Abstract
The communities of bacteria that assemble around marine microphytoplankton are predictably dominated by Rhodobacterales, Flavobacteriales, and families within the Gammaproteobacteria. Yet whether this consistent ecological pattern reflects the result of resource-based niche partitioning or resource competition requires better knowledge of the metabolites linking microbial autotrophs and heterotrophs in the surface ocean. We characterized molecules targeted for uptake by three heterotrophic bacteria individually co-cultured with a marine diatom using two strategies that vetted the exometabolite pool for biological relevance by means of bacterial activity assays: expression of diagnostic genes and net drawdown of exometabolites, the latter detected with mass spectrometry and nuclear magnetic resonance using novel sample preparation approaches. Of the more than 36 organic molecules with evidence of bacterial uptake, 53% contained nitrogen (including nucleosides and amino acids), 11% were organic sulfur compounds (including dihydroxypropanesulfonate and dimethysulfoniopropionate), and 28% were components of polysaccharides (including chrysolaminarin, chitin, and alginate). Overlap in phytoplankton-derived metabolite use by bacteria in the absence of competition was low, and only guanosine, proline, and N-acetyl-d-glucosamine were predicted to be used by all three. Exometabolite uptake pattern points to a key role for ecological resource partitioning in the assembly marine bacterial communities transforming recent photosynthate.
Subject terms: Microbial ecology, Bacteria
Introduction
The marine dissolved organic carbon (DOC) reservoir plays two critical roles in the global carbon cycle. The first is as a long-term repository of carbon nearly equal in magnitude to the inorganic carbon stored in the atmosphere. This role is fulfilled by ~660 Pg C of refractory organic compounds that accumulate in seawater, some with lifetimes of many thousands of years [1]. The second is as the primary source of substrates for heterotrophic marine microbes. This role is fulfilled by ~0.2 Pg of highly bioreactive organic compounds that are rapidly cycled by bacterioplankton [2], some with turnover times on the order of hours [3–5]. Recent chemical analysis of bulk marine DOC concentrated from seawater has shown it to be conservatively composed of tens of thousands of distinct organic compounds [6, 7]. Yet chemical analysis of the bioreactive subset of marine DOC has thus far been dominated by targeted analysis of a limited number of core metabolites, such as amino acids, sugars, and select biopolymers [8–11]. Untargeted chemical approaches have lagged behind, plagued by low concentrations, similar physical properties of salt and polar metabolites, and short lifetimes of component metabolites. Thus many critical but largely invisible chemical connections between marine microbes remain unstudied.
The primary source of labile metabolites for surface ocean bacteria is phytoplankton. It is estimated that these microbial autotrophs release 10–20% of net primary production (NPP) into the marine DOC pool in coastal systems [12] and up to 40% in oligotrophic systems [13], although this percentage varies across species and growth conditions [14, 15]. Mechanisms by which metabolites are released from phytoplankton cells range from passive leakage of small and uncharged molecules [16], to active exudation related to redox balance, defense, and signaling [17–19], to photosynthetic overflow due to stoichiometric imbalance [15]. Labile organic compounds also become available by phytoplankton cell death from processes such as viral lysis, protist grazing, and senescence [20–22]. Once metabolites are released or excreted from phytoplankton cells, heterotrophic bacteria play the dominant role in their transformation.
Three marine bacterial taxa have consistently been found associated with microphytoplankton in natural marine environments and phytoplankton cultures, and are thought to dominate processing of recently-fixed carbon using genes that allow them to quickly respond to transient nutrient pulses [23]. Members of these three groups, the Rhodobacterales, Gammaproteobacteria, and Flavobacteriales, appear to specialize on different components of bioreactive DOC [11, 23–25]. Rhodobacterales typically play a prominent role in processing low molecular weight (LMW) phytoplankton-derived metabolites [26, 27], Flavobacteriales largely transform high molecular weight carbohydrate polymers [28, 29], and several copiotrophic Gammaproteobacteria families utilize compounds from both classes [30, 31]. Here, we address the ecological basis of this widespread taxonomic pattern by generating metabolite uptake profiles for three model bacterial species, one from each of the major phytoplankton-associated groups, when growing on a microphytoplankton exometabolite pool in the absence of competition from other bacteria.
Pairwise co-culture systems were established with the diatom Thalassiosira pseudonana as the sole source of substrates for Ruegeria pomeroyi DSS-3 (Rhodobacterales), Stenotrophomonas sp. SKA14 (Xanthomonadales), or Polaribacter dokdonensis MED152 (Flavobacteriales). T. pseudonana was selected as the autotrophic member of the model systems because as a group, marine diatoms contribute ~20% of global NPP [32]. The heterotrophic bacterial strains were selected because they have high identity to 16S rRNA genes from phytoplankton cultures or flow-sorted phytoplankton cells, with percent similarities as high as 99.6% for R. pomeroyi [33–36], 98.8% for Stenotrophomonas sp. SKA14 [33], and 97.2% for P. dokdonensis [35, 37]. To overcome some challenges of direct chemical analysis of low-concentration compounds in seawater, we used two biological vetting approaches that highlighted metabolites most likely to be important in phytoplankton-bacteria carbon exchange. In the first, expression patterns of bacterial transporters and catabolic genes were used to identify the cellular machinery for carbon acquisition activated by each strain when growing in co-culture. In the second, drawdown of diatom-derived exometabolites from the co-culture media was used to distinguish compounds decreasing in the presence of bacteria. Metabolite concentrations were measured using liquid chromatography-mass spectrometry (LC-MS) and heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectroscopy. Both biological vetting strategies relied on bacterial activity to spotlight compounds within a complex pool of dilute metabolites that were likely supporting bacterial heterotrophy.
Methods
Co-cultures
Three bacterial strains were introduced individually into 7-day diatom co-cultures. Samples were collected after 8, 24, and/or 48 h and analyzed for bacterial response via transcriptomics (to measure regulation changes) or via mass spectrometry (MS) or NMR (to measure metabolite drawdown). To initiate co-cultures, T. pseudonana 1335 (National Center for Marine Algae) was grown axenically in organic carbon-free medium L1 +Si [38] in 1900-ml vented polystyrene tissue culture flasks at 18 °C under 16 h light at 160 µmol photons m−2 s−1 and 8 h dark. For samples used in NMR analysis, 13C bicarbonate was used to make the L1 medium. After the diatom cultures had been growing for 7 days, bacteria pre-grown in YTSS medium were washed five times in sterile L1 medium and inoculated into three replicate diatom cultures at ~106 cells ml−1; three flasks remained uninoculated. Following incubation in the light for 8 h, T. pseudonana cells were removed by pre-filtration through 2.0-µm-pore-size filters, and bacteria were collected on 0.2-µm-pore-size filters. Filters were immediately flash frozen in liquid nitrogen and stored at −80 °C until processing and the filtrate was stored frozen for subsequent chemical analysis. Filtrate was also obtained 24 and 48 h after bacterial inoculation for chemical analysis. The control for transcriptome analysis was established by growing bacteria in a defined glucose medium (L1 medium +Si with 2.5 mM glucose); this masked signals of glucose utilization in the co-cultures but provided baseline transcriptomes of actively growing bacteria. Bacterial strains were similarly inoculated into the control medium, collected on 0.2 pore-size filters after 8 h, and flash frozen. Detailed methods are given in Supplementary File 1.
Cell counts
Culture samples were fixed to a final concentration of 1% glutaraldehyde, incubated at 4°C for 20 min, and stored at −80 °C. Just prior to analysis, an internal standard of 5-μm fluorescent beads was added (Spherotech, Lake Forest, IL, USA), followed by staining for 15 min with SYBR Green I (final concentration 0.75X; Life Technologies, Waltham, MA, USA). Samples were analyzed on an Agilent Quanteon flow cytometer (Acea, Biosciences Inc., San Diego CA). Fluorescence was detected with a 405 nm laser using a 530/30 bandpass filter for SYBR Green (bacteria) and a 695/40 bandpass filter for chlorophyll a. There was no bacterial contamination of axenic cultures based on scattergrams from flow cytometry and aliquots from axenic cultures spread onto YTSS plates.
RNA-seq analysis
Filters were incubated in SDS (0.6% final concentration) and proteinase K (120 ng μl–1 final concentration). RNA was extracted from duplicates of each treatment by adding an equal volume of acid phenol:chloroform:isoamyl-alcohol, followed by shaking, centrifugation, collection of the supernatant, and addition of an equal volume of chloroform:isoamyl-alcohol. RNA was recovered from the supernatant, treated to remove rRNA, and sequenced on an HiSeq Illumina 2500. Quality control of the 207 million 50-bp reads was performed using the FASTX toolkit. Reads aligning to rRNA were removed and remaining reads were mapped to the bacterial genomes. Genes with differential expression were determined with DESeq2 [39], and are available in Supplementary Tables S1, S2, and S3. The dbCAN web resource was used for identification of carbohydrate-active enzyme annotations [40]. Raw RNA-seq data are available in the NCBI SRA BioProject database under accession PRJNA448168. Detailed methods are given in Supplementary File 1. Microbial genome sequences are available at NCBI RefSeq under accession numbers ASM14940v2 (T. pseudonana CCMP1335), ASM1196v2 (R. pomeroyi DSS-3), ASM15857v1 (Stenotrophomonas sp. SKA14), and ASM15294v2 (P. dokdonensis MED152).
Mass spectrometry analysis
Chemical analysis was conducted on filtered spent media from the co-cultures and axenic T. pseudonana culture using an uninoculated L1 as the medium blank. For MS analysis, 8, 24, and 48 h co-culture spent media were analyzed. Metabolites were derivatized with benzoyl chloride [41] by modification of methods from Oehlke et al. [42] and Wong et al. [43], extracted using a solid phase resin (Agilent, Bond Elut PPL), and analyzed using ultra high performance liquid chromatography coupled with electrospray ionization and tandem MS with modifications to Kido Soule et al. [44]. Metabolite peak areas were selected and integrated using Skyline [45, 46] (Fig. S1). MS metabolites were evaluated statistically in MATLAB by comparing adjusted sample concentrations grouped across time points using a one-way ANOVA (α = 0.05). Post hoc Dunnett’s test was used to compare each co-culture to the axenic T. pseudonana treatment, and p values were adjusted for multiple comparisons. Outliers were defined as values that exceeded three scaled median absolute deviations and were excluded from statistical analysis. All MS data are available at MetaboLights under accession number MTBLS1751. Detailed methods are given in Supplementary File 1.
NMR analysis
For NMR analysis, 5 ml of 48 h co-culture spent medium was lyophilized, homogenized dry using 5 × 3.5 mm glass beads, reconstituted in dimethyl-sulfoxide-d6, and re-homogenized. The supernatant was analyzed using two-dimensional HSQC NMR (Bruker 800 MHz NEO with 1.7 mm cryoprobe) using acquisition parameters modified from a hsqcetgpsisp2.2 pulse program (TopSpin V4.0.6). The indirect 13C (f1) dimension had a spectral width of 90.0027 ppm, 128 data points, and a carrier frequency of 45 ppm. The direct 1H dimension (f2) had a spectral width of 13.0255 ppm, 4166 data points, and a carrier frequency of 3.691 ppm. Spectra were processed in MNOVA and transformed spectra were auto-phased, baseline-corrected, and referenced along f1 and f2 to DSS-d6 (0.0, 0.0 ppm). All peaks above noise were manually integrated (Fig. S2). A MATLAB workflow was used to normalize, scale, and analyze spectral features. p values were calculated for all peak integrals using the [ttest] function in MATLAB for sample pairs. False discovery rates and q values were calculated using MATLAB built-in function [mafdr]. Raw data, peak lists, and analysis scripts are available at MetaboLights under accession MTBLS1544. Detailed methods are given in Supplementary File 1.
Results
Identification of ecologically relevant exometabolites was carried out in co-culture systems in which marine phytoplankter T. pseudonana CCMP1335 served as the sole carbon source for three bacterial strains. R. pomeroyi DSS-3, Stenotrophomonas sp. SKA14, and P. dokdonensis MED152 were individually inoculated into a T. pseudonana culture that had accumulated exometabolites over 7 days. Bacterial gene expression in response to compounds in the exometabolite pool was identified after 8 h by comparison to gene expression in a single-substrate (glucose) control. Drawdown of T. pseudonana exometabolites in the co-culture spent medium was analyzed after 8, 24, and 48 h by LC-MS (Table S4) [41] and after 48 h incubations by 2D HSQC NMR (Holderman et al. in prep.). The control for the chemical analyses was spent medium from axenic T. pseudonana cultures. Microscopic observations did not reveal close physical attachment of the bacteria to diatom cells, although P. dokdonensis appeared associated with diatom extracellular polysaccharides after 48 h in co-culture. There was no evidence of altered growth rates by the diatom in the presence of the bacteria (Kruskal Wallis, p = 0.30, n = 3, T = 253 h; Fig. 1).
Fig. 1. Cell numbers in co-culture experiments after inoculation of bacterial strains into T. pseudonana cultures at 168 h (day 7; gray dashed line).
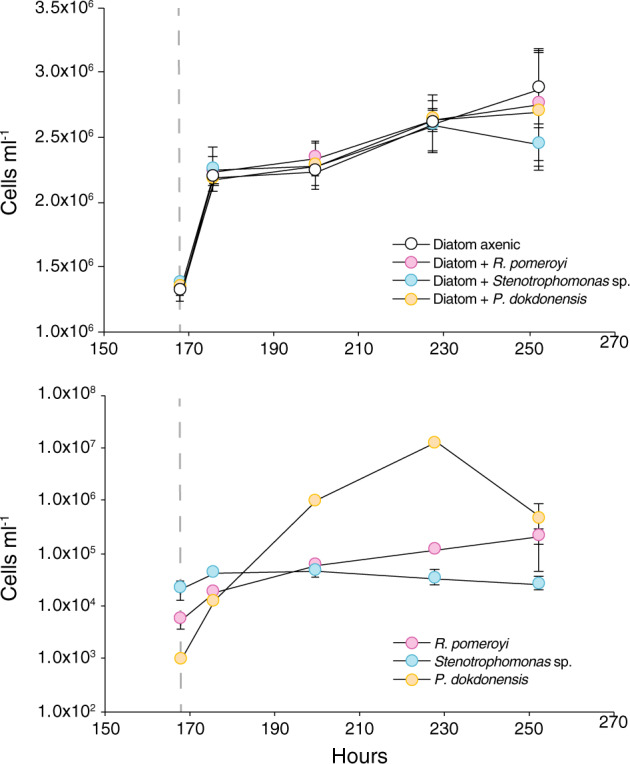
Top: growth of T. pseudonana under axenic conditions and in cultures with each bacterial strain. Bottom: growth of bacterial strains in co-culture with T. pseudonana. n = 3; error bars represent ±1 standard deviation.
Ruegeria pomeroyi DSS-3 metabolite utilization
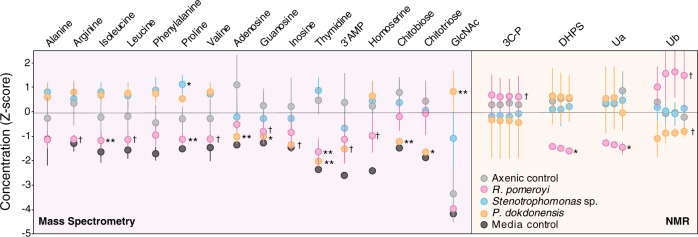
R. pomeroyi transporters that were enriched in the co-cultures compared to the glucose control included ABC and TRAP transporters, both employing dedicated solute binding proteins that recognize substrates with high affinity, as well as permeases that depend largely on diffusion. Five enriched transporters had hypothesized annotations for amino acid uptake (Fig. 2 and Table S5). Spent medium drawdown patterns were consistent with this, and specifically indicated that concentrations of arginine and the branched chain amino acids valine, leucine, and isoleucine were lower in co-cultures with R. pomeroyi compared to axenic T. pseudonana cultures (Fig. 3). Nucleoside uptake was also indicated by spent medium drawdown, with significantly lower concentrations of guanosine and thymidine (Figs. 3 and S1).
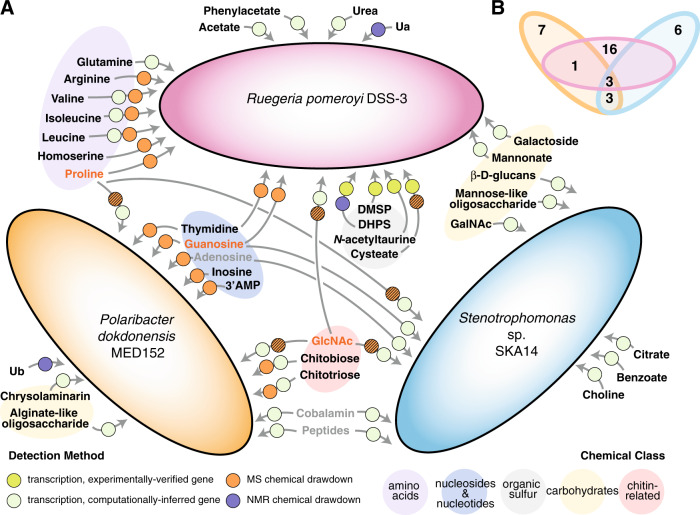
Fig. 2. Metabolites involved in autotroph-heterotroph carbon transfer for three model marine bacteria when co-cultured with T. pseudonana.
A. Summary of results from exometabolite chemical analysis and bacterial gene expression. Black font indicates metabolites unique to a single strain; gray font indicates metabolites shared by two strains; and orange font indicates metabolites shared by three strains. Circles specify the detection method(s), with hatched symbols indicating a mismatch between expression and drawdown methods. Metabolites Ua and Ub are unidentified compounds detected by 13C-HSQC NMR. B Venn diagram summary of unique and shared metabolites.
Fig. 3. Bacterial drawdown of diatom exometabolites compared to axenic diatom cultures based on analysis of spent media by targeted LC-ESI-MS (left) and 13C-HSQC NMR (right).
Data are Z scores of adjusted concentration (MS analysis) and scaled area (NMR analysis). Grouped data points indicate linked carbon atoms in NMR analysis. See Fig. S1 for time course of MS data and Fig. S2 for contour plots of NMR features. n = 3–7. ** = treatments significantly different from the axenic diatom cultures at adjusted p ≤ 0.01; * = treatments different from the axenic diatom cultures at p ≤ 0.05; † = treatments different from the axenic diatom cultures at p < 0.10.
Organic sulfur substrates indicated by transcript enrichment included 2,3,-dihydroxypropane-1-sulfonate (DHPS) and cysteate and N-acetyltaurine each containing a nitrogen atom as well (Tables 1 and S5). For all of these, the transporter genes have been verified experimentally in R. pomeroyi (Table 1). In the case of DHPS, spent medium drawdown analysis further indicated bacterial consumption based on significantly lower concentrations in the co-cultures. Drawdown analysis also indicated homoserine consumption (Fig. 3). Although the transporter for dimethylsulfoniopropionate (DMSP) has not yet been identified in the R. pomeroyi genome, enrichment of transcripts for functionally verified genes dmdA (DMSP demethylase) [47] and acuI (acrylyl-CoA reductase) [48] (Tables 1 and S5) suggested uptake of this organic sulfur metabolite.
Table 1.
Metabolites hypothesized to support bacterial heterotrophy in diatom co-cultures based on significant changes in bacterial gene expression (adjusted p < 0.05) or metabolite concentrations in spent media (adjusted p < 0.10) for three bacterial strains, and the methodological approach providing support.
| Substrate | R. pomeroyi | Stenotrophomonas | P. dokdonensis | References |
|---|---|---|---|---|
| Organic nitrogen | ||||
| 3′ AMP | M | |||
| Adenosine | G | M | ||
| Arginine | M | |||
| Chitobiose | M, G | |||
| Chitotriose | M, G | |||
| Choline | G | |||
| N-acetyl-d-galactosamine | Ga | [53] | ||
| N-acetyl-d-glucosamine | G | G | G | |
| Glutamine | G | |||
| Guanosine | M | G | M | |
| Homoserine | M | |||
| Inosine | M | |||
| Isoleucine | M, G | |||
| Leucine | M, G | |||
| Peptides | G | G | ||
| Proline | M | G | G | |
| Thymidine | M | M | ||
| Urea | G | |||
| Valine | M, G | |||
| Organic nitrogen and sulfur | ||||
| Cysteate | Ga | [98] | ||
| N-acetyltaurine | Ga | [99] | ||
| Organic sulfur | ||||
| DHPS | M, Ga | [100] | ||
| DMSP | Ga | [47, 48] | ||
| Carbon only | ||||
| Acetate | G | |||
| Alginate-like oligosaccharide | G | |||
| β-D-Glucans | G | |||
| Benzoate | G | |||
| Chrysolaminarin | G | |||
| Citrate | G | |||
| Mannonate | G | |||
| Mannose-like oligosaccharide | G | |||
| Phenylacetate | G | |||
| Galactoside | G | |||
| Others | ||||
| Cobalamin | G | G | ||
| Ua | M | |||
| Ub | M | |||
Metabolites Ua and Ub are unidentified compounds detected by 13C-HSQC NMR (Fig. S2).
M uptake suggested from metabolomics analysis, G uptake suggested from gene expression.
aGene function is experimentally verified.
For five enriched transporter systems, computationally inferred target substrates included urea, phenylacetate, mannonate, acetate, and a galactoside. Transcriptome analysis indicated uptake of six additional metabolites by transporters with general (e.g., sugar and dicarboxylate) or no substrate annotation (Fig. 2 and Table S5). Drawdown analysis revealed a lower concentration of one unidentified metabolite (designated Ua). In two cases there were higher concentrations of compounds in the co-culture spent media (phosphate-containing three carbon compound 3C-P and Ub; Fig. 3), suggesting either release of these unidentified metabolites by R. pomeroyi or enhanced export by T. pseudonana in the presence of the bacterium. Overall, the transcriptome and spent medium analyses of the T. pseudonana–R. pomeroyi co-culture indicated that at least 20 components of the diatom exometabolite pool were used by this bacterium, 13 of which were organic nitrogen molecules and 4 of which were organic sulfur molecules.
Stenotrophomonas sp. SKA14 metabolite utilization
Of the 23 organic compound transporters significantly enriched in the Stenotrophomonas sp. SKA14 transcriptome, six are TonB-dependent transporters (TBDTs) functioning in the outer membrane to bring molecules into the periplasm using energy from TonB proteins located in the inner membrane (Table S6); subsequent passage of substrates into the cytoplasm is typically carried out by permeases or other types of inner membrane transporters. Among the enriched TBDTs, three are annotated as cobalamin/B12 receptors and likely involved in vitamin acquisition. Other types of enriched transporters indicate uptake of peptides (major facilitator superfamily (MFS) and oligopeptide family transporters), amino acids (two permeases, a serine/alanine/glycine transporter), and benzoate (Fig. 2 and Table S6). However, there were no decreases in amino acids in the spent medium compared to the T. pseudonana axenic control. An actual increase in proline and phenylalanine concentrations (Fig. 3) suggested either release of these metabolites by Stenotrophomonas or their enhanced export by T. pseudonana when Stenotrophomonas was present. Transporters for purines (MFS transporter) and nucleosides (NupC) were upregulated in co-culture (Table 1).
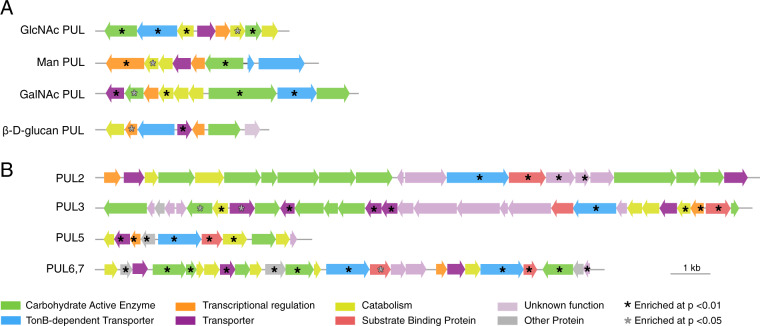
Carbohydrate utilization was important in the Stenotrophomonas co-cultures and accomplished largely via polysaccharide utilization loci (PULs), genomic regions that provide Gammaproteobacteria [7] and Flavobacteriales [49] with the capacity to hydrolyze polysaccharide structures while retaining the resulting monomers in the periplasm until passage inside the cell. Both TBDTs and carbohydrate degrading enzymes (CAZymes) are common in these regions. Among the four PULs containing enriched genes in the Stenotrophomonas co-culture transcriptome is a likely N-acetyl-d-glucosamine (GlcNAc) PUL (Fig. 4) containing an enriched glycoside hydrolase (GH) that acts on N-acetylglucosides (GH-20), a glucokinase that phosphorylates glucose, a sugar membrane permease, and several GlcNAc catabolic enzymes (such as nagA) [50, 51]. The two chitinases also present in this PUL were not enriched, a pattern suggesting that Stenotrophomonas sp. SKA14 was targeting GlcNAc or chitin oligosaccharides in the T. pseudonana exometabolite pool. The hypothesized mannose oligosaccharide PUL system includes two GHs annotated as a β-mannosidase (GH2) and α-1,2-mannosidase (GH92). The likely N-acetyl-d-galactose PUL (GalNAc PUL) has a gene content consistent with activity toward galactose-containing carbohydrates [52–54], and is homologous to an operon in Stenotrophomonas maltophilia K279a verified experimentally to catabolize N-acetyl-d-galactosamine [53]. Significantly enriched genes in the co-culture transcriptome (Fig. 4) included an MFS transporter that brings N-acetyl-d-galactosamine through the inner membrane (AgaP), a tagatose-biphosphate aldolase (AgaY) that carries out the final step in N-acetyl-d-galactosamine catabolism, and a GH containing a carbohydrate binding module with activity against substrates such as galactose oligosaccharides, galactomannans, and galactolipids (Table S6). Finally, a likely 1,3/1,4-β-d-glucan PUL contains an upregulated exo 1,3/1,4-β-d-glucan glucohydrolase (GH3) predicted to hydrolyze residues of β-d-glucans [55].
Fig. 4. Polysaccharide utilization loci (PULs) containing genes enriched in diatom co-cultures relative to glucose controls.
A Stenotrophomonas sp. SKA14. B Polaribacter dokdonensis MED152. Gene annotations and fold-change data are given in Tables S6 and S7.
There were also seven carbohydrate-active enzymes located outside PUL regions among the enriched Stenotrophomonas sp. genes (Table S6), two of which have computationally derived annotations for utilization of choline and chitin. Overall, transcriptomes and drawdown assays hypothesized at least 12 diatom metabolites serving as substrates for Stenotrophomonas in the co-cultures (Table 1), 7 of which contain nitrogen and 4 of which are carbohydrates.
Polaribacter dokdonensis MED152 metabolite utilization
In the co-culture transcriptomes of P. dokdonensis, components of 13 organic molecule transporters were significantly enriched compared to the glucose control (DeSeq2; p < 0.01) (Fig. 2 and Table S7). Six of these are TBDTs, most located in PULs with carbohydrate utilization functions but one annotated for cobalamin uptake. Of the seven other transporters, one is a general MFS transporter with no substrate information, and one is annotated as a putative amino acid transporter. Exometabolite analysis showed no evidence of amino acid drawdown in the P. dokdonensis co-culture, but did indicate decreases in concentrations of inosine, adenosine, thymidine, and guanosine (Fig. 3). Peaks putatively assigned to unidentified metabolite Ub were also drawn down in the P. dokdonensis co-culture relative to axenic T. pseudonana.
Carbohydrate utilization via PULs was also important in the P. dokdonensis co-cultures. PUL2 (designations from the CAZy database) [55] (Fig. 4) is predicted to degrade chrysolaminarin, a glucose storage glucan synthesized by diatoms [56, 57] that is ubiquitous in ocean organic matter pools [58]. The enriched substrate binding proteins (susD) is an ortholog of a Flavobacterales gene demonstrated to bind laminarin [59], a storage glucan similar to chrysolaminarin found in brown macroalgae. For PUL3 of P. dokdonensis (Fig. 4), we hypothesize a function in transport and catabolism of fucose-like or maltooligosaccharide-containing compounds based on inner membrane transporters (Table S7). PUL5 was the most enriched PUL in the P. dokdonensis transcriptome, and we predict it transports and catabolizes GlcNAc in the form of chitin monomers or oligomers. This PUL aligns modularly with a Bacteroidaceae PUL in a bacterium confirmed to grow on GlcNAc [60], and the enriched MFS transporter (Table S7) is an ortholog of a gene in the GlcNAc PUL of a Stenotrophomonas species. Genes encoding chitin hydrolysis were not found in the P. dokdonensis genome, suggesting this bacterium is only able to use monomers and oligomers of chitin. Bacterial drawdown analysis agreed with this annotation by indicating significantly lower concentrations of chitobiose and chitotriose in the co-culture spent medium (Fig. 3), but concentrations of GlcNAc were higher. PUL6 and PUL7 are co-located (Fig. 4) and we predict they target glycans containing guluronic acid and mannuronic acid, the two primary monomers of alginate. While alginate has not been reported as a component of T. pseudonana polysaccharides, guluronic acid is present in diatom cell walls [61] and the P. dokdonensis PUL6 is highly syntenic with those of previously described alginate-catabolizing Flavobacteriales [62–64]. The enriched components of PUL6 and PUL7 include a poly(β-d-mannuronate) lyase and two alginate lyases, also supporting a function for this genomic region in utilization of an alginate-like polysaccharide (Table S7). Nine enriched CAZymes that were not associated with PULs included a gene with catabolic activity against β-1,3 glucans, a gene with activity toward various β-1,3 and β-1,4 glucans, and an alginate lyase. A total of 14 possible substrates were identified for P. dokdonensis, 10 of which were organic nitrogen compounds and 5 of which were carbohydrates (Table 1).
Discussion
Metabolites passed between marine microbes play key roles in ocean ecosystems, influencing bacterial community assembly and diversity [65, 66], mediating competition, antagonism, and mutualisms [25], and serving as currencies of carbon flux. The last of these roles is a central function in global carbon cycling by which ~50% of marine photosynthate is transferred from phytoplankton to heterotrophic bacteria via the labile DOC pool [1, 67, 68]. There are two primary reasons why little is known about the metabolites linking marine phytoplankton and bacteria [69]. One is that the most biologically reactive components of marine DOC do not accumulate and therefore account for an extremely small fraction of the total reservoir [1]; and the other is that direct chemical analysis, especially in a seawater matrix, is challenging unless the identity of a compound is already known [70].
Our approach delineated clear differences in resource use patterns by bacteria individually inoculated into identical exometabolite pools formed during exponential through stationary growth phases of a diatom. Of the 36 molecules (Fig. 2) covering chemical classes ranging from amino acids to organic sulfur compounds, monosaccharides, oligosaccharides, nucleosides, and organic acids, 29 (80%) were targeted for uptake up by only one of the three representative bacterial strains. Rhodobacterales member R. pomeroyi was hypothesized to uniquely utilize four amino acids, the organic sulfur compounds DHPS, cysteate, DMSP, and N-acetyltaurine, the carboxylic acids acetate and phenylacetate, two carbohydrates, and urea, suggesting a substrate suite that is diverse, dominated by LMW compounds, and distinguished by a focus on organic sulfur metabolites. Gammaprotebacteria member Stenotrophomonas sp. SKA14 was hypothesized to uniquely utilize three carbohydrates plus benzoate, citrate, and choline. Flavobacteriales member P. dokdonensis uniquely utilized the chitin oligomers chitobiose and chitotriose, the carbohydrate storage compound chrysolaminarin, an alginate-like carbohydrate, 3′AMP, and the nucleoside inosine. The latter two bacteria had substrate use linked to polymeric and oligomeric carbohydrates that, particularly in the case of P. dokdonensis, is consistent with significant roles in aging blooms [11, 23]. Low overlap in the utilization of labile organic matter has been proposed to explain the predictable co-occurrence of Rhodobacterales, Gammaproteobacteria, and Flavobacteriales with microphytoplankton in the surface ocean [11, 23, 27, 71, 72]. Our analyses concur that substrate overlap is low among these groups when processing a natural pool of phytoplankton-derived molecules. Furthermore, they point to resource-based niche partitioning of the available resources, rather than competition for them, as the underlying ecological explanation.
Seven of the 36 metabolites, however, were targeted for uptake by more than one bacterial strain: proline, GlcNAc, peptides, cobalamin, and the nucleosides adenosine, guanosine, and thymidine. All of these are organic nitrogen compounds and one is also a vitamin, suggesting these may be resources for which there is competition among the marine bacteria growing at the expense of diatom exometabolites. Proline, guanosine, and GlcNAc were targeted for uptake by all three strains, with the availability of GlcNAc possibly linked to the chitin present in diatom cell walls [73]. Organic nitrogen compounds were also common in the non-shared metabolites, making up ~45% of metabolites.
Previous studies of endometabolite composition of diatom cells reported some of the same compounds identified here in the T. pseudonana exometabolome (Table 1). These include the amino acids glutamine, arginine, valine, isoleucine, leucine, and proline [74, 75], the organic sulfur compounds DMSP and DHPS [76, 77], choline [74, 78], galactose [79], and chrysolaminarin [77]. Previous studies have also shown chemotaxis by marine bacteria to several compounds identified in the T. pseudonana exometabolome. These include valine, proline, DMSP, and GlcNAc [80, 81]. Evidence that these compounds are present in diatom cells, function as attractants in chemotaxis, and are taken up differentially by heterotrophic bacteria suggests they play roles in the chemical ecology of bacterial community assembly. Niche dimensions unrelated to carbon and nutrient acquisition were not examined here, but may also influence ecological differentiation among co-existing bacterial species in the surface ocean; such factors include physical conditions [82], growth kinetics [83], antimicrobial defenses [84], and death processes through viral and protist grazing [85].
Some metabolites had different patterns in gene expression compared to drawdown. Examples are the enrichment of transcripts from the experimentally verified cysteate transporter in R. pomeroyi (Tables 1 and S5) but no evidence of cysteate release by axenic diatoms; and enrichment of transcripts from possible proline transporters in both Stenotrophomonas sp. and P. dokdonensis (Tables 1, S6, and S7) but no evidence of proline drawdown for either (Fig. 3). Differential exometabolite release by the diatom in axenic versus co-culture (i.e., different composition or rate of exometabolite release in the presence versus absence of bacteria) could explain these methodological mismatches. There is growing evidence that marine phytoplankton can detect the presence of bacteria, including a potential recognition cascade invoked by T. pseudonana when growing with R. pomeroyi [86]. Other scenarios that could lead to mismatches between these methodological approaches include lower bacterial drawdown rates than diatom release rates, and bacterial metabolite release. A potential example of the last scenario is the accumulation of GlcNAc in the P. dokdonensis co-culture but not the axenic diatom culture. In this case, Polaribacter hydrolysis of chitobiose and chitotriose (Fig. 3) may be outpacing its uptake of the hydrolysis product GlcNAc [87].
The interpretation of gene expression patterns used here assumed that upregulation of transport and catabolic genes was stimulated by the availability of a substrate. This is based on the unfavorable energetics of across-the-board synthesis of transporter systems that may not yield benefits [88], as well as previous observations of transporter expression changes when the substrates available to heterotrophic marine bacteria are manipulated [26, 89, 90]. Other regulatory strategies are possible, however, such as constitutive regulation, posttranscriptional regulation, or co-regulation, and these may not be detected in this study or could mislead interpretation. Finally, gene expression interpretations assume that computationally inferred annotations of transporter and catabolic genes in the bacterial genomes are generally correct, which is not always the case [91]. Across the three bacterial genomes, half the upregulated transporters have either general annotations or no annotations regarding their target substrates (Tables S5, S6, and S7), emphasizing the limitation of comparative genome analysis alone to address bacterial resource dimensions.
The role of resource competition in determining bacterial community assembly has been explored recently both in experimental systems and metabolic models [66, 92–94]. Yet current knowledge of microbial metabolites is hindered by barriers to capturing and identifying microbial products, and further exacerbated by the difficulty of microbial gene annotation. Thus it is challenging to go beyond generic products of central metabolism in addressing resource-based bacterial niche dimensions. Indeed, metabolites of noncore processes that are by and large missing from experiments and models may be the compounds most likely to support resource-based niche differentiation. For example, DHPS release by marine diatoms (Fig. 2) was mentioned only two times in the oceanographic literature [95, 96] before it emerged as a major resource for certain taxa of heterotrophic bacteria [26, 97]. Two types of information are critical for improving understanding of bacterial resource partitioning. The first is a chemical database that captures the diversity of microbial metabolites. The second is experimental annotation of bacterial transporter genes that hold important clues about resource-based niche dimensions in the surface ocean [69].
High turnover rates and low concentrations make identification of the labile organic matter released by phytoplankton problematic. Here we used biological vetting based on bacterial activity to identify the molecules most likely to shape heterotrophic bacterial communities reliant on recent photosynthate. Progress in closing the knowledge gap of marine metabolites will enable new insights into the transfer of carbon between major ocean reservoirs.
Supplementary information
Acknowledgements
The authors thank S. Roth, J. Nelson, J. Schreier, B. Nowinski, C. Smith, S. Sharma, M. Uchimiya, M. Olofsson, H. Panagos, H. Fu, and M. Kido Soule for advice and assistance, and J. Pinhassi for bacterial strains. This work was supported by grants from the Gordon and Betty Moore Foundation (5503) and the National Science Foundation (IOS-1656311) to MAM, ASE, and EBK, and by the Simons Foundation grant 542391 to MAM within the Principles of Microbial Ecosystems (PriME) Collaborative.
Author contributions
MAM, EBK, and ASE conceptualized the study. FXF-G, ASE, EBK, and MAM designed the study. BW, NRH, ASE, and EBK designed and developed chemical methodologies. FXF-G, BW, and NRH generated and analyzed data. FXF-G, BW, NRH, and MAM wrote the paper with input from all authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Frank Xavier Ferrer-González, Brittany Widner, Nicole R. Holderman
Supplementary information
The online version of this article (10.1038/s41396-020-00811-y) contains supplementary material, which is available to authorized users.
References
- 1.Hansell DA. Recalcitrant dissolved organic carbon fractions. Ann Rev Mar Sci. 2013;5:421–45. doi: 10.1146/annurev-marine-120710-100757. [DOI] [PubMed] [Google Scholar]
- 2.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–6. [Google Scholar]
- 3.Simon M, Rosenstock B. Different coupling of dissolved amino acid, protein, and carbohydrate turnover to heterotrophic picoplankton production in the Southern Ocean in austral summer and fall. Limnol Oceanogr. 2007;52:85–95. [Google Scholar]
- 4.Suttle CA, Chan AM, Fuhrman JA. Dissolved free amino acids in the Sargasso Sea: uptake and respiration rates, turnover times, and concentrations. Mar Ecol Prog Ser. 1991;70:189–99. [Google Scholar]
- 5.Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep Sea Res II Top Stud Oceanogr. 2002;49:3017–38. [Google Scholar]
- 6.Hertkorn N, Benner R, Frommberger M, Schmitt-Kopplin P, Witt M, Kaiser K, et al. Characterization of a major refractory component of marine dissolved organic matter. Geochim Cosmochim Acta. 2006;70:2990–3010. [Google Scholar]
- 7.Koch H, Duerwald A, Schweder T, Noriega-Ortega B, Vidal-Melgosa S, Hehemann J-H, et al. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 2019;13:92. doi: 10.1038/s41396-018-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson RL, Sunda WG. Utilization of amino acids by planktonic marine bacteria: importance of clean technique and low substrate additions 1, 2. Limnol Oceanogr. 1984;29:258–74. [Google Scholar]
- 9.Fuhrman JA, Ferguson RL. Nanomolar concentrations and rapid turnover of dissolved free amino acids in seawater: agreement between chemical and microbiological measurements. Mar Ecol Prog Ser. 1986;33:237–42. [Google Scholar]
- 10.Hodson R, Azam F, Carlucci A, Fuhrman J, Karl D, Holm-Hansen O. Microbial uptake of dissolved organic matter in McMurdo Sound, Antarctica. Mar Biol. 1981;61:89–94. [Google Scholar]
- 11.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–11. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 12.Thornton DC. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur J Phycol. 2014;49:20–46. [Google Scholar]
- 13.Fogg GE. The ecological significance of extracellular products of phytoplankton photosynthesis. Bot Mar. 1983;26:3–14. [Google Scholar]
- 14.Mühlenbruch M, Grossart HP, Eigemann F, Voss M. Mini‐review: phytoplankton‐derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ Microbiol. 2018;20:2671–85. doi: 10.1111/1462-2920.14302. [DOI] [PubMed] [Google Scholar]
- 15.Obernosterer I, Herndl GJ. Phytoplankton extracellular release and bacterial growth: dependence on the inorganic N:P ratio. Mar Ecol Progr Ser. 1995;116:247–57. [Google Scholar]
- 16.Flynn KJ, Clark DR, Xue Y. Modeling the release of dissolved organic matter by phytoplankton 1. J Phycol. 2008;44:1171–87. doi: 10.1111/j.1529-8817.2008.00562.x. [DOI] [PubMed] [Google Scholar]
- 17.Seymour JR, Simó R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science. 2010;329:342–5. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 18.Strom S, Wolfe G, Slajer A, Lambert S, Clough J. Chemical defense in the microplankton II: Inhibition of protist feeding by β‐dimethylsulfoniopropionate (DMSP) Limnol Oceanogr. 2003;48:230–7. [Google Scholar]
- 19.Wolfe GV, Steinke M, Kirst GO. Grazing-activated chemical defence in a unicellular marine alga. Nature. 1997;387:894. [Google Scholar]
- 20.Bidle KD. The molecular ecophysiology of programmed cell death in marine phytoplankton. Ann Rev Mar Sci. 2015;7:341–75. doi: 10.1146/annurev-marine-010213-135014. [DOI] [PubMed] [Google Scholar]
- 21.Caron DA, Hutchins DA. The effects of changing climate on microzooplankton grazing and community structure: drivers, predictions and knowledge gaps. J Plank Res. 2013;35:235–52. [Google Scholar]
- 22.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 23.Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–98. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 24.Sapp M, Schwaderer AS, Wiltshire KH, Hoppe H-G, Gerdts G, Wichels A. Species-specific bacterial communities in the phycosphere of microalgae? Micro Ecol. 2007;53:683–99. doi: 10.1007/s00248-006-9162-5. [DOI] [PubMed] [Google Scholar]
- 25.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–84. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landa M, Burns AS, Roth SJ, Moran MA. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 2017;11:2677. doi: 10.1038/ismej.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 28.Avcı B, Krüger K, Fuchs BM, Teeling H, Amann RI. Polysaccharide niche partitioning of distinct Polaribacter clades during North Sea spring algal blooms. ISME J. 2020;14:1369–83. [DOI] [PMC free article] [PubMed]
- 29.Kirchman DL. The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 30.Pedler BE, Aluwihare LI, Azam F. Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc Nat Acad Sci. 2014;111:7202–7. doi: 10.1073/pnas.1401887111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang K, Jiao N, Liu K, Zhang Y, Li S. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PloS ONE. 2012;7:e41204. [DOI] [PMC free article] [PubMed]
- 32.Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob Biogeochem Cycles. 1995;9:359–72. [Google Scholar]
- 33.Alavi M, Miller T, Erlandson K, Schneider R, Belas R. Bacterial community associated with Pfiesteria‐like dinoflagellate cultures. Environ Microbiol. 2001;3:380–96. doi: 10.1046/j.1462-2920.2001.00207.x. [DOI] [PubMed] [Google Scholar]
- 34.Amin S, Hmelo L, Van Tol H, Durham B, Carlson L, Heal K, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 35.Behringer G, Ochsenkühn MA, Fei C, Fanning J, Koester JA, Amin SA. Bacterial communities of diatoms display strong conservation across strains and time. Front Microbiol. 2018;9:659. doi: 10.3389/fmicb.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJ. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol Ecol. 2004;47:345–57. doi: 10.1016/S0168-6496(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 37.Hold GL, Smith EA, Rappë MS, Maas EW, Moore ER, Stroempl C, et al. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol Ecol. 2001;37:161–73. [Google Scholar]
- 38.Guillard R, Hargraves P. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–6. [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widner B, Kido Soule MCK, Ferrer-González FX, Moran MA, Kujawinski EB. Superior and novel detection of small, polar metabolites in saline samples using pre-extraction benzoyl chloride derivatization and Ultra-High Performance Liquid Chromatography Tandem Mass Spectrometry (UHPLC MS/MS). chemRXiv. 2020. 10.26434/chemrxiv.12915488.v1. [DOI] [PubMed]
- 42.Oehlke J, Brudel M, Blasig IE. Benzoylation of sugars, polyols and amino acids in biological fluids for high-performance liquid chromatographic analysis. J Chromatogr B Biomed Sci Appl. 1994;655:105–11. doi: 10.1016/0378-4347(94)00067-0. [DOI] [PubMed] [Google Scholar]
- 43.Wong J-MT, Malec PA, Mabrouk OS, Ro J, Dus M, Kennedy RT. Benzoyl chloride derivatization with liquid chromatography–mass spectrometry for targeted metabolomics of neurochemicals in biological samples. J Chromatogr A. 2016;1446:78–90. doi: 10.1016/j.chroma.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kido Soule MC, Longnecker K, Johnson WM, Kujawinski EB. Environmental metabolomics: analytical strategies. Mar Chem. 2015;177:374–87. [Google Scholar]
- 45.Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, MacCoss MJ. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom Rev. 2017;39:229–44. doi: 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henderson CM, Shulman NJ, MacLean B, MacCoss MJ, Hoofnagle AN. Skyline performs as well as vendor software in the quantitative analysis of serum 25-hydroxy vitamin D and vitamin D binding globulin. Clin Chem. 2018;64:408–10. doi: 10.1373/clinchem.2017.282293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–52. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 48.Todd JD, Curson AR, Sullivan MJ, Kirkwood M, Johnston AW. The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PloS ONE. 2012;7:e35947. [DOI] [PMC free article] [PubMed]
- 49.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199:e00860–16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boulanger A, Déjean G, Lautier M, Glories M, Zischek C, Arlat M, et al. Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J Bacteriol. 2010;192:1487–97. doi: 10.1128/JB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenbeis S, Lohmiller S, Valdebenito M, Leicht S, Braun V. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J Bacteriol. 2008;190:5230–8. doi: 10.1128/JB.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkkötter A, Shakeri-Garakani A, Lengeler JW. Two class II D-tagatose-bisphosphate aldolases from enteric bacteria. Arch Microbiol. 2002;177:410–9. doi: 10.1007/s00203-002-0406-6. [DOI] [PubMed] [Google Scholar]
- 53.Leyn SA, Gao F, Yang C, Rodionov DA. N-Acetylgalactosamine utilization pathway and regulon in Proteobacteria, genomic reconstruction and experimental characterization in Shewanella. J Biol Chem. 2012;287:28047–56. doi: 10.1074/jbc.M112.382333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodionov DA, Yang C, Li X, Rodionova IA, Wang Y, Obraztsova AY, et al. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genomics. 2010;11:494. doi: 10.1186/1471-2164-11-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beattie A, Hirst E, Percival E. Studies on the metabolism of the Chrysophyceae. Comparative structural investigations on leucosin (chrysolaminarin) separated from diatoms and laminarin from the brown algae. Biochem J. 1961;79:531. doi: 10.1042/bj0790531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Størseth TR, Hansen K, Reitan KI, Skjermo J. Structural characterization of β-D-(1→ 3)-glucans from different growth phases of the marine diatoms Chaetoceros mülleri and Thalassiosira weissflogii. Carbohydr Res. 2005;340:1159–64. doi: 10.1016/j.carres.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 58.Becker S, Tebben J, Coffinet S, Wiltshire K, Iversen MH, Harder T, et al. Laminarin is a major molecule in the marine carbon cycle. Proc Nat Acad Sci. 2020;117:6599–607. [DOI] [PMC free article] [PubMed]
- 59.Unfried F, Becker S, Robb CS, Hehemann J-H, Markert S, Heiden SE, et al. Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J. 2018;12:2894. doi: 10.1038/s41396-018-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gügi B, Le Costaouec T, Burel C, Lerouge P, Helbert W, Bardor M. Diatom-specific oligosaccharide and polysaccharide structures help to unravel biosynthetic capabilities in diatoms. Mar Drugs. 2015;13:5993–6018. doi: 10.3390/md13095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, et al. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae‐associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol. 2016;18:4610–27.. doi: 10.1111/1462-2920.13584. [DOI] [PubMed] [Google Scholar]
- 63.Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, et al. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 2014;8:1492. doi: 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, et al. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol. 2013;79:6813–22. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun. 2016;7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azam F, Fenchel T, Field J, Grey J, Meyer-Reil L, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–63. [Google Scholar]
- 68.Cole JJ, Findlay S, Pace ML. Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar Ecol Progr Ser. 1988;43:1–10. [Google Scholar]
- 69.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, et al. Deciphering ocean carbon in a changing world. Proc Nat Acad Sci. 2016;113:3143–51. doi: 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kujawinski EB. The impact of microbial metabolism on marine dissolved organic matter. Ann Rev Mar Sci. 2011;3:567–99. doi: 10.1146/annurev-marine-120308-081003. [DOI] [PubMed] [Google Scholar]
- 71.Larsbrink J, Zhu Y, Kharade SS, Kwiatkowski KJ, Eijsink VG, Koropatkin NM, et al. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol Biofuels. 2016;9:260. doi: 10.1186/s13068-016-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathieu S, Touvrey-Loiodice M, Poulet L, Drouillard S, Vincentelli R, Henrissat B, et al. Ancient acquisition of “alginate utilization loci” by human gut microbiota. Sci Rep. 2018;8:8075. doi: 10.1038/s41598-018-26104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 74.Azizan A, Ahamad Bustamam MS, Maulidiani M, Shaari K, Ismail IS, Nagao N, et al. Metabolite profiling of the microalgal diatom Chaetoceros calcitrans and correlation with antioxidant and nitric oxide inhibitory activities via 1H NMR-based metabolomics. Mar Drugs. 2018;16:154. doi: 10.3390/md16050154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hellebust JA. Excretion of some organic compounds by marine phytoplankton 1. Limnol Oceanogr. 1965;10:192–206. [Google Scholar]
- 76.Boroujerdi AF, Lee PA, DiTullio GR, Janech MG, Vied SB, Bearden DW. Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance. Anal Bioanal Chem. 2012;404:777–84. doi: 10.1007/s00216-012-6169-2. [DOI] [PubMed] [Google Scholar]
- 77.Iglesias MJ, Soengas R, Probert I, Guilloud E, Gourvil P, Mehiri M, et al. NMR characterization and evaluation of antibacterial and antiobiofilm activity of organic extracts from stationary phase batch cultures of five marine microalgae (Dunaliella sp., D. salina, Chaetoceros calcitrans, C. gracilis and Tisochrysis lutea) Phytochemistry. 2019;164:192–205. doi: 10.1016/j.phytochem.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 78.Jeong K-S, Jeong K-Y, Hong Y-S, Kim D-K, Oh H-J, Chang K-H. Application of nuclear magnetic resonance for analyzing metabolic characteristics of winter diatom blooms. J Plank Res. 2020;42:31–9. [Google Scholar]
- 79.Scholz B, Liebezeit G. Compatible solutes in three marine intertidal microphytobenthic Wadden Sea diatoms exposed to different salinities. Eur J Phycol. 2012;47:393–407. [Google Scholar]
- 80.Bell W, Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull. 1972;143:265–77. [Google Scholar]
- 81.Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol. 2004;70:4692–701. doi: 10.1128/AEM.70.8.4692-4701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–7. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 83.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kinkel LL, Schlatter DC, Xiao K, Baines AD. Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. ISME J. 2014;8:249–56. doi: 10.1038/ismej.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirchman DL. Phytoplankton death in the sea. Nature. 1999;398:293–4. [Google Scholar]
- 86.Durham BP, Dearth SP, Sharma S, Amin SA, Smith CB, Campagna SR, et al. Recognition cascade and metabolite transfer in a marine bacteria‐phytoplankton model system. Environ Microbiol. 2017;19:3500–13. doi: 10.1111/1462-2920.13834. [DOI] [PubMed] [Google Scholar]
- 87.Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, et al. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol. 2019;29:1528–35. doi: 10.1016/j.cub.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 88.Ferraris RP, Diamond JM. Specific regulation of intestinal nutrient transporters by their dietary substrates. Ann Rev Physiol. 1989;51:125–41. doi: 10.1146/annurev.ph.51.030189.001013. [DOI] [PubMed] [Google Scholar]
- 89.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Nat Acad Sci. 2010;107:16420–7. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–27. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salzberg SL. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019;20:92. doi: 10.1186/s13059-019-1715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, et al. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 93.Fu H, Uchimiya M, Gore J, Moran MA. Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc Nat Acad Sci. 2020;117:3656–62. [DOI] [PMC free article] [PubMed]
- 94.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Nat Acad Sci. 2015;112:6449–54. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benson A, Lee R. The sulphoglycolytic pathway in plants. Biochem J. 1972;128:29P. doi: 10.1042/bj1280029pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Busby WF. Sulfoporpanedial and cysteinolic acid in the diatom. Biochim Biophys Acta. 1966;121:160–1. doi: 10.1016/0304-4165(66)90360-6. [DOI] [PubMed] [Google Scholar]
- 97.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Nat Acad Sci. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Landa M, Burns AS, Durham BP, Esson K, Nowinski B, Sharma S, et al. Sulfur metabolites that facilitate oceanic phytoplankton–bacteria carbon flux. ISME J. 2019;13:2536–50. [DOI] [PMC free article] [PubMed]
- 99.Denger K, Lehmann S, Cook AM. Molecular genetics and biochemistry of N-acetyltaurine degradation by Cupriavidus necator H16. Microbiology. 2011;157:2983–91. doi: 10.1099/mic.0.048462-0. [DOI] [PubMed] [Google Scholar]
- 100.Mayer J, Huhn T, Habeck M, Denger K, Hollemeyer K, Cook AM. 2, 3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology. 2010;156:1556–64. doi: 10.1099/mic.0.037580-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.