Abstract
RNase III DROSHA is upregulated in multiple cancers and contributes to tumor progression by hitherto unclear mechanisms. Here, we demonstrate that DROSHA interacts with β-Catenin to transactivate STC1 in an RNA cleavage-independent manner, contributing to breast cancer stem-like cell (BCSC) properties. DROSHA mRNA stability is enhanced by N6-methyladenosine (m6A) modification which is activated by AURKA in BCSCs. AURKA stabilizes METTL14 by inhibiting its ubiquitylation and degradation to promote DROSHA mRNA methylation. Moreover, binding of AURKA to DROSHA transcript further strengthens the binding of the m6A reader IGF2BP2 to stabilize m6A-modified DROSHA. In addition, wild-type DROSHA, but not an m6A methylation-deficient mutant, enhances BCSC stemness maintenance, while inhibition of DROSHA m6A modification attenuates BCSC traits. Our study unveils the AURKA-induced oncogenic m6A modification as a key regulator of DROSHA in breast cancer and identifies a novel DROSHA transcriptional function in promoting the BCSC phenotype.
Subject terms: Cancer stem cells, Breast cancer, Mechanisms of disease
Introduction
DROSHA processes primary miRNAs into precursor miRNAs. Accumulated evidence further reveals that DROSHA directly regulates diverse RNA metabolism across the transcriptome, including post-transcriptional control of RNA stability,1 alternative splicing,2 3′-end processing3 and transcriptional termination.4 Pathologically, DROSHA is aberrantly expressed in multiple cancer types,5 and contributrun 0tes to cancer progression through its miRNA processing and RNA cleavage functions.6,7 A recent study reports that DROSHA specifically binds proximal promoter regions to induce gene transcription through complexing with CBP80 and RNA Polymerase II.8 Yet, how this transcriptional role of DROSHA promotes tumor development and the underlying mechanisms involved are much less well understood. In addition, previous studies have revealed that DROSHA is transcriptionally regulated by key oncogenes in cancers. For instance, c-Myc directly binds to the E-box of DROSHA promoter to activate its transcription in B lymphoma cells and lung cancer cells.9 However, the regulation of DROSHA mRNA stability during tumor development remains unknown.
m6A is a highly prevalent modification in mRNA10 and is recognized by m6A-binding proteins (readers) that determines mRNA fate, through modulating their splicing, translation, or stability.11 m6A modification is catalyzed by a core METTL3-METTL14-WTAP m6A methyltransferase complex (writer) and removed by two key eraser enzymes (FTO and ALKBH5), and these factors are essential for cancer initiation and progression.12 For example, ALKBH5 demethylates FOXM1 nascent transcripts and enhances FOXM1 expression via lncRNA FOXM1-AS to promote tumorigenicity of glioblastoma stem-like cells.13 METTL14, negatively regulated by transcription factor SPI1, elevates the expression of oncogenes MYB and MYC via m6A modification to stimulate acute myeloid leukemia (AML) development and leukemia stem cell (LSC) self-renewal.14 Moreover, the m6A reader YTHDF2 represses mRNA stability of methylated tumor suppressor SOCS2 to promote tumor growth and lung metastasis in hepatocellular carcinoma.15 In contrast, newly identified m6A reader proteins (IGF2BP1-3) also play oncogenic roles in cancers by stabilizing methylated mRNA of oncogenic targets (especially, MYC).16 Thus, whether m6A modification regulates DROSHA mRNA stability to promote tumorigenicity is worth investigating.
The Aurora kinase A (AURKA) oncogene is overexpressed in a variety of tumors and plays multiple roles in cancer development.17 Our recent study also demonstrates that AURKA displays a nuclear kinase-independent function in transactivating the MYC promoter in cooperation with hnRNP K, contributing to breast cancer stem cell traits.18 Here, our data show that DROSHA interacting with β-Catenin transactivates the stemness gene STC1 in a non-canonical fashion to promote BCSC properties. Moreover, DROSHA mRNA stability is enhanced by m6A modification, which is in turn mediated by the AURKA-METTL14 axis and stabilized by AURKA-IGF2BP2 complex. Depletion of DROSHA m6A modification significantly suppresses BCSC phenotype. Altogether, these findings reveal the AURKA-mediated m6A modification as a key regulator of DROSHA expression, and define a transactivating role of DROSHA in BCSC maintenance.
Results
A non-canonical function of DROSHA in promoting STC1 transcription
As cancer stem-like cells contribute to tumorigenesis,19 we firstly examined the role of DROSHA in BCSC maintenance. A significant upregulation of both the DROSHA mRNA and protein levels, which were associated with enrichment of stemness factors including MYC, POU5F1, and NANOG (Supplementary information, Fig. S1a, b), was determined in BCSC-enriched spheroids. In breast cancer SK-BR-3 cells with overexpression of wild-type (WT) DROSHA, the proportion of ALDH+ cells were increased (Supplementary information, Fig. S1c, d). Meanwhile, forced expression of DROSHA significantly promoted sphere formation ability in replating sphere formation (Supplementary information, Fig. S1e, f) and extreme limiting dilution assays (Supplementary information, Fig. S1g). In contrast, depletion of DROSHA dramatically inhibited sphere formation abilities (Supplementary information, Fig. S1h–j). Moreover, silencing of DROSHA suppressed tumor growth and tumorigenesis in primary tumor xenograft and secondary limited dilution xenograft assays (Supplementary information, Fig. S1k–m). Clinically, high DROSHA expression predicted an inferior overall survival in breast cancer patients (n = 1076), suggesting a significantly unfavorable prognosis and shorter patient lifespan (Supplementary information, Fig. S1n).
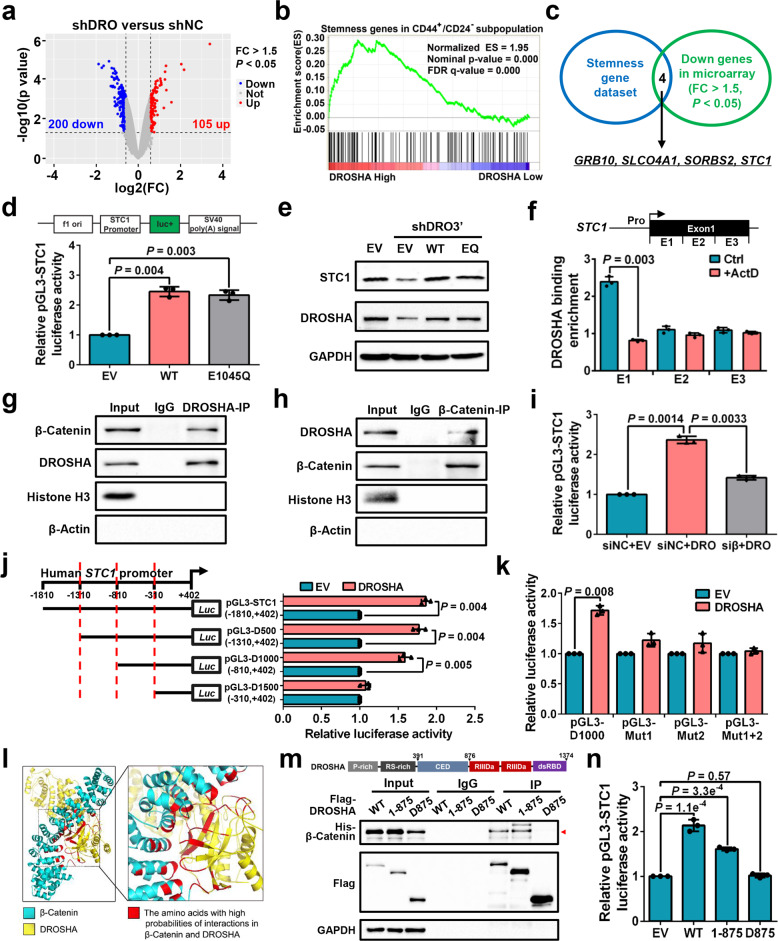
Next, we analyzed the differentially expressed genes (DEGs) in microarray data, comparing DROSHA-knockdown with control MDA-MB-231 cells. Two hundred genes were downregulated among the DEGs upon DROSHA knockdown (Fig. 1a). We then performed gene set enrichment analysis (GSEA) and found that the upregulated genes of DROSHA High groups in microarray data are also enriched in the CD44+/CD24− BCSC subpopulations (Fig. 1b), indicating that besides its role in RNA cleavage, DROSHA transcriptional activity also contributes to BCSC stemness. We thus overlapped those downregulated genes from DROSHA-knockdown microarray data with the stemness genes.20,21 The overlapped genes from these two groups include GRB10, SLCO4A1, SORBS2, and STC1 (Fig. 1c). To validate these candidates, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed and showed STC1, GRB10, and SLCO4A1 mRNA levels were downregulated and upregulated, respectively, in the DROSHA-deficient and DROSHA-overexpressing cells (Supplementary information, Fig. S2a, b). Ablation of DROSHA also decreased the protein expression of STC1, GRB10 and SLCO4A1 (Supplementary information, Fig. S2c, d). Both STC1 and GRB10 mRNA levels were enriched in sphere cells compared with non-spheroid cells (Supplementary information, Fig. S2e). Specifically, depletion of STC1, but not GRB10, SLCO4A1 or SORBS2, blocked DROSHA-deficiency-mediated ALDH+ subpopulation reduction in MDA-MB-231 cells (Supplementary information, Fig. S2f, g), indicating that STC1 might be critical for DROSHA-regulated BCSCs. High expression of STC1 denoted a poor prognosis for breast cancer patients (Supplementary information, Fig. S2h). To further confirm that DROSHA transactivates STC1, forced expression of WT or cleavage function mutation (E1045Q) DROSHA22 enhanced luciferase activity of the STC1 promoter reporter (Fig. 1d). Overexpression of WT or E1045Q DROSHA also rescued the STC1 expression in endogenous DROSHA-knockdown cells (Fig. 1e). As DROSHA can bind to proximal promoter regions of human protein-coding genes in a transcription-dependent manner,8 we also found that DROSHA can bind to the STC1 Exon 1 (E1) dependent on STC1 transcription by chromatin immunoprecipitation (ChIP) assay (Fig. 1f; Supplementary information, Fig. S2i).
Fig. 1. A non-canonical function of DROSHA in promoting STC1 transcription.
a Volcano plots displaying DEGs in microarray data comparing shDROSHA with shNC MDA-MB-231 cells. The numbers of significantly variant genes (FC > 1.5, P < 0.05) were shown. Vertical dashed lines indicate cut-off of FC (1.5), whereas the horizontal dashed lines indicate cut-off of P value (0.05). FC, fold change. b Upregulated DEGs in shNC group of microarray data were subjected to GSEA using the gene expression signature that was upregulated in the CD44+/CD24− signature acquired from a public database (GSE7513). c Overlapping stemness gene dataset and downregulated genes in shDROSHA cells from microarray data (FC > 1.5, P < 0.05), common genes were listed. d Relative luciferase activity of pGL3-STC1 in HEK293T cells with forced expression of WT and E1045Q DROSHA. e Western blotting showing STC1 and DROSHA expression in MDA-MB-231 cells with endogenous DROSHA knockdown and forced expression of WT and E1045Q DROSHA. f ChIP was performed to quantify the binding of DROSHA to STC1 in MDA-MB-231 cells with or without 5 μg/mL actinomycin D (ActD). g, h Co-IP assay to detect the interaction between DROSHA and β-Catenin in the nuclear extraction of MDA-MB-231 cells. Histone H3 as nuclear internal control and β-Actin as cytoplasmic internal control. i Relative luciferase activity of pGL3-STC1 in HEK293T cells with knockdown of β-Catenin and forced expression of DROSHA. siβ, siRNA against β-Catenin. j, k Relative luciferase activity of STC1 truncated promoters (j) and mutant promoters (k) in HEK293T cells with overexpression of DROSHA. l The simulated interaction diagram of β-Catenin and DROSHA. m Ability of the indicated Flag-labeled DROSHA derivatives to co-immunoprecipitate His-tagged β-Catenin defined by immunoblotting in HEK293T cells. n Relative luciferase activity of pGL3-STC1 in HEK293T cells with overexpression of the indicated DROSHA derivatives. Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test and P < 0.05 is considered statistically significant.
We next explored how DROSHA activates STC1 transcription, sought to identify DROSHA-interacting proteins using mass spectrometry analysis (Supplementary information, Table S2) and searched for the regulatory proteins of STC1 promoter with the assistance of software JASPAR. Comparing the candidates from the two analyses, a transcriptional factor complex, β-Catenin/TCF4, was identified (Supplementary information, Fig. S2j). Indeed, we confirmed the interaction between DROSHA and β-Catenin in the nucleus (Fig. 1g, h). Silencing of β-Catenin also significantly decreased the pre-mRNA, mRNA and protein levels of STC1 in DROSHA-overexpressing cells (Supplementary information, Fig. S2k, l). In addition, depletion of β-Catenin markedly reversed DROSHA-increased luciferase activity of the STC1 promoter reporter, indicating that β-Catenin is critical for the transcription of STC1 by DROSHA (Fig. 1i; Supplementary information, Fig. S2m). We next determined the region of STC1 promoter that is required for DROSHA-activated STC1 transcription and deletion analysis showed that DROSHA failed to activate STC1 promoter when the -810 to -310 region of STC1 promoter was removed (Fig. 1j). As two putative β-Catenin/TCF4 binding motifs are located within the -810 to -310 region of the STC1 promoter, we constructed three binding motif mutant reporters (Mut1, Mut2 and Mut1 plus Mut2) to perform luciferase assay (Supplementary information, Fig. S2n). Forced expression of DROSHA promoted luciferase activity of the pGL3-D1000 reporter, but had no effects on three mutant reporters (Fig. 1k). Further corroborating the mass spectrometry results, molecular dynamics (MD) simulation revealed 43 combinations of high interaction possibilities between amino acids of DROSHA CED domain (390–875 amino acids) and β-Catenin (Supplementary information, Fig. S2o), with the amino acids responsible for the DROSHA and β-Catenin interaction (red color) displaying spatial proximity (Fig. 1l; Supplementary information, Movie S1). As expected, only N-terminal 875 amino acids of DROSHA (1–875), but not the C-terminus of DROSHA (D875), interacted with His-β-Catenin (Fig. 1m). Importantly, DROSHA D875 failed to transactivate STC1 (Fig. 1n). Together, these data support a role of the CED domain of DROSHA in the recognition of β-Catenin to promote STC1 transcription.
DROSHA-STC1 axis promotes breast cancer stem-like cell properties
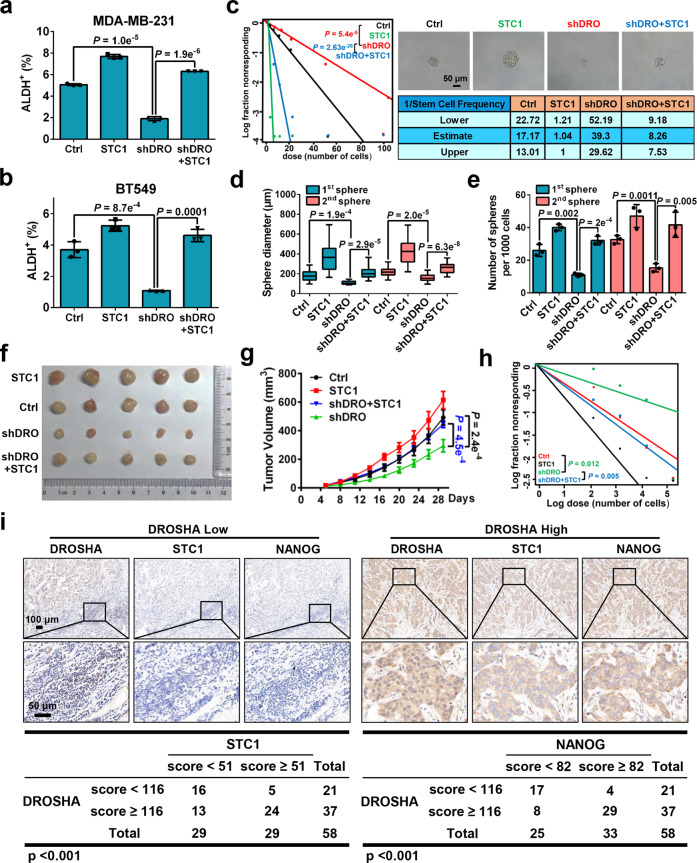
We next verified the role of DROSHA-STC1 axis in promoting BCSC properties. Overexpression of STC1 elevated ALDH+ subpopulations in DROSHA repressed breast cancer cells (Fig. 2a, b; Supplementary information, Fig. S3a, b). In addition, forced expression of STC1 restored sphere formation ability in extreme limiting dilution assays (Fig. 2c; Supplementary information, Fig. S3c). Consistently, overexpressing STC1 significantly increased the diameter and number of spheres in DROSHA-deficient cells using replating sphere formation assay (Fig. 2d, e). Next, MDA-MB-231 cells with DROSHA stable knockdown or control cells with overexpression of STC1 were injected into nude mice. Mice inoculated with DROSHA-deficient cells with overexpression of STC1 was shown to form larger tumor masses than the mice injected with DROSHA-deficient cells (Fig. 2f, g). Overexpression of STC1 also rescued DROSHA knockdown-mediated inhibition in tumorigenesis in secondary limited dilution xenograft assays (Fig. 2h; Supplementary information, Fig. S3d). Clinically, mRNA levels of DROSHA and STC1 were highly expressed in 21 breast tumor specimens compared with paired normal breast specimens (Supplementary information, Fig. S3e). As NANOG acts as a BCSC marker,23 we then performed immunohistochemistry (IHC) staining of DROSHA, STC1 and NANOG, to determine their correlations in breast tumor tissues. DROSHA expression positively correlated with the expression of STC1 and NANOG was shown in breast tumor tissues (Fig. 2i). Hence, our data reveal that DROSHA-STC1 axis has a crucial role in promoting BCSC phenotype.
Fig. 2. DROSHA-STC1 axis promotes BCSC properties.
a, b ALDH+ populations were analyzed in MDA-MB-231 (a) and BT549 (b) cells with DROSHA knockdown and forced expression of STC1. shDRO, shRNA against DROSHA. STC1, overexpression of STC1. c Left: ELDA was performed in MDA-MB-231 cells with DROSHA knockdown and forced expression of STC1. Top right: The representative sphere images are shown. Scale bars, 50 μm. Bottom right: Stemness frequency illustration of the cells with the upper and lower 95% confidence intervals meaning that the frequency of one stem cell in cancer cells. Spheres were counted from 24 replicate wells. d, e Replating sphere formation was performed in MDA-MB-231 cells with DROSHA knockdown and forced expression of STC1. The diameter (d) and number (e) of spheres were quantified. Spheres were counted from three replicate wells. The diameter and number of each experiment represent the total count of three replicate wells. f, g Immunodeficient mice (n = 5, biological replicates) were subcutaneously inoculated with MDA-MB-231 cells with forced expression of STC1 and DROSHA silencing (f), and tumor volumes were monitored (g). Ctrl, control cells. h Secondary limited dilution xenograft was performed by plating gradually decreasing numbers of primary xenografted tumor cells into immunodeficient mice (n = 5, biological replicates) and calculated with ELDA analysis. i Representative images of DROSHA, STC1 and NANOG IHC staining in breast tumor specimens (n = 58, biological replicates). Scale bars, 100 μm or 50 μm. Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test in a, b, e, g, ANOVA test in d, χ2 test in c, h, Pearson’s correlation test in i. P < 0.05 is considered statistically significant.
DROSHA mRNA is stabilized by m6A deposition
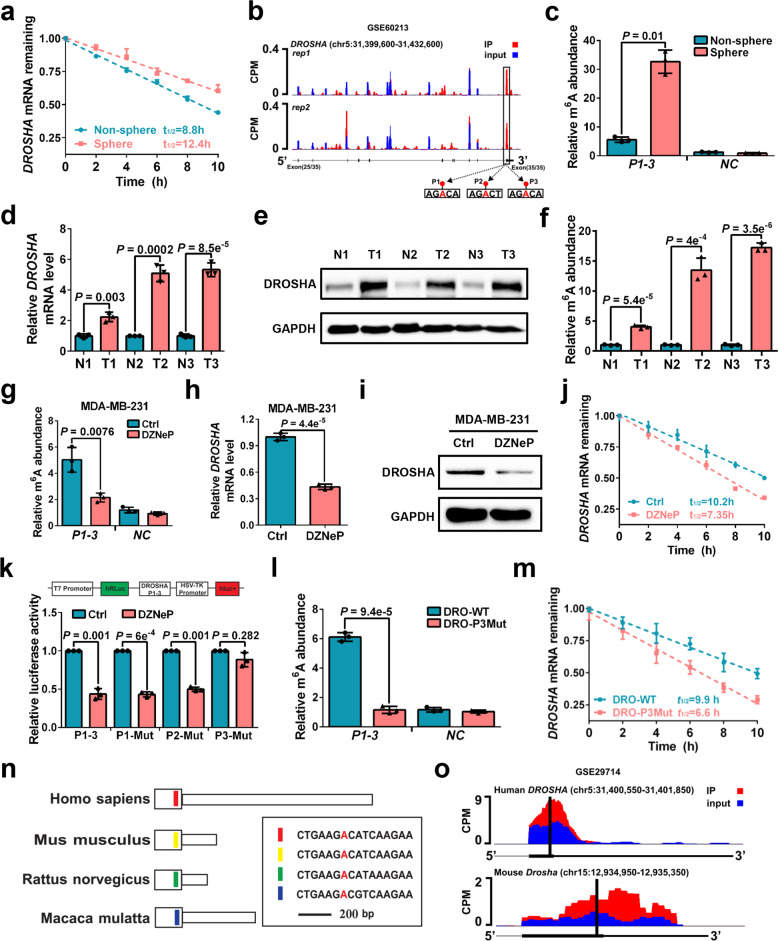
We further investigated how DROSHA is upregulated in BCSCs. As c-Myc could promote DROSHA transcription in B lymphoma cells and lung cancer cells,9 overexpression of MYC was shown to have no effects on DROSHA promoter activity, DROSHA mRNA level or DROSHA protein expression in breast cancer cells (Supplementary information, Fig. S4a–c). Moreover, there were no significant differences in DROSHA promoter activity and DROSHA pre-mRNA level between sphere cells and non-sphere cells (Supplementary information, Fig. S4d, e). In contrast, DROSHA mRNA exhibited a longer half-life in sphere cells compared to non-sphere cells (Fig. 3a; Supplementary information, Fig. S4f), suggesting that mRNA stability is responsible for high expression of DROSHA in BCSCs. By analyzing the m6A methylated RNA immunoprecipitation sequencing (meRIP-seq) data available for breast cancer cells, we found that abundant m6A peaks were accumulated near DROSHA mRNA stop codon including three putative m6A motifs (P1-P3) (Fig. 3b). meRIP-qPCR data further confirmed that m6A methylation had a higher abundance near the DROSHA mRNA stop codon in sphere cells compared to non-sphere cells (Fig. 3c). The mRNA expression, protein expression and m6A methylation of DROSHA were also much higher in breast tumor specimens compared to the paired normal breast specimens (Fig. 3d–f).
Fig. 3. DROSHA mRNA is stabilized by m6A deposition.
a Stability of DROSHA mRNA in non-sphere and sphere of MDA-MB-231 cells. mRNA levels were quantified by RT-qPCR. b UCSC Genome Browser plot containing tracks for m6A-seq IP reads (red) and input reads (blue) at the DROSHA locus by analyzing meRIP-seq data (GSE60213). The sequence of m6A motif near DROSHA stop codon is highlighted in three black boxes. c meRIP-qPCR was used to quantify relative DROSHA P1-3 m6A levels in non-sphere or sphere of MDA-MB-231 cells. NC, negative control, the primer of DROSHA non-m6A segment. d, e Relative mRNA (d) and protein (e) levels of DROSHA were determined between three breast tumor specimens and paired adjacent normal breast specimens. f meRIP-qPCR was used to quantify relative DROSHA m6A levels in three breast tumor specimens and paired adjacent normal breast specimens. g meRIP-qPCR was used to quantify relative DROSHA P1-3 m6A levels in MDA-MB-231 cells treated with DZNeP. h, i Relative mRNA (h) and protein (i) levels of DROSHA were determined in MDA-MB-231 cells treated with DZNeP. j Stability of DROSHA mRNA in MDA-MB-231 cells treated with DZNeP. k Relative luciferase activity of the DROSHA P1-3, P1-Mut, P2-Mut or P3-Mut reporter in HEK293T cells treated with DZNeP (n = 3, biological replicates). l meRIP-qPCR was used to quantify relative DROSHA P1-3 m6A levels in MDA-MB-231 cells with forced expression of DRO-WT and DRO-P3Mut. m Stability of DROSHA mRNA in MDA-MB-231 cells by forced expression of DRO-WT and DRO-P3Mut. n Diagram of the last exon of DROSHA genes from various organisms including coding sequence (large box) and 3’UTR. The conserved sequences are indicated by colored boxes. o m6A-seq reads cluster at the same distinct regions of DROSHA in both human brain RNA (top) and mouse brain RNA (bottom) by analyzing meRIP-seq data (GSE29714). Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test and P < 0.05 is considered statistically significant.
As 3-deazaneplanocin A (DZNeP) or methionine deprivation has been reported to inhibit RNA methylation,24,25 dot blot showed that DZNeP treatment or methionine deprivation significantly decreased mRNA m6A methylation in breast cancer cells (Supplementary information, Fig. S4g, h). After treating cells with DZNeP treatment or methionine depletion medium, we found that the m6A modification of DROSHA decreased dramatically (Fig. 3g; Supplementary information, Fig. S4i). Both the DROSHA mRNA and protein were downregulated in DZNeP-treated or methionine-depleted cells (Fig. 3h, i; Supplementary information, Fig. S4j, k). Furthermore, accelerated DROSHA mRNA decay upon DZNeP treatment or methionine depletion was observed in MDA-MB-231 cells (Fig. 3j; Supplementary information, Fig. S4l). DZNeP or methionine depletion also reduced luciferase activity of WT m6A motif DROSHA (P1-3), P1 m6A motif mutant DROSHA (P1-Mut) and P2 m6A motif mutant DROSHA (P2-Mut) reporters, but not that of the P3 m6A motif mutant DROSHA (P3-Mut) reporter (Fig. 3k; Supplementary information, Fig. S4m), suggesting that m6A modification of DROSHA mRNA at P3 is critical for DROSHA mRNA stability. We next confirmed that m6A abundance was decreased in MDA-MB-231 cells after the P3 m6A motif of DROSHA mRNA was mutated (Fig. 3l). Indeed, ectopic co-expression studies of WT DROSHA (DRO-WT) and P3 mutant DROSHA (DRO-P3Mut) in endogenous DROSHA-deficient cells showed that the P3 m6A motif mutation remarkably accelerated DROSHA mRNA decay (Fig. 3m) and decreased DROSHA protein expression (Supplementary information, Fig. S4n). Moreover, examination of the sequence of DROSHA transcript revealed that the P3 m6A motif of DROSHA mRNA is highly conserved across species (Fig. 3n). The m6A peaks were found to be enriched near DROSHA P3 motif in both human and mouse brain tissues by analyzing published meRIP-seq data (Fig. 3o). Similar results were also detected in hepatocellular carcinoma and lung cancer cells (Supplementary information, Fig. S4o). Thus, these findings demonstrate that BCSC-enriched m6A modification deposited on DROSHA transcript plays a crucial role in DROSHA mRNA stability in breast cancer cells.
AURKA-stabilized METTL14 maintains DROSHA transcript via m6A deposition
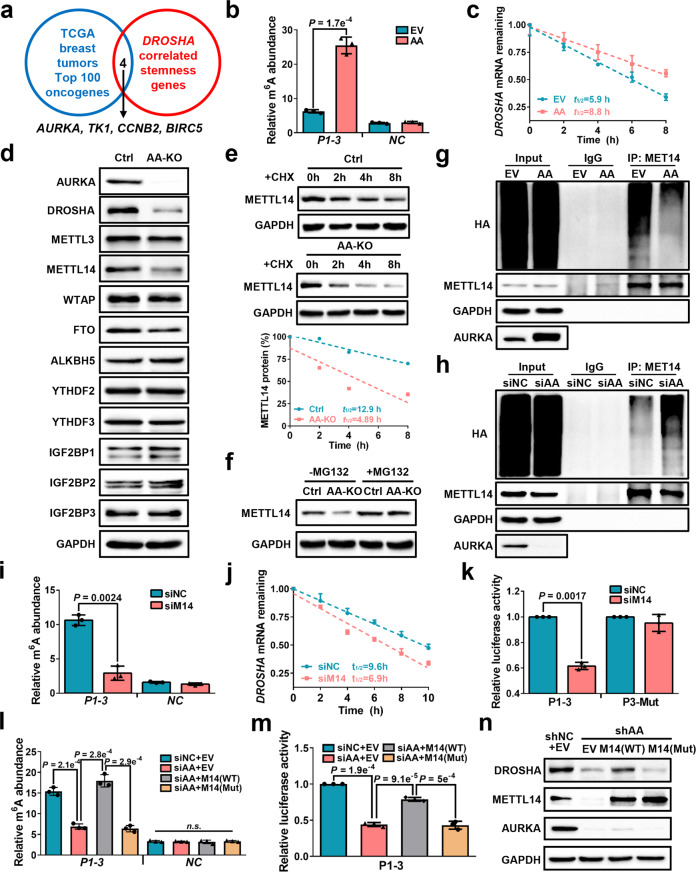
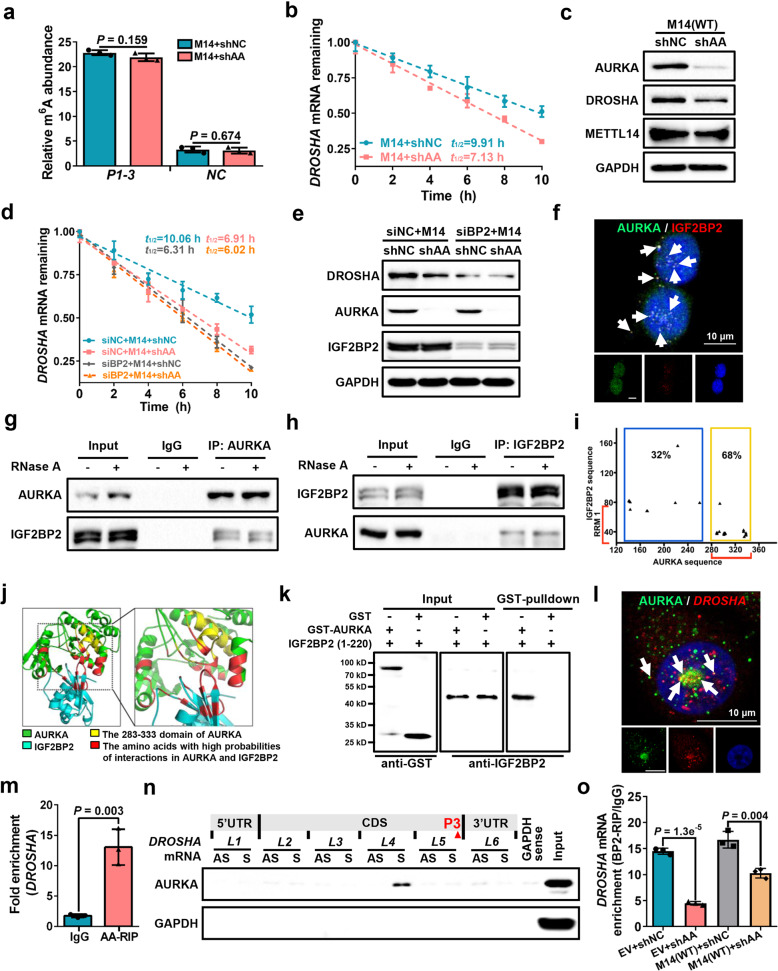
We next explored the key oncogenes that control DROSHA mRNA expression via m6A modification in BCSCs. To this end, we overlapped the top 100 highly expressed oncogenes from TCGA breast tumors with DROSHA-correlated stemness genes (Supplementary information, Table S3), and identified four oncogenes, including AURKA, TK1, CCNB2 and BIRC5 (Fig. 4a). Compared to non-sphere cells, the mRNA levels of AURKA and CCNB2 were significantly enriched in sphere cells (Supplementary information, Fig. S5a). Importantly, depletion of AURKA, but not TK1, CCNB2 or BIRC5, significantly decreased luciferase activity of the DROSHA P1-3 reporter, but not that of the P3-Mut reporter (Supplementary information, Fig. S5b, c), indicating that AURKA might act as a key regulator of DROSHA. We further determined whether AURKA regulates DROSHA m6A modification and mRNA stability. As expected, overexpression or ablation of AURKA increased or reduced DROSHA mRNA m6A methylation, respectively (Fig. 4b; Supplementary information, Fig. S5d), and forced expression of AURKA decelerated DROSHA mRNA decay in breast cancer cells (Fig. 4c; Supplementary information, Fig. S5e). Moreover, AURKA enhanced luciferase activity of P1-3, P1-Mut and P2-Mut reporters, but not the P3-Mut reporter (Supplementary information, Fig. S5f).
Fig. 4. AURKA-stabilized METTL14 maintains DROSHA transcript via m6A deposition.
a Comparison of DROSHA-correlated stemness genes with highly expressed oncogenes in TCGA breast tumor (top 100), common genes were listed. b meRIP-qPCR was used to determine relative DROSHA P1-3 m6A levels in AURKA-overexpressing SK-BR-3 cells. EV, empty vector, AA, overexpression of AURKA. c Stability of DROSHA mRNA in AURKA-overexpressing SK-BR-3 cells. d The expression of indicated proteins was detected in AURKA-knockout MDA-MB-231 cells by western blotting. AA-KO, knockout of AURKA. e METTL14 expression was detected in AURKA-knockout MDA-MB-231 cells treated with cycloheximide (CHX, 200 μg/mL). METTL14 proteins were quantified by densitometry and plotted as a scatter diagram at the bottom. f METTL14 expression was detected in AURKA-knockout MDA-MB-231 cells treated with MG132 (10 μM) for 8 h. g, h HEK293T cells were transfected with indicated constructs (g) or siRNAs (h) followed by co-transfection with HA-Ub and METTL14 constructs. Cells were treated with MG132 (10 μM) for 8 h before collection. The whole cell lysate was subjected to immunoprecipitation with METTL14 antibody and western blotting with anti-HA antibody to detect ubiquitylated METTL14. i meRIP-qPCR was performed to verify relative DROSHA P1-3 m6A levels in METTL14-knockdown MDA-MB-231 cells. j Stability of DROSHA mRNA in METTL14-knockdown MDA-MB-231 cells. k Relative luciferase activity of P1-3 and P3-Mut in HEK293T cells with METTL14 knockdown. l Relative DROSHA P1-3 m6A levels were detected in AURKA-deficient MDA-MB-231 cells with or without overexpression of indicated METTL14. m Relative luciferase activity of P1-3 in AURKA-deficient HEK293T cells with or without indicated overexpression of METTL14. n DROSHA, METTL14 and AURKA protein expression was identified in AURKA-deficient MDA-MB-231 cells with or without overexpression of indicated METTL14. shAA, shRNA against AURKA. Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test and P < 0.05 is considered statistically significant.
We next investigated how AURKA upregulates DROSHA m6A modification, as our previous study revealed that AURKA acted as a co-transcription factor to transactivate MYC.18 Overexpression and depletion of AURKA increased and decreased the mRNA and protein levels of MYC, respectively (Supplementary information, Fig. S5g–j), while knockdown or overexpression of AURKA had little effects on mRNA levels of the m6A methyltransferases and demethylases (Supplementary information, Fig. S5k, l). Furthermore, we found that the protein expression of core methyltransferase METTL14, but not other methyltransferases, demethylases and readers, was consistently downregulated in AURKA-knockout cells and upregulated in AURKA-overexpressing cells (Fig. 4d; Supplementary information, Fig. S5m). Inhibition of AURKA kinase activity also displayed no effects on METTL14 expression (Supplementary information, Fig. S5n), indicating AURKA regulates METTL14 expression in a kinase-independent manner. After treatment with the protein synthesis inhibitor cycloheximide (CHX), knockout of AURKA shortened the half-life of endogenous METTL14 protein (Fig. 4e), whereas overexpression of AURKA resulted in the prolonged half-life of endogenous METTL14 protein (Supplementary information, Fig. S5o). We next treated cells with the proteasome inhibitor MG132 in the absence of AURKA. MG132 enhanced METTL14 expression, whereas AURKA knockout did not decrease DROSHA level in the presence of MG132 treatment (Fig. 4f), suggesting that AURKA stabilizes METTL14 protein expression through inhibition of the proteasome-dependent degradation pathway. Moreover, we found that overexpression of AURKA reduced METTL14 ubiquitylation (Fig. 4g), whereas knockdown of AURKA increased METTL14 ubiquitylation (Fig. 4h). These data propose that AURKA protects METTL14 protein from ubiquitylation-degradation to promote DROSHA m6A modification.
Next, we found that ablation of METTL14 significantly decreased DROSHA expression in breast cancer cells (Supplementary information, Fig. S6a, b). The available m6A-sequence data revealed that depletion of METTL14 reduced m6A peak near the DROSHA P3 motif in various cancer cells (Supplementary information, Fig. S6c, d). Similar reduction of DROSHA mRNA m6A methylation was observed in the METTL14-knockdown MDA-MB-231 cells compared to controls in meRIP-qPCR assays (Fig. 4i). Accelerated DROSHA mRNA decay upon silencing METTL14 was confirmed in MDA-MB-231 cells (Fig. 4j). Furthermore, silencing of METTL14 reduced luciferase activity of the P1-3 reporter, but not that of the P3-Mut reporter in MDA-MB-231 cells (Fig. 4k). Moreover, R298P mutation (R298P) significantly reduced the m6A methylation activity of METTL14 (Supplementary information, Fig. S6e, f), overexpression of WT METTL14 promoted luciferase activity of the P1-3 reporter but not P3-Mut reporter, while the R298P METTL14 had no effect on luciferase activity of either DROSHA reporters (Supplementary information, Fig. S6g). Clinically, METTL14 mRNA and protein were highly expressed in breast tumor compared with paired normal breast specimens (Supplementary information, Fig. S6h, i). In addition, WT, but not R298P, METTL14 rescued DROSHA mRNA m6A level in AURKA-deficient cells (Fig. 4l). Importantly, WT, but not R298P, METTL14 partially restored luciferase activity of the P1-3 DROSHA (Fig. 4m) as well as DROSHA expression (Fig. 4n) following AURKA silencing. Taken together, these results reveal that AURKA enhances DROSHA mRNA stability partially via METTL14-dependent m6A deposition in breast cancer cells.
AURKA strengthens IGF2BP2 binding to m6A for DROSHA transcript stabilization
To further explore other mechanisms by which AURKA regulated DROSHA mRNA stability independent of the METTL14-regulated m6A change, METTL14-overexpressing cells were established to determine whether a steady DROSHA m6A state could be maintained following AURKA depletion. Indeed, AURKA knockdown had no effects on DROSHA m6A levels in METTL14-overexpressing cells (Fig. 5a). However, ablation of AURKA attenuated DROSHA mRNA stability and decreased DROSHA expression in METTL14-overexpressing cells (Fig. 5b, c). As insulin-like growth factor 2 mRNA-binding proteins 1, 2 and 3 (IGF2BP1-3) have recently been reported as m6A readers to recognize and stabilize m6A modified transcripts,16 we hypothesized that IGF2BP1-3 mediate AURKA-enhanced DROSHA stability in an METTL14-independent manner. We found that depletion of IGF2BP2, but not IGF2BP1 or 3, substantially reduced AURKA-regulated DROSHA mRNA stability in METTL14-overexpressing cells (Fig. 5d; Supplementary information, Fig. S7a–c). Ablation of IGF2BP2 also restricted AURKA-regulated DROSHA protein expression in METTL14-overexpressing cells (Fig. 5e). In addition, binding of IGF2BP2 to DROSHA transcript was enriched in sphere cells compared to non-sphere cells (Supplementary information, Fig. S7d). Both the mRNA and protein levels of IGF2BP2 showed no significant differences between breast tumor specimens and the paired normal breast specimens (Supplementary information, Fig. S7e, f).
Fig. 5. AURKA strengthens IGF2BP2 binding to m6A for DROSHA transcript stabilization.
a meRIP-qPCR was performed to verify relative DROSHA P1-3 m6A levels in METTL14-overexpressing MDA-MB-231 cells with or without AURKA-deficiency. b Stability of DROSHA mRNAs were identified in METTL14-overexpressing MDA-MB-231 cells with or without AURKA-deficiency. c The protein expression of AURKA, DROSHA and METTL14 was identified in METTL14-overexpressing MDA-MB-231 cells with or without AURKA deficiency. d Stability of DROSHA mRNAs was evaluated in METTL14-overexpressing plus IGF2BP2-knockdown (siIGF2BP2) or siNC MDA-MB-231 cells with or without AURKA deficiency. e DROSHA, AURKA and IGF2BP2 proteins were detected by western blotting in METTL14-overexpression plus siIGF2BP2 or siNC MDA-MB-231 cells with or without AURKA deficiency. f The representative image of co-staining of AURKA protein (green) and IGF2BP2 protein (red) were observed in MDA-MB-231 cells. The nucleus was stained by DAPI (blue). Scale bars, 10 μm. g, h Co-IP assay to identify the endogenous interaction between AURKA and IGF2BP2 in MDA-MB-231 cells. Cell extracts were untreated (–) or treated (+) with RNase A (50 mg/mL). i The dot plot represents the amino acids in orange box located in the nucleotide 283–333 region of the AURKA sequence and the RRM region of the IGF2BP2 sequence. j The simulated interaction diagram of AURKA and IGF2BP2. k Direct interaction between GST-AURKA fusion protein and recombinant IGF2BP2 (1–220) protein was determined by in vitro interaction assay. l The representative image of co-staining (indicated by white arrows) of AURKA protein (green) and DROSHA mRNA (red) observed in the nucleus (blue). Scale bars, 10 μm. m Levels of AURKA-binding to DROSHA mRNA were determined by RIP assay in MDA-MB-231 cells. n Proteins in MDA-MB-231 cells pulled down by the indicated biotin-RNAs were analyzed with AURKA antibody. AS, antisense; S, sense. o Levels of IGF2BP2 binding to DROSHA mRNA were determined by RIP assay with or without AURKA-deficiency in control MDA-MB-231 cells or METTL14-overexpressing MDA-MB-231 cells. Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test and P < 0.05 is considered statistically significant.
We next investigated how IGF2BP2 mediated AURKA-enhanced DROSHA mRNA stability. The mass spectrometry result of AURKA-interacting proteins indicates that AURKA can interact with IGF2BP2 (Supplementary information, Table S4). As expected, AURKA and IGF2BP2 were colocalized in breast cancer cells (Fig. 5f). Co-immunoprecipitation (co-IP) results confirmed the cellular interaction between AURKA and IGF2BP2 (Fig. 5g, h). Furthermore, MD simulation unveiled 33 combinations of high interaction possibilities between amino acids of AURKA and IGF2BP2 (Fig. 5i), with the amino acids responsible for the AURKA and IGF2BP2 interaction (red color) displaying spatial proximity (Fig. 5j; Supplementary information, Movie S2). Direct interaction between AURKA and IGF2BP2 (1–220 amino acids) purified proteins was determined using in vitro interaction assay (Fig. 5k). In addition, co-localization of AURKA protein and DROSHA mRNA was observed in breast cancer cells (Fig. 5l; Supplementary information, Fig. S7g), which indicates that AURKA protein can bind to DROSHA mRNA. To verify this, we confirmed the binding of AURKA to DROSHA transcript by RNA immunoprecipitation (RIP) assay (Fig. 5m; Supplementary information, Fig. S7h). RNA pull-down results showed that amongst all antisense and sense RNA fragments, only the DROSHA L4 sense fragment pulled down AURKA protein from breast cancer cell lysates (Fig. 5n; Supplementary information, Fig. S7i).
As AURKA interacts with IGF2BP2 in an RNA-independent manner (Fig. 5g, h) and predominantly localizes in the nucleus in breast cancer cells,18 we evaluated whether AURKA promotes nuclear translocation of IGF2BP2 to recognize m6A modification. However, depletion of AURKA did not alter the cytoplasmic and nuclear distribution of IGF2BP2 (Supplementary information, Fig. S7j). Moreover, the IGF2BP2-binding DROSHA P3 m6A motif is located in L5 region near the AURKA-binding L4 region, we reasoned that AURKA reinforced the binding of IGF2BP2 to the m6A modification to stabilize DROSHA transcript. To validate this hypothesis, we performed AURKA knockdown and found decreased co-localization between IGF2BP2 protein and DROSHA mRNA in the nucleus (Supplementary information, Fig. S7k). Importantly, depletion of AURKA decreased m6A levels in control cells but not in METTL14-overexpressing cells (Supplementary information, Fig. S7l), and diminished the capacity of IGF2BP2 binding to DROSHA mRNA in control cells (decreased by ~3.3 fold) more than in METTL14-overexpressing cells (decreased by ~1.7 fold) (Fig. 5o). In contrast, IGF2BP2 knockdown had little effect on the capacity of AURKA binding to DROSHA mRNA (Supplementary information, Fig. S7m). Altogether these findings confirmed that AURKA strengthens the binding of IGF2BP2 to the m6A-modified transcript to stabilize DROSHA mRNA.
Suppression of DROSHA m6A modification attenuates BCSC traits
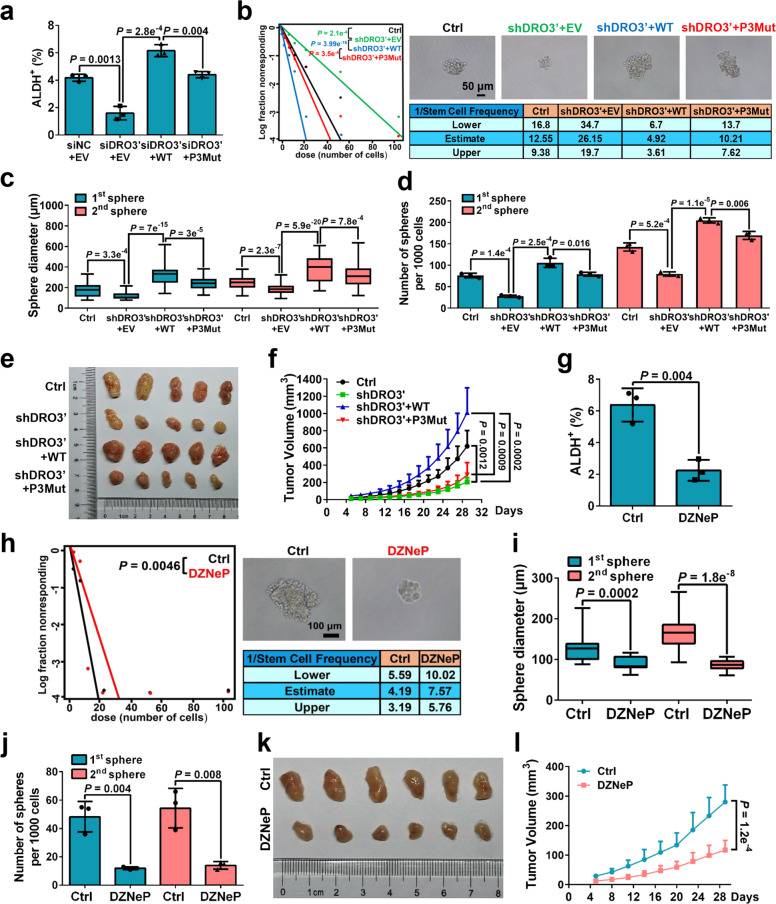
We next evaluated the effects of DROSHA m6A modification on BCSC phenotype. Overexpression of DRO-WT, but not the DRO-P3Mut, substantially rescued the ALDH+ subpopulations in endogenous DROSHA-knockdown cells (Fig. 6a; Supplementary information, Fig. S8a, b). Consistently, forced expression of DRO-WT rather than DRO-P3Mut remarkably restored sphere formation ability in endogenous DROSHA-depleted cells using extreme limiting dilution assays and replating sphere formation assays (Fig. 6b–d). Endogenous DROSHA stably knocked down MDA-MB-231 cells or control cells with forced expression of DRO-WT or DRO-P3Mut were injected into nude mice (n = 5). Mice inoculated with overexpression of DRO-WT, but not DRO-P3Mut cells significantly increased the volume of tumor masses compared to the mice injected with DROSHA-deficient cells (Fig. 6e, f; Supplementary information, Fig. S8c). Furthermore, we found that the proportion of ALDH+ populations was decreased in breast cancer cells with DZNeP treatment (Fig. 6g; Supplementary information, Fig. S8d). DZNeP also inhibited sphere formation ability in extreme limiting dilution assays (Fig. 6h). Similarly, treatment with DZNeP significantly decreased the diameter and number of spheres in breast cancer cells using replating sphere formation assay (Fig. 6i, j; Supplementary information, Fig. S8e, f). In addition, MDA-MB-231 cells were injected into nude mice treated with DZNeP each day during the tumor measurement. Mice treated with DZNeP evidently formed smaller tumor masses compared to the mice injected with normal saline (Fig. 6k, l). Taken together, these results reveal that blockage of DROSHA m6A modification significantly inhibits BCSC phenotype.
Fig. 6. Suppression of DROSHA m6A modification attenuates BCSC traits.
a ALDH+ populations were analyzed following endogenous DROSHA knockdown and forced expression of WT or P3Mut DROSHA in MDA-MB-231 cells. siDRO3′, siRNA against DROSHA 3′UTR. b Left: ELDA was performed in MDA-MB-231 cells with endogenous DROSHA knockdown and forced expression of WT or P3Mut DROSHA. Top right: The representative sphere images are shown. Scale bars, 50 μm. Bottom right: Stemness frequency illustration of the cells with the upper and lower 95% confidence intervals meaning that the frequency of one stem cell in cancer cells. shDRO3′, shRNA against DROSHA 3′UTR. Spheres were counted from 24 replicate wells. c, d Replating sphere formation was performed in MDA-MB-231 cells with endogenous DROSHA knockdown and forced expression of indicated DROSHA. The diameter (c) and number (d) of spheres were quantified. Spheres were counted from three replicate wells. The diameter and number of each experiment represent the total count of three replicate wells. e, f Immunodeficient mice (n = 5, biological replicates) were subcutaneously inoculated with MDA-MB-231 cells with endogenous DROSHA knockdown and forced expression indicated DROSHA (e), and tumor volumes were monitored (f). g ALDH+ populations were analyzed in MDA-MB-231 cells treated with DZNeP. h Left: ELDA was addressed in MDA-MB-231 cells treated with DZNeP. Top right: The representative sphere images are shown. Scale bars, 100 μm. Bottom right: Stemness frequency illustration of the cells with the upper and lower 95% confidence intervals meaning that the frequency of one stem cell in cancer cells. Spheres were counted from twenty-four replicate wells. i, j Replating sphere formation was performed in MDA-MB-231 cells treated with DZNeP. The diameter (i) and number (j) of spheres were quantified. Spheres were counted from three replicate wells. The diameter and number of each experiment represent the total count of three replicate wells. k, l Immunodeficient mice (n = 6, biological replicates) were subcutaneously inoculated with MDA-MB-231 cells treated with DZNeP (k), and tumor volumes were measured (l). Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test in a, d, f–g, j, l, ANOVA test in c and i, χ2 test in b, h. P < 0.05 is considered statistically significant.
Discussion
DROSHA, a catalytic subunit of the microprocessor complex, has been shown to be dysregulated during tumorigenesis.6 However, the aberrant regulatory network of DROSHA in promoting BCSC property remains elusive. Here, we demonstrate a non-canonical function of DROSHA to transactivate STC1 through interacting with β-Catenin in BCSCs. Oncogenic AURKA enhances m6A-dependent DROSHA mRNA stability through (1) stabilizing METTL14 by inhibition of ubiquitylation-mediated degradation and (2) strengthening IGF2BP2 to stabilize m6A-modified DROSHA transcript. Indeed, WT DROSHA, but not an m6A methylation-deficient mutant, enhance BCSC stemness, whereas inhibition of DROSHA m6A modification suppresses tumor growth. Clinically, both DROSHA and STC1 predict adverse prognosis of breast cancer patients. Moreover, DROSHA m6A level is positively correlated with the expression of DROSHA and METTL14 enriched in breast tumor tissues. Thus, we unveil the AURKA-induced oncogenic m6A modification in stabilizing DROSHA transcript and identify a novel DROSHA transcriptional function in promoting BCSC phenotype.
The regulation of miRNAs by DROSHA is widely involved in tumor development5. Interestingly, our GSEA reveals that positively regulated targets by DROSHA are enriched in the CD44+/CD24− BCSC subpopulations, indicating that besides its conventional role in RNA processing, DROSHA transcriptionally enhances stemness factors to maintain BCSCs. Previous studies have shown that DROSHA binding with promoter-proximal regions to regulate human gene transcription by associating with the RNA-binding protein CBP80 and RNA Polymerase II.8 In this study, DROSHA interacts with β-Catenin to promote STC1 transcription in breast cancer cells. The CED domain (390–875 amino acids) of DROSHA is responsible for its interaction with β-Catenin to transactivate STC1, which represents a novel mechanism of the transcriptional function for DROSHA. In addition, STC1 has recently been shown to promote stem-like traits in glioblastoma cells.26 Consistently, STC1 functions downstream of DROSHA to maintain BCSCs, as overexpression of STC1 rescues the DROSHA-repressed BCSC phenotype.
DROSHA mRNA is transcriptionally regulated by c-Myc and E2F1 to promote miRNA processing in B lymphoma and lung cancer cells,9,27 whereas the potential mechanism by which DROSHA mRNA stability is regulated has hitherto remained unknown. Accumulated studies reveal that m6A modification can either enhance or attenuate mRNA stability dependent on the type of m6A reader protein11. YTHDF2 targets thousand m6A-modified transcripts to promote mRNA decay.28 Conversely, IGF2BPs selectively recognize m6A-modified transcript to promote mRNA stability.16 In our study, m6A modification is enriched near the DROSHA mRNA stop codon, consistent with the findings that DROSHA expression is much higher in breast tumor tissues. Inhibition of m6A modification or mutation of the m6A site attenuates the stability of DROSHA mRNA, which uncovers a novel regulatory mechanism of DROSHA mRNA stability. Furthermore, RNA m6A methylation has been shown to be required for cancer initiation and progression in numerous solid tumors and acute myeloid leukemia.29,30 As DZNeP has been reported as an RNA methylation inhibitor in both normal and cancer cells,24 we find that DZNeP inhibits DROSHA m6A methylation and effectively attenuates BCSC traits. On the other hand, DZNeP as the first nonspecific EZH2 inhibitor has been used to mediate several antitumor activities including inhibition of biliary tract cancer stem cells through repression of PRC2 and removal of H3K27me3 marks,31,32 which implies another possibility that DZNeP might suppress BCSC properties through inhibition of histone methylation.
The mechanistic roles of m6A methyltransferases have been substantially investigated during cancer progression. For example, METTL14 elevates the expression of oncogenic MYB and MYC via m6A modification to promote AML development and LSC self-renewal14. Reductions in m6A methylation due to either METTL14 mutation or reduced expression of METTL3 promote tumorigenicity through activation of the AKT pathway.33 Although recent study reveals that SUMOylation of METTL3 is critical for m6A methyltransferase activity in lung cancer,34 how the m6A methyltransferase METTL14 is controlled during cancer progression has remained enigmatic. Previously, we show that AURKA locates preferentially into nucleus to transactivate MYC in a kinase-independent manner.18 In the present work, we find that inhibition of AURKA kinase activity has no impacts on METTL14 expression. Yet, we uncover a novel role of the oncogenic AURKA in stabilizing METTL14 protein through suppressing ubiquitylation-dependent degradation, another kinase-independent role of AURKA in regulating protein stability.
The presence or absence of association partners might account for the variable distribution of m6A readers, such as YTHDF2, contributing to different biological functions.35 How diverse readers achieve selectivity toward certain m6A sites or certain m6A-modified transcripts remains elusive. One likely scenario is that readers may be localized to different regions of mRNA through interacting with other RNA binding proteins (RBPs) that recognize distinct features of the RNA.36 In concordance, we find that the RBP-like AURKA binds to DROSHA transcript near the DROSHA m6A site and strengthens the binding of IGF2BP2 to m6A modification for DROSHA stability. We speculate that AURKA likely acts as a ‘locker’ or ‘harness’ to strengthen IGF2BP2 binding to the m6A modification and subsequently stabilize the DROSHA transcript. Similar notion was raised in a recent study revealing that SMAD2/3 promotes binding of the m6A methyltransferase complex METTL3-METTL14-WTAP to a subset of transcripts involved in early cell fate decisions.37 In addition, the KH3-4 domains are critical for the binding of IGF2BPs to m6A-modified RNAs. However, KH3-4 peptides alone showed poor selectivity and affinity for m6A RNA compared with full-length proteins, indicating that other mechanisms may be involved in the recognition of m6A by the KH domains of IGF2BPs.16 Thus, it is possible that AURKA directly interacting with N-terminal RRM domains of IGF2BP2 might further alter the conformation of KH3-4 domains, leading to preferential recognition of m6A. This hypothesis is worth further investigation in the future study. Hence, this dual function of the oncogenic AURKA represents a novel and intriguing mechanism of oncogenic m6A epigenetic modification in BCSC maintenance.
Collectively, our results reveal that DROSHA interacts with β-Catenin to transactivate STC1 in a non-canonical fashion, contributing to BCSC phenotype. The AURKA/METTL14/IGF2BP2-regulated oncogenic m6A modification enhances the mRNA stability of DROSHA to maintain its high expression levels in BCSCs. Targeting m6A modification of DROSHA represents a novel therapeutic approach for breast malignancies (Supplementary information, Fig. S8g).
Materials and methods
Cell culture and breast tissue specimens
The human breast cancer cell lines, MDA-MB-231 (HTB-26), SK-BR-3 (HTB-30), BT549 (HTB-122) and MCF-7 (HTB-22) purchased from American Type Culture Collection (ATCC) were, respectively, cultured in Leibovitz’s L-15 Medium (Gibco, 11415056), McCoy’s 5A Modified Medium (Gibco, 16600082), RPMI-1640 Medium (Gibco, 11876093) containing 0.023 IU/mL insulin (Sigma, 91077C) and Eagle’s Minimum Essential Medium (Gibco, 11095080) containing 0.01 mg/mL human recombinant insulin (Sigma) with 10% fetal bovine serum (FBS, Gibco, 10100147), 0.1% penicillin-streptomycin (Thermo, 15140122) and anti-mycoplasma reagent SaveIt (Hanbio, HB-SV1000). HEK293T (CRL-11268) cells from ATCC were grown in dulbecco’s modified eagle medium (Gibco, 10569044) containing 10% FBS, 0.1% penicillin-streptomycin (Thermo) and SaveIt. All cells were maintained at 37 °C in a humidified 5% CO2 atmosphere except MDA-MB-231 cultured in atmospheric air and were not cultured continuously for more than 3 months. Cryopreservation of all cell lines were using CELLSAVING (New Cell & Molecular Biotech, C40100). All cell lines were authenticated by STR profiling and tested for mycoplasma contamination with the method of PCR. Petri dishes and cell culture plates were purchased from Guangzhou Jet Bio-Filtration Co., Ltd.
Twenty-one pairs of breast tumor specimens and conjugate breast normal specimens were obtained from the same patients undergoing surgery, following informed consent from patients and approved by the Institutional Ethics Review Board of the Second Affiliated Hospital of Dalian Medical University (No. 2019-028). The clinical information of breast cancer patients was listed in Supplementary information, Table S5.
Transfection, lentivirus package and stable cell line generation
Transient transfection with siRNAs (GenePharma, Suzhou, China) or plasmids was performed using Lipofectamine 3000 (Lipo3000, Invitrogen, L3000015) according to the manufacturers’ protocols. Lentivirus was packaged in HEK293T cells using the second-generation packaging system plasmids psPAX2 (Addgene plasmid, #12260) and pMD2.G (Addgene plasmid, #12259). HEK293T cells were seeded in a 6-well plate to allow 80% confluency in the next day. Lentiviral plasmid (pLVX-STC1 or pLVX-METTL14), psPAX2 and pMD2.G were co-transfected into HEK293T cells using Lipo3000. Supernatants were collected every 24 h between 48 and 72 h after transfection and the viral titer was determined. Lentiviral-mediated short hairpin RNA (shRNA) directed against DROSHA were purchased from GenePharma, Suzhou, China. All siRNA and shRNA sequences were listed in Supplementary information, Table S1.
For lentiviruses infection, cells were infected and subsequently selected in the presence of 2 μg/mL puromycin (Sigma, P8833) for over 72 h. MDA-MB-231 (Tet-on shAURKA) cells was achieved in our previous study.18
To construct the AURKA-knockout cell line, we used one vector CRISPR-Cas9 genome editing system (lentiCRISPR v2, Addgene #52961) to generate an AURKA knockout cell line in MDA-MB-231 cells following the manufacturer’s protocols.38 The gRNA sequence targeting AURKA was 5′-TGAGTCACGAGAACACGTTT-3′.
Plasmid construction
DRO-WT was subcloned into pcDNA6b vector. WT, amino acid truncation (1–875 and 876–1374, D875) DROSHA. WT β-Catenin was subcloned into pcDNA6b vector with His tag. m6A-enriched fragment of DROSHA (P1-3) was subcloned into psiCHECK2 vector. WT METTL14 and WT STC1 were subcloned into pLVX-DsRed-Monomer-N1. WT and truncated 500, 1000, 1500 bp (D500, D1000, D1500) promoters of STC1 were subcloned into pGL3-Basic vectors. STC1 and DROSHA promoters were subcloned into pLVX-OS-dGFP vectors. Plasmids of psiCHECK2-DROSHA (P1-Mut, P2-Mut and P3-Mut), pLVX-METTL14 (R298P, catalytic mutant), pcDNA6-DROSHA (E1045Q, RNase III domain mutant), pcDNA6-DROSHA (P3Mut, m6A site mutant), pGL3-STC1 (Mut1, Mut2 and Mut1 plus Mut2, predicted TCF-bind sites mutant) were generated by site-directed mutagenesis using PCR. WT AURKA was subcloned into pGEX-6P-1 vector with His tag. All the primers used for plasmid construction are listed in Supplementary information, Table S1. pcDNA6b-Flag-AURKA was established in our previous study.18 pcDNA6b-MYC was established in our previous study.39 HA-Ub was a gift from Prof. Lingqiang Zhang lab (Beijing Institute of Lifeomics, Beijing, China).
RNA extraction and RT-qPCR
Total RNAs were extracted using AG RNAex Pro Reagent (AG21101, ACCURATE BIOTECHNOLOGY, HUNAN) or RNAiso Plus (TaKaRa, 9109) according to the manufacturer’s instructions, which were used to generate cDNA using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, AE311-02) or Evo M-MLV RT Kit with gDNA Clean for qPCR (AG11705, ACCURATE BIOTECHNOLOGY, HUNAN). RT-qPCR using ChamQTM Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd, Q711-02) was performed on a Real-time PCR System (Agilent Technologies Stratagene Mx3005P). ACTB or GAPDH was used as an internal control to normalize RNA expression. The primers used in RT-qPCR were shown in Supplementary information, Table S1.
m6A dot blot
Total RNAs were extracted with RNAiso Plus (TaKaRa). Then, intact mRNAs were purified from total RNAs using Dynabeads mRNA purification kit (Ambion, 61006). The indicated amount of purified mRNA was denatured in 10 μL volume of RNA incubation buffer (50% formamide, 2.5% formaldehyde and 0.5× MOPS) at 55 °C for 15 min, followed by chilling on ice. RNA samples were applied to Amersham Hybond-N+ membrane (GE Healthcare, RPN303B) with a Bio-Dot Apparatus (Bio-Rad). After UV crosslinking, the membrane was stained with 0.02% methylene blue (MB) in 0.3 M sodium acetate. The membrane was then washed with 1× PBST buffer, blocked with 5% non-fat milk in PBST, and incubated with anti-m6A antibody (Synaptic Systems, #202003) overnight at 4 °C. After incubating with Goat anti-Rabbit IgG (H + L) Secondary Antibody (Thermo Fisher Scientific, 31460), the membrane was incubated using WesternBright ECL kit (Advansta, K-12045-D50) for chemiluminescent reading on ChemiDoc System (Bio-Rad).
Met depletion and DZNeP treatment
For Met depletion, cells were washed twice by Dulbecco’s Phosphate Buffer Saline (0.137 M NaCl, 2.7 mM KCl, 1.1 mM KH2PO4, 0.5 mM MgCl2, 8.1 mM Na2HPO4, 0.9 mM CaCl2, pH 7.4), and then supplemented with Met-free media (Thermo Fisher, 21013024) to decrease m6A level. For DZNeP treatment, cells were managed with 10 μM DZNeP (Selleck, S7120) to inhibit m6A modification. In animal model, the mice were subcutaneously injected with DZNeP (8 mg/kg per mice) in each two days.
RNA stability assay
Breast cancer cells were transfected with plasmids or treated by DZNeP or methionine deprivation for 24 h. Then, cells were treated with 5 μg/mL actinomycin D (ActD, Sigma, CA1201) and collected at the indicated time points. The total RNAs were extracted by RNAiso Plus (TaKaRa) and analyzed by RT-qPCR.
Western blotting
Cells were lysed using RIPA buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 1% Triton X-100, 0.5% Sodium Deoxycholate, 0.1% SDS, 5 mM EDTA) with protease inhibitor cocktail (MedChemExpress, HY-K001) for 30 min. After centrifugation, supernatant was collected and boiled with 6× loading buffer. Equal amounts of proteins were loaded and separated by SDS-PAGE, transferred to nitrocellulose membranes (Millipore, HATF00010), incubated with primary and secondary antibodies. The membrane was incubated using WesternBright ECL kit for chemiluminescent reading on Bio-Rad. Here are antibodies used for western blotting: DROSHA (3364S), C-MYC (9402S), Phospho-Aurora A (T288) (3079S), Histone H3 (9715S), FTO (14386S) and GST (2625S) were purchased from Cell Signaling Technology (CST) in MA, USA. OCT4 (GTX101497) and NANOG (GTX100863) were from GeneTex in Southern California, USA. GAPDH (60004-1-ZJ), Beta-Actin (66009-1-Ig), GRB10 (23591-1-AP), SLCO4A1 (26399-1-AP), SORBS2 (24643-1-AP), METTL3 (15073-1-AP), METTL14 (26158-1-AP), WTAP (60188-1-IG), YTHDF2 (24744-1-AP), YTHDF3 (25537-1-AP), IGF2BP1 (14642-1-AP), IGF2BP2 (11601-1-AP), IGF2BP3 (22803-1-AP) and HA Tag (66006-2-Ig) were from Proteintech Group in Wuhan, China. β-Catenin (06-734), Aurora A (07-648) and ALKBH5 (ABE547) were from Millipore in MA, USA. STC1 (ab229477) was from Abcam in Cambridgeshire, UK. PARP (sc-8007) was from Santa Cruz Biotechnology in CA, USA. Goat anti-Rabbit IgG (H + L) Secondary Antibody (31460) and Goat anti-Mouse IgG (H + L) Secondary Antibody (31430) were purchased from Thermo Fisher Scientific, MA, USA.
meRIP-seq data analysis
meRIP-seq data were analyzed according to the protocol described previously.40 In brief, reads underwent a set of QC checks with FastQC and Fastp, then were mapped to the reference genome (hg19 and mm10) with STAR. Mapped reads were provided as input for Deeptools and MACS2, which identified m6A peaks adapted for visualization on the UCSC genome browser. The meRIP-seq data of GSE60213, GSE29714, GSE90642 and GSE54365 were used in this study.
mRNA purification and meRIP-qPCR
Total RNAs were extracted with RNAiso Plus (TaKaRa). Then, intact mRNAs were purified from total RNAs using Dynabeads mRNA purification kit (Ambion, 61006). For meRIP, the procedure was described previously.41 In brief, purified mRNAs (5 μg) were digested by DNase I (M0303, NEB) and then fragmented into around 200-nt fragments by incubation at 95 °C for 25 s in RNA Fragmentation Reagents (Ambion, AM8740), followed by standard ethanol precipitation and collection. Anti-m6A antibody (10 μg antibody for 5 μg mRNAs; Synaptic Systems) was incubated with 40 μL Protein A beads (Sigma, P9424) in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl, pH 7.4) for 2 h at room temperature. The fragmented mRNAs (5 μg) were incubated with the prepared antibody-bead mixture for 4 h at 4 °C. By washing three times, bound RNA was eluted from the beads with 0.5 mg/mL N6-methyladenosine (BERRY & ASSOCIATES, P3732) in IPP buffer. The eluted RNA was extracted by Enol:Chloroform:Isoamylol (pH < 5.0, Solarbio life science, P1025) and then generated to cDNA using 5× All-In-One RT MasterMix (ABM, G490). The enrichment of m6A was quantified by qPCR. The sequences of qPCR primers are listed in Supplementary information, Table S1.
Dual-luciferase reporter assay
To measure the gene transcriptional activation, pGL3-STC1 (WT, D500, D1000, D1500, Mut1, Mut2 and Mut1 plus Mut2) reporter plasmids (Fluc) and renilla luciferase (Rluc) control plasmids (pRL-TK) were co-transfected with other plasmids or siRNAs in HEK293T cells. After 24 h, Fluc and Rluc activities were measured with the Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer’s instructions. The relative luciferase activity was calculated by dividing Fluc by Rluc and normalized to individual control for each assay.
To determine the RNA stability, psiCHECK2-DROSHA (WT, P1-Mut, P2-Mut and P3-Mut) reporter plasmids were co-transfected with other plasmids, siRNAs or treated by DZNeP or methionine deprivation. The activities of the renilla and firefly luciferases were quantified with the Dual-Luciferase Reporter Assay System after 24 h. The relative luciferase activity was calculated by dividing Rluc by Fluc and normalized to individual control for each assay.
Fluorescence in situ hybridization-immunofluorescence
Cells were mounted on the Circle Microscope Cover Glass (NEST, 801009). After fixation with 3.6% paraformaldehyde (Solarbio, P1110) and 10% acetic acid (Kermel, 6762007) for 15 min at room temperature, cells were pre-hybridized at 55 °C for 2 h and hybridized overnight at 55 °C in a humidified chamber with DIG-labeled UTP (Roche, 11277073910) nucleic acid probes. Post-hybridized glasses were stringently washed with 50% formamide/2× SSC (20× SSC, 3.0 M NaCl, 0.3 M sodium citrate, pH 7.0). The hybridization was visualized using anti-digoxygenin (1:100, Roche) and Alexa FlourTM 555 donkey anti-sheep IgG (1:200, Thermo Fisher Scientific). The protein was visualized using anti-AURKA antibody (1:100, Abcam) or anti-IGF2BP2 antibody (1:100, Proteintech Group) and Alexa FlourTM 488 donkey anti-rabbit IgG (1:200, Thermo Fisher Scientific). Nuclei were counterstained with DAPI. Image acquisition and analysis were addressed in the confocal microscope (Leica, TCS SP5II).
DROSHA probes were transcripted from DROSHA PCR fragments with T7 promoter in vitro with MEGAscript™ T7 Transcription Kit (Thermo Fisher Scientific, AM1333) and purified with RNAiso Plus (TaKaRa). Primers used to prepare DROSHA PCR fragments were listed in Supplementary information, Table S1.
RIP assay
Endogenous RIP studies were performed using the MagnaRIP Kit according to the manufacturer’s protocol (Millipore, 17-700). Briefly, cells were lysed with RIP lysis buffer for 30 min on ice. The whole-cell lysates were incubated at 4 °C overnight with magnetic protein A-protein G beads coupled with 5 µg of either normal IgG antibody (Millipore) or AURKA antibody (Sigma, A1231), IGF2BP2 antibody (Proteintech, 11601-1-AP). Beads were then washed three times and incubated with proteinase K buffer (30 min at 55 °C). The co-precipitated RNAs were extracted with Enol:Chloroform:Isoamylol (pH < 5.0, Solarbio life science) and detected by RT-qPCR. The primers were listed in Supplementary information, Table S1.
Cytoplasmic and nuclear protein extraction
Cytoplasmic extracts were collected by resuspending the cell pellets in hypotonic buffer (20 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2) supplemented with protease inhibitor (cocktail) and then were incubated on ice for 15 min. Next, 0.9% NP-40 (final concentration) was added with vortexing for 10 s and then were centrifuged for 10 min at 3000 rpm at 4 °C. The supernatants were transferred to new prechilled Ep tube. The residual pellets were washed with hypotonic buffer for 3 times and the supernatants were discarded thoroughly. To achieve nuclear extraction, the nuclear pellets were treated with nuclear extraction buffer (100 mM Tris-HCl, pH 7.4, 2 mM Na3VO4, 100 mM NaCl, 1% TritonX-100, 1 mM EGTA, 0.1% SDS, 1 mM NaF, 20 mM Na4P2O7). After vortexing at every 10 min for 30 min at 4 °C, the lysates were centrifuged at 14,000 × g for 30 min at 4 °C. Supernatants after this spin contained the nuclear protein fraction.
Protein co-IP and LC-MS/MS analysis
Cells grown in 15-cm dishes at 70%–80% were lysed with co-IP buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethyl sulfonyl fluoride, protease cocktail), and collected by cell scrapers. Pull-downs were carried out in RNase A untreated lysates, except Fig. 5g, h, where 50 mg/mL RNase A was used to treat cell lysates during the pull-down procedure. Equal volume of lysates was respectively incubated with AURKA antibody (Sigma, A1231), IGF2BP2 antibody (Proteintech, 11601-1-AP), β-Catenin antibody (Millipore, 06-734), DROSHA antibody (CST, 3364 S), Flag-Tag antibody (Proteintech, 20543-1-AP), His-Tag antibody (CST, 12698), METTL14 antibody (Abcam, 252562) and the corresponding mouse IgG antibody (CST, 7076) or rabbit IgG antibody (CST, 7074). After applying a centrifugation, proteins associated with Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology, SC-2003) were washed three times and analyzed by western blotting.
For mass spectrometry analysis, samples from immuno-enrichment with AURKA, DROSHA or IgG antibodies were loaded onto separated lanes of a 10% SDS polyacrylamide gel, then electrophoresed for 20 min. The entire gel region containing the proteins was cut from the gel and subjected to in-gel trypsin digestion and subsequent recovery of peptides as described previously.42 LTQ-Orbitrap Elite (Thermo Scientific) coupled with Dionex UltiMate 3000 RSLCnano system (Thermo Scientific) was used for all proteomic analyses. The MS data were analyzed by Q-Exactive mass spectrometer and searched against the Uniprot human database by using the MaxQuant (version 1.6.0.1) software for peptide and protein identifications. A summary of the identified proteins can be found in Supplementary information, Tables S2 and S4.
Recombinant protein purification and in vitro interaction assay
AURKA protein was purified from BL21 E. coli that was transformed with pGEX6P1-GST-AURKA-His. Bacteria expressing GST-AURKA or GST were then lysed by sonication with 1 mg/mL lysozyme in PBS and 200 mM KCl. GST proteins were purified via magnetic glutathione affinity (V8611, Promega Corporation) and eluted in 50 mM Tris (pH 8.0) with 10 mM glutathione. Commercial recombinant human protein IGF2BP2 (1-220) was purchased from Novoprotein Scientific Inc (Q9Y6M1).
For in vitro interaction assay, recombinant IGF2BP2 (1-220) protein (5 μg) was incubated with 30 μL magnetic glutathione particles adsorbed with GST-AURKA (5 μg) or GST (1.8 μg) fusion proteins in a total volume of 300 μL binding buffer (20 mM HEPE, pH 7.5, 200 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 1 mM dithiothreitol, 0.05% Triton X-100) for 3 h at 4 °C. The magnetic glutathione particles were then washed by 3 times with the binding buffer. The bound proteins were eluted with 5 mM glutathione/10 mM Tris (pH 8.0) and subjected to western blotting.
RNA pull-down assay
Biotinylated transcripts were transcripted from DROSHA PCR fragments with T7 promoter using MEGAscript T7 kit (Ambion, AM1333) with biotin-16-UTP (Ambion, AM8452) and then purified by RNAiso Plus (TaKaRa). Whole cell lysates (1 mg per sample) were incubated with 5 μg of purified biotinylated transcripts for 1 h at 25 °C. Complexes were isolated with Streptavidin Agarose (Invitrogen, SA10004). The co-precipitated proteins were detected by western blotting. Primers used to prepare PCR fragments were listed in Supplementary information, Table S1.
RNA-seq
Two replicate samples from MDA-MB-231 (Tet-on shAURKA) treated with or without doxycycline for 72 h were submitted to Novogene company (Beijing, China). Total RNAs were extracted according to RNeasy plus kit (Qiagen, 74104). Libraries were prepared according to the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB) following the manufacturer’s recommendations. All samples were sequenced by illumina Hiseq platform with pair end 125 bp read length. The RNA-seq data were mapped to the reference genome hg19 using the software Hisat2 v2.0.5 based on the default parameters. DEGs were analyzed using DEseq2.43 Genes with an adjusted P value < 0.05 and FC > 1.5 were assigned as differentially expressed. Data accession: all the raw data have been deposited in the GEO under GSE128428.
Microarray
Two replicate RNA samples of MDA-MB-231 shDROSHA cells and shNC cells were extracted by RNAiso Plus and submitted to the Gene Tech Company Limited (Shanghai, China) for labeling and hybridization for 16 h at 45 °C using Affymetrix Clariom D. Microarray scans were obtained with a GeneChip Scanner 3000 7 G (Affymetrix) using the default settings. Data were normalized with the Robust Multichip Analysis (RMA) algorithm using default analysis settings and some additional median/quantile normalization. We eliminated all probes with a means < 6.0 and standard deviation < 1.0 to filter the number of probes. Then, we filter the genes with a FC > 1.5 and P value < 0.05. Data accession: all the raw data have been deposited in the GEO under GSE125705.
IHC and scoring
The tissue microarray was obtained from OUTDO BIOTECH (HBreD030CS01, HBreD030PG04). Antigen retrieval was performed by heating the sample in EDTA buffer (pH 8.0) in a microwave oven for 15 min. The slides were stained for 25 min at room temperature (RT). Histological detection of DROSHA (ab183732, Abcam), STC1 (ab229477, Abcam) or NANOG (GTX100863, GeneTex) was performed using EnVision Detection Systems. The IHC staining was quantified as the H-score, which has been validated for breast cancer IHC staining.44 The images were acquired using a Nuance EX multispectral imaging system (PerkinElmer) under identical conditions; two fields of each sample in the tissue microarray were acquired using a 20× objective. Scoring was performed using inForm software (PerkinElmer). Typical images corresponding to negative (four images, scored as ‘0’), weak (four images, scored as ‘1’), intermediate (four images, scored as ‘2’) and strong (four images, scored as ‘3’) brown staining were selected by two independent experienced pathologist for software training. During training, an algorithm, which helps the software to distinguish between the cancer and non-cancer tissues, and distinguish between the nuclear and cytoplasmic regions, and sets the thresholds for 0–3 staining, was optimized. Next, the optimized algorithm was used to perform scoring of the other samples. The H-score (between 0 and 300) for each sample was calculated in the following way: (% of cells stained at intensity 1 × 1) þ (% of cells stained at intensity 2 × 2) þ (% of cells stained at intensity 3 × 3). The receiver operating characteristic curve analysis was used to select the cut-off point for each variable.
GSEA
The gene set enrichment analysis software (GSEA v2.1.0) was used to identify the molecular phenotype of downregulated genes in shDROSHA microarray data. The gene expression signatures were acquired from published microarray dataset.45 Gene sets with a nominal P value < 0.05 and FC > 1.25 were considered significant hits.
Molecular dynamics (MD) simulation
The crystallographic structure of β-Catenin (PDB ID: 3TX7) was obtained from RCSB Protein Data Bank. The 3D structure of DROSHA was predicted using I-TASSER Server.46 The conformation of the complexes formed between β-Catenin and DROSHA were predicted using Zdock program. MD simulations were performed by Gromacs 5.1.5 with Amber99sb force field. After MD simulation, the molecular mechanics energies combined with the Poisson-Boltzmann and surface area continuum solvation (MM/PBSA) methods were employed to evaluate the binding free energy of β-Catenin with DROSHA.18 The interaction residues between these two proteins were analyzed using LIGPLOT (v.4.4.2).47
The crystallographic structure of AURKA (PDB ID: 5G1X) was obtained from RCSB Protein Data Bank. The 3D structure of IGF2BP3 RRM domain (amino acid: 1–157) was predicted using existing crystal structure of IGF2BP2 (PDB ID: 6FQ1) as template by Swiss Model Server.48 The conformation of the complexes formed between AURKA and IGF2BP2 were predicted using Zdock program and all of the MD simulations were performed by Gromacs 5.1.5 with Amber99sb force field. After MD simulation, the molecular mechanics energies combined with the Poisson-Boltzmann and surface area continuum solvation (MM/PBSA) methods were employed to evaluate the binding free energy of AURKA with IGF2BP2.18 The interaction residues between these two proteins were analyzed using LIGPLOT (v.4.4.2).47
ALDH+ cell staining
The ALDEFLUOR assay was carried out according to the manufacturer’s guidelines (STEMCELL Technologies, 01700). Briefly, cells were suspended in ALDEFLUOR assay buffer containing an ALDH substrate, BODIPY-aminoacetaldehyde (BAAA) and incubated for 30 min at 37 °C. To distinguish between ALDH+ and ALDH− cells, a fraction of cells was incubated with a 10-fold excess of an ALDH inhibitor, Diethylamino-benzaldehyde. The result in fluorescence intensity of ALDH+ cells was analyzed by flow cytometer (Beckman Coulter, CytoFLEX).
Sphere formation assay
For replating sphere formation, cells were seeded in 6-well ultralow attachment plates (Corning, 3471) at a density of 1 × 103 cells/well with sphere medium. After 7 days, the number and diameter of tumor spheres were calculated with the inverted microscope (Olympus, DP73). Then, cells were digested with trypsin and replanted into 6-well ultralow attachment plates at a density of 1 × 103 cells/well. After another 7 days, the number and diameter of spheres were calculated.
For Extreme limiting dilution assay (ELDA), cells were seeded into 96-well ultralow attachment plates (Corning, 3474) with sphere medium at density of 1, 5, 10, 20, 50, 100 cells/well (24 wells per cell density). After 7 days, positive (sphere formation) well numbers in each group were uploaded and calculated in the ELDA website.49
Sphere medium includes DMEM/F12 (Gibco, C11330500BT), 20 ng/mL basic fibroblast growth factor (Petrotech, 100-18B), 20 μL/mL B27 (Thermo Fisher Scientific, 17504044) and 20 ng/mL epidermal growth factor (Petrotech, 100-05).
Animal studies
Female BALB/c nude mice (4–6 weeks, Beijing Vital River Laboratory Animal Technology Co., Ltd) subcutaneous were injected with the indicated MDA-MB-231 cells (105 cells/0.1 mL/mouse). The tumor volumes were measured by a calliper once every three days and estimated using the formula = 0.5 × a × b2 (a and b were the long and short diameter of the tumors, respectively). Mice were sacrificed after about four weeks and the xenografted tumors were immediately dissected to take photos.
For secondary limited dilution xenograft, primary xenografted tumors were digested with both collagenase I (Sigma, C0130) and hyaluronidase (Sigma, H1136). Serially diluted single cell suspensions (105, 104, 103, 102) were subcutaneously injected into nude mice (4–6 weeks old). After about four weeks, tumor formation ability was calculated using the Extreme Limiting Dilution Analysis software.49
All animal studies were approved by the Institute Animal Care and Use Committee (IACUC) of Dalian Medical University (No. AAE18017) and carried out in accordance with established institutional guidelines and approved protocols.
ChIP assay
ChIP Assay was conducted with ChIP-IT Express Chromatin Immunoprecipitation Kits (Active Motif). In short, fix 1 × 107 cells with fixation solution for 10 min at RT, then wash cells with ice-cold 1× PBS and rock the plate for 5 s and discard PBS. Stop fixation reaction with Glycine Stop-fix Solution and rock the plate at RT for 5 min. Scrape cells with ice-cold cell Scraping Solution including PMSF (final concentration is at 0.6 mM) and centrifuge cells at 2500 rpm for 10 min at 4 °C. Resuspend cells with 1 mL ice-cold Lysis Buffer for 30 min. Transfer cells to an ice-cold dounce homogenizer and dounce with 10 stokes to aid in nuclei release. Shear the DNA with the sonicator. Store 10 μL chromatin as “Input DNA” at –20 °C. The other chromatin was immunoprecipitated with 2 μg DROSHA antibody (3364S, CST) or corresponding rabbit IgG antibody (CST, 7074) and protein A/G magnetic beads at 4 °C overnight. Wash beads with ChIP Buffer I and ChIP Buffer II. Elute immune complex with 50 μL Elution Buffer AM2. Add 50 μL Reverse Cross-linking Buffer to elute chromatin. Incubate chromatin with 2 μL Proteinase K at 37 °C for 1 h and get DNA with phenol and phenol/chloroform extractions. The specific primers used for PCR targeting STC1 promoter were listed in Supplementary information, Table S1.
Software
JASPAR was used to predict the potential binding of transcription factors (TFs) on the STC1 “core promoter” region, from +402 to –1810 relative to the transcription start site (TSS).50 We identified top 100 highly expressed genes in breast tumors with the assistant of Gene Expression Profiling Interactive Analysis (GEPIA) with default setting.51 We also identified DROSHA correlated genes in breast cancer (METABRIC dataset) with the assistant of cBioPortal.52 The m6A site prediction was performed by “RMBase v2.0”.53 Hazardous ratios (HRs) for tumor cohorts of breast cancer were determined by the Kaplan–Meier (KM) plotter online tool using best cutoff analyses and the multigene classifier.54 The overall survival (OS) of breast cancer patients were analyzed using a Kaplan–Meier survival plot.
Statistical analysis
Statistical comparisons were performed by using Student’s t-test (two-tailed unpaired), ANOVA test or χ2 test as indicated in the figure legends. Statistical analysis for correlation was performed using two-tailed Pearson’s correlation test. Data were presented as means ± SD, P < 0.05 was considered significant.
Supplementary information
Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFA0110300 to Q.Liu and 2017YFA0505600-04 to Q.Liu), the National Natural Science Foundation of China (81820108024 to Q.Liu, 81630005 to Q.Liu, 81972786 to J.X., 81573025 to Q.Liu, 81703062 to L.H. and 81703091 to F.A.), Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (IRT_17R15), Innovative Research Team in University of Liaoning (LT2017001 to Q.Liu), the Natural Science Foundation of Liaoning (2019-BS-081 to F.P.), the “Seedling cultivation” program for young scientific and technological talents of Liaoning (LZ2019067 to B.C. and 2020 to F.P.), the program for climbing Scholars of Liaoning, the Science and Technology Innovation Foundation of Dalian (2020JJ25CY008 to Q.Liu), Dalian High-level Talent Innovation Program (2016RD12 to Q.Liu), International Scientific and Technological Cooperation of Dalian (2015F11GH095 to Q.Liu), the Natural Science Foundation of Guangdong (2016A030311038 and 2017A030313608 to Q.Liu, 2017A020215098 to Z.W.), the Science and Technology Planning Project of Guangzhou (201804020044 to Q.Liu). E.W-F.L.’s work is supported by MRC (MR/N012097/1), CRUK (C37/A12011; C37/18784), Breast Cancer Now (2012MayPR070; 2012NovPhD016; 2014NovPhD326). The authors thank Prof. Yungui Yang (Beijing Institute of Genomics, CAS) for scientific advice and technical assistance, especially meRIP-seq data analysis and the method of meRIP-qPCR assay. The authors thank Prof. Lingqiang Zhang (Beijing Institute of Lifeomics, Beijing, China) for a gift of HA-Ub plasmid.
Author contributions
Q.Liu, J.X and F.P. conceived and designed the entire project. Q.Liu, F.P., J.X., B.H. and Z.W., designed and supervised the research. F.P., B.C., Q.Liang, S.Z., H.Zou, M.L., H.Zhao, Y.M., J.H. and J.Z. performed the experimental data analyses and/or experimental planning. H.Zou, Y.L., J.L. and Z.L., performed the transcriptome-wide and meRIP-seq data analyses. B.L., S.L., J.C. and F.W. performed the proteomic analyses by LC-MS. P.C. and Z.S. performed MD simulation. F.A. drafted the working model. L.X. collected breast tumor specimens and conjugated normal breast specimens. Q.Liu, J.X., F.P., B.C. and E.W-F.L. contributed reagents/analytic tools and/or grant support. F.P., B.C., Q.Liu, J.X., B.H., Q.Liang, S.Z., Y.Z. and E.W-F.L. wrote and revised the manuscript. All authors discussed the results and commented on the manuscript.
Data availability
The RNA-seq and microarray data generated by this study have been deposited in the GEO database under the accession number GSE128428 (RNA-seq) and GSE125705 (microarray). Human breast tumor gene expression and clinical data were derived from TCGA Research Network (http://cancergenome.nih.gov/). Previously published meRIP-seq data and microarray data were re-analyzed. They are available under accession codes: GSE60213, GSE29714, GSE90642 and GSE54365 for meRIP-seq; GSE7513 and GSE15192 for microarray.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Fei Peng, Jie Xu, Bai Cui, Qilan Liang, Sai Zeng, Bin He
Contributor Information
Jie Xu, Email: xujie@dmu.edu.cn.
Quentin Liu, Email: liuq9@mail.sysu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-00397-2.
References
- 1.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melamed Z, et al. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol. Cell. 2013;50:869–881. doi: 10.1016/j.molcel.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Nepal C, et al. Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs. Nucleic Acids Res. 2016;44:3070–3081. doi: 10.1093/nar/gkv1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagschal A, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hata A, Kashima R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016;51:121–134. doi: 10.3109/10409238.2015.1117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu WT, et al. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018;9:532. doi: 10.1038/s41467-018-02893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gromak N, et al. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013;5:1499–1510. doi: 10.1016/j.celrep.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao X, Gao P, Wu M. c-Myc modulates microRNA processing via the transcriptional regulation of Drosha. Sci. Rep. 2013;3:1942. doi: 10.1038/srep01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X, Su R, Feng X, Wei M, Chen J. Role of N6-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan M, et al. Aurora-A kinase: a potent oncogene and target for cancer therapy. Med. Res. Rev. 2016;36:1036–1079. doi: 10.1002/med.21399. [DOI] [PubMed] [Google Scholar]
- 18.Zheng F, et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat. Commun. 2016;7:10180. doi: 10.1038/ncomms10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke MF. Clinical and therapeutic implications of cancer stem cells. N. Engl. J. Med. 2019;380:2237–2245. doi: 10.1056/NEJMra1804280. [DOI] [PubMed] [Google Scholar]
- 20.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 21.Seita J, et al. Gene expression commons: an open platform for absolute gene expression profiling. PloS One. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J. Biol. Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Pendleton KE, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Stanniocalcin-1 augments stem-like traits of glioblastoma cells through binding and activating NOTCH1. Cancer Lett. 2018;416:66–74. doi: 10.1016/j.canlet.2017.11.033. [DOI] [PubMed] [Google Scholar]
- 27.Cao JX, et al. E2F1-regulated DROSHA promotes miR-630 biosynthesis in cisplatin-exposed cancer cells. Biochem. Biophys. Res. Commun. 2014;450:470–475. doi: 10.1016/j.bbrc.2014.05.138. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu Y, Wan A, Lin Z, Lu X, Wan G. N6-methyladenosine modification: a novel pharmacological target for anti-cancer drug development. Acta Pharm. Sin. B. 2018;8:833–843. doi: 10.1016/j.apsb.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu LP, Cheng Y, Kharas MG. The Biology of m6A RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 2019;9:25–33. doi: 10.1158/2159-8290.CD-18-0959. [DOI] [PubMed] [Google Scholar]
- 31.Tan J, et al. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayr C, et al. 3-deazaneplanocin A may directly target putative cancer stem cells in biliary tract cancer. Anticancer Res. 2015;35:4697–4705. [PubMed] [Google Scholar]
- 33.Liu J, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y, et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46:5195–5208. doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertero A, et al. The SMAD2/3 interactome reveals that TGFbeta controls m6A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui B, et al. Stress-induced epinephrine enhances lactate dehydrogenase A and promotes breast cancer stem-like cells. J. Clin. Invest. 2019;129:1030–1046. doi: 10.1172/JCI121685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 41.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Detre S, Saclani Jotti G, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng L, et al. De novo molecular design of a novel octapeptide that inhibits in vivo melanogenesis and has great transdermal ability. J. Med. Chem. 2018;61:6846–6857. doi: 10.1021/acs.jmedchem.8b00737. [DOI] [PubMed] [Google Scholar]
- 48.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xuan JJ, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46:D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanczky A, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and microarray data generated by this study have been deposited in the GEO database under the accession number GSE128428 (RNA-seq) and GSE125705 (microarray). Human breast tumor gene expression and clinical data were derived from TCGA Research Network (http://cancergenome.nih.gov/). Previously published meRIP-seq data and microarray data were re-analyzed. They are available under accession codes: GSE60213, GSE29714, GSE90642 and GSE54365 for meRIP-seq; GSE7513 and GSE15192 for microarray.