Abstract
Osimertinib (AZD9291) has been widely used for the treatment of EGFR mutant non-small cell lung cancer. However, resistance to osimertinib is inevitable. In this study we elucidated the molecular mechanisms of resistance in osimertinib-resistant NCI-H1975/OSIR cells. We showed that NCI-H1975/OSIR cells underwent epithelial–mesenchymal transition (EMT), which conferred sensitivity to the GPX4 inhibitor 1S, 3R-RSL3 to induce ferroptotic cell death. The EMT occurrence resulted from osimertinib-induced upregulation of TGFβ2 that activated SMAD2. On the other hand, we revealed that NCI-H1975/OSIR cells were highly dependent on NF-κB pathway for survival, since treatment with the NF-κB pathway inhibitor BAY 11–7082 or genetic silence of p65 caused much greater cell death as compared with the parental NCI-H1975 cells. In NCI-H1975 cells, osimertinib activated NF-κB pathway, evidenced by the increased p65 nuclear translocation, which was abolished by knockdown of TGFβ2. In the cancer genome atlas lung adenocarcinoma data, TGFB2 transcript abundance significantly correlated with EMT-associated genes and NF-κB pathway. In addition, coexistence of EMT and activation of NF-κB pathway was observed in several NCI-H1975/OSIR clones. These findings shed new light on distinct roles of TGFβ2 in osimertinib-resistant cells and provide new strategies for treatment of this resistant status.
Keywords: EGFR mutant non-small cell lung cancer, osimertinib resistance, TGFβ2, epithelial-mesenchymal transition, NF-κB
Introduction
Osimertinib (AZD9291) is a third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) [1]. Currently, it is used for the treatment of non-small cell lung cancer (NSCLC) patients with EGFR-sensitizing mutations (exon 19 deletions or L858R mutations) and EGFR T790M mutation. Compared with first-generation EGFR TKIs (gefitinib and erlotinib), osimertinib significantly improves the overall survival of previously untreated and advanced EGFR mutant NSCLC patients [2]. With widespread clinical usage of osimertinib, the emergence of acquired resistance to osimertinib with highly heterogeneous mechanisms has been reported [3]. T790M is lost in more than half of osimertinib-resistant NSCLC patients. In T790M-maintaining patients, EGFR C797 and L792 mutations are the most common resistance mechanisms [4, 5]. Besides, activation of bypass signaling pathways independent of EGFR, e.g., MET amplification and FGFR amplification contributes to osimertinib resistance [4]. In addition, phenotypic transformation is observed in osimertinib-resistant patients, such as those with small cell lung cancer (SCLC) transformation [6]. However, the osimertinib resistance mechanisms in ~40% of patients remain unknown [7].
Epithelial–mesenchymal transition (EMT) refers to the transformation of cells from an epithelial phenotype to a mesenchymal phenotype. It has been generally confirmed that cancer cells undergoing EMT show increased motility and invasiveness, resulting in the dissemination of cancer cells to distant sites as well as the formation of metastases [8, 9]. In addition, tumor cells in the mesenchymal state become resistant to apoptosis and anticancer drugs [8]. EMT is induced by various cytokines, including epidermal growth factor, hepatocyte growth factor, and transforming growth factor beta (TGFβ). Classically, the induction of EMT is mediated by TGFβ/SMAD signaling [10]. All three isoforms of TGFβ (TGFβ1, TGFβ2, and TGFβ3) induce EMT by activating TGFβ receptor-SMAD2/3 signaling in cancer cells [11]. In addition to cytokines, nuclear factor-κB (NF-κB) regulates EMT-associated genes and is critical to the induction and maintenance of EMT [12, 13].
The NF-κB heterodimer functions as a transcriptional factor. The inhibitory protein IκB binds and sequesters the NF-κB dimer in the cytoplasm. Once IκB is phosphorylated and dissociated from NF-κB, the NF-κB dimer translocates into the nucleus to promote gene transcription [14]. NF-κB exists in almost all cell types and is involved in the regulation of inflammation and the immune response. In addition, NF-κB plays a very important role in cancer cell proliferation and cell survival as well as drug resistance [15, 16]. However, its role in osimertinib resistance is still elusive.
Previously, we established an osimertinib-resistant NCI-H1975/OSIR cell line [17]. The resistant cells were resistant to EGFR TKIs and exhibited EMT-like features with elongated spindle shapes, accompanied by enhanced cell migration and invasion abilities [17]. Based on the RNA-seq analysis, we demonstrated the cooccurrence of EMT and NF-κB activation in osimertinib-resistant cells, which was mediated by TGFβ2 and resulted in sensitivity to glutathione peroxidase 4 (GPX4) and NF-κB inhibitors. Collectively, this study highlights the distinct roles of TGFβ2 in osimertinib-resistant cells and provides new strategies to address this resistance.
Materials and methods
Reagents
Osimertinib, 1S, 3R-RSL3 (RSL3), ferrostatin-1 (Fer-1), and SB525334 were purchased from Selleck Chemicals (Houston, USA). Hoechst 33342, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), deferoxamine mesylate salt (DFO), nordihydroguaiaretic acid (NDGA), and dimethyl sulfoxide (DMSO) were derived from Sigma (St Louis, USA). Vitamin E (VE) and BAY 11–7082 were purchased from Beyotime Biotechnology (Shanghai, China).
Cell culture
Human NSCLC NCI-H1975 cells (EGFR L858R/T790M mutation) were derived from the Shanghai Cell Bank (Shanghai, China). Osimertinib-resistant NCI-H1975/OSIR cells were established in our previous study [17]. The monoclonal cells (OSIR/C1, OSIR/C2, OSIR/C3, and OSIR/C4) were generated by limiting dilution to ensure that one well contained no cells or one cell in a 96-well plate. All the cells were cultured in RPMI-1640 containing 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 units/mL penicillin and were grown at 37 °C with 5% CO2.
RNA extraction and RNA-seq
Total RNA was extracted from NCI-H1975 and NCI-H1975/OSIR cells in triplicate by using TRIzol reagent (Life Technologies, Shanghai, China) following the manufacturer’s instructions. RNA-seq was performed by Omigen (Hangzhou, China). Briefly, the reads were first mapped to the UCSC Genome Browser (GRCh37/hg19) transcript set using STAR version 2.4.1d, and the gene expression level was quantified by using Partek E/M annotation model. Gene expression levels were normalized to the total counts (available at 10.6084/m9.figshare.11653641.v1). Differentially expressed genes were identified using the differential gene expression algorithm in Partek Genomics Suite. Genes showing altered expression with FDR < 0.05 and more than twofold changes were considered differentially expressed. The heatmap of differentially expressed genes was generated by MeV version 4.9.0. Gene Set Enrichment Analysis (GSEA) version 3.0 was employed to enrich the canonical pathway of interest.
Quantitative real-time PCR (qRT-PCR)
QRT-PCR assays were performed as described previously [18]. Primers for PCR were synthesized by Invitrogen Life Technologies (Shanghai, China). The primers utilized to amplify the genes of interest were listed in Table 1. GAPDH or ACTB was used as the internal control, and the relative gene expression was calculated by 2−ΔΔCT. All experiments were performed at least in triplicate.
Table 1.
Primers and siRNAs sequences.
| Genes | Analysis | Sequences |
|---|---|---|
| CDH1 | qRT-PCR |
Forward: 5′-GCCTCCTGAAAAGAGAGTGGAAG-3′ Reverse: 5′-TGGCAGTGTCTCTCCAAATCCG-3′ |
| VIM | qRT-PCR |
Forward: 5′-CCCTCACCTGTGAAGTGGAT-3′ Reverse: 5′-GCTTCAACGGCAAAGTTCTC-3′ |
| ZEB1 | qRT-PCR |
Forward: 5′-GATGATGAATGCGAGTCAGATGC-3′ Reverse: 5′-ACAGCAGTGTCTTGTTGTTGTAG-3′ |
| ZEB2 | qRT-PCR |
Forward: 5′-CAAGAGGCGCAAACAAGC-3′ Reverse: 5′-GGTTGGCAATACCGTCAT-3′ |
| TGFB1 | qRT-PCR |
Forward: 5′-GAGCCCTGGACACCAACTATT-3′ Reverse: 5′-AGGTCCTTGCGGAAGTCAAT-3′ |
| TGFB2 | qRT-PCR |
Forward: 5′-CCCCACATCTCCTGCTAATGT-3′ Reverse: 5′-AGGCAGCAATTATCCTGCAC-3′ |
| TGFB3 | qRT-PCR |
Forward: 5′-ACTGTCCATGTCACACCTTTCA-3′ Reverse: 5′-GCCATGGTCATCCTCATTGT-3′ |
| GAPDH | qRT-PCR |
Forward: 5′-GCGACACCCACTCCTCCACCTTT-3′ Reverse: 5′-TGCTGTAGCCAAATTCGTTGTCATA-3′ |
| ACTB | qRT-PCR |
Forward: 5′-AGCGAGCATCCCCCAAAGTT-3′ Reverse: 5′-GGGCACGAAGGCTCATCATT-3′ |
| TGFB2 | siRNA |
Sense: 5′-GCGGCCUAUUGCUUUAGAATT-3′ Antisense: 5′-UUCUAAAGCAAUAGGCCGCTT-3′ |
| RELA (p65)-#1 | siRNA |
Sense: 5′-GAUUGAGGAGAAACGUAAATT-3′ Antisense: 5′-UUUACGUUUCUCCUCAAUCTT-3′ |
| P65-#2 | siRNA |
Sense: 5′-GAUCAAUGGCUACACAGGATT-3′ Antisense: 5′-UCCUGUGUAUCCAUUGAUCTT-3′ |
| Negative control (NC) | siRNA |
Sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ Antisense: 5′-ACGUGACACGUUCGGAGAATT-3′ |
Western blot assay
Western blot assays were conducted as previously described [18]. The primary antibodies against the proteins of interest were as follows: p-EGFR (Y1068) (Cell Signaling Technology (CST), Beverly, USA, #3777), EGFR (CST, #2232), CDH1 (CST, #3195), vimentin (VIM) (Becton, Dickinson and Company (BD), Franklin Lakes, USA, 550513), ZEB1 (CST, #3396), ZEB2 (Santa Cruz Biotechnology (SCBT), Dallas, USA, sc-271984), GAPDH (CST, #5174), p-SMAD2 (S465/467) (CST, #3108), SMAD2 (CST, #5339), p-IKKα/β (S176/180) (CST, #2697), p-IκBα (S32) (CST, #2859), p65 (CST, #8242), TGFβ2 (Abcam, Cambridge, UK, ab36495), and ACTB (CST, #3700).
Immunofluorescence
The immunofluorescence assay was performed following the manufacturer’s instructions. Briefly, the cells were washed with PBS three times, fixed with 4% paraformaldehyde for 20 min, and permeated with 0.5% Triton X-100 in PBS for 20 min at room temperature. Then, the cells were blocked with 0.5% BSA containing 0.2% Triton X-100 in PBS for 1 h, followed by incubation with primary antibodies against p65 (CST, #8242) overnight at 4 °C. After washing with PBS three times, the cells were incubated with anti-rabbit IgG (H + L), F(ab’)2 fragment (Alexa Fluor® 488 Conjugate) (CST, #4412) at room temperature for 45 min in the dark. After that, the cells were stained with Hoechst 33342 for 10 min and photographed by using a Leica TCS SP8 microscope (Solms, Germany).
MTT assay
The MTT assay was conducted as described in our previous study [18]. Briefly, the supernatant of cultured cells was discarded, and 100 μL of MTT solution within medium (1 mg/mL) was added into each well and incubated at 37 °C for 4 h. After that, the supernatant was discarded again, followed by the addition of 100 μL of DMSO. Then, the plate was shaken for 10 min in the dark to solubilize the formazan. The absorbance at 570 nm was measured by using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, USA).
Propidium iodide (PI) staining assay
The NCI-H1975/OSIR cells were seeded on 12-well plates at 80,000 cells per well overnight. Then, the cells were treated with RSL3 in combination with ferroptosis-associated inhibitors Fer-1, DFO, NDGA, and VE for 12 h. After rinsing with PBS, the cells were incubated with 20 μg/mL PI for 10 min at room temperature and then photographed with an IX73 inverted microscope system (Olympus, Tokyo, Japan).
Lactate dehydrogenase (LDH) release assay
The NCI-H1975/OSIR cells were seeded on 96-well plates at 5000 cells per well for 24 h. Then, the cells were treated with RSL3 in combination with Fer-1, DFO, NDGA, and VE for another 12 h. LDH release assay was conducted according to the manufacturer’s instructions and performed as described previously [19].
Lipid peroxidation
NCI-H1975/OSIR cells were seeded in six-well plates at 200,000 cells per well for 24 h. The cells were then incubated with RSL3 in combination with Fer-1, DFO, NDGA, and VE for another 12 h. After washing with PBS, the cells were stained with 5 μM BODIPY™ 581/591 C11 (Thermo Fisher Scientific, Waltham, USA) for 20 min at 37 °C. Then, the cells were collected by centrifugation at 800 rpm for 5 min. The lipid peroxidation was analyzed by using flow cytometry (Becton Dickinson FACS Canto, Franklin Lakes, USA), and data analysis was conducted by using FlowJo VX (TreeStar, Ashland, USA).
SiRNA transfection
Specific siRNAs targeting genes of interest were purchased from Shanghai GenePharma (Shanghai, China). Cells were transfected with siRNAs by using Lipofectamine® 2000 Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocols. The sequences used to knock down the indicated genes were listed in Table 1.
Detection of TGFβ2 by enzyme-linked immunosorbent assay (ELISA)
NCI-H1975 cells were seeded on 60-mm dishes at 450,000 cells and incubated overnight. Then, the cells were treated with osimertinib (125 nM) for 48 h. After that, the supernatants were harvested to detect TGFβ2 secretion using the TGF beta-2 Human ELISA Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Gene expression profiling
Lung adenocarcinoma (The Cancer Genome Atlas (TCGA), Provisional) data were downloaded from the cBioPortal website (https://www.cbioportal.org/) [20, 21]. TGFB2 mRNA expression Z-scores (RNA-Seq V2 RSEM) below 0 were considered to have low expression, and those with Z-scores above 0 were considered to have high expression. The significance of p65 (S536) between the high and low TGFB2 levels was analyzed by using Student’s unpaired t test. The analysis of the correlation of TGFB2 and EMT marker mRNA expression was assessed by calculating Pearson correlation coefficients.
Statistical analysis
The significance between two groups was analyzed by using Student’s unpaired t test. The correlation analysis was performed by using Pearson correlation coefficients. All data analysis was conducted by using GraphPad Prism software 6 (GraphPad Software, Inc., San Diego, USA). All experiments were performed at least in triplicate, and each value represents the mean ± SD. A P value < 0.05 represents statistical significance.
Results
NCI-H1975/OSIR cells undergo EMT
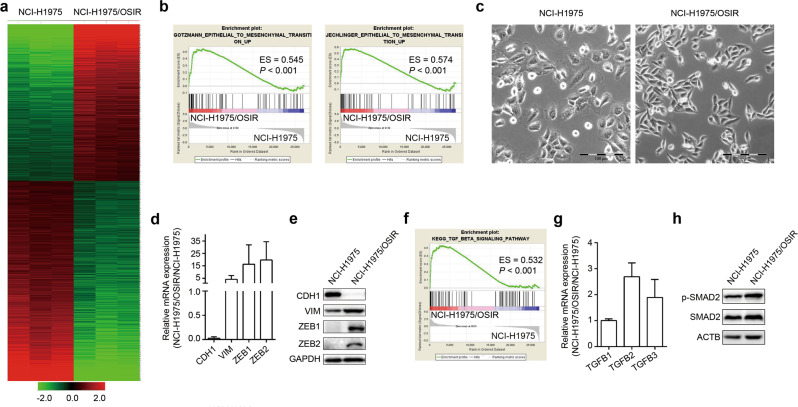
In 2016, we established osimertinib-resistant NCI-H1975/OSIR cells and observed that the cells exhibited EMT-like features with elongated spindle shapes [17]. To further explore the osimertinib resistance mechanism in NCI-H1975/OSIR cells, we performed RNA-seq to compare the differentially expressed genes between NCI-H1975 and resistant cells. As shown in Fig. 1a, 715 upregulated and 911 downregulated genes (fold change > 2, FDR < 0.05) were obtained. GSEA and morphology observations implied that the resistant cells underwent EMT (Fig. 1b, c). As expected, compared with the parental NCI-H1975 cells, the resistant cells showed downregulation of the epithelial marker CDH1 and upregulation of the mesenchymal marker VIM as well as EMT-associated transcriptional factors ZEB1 and ZEB2 both at the mRNA and protein levels (Fig. 1d, e), indicating that EMT does occur in NCI-H1975/OSIR cells.
Fig. 1. NCI-H1975/OSIR cells undergo EMT.
a Heatmap of differentially expressed genes between NCI-H1975 and NCI-H1975/OSIR cells (fold change > 2, FDR < 0.05). b GSEA of EMT signatures based on differentially expressed genes between NCI-H1975 and NCI-H1975/OSIR cells. ES represents enrichment score. c The morphology of NCI-H1975 and NCI-H1975/OSIR cells. Scale bar: 100 μm. d The mRNA levels of EMT-related markers in NCI-H1975 and NCI-H1975/OSIR cells were detected by qRT-PCR. e The protein levels of EMT-related markers were determined by Western blotting of NCI-H1975 and NCI-H1975/OSIR cells. f Enrichment of the TGFβ signaling pathway by GSEA according to the RNA-seq data. g The mRNA levels of TGFB1, TGFB2, and TGFB3 were analyzed by qRT-PCR. h The protein levels of p-SMAD2 and SMAD2 were evaluated by Western blotting of NCI-H1975 and NCI-H1975/OSIR cells.
On the basis of GSEA analysis, the TGFβ signaling pathway was enriched in osimertinib-resistant cells, suggesting that TGFβ mediates EMT in our model (Fig. 1f). There are three isoforms of TGFβ, i.e., TGFβ1, TGFβ2, and TGFβ3 [11]. To our surprise, the mRNA levels of TGFB2 and TGFB3, but not TGFB1, which is the classical inducer of EMT for in vitro studies [22, 23], were increased in NCI-H1975/OSIR cells compared with the parental cells (Fig. 1g). In addition, osimertinib-resistant cells showed an increase in phosphorylated SMAD2 (Fig. 1h), implying that TGFβ2 or TGFβ3 activates SMAD2 to induce EMT.
Osimertinib-induced activation of SMAD2 is mediated by TGFβ2
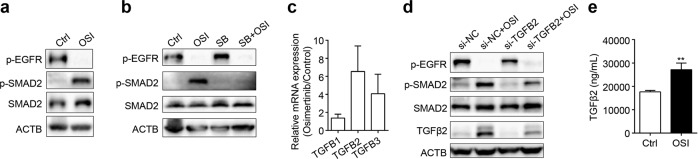
Intriguingly, treatment with osimertinib for 48 h increased phosphorylated SMAD2 in NCI-H1975 cells (Fig. 2a). The potent TGFβRI inhibitor SB525334 abolished osimertinib-induced SMAD2 activation (Fig. 2b), demonstrating that osimertinib-induced activation of SMAD2 may be triggered by TGFβ. Furthermore, osimertinib obviously upregulated the mRNA levels of TGFB2 and TGFB3 in NCI-H1975 cells (Fig. 2c). We supposed that osimertinib-induced activation of SMAD2 is possibly due to an increase in TGFβ2 or TGFβ3. For this purpose, we used specific siRNAs to silence TGFB2 or TGFB3. Knockdown of TGFB2 largely abolished osimertinib-induced SMAD2 activation (Fig. 2d), while silencing TGFB3 further increased the phosphorylation of SMAD2 (data not shown). Furthermore, osimertinib increased the secretion of TGFβ2 into the cell culture medium (Fig. 2e). Thus, osimertinib-induced activation of SMAD2 is mediated, at least in part, by TGFβ2 and gradually contributes to EMT.
Fig. 2. Osimertinib-induced activation of SMAD2 is mediated by TGFβ2 in NCI-H1975 cells.
a Western blot assay was utilized to detect the indicated proteins after treatment with osimertinib (OSI) for 48 h in NCI-H1975 cells. Control Ctrl. b The indicated protein levels were determined by Western blotting after cotreatment with the TGFβRI inhibitor SB525334 (SB, 1 μM) and OSI (125 nM) for 48 h. c NCI-H1975 cells were treated with OSI for 48 h, and then the mRNA levels of TGFB1, TGFB2, and TGFB3 were measured by qRT-PCR. d The siRNAs were used to knockdown TGFB2 for 24 h, followed by treatment with OSI for another 48 h. Then, the indicated proteins were detected by Western blot. e The secretion of TGFβ2 was detected by ELISA after treatment with OSI for 48 h in NCI-H1975 cells. **P < 0.01.
NCI-H1975/OSIR cells are sensitive to the GPX4 inhibitor RSL3, a ferroptosis inducer
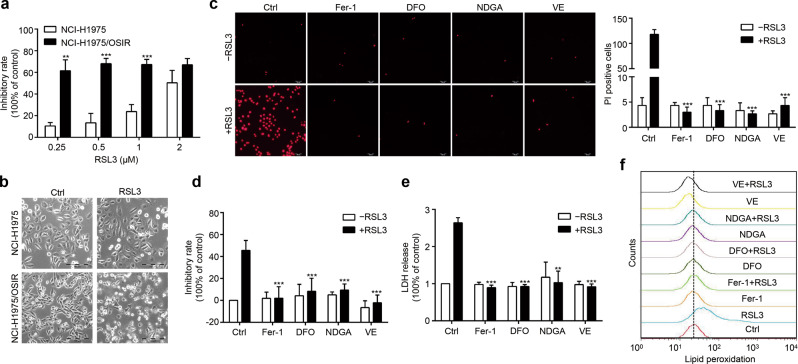
Therapy-resistant states of cancer cells, including high-mesenchymal cell state like EMT, are dependent on the lipid peroxidase GPX4 to protect against ferroptotic cell death (ferroptosis), which indicates that these cells are sensitive to GPX4 inhibitor-induced ferroptosis [24]. As mentioned above, NCI-H1975/OSIR cells undergo EMT, which may result in sensitivity to GPX4 inhibitors. Here, NCI-H1975 and NCI-H1975/OSIR cells were treated with the GPX4 inhibitor RSL3, and we found that NCI-H1975/OSIR cells were more sensitive to RSL3 (Fig. 3a). At a concentration of 0.5 μM, RSL3 caused substantial cell death in the resistant cells but not in the parental NCI-H1975 cells (Fig. 3b); these results were further confirmed by detection of PI-positive cells and LDH release (features of ferroptosis) in NCI-H1975/OSIR cells (Fig. 3c–e). Ferroptosis is an iron-dependent nonapoptotic cell death caused by lipid peroxidation, which is enhanced by lipoxygenase-catalyzed lipid hydroperoxide generation [25]. As expected, all these effects caused by RSL3 were totally abolished by cotreatment with ferroptosis-related inhibitors, including lipophilic antioxidants Fer-1, iron chelator DFO, and lipoxygenase inhibitors NDGA and VE (Fig. 3c–e), indicating that RSL3 triggers canonical ferroptosis in resistant cells. Since ferroptosis is mainly due to the production of lipid peroxidation, we further determined whether RSL3 caused this effect in our model. As shown in Fig. 3f, RSL3 induced lipid peroxidation by BODIPY staining in the resistant cells, which was totally rescued by cotreatment with ferroptosis-related inhibitors but not in NCI-H1975 cells (data not shown). Collectively, these results indicate that GPX4 inhibitor-induced ferroptosis is a vulnerability for NCI-H1975/OSIR cells.
Fig. 3. GPX4 inhibitor RSL3 induces ferroptosis in NCI-H1975/OSIR cells.
a NCI-H1975 and NCI-H1975/OSIR cells were treated with various concentrations of RSL3 for 24 h, and then the sensitivities were determined by MTT assay. b Morphological changes in NCI-H1975 and NCI-H1975/OSIR cells after treatment with RSL3 (0.5 μM) for 12 h. Scale bar: 100 μm. c After cotreatment with RSL3 and ferroptosis-related inhibitors, including lipophilic antioxidants ferrostatin-1 (Fer-1, 2 μM), iron chelator deferoxamine mesylate salt (DFO, 100 μM), lipoxygenase inhibitors nordihydroguaiaretic acid (NDGA, 5 μM) and vitamin E (VE, 100 μM), for 12 h, NCI-H1975/OSIR cells were stained with PI for 10 min and photographed with a confocal laser scanning microscope. The number of PI-stained cells was counted in each group. Scale bar: 500 μm. d The inhibitory rates were evaluated by MTT assay after cotreatment with RSL3 and ferroptosis-related inhibitors for 12 h in NCI-H1975/OSIR cells. e After coincubation with RSL3 and ferroptosis-related inhibitors for 12 h, cytotoxicity was analyzed by LDH release assay. f NCI-H1975/OSIR cells were cotreated with RSL3 and ferroptosis-related inhibitors for 3 h, and then lipid peroxidation was determined by staining with BODIPY (5 μM) for 20 min. **P < 0.01, ***P < 0.001.
Resistant cells are dependent on the NF-κB pathway
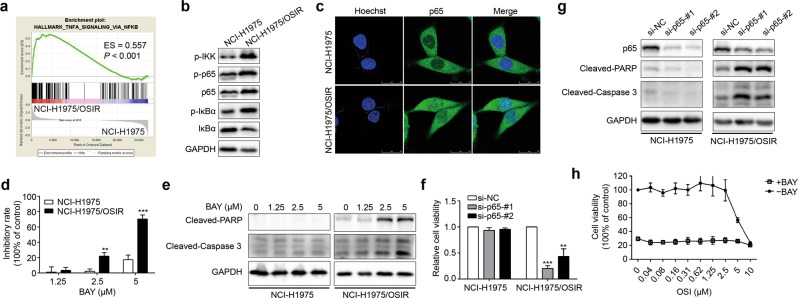
According to the GSEA of the RNA-seq data, the NF-κB pathway was enriched in the resistant cells (Fig. 4a). Compared with NCI-H1975 cells, the resistant cells showed an increase in phosphorylated IKK and IκBα as well as p65 (Fig. 4b). Moreover, massive nuclear localization of p65 was observed by immunofluorescence assay in the resistant cells (Fig. 4c). Accordingly, these results illustrate that the NF-κB pathway is activated in osimertinib-resistant cells.
Fig. 4. The resistant cells are addictive to the NF-κB pathway.
a Enrichment of the NF-κB pathway by GSEA according to RNA-seq data. b Western blot assay was used to evaluate the protein levels of the NF-κB pathway. c The expression and localization of p65 were analyzed by immunofluorescence in NCI-H1975 and NCI-H1975/OSIR cells. Scale bar: 25 μm. d NCI-H1975 and NCI-H1975/OSIR cells were treated with various concentrations of BAY 11–7082 (BAY, 1.25, 2.5, 5 μM) for 48 h, and then the sensitivities were evaluated by MTT assay. e The protein levels of cleaved PARP and cleaved caspase-3 were determined by Western blot after treatment with BAY (1.25, 2.5, 5 μM) for 24 h in NCI-H1975 and NCI-H1975/OSIR cells. f After silencing p65 with two specific siRNAs for 72 h, the cell viabilities of NCI-H1975 and NCI-H1975/OSIR cells were detected by MTT. g After silencing p65 with two specific siRNAs for 72 h in NCI-H1975 and NCI-H1975/OSIR cells, the protein levels of cleaved PARP and cleaved caspase-3 were determined by Western blotting. h NCI-H1975/OSIR cells were cotreated with OSI and BAY (5 μM) for 48 h, and cell viability was measured by MTT assay. **P < 0.01, ***P < 0.001.
We further explored the function of the NF-κB pathway in NCI-H1975/OSIR cells. The osimertinib-resistant cells showed more sensitivity to the NF-κB pathway inhibitor BAY 11–7082 than NCI-H1975 cells (Fig. 4d). BAY 11–7082 treatment triggered apoptosis as indicated by increased cleavage of caspase-3 and its substrate PARP in the resistant cells, whereas it caused slight growth inhibition without inducing apoptosis in NCI-H1975 cells (Fig. 4e). Furthermore, knockdown of p65 by two specific siRNAs in the resistant cells caused almost total cell death accompanied by an increase in caspase-3 and PARP cleavage, which was not obviously observed in NCI-H1975 cells (Fig. 4f, g). However, BAY 11–7082 did not enhance the sensitivity of NCI-H1975/OSIR cells to osimertinib, likely because the cells were too sensitive to BAY 11–7082 (Fig. 4h). Since EGFR expression was obviously decreased in the resistant cells, as we reported previously [17], NCI-H1975/OSIR cells might be dependent upon the NF-κB pathway for their survival, while NCI-H1975 cells are dependent upon EGFR.
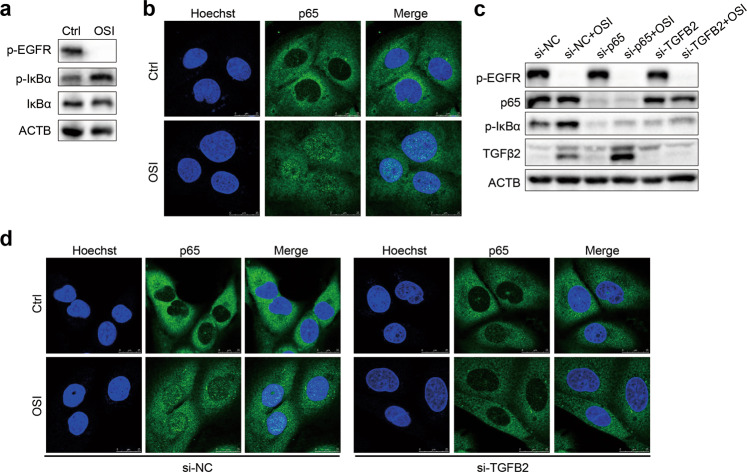
Osimertinib induces activation of the NF-κB pathway via TGFβ2
Since the resistant cells were dependent on the NF-κB pathway, we further explored whether osimertinib affected the NF-κB pathway in the parental cells. As expected, osimertinib obviously increased the phosphorylation of IκBα and the nuclear translocation of p65 (Fig. 5a, b), indicating that osimertinib triggers the activation of the NF-κB pathway in NCI-H1975 cells. TGFβ2 constitutively activates the NF-κB pathway for the survival of some tumor cells [26]. In addition to upregulation of TGFB2 at the mRNA level (Fig. 1f), we also found that osimertinib increased the secretion of TGFβ2 in cell culture medium (Fig. 2e). Thus, the effect of TGFβ2 on the activation of the NF-κB pathway caused by osimertinib was further investigated. To this end, TGFB2 was silenced by using a specific siRNA before osimertinib treatment. Knockdown of TGFB2 abrogated osimertinib-induced phosphorylation of IκBα in NCI-H1975 cells (Fig. 5c). Furthermore, genetic silencing of TGFB2 obviously reduced osimertinib-triggered nuclear accumulation of p65 (Fig. 5d), demonstrating that osimertinib-induced TGFβ2 may activate the NF-κB pathway.
Fig. 5. Osimertinib induces activation of the NF-κB pathway by TGFβ2.
a The protein levels of p-EGFR, p-IκBα, IκBα, and ACTB were determined by Western blotting after treatment with OSI for 48 h in NCI-H1975 cells. b After treatment with OSI for 48 h in NCI-H1975 cells, the expression and localization of p65 were analyzed by immunofluorescence. Scale bar: 25 μm. c The indicated proteins were evaluated by Western blotting after knockdown of p65 and TGFB2 for 24 h, followed by treatment with OSI for another 48 h. d Immunofluorescence assay was performed to analyze the expression and localization of p65 in NCI-H1975 cells with silenced TGFB2 for 24 h, followed by treatment with OSI for another 48 h. Scale bar: 25 μm.
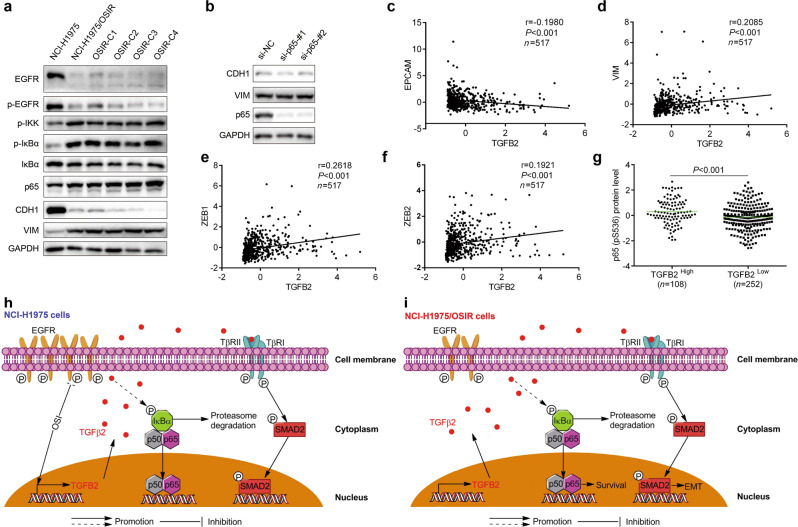
Coexistence of activation of the NF-κB pathway and EMT in NCI-H1975/OSIR clones
To determine whether cancer clonal heterogeneity existed in our osimertinib-resistant model, we selected several monoclones from NCI-H1975/OSIR cells. Consistent with the decrease in EGFR in the resistant cell population, as we previously reported [17], the EGFR protein level was obviously reduced in all tested monoclones. Importantly, the clones showed the coexistence of activation of the NF-κB pathway and EMT (Fig. 6a), suggesting that they are the main resistance mechanisms of NCI-H1975/OSIR cells. Accordingly, low heterogeneity of the resistance mechanism existed in NCI-H1975/OSIR cells. It has been shown that NF-κB mediates the development of EMT [12, 13]. We wondered whether the relationship existed in our model. To this end, we silenced p65 to determine the changes in EMT-related markers in the resistant cells. However, knockdown of p65 failed to change the protein levels of EMT markers (Fig. 6b), implying that NF-κB may not regulate EMT in resistant cells.
Fig. 6. Cooccurrence of EMT and NF-κB pathway activation in resistant cells.
a The indicated proteins were detected by Western blot assay in NCI-H1975 cells, NCI-H1975/OSIR cells and several NCI-H1975/OSIR monoclones. b The indicated proteins were detected by Western blot assay after knockdown of p65. The correlations between TGFB2 and EPCAM (c), VIM (d), ZEB1 (e), and ZEB2 (f) were analyzed by calculating the Pearson correlation coefficients. g The difference in p65 (pS536) protein levels between the TGFB2 high group and the TGFB2 low group was analyzed by using Student’s unpaired t test. h Schematic representation of the proposed mechanisms of OSI resistance in NCI-H1975 cells. OSI-induced TGFβ2-mediated activation of NF-κB and SMAD2. i Schematic representation of the proposed mechanisms of OSI resistance in NCI-H1975/OSIR cells. OSI-resistant cells exhibited NF-κB pathway activation concurrent with EMT.
To determine the relationship between TGFB2, EMT markers, and the NF-κB pathway in human samples, we interrogated lung adenocarcinoma-TCGA data through the cBioPortal website (https://www.cbioportal.org/) [20, 21]. Here, we found that the mRNA levels of TGFB2 and epithelial marker EPCAM were negatively correlated (Fig. 6c). TGFB2 mRNA level was also positively correlated with the expression levels of VIM, ZEB1, and ZEB2 (Fig. 6d–f), indicating the role of TGFB2 in EMT development. Samples with high TGFB2 expression showed significantly high protein levels of p65 (pS536), which is positively correlated with NF-κB pathway activation; the opposite trends were also observed in samples with low TGFB2 expression (Fig. 6g), demonstrating the effect of TGFB2 on the activation of the NF-κB pathway.
Discussion
We demonstrated for the first time the simultaneous presence of NF-κB pathway activation and EMT in osimertinib-resistant cells. On the one hand, osimertinib-induced upregulation of TGFβ2 may mediate EMT in NCI-H1975/OSIR cells. On the other hand, the resistant cells were addictive to the NF-κB pathway, independent of EGFR, caused gradually by osimertinib-induced TGFβ2 expression (Fig. 6h, i), pointing out important roles of TGFβ2, but not other isoforms of TGFβ, in osimertinib-resistant NSCLC cells.
Currently, osimertinib has been approved as a first-line treatment for EGFR mutant NSCLC. However, resistance to osimertinib is inevitable in the clinic [3]. Resistance mechanisms have been widely explored, including EGFR-dependent and -independent resistance [3, 4]. The development of EGFR-dependent resistance is mainly due to EGFR C797S mutation or loss of its T790M mutation [4, 5]. Based on Sanger DNA sequencing of the EGFR kinase domain, we found that NCI-H1975/OSIR cells still had the EGFR T790M mutation without the C797S mutation or other kinase domain mutations (data not shown). Our previous study showed decreased EGFR expression in NCI-H1975/OSIR cells relative to NCI-H1975 cells [17], which was further demonstrated by two other studies [27, 28]. Niederst et al. reported that EGFR TKI-resistant NSCLC patients transformed to SCLC accompanied by decreased EGFR expression, which resulted in insensitivity to EGFR TKIs [29, 30]. Clinical data showed that SCLC transformation was reported in EGFR mutant patients after progression on osimertinib [6], indicating that a decrease in EGFR as a resistance mechanism may exist in osimertinib-resistant patients. Recently, AURKA and PKCδ have been identified as key factors that mediate osimertinib resistance with no additional EGFR mutation, and inhibition of AURKA and PKCδ resensitized resistant cells to osimertinib [31, 32]. However, it seems that they do not contribute to osimertinib resistance in NCI-H1975/OSIR cells (data not shown), demonstrating the diversity of osimertinib resistance mechanisms.
According to the RNA-seq analysis, NCI-H1975/OSIR cells underwent EMT with activation of SMAD2 (Fig. 1b–h). EMT occurred in EGFR TKI-resistant NSCLC patients [33] and was reported as a resistance mechanism of EGFR TKIs [34–36], implying that it confers osimertinib resistance in our model. In NCI-H1975 cells, osimertinib-induced activation of SAMD2, which was blocked by TGFβRI inhibition (Fig. 2b), indicating that osimertinib triggers activation of TGFβ/SMAD2 signaling, which may further cause osimertinib resistance. Previous studies showed that TGFβ was necessary for EGFR TKI resistance and the development of EMT [37–39]. In our study, osimertinib-increased TGFβ2-mediated activation of SMAD2 (Fig. 2c, d), which also showed high expression in the resistant cells relative to NCI-H1975 cells (Fig. 1g, h). It has been demonstrated that TGFβ2 is involved in the development of EMT in several EGFR TKI resistance models [37–39]. Furthermore, the abundance of TGFB2 significantly correlated with EMT-related markers in patient samples (Fig. 6c–f), supporting the important role of TGFβ2 in the occurrence of EMT and osimertinib resistance. However, the underlying mechanism by which osimertinib increases TGFB2 needs to be further investigated.
Cancer cells acquire dependence on critical signals, including oncogene addiction, for their survival. Drugs that combat the oncogene addiction of cancer cells, such as EGFR and ALK, have been successfully developed in the clinic [40]. The parental NCI-H1975 cells were dependent on the oncogene EGFR for survival. However, EGFR expression was decreased in the osimertinib-resistant NCI-H1975/OSIR cells (Fig. 6a). In ALK-mutated and MYCN-amplified neuroblastoma cells that developed resistance to ALK inhibition, Debruyne et al. found that the resistant cells lost ALK activation and MYCN expression accompanied by a switch in cellular dependence from MYCN to BORIS [41]. This suggests that NCI-H1975/OSIR cells rely on other signals instead of EGFR for survival. On the basis of GSEA analysis, the NF-κB pathway was activated in the resistant cells. The activation of this pathway is a resistance mechanism of other EGFR TKIs, such as gefitinib and erlotinib, in both in vitro and in vivo models [42–44]. However, the role of NF-κB in osimertinib resistance remains unclear. In our model, we showed that the resistant cells were addictive to the NF-κB pathway for their survival instead of the addiction gene EGFR in NCI-H1975 cells (Fig. 4d–f), providing a therapeutic target for this type of resistance. TGFβ2-mediated activation of the NF-κB pathway, which has also been validated in lung adenocarcinoma samples (Fig. 6g), is critical for survival and resistance to apoptosis in some cancers [26]. We further illustrated that osimertinib-induced TGFβ2 activated the NF-κB pathway in NCI-H1975 cells, which may lead gradually to dependence on NF-κB in the resistant cells. In addition, the resistant population cells and several monoclones showed the coexistence of activation of the NF-κB pathway and EMT as resistance mechanisms (Fig. 6a). Moreover, we wondered whether there are any links between the NF-κB pathway and EMT in our model. Although NF-κB is critical for the induction and maintenance of EMT [12], it seems that NF-κB does not regulate the occurrence of EMT in NCI-H1975/OSIR cells (Fig. 6b).
Preclinical studies have shown that EMT is dispensable for cancer metastasis but contributes to drug resistance [45, 46]. Our previous study indicated that NCI-H1975/OSIR cells with EMT were resistant to EGFR TKIs [17]. In addition, clinical data showed that EGFR mutant NSCLC patients were less responsive to immunotherapy [47]. Thus, it seems that no effective strategy could be utilized to treat the resistant cells. The lipid peroxidase GPX4 protected therapy-resistant cancer cells including EMT from ferroptosis [24]. In addition, CDH1 negatively regulated ferroptosis, and the loss of CDH1, a marker of EMT, made the cells more sensitive to ferroptosis inducers [48]. As expected, NCI-H1975/OSIR cells underwent EMT with loss of CDH1, showing more sensitivity to the GPX4 inhibitor to induce ferroptosis than NCI-H1975 cells (Fig. 3a). These findings provide a GPX4 inhibitor as a druggable vulnerability for osimertinib-resistant NSCLC cells with EMT. Since the resistant cells were deadly dependent on the NF-κB pathway, we illustrated that the small molecular inhibitor or genetic suppression of the NF-κB pathway triggered apoptosis in NCI-H1975/OSIR cells (Fig. 4d–f), providing another strategy for the treatment of resistant cells. Of note, the baseline cleaved PARP level was higher in NCI-H1975/OSIR cells than in NCI-H1975 cells (Fig. 4e). It seems that the resistant cells are prone to apoptosis, whereas the reason is not clear. Taken together, we identified that targeting EMT and the NF-κB pathway are potent strategies against osimertinib-resistant cells, which may represent a subset of osimertinib-resistant NSCLC patients in the clinic. In addition, TGFβ2 induced EMT and activated the NF-κB pathway in our model; thus, therapeutics with TGFβ2 antisense oligonucleotides or TGFβ receptor inhibitors [11], some of which have been evaluated in clinical trials, may also be promising strategies for treatment.
Collectively, we show for the first time that the critical roles of TGFβ2, but not other isoforms of TGFβ, contribute to osimertinib resistance. Importantly, targeting GPX4 against EMT or inhibiting the NF-κB pathway are treatment strategies for osimertinib-resistant NSCLC cells.
Acknowledgements
This study was funded by University of Macau (File no. MYRG2018-00165-ICMS and MYRG2016-152-ICMS), National Natural Science Foundation of China (81973516) and The Science and Technology Development Fund, Macau SAR (File no. 176/2017/A3). The results of gene expression profiling shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions
XMJ and JJL designed the research. XMJ, YLX, LWY, MYH, ZHY, and MXS conducted the experiments. XMJ and LLZ contributed to data analysis. XMJ and JJL wrote the paper. JJL, XPC, RDY, and HZ revised the paper. All authors provided final approval of the paper.
Data availability
Datasets used and/or analyzed during the current study are deposited at 10.6084/m9.figshare.11653641.v1 or from the cBioPortal website.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xiao-ming Jiang, Yu-lian Xu
References
- 1.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–40. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020;382:41–50. doi: 10.1056/NEJMoa1913662. [DOI] [PubMed] [Google Scholar]
- 3.Tang ZH, Lu JJ. Osimertinib resistance in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Lett. 2018;420:242–6. doi: 10.1016/j.canlet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Le X, Puri S, Negrao MV, Nilsson MB, Robichaux J, Boyle T, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res. 2018;24:6195–203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–34. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–39. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121:725–37. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci F, Stassi G, De Maria R. Epithelial–mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15:311–25. doi: 10.1038/nrd.2015.13. [DOI] [PubMed] [Google Scholar]
- 9.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 10.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colak S, Ten Dijke P. Targeting TGF-beta signaling in cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, et al. NF-kappaB is essential for epithelial–mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Investig. 2004;114:569–81. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pires BR, Mencalha AL, Ferreira GM, de Souza WF, Morgado-Diaz JA, Maia AM, et al. NF-kappaB is involved in the regulation of EMT genes in breast cancer cells. PLoS ONE. 2017;12:e0169622. doi: 10.1371/journal.pone.0169622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–83. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 17.Tang ZH, Jiang XM, Guo X, Fong CM, Chen X, Lu JJ. Characterization of osimertinib (AZD9291)-resistant non-small cell lung cancer NCI-H1975/OSIR cell line. Oncotarget. 2016;7:81598–610. doi: 10.18632/oncotarget.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang XM, Xu YL, Huang MY, Zhang LL, Su MX, Chen X, et al. Osimertinib (AZD9291) decreases programmed death ligand-1 in EGFR-mutated non-small cell lung cancer cells. Acta Pharmacol Sin. 2017;38:1512–20. doi: 10.1038/aps.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu XH, Liu QY, Li T, Liu JL, Chen X, Huang L, et al. Garcinone E induces apoptosis and inhibits migration and invasion in ovarian cancer cells. Sci Rep. 2017;7:10718. doi: 10.1038/s41598-017-11417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo W, Liu X, Sun W, Lu JJ, Wang Y, Chen X. Toosendanin, a natural product, inhibited TGF-beta1-induced epithelial–mesenchymal transition through ERK/Snail pathway. Phytother Res. 2018;32:2009–20. doi: 10.1002/ptr.6132. [DOI] [PubMed] [Google Scholar]
- 23.Zhang LL, Jiang XM, Huang MY, Feng ZL, Chen X, Wang Y, et al. Nagilactone E suppresses TGF-beta1-induced epithelial–mesenchymal transition, migration and invasion in non-small cell lung cancer cells. Phytomedicine. 2019;52:32–9. doi: 10.1016/j.phymed.2018.09.222. [DOI] [PubMed] [Google Scholar]
- 24.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–7. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–50. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu T, Burdelya LG, Swiatkowski SM, Boiko AD, Howe PH, Stark GR, et al. Secreted transforming growth factor beta2 activates NF-kappaB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci USA. 2004;101:7112–7. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Zhao X, He D, Wang J, Li W, Liu Y, et al. Loss of EGFR confers acquired resistance to AZD9291 in an EGFR-mutant non-small cell lung cancer cell line with an epithelial–mesenchymal transition phenotype. J Cancer Res Clin Oncol. 2018;144:1413–22. doi: 10.1007/s00432-018-2668-7. [DOI] [PubMed] [Google Scholar]
- 28.Lopez Sambrooks C, Baro M, Quijano A, Narayan A, Cui W, Greninger P, et al. Oligosaccharyltransferase inhibition overcomes therapeutic resistance to EGFR tyrosine kinase inhibitors. Cancer Res. 2018;78:5094–106. doi: 10.1158/0008-5472.CAN-18-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–72. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PC, Fang YF, Yamaguchi H, Wang WJ, Chen TC, Hong X, et al. Targeting PKCdelta as a therapeutic strategy against heterogeneous mechanisms of EGFR inhibitor resistance in EGFR-mutant lung cancer. Cancer Cell. 2018;34:954–69. doi: 10.1016/j.ccell.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–8. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGowan M, Kleinberg L, Halvorsen AR, Helland A, Brustugun OT. NSCLC depend upon YAP expression and nuclear localization after acquiring resistance to EGFR inhibitors. Genes Cancer. 2017;8:497–504. doi: 10.18632/genesandcancer.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suda K, Murakami I, Yu H, Kim J, Tan AC, Mizuuchi H, et al. CD44 facilitates epithelial-to-mesenchymal transition phenotypic change at acquisition of resistance to EGFR kinase inhibitors in lung cancer. Mol Cancer Ther. 2018;17:2257–65. doi: 10.1158/1535-7163.MCT-17-1279. [DOI] [PubMed] [Google Scholar]
- 36.Poh ME, Liam CK, Rajadurai P, Chai CS. Epithelial-to-mesenchymal transition (EMT) causing acquired resistance to afatinib in a patient with epidermal growth factor receptor (EGFR)-mutant lung adenocarcinoma. J Thorac Dis. 2018;10:E560–E3. doi: 10.21037/jtd.2018.06.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiagarajan PS, Wu X, Zhang W, Shi I, Bagai R, Leahy P, et al. Transcriptomic-metabolomic reprogramming in EGFR-mutant NSCLC early adaptive drug escape linking TGFbeta2-bioenergetics-mitochondrial priming. Oncotarget. 2016;7:82013–27. doi: 10.18632/oncotarget.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serizawa M, Takahashi T, Yamamoto N, Koh Y. Combined treatment with erlotinib and a transforming growth factor-beta type I receptor inhibitor effectively suppresses the enhanced motility of erlotinib-resistant non-small-cell lung cancer cells. J Thorac Oncol. 2013;8:259–69. doi: 10.1097/JTO.0b013e318279e942. [DOI] [PubMed] [Google Scholar]
- 39.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–40. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivanco I. Targeting molecular addictions in cancer. Br J Cancer. 2014;111:2033–8. doi: 10.1038/bjc.2014.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debruyne DN, Dries R, Sengupta S, Seruggia D, Gao Y, Sharma B, et al. BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells. Nature. 2019;572:676–80. doi: 10.1038/s41586-019-1472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu CF, Chang YW, Kuo KT, Shen YS, Liu CY, Yu YH, et al. NF-kappaB-driven suppression of FOXO3a contributes to EGFR mutation-independent gefitinib resistance. Proc Natl Acad Sci USA. 2016;113:E2526–35. doi: 10.1073/pnas.1522612113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–6. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 2015;11:98–110. doi: 10.1016/j.celrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee CK, Man J, Lord S, Cooper W, Links M, Gebski V, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol. 2018;4:210–6. doi: 10.1001/jamaoncol.2017.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, et al. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572:402–6. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets used and/or analyzed during the current study are deposited at 10.6084/m9.figshare.11653641.v1 or from the cBioPortal website.