Abstract
Histamine receptors play important roles in various pathophysiological conditions and are effective targets for anti-allergy treatment, however the mechanism of receptor activation remain elusive. Here, we present the cryo-electron microscopy (cryo-EM) structure of the human H1R in complex with a Gq protein in an active conformation via a NanoBiT tethering strategy. The structure reveals that histamine activates receptor via interacting with the key residues of both transmembrane domain 3 (TM3) and TM6 to squash the binding pocket on the extracellular side and to open the cavity on the intracellular side for Gq engagement in a model of “squash to activate and expand to deactivate”. The structure also reveals features for Gq coupling, including the interaction between intracellular loop 2 (ICL2) and the αN-β junction of Gq/11 protein. The detailed analysis of our structure will provide a framework for understanding G-protein coupling selectivity and clues for designing novel antihistamines.
Subject terms: Cryoelectron microscopy, Hormone receptors
Histamine receptors are effective targets for allergy treatments and antihistamines are the first choice of many allergic disorders, but the exact mechanism of agonist binding and receptor activation remain unknown. Here, the authors present the cryo-EM structure of histamine-bound H1R/Gq complex and propose a mechanism of ligand induced receptor activation.
Introduction
Histamine is a biogenic amine that mediates a variety of pathophysiological responses and signaling events through the binding of histamine receptors, members of the class A G-protein-coupled receptor (GPCR) superfamily1. There are four types of histamine receptors, H1R, H2R, H3R, and H4R2. H1R and H2R are validated targets for the treatment of allergies and forms of gastric acid-related conditions, while H3R and H4R have a clinical potential for dementia, asthma, inflammatory bowel disease, and rheumatoid arthritis1,3–5. Histamine binding of receptor recruits heterotrimeric G-protein and triggers downstream signaling cascade. The H1R is mainly coupled with Gq protein that activates phospholipase C to increases inositol phosphates and intracellular calcium level, H2R couples to Gs protein to stimulate cAMP production, H3R and H4R signals through Gi/o proteins2.
Histamine has an active role in allergy and anaphylaxis and is mainly mediated by H1R3. The earliest use of antihistamines for allergic disorders can be traced back to early 1950s and over more than half century antihistamines remain as the first choice for many allergic disorders, such as allergic rhinitis, hay fever, and urticarial3. Early antihistamines can easily penetrate the blood-brain barrier and have low receptor selectivity, therefore have considerable side effects such as sedation, dry mouth, and arrhythmia6,7. The later introduction of carboxyl moiety and protonated amine improve receptor selectivity and significantly reduce side effects associated with brain permeability. However, even the second- or third-generation antihistamine, such as Certirizine (Zyrtec), Loratidine (Claritin), and Fexofenadine (Allegra) still have some unwanted side effects, such as drowsiness, dizziness, and headache1,8,9. The most successful antihistamine design is bulky molecules with a basic amino group which is very different from the simple imidazole ring and ethylamine side chain of histamine3, yet the mechanism by which those bulky antihistamines block the H1R signaling is still unknown.
An early study has revealed the structure of H1R bound to the first generation of antihistamine, doxepin, in an inactive conformation10. The structure revealed that the amine moiety of doxepin forms a salt bridge with a strictly conserved D1073.32 and the bulky tricyclic dibenzooxepin ring sets in a hydrophobic pocket form by conserved residues of TM 3, 5 and 6. The study also explained the improvement of specificity of the second generation of antihistamine via the docking method. However, lacking the active conformation of the receptor, the paper did not articulate a clear mechanism by which those inverse agonists inactivate receptor. Limited structural information of the receptor, particularly the lack of precise agonist binding information and the active conformation of receptor, hampered the development of novel antihistamine that may shut down the receptor activity more effectively and have less side effects.
GPCRs primarily couple to 4 major Gα families, Gs, Gi/o, Gq/11, and G12/13, that dictate different signaling cascade11. Considering the large number of receptors (more than 800) and the limited number of G protein (16 of 4 families)12, a general question is whether there is a selective barcode for receptor/G-protein recognition. A number of studies have suggested that the engagement of G-protein to receptor is more complicated than previous envisioned13–15, and there is no simple primary or second structural pattern on receptor/G-protein recognition. A barcode, if exists, it must lie in the tertiary or even the quaternary structure of receptor/G-proteins complex. A number of GPCR/Gs and GPCR/Gi complexes have been solved by cryo-EM, however, there is very few of Gq/11-coupled receptor complex structure available. The muscarinic acetylcholine receptor 1 (M1R)/G11 complex structure first revealed some distinct features for the Gq/11-coupled receptors, including an extended TM5 and a receptor c-tail/G-protein interaction16. Most recently, a 5-HT2A serotonin receptor (HTR2A)/mini-Gq protein complex structure was solved17. It is imaginable that with more Gq-coupled receptor complex structures being solved, a pattern of receptor/Gq engagement can be found.
Here, we present a cryo-EM structure of H1R in complex with a N-terminal engineered Gq protein by a NanoBiT tethering strategy. The structure reveals the mechanism of ligand-induced receptor activation in a model of “squash to activate and expand to deactivate” (Supplementary Fig. 1). Our structure analysis also unravels some distinct features of H1R/Gq engagement. These findings could help to understand the Gq coupling selectivity and provide clues for designing novel antihistamines that can more specifically block histamine signaling and have fewer side effects.
Results
The overall structure of H1R/Gq complex
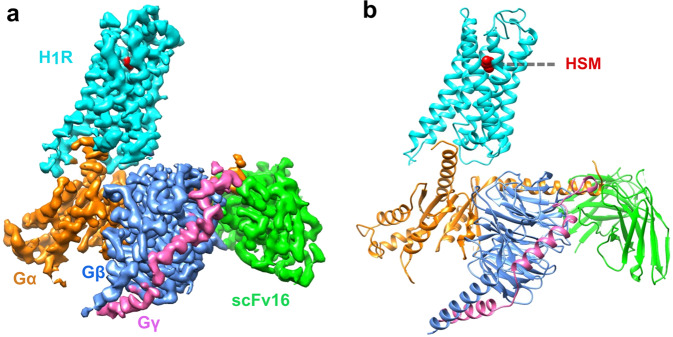
To facilitate the cryo-EM structure solving, we use an engineered Gq protein (GqiN) in which the N-terminus (residue 1–32) of Gq was replaced by the N-terminus (residue 1–28) of Gi protein to render the protein binding ability to the scFv16 antibody that has been successfully used in solving numerous receptor/G-protein complexes16,18–20, including the M1R/G11 complex. Also the long ICL3 of H1R (residue 224–401) was deleted to improve protein expression and folding. The initial attempt to get a stable complex via co-expression of the receptor, GqiN and scFV16 was not very successful. We then adopted to the NanoBiT tethering strategy21 in which the C-terminus of H1R was fused to the large part of NanoBiT (LgBiT), and the C-terminus of Gβ was fused to the renovated 13-amino acid peptide of NanoBiT (HiBiT) (Supplementary Fig. 2). This strategy has been successfully used in solving several GPCR/G-protein complexes structures, including the vasoactive intestinal polypeptide receptor21 (VIP1R)/Gs and growth hormone-releasing hormone receptor/Gs complexes22. Indeed, the NanoBiT tethering strategy greatly improves the composition of the complex (Supplementary Fig. 3), and the structure was solved by the single-particle cryo-EM analysis of the H1R-LgBiT/GαqiN/Gβ-HiBiT/Gγ/scFv16 complex at 3.64 Å resolution (Methods and Supplementary Figs. 4–6). The overall reconstruction of the H1R/Gq complex is shown in Fig. 1a with the resulting model in Fig. 1b. The global arrangement of the complex is similar to other GPCR/G-protein complexes with the Gα protein engaging with the intracellular side of the receptor. Local resolution analysis shows that the WD40 repeat domain of Gβ and the core transmembrane domain of the receptor have the highest resolution, the extracellular part of receptor, the border of Gα RAS domain and the N-terminus of Gβγ have the relative lower resolution (Supplementary Fig. 5a). The alpha helical domain of Gα cannot be resolved in our analysis due to its high flexibility. We also observed trace amount of density of the NanoBiT complex formed by the LgBiT and HiBiT (Supplementary Fig. 7), however, the local resolution much worse than 6 Å, we therefore omit it in the later structural analysis. Previous VIP1R study has shown that the fusion of LgBiT to receptor has almost no effect on the function of the receptor. We also did a nuclear factor of activated T-cells response element (NFAT-RE) reporter assay, a well-established reporter assay for the Gq/11 signaling pathway23, showing that the fusion of LgBiT to H1R only marginally affects receptor activity (Supplementary Fig. 2c). Because of the NanoBiT density is far from the receptor/Gq interface (Supplementary Fig. 7) and the fusion of LgBiT do not affect receptor’s response to histamine, we reason that the complex structure of H1R-LgBiT/GαqiN/Gβ-HiBiT/Gγ represents the complex structure of H1R/Gq, and hence use H1R/Gq to represent the complex in this study. The transmembrane helical bundle has strong electron signal and the histamine ligand is well resolved in the ligand-binding pocket (discussed in detail in the following section).
Fig. 1. Overall structure of the H1R/Gq complex.
a Orthogonal views of the cryo-EM density map of the H1R/Gq complex. b Model of the complex in same view and color scheme as shown in (a). HSM: histamine.
Histamine binding and the ligand-binding pocket
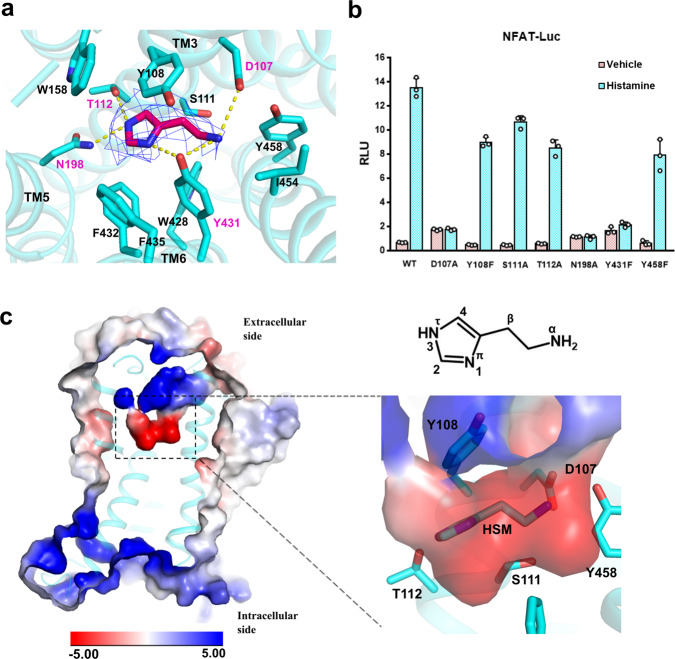
The native ligand histamine is well resolved in the ligand-binding pocket (Fig. 2a). The structure shows that all three histamine nitrogen atoms establish hydrogen bonds with the surrounding polar residues from TM3, TM5, and TM6. The primary amino group of histamine (Nα) forms hydrogen bonds with the D1073.32 of TM3 and Y4316.51 of TM6, the 1 position nitrogen atom (Nπ) of the imidazole ring form a hydrogen bond with Y4316.51 of TM6, and the 3 position nitrogen atom (Nτ) forms hydrogen bonds with N1985.46 of TM5 and T1123.37 of TM3 (Fig. 2a). Those key positions are conserved in the histamine receptor family (Supplementary Fig. 8) and the interactions are confirmed by mutation experiment on the NFAT-RE reporter assay. Single mutation of D107A, N198A, and Y431F totally abolish histamine-induced receptor activation (Fig. 2b), suggesting that they play a key role in ligand binding. The T112A mutation only partially decreases the ligand-induced receptor activity, this may be due to the factor that the Nτ atom also forms hydrogen bond with the N1985.46 of TM5. On the other hand, the Y108F and S111A mutations do not have a detrimental effect on ligand-induced receptor activation, indicating that they do not participate the forming of the hydrogen bond network with the ligand. Interestingly those two residues are also not conserved in the histamine receptor family (Supplementary Fig. 8). The periphery of the ligand-binding pocket is surrounded by hydrophobic residues, including W4286.48, F4326.52, F4356.55, and W1584.56 (Fig. 2a). To investigate whether those non-polar bulky residues also contribute to receptor activation through hydrophobic or van der Waals interactions, we mutate those bulky residues (including two polar bulky residues that do not form hydrogen bond with histamine, Y1083.33 and Y4587.43) to small non-polar residues and test them in the NFAT-RE reporter assay. The data show that mutation of W158A and W428A totally abolish receptor activation, and F435A severely affects receptor activity (Supplementary Fig. 9), suggesting that they may play a crucial role in defining the ligand-binding pocket and providing some necessary hydrophobic interaction to support the correct ligand binding. Particularly, the W4286.48 of the conserved CWxP motif is the “toggle-switch” of receptor, plays a key role in switching receptor from inactive state to active state. The Y108V and Y458A mutations retain about half of receptor activity, and the F432A and I454A mutations are almost as active as the wild-type receptor, suggesting those residues do not directly participate ligand binding. An electrostatic potential calculated and analyzed by the APBS Eletrostatics PyMol Plugin shows a negative charged pocket setting on the up-middle (toward the extracellular side) of the receptor (Fig. 2c). The pocket is formed by D1073.32, Y4587.43, Y4316.51, N1985.46, T1123.37, Y1083.33, S1113.36, and histamine sets right middle of the pocket.
Fig. 2. Histamine binding and the ligand-binding pocket.
a The ligand-binding pocket of histamine. The histamine is shown in red in the middle of the pocket, density map of histamine (blue mesh) is set at contour level of 3.0. Surrounding residues within 4.1 Å of histamine are shown in sticks and colored in cyan. Hydrogen bonds are marked as yellow dash line. b A NFAT-RE reporter assay of mutations of key residues in the ligand-binding pocket. Histamine, 10 µM; data are presented as mean values ± SD; n = 3 independent samples. RLU, relative luciferase unit. WT, wild-type. c The electrostatic potential surface of H1R and the histamine binding pocket. The chemical structure of histamine is shown on the up-right panel.
Active H1R vs inactive H1R
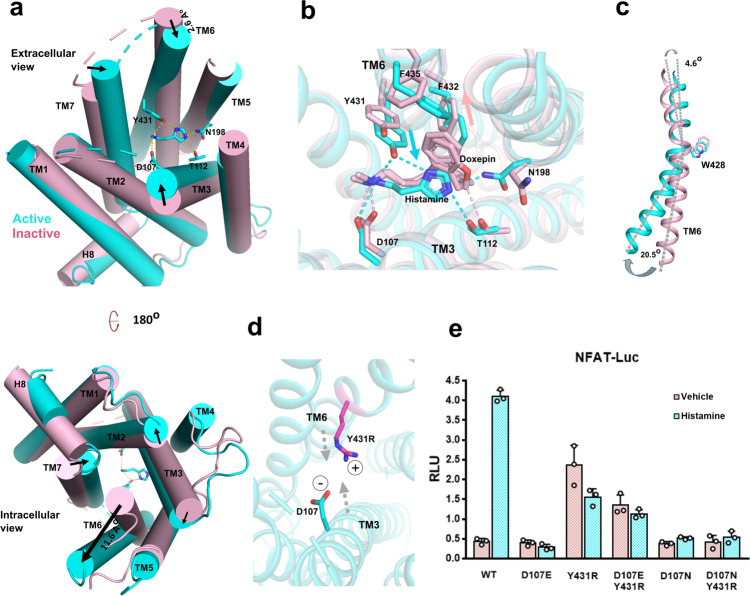
Compared to the inactive conformation of doxepin-bound H1R10 (Cα root-mean-square deviation of 1.235 Å), the biggest difference is the squash of ligand-binding pocket in the active conformation. Calculating by the CASTp 3.0 server24, the solvent-accessible volume of doxepin-bound H1R is 249 Å3, and the histamine-bound H1R is 79 Å3 (Supplementary Fig. 10). A close look at the ligand-binding sites shows that in the active conformation, the agonist histamine forms hydrogen bonds with D1073.32, T1123.37 of TM3 on one side, and forms hydrogen bond with the Y4316.51 of TM6 on the other side, acting as a magnet to pull the extracellular half of TM6 toward TM3 (Fig. 3a). Although the inverse agonist doxepin also forms hydrogen bond with the conserved D1073.32 of TM3, its hydrophobic tricyclic dibenzooxepin ring cannot form a hydrogen bond with Y4316.51 of TM6, in fact the bulky tricyclic dibenzooxepin ring push F4326.52 and F4356.55 of TM6 away from TM3 (Fig. 3b). It is conceivable that the contraction of the extracellular binding pocket can lead to the expansion of the intracellular end, thus to open the hydrophobic cavity of the intracellular side for transducer engagement, as seen in β2 adrenergic receptor (β2-AR)/Gs, M1R/G11 and cannabinoid receptor 1 (CB1)/Gi complex16,25,26. Indeed, we see an inward displacement of the extracellular part of TM6 (2.6 Å measured by the Cα of A4396.59) and a dramatic outward displacement of the intracellular part of TM6 (11.6 Å as measured by the Cα of N4086.28) upon ligand binding (Fig. 3a). The conserved W4286.48 serves as a pivot point for these movements, where the “up” (extracellular) side move inward (4.6 degree), and the “down” (intracellular) side move outward (20.5 degree) (Fig. 3c). We also see the coordinated movements of TM3, TM2, and TM7 in squashing the binding pocket and opening the hydrophobic cavity of the intracellular side (Fig. 3a). Together, those movements render receptor in an active conformation, enabling the engagement of the αH5 of Gαq. Based on those observations, we postulate that the key mechanism for receptor activation is to squash the ligand-binding pocket via the hydrogen bonds formed by histamine with key residues of TM3 and TM6. To test the hypothesis, we have mutated the Y4316.51, the key residue of TM6 that forms hydrogen bond with the Nπ of histamine, to the positive charged residue R, rendering it ability to form a salt bridge with the negative charged residue D1073.32 to mimic the effect of histamine to pull TM6 toward TM3 (Fig. 3d). In the NFAT-RE reporter assay, the Y431R shows high basal activity (Fig. 3e). To prove the high basal activity is caused by the salt bridge interaction, we mutated D1073.32 to N. The data show that the combination of Y431R/D107N or D107N alone has almost no basal activity. To further prove the lost basal activity of the Y431R/D107N is due to the loss of salt-bridge interaction, we also mutate D1073.32 to E which retains the ability to form a salt bridge with Y431R. The data shows while D107E has no basal activity as the D107N, the combination of Y431R/D107E retains the most basal activity of Y131R (Fig. 3e), suggesting the high basal activity indeed comes from the salt-bridge interaction between TM3 and TM6. We also obtained similar results when Y4316.51 is mutated to K (Supplementary Fig. 11). Taken together, we propose a model of “squash to activate and expand to deactivate” for H1R action (Supplementary Fig. 1) where agonist activates receptor via forming hydrogen bonds with TM3 and TM6 to squash ligand-binding pocket on the extracellular side and open the cavity for G-protein engagement on the intracellular side; on the other hand, antihistamine (inverse agonist) use its bulky group to expand the ligand-binding pocket on the extracellular side and to close the G-protein binding cavity on the intracellular side to shut down receptor signaling. Indeed, a comparison of doxepin-bound H1R with histamine-bound H1R shows that the bulky tricyclic dibenzooxepin ring of doxepin pushes F4326.52 and F4356.55 of TM6 away from TM3. It is also very interesting to find that the most successful antihistamines are those with bulky ring group, such as fexofenadine (Allergra), loratadine (Claritin), and certirizine (Zyrtec) (Supplementary Fig. 12), further supporting the idea of “expand to deactivate and squash to activate”. To examine whether this is a specific activation mechanism for H1R, or it is common mechanism for monoamine GPCRs, we compared the size of agonist and antagonist binding pockets of H1R with β2-AR, dopamine receptor DRD2 and serotonin receptor HTR2A, members of the monoamine GPCRs (Supplementary Fig. 13). The side-by-side comparisons show that the ligand-binding pocket of H1R condenses most dramatically upon agonist binding, the β2-AR ligand-binding pocket slightly shrinks while there is no significant change in the size of DRD2 pocket upon agonist binding, and HTR2A’s pocket actually increase upon agonist binding. Those data suggest that the “squash to activate and expand to deactivate” might be a more specific model for H1R action.
Fig. 3. Activation of H1R by the binding of histamine.
a A comparison of histamine-bound active H1R and the inverse agonist doxepin-bound inactive H1R (PDB 3RZE). The helices are shown in cylinders. Active receptor is shown in cyan and inactive receptor is shown in light pink. Up-panel is the extracellular side view and lower-panel is the intracellular side view. Arrows mark the movement of the designate part of receptor upon histamine binding. b A comparison of the agonist (histamine) binding pocket (cyan) with the inverse agonist (doxepin) binding pocket (light pink). The cyan arrow marks the movements of key residues on TM6 upon agonist (histamine) binding and the light pink arrow marks the movements of key residues on TM6 upon inverse agonist (doxepin) binding. c The movements of TM6 upon receptor activation. The W4286.48 severs as a pivot between the upside (extracelluar) movement and the downside movement of TM6. d A simple model (for illustration only) of a designed salt bridge interaction between the Y431R mutation of TM6 and with the negative charge D1073.32 of TM3. e A NFAT-RE reporter assay of the Y4316.51 and D1073.32 mutations. Histamine, 10 µM; data are presented as mean values ± SD; n = 3 independent samples. RLU, relative luciferase unit.
We also look at how the activation signal on the extracellular side is transmitted to the intracellular side via examining the conformation changes on the conserved DRY, CWxP, NPxxY, and PIF motifs. Those motifs lie on the intracellular half of receptor and are usually associated with locking receptor in the inactive state (Supplementary Fig. 14). The CWxP motif of TM3 lies in the hinge region of receptor where the extracellular side signal is transmitted to the intracellular side of receptor. In receptor activation, the binding of agonist ligand histamine triggers the rotameric switch of W4286.48 and the concomitant side chain rotation of Y4316.51, F4326.52, F4246.44 and F1995.47, initiating the rotation of TM6. Mutation of W4286.48 to A totally abolishes receptor activity (Supplementary Fig. 9b), indicating the importance of this residue in the switch of receptor activity. The DRY motif localizes on the lower part of TM3. In the inactive state, the Y1253.50 of DRY motif interacts with both A4136.33 and Q4166.36 of TM6 to lock receptor in an inactive conformation. Upon receptor activation, the interaction is broken as TM6 sways away from inside to outside (Supplementary Fig. 14d). The N4647.49 of the NPxxY motif on TM7 associates with D732.50 in both active and inactive states, however, upon receptor activation, there is 8.2 Å displacement of Y4687.53 toward the center of the cavity when TM6 sways away, the aromatic ring of Y4687.53 also tilts about 40 degrees to form new contact with residue V1183.43, L1213.46, and R1253.50 of TM3 (Supplementary Fig. 14e). In the PIF motif, the most significant change is the side chain displacement of F4246.44 upon receptor activation, similar to the observation in the receptor activation of the HTR2A/mini-Gq complex17.
The engagement of Gq to H1R
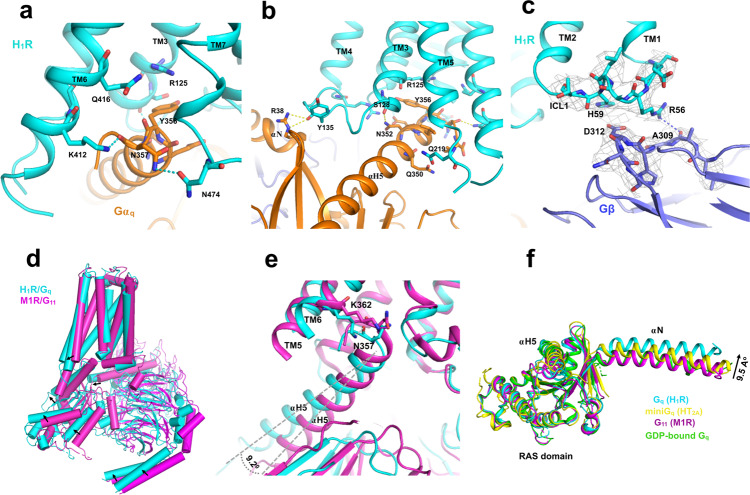
The engagement of Gq to H1R is mainly maintained by key interaction from TM6, TM5, TM3, ICL2, and TM7-H8 kink region. The K4126.32 of TM6 makes a key interaction with the carboxy group of N357G.H5.24 of the αH5 (Fig. 4a). The side chain of N352G.H5.19 in αH5 forms a hydrogen bond with the backbone carbonyl of S1283.53 in TM3, while the last residue of αH5, Y356G.H5.23, is blocked by the R1253.50 to prevent further intrusion (Fig. 4b). The ICL2/αN-β1 junction interaction has been reported in M1R/G11 complex16, we also observe a similar interaction in the H1R/Gq complex but with a different pattern where the backbone carbonyl of Y135ICL2 forms a hydrogen bond with the αN-β1 junction. In TM7-H8 kink region, we observe a close contact of N4748.49 with the N357G.H5.24 of αH5 (Fig. 4a). Residue L133 in ICL2 is highly conserved in GPCR family and has been implicated play an important role in Gs and Gq coupling. Similar to the HTR2A/mini-Gq complex17, we found that the hydrophobic residue L133 ICL2 intrudes into a hydrophobic “pocket” formed by L40, V199, F201, F341 and I348 of Gαq (Supplementary Fig. 15). Mutation studies has suggested that this hydrophobic interaction play a crucial role in stabling the receptor/Gq complex17. Interestingly, we also observe a patch of interactions between the ICL1 and the Gβ subunit where the side chain of R56ICL1 forms a hydrogen bond with the backbone carbonyl of A309 of Gβ, and the H59ICL1 is in close contact with the negative charged D312 of Gβ (Fig. 4c).
Fig. 4. The engagement Gq to H1R.
a The interaction between the receptor and the αH5, viewing from the TM6, TM7-H8 front angle. The receptor is colored in cyan and Gαq is colored in orange. b The interaction between the receptor and Gαq, viewing from the TM5, TM3, and ICL2 front angle. c the ICL2/Gβ interaction. The receptor is colored in cyan and Gβ is colored in slate. Density map of designated region is set to contour level of 3.0. d An overall comparison of the H1R/Gq complex with the M1R/G11 complex (PDB 6OIJ). e A comparison of the engagement of αH5 to receptor between the H1R/Gq complex with the M1R/G11 complex. f A comparison of the αN displacements of Gq/11 in receptor engagement. M1R/G11 (PDB 6OIJ), HT2AR/mini-Gq (PDB 6WHA) and GDP-bound Gq (PDB 3AH8).
The overall engagement of H1R/Gq is similar to M1R/G11 (Fig. 4d), however, distinct features were observed. Aligned with receptors, a comparison of M1R/G11 with H1R/Gq shows a clear difference in the orientation and position of G protein relative to receptor. The insertion of αH5 into H1R is less deeper (about 1/2 helix) than the G11 in M1R (Fig. 4e). Also the αH5 of Gq shows a 9.2 degree outward displacement in the H1R/Gq complex, we also observe the coordinated displacements on Gα RAS domain and Gβγ core domain (Fig. 4d). However, the largest observed displacement is the translational displacement of αN (9.5 Å as measured by the Cα of A7 between Gq and G11) when aligned with the core of RAS domain which seldom changes shape in receptor engagement (Fig. 4f). The αN of the H1R/Gq complex shows a dramatic outward translational displacement compared to that of the M1R/G11 and HTR2A/mini-Gq, as well as the GDP-bound inactive Gq protein27. While the interaction between ICL2 and αN-β1 junction seems to be a common feature of the Gq/11 coupled receptor, H1R use a different pattern to interact the junction in which the backbone carbonyl of Y135ICL2 interacts with the side chain of R38 of Gq (Fig. 4b and Supplementary Fig. 16a), instead of the “head to toe” pattern observed in M1R/G11 and HTR2A/mini-Gq where the side chain of R134ICL2 interact with the backbone carbonyl of R37, and the side chain of R37 interact with the backbone carbonyl of R134ICL2 in the M1R/G11 complex (for HTR2A/mini-Gq, the pattern is R185ICL2/R32) (Supplementary Fig. 16). We speculate that the interaction difference may contribute to the large displacement of αN in the H1R/Gq complex. In addition, we also find the TM7-H8 kink/αH5 interaction might be a common feature for Gq/11 coupling receptors. In H1R/Gq complex, the N4748.49 interacts with the N357G.H5.24 of αH5 (Fig. 4a and Supplementary Fig. 17). In HTR2A/mini-Gq complex, the N3848.47 interacts with N244 of the mini-Gq. Similarly in M1R/G11, the N4228.47 of M1R is in a close contact with E355G.H5.22 and N357G.H5.24 of the αH5.
Comparison with GPCR-Gs and -Gi complexes
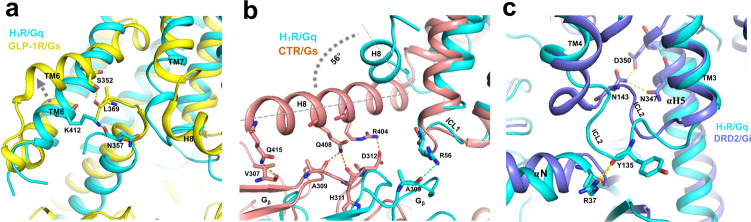
The structure of H1R/Gq, M1R/G11, and HTR2A/mini-Gq complex, enable us to do a comparison of Gq-coupled with Gs-coupled and Gi-coupled receptor complex. Since Gs complex structures predominantly come from Class-B GPCR/Gs complex, we first compare with the class B GPCR. It has been reported that the Gs family has the most pronounced outward displacement of TM6 in receptor activation. Compared to Glucagon-like peptide 1 receptor (GLP-1R)/Gs complex28, the TM6 of GLP-1R shows a 6.0 Å more outward displacement than H1R (Fig. 5a). Similar displacement of TM6 was observed on parathyroid hormone receptor-1 (PTH1R)29 and calcitonin receptor (CTR)30. We also noticed a different pattern of TM6/αH5 interaction. Instead of using the positive charged residue (K4126.32 of H1R and K3626.32 of M1R) for αH5 interaction, the Gs-coupled receptor use serine or threonine (S3526.41 of GLP-1R and S4096.41 of PTH1R) for αH5 interaction (Fig. 5a). It has been reported that the Gβ subunit participates receptor interaction in class B GPCR/Gs complex22,28–30, since we discovered an interaction between ICL1 and Gβ, we compared H1R with class B GPCRs. We noticed a significant difference in the length and orientation of helix 8, the class B GPCRs have a much longer H8 than the Gq-coupling receptor (H1R, M1R and HTR2A). Most notably, a H8 of Gs-coupled class B GPCR (CTR) shows a 56 degrees of tilt toward the Gβ subunit side, which explains the general observation of H8/Gβ interaction in this family (Fig. 5b). For instance, the Q4158.67, Q4088.6 and R4048.56 of calcitonin receptor (CTR) make contact with V307, A309, and D312 of Gβ, respectively. On the other hand, H1R use its ICL1 (R56ICL1) to interact the Gβ subunit (A309) (Figs. 4c, 5b). The role of H8/Gβ interaction of the class B GPCR is generally believed to be related to receptor stability but not the receptor/G-protein coupling as mutations show little effect on the signaling28,30. Consistent with this, mutation of R56ICL1 and H59ICL1 of the H1R to alanine show no effect on the NFAT-RE reporter assay (Supplementary Fig. 18).
Fig. 5. Comparisons of Gq-coupled H1R with Gs- and Gi-coupled receptors.
a A comparison of the αH5 engagement between GLP-1R/Gs (PDB 5VAI) and the H1R/Gq complex. The dashed arrow shows the additional outward displacement of the TM6 of GLP-1R. b A comparison of the Gβ interactions between the CTR/Gs (PDB 5UZ7). c A comparison of the ICL2 interaction between the DRD2/Gi (PDB 6VMS) and the H1R/Gq complex.
We then compare the class A Gs-coupled receptors with H1R. There are only two class A GPCR/Gs complex structures (β2-AR and β1-AR) and two class A GPCR/mini-Gs (GPR52 and A2AR) complex structures available. We first compare with Gs complexes. Similar to class B GPCR, the outward displacement of TM6 is more pronounced in β2-AR and β1-AR than H1R (Supplementary Fig. 19a). The TM6/αH5 interaction pattern is similar to class B GPCR, β2-AR and β1-AR use threonine (T2746.36 and T2916.36, respectively) for αH5 interaction while H1R use positive charged residue (K4126.32) for αH5 interaction (Supplementary Fig. 19a). We also noticed a subtle difference of the TM3/αH5 interaction. For instance, β2-AR use both the last two residues (I1353.54, T1363.55) of TM3 to interact with the αH5 of Gs (Q384G.H5.5.16 and R380G.H5.5.12) while the Gq-coupled receptor only use the second last residue of TM3 (S1283.53 of H1R and S1263.53 of M1R) to interact the αH5 (N352G.H5.5.19 and Y356G.H5.5.23) (Supplementary Fig. 19b). For the mini-Gs/Receptor complexes, a distinct feature is the TM5/α H5 interaction (H2335.69 of GPR52 interact with Q384 of mini-Gs, and Q2075.63 of A2AR interact with Q374 of mini-Gs) which has not been seen in H1R or M1R (Supplementary Fig. 20).
Compared to the Gi-coupled receptors, the difference is much subtle, the most noticeable difference is the ICL2 interaction. The Gq-coupled receptors seem uniquely use the ICL2 to interact with the αN-β1 junction of Gα (Fig. 5c and Supplementary Fig. 16a), while the Gi-coupled receptors dominantly use the ICL2-TM4 junction to interact with the αH5. For instance, the D2 dopamine receptor (DRD2) use the N143ICL2 to interact with the D350G.H5.22 of the αH5 of Gi19 (Fig. 5c), while H1R use the Y135ICL2 to interact with the R37 of the αN of Gq. Similarly, CB126 use P221ICL2, CB231 use K142ICL2 and μ-opioid receptor (µOR)20 use R179ICL2 to interact with the αH5 of Gi (Supplementary Fig. 21), and M1R use R134ICL2 to interact the R37 of the αN of G11 (Supplementary Fig. 16a). On the G protein side, compared with Gs and Gi-coupled receptor complexes, the αN of H1R/Gq complex displays the largest outward translational displacement (Supplementary Fig, 22) as seen in the Gq-coupled receptor complex (Fig. 4f). Mutations of key residues of H1R (Y135A, K137A) that interacts with the αN-β1 junction of Gq shows no severe effect on receptor activity on the NFAT-RE reporter assay (Supplementary Fig. 16b), suggesting that function of the large translational displacement of αN in H1R/Gq complex may not directly relate to receptor coupling.
Discussion
In this study, we use the NanoBiT strategy to solved the structure of histamine-bound H1R in complex with Gq via single-particle analysis of cryo-EM. To facilitate receptor expression and folding, we use a ICL3 truncation version of receptor (224-401) for structure study. It has been reported that the ICL3 contributes to the recognition and coupling of GPCRs with Gs, Gi, Gq and G12/1313,14, we therefore ask whether the ICL3 deletion will affect receptor activation. In a NFAT-RE reporter assay, the ICL3 deleted receptor responds ligand well, however, comparing the dose-response curve, the ICL3 truncation shows 10 folders higher of EC50 than wild-type receptor (118.6 nM for wild type, 1651 nM for ICL3 truncation, Supplementary Fig. 23), suggesting that the ICL3 does contribute receptor activation and Gq coupling. We suspected that the flexible region of ICL3 may interact with αH5 and the Ras domain of the Gαq that faces the receptor side, and those interactions may stabilize the receptor/Gq complex, the detail of those interaction is worthy of future investigation for a comprehensive understanding of Gq coupling. In summary, we have revealed the active conformation of H1R in complex with Gq and presented a model of “squash to activate and expand to deactivate” for H1R action. Our findings, including both the mechanistic insights and the featured observations of the Gq coupling will benefit both the understanding of Gq signaling and the rational design of novel antihistamines.
Methods
Constructs
The human H1R gene was subcloned into the pFastBac plasmid with a HA-signal peptide sequence on its N-terminus and the LgBiT fused to its c-terminus followed by a Tobacco etch virus (TEV) cutting site and 2 fused maltose-binding proteins to facilitate protein expression and purification. A 12 amino sequence of GASGASGASGAS sequence was inserted between the receptor and LgBiT. The first 28 residues (residue 1-28) and the ICL3 loop (residue 224–401) of H1R were chopped off to increase protein expression and folding. The HiBiT was fused to the c-terminus of human Gβ1 and cloned into pFastBac plasmid as described in the VIP1R paper21. The N-terminus (residue 1-32) of human Gαq was replaced by the N-terminus of Gi (residue 1–28) and subcloned into pFastBac plasmid. The wild-type human Gγ2 was cloned into pFastBac plasmid. The scFv16 that encodes the single-chain variable fragment of mAb16 was subcloned into pFastBac plasmid.
Expression and purification of H1R/Gq complex
Bacmid preparation and virus production were performed according to the Bac-to-Bac baculovirus system manual (Gibco, Invitrogen). For expression, the Spodoptera frugiperda (Sf9) cells at density of 2 × 106 cells per ml were co-infected with baculovirus encoding the H1R-LgBiT-tev-2MBP, GqiN, Gβ, Gγ and scFv16 protein at a ratio of 1:100 (virus volume vs cells volume). Cells were harvest 48 h after infection. Cell pellets were resuspended in 20 mM Hepes buffer (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 20 mM KCl, 5 mM CaCl2, and homogenized by douncing ~30 times. Apyase was added to the lysis at a final concentration of 0.5 mU/ml. To keep the complex stable, histamine was added at a final concentration of 100 µM all through the purification procedure. The lysate was incubated at room temperature for 1 h with flipping. Then, n-dodecyl-β-D-maltoside (DDM) was added at the final concentration of 0.5% to solubilize the membrane at 4 °C for 2 h. Then the lysis was ultracentrifuged at 56,000g (45,000 rpm) at 4 °C for 40 min. The supernatant was collected and incubated with amylose column for 2 h. The amylose column was washed with a buffer of 25 mM Hepes (pH 7.5), 200 mM NaCl and 0.02% DDM, and 0.004% cholesteryl hemi-succinate (CHS), then eluted with the same buffer plus 10 mM maltose. The elution was concentrated and processed with home-made TEV for overnight at 4 °C. Then the digest was separated on a Superdex 200 Increase 10/300 GL (GE health science) gel infiltration column with a buffer of 25 mM Hepes (pH 7.5), 200 mM NaCl, and 0.1% digitonin (Biosynth). The peak corresponding to the H1R/Gq complex was concentrated at about 10 mg/ml and snap frozen for later cryo-EM grid preparation.
Grid preparation and cryo-EM data collection
Three microlitters of H1R/Gq complex sample at ~10 mg/ml was applied to a glow-charged quantifoil R1.2/1.3 CuRh holey carbon grids (Quantifoil GmbH). The grids were vitrified in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific) instrument in the setting of blot force of 10, blot time of 5 s, humidity of 100%, temperature of 6 °C. Grids were first screened on a FEI 200 kV Arctica transmission electron microscope (TEM) and grids with evenly distributed thin ice and promising grids were transferred to a FEI 300 kV Titan Krios TEM equipped with a Gatan Quantum energy filter and a spherical corrector for data collection. Images were taken by a Gatan K3 direct electron detector at magnitude of 64,000, super-resolution counting model at pixel size of 0.54 Å, the energy filter slit was set to 20 eV. Each image was dose-fractionated in 32 frames using a total exposure time of 2.56 s at a dose rate of 1.56 e/Å2/s (total dose 50 e/Å2). All image stacks were collected by the EPU program of FEI, nominal defocus value varied from 1.2 to 2.0 µm.
Data processing
Raw movies at a size of 0.54 Å were binned once to generate a pixel size of 1.08 Å and then motion-corrected using MotionCor232, followed by CTF estimation using CTFFIND 4.133. Particles were picked from the micrographs using crYOLO34. Then the picked particles (about 2.4 million) were extracted by RELION35,36 (version 3.1) and subjected to 2 rounds of reference-free 2D classification in RELION. About 650,000 particles were selected and the initial model was generated by cryoSPARC37 ab initio. Then the model was used as reference in RELION 3D classification. Classes showed clear secondary structure features were select for a 3D refinement in RELION, followed by a Baysian polishing38 implemented in RELION. Then the polished particles were subject to one round of 2D classification to get rid of the irregular particles that may not contribute to the high resolution 3D reconstitution. Then, this was followed by a 3D refinement and a CTF refinement implemented in RELOIN. The CTF refined particles were subjected to a 3D classification with fine angular sampling which yields a promising class of 169,241 particles. Then the particles were transferred to cryoSPARC and followed a Non-uniform Refinement which yields a map of 3.64 Å based on the gold standard Fourier Shell Correlation (FSC) = 0.143 criterion. Later Phenix real_space-refinement show a resolution of 3.3 Å at 0.143. Local resolution estimations were performed using either RELION or cryoSPARC.
Model building
The crystal structures of human doxepin-bound H1R10 (PDB 3RZE) and the G11 protein complex from the M1R/G11 (PDB 6OIJ)16 were used as initial models for model rebuilding and refinement against the electron microscopy map. All models were docked into the electron microscopy density map using UCSF Chimera39. The resulting model was subjected to iterative manual adjustment using Coot40, followed by a rosetta cryoEM refinement41 at relax model and Phenix real_space refinement42. The model statistics were validated using MolProbity43. Structural figures were prepared in UCSF Chimera39 and PyMOL (https://pymol.org/2/). The statistics for data collection and refinement are included in Supplementary Table 1.
The NFAT-reporter assay
The nuclear factor of activated T-cells response element (NFAT-RE) reporter assay was performed according to the luciferase reporter assay for deciphering GPCR pathways paper23 and Promega instruction. Briefly, AD293 cells were split into 24 well plates at a density of 40,000 per well. After one day of growth on 37 °C at 5% CO2, cells (per well) were transfected with 100 ng of NFAT-RE-Luc, 10 ng of pcDNA3-H1R wild-type or mutations, 10 ng of phRGtkRenilla plasmids by X-tremeGENE HP (Roche) at a ratio 3:1 to DNA amount. 16 h after transfection, cells were induced by histamine at 10 µM or vehicle. Six hours after induction, cells were harvested and lysed by addition of 1× Passive Lysis Buffer (Promega), and luciferase activity was assessed by the Dual-Glo Luciferase system (Promega). Data were plotted as firefly luciferase activity normalized to Renilla luciferase activity in Relative Luciferase Units (RLU).
Structure and sequence comparison
The calculation of the pocket volume was done by the CASTp 3.0 sever24. Sequence alignment by the Clustal Omega44 sever and the representation of sequence alignment was generated using the ESPript45 website (http://espript.ibcp.fr). The generic residue numbering of GPCR is based on the GPCRdb46 (https://gpcrdb.org/).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors thank the Cryo-EM facility of Harbin Institute of Technology and the Cryo-EM core of the Shuimu Bioscience (Beijing) for sample screening and data collection. We thank the Startup Funds of HIT Center for Life Science. We thank the National Natural Science Foundation of China (32070048 to Y.H.). We thank Dr. Zhiwei Huang for suggestion and support of this project.
Author contributions
Y.H. conceived the project and design the experiments. R.X. made the expression constructs, purified the proteins and assembled the complex. N.W. made mutation constructs and performed the functional assays. Z.X. purified the proteins and prepared the grids. Y.L. initial the project and made early constructs. J.S. made some mutation constructs. Y.H., A.Z., and C.G. collected the data. Y.H. solved the structure and wrote the manuscript. All authors contributed to data interpretation and preparation of the manuscript.
Data availability
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper. Structural data have been deposited with the PDB (accession number 7DFL), and maps have been deposited with the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-30665.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks So Iwata and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ruixue Xia, Na Wang, Zhenmei Xu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-22427-2.
References
- 1.Monczor F, Fernandez N. Current knowledge and perspectives on histamine H1 and H2 receptor pharmacology: functional selectivity, receptor crosstalk, and repositioning of classic histaminergic ligands. Mol. Pharm. 2016;90:640–648. doi: 10.1124/mol.116.105981. [DOI] [PubMed] [Google Scholar]
- 2.Panula P, et al. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine receptors. Pharm. Rev. 2015;67:601–655. doi: 10.1124/pr.114.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons ME, Ganellin CR. Histamine and its receptors. Br. J. Pharm. 2006;147:S127–S135. doi: 10.1038/sj.bjp.0706440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung-Leung WP, Thurmond RL, Ling P, Karlsson L. Histamine H4 receptor antagonists: the new antihistamines? Curr. Opin. Investig. Drugs. 2004;5:1174–1183. [PubMed] [Google Scholar]
- 5.Leurs R, Bakker RA, Timmerman H, de Esch IJ. The histamine H3 receptor: from gene cloning to H3 receptor drugs. Nat. Rev. Drug Discov. 2005;4:107–120. doi: 10.1038/nrd1631. [DOI] [PubMed] [Google Scholar]
- 6.Yanai K, Tashiro M. The physiological and pathophysiological roles of neuronal histamine: an insight from human positron emission tomography studies. Pharm. Ther. 2007;113:1–15. doi: 10.1016/j.pharmthera.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Fein MN, Fischer DA, O’Keefe AW, Sussman GL. CSACI position statement: Newer generation H1-antihistamines are safer than first-generation H1-antihistamines and should be the first-line antihistamines for the treatment of allergic rhinitis and urticaria. Allergy Asthma Clin. Immunol. 2019;15:61. doi: 10.1186/s13223-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naqvi, A. & Gerriets, V. Cetirizine. In StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. (2021).
- 9.Tashiro M, et al. Dose dependency of brain histamine H(1) receptor occupancy following oral administration of cetirizine hydrochloride measured using PET with [11C]doxepin. Hum. Psychopharmacol. 2009;24:540–548. doi: 10.1002/hup.1051. [DOI] [PubMed] [Google Scholar]
- 10.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjarnadottir TK, et al. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–273. doi: 10.1016/j.ygeno.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell. 2019;177:1933–1947.e1925. doi: 10.1016/j.cell.2019.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flock T, et al. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding SD, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda S, Qu Q, Robertson MJ, Skiniotis G, Kobilka BK. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science. 2019;364:552–557. doi: 10.1126/science.aaw5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell. 2020;182:1574–1588.e1519. doi: 10.1016/j.cell.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda S, et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 2018;9:3712. doi: 10.1038/s41467-018-06002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin J, et al. Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. Nature. 2020;584:125–129. doi: 10.1038/s41586-020-2379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehl A, et al. Structure of the micro-opioid receptor-Gi protein complex. Nature. 2018;558:547–552. doi: 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan J, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 2020;11:4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou F, et al. Structural basis for activation of the growth hormone-releasing hormone receptor. Nat. Commun. 2020;11:5205. doi: 10.1038/s41467-020-18945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Z, et al. Luciferase reporter assay system for deciphering GPCR pathways. Curr. Chem. Genomics. 2010;4:84–91. doi: 10.2174/1875397301004010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna Kumar K, et al. Structure of a signaling cannabinoid receptor 1-G protein complex. Cell. 2019;176:448–458.e412. doi: 10.1016/j.cell.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura A, et al. Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc. Natl Acad. Sci. USA. 2010;107:13666–13671. doi: 10.1073/pnas.1003553107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao LH, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364:148–153. doi: 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang YL, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing C, et al. Cryo-EM structure of the human cannabinoid receptor CB2-Gi signaling complex. Cell. 2020;180:645–654.e613. doi: 10.1016/j.cell.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr D. Struct. Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife7, 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed]
- 37.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 38.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Wang, R. Y. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife5, 10.7554/eLife.17219 (2016). [DOI] [PMC free article] [PubMed]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madeira F, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandy-Szekeres G, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper. Structural data have been deposited with the PDB (accession number 7DFL), and maps have been deposited with the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-30665.