Abstract
Recent empirical studies have demonstrated that speciation with gene flow is more common than previously thought. From a conservation perspective, the potential negative effects of hybridization raise concerns on the genetic integrity of endangered species. However, introgressive hybridization has also been growingly recognized as a source of diversity and new advantageous alleles. Carbonell’s wall lizard (Podarcis carbonelli) is an endangered species whose distribution overlaps with four other congeneric species. Our goal here was to determine whether P. carbonelli is completely reproductively isolated from its congeners and to evaluate the relevance of hybridization and interspecific gene flow for developing a conservation plan. We used restriction site associated DNA (RAD) sequencing to discover SNPs in samples from four contact zones between P. carbonelli and four other species. Principal component analysis, multilocus genotype assignment and interspecific heterozygosity suggest incomplete reproductive isolation and ongoing gene flow between species. However, hybridization dynamics vary across all pairs, suggesting complex interactions between multiple intrinsic and extrinsic barriers. Despite seemingly ubiquitous interspecific gene flow, we found evidence of strong reproductive isolation across most contact zones. Instead, indirect effects of hybridization like waste of reproductive effort in small isolated populations may be more problematic. Our results highlight the need to further evaluate the consequences of introgression for P. carbonelli, both on a geographic and genomic level and included in a comprehensive and urgently needed conservation plan. Besides, those findings will add important insights on the potential effects of hybridization and introgression for endangered species.
Subject terms: Population genetics, Speciation
Introduction
While speciation is often defined as the evolution of reproductive isolation (Mayr 1942; Coyne and Orr 2004), recent empirical and theoretical work has demonstrated that distinct species can persist in the presence of gene flow. This phenomenon may occur by sympatric divergence with gene flow (primary gene flow; Niemiller et al. 2008; Martin et al. 2013), or more commonly is the result of allopatric speciation followed by secondary contact (secondary gene flow; Tarroso et al. 2014; Grossen et al. 2016). Many species are maintained in nature despite some level of gene exchange, either coexisting in sympatry due to niche and/or phenotypic divergence (e.g., Whittemore and Schaal 1991; Milne et al. 1999; Neaves et al. 2010; Hochkirch and Lemke 2011); or replacing each other abruptly at narrow contact zones (e.g., Szymura and Barton 1986; Irwin et al. 2009; Tarroso et al. 2014; Grossen et al. 2016). Thus, it has been increasingly accepted that speciation can occur without complete reproductive isolation (Mallet 2008; Pinho and Hey 2010) and complete reproductive isolation is not a pre-requisite for most current species definitions such as the Evolutionary Species Concept (Simpson 1961) or even for most current versions of the Biological Species Concept originally proposed by Mayr (1940, 1942). This perspective calls for a new vision on gene flow in fields like evolutionary biology, ecology and conservation biology (VonHoldt et al. 2018).
The ultimate fate of two lineages that come into secondary contact depends on the strength of the barriers to gene flow. In empirical studies, three key questions arise when closely related lineages meet: (i) do they hybridize (i.e., is there interspecific mating forming F1 hybrids)? (ii) does this lead to introgression (i.e., occurrence of backcrosses leading to persistence of gene flow)? (iii) are there intrinsic or extrinsic barriers to gene flow that “protect” each genome from complete “invasion” by the other gene pool? Answering these questions will inform on the future evolutionary trajectories of the lineages involved (Wu 2001), i.e., whether they are likely to persist as distinct lineages or to fuse back in the future. In this context the amount of gene flow depends, among other factors, on the existence and efficiency of pre-mating barriers and mate choice, the genomic architecture of differentiation, the strength of selection of loci combinations in each genomic background, and on whether or not such combinations are involved in epistatic interactions (Wu 2001).
These questions are particularly relevant for conservation biology. Gene flow and introgression are processes that have recently become more important as human-mediated species translocations and range modifications bring into contact previously allopatric species or lineages that can still hybridize (e.g., Rubidge and Taylor 2005; Ayres et al. 2008; Senn and Pemberton 2009; VonHoldt et al. 2018). Distinct views on the evolutionary consequences of hybridization often conflict (Butlin and Ritchie 2013; Sætre 2013), which may hamper management decisions (Allendorf et al. 2001; Wayne and Shaffer 2016; VonHoldt et al. 2018). On the one hand, hybridization is often regarded as having negative effects on the conservation of endangered species because introgression may decrease divergence (Seehausen et al. 2008), impede reproductive isolation (Owens and Samuk 2019) and ultimately lead to genetic swamping (e.g., Rhymer et al. 1994; Roberts et al. 2010). Other negative effects of hybridization include the waste of reproductive effort in the generation of inviable or maladapted hybrid offspring (Lepais et al. 2009; Beatty et al. 2010) or the loss of locally adapted alleles (Bourret et al. 2011). In extreme cases, these situations may contribute to extinction (Todesco et al. 2016), particularly in species that are already rare or affected by other threats. On the other hand, hybridization and introgression have been growingly recognized as a source of genetic novelty, as they may increase diversity and facilitate the acquisition of advantageous alleles through adaptive introgression (Anderson et al. 2009; Whitney et al. 2010; Becker et al. 2013; Leroy et al. 2019) or prevent inbreeding depression (Johnson et al. 2010), thus enhancing the resilience of endangered populations (Tompkins et al. 2006) and even triggering biological diversification (Lamichhaney et al. 2016). This would suggest that the effects of hybridization on endangered species may sometimes be positive in conservation frameworks.
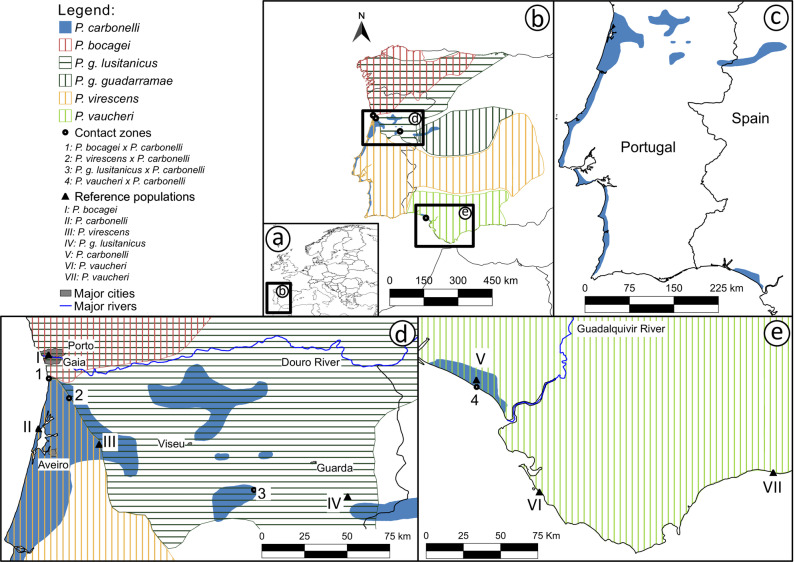
Carbonell’s wall lizard (Podarcis carbonelli) is a small lacertid lizard belonging to the Podarcis hispanicus species complex, a monophyletic group of genetically divergent but ecologically and morphologically similar species inhabiting the Iberian Peninsula and North Africa (Harris and Sá-Sousa 2002; Kaliontzopoulou et al. 2012b; Kaliontzopoulou et al. 2011). Natural hybridization and introgression have been documented in Podarcis (Capula 1993, 2002), and the P. hispanicus complex makes no exception (Pinho et al. 2009; Renoult et al. 2009) despite ancient diversification with speciation events within the complex ranging from ~2.5 to 10 million years ago (Kaliontzopoulou et al. 2011). Most species of the P. hispanicus complex are parapatric and replace each other abruptly, a pattern that is likely mediated by the interaction between suitable ecological conditions and competitive exclusion (Caeiro-Dias et al. 2018). P. carbonelli, on the contrary, is sympatric with at least one other species of the P. hispanicus complex in most parts of its range (Podarcis vaucheri, Podarcis guadarramae, or Podarcis virescens) and meets Podarcis bocagei across a narrow contact zone (Carretero et al. 2002; Sá-Sousa and Harris 2002; Pinho et al. 2009; Caeiro-Dias et al. 2018, see Fig. 1). At this contact zone the two species are known to hybridize, but nothing is known about the level of reproductive isolation between P. carbonelli and the other species.
Fig. 1. Distribution of each Podarcis species in Iberian Peninsula, the location of sampled contact zones and reference populations analyzed.
a Map of Europe with location of the study area boxed; b Distribution of the four Podarcis species (including both Podarcis guadarramae subspecies). Black dots represent the location of the contact zones; c Map highlighting P. carbonelli distribution; Zoom-in of the Northwest (d) and Southern (e) Iberian Peninsula showing the detailed locations of the contact zones and the reference populations analyzed (black triangles).
Remarkably, P. carbonelli has a highly fragmented distribution, throughout western Portugal, west-central Spain, and very small areas of southwestern Spain (Sá-Sousa 2008, see Fig. 1c) which has likely been shaped by a significant range reduction due to climatic changes after the last glacial maximum (Sá-Sousa 2001; Sillero and Carretero 2013), resulting in moderate levels of population substructure (Pinho et al. 2007, 2011). Several field studies have detected a trend of declining population sizes in recent years (Sillero et al. 2012, 2014), which, together with P. carbonelli’s limited geographic range and population fragmentation, led to the species categorization as endangered (EN) by IUCN (Sá-Sousa et al. 2009). Hybridization could, in theory, add to the existing threats to the species and the worrisome conservation status of P. carbonelli calls for an evaluation of the reproductive isolation as well as the amount and consequences of hybridization between the species and its congeners. To do so, we collected genetic samples from areas of syntopy and analyzed patterns of hybridization and introgression between P. carbonelli and other four co-distributed wall lizard species, using single nucleotide polymorphisms (SNPs) detected with double digest RAD sequencing (ddRAD, Peterson et al. 2012). We sought to answer the following questions: (1) does P. carbonelli hybridize with conspecific co-occurring species? (2) if hybridization occurs, does it lead to interspecific gene flow or is it restricted to F1 hybrids? (3) is the strength of reproductive barriers similar across species pairs? and (4) considering that P. carbonelli contacts with at least one other Podarcis species across most of its distribution range, can hybridization threaten its persistence, and should it be taken into account when devising conservation plans?
Material and methods
Sampling
Samples were collected between spring and autumn of 2013 in four contact zones between P. carbonelli and one of four other Podarcis species. In all of the contact zones the two species were found in strict syntopy. The sampling scheme was aimed at capturing all the individuals encountered, avoiding bias of age, sex or species. Lizards were captured with a noose, which is harmless, and kept in individual cloth bags until they were processed. All adults were identified to species-level in the field based upon a combination of coloration, head shape and behavior, but most juveniles were not possible to identify. Each sample was geo-referenced and photographed. A small tail tip was collected and immediately stored in 96% ethanol for subsequent DNA extraction. Animals were released the same day at the place of capture. We added reference individuals from nearby populations outside the contact zones (Fig. 1d, e) retrieved from the tissue collections of the CIBIO-InBio, Portugal, and EPHE-CEFE, France (BEV collection). Because of the scarcity of P. carbonelli in inland locations, we could not obtain reference populations for each of the three contact zones in Northwest Iberia and we therefore used the same P. carbonelli population (II in Fig. 1d) as reference for the contact zones 1 (with P. bocagei), 2 (with P. virescens) and 3 (with P. g. lusitanicus). These three contact zones are all geographically close (Fig. 1d) and given the low level of population differentiation reported for this part of the range (Pinho et al. 2011), this should not be an issue. Reference population II comes from an area where P. virescens is present but where we did not detect any syntopy (pers. obs.). Similarly, the southern reference population (V in Fig. 1e), used for the contact zone 4 (with P. vaucheri), comes from an area of sympatry with this species but is composed of individuals collected far from the actual contact zone and in habitats where P. vaucheri have never been observed (pers. obs., see also Discussion section). Sampling information for each contact zone and reference population is provided in Table 1 and detailed information about each sample is available in Supplementary Information Table S1.
Table 1.
Number of individuals analyzed in each contact zone (CZ) and detailed information about each dataset.
| CZ | Data set | Nr of individuals analyzed | Nr of loci | |||
|---|---|---|---|---|---|---|
| Total in CZ | Ref Pc. | Ref Sp. | Total | |||
| 1 (bocagei) | Complete | 115 | 23 | 20 | 158 | 19233 |
| Diagnostic | 114 | 23 | 20 | 157 | 1786 | |
| 2 (virescens) | Complete | 57 | 10 | 7 | 74 | 5207 |
| Diagnostic | 58 | 10 | 7 | 75 | 322 | |
| 3 (lusitanicus) | Complete | 61 | 21 | 18 | 100 | 727 |
| Diagnostic | 65 | 21 | 18 | 104 | 32 | |
| 4 (vaucheri) | Complete | 63 | 10 | 9 | 82 | 3549 |
| Diagnostic | 63 | 10 | 9 | 82 | 56 | |
CZ is the number of contact zone (as in Fig. 1), followed by the lineage (other than P. carbonelli) involved in the CZ; “Total in CZ” is the total number of individual samples in the contact zones; Ref Pc. and Ref Sp. are the number of sample in the reference populations for P. carbonelli and the other species, respectively, used for each contact zone.
The syntopy area between P. bocagei and P. carbonelli (contact zone 1) is located in a narrow coastal dune stripe about 450 meters wide with dune scrub vegetation north of the locality of Espinho (Aveiro District, Portugal, see Carretero et al. 2002). Across the area where both species were sampled in syntopy no obvious ecological segregation was identified. Podarcis virescens and P. carbonelli (contact zone 2) were found in the same walls of the castle of Santa Maria da Feira (Aveiro District, Portugal) clearly occurring in the exact same places. Although P. carbonelli is found in sympatry with the two currently recognized subspecies of P. guadarramae (P. g. guadarramae and P. g. lusitanicus; Geniez et al. 2014), only P. g. lusitanicus was used in this study (see Fig. 1 for details on geographic distributions). Specimens of syntopic P. g. lusitanicus with P. carbonelli (contact zone 3) come from a 500-meter-long area in Vale do Rossim (Serra da Estrela Natural Park, Guarda District, Portugal), an area dominated by sparse pine trees with relatively dense scrub cover with open patches and rocky outcrops. While P. g. lusitanicus was collected mostly on rocks, P. carbonelli was always found on the ground. P. vaucheri specimens in syntopy with P. carbonelli (contact zone 4) were found along a very narrow strip of a few tens of meters in the far northwest of the coastal village of Matalascañas (Huelva Province, Spain). Both species inhabited distinct micro-habitats, P. vaucheri human-made structures and P. carbonelli only semi-natural dune environments with pine trees and scrub vegetation. Individuals were collected along a 500 meters stripe comprising both environment types.
RAD sequencing, data filtering and SNP calling
We obtained ddRAD sequence data using modifications to protocols from Parchman et al. (2012), Peterson et al. (2012) and Purcell et al. (2014). The complete protocol is described by Brelsford et al. (2016). The main steps were the digestion of genomic DNA with the restriction enzymes SbfI and MseI, ligation of barcoded adapters to restriction sites, amplification of each individual sample in four independent separate PCR reactions, pooling of all PCR products and fragment selection between 400 and 500 bp using a 2.5% agarose gel. Samples used in this work were included in two separate libraries constructed following the same protocol. Both libraries included other samples not used in this study. One had a total of 329 samples and included samples from contact zone 1. This library was sequenced on eight Illumina® (San Diego, CA, USA) HiSeq 2000 lanes in the Lausanne Genetic Technology Facility (Lausanne, Switzerland), with single-end 100 bp reads. The datasets for contact zones 2, 3, and 4 were obtained from a library containing a total of 665 samples that was sequenced on two Illumina® HiSeq 2000 lanes at the Lausanne Genetic Technology Facility (Lausanne, Switzerland) and on four Illumina® HiSeq 1500 lanes at the CIBIO Next Generation Sequencing Platform (Vairão, Portugal), also with single-end 100 bp reads.
We demultiplexed individual raw reads using the process_radtags module of Stacks version 2.2 (Catchen 2013) allowing one mismatch per barcode, removing reads containing adapter sequence, reads with uncalled bases and reads that failed the Illumina® ‘chastity’ filter. We then tested the optimal de novo assembly parameters for our data set following the protocol described in Rochette and Catchen (2017) adapted to Stacks version 2.2, prior to the final de novo read alignment. For this test we used the samples of one contact zone only (P. carbonelli × P. vaucheri), running consecutively ustacks (build loci), cstacks (create a catalog of loci), sstacks (match individual samples against the catalog), tsv2bam (transpose data), gstacks (align each read to a locus and call SNPs) and populations (SNP filtering and output data) units. We performed separate runs varying the number of mismatches allowed between reads within individuals (M) and between individuals (n). As suggested by Rochette and Catchen (2017), we varied M and n between 1 and 9 and kept M = n, while the minimum depth of coverage to create a stack (m) was kept constant at 3, the ustacks default. Both in these prior tests and in the final analyses we used a bounded SNP model (--model_type bounded option) in ustacks, with an upper bound for the error rate of 0.1. Other than the mentioned changes to test the parameter values, we used default settings for all other steps of the pipeline. After completion of the pipeline for the 9 different sets of parameter values, we analyzed the numbers of loci shared by at least 80% of the samples and the distribution of variable sites within loci for the range of tested values. Based on this analysis we chose to retain M = n = 5 for the final de novo read alignment and SNP calling procedure, which was conducted separately for each contact zone and their respective reference populations.
The populations module from Stacks was used to filter out potential paralog loci by removing the resulting variants with higher than 0.7 maximum observed heterozygosity across all samples. Subsequently, vcftools version 0.1.15 (Danecek et al. 2011) was used to discard SNPs with depth coverage less than 8, SNPs with alleles with minimum frequency lower than 0.05, and present in less than 80% of the loci for each dataset. We then performed two additional filtering steps using a custom Python script (available at https://github.com/catpinho/filter_RADseq_data): i) we removed loci exhibiting more than 8 SNPs per RAD tag; ii) we kept only one SNP per locus, choosing the SNPs maximizing frequency differences between species. For each contact zone we then kept two separate datasets: the complete dataset with all filtered SNPs; and the diagnostic dataset including only loci with fixed differences between reference populations but excluding the loci with private alleles, i.e., loci with alleles that were not found in the contact zone. In each dataset and subset (see details below), individuals with more than 35% of missing data for each locus were discarded. Library construction, sequencing, demultiplexing and data filtering (except data subsets) were repeated independently for ~ 6% of the samples to evaluate replicability.
Genetic characterization of the contact zones and admixture analysis
The genomic variability among individuals was visualized by performing principal component analyses (PCAs) on the complete dataset for each contact zone separately using the adegenet R package version 2.0.1 (Jombart 2008; Jombart and Ahmed 2011). We then used Structure version 2.3.4 (Pritchard et al. 2000) to evaluate the proportion (Q) of each individual’s genome originating from each of the parental species and its 90% posterior probability intervals (CI), including the reference populations. For clarity we will refer to the proportion of P. carbonelli as QC in each contact zone, as it is the common element to all. The proportion of assignment of each individual to the other species in a particular contact zone is 1 – QC. The distributions of Qc scores in 10 classes of equal size for each contact zone are also reported.
The number of loci is variable between contact zones and this influences the ability of Structure to estimate the CI (confirmed by comparing preliminary Structure results with different number of loci – results not shown). Thus, the proportion of admixed/parental individuals across contact zones are not comparable based on Qc CI if we use largely different number of SNPs. We ran the final Structure analysis with the same number of SNP’s for all contact zones, to make the analysis across contact zones more comparable. We did this by selecting a random subset of 727 SNPs, the same number of SNPs as in the contact zone with the lowest number of markers (contact zone 3). Comparison of preliminary runs with five different random subsets for each of the three contact zones showed no substantial disparities (results not shown). We thus chose randomly one of the five subsets to present the results. We ran Structure for K = 2 for each contact zone, as we were only interested in detecting admixture between each pair of species, using the complete datasets for each of the contact zones. Runs were performed five times independently with one million repetitions and a burn-in of 250 000. We used a model assuming admixture and independent allelic frequencies using a prior of individual ancestry of 0.5, as recommended for unbalanced sample sizes by Wang (2017). The 90% Qc CI were estimated. Structure Harvester web version 0.6.94 (Earl and VonHoldt 2012) was used to visualize the likelihood of the data for each contact zone. The run that maximized the likelihood was retained and is presented in the Results. We opted for this approach, instead of concatenating the five runs for each dataset, as CI estimations are used in further analyses.
Individuals from reference populations were then used to calculate the hybrid index (HI) of each individual from the admixed population using the R package Introgress version 1.2.3 (Gompert, Alex Buerkle 2010). P. carbonelli parental individuals were set to have an HI of 1, and the other species were set to an HI of 0 for each contact zone. The proportion of loci in an admixed individual’s genome with alleles inherited from both parental species, i.e., interspecific heterozygosity (He), was calculated for each admixed individual using Introgress. This method for calculating interspecific heterozygosity assumes that parental allele frequencies are known. Therefore, the same individuals used as parentals for HI estimation were also used to calculate He. A triangle plot can represent the relationship between the HI and He (Fitzpatrick 2012). He ranges from 0 to 1, where values of 1 are interpreted as “perfect” F1 hybrids and values lower than 1 indicate later generation hybrids that have either backcrossed with the parentals (overlapping the lines of the triangle plot) and/or with other hybrids (below the triangle plot lines; Fitzpatrick 2012). This analysis requires that loci are fully diagnostic between species or at least exhibit large differences for a good approximation. Here we performed this analysis by restricting the SNPs just to loci with fixed differences between reference populations and excluding loci with private alleles from each population (diagnostic dataset).
To assess whether or not the strength of reproductive isolation between all the species pairs is similar, we tested if the overall genotypic hybrid composition was effectively distinct across contact zones, using Fisher’s exact tests. We tested the global independence of overall admixed genotype composition among the four contact zones, against the alternative hypothesis that the proportion of admixed genotypes is similar across contact zones, by evaluating the frequency of occurrence of two categories: “parental genotypes” (individuals with 90% CI of Qc overlapping 0 or 1) and “admixed genotypes” (individuals with 90% CI of Qc non-overlapping 0 or 1). Additionally, we tested if recent hybridization events have different contributions to each contact zone. To do so we performed a similar global Fisher’s exact test, but in this case, we considered the frequencies of recently admixed genotypes (QC between 0.4 and 0.6) against parental genotypes and later generation backcrosses (genotypes with Qc ≥ 0.6 and Qc ≤ 0.4). In the case that the global test was statistically significant we further performed pairwise Fisher’s exact tests to identify the contact zone pairs that present differences in genotype composition. All tests were performed with R package stats version 3.3.3 (R Core Team 2017) and for pairwise tests we applied a Bonferroni correction for multiple comparisons.
Results
Data filtering and SNP calling
Characteristics of the final datasets (complete and diagnostic), after removing loci with depth coverage <8 and missing data >20% and removing individuals with more than 35% of missing data are summarized in Table 1. Across complete and diagnostic datasets, mean coverage by individuals ranged from 28 to 47 and by loci from 28 to 44 (see detailed results in Supplementary Information Table S2). The analysis of replicate samples showed high levels (>99%) of multilocus genotype replicability.
Podarcis bocagei × Podarcis carbonelli
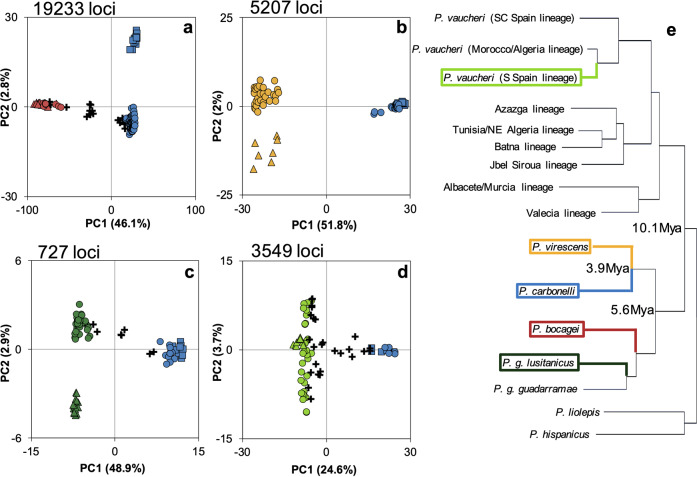
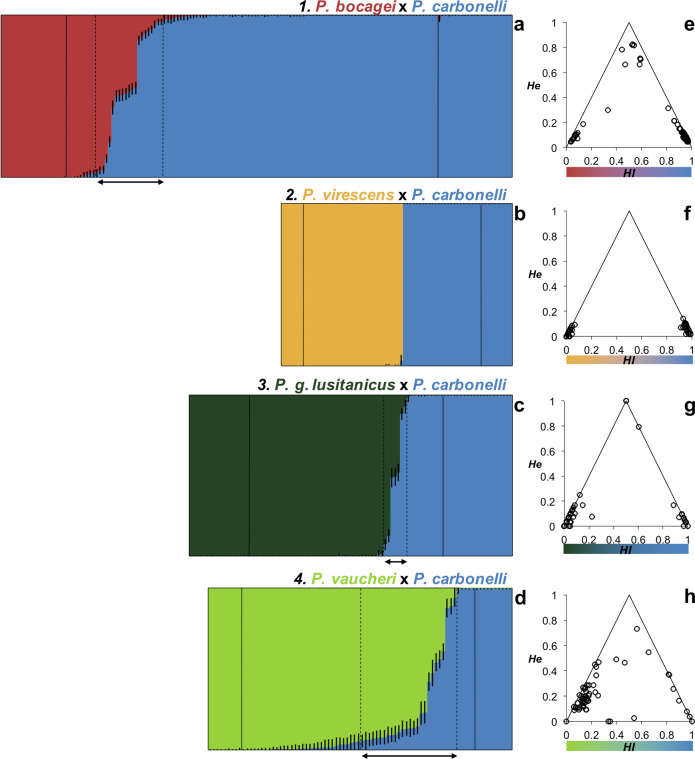
The PCA analysis based on the 19233 SNPs separated two groups: PC1 explained 46.1% of the variance and separated P. bocagei from P. carbonelli, broadly matching the field identification of each individual (Fig. 2a) but with several individuals in-between. PC2 explained 2.8% of the variance and is due to variation between P. carbonelli populations. We found 7.8% of P. bocagei and 73.9% of P. carbonelli “parental genotypes” while 18.2% of the individuals were identified as “admixed genotypes” (according to the Structure CI criterion; Fig. 3a). Most individuals had Qc values close to the extreme values (Supplementary Information Fig. S1a) as expected in bimodal hybrid zones. When plotting He against the HI (Fig. 3b) the results were concordant with Structure and PCA analysis, where most individuals showed at least some degree of admixture (He ranges between 0.03 and 0.82) and several parental individuals had residual levels of admixture indicating the existence of gene flow between the two species.
Fig. 2. Principal Component Analysis (PCA) of SNP variation.
PCA for a P. bocagei (red)×P. carbonelli (blue); b P. virescens (yellow)×P. carbonelli; c P. guadarramae lusitanicus (dark green)×P. carbonelli; and d P. vaucheri (light green)×P. carbonelli contact zones; the variation explained by each axis (PC) is represented as percentage; circles represent individuals in syntopy with Structure 90% CI overlapping 0 or 1, crosses denote individuals with Structure 90% CI non-overlapping 0 or 1, squares the P. carbonelli reference population and triangles the reference population of the other species; individuals were colored after Structure results; e schematic representation of phylogenetic relationships of all Podarcis hispanicus complex lineages highlighting the divergence times between species from contact zones analyzed in this study (see Kaliontzopoulou et al. 2011).
Fig. 3. Analysis of genetic variability and admixture performed with SNP data.
Results for a and e P. bocagei (red)×P. carbonelli (blue); b and f P. virescens (yellow)×P. carbonelli; c and g P. g. lusitanicus (dark green)×P. carbonelli; and d and h P. vaucheri (light green)×P. carbonelli contact zones. Plots on the left (a–d) show the results from individual multilocus genotype clustering analysis performed with Structure based on the runs with the 727 loci datasets; each individual is represented as a horizontal line partitioned into the 2 colored segments (K=2) and each segment length is proportional to the assignment to one of the two species. Horizontal black lines delimit individuals in contact zone (in the center) from the reference populations and between the horizontal dashed lines are the individuals whose 90% CI (back segments in each individual) do not overlap 0 or 1. Plots on the right (e–h) show distribution of individual hybrid index (HI) and interspecific heterozygosity (He) in the contact zones based on diagnostic loci between reference populations; in each contact zone P. carbonelli reference individuals were set to have a HI of 1, and the other species was set to an HI of 0.
Podarcis virescens × Podarcis carbonelli
The PCA showed two clusters of individuals based on the 5207 SNPs analyzed (Fig. 2b). PC1 explained 51.8% of the variation, separating the two species while PC2 explained 2% of the variation and captured mostly intraspecific variability in P. virescens. The assignment analysis (Fig. 3c) and the distribution of Qc values (Supplementary Information Fig. S1b) were concordant with the results of the PCA. None of the individuals fell into the category of “admixed genotypes” and only 10.5% of the individuals had Qc values slightly deviating from 0 or 1 suggesting a residual level of admixture. The HI distributions were concordant with previous results and the estimations of He were close to zero (Fig. 3d). Therefore, no recent hybridization was detected in this contact zone.
Podarcis guadarramae lusitanicus × Podarcis carbonelli
The PCA revealed two main groups of samples based on 727 SNPs (48.9% of the variance explained by PC1) showing that the two species in the contact zone are clearly separable. PC2 explains 2.9% of the variation and captures mostly the intraspecific variability within P. g. lusitanicus. Several individuals were in an intermediate position between both groups along PC1, indicating recent hybridization (Fig. 2c). Based on Structure results 11.5% of the individuals with QC between 0.07 and 0.93 were considered “admixed genotypes”, confirming some degree of admixture, while 67.2% were assigned to P. g. lusitanicus and 21.3% to P. carbonelli (Fig. 3e), showing that most individuals had a Qc value close to zero or one (Supplementary Information Fig. S1c). Estimates of He were low for most individuals with HI close to 0 and 1 (Fig. 3f). Two individuals perfectly matched the predictions for F1 hybrids and one individual with QC = 0.52 had HI = 0.6 and He = 0.79, suggesting a backcross between an individual with mixed ancestry and a P. carbonelli “parental genotype”.
Podarcis vaucheri × Podarcis carbonelli
The first axis (PC1) of the PCA based on 3549 SNPs explained 24.6% of the variance (Fig. 2d) and, separating P. vaucheri from P. carbonelli, but with several individuals between the two groups. A large amount of variation among P. vaucheri individuals is obvious along PC2 (3.7% of the variance). Structure results showed that 7.9% of the individuals were assigned to P. carbonelli “parental genotypes” and 50.8% to P. vaucheri (Fig. 3g) but a large proportion of individuals (41.3%) showed some degree of admixture (Supplementary Information Fig. S1d), highlighting the permeability of the genomes to gene flow. Furthermore, the presence of individuals with high He revealed the occurrence of contemporary admixture (Fig. 3h). Because we used parental individuals from outside of the contact zone (virtually “pure”) to estimate heterozygosity and several parental individuals in the population of contact had residual levels of admixture (Fig. 3g), “perfect” F1 hybrids were not expected to be found, as in the contact zone 1. Surprisingly, two individuals in the contact zone 4 had He = 0 but HI ~ 0.35 and another individual had He = 0.02 with HI = 0.54 (Fig. 3h). Such results may be explained by poor estimates of heterozygosity due to the lower average coverage for these individuals compared to the others (data not shown). In this contact zone there were only four individuals, including these three, with depth of coverage lower or equal to 20x.
Comparison between contact zones
The total proportion of admixed genotypes, i.e., genotypes whose Qc 90% CIs do not overlap 0 or 1, ranged between 0 and 41% across different contact zones (Fig. 4a). In contact zone 4, the proportion of admixed individuals (41%) was almost half of the total genotypes. The global test revealed statistical differences among contact zones (Supplementary Information Table S3). When we compared the proportions of admixed genotypes between all contact zones, five comparisons were significant and we could reject the null hypothesis that the proportions are similar (p value < 0.008, the corrected significance level after employing the Bonferroni correction). The comparison between contact zones 1 and 3 (with 25% and 13% of admixed genotypes, respectively) did not differ statistically from each other (Supplementary Information Table S3).
Fig. 4. Overall genotypic composition for each contact zone.
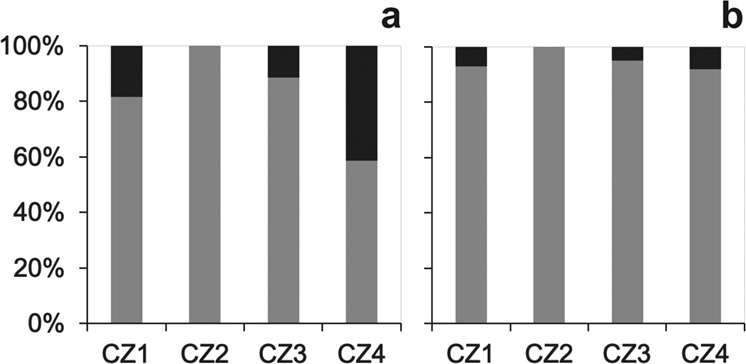
a Proportion of parental genotypes (90% CI of QC that overlap 0 or 1), represented in gray, and hybrid genotypes (90% CI of QC that do not overlap 0 or 1), represented in black, for each contact zone. QC results from the clustering analysis, as in Fig. 2. Fisher exact test statistically significant (p=8.29×10−9). b Proportion of parental and later generation hybrid genotypes resulting from recurrent backcrosses (QC≥0.6 and ≤ 0.4; gray) and earlier generation hybrid genotypes, (QC<0.6 and QC>0.4; black) in each contact zone. Fisher exact test not statistically significant (p=0.12). CZ1: P. bocagei×P. carbonelli, CZ2: P. virescens×P. carbonelli, CZ3: P. guadarramae lusitanicus×P. carbonelli, CZ4: P. vaucheri×P. carbonelli.
Our results show that in three out of the four contact zones studied recent hybridization events (identified by the presence of individuals showing QC between 0.4 and 0.6 and high interspecific heterozygosity) are relatively rare; evidence for recent admixture was completely absent in the other contact zone (Fig. 4b). Recently admixed genotypes (Qc between 0.4 and 0.6) made up between 0 and 8% of the admixed genotypes across the analyzed contact zones (Fig. 4b). When we compared the number of recent hybrid genotypes vs. parental and later-generation admixed genotypes, the differences across contact zones were not significant and thus we could not reject the null hypothesis that the proportions of recently admixed genotypes in each contact zone were similar (Supplementary Information Table S3).
Discussion
Our results provide evidence that P. carbonelli, a species co-occurring across most of its distribution range with at least another congener, can maintain its genetic identity in extensive sympatry in spite of incomplete reproductive isolation with the species it coexists with: in three of the four contact zones investigated we found that recent admixture has occurred, and that it is not restricted to F1 hybrids. The markedly bimodal distribution of genotypes in all contact zones suggests strong barriers to gene flow, however, even though the strength of reproductive barriers varies between species pairs. Interspecific gene flow is thus an important feature in the evolutionary history of P. carbonelli.
Species validity and persistence in the presence of gene flow
The four species studied here are indubitably distinct, valid species under all current species concepts, with divergence times based on mtDNA ranging between 3.9 Mya (P. virescens and P. carbonelli) and 10.1 Mya (P. vaucheri and P. carbonelli; Fig. 2e; also see Kaliontzopoulou et al. 2011), most of them with distinct morphology (Kaliontzopoulou et al. 2012b) and realized climatic niches (Caeiro-Dias et al. 2018). They also maintain highly distinct nuclear genomes in spite of multiple opportunities for hybridization, as evidenced by the PCAs on SNP data that reveal much higher levels of between- than within-species variation. Nevertheless, we found evidence for recent hybridization in three contact zones and our data suggest later generation hybrids and/or signal from older introgression events in the four contact zones (Fig. 3), indicating interspecific gene flow and a lack of complete reproductive isolation. As P. carbonelli is sympatric across all of its distributional range with other Podarcis species, these results suggest that hybridization is likely to occur across numerous populations of this species.
The overall deep divergence between our species (Kaliontzopoulou et al. 2011) and the strong bimodality of their hybrid zones in spite of the presence of introgressed individuals, other than first generation hybrids, is consistent with the notion that these species are in the late stages of speciation, where speciation is already well advanced but reproductive isolation is still incomplete. The bimodality observed for the hybrid zones is likely not the result of the recent establishment of these contact zones (i.e., few generations scale), as evidenced by the presence of a large proportion of individuals with some degree of admixture in contact zones 1 and 4 (Fig. 3b, h) and by the presence of individuals with residual levels of admixture in all four contact zones. In addition, hybridization is known to have occurred between P. bocagei and P. carbonelli (contact zone 1) for several generations (Pinho et al. 2009) and all the syntopic populations sampled for this study were known prior to sampling, sometimes for many years (pers. obs.).
The occurrence of regular gene flow between sympatric species at levels that do not reverse species divergence have been increasingly reported (Steeves et al. 2010; Palma‐Silva et al. 2011; McIntosh et al. 2014). Our results support the notion that introgression may be a regular component in the evolutionary history among the species of the P. hispanicus complex (Pinho et al. 2008; Renoult et al. 2009) and an integral part of the speciation process in general, not only in its initial stages but also in later stages.
Intrinsic and extrinsic factors potentially influencing hybridization dynamics
Regular interspecific gene flow, even when rare, leads to extensive admixture over time and several generations of rare introgression would quickly remove all parental genotypes in the absence of post-zygotic isolation (Arnold et al. 1999). Therefore, post-zygotic barriers, such as high mortality or reduced fertility of highly admixed individuals, must be acting in these contact zones. In contact zone 1, hybrids do not have lower fertility than either parental species (Pinho et al. 2009) but the fitness of their progeny has not been evaluated. The rarity of F1 hybrids relative to pure genotypes may also be the result of pre-zygotic barriers preventing gene flow. For example, in half contact zones studied here, syntopy is accompanied by some degree of ecological segregation, also previously verified between P. bocagei and P. g. lusitanicus (Kaliontzopoulou et al. 2012a; Gomes et al. 2016). Chemically-mediated candidate species recognition systems were also found between several species of the complex (Barbosa et al. 2005, 2006; Gabirot et al. 2010, 2012), but actual validation of their effectiveness in natural conditions is entirely lacking. Deciphering the relative contribution of these or other pre- and post-zygotic barriers would require finer experimental dissection of each contact zone, but it is clear that post-zygotic barriers are crucial in maintaining P. carbonelli as a distinct species.
Why do levels of interspecific gene flow vary across contact zones? One would expect to detect increased hybridization levels between species with closer evolutionary relationships, as genetic divergence is positively associated with reproductive isolation (Pereira et al. 2011; Sánchez‐Guillén et al. 2014; Harvey et al. 2017). Remarkably, a higher level of admixture between P. carbonelli was found with the most phylogenetically divergent species (P. vaucheri) while we found almost no admixture with the most closely related species (P. virescens; Figs. 2 and 3; see also Kaliontzopoulou et al. 2011 for phylogenetic relationships). Consequently, the level of reproductive isolation does not seem to be explained by the degree of divergence in this system. The amount of reproductive isolation between P. carbonelli and the species it coexists with seems to depend more on other factors rather than on genetic divergence. In other systems, ecology (Funk et al. 2006) or phenotype (Stelkens and Seehausen 2009) were found to influence reproductive isolation.
While genetic divergence does not seem to determine reproductive isolation, the extent and duration of sympatry appears associated with the level of admixture in our system. The two species with the most restricted introgression are P. virescens and P. carbonelli (contact zone 2), which are the species pair with the largest area of total overlap across their distribution areas. On the contrary, P. bocagei and P. carbonelli (contact zone 1) are allopatric and only come into contact at a very narrow hybrid zone, where admixture is high. The two species with the highest level of admixture are P. vaucheri and P. carbonelli (contact zone 4), which are only marginally sympatric and are naturally segregated by their distinct use of microhabitats. In contact zone 4, P. vaucheri was observed exclusively in human-made structures while P. carbonelli inhabited semi-natural environments outside the village and the syntopy between them was restricted to the edge of the town. In the Doñana area (where Matalascañas is located), P. vaucheri seems to be restricted to man-made structures such as buildings, bridges, concrete walls, and was never observed in the sandy semi-natural habitats inhabited by P. carbonelli (pers. obs). Given that Matalascañas is a village where development for tourism is relatively recent, occurring mostly after 1972 (IECA 2014), and that elsewhere in the Doñana area we did not detect areas of syntopy, the two species may have only been in contact for a short time (several tens of generations). Human-mediated habitat modifications have been widely identified as factors promoting geographic contact between species and thus contributing to hybridization when pre-mating barriers are incomplete (Levin et al. 1996; Allendorf et al. 2001). Given the apparent negative correlation between the geographic extent, duration of sympatry and the levels of hybridization, our results are in line with the idea that reinforcement is important for incompletely isolated species to persist in sympatry (Bímová et al. 2011; Yukilevich 2012; Hudson and Price 2014). Further research needs to be done to formally test this hypothesis.
Hybridization may increase as a result of an imbalance in species frequencies, as the rarity of one species promotes opportunities for interspecific mating (Burgess et al. 2005; Lepais et al. 2009; Beatty et al. 2010). We detected hybridization in the three contact zones where the proportions of both species are highly imbalanced. The contact zone with the lowest interspecific gene flow (contact zone 2) had, apparently, a more equally balanced abundance of the two species in contact (Fig. 2b). This hypothesis remains to be tested but if it is valid the declining trend of P. carbonelli (Sillero et al. 2012, 2014) may make it more susceptible to hybridization when it comes into contact with more abundant species.
Outcomes of hybridization for P. carbonelli
Hybridization has long been emphasized as a threat to species, endangered (Milián-García et al. 2015; Vuillaume et al. 2015) or not (Rhymer and Simberloff 1996; Allendorf et al. 2001; Wolf et al. 2001; Wayne and Shaffer 2016). In our system, the level of reproductive isolation detected between P. carbonelli and its sympatric congeners seems strong enough to prevent global genetic swamping (see Steeves et al. 2010 for a similar conclusion in the critically endangered bird Himantopus novaezelandiae). However interspecific mating per se may have negative effects, such as wasted reproductive effort in small populations that coexist with more abundant congeneric species (Burgess et al. 2005; Lepais et al. 2009; Beatty et al. 2010). This is particularly relevant for the southern-most P. carbonelli population that came into contact with P. vaucheri (contact zone 4), more recently as well as, some of the small fragmented populations in the north that come into contact with P. g. lusitanicus. Alternatively, interspecific gene flow may be a source of favorable alleles or allelic combinations (Anderson et al. 2009; Whitney et al. 2010; Becker et al. 2013; Leroy et al. 2019) and a potentially important contributor to the origins of evolutionary novelty. Our data does not reveal if alleles crossed the species boundaries beyond the contact zones but given the variable levels of incomplete reproductive isolation between P. carbonelli and other co-occurring Podarcis species this is a topic worth studying in the near future.
At this stage, making detailed recommendations for conservation would be premature, but monitoring the amount of hybridization should certainly be part of any forthcoming management plan, particularly for the Andalusian P. carbonelli population that has recently been affected following human-mediated habitat changes. Most management plans now include a research program, given our findings, we highlight the need for these research programs to: (1) extend the evaluation of hybridization and interspecific gene flow across the P. carbonelli distribution; (2) investigate the extent of geographic and genomic introgression; (3) correlate the levels of interspecific gene flow with intrinsic and extrinsic factors that can modulate it; (4) look for traces of adaptive introgression from other members of the P. hispanicus complex. This will allow us to better understand which factors are involved in reproductive isolation as well as the evolutionary and conservation consequences of hybridization.
Supplementary information
Acknowledgements
We thank E. Pérez, M. A. Carretero, C. Rato, A. Perera, V. Gomes, N. Sillero, A. Crotini and A. Žagar for helping with sample collection. We also thank B. Gernster for the manuscript revision. This study benefited from the Montpellier Bioinformatics Biodiversity platform supported by the LabEx CeMEB, an ANR “Investissements d’avenir” program (ANR-10-LABX-04-01). GCD was supported by a PhD grant (SFRH/BD/89750/2012), and AK and CP by IF contracts (IF/00641/2014/CP1256/CT0008 and IF/01597/2014/CP1256/CT0009, respectively) under the Programa Operacional Potencial Humano—Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese Ministério da Educação e Ciência and PAC by the ANR grant ANR-19-CE02-0011 – IntroSpec. Support was also provided by national funds through FCT projects: PTDC/BIA-BEC/102179/2008 – FCOMP-01-0124-FEDER-007062, under FEDER COMPETE funds; PTDC/BIA-EVL/28090/2017 – POCI-01-0145-FEDER-028090 and PTDC/BIA-EVL/30288/2017 – NORTE-01-0145-FEDER-30288 co-funded by NORTE2020 through Portugal 2020 and FEDER Funds. Specimens for this study were captured and handled under permit numbers 119 to 124/2013/CAPT and 486 to 490/2014/CAPT by Instituto da Conservação da Natureza e das Florestas (ICNF, Portugal) and permit 201318800014253 by Junta de Andalucía, Consejería de Agricultura, Pesca y Medio Ambiente (Spain).
Data availability
Raw sequence reads from ddRAD-seq are deposited in the NCBI Sequence Read Archive (SRA). The BioProject accession number is PRJNA665746. VCF files with the complete datasets for each contact zone have been deposited on DRYAD (10.5061/dryad.k0p2ngf51).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Associate editor: Lounès Chikhi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pierre-André Crochet, Catarina Pinho
Supplementary information
The online version of this article (10.1038/s41437-020-00386-6) contains supplementary material, which is available to authorized users.
References
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- Anderson TM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Bulger MR, Burke JM, Hempel AL, Williams JH. Natural hybridization: how low can you go and still be important? Ecology. 1999;80:371–381. [Google Scholar]
- Ayres DR, Grotkopp E, Zaremba K, Sloop CM, Blum MJ, Bailey JP, et al. Hybridization between invasive Spartina densiflora (Poaceae) and native S. foliosa in San Francisco Bay, California, USA. Am J Bot. 2008;95:713–719. doi: 10.3732/ajb.2007358. [DOI] [PubMed] [Google Scholar]
- Barbosa D, Desfilis E, Carretero MA, Font E. Chemical stimuli mediate species recognition in Podarcis wall lizards. Amphib-reptil. 2005;26:257–263. doi: 10.1007/s10886-006-9072-5. [DOI] [PubMed] [Google Scholar]
- Barbosa D, Font E, Desfilis E, Carretero MA. Chemically mediated species recognition in closely related Podarcis wall lizards. J Chem Ecol. 2006;32:1587–1598. doi: 10.1007/s10886-006-9072-5. [DOI] [PubMed] [Google Scholar]
- Beatty GE, Philipp M, Provan J. Unidirectional hybridization at a species’ range boundary: implications for habitat tracking. Divers Distrib. 2010;16:1–9. [Google Scholar]
- Becker M, Gruenheit N, Steel M, Voelckel C, Deusch O, Heenan PB, et al. Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nat Clim Chang. 2013;3:1039. [Google Scholar]
- Bímová BV, MacHolán M, Baird SJE, Munclinger P, Dufková P, Laukaitis CM, et al. Reinforcement selection acting on the European house mouse hybrid zone. Mol Ecol. 2011;20:2403–2424. doi: 10.1111/j.1365-294X.2011.05106.x. [DOI] [PubMed] [Google Scholar]
- Bourret V, O’reilly PT, Carr JW, Berg PR, Bernatchez L. Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees. Heredity. 2011;106:500. doi: 10.1038/hdy.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Dufresnes C, Perrin N. High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity. 2016;116:177. doi: 10.1038/hdy.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess KS, Morgan M, Deverno L, Husband BC. Asymmetrical introgression between two Morus species (M. alba, M. rubra) that differ in abundance. Mol Ecol. 2005;14:3471–3483. doi: 10.1111/j.1365-294X.2005.02670.x. [DOI] [PubMed] [Google Scholar]
- Butlin RK, Ritchie MG. Pulling together or pulling apart: hybridization in theory and practice. J Evol Biol. 2013;26:294–298. doi: 10.1111/jeb.12080. [DOI] [PubMed] [Google Scholar]
- Caeiro-Dias G, Luís C, Pinho C, Crochet PA, Sillero N, Kaliontzopoulou A. Lack of congruence of genetic and niche divergence in Podarcis hispanicus complex. J Zool Syst Evol Res. 2018;56:479–492. [Google Scholar]
- Capula M. Natural hybridization in Podarcis sicula and P. wagleriana (Reptilia: Lacertidae) Biochem Syst Ecol. 1993;21:373–380. [Google Scholar]
- Capula M. Genetic evidence of natural hybridization between Podarcis sicula and Podarcis tiliguerta (Reptilia: Lacertidae) Amphib-reptil. 2002;23:313–321. [Google Scholar]
- Carretero MA, Sá-Sousa P, Barbosa D, Harris DJ, Pinho C. Sintopía estricta entre Podarcis bocagei y P. carbonelli. Bol Asoc Herpetol Esp. 2002;13:20–24. [Google Scholar]
- Catchen JM. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland
- Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Fitzpatrick BM. Estimating ancestry and heterozygosity of hybrids using molecular markers. BMC Evol Biol. 2012;12:131. doi: 10.1186/1471-2148-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc Natl Acad Sci USA. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabirot M, Castilla AM, López P, Martín J. Differences in chemical signals may explain species recognition between an island lizard, Podarcis atrata, and related mainland lizards, P. hispanica. Biochem Syst Ecol. 2010;38:521–528. [Google Scholar]
- Gabirot M, Lopez P, Martín J (2012) Differences in chemical sexual signals may promote reproductive isolation and cryptic speciation between Iberian wall lizard populations. Int J Evol Biol 2012:698520 [DOI] [PMC free article] [PubMed]
- Geniez P, Sá-Sousa P, Guillaume CP, Cluchier A, Crochet PA. Systematics of the Podarcis hispanicus complex (Sauria, Lacertidae) III: valid nomina of the western and central Iberian forms. Zootaxa. 2014;3794:1–51. doi: 10.11646/zootaxa.3794.1.1. [DOI] [PubMed] [Google Scholar]
- Gomes V, Carretero MA, Kaliontzopoulou A. The relevance of morphology for habitat use and locomotion in two species of wall lizards. Acta Oecol. 2016;70:87–95. [Google Scholar]
- Gompert Z, Alex Buerkle C. INTROGRESS: a software package for mapping components of isolation in hybrids. Mol Ecol Resour. 2010;10:378–384. doi: 10.1111/j.1755-0998.2009.02733.x. [DOI] [PubMed] [Google Scholar]
- Grossen C, Seneviratne SS, Croll D, Irwin DE. Strong reproductive isolation and narrow genomic tracts of differentiation among three woodpecker species in secondary contact. Mol Ecol. 2016;25:4247–4266. doi: 10.1111/mec.13751. [DOI] [PubMed] [Google Scholar]
- Harris DJ, Sá-Sousa P. Molecular phylogenetics of Iberian wall lizards (Podarcis): is Podarcis hispanica a species complex? Mol Phylogenet Evol. 2002;23:75–81. doi: 10.1006/mpev.2001.1079. [DOI] [PubMed] [Google Scholar]
- Harvey MG, Seeholzer GF, Smith BT, et al. Positive association between population genetic differentiation and speciation rates in New World birds. Proc Natl Acad Sci USA. 2017;114:6328–6333. doi: 10.1073/pnas.1617397114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochkirch A, Lemke I. Asymmetric mate choice, hybridization, and hybrid fitness in two sympatric grasshopper species. Behav Ecol Sociobiol. 2011;65:1637–1645. [Google Scholar]
- Hudson EJ, Price TD. Pervasive reinforcement and the role of sexual selection in biological speciation. J Hered. 2014;105:821–833. doi: 10.1093/jhered/esu041. [DOI] [PubMed] [Google Scholar]
- IECA (2014) El tiempo vuela… Matalascañas 1956-2011. Instituto de Estadística y Cartografía de la Junta de Andalucía. (https://www.juntadeandalucia.es/institutodeestadisticaycartografia/blog/2014/04/matalascanas-1956-2011/; acceded at 03-08-2019)
- Irwin DE, Brelsford A, Toews DPL, MacDonald C, Phinney M. Extensive hybridization in a contact zone between MacGillivray’s warblers Oporornis tolmiei and mourning warblers O. philadelphia detected using molecular and morphological analyses. J Avian Biol. 2009;40:539–552. [Google Scholar]
- Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden, et al. (2010) Genetic restoration of the Florida panther. Science 329:1641–1645 [DOI] [PMC free article] [PubMed]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliontzopoulou A, Adams DC, van der Meijden A, Perera A, Carretero MA. Relationships between head morphology, bite performance and ecology in two species of Podarcis wall lizards. Evol Ecol. 2012;26:825–845. [Google Scholar]
- Kaliontzopoulou A, Carretero MA, Llorente GA. Morphology of the Podarcis wall lizards (Squamata: Lacertidae) from the Iberian Peninsula and North Africa: patterns of variation in a putative cryptic species complex. Zool J Linn Soc. 2012;164:173–193. [Google Scholar]
- Kaliontzopoulou A, Pinho C, Harris DJ, Carretero MA. When cryptic diversity blurs the picture: a cautionary tale from Iberian and North African Podarcis wall lizards. Biol J Linn Soc. 2011;103:779–800. [Google Scholar]
- Lamichhaney S, Han F, Berglund J, Wang C, Almén MS, Webster MT, Grant BR, et al. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science. 2016;352:470–474. doi: 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, Kremer A, et al. Species relative abundance and direction of introgression in oaks. Mol Ecol. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Levin DA, Francisco‐Ortega J, Jansen RK. Hybridization and the extinction of rare plant species. Conserv Biol. 1996;10:10–16. [Google Scholar]
- Leroy T, Louvet J-M, Lalanne C, Le Provost G, Labadie K, Aury JM, et al. Adaptive introgression as a driver of local adaptation to climate in European white oaks. N. Phytol. 2019;226:1171–1182. doi: 10.1111/nph.16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E (1942) Systematics and the origin of species, from the viewpoint of a zoologist. Columbia University Press, New York
- Mayr E. Speciation phenomena in birds. Am Nat. 1940;74:249–278. [Google Scholar]
- McIntosh EJ, Rossetto M, Weston PH, Wardle GM. Maintenance of strong morphological differentiation despite ongoing natural hybridization between sympatric species of Lomatia (Proteaceae) Ann Bot. 2014;113:861–872. doi: 10.1093/aob/mct314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milián-García Y, Ramos-Targarona R, Pérez-Fleitas E, et al. Genetic evidence of hybridization between the critically endangered Cuban crocodile and the American crocodile: implications for population history and in situ/ex situ conservation. Heredity. 2015;114:272. doi: 10.1038/hdy.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RI, Abbott RJ, Wolff K, Chamberlain DF. Hybridization among sympatric species of Rhododendron (Ericaceae) in Turkey: morphological and molecular evidence. Am J Bot. 1999;86:1776–1785. [PubMed] [Google Scholar]
- Neaves LE, Zenger KR, Cooper DW, Eldridge MDB. Molecular detection of hybridization between sympatric kangaroo species in south-eastern Australia. Heredity. 2010;104:502. doi: 10.1038/hdy.2009.137. [DOI] [PubMed] [Google Scholar]
- Niemiller ML, Fitzpatrick BM, Miller BT. Recent divergence with gene flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene genealogies. Mol Ecol. 2008;17:2258–2275. doi: 10.1111/j.1365-294X.2008.03750.x. [DOI] [PubMed] [Google Scholar]
- Owens GL, Samuk K (2020) Adaptive introgression during environmental change can weaken reproductive isolation. Nat Clim Change 10:58–62
- Palma‐Silva C, Wendt T, Pinheiro F, Barbará T, Fay MF, Cozzolino S, et al. Sympatric bromeliad species (Pitcairnia spp.) facilitate tests of mechanisms involved in species cohesion and reproductive isolation in Neotropical inselbergs. Mol Ecol. 2011;20:3185–3201. doi: 10.1111/j.1365-294X.2011.05143.x. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle CA. Genome‐wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol. 2012;21:2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x. [DOI] [PubMed] [Google Scholar]
- Pereira RJ, Monahan WB, Wake DB. Predictors for reproductive isolation in a ring species complex following genetic and ecological divergence. BMC Evol Bio. 2011;11:194. doi: 10.1186/1471-2148-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho C, Harris DJ, Ferrand N. Comparing patterns of nuclear and mitochondrial divergence in a cryptic species complex the case of Iberian and North African wall lizards (Podarcis, Lacertidae) Biol. J Linn Soc Lond. 2007;91:121–133. [Google Scholar]
- Pinho C, Harris DJ, Ferrand N. Non-equilibrium estimates of gene flow inferred from nuclear genealogies suggest that Iberian and North African wall lizards (Podarcis spp.) are an assemblage of incipient species. BMC Evol Biol. 2008;8:63. doi: 10.1186/1471-2148-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho C, Hey J. Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst. 2010;41:215–230. [Google Scholar]
- Pinho C, Kaliontzopoulou A, Carretero MA, Harris DJ, Ferrand N. Genetic admixture between the Iberian endemic lizards Podarcis bocagei and Podarcis carbonelli: evidence for limited natural hybridization and a bimodal hybrid zone. J Zool Syst Evol Res. 2009;47:368–377. [Google Scholar]
- Pinho C, Kaliontzopoulou A, Harris DJ, Ferrand N. Recent evolutionary history of the Iberian endemic lizards Podarcis bocagei (Seoane, 1884) and Podarcis carbonelli Pérez-Mellado, 1981 (Squamata: Lacertidae) revealed by allozyme and microsatellite markers. Zool J Linn Soc. 2011;162:184–200. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell J, Brelsford A, Wurm Y, Perrin N, Chapuisat M. Convergent genetic architecture underlies social organization in ants. Curr Biol. 2014;24:2728–2732. doi: 10.1016/j.cub.2014.09.071. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna,Austria. https://www.R-project.org/
- Renoult JP, Geniez P, Bacquet P, Benoit L, Crochet PA. Morphology and nuclear markers reveal extensive mitochondrial introgressions in the Iberian Wall Lizard species complex. Mol Ecol. 2009;18:4298–4315. doi: 10.1111/j.1365-294X.2009.04351.x. [DOI] [PubMed] [Google Scholar]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu Rev Ecol Syst. 1996;27:83–109. [Google Scholar]
- Rhymer JM, Williams MJ, Braun MJ (1994) Mitochondrial analysis of gene flow between New Zealand mallards (Anas platyrhynchos) and grey ducks (A. superciliosa). Auk 111:970–978
- Roberts DG, Gray CA, West RJ, Ayre DJ. Marine genetic swamping: hybrids replace an obligately estuarine fish. Mol Ecol. 2010;19:508–520. doi: 10.1111/j.1365-294X.2009.04501.x. [DOI] [PubMed] [Google Scholar]
- Rochette NC, Catchen JM. Deriving genotypes from RAD-seq short-read data using Stacks. Nat Protoc. 2017;12:2640. doi: 10.1038/nprot.2017.123. [DOI] [PubMed] [Google Scholar]
- Rubidge EM, Taylor EB. An analysis of spatial and environmental factors influencing hybridization between native westslope cutthroat trout (Oncorhynchus clarki lewisi) and introduced rainbow trout (O. mykiss) in the upper Kootenay River drainage, British Columbia. Conserv Genet. 2005;6:369–384. [Google Scholar]
- Sá-Sousa P. Comparative chorology between Podarcis bocagei and P. carbonellae (Sauria: Lacertidae) in Portugal. Rev Esp Herpetol. 2001;15:85–97. [Google Scholar]
- Sá-Sousa P (2008) Podarcis carbonelli. In: Loureiro A, Ferrand de Almeida N, Carretero MA, Paulo OS (eds.), Atlas dos Anfíbios e Répteis de Portugal. Instituto da Conservação da Natureza e da Biodiversidade, Lisboa. pp. 152–153
- Sá-Sousa P, Pérez-Mellado V and Martínez-Solano I (2009) Podarcis carbonelli. The IUCN Red List of Threatened Species 2009: e.T61545A12512496. 10.2305/IUCN.UK.2009.RLTS.T61545A12512496.en. Acceded 29 May 2019
- Sá-Sousa P, Harris DJ. Podarcis carbonelli Perez-Mellado, 1981 is a distinct species. Amphib-reptil. 2002;23:459–468. [Google Scholar]
- Sætre G. Hybridization is important in evolution, but is speciation? J Evol Biol. 2013;26:256–258. doi: 10.1111/jeb.12005. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Guillén RA, Córdoba‐Aguilar A, Cordero‐Rivera A, Wellenreuther M. Genetic divergence predicts reproductive isolation in damselflies. J Evol Biol. 2014;27:76–87. doi: 10.1111/jeb.12274. [DOI] [PubMed] [Google Scholar]
- Seehausen OLE, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol. 2008;17:30–44. doi: 10.1111/j.1365-294X.2007.03529.x. [DOI] [PubMed] [Google Scholar]
- Senn HV, Pemberton JM. Variable extent of hybridization between invasive sika (Cervus nippon) and native red deer (C. elaphus) in a small geographical area. Mol Ecol. 2009;18:862–876. doi: 10.1111/j.1365-294X.2008.04051.x. [DOI] [PubMed] [Google Scholar]
- Sillero N, Argaña E, Matos C, et al. Unexpected low population levels of Podarcis carbonelli in southern Salamanca (Spain) Bol Asoc Herpetol Esp. 2012;23:69–73. [Google Scholar]
- Sillero N, Carretero MA. Modelling the past and future distribution of contracting species. The Iberian lizard Podarcis carbonelli (Squamata: Lacertidae) as a case study. Zool Anz. 2013;252:289–298. [Google Scholar]
- Sillero N, Leerschool T, Hollanders M, Beukema W. No more Podarcis carbonelli in its type locality? Herpetol Notes. 2014;7:751–754. [Google Scholar]
- Simpson GG. Principles of animal taxonomy. New York: Columbia University Press; 1961. [DOI] [PubMed] [Google Scholar]
- Steeves TE, Maloney RF, Hale ML, Tylianakis JM, Gemmell NJ. Genetic analyses reveal hybridization but no hybrid swarm in one of the world’s rarest birds. Mol Ecol. 2010;19:5090–5100. doi: 10.1111/j.1365-294X.2010.04895.x. [DOI] [PubMed] [Google Scholar]
- Stelkens RB, Seehausen O. Phenotypic divergence but not genetic distance predicts assortative mating among species of a cichlid fish radiation. J Evol Biol. 2009;22:1679–1694. doi: 10.1111/j.1420-9101.2009.01777.x. [DOI] [PubMed] [Google Scholar]
- Szymura JM, Barton NH. Genetic analysis of a hybrid zone between the fire‐bellied toads, Bombina bombina and B. variegata, near Cracow in southern Poland. Evolution. 1986;40:1141–1159. doi: 10.1111/j.1558-5646.1986.tb05740.x. [DOI] [PubMed] [Google Scholar]
- Tarroso P, Pereira RJ, Martínez-Freiría F, Godinho R, Brito JC. Hybridization at an ecotone: ecological and genetic barriers between three Iberian vipers. Mol Ecol. 2014;23:1108–1123. doi: 10.1111/mec.12671. [DOI] [PubMed] [Google Scholar]
- Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, et al. (2016) Hybridization and extinction. Evol Appl 9: 892–908 [DOI] [PMC free article] [PubMed]
- Tompkins DM, Mitchell RA, Bryant DM. Hybridization increases measures of innate and cell‐mediated immunity in an endangered bird species. J Anim Ecol. 2006;75:559–564. doi: 10.1111/j.1365-2656.2006.01076.x. [DOI] [PubMed] [Google Scholar]
- VonHoldt BM, Brzeski KE, Wilcove DS, Rutledge LY. Redefining the role of admixture and genomics in species conservation. Conserv Lett. 2018;11:e12371. [Google Scholar]
- Vuillaume B, Valette V, Lepais O, Grandjean F, Breuil M. Genetic evidence of hybridization between the endangered native species Iguana delicatissima and the invasive Iguana iguana (Reptilia, Iguanidae) in the Lesser Antilles: management implications. PloS One. 2015;10:e0127575. doi: 10.1371/journal.pone.0127575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. The computer program Structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour. 2017;17:981–990. doi: 10.1111/1755-0998.12650. [DOI] [PubMed] [Google Scholar]
- Wayne RK, Shaffer HB. Hybridization and endangered species protection in the molecular era. Mol Ecol. 2016;25:2680–2689. doi: 10.1111/mec.13642. [DOI] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. N. Phytol. 2010;187:230–239. doi: 10.1111/j.1469-8137.2010.03234.x. [DOI] [PubMed] [Google Scholar]
- Whittemore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DE, Takebayashi N, Rieseberg LH. Predicting the risk of extinction through hybridization. Conserv Biol. 2001;15:1039–1053. [Google Scholar]
- Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
- Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in. Drosoph Evol. 2012;66:1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads from ddRAD-seq are deposited in the NCBI Sequence Read Archive (SRA). The BioProject accession number is PRJNA665746. VCF files with the complete datasets for each contact zone have been deposited on DRYAD (10.5061/dryad.k0p2ngf51).