dos Santos Correia MT, de Carvalho E, Coelho L. The genus Aeromonas: a general approach. Microb pathogenesis. 2019;130:81–94. doi: 10.1016/j.micpath.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 57.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–36. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol. 2014;94:1361–74. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio. 2016;7:e01055–16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, et al. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell host microbe. 2018;24:296–307.e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsbrink J, McKee LS. Bacteroidetes bacteria in the soil: glycan acquisition, enzyme secretion, and gliding motility. Adv Appl Microbiol. 2020;110:63–98. doi: 10.1016/bs.aambs.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–35. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 63.Ganz J, Melancon E, Eisen JS. Zebrafish as a model for understanding enteric nervous system interactions in the developing intestinal tract. Methods cell Biol. 2016;134:139–64. doi: 10.1016/bs.mcb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–86. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Yan Q, van der Gast CJ, Yu Y. Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS ONE. 2012;7:e30603. doi: 10.1371/journal.pone.0030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–62. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers GB, Hoffman LR, Carroll MP, Bruce KD. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2013;21:271–6. doi: 10.1016/j.tim.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–64. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shafquat A, Joice R, Simmons SL, Huttenhower C. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol. 2014;22:261–6. doi: 10.1016/j.tim.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shade A, Peter H, Allison SD, Baho DL, Berga M, Burgmann H, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–43. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 73.Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, et al. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology. 2009;137:1757–67.e1. doi: 10.1053/j.gastro.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 74.Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–34. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 75.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–23. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 76.Shabgah AG, Navashenaq JG, Shabgah OG, Mohammadi H, Sahebkar A. Interleukin-22 in human inflammatory diseases and viral infections. Autoimmun Rev. 2017;16:1209–18. doi: 10.1016/j.autrev.2017.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequences generated for the study can be found in the NCBI Short Read Archive under BioProject No. PRJNA649696. Bacterial genome sequences obtained in the present study are available at the European Nucleotide Archive with the project number PRJEB36872, under accession numbers ERS4385993 (Aeromonas veronii 1); ERS4386000 (Aeromonas veronii 2); ERS4385996 (Aeromonas caviae); ERS4385998 (Chryseobacterium massiliae); ERS4385999 (Phyllobacterium myrsinacearum); ERS4406247 (Pseudomonas sediminis); ERS4385994 (Pseudomonas mossellii) ERS4386001 (Pseudomonas nitroreducens); ERS4385997 (Pseudomonas peli); ERS4385995 (Stenotrophomas maltophilia).