B cell development in the bone marrow is critical for producing numerous B cells that play an essential role in the adaptive immune response. B cells develop from common lymphoid progenitors (CLPs), which then differentiate through a series of stages, which include prepro-B cells, pro-B cells, pre-B cells, immature B cells, and mature B cells.1 Within the pro-B stage, cells undergo proliferation to expand the pro-B cell pool and complete the recombination of the VH to DHJH segments to form the μ chain. However, the mechanisms involved in pro-B cell development are not fully understood. There have been several studies about the critical role of ubiquitination modifications in B cell development.2,3 Here, we report that DNA damage binding protein 1 (DDB1), an adaptor protein in CUL4-ring E3 ligases that is highly expressed in a variety of highly proliferative precursor cells,4 is involved in regulating pro-B cell development.
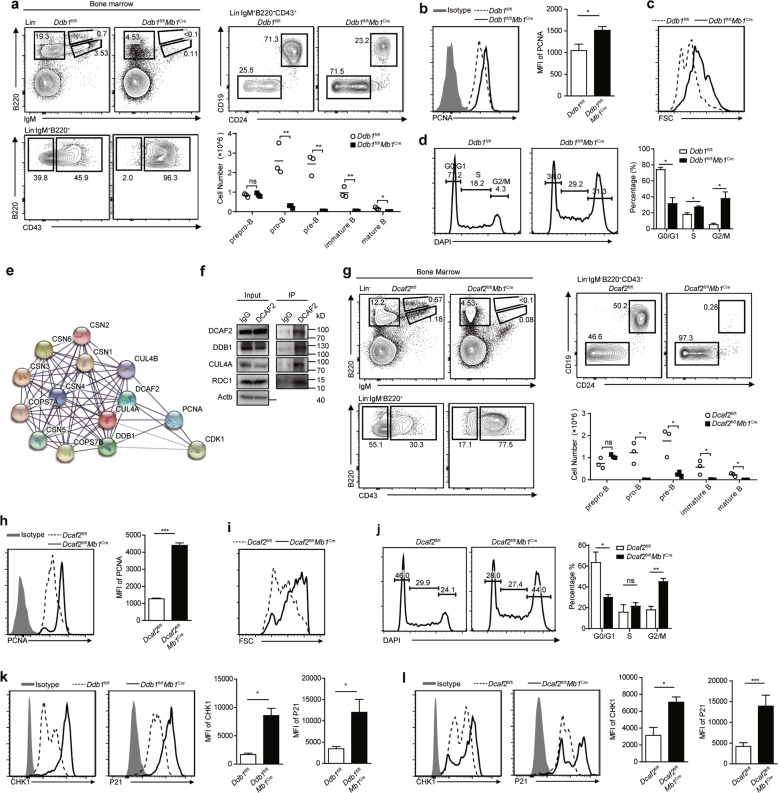
We initially examined DDB1 expression in B cell precursors at different stages. Pro-B cells expressed the highest amount of DDB1 (Fig. S1a), indicating that this protein might be involved in B lymphocytopoiesis. To further explore the role of DDB1 in B cell development, we crossed Ddb1fl/fl mice with Mb1Cre mice to delete DDB1 from late-stage prepro-B cells. The efficiency of DDB1 deletion was verified (Fig. S1b). Strikingly, B cells in the spleen, lymph nodes and peripheral blood were severely decreased in DDB1-deficient mice (Fig. S1c). Few serum immunoglobulins were detectable in DDB1-deficient mice (Fig. S1d). Then, B cell subsets at different developmental stages in the bone marrow were analyzed. The number of cells in the pro-B stage decreased (Fig. 1a). Together, these data demonstrated that DDB1 disruption severely impaired B cell development at the pro-B cell stage.
Fig. 1.
The DDB1-DCAF2 complex is required for B cell development because it controls cell cycle progression. a Prepro-B (Lin−IgM−B220+CD43+CD24−CD19−), pro-B (Lin−IgM−B220+CD43+CD24+CD19+), pre-B (Lin−IgM−B220+CD43−), immature B (Lin−IgM+B220low), and mature B (Lin−IgM+B220hi) cells from the indicated mice were analyzed by flow cytometry (n = 3). The absolute numbers of B cells at different stages are shown in the column chart. b PCNA staining of pro-B cells. MFI of PCNA is shown on the right. c The size of pro-B cells from the indicated mice. d Cell cycle analysis of pro-B cells by DAPI staining. The percentage of each cell phase is shown on the right. e Ba/F3 cells expressing Flag or Flag-DCAF2 were subjected to immunoprecipitation, and proteins that were pulled down were identified by mass spectrometry. A protein-protein interaction network was generated with the identified proteins by using the STRING database (https://string-db.org). f Coimmunoprecipitation of CUL4A, DDB1 and ROC1 with DCAF2 in Ba/F3 cells. Each experiment was independently repeated at least three times. g Flow cytometry analysis of B cell subsets in the bone marrow from the indicated mice (n = 3). The absolute numbers of B cells at different stages are shown in the column chart. h PCNA staining of pro-B cells from wild-type and DCAF2-deficient mice. MFI of PCNA is shown on the right (n = 3). i The size of pro-B cells was determined by forward scatter (FSC) signals. j Cell cycle analysis of pro-B cells of the indicated genotypes by DAPI staining. The percentage of each cell phase is shown on the right. k The protein levels of CHK1 and P21 in pro-B cells from DDB1-deficient mice and wild-type littermates were analyzed by flow cytometry. MFI of each protein was determined and is shown on the right (n = 3). l The protein levels of CHK1 and P21 in pro-B cells from DCAF2-deficient mice and wild-type littermates were analyzed by flow cytometry. MFI of each protein was determined and is shown on the right (n = 3). Bars represent the mean ± SD. Data are representative of at least three independent experiments. Statistical analysis was performed using two-tailed, unpaired Student’s t test. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001
Within the pro-B stage, IL-7 signaling provides survival and proliferation signaling and regulates the V-DJ recombination of immunoglobulin heavy chain.5 However, DDB1 deletion did not induce obvious defects in IL-7R expression in pro-B cells (Fig. S2a). During the pro-B stage, only cells with successfully rearranged V-DJ segments enter the next stage. PAX5 is an important regulator of VH-DHJH recombination. DDB1 deletion did not affect PAX5 expression in pro-B cells (Fig. S2b). The recombination of DH to JH and proximal VH segments (VH7183 and VHQ52 to DHJH) was intact, while the recombination of distal VH segments (VHGam3.8, VH3609, and VHJ558 to DHJH) was slightly diminished in DDB1-deficient pro-B cells (Fig. S2c). This finding might explain the modest decrease in the intracellular μ chain in DDB1-deficient pro-B cells (Fig. S2d). It was unclear whether the loss of pro-B cells was resulted from elevated apoptosis. We performed Annexin V staining and found that the percentage of Annexin V-positive pro-B cells was comparable between wild-type and DDB1-deficient mice (Fig. S2e). Therefore, the severe loss of pro-B cells in DDB1-deficient mice was not caused by altered IL-7R expression, V-DJ recombination of IgH or cell apoptosis.
The proliferation of pro-B cells following VH-DHJH rearrangement is crucial for the maintenance of the B cell pool. Next, we examined whether the loss of pro-B cells was caused by proliferation defects. Thus, we detected the proliferation markers proliferating cell nuclear antigen (PCNA) and Ki67. DDB1-deficient pro-B cells expressed more PCNA and Ki67 (Fig. 1b, Fig. S3a). Consistently, pro-B cells without DDB1 were larger (Fig. 1c). Cell cycle analysis showed that DDB1 deficiency led to G2/M-phase arrest in pro-B cells, with a slight block in S phase (Fig. 1d). Western blot analysis also showed increased expression of cyclin B, a G2/M cyclin, in DDB1-deficient pro-B cells, while G1/S cyclins, including cyclin A and cyclin E, remained unaltered (Fig. S3b). DDB1 deletion in the mouse brain and lens has been reported to cause DNA damage accumulation in proliferating cells.4 We stained cells with antibodies against histone H2AX phosphorylated at serine 139 (γH2AX) to assess DNA damage. We found more γH2AX accumulation in DDB1-deficient pro-B cells (Fig. S3c). Thus, we proposed that DDB1 deficiency led to the severe loss of pro-B cells mainly by preventing pro-B cells from going through the G2/M phase.
DDB1 interacts with DDB1-CUL4 associating factors (DCAFs), and more than ninety DCAFs have been identified.6 The ImmGen database showed that among the most studied DCAFs in mice, DCAF2 was relatively highly expressed in B cell progenitors (Fig. S4a). In addition, DCAF2 is important for maintaining normal cell cycle progression.7 The expression profile of DCAF2 in B cell subsets showed that the highest abundance of DCAF2 was found in pro-B cells (Fig. S4b), which is similar to the DDB1 expression pattern (Fig. S1a). To determine whether DCAF2 interacted with DDB1 in pro-B cells, we performed immunoprecipitation using Ba/F3 cells, and immunoprecipitated products were analyzed by mass spectrometry. The results showed that DDB1 and CUL4A were enriched in the Flag-DCAF2 group (Fig. 1e). Regulators of CRL4 E3 ligase, such as PCNA and the constitutive photomorphogenesis 9 signalosome, were enriched in the Flag-DCAF2 group as well (Fig. 1e). In addition, endogenous DCAF2 interacted with endogenous components of CRL4, such as CUL4A, DDB1 and ROC1 (Fig. 1f). These data suggested that DDB1 interacted with DCAF2 to form a complex in pro-B cells.
Then, we crossed Dcaf2fl/fl mice with Mb1Cre mice. Peripheral lymphoid organs showed few B cells, and serum immunoglobulins were undetectable in DCAF2-deficient mice (Fig. S5a, b). In addition, B cell development was severely blocked in the pro-B stage (Fig. 1g). Further analysis showed that DCAF2-deficient pro-B cells displayed similar defects as DDB1-deficient pro-B cells: increased PCNA levels, larger cell sizes, G2/M-phase arrest and more DNA damage (Fig. 1h-j and Fig. S6a). Moreover, DCAF2 loss did not affect the apoptosis of pro-B cells (Fig. S6b). These data suggested that the reduction in pro-B cells in DCAF2-deficient mice was due to G2/M retention, as observed in DDB1-deficient mice.
We tested the expression of three reported targets of DCAF2 in DDB1- or DCAF2-deficient pro-B cells8–10 and found that either DDB1 or DCAF2 deletion resulted in the abnormal accumulation of CHK1, P21, and SET8 in pro-B cells (Fig. 1k, l and Fig. S7a, b). Together, these results indicated that the DDB1-DCAF2 complex was involved in the proper degradation of cell cycle-related proteins such as CHK1, P21, and SET8, which induced pro-B cells to progress through the cell cycle.
In summary, we found that CRL4-DDB1-DCAF2 E3 ligase acts as a new player in B cell lymphopoiesis by regulating the proper G2/M phase exit of pro-B cells. These findings will help to provide a better understanding of the regulatory network of B cell development.
Supplementary information
DDB1-DCAF2 supplementary materials and methods
Acknowledgements
We thank Dr. Xiaolong Liu (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for his generous gifts of cell lines and plasmids. We thank Yingying Huang, Yanwei Li, and Jiajia Wang (Core Facilities, School of Medicine, Zhejiang University) for helping with cell sorting; Yu Zhang and Huihui Jin (Laboraty Animal Center, Zhejiang University) for feeding the mice; Xuliang Zhang, Yanxia Ding (Laboraty Animal Center, Zhejiang University), Xiaoyu Meng, and Xiaoqian Liu (Institute of Immunology, Zhejiang University School of Medicine) for the animal study; and Wenqiang Cao and Zhaoyuan hui (Institute of Immunology, Zhejiang University School of Medicine) for improving the writing. This work was supported in part by grants from the National Natural Science Foundation of China (81830006, 31670887, 31870874, and 31800734), Zhejiang Provincial Key Project of Research and Development (2019C03043), Zhejiang Natural Science Foundation (LQ16H030003), and Zhejiang Science and Technology Program (2017C37117 and 2017C37170).
Author contributions
Lie Wang, Zhonghui Xue, and Jing Guo designed the study. Zhonghui Xue, Jing Guo, Ruoyu Ma, Lina Zhou, and Yixin Guo performed the experiments and data analysis. Lie Wang, Zhonghui Xue, and Jing Guo wrote the paper. Yong Cang and Hengyu Fan provided the reagents. Jian Chen and Wenbin Qian provided expertise and advice. Lie Wang supervised the project.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zhonghui Xue, Jing Guo
Supplementary information
The online version of this article (10.1038/s41423-020-0390-2) contains supplementary material.
References
- 1.Wang W, et al. MEF2C protects bone marrow B-lymphoid progenitors during stress haematopoiesis. Nat. Commun. 2016;7:12376. doi: 10.1038/ncomms12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, et al. The endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 controls a critical checkpoint in B cell development in mice. J. Biol. Chem. 2018;293:12934–12944. doi: 10.1074/jbc.RA117.001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, et al. The E3 ubiquitin ligase Itch is required for B-cell development. Sci. Rep. 2019;9:421. doi: 10.1038/s41598-018-36844-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J, et al. The CUL4-DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. Elife. 2015;4:e07539. doi: 10.7554/eLife.07539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark MR, et al. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochemical Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas T, Dutta A. CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huh J, Piwnica-Worms H. CRL4(CDT2) targets CHK1 for PCNA-independent destruction. Mol. Cell. Biol. 2013;33:213–226. doi: 10.1128/MCB.00847-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishitani H, et al. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbas T, et al. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DDB1-DCAF2 supplementary materials and methods