Abstract
Following human occupation, the house mouse has colonised numerous islands, exposing the species to a wide variety of environments. Such a colonisation process, involving successive founder events and bottlenecks, may either promote random evolution or facilitate adaptation, making the relative importance of adaptive and stochastic processes in insular evolution difficult to assess. Here, we jointly analyse genetic and morphometric variation in the house mice (Mus musculus domesticus) from the Orkney archipelago. Genetic analyses, based on mitochondrial DNA and microsatellites, revealed considerable genetic structure within the archipelago, suggestive of a high degree of isolation and long-lasting stability of the insular populations. Morphometric analyses, based on a quantification of the shape of the first upper molar, revealed considerable differentiation compared to Western European populations, and significant geographic structure in Orkney, largely congruent with the pattern of genetic divergence. Morphological diversification in Orkney followed a Brownian motion model of evolution, suggesting a primary role for random drift over adaptation to local environments. Substantial structuring of human populations in Orkney has recently been demonstrated, mirroring the situation found here in house mice. This synanthropic species may thus constitute a bioproxy of human structure and practices even at a very local scale.
Subject terms: Evolutionary ecology, Population genetics
Introduction
Islands are well-known “laboratories of evolution” suited to investigation of processes of divergence (Berry 1996). The paradigm examples of insular evolution involve dramatic changes in body size, with dwarfism of large species and gigantism of small species (Lomolino 1985), but morphological differentiation and radiation are also frequent [e.g. (Losos and Ricklefs 2009)]. Such cases of extreme evolution are mostly ascribed to adaptation to local ecological conditions, in particular the release from predation and interspecific competition (Lomolino et al. 2012). Random evolution is also an important driver of morphological differentiation on islands, because of the large effect of genetic drift in the small founding propagules and in the subsequent isolated populations (Sendell-Price et al. 2020). A way to disentangle adaptive and random factors, and hence to better assess the contextual driving forces of insular evolution, may be to consider the relationship between genetic and morphological markers: random morphological evolution should be coupled to, and adaptive evolution uncoupled from, neutral genetic divergence (Polly 2004; Renaud et al. 2007).
The Orkney archipelago, lying close to the northern coast of mainland Scotland, is well-known for its wealth of Neolithic sites. By this period, ca. 5000 years BP, the long-tailed field mouse (Apodemus sylvaticus) and the common vole (Microtus arvalis) had been introduced to the archipelago (Romaniuk et al. 2016). The Orkney vole is recognisable by its large size and characteristic tooth shape (Cucchi et al. 2014). Localised morphological diversification, echoed in the genetic structure, also occurred between islands of the archipelago (Martínková et al. 2013). The congruent patterns of divergence in molar shape and neutral molecular markers suggest a primary role for drift in the process of Orkney vole morphological evolution (Cucchi et al. 2014).
The Western European house mouse (Mus musculus domesticus) was first introduced to Orkney about 4000 years later. The Orkney islands had a central position within the Norwegian Viking kingdom, which was active from the late 8th to the 11th centuries AD. The house mouse was introduced as an unintentional stowaway during this period of intense maritime traffic (Searle et al. 2009). Despite the relatively short period of time since their introduction to Orkney, house mice display considerable diversity in tooth shape on the archipelago (Ledevin et al. 2016). The house mouse arrived on Orkney around the same time as potential predators such as the domestic cat, the red fox and the black rat (Cucchi et al. 2014). It was therefore confronted by ecological conditions similar to those encountered on the continent, a situation that is less prone to drive accelerated morphological divergence than would be the case when arriving in an insular location with a depauperate fauna (van der Geer et al. 2013). This should have mitigated the role of adaptation in the evolution of Orkney mice compared with continental relatives. As a synanthropic species, house mice are frequently translocated by humans (Cucchi 2008; García-Rodríguez et al. 2018), possibly limiting the impact of founder effect and subsequent isolation.
Samples from house mouse populations on different Orkney islands were investigated for variation in mitochondrial DNA and at microsatellite loci, together with a geometric morphometric analysis of their molar shape. The genetic and morphological diversity of Orkney mice, and its relationship with Western Europe, were assessed, with particular emphasis on the comparison between morphological and molecular evolution. The main aims were firstly to identify the degree of differentiation between mice from Orkney and the continent, and between mice from within the Orkney archipelago, and secondly to determine the relative role of adaptation and drift in this differentiation. An accelerated morphological evolution on Orkney, compared to the genetic divergence, would point to a prime role of adaptation (Renaud et al. 2007), likely to occur in the peculiar insular conditions (Millien 2006). In contrast, a primary role for drift would be indicated by morphological evolution paralleling genetic divergence, according to a Brownian model of evolution.
Materials and methods
Sampling
Mice were trapped during two field trips to the Orkney archipelago (Fig. S1). The islands of Eday, Faray, Papa Westray, Sanday and Westray were sampled in 1992, and Papa Westray, Burray, South Ronaldsay and Mainland were sampled in 2012 (Fig. S1B; Table S1). The sole mouse from Burray was grouped with mice from South Ronaldsay because the two islands are physically connected by a narrow isthmus.
Most mice were kept for several months in captivity before sacrifice (Ganem 1998; Souquet et al. 2019). All mice were sacrificed according to the directive 2010/63/UE of the European Parliament on the protection of animals used for scientific purposes. Skulls were manually prepared and stored at the Institut des Sciences de l’Evolution (Montpellier, France).
In total, 303 mice from Orkney were included in the morphometric analyses, 279 mice were genotyped at 19 microsatellite loci, and 79 mice were sequenced for the mitochondrial D-loop.
Comparison between Orkney and continental populations
For the comparison of Orkney mice with continental populations from Western Europe, our 79 D-loop sequences were combined with sequences retrieved from GenBank. The first dataset included D-loop sequences from various origins in order to insert Orkney mice into a large phylogeographic context. A second dataset was compiled with sequences from the same localities as the morphometric sampling (Fig. S1A, Table S2).
The corresponding morphometric analysis included 593 mice from Orkney, the adjacent Scottish mainland and various continental populations (Fig. S1A; Table S2). Since body weight data were only available for a few continental populations, and given that most of the Orkney mice aged in laboratory conditions, a comparison of body size between continental and Orkney mice was not performed.
Genetic and morphometric variation in Orkney
A second set of analyses were devoted to the geographic structure within the Orkney archipelago; three markers were considered. (1) D-loop sequences. (2) Microsatellite data. (3) A morphometric analysis of molar shape, focused on mice that were also genotyped for microsatellites. This dataset included 268 mice (Table 1, Table S1). Given its extent, the island of Mainland was divided into discrete areas: North West (NW), North Central (NC), North East (NE), Central A5 and A7 localities (C57), Kirkwall (KIRK), South East (SE) and Deerness (DE).
Table 1.
Number of house mice trapped on Orkney islands and included in the different analyses, genetic diversity measures based on the D-loop and 19 microsatellite loci datasets and island size.
| Area (km2) | NDloop | NH | h ± SD | π ± SD | Nmic | A (range) | Ho | He | Nmor | Nmm | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Orkney (all islands) | 990 | 146 | 17 | 0.8913 ± 0.0091 | 0.004191 ± 0.002360 | 279 | 10.32 (5–18) | 0.39 | 0.68 | 303 | 268 |
| Mainland | 523.25 | 84 | 10 (5) | 0.8371 ± 0.0180 | 0.003505 ± 0.002040 | 184 | 7.89 (3–18) | 0.43 | 0.61 | 179 | 174 |
| South Ronaldsay | 49.8 | 11 | 3 (1) | 0.6364 ± 0.0895 | 0.001451 ± 0.001112 | 25 | 4.16 (1–7) | 0.47 | 0.54 | 27 | 25 |
| Burray | 9.03 | 1 | 1 (1) | 1 | 0 | 1 | 1.16 (1–2) | 1 | 1 | ||
| Eday | 27.45 | 12 | 3 (1) | 0.4394 ± 0.1581 | 0.002675 ± 0.001765 | 17 | 2.89 (1–8) | 0.34 | 0.4 | 37 | 16 |
| Faray | 1.8 | 6 | 1 (0) | 0 | 0 | 12 | 1.21 (1–2) | 0.23 | 0.21 | 12 | 12 |
| North Ronaldsay | 6.9 | 2 | 2 (1) | 1.0000 ± 0.5000 | 0.009101 ± 0.009653 | – | – | – | – | – | – |
| Papa Westray | 9.18 | 16 | 2 (2) | 0.5250 ± 0.0546 | 0.000598 ± 0.000590 | 24 | 3 (1–6) | 0.32 | 0.36 | 24 | 24 |
| Sanday | 50.43 | 7 | 1 (0) | 0 | 0 | 7 | 2.21 (1–5) | 0.42 | 0.48 | 14 | 7 |
| Westray | 47.13 | 7 | 3 (0) | 0.6667 ± 0.1598 | 0.003041 ± 0.002104 | 9 | 4.26 (1–7) | 0.37 | 0.61 | 9 | 9 |
NDloop number of D-loop sequenced, k mean number of paiwise difference, NH number of haplotype, private haplotypes are indicated within brackets, h haplotype diversity, π nucleotide diversity, Nmic number of mice scored at 19 microsatellites, A mean number of alleles per locus with the range of allele number in brackets, (Ho) observed and (He) expected heterozygosities, Nmor morphometrics, Nmm match microsatellites + morphometrics.
Molecular analyses
Data acquisition
DNA was extracted, amplified and aligned using standard protocols (Supplementary Text). The 79 new D-loop sequences were submitted to EMBL: accession numbers LR862585–LR862663.
Nineteen microsatellite loci were selected based on previous studies (Britton-Davidian et al. 2017; Hardouin et al. 2010); genotyping and scoring were done according to standard procedures (Supplementary Text). In total, 279 mice from the Orkney archipelago were successfully genotyped.
Phylogenetic analyses of D-loop sequences (for details, see Supplementary Text)
Three different datasets were analysed.
Inserting Orkney mice within the phylogeny of the house mouse. The corresponding dataset included 1812 D-loop sequences of Mus musculus domesticus and two sequences each of M. musculus castaneus and M. musculus musculus used as outgroups. The final alignment comprised 728 haplotypes and 898 sites. The phylogenetic tree was reconstructed using MrBayes (Ronquist et al. 2012); robustness of the nodes was estimated with posterior probabilities (PP).
A D-loop dataset for comparison with the morphometric analysis at the European scale. This dataset was designed to include only sequences matching the morphometric sampling (Table S2). It comprised 414 D-loop sequences and 879 positions. A neighbour-joining phylogeny was reconstructed based on p-distance estimated with MEGA 7 (Kumar et al. 2016).
D-loop-based genetic structure on the Orkney archipelago. The 79 new D-loop sequences were combined with 67 sequences retrieved from previous studies. The final alignment comprised 146 sequences and 879 nucleotides. Haplotypes and nucleotide diversity indices were determined for each island with DNAsp v6 (Rozas et al. 2017) and Arlequin 3.5 (Excoffier and Lischer 2010). A haplotype network was inferred using the median-joining algorithm as implemented in POPART (Leigh and Bryant 2015). As three of the 17 haplotypes were characterised by indels that are not taken into account by PopART, we coded these indels as new sites with A(deletion)/T(insertion) at the end of the alignment.
Population genetic analyses based on microsatellite data
Genetic diversity
The genetic diversity within each island of the Orkney archipelago was characterised, using Arlequin 3.5 (Excoffier and Lischer 2010), by the mean number of alleles per locus (A), expected (He) and observed (Ho) heterozygosities. The differentiation between Orkney islands and populations of Mainland was estimated using pairwise Fst. Significance was tested using 1000 permutations.
Population structure
The microsatellite data were first analysed using STRUCTURE 2.3.4 (Pritchard et al. 2000). This method failed to provide consistent results, providing K estimates that greatly varied between each run. Therefore, we used a discriminant analysis of principal components (DAPC) (Jombart et al. 2010). Isolation by distance (IBD) on molecular distances was tested at the scale of the whole archipelago, and on Mainland only. The distance between locations “as the crow flies” (i.e. in straight line) was used in both cases, because mice may have benefitted from human-mediated transport on both land and sea.
Data acquisition for morphometrics
The first upper molar (UM1) is known to be highly evolvable in the house mouse, especially on islands (Renaud et al. 2011), and was therefore chosen as the character of interest. Three-dimensional methods have been developed to characterise its occlusal geometry (Ledevin et al. 2016). However, an outline-based 2D approach has been chosen here, for two reasons. Firstly, the 2D approach provides a good approximation of the 3D signal (Ledevin et al. 2016), and because of its reduced cost compared to 3D analyses based on µCT scans, it allowed us to include all the available specimens in the morphometric study. In addition, the 3D geometry of the molar is strongly impacted by wear. Most Orkney mice were kept in the laboratory for some time, allowing them to grow older than in the field that caused an increase in wear of the teeth. The 2D outline of the occlusal surface is measured low on the crown, mitigating this issue (Renaud et al. 2017).
The UM1 outline was thus described using 64 points sampled at equal curvilinear distance using the Optimas software. The maximum length of the tooth was automatically extracted from this dataset. The starting point was positioned at the most anterior part of the tooth.
Morphometric analyses
Molar size
The relationship between molar length and body weight was tested using a linear regression on the subset of mice that were trapped on Orkney in 2012 and sacrificed at capture. Mice subsequently bred in captivity were not considered in this analysis. Differences in molar length between continental, Scottish and Orkney mice were tested using an analysis of variance (ANOVA) followed by pairwise Tukey post hoc tests.
Procrustes superimposition
The points along the outline were analysed as sliding semi-landmarks (Cucchi et al. 2013) using a Generalised Procrustes Analysis (GPA) standardising size, position and orientation while retaining the geometric relationships between specimens. During the superimposition, semi-landmarks were allowed to slide along their tangent vectors until their positions minimised the shape difference between specimens, the criterion being bending energy. Because the first point was only defined as a maximum of curvature, some slight offset might occur between specimens. It was therefore considered as a semi-landmark and allowed to slide between the last and second point. Two superimpositions were performed, one including the total dataset, and the other focusing on Orkney mice. Differences between groups were tested using Procrustes ANOVA (10,000 permutations). The GPA and the Procrustes ANOVA were performed using the R package geomorph (Adams and Otarola-Castillo 2013).
Multivariate analyses and statistics
Principal component analyses (PCA) on the aligned coordinates were used to visualise the pattern of total variance and to reduce the dimensionality of the data, by retaining only PCs > 1% of variance in the subsequent analyses. Relationships between groups were further investigated using between-group PCA (bgPCA). While the PCA is an eigenanalysis of the total variance–covariance in the dataset, the bgPCA analyses the variance–covariance between group means, weighted by the sample size of each group. Relationships between groups were also visualised using unrooted neighbour-joining trees based on Euclidean distances between group means.
Comparison between morphometric and genetic data
The match between the genetic and morphometric structure was investigated at different scales and using different complementary approaches. (1) The relationship between the microsatellite and morphometric datasets was visualised using a co-inertia analysis. This approach aims to find orthogonal vectors (co-inertia axes) maximising the covariance between the two datasets (Dolédec and Chessel 1994), allowing their projection in a common space. (2) The relationship between the microsatellite and morphometric datasets was tested using Protests (Peres-Neto and Jackson 2001) and RV tests (Escoufier 1973), the significance being based on 10,000 permutations. These tests were performed on the complete datasets (all microsatellite data vs aligned coordinates) and using a reduction of dimensionality, retaining only PC axes explaining more than 1% of variance in each case. For that purpose, the microsatellite dataset was analysed using a PCA. (3) The degree of the phylogenetic signal in a morphometric dataset can be estimated by comparison with a reference phylogenetic tree (statistics Kmult) (Adams 2014). This approach compares the observed morphometric dataset to the expectation of evolution along the tree under a Brownian motion model, significance being assessed by permuting the shape data among the tips of the phylogeny. The significance was assessed based on 10,000 permutations. Two reference phylogenies were used to address two different geographic scales. First, the neighbour-joining tree based on the D-loop p-distances was used to compare Orkney and continental European populations. The match between morphometric and genetic sampling was only possible at the level of the locality (or even neighbouring area) and group means had to be considered. We also used this phylogeny to test for differences in the net rates of morphological evolution between Orkney, Scotland and the continent. For this test, the net rate of shape evolution for each group in the multidimensional space is calculated under a Brownian motion model of evolution, and a ratio of rates is obtained (Adams 2014). Second, the neighbour-joining tree based on the microsatellite distances was used to investigate the morphological diversification within Orkney. For this dataset, the match between genetic and morphometric variation was assessed at the individual level.
The PCA, bgPCA, co-inertia analyses and the RV tests were performed using the R package ade4 (Dray and Dufour 2007). Protests were performed using the R package vegan (Oksanen et al. 2013). Kmult tests were performed using the R package geomorph (Adams and Otarola-Castillo 2013).
Results
Continental vs Orkney mice: phylogeny and morphometrics
D-loop-based phylogeny
In the phylogenetic tree reconstructed with the 728 haplotypes (Fig. S2), most of the sequences from Orkney belong to the well-supported (PP = 0.9) and previously defined “Orkney lineage” or clade F (Jones et al. 2011b; Searle et al. 2009). The two other sequences are in clade E (Jones et al. 2011b) or at the base of the phylogeny.
Molar-size differences between populations
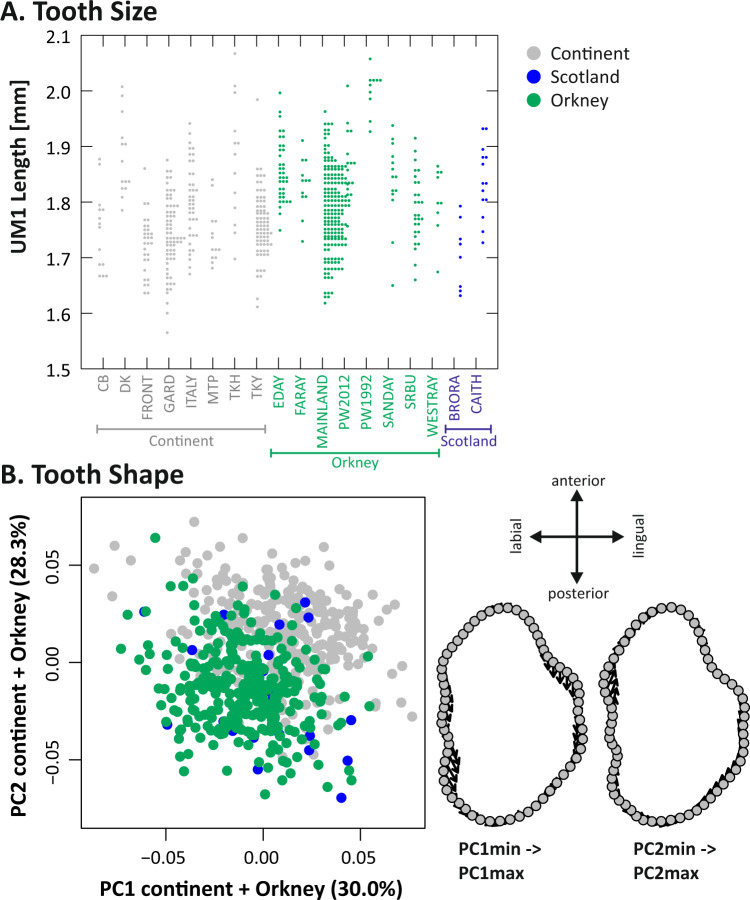
Molar length was not related to body weight in the subset of 75 mice sacrificed at capture on Mainland, Papa Westray and South Ronaldsay (P = 0.837). Orkney, continental European and Scottish mice differed in molar length (ANOVA: P < 0.0001). Orkney mice had longer molars (average length ± standard deviation: 1.82 ± 0.09 mm) than their continental relatives (1.77 ± 0.08 mm) (Tukey: P < 0.0001). Scottish mice had molars of intermediate length (1.79 ± 0.08 mm), and were therefore different neither from the continental ones (P = 0.4081) nor the Orkney ones (P = 0.3656). The range of within-population variation was large (Fig. 1a). Populations with long molars occurred both on the continent (Denmark [DK] and Brittany [TKH]) and Orkney (Eday, Faray and Papa Westray).
Fig. 1. Molar size and shape in continental, Scottish and Orkney mice.
a Variation in tooth length among populations. b Morphospace describing molar shape in continental (grey), Scottish (blue) and Orkney (green) mice. The first two axes of a PCA on the aligned coordinates are displayed. Tooth-shape changes from the minimum to the maximum scores along PC1 and PC2 are depicted to the right.
Molar-shape differentiation between the continent and Orkney
Molar-shape differentiation between populations from the continent, Scotland and Orkney, was highly significant (Procrustes ANOVA: P = 0.0001). The differentiation of molar shape between mice from Orkney and those from western continental Europe was expressed both along PC1 (30% of variance) and PC2 (28.3%) (Fig. 1b). Molars from Orkney mice were as widely distributed in the morphospace as those of continental mice, with parallel variation in Orkney and the continent mostly expressed along PC1. This axis described an elongation of the anterior part of the UM1. Tooth-shape variations along PC2 were more localised, and mostly involved the labial anterior cusp. Molars from Scottish mainland mice were mostly within the range of molars from Orkney mice.
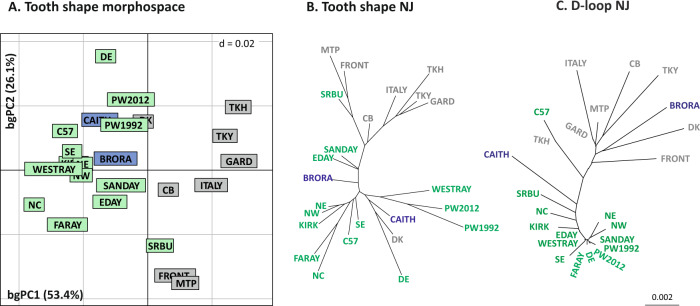
Considering a bgPCA, the difference between localities corresponded to 32.3% of the total variance (Fig. 2a). The first axis (53.4%) differentiated continental from Scottish localities, including those in Orkney. The diversification within continental Europe and within Orkney was expressed on bgPC2 (26.1%). The two groups from Papa Westray (1992 and 2012) closely resembled each other. Two localities departed from the pattern separating Orkney from continental populations: Denmark [DK] grouped with Orkney, while South Ronaldsay grouped with continental populations (Fig. 2a, b).
Fig. 2. Morphometric and genetic between-group differentiation.
a Differentiation of the groups on the first two axes of a between-group PCA on the aligned coordinates. b Neighbour-joining tree based on morphometric distances between the group means. c Neighbour-joining tree based on average molecular p-distances calculated between the groups. Continental Western Europe in grey, Scotland in blue and Orkney in green.
The between-group morphometric distance matrix was only weakly correlated (R = 0.2251, P = 0.0256) to the D-loop p-distance matrix. A reduced multivariate morphometric dataset (first ten PCs totalling 93% of variance) was compared with the D-loop phylogenetic tree (Fig. 2c), showing that tooth shape did not evolve according to Brownian motion (Kmult = 0.00716, P = 0.7418).
Differences of evolutionary rates between groups were tested further. The difference between continental Europe + mainland Scotland vs Orkney was not significant (P = 0.2178), despite very different average evolutionary rates (continental Europe + mainland Scotland = 0.022 vs Orkney = 3.889), possibly because of high heterogeneity within groups. Considering four groups (Orkney Mainland, small Orkney islands, mainland Scotland and the continent), the difference in evolutionary rates was significant (P = 0.0465; evolutionary rates for continental Europe = 0.025, mainland Scotland = 0.011, Orkney Mainland = 3.727 and small Orkney islands = 4.051).
Genetic structure in Orkney
Population structure based on the D-loop
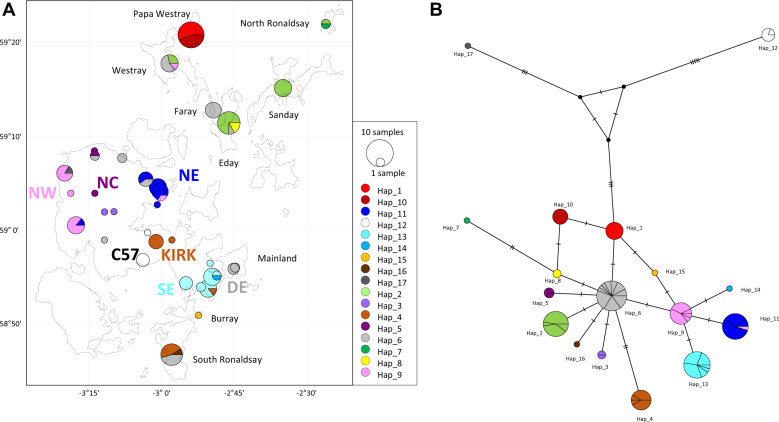
Seventeen haplotypes were distinguished among the 146 D-loop sequences from the Orkney archipelago. Fifteen belonged to the “Orkney lineage” (Fig. S2) and among them, 11 were restricted to only one of the Orkney islands (Fig. 3a; Table 1).
Fig. 3. Population structure in Orkney based on the D-loop.
a Distribution of the 17 haplotypes sampled in Orkney. b Network of the haplotypes present in Orkney. The colours correspond to the different haplotypes.
In the network (Fig. 3b), 14 haplotypes are grouped together around a central haplotype (hap_6), which is present in several localities on Mainland and on three other Orkney islands. The remaining two haplotypes are only distantly related to each other and to the other Orkney haplotypes. All the haplotypes in the “Orkney lineage” are only separated by one or two mutational steps. Most of them are restricted to one or a few localities. Therefore, each island, and each group identified in Mainland, is characterised by its own haplotype composition.
Microsatellite analyses
The 19 loci showed high levels of polymorphism with 5–18 (average 10.32) alleles per locus, and a total of 196 alleles across all sampling locations (Table 1). The different islands displayed low-to-moderate levels of observed (Ho) and expected (He) heterozygosity ranging from 0.23 to 0.47 for the former and from 0.21 to 0.61 for the latter (Table 1). Fst values between islands were high (Fst = 0.21–0.82, Table S3) and highly significant (P < 0.001). The populations on Mainland were genetically structured, as indicated by high Fst values (mean = 0.27, Table S4). The comparisons were significant (P < 0.01) in most cases, except for localities sampled by a single specimen (Table S4).
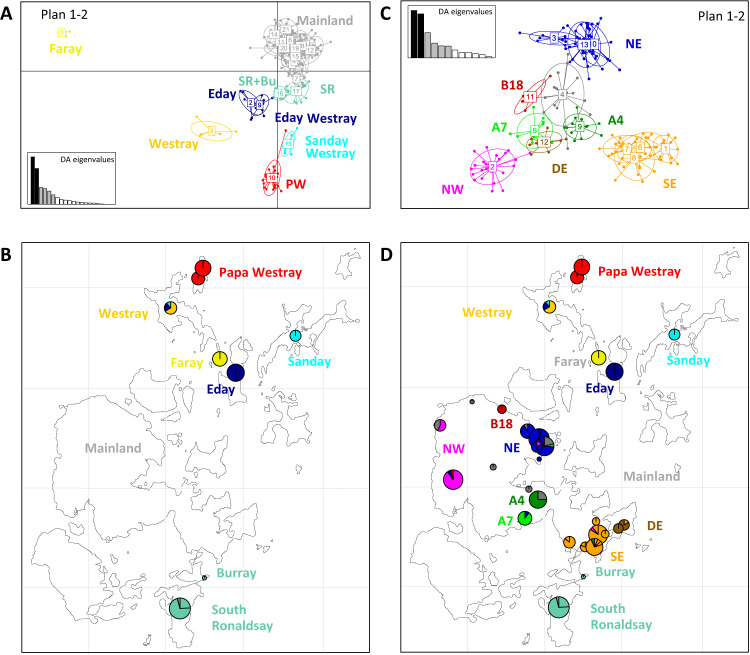
The DAPC analyses demonstrated that the populations were highly structured at the scale of the Orkney archipelago (Fig. 4a, b, 21 clusters) and at the scale of Mainland alone (Fig. 4c, d, 13 clusters). Within Orkney, the different islands clearly separated from each other on the plane defined by the first two axes of the DAPC (Fig. 4a). The first axis separates Faray, Westray and to a lesser extent, Eday from the other islands. The Mainland, South Ronaldsay, Eday, Sanday and Papa Westray populations are spread along the second axis, each being well separated from the other islands. The Papa Westray population did not change over time. A focus on Mainland (Fig. 4c, d) revealed a strong geographic structure very similar to the one that was found with the D-loop (Fig. 3a), supporting the grouping of populations into differentiated regions. The only notable discrepancy concerns mice from the localities in the central part of Mainland (A5 and A7). They harboured a D-loop haplotype that did not belong to the “Orkney lineage”, suggesting an import from elsewhere. However, the mice from these localities group with those from other Mainland localities with respect to the microsatellite data, showing no evidence of an allochthonous genetic signature.
Fig. 4. DAPC analysis of the microsatellite data.
a At the level of the Orkney archipelago. b Projection of the clusters present on the small islands. c At the level of Mainland alone. d Projection of the 21 clusters present in the whole archipelago. In a and c, the number of the clusters resulting from the DAPC analysis is indicated at the centre of the corresponding group.
Finally, IBD was stronger within the Orkney archipelago (R2 = 0.135, P < 0.001) than within Mainland (R2 = 0.023, P = 0.03).
Relationship between microsatellite and morphometric data
Orkney archipelago
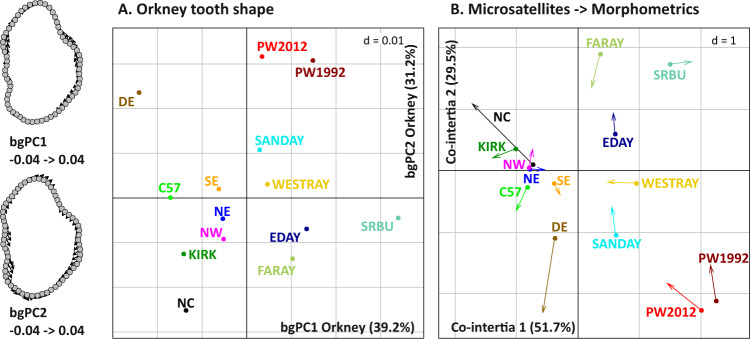
Morphometric and microsatellite data were available for 268 specimens from Orkney. Molar-shape differences were significant between populations (Procrustes ANOVA: P = 0.0001) and explained 25.8% of the total variance, based on the bgPCA. The first bgPCA axis tended to separate small islands from Mainland (Fig. 5a). Papa Westray samples from 1992 to 2012 were close to each other. Both diverged from the populations of the other small Orkney islands along bgPC2, due to an anterior elongation of the tooth. Mainland groups were relatively close to each other.
Fig. 5. Morphometric differentiation in Orkney and comparison with microsatellites.
a Molar-shape differentiation on Orkney, based on a between-group PCA on the aligned coordinates of the molar outline. To the left, visualisation of the shape changes along the axes. b Co-inertia analysis between microsatellite and morphometric datasets. The arrows indicate the change in topology going from microsatellites to morphometrics. Colour code as in Fig. 4d.
The congruence between microsatellite and morphometric data was visualised using a co-inertia analysis (Fig. 5b). The overall match was good (short arrows), with populations from the small islands tending to differentiate less in tooth shape than expected based on the microsatellites (arrows pointing to each other, as exemplified for Faray, Westray, Sanday and Papa Westray). Mainland populations NC and DE tended to differentiate more in tooth shape than for the microsatellites (diverging arrows).
Microsatellites and morphometric datasets were compared based on between-group (N = 14) and individual (N = 268) estimates, using several tests (Protest, RV and Kmult) and using all variables and reduced datasets. The results were congruent overall (Table S5; only the results based on Kmult test on the total datasets, i.e. 196 variables for the microsatellites and 128 aligned coordinates for the morphometric analysis, are provided in the text). Genetic and morphometric datasets appeared significantly correlated (all specimens, PKmult = 0.0003).
Local scale on Mainland
There was a significant difference in tooth shape between the regions of Orkney Mainland, as determined from the microsatellite analysis (N = 174, Procrustes ANOVA: P = 0.0001). Microsatellite and morphometric structures were significantly related (PKmult = 0.0001).
Lastly, the analysis was focused on two sets of neighbouring farms. Northeast Mainland and Southeast Mainland were selected because of the good sample sizes and high genetic homogeneity at these locations.
For Northeast Mainland (N = 56), molar shape differed between neighbouring farms (Procrustes ANOVA: P = 0.0122), and the morphometric differentiation was related to the phylogenetic signal (PKmult = 0.0468).
The pattern was similar in Southeast Mainland (N = 50). Molar shape differed between neighbouring populations (Procrustes ANOVA: P = 0.0231), and morphometric divergence followed a Brownian motion along the phylogeny (PKmult = 0.0020).
Discussion
A low genetic diversity in a remote northern archipelago
The present data confirm the dominance of the “Orkney lineage” (Searle et al. 2009) on the archipelago, attributed to an introduction by the Vikings (Searle et al. 2009). Twelve of the 15 D-loop haplotypes belonging to this clade are endemic to the Orkney archipelago. The three haplotypes found outside Orkney probably represent ancestral diversity, whereas the endemic ones may be the result of local differentiation (Searle et al. 2009). This dominance of a single haplogroup can be interpreted as the consequence of the first invading population being resilient to subsequent invasion, as documented in Kerguelen (Hardouin et al. 2010) and Madeira (Günduz et al. 2001). In contrast, a high genetic diversity was found in more meridional locations such as the Canary islands, Aeolian archipelago and the Azores (Bonhomme et al. 2011; Gabriel et al. 2015; Solano et al. 2013); Cyprus displays the highest mitochondrial diversity of any island population, with 9 of the 11 described haplogroups present (García-Rodríguez et al. 2018).
The contrast between high genetic diversity in meridional islands and moderate-to-low diversity in Orkney and other high-latitude islands is also evident from microsatellite data (Gabriel et al. 2013; Hardouin et al. 2010; Jones et al. 2011a; 2012); Cyprus here again displays the highest diversity (García-Rodríguez et al. 2018).
The genetic diversity in meridional islands reflects multiple colonisation events that can be ascribed to their complex human history, especially in the case of the Mediterranean islands such as Cyprus, which was colonised as early as the Neolithic, and experienced a considerable volume of sea traffic over several millennia (Cucchi et al. 2020; 2005; García-Rodríguez et al. 2018; Solano et al. 2013). In contrast, Orkney has been relatively isolated since the Viking period, as confirmed by human genetics, with the human population of Orkney differing substantially from other British populations, and including an important contribution from Norway in its ancestry (Leslie et al. 2015). The resilience of house mouse populations to later invasion is not complete, however, as shown by two haplotypes (six mice) belonging to other clades that are mostly found in localities of mainland Britain (Searle et al. 2009), tracing more recent exchanges between Britain and Orkney. Assimilation nevertheless occurred, since these mice display a typical Orkney microsatellite signature.
Orkney mice: an initial adaptive step?
Despite the morphological diversity among the islands of the archipelago [(Ledevin et al. 2016), this study], the first-order morphological signal is a divergence of all Orkney mice from continental ones. This idiosyncratic Orkney molar shape echoes what has been found for mandible shapes (Souquet et al. 2019). This result was expected, given the genetic homogeneity of Orkney mice, and yet, molar-shape evolution appeared to be weakly related to neutral genetic evolution. This relative decoupling may be due to accelerated evolution on Scottish mainland and Orkney, an effect that likely increases in smaller and presumably, more isolated islands. A component of adaptation to northern environment may further contribute to the divergence of Orkney mice, since mice from Northern Scotland and Denmark present morphological similarities with Orkney mice, despite their different haplotypic signatures and translocation history (Searle et al. 2009).
Beyond shape differentiation, an increase in body size is expected for insular small mammals (Lomolino 1985; 2005). Body-size response was difficult to assess here because most of the Orkney mice were kept for a while in laboratory conditions, allowing them to grow older and larger. Molar size is considered to be a good proxy of body size at a broad taxonomic scale (Gingerich et al. 1982), but not at a population level, because the first molar erupts early after birth and is therefore not affected by subsequent growth (Renaud et al. 2017). As a consequence, the increase in molar size observed on some Orkney islands is probably not related to differences in body size, and the larger molar size of Orkney mice may not be indicative of their larger body size compared to mice from the continent.
A strong genetic structure within the Orkney archipelago
Beyond the typical Orkney signature, molar shape diversified within Orkney. The population structure based on mitochondrial and microsatellite data is highly congruent with this morphological diversification. Fst values are high for an intraspecific structure, but are similar to what has been observed for other insular house mouse populations, such as those in the Azores, Madeira and Faroe (Gabriel et al. 2013; Jones et al. 2011a). Divergence between populations from the different Orkney Islands was also found for Orkney voles (Cucchi et al. 2014). The fragmentation of the archipelago obviously constitutes a barrier to human and animal exchanges between islands.
The population structure observed on Orkney Mainland is more intriguing, since no visible geographic barriers divide the landscape, except perhaps for the narrow isthmus connecting the Deerness peninsula. Accordingly, no genetic structure was observed for Mainland Orkney voles (Martínková et al. 2013). House mice are supposed to be readily translocated by people, even at a local scale, to an extent that will erase any geographic structure on an island as large as Cyprus (García-Rodríguez et al. 2018). However, on Mainland Orkney, they appear to have accumulated more geographic structure than voles, within a much shorter time span. This small-scale geographic structure may firstly be due to the cool and wet climate of Orkney, which may dissuade mice from foraging extensively outdoors, although Orkney mice could occasionally be trapped in fields during summer time. Secondly, house mice are at a competitive disadvantage to wood mice (A. sylvaticus) beyond the vicinity of human buildings and activity (Berry and Tricker 1969; Fairley and Smal 1987). This could contribute to their restricted overland dispersal in Mainland, given that wood mice were introduced there several millennia before the arrival of house mice (e.g. (Romaniuk et al. 2016)). Indeed, competition with the wood mouse appeared to be an important factor influencing tooth-shape diversification of house mice from the islands of Orkney (Ledevin et al. 2016).
The congruence of the microsatellite signal with the more slowly evolving mitochondrial genetic variation underlines the long-lasting stability of this geographic structure. A dynamic of local extinction and recolonisation is typical for synanthropic house mouse populations that are known to function in small demes, with groups of related individuals structured at a very small geographic scale of only a few metres (Pocock et al. 2004). In this process, human-mediated translocations can erase any geographic structure; however, our results suggest that there have been few such exchanges across Mainland. This may indeed be related to local human practices, since people in the north-eastern, north-western and south-eastern parts of Mainland Orkney appear to be genetically differentiated (Gilbert et al. 2019).
The congruence of human and house mouse phylogeography is well-known (Searle et al. 2009), and the house mouse has consequently been considered as a bioproxy to infer past human long-distance travels (Jones et al. 2013). Our data suggest that the structure of house mouse populations may even reflect human spatial organisation and social practices at a surprisingly small local level.
Isolation and fragmentation as drivers of morphological diversification
A strong and small-scale genetic structure therefore resulted from behavioural patterns of both mice and men. This genetic structure is tightly mirrored in tooth shape, which displays a high disparity among Orkney islands and even within Mainland. An analysis of the molar row topography, however, suggested that the most extreme Orkney phenotypes were not functionally advantageous and may even be the result of a relaxation of functional demands in the insular environment (Renaud et al. 2018; Souquet et al. 2019).
The morphological evolution on Orkney thus appears to be largely neutral and to relate to isolation between populations. It is notable that isolation occurred even between neighbouring groups of farms, triggering differences in upper molar shape even at this very small geographic scale because of developmental properties favouring rapid evolution (Hayden et al. 2020).
Conclusions
On the continent, frequent translocations of mice, associated with local extinctions, are likely continually reshuffling genetic composition and consequently erasing local morphological divergence, resulting in a rather homogeneous molar tooth morphology (Ledevin et al. 2016; Renaud et al. 2017). The ability of house mice to rapidly evolve in small fragmented populations may fuel genetic and morphological diversity, providing ample variation for the action of selection, and possibly contributing to the success of the house mouse as a worldwide invasive species. Nevertheless, while adaptation to local conditions might have initially contributed to the evolution of Orkney mice, our data suggest that drift is the primary driver sustaining morphological disparity in these fragmented and isolated populations. Whether or not this is the case, the genetic structure was clearly mirrored by the morphological evolution, demonstrating that molar shape is a useful marker of evolution at very short timescales.
Supplementary information
Acknowledgements
We warmly thank Josette Catalan and Annie Orth for their participation to the 2012 field trip to Orkney, Jeremy Searle for original collection of specimens from northern Scotland and all the people from Orkney who kindly made possible the extensive trapping campaigns. The paper benefited from the constructive remarks of three anonymous reviewers and Stuart J.E. Baird. This work was performed using the computing facilities of the CC LBBE/PRABI. It was supported by the ANR Bigtooth (ANR-11-BSV7-008). This is a publication ISEM 2020-216.
Data availability
The DNA sequences are deposited in EMBL, accessions LR862585–LR862663. Sampling locations are indicated in Table S1. Morphological data and microsatellite genotypes are deposited in Dryad: 10.5061/dryad.nvx0k6dqm.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Associate editor: Bastiaan Star
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-020-00368-8) contains supplementary material, which is available to authorized users.
References
- Adams DC. A generalized K statistic for estimating phylogenetic signal from shape and other high-dimensional multivariate data. Syst Biol. 2014;63(5):685–697.. doi: 10.1093/sysbio/syu030. [DOI] [PubMed] [Google Scholar]
- Adams DC, Otarola-Castillo E. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods Ecol Evol. 2013;4:393–399.. [Google Scholar]
- Berry RJ. Small mammal differentiation on islands. Philos Trans R Soc Lond B. 1996;351(1341):753–764.. doi: 10.1098/rstb.1996.0070. [DOI] [PubMed] [Google Scholar]
- Berry RJ, Tricker BJK. Competition and extinction: the mice of Foula, with notes on those of Fair Isle and St Kilda. J Zool Lond. 1969;158:247–265.. [Google Scholar]
- Bonhomme F, Orth A, Cucchi T, Rajabi-Maham H, Catalan J, Boursot P, et al. Genetic differentiation of the house mouse around the Mediterranean basin: matrilineal footprints of early and late colonization. Proc R Soc Lond Biol Sci. 2011;278:1034–1043.. doi: 10.1098/rspb.2010.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J, Caminade P, Davidian E, Pagès M. Does chromosomal change restrict gene flow between house mouse populations (Mus musculus domesticus)? Evidence from microsatellite polymorphisms. Biol J Linn Soc. 2017;122(1):224–240.. [Google Scholar]
- Cucchi T. Uluburun shipwreck stowaway house mouse: molar shape analysis and indirect clues about the vessel’s last journey. J Archaeol Sci. 2008;35:2953–2959.. [Google Scholar]
- Cucchi T, Barnett R, Martinkova N, Renaud S, Renvoisé E, Evin A, et al. The changing pace of insular life: 5000 years of microevolution in the Orkney vole (Microtus arvalis orcadensis) Evolution. 2014;68(10):2804–2820.. doi: 10.1111/evo.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchi T, Kovács ZE, Berthon R, Orth A, Bonhomme F, Evin A, et al. On the trail of Neolithic mice and men towards Transcaucasia: zooarchaeological clues from Nakhchivan (Azerbaijan) Biol J Linn Soc. 2013;108:917–928.. [Google Scholar]
- Cucchi T, Papayianni K, Cersoy S, Aznar-Cormano L, Zazzo A, Debruyne R, et al. Tracking the Near Eastern origins and European dispersal of the western house mouse. Sci Rep. 2020;10:8276.. doi: 10.1038/s41598-020-64939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchi T, Vigne J-D, Auffray J-C. First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the Western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol J Linn Soc. 2005;84:429–445.. [Google Scholar]
- Dolédec S, Chessel D. Co-inertia analysis: an alternative method for studying species-environment relationships. Freshw Biol. 1994;31(3):277–294.. [Google Scholar]
- Dray S, Dufour A-B. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20.. [Google Scholar]
- Escoufier Y. Le traitement des variables vectorielles. Biometrics. 1973;29(4):751–760.. [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3. 5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567.. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fairley JS, Smal CM. Feral house mice in Ireland. Ir Naturalists’ J. 1987;22(7):284–290.. [Google Scholar]
- Gabriel SI, Mathias MDL, Searle JB. Genetic structure of house mouse (Mus musculus Linnaeus 1758) populations in the Atlantic archipelago of the Azores: colonization and dispersal. Biol J Linn Soc. 2013;108(4):929–940.. [Google Scholar]
- Gabriel SI, Mathias ML, Searle JB. Of mice and the ‘Age of Discovery’: the complex history of colonization of the Azorean archipelago by the house mouse (Mus musculus) as revealed by mitochondrial DNA variation. J Evolut Biol. 2015;28(1):130–114.. doi: 10.1111/jeb.12550. [DOI] [PubMed] [Google Scholar]
- Ganem G. Behavioural and physiological characteristics of standard and chromosomally divergent populations of house mice from the Orkney archipelago (Scotland) Acta Theriol. 1998;43(1):23–38.. [Google Scholar]
- García-Rodríguez O, Andreou D, Herman JS, Mitsainas GP, Searle JB, Bonhomme F, et al. Cyprus as an ancient hub for house mice and humans. J Biogeogr. 2018;45:2618–2630.. [Google Scholar]
- Gilbert E, O’Reilly S, Merrigan M, McGettigan D, Vitart V, Joshi PK, et al. The genetic landscape of Scotland and the Isles. Proc Natl Acad Sci USA. 2019;116(38):201904761.. doi: 10.1073/pnas.1904761116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich PD, Smith BH, Rosenberg K. Allometric scaling in the dentition of primates and prediction of body weight from tooth size in fossils. Am J Phys Anthropol. 1982;58:81–100.. doi: 10.1002/ajpa.1330580110. [DOI] [PubMed] [Google Scholar]
- Günduz İ, Auffray J-C, Britton-Davidian J, Catalan J, Ganem G, Ramalhinho MG, et al. Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol Ecol. 2001;10:2023–2029.. doi: 10.1046/j.0962-1083.2001.01346.x. [DOI] [PubMed] [Google Scholar]
- Hardouin E, Chapuis J-L, Stevens MI, van Vuuren JB, Quillfeldt P, Scavetta RJ, et al. House mouse colonization patterns on the sub-Antarctic Kerguelen Archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evolut Biol. 2010;10:325.. doi: 10.1186/1471-2148-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden L, Lochovska L, Sémon M, Renaud S, Delignette-Muller M-L, Vicot M, et al. Developmental variability channels mouse molar evolution. eLife. 2020;9:e50103.. doi: 10.7554/eLife.50103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11(1):94.. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EP, Eager HM, Gabriel SI, Jóhannesdóttir F, Searle JB. Genetic tracking of mice and other bioproxies to infer human history. Trends Genet. 2013;29(5):298–308.. doi: 10.1016/j.tig.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Jones EP, Jensen J-K, Magnussen E, Gregersen N, Hansen HS, Searle JB. A molecular characterization of the charismatic Faroe house mouse. Biol J Linn Soc. 2011;102:471–482.. [Google Scholar]
- Jones EP, Jóhannesdóttir F, Gündüz İ, Richards MB, Searle JB. The expansion of the house mouse into north-western Europe. J Zool Lond. 2011;283(4):257–268.. [Google Scholar]
- Jones EP, Skirnisson K, McGovern T, Gilbert M, Willerslev E, Searle JB. Fellow travellers: a concordance of colonization patterns between mice and men in the North Atlantic region. BMC Evolut Biol. 2012;12(1):35.. doi: 10.1186/1471-2148-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874.. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledevin R, Chevret P, Ganem G, Britton-Davidian J, Hardouin EA, Chapuis J-L, et al. Phylogeny and adaptation shape the teeth of insular mice. Proc R Soc Lond Biol Sci. 2016;283:20152820.. doi: 10.1098/rspb.2015.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9):1110–1116.. [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, et al. The fine-scale genetic structure of the British population. Nature. 2015;519(7543):309–314.. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomolino MV. Body size of mammals on islands: the island rule reexamined. Am Naturalist. 1985;125:310–316.. [Google Scholar]
- Lomolino MV. Body size evolution in insular vertebrates: generality of the island rule. J Biogeogr. 2005;32:1683–1699.. [Google Scholar]
- Lomolino MV, Sax DF, Palombo MR, van der Geer AA. Of mice and mammoths: evaluations of causal explanations for body size evolution in insular mammals. J Biogeogr. 2012;39:842–854.. [Google Scholar]
- Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457(7231):830–836.. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- Martínková N, Barnett R, Cucchi T, Struchen R, Pascal M, Pascal M, et al. Divergent evolutionary processes associated with colonization of offshore islands. Mol Ecol. 2013;22:5205–5220.. doi: 10.1111/mec.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millien V. Morphological evolution is accelerated among island mammals. PLoS Biol. 2006;4(10):e321.. doi: 10.1371/journal.pbio.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. (2013) Vegan: Community Ecology Package. R Package Version. 2.0-10. CRAN.
- Peres-Neto PR, Jackson DA. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia. 2001;129(2):169–178.. doi: 10.1007/s004420100720. [DOI] [PubMed] [Google Scholar]
- Pocock MJO, Searle JB, White PCL. Adaptations of animals to commensal habitats: population dynamics of house mice Mus musculus domesticus on farms. J Anim Ecol. 2004;73:878–888.. [Google Scholar]
- Polly PD. On the simulation of the evolution of morphological shape: multivariate shape under selection and drift. Palaeontol Electron. 2004;7(2):7A:28.. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959.. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Chevret P, Michaux J. Morphological vs. molecular evolution: ecology and phylogeny both shape the mandible of rodents. Zool Scr. 2007;36:525–535.. [Google Scholar]
- Renaud S, Hardouin EA, Quéré J-P, Chevret P. Morphometric variations at an ecological scale: seasonal and local variations in feral and commensal house mice. Mamm Biol. 2017;87:1–12.. [Google Scholar]
- Renaud S, Ledevin R, Souquet L, Gomes Rodrigues H, Ginot S, Agret S, et al. Evolving teeth within a stable masticatory apparatus in Orkney mice. Evolut Biol. 2018;45(4):405–424.. [Google Scholar]
- Renaud S, Pantalacci S, Auffray J-C. Differential evolvability along lines of least resistance of upper and lower molars in island house mice. PLoS One. 2011;6(5):e18951.. doi: 10.1371/journal.pone.0018951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk AA, Shepherd AN, Clarke DV, Sheridan AJ, Fraser S, Bartosiewicz L, et al. Rodents: food or pests in Neolithic Orkney. R Soc Open Sci. 2016;3(10):160514.. doi: 10.1098/rsos.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Pvd Mark, Ayres D, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–3302.. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Searle JB, Jones CS, Gündüz İ, Scascitelli M, Jones EP, Herman JS, et al. Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proc R Soc Lond Biol Sci. 2009;276:201–207.. doi: 10.1098/rspb.2008.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendell-Price AT, Ruegg KC, Clegg SM. Rapid morphological divergence following a human-mediated introduction: the role of drift and directional selection. Heredity. 2020;124:535–549.. doi: 10.1038/s41437-020-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano E, Franchini P, Colangelo P, Capanna E, Castiglia R. Multiple origins of the western European house mouse in the Aeolian Archipelago: clues from mtDNA and chromosomes. Biol Invasions. 2013;15(4):729–739.. [Google Scholar]
- Souquet L, Chevret P, Ganem G, Auffray J-C, Ledevin R, Agret S, et al. Back to the wild: does feralization affect the mandible of non-commensal house mice (Mus musculus domesticus)? Biol J Linn Soc. 2019;126:471–486.. [Google Scholar]
- van der Geer AA, Lyras GA, Lomolino MV, Palombo MR, Sax DF. Body size evolution of palaeo-insular mammals: temporal variations and interspecific interactions. J Biogeogr. 2013;40:1440–1450.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences are deposited in EMBL, accessions LR862585–LR862663. Sampling locations are indicated in Table S1. Morphological data and microsatellite genotypes are deposited in Dryad: 10.5061/dryad.nvx0k6dqm.