Abstract
One of the hallmarks of live cells is the asymmetric distribution of lipids across their plasma membrane. Changes in this asymmetry due to lipid “scrambling” result in phosphatidylserine exposure at the cell surface that is detected by annexin V staining. This alteration is observed during cell death processes such as apoptosis, and during physiological responses such as platelet degranulation and membrane repair. Previous studies have shown that activation of NK cells is accompanied by exposure of phosphatidylserine at the cell surface. While this response was thought to be indicative of ongoing NK cell death, it may also reflect the regulation of NK cell activation in the absence of cell death. Herein, we found that NK cell activation was accompanied by rapid phosphatidylserine exposure to an extent proportional to the degree of NK cell activation. Through enforced expression of a lipid scramblase, we provided evidence that activation-induced lipid scrambling in NK cells is reversible and does not lead to cell death. In contrast, lipid scrambling attenuates NK cell activation. This response was accompanied by reduced cell surface expression of activating receptors such as 2B4, and by loss of binding of Src family protein tyrosine kinases Fyn and Lck to the inner leaflet of the plasma membrane. Hence, lipid scrambling during NK cell activation is, at least in part, a physiological response that reduces the NK cell activation level. This effect is due to the ability of lipid scrambling to alter the distribution of membrane-associated receptors and kinases required for NK cell activation.
Keywords: NK cell activation, Phosphatidylserine exposure, Lipid scrambling, TMEM16F, Signaling
Subject terms: Signal transduction, Innate lymphoid cells
Introduction
Eukaryotic cells are enveloped by a lipid bilayer, the plasma membrane, that serves as a physical barrier between the extracellular environment and the intracellular milieu.1–3 The plasma membrane also acts as a sensor of cues in the extracellular environment, enabling the initiation of signals that are propagated to the intracellular space.1,2 One of the most notable features of the plasma membrane is the asymmetrical distribution of its lipids.3–5 The phospholipids phosphatidylserine (PS) and phosphatidylethanolamine (PE) are concentrated in the inner layer of the plasma membrane, while phosphatidylcholine and sphingolipids are mostly positioned in the outer layer.2,3
Lipid asymmetry in the plasma membrane is a hallmark of living cells.6–12 The loss of lipid asymmetry leads to exposure of PS at the cell surface, which occurs early during death processes such as apoptosis, necroptosis and pyroptosis.6,13–15 In this context, exposed PS acts as a ligand for prophagocytic receptors expressed on macrophages that trigger phagocytosis.13,14,16 The loss of plasma membrane lipid asymmetry is also seen in death-independent physiological processes such as platelet activation and pore-induced membrane repair.17,18 In these settings, it appears that the loss of asymmetry increases the plasticity of the plasma membrane, thereby promoting degranulation and repair.
As PS and PE are acidic phospholipids, their asymmetric distribution renders the inner leaflet of the plasma membrane negatively charged (i.e., acidic). This negative charge is important for several intracellular signaling molecules bearing stretches of positively charged (i.e., basic) sequences localizing to and functioning at the plasma membrane.19 These molecules include a variety of cell surface receptors, Src family protein tyrosine kinases (PTKs) and GTP-binding (G) proteins.20–22 Recent data suggested that reducing the negative charge of the inner leaflet of the plasma membrane through the loss of lipid asymmetry can compromise the targeting of at least some of these proteins to the membrane, thereby altering cellular signaling and functions.19,23
The maintenance of lipid asymmetry of the plasma membrane is achieved by three categories of enzymes: flippases, which transport phospholipids from the outer layer to the inner layer; floppases, which move phospholipids from the inner layer to the outer layer; and scramblases, which randomly redistribute phospholipids in the lipid bilayer in a process named lipid scrambling.4 Compared to the relatively slow energy-dependent lipid turnover mediated by flippases and floppases, lipid scrambling mediated by scramblases is a faster energy-independent mechanism that disrupts lipid asymmetry. The caspase-activated lipid scramblase Xkr8 is critical for lipid randomization during apoptosis,24,25 whereas the calcium-activated lipid scramblase TMEM16F (also known as ANO6) mediates lipid randomization during platelet activation and pore-induced membrane repair.17,18,26
NK cells are immune cells that play critical roles in innate immunity against cancer cells and virus-infected cells. Their activity is significantly mediated in part through direct cytotoxicity due to the release of cytotoxic granules toward target cells. The activation of NK cells is controlled by the balance between the signals delivered by a broad range of activating receptors and inhibitory receptors, which are engaged by their ligands being expressed or not expressed on target cells. Activating receptors include NKG2D, DNAM-1, and 2B4.27 Previous studies have shown that NK cell activation is also accompanied by increased PS exposure on the cell surface. It was suggested that this alteration is indicative of imminent NK cell death and is part of a mechanism aimed at terminating NK cell activation. However, considering that PS exposure can occur under physiological conditions independent of cell death, it is also plausible that lipid scrambling during NK cell activation repairs the plasma membrane after granule release. Alternatively, it may be involved in controlling the activity of the NK cell signaling apparatus to either enhance or suppress NK cell activation.
Herein, we found that PS exposure during NK cell activation occurs in a manner proportional to the level of NK cell activation. By augmenting lipid scrambling in NK cells with the lipid scramblase TMEM16F as a tool, we observed that lipid scrambling was reversible and not indicative of NK cell death. In contrast, lipid scrambling resulted in decreased signaling by the activating receptor 2B4 and diminished NK cell-mediated cytotoxicity toward target cells. These effects correlated with a decrease in the cell surface expression of 2B4 and a loss of the association of Src-related PTKs Fyn and Lck with the inner leaflet of the plasma membrane. These data argue that activation-induced lipid scrambling in NK cells is, at least in part, aimed at limiting NK cell activation because it influences the spatial distribution of the signaling apparatus that initiate NK cell activation without causing NK cell death.
Results
Lipid scrambling in NK cells correlates with the level of NK cell activation
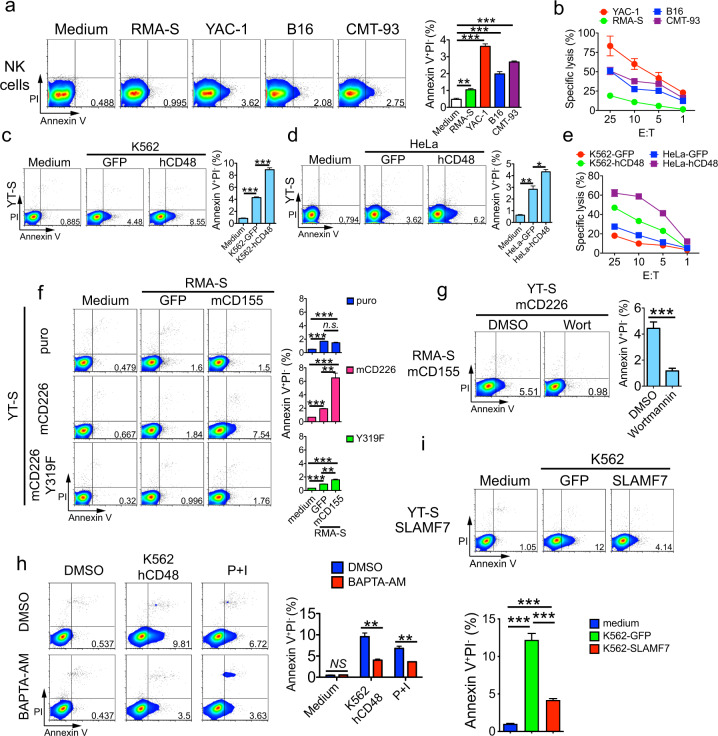
To determine which conditions influence lipid scrambling in the plasma membrane during NK cell activation, interleukin (IL)-2-activated mouse NK cells were stimulated with various tumor target cells, either hematopoietic (YAC-1 thymoma and RMA-S lymphoma cells) or nonhematopoietic (B16 melanoma and CMT-93 rectal carcinoma cells), in vitro. PS exposure in response to the targets was then measured by staining cells with annexin V, which binds PS. In parallel, cells were stained with propidium iodide (PI), a DNA-binding dye that enters cells when the integrity of the plasma membrane is compromised. Furthermore, cytotoxicity toward target cells was assessed in a 51Cr release assay. Compared to NK cells stimulated with medium alone, a small fraction (~1–3%) of NK cells activated by tumor target cells displayed PS exposure (Fig. 1a). However, they remained negative for PI. These effects were seen with all target cells. The extent of PS exposure on NK cells correlated with the extent of cytotoxicity toward the targets (Fig. 1b).
Fig. 1.
Plasma membrane lipid scrambling during NK cell activation by tumor target cells. a Annexin V and PI staining was performed on IL-2-activated mouse NK cells incubated with tumor target cells. NK cells without target cells (medium alone) were used as controls. Representative dot plots are shown on the left, and the statistics of annexin V+PI− populations for 4 independent experiments are shown on the right. PI propidium iodide. b NK cell cytotoxicity induced in YAC-1, RMA-S, B16 and CMT-93 cells was assessed by 51Cr release assay. The data are representative of three independent experiments. Annexin V and PI staining was performed on YT-S cells activated or not with K562 (c) or HeLa (d) target cells, which express green fluorescent protein (GFP) alone or in combination with human CD48 (hCD48). Representative dot plots are shown on the left, and the statistics for three independent experiments are shown on the right. e YT-S cell cytotoxicity induced in K562 and HeLa cells expressing GFP or human CD48. The data are representative of two independent experiments. f Annexin V and PI staining was performed on YT-S cells expressing an empty vector alone (puromycin resistance marker; puro) or with wild-type mouse CD226 (mCD226) or the mCD226 tyrosine 319-to-phenylalanine 319 (Y319F) mutant, incubated or not with RMA-S expressing GFP alone or with mCD155. Representative dot plots are shown on the left, and the statistics for three independent experiments are shown on the right. g Annexin V and PI staining was performed on YT-S cells expressing mCD226 incubated with RMA-S cells expressing mCD155 with or without wortmannin [wort, 1 μM; dissolved in dimethyl sulfoxide (DMSO)]. DMSO alone was used as the control. Representative dot plots are shown on the left, and the statistics for four independent experiments are depicted on the right. h Annexin V and PI staining was performed on YT-S cells stimulated or not with K562 cells expressing hCD48, with or without 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester) (BAPTA-AM; 100 μM in DMSO). Phorbol myristate acetate (PMA; 100 ng/ml) plus ionomycin (1 μM) (P + I) was used as a control. Representative dot plots are shown on the left, and the statistics for three independent experiments are shown on the right. i Annexin V and PI staining was performed on YT-S cells expressing empty vector (puro) or mSLAMF7 and incubated or not with K562 cells expressing GFP alone or with mSLAMF7. Representative dot plots are shown at the top, and statistics for three independent experiments are shown at the bottom. NS not significant; *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t tests). The data are presented as the means ± s.e.m
The latter observation suggested that the extent of PS exposure during NK cell activation might correlate positively with the level of NK cell activation. To address this possibility more rigorously, we examined PS exposure in the human NK cell line YT-S, which was activated by target cells expressing ligands for specific activating NK cell receptors. These various YT-S derivatives, as well as the targets selected, were previously reported.28–30 In the presence of target cells expressing CD48, the ligand for the activating receptor 2B4 expressed on parental YT-S cells, YT-S cells displayed greater PS exposure, when compared to YT-S cells incubated with target cells expressing the green fluorescent protein (GFP) selection marker alone (Fig. 1c, d). This outcome was observed with both K562 erythroleukemia target cells and HeLa cervical carcinoma target cells. As expected in this system, the cytotoxicity induced by the K562 and HeLa cells bearing CD48 was greater than that induced by target cells expressing an empty GFP vector (Fig. 1e).
We also ascertained PS exposure on YT-S cells expressing or not expressing the activating receptor CD226 (also known as DNAM-1). The YT-S derivatives expressed the murine (m) version of CD226, as described elsewhere.29 In the presence of RMA-S target cells expressing mouse CD155, the ligand of CD226, mCD226-expressing YT-S cells exhibited a marked increase in PS exposure, compared to YT-S cells incubated with RMA-S target cells that lacked mCD155 (Fig. 1f). This effect was not apparent in YT-S cells lacking mCD226. It was also not evident in derivatives of YT-S cells expressing a mutant form of mCD226 (tyrosine-to-phenylalanine at position 319; Y319F) that is unable to trigger intracellular signals that enhance NK cell activation.29 A similar abrogation of the impact of CD226 was observed when mCD226 YT-S cells were pretreated with wortmannin, a pharmacological inhibitor of phosphatidylinositol 3′ kinase (PI3′K), a key intracellular effector of CD226 (Fig. 1g). The impact of the calcium chelator BAPTA-AM on PS exposure during NK cell activation was also analyzed. The addition of BAPTA-AM, but not medium alone, resulted in a decrease in PS exposure on YT-S cells in response to target cells expressing CD48 during a cytotoxicity assay (Fig. 1h).
Finally, we tested the impact of the engagement of the homotypic receptor SLAMF7. As YT-S cells do not express the adapter EAT-2, SLAMF7 acts as an inhibitory receptor in these cells upon engagement by its ligand, SLAMF7, which is expressed on target cells.30 When incubated with K562 target cells expressing SLAMF7, YT-S cells expressing SLAMF7 displayed reduced PS exposure compared to YT-S cells incubated with the target cells expressing GFP alone (Fig. 1i).
Hence, during NK cell activation, loss of lipid asymmetry in the plasma membrane, as reflected by PS exposure, positively correlated with the level of NK cell activation. It was not associated with increased permeability to PI, implying that it was not involved in NK cell death triggered by NK cell activation.
Lipid scramblase TMEM16F enhances PS exposure in a reversible manner during NK cell activation
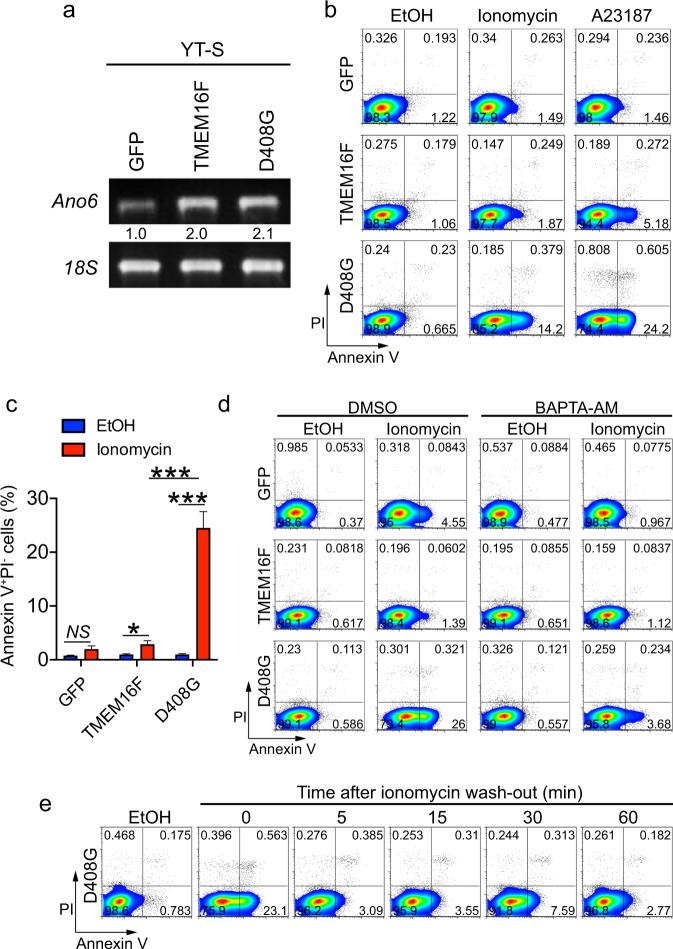
To determine how lipid scrambling influences NK cell activation, the effects of enforced expression of the calcium-activated lipid scramblase TMEM16F were studied in YT-S NK cells. We postulated that an enhancement of the activity of TMEM16F might augment lipid scrambling during NK cell activation, thereby showing the impact of this modification on NK cell functions. Two versions of TMEM16F were used: wild-type TMEM16F and the calcium-hypersensitive aspartic acid 408-to-glycine 408 (D408G) TMEM16F mutant.26 Human proteins were used. Overexpression of human TMEM16F in these cells, compared to in control cells expressing GFP alone, was confirmed at the RNA level by real-time PCR (Fig. 2a).
Fig. 2.
Lipid scramblase TMEM16F causes transient and reversible PS exposure on NK cells. a Expression of human TMEM16F-encoding RNA (Ano6) in YT-S cells expressing GFP alone or in combination with wild-type TMEM16F or the aspartate 408-to-glycine 408 (D408G) TMEM16F mutant was measured by RT-PCR. 18S RNA was used as control. b, c Annexin V and PI staining was performed on YT-S cells expressing GFP alone or in combination with wild-type TMEM16F or D408G TMEM16F that were treated or not for 5 min with the calcium ionophore ionomycin [10 μM; in ethanol (EtOH)] or A23187 (10 μM in EtOH). Representative dot plots are shown in (b), while the statistics for seven independent experiments are shown in (c). d Same as (b), except that the cells were incubated or not with BAPTA-AM (100 μM). The data are representative of two independent experiments. e Same as (b), except that ionomycin was washed out, and the cells were incubated for the indicated periods at 37 °C in the culture medium. Representative of two independent experiments. NS not significant; *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t tests). The data are presented as the means ± s.e.m
Compared to control cells expressing GFP alone, YT-S cells overexpressing wild-type TMEM16F exhibited only a small increase in PS exposure in response to the calcium ionophores ionomycin or A23187 (Fig. 2b, c). In contrast, cells expressing D408G TMEM16F displayed markedly increased PS exposure upon treatment with calcium ionophores. No effect was seen in the absence of the ionophores. There was little or no concomitant increase in PI staining, implying that membrane integrity was preserved. The augmentation of PS exposure in D408G TMEM16F-expressing cells was blocked by the calcium chelator BAPTA-AM, supporting the idea that these increases in PS exposure were calcium-dependent (Fig. 2d). Moreover, it was rapidly (within 5 min) reversible after washing away ionomycin (Fig. 2e).
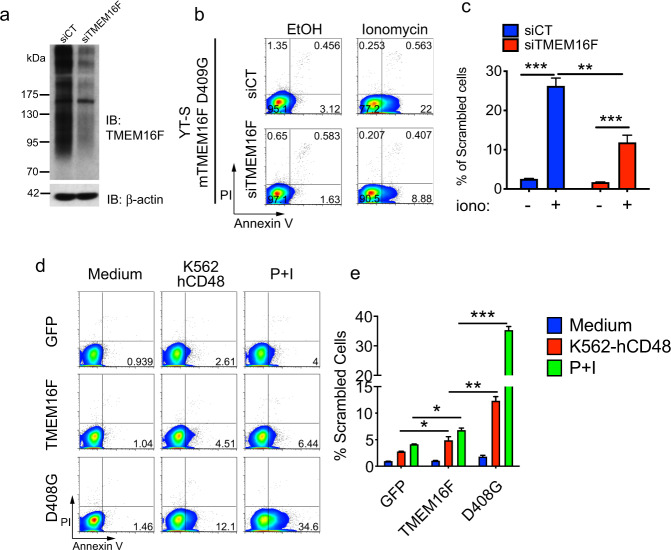
To ensure that these effects were not indirect consequences of sustained overexpression of TMEM16F, we used YT-S cells stably expressing the mouse variant (D409G) of the activated TMEM16F mutant that were then transduced with small interfering (si) RNAs targeting mouse TMEM16F. The choice of mouse TMEM16F was made for this experiment because the only antibodies available against TMEM16F recognized the mouse protein, not the human protein, which had instead been used for the studies described in Figs. 1 and 2. An anti-TMEM16F immunoblot demonstrated that the expression of the mouse TMEM16F protein was downregulated by the specific siRNA but not by a nontargeting siRNA (Fig. 3a). Notably, TMEM16F migrated as multiple ill-defined bands in the gel, in keeping with previous reports.18,26 This is presumably because multiple glycoforms of TMEM16F exist. Concurrent with the reduction of TMEM16F expression by the siRNA, PS exposure induced by ionomycin was reduced (Fig. 3b, c).
Fig. 3.
Enhanced PS exposure during NK cell activation is TMEM16F-dependent. a Anti-TMEM16F immunoblot (IB) of total cell lysates from YT-S cells expressing mouse D409G TMEM16F transfected either with control nontargeting siRNAs (siCT) or with siRNAs against mouse TMEM16F (siTMEM16F). β-actin was used as the loading control. b, c Annexin V and PI staining was performed on the cells described in (a) treated or not with ionomycin. Representative dot plots are shown in (b), while the statistics for four independent experiments are shown in (c). d, e Annexin V and PI staining was conducted with YT-S cells expressing GFP alone or in combination with wild-type TMEM16F or the aspartate 408-to-glycine 408 (D408G) TMEM16F mutant that were incubated or not with K562 cells expressing hCD48 or the combination of PMA (100 ng/ml) plus ionomycin (1 μM) (P + I). Representative dot plots are shown in (d), while statistics for three independent experiments are shown in (e). *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t tests). The data are presented as the means ± s.e.m
Next, we tested whether TMEM16F also influenced PS exposure when NK cells were activated by tumor target cells. Compared to the expression of GFP alone, overexpression of wild-type TMEM16F caused a small increase in the exposure of PS on YT-S cells in response to CD48-expressing K562 target cells, in contrast to the effect of medium alone (Fig. 3d, e). A much more robust increase was observed when YT-S cells expressed D408G TMEM16F than when they expressed wild-type TMEM16F. Similar effects were observed when cells were stimulated with a combination of phorbol myristate acetate (PMA) and ionomycin, two pharmacological agonists of NK cell activation.
Therefore, the expression of an activated version of the calcium-activated lipid scramblase TMEM16F enhanced lipid scrambling, causing PS exposure in response to calcium ionophores, as well as during NK cell activation by target cells. This effect was rapidly reversible and not associated with appreciable NK cell death.
Lipid scrambling reduces NK cell cytotoxicity and 2B4 signaling
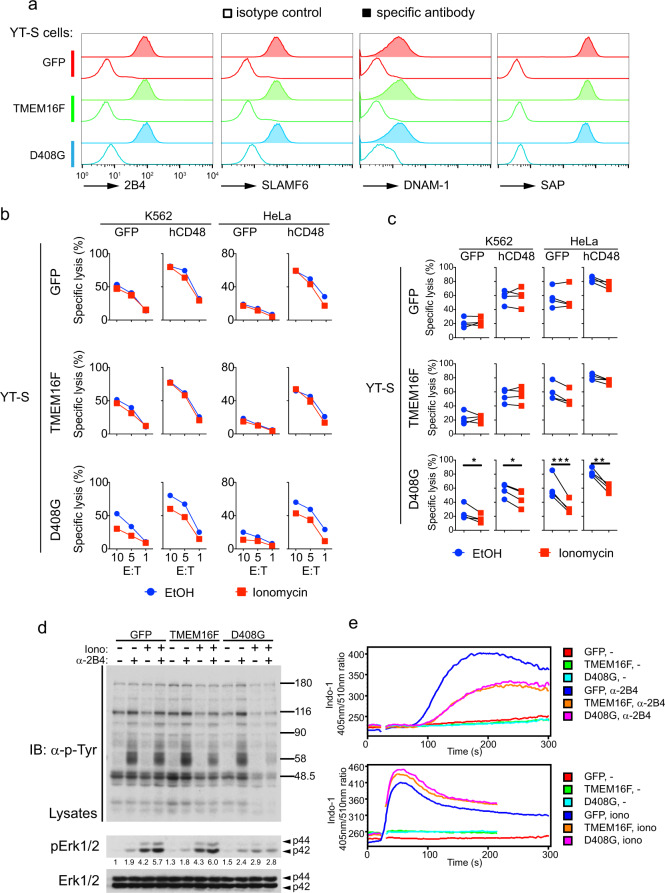
Next, we examined whether the increased lipid scrambling induced by TMEM16F was associated with altered NK cell activation. To this end, the YT-S derivatives were incubated with K562 or HeLa cells expressing or not expressing CD48, in the presence or absence of ionomycin to activate scrambling by TMEM16F. Target cell killing was measured using a chromium release assay. All YT-S derivatives expressed equivalent amounts of various markers, including 2B4 and SAP, the intracellular adapter that couples 2B4 to Fyn kinase (Fig. 4a). In general, the cytotoxicity displayed by these YT-S derivatives toward targets expressing CD48 was greater than that toward targets lacking CD48, in keeping with the activating function of 2B4 in this system (Fig. 4b, c).
Fig. 4.
TMEM16F-triggered lipid scrambling reduces NK cell cytotoxicity and 2B4 signaling. a Expression of membrane receptors 2B4, SLAMF6, and DNAM-1, as well as adapter SAP, was determined by flow cytometry of YT-S cells expressing empty vector (GFP), wild-type TMEM16F, or the D408G TMEM16F mutant. For detection of SAP, the cells were permeabilized. Open histogram, isotype control antibody; filled histogram, specific antibody. Representative histograms of two independent experiments. b, c YT-S cells expressing GFP alone or in combination with wild-type TMEM16F or D408G TMEM16F were incubated for 6 h with K562 or HeLa cells expressing hCD48 in the presence or in the absence of ionomycin (1 μM) to activate TMEM16F. Cytotoxicity was measured by 51Cr release at the indicated effector:target (E:T) ratios. The extent of the cell lysis was indicated as the percentage of maximum 51Cr release. The data are presented as mean ± s.d. of duplicate samples. Representative cytotoxicity assays are shown in (b), while statistics for four independent experiments are depicted in (c). d The indicated YT-S derivatives were preincubated for 5 min at room temperature in the presence or in the absence of ionomycin (10 μM). They were then stimulated or not for 5 min at 37 °C with anti-2B4 antibodies C1.7 and the relevant secondary anti-mouse antibody. Total cell lysates were probed by immunoblotting with anti-phosphotyrosine (p-Tyr; top), anti-phospho-Erk1/2 (pErk1/2; middle) or anti-Erk1/2 (bottom) antibodies. The levels of phospho-Erk1/2 were measured and normalized according to the total Erk1/2 level. Representative blots of three independent experiments. e Calcium influx in the indicated YT-S cells stimulated or not with anti-2B4 antibodies. Calcium influx was determined by the indicated fluorescence ratio for Indo-1. Ionomycin served as the positive control. Representative histograms of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t tests). The data are presented as the means ± s.e.m
Compared to YT-S cells expressing GFP alone, YT-S cells overexpressing wild-type TMEM16F did not display altered cytotoxicity toward the targets (Fig. 4b, c). This was true whether or not cells were incubated with ionomycin, which enhances TMEM16F-dependent scrambling (Figs. 4b, c and 2a). In striking contrast, however, YT-S cells expressing D408G TMEM16F exhibited markedly reduced cytotoxicity toward the two targets in the presence of ionomycin compared to vehicle alone (Fig. 4b, c). This was seen whether the targets expressed CD48 or not.
While the activating receptor(s) that mediate the killing of K562 or HeLa cells lacking CD48 by YT-S cells is (are) not known, the increase in cytotoxicity observed in targets expressing CD48 is mediated by 2B4. To ascertain the molecular mechanism by which augmented lipid scrambling leads to decreased NK cell-mediated cytotoxicity, the impact of TMEM16F on 2B4-triggered intracellular signals was studied. The most proximal outcome of the signal triggered by 2B4 is protein tyrosine phosphorylation, which is initiated via SAP by the Src-related PTK Fyn. Thus, YT-S derivatives were stimulated with agonistic anti-2B4 antibodies in the presence or in the absence of ionomycin to induce scrambling, and overall protein tyrosine phosphorylation was monitored. Consistent with the results of PS exposure (Fig. 2), YT-S cells overexpressing wild-type TMEM16F showed slightly reduced protein tyrosine phosphorylation in response to anti-2B4 antibodies plus ionomycin compared to control YT-S cells (Fig. 4d). A much more pronounced decrease was observed in YT-S cells bearing D408G TMEM16F. All 2B4-regulated substrates were affected by this diminution.
We also examined other downstream 2B4-induced signals, particularly calcium flux and Erk activation. Using a flow cytometry-based assay, we observed that YT-S cells overexpressing wild-type TMEM16F or D408G TMEM16F exhibited reduced anti-2B4-induced calcium flux in comparison to the flux in the control YT-S cells (Fig. 4e). No difference was seen when calcium flux was triggered by a high concentration of ionomycin, which induces calcium influx independent of 2B4 or protein tyrosine phosphorylation. Similar effects were observed with Erk activation, which was assayed by immunoblotting with a specific antibody recognizing the activated phosphorylated form of Erk (Fig. 4d).
Thus, augmented lipid scrambling leading to increased PS exposure was associated with reduced NK cell-mediated cytotoxicity and with compromised 2B4-triggered protein tyrosine phosphorylation, calcium flux and Erk activation.
Loss of surface 2B4 and membrane dissociation of Src kinases during lipid scrambling
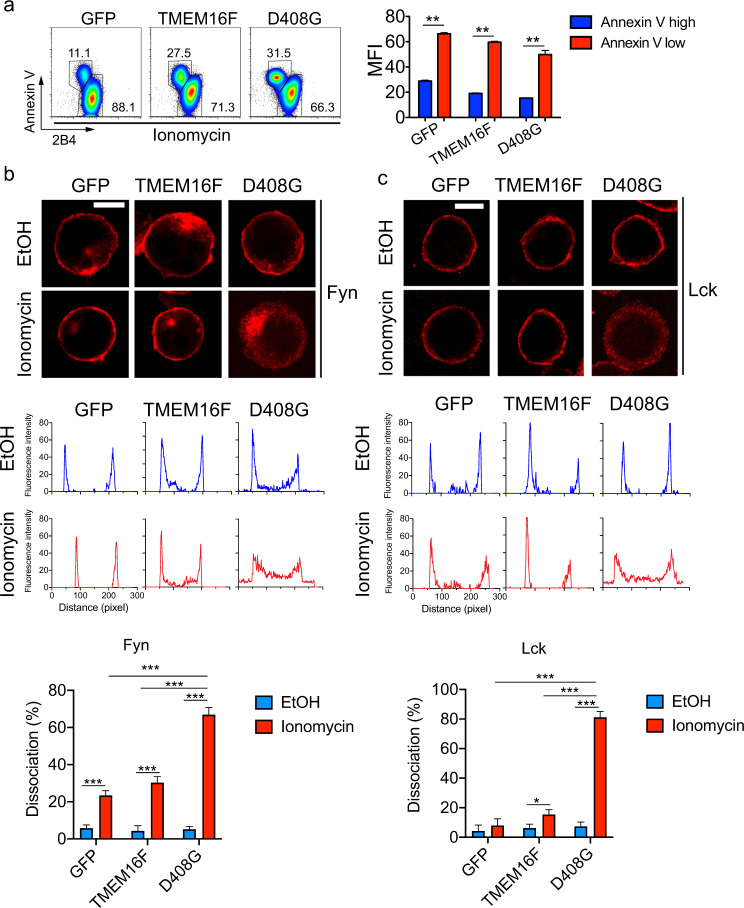
To assess how lipid scrambling attenuated 2B4-mediated signals and cytotoxicity at the molecular level, the effects of lipid scrambling on 2B4 expression at the cell surface and on the membrane association of Fyn were examined. Using flow cytometry to detect cell surface 2B4, we found that the subset of YT-S cells displaying increased annexin V staining in response to ionomycin also showed reduced levels of 2B4, by ~50–70% compared to cells not showing augmented annexin V staining (Fig. 5a). The proportion of annexin V+2B4lo cells was higher in YT-S derivatives expressing wild-type TMEM16F or D408G TMEM16F than in control cells. No difference in 2B4 expression between the various YT-S derivatives was observed in the absence of ionomycin (Fig. 4a).
Fig. 5.
Loss of surface 2B4 and membrane dissociation of Src kinases during lipid scrambling. a The intensity of the 2B4 and annexin V staining was analyzed on YT-S derivatives treated with ionomycin (10 μM). Representative dot plots are on the left, while the statistics of the mean fluorescent intensity (MFI) of 2B4 in the annexin Vhigh versus the annexin Vlow populations from three independent experiments are depicted on the right. Cellular localization of Fyn (b) and Lck (c) in YT-S derivatives treated or not with ionomycin (10 μM) for 5 min at RT was determined by staining with the relevant antibodies and confocal microscopy. Representative images are shown in the top two panels, while profiles of the fluorescence as measured by ZEN software are shown in the middle two panels, and the statistics for multiple cells are shown in the bottom two panels. The data are from seven pictures for each condition, with 20–50 cells in each picture. The data are representative of two experiments. Scale bar is 10 μm. *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t tests). The data are presented as the means ± s.e.m. The data are related to Fig. S1
Next, we studied the intracellular localization of Fyn using confocal microscopy. Lck, another Src family kinase expressed in NK cells, was analyzed in parallel. Staining with anti-Fyn antibodies showed that in control cells expressing GFP alone, Fyn was primarily located at the plasma membrane (Fig. 5b; Fig. S1). This was true whether cells were treated or not with ionomycin. Some staining was also seen in intracellular organelles that presumably represent endosomes, as described for the localization of Fyn in other cell types.31 Similar findings were obtained with cells overexpressing wild-type TMEM16F. In contrast, cells expressing D408G TMEM16F displayed localization of Fyn that was primarily (~60%) cytosolic in the presence of ionomycin, differing from that of cells treated with vehicle alone. Analogous effects were seen for Lck, although, in contrast to Fyn, Lck did not show any endosomal distribution (Fig. 5c).
Therefore, augmented lipid scrambling was accompanied by reduced staining of 2B4 on the cell surface and by loss of the association of Src kinases Fyn and Lck with the inner leaflet of the plasma membrane.
Specific plasma membrane remodeling induced by lipid scrambling
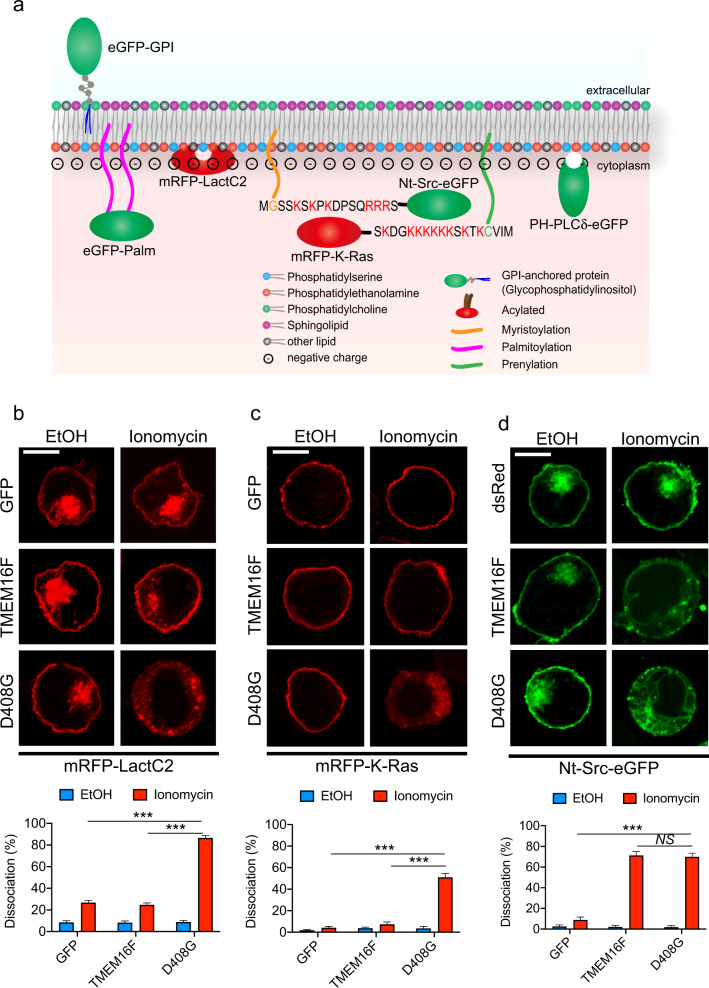
As mentioned above, the acidic phospholipids PS and PE are normally concentrated at the inner leaflet of the plasma membrane, where they are believed to provide electrostatic anchors for molecules such as Src family kinases and G-proteins, which possess complementary stretches rich in basic residues.19–23 To ascertain the extent and specificity of the changes in the plasma membrane caused by lipid scrambling, the distribution of previously described genetically encoded membrane-bound fluorescent probes was assessed.19,23
YT-S cells were stably transfected with cDNAs encoding a reporter protein [enhanced (e) GFP or monomeric red fluorescent protein (mRFP)], fused to the following sequences (Fig. 6a): (1) the C2 domain of lactadherin that directly binds inner membrane PS in a manner analogous to annexin V (mRFP-LactC2); (2) the carboxy (C)-terminal region of K-Ras, containing nine basic residues that bind to inner membrane acidic phospholipids via electrostatic charges and containing a site of farnesylation (mRFP-K-Ras); (3) the amino (N)-terminal region of Src, bearing six basic residues that also interact with internal membrane acidic phospholipids, in addition to a myristoylation signal (Nt-Src-eGFP); (4) palmitoylation and acylation sequences from H-Ras that bind the inner plasma membrane via hydrophobic interactions (eGFP-Palm); (5) a glycosylphosphatidylinositol (GPI) anchor that associates with the outer plasma membrane through hydrophobic interactions (eGFP-GPI); and (6) the pleckstrin homology (PH) domain of phospholipase C (PLC) δ that binds the inner plasma membrane via phosphatidylinositol-4,5-biphosphate (PIP2) (PH-PLCδ-eGFP).
Fig. 6.
The loss of inner membrane acidic charges induced by lipid scrambling. a A depiction of the fluorescent probes used in Figs. 6 and 7 is shown. Cellular localization of mRFP-LactC2 (b), mRFP-K-Ras (c) and Nt-Src-eGFP (d) in YT-S derivatives treated or not with ionomycin (10 μM) for 5 min at RT was determined by confocal microscopy. Representative images are shown at the top, while the statistics for multiple cells are shown at the bottom. The data are from 7 (b, c) or 10 (d) pictures for each condition, with 20–50 cells in each picture. The data are representative of two experiments. The scale bar is 10 μm in all the images. NS, not significant; ***p < 0.001 (two-tailed Student’s t tests). Data are means ± s.e.m
The resulting YT-S derivatives were treated or not with ionomycin to trigger lipid scrambling, and the association of the probes with the plasma membrane was assessed. Similar to the effect on Fyn and Lck (Fig. 5b–e), ionomycin disrupted the association of mRFP-LactC2 and mRFP-K-Ras with the plasma membrane (Fig. 6b, c). These effects were observed in YT-S cells expressing D408G TMEM16F but not in cells overexpressing wild-type TMEM16F or GFP alone. Ionomycin also triggered the loss of membrane binding of Nt-Src-eGFP, and this effect was seen in cells expressing D408G or overexpressing wild-type TMEM16F but not in cells containing GFP alone (Fig. 6d).
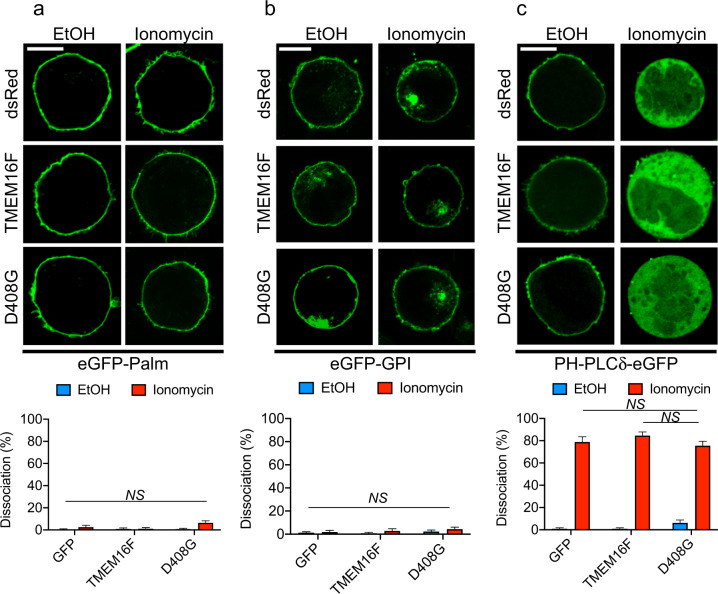
In contrast, ionomycin had no impact on the membrane association of eGFP-Palm and eGFP-GPI, which bind the inner and outer leaflets of the plasma membrane via hydrophobic interactions, respectively (Fig. 7a, b). Nonetheless, ionomycin provoked marked dissociation of PH-PLCδ-eGFP from the plasma membrane (Fig. 7c). This effect was seen in all cells, including cells expressing GFP alone, implying that it was not related to lipid scrambling. Ionomycin is known to activate calcium-dependent PLCs, which deplete PIP2, the ligand for this probe, from the plasma membrane.
Fig. 7.
Specificity of plasma membrane remodeling induced by lipid scrambling. Same as Fig. 6, except that a different set of probes was introduced to YT-S cells expressing Discosoma (ds) Red alone, or in combination with wild-type or D408G TMEM16F, were used. a eGFP-Palm; b eGFP-GPI; c PH-PLCδ-eGFP. Data are from ten pictures for each condition, with 20–50 cells per picture. The data are representative of two experiments. The scale bar is 10 μm in all the images. NS not significant, two-tailed Student’s t tests. The data are presented as the means ± s.e.m
Thus, lipid scrambling was associated with the loss of PS at the inner leaflet of the plasma membrane, as revealed by the mRFP-LactC2 probe. It was also accompanied by overall loss of acidic charges in the inner leaflet of the plasma membrane, as shown by the dissociation of the mRFP-K-Ras and Nt-Src-eGFP probes. However, there was no effect on the hydrophobicity of the plasma membrane, as indicated by the eGFP-GPI and eGFP-Palm probes.
Discussion
The loss of plasma membrane asymmetry, leading to PS exposure at the cell surface, has been observed in two types of situations. First, it is typical of cell death processes such as apoptosis, pyroptosis, and necroptosis.6,15,16 In this context, it is a prelude to cell death, and it triggers phagocytosis by macrophages.13,14 Second, the loss of plasma membrane asymmetry is seen in physiological situations, independent of cell death. These settings include platelet activation,4,17 during which loss of plasma membrane asymmetry is needed for platelet degranulation and hemostasis, and in pore-induced membrane repair,18 which maintains plasma membrane integrity after damage is caused by pore-forming agents such as bacterial toxins.
Previous studies showed that NK cells displayed an enhancement of PS exposure on their surface during activation. For instance, activated NK cells from mice infected with mouse cytomegalovirus exhibited increased annexin V staining and higher levels of activation markers CD69 and CD25.32 Augmented staining was also observed for activated caspases. In another report, the extent of NKG2D-mediated activation of mouse NK cells was shown to correlate positively with PS exposure at the NK cell surface.33 This correlation was accompanied by increased PI staining, which is indicative of increased membrane permeability. In light of these and related findings, it has been assumed that activation-induced PS exposure on NK cells is a reflection of ongoing NK cell death, which terminates NK cell activation. However, as PS exposure, in addition to caspase activation and PI staining, can be detected in immune cells that are not dying4,17,18, it is unclear whether the loss of plasma membrane asymmetry in NK cells is always a marker of ongoing cell death;34,35 our data indicate that this loss of asymmetry is not necessarily a harbinger of cell death.
Herein, we confirmed previous findings showing that activation of NK cells is accompanied by increased PS exposure at the cell surface. This was shown with normal mouse NK cells or with the human NK cell line YT-S. Using a series of YT-S derivatives and genetically modified target cells, we established that the extent of PS exposure during NK cell activation was proportional to the level of NK cell activation. Furthermore, PS exposure was not accompanied by any appreciable increase in PI staining, one of the early markers of cell death. Thus, in our studies, lipid scrambling during NK cell activation did not lead to appreciable NK cell death.
To probe more directly the impact of lipid scrambling on NK cell activation, we took advantage of the lipid scramblase TMEM16F, which can be activated in response to an increase in intracellular calcium.17,18,26 The D408G mutation renders TMEM16F hypersensitive to calcium. By overexpressing wild-type TMEM16F or, to a greater extent, expressing D408G TMEM16F, we found that an increase in lipid scramblase activity in YT-S cells resulted in augmented PS exposure. This effect was seen in response to the calcium ionophores A23187 and ionomycin, tumor target cells and PMA plus ionomycin. Importantly, as was the case in normal NK cells, the response was independent of augmented PI staining. Moreover, it was rapidly reversed when ionomycin was removed. Thus, TMEM16F augmented the loss of plasma membrane asymmetry during NK cell activation with a dearth of detectable cell death.
Further analyses showed that the increase in lipid scrambling evoked by TMEM16F was accompanied by reduced NK cell-mediated cytotoxicity toward tumor cells. This reduction was observed whether or not target cells expressed CD48, suggesting that it involved one or more activating NK cell receptors, including 2B4. Using 2B4 as a model activating receptor, we discovered that enhanced lipid scrambling resulted in severely compromised 2B4-mediated signaling. This was true for overall protein tyrosine phosphorylation, calcium flux and Erk activation, which are known to be initiated by Fyn in this setting and to be essential for 2B4-dependent cytotoxicity. Confocal microscopy studies showed that lipid scrambling was associated with the loss of Fyn binding to the inner leaflet of the plasma membrane. A similar effect was seen for Lck. There was also a loss of 2B4 expression at the cell surface. Thus, altogether, these data implied that augmented lipid scrambling compromised NK cell activation by altering the signaling machinery involved in the initiation of NK cell activation.
Studies using a series of fluorescent probes indicated that the impact of augmented lipid scrambling on the interactions of cellular proteins with the plasma membrane was specific. Probes containing polybasic stretches, such mRFP-K-Ras and Nt-Src-eGFP, displayed the loss of membrane binding in response to augmented scrambling, supporting the idea that they interact with inner membrane acidic phospholipids such as PS and PE. However, other probes, such as eGFP-Palm and eGFP-GPI, which bind the plasma membrane through hydrostatic interactions, were not affected. Consequently, lipid scrambling caused a loss of membrane binding specific for molecules with polybasic stretches but not for proteins binding the membrane through alternative mechanisms.
Intriguingly, the effects of the overexpression of wild-type TMEM16F and the expression of the activated version of TMEM16F (D408G) were not identical. In general, the impact of wild-type TMEM16F was much milder than that of D408G TMEM16F. This outcome was evident with ionomycin-induced PS exposure, target cell-induced PS exposure, 2B4-triggered protein tyrosine phosphorylation, membrane dissociation of Fyn and Lck, loss of membrane binding of the LactC2 or K-Ras probes, and reduced target cell cytotoxicity. However, other effects of wild-type TMEM16F and D408G TMEM16F were nearly identical, as was the case for 2B4-induced calcium flux and the loss of membrane binding of the Nt-Src probe. Given that the Nt-Src probe possesses only six basic residues, compared to nine for the K-Ras probe, it is possible that these differential effects are related to the extent of lipid scrambling triggered by wild-type versus D408G TMEM16F. Perhaps the interactions of the Nt-Src probe or of the machinery involved in 2B4-evoked calcium flux with the plasma membrane were less stable or located in different membrane domains such that they might be more easily reversed by enhanced lipid scrambling.
The identity of the lipid scramblase(s) responsible for activation-induced lipid scrambling in NK cells remains to be clarified. We did not detect reduced PS exposure in activated TMEM16F-deficient NK cells, implying that TMEM16F is not implicated or essential for this response (Fig. S2a, b). Notably, however, compared to thymocytes, NK cells express lower levels of TMEM16F (www.immgen.org). Another candidate scramblase is TMEM16K (ANO10), which is also expressed in NK cells (www.immgen.org). However, recent reports have suggested that this scramblase is primarily localized to the endoplasmic reticulum, not the plasma membrane.36 Thus, one or more currently unidentified scramblases are presumably implicated. Given that PS exposure during NK cell activation in the absence of TMEM16F enforced expression was reduced by the calcium chelator BAPTA-AM, it is plausible that these scramblase(s) are also activated by calcium. Future studies will be needed to resolve this issue.
In addition to NK cells, PS exposure has been reported during the activation of other immune cell types. For example, it was reported that a subset of activated CD4+ T cells express high levels of PS at their surface.9 It was proposed that PS exposure in this setting influenced T-cell migration. In another study, it was suggested that TMEM16F is a negative regulator of T-cell activation and prevents T-cell exhaustion.37 Although there was no documentation of TMEM16F-dependent PS exposure in this setting, an effect on multivesicular body formation in endosomes was shown. In addition, engagement of the antigen receptor on B cells was reported to lead to PS exposure.8 Once again, the impact of this modification on B-cell functions was not determined. Considering our findings, it is plausible that activation-induced lipid scrambling occurs in all immune cells and that it is broadly aimed at terminating immune cell activation.
Interestingly, a recent report suggested that in activated CD8+ T cells, localized PS exposure at the immunological synapse prevents self-killing of activated T cells.34 It was proposed that this protective effect was related in part to the quenching of perforin by localized PS. Considering our data with NK cells, it is possible that this impact was also due to a negative feedback effect of lipid scrambling to prevent excessive T-cell activation. Furthermore, it may relate to the ability of lipid scrambling to facilitate T-cell membrane repair following self-damage by perforin, as we reported for other pore-forming agents.18 Hence, the protective influence of lipid scrambling in activated T cells may be multifactorial. The same may be true for NK cells.
In summary, our results show that NK cell activation is accompanied by a loss of lipid asymmetry in the plasma membrane, seemingly attenuating NK cell activation responses. These data also imply that the entire machinery maintaining lipid asymmetry, that is, scramblases, flippases, and floppases, is collectively critical for securing normal cell signaling and functions in NK cells. Considering this homeostatic role, it is attractive to speculate that alterations in the activity of these scramblases, flippases or floppases, leading to loss of lipid asymmetry, can affect the genesis of conditions where NK cell responses are modified or compromised, including NK cell education, anergy, tolerance and exhaustion. Similar activities may be evident in other immune cell types, including T cells and B cells.
Materials and methods
Mice and cells
C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). IL-2-activated NK cells (lymphokine-activated killer cells; LAK cells) were produced as described elsewhere.38 TMEM16F-KO mice were previously described.18 Briefly, splenic NK cells were first enriched with a negative selection kit (StemCell Technologies) and then cultured for 5 days in complete RPMI 1640 medium supplemented with mouse IL-2 (1000 U/ml) (PeproTech). YT-S, YAC-1, B16, RMA-S, CMT-93, HeLa, and K562 cells were described previously.38 YT-S derivatives expressing mouse CD226 (wild-type or Y319F) or mouse SLAMF7 or neither and HeLa and K562 cells expressing human CD48 or mouse SLAMF7 or neither, and RMA-S cells expressing mouse CD155 or not, were described elsewhere.28–30 All cell lines were cultured under the conditions recommended by the providers. Animal experimentation was conducted according to the guidelines of the Canadian Council for Animal Care and approved by the Animal Care Committee of the Institut de recherches cliniques de Montréal. It was also approved by the Animal Ethics Committee of Huazhong University of Science and Technology.
Plasmids and siRNAs
cDNAs encoding mouse or human TMEM16F and their mutants were cloned into the pFB-GFP retroviral vector. Plasmids encoding mRFP-LactC2, mRFP-K-Ras, Nt-Src-eGFP, eGFP-GPI, eGFP-Palm, and PH-PLCδ-eGFP were kindly provided by Dr. Sergio Grinstein (Toronto, Ontario, Canada).19,23 Cells were transduced with the indicated cDNAs using retrovirus-mediated gene transfer. siRNAs targeting mouse TMEM16F-encoding RNAs (GS105722) and Allstars negative control siRNAs (SI03650318) were purchased from Qiagen. siRNAs were electroporated into cells using an Amaxa Nucleofector kit V (Lonza). The human Ano6 and 18S real-time PCR primers used were as follows: Ano6: forward, TGTCCCCGATTTGGGATCACT; reverse, CGTATGCTTGTCTTTTCCTCCT. 18S: forward, CAGCCACCCGAGATTGAGCA; reverse, TAGTAGCGACGGGCGGTGTG.
Annexin V, antibodies, and other reagents
PS externalization at the cell surface was detected by staining with annexin V (BioLegend) in the presence of PI to identify cells with altered membrane permeability (Sigma). Antibodies against human 2B4 (C1.7) were obtained from BioLegend. Western blot antibodies against β-actin (C4) were obtained from Santa Cruz. Antibodies against Fyn, Lck, SAP, SLAMF6, DNAM-1, and mouse TMEM16F were generated in our laboratory and described elsewhere.18,39,40 Anti-phosphotyrosine antibody (4G10) was from Millipore. Antibodies against phosphorylated Erk (E10) and p44/42 MAPK (9102) were purchased from Cell Signaling Technology. Ionomycin, A23197, DMSO, wortmannin, BAPTA-AM, and PMA were obtained from Sigma. Under the conditions used, ionomycin had no appreciable toxic effects on NK cells or target cells (data not shown).
Flow cytometry, calcium flux, and immunoblotting
Flow cytometry, a calcium flux assay and immunoblotting were performed as described elsewhere.18,38 Immunoblots were quantified by ImageJ and normalized for total Erk1/2.
PS exposure
To study PS exposure during NK cell activation, NK cells were incubated for 4 h at 37 °C in complete medium containing target cells at an effector:target ratio of 1:1 or containing no target cells. PS exposure triggered by calcium ionophores was studied as described elsewhere.26 Briefly, after washing cells at room temperature (RT) in phosphate-buffered saline, cells were treated or not for 5 min with ionomycin (10 μM) or A23187 (10 μM) in calcium-free HBSS buffer. After the indicated treatments, the cells were pelleted and stained with annexin V and PI and then subjected to flow cytometry (CyAn, Dako).
NK cell cytotoxicity and degranulation
The 51Cr release and CD107a exposure assays were performed as described previously.38 Briefly, for 51Cr release, NK cells were incubated for 6 h at 37 °C with target cells preloaded with 51Cr at the indicated effector:target ratios. Supernatants were then assayed for 51Cr release. For CD107a exposure, mice were injected with poly(I:C) (200 μg per mouse) intraperitoneally, and splenocytes were isolated 48 h later. Then, the splenocytes were cocultured for 6 h with target cells in the presence of an anti-CD107a antibody. NK cells were gated as TCRβ−NK1.1+.
Antibody-mediated stimulation of 2B4
Protein tyrosine phosphorylation, Erk activation, and calcium influx induced by antibody-mediated stimulation of 2B4 were measured as outlined in a previous report.38
Confocal microscopy
To assess the localization of the fluorescent probes, cells were treated or not with ionomycin as described above for flow cytometry. After treatment, the cells were fixed in a solution containing freshly prepared 2% (vol:vol) paraformaldehyde and directly mounted onto cover slides for staining with rabbit anti-Fyn or anti-Lck antibody. Goat anti-rabbit IgG-Alexa Fluor 594 was used as the secondary antibody (Invitrogen). Data were acquired using an LSM-710 laser-scanning microscope (Carl Zeiss). The results were further analyzed by ImageJ (NIH). In the absence of ionomycin, fluorescent probes, as well as Fyn and Lck, were primarily attached to the plasma membrane, forming a tight and thin layer at the periphery of the cell. In contrast, upon dissociation from the plasma membrane, diffuse cytoplasmic fluorescence was observed. The “dissociation ratio” was calculated as the number of cells with diffuse cytoplasmic fluorescence compared to the total number of cells in each picture. Typically, pictures contained 20–50 cells, and 7–10 pictures of each sample were counted, for a total of 140–500 cells per condition. The fluorescence profile was quantified by ZEN software (Carl Zeiss).
Statistical analyses
Prism software V7 was used for the statistical analysis.
Supplementary information
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) MT-14429, MOP-82906, and FDN-143338 to A.V., and NSFC-31870863 from the National Natural Science Foundation of China to N.W. N.W. was supported by a Postdoctoral Fellowship from Fonds de recherche du Québec Santé (FRQS). A.V. is the Canada Research Chair on Signaling in the Immune System.
Competing interests
A.V. received a contract from Bristol Myers-Squibb to study the mechanism of action of the anti-SLAMF7 monoclonal antibody elotuzumab in multiple myeloma. He was also a consultant for Boehringer-Ingelheim on the topic of the SIRPα-CD47 blockade in anticancer immunotherapy. The authors declare no competing interests.
Footnotes
These authors contributed equally: Ning Wu, Hua Song
Contributor Information
Ning Wu, Email: wun@hust.edu.cn.
André Veillette, Email: andre.veillette@ircm.qc.ca.
Supplementary information
The online version of this article (10.1038/s41423-020-00600-9) contains supplementary material.
References
- 1.Sunshine H, Iruela-Arispe ML. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017;28:408–413. doi: 10.1097/MOL.0000000000000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 4.Bevers EM, Williamson PL. Getting to the outer leaflet: physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 2016;96:605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: a matter of life and death. Annu. Rev. Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 6.Gong YN, et al. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell. 2017;169:286–300 e216. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann B, et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–222. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- 8.Dillon SR, Mancini M, Rosen A, Schlissel MS. Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J. Immunol. 2000;164:1322–1332. doi: 10.4049/jimmunol.164.3.1322. [DOI] [PubMed] [Google Scholar]
- 9.Elliott JI, et al. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat. Cell Biol. 2005;7:808–816. doi: 10.1038/ncb1279. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K, et al. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood. 2006;108:4094–4101. doi: 10.1182/blood-2006-03-011742. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Poole K, Goyette J, Gaus K. Introducing membrane charge and membrane potential to T cell signaling. Front Immunol. 2017;8:1513. doi: 10.3389/fimmu.2017.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rival CM, et al. Phosphatidylserine on viable sperm and phagocytic machinery in oocytes regulate mammalian fertilization. Nat. Commun. 2019;10:4456. doi: 10.1038/s41467-019-12406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagata S, Segawa K. Sensing and clearance of apoptotic cells. Curr. Opin. Immunol. 2020;68:1–8. doi: 10.1016/j.coi.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Segawa K, Nagata S. An apoptotic ‘Eat Me’ signal: phosphatidylserine exposure. Trends Cell Biol. 2015;25:639–650. doi: 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Ruhl S, et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 2018;362:956–960. doi: 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- 16.Nagata S. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 2018;36:489–517. doi: 10.1146/annurev-immunol-042617-053010. [DOI] [PubMed] [Google Scholar]
- 17.Fujii T, Sakata A, Nishimura S, Eto K, Nagata S. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc. Natl Acad. Sci. USA. 2015;112:12800–12805. doi: 10.1073/pnas.1516594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu N, et al. Critical role of lipid scramblase TMEM16F in phosphatidylserine exposure and repair of plasma membrane after pore formation. Cell Rep. 2020;30:1129–1140 e1125. doi: 10.1016/j.celrep.2019.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung T, et al. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell VB, Rossjohn J, Wakelam MJ. Phospholipid signaling in innate immune cells. J. Clin. Investig. 2018;128:2670–2679. doi: 10.1172/JCI97944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resh MD. Myristylation and palmitylation of Src family members: the fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-X. [DOI] [PubMed] [Google Scholar]
- 22.Resh MD. Targeting protein lipidation in disease. Trends Mol. Med. 2012;18:206–214. doi: 10.1016/j.molmed.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 24.Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki J, Imanishi E, Nagata S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 2014;289:30257–30267. doi: 10.1074/jbc.M114.583419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 27.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo H, et al. Deletion of Slam locus in mice reveals inhibitory role of SLAM family in NK cell responses regulated by cytokines and LFA-1. J. Exp. Med. 2016;213:2187–2207. doi: 10.1084/jem.20160552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. DNAM-1 controls NK cell activation via an ITT-like motif. J. Exp. Med. 2015;212:2165–2182. doi: 10.1084/jem.20150792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 2009;10:297–305. doi: 10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 31.Sandilands E, Brunton VG, Frame MC. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J. Cell Sci. 2007;120:2555–2564. doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- 32.Stacey MA, Marsden M, Wang EC, Wilkinson GW, Humphreys IR. IL-10 restricts activation-induced death of NK cells during acute murine cytomegalovirus infection. J. Immunol. 2011;187:2944–2952. doi: 10.4049/jimmunol.1101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura K, et al. Fratricide of natural killer cells dressed with tumor-derived NKG2D ligand. Proc. Natl Acad. Sci. USA. 2013;110:9421–9426. doi: 10.1073/pnas.1300140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudd-Schmidt JA, et al. Lipid order and charge protect killer T cells from accidental death. Nat. Commun. 2019;10:5396. doi: 10.1038/s41467-019-13385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zech T, et al. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petkovic M, Oses-Prieto J, Burlingame A, Jan LY, Jan YN. TMEM16K is an interorganelle regulator of endosomal sorting. Nat. Commun. 2020;11:3298. doi: 10.1038/s41467-020-17016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, et al. Scramblase TMEM16F terminates T cell receptor signaling to restrict T cell exhaustion. J. Exp. Med. 2016;213:2759–2772. doi: 10.1084/jem.20160612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu N, et al. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat. Immunol. 2016;17:387–396. doi: 10.1038/ni.3369. [DOI] [PubMed] [Google Scholar]
- 39.Lemay S, Davidson D, Latour S, Veillette A. Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Mol. Cell Biol. 2000;20:2743–2754. doi: 10.1128/MCB.20.8.2743-2754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veillette A, Thibaudeau E, Latour S. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J. Biol. Chem. 1998;273:22719–22728. doi: 10.1074/jbc.273.35.22719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.