Abstract
Microorganisms in marine sediments play major roles in marine biogeochemical cycles by mineralizing substantial quantities of organic matter from decaying cells. Proteins and lipids are abundant components of necromass, yet the taxonomic identities of microorganisms that actively degrade them remain poorly resolved. Here, we revealed identities, trophic interactions, and genomic features of bacteria that degraded 13C-labeled proteins and lipids in cold anoxic microcosms containing sulfidic subarctic marine sediment. Supplemented proteins and lipids were rapidly fermented to various volatile fatty acids within 5 days. DNA-stable isotope probing (SIP) suggested Psychrilyobacter atlanticus was an important primary degrader of proteins, and Psychromonas members were important primary degraders of both proteins and lipids. Closely related Psychromonas populations, as represented by distinct 16S rRNA gene variants, differentially utilized either proteins or lipids. DNA-SIP also showed 13C-labeling of various Deltaproteobacteria within 10 days, indicating trophic transfer of carbon to putative sulfate-reducers. Metagenome-assembled genomes revealed the primary hydrolyzers encoded secreted peptidases or lipases, and enzymes for catabolism of protein or lipid degradation products. Psychromonas species are prevalent in diverse marine sediments, suggesting they are important players in organic carbon processing in situ. Together, this study provides new insights into the identities, functions, and genomes of bacteria that actively degrade abundant necromass macromolecules in the seafloor.
Subject terms: Microbial ecology, Environmental microbiology
Introduction
The majority of marine sediments that underlie the Earth’s oceans are dominated by heterotrophic microorganisms, which are primarily sustained by “pelagic-benthic” coupling [1]. This is driven by a constant supply of organic matter from planktonic organisms that thrive in the overlying water column and settle with particulate aggregates to the seafloor after their death [2]. The amount and composition of organic matter that reaches the seafloor is strongly dependent on water depth, whereby continental shelves and shallow sediments generally receive greater inputs compared to deep-sea sediments [3]. In cold polar regions with lower microbial activities in the water column [4], a large fraction of the phytodetritus from spring bloom-like events reaches the underlying sediments [5]. Additionally, organic matter can originate from autochthonous microbial production [6] and from land-derived sources, which can be substantial in areas close to land and rivers, as well as polar regions subjected to melting [2, 7].
Organic matter in marine sediments is composed of an immense diversity of macromolecules, of which a substantial fraction can be enzymatically degraded and utilized as nutrient and energy sources by microorganisms [2]. Proteins and lipids typically constitute 10% and 5–10% of the organic matter found in marine sediments, respectively [8, 9]. Proteins, peptides, and amino acids are important sources of nitrogen [10], particularly in sediment habitats containing limited amounts of inorganic nitrogen sources [11]. Lipids generally consist of an alkyl chain that is ester- or ether-bound to a polar head group such as phospho-glycerol, or a glycerol that is glycosidically bound to a sugar moiety. They are major components of phytoplankton biomass [12, 13], of which the less labile fraction survives degradation in the water column [14] and is degraded by microorganisms in the underlying sediments [15]. Long-chain fatty acids released from lipid hydrolysis are energy-rich compounds [16], and can be expected to be favorable substrates for microorganisms.
Most macromolecules are too large to be directly imported into cells and must therefore be degraded, at least partially, outside the cells [17]. Hydrolysis of macromolecules via the activity of extracellular enzymes secreted by microorganisms is the rate limiting step during organic matter mineralization in marine sediments [17, 18]. Microorganisms utilize a compositionally and functionally large diversity of extracellular hydrolases to facilitate the extracellular breakdown of macromolecules [19]. For example, peptidases in sediments of Arctic fjords in Svalbard had greater substrate ranges for different peptides and much higher activities (tens to hundreds of thousands nmol l−1 h−1) than peptidases in the water column [20]. This showed that sediments act as important biogeochemical hotspots for processing organic macromolecules. Interestingly, peptidase activity was negatively correlated to amounts of phytodetritus inputs, whereas lipase activity was positively correlated [11]. This indicated that peptidases are excreted in times of nutrient limitation, whereas lipases are excreted when organic matter availability is high [11].
The microbial degradation of organic macromolecules in anoxic marine sediments is a complex inter-species process involving “primary degraders” that break down larger macromolecules into oligomers and monomers for fermentation to alcohols, lactate, and/or short-chain volatile fatty acids (VFAs), which are then mineralized to CH4, CO2 and/or H2 [21]. “Terminal oxidizers” of fermentation products prevalent in marine sediments, such as sulfate-reducing microorganisms (SRM), have been relatively well studied [22–27]. On the other hand, the key microbial players responsible for the primary hydrolysis of different types of organic matter and macromolecules are poorly understood. Most studies are based on predictions from genomic analyses [28–31], whereas experimental evidence linking identities to functions are lacking.
In the present study, we investigated the identities, genomic features, and ecological interactions of bacterial taxa that may play important roles in protein and lipid macromolecule degradation in subarctic marine sediments. We performed laboratory experiments whereby sulfidic arctic marine sediments were incubated in microcosms under cold (4 °C), anoxic conditions, and supplemented with either 13C-labeled proteins or 13C-labeled lipids. Incubations were performed at 4 °C to simulate temperature conditions of deep marine waters and seafloor, which are permanently cold [32]. These microcosms were also incubated under conditions where sulfate reduction was specifically inhibited, in order to elucidate the roles of SRM. Catabolism and assimilation of 13C-labeled substrates by the sediment bacteria were investigated by DNA-based stable isotope probing (DNA-SIP) and amplicon sequencing of 16S rRNA genes. The degradation of organic matter was monitored by measuring concentrations of VFAs and sulfate. Genome-resolved metagenomic analyses were used to predict secreted hydrolytic enzymes and reconstruct organic matter degradation pathways encoded by the taxa identified by DNA-SIP. This revealed (1) the identities and foraging strategies of several key protein- and lipid-consuming bacteria in marine sediments, (2) that niche partitioning among Psychromonas subspecies was based on differential utilization of proteins and lipids, and (3) that SRM of the family Desulfobacteraceae mainly utilized the macromolecule degradation intermediates, i.e., VFAs.
Materials and methods
Sediment incubations
Sediment slurries were produced with sediment obtained from intact Rumohr cores (0–30 cm below seafloor) from Greenland (Nuuk fjord “station 3,” water depth 498 m, August 2013, 64°26′45″N, 52°47′39″W) [33], mixed 1:1 (v/v) with anoxic artificial seawater [34] containing ~28 mM sulfate. Intact sediments were stored for ~5 weeks at 4 °C prior to the incubations. For each microcosm, 40 ml of this slurry was distributed into 250 ml serum vials under anoxic conditions in an anoxic glovebox (nitrogen atmosphere containing ~2% hydrogen and 10% CO2). Microcosms were sealed with thick butyl rubber stoppers and crimped, and flushed with N2 after removing from the anoxic glovebox. All subsequent subsampling was also performed within the anoxic glovebox, with microcosms placed on ice packs to minimize warming of incubations. Triplicate microcosms were either supplemented with a single dose of: (1) 300 µg C g−1 13C-labeled Algal lipid mixture-99 atom% 13C (ISOTEC, Sigma-Aldrich); (2) 300 µg C g−1 13C-labeled Algal crude protein extract-98 atom% 13C (ISOTEC, Sigma-Aldrich), (3) 300 µg C g−1 13C-labeled Algal lipid mixture (as above) and the sulfate reduction inhibitor molybdate (28 mM; [35]); (5) 300 µg C g−1 13C-labeled Algal protein mixture (as above) and molybdate (28 mM); or (6) were left without supplementation (no-substrate controls). Concentrations of substrates added to microcosms were chosen on the basis of previous research that showed bioavailable (hydrolysable) proteins up to 335 µg g−1 of deep-sea surface sediments [36]. Although no information for bioavailable lipids in sediments was available, we considered that they may be in a similar range because concentrations of total lipids and proteins are typically within similar ranges (at least within the same order of magnitude) in various sediments [37]. The microcosms were incubated at 4 °C and were sampled after 2, 5, 10, 17, 25, and 48 days of incubation. Sediment slurry samples were taken for analysis of VFAs and sulfate (1 ml), as well as for DNA-based analyses (0.5 ml). Samples were stored on precooled ice packs in the anoxic glovebox and were transferred immediately to the −80 °C freezer after sampling. Sampling, processing and molecular biological analyses of sediment samples from Svalbard, Norway, are presented as Supplementary information.
Determination of sulfate and volatile fatty acid concentrations
Sulfate concentrations in interstitial water samples of microcosms were determined by capillary electrophoresis (P/ACETM MDQ molecular characterization system, Beckman Coulter) with the CEofixTM anions 5 kit (Analis). Standards were produced by dissolving known concentrations of Na2SO4 in artificial seawater. The concentrations of VFAs were determined as follows: samples were defrosted, vortexed, and centrifuged at 10,000 rpm for 10 min. 100 µl of the supernatant was diluted 1:10 (v/v) in Milli-Q water. Syringe filters (Acrodisc IC grade filter, d = 13 mm, Suprapor® membrane with 0.2 µm pore size) were first rinsed with 10 ml Milli-Q water followed by 0.5 ml of diluted sample to minimize further dilution with the rinsing water. Another 0.5 ml of sample was then filtered and measured by two-dimensional ion chromatography-mass spectrometry (IC-IC-MS; Dionex ICS-3000 coupled to an MSQ Plus™, both Thermo Scientific), equipped with an Ion Pack™ AS 24 as the first column to separate bulk VFAs from chloride, and an Ion Pack™ AS 11 HC as the second column to separate individual VFAs [38].
DNA extraction
For DNA-SIP gradients and PCR-based amplicon sequencing, DNA was extracted using a combination of bead beating, cetyltrimethylammonium bromide-containing buffer, and phenol-chloroform extractions. Sediment slurry (0.5 ml) was added to Lysing Matrix E tubes (MP Biomedicals) and was suspended in 0.675 ml cetyltrimethylammonium bromide-containing extraction buffer, as described previously [39]. The tubes were placed on a rotary shaker (200 rpm) and incubated at 37 °C for 30 min. The samples were supplemented with 75 µL of sodium dodecyl sulfate (20% w/v) and were then incubated for 1 h at 65 °C (tubes were inverted every 20 min). Samples were subjected to two rounds of bead beating with a speed setting 6 for 30 s using a FastPrep®-24 bead beater (MP Biomedicals). In between the beat beating steps, samples were cooled on ice. Debris was pelleted by centrifugation at 6000 × g for 10 min at 25 °C and supernatant was collected. An equal volume of phenol:chloroform:isoamyl (25:24:1) alcohol (ROTH) was added to the supernatant and tubes were repeatedly inverted, followed by centrifugation at 16,000 × g for 10 min at 25 °C. The aqueous phase (upper layer) was transferred into a clean 1.5 ml tube, supplemented with 0.6 volume of 2-propanol, and incubated for 1 h at 4 °C to precipitate DNA. The precipitate was then pelleted by centrifugation at 16,000 × g for 30 min at 4 °C. DNA pellets were washed with 250 µL of 70% (v/v) ethanol, air-dried, and resuspended in 50 µL Tris buffer (10 mM Tris-HCl (pH 8.0)). For metagenome sequencing, DNA was extracted from day 0 and from individual sediment microcosm samples at days 5, 17, and 25 (for both protein and lipid-amended treatments) using the PowerSoil DNA Isolation Kit (MoBio) and following manufacturers’ protocol.
DNA-SIP gradients
CsCl gradients with 5 µg DNA were prepared in a temperature controlled room at 23 °C according to a previously published protocol [40]. The gradient mixtures were added to ultracentrifuge tubes (Beckman Coulter) and were centrifuged in an Optima L-100XP ultracentrifuge (Beckman Coulter) using the VTi 90 rotor for >48 h at 146286 rcf (44,100 rpm) at 20 °C. After centrifugation, gradients were fractionated into 250 µl fractions by puncturing the bottom of the tube with a sterile needle and adding water to the gradient tube at the top using a sterile needle and a syringe pump (World Precision Instruments). The density of collected fractions was determined by using a digital refractometer (AR 200, Reichert Analytical Instrument) at 23 °C. The DNA-SIP fractions were considered heavy at densities >1.726 g ml−1 and light at densities < 1.720 g ml−1 (Supplementary Fig. S1). Afterwards, DNA was precipitated with 500 µL sterile PEG 6000 (30% polyethylene glycol 6000 and 1.6 M NaCl) and 1 µL of glycogen (5 µg ml−1) and subsequently purified as described previously [40]. Bacterial DNA in SIP fractions was quantified by quantitative PCR (qPCR) using the primers 341F 5′-CCT ACG GGA GGC AGC AG-3′ and 534R 5′-ATT ACG GCG GCT GCT GGC A-3′. The 20 µL qPCR mix contained 1× IQTM SYBR Green Supermix (BIO-RAD), 0.25 µM of each primer, and 1 µL of 1:10 diluted DNA from individual gradient fractions. The program used for thermal cycling on the iCycler thermal cycler (Bio-Rad) consisted of: 3 min at 95 °C, followed by 39 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 39 s, and was followed by a melting curve from 60 °C to 95 °C by increments of 0.5 °C every 5 s.
Amplicon sequencing and analysis
Barcoded 16S rRNA gene amplicons for Illumina MiSeq sequencing were produced using a previously established two-step PCR approach [41, 42]. The primers were based on U519F 5′-CAG CMG CCG CGG TAA TWC-3′ and 802R 5′-TAC NVG GGT ATC TAA TCC-3′. Primer coverage was checked using TestProbe against SILVA v138 [43]. Raw reads were processed as described previously [41, 42]. The identity threshold used for OTU clustering was 97% identity. Sub-OTU diversity at single-nucleotide resolution in the amplicon data was investigated using cluster-free filtering [44]. Statistical significance of differences in OTU relative abundances between treatments was determined using DESeq2 (version 1.10.1) [45] in the R software environment http://www.r-project.org/index.html. Additionally, an OTU was considered to be 13C-enriched when it was significantly more abundant in heavy SIP fractions compared to light fractions obtained from the same incubation [46], which was also determined using DESeq2. The sequence abundance of each OTU in the “heavy” part (>1.726 g ml−1) of the DNA-SIP gradient (numerator) was compared to sequence abundance in the “light” part (<1.720 g ml−1) of the DNA-SIP gradient (denominator). Specific samples used for comparisons are indicated in Supplementary Fig. S1. Only OTUs that had ≥5 reads and that were present in ≥5 of the 32 gradient fraction sample data sets were considered for comparisons by DESeq2 analyses. Results were extracted with the command:
results(cooksCutoff = FALSE, altHypothesis = “greater”)
and were considered statistically significant if: (1) the false-discovery-rate (FDR)-adjusted p value was below 0.1, and (2) the respective OTU was not significantly enriched in the heavy fractions of the DNA-SIP gradient from the no-substrate incubations. OTU counts were “rlog” transformed with DESeq2 and heatmaps of enriched OTUs were created with R software package pheatmap (version 1.0.8).
The presence of 16S rRNA gene sequences related (>97% identity) to specific organisms of interest in publicly available amplicon-derived data sets was determined using the IMNGS server [47], with 100 bp overlap as minimum.
Metagenome sequencing and differential coverage binning
Metagenome libraries were produced with the Nextera XT DNA Library Preparation Kit (Illumina) and sequencing was performed at the Vienna Biocenter Core Facilities Next Generation Sequencing facility (Vienna, Austria) on an Illumina Hiseq2500 using HiSeq V4 chemistry with the 125 bp paired-end mode. Reads were end trimmed at the first base with a q score below 10 and reads with <50 bp were removed. For assembly, sequence coverage was normalized across samples using bbnorm with an average read depth of 100 and a minimum read depth of 3 (BBmap version 33.57 http://sourceforge.net/projects/bbmap/). Normalized read files were assembled with IDBA-UD [48] and SPAdes [49] using default parameters. Reads of each sample (non-normalized) were mapped to each assembly using BWA [50], and coverage information was obtained using SAMtools [51]. Differential coverage binning was performed with MetaBAT [52], MaxBin [53], and CONCOCT [54]. Following the binning programs default parameters, only contigs with a minimum length of 2500 and 1000 bp were used for binning with MetaBAT and MaxBin/CONCOCT, respectively. The resulting metagenome-assembled genomes (MAGs) generated from different assemblies and binning programs were aggregated with DASTool [55], and de-replicated with dRep using default parameters [56]. MAGs that could be linked to 13C-incorporating OTUs during protein and lipid hydrolysis by 16S rRNA gene sequence identity were further analyzed. Selected MAGs were evaluated for completion and contamination using CheckM [57], and classified using the Genome Taxonomy Database (GTDB) release 86 [58] and GTDB-Tk version 0.1.3 [59]. For a genome-based phylogenetic analysis of MAGs, the genome of the type strain Psychrilyobacter atlanticus HAW-EB21 and closely related reference genomes, was constructed from the concatenated marker protein alignment produced by CheckM [57]. The phylogenetic tree was built from this concatenated marker alignment with IQ-TREE using automatic substitution model selection (LG + F + I + G4) [60] and ultrafast bootstrap approximation with 1000 replicates [61]. The tree was visualized with iTOL [62]. Average nucleotide identities (ANI) and average amino acid identities (AAI) of selected MAGs and closely related reference genomes were determined using all-vs-all FastANI 1.1 with default parameters [63] and CompareM 0.0.23 (aai_wf) with default parameters (https://github.com/dparks1134/CompareM), respectively.
Genome annotation
MAGs were annotated with the MicroScope annotation platform [64] and using the RAST server [65]. The annotations of proteins were confirmed by DIAMOND searches [66] against the NCBI-nr database (e value 10−5), hidden Markov model-based searches using InterProScan [67] with the databases Pfam-A [68] and TIGRFAM [69], and online BLASTP searches against the UniProt database [70]. Possible hydrolytic enzymes were examined for signal peptide sequences that facilitate secretion from the cytoplasm and transmembrane helices using the Phobius online server [71] and using PSORTb version 3.0 [72]. Furthermore, peptidases, lipases/esterases, and glycoside hydrolases were additionally compared to the databases MEROPS [73], ESTHER [74], and CAZY [75], respectively, using DIAMOND searches (e value > 10−5, identity ≥ 30%). Furthermore, searches were made against the NCBI-nr database using BLASTP [76] for certain proteins of interest. The selection of potentially catabolic peptidases encoded by genomes and MAGs was based on two recent studies about extracellular peptidases in marine sediments [29, 77]. In addition, non-peptidase homologs and peptidases with regulatory functions, e.g., peptidases involved in membrane protein remodeling, as indicated by MEROPS database descriptions, were not considered (Supplementary Table S1). Potentially catabolic lipases/esterases were selected based on the ESTHER database descriptions (Supplementary Table S1).
Phylogenetic analyses of 16S rRNA gene sequences
16S rRNA gene sequences were aligned and classified with the SINA aligner [78], using the SILVA database release 128 [43]. Full-length sequences of close relatives of 16S rRNA OTUs were extracted from the SILVA database and combined with the aligned 16S rRNA genes sequences that were obtained from MAGs. A reference tree was then constructed using FastTree [79]. Subsequently, 16S rRNA OTUS and partial 16S rRNA genes from MAGs were added to the reference tree using the EPA algorithm [80] in RAxML [81]. Trees were visualized in iTOL [62].
Statistical analyses
Statistics for comparisons of VFAs and sulfate in treatment series versus controls were determined with Student’s t tests using the function t.test() in the R software environment.
Results
Sulfate removal and volatile fatty acid turnover during protein and lipid degradation
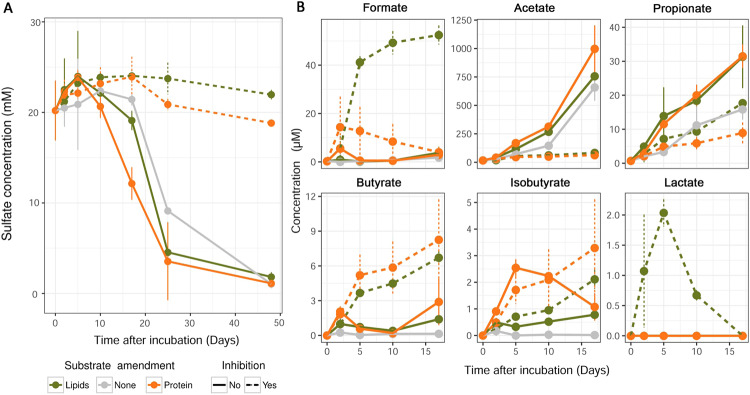
Sulfate was largely turned over after 48 days in all incubation treatments (down to 0.6–2.4 mM), except in incubations where sulfate reduction was inhibited by molybdate, where it remained between 18.1 and 24.2 mM (Fig. 1A). Sulfate turnover was fastest between days 5 and 25, and was stimulated by the additions of proteins or lipids. Of all measured VFAs, acetate was the most prominent, reaching concentrations of over 900 µM in protein-amended microcosms on day 17. Formation of the next most abundant VFA, formate, peaked at around 50 µM in lipid/molybdate-amended microcosms. Supplementation of proteins to the incubations resulted in significantly higher concentrations of acetate from days 2 to 5 compared to no-substrate controls (Supplementary Table S2). While the stimulation of acetate production from lipid additions was noticeably higher than in no-substrate controls, the differences were not statistically significant (Supplementary Table S2). Significantly more propionate, butyrate and isobutyrate were produced in protein- and lipid-amended microcosms, as compared to the no-substrate controls (mainly between 2 and 5 days) (Fig. 1B and Supplementary Table S2). Molybdate-inhibited incubations supplemented with proteins or lipids showed increased accumulation of formate, butyrate, and isobutyrate, but decreased production of acetate and propionate.
Fig. 1. Concentration profiles of sulfate and fermentation products in subarctic marine sediment microcosms.
Concentrations of sulfate (A) and volatile fatty acids (B) were determined in anoxic incubations with 13C-protein or 13C-lipids, and with or without the sulfate reduction inhibitor molybdate. Control microcosms were incubated without organic substrates and without molybdate.
13C-labeling of OTUs and sub-OTUs during protein and lipid degradation
To evaluate incorporation of 13C from the added 13C-labeled proteins or lipids into the DNA of microorganisms, bacterial 16S rRNA genes were quantified by qPCR across individual SIP fractions (Supplementary Fig. S1). Higher relative copy numbers were detected in the “heavy” fractions of the gradients (i.e., densities >1.726 g ml−1) from both 13C-protein and 13C-lipid incubations relative to corresponding no-substrate controls at days 5 and 10. This indicated 13C-uptake and incorporation into DNA at these early time points. At days 17 and 25, only very small or no differences in relative 16S rRNA gene copy numbers between gradients from the 13C-substrate incubations and the no-substrate control incubations were observed.
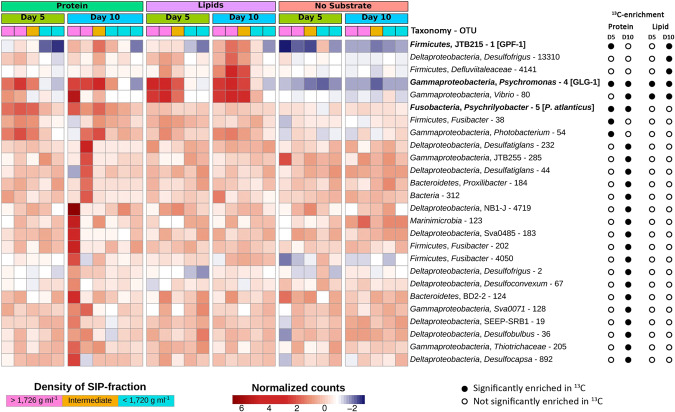
Bacterial community analysis of SIP fractions by 16S rRNA gene amplicon sequencing identified five OTUs that affiliated with various taxa, i.e., Clostridia-JTB215 OTU 1, Psychromonas OTU 4, Psychrilyobacter OTU 5, Fusibacter-related OTU 38, and Photobacterium OTU 54, were significantly enriched in heavy SIP fractions from the 13C-protein incubations at day 5 (Fig. 2A and Supplementary Table S3). At day 10, another 19 OTUs were significantly enriched in heavy SIP fractions from incubations with 13C-protein amendments (Fig. 2A). These were affiliated with the classes Deltaproteobacteria (OTUs 2, 19, 36, 44, 67, 183, 232, 892, 4719), Gammaproteobacteria (OTUs 80, 128, 205, 285) and the phyla Bacteroidetes (OTUs 124 and 184), Firmicutes (OTUs 202 and 4050), and Marinimicrobia (OTU 123), or were unclassified (OTU 312) (Supplementary Fig. S2).
Fig. 2. 16S rRNA gene OTUs enriched in heavy 13C-DNA-SIP gradient fractions.
Heatmaps showing the relative abundance of 16S rRNA gene OTUs in DNA-SIP gradient fractions from anoxic sediment incubations amended with 13C-proteins or 13C-lipids and from unamended control incubations. Significant (FDR-adjusted p value was <0.1) 13C-enrichment was determined by differential abundance analysis between high density (13C-enriched, >1.726 g ml−1, pink blocks) and low density (13C-free, <1.720 g ml−1, turquoise blocks) DNA, and is indicated by filled circles. 16S rRNA genes that were matched to 16S rRNA gene sequences of genomes/MAGs are indicated in bold.
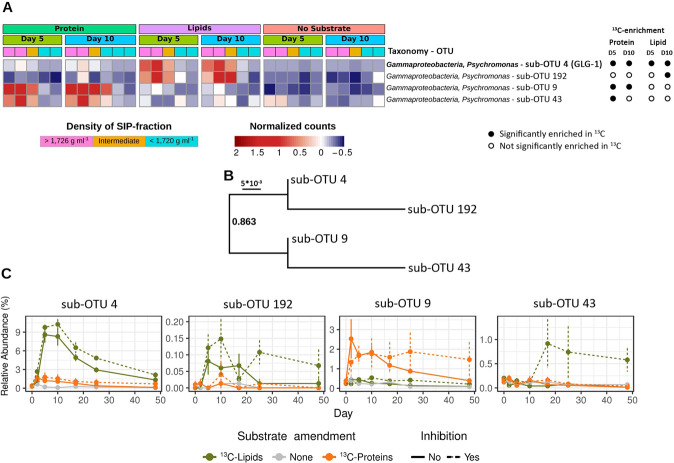
In SIP gradients from 13C-lipid-amended incubations at day 5, significant 13C-enrichment was revealed for only two gammaproteobacterial OTUs, i.e., Psychromonas OTU 4 and Vibrio OTU 80 (Fig. 2). An analysis of the microdiversity within Psychromonas OTU 4 identified four Psychromonas sub-OTUs (sub-OTUs 4, 192, 9, and 43) that responded differently in incubations with protein or lipid amendments. The sub-OTUs 4 and 192 were significantly enriched in heavy fractions of 13C-lipid incubations (Fig. 3A) and were more closely related to each other than to sub-OTUs 9 and 43 (Fig. 3B). Furthermore, the relative abundance of sub-OTUs 4 and 192 increased in the microcosms from 13C-lipid incubations compared to no-substrate control microcosms (Fig. 3C). In contrast, the sub-OTUs 9 and 43 were significantly enriched in heavy SIP fractions from 13C-protein incubations, and sub-OTU 9 increased in relative abundance in the microcosms from 13C-protein incubations compared to no-substrate control microcosms (Fig. 3C). No sub-OTU microdiversity was detected in any other OTU that was significantly enriched in 13C. At day 10, we identified three additional OTUs enriched in the heavy SIP fractions from 13C-lipid incubations that were affiliated with the phylum Firmicutes (OTUs 1 and 4141) and the class Deltaproteobacteria (OTU 13310) (Fig. 2A).
Fig. 3. Differential 13C-labeling and response of Psychromonas sub-OTUs in protein- and lipid-amended sediment microcosms.
A Heatmap showing the relative abundance of four sub-OTUs of Psychromonas 16S rRNA OTU 4 in DNA-SIP gradients. Significant 13C-enrichment was determined by differential abundance analysis between high density (13C-enriched) and low density (12C-enriched) DNA-SIP fractions. Sub-OTU 4, which was identical to the 16S rRNA gene sequence of Psychromonas MAG GLG-1, is indicated in bold. B Phylogeny of Psychromonas sub-OTUs. The sequences were aligned with MAFFT [89] and the tree was calculated with FastTree [79]. C Changes in relative abundance of Psychromonas sub-OTUs in protein-amended, lipid-amended, and unamended sediment microcosms with or without the sulfate reduction inhibitor molybdate.
We also compared the relative abundances of 16S rRNA OTUs among the protein- or lipid-amended microcosms versus no-substrate control microcosms over time. Here, we restricted the analysis to OTUs (n = 26) that were enriched in 13C from the protein- and lipid-amended treatments (described above). Between days 5 and 10, ten OTUs (1, 2, 4, 5, 19, 54, 80, 285, 4141, and 13310) significantly increased in relative abundances in substrate-amended incubations as compared to the no-substrate controls (Supplementary Fig. S3A and Supplementary Table S2). Most notably, several OTUs showed comparably large differences in relative abundances between amended and no-substrate controls, and consequently had high relative abundances among the overall communities. For instance, Psychrilyobacter OTU 5 reached relative abundances of around 9–10% after 2–5 days in protein-amended microcosms, while staying around 3–4% in no-substrate controls. Psychromonas OTU 4 reached relative abundances of around 15–16% between 5 and 10 days after lipid amendments, while also reaching around 5–7% in the protein-amended microcosms from days 2 to 10. In contrast, Psychromonas OTU 4 stayed below 2.5% in the no-substrate controls over time. The relatively fast (<10 days) and clear increases in relative abundances of these OTUs due to substrate additions therefore provided evidence for their direct involvement in substrate utilization for growth. On the contrary, the Clostridia-JTB215 OTU 1 also showed large increases in relative abundances from both protein and lipid amendments versus no-substrate controls, however, the response was relatively delayed, i.e., such increases developed mainly after 10 days.
Relative abundances of OTUs 1, 2, 67, 4050, and 4141 were significantly lower in microcosms with the sulfate-reduction-inhibitor molybdate than in microcosms without molybdate. This suggests that these OTUs are SRM or dependent on SRM activity.
Genomic evidence for extracellular hydrolysis of proteins and lipids
Samples from day 0, and from days 5, 17, and 25 from the lipid- and protein-amended microcosms were selected for metagenome sequencing. The aim was to recover genomes of organisms that were 13C-labeled from protein or lipid treatments, and to specifically analyze their catabolic potentials for these macromolecules. Overall, nine MAGs were recovered with >80% completeness and <5% contamination. Two had 16S rRNA sequences identical to bacterial OTUs that incorporated 13C from protein or lipid hydrolysis, i.e., Psychromonas MAG GLG-1 and Clostridia MAG GPF-1 (Supplementary Fig. S2). All MAGs that were affiliated with the genus Psychrilyobacter were very incomplete (<20%) (data not shown). Therefore, we analyzed the publicly available genome of the type strain P. atlanticus HAW-EB21. This strain was isolated from marine sediments [82], and had a 16S rRNA sequence that was identical to Psychrilyobacter OTU 5 that was labeled from protein amendments (Supplementary Fig. S2). A Desulfoluna MAG GLD-1 was also analyzed in detail because it encoded potential to degrade VFAs and/or lipids, although it could not be confidently linked to any 16S rRNA OTU sequence.
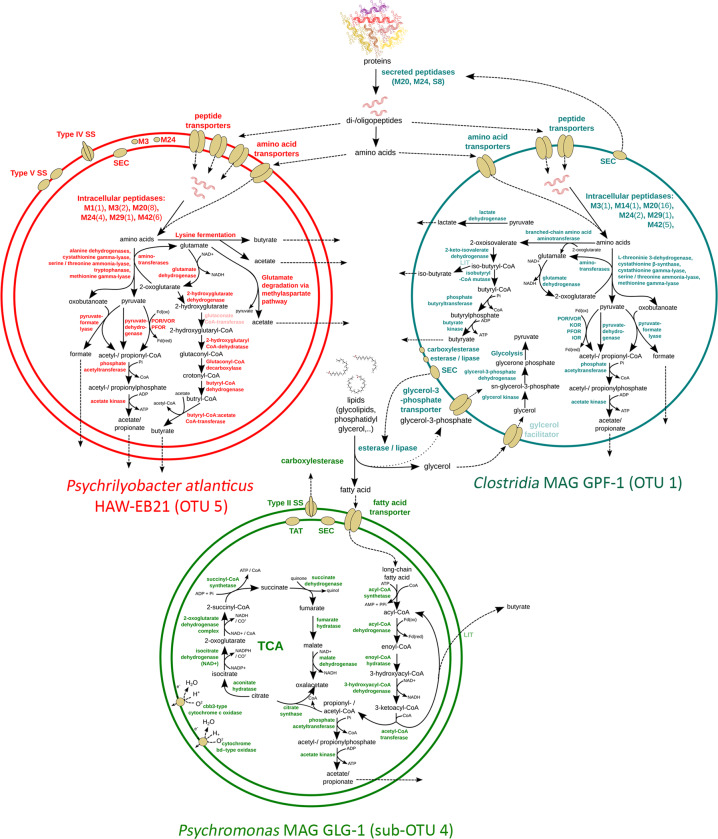
Psychromonas MAG GLG-1 was most closely related to Psychromonas aquimarina ATCC BAA-1526 (ANI 80.7% of 41.8% aligned) (Supplementary Table S4) and encoded a 16S rRNA sequence that was identical to Psychromonas sub-OTU 4 (Supplementary Fig. S2). Clostridia MAG GPF-1 was only distantly related to any publicly available genome (Supplementary Fig. S4), with an AAI of only 53.4% (of 37% aligned) (ANI was too low to calculate) to the most-related genome of Caloranaerobacter azorensis DSM 13643 (Supplementary Table S4). A short 16S rRNA fragment (92 bp) was retrieved from Clostridia MAG GPF-1 and was linked to the 16S rRNA OTU 1 by phylogenetic placement into a reference tree (Supplementary Fig. S2). The genomes/MAGs of putative protein and/or lipid degraders were subsequently analyzed for encoded capabilities to degrade proteins (P. atlanticus corresponding to 16S rRNA OTU 5), lipids (Psychromonas MAG GLG-1 corresponding to sub-OTU 4) or both macromolecules (Clostridia MAG GPF-1 corresponding to 16S rRNA OTU 1).
The genome of P. atlanticus HAW-EB21 encodes a variety of peptidases (n = 24), which were likely involved in the breakdown of peptides for further catabolism of amino acids (Supplementary Table S5). Two of these peptidases (i.e., M3 and M24) encoded signal peptides for translocation across the cytoplasmic membrane (Fig. 4). Overall, the genome of P. atlanticus encodes an array of predicted peptide (n = 19) and amino acid transporters (n = 38), especially when compared to the MAGs of the putative lipid degraders Psychromonas MAG GLG-1 (peptide transporter, n = 6; amino acid transporter, n = 14) (Supplementary Table S5). Furthermore, the genome of P. atlanticus was encoded for proton-dependent peptide transportation (Supplementary Table S5).
Fig. 4. Genome-inferred metabolic models of protein- and lipid-degrading bacteria in arctic marine sediment.
Schematic depiction of predicted metabolic properties of Psychrilyobacter atlanticus, Clostridia MAG GPF-1, and Psychromonas MAG GLG-1, which correspond to the 16S rRNA OTUs 5, 1, and sub-OTU 4, respectively (indicated in parenthesis). The models indicate macromolecule degradation processes and enzymes that may explain the observed biogeochemical patterns and the 13C-enrichment of corresponding 16S rRNA OTUs in the DNA-SIP incubations. Enzymes that are indicated in pale color were not found in the respective genome/MAG. SS secretion system, TCA tricarboxylic acid cycle, TAT twin-arginine translocation pathway, SEC Sec secretion system; LIT process has physiological evidence but the involved enzymes are not identified yet.
The lipid-metabolizing capability of Psychromonas MAG GLG-1 was supported by the presence of genes for an extracellular phospholipase D/alkaline phosphatase and a carboxylesterase with a signal peptide sequence, which corresponded to the ESTHER family carboxylesterase type B (Supplementary Table S5). The long-chain fatty acids that resulted from extracellular degradation of acyl chain esters were likely taken-up by cells via two predicted long-chain fatty acid transporters (Supplementary Table S5). Predicted glycerol transporters were not encoded in the genome of Psychromonas MAG GLG-1. Because our study revealed Psychromonas spp. to be potentially important lipid and protein degraders, we also investigated available genomes and/or MAGs (n = 16) from publicly available databases in order to examine the prevalence of the encoded capacity to utilize extracellular proteins or lipids via extracellular hydrolases (Supplementary Fig. S5 and Supplementary Table S6). This identified predicted secreted phospholipases in most genomes (n = 10), while predicted secreted proteases/peptidases were restricted to four genomes (Supplementary Fig. S5 and Supplementary Table S6).
Clostridia MAG GPF-1 encoded the largest number of peptidases (n = 29) of all analyzed MAGs/genomes (Supplementary Table S5). Three of these peptidases (M20, M24, and S8 peptidase families) have predicted secretion signal peptides and might be excreted from the cytoplasm via the SEC pathway (Fig. 4). Similar to P. atlanticus, Clostridia MAG GPF-1 encoded a broad array of predicted peptide (n = 14) and amino acid (n = 24) transporters. In accordance with the DNA-SIP results, Clostridia MAG GPF-1 also has the potential for catabolic breakdown of lipids. The MAG encoded one potentially secreted catabolic esterase/lipase that correspond to the ESTHER family Bacterial_EstLip_FamX, and two predicted membrane-bound esterases/lipases that correspond to the ESTHER families Bacterial_EstLip_Fam and carboxylesterase, type B (Supplementary Table S5). The presence of genes for esterases/lipases in the genome of Clostridia MAG GPF-1 indicated the potential of this MAG to hydrolyze ester bonds between glycerol and fatty acids. Because genes encoding for known fatty acid transport and catabolism were absent from the genome of Clostridia MAG GPF-1, this bacterium might instead utilize glycerol. This was supported by an encoded glycerol-3-phosphate transporter and a putative glycerol facilitator protein, which were also encoded directly adjacent to a glycerol kinase, which catalyses the first step of the cytoplasmic catabolism of this substrate (Supplementary Table S5).
Intracellular catabolism of protein and lipid degradation intermediates
The genome of P. atlanticus encoded 22 cytoplasmic peptidases with possible catabolic roles (Supplementary Table S5). In comparison, the genomes of the putative lipid degrader, i.e., Psychromonas MAG GLG-1 encoded only seven intracellular peptidases. In this study, we focused on amino acid degradation processes that form glutamate and/or keto-acids for central carbon metabolism and that could therefore explain incorporation of 13C-carbon from the 13C-labeled proteins into DNA, as well as the development of VFAs in our incubations (Fig. 4). The genome of P. atlanticus encoded several aminotransferases and other amino acid degrading enzymes (i.e., alanine dehydrogenases, cystathionine gamma-lyase, serine/threonine ammonia-lyase, tryptophanase, and methionine gamma-lyase) (Supplementary Table S5). These can convert amino acids to the keto-acids pyruvate and oxobutanoate, which could then be fermented to formate, and either acetate or propionate, respectively (Fig. 4).
Psychromonas MAG GLG-1 encoded all enzymes required for beta-oxidation of fatty acids, including a putative multienzyme complex that contains the two enzymes enoyl-CoA hydratase/isomerase and 3-hydroxyacyl-CoA dehydrogenase (Supplementary Table S5). Through beta-oxidation, even- and odd-chain fatty acids can be converted to propionyl- and acetyl-CoA, which can be further fermented to acetate and propionate, respectively (Fig. 4). In comparison to P. atlanticus and Clostridia MAG GPF-1, which encoded strict fermentative metabolisms, Psychromonas MAG GLG-1 also encoded a complete oxidative TCA cycle, which enables a respiratory conversion of acetyl-CoA to CO2 (Fig. 4). When oxygen is available, Psychromonas MAG GLG-1 might use one of the two encoded high affinity oxidases for respiration, i.e., cbb3-type cytochrome c oxidase and cytochrome bd-type oxidase (Supplementary Table S5). In oxygen-depleted sediments like in our microcosm incubations, Psychromonas GLG-1 might ferment, or may respire using nitrate (dissimilatory nitrate reduction to ammonium) or fumarate as terminal electron acceptors (Supplementary Table S5). Genes necessary for the intracellular degradation of glycerol were absent from Psychromonas MAG GLG-1 (Supplementary Table S5). Furthermore, despite the presence of genes for several extracellular glycosyl hydrolases in the genome of Psychromonas MAG GLG-1 and a complete glycolysis pathway (Supplementary Table S5), enzymes for feeding galactose or glycerol (subcomponents of Spirulina-derived galactosyl diglycerides), into glycolysis, were not identified.
The Clostridia MAG GPF-1 encoded the largest set of intracellular peptidases (n = 26) of all investigated genomes (Supplementary Table S5). The fermentation of amino acids by Clostridia GPF-1 to propionate, acetate, or formate, i.e., via aminotransferases and other amino acid degrading enzymes (i.e., L-threonine 3-dehydrogenase, cystathionine β-synthase, cystathionine gamma-lyase, serine/threonine ammonia-lyase, and methionine gamma-lyase), was mostly similar to the predicted routes used by P. atlanticus, although genes for lysine fermentation were not present and the glutamate degradation pathways were incomplete (Supplementary Table S5). In addition, the Clostridia MAG GPF-1 was the only MAG that encoded enzymes that initiate valine degradation, i.e., two branched-chain amino acid transferases and the gene 2-keto-isovalerate dehydrogenase that is part of the branched-chain α-keto acid dehydrogenase complex (Supplementary Table S5). Besides the potential for amino acid degradation, Clostridia MAG GPF-1 also encoded all genes necessary for the conversion of glycerol and glycerol-3-phosphate to pyruvate (Supplementary Table S5), which may help explain with the 13C-enrichment of 16S rRNA OTU 1 in 13C-lipid-amended incubations (Fig. 2).
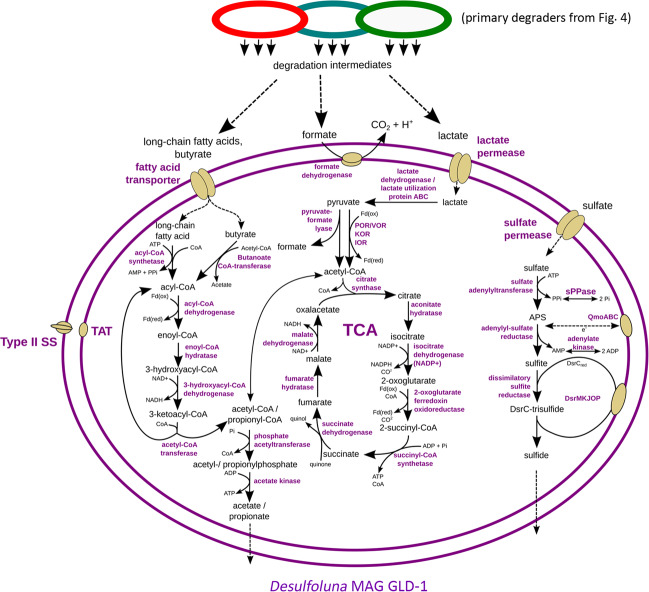
A putative sulfate-reducing Desulfoluna MAG GLD-1 with potential to use VFAs and long-chain fatty acids
The deltaproteobacterial Desulfoluna MAG GLD-1 encoded the capacity to utilize various VFAs and potentially lipids/long-chain fatty acids. In addition to several acyl-CoA synthetases, it encoded a butanoate CoA-transferase that enables conversion of butyrate to butyryl-CoA, which can then be funneled into beta-oxidation (Fig. 5). For lactate degradation, GLD-1 encoded several lactate dehydrogenases and a lactate utilization operon (Supplementary Table 5). Desulfoluna MAG GLD-1 also encoded a formate dehydrogenase enzyme complex (Supplementary Table 5) to oxidize formate to CO2 and H2 (Fig. 5). Furthermore, the potential to utilize lipids and long-chain fatty acids was indicated by genes for an outer membrane phospholipase A, a long-chain fatty acid transporter, and multiple long-chain fatty acid CoA ligases (Supplementary Table 5), all with high similarity to homologs from other known long-chain fatty acid- and lipid-degrading SRM. The Desulfoluna MAG GLD-1 encoded the complete pathway for dissimilatory sulfate reduction (Fig. 5).
Fig. 5. Genome-inferred metabolic model of Desulfoluna MAG GLD-1.
SS secretion system, TCA tricarboxylic acid cycle, TAT twin-arginine translocation system.
Environmental distributions of key taxa
To explore fine-scale depth distributions of the main primary-hydrolyzing Psychromonas and Psychrilyobacter species in sulfidic arctic marine sediments, we examined 16S rRNA-gene and -cDNA derived sequence data sets from sediments of Smeerenburgfjorden, Svalbard (Supplementary Fig. S6). This showed that Psychromonas sequences reached 1.7% and 1.9% relative abundances in 16S rRNA-gene and -cDNA libraries in 0–1 centimetres below seafloor (cmbsf) samples from Station GK, respectively. In a nearby core from Station J, they also reached 1.2% relative abundances in both 16S rRNA-gene and -cDNA libraries in 0–1 cmbsf samples. At both sites, relative abundances of Psychromonas 16S rRNA-genes and -transcripts were very low under 3 cmbsf (Supplementary Fig. S6). Sequences from the Psychrilyobacter reached only 0.3% in 0–1 cmbsf from Station J samples and comprised maximally only 0.05% of the 16S rRNA transcripts in the same sample (results not shown). Psychrilyobacter sequences were under 0.03% in all other samples deeper than 1 cmbsf. Furthermore, to gauge their prevalence in other marine sediments, we examined sequences that were similar (>97% identity) to the Psychromonas and Psychrilyobacter OTUs recovered in this study to sequences in publicly available 16S rRNA gene data sets. This showed that sequences related to these organisms were widely distributed in sediment sites around the world’s oceans (Supplementary Fig. S7). For example, Psychromonas sequences reached a maximum of 4% relative abundance in seagrass-associated sediments (Nantucket Sound, USA).
Discussion
Proteins and lipids comprise a significant fraction (up to 10% each) of organic matter, and are especially abundant and available in marine surface sediments that receive fresh organic matter from the water column [8, 36, 83]. They therefore represent major nutrient and energy sources for sediment microbiomes, although knowledge of the identities of microorganisms that actively degrade these macromolecules is lacking. The experimental SIP work presented here is important because previous metagenomic studies [31] only indicate potential to use certain macromolecules, but do not reveal which microorganisms actively perform such processes and to what extent. Our experiment showed that supplemented proteins and lipids were actively utilized by bacteria within 5 days of incubation. Both protein and lipid amendments induced rapid accumulation of several VFAs, demonstrating primary fermentation of both macromolecules. DNA-SIP further showed that 13C-carbon from protein and lipid catabolism was incorporated into the DNA of specific taxa within 5 days. This was also paralleled by sharp increases in the relative abundances by many of the same taxa in the corresponding microcosms. We therefore reasoned that taxa that were 13C-labeled and stimulated to higher relative abundances (>1%) within the first 5 days represented the main primary hydrolysers and fermenters of proteins or lipids. Thus, the following discussion especially focuses on these primary-hydrolyzing taxa.
Overall, more diverse OTUs incorporated 13C-carbon from proteins compared to lipids, especially after 10 days of incubation, possibly due to a combination of: (1) higher proportions and bioavailability of proteins in sediments [8, 9], (2) greater nutritional value of proteins (e.g., nitrogen), and/or (3) a greater biodegradability of proteins as compared to lipids [84]. Sediment microorganisms are also known to efficiently salvage organic nitrogen sources such as amino acids in times of low nutrient supply [11]. Therefore, some microorganisms may have salvaged free peptides or amino acids released from the primary hydrolysers and became 13C-labeled.
The Psychromonas 16S rRNA gene OTU 4 was notable in this study because our DNA-SIP results showed that it was a prominent degrader of both lipids and proteins. Interestingly, we could distinguish four sub-OTUs that each had distinct preferences for either lipids or proteins. Thus, this demonstrated that closely related populations of Psychromonas in marine sediments can have very different preferences for macromolecules that require completely different catabolic machinery. From the metagenomic analyses, the Psychromonas MAG GLG-1, which represented the lipid-degrading sub-OTU 4 population, indeed had gene content that supported its capacity to digest lipids and catabolize fatty acids. For instance, it encoded a secreted esterase/lipase to digest lipids from the extracellular environment, as well as long-chain fatty acid transporters and a beta-oxidation pathway to import and catabolize the fatty acids (Fig. 4). Although we did not recover any Psychromonas MAGs representative of the protein-degrading populations, comparative genomic analyses of publicly available Psychromonas genomes and MAGs showed that predicted secreted lipases and/or peptidases are common among this genus (Supplementary Fig. S5). Diverse members of this genus therefore have unique potentials to utilize lipids or proteins. An alternative possibility is that the Psychromonas sub-OTU 9 population may have become 13C-labeled after taking-up free amino acids released by other primary degraders. Yet if this was the case, one could expect that many other taxa would have been 13C-labeled at day 5, however, this was not apparent. It therefore points to the direct involvement of the Psychromonas sub-OTU 9 in the primary degradation of proteins.
Our bacterial community profiling showed that Psychromonas were relatively abundant and active (based on 16S rRNA expression) in shallow sediments (<3 cmbsf) from arctic Svalbard. Psychromonas were previously shown to be highly correlated with fluxes of fresh phytodetritus to surface sediments of the arctic Laptev Sea [85]. Together with our experimental and genomic analyses, this suggests that Psychromonas play an active role in the turnover of “fresh” and labile organic matter delivered to surface marine sediments from the water column. This is in line with previous experiments that showed Psychromonas were important players in the initial degradation of whole Spirulina (cyanobacterial) necromass in arctic marine sediment [23]. This may be explained by the high amounts of protein in such necromass. Our study therefore suggests that Psychromonas populations thrive in surface sediments by utilizing abundant macromolecules that represent fresh and labile organic matter, e.g., lipids and/or proteins.
We further showed that Psychrilyobacter OTU 5 was one of the most prominent protein and/or amino acid degraders in our experiment. Members of this genus were also shown to be important degraders of whole Spirulina necromass in marine sediment experiments [23, 34]. Here, we therefore deciphered their role in bulk organic matter degradation by showing that they are efficient degraders of detrital protein/peptides. Correspondingly, genomic analyses showed that the P. atlanticus type strain HAW-EB21 [82], which had 100% 16S rRNA sequence identity to OTU 5, encoded various routes for the degradation of peptides and for the subsequent fermentation of amino acids (Fig. 4) [82]. Their overall contributions to protein degradation in marine sediments may, however, be less than the Psychromonas because they were not abundant members of most sedimentary microbiomes analyzed.
The Clostridia MAG GPF-1 (16S rRNA OTU 1) had <54% AAI to any available genome, and therefore our data represent the first insights into the potential metabolism of this uncultured group. Overall, our analyses suggested that these organisms are versatile organic molecule utilizers. The Clostridia OTU 1 population was 13C-labeled in SIP analyses from both 13C-protein and -lipid incubations, and its relative abundances were also increased by the additions of both substrates, although to a much lower extent than for Psychromonas or Psychrilyobacter. Our genomic analyses indicated the potential to utilize peptides, amino acids, and glycerol (Fig. 4), thereby highlighting its versatility. Furthermore, OTU 1 also increased in no-substrate controls over time, suggesting it either utilized natural organic matter in the microcosms, or fermentation products produced from the degradation of the natural organic matter by other members of the community.
In addition to the relatively abundant taxa described above that were 13C-enriched and that could be linked to genomes or MAGs, we also identified 13C-enrichment in several low abundance taxa (<1% relative abundances), i.e., members of the genera Photobacterium (OTU 54), Vibrio (OTU 80), and Fusibacter-related OTU 38. These taxa may have therefore played a less prominent role in the primary hydrolysis of the macromolecules and are therefore discussed briefly in the Supplementary Discussion.
After the main primary degradation processes and increased formation of fermentation products (i.e., VFAs) in the first 5 days, ongoing depletion of sulfate and inhibition of sulfate depletion by molybdate indicated SRM were active. Various taxa including Deltaproteobacteria related to known SRM became 13C-labeled at day 10, which suggested transfer of carbon from the added macromolecules to SRM. This was likely via oxidation of the VFAs by the SRM, because deltaproteobacterial SRM are known to be important oxidizers of fermentation products in marine sediments [33, 86, 87]. A deltaproteobacterial Desulfoluna MAG GLD-1 recovered in this study supported previous observations because it encoded capacity to use various fermentation products, including butyrate, lactate, and formate (Supplementary Fig. 5). The detection of more 13C-labeled putative SRM of the Deltaproteobacteria from protein incubations than from lipid incubations may have reflected the more diverse taxa involved in primary degradation of the proteins, e.g., through more diverse metabolic interactions. We also hypothesized that the additional nitrogen supplied by the proteins may have facilitated the growth of more diverse Deltaproteobacteria, since none of the VFAs appeared to be limiting in either protein or lipid treatments.
In conclusion, this study provided new insights into the identities and functions of various protein- and lipid-degrading bacteria in cold marine sediments, and indicates that Psychromonas spp. are prominent and active players in the utilization of fresh detrital protein and/or lipid macromolecules. The activity of these primary hydrolysers also facilitates transfer of carbon and energy between trophic levels within the sediment microbial community. Future work could specifically examine the roles of Archaea, because our PCR primer sets only matched about 56% of Archaea in the SILVA database [43], and therefore they may have been underrepresented in our analyses. Archaea could be important, as Bathyarchaeota members in marine sediments encode the potential for protein or amino acid degradation in their genomes [28], and may be more important in deeper sediments where they often become more abundant and microbial activity is low [88].
Supplementary information
Acknowledgements
The authors would like to thank the crew of the R/V Sanna 2013 summer sampling campaign, which was funded by the Arctic Research Centre, Aarhus University, and Kasper Kjeldsen, Hans Røy, and Marit-Solveig Seidenkranz for providing the sediment samples. We thank captain (Stig Henningsen) and first mate of MS Farm for their assistance during sampling of Svalbard sediments in 2017, as well as the Kings Bay Marine Laboratory and the AWIPEV Arctic Research Base in Ny-Ålesund for assistance and laboratory space. Furthermore, the authors want to thank Carmen Czepe from the Vienna Biocenter Core Facilities Next Generation Sequencing facility for sequencing of metagenomic libraries on the Illumina Hiseq2500 instrument. This work was financially supported by the Austrian Science Fund (P29426-B29 to KW; P25111-B22 to AL).
Data availability
All sequence data sets and metagenome-assembled genomes from the DNA-SIP experiments are available under the NCBI-Genbank Bioproject PRJNA609450. DNA-SIP amplicon sequencing data sets are available in the NCBI-Genbank Sequence Read Archive under BioSample accession SAMN14253696 and SRA accessions SRR11221408-SRR11221561. Metagenomic sequence reads are available in the NCBI-Genbank Sequence Read Archive under BioSample accession numbers SAMN14421543–SAMN14421548. Metagenome-assembled genomes are available in the NCBI-Genbank under BioSample accession numbers SAMN14421524–SAMN14421532. Annotated MAGs and genomes from the MicroScope annotation platform for key protein- and lipid-degrading organisms, and Psychrilyobacter atlanticus DSM 19335, are publicly available in the MaGe-Microscope server (https://mage.genoscope.cns.fr/). Amplicon sequencing data sets from Svalbard sediments are available under NCBI-Genbank Bioproject PRJNA623111 and the BioSample accession numbers SAMN14538997–SAMN14539076.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Claus Pelikan, Kenneth Wasmund
Contributor Information
Kenneth Wasmund, Email: kenneth.wasmund@univie.ac.at.
Alexander Loy, Email: alexander.loy@univie.ac.at.
Supplementary information
The online version of this article (10.1038/s41396-020-00817-6) contains supplementary material, which is available to authorized users.
References
- 1.Hop H, Pearson T, Hegseth EN, Kovacs KM, Wiencke C, Kwasniewski S, et al. The marine ecosystem of Kongsfjorden, Svalbard. Polar Res. 2002;21:167–208. [Google Scholar]
- 2.Arndt S, Jørgensen BB, LaRowe DE, Middelburg JJ, Pancost RD, Regnier P. Quantifying the degradation of organic matter in marine sediments: a review and synthesis. Earth-Sci Rev. 2013;123:53–86. [Google Scholar]
- 3.Dunne JP, Sarmiento JL, Gnanadesikan A. A synthesis of global particle export from the surface ocean and cycling through the ocean interior and on the seafloor. Glob Biogeochem Cycles. 2007;21:1–16. [Google Scholar]
- 4.Christian JR, Karl DM. Bacterial ectoenzymes in m`arine waters: activity ratios and temperature responses in three oceanographic provinces. Limnol Oceanogr. 1995;40:1042–9. [Google Scholar]
- 5.Fabiano M, Pusceddu A. Total and hydrolizable particulate organic matter (carbohydrates, proteins and lipids) at a coastal station in Terra Nova Bay (Ross Sea, Antarctica) Polar Biol. 1998;19:125–32. [Google Scholar]
- 6.Bradley JA, Amend JP, LaRowe DE. Necromass as a limited source of energy for microorganisms in marine sediments. J Geophys Res Biogeosci. 2018;123:577–90. [Google Scholar]
- 7.Wehrmann LM, Formolo MJ, Owens JD, Raiswell R, Ferdelman TG, Riedinger N, et al. Iron and manganese speciation and cycling in glacially influenced high-latitude fjord sediments (West Spitsbergen, Svalbard): evidence for a benthic recycling-transport mechanism. Geochim Cosmochim Acta. 2014;141:628–55. [Google Scholar]
- 8.Burdige DJ. Preservation of organic matter in marine sediments: controls, mechanisms, and an imbalance in sediment organic carbon budgets? Chem Rev. 2007;107:467–85. doi: 10.1021/cr050347q. [DOI] [PubMed] [Google Scholar]
- 9.Hedges JI, Oades JM. Comparative organic geochemistries of soils and marine sediments. Org Geochem. 1997;27:319–61. [Google Scholar]
- 10.McCarthy M, Pratum T, Hedges J, Benner R. Chemical composition of dissolved organic nitrogen in the ocean. Nature. 1997;390:150–4. [Google Scholar]
- 11.Vetter YA, Deming JW. Extracellular enzyme activity in the Arctic Northeast Water polynya. Mar Ecol Prog Ser. 1994;114:23–34. [Google Scholar]
- 12.Parsons TR, Stephens K, Strickland JDH. On the chemical composition of eleven species of marine phytoplankters. J Fish Res Board Can. 1961;18:1001–16. [Google Scholar]
- 13.Hudson BJ, Karis IG. The lipids of the alga Spirulina. J Sci Food Agric. 1974;25:759–63. doi: 10.1002/jsfa.2740250703. [DOI] [PubMed] [Google Scholar]
- 14.Wakeham SG, Lee C, Farrington JW, Gagosian RB. Biogeochemistry of particulate organic matter in the oceans: results from sediment trap experiments. Deep Sea Res A. 1984;31:509–28. [Google Scholar]
- 15.Harvey HR, Rodger Harvey H, Fallon RD, Patton JS. The effect of organic matter and oxygen on the degradation of bacterial membrane lipids in marine sediments. Geochim Cosmochim Acta. 1986;50:795–804. [Google Scholar]
- 16.Sousa DZ, Smidt H, Alves MM, Stams AJM. Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol Ecol. 2009;68:257–72. doi: 10.1111/j.1574-6941.2009.00680.x. [DOI] [PubMed] [Google Scholar]
- 17.Meyer-Reil L-A. Ecological aspects of enzymatic activity in marine sediments. Brock/Springer Series in Contemporary Bioscience; Springer New York New York, NY 1991. p. 84–95.
- 18.Beulig F, Røy H, Glombitza C, Jørgensen BB. Control on rate and pathway of anaerobic organic carbon degradation in the seabed. Proc Natl Acad Sci USA. 2018;115:367–72. doi: 10.1073/pnas.1715789115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnosti C. Microbial extracellular enzymes and the marine carbon cycle. Ann Rev Mar Sci. 2011;3:401–25. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 20.Arnosti C. Contrasting patterns of peptidase activities in seawater and sediments: an example from Arctic fjords of Svalbard. Mar Chem. 2015;168:151–6. [Google Scholar]
- 21.Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6:441–54. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 22.Webster G, Watt LC, Rinna J, Fry JC, Evershed RP, Parkes RJ, et al. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ Microbiol. 2006;8:1575–89. doi: 10.1111/j.1462-2920.2006.01048.x. [DOI] [PubMed] [Google Scholar]
- 23.Müller AL, Pelikan C, de Rezende JR, Wasmund K, Putz M, Glombitza C, et al. Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ Microbiol. 2018;20:2927–40. doi: 10.1111/1462-2920.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoblauch C, Sahm K, Jørgensen BB. Psychrophilic sulfate-reducing bacteria isolated from permanently cold arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int J Syst Bacteriol. 1999;49:1631–43. doi: 10.1099/00207713-49-4-1631. [DOI] [PubMed] [Google Scholar]
- 25.Sahm K, Knoblauch C, Amann R. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine Arctic sediments. Appl Environ Microbiol. 1999;65:3976–81. doi: 10.1128/aem.65.9.3976-3981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Na H, Lever MA, Kjeldsen KU, Schulz F, Jørgensen BB. Uncultured desulfobacteraceae and crenarchaeotal group C3 incorporate 13C-acetate in coastal marine sediment. Environ Microbiol Rep. 2015;7:614–22. doi: 10.1111/1758-2229.12296. [DOI] [PubMed] [Google Scholar]
- 27.Wasmund K, Mußmann M, Loy A. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep. 2017;9:323–44. doi: 10.1111/1758-2229.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–8. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 29.Zinke LA, Glombitza C, Bird JT, Røy H, Jørgensen BB, Lloyd KG, et al. Microbial organic matter degradation potential in Baltic Sea sediments influenced by depositional conditions and in situ geochemistry. Appl Environ Microbiol. 2018;85:e02164–18.. doi: 10.1128/AEM.02164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orsi WD, Richards TA, Francis WR. Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol. 2018;3:32–7. doi: 10.1038/s41564-017-0047-9. [DOI] [PubMed] [Google Scholar]
- 31.Baker BJ, Lazar CS, Teske AP, Dick GJ. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome. 2015;3:14. doi: 10.1186/s40168-015-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer T, Levitus S, Garcia H, Locarnini RA, Stephens C, Antonov J. Objective analyses of annual, seasonal, and monthly temperature and salinity for the World Ocean on a 0.25 grid. Int J Climatol. 2005;25:931–45. [Google Scholar]
- 33.Glombitza C, Jaussi M, Røy H, Seidenkrantz M-S, Lomstein BA, Jørgensen BB. Formate, acetate, and propionate as substrates for sulfate reduction in sub-arctic sediments of Southwest Greenland. Front Microbiol. 2015;6:846. doi: 10.3389/fmicb.2015.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graue J, Engelen B, Cypionka H. Degradation of cyanobacterial biomass in anoxic tidal-flat sediments: a microcosm study of metabolic processes and community changes. ISME J. 2012;6:660–9. doi: 10.1038/ismej.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newport PJ, Nedwell DB. The mechanisms of inhibition of Desulfovibrio and Desulfotomaculum species by selenate and molybdate. J Appl Bacteriol. 1988;65:419–23. [Google Scholar]
- 36.Danovaro R, Dell’Anno A, Fabiano M. Bioavailability of organic matter in the sediments of the Porcupine Abyssal Plain, northeastern Atlantic. Mar Ecol Prog Ser. 2001;220:25–32. [Google Scholar]
- 37.Pusceddu A, Dell’Anno A, Fabiano M, Danovaro R. Quantity and bioavailability of sediment organic matter as signatures of benthic trophic status. Mar Ecol Prog Ser. 2009;375:41–52. [Google Scholar]
- 38.Glombitza C, Pedersen J, Røy H, Jørgensen BB. Direct analysis of volatile fatty acids in marine sediment porewater by two-dimensional ion chromatography-mass spectrometry. Limnol Oceanogr Methods. 2014;12:455–68. [Google Scholar]
- 39.Dumont MG, Radajewski SM, Miguez CB, McDonald IR, Murrell JC. Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and metagenomic analysis. Environ Microbiol. 2006;8:1240–50. doi: 10.1111/j.1462-2920.2006.01018.x. [DOI] [PubMed] [Google Scholar]
- 40.Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 41.Pelikan C, Herbold CW, Hausmann B, Müller AL, Pester M, Loy A. Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics. Environ Microbiol. 2016;18:2994–3009. doi: 10.1111/1462-2920.13139. [DOI] [PubMed] [Google Scholar]
- 42.Herbold CW, Pelikan C, Kuzyk O, Hausmann B, Angel R, Berry D, et al. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front Microbiol. 2016;6:731. doi: 10.3389/fmicb.2015.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orsi WD, Smith JM, Liu S, Liu Z, Sakamoto CM, Wilken S, et al. Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 2016;10:2158–73. doi: 10.1038/ismej.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, et al. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 49.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 54.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 55.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 59.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019 Nov 15:btz848. 10.1093/bioinformatics/btz848. Epub ahead of print. PMID: 31730192. [DOI] [PMC free article] [PubMed]
- 60.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 63.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017;45:D517–28.. doi: 10.1093/nar/gkw1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 67.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2013;42:D222–30.. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–3. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kall L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:W429–32.. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–15. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rawlings ND. MEROPS: the peptidase database. Nucleic Acids Res. 2000;28:323–5. doi: 10.1093/nar/28.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–9. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–95. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed]
- 76.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steen AD, Kevorkian RT, Bird JT, Dombrowski N, Baker BJ, Hagen SM, et al. Kinetics and identities of extracellular peptidases in subsurface sediments of the White Oak River Estuary, North Carolina. Appl Environ Microbiol. 2019;85:e00102–19. [DOI] [PMC free article] [PubMed]
- 78.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao J-S, Manno D, Hawari J. Psychrilyobacter atlanticus gen. nov., sp. nov., a marine member of the phylum Fusobacteria that produces H2 and degrades nitramine explosives under low temperature conditions. Int J Syst Evol Microbiol. 2009;59:491–7. doi: 10.1099/ijs.0.65263-0. [DOI] [PubMed] [Google Scholar]
- 83.Hedges JI, Oades JM. Comparative organic geochemistries of soils and marine sediments. Org Geochem. 1997;27:319–61. [Google Scholar]
- 84.Wakeham SG, Canuel EA. Degradation and preservation of organic matter in marine sediments. In: The handbook of environmental chemistry; Springer Berlin Heidelberg Berlin, Heidelberg 2006. p. 295–321.
- 85.Bienhold C, Boetius A, Ramette A. The energy–diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 2011;6:724–32. doi: 10.1038/ismej.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Finke N, Vandieken V, Jørgensen BB. Acetate, lactate, propionate, and isobutyrate as electron donors for iron and sulfate reduction in Arctic marine sediments, Svalbard. FEMS Microbiol Ecol. 2007;59:10–22. doi: 10.1111/j.1574-6941.2006.00214.x. [DOI] [PubMed] [Google Scholar]
- 87.Glombitza C, Egger M, Røy H, Jørgensen BB. Controls on volatile fatty acid concentrations in marine sediments (Baltic Sea) Geochim Cosmochim Acta. 2019;258:226–41. [Google Scholar]
- 88.Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 2012;6:1949–65. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data sets and metagenome-assembled genomes from the DNA-SIP experiments are available under the NCBI-Genbank Bioproject PRJNA609450. DNA-SIP amplicon sequencing data sets are available in the NCBI-Genbank Sequence Read Archive under BioSample accession SAMN14253696 and SRA accessions SRR11221408-SRR11221561. Metagenomic sequence reads are available in the NCBI-Genbank Sequence Read Archive under BioSample accession numbers SAMN14421543–SAMN14421548. Metagenome-assembled genomes are available in the NCBI-Genbank under BioSample accession numbers SAMN14421524–SAMN14421532. Annotated MAGs and genomes from the MicroScope annotation platform for key protein- and lipid-degrading organisms, and Psychrilyobacter atlanticus DSM 19335, are publicly available in the MaGe-Microscope server (https://mage.genoscope.cns.fr/). Amplicon sequencing data sets from Svalbard sediments are available under NCBI-Genbank Bioproject PRJNA623111 and the BioSample accession numbers SAMN14538997–SAMN14539076.