Abstract
Sterol homeostasis is tightly controlled by molecules that are highly conserved from yeast to humans, the dysregulation of which plays critical roles in the development of antifungal resistance and various cardiovascular diseases. Previous studies have shown that sterol homeostasis is regulated by the ubiquitin–proteasome system. Two E3 ubiquitin ligases, Hrd1 and Doa10, are known to mediate the proteasomal degradation of 3-hydroxy-3-methylglutaryl-CoA reductase Hmg2 and squalene epoxidase Erg1 with accumulation of the toxic sterols in cells, but the deubiquitinases (DUBs) involved are unclear. Here, we screened for DUBs responsible for sterol homeostasis using yeast strains from a DUB-deletion library. The defective growth observed in ubp3-deleted (ubp3Δ) yeast upon fluconazole treatment suggests that lack of Ubp3 disrupts sterol homeostasis. Deep-coverage quantitative proteomics reveals that ergosterol biosynthesis is rerouted into a sterol pathway that generates toxic products in the absence of Ubp3. Further genetic and biochemical analysis indicated that Ubp3 enhances the proteasome's ability to degrade the ergosterol biosynthetic enzymes Erg1 and Erg3. The retardation of ergosterol enzyme degradation in the ubp3Δ strain resulted in the severe accumulation of the intermediate lanosterol and a branched toxic sterol, and ultimately disrupted sterol homeostasis and led to the fluconazole susceptibility. Our findings uncover a role for Ubp3 in sterol homeostasis and highlight its potential as a new antifungal target.
Keywords: sterol homeostasis, lanosterol, toxic sterol, ubp3, fluconazole susceptibility, ergosterol synthesis enzymes, protein stability
Abbreviations: CHX, cycloheximide; DUBs, deubiquitinases; FC, fold change; HB, histidine and biotin; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; SC, synthetic complete; SILAC, stable isotope labeling by amino acids in cell culture; TAP, tandem affinity purification
Sterols, such as the cholesterol found in animals and the ergosterol in fungi, are essential for sustaining the structure and properties of cellular membranes, such as fluidity and rigidity (1, 2). The intermediate and the final products in cholesterol/ergosterol biosynthetic pathway are precursors of many steroid hormones or terpenoids that are essential for cell survival. A large number of studies have shown that the dysregulation of sterol homeostasis can lead to the development of cardiovascular diseases in humans and antifungal resistance in fungi (2, 3, 4).
In fungi, the biosynthesis of ergosterol is tightly regulated by a cascade of enzymes encoded by a group of ergosterol biosynthetic enzymes (Erg genes). These enzymes can be assigned to three pathways: the “early” mevalonate pathway, the “late” ergosterol pathway, and the “alternate” branched toxic sterol pathway (5, 6). Enzymes of the mevalonate pathway control the conversion of acetyl-CoA to farnesyl pyrophosphate. Among them, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductases Hmg1 and Hmg2 are rate-limiting enzymes (3). Enzymes of the ergosterol pathway are responsible for the biosynthesis of ergosterol from squalene, and some of them are chosen as targets for antifungal drug designing. Among them, the squalene epoxidase Erg1 and the lanosterol 14-alpha-demethylase Erg11 are rate-limiting enzymes. Azoles, such as fluconazole, can inhibit the activity of Erg11 and redirect the metabolic flux to the toxic sterol pathway (5, 7). Byproducts of this pathway then inhibit fungi by disrupting the plasma membrane and affecting the membrane permeability (8, 9). As for the alternative pathway, the C-5 sterol desaturase (Erg3) is known to mediate the production of toxic sterol 14α-methylergosta 8,24(28)-dien-3β,6α-diol (10). Previous studies have indicated that loss of function of both Erg3 and Erg11 enables fungi to become tolerant to azoles by effectively blocking the synthesis of toxic sterols (4, 9, 11, 12).
The sophisticated and precise regulation of ergosterol biosynthesis requires the combined actions of transcription, translation, and post-translational modifications and plays an important role in maintaining cellular sterol homeostasis (3, 13, 14, 15, 16, 17). In fungi, transcriptional factors Upc2 and Ecm22 regulate the transcription of most of Erg genes (17, 18). The methylation of histone H3 lysine 4 (H3K4) has also been shown to impact the gene expression of HMG1, HMG2, and ERG11 (16, 19). At protein level, a key mechanism in sterol homeostasis involves the ubiquitination-dependent proteasomal degradation of Erg enzymes in a sterol-dependent manner (20, 21). The overaccumulation of geranylgernayl pyrophosphate and lanosterol would feedback to induce the proteasomal degradation of Hmg2 and Erg1, respectively. The E3 ubiquitin ligases, Hrd1 and Doa10, control the sterol-dependent degradation of Hmg2 and Erg1, respectively, through the endoplasmic reticulum–associated protein degradation pathway (13, 14, 15, 21, 22). However, the deubiquitination machinery and the identity of deubiquitinases (DUBs) for sterol homeostasis remain unclear.
Herein, we identified a novel DUB, namely Ubp3, that promotes the degradation of Erg enzymes in yeast through enhancing the ability of proteasome. In ubp3Δ yeast, an increased level of Erg enzyme triggers the accumulation of intermediate lanosterol and toxic sterols, rendering the strain sensitive to fluconazole. Our finding shed new light on the role of Ubp3 in sterol homeostasis and its potential as an antifungal therapeutic target.
Results
DUB Ubp3 is required for fluconazole resistance
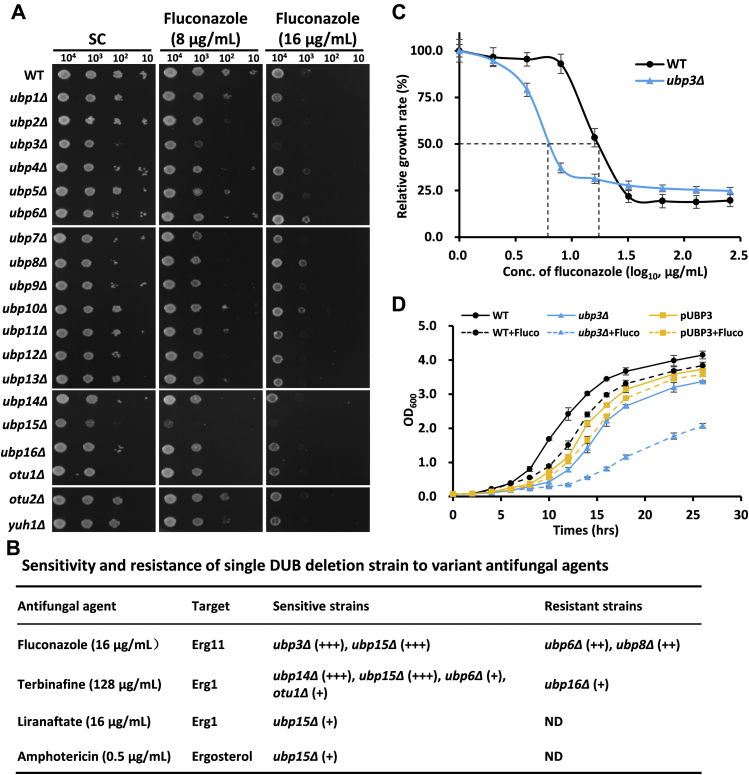
In yeast, there are 21 DUBs that belong to five families (23). To identify DUBs responsible for regulating sterol homeostasis, we systematically compared susceptibility of 19 single DUB deletion strains, excluding the newly identified MIY1 (24) and the essential RPN11 (25), to the antifungal agent fluconazole, which disrupted the sterol homeostasis. On synthetic complete (SC) media, the growth of ubp3Δ and ubp15Δ strains was slightly poorer than that of WT. No obvious effects were observed in other strains (Fig. 1A, left). In the presence of fluconazole stress that blocked the activity of Erg11, ubp3Δ and ubp15Δ strains exhibited serious growth defect when compared with WT, indicating that these DUBs might be involved in regulating sterol homeostasis (Fig. 1A, middle and right). Among them, the growth defect of ubp3Δ was most severe (Fig. 1B). We also tried replacing fluconazole with three other antifungal agents, including terbinafine and liranaftate, that target Erg1 and amphotericin B that bind with ergosterol (8). The results showed that ubp15Δ responded to all three antifungal agents at varying degrees and ubp14Δ was sensitive to terbinafine (Fig. S1). However, ubp3Δ was insensitive to all three antifungal agents (Fig. 1B and Fig. S1), suggesting its specificity to fluconazole.
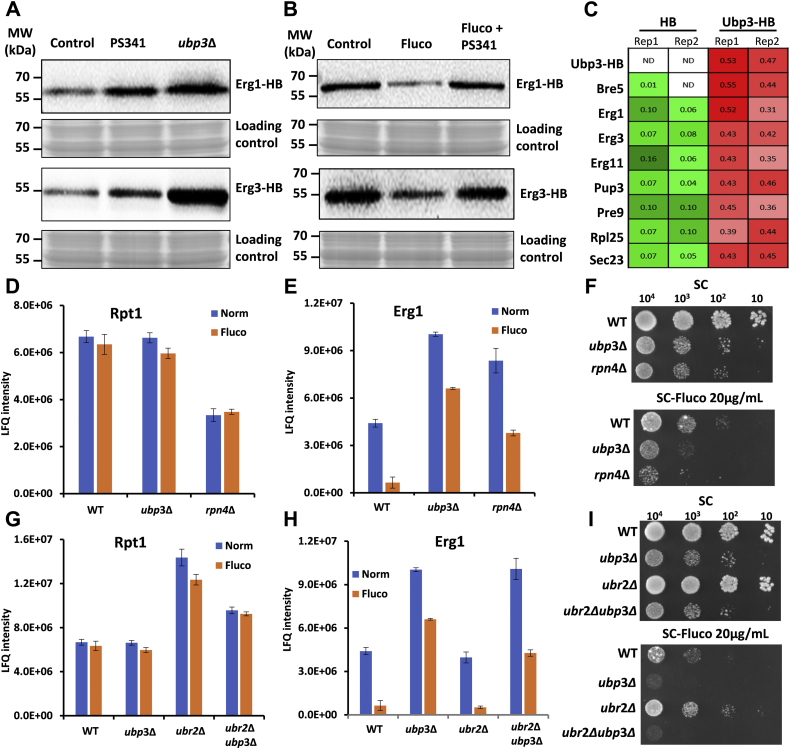
Figure 1.
Ubp3 is required for the tolerance to fluconazole.A, WT and 19 DUB deletion yeast strains were plated on standard synthetic complete (SC) media, SC containing 8 and 16 μg/ml fluconazole. B, sensitive and resistant phenotypes to several antifungal agents. “+” stands for the sensitive or resistant degree. C, the IC50 curve of WT and ubp3Δ to fluconazole in SC media. Three biological replicates were performed. D, growth curve of WT, ubp3Δ, and Ubp3-replenished (pUBP3) strains with or without fluconazole (Fluco). Three biological replicates were performed. DUB, deubiquitinase.
The IC50 value of ubp3Δ was 5.7 μg/ml, which was about one-third of that (18.2 μg/ml) of WT strain (Fig. 1C). The growth curves in SC media with or without fluconazole also verified the growth defect in ubp3Δ strain, and the phenotype was rescued after Ubp3 was re-expressed in ubp3Δ (Fig. 1D). These results supported that Ubp3 was required for fluconazole resistance and might be involved in maintaining the sterol homeostasis in yeast cells.
Loss of Ubp3 rewires ergosterol biosynthetic pathway
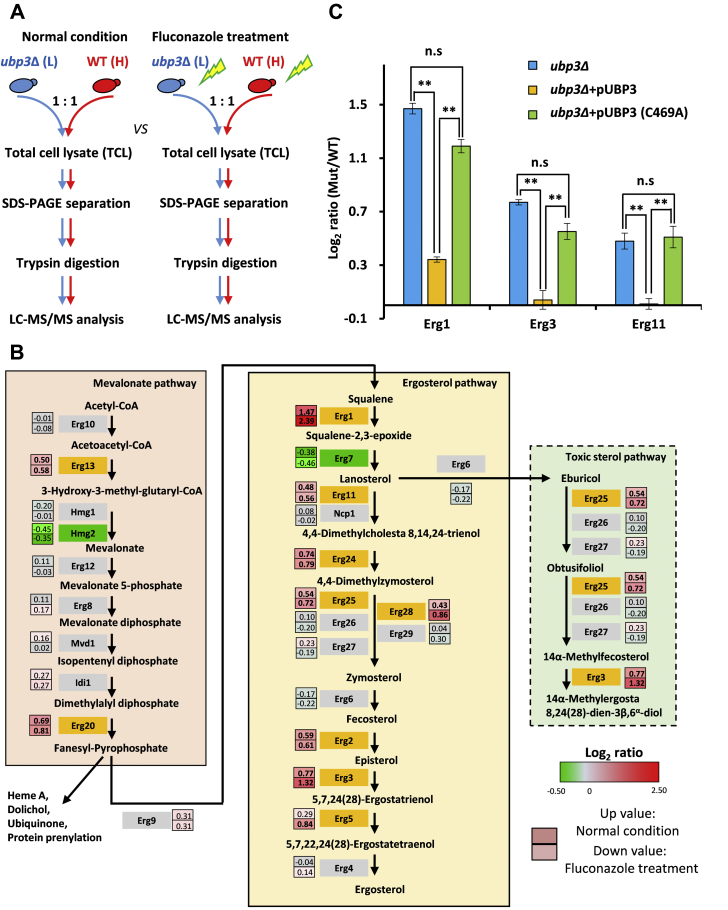
Previous studies have shown that the proteomes of WT and ubp3Δ differ by 30% (26, 27). This makes the study of fluconazole resistance in ubp3Δ particularly challenging, as most of the changes in proteome might indirectly affect the sensitivity of the strains to fluconazole stress. To reduce experimental errors, we used a stable isotope labeling by amino acids in cell culture (SILAC) label-swap approach that combined high-resolution peptide fractionation with off-line high-pH LC and LC–MS/MS platform to compare the proteome difference between WT and ubp3Δ before and after fluconazole treatment (Fig. 2A).
Figure 2.
Loss of Ubp3 perturbed the ergosterol biosynthesis pathway.A, SILAC-based quantitative proteomics to compare the proteome of WT and ubp3Δ strains with or without 32 μg/mL fluconazole treatment. Label-swapped experiments (forward and reverse) were performed for normal and fluconazole condition. B, ergosterol biosynthesis enzyme quantification between WT and ubp3Δ. The average value of log2 ratio (ubp3Δ/WT) of forward and reverse data set was in the bar box. Top value was for normal condition, and bottom value was for fluconazole treatment. Upregulated values were marked as red, and the corresponding enzymes were filled with orange. Unchanged value and its enzyme were marked with gray. Down value and its enzyme were marked with green. C, Ubp3 with full deubiquitinase activity was required for maintaining the Erg1, Erg3, and Erg11 level in cell. The log2 ratios of ubp3Δ, pUBP3, and C469A to WT were compared, respectively. The results represent the means (±SD) of forward and reverse experiments. ∗∗p < 0.005. ns, no significance; SILAC, stable isotope labeling by amino acids in cell culture.
Our deep-coverage proteomics platform (28) allowed us to quantify all the 25 Erg enzymes in the ergosterol pathway (Fig. 2B and Table S3). Surprisingly, deletion of UBP3 led to accumulation of nearly half of the enzymes involved in ergosterol biosynthetic pathway, especially those in the late ergosterol and toxic sterol pathways (Fig. 2B). Among them, 10 enzymes showed significant upregulation, whereas only two showed slight downregulation (Fig. 2B). The difference in protein abundance of these upregulated Erg enzymes between ubp3Δ and WT was further increased in the presence of fluconazole, indicating that Ubp3 plays a critical role in regulating the stability of the enzymes in the ergosterol biosynthesis pathways. Erg1, the first enzyme of ergosterol pathway (late), showed the largest accumulation in ubp3Δ (Fig. 2B), and our observation was further supported by tryptic peptide intensity measurements (Fig. S2). The loss of Ubp3 also resulted in the accumulation of Erg11, an enzyme in the bifurcation node of the ergosterol pathway (late) and toxic sterol pathways, which disagrees with the previous study that its upregulation contributed to fluconazole resistance (29).
To examine if the accumulation of Erg enzymes was directly regulated by Ubp3, we picked three key Erg enzymes of ergosterol and branched toxic sterol pathways, namely Erg1, Erg3, and Erg11, for further investigation. The levels of these Erg enzymes in ubp3Δ, Ubp3 complementary strain, and the catalytically inactive Ubp3 (C469A) complementary strain were compared using SILAC (Fig. 2A and Fig. S3). The results showed that these three enzymes were downregulated in the Ubp3 complementary strain but not in the Ubp3–C469A (Fig. 2C). Quantitative RT-PCR results showed little difference in the relative mRNA levels of ERG1, ERG3, and ERG11 between ubp3Δ and WT strains, regardless of fluconazole treatment (Fig. S4). These results suggest that the abundance of Erg enzymes is unlikely to be regulated on the mRNA level. Thus, in contrast to the general notion that DUBs stabilize proteins (30), Ubp3 might play an unexpected role in promoting the degradation of Erg enzymes in a deubiquitination activity–dependent manner.
Ubp3 is required for fluconazole-induced feedback degradation of the Erg enzymes
To confirm the regulation of Ubp3 on the stability of Erg enzymes, the turnover rates of Erg enzymes in WT and ubp3Δ were measured using pulse-chase SILAC approach (21, 31) (Fig. 3A). The result showed that the degradative rates of accumulated Erg enzymes, such as Erg1, Erg3, and Erg11, in ubp3Δ cells were significantly lower than those of WT (Fig. 3, B and C). In contrast, deletion of Ubp3 had no effect on the half-lives or abundance of other enzymes in this pathway, such as Erg9, Erg10, and Erg27 (Figs. 2B and 3C). We also validated this phenomenon with a His6-biotin–tagged Erg1 and Erg3 in pdr5Δ background (Fig. 3, D and E). These results suggest that Ubp3 promotes the degradation of a subset of Erg enzymes under normal condition.
Figure 3.
Ubp3 promoted the degradation of specific Erg enzymes in ergosterol biosynthesis pathway.A, workflow for the protein turnover measurements based on SILAC-based quantitative proteomics. WT and ubp3Δ were labeled completely and then transferred into light media and harvested as the time indicated. B, Erg1 degradation in WT and ubp3Δ strains. The results represent the means (±SD) of two repetitions. C, half-life fold change of the indicated proteins in ubp3Δ compared with WT strains. The asterisk stands for significant change in proteins. D, the degradation of Erg1 and Erg3 was monitored after inhibition of protein synthesis by 200 μg/mL cycloheximide in WT and ubp3Δ cells in the pdr5Δ background. Whole-cell extracts were analyzed by SDS-PAGE and Western blotting. Erg1 and Erg3 were detected with the antibody of the biotin tag labeled on their C-terminal region. Ponceau S staining was used as loading control. SILAC, stable isotope labeling by amino acids in cell culture.
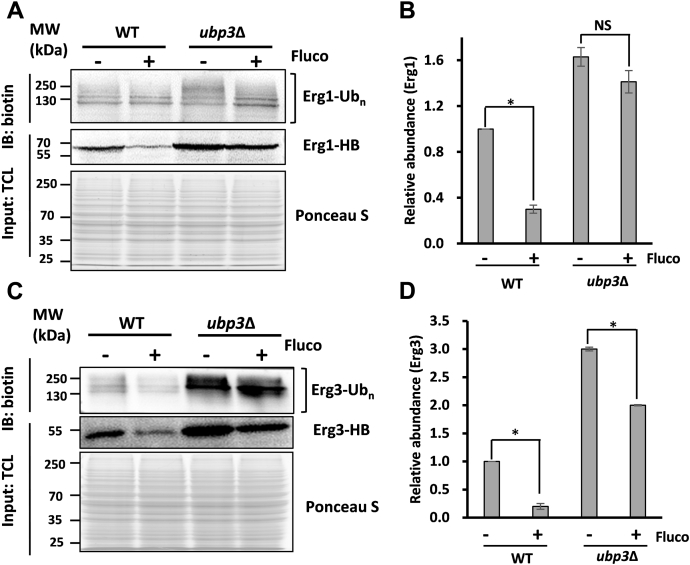
Previous study has suggested that the sterol homeostasis is regulated by a feedback inhibition in an ubiquitination-dependent degradation manner (20). Lanosterol accumulation induced by fluconazole could enhance the degradation of its upstream enzyme Erg1, which in turn reduces the biosynthesis of lanosterol to maintain the sterol homeostasis (14). To test the hypothesis that Ubp3 promotes the degradation of Erg enzymes in the presence of fluconazole, the abundances of the key Erg enzymes were compared before and after fluconazole treatment by Western blotting (Fig. 4). As expected, fluconazole treatment resulted in a significant decrease of Erg1 in WT strain, which reflected the feedback signal. Similar decrease was observed in phenotype of Erg3 protein levels in WT implying the same regulatory machinery for Erg1 and Erg3 (Fig. 4, C and D). Fluconazole treatment might divert lanosterol into the biosynthesis of toxic sterols, in which Erg3 acts as the key enzyme (5). We speculated that the accumulation of toxic sterol could also induce the feedback signal to increase the degradation of its upstream enzyme Erg3. This is a novel feedback signal that was induced by the toxic sterol besides lanosterol and geranylgernayl pyrophosphate.
Figure 4.
Fluconazole-induced feedback degradation of Erg enzymes was blocked upon loss of Ubp3.A, the abundance of Erg1-HB was detected after fluconazole (Fluco) treatment in WT and ubp3Δ cells. Ubiquitinated forms of Erg1 were enriched based on the HB tag on its C terminal and form equal amounts of whole-cell lysates of the indicated four strains. Whole-cell lysates and their ubiquitinated forms were analyzed by SDS-PAGE and Western blotting. Erg1 was probed with anti-streptavidin antibody. The Ponceau S staining was taken as loading control. A representative image of two independent experiments was shown. B, the relative gray values of the Erg1 itself in A. The ratios were based on the gray value of WT without fluconazole treatment. The asterisk stands for significant change in two independent experiments. C, the abundance of Erg3-HB and its ubiquitinated forms after fluconazole treatment as described in panel A. D, the relative gray values of Erg3 itself as described in C. NS, no significance.
Moreover, the accelerated degradation of Erg1 and Erg3, where both were arrested in ubp3Δ cells (Fig. 4, A–D), suggested that Ubp3 might participate in this feedback inhibition by facilitating their degradation. The results are consistent with those obtained using SILAC-based quantitative proteomics (Fig. 2A and Fig. S5). These results further support the large ratios of Erg1 and Erg3 between ubp3Δ and WT strains treated with fluconazole (Fig. 2B). In addition, the absence of Ubp3 also significantly upregulated the ubiquitinated forms of Erg1 and Erg3 regardless of fluconazole incubation or not (Fig. 4, A and C) but did not increase their unstability to decrease the whole amount of Erg enzymes. These results indicated that the loss of Ubp3 did not block the ubiquitination process of Erg proteins but did affect the degradation process of its ubiquitinated form.
Given that both Hrd1 and Doa10 were required for the accelerated degradation of key Erg enzymes induced by sterol intermediates (14, 15, 20), we examined if Ubp3 had any effects on the abundance of these two E3 ligases. Our data from the SILAC-based quantitative proteomics showed that lack of Ubp3 had no impact on the levels of Hrd1 and Doa10, which were not changed by fluconazole as well (Fig. 2A and Fig. S6). Therefore, the absence of Ubp3 could not affect the abundance of key Erg enzymes through altering the protein level of E3s, which mediated their degradation. We thus speculated that Ubp3 was required for the feedback control of the degradation of Erg1 and Erg3 induced by fluconazole treatment, and loss of Ubp3 inhibited their degradation and blocked the feedback signal.
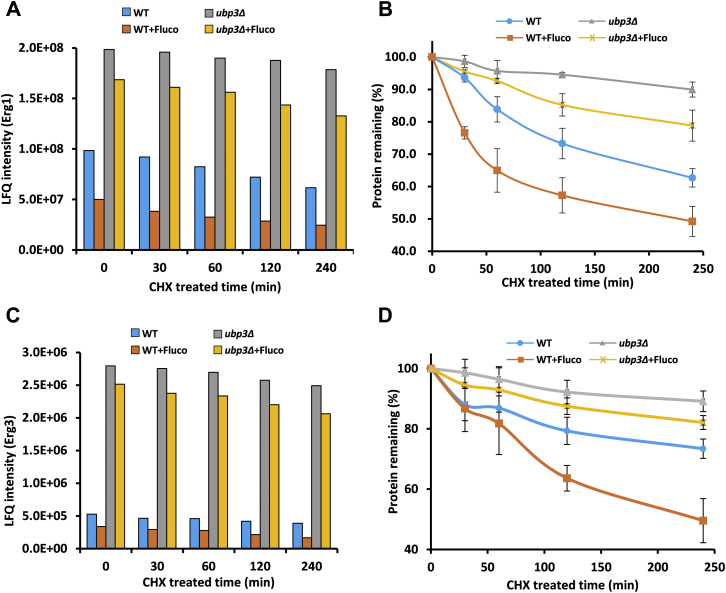
We also measured protein turnover rates of Erg1 and Erg3 in WT and ubp3Δ without or with fluconazole treatment after protein translation is blocked by cycloheximide (CHX) (32). Harvested yeast cells at indicated time points after CHX incubation, lysed and took equal amounts of total cell lysates for SDS-PAGE separation. The slices that contained Erg1 and Erg3 based on the molecular weight were cut for further LC–MS/MS analysis (Fig. S7A). We found that fluconazole treatment promoted the degradation of Erg1 and Erg3 in WT but not in ubp3Δ (Fig. 5, A–D). Therefore, Ubp3 was required for the feedback signal by accelerating the degradation of Erg1 and Erg3 during fluconazole stimulation.
Figure 5.
Loss of Ubp3 defects the feedback degradation of Erg enzyme response to fluconazole stress.A, the LFQ intensity of endogenous Erg1 with time course after CHX treated. The WT and ubp3Δ strains were cultured once the optical density was 0.5 (OD600) and treated with or without 32 μg/mL fluconazole for 2 h. Then the cells were treated with CHX as the time presented. Whole-cell extracts were analyzed by SDS-PAGE and LC–MS/MS. B, Erg1 half-life analysis by CHX chase and LC–MS/MS. The percent of protein remaining was normalized to the 0 point. Two independent experiments were performed. C, LFQ intensity of Erg3 as mentioned in panel A. D, half-life analysis of Erg3 as mentioned in panel B. CHX, cycloheximide; LFQ, label-free quantification.
Ubp3 enhances proteasomal degradation of Erg enzymes
To investigate the physiological role of proteasome on these Erg enzymes, we analyzed the levels of these proteins after proteasome inhibition. Similar to ubp3Δ, treatment with PS341, an inhibitor of proteasome, led to significant accumulations in both Erg1 and Erg3 in WT cells, indicating their proteasomal degradations (Fig. 6A). The fluconazole-induced degradation of Erg1 and Erg3 was blocked by PS341 treatment in WT strain (Fig. 6B), suggesting that fluconazole promotes their degradations through proteasomes. Similar accumulation phenomenon of Erg1 and Erg3 in PS341-treated WT yeast and ubp3Δ, with and without fluconazole treatment, implied that Ubp3 absence disturbed their proteasomal degradation.
Figure 6.
Ubp3 located to the upstream of proteasome to facilitate the degradation of Erg enzymes.A, the degradation of Erg proteins was delayed upon PS341 treatment or Ubp3 absence. The Erg1 and Erg3 were expressed with HB tag in WT and ubp3Δ strains and detected with biotin antibody. The WT yeast was treated without or with 50 μM PS341 for 4 h. Equal amount of whole-cell lysates was loaded for analysis. B, the abundances of Erg1 and Erg3 were detected after fluconazole and combined fluconazole with PS341 treatment in WT. The yeast cell was pretreated with 50 μM PS341 for 15 min and then added with 32 μg/ml fluconazole for 4 h. The target protein was probed with anti-streptavidin antibody. C, proportion quantification of the interactors with Ubp3. The proportion was based on the sum iBAQ of each protein identified in the HB and UBP3-HB experiments. D and E, the LFQ intensity of Rpt1 (subunit of proteasome) (D) or Erg1 (E) in WT, ubp3Δ, and rpn4Δ strains before and after 32 μg/ml fluconazole incubation. Equal amounts of whole-cell extracts were analyzed by SDS-PAGE and LC–MS/MS. The value represented the means (±SD) of two technical replication quantitative results. F, serial dilutions of WT, ubp3Δ, and rpn4Δ strains spotted onto synthetic complete (SC) (up) and SC containing 20 μg/ml fluconazole (bottom). The plates were incubated at 30 °C for 72 h. G and H, the LFQ intensity of Rpt1 (G) or Erg1 (H) in WT, ubp3Δ, ubr2Δ, and ubp3Δubr2Δ strains before and after 32 μg/ml fluconazole treatment. The value represents the means (±SD) of two quantitative results. I, serial dilutions of WT, ubp3Δ, ubr2Δ, and ubp3Δ ubr2Δ strains spotted onto SC (up) and SC containing 20 μg/ml fluconazole (bottom). The plates were incubated at 30 °C for 72 h. iBAQ, intensity-based absolute quantitation. ND, no detection.
To clarify the relationship among Ubp3, Erg enzymes, and proteasome, the protein–protein interactions among Ubp3, the two Erg enzymes, and proteasome were examined. Ubp3 that was either C-terminally tagged with tandem 6× histidine and biotin (Ubp3-HB) or free of HB (control) was expressed in ubp3Δ (Fig. S8A and Table S1). After subjecting total cell lysates to tandem affinity purification (TAP) under native conditions, the purified Ubp3 interactome was analyzed by LC–MS/MS (Fig. S8B). As expected, Bre5, a cofactor of Ubp3 (33, 34, 35), was enriched significantly (Fig. 6C). Some of the known substrates of Ubp3, such as Sec23 and Rpl25 (33, 36), were also enriched (Fig. 6C), suggesting the high quality of our samples. The enrichment of Erg1 and Erg3 in Ubp3-HB sample was also significantly higher than those in the HB control (Fig. 6C), suggesting direct or indirect interaction between Ubp3 and Erg1 and Erg3. Except for these proteins, the subunits of proteasome, such as Pup3 and Pre9 (37), were also enriched by Ubp3 TAP, suggesting that Ubp3 interacts with the proteasome as well (Fig. 6C). Our result supported a previous finding by Marmorstein group that Ubp3 could interact with subunits of proteasome both in vivo and in vitro (38). To further verify the protein–protein interaction, HB-tagged Erg1 and Erg3 at their carboxyl termini were expressed in WT strain, respectively, followed by TAP through the same HB tag (Fig. S8, C and D). The enrichment of Erg1 and Erg3 was more than that of their respective controls (Fig. S8E). Same as these bait Erg proteins, the Ubp3/Bre5 complex was also greatly enriched (Fig. S8E).
In general, Ubp3 has been known to prevent its substrates from proteasome degradation by removing the polyubiquitin chains modified on them (30, 33, 39), which was contradictory to our results. To understand the role of Ubp3 in the proteasomal degradation of Erg enzymes, the abundance of proteasomes was reduced by mutating the proteasome transcriptional factor RPN4 (40). Erg1 was increased in rpn4Δ, similar to that in ubp3Δ under the normal condition (Fig. 6E, blue bars). The amounts of proteasome, as indicated by a proteasomal subunit, Rpt1, remain unchanged between normal condition and fluconazole treatment in WT, rpn4Δ, and ubp3Δ cells, respectively (Fig. 6D). However, the rpn4Δ and ubp3Δ cells showed a delayed degradation of Erg1 and susceptibility to fluconazole as compared with WT (Fig. 6, E and F). These results support that either knockout of Ubp3 or reduced proteasome level can inhibit the feedback degradation of Erg enzymes upon fluconazole stress.
Conversely, we increased the amounts of proteasomes by removing Ubr2, an E3 ligase that mediates the proteasomal degradation of Rpn4 (41, 42) (Fig. 6G). The result showed that there was a slight increase in the degradation of Erg1 in ubr2Δ cells as compared with WT (Fig. 6H). Consistently, the enhancement of proteasome amounts slightly accelerated feedback degradation of Erg1 and tolerance of ubr2Δ to fluconazole (Fig. 6, H and I). However, both fluconazole-induced degradation of Erg1 and the resistance to fluconazole were lost when UBP3 was knocked out in ubr2Δ cells (Fig. 6, H and I), despite that there were more proteasomes in these cells than WT cells (Fig. 6G). These results support that Ubp3 is involved in proteasomal degradation of Erg enzymes.
Disrupting the feedback degradation of key Erg enzymes in ubp3Δ resulted in toxic sterol accumulation and fluconazole susceptibility
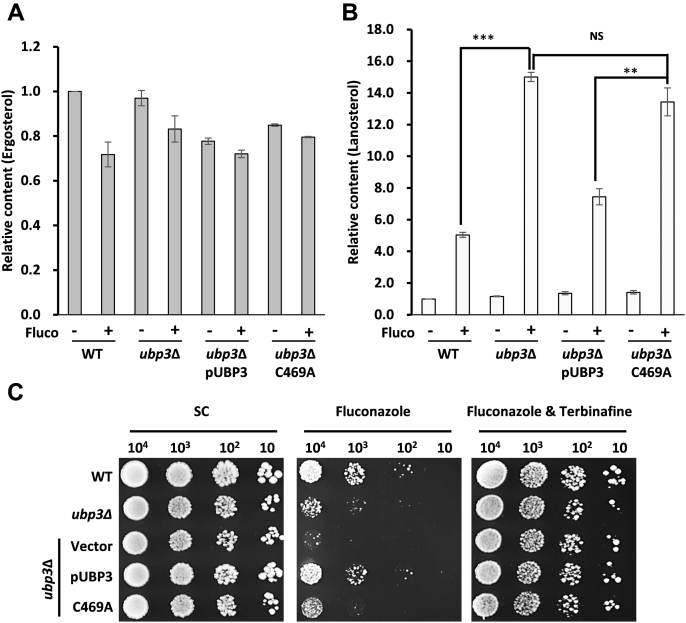
The upregulation of toxic sterol pathway by fluconazole induced a feedback suppression mechanism by which increased Erg1 and Erg3 degradation leads to reduced biosynthesis of these sterols (14, 20), which serves as a protective mechanism for yeast by developing resistance to azoles. We hypothesized that the loss of function of Ubp3 would block the feedback inhibition to accelerate the degradation of Erg1 and Erg3 that eventually led to a significant accumulation of lanosterol for toxic sterol biosynthesis. To test this idea, the cellular levels of the intermediate product lanosterol and the final product ergosterol were analyzed before and after fluconazole treatment in WT, ubp3Δ, Ubp3 complementary strain, and the Ubp3 catalytically inactive mutant (C469A) complementary strain. Unsurprisingly, the cellular level of ergosterol was similar in all these four strains and slightly decreased after fluconazole treatment (Fig. 7A). In normal condition, the cellular level of lanosterol in all these four strains remained the same (Fig. 7B). However, the lanosterol level was greatly increased by over 4 times in both WT and ubp3Δ under the treatment of fluconazole (Fig. 7B), possibly because of the blockage of ergosterol synthesis through inhibiting Erg11 by fluconazole. Furthermore, the accumulation of lanosterol in ubp3Δ was two fold higher than that in WT strain (Fig. 7B), indicating that an enhanced metabolic flux was redirected to toxic sterol biosynthesis in ubp3Δ. Complementary WT but not C469A Ubp3 restored the level of lanosterol to that in WT (Fig. 7B), suggesting that the catalytic activity of Ubp3 is required. Altogether, we propose that loss of feedback degradation of Erg1 and Erg3 would lead to the accumulation of lanosterol in ubp3Δ cell and subsequently increases the production of toxic sterol through branched toxic sterol pathway and fluconazole sensitivity. The growth spots analysis showed that re-expression of WT but not C469A Ubp3 rescued the growth defect and restored fluconazole resistance (Fig. 7C, middle), probably through reducing the expression of the Erg enzymes and the level of lanosterol (Figs. 2C and 7B). Thus, the feedback regulation is dependent on the deubiquitination activity of Ubp3.
Figure 7.
Ubp3 was essential for maintaining cellular sterol homeostasis when exposed to fluconazole.A, relative abundance of ergosterol stoichiometry in WT, ubp3Δ, and its replenishing UBP3 (pUBP3) and catalytic site mutant (C469A) strains with fluconazole (Fluco) treatment or not. The values were normalized based on that of WT without fluconazole and represented as the means (±SD) of three biological repetitions. B, relative abundance of lanosterol stoichiometry as mentioned in panel A. ∗∗∗p < 0.001; ∗∗p < 0.005; NS, no significance. C, serial dilutions of WT, ubp3Δ, and its derivated strains replenishing control empty vector, UBP3 (pUBP3), and catalytic site mutant (C469A) strains spotted onto synthetic complete (SC) (left), SC containing 16 μg/ml fluconazole (middle), or fluconazole plus 32 μg/ml terbinafine (right). The plates were incubated at 30 °C for 72 h.
To further validate this hypothesis, we inhibited the biosynthesis of lanosterol and toxic sterol through the inhibition of Erg1 by terbinafine (43, 44). The presence of terbinafine alone did not impact the viability of these strains (Fig. S9). Indeed, ubp3Δ and C469A re-expressed yeast cells treated with additional terbinafine were no longer sensitive to fluconazole and grew normally as WT (Fig. 7C, right). These results further confirmed that the overaccumulation of lanosterol and toxic sterol in ubp3-deficency cell was the reason for its susceptibility to fluconazole. Therefore, Ubp3-dependent degradation of Erg1 and Erg3 reduced the cellular level of intermediate lanosterol, which prevented the accumulation of toxic sterol and maintained sterol homeostasis (Fig. 8).
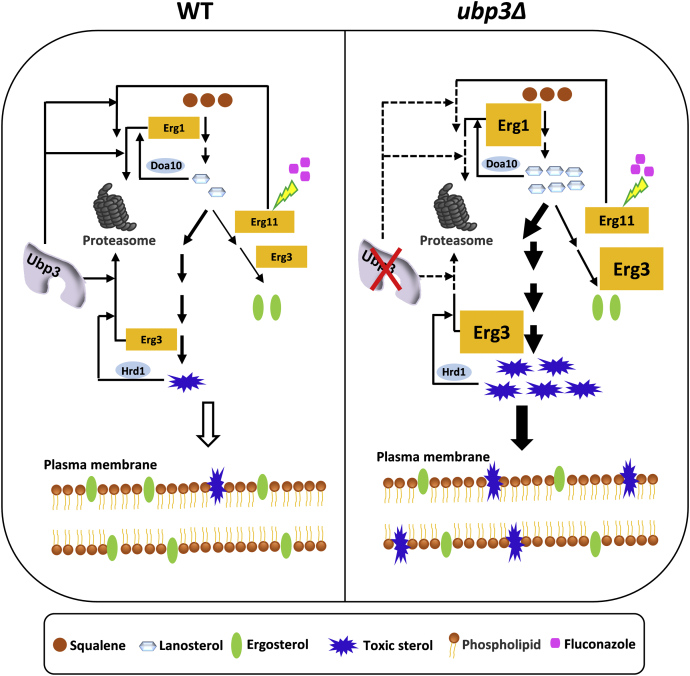
Figure 8.
Model of Ubp3 for facilitating Erg enzyme degradation to maintain sterol homeostasis and fluconazole tolerance. Normally, inhibiting the activity of Erg11 with fluconazole would block the ergosterol biosynthesis pathway and activate alternate toxic pathway in WT strain. With the accumulation of lanosterol and toxic sterol, the feedback proteasomal degradation of Erg1 and Erg3 by E3 ligases Doa10 and Hrd1 is benefited to sterol homeostasis and fluconazole tolerance. Herein, Ubp3 was the key enhancer for the feedback degradation system to reduce the key Erg enzymes and maintain the sterol homeostasis. In ubp3Δ strain, the feedback degradation of Erg1 and Erg3 after fluconazole treatment was disrupted and further resulted into the huge accumulation of lanosterol and toxic sterol, leading to the sensitive phenotype.
Discussion
The ubiquitin and proteasome signaling pathways have been known to regulate sterol homeostasis through balancing the abundance of Erg1 and Hmg2. Sterol-dependent degradation of Erg1 requires yeast Doa10 and its homolog mammalian Teb4 (14, 20). However, little is known about the role of deubiquitination in the regulation of key enzymes for ergosterol biosynthesis. Our study identified several DUBs whose deletion increased the sensitivity to antifungal agents. We further showed that one of these DUBs, Ubp3, protects yeast from sensitivity to antifungal agents through maintaining cellular sterol homeostasis.
In previous genetic screen and haploinsufficiency profiling–homozygous deletion profiling studies (45, 46), Ubp3 was identified as potential target of antifungal agents. However, the mechanism how Ubp3 confers antifungal resistance and sterol homeostasis has been ambiguous. Taking advantage of the deep-coverage proteomics platform established in the laboratory (28), we were able to identify and precisely quantify all enzymes in the ergosterol pathway (Fig. 2B). Our study also revealed that Ubp3 deletion increased the Erg enzyme levels (Fig. 2B). Fluconazole treatment augmented the disparity of Erg enzyme levels between WT and ubp3Δ strains and diverted the ergosterol pathway to toxic sterol pathway, indicating the importance of Ubp3 in sterol homeostasis (Fig. 2B). In combination of protein–protein interaction and half-life analysis of the proteome using SILAC pulse-chase quantification and translation inhibitor, we proved that Ubp3 facilitated the feedback proteasomal degradation of several key Erg enzymes, including Erg1 and Erg3, to maintain sterol homeostasis and fluconazole resistance.
As a DUB, Ubp3 is expected to inhibit the proteolysis of its substrates by removing the ubiquitin on these proteins, therefore preventing their delivery to proteasomes (30, 33). Interestingly, our study has shown that Ubp3 promotes the proteasomal degradation of Erg enzymes in a deubiquitination activity–dependent manner, indicating dual function of this DUB. A previous research also observed the completely opposite roles of Ubp3-dependent deubiquitination in regulating replicative life span and heat resistance, by either promoting or inhibiting the 26S proteasome-dependent degradation of damaged proteins (47). They proposed that promotion or inhibition of protein degradation by Ubp3 depends on what “state” the protein remains. If the presence of the proteins benefits the cells, Ubp3 could remove the polyubiquitin chains to keep them from degradation for reuse. On the other hand, Ubp3 could also enhance the degradation of the proteins if they are toxic to cells. Surprisingly, in our study, the abundance of the enzymes seems depending on the toxic sterol metabolites, which might instruct Ubp3 to accelerate the degradation of its upstream enzymes. In another word, Ubp3 could recognize and then promote the proteasomal degradation of Erg enzymes by interacting with Erg enzymes and proteasome simultaneously, upon fluconazole treatment (Figs. 4, A and C and 6C). The precise mechanism of how Ubp3 senses the fluctuation of the sterol levels needs further study.
In addition, researchers proposed a hypothesis that Ubp3 acts as a cofactor of proteasome and removes the polyubiquitin chains from its substrates to facilitate the substrates to enter into the catalytic particle (20S) of proteasome for proteolysis (38, 48). In our study, deubiquitination activity of Ubp3 is required for the proteasomal degradation rate of Erg enzymes, suggesting that removal of ubiquitin from the Erg enzymes is required for their degradations. It was likely that the Erg enzymes may be easier to be delivered into proteasomes by the direct interaction between Ubp3 and its substrates. This model needs further validations.
On the other hand, further research is needed to understand how Ubp3 chooses to protect or accelerate the degradation of a substrate. In our opinion, Ubp3 might prevent the substrates from degradation when Ubp3 interacts with these substrates indirectly through Bre5 (33, 35, 49). However, Ubp3 promotes the degradation of its substrates when it interacts with proteasome and the ubiquitinated substrates directly (38, 48).
Experimental procedures
Yeast strains, plasmids, and growth conditions
All yeast strains used in this study were derived from Saccharomyces cerevisiae JMP024 (50). Yeast strains and protocols are described in Table S1 and the Supporting information.
Yeast susceptibility to antifungal agents
Yeast cells were cultured in yeast extract peptone dextrose to exponential phase and then washed with distilled water. Equal amounts of cells were spotted on standard SC and SC-containing antifungal agents. The plates were incubated at 30 °C for about 72 h. The IC50 of yeast to fluconazole was measured as described (51). The IC50 was calculated using GraphPad Prism 5 software (GraphPad Software).
Protein identification and quantification in SILAC
For SILAC labeling, WT (JMP024) and ubp3Δ strains (PXL007) (Table S1) were cultured in SC containing Arg (20 mg/l) and Lys (30 mg/l), and the heavy media contained equal molar amounts of [13C6 15N4] arginine (+10.0083 Da) and [13C6] lysine (+6.0201 Da) from Cambridge Isotope Laboratories. The cells were harvested with or without 32 μg/mL fluconazole treatment for 4 h. Differentially labeled WT and ubp3Δ cells were equally mixed and then lysed under denatured condition. Total cell lysates were subjected to SDS-PAGE separation, trypsin in-gel digestion overnight, and analyzed by an ultraperformance LC–MS/MS platform of hybrid LTQ-Orbitrap Velos (Thermo Fisher Scientific) (50, 52). Eluting peptides were analyzed using the mass spectrometer as described before (53).
All the raw files were searched by MaxQuant (version 1.5.30) against the target-decoy protein FASTA file from Saccharomyces genome database (version released in 2016.02; 6717 entries, Saccharomyces genome database; http://www.yeastgenome.org). For SILAC-based quantification, fold change (FC) and p value were both used to calculate the significance of amount of changed proteins. Only proteins with more than 1.4-FC (log2FC at ±0.50) and p value <0.05 in both forward and reverse experiments were considered as significantly changed proteins. The detailed protocols were described in the Supporting information.
TAP strategy
A two-step TAP approach was used to obtain high-purity protein as previously described (54). Yeast cells were harvested during log phase and lysed with glass beads in native condition. The target protein was enriched onto a nickel column followed by second-step purification with streptavidin beads. Finally, the protein was eluted by boiling in 2× SDS-PAGE loading buffer at 80 °C for 10 min. Affinity-purified protein and its interactors were subjected to LC–MS/MS analysis.
Quantitative RT-PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). To eliminate genomic DNA contamination, an additional DNase treatment was performed with RNase-free DNase I (Promega). About 200 ng of RNA was reverse transcribed into complementary DNA in a 10-μl reaction mixture using the Revertra Ace qPCR RT Kit (TOYOBO). The complementary DNAs were then used as templates for PCR. mRNA level of each gene was determined by quantitative real-time PCR using SYBR Green Master Mix (TOYOBO). 18S recombinant DNA was used as the internal control. The primer sequences used in this study are shown in Table S2.
Protein turnover profiling
We applied two methods to analyze the dynamic degradation process of the proteome (21, 50). In the first method, yeast cells were completely labeled in SC heavy media and then transferred into new fresh SC light media when the optical density was 0.6 (OD600). Yeasts were harvested at 0, 15, 30, 60, and 120 min, lysed, and digested in gel with trypsin overnight. The peptides were desalted with C18 Sep-Pak SPE cartridge (Waters) and vacuum evaporated. The resulting peptides were analyzed by LC–MS/MS. In the second method, yeast cells were cultured in SC light media until the optical density was 0.8 and treated with 200 μg/mL CHX to inhibit protein synthesis. Equal amounts of cells were harvested at 0, 30, 60, 120, and 240 min and lysed at denatured condition. Equal amounts of resulting peptides were loaded for LC–MS/MS analysis.
Mass spectrometric lipid analysis
Quantification of sterol species was performed based on the previously described (55, 56). First, yeast cells were lysed by vigorous vortex mixing in 150 mM NH4HCO3. The precipitated lipid sample was extracted with 990 μl chloroform/methanol (17:1, v/v) for 120 min. The organic phase was collected, vacuum evaporated, and analyzed on an ultraperformance LC–MS/MS platform of QTRAP 4500 mass spectrometer (AB Sciex). Lipid species were identified and quantified using MSFileReader (Thermo Fisher Scientific), ALEX software, and Orange software 2.6.
Data availability
All analyzed data sets used for this study are referenced in the text and included in the supporting information, including methods and results, nine supporting figures, and three supporting tables.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Drs Guanghua Huang, Lilin Du, and Cong Xu for gracious gifts of their reagents and strains. We thank Drs Guangwei Du and Hua Mo for the kindness advice for our article. This study was funded by the Ministry of Science and Technology (2017YFA0505002, 2017YFA0505100, and 2016YFA0501300), the National Natural Science Foundation of China (32071431, 31700723, 31670834, 31870824, and 91839302), the Innovation Foundation of Medicine (BWS17J032, AWS17J008, 20SWAQX34, and 19SWAQ17), National Megaprojects for Key Infectious Diseases (2018ZX10302302), Guangzhou Science and Technology Innovation & Development Project (201802020016), the Unilevel 21th Century Toxicity Program (MA-2018-02170N), the Foundation of State Key Lab of Proteomics (SKLP-K201704 and SKLP-Y201901), the China Postdoctoral Science Foundation (2019M664016), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-017), and the Beijing-Tianjin-Hebei Basic Research Cooperation Project (J200001).
Author contributions
P. X., Q. L., and Y. L. designed research; Q. L., Y. L., F. W., Z. L., H. L., and Y. G. performed research; Q. L., Y. L., and Y. W. analyzed the data; Z. Z. analyzed the content of sterol; P. X., Q. L., Y. L., and J. W. wrote the article; and Z. D. and F. H. provided vital advice.
Edited by George DeMartino
Footnotes
This article contains supporting information.
Contributor Information
Yanchang Li, Email: liyanchang1017@163.com.
Junzhu Wu, Email: wujunzhu@whu.edu.cn.
Ping Xu, Email: xuping@ncpsb.org.cn.
Supporting information
References
- 1.Brown M.S., Goldstein J.L. Cholesterol feedback: From Schoenheimer's bottle to Scap's MELADL. J. Lipid Res. 2009;50 Suppl:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein J.L., Deboseboyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Espenshade P.J., Hughes A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 4.Sanglard D., Ischer F., Parkinson T., Falconer D., Bille J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 2003;47:2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya S., Esquivel B.D., White T.C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio. 2018;9 doi: 10.1128/mBio.01291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 7.Mercer E.I. The biosynthesis of ergosterol. Pestic. Sci. 1984;15:133–155. [Google Scholar]
- 8.Cowen L.E., Steinbach W.J. Stress, drugs, and evolution: The role of cellular signaling in fungal drug resistance. Eukaryot. Cell. 2008;7:747–764. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodedova M., Sychrova H. Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daum G., Lees N.D., Bard M., Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Kelly S.L., Lamb D.C., Kelly D.E., Manning N.J., Loeffler J., Hebart H., Schumacher U., Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 12.Yan L., Zhang J., Li M., Cao Y., Xu Z., Cao Y., Gao P., Wang Y., Jiang Y. DNA microarray analysis of fluconazole resistance in a laboratory Candida albicans strain. Acta Biochim. Biophys. Sin. (Shanghai) 2008;40:1048–1060. doi: 10.1111/j.1745-7270.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 13.Burg J.S., Espenshade P.J. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 2011;50:403–410. doi: 10.1016/j.plipres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foresti O., Ruggiano A., Hannibal-Bach H.K., Ejsing C.S., Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife. 2013;2 doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzies S.A., Volkmar N. The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1. Elife. 2018;7 doi: 10.7554/eLife.40009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.South P.F., Harmeyer K.M., Serratore N.D., Briggs S.D. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1016–1025. doi: 10.1073/pnas.1215768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H., Tong J., Lee C.W., Ha S., Eom S.H., Im Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015;6:1–13. doi: 10.1038/ncomms7129. [DOI] [PubMed] [Google Scholar]
- 18.Vik A., Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett M.K., Osborne T.F. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: Increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatsukasa K., Okumura F., Kamura T. Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: Lessons from yeast. Crit. Rev. Biochem. Mol. Biol. 2015;50:489–502. doi: 10.3109/10409238.2015.1081869. [DOI] [PubMed] [Google Scholar]
- 21.Christiano R., Nagaraj N., Frohlich F., Walther T.C. Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep. 2014;9:1959–1965. doi: 10.1016/j.celrep.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampton R.Y., Gardner R.G., Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mevissen T.E., Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 24.Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., Hofmann K., Kulathu Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Lan Q., Gao Y., Xu C., Xu Z., Wang Y., Chang L., Wu J., Deng Z., He F., Finley D., Xu P. Ubiquitin linkage specificity of deubiquitinases determines cyclophilin nuclear localization and degradation. iScience. 2020;23:100984–100998. doi: 10.1016/j.isci.2020.100984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isasa M., Rose C.M., Elsasser S., Navarrete-Perea J., Paulo J.A., Finley D.J., Gygi S.P. Multiplexed, proteome-wide protein expression profiling: Yeast deubiquitylating enzyme knockout strains. J. Proteome Res. 2015;14:5306–5317. doi: 10.1021/acs.jproteome.5b00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulsen J.W., Madsen C.T., Young C., Kelstrup C.D., Grell H.C., Henriksen P., Juhl-Jensen L., Nielsen M.L. Comprehensive profiling of proteome changes upon sequential deletion of deubiquitylating enzymes. J. Proteomics. 2012;75:3886–3897. doi: 10.1016/j.jprot.2012.04.055. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Dammer E.B., Gao Y., Lan Q., Villamil M.A., Duong D.M., Zhang C., Ping L., Lauinger L., Flick K., Xu Z., Wei W., Xing X., Chang L., Jin J. Proteomics links ubiquitin chain topology change to transcription factor activation. Mol. Cell. 2019;76:126–137.e127. doi: 10.1016/j.molcel.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard D., Coste A., Ferrari S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009;9:1029–1050. doi: 10.1111/j.1567-1364.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- 30.Kvint K., Uhler J.P., Taschner M.J., Sigurdsson S., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol. Cell. 2008;30:498–506. doi: 10.1016/j.molcel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 31.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 32.Edwards P.A., Gould R.G. Turnover rate of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase as determined by use of cycloheximide. J. Biol. Chem. 1972;247:1520–1524. [PubMed] [Google Scholar]
- 33.Cohen M., Stutz F., Belgareh N., Haguenauer-Tsapis R., Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- 34.Li K., Ossareh-Nazari B., Liu X., Dargemont C., Marmorstein R. Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J. Mol. Biol. 2007;372:194–204. doi: 10.1016/j.jmb.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K., Zhao K., Ossareh-Nazari B., Da G., Dargemont C., Marmorstein R. Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor. J. Biol. Chem. 2005;280:29176–29185. doi: 10.1074/jbc.M502975200. [DOI] [PubMed] [Google Scholar]
- 36.Ossareh-Nazari B., Bonizec M., Cohen M., Dokudovskaya S., Delalande F., Schaeffer C., Van Dorsselaer A., Dargemont C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finley D., Chen X., Walters K.J. Gates, channels, and switches: Elements of the proteasome machine. Trends Biochem. Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao P., Smerdon M.J. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J. Biol. Chem. 2010;285:37542–37550. doi: 10.1074/jbc.M110.170175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chew B.S., Siew W.L., Xiao B., Lehming N. Transcriptional activation requires protection of the TATA-binding protein Tbp1 by the ubiquitin-specific protease Ubp3. Biochem. J. 2010;431:391–399. doi: 10.1042/BJ20101152. [DOI] [PubMed] [Google Scholar]
- 40.Xie Y., Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Mao X., Ju D., Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 42.Ju D., Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and-independent. J. Biol. Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- 43.Klobucníková V., Kohút P., Leber R., Fuchsbichler S., Schweighofer N., Turnowsky F., Hapala I. Terbinafine resistance in a pleiotropic yeast mutant is caused by a single point mutation in the ERG1 gene. Biochem. Biophys. Res. Commun. 2003;309:666–671. doi: 10.1016/j.bbrc.2003.08.051. [DOI] [PubMed] [Google Scholar]
- 44.Leber R., Fuchsbichler S., Klobucníková V., Schweighofer N., Pitters E., Wohlfarter K., Lederer M., Landl K., Ruckenstuhl C., Hapala I., Turnowsky F. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2003;47:3890–3900. doi: 10.1128/AAC.47.12.3890-3900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mor V., Rella A., Farnoud A.M., Singh A., Munshi M., Bryan A., Naseem S., Konopka J.B., Ojima I., Bullesbach E., Ashbaugh A., Linke M.J., Cushion M., Collins M., Ananthula H.K. Identification of a new class of antifungals targeting the synthesis of fungal sphingolipids. mBio. 2015;6 doi: 10.1128/mBio.00647-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Fang Y., Jaiseng W., Hu L., Lu Y., Ma Y., Furuyashiki T. Characterization of tamoxifen as an antifungal agent using the yeast Schizosaccharomyces pombe model organism. Kobe J. Med. Sci. 2015;61:54–63. [PubMed] [Google Scholar]
- 47.Öling D., Eisele F., Kvint K., Nyström T. Opposing roles of Ubp3-dependent deubiquitination regulate replicative life span and heat resistance. EMBO J. 2014;33:747–761. doi: 10.1002/embj.201386822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Zhu M., Ayalew M., Ruff J.A. Down-regulation of Pkc1-mediated signaling by the deubiquitinating enzyme Ubp3. J. Biol. Chem. 2008;283:1954–1961. doi: 10.1074/jbc.M705682200. [DOI] [PubMed] [Google Scholar]
- 49.Bilsland E., Hult M., Bell S.D., Sunnerhagen P., Downs J.A. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair. 2007;6:1471–1484. doi: 10.1016/j.dnarep.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agarwal A.K., Rogers P.D., Baerson S.R., Jacob M.R., Barker K.S., Cleary J.D., Walker L.A., Nagle D.G., Clark A.M. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 52.Xiao W., Zhang J., Wang Y., Liu Z., Wang F., Sun J., Chang L., Xia Z., Li Y., Xu P. Ac-LysargiNase complements trypsin for the identification of ubiquitinated sites. Anal. Chem. 2019;91:15890–15898. doi: 10.1021/acs.analchem.9b04340. [DOI] [PubMed] [Google Scholar]
- 53.Gao Y., Li Y., Zhang C., Zhao M., Deng C., Lan Q., Liu Z., Su N., Wang J., Xu F., Xu Y., Ping L., Chang L., Gao H., Wu J. Enhanced purification of ubiquitinated proteins by engineered tandem hybrid ubiquitin-binding domains (ThUBDs) Mol. Cell. Proteomics. 2016;15:1381–1396. doi: 10.1074/mcp.O115.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: Application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Della Corte A., Chitarrini G., Di Gangi I.M., Masuero D., Soini E., Mattivi F., Vrhovsek U. A rapid LC-MS/MS method for quantitative profiling of fatty acids, sterols, glycerolipids, glycerophospholipids and sphingolipids in grapes. Talanta. 2015;140:52–61. doi: 10.1016/j.talanta.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Ejsing C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyzed data sets used for this study are referenced in the text and included in the supporting information, including methods and results, nine supporting figures, and three supporting tables.