Abstract
The overwhelming taxonomic diversity and metabolic complexity of microorganisms can be simplified by a life-history classification; copiotrophs grow faster and rely on resource availability, whereas oligotrophs efficiently exploit resource at the expense of growth rate. Here, we hypothesize that community-level traits inferred from metagenomic data can distinguish copiotrophic and oligotrophic microbial communities. Moreover, we hypothesize that oligotrophic microbial communities harbor more unannotated genes. To test these hypotheses, we conducted metagenomic analyses of soil samples collected from copiotrophic vegetated areas and from oligotrophic bare ground devoid of vegetation in an arid-hyperarid region of the Sonoran Desert, Arizona, USA. Results supported our hypotheses, as we found that multiple ecologically informed life-history traits including average 16S ribosomal RNA gene copy number, codon usage bias in ribosomal genes and predicted maximum growth rate were higher for microbial communities in vegetated than bare soils, and that oligotrophic microbial communities in bare soils harbored a higher proportion of genes that are unavailable in public reference databases. Collectively, our work demonstrates that life-history traits can distill complex microbial communities into ecologically coherent units and highlights that oligotrophic microbial communities serve as a rich source of novel functions.
Subject terms: Microbial ecology, Community ecology
Introduction
Microorganisms represent the vast majority of organisms and possess enormous functional versatility [1, 2]. They are essential to a multitude of biogeochemical cycles and have a profound role in ecosystem functioning [3]. This tremendous complexity poses a daunting challenge to building a general understanding of the ecological role of microbes. Such generalizations could be achieved if we simplify the complexity by categorizing the microbes into ecological groups on the basis of shared morphological, physiological, or life-history traits [4, 5]. A classic life-history classification of microbes is the copiotroph-oligotroph dichotomy. Copiotrophs grow faster and rely on resource availability, whereas oligotrophs efficiently exploit resource at the expense of growth rate [6]. Therefore, these distinct life-history strategies represent a fundamental trade-off between growth rate and resource use efficiency. This life-history framework enables us to directly link microbial performance to environmental regimes; copiotrophs grow faster in resource-rich environments, whereas oligotrophs grow slower in resource-poor environments [7, 8].
Deciphering the genomic underpinnings of microbial life-history strategies requires a consensus on genomic traits that distinguish copiotrophs and oligotrophs. One of the most often invoked traits is copy number of ribosomal RNA (rRNA) genes [9]. Copiotrophic bacteria are hypothesized to encode more rRNA copies in their genomes to promote the higher ribosomal content required by rapid growth [10]. Fast-growing copiotrophs are also predicted to enhance the usage of synonymous codons (i.e., different codons encoding the same amino acid) in their ribosomal genes because they underwent translational selection, leading to greater codon usage bias in the ribosomal genes of copiotrophs [11]. Thus, greater codon usage bias is typically found to be associated with higher maximum growth rate [12], a pivotal trait representing the ability of copiotrophs to grow rapidly in response to resource pulses. With the advances in shotgun metagenomic sequencing, community-level measurements of these life-history traits have been developed to improve our perception of the ecological strategies of complex microbial communities [12, 13]. We hypothesize that these traits can serve as important life-history traits to facilitate the differentiation of copiotrophic and oligotrophic microbial communities. Such knowledge will provide a rigorous framework for trait-based understanding of microbial communities.
Copiotrophs and oligotrophs differ in a myriad of functional genes encoding metabolic pathways, environmental sensing, substrate uptake mechanisms, among others [14]. These functional attributes provide critical insights into the genetic mechanisms underlying the adaptive responses of copiotrophs and oligotrophs to resource availability. However, a considerable fraction of genes remains unannotated [15]. The unannotated genes can account for ~80% of the total gene pool in microbial communities [16]. The root of this limitation is that functional annotation relies on reference databases (e.g., [17]), which are mostly based on cultured organisms and heavily biased toward a relatively small set of model organisms. However, most bacteria are still uncultured [18] and we do not even have cultivated isolates and genomic information for the most cosmopolitan bacteria [19]. The massive incompleteness of reference genome databases hinders our ability to chart the full extent of functional capacities of copiotrophic and oligotrophic microbial communities, and our inability to characterize the unannotated genes might obscure important ecological patterns [20]. Given that oligotrophs are known to be challenging to culture due to their small sizes and slow growth [21, 22], we hypothesize that oligotrophic microbial communities harbor a higher proportion of unannotated genes.
We contend that hyperarid ecosystems provide a unique opportunity to test our hypotheses within a single environment. Arid landscapes are typically characterized by vegetation patchiness, in which bare ground is interspersed with sparse vegetation spots [23]. Soils beneath vegetation patches show higher nutrient levels and water storage capacity [24]. In contrast, soils in adjacent bare ground devoid of vegetation are exposed to higher rates of soil erosion, higher temperature and more intense radiation, leading to nutrient depletion [25]. In this study, we conducted metagenomic analyses of 24 soil samples collected from four sites (each site had three samples beneath vegetation patches and three samples in bare ground) in western Arizona of the Sonoran Desert (Fig. S1). By calculating average 16S rRNA copy number, codon usage bias in ribosomal genes and maximum growth rate using metagenomic data, we would expect that these trait values are higher in copiotrophic vegetated soils than oligotrophic bare soils. Moreover, we would expect that microbial communities in bare soils harbor more unannotated genes.
Materials and methods
Study sites and sampling
The study was conducted in western Sonoran Desert, Arizona, USA; a region in which vegetation density is controlled by increasing aridity [26]. In October 2017, soil samples were collected from four sites along a 77 km north-south transect in the Sonoran Desert (Fig. S1). The transect traversed an arid-hyperarid region with aridity index ranging from 0.041 to 0.052 (Kushwaha et al. unpublished data). At each site, three 30-m transects were established. From each transect, two composite soil samples (each consists of three samples at 0–20 cm depth, and they were collected from equidistant locations along the transect) under vegetation patches and from bare ground area devoid of vegetation were collected. Biological crusts were not present in any of the locations sampled. The shrub species Larrea tridentata and Ambrosia dumosa comprised the dominant vegetation (~70%). A total of 24 soil samples were collected. Samples were sealed in sterilized plastic bags, and they were immediately transported on ice to the laboratory. Soil samples were sieved through 2-mm mesh and homogenized, and any visible living plant material (e.g., roots) was removed. For each sample, a subsample was placed into a sterilized tube and stored at −80 °C until DNA extraction, and the remaining soils were used for soil physiochemical analyses.
Molecular analyses and sequencing
Total soil genomic DNA was extracted using a Fast DNA SPIN for Soil Kit™ (MP Biomedicals, Solon, OH) and further purified using a DNeasy PowerClean Pro Cleanup Kit (Qiagen, Hilden, Germany) according to the manufacturers’ instructions. To assess the taxonomic profiles of bacterial and archaeal communities, the V4 region of the 16S rRNA gene was sequenced using Illumina MiSeq platform (Supplementary methods). The amplicon sequence variants (16S rRNA phylotypes hereafter) were obtained using DADA2 pipeline (Supplementary methods; [27]).
To assess the functional attributes of microbial communities (Supplementary methods), total genomic DNA was fragmented and ligated to Illumina adapters using the QIAseq FX DNA Library Kit (Qiagen, Hilden, Germany). The quality and quantity of all libraries were determined with Agilent 4150 TapeStation DNA bioanalyzer. All samples were shotgun-sequenced on a 2 × 150 bp Illumina NextSeq550 platform at the Microbiome Core, Steele Children Research Center, University of Arizona.
Metagenomic data analyses
Raw reads were preprocessed by removing adapters with Cutadapt v. 2.1 [28]. Reads shorter than 50 bp and low-quality bases were removed using Trimmomatic v. 0.38 [29]. A total of 69,333,926 to 356,676,854 reads per sample remained after quality trimming (Table S1). Reads from each sample were de novo assembled using Megahit v. 1.1.4 [30], with the k-mer length increasing from 21 to 141 in steps of 20. The contig N50 ranged from 715 to 964 bp among samples (Table S1). Protein-coding genes were predicted on assembled contigs longer than 500 bp using Prodigal v. 2.6 [31]. After discarding genes shorter than 100 bp, genes from all samples were clustered at ≥95% identity and ≥90% overlap with MMseqs2 [32]. This resulted in a catalog containing 32,930,898 non-redundant genes. Paired-end reads of each sample were mapped to the gene catalog using BWA v. 0.7.16 [33]. Functional information of genes was annotated by comparison to Kyoto Encyclopedia of Genes and Genomes (KEGG) database using GhostKOALA because KEGG database is one of the most widely used reference databases for pathway mapping [34]. Moreover, genes were annotated with the eggNOG database using eggNOG-mapper [35, 36].
Calculation of life-history traits
For each soil sample, we calculated a set of traits from metagenomic data to shed light on the microbial life-history strategies. First, we estimated the average 16S rRNA copy number and average genome size as described in Pereira-Flores et al. [13]. Briefly, the number of genomes was first estimated as the mean coverage of the 35 single-copy genes (Supplementary methods). The average 16S rRNA copy number was estimated as the coverage of 16S rRNA genes divided by the number of genomes. The average genome size was estimated as the number of base pairs divided by the number of genomes. Second, we calculated the codon usage bias in ribosomal genes and minimum generation time (hours, h) as described in Vieira-Silva & Rocha [12]. Briefly, the ribosomal genes were retrieved by blasting on a database of ribosomal proteins of all sequenced genomes [12]. The codon usage bias in each ribosomal gene was calculated using effective number of codons (ENC′) [37]. The codon usage bias of a community was calculated as the inverse mean ENC′ of all ribosomal genes. The minimum generation time was predicted using codon usage bias as described in Vieira-Silva & Rocha [12] (available at https://galaxy.pasteur.fr/?tool_id=toolshed.pasteur.fr%2Frepos%2Fkhillion%2Fgrowthpred%2Fgrowthpred%2F1.07&version=1.07&__identifer=unl8nld8ngn). We then calculated the maximum growth rate as the inverse of minimum generation time (h−1) [38]. Third, we calculated the GC content and the variance of GC content of the quality-filtered reads as described in Barberán et al. [39].
Statistical analyses
Statistical analyses were implemented in R [40]. To test the differences in microbial life-history traits between bare and vegetated soils, we used linear mixed-effects models to fit life-history trait values as a function of soil environment (i.e., bare and vegetated soils), using lme4 and lmerTest packages. Soil environment was used as a fixed effect, and site was treated as a random effect. R2-value of the fixed effect was estimated following Nakagawa and Schielzeth [41]. We used a negative binomial distribution model as implemented in DESeq2 package [42] to identify KEGG pathways that were significantly abundant in bare or vegetated soils. P-values for multiple testing were corrected using the Benjamini–Hochberg method [43]. To explore the potential sources of unannotated genes in bare and vegetated soils, we examined the correlation between the proportion of genes without KEGG annotation and the proportion of phylotypes that were unclassified at different taxonomic levels (i.e., phylum, class, order, family, genus and species). To examine whether our results are robust to the use of functional annotation database, we calculated the proportion of genes without eggNOG annotation and repeated the correlation analyses.
Results and discussion
Life-history traits
The contents of total organic carbon, dissolved organic carbon, dissolved nitrogen and bioavailable phosphorus were significantly higher in vegetated than bare soils (Fig. S2). The higher soil nutrient availability in vegetated areas is mainly ascribed to the effects of desert plants such as leaf litter and root exudates on soil nutrients, which is known as the fertility island effect [44, 45]. In contrast, this effect is absent in bare ground devoid of vegetation. In particular, the low nitrogen availability in bare ground is considered to be a primary limiting factor to ecosystem productivity in arid ecosystems [46]. Therefore, our results indicate that vegetated soils represent a copiotrophic environment and bare soils represent an oligotrophic environment.
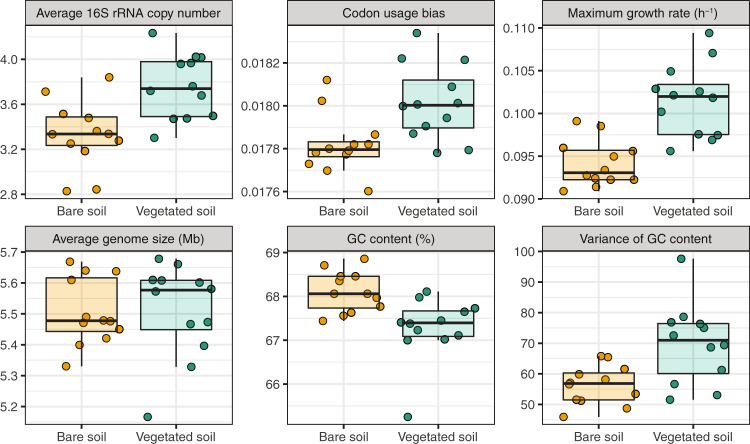
Microbial communities in vegetated soils had a higher average 16S rRNA gene copy number than those in bare soils (linear mixed-effects model, F1,19 = 12.91, P = 0.002, R2 = 0.36; Fig. 1), suggesting that soil microbes in vegetated areas have a higher cellular ribosome content than microbes in bare soils. In addition, microbes beneath vegetation patches exhibited a greater codon usage bias in their ribosomal genes (F1,19 = 10.40, P = 0.004, R2 = 0.28; Fig. 1). The greater codon usage bias in vegetated soils represented a differential preference of the ribosomal genes’ codons because ribosomal genes are highly expressed during fast growth [12]. As a result, we found that maximum growth rate was higher for microbial communities in vegetated soils than those in bare soils (F1,19 = 28.30, P = 3.91 × 10−5, R2 = 0.53; Fig. 1). Result of these trait comparisons suggest that high-resource conditions in vegetated soils favored copiotrophic microbial communities and bare soils selected for oligotrophic microbial communities adapted to resource scarcity. Consistent with our hypothesis, these life-history traits play pivotal roles in distinguishing copiotrophic and oligotrophic microbial communities.
Fig. 1. Microbial life-history traits.
Box plots show the comparisons of life-history trait values between bare and vegetated soils. The medians in these box plots are as follows (bare soil vs vegetated soil): average 16S rRNA copy number (3.34 vs 3.74), codon usage bias (0.0178 vs 0.0180), maximum growth rate (0.093 h−1 vs 0.102 h−1), average genome size (5.48 Mb vs 5.58 Mb), GC content (68.06% vs 67.39%), variance of GC content (56.84 vs 70.96).
Average genome size was not significantly different between microbial communities in bare and vegetated soils (F1,19 = 0.01, P = 0.91, R2 = 0.001; Fig. 1). Most studies in aquatic habitats showed that oligotrophs tend to have smaller genomes [14, 47], whereas a grassland study demonstrated that soil copiotrophs have smaller genomes [48]. The mixed evidence provided by previous studies and our observation might reflect a habitat difference of selective pressure on genome size, different methods of genome size estimation, or a disparity between community and species/genome level comparison. Moreover, microbial communities in bare soils had a higher GC content (F1,19 = 15.57, P = 0.0009, R2 = 0.31; Fig. 1). A higher genomic GC content can enhance the thermotolerance of a microorganism because the GC base pair is known for its high thermal stability [49, 50]. This thermotolerance ability is particularly relevant for oligotrophs in bare soils because they are exposed to higher radiation and temperature [44]. The higher GC content in bare soils was also mirrored by the greater relative abundance of Actinobacteria (Fig. S3). Members of Actinobacteria represent a well-defined clade of GC-rich taxa, and their exclusive tolerance to desiccation facilitates their widespread distribution in desert soils [51]. Furthermore, bare soil microbial communities had a lower variance of genomic GC content (F1,19 = 16.97, P = 0.0006, R2 = 0.30; Fig. 1), suggesting that the stressful environment in bare soils enriched microbes with similar GC contents and limited their variability.
Trait-based approaches have elicited recent interest in microbial ecology, but the complexities of microbial traits and trait measurements make the integration and generalization among trait-based microbial studies challenging [52]. Moreover, it has been well-recognized that microbes can be differentiated into broad ecological categories according to a copiotroph-oligotroph framework [7]. On the one hand, average 16S rRNA copy number, codon usage bias in ribosomal genes and maximum growth rate can serve as universal and mechanistic life-history traits to classify microbial communities as fast-growing copiotrophic communities or slow-growing oligotrophic communities. More generally, these life-history traits can extend the copiotroph-oligotroph dichotomy to a continuum running from copiotrophic to oligotrophic microbial communities, which is analogous to the “fast-slow” plant economics spectrum [53]. We advocate that grounding such a trait-based approach into an established life-history framework would help to develop a more mechanistic picture of microbial responses to changing environmental conditions. For example, recent research has shown that shifts in microbial growth-related traits can explain successional trajectories associated with changes in resource availability [54, 55]. On the other hand, average genome size, GC content, and GC variance represent habitat-specific properties of copiotrophic and oligotrophic communities. For example, GC content and GC variance reflect thermotolerance in desert soils. It is worth noting, however, that these traits do not take into account the abundance of particular species or the intraspecies trait variance, a limitation that should be addressed in future studies. Despite this limitation, these traits facilitate the comparisons of microbial life-history strategies across contrasting environments and hold great promise for distilling complex microbial communities into important ecological attributes.
Functional attributes
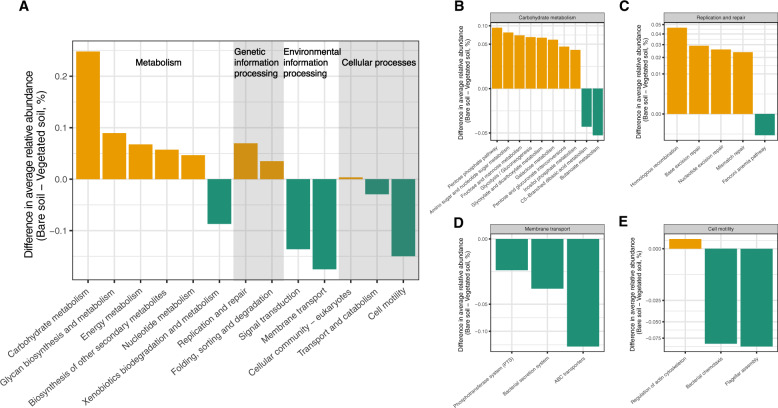
Microbial communities with contrasting life-history strategies exhibited different functional attributes (Fig. 2). Bare soil microbes had greater relative abundances of genes associated with metabolism functions (Fig. 2a), suggesting that metabolic versatility is an essential trait of oligotrophic microbial communities. This metabolic versatility likely reflects an important adaptive strategy of oligotrophs in coping with resource scarcity. For example, although plant-derived carbon inputs are rare in bare soils, bare soil microbes had more abundant genes associated with metabolizing a broad range of saccharides including pentose, amino sugars, nucleotide sugars, among others (Fig. 2b). This observation is in line with a previous study showing that oligotrophs might be capable of scavenging a broad range of carbon substrates [56]. Moreover, microbes in bare soils had more abundant genes encoding homologous recombination, base excision repair, nucleotide excision repair, and mismatch repair (Fig. 2c), likely representing stress-tolerant traits enabling oligotrophs to maintain genome integrity by preventing radiation-induced DNA damage in bare soils [44]. We further found that maximum growth rate was negatively associated with relative abundances of genes involved in DNA repair (Fig. S4). This result reflects a trade-off between growth rate and resistance [57], where oligotrophs allocate more resources into stress-resistance at the expense of growth rates [58].
Fig. 2. Functional attributes of microbial communities.
Bar plots represent the differences in average relative abundances of KEGG pathways between bare and vegetated soils. Only those KEGG pathways that were significantly more abundant in bare soils (yellow) or vegetation soils (green) according to DESeq2 analysis (P < 0.05 after FDR correction) are shown.
Microbial communities in vegetated soils had greater relative abundances of genes involved in environmental information processing (Fig. 2a). The inflated relative abundances of genes encoding ATP-binding cassette transporters (ABC-transporters) and transporters in the phosphotransferase system (PTS; Fig. 2d) would facilitate copiotrophs to uptake readily available extracellular compounds in soils or plant root exudates [59]. The enrichment of genes encoding two-component system (Fig. S5) might facilitate copiotrophs to sense and respond to changes in environmental conditions. In addition, a larger proportion of genes encoding flagellar assembly and chemotaxis (Fig. 2e) might represent motility-enabling traits of copiotrophs in vegetated soils. While cell motility is widely suggested as an important trait for copiotrophs in aquatic habitats [14], it might be important for copiotrophs in vegetated soils to be recruited as members of microbial communities in rhizosphere [60].
Potential sources of unannotated genes
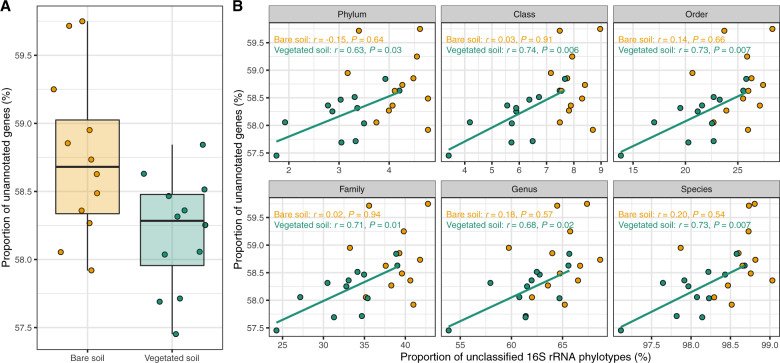
Results of KEGG annotation showed that the proportion of unannotated genes ranged from 57 to 60% among samples (Fig. 3a), suggesting that a substantial fraction of functional attributes remain poorly described in desert soil microbes. The proportion of unannotated genes was higher for microbial communities in bare soils than those in vegetated soils (F1,19 = 6.97, P = 0.01, R2 = 0.23; Fig. 3a, see a consistent pattern for genes without eggNOG annotation in Fig. S6). In addition, bare soils harbored a higher proportion of 16S rRNA phylotypes that were unclassified at different taxonomic levels (Fig. S7). These results can be attributed to the fact that oligotrophs are less readily culturable than copiotrophs, and thus their taxonomic and functional information are underrepresented in current reference databases [61]. Therefore, bare soils serve as a reservoir for uncharacterized microbial taxonomic and functional diversity in arid ecosystems. In support of our hypothesis, these results indicate that oligotrophic microbial communities harbor more unannotated genes than copiotrophic microbial communities.
Fig. 3. Potential sources of unannotated genes.
For each sample, the proportion of unannotated genes was measured as the number of genes without KEGG annotation divided by the total number of genes, the proportions of unclassified 16S rRNA phylotypes at different taxonomic levels were measured as the number of unclassified 16S rRNA phylotypes divided by the total number of 16S rRNA phylotypes. a Comparison of the proportion of genes without KEGG annotation in bare (median = 58.68%) and vegetated soils (median = 58.28%). b Relating the proportion of genes without KEGG annotation to the proportion of unclassified phylotypes at different taxonomic levels. Results of Pearson correlations are shown.
Experimental characterization of gene-function relationship is the most promising way to tackle the unannotated genes, but it is labor-intensive [62]. The development of a strategy to identify which taxa the unannotated genes most likely arise from can provide practical hints on prioritizing microbial members for experimental investigations. In this study, we found that the primary sources of unannotated genes were different in copiotrophic and oligotrophic microbial communities (Fig. 3b). For the copiotrophic communities in vegetated soils, we found that the proportion of unannotated genes was positively related to the proportion of unclassified 16S rRNA phylotypes at different taxonomic levels (Fig. 3b, Fig. S6), suggesting that the unannotated genes in copiotrophs might originate from unclassified copiotrophs. In other words, taxonomically known copiotrophs are most likely to have available genomic information. While we do not exclude the possibility that there are some copiotrophs that are taxonomically known but have unavailable reference genomes, our results suggest that the most productive way to obtain novel genes of copiotrophs is to culture isolates of taxonomically unknown copiotrophs and investigate their gene functions [63]. In contrast, in the oligotrophic communities in bare soils we found that the proportion of unannotated genes was not related to the proportion of unclassified 16S rRNA phylotypes (Fig. 3b, Fig. S6). A plausible explanation is that the unannotated genes in oligotrophs come from two sources: (i) oligotrophs that are taxonomically unknown, and (ii) oligotrophs that are taxonomically known but have unavailable genomic information. Thus, the novel genes of oligotrophs are hiding in plain sight: we should preferentially investigate the genes of taxonomically known oligotrophs in hand. These correlational analyses provide a preliminary strategy for unraveling unannotated genes, more complicated computation approaches are required to prioritize microbial lineages that enrich unannotated genes.
Conclusions and perspectives
Metagenomics has greatly expanded our ability to characterize the functional capacities of microorganisms, but it remains limited in providing new ecological insights. We demonstrated that multiple ecologically informed traits inferred from metagenomic data can advance the trait-based characterization of microbial communities within a copiotroph-oligotroph framework. This trait-based classification opens new opportunities for reducing the complexity of microbial communities. For example, microbial communities that vary across different geographic regions can be simplified by the classification of community-level ecological strategies [64]. In addition, we showed that oligotrophic microbial communities have a higher proportion of functionally unknown genes than copiotrophic microbial communities. We provided a preliminary strategy for unraveling these unknown genes. In copiotrophic communities, the priority should be placed on taxonomically unknown phylotypes, whereas in oligotrophic communities the priority should be placed on phylotypes that are taxonomically known. With the increasing amount of metagenomic data being generated across a wide range of ecosystems, an important avenue for future work will be to facilitate the global integration and generalization of our results.
Supplementary information
Acknowledgements
We thank members of the Barberán lab and three anonymous reviewers for providing constructive comments on this manuscript. We acknowledge the help of Bradley Butterfield with site selection. We acknowledge the help of Daniel Laubitz with shotgun metagenomic sequencing. An allocation of computer resource from the UA Research Computing High Performance Computing (HPC) at the University of Arizona is gratefully acknowledged. The research was supported by the National Institute of Environmental Health Sciences grant P42ES004940 and University of Arizona’s Research, Discovery, and Innovation Accelerate for Success Grant 2018-2019. The research was also supported by USDA-NIFA grant 2017-68005-26867, Sustainable Bioeconomy for Arid Lands. Opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of US Department of Agriculture.
Author contributions
JWN, RMM, and AB conceived the research; YC and PK performed laboratory analyses; YC analyzed the data and, along with AB, led the writing of the manuscript; All authors contributed critically to manuscript writing and revisions.
Data availability
The sequence data have been deposited in the NCBI Sequence Read Archive under BioProject accession code PRJNA664514.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongjian Chen, chenyj@email.arizona.edu.
Albert Barberán, barberan@email.arizona.edu.
Supplementary information
The online version of this article (10.1038/s41396-020-00803-y) contains supplementary material, which is available to authorized users.
References
- 1.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 2.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardgett RD, van der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515:505–11. doi: 10.1038/nature13855. [DOI] [PubMed] [Google Scholar]
- 4.Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–43. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 5.Martiny JBH, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: a phylogenetic perspective. Science. 2015;350:aac9323. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 6.Koch AL. Oligotrophs versus copiotrophs. BioEssays. 2001;23:657–61. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- 7.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–64. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 8.Ho A, Di Lonardo DP, Bodelier PLE. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol Ecol. 2017;93:fix006. doi: 10.1093/femsec/fix006. [DOI] [PubMed] [Google Scholar]
- 9.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–33. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roller BRK, Stoddard SF, Schmidt TM. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol. 2016;1:1–7. doi: 10.1038/nmicrobiol.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botzman M, Margalit H. Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 2011;12:R109. doi: 10.1186/gb-2011-12-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieira-Silva S, Rocha EPC. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 2010;6:e1000808. doi: 10.1371/journal.pgen.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira-Flores E, Glöckner FO, Fernandez-Guerra A. Fast and accurate average genome size and 16S rRNA gene average copy number computation in metagenomic data. BMC Bioinforma. 2019;20:453. doi: 10.1186/s12859-019-3031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–33. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman SK, Avila-Herrera A, Nayfach S, Pollard KS. A most wanted list of conserved microbial protein families with no known domains. PLoS ONE. 2018;13:e0205749. doi: 10.1371/journal.pone.0205749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galand PE, Pereira O, Hochart C, Auguet JC, Debroas D. A strong link between marine microbial community composition and function challenges the idea of functional redundancy. ISME J. 2018;12:2470–8. doi: 10.1038/s41396-018-0158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steen AD, Crits-Christoph A, Carini P, DeAngelis KM, Fierer N, Lloyd KG, et al. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–30. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–5. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 20.Jaroszewski L, Li Z, Krishna SS, Bakolitsa C, Wooley J, Deacon AM, et al. Exploration of uncharted regions of the protein universe. PLoS Biol. 2009;7:e1000205. doi: 10.1371/journal.pbio.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giovannoni S, Stingl U. The importance of culturing bacterioplankton in the ‘omics’ age. Nat Rev Microbiol. 2007;5:820–6. doi: 10.1038/nrmicro1752. [DOI] [PubMed] [Google Scholar]
- 22.Barberán A, Caceres Velazquez H, Jones S, Fierer N. Hiding in plain sight: Mining bacterial species records for phenotypic trait information. mSphere. 2017;2:e00237–17. doi: 10.1128/mSphere.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguiar MR, Sala OE. Patch structure, dynamics and implications for the functioning of arid ecosystems. Trends Ecol Evol. 1999;14:273–7. doi: 10.1016/s0169-5347(99)01612-2. [DOI] [PubMed] [Google Scholar]
- 24.Schlesinger WH, Raikes JA, Hartley AE, Cross AF. On the spatial pattern of soil nutrients in desert ecosystems. Ecology. 1996;77:364–74. [Google Scholar]
- 25.Maestre FT, Bautista S, Cortina J, Bellot J. Potential for using facilitation by grasses to establish shrubs on a semiarid degraded steppe. Ecol Appl. 2001;11:1641–55. [Google Scholar]
- 26.Butterfield BJ, Betancourt JL, Turner RM, Briggs JM. Facilitation drives 65 years of vegetation change in the Sonoran Desert. Ecology. 2010;91:1132–9. doi: 10.1890/09-0145.1. [DOI] [PubMed] [Google Scholar]
- 27.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. [Google Scholar]
- 29.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 31.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinegger M, Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35:1026–8. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–14. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–4. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- 38.Vieira-Silva S, Falony G, Darzi Y, Lima-Mendez G, Yunta RG, Okuda S, et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat Microbiol. 2016;1:1–8. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 39.Barberán A, Fenández-Guerra A, Bohannan BJ, Casamayor EO. Exploration of community traits as ecological markers in microbial metagenomes. Mol Ecol. 2012;21:1909–17. doi: 10.1111/j.1365-294X.2011.05383.x. [DOI] [PubMed] [Google Scholar]
- 40.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2018. https://www.R-project.org/.
- 41.Nakagawa S, Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol. 2013;4:133–42. [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 44.Goberna M, Navarro‐Cano JA, Valiente‐Banuet A, García C, Verdú M. Abiotic stress tolerance and competition‐related traits underlie phylogenetic clustering in soil bacterial communities. Ecol Lett. 2014;17:1191–201. doi: 10.1111/ele.12341. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez-Echeverría S, Lozano YM, Bardgett RD. Influence of soil microbiota in nurse plant systems. Funct Ecol. 2016;30:30–40. [Google Scholar]
- 46.Yahdjian L, Gherardi L, Sala OE. Nitrogen limitation in arid-subhumid ecosystems: a meta-analysis of fertilization studies. J Arid Environ. 2011;75:675–80. [Google Scholar]
- 47.Giovannoni SJ, Thrash JC, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL, et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci USA. 2015;112:10967–72. doi: 10.1073/pnas.1508382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musto H, Naya H, Zavala A, Romero H, Alvarez-Valín F, Bernardi G. Correlations between genomic GC levels and optimal growth temperatures in prokaryotes. FEBS Lett. 2004;573:73–7. doi: 10.1016/j.febslet.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 50.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–74. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neilson JW, Quade J, Ortiz M, Nelson WM, Legatzki A, Tian F, et al. Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles. 2012;16:553–66. doi: 10.1007/s00792-012-0454-z. [DOI] [PubMed] [Google Scholar]
- 52.Lajoie G, Kembel SW. Making the most of trait-based approaches for microbial ecology. Trends Microbiol. 2019;27:814–23. doi: 10.1016/j.tim.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Reich PB. The world-wide ‘fast-slow’ plant economics spectrum: a traits manifesto. J Ecol. 2014;102:275–301. [Google Scholar]
- 54.Nemergut DR, Knelman JE, Ferrenberg S, Bilinski T, Melbourne B, Jiang L, et al. Decreases in average bacterial community rRNA operon copy number during succession. ISME J. 2016;10:1147–56. doi: 10.1038/ismej.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortiz-Álvarez R, Fierer N, de Los Ríos A, Casamayor EO, Barberán A. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J. 2018;12:1658–67. doi: 10.1038/s41396-018-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song H-K, Song W, Kim M, Tripathi BM, Kim H, Jablonski P, et al. Bacterial strategies along nutrient and time gradients, revealed by metagenomic analysis of laboratory microcosms. FEMS Microbiol Ecol. 2017;93:fix114. doi: 10.1093/femsec/fix114. [DOI] [PubMed] [Google Scholar]
- 57.Ferenci T. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 2016;24:209–23. doi: 10.1016/j.tim.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 58.Gray DA, Dugar G, Gamba P, Strahl H, Jonker MJ, Hamoen LW. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat Commun. 2019;10:890. doi: 10.1038/s41467-019-08719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trivedi P, Anderson IC, Singh BK. Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 2013;21:641–51. doi: 10.1016/j.tim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Müller DB, Vogel C, Bai Y, Vorholt JA. The plant microbiota: systems-level insights and perspectives. Annu Rev Genet. 2016;50:211–34. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- 61.Brewer TE, Aronson EL, Arogyaswamy K, Billings SA, Botthoff JK, Campbell AN, et al. Ecological and genomic attributes of novel bacterial taxa that thrive in subsurface soil horizons. MBio. 2019;10:e01318–19. doi: 10.1128/mBio.01318-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature. 2018;557:503–9. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 63.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pascual-García A, Bell T. Community-level signatures of ecological succession in natural bacterial communities. Nat Commun. 2020;11:1–1. doi: 10.1038/s41467-020-16011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data have been deposited in the NCBI Sequence Read Archive under BioProject accession code PRJNA664514.