Abstract
Microbial communities are comprised of many species that coexist on small spatial scales. This is difficult to explain because many interspecies interactions are competitive, and ecological theory predicts that one species will drive the extinction of another species that competes for the same resource. Conversely, evolutionary theory proposes that natural selection can lead to coexistence by driving competing species to use non-overlapping resources. However, evolutionary escape from extinction may be slow compared to the rate of competitive exclusion. Here, we use experimental co-cultures of Escherichia coli and Saccharomyces cerevisiae to study the evolution of coexistence in species that compete for resources. We find that while E. coli usually outcompetes S. cerevisiae in co-culture, a few populations evolved stable coexistence after ~1000 generations of coevolution. We sequenced S. cerevisiae and E. coli populations, identified multi-hit genes, and engineered alleles from these genes into several genetic backgrounds, finding that some mutations modified interactions between E. coli and S. cerevisiae. Together, our data demonstrate that coexistence can evolve, de novo, from intense competition between two species with no history of coevolution.
Subject terms: Molecular evolution, Microbial ecology
Introduction
Competition between microbial species is probably more common than cooperation [1]. Can two species that compete for the same resource evolve to coexist? Theory predicts that if competition between species is strong, then one species is likely to drive the other extinct [2]. However, there are mechanisms that can promote coexistence—equalising forces that reduce the fitness differences between competing species, and stabilising forces that reduce negative interspecific interactions [3]. Equalising mechanisms act to reduce the rate of competitive exclusion so that, even if species depend on the same resources, they can coexist neutrally for extended periods [4–6]. Alternatively, stabilising mechanisms promote coexistence by reducing interspecific interactions, for example, by niche differentiation [4, 7]. Recent work has shown that evolutionary and ecological processes act over similar timescales in a process known as eco-evolutionary feedback [8–12]. Evidence for eco-evolutionary feedback has been found in a wide variety of model systems, ranging from laboratory populations of Bacillus subtilis [11], Caenorhabditis elegans and its parasite Bacillus thuringiensis [13], Drosophila melanogaster [14], the plant species Lemna minor and Spirodela polyrhiza [15] and island populations of Anolis lizards [12]. The prevalence of eco-evolutionary feedback among these model systems suggest these coexistence mechanisms may be able to evolve in newly formed communities comprised of competing species.
Evolution experiments with populations of microbes have shown that rates of adaptation are sufficient for multiple coexisting ecotypes to evolve from a single progenitor during a laboratory experiment [16–20]. A well characterised example is the evolution of P. fluorescens SBW25 in spatially structured microcosms, where intraspecific competition drives the evolution and maintenance of at least two ecotypes [19, 20]. Experimental adaptive radiation has also been observed in the microbial eukaryote S. cerevisiae, which rapidly evolves two, coexisting, ecotypes in non-shaking microwell plates [21]. In both cases, strong intraspecific competition and the availability of alternative niches in heterogenous environments provided the opportunity for the evolution of novel ecotypes.
There is also an evidence of eco-evolutionary feedback mechanisms at work in homogenous environments [12, 15, 22, 23]. In one well-known example, E. coli populations propagated in environments with glucose as the sole carbon source can evolve to secrete high levels of acetate, a by-product of fermentation. This acetate waste product can be exploited as a carbon source by a second ecotype that evolves during the experiment [24, 25]. In other experiments with E. coli, the loss of the capacity to produce an amino acid was found to be advantageous, as long as there was enough of that amino acid being produced by other E. coli cells in the population [26]. Partially engineered microbial models have also been used to study the relationship between consumers and producers of specific metabolites [27–30]. These systems lead to the evolution of mutualisms, as costly excretions can be exploited as a resource when a mutualistic partner can offer something in return [31–33]. Studies of naturally co-occurring bacteria often find the presence of mutualisms that avoid niche overlap in complex systems [1, 34–37]. These studies of multi-species communities provide many insights into coevolution and multi-species interactions, however, since these interspecies interactions are pre-existing, it is difficult to trace the evolutionary origins of coexistence.
Taken together, these evolution experiments have shown that diversification frequently evolves in single species evolution experiments. However, the outcome for two or more evolving species in co-culture is less certain. Theory predicts that coexistence is more likely to evolve when the initial fitness difference between species is small [4, 38], and two ecotypes evolved from the same species are much more likely to have growth rates compatible with coexistence than two competing species. While there have been experimental studies of coevolving species, most have focused on species with established relationships, such as phage and their bacterial hosts [39, 40] or natural and synthetic bacterial communities that already coexist [35, 37, 41]. These, and studies of coevolving bacteria, have found that the presence of multiple species can drastically alter the outcomes of evolution [35]. In this study, we explore the evolutionary origins of coexistence by co-culturing two species, a prokaryote (E. coli) and a eukaryote (S. cerevisiae), within a homogenous environment. These two species have been well characterised and have a suite of genetic and genomic resources. Since these two organisms do not have a history of coexistence in nature [42, 43], and are competitively mismatched in the lab, this allows us to explore the genetic basis of the eco-evolutionary mechanisms that lead to the de novo evolution of coexistence.
Results
E. coli is competitively superior to S. cerevisiae in co-culture conditions
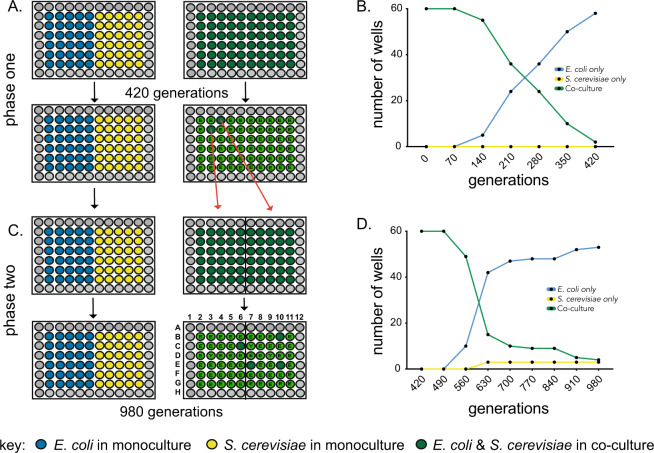
To study the evolutionary origins of coexistence, we propagated replicate populations of E. coli and S. cerevisiae in either monoculture or co-culture treatment conditions. We used a nutrient-rich complex growth medium with glucose (4%) and yeast extract, and incubated cultures with rapid shaking to evenly distribute nutrients and reduce spatial structure (“Methods”). The monoculture treatment consisted of either S. cerevisiae or E. coli grown in isolation from the other species. In the co-culture treatment, S. cerevisiae and E. coli were propagated together in the same vessel (Fig. 1A). We tracked the presence of both species in each population to monitor rates of competitive exclusion. In the first phase of the experiment (420 generations, Fig. 1), E. coli tended to outcompete S. cerevisiae, until only two of the initial 60 replicate co-culture populations contained both species (Fig. 1B). This confirmed that, in the co-culture conditions of this experiment, E. coli has a superior competitive fitness to S. cerevisiae. For the second phase of the experiment, the two co-culture wells remaining after 420 generations were used to found 60 new replicate co-culture populations (30 each). After these were propagated for a further 560 generations, four of the 60 co-culture populations maintained both E. coli and S. cerevisiae, while S. cerevisiae had fixed in three populations and E. coli had fixed in the remaining 53 cultures (Fig. 1C, D). Altogether, the co-culture and monoculture populations were propagated for around 980 generations. Each of the 60 monoculture populations in phase two were founded by transfer directly from the 60 monocultures from phase one (Fig. 1B).
Fig. 1. Experimental co-evolution of E. coli and S. cerevisiae.
A Replicate populations of E. coli and S. cerevisiae were propagated as either monoculture or co-culture for 420 generations (phase one). B The species composition of 60 co-culture replicate populations were tracked with S. cerevisiae going extinct in 58/60 wells. C These two persisting co-cultures were each used to found 30 replicate populations and propagated for 580 generations (phase two) and D the species composition of the co-cultures tracked. Letters inside green circles indicate that either E. coli (E) or S. cerevisiae (Y) has fixed in that co-culture. Alphanumeric well positions (e.g. “C6”) are used to refer to specific populations.
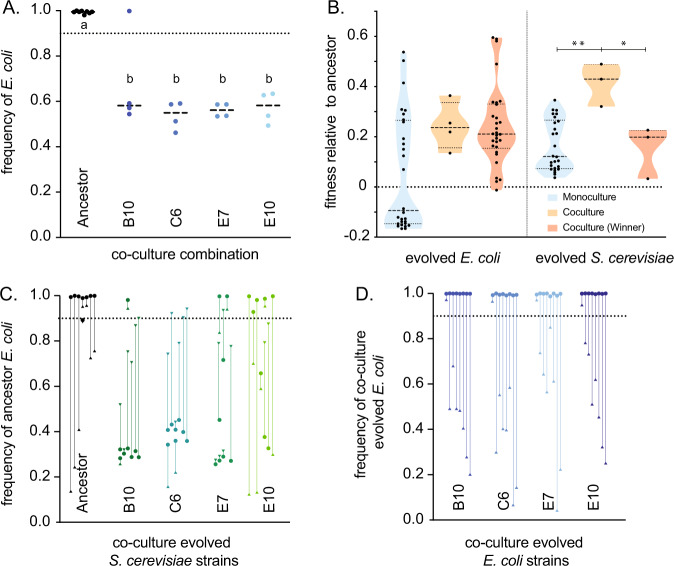
We set up replicate cultures of the four co-cultures (B10, C6, E7, and E10) that remained after 980 generations to determine the new equilibrium frequencies of E. coli and S. cerevisiae. We tracked the frequencies of E. coli and S. cerevisiae for 70 generations, to determine whether the two species could persist in co-culture. While E. coli had fixed in nearly all ancestral co-cultures (fE. coli = 0.994, CI ±0.005), the evolved co-cultures maintained an equilibrium at around 60% E. coli (fE. coli = 0.58, CI ±0.059), significantly different from the equilibrium frequency of the E. coli and S. cerevisiae ancestors (post-hoc t-test, p < 1 × 10−4) (Fig. 2A). Since the E. coli ancestor nearly always outcompeted the S. cerevisiae ancestor, these data show that the co-culture evolved E. coli/S. cerevisiae pairs were able to persist in co-culture for significantly longer than the pair of ancestral strains.
Fig. 2. The evolution of stable E. coli - S. cerevisiae co-cultures.
A The equilibrium frequency of S. cerevisiae and E. coli co-culture for ancestral (black dots) and co-culture evolved strains (coloured dots) expressed as the frequency of E. coli (fE. coli). B The fitness of evolved monoculture populations (blue), populations that persisted in co-culture for 980 generations (orange), and “winners”, populations that outcompeted its opposite species in co-culture during the second phase (generation 420–980) of the evolution experiment. E. coli strains are shown in the left panel and S. cerevisiae shown on the right panel. Stars show significant differences between individual comparisons using Bonferroni-corrected Mann–Whitney U tests. C The final equilibrium frequency of ancestral E. coli co-cultured with ancestor and co-culture evolved S. cerevisiae populations after an invasion assay; and D ancestral S. cerevisiae with co-culture evolved E. coli. An equilibrium frequency of “1” means that E. coli has driven S. cerevisiae extinct, and 0.5 means that E. coli and S. cerevisiae are equally represented in stable co-culture. Arrows start at the inoculation frequency for each of the replicate populations. Capitalised letters show different groups as designated by Tukey’s post-hoc test.
The evolution of co-culture specific adaptations in S. cerevisiae and E. coli
Previous evolution experiments have found that coevolution can speed rates of adaptation. We carried out competition assays between the evolved co-culture populations and their corresponding ancestor. S. cerevisiae that evolved in co-culture adapted more rapidly to the growth medium than S. cerevisiae that evolved in monoculture (Mann–Whitney test, p = 1.1 × 10−3) (Fig. 2B). Fitness assays showed that while both the winning and co-culture E. coli were significantly fitter than the ancestor strain (post-hoc t-test, p < 0.01), neither were significantly fitter than monoculture E. coli (post-hoc t-test, p > 0.05) (Fig. 2B). These results suggest that only co-cultured S. cerevisiae that adapted faster than average had the potential to persist in co-culture with E. coli.
Ecologically stable coexistence is defined by the ability of each species to invade from rare. To test whether the genetic changes that evolved in S. cerevisiae were sufficient for E. coli/S. cerevisiae coexistence, we carried out reciprocal invasion assays of ancestral E. coli against evolved and ancestral S. cerevisiae. We founded cultures with a range of starting frequencies for each species, ranging between 9:1 and 1:9 (E. coli:S. cerevisiae) (Fig. 2C, D). We found strong genotype-dependent effects across different combinations of S. cerevisiae and E. coli regardless of the initial frequency ratios (Fig. 2C, D). Ancestral E. coli could form stable co-culture with two of the evolved co-cultured S. cerevisiae, B10 and C6, (Fig. 2C) with ancestral E. coli comprising around 30–40% of the co-culture. This was lower than the frequency of E. coli in evolved co-cultures (50–60% E. coli). Conversely, ancestral S. cerevisiae was not able to form stable co-culture with any of the co-culture evolved E. coli (Fig. 2D). These data show that the evolutionary changes in co-culture evolved S. cerevisiae were necessary and sufficient for coexistence with E. coli. However, since co-culture evolved E. coli attained higher frequencies in co-culture than ancestral E. coli, this suggests that there was also further evolutionary changes in co-culture evolved E. coli.
Parallel evolution suggests co-culture specific adaptation in E. coli but not S. cerevisiae
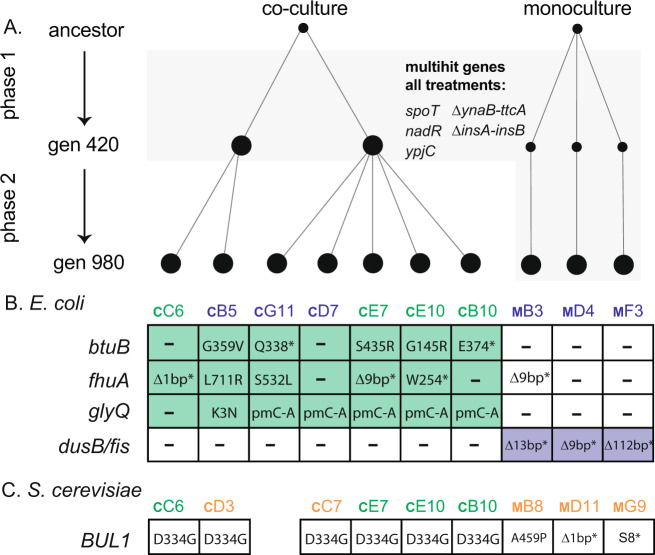
To determine the genetic causes of adaptation, we sequenced S. cerevisiae and E. coli populations taken from monoculture and co-culture treatments (Fig. 3, Supplementary Data file). The evolution of mutations in the same gene or genetic pathway across independent replicate populations is evidence for the action of natural selection. We looked for multi-hit genes, or genes that were mutated across all replicate populations for a given treatment. In S. cerevisiae, we identified 146 mutations across 9 sequenced populations. However, no genes stood out as co-culture specific. We identified a single multi-hit gene, BUL1, that had evolved distinct nonsynonymous substitutions in both co-culture and monoculture populations. In E. coli, we found a clear delineation between the mutations that fixed in co-culture and those that fixed in monoculture populations (Fig. 3). We identified mutations across all evolved E. coli populations (monoculture and co-culture) that have been described in previous evolution experiments with E. coli MG1655. Indel mutations between pryE and rph known to restore a fitness defect in MG1655 were found in 11 of the 12 populations that we sequenced. Mutations in spoT and nadR are known to increase fitness in evolution experiments with glucose as the sole carbon source, and fixed in every E. coli population. Other mutational differences evolved that were specific to the monoculture and co-culture treatments, respectively (Fig. 3). Each monoculture E. coli population that we sequenced had a unique mutation that disrupted the dusB-fis operon. Co-culture populations showed parallelism in three genes, glyQ, btuB, and fhuA (Supplementary Data). glyQ is an essential gene involved in tRNA synthesis, attaching glycine to its relevant tRNA for subsequent protein construction. Interestingly, btuB and fhuA are TonB-dependent transmembrane transporters of vitamin B12 (btuB) and ferrichrome siderophores (fhuA), respectively. Both are also implicated in the uptake of a range of toxins and antibiotics produced by microbes and is a receptor for a range of bacteriophage. The btuB and fhuA genes both contain a variety of nonsynonymous mutations, including missense and nonsense mutations across the four co-cultured populations sequenced (Fig. 3), indicating selection for loss of function in these genes.
Fig. 3. The genetic causes of co-culture and monoculture specific evolution.
A E. coli and S. cerevisiae populations from co-cultures were sequenced after 420 (two populations) and 980 generations (E. coli = seven and S. cerevisiae = six populations) and monoculture populations were sequenced after 980 generations (three populations) (Supplementary Data). In E. coli, mutations in five genes were found to be at high frequencies in all populations (A, grey shaded area). B Six independent mutations were found across the co-culture evolved populations in btuB and fhuA and two in glyQ. Population E10 had two mutations in both btuB and fhuA. All three monoculture populations had unique mutations in the dusB/fis operon. C BUL1 was mutated in all sequenced S. cerevisiae populations. Coloured text indicates the species present at generation 980. E. coli and S. cerevisiae (green text), E. coli only (blue text), S. cerevisiae only (orange text). Alphanumeric population codes (e.g., cC6) denote whether the population was evolved in coevolution (C) or monoculture (M) conditions and the position in the 96-well plate. “pm” indicates promoter, “*” indicates early stop or nonsense mutations. “–” indicates that no mutation was discovered in that gene.
BUL1 mutations in S. cerevisiae and glyQ mutations in E. coli are beneficial in monoculture growth conditions
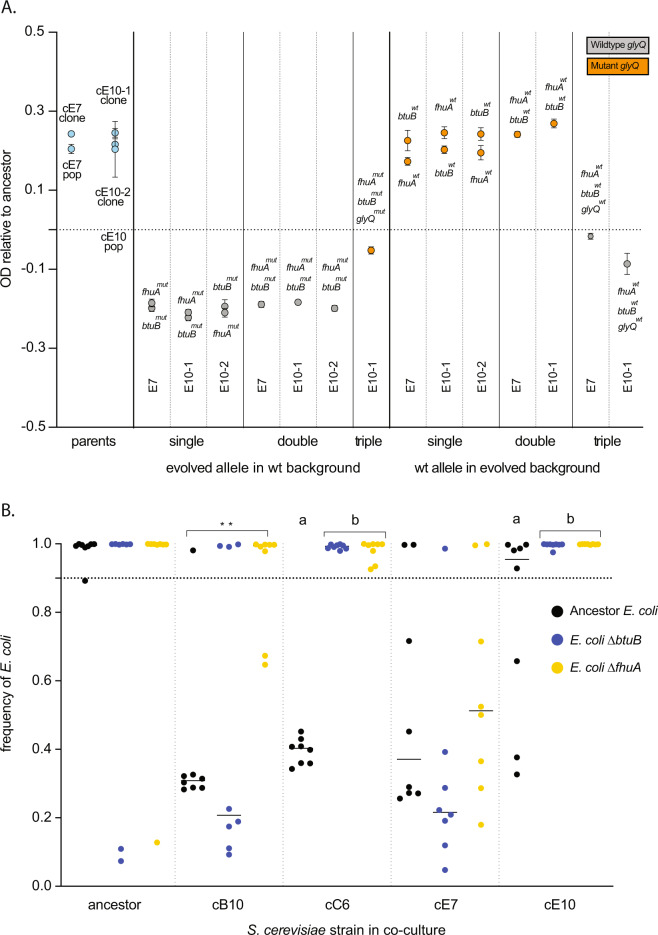
To measure the phenotypic effect of mutation of our single multi-hit S. cerevisiae gene, we engineered an allele that evolved in BUL1 into the S. cerevisiae genetic background, and found an increase in growth rate that matched the increase in growth rate observed in the co-culture evolved yeast (post-hoc t-test, p < 1 × 10−4) (Fig. 4). To test the effect of the mutations discovered in E. coli multi-hit genes, we engineered deletions of fhuA and btuB into an ancestral strain of E. coli and reconstructed the evolved mutations of three clones from two of the coevolved populations that had occurred in the btuB, fhuA, and glyQ. Where possible, we created single, double, and triple mutant combinations to measure combinatorial effect of mutations, although we had difficulty engineering glyQ (an essential gene) mutations into some genetic backgrounds. In order to check whether we had missed any mutations that could impact fitness, we also engineered the wild-type version of alleles for these three genes into evolved clones. In total, this was 22 separately engineered clones with different mutations in a range of combinations. We measured the growth of each of these mutants compared to the ancestral strains and found that glyQ mutations could account for a significant proportion of fitness increase in E. coli (post-hoc t-test, p < 1 × 10−4) (Fig. 5A). To check whether other genes aside from the three we checked were important for fitness, we replaced mutant alleles in the evolved strains with wild-type alleles. This revealed that while a mutant glyQ allele was necessary for high fitness it was not sufficient, suggesting the importance of other mutations or epistatic interactions that we have not identified (Fig. 5A). Interestingly, mutations in fhuA and btuB reduced growth compared to the ancestral strain (post-hoc t-test, p < 1 × 10−4) (Fig. 5A) but did not have a significant effect on competitive fitness (Supplementary Fig. 1).
Fig. 4. Amino acid substitutions from BUL1 (D334G), YFR018C (L7F), and TIF35 (V272L) were engineered into the ancestral genetic background of S. cerevisiae (“Methods”).
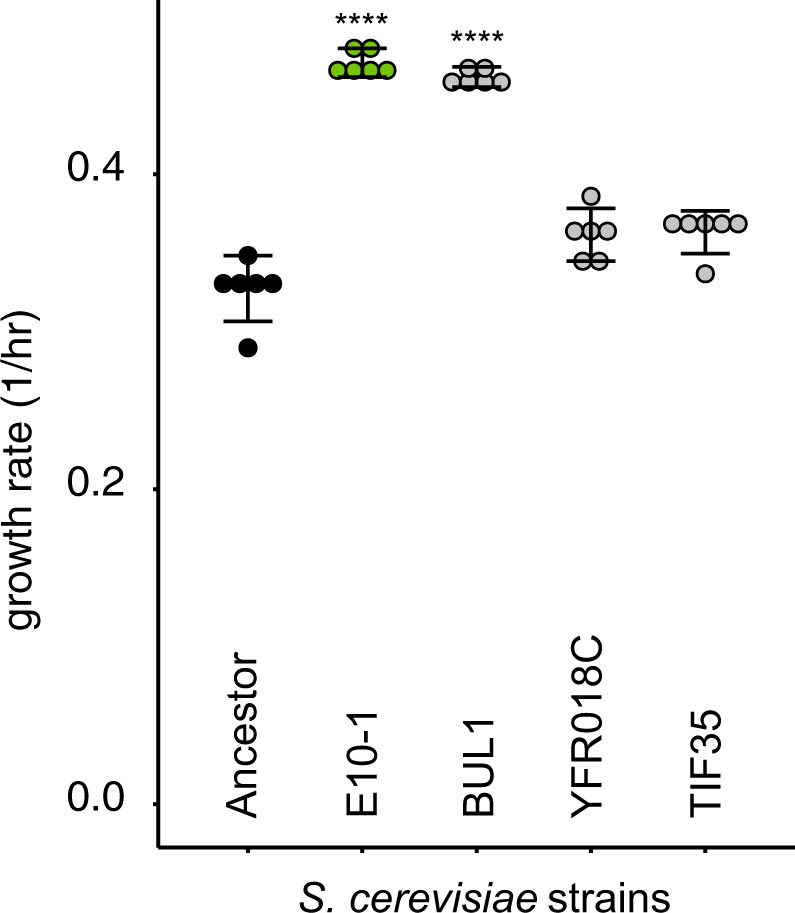
Growth rates were measured for the S. cerevisiae ancestor, the co-culture evolved S. cerevisiae clone E10-1 and the engineered three mutants. Stars indicate a significant difference from the ancestor (Bonferroni-corrected post-hoc t-tests).
Fig. 5. The genetic causes of E. coli adaptation.
A Mutations in the essential gene glyQ cause the improved growth of evolved E. coli compared to ancestor E. coli. Growth was measured as OD600 attained after 24 h of incubation at experimental conditions. Each dot represents a distinct engineered clone, where either one, two, or three alleles were altered. In one example, the two grey dots in the column indicated “evolved allele in wt background, single, E7” denote two separate strains where the ancestor E. coli genotype was engineered to carry either the fhuA or btuB mutant alleles that evolved in the E7 co-culture E. coli. For a second example, the single grey dot in the right-most column indicated “wt allele in evolved background, triple, E10-1” denotes engineering of an E. coli clone from the E10 co-culture evolved population. Here three mutant alleles (fhuA, btuB, and glyQ) were replaced with the wild-type alleles. The growth of each measured strain is plotted relative to the ancestor, which is set at 0, (the dotted line), all measured strains were judged significantly different from the ancestor using bonferroni-corrected post-hoc t-tests, p < 0.001. Error bars represent standard error of the mean (SEM). B The final equilibrium frequency of ancestral and mutant E. coli co-cultured with ancestor and co-culture evolved S. cerevisiae populations after a reciprocal invasion assay (B). The equilibrium frequency of the ancestor E. coli when co-cultured with each of these S. cerevisiae populations is shown (black dots). Also shown are mutant E. coli engineered to contain either a btuB (blue dots) or fhuA mutation (yellow dots). Each dot represents a single replicate co-culture experiment. Stars show significant differences between individual comparisons; lower case letters show significantly different groups as designated by the Tukey’s post-hoc test.
Mutations in E. coli genes fhuA and btuB alter co-culture dynamics with S. cerevisiae
We tested whether engineered E. coli strains could invade ancestral and co-culture evolved S. cerevisiae (Fig. 5B, Supplementary Fig. 2). We found that mutations that disrupt fhuA and btuB were sufficient to change the equilibrium frequency of co-cultures. The capacity to reduce community stability was dependent on both the gene that had been ablated, and the S. cerevisiae genotype (Fig. 5B). For instance, co-culture-evolved S. cerevisiae B10 formed stable co-cultures with the E. coli ancestor from a range of starting frequencies, and similarly stable co-cultures with the E. coli btuB mutant (post-hoc t-test, p > 0.05). However, co-culture-evolved S. cerevisiae B10 was less able to coexist with the fhuA mutant (post-hoc t-test, p = 4.5 × 10−3). Notably, the evolved B10 co-culture E. coli did not have a mutation in fhuA. When the mutant E. coli is paired with evolved S. cerevisiae C6, stability is completely disrupted for btuB and fhuA so that different equilibria are reached (post-hoc t-test, p < 1 × 10−4). Even though the E. coli mutations do not increase competitive fitness against the E. coli ancestor, they were able to disturb mechanisms of coexistence and increase the capacity for E. coli to competitively exclude co-culture evolved S. cerevisiae.
E. coli and S. cerevisiae evolved improved performance on spent media
We tested the idea that S. cerevisiae and E. coli had evolved to occupy different niches by carrying out growth experiments in spent medium. If S. cerevisiae can grow in medium that has been exhausted by a culture of E. coli (and the E. coli cells subsequently removed), this would suggest that S. cerevisiae can utilise a resource in the growth medium that cannot be accessed by E. coli or that E. coli produces a resource that can be utilised by S. cerevisiae. We found that ancestral E. coli and S. cerevisiae could not grow in medium previously exhausted by their own species, but both could grow on the spent medium of the other (Supplementary Figs. 3 and 4). After 980 generations of evolution, all of the co-culture evolved E. coli had an improved growth rate on the spent medium of its evolved partner S. cerevisiae strain (Fig. 6A), having a shorter lag phase than ancestor E. coli growing on S. cerevisiae spent medium and of which, three had evolved an increased carrying capacity (Supplementary Fig. 5). When we tested co-culture evolved S. cerevisiae on the spent medium of the E. coli they had coevolved with, only two of the co-culture-evolved S. cerevisiae had higher OD and none had a shorter lag phase (Supplementary Fig. 6).
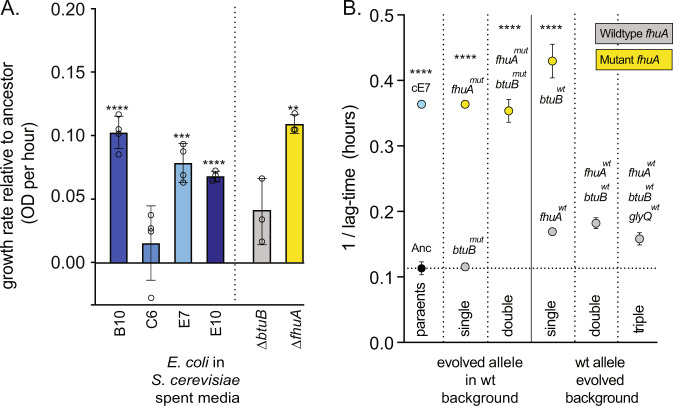
Fig. 6. Mutations in fhuA cause co-culture specific adaptation.
A The co-culture evolved E. coli, and ΔfhuA mutant, have improved growth in spent medium relative to the E. coli ancestor. Each E. coli population was grown on spent medium from its respective co-culture S. cerevisiae. E. coli ΔbtuB and ΔfhuA mutants were grown on spent medium from a culture from the co-culture evolved “E7” population of S. cerevisiae. Error bars represent standard error of the mean (SEM). Stars show significant differences between strain growth in ancestor and evolved spent medium using post-hoc t-tests. B The change in lag-time for ancestral, coevolved, mutant and wild-type revertant strains when grown in the presence of the FhuA-targeting antibiotic albomycin. Error bars represent standard deviation (SD).
We next tested whether btuB or fhuA mutations in E. coli conferred a growth advantage in spent medium from S. cerevisiae (Fig. 6). We found that the fhuA mutant had a significantly increased growth rate (post-hoc t-test, p = 6 × 10−4) (Fig. 6A); while btuB was not significantly different to the ancestor (post-hoc t-test, p = 0.56).
Evolved mutations in fhuA cause resistance to the antibiotic albomycin
While the selective benefit of glyQ is apparent in monoculture (Fig. 5A), the effects of fhuA and btuB mutations are specific to co-culture with E. coli and spent medium. The primary role of the FhuA and BtuB outer membrane transporters is to import ferrichrome and vitamin B12. However, both are also implicated in the transport of bacterial toxins (bactericins), are binding sites for bacteriophage, and FhuA imports the antibiotics albomycin and rifamycin. BtuB and FhuA depend on the periplasmic membrane protein TonB to provide energy for transport, and compete with five other E. coli outer membrane transporters for access to TonB. We hypothesised that the mutations in btuB and fhuA could prevent the binding of the cognate substrate. To test whether fhuA mutations are altering the potential for taking up substrates, we measured the effect of an antibiotic, albomycin, that depends on binding fhuA for entry into the cell (Fig. 6B). We found that all of the evolved variants of fhuA were sufficient to confer resistance to albomycin (post-hoc t-test, p < 1 × 10−4) (Fig. 6B). This suggests that the loss of fhuA substrate binding is important for improved growth of E. coli in co-culture with S. cerevisiae.
Discussion
Competitive exclusion sets a time limit for the evolution of coexistence
While previous evolution experiments have shown that how a single species can diversify into multiple coexisting ecotypes, the evolution of coexistence in this experiment is a relatively rare outcome (Fig. 1). In E. coli, 9 of 12 populations evolved coexisting ecotypes for tens of thousands of generations during a 61,500 generation experiment [44]. In P. fluorescens, the Wrinkly Spreader type evolves within a week whenever grown in non-shaken conditions [19]. Similarly, in a 1000 generations experiment with S. cerevisiae, 100 out of 1000 populations evolved an adherent type that coexisted with the progenitor ecotype [21, 45].
One factor working against the evolution of coexistence is the time limit set by the rate of competitive exclusion. Evolution experiments in a range of species have shown how ecological change can occur fast enough to change the selective conditions experienced by a population or community, and therefore drive further evolution [8, 10, 12, 15]. However, even rapid evolution of the more poorly adapted species does not always lead to stable coexistence [14]. In experiments founded from a single species, there is no time limit on evolutionary change in favour of coexistence, because a novel ecotype always has the potential to evolve via spontaneous mutation [44]. By contrast, in this experiment, S. cerevisiae must evolve the characteristics necessary to promote coexistence before it is driven extinct by E. coli. In other words, interspecific competition is much stronger than intraspecific competition. This constraint might be expected to limit the potential for stable microbial communities to evolve from competing species. However, evolution in natural microbial communities might not operate under such constraints. For instance, competitive exclusion of one species will only cause local or temporary extinction of the weaker species. Future immigration events can provide further opportunities for the two species to co-habit, and co-evolve the mechanisms vital to coexistence. This suggests that the propensity for competing species to evolve coexistence may be increased by the presence of reservoir populations, potentially in stratified sub-sections of the environment, to provide multiple “trials” for the evolution of coexistence.
Genetic evolution in both S. cerevisiae and E. coli underpins coexistence
Our fitness measurements showed that the few S. cerevisiae that evolved to persist in co-culture with E. coli had a higher competitive fitness than monoculture evolved S. cerevisiae. E. coli on the other hand had similar increases in fitness in monoculture and co-culture (Fig. 2). While we did not find any co-culture specific genetic changes in S. cerevisiae, we did find one change consistent to all populations, suggesting that S. cerevisiae adaptations may have caused general increases in growth rate that were not dependent on the presence of E. coli. Crucially, co-culture evolved S. cerevisiae formed stable co-cultures with ancestral E. coli, while co-culture evolved E. coli rapidly outcompeted ancestral S. cerevisiae Fig. 2C, D). This result suggests that the genetic changes in co-culture evolved S. cerevisiae were sufficient for coexistence.
Despite this result, we found strong evidence of co-culture specific adaptation in E. coli populations. All of the co-culture evolved E. coli had significantly improved growth in S. cerevisiae spent medium. Our genome sequencing experiments also showed that a distinct set of E. coli genes were targets for selection in co-culture (Fig. 3). The engineered substitutions of mutant and wild-type glyQ alleles into wild-type and evolved E. coli showed that mutations in glyQ, and probably another unknown locus (Fig. 5A), were responsible for increased E. coli growth in unspent medium. However, engineered disruptions of fhuA and btuB genes in E. coli did not improve growth, or increase fitness compared to the ancestor E. coli (Fig. 5A).
Why then were these mutations so strongly selected in all co-culture populations? We found that deleting fhuA and btuB altered the outcomes of invasion assays with ancestral and evolved S. cerevisiae (Fig. 5B). Moreover, the fhuA mutant had significantly improved growth in S. cerevisiae’s spent growth medium (Fig. 6A). Our experimental evidence supports that the selective pressure driving the fixation of btuB and fhuA mutations emerged from interactions with S. cerevisiae.
S. cerevisiae persistence in co-culture led to rapid co-culture specific E. coli evolution
The evolution of stable co-culture depends strongly on the genotype of both E. coli and S. cerevisiae. We expected rapid adaptation in S. cerevisiae in order to ameliorate or avoid direct interactions with E. coli that reduce fitness. However, it was surprising that E. coli evolved so many co-culture specific genetic changes, since E. coli was the fitter of the two species initially. We propose that in the rare cases where S. cerevisiae adapted quickly enough to coexist with E. coli, S. cerevisiae imposed strong selective pressures on E. coli to access new nutrients or avoid suppression, driving co-culture specific adaptations in E. coli and subsequently stabilising coexistence. This could explain why S. cerevisiae adaptations were general to both treatments of this experiment, but E. coli has multiple co-culture specific adaptations. Whole-genome sequencing in S. cerevisiae revealed that despite the discovery of many mutations across the genome, very few were in the same multi-hit genes (Supplementary Data). This may be because of the broad number of mutational targets available for S. cerevisiae to improve growth in high glucose medium. While our growth assays with engineered BUL1 mutants confirm the presence of a beneficial mutation in evolved yeast populations, future work will determine any other genetic causes of adaptation in the S. cerevisiae populations.
Molecular causes for the evolution of coexistence
Previous studies have found that microbial communities growing in rich medium are likely to produce toxic compounds that inhibit each other’s growth [46]. Since mutations in fhuA and btuB are known to provide resistance to bacteriocins [47], it is possible that the mutations discovered in this study provide resistance to a secreted product of S. cerevisiae. However, while some S. cerevisiae strains secrete antimicrobial peptides [48], our data do not suggest any candidates for such a secreted molecule. A second explanation is related to the role of the FhuA and BtuB proteins as two of seven membrane transporters that depend on TonB to provide the energy for substrate transport [49]. TonB dependent transporters compete for access to TonB [50], and we found that the mutations that evolved in our study render FhuA insensitive to its cognate substrate, and therefore unable to activate TonB. The inactivation of FhuA and BtuB would increase availability of TonB for other TonB-dependent transporters. If one or more of these transporters is in high use due to co-culture with S. cerevisiae, this would provide a selective advantage. In future work, we plan to carry out experiments to resolve these two hypotheses.
Conclusions
Here we have shown that a prokaryote and a eukaryote, that are mismatched in co-culture and are not known to co-habit in nature, can evolve to coexist. The adaptive changes discovered in E. coli reinforce that evolution in the context of a community can be dramatically different to evolution in isolation. Most studies of microbial evolution, molecular biology, and antibiotic resistance have been carried out in monocultures; microbiology in co-culture could yield new insights into each of these areas of research.
Methods
Strains
The E. coli strain used to found the evolution experiment is isogenic with the laboratory strain E. coli MG1655 K-12 F– λ– ilvG– rfb-50 rph-1. The S. cerevisiae ancestor was haploid, non-recombining S. cerevisiae strain R1158 with genotype trp1::Hph URA::CMV-tTA MATa his3-1 leu2-0met15-0 [51].
Medium composition
All strains were grown within a defined medium supplemented with 4% glucose, also called high-glucose medium (HGM), described in previous work [52].
Long-term evolution
To set up the evolution experiment, single clones of MG1655 and R1158 were grown to saturation in high-glucose medium, diluted 1:210 and used to seed 60 replicate co-culture populations in a single 96-well plate, as well as 30 replicate populations for each species in monoculture in a separate 96-well plate. The cultures were evolved through 420 generations (42 daily cycles) of growth and dilution in high-glucose medium at 28 °C and 750 RPM. Every 24 h, the populations were diluted 1:210 by serially diluting 1:25 (4 μL into 125 μL) × 1:25 (4 μL into 125 μL) into new high-glucose medium. All dilutions were performed using the Hamilton96 Liquid Handler with the CO-RE 96 Probe Head. After every 70 generations, populations were mixed with 50 μL of 75% glycerol and archived at −80 °C. At 420 generations, single clones of each co-culture species were extracted from wells C3 and B4 of the co-culture 96-well plate by plating onto selective agar, grown to saturation in high-glucose medium at 28 °C, and used to found 60 replicate co-culture populations in a new 96-well plate. All monoculture population lineages continued to be propagated after 420 generations. Both monoculture and co-culture populations were evolved for a further 580 generations (58 daily cycles) under the same conditions, for a total of ~1000 generations of growth (100 daily cycles).
Stable frequency assay
Single colonies of each ancestor were grown on YPD agar, and one colony was picked from each ancestral population. These individual colonies were grown to saturation in 3 mL of defined supplemented medium. Ancestral E. coli and ancestral S. cerevisiae were mixed together and propagated together under long-term evolution conditions as outlined above for 70 generations, along with population samples of each co-culture at 1000 generations. In conjunction with the continual growth and dilution of evolving cultures, populations were also diluted 1:10 and 1:100 (13 μL in 119 μL × 13 μL in 119 μL) into 1× phosphate-buffered saline for measurement of total cell count using flow cytometry. Nine microliters samples were taken from each population for cell counts, with a maximum total count of 30,000 events.
Competitive fitness assays
For fitness assays of evolved E. coli populations, we selected 35 co-culture populations and 30 monoculture populations to measure their fitness relative to a GFP-expressing reference E. coli strain. For fitness assays of evolved S. cerevisiae populations, we selected all remaining co-culture populations (seven total) and 30 monoculture populations to measure their fitness relative to a GFP-expressing reference S. cerevisiae strain.
Experimental and reference strains were grown to saturation in separate 15 mL falcon tubes. If the strains to be tested were coming from the −80 °C freezer, strains were passaged further by diluting 1:25 in a 96-well plate to re-acclimatise to the appropriate medium. Experimental and reference strains were mixed 25 μL:75 μL (1:3) manually, before propagating for 20 generations (two daily cycles) using the Hamilton96 Liquid Handler. In conjunction with the continual growth and dilution of evolving cultures, populations were also diluted each day 1:10 and 1:100 (13 μL in 119 μL × 13 μL 119 μL) into 1× phosphate buffered saline for measurement of total cell count by flow cytometry using a LSR Fortessa X20a. Nine microliters samples were taken from each population for cell counts, with a minimum count of 10,000 events, and a maximum total count of 30,000 events.
For fitness assays of mutant E. coli populations, the same competitive fitness assay protocol was used, but with the following changes. Instead of population samples, clonal samples of ancestral, mutant, and coevolved populations were taken from single colonies grown on YPD agar.
Whole-genome sequencing
For sequencing clones, we used single colonies grown on YPD agar and picked one colony from each population to sequence at two time points. These individual colonies were grown to saturation in 3 mL of defined supplemented medium and total genomic DNA was isolated for each sample the GenElute bacterial DNA prep kit (Sigma-Aldrich). DNA concentrations were measured on a Qubit analyser and sent to Genewiz (NJ, USA) for library preparation and short-read sequencing on a Hiseq X-10 (illumina). Upon receipt of fastQ files, variants were called using Breseq v.0.33.2, using default parameters [53]. Each E. coli sample was aligned to the complete and annotated MG1655 genome sourced from NCBI, and each S. cerevisiae sample was aligned to the complete and annotated R1158 genome sourced from NCBI. Each sample was sequenced to a depth of ~100× coverage for E. coli and ~50× coverage for S. cerevisiae. Outputs were aligned together using the gdtools. COMPARE function within breseq used to determine which mutations were unique or shared across monoculture and co-culture populations. Single nucleotide polymorphisms at or above 40% frequency were used for analysis of genomic alterations.
S. cerevisiae growth assay
Single colonies were grown on YPD agar, and one colony was picked from ancestral, evolved E10 and mutant populations. These individual colonies were grown in 10 mL non-shaking HGM for 24 h [54]. All strains were standardised to 0.01 OD600 using HGM as diluent and placed into 96-well microplates (n = 6). Growth of each population monitored by tracking optical density (OD) of cultures in a plate reader for 24 h, with readings taken every 10 min. The resultant growth data were fitted to logistic curves using the R package growthcurver to calculate growth rates, r, of each strain [55].
Invasion assay
Single colonies were grown on YPD agar, and one colony was picked from each ancestral, evolved and mutant population. These individual colonies were grown to saturation in 3 mL of defined supplemented medium. E. coli and S. cerevisiae were mixed together at different volumetric ratios and correlated with total cell count using flow cytometry. From this a range of ratios was created by mixing E. coli and S. cerevisiae together in different volumetric ratios across 3, 96-well plates, ranging from 9:1 to 1:9 (E. coli:S. cerevisiae). This initial mix plate then underwent long-term evolution transfer conditions as outlined above, except instead of evolving populations for 1000 generations (100 daily cycles), populations were evolved for 140 generations (14 daily cycles). In conjunction with the continual growth and dilution of evolving cultures, populations were also diluted 1:10 and 1:100 (13 μL in 119 μL × 13 μL in 119 μL) into 1× phosphate-buffered saline for measurement of total cell count using flow cytometry. Nine microliters samples were taken from each population for cell counts, with a maximum total count of 30,000 events.
Spent medium assay
To generate spent medium for ancestral samples, single clones of ancestral MG1655 and ancestral R1158 were grown to saturation in 3 mL of high-glucose medium at 28 °C over 24 h. Cells were removed from overnight cultures via centrifugation (10,000 rpm for 2 min) and supernatant removed. Cells were washed in phosphate-buffered saline via centrifugation before being resuspended in 3 mL phosphate-buffered saline. Supernatant was filter sterilised through a syringe filter with a pore size of 0.2 μm. Filtered spent medium was then added to a 96-well plate, with 128 μL per well. Four microliters of resuspended cells was then added to spent medium. Each species was added to both its own species spent medium and the opposite species spent medium in four replicates each. Growth of each population monitored by tracking OD of cultures in plate reader for 24 h, with readings taken every 20 min. Growth conditions were the same as long-term evolution experiment. Maximum OD, time to exponential phase (defined as OD greater than 0.1), and maximum growth rate (calculated over five data points) recorded.
For spent medium assays with evolved population samples, mixed population samples of each evolved population were grown in 3 mL of defined medium overnight, with antibiotics to remove either S. cerevisiae (cycloheximide) or E. coli (tetracycline). Ten microliters of culture then plated onto non-selective agar and incubated. Multiple colonies from agar plate picked from plate and regrown in 3 mL of defined medium without antibiotics. The same sterilisation and filtration procedure was followed to generate spent medium and resuspended cells. Each evolved strain was grown in ancestral and evolved spent medium of both MG1655 and R1158.
For spent medium assays of E. coli mutant strains, single colonies of each mutant selected was grown overnight in 3 mL of high-glucose medium at 28 °C over 24 h, before being centrifuged, washed and resuspended as outlined above. The same sterilisation and filtration procedure was followed to generate spent medium and resuspended cells. Each E. coli mutant strain was grown in ancestral and evolved spent medium of R1158.
Albomycin growth assays
Both population and clonal samples of evolved strains were grown overnight in 3 mL of high-glucose medium at 28 °C over 24 h, as well as single clones of each mutant and wild-type revertant strains. Overnight cultures were diluted 1:210 by serially diluting 1:25 (4 μL into 125 μL) × 1:25 (4 μL into 125 μL) into new high-glucose medium in a 96-well plate. Growth of each population monitored by tracking OD of cultures in plate reader for 24 h, with readings taken every 20 min. Maximum OD, time to exponential phase (defined as OD greater than 0.1), and maximum growth rate (calculated over 5 data points) recorded.
For growth curves with additional albomycin, the same protocol was used as outlined above, with the following changes. Overnight cultures were diluted 20-fold in 1 mL of PBS (50 μL into 950 μL) before a further 50-fold dilution into 200 μL of high-glucose medium with 50 mg/mL albomycin.
Statistical methods
All statistical tests conducted were one way ANOVA with Bonferroni-corrected post-hoc t-tests (p values <0.05*, 0.01**, 0.001***, and 0.0001****) or tukey’s post-hoc tests (groups shown by letters), except where otherwise specified.
E. coli strain modifications
Strain constructions were carried out using CRISPR-Cas for E. coli, adopted from Jiang et al. [56]. Both pTargetF (guide RNA expressing, Plasmid #62226) and pCas (Cas9 expressing, Plasmid #62225) plasmids were obtained from Addgene. E. coli Top10 cells were used as a cloning host and genetic changes were introduced into the ancestral MG1655 strain. Further, we have swapped mutant alleles with the wild-type alleles (from the ancestor) in the selected evolved clones.
Modification of 20 N sequence in pTargetF
Target sequences were chosen for each gene using the online tool CCTop [57] (Supplementary Table 1). Plasmid modifications were carried out in three steps. Step 1 was the removal of 20N sequence from pTargetF. Plasmid pTargetF was cut with SpeI and PshAI restriction enzymes, then the fragment containing the backbone of the plasmid without 20N sequence was gel purified. Step 2 was the construction of DNA fragments with new 20 N sequence. PCR fragments with 20N target sequences for each gene were generated in three PCR reactions. First, PCR fragment was amplified using pTargetF-F1 and pTargetF-R1 primers (R1 primers have reverse complement of 20N sequence specific to each gene at the 5′-end) (see Supplementary Table 2 for primers). Second, PCR reactions used pTargetF-F2, with 20N sequence for each gene at the 5′-end, and pTargetF-R2 primers. These two PCR fragments were gel purified and used as templates in the third PCR with the outer primers (pTargetF-F1 and pTargetF-R2) to generate a PCR fragment with a 20N sequence. Later, the third PCR fragments were also cut with SpeI and PshAI restriction enzymes to generate complementary ends to the plasmid backbone. Step 3 involved insertion of plasmid with 20N target sequence into cloning host cells. Plasmid backbone from step 1 and DNA fragments with the new 20N sequences from step 2 were ligated using instant Sticky-end Ligase Master Mix (NEB) and electroporated with 50 μL electro-competent Top10 cells using 1-mm Gene Pulser cuvette (Bio-Rad), at 2.5 kV. Cells were recovered at 37° for 1 h before being spread onto LB agar plates containing spectinomycin (50 mg/L). Sanger sequencing confirmed the presence of colonies with the modified pTargetF plasmids, namely; trpF pTargetF, tonB pTargetF, fhuA pTargetF, btuB1 pTargetF, btuB2 pTargetF, and ampR pTargetF.
Repair DNA constructions
CRISPR engineering requires a donor “repair” DNA that the cell will use to repair the cut target side, thereby introducing the desired DNA sequenced change.
eGFP strain
A 50 base pair cloning strategy was used for the construction of GFP-containing fragment. The trpF gene was targeted (with trpF pTargetF) as a GFP insertion site. GFPmut2 DNA was amplified from pMGS053 plasmid using GFP-F and GFP-R primers (see Supplementary Table 2). The resultant DNA fragment had 50 base pair homologies at both 5′ and the 3′-ends corresponding to the upstream and downstream of 20 N sequence in trpF gene.
fhuA mutants
The fhuA mutant allele swap was generated by swapping this gene with AmpR cassette, then swapping ampR cassette with the exact mutations seen in the evolution experiment. In the first stage, fhuA pTargetF plasmid was used to direct Cas9 enzyme to introduce double strand breaks in fhuA gene. Then, fhuA ampR cassette DNA fragment was used as repair DNA. This DNA fragment was constructed with 400 to 600 base pair homologies of upstream and downstream of fhuA gene. ampR gene (including its promoter and terminator sequences) was amplified from pUC19 plasmid using ampR-F and ampR-R primers. 5′-end of these primers were designed to generate homology (about 20 bp) to upstream and downstream of fhuA. Upstream sequence was amplified with fhuA-F1 and fhuA-R1 and downstream sequence was amplified with fhuA-F2 and fhuA-R2 primers (Supplementary Table 2) using MG1655 DNA as a template. Finally, overlap PCR method was used to join the three DNA fragments. This stage results in a knock-out mutant. In the second stage, ampR cassette was targeted by Cas9 enzyme (with ampR pTargetF) and swapped with repair DNA containing exact mutations for fhuA. A colony PCR was used to amplify repair DNA using evolved clones as template DNA and fhuA-F1, fhuA-R2 primer pairs (see Supplementary Table 2).
btuB mutants
A deletion strategy in two stages was used for the btuB mutant construction. In the first stage, Cas9 enzyme was directed to a site in the btuB gene with btuB1 pTargetF plasmid. Then, a deletion repair DNA fragment was used to introduce the deletion. This DNA fragment was generated with three PCR reactions. The first PCR step used btuB-F1 and btuB-R1 primers (Supplementary Table 2) and MG1655 template DNA resulting in a DNA fragment containing upstream and downstream of start codon of btuB. The second step used btuB-F2 (containing the forward sequence of btuB-R1 at 5′-end to generate homology between the two PCR fragments) and btuB-R2 primers (Supplementary Table 2) to amplify upstream of btuB stop codon (making sure murl start codon is undisturbed) and downstream sequence. The third step used these two PCR fragments as template DNA and btuB-F1 and btuB-R2 primers to generate a repair DNA fragment resulting in a 1386 bp deletion in the middle section of btuB gene. In the second stage, a 20 N sequence targeted the location where the two PCR fragments fused (see above) with the btuB2 pTargetF. Then a DNA fragment was amplified using btuB-F1, btuB-R2 primer pair with an evolved clone as template and used as a repair DNA to introduce btuB mutations from the evolution experiment to ancestral MG1655 cells.
glyQ mutants
glyQ is an essential gene and its modification requires a different two stage CRISPR strategy as its inactivation results in cell death. In the first stage, ampR cassette was carefully inserted upstream of glyQ promoter, leaving the promoter undisturbed. No pTargetF plasmid was used to introduce double strand breaks at this location [58]. A glyQ ampR cassette DNA fragment was generated with a four step PCR protocol. In the first step, upstream of the promoter was amplified using glyQ-F1 and glyQ-R1 primers (Supplementary Table 2). In the second step glyQ-F2 and glyQ-R2 primers were used to amplify the region that includes few bases upstream of the promoter, glyQ start codon and partial glyQ sequence following downstream of the start codon. ampR cassette was then amplified from pUC19 plasmid using glyQ-amp-F and glyQ-amp-R primers in the third step (Supplementary Table 2). Finally, the resulting three PCR fragments were merged in a fusion PCR using glyQ-F1 and glyQ-R2 primers. In the second stage of CRISPR, ampR cassette was targeted with ampR pTargetF and the cassette was deleted with the insertion of the new repair DNA (amplified with glyQ-F1, glyQ-R2 primer pair and evolved clone template DNA) containing the evolved glyQ promoter mutation.
Wild-type allele revertants
The same two stage CRISPR strategy was used to swap evolved alleles with the ancestral alleles in the evolved E. coli clones: E10-1, E10-2, and E7 for fhuA, btuB, and glyQ genes. In the first stage, evolved alleles were swapped with the ampR cassette, before being swapped with the wild-type ancestral allele in the second stage. For glyQ this ampR cassette was inserted in front of the glyQ promoter region.
CRISPR-Cas system transformations
Ancestral MG1655 cells were first transformed with pCas plasmid via electroporation. Cells harbouring pCas grown overnight before a 1 in 100 v/v dilution in medium including kanamycin (50 mg/L). After allowing them to grow for 2 h, λ-Red recombination genes were induced with arabinose (10 mM final concentration) addition. Cells were allowed to grow for another hour and made electro-competent as described previously [59]. Fifty microliters of cells mixed with 100 ng of modified pTargetF plasmids and 1–5 μg of repair DNAs in a 1-mm Gene Pulser cuvettes (Bio-Rad) at 2.5 kV. Cells resuspended in 1 mL of LB medium and allowed to recover at 28 °C for 1 h before being spread onto LB agar plates containing kanamycin (50 mg/L) and spectinomycin (50 mg/L) and incubated at 28 °C overnight. The presence of mutations confirmed with colony PCR and Sanger sequencing.
S. cerevisiae strain modifications
Strain constructions were carried out using CRISPR-Cas for S. cerevisiae, following methods from DiCarlo et al. [60] and Biot-Pelletier and Martin [61]. Both p426-SNR52p-gRNA.CAN1.Y-SUP4t (guide RNA expressing, Plasmid #43803) and p414-TEF1p-Cas9-CYC1t (Cas9 expressing, Plasmid #43802) plasmids were obtained from Addgene. E. coli Top10 cells were used as a cloning host and genetic changes were introduced into the ancestral strain R1158.
As R1158 grows in synthetic media lacking uracil, URA3 marker was changed to a LEU2 marker in p426 plasmid as follows. First, the LEU2 marker was amplified from p415-GalL-Cas9-CYC1t (Addgene, Plasmid #43804) using p415-Leu-F and p415-Leu-R primers (Supplementary Table 2). Second, the backbone of p426 plasmid was amplified using p426-minus-Ura-F and p426-minus-Ura-R primers. Third, we have assembled first two PCR fragments using Circular Polymerase Extension Cloning (CPEC) [62]. Finally, 5 µL of CPEC product was electroporated with 50 µL electro-competent Top10 cells using 1-mm Gene Pulser cuvette (Bio-Rad), at 2.5 kV. Cells were recovered at 37 °C for 1 h before spreading onto LB agar plates containing ampicillin (50 mg/L). The resultant plasmid was called p426-LEU2.
Modification of 20 N sequence in p426-LEU2
Target sequences were chosen for each gene using the online tool CCTop [57] (Supplementary Table 1). Plasmid modifications were carried out in three steps. Step 1 involved the amplification of the backbone of the plasmid without 20 N sequence using p426-F1-reversed and p426-R2-reversed primers (Supplementary Table 2). Step 2 was the construction of DNA fragments with new 20 N sequence. PCR fragments with 20 N target sequences for each gene were generated in three PCR reactions. First PCR used p426-F1 and p426-R1 primers (R1 primers have reverse complement of 20 N sequence specific to each gene at the 5′-end (see Supplementary Table 2 for primers). Second PCR reactions used p426-F2, with 20 N sequence for each gene at the 5′-end, and p426-R2 primers. These two PCR fragments were gel purified and used as templates in the third PCR with the outer primers (p426-F1 and p426-R2) to generate a PCR fragment with a 20 N sequence. In the 3rd step, PCR fragments from step 1 and step 2 were assembled using CPEC [62] and electroporated into Top10 cells and confirmed with Sanger sequencing.
Repair DNA constructions
BUL1 and YFR018C mutants
Mutant alleles for BUL1 and YFR018C were generated in two successive CRISPR events. In the first event, a repair DNA containing 20 N stuffer sequence was inserted [61]. The stuffer repair DNA was generated in three PCR steps. The first PCR used gene-F1 and gene-R1 primers to amplify upstream of the stuffer insertion location. The second PCR used gene-F2 and gene-R2 primers to amplify downstream of the insertion location. Stuffer sequence was used to generate the required homology between the first and the second DNA fragments and the two fragments were joined in the 3rd PCR using gene-F1 and gene-R2 primers. Finally, the stuffer sequence was targeted using stuffer-p426-LEU2 plasmid and swapped with the new repair DNA having exact mutations seen in the evolution experiment. Repair DNA was amplified with a colony PCR from the evolved clones using primers gene-F1 and gene-R2.
TIF35 mutant
TIF35 is an essential gene, requiring a one step CRISPR event. We chose a 20N target sequence such that NGG PAM sequence is the mutation site in the evolved clone. We have targeted this 20N sequence in the ancestor using TIF35-p426-LEU2 plasmid and provided the repair DNA from the evolved clones, amplified with using TIF35-F1 and TIF35-R1 primers.
All transformation events (plasmids and repair DNAs) were carried out using a standard lithium acetate transformation method [63]. The presence of mutations confirmed with colony PCR and Sanger sequencing.
Supplementary information
Acknowledgements
MJM was supported by ARC Discovery Grant (DP180102161) and an ARC Future Fellowship (FT170100441).
Author contributions
MJM conceived the study, JNB, ALS, JMV, TDA and MJM designed and carried out experiments, JNB, ALS, LCW and MJM analysed the data. JNB and MJM wrote the paper. All authors have read and approved the final manuscript.
Data availability statement
Raw sequencing reads have been deposited in the NCBI Biosample database with Bioproject ID: PRJNA668197
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jake N. Barber, Aysha L. Sezmis
Supplementary information
The online version of this article (10.1038/s41396-020-00810-z) contains supplementary material, which is available to authorized users.
References
- 1.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–50. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Gause GF. Experimental analysis of Vito Volterra’s mathematical theory of the struggle for existence. Science. 1934;79:16–7. doi: 10.1126/science.79.2036.16-a. [DOI] [PubMed] [Google Scholar]
- 3.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66. [Google Scholar]
- 4.Lankau RA. Rapid evolutionary change and the coexistence of species. Annu Rev Ecol Evol Syst. 2011;42:335–54. [Google Scholar]
- 5.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–41. [Google Scholar]
- 6.Hubbell SP. Neutral theory and the evolution of ecological equivalence. Ecology. 2006;87:1387–98. doi: 10.1890/0012-9658(2006)87[1387:ntateo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.MacArthur R, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Nat. 1967;101:377–85. [Google Scholar]
- 8.Bailey JK, Hendry AP, Kinnison MT, Post DM, Palkovacs EP, Pelletier F, et al. From genes to ecosystems: an emerging synthesis of eco-evolutionary dynamics. N Phytol. 2009;184:746–9. doi: 10.1111/j.1469-8137.2009.03081.x. [DOI] [PubMed] [Google Scholar]
- 9.Baquero F, Blázquez J. Evolution of antibiotic resistance. Trends Ecol Evol. 1997;12:482–7. doi: 10.1016/s0169-5347(97)01223-8. [DOI] [PubMed] [Google Scholar]
- 10.Grant PR, Grant BR. Evolution of character displacement in Darwin’s finches. Science. 2006;313:224–6. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 11.Koeppel AF, Wertheim JO, Barone L, Gentile N, Krizanc D, Cohan FM. Speedy speciation in a bacterial microcosm: new species can arise as frequently as adaptations within a species. ISME J. 2013;7:1080–91. doi: 10.1038/ismej.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart YE, Campbell T, Hohenlohe P, Reynolds RG, Revell L, Losos J. Rapid evolution of a native species following invasion by a congener. Science. 2014;346:463–6. doi: 10.1126/science.1257008. [DOI] [PubMed] [Google Scholar]
- 13.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci USA. 2010;107:7359–64. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pimentel D, Feinberg EH, Wood PW, Hayes JT. Selection, spatial distribution, and the coexistence of competing fly species. Am Nat. 1965;99:97–109. [Google Scholar]
- 15.Hart SP, Turcotte MM, Levine JM. Effects of rapid evolution on species coexistence. Proc Natl Acad Sci USA. 2019;116:2112–7. doi: 10.1073/pnas.1816298116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA. 2012;110:250–9. doi: 10.1073/pnas.1207025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner CB, Marshall CW, Cooper VS. Parallel genetic adaptation across environments differing in mode of growth or resource availability. Evolution Lett. 2018;2:355–67. doi: 10.1002/evl3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elena SF, Lenski RE. Microbial genetics: evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 19.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson GC, Bertels F, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. V. Insight into the niche specialist fuzzy spreader compels revision of the model Pseudomonas radiation. Genetics. 2013;195:1319–35. doi: 10.1534/genetics.113.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frenkel EM, McDonald MJ, Van Dyken JD, Kosheleva K, Lang GI, Desai MM. Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proc Natl Acad Sci USA. 2015;112:11306–11. doi: 10.1073/pnas.1506184112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuppinger-Dingley D, Schmid B, Petermann JS, Yadav V, De Deyn GB, Flynn DF. Selection for niche differentiation in plant communities increases biodiversity effects. Nature. 2014;515:108. doi: 10.1038/nature13869. [DOI] [PubMed] [Google Scholar]
- 23.Le Gac M, Plucain J, Hindré T, Lenski RE, Schneider D. Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2012;109:9487–92. doi: 10.1073/pnas.1207091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenzweig RF, Sharp R, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–17. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–58. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Souza G, Kost C. Experimental evolution of metabolic dependency in bacteria. PLoS Genet. 2016;12:e1006364. doi: 10.1371/journal.pgen.1006364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci USA. 2014;111:17941–6. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallus Y, Miller JH, Libby E. Paradoxes in leaky microbial trade. Nat Commun. 2017;8:1361. doi: 10.1038/s41467-017-01628-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller EE, Faust K, Widder S, Herold M, Arbas SM, Wilmes P. Using metabolic networks to resolve ecological properties of microbiomes. Curr Opin Syst Biol. 2018;8:73–80. [Google Scholar]
- 30.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1:0109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 31.Harcombe WR, Betts A, Shapiro JW, Marx CJ. Adding biotic complexity alters the metabolic benefits of mutualism. Evolution. 2016;70:1871–81. doi: 10.1111/evo.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harcombe WR, Chacón JM, Adamowicz EM, Chubiz LM, Marx CJ. Evolution of bidirectional costly mutualism from byproduct consumption. Proc Natl Acad Sci USA. 2018;115:12000–4. doi: 10.1073/pnas.1810949115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrell JM, Brown SP. Evolution of bacterial trade in a two-species community. Proc Natl Acad Sci USA. 2018;115:11874–5. doi: 10.1073/pnas.1817344115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris JJ. Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 2015;31:475–82. doi: 10.1016/j.tig.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–64. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adler PB, HilleRisLambers J, Levine JM. A niche for neutrality. Ecol Lett. 2007;10:95–104. doi: 10.1111/j.1461-0248.2006.00996.x. [DOI] [PubMed] [Google Scholar]
- 39.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc R Soc Lond Ser B: Biol Sci. 2002;269:931–6. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccardi P, Vessman B, Mitri S. Toxicity drives facilitation between 4 bacterial species. Proc Natl Acad Sci USA. 2019;116:15979–84. doi: 10.1073/pnas.1906172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goddard MR, Greig D. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 2015;15:fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Elsas JD, Semenov AV, Costa R, Trevors JT. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 2011;5:173–83. doi: 10.1038/ismej.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM. The dynamics of molecular evolution over 60,000 generations. Nature. 2017;551:45. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–61. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratzke C, Barrere J, Gore J. Strength of species interactions determines biodiversity and stability in microbial communities. Nat Ecol Evol. 2020;4:376–83. doi: 10.1038/s41559-020-1099-4. [DOI] [PubMed] [Google Scholar]
- 47.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 48.Branco P, Francisco D, Chambon C, Hebraud M, Arneborg N, Almeida MG, et al. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl Microbiol Biotechnol. 2014;98:843–53. doi: 10.1007/s00253-013-5411-y. [DOI] [PubMed] [Google Scholar]
- 49.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howard SP, Herrmann C, Stratilo CW, Braun V. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J Bacteriol. 2001;183:5885–95. doi: 10.1128/JB.183.20.5885-5895.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 52.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33:377. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genom. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall BG, Acar H, Nandipati A, Barlow M. Growth rates made easy. Mol Biol Evol. 2014;31:232–8. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 55.Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016;17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–14. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Cheng Q-X, Liu A-M, Zhao G-P, Wang J. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol. 2017;8:812. doi: 10.3389/fmicb.2017.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biot-Pelletier D, Martin VJ. Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9. J Biol Eng. 2016;10:6. doi: 10.1186/s13036-016-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads have been deposited in the NCBI Biosample database with Bioproject ID: PRJNA668197